Abstract
Background
Common fetal aneuploidies include Down syndrome (trisomy 21 or T21), Edward syndrome (trisomy 18 or T18), Patau syndrome (trisomy 13 or T13), Turner syndrome (45,X), Klinefelter syndrome (47,XXY), Triple X syndrome (47,XXX) and 47,XYY syndrome (47,XYY). Prenatal screening for fetal aneuploidies is standard care in many countries, but current biochemical and ultrasound tests have high false negative and false positive rates. The discovery of fetal circulating cell‐free DNA (ccfDNA) in maternal blood offers the potential for genomics‐based non‐invasive prenatal testing (gNIPT) as a more accurate screening method. Two approaches used for gNIPT are massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS).
Objectives
To evaluate and compare the diagnostic accuracy of MPSS and TMPS for gNIPT as a first‐tier test in unselected populations of pregnant women undergoing aneuploidy screening or as a second‐tier test in pregnant women considered to be high risk after first‐tier screening for common fetal aneuploidies. The gNIPT results were confirmed by a reference standard such as fetal karyotype or neonatal clinical examination.
Search methods
We searched 13 databases (including MEDLINE, Embase and Web of Science) from 1 January 2007 to 12 July 2016 without any language, search filter or publication type restrictions. We also screened reference lists of relevant full‐text articles, websites of private prenatal diagnosis companies and conference abstracts.
Selection criteria
Studies could include pregnant women of any age, ethnicity and gestational age with singleton or multifetal pregnancy. The women must have had a screening test for fetal aneuploidy by MPSS or TMPS and a reference standard such as fetal karyotype or medical records from birth.
Data collection and analysis
Two review authors independently carried out study selection, data extraction and quality assessment (using the QUADAS‐2 tool). Where possible, hierarchical models or simpler alternatives were used for meta‐analysis.
Main results
Sixty‐five studies of 86,139 pregnant women (3141 aneuploids and 82,998 euploids) were included. No study was judged to be at low risk of bias across the four domains of the QUADAS‐2 tool but applicability concerns were generally low. Of the 65 studies, 42 enrolled pregnant women at high risk, five recruited an unselected population and 18 recruited cohorts with a mix of prior risk of fetal aneuploidy. Among the 65 studies, 44 evaluated MPSS and 21 evaluated TMPS; of these, five studies also compared gNIPT with a traditional screening test (biochemical, ultrasound or both). Forty‐six out of 65 studies (71%) reported gNIPT assay failure rate, which ranged between 0% and 25% for MPSS, and between 0.8% and 7.5% for TMPS.
In the population of unselected pregnant women, MPSS was evaluated by only one study; the study assessed T21, T18 and T13. TMPS was assessed for T21 in four studies involving unselected cohorts; three of the studies also assessed T18 and 13. In pooled analyses (88 T21 cases, 22 T18 cases, eight T13 cases and 20,649 unaffected pregnancies (non T21, T18 and T13)), the clinical sensitivity (95% confidence interval (CI)) of TMPS was 99.2% (78.2% to 100%), 90.9% (70.0% to 97.7%) and 65.1% (9.16% to 97.2%) for T21, T18 and T13, respectively. The corresponding clinical specificity was above 99.9% for T21, T18 and T13.
In high‐risk populations, MPSS was assessed for T21, T18, T13 and 45,X in 30, 28, 20 and 12 studies, respectively. In pooled analyses (1048 T21 cases, 332 T18 cases, 128 T13 cases and 15,797 unaffected pregnancies), the clinical sensitivity (95% confidence interval (CI)) of MPSS was 99.7% (98.0% to 100%), 97.8% (92.5% to 99.4%), 95.8% (86.1% to 98.9%) and 91.7% (78.3% to 97.1%) for T21, T18, T13 and 45,X, respectively. The corresponding clinical specificities (95% CI) were 99.9% (99.8% to 100%), 99.9% (99.8% to 100%), 99.8% (99.8% to 99.9%) and 99.6% (98.9% to 99.8%). In this risk group, TMPS was assessed for T21, T18, T13 and 45,X in six, five, two and four studies. In pooled analyses (246 T21 cases, 112 T18 cases, 20 T13 cases and 4282 unaffected pregnancies), the clinical sensitivity (95% CI) of TMPS was 99.2% (96.8% to 99.8%), 98.2% (93.1% to 99.6%), 100% (83.9% to 100%) and 92.4% (84.1% to 96.5%) for T21, T18, T13 and 45,X respectively. The clinical specificities were above 100% for T21, T18 and T13 and 99.8% (98.3% to 100%) for 45,X. Indirect comparisons of MPSS and TMPS for T21, T18 and 45,X showed no statistical difference in clinical sensitivity, clinical specificity or both. Due to limited data, comparative meta‐analysis of MPSS and TMPS was not possible for T13.
We were unable to perform meta‐analyses of gNIPT for 47,XXX, 47,XXY and 47,XYY because there were very few or no studies in one or more risk groups.
Authors' conclusions
These results show that MPSS and TMPS perform similarly in terms of clinical sensitivity and specificity for the detection of fetal T31, T18, T13 and sex chromosome aneuploidy (SCA). However, no study compared the two approaches head‐to‐head in the same cohort of patients. The accuracy of gNIPT as a prenatal screening test has been mainly evaluated as a second‐tier screening test to identify pregnancies at very low risk of fetal aneuploidies (T21, T18 and T13), thus avoiding invasive procedures. Genomics‐based non‐invasive prenatal testing methods appear to be sensitive and highly specific for detection of fetal trisomies 21, 18 and 13 in high‐risk populations. There is paucity of data on the accuracy of gNIPT as a first‐tier aneuploidy screening test in a population of unselected pregnant women. With respect to the replacement of invasive tests, the performance of gNIPT observed in this review is not sufficient to replace current invasive diagnostic tests.
We conclude that given the current data on the performance of gNIPT, invasive fetal karyotyping is still the required diagnostic approach to confirm the presence of a chromosomal abnormality prior to making irreversible decisions relative to the pregnancy outcome. However, most of the gNIPT studies were prone to bias, especially in terms of the selection of participants.
Keywords: Female; Humans; Pregnancy; Aneuploidy; Cell-Free Nucleic Acids; Cell-Free Nucleic Acids/blood; Chromosome Disorders; Chromosome Disorders/diagnosis; Chromosome Disorders/genetics; Disorders of Sex Development; Disorders of Sex Development/diagnosis; Disorders of Sex Development/genetics; Fetal Diseases; Fetal Diseases/diagnosis; Fetal Diseases/genetics; High-Throughput Nucleotide Sequencing; High-Throughput Nucleotide Sequencing/methods; Pregnancy, High-Risk; Prenatal Diagnosis; Prenatal Diagnosis/methods
Plain language summary
Accuracy of gNIPT for identifying genetic abnormalities in unborn babies
What is the issue?
How accurate is the new test (genomics‐based non‐invasive prenatal testing (gNIPT)) for detecting abnormal chromosome number in an unborn baby's genetic material (DNA) found in the mother's blood? We assessed the accuracy for the screening of Down syndrome (trisomy 21), Edward syndrome (trisomy 18), Patau syndrome (trisomy 13), Turner syndrome (45,X), Klinefelter syndrome (47,XXY), Triple X syndrome (47,XXX) and 47,XYY syndrome. There are different methods in use for gNIPT. We assessed MPSS (massively parallel shotgun sequencing) that tests whole DNA and TMPS (targeted massively parallel sequencing) that tests targeted DNA.
Background
There are 46 chromosomes (23 pairs) in humans. Abnormal numbers of chromosomes can cause genetic disorders for which there are no cures. Having an extra chromosome is called trisomy and an excess (or less) of sexual chromosome is called sex chromosome abnormality (SCA). The most common trisomy is Down syndrome which occurs in about one in 1000 babies. Children with Downs have slow growth, characteristic facial features and mild to moderate intellectual disability, with some requiring specialist education later in life. However, the symptoms vary from mild to severe so that some infants lead relatively normal lives. The other trisomy or SCA conditions have varying degrees of disability but the chance of a baby being affected is much less.
Current screening tests for these conditions require confirmation if the baby has the condition or not and for this an invasive test like amniocentesis is used. Amniocentesis is where fetal cells that float in the fluid surrounding the unborn baby are collected by putting a fine needle through the mother’s abdomen and collecting the fluid. Alternatively, tissue can be collected from the placenta (chorionic villus sampling (CVS)). With these invasive tests, pregnant women are exposed to a higher chance of losing their baby even if the baby is unaffected by Down syndrome. So, this invasive test is only offered to women who are thought to have a higher chance of having an affected unborn baby
What we did
We looked for studies that included women of any age, ethnicity and gestational age who were carrying either a single baby or more than one. We searched for studies (up to July 2016) that assessed the accuracy of the new test.
What we found
We found 65 studies with a total of 86,139 pregnant women, including 3141 affected pregnancies. Forty‐two studies (65%) enrolled pregnant women with a high chance of having babies with abnormal chromosome number. Forty‐eight (74%) studies included only women with a singleton pregnancy. Forty‐four studies (68%) used MPSS and 21 studies (32%) used TMPS.
gNIPT seems to be accurate for screening unborn babies (either singletons or twins), especially for detecting Down syndrome, trisomy 18 and trisomy 13. However, there were some problems with how the studies were conducted which makes us cautious about our findings. This may result in gNIPT appearing to perform better than it really does.
Other important information to consider
gNIPT method appears to perform well in identifying unborn babies with abnormal number of chromosomes. However, when a gNIPT detects an abnormal chromosome number, then a confirmation using invasive tests (like amniocentesis or CVS) is still needed before pregnancy‐related decisions can be made.
It is important that pregnant women are given full information on the possible health problems that might arise for babies affected by an additional chromosome. For example, with Down syndrome though some children have considerable disability, others can lead relatively normal lives. In addition, in this review most studies enrolled pregnant women with increased chance of having babies with abnormal chromosome number, so our findings do not directly apply to general populations of pregnant women.
Summary of findings
Summary of findings 1. Summary characteristics of included studies.
| Summary characteristics of included studies | |
| Review question | What is the diagnostic accuracy of massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS) using circulating cell‐free DNA (ccfDNA) in maternal blood for the detection of common fetal aneuploidies (T21, T18, T13, 45,X, 47,XXY, 47,XXX and 47,XYY) in pregnant women according to their prior risk of fetal aneuploidy? |
| Importance (rationale) | These new genomics‐based non‐invasive prenatal testing (gNIPT) approach report higher sensitivity and lower false positive rate than traditional screening tests. gNIPT is already advertised and marketed. How gNIPT should be used in clinical practice should be assessed in order to provide a framework for its use. |
| Study design | There were 40 prospective cohort studies, 8 retrospective cohort studies, 16 case‐control studies and 1 prospective and retrospective cohort study. |
| Population | Pregnant women of any age, ethnicity and gestational age, with singleton or multifetal pregnancy who had a screening test for fetal aneuploidy using gNIPT and received a reference standard. 42 studies enrolled pregnant women selected at high risk of fetal aneuploidy, 5 enrolled unselected pregnant women undergoing aneuploidy screening and 18 enrolled pregnant women from a mixed‐risk population of fetal aneuploidy. 48 studies included only women with singleton pregnancy, 5 included only multifetal pregnancies, 4 included either type of pregnancy and 8 did not report type of pregnancy. 10 studies included only women in the first trimester (15 weeks or less), 21 studies included women in the first 2 trimesters (29 weeks or less), 24 studies included women in the 3 trimesters (42 weeks or less) and 10 studies (15%) did not report gestational age. |
| Index tests | gNIPT by MPSS (44 studies) or TMPS (21 studies), including 5 studies that compared a gNIPT with a traditional screening test. 37 studies were industry‐funded or were written by 1 or more authors affiliated with a company who sells gNIPT. 22 studies were not reported to be funded by industry but samples were sequenced and analysed by a commercial laboratory. 3 studies had no links with industry. |
| Target conditions | 36 studies reported results for only autosomes (T21, T18, T13), 4 for only SCA (45,X, 47,XXY, 47,XXX and 47,XYY), and 25 for both autosomes and SCA. |
| Reference standard | Fetal karyotyping performed on cells obtained from chorionic villi sampling, amniotic fluid, placental tissue, a fetus lost by miscarriage or other equivalent and recognised methods on the same materials for autosomes and SCA. If fetal karyotyping was not performed, we used neonatal clinical examination or medical records from birth (for autosomes only). Only 1 reference standard was used for all pregnant women included in 36 studies while multiple reference standards were used in 29 studies. |
| Risk of bias | The QUality Assessment of Diagnostic Accuracy Studies (QUADAS‐2) tool was used to assess the methodological quality of included studies. No study was assessed as being at low risk of bias across all domains. For the patient selection domain, no study was assessed as being at low risk of bias. For the index test, reference standard and flow and timing domains, the risk of bias was low for 94%, 77% and 23% of studies, respectively. |
| Applicability concerns | Applicability was of low concern for all studies in the index test and reference standard domains because the studies matched the review question. In the patient selection domain, 47 (71%) studies were judged to be of low applicability concern because they included pregnant women matching the review question. |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13.
Summary of findings 2. Performance of gNIPT for detection of T21.
| Performance of gNIPT for detection of T21 | |||||||
| Test strategy |
Number of studies |
Number of affected pregnancies (Number of unaffected pregnancies)a |
Sensitivity % (95% CI) |
Specificity % (95% CI) |
Median prevalenceb % (range) |
Missed cases (FN)c |
False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 8 (1733) | 100 (67.6 to 100) | 100 (99.8 to 100) | 0.46 (0.24 to 5.21) |
0 | 0 |
| TMPS | 4 | 88 (20,679) | 99.2 (78.2 to 100) | 100 (> 99.9 to 100) | 4 | 0 | |
| Traditional screening teste | 1 | 38 (15,803) | 78.9 (63.7 to 88.9) | 94.6 (94.2 to 94.9) | 97 | 5375 | |
| Implications |
|
||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 30 | 1048 (15,937) | 99.7 (98.0 to 100) | 99.9 (99.8 to 100) | 4.95 (0.44 to 27.66) |
15 | 95 |
| TMPS | 6 | 246 (4380) | 99.2 (96.8 to 99.8) | 100 (99.8 to 100) | 40 | 0 | |
| Difference between MPSS and TMPS | 0.53 (‐0.73 to 1.78) | ‐0.03 (‐0.11 to 0.04) | NA | ||||
| Implications |
|
||||||
MPSS: massively parallel shotgun sequencing, NA; not applicable, TMPS: targeted massively parallel sequencing, T21: trisomy 21.
aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered.
cMissed cases per 100,000 tested. FN: false negatives.
dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman.
eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test.
Summary of findings 3. Performance of gNIPT for detection of T18.
| Performance of gNIPT for detection of T18 | |||||||
| Test strategy |
Number of studies |
Number of affected pregnancies (Number of unaffected pregnancies)a |
Sensitivity % (95% CI) |
Specificity % (95% CI) |
Median prevalenceb % (range) |
Missed cases (FN)c |
False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 2 (1739) | 100 (34.3 to 100) | 99.9 (99.7 to 100) | 0.11 (0.06 to 0.36) |
0 | 100 |
| TMPS | 3 | 22 (20,553) | 90.9 (70.0 to 97.7) | 100 (99.9 to 100) | 10 | 0 | |
| Traditional screening teste | 1 | 10 (15,831) | 80.0 (49.0 to 94.3) | 99.7 (99.6 to 99.8) | 22 | 300 | |
| Implications |
|
||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 28 | 332 (16,180) | 97.8 (92.5 to 99.4) | 99.9 (99.8 to 100) | 1.46 (0.22 to 17.02) |
32 | 99 |
| TMPS | 5 | 112 (4010) | 98.2 (93.1 to 99.6) | 100 (99.8 to 100) | 26 | 0 | |
| Difference between MPSS and TMPS | ‐0.41 (‐4.11 to 3.28) | ‐0.06 (‐0.14 to 0.03) | NA | ||||
| Implications |
|
||||||
MPSS: massively parallel shotgun sequencing, NA: not applicable, TMPS: targeted massively parallel sequencing, T18: trisomy 18.
aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered.
cMissed cases per 100,000 tested. FN: false negatives.
dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman.
eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test.
Summary of findings 4. Performance of gNIPT for detection of T13.
| Performance of gNIPT for detection of T13 | |||||||
| Test strategy |
Number of studies |
Number of affected pregnancies (Number of unaffected pregnancies)a |
Sensitivity % (95% CI) |
Specificity % (95% CI) |
Median prevalenceb % (range) |
Missed cases (FN)c |
False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 1 (1740) | 100 (20.7 to 100) | 100 (99.8 to 100) | 0. 12 (0.01 to 0.52) |
0 | 0 |
| TMPS | 3 | 8 (14,154) | 65.1 (9.16 to 97.2) | 100 (99.9 to 100) | 41 | 0 | |
| Traditional screening teste | 1 | 2 (11,183) | 50.0 (9.45 to 90.5) | 99.7 (99.6 to 99.8) | 59 | 300 | |
| Implications |
|
||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 20 | 128 (13,810) | 95.8 (86.1 to 98.9) | 99.8 (99.8 to 99.9) | 1.09 (0.04 to 3.54) |
46 | 198 |
| TMPS | 2 | 20 (293) | 100 (83.9 to 100)f | 100 (98.7 to 100)f | 0 | 0 | |
| Implications |
|
||||||
MPSS: massively parallel shotgun sequencing, NA: not applicable, TMPS: targeted massively parallel sequencing, T13: trisomy 13.
aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered.
cMissed cases per 100,000 tested. FN: false negatives.
dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman.
eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test.
fSimple pooling used to obtain summary estimates of sensitivity, specificity or both.
Summary of findings 5. Performance of gNIPT for detection of 45,X.
| Performance of gNIPT for detection of 45,X | |||||||
| Test strategy |
Number of studies |
Number of affected pregnancies (Number of unaffected pregnancies)a |
Sensitivity % (95% CI) |
Specificity % (95% CI) |
Median prevalenceb % (range) |
Missed cases (FN)c |
False positives (FP)d |
| Selected high‐risk pregnant women | |||||||
| MPSS | 12 | 119 (7440) | 91.7 (78.3 to 97.1) | 99.6 (98.9 to 99.8) | 1.04 (0.27 to 18.58) |
86 | 396 |
| TMPS | 4 | 79 (985) | 92.4 (84.1 to 96.5) | 99.8 (98.3 to 100) | 79 | 198 | |
| Difference between MPSS and TMPS | ‐0.74 (‐11.1 to 9.60) | ‐0.23 (‐0.82 to 0.36) | NA | ||||
| Implications |
|
||||||
45,X: Turner syndrome, MPSS: massively parallel shotgun sequencing, NA: not applicable, TMPS: targeted massively parallel sequencing.
aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered.
cMissed cases per 100,000 tested. FN: false negatives.
dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman.
Summary of findings 6. Performance of gNIPT for detection of autosomes aneuploidies (T21, T18 and T13 combined).
| Performance of gNIPT for detection of autosomes aneuploidies (T21, T18 and T13 combined) | |||||||
| Test strategy |
Number of studies |
Number of affected pregnancies (Number of unaffected pregnancies)a |
Sensitivity % (95% CI) |
Specificity % (95% CI) |
Median prevalenceb % (range) |
Missed cases (FN)c |
False positives (FP)d |
| Unselected pregnant women | |||||||
| MPSS | 1 | 11 (1730) | 100 (74.1 to 100) | 99.9 (99.7 to 100) | 0,63 (0.32 to 5.73) |
0 | 99 |
| TMPS | 4 | 118 (20,649) | 94.9 (89.1 to 97.7) | 99.9 (99.8 to 99.9) | 32 | 99 | |
| Traditional screening teste | 4 | 120 (22,247) | NDf | ND | |||
| Implications |
|
||||||
| Selected high‐risk pregnant women | |||||||
| MPSS | 32 | 1508 (15,797) | 98.8 (97.2 to 99.5) | 99.9 (99.7 to 100) | 5.85 (0.67 to 46.81) |
70 | 94 |
| TMPS | 7 | 378 (4282) | 98.9 (97.2 to 99.6) | 99.9 (99.8 to 100) | 64 | 94 | |
| Difference between MPSS and TMPS | ‐0.11 (‐1.58 to 1.35) |
‐0.08 (‐0.22 to 0.07) |
NA | ||||
| Implications |
|
||||||
MPSS: massively parallel shotgun sequencing, NA: not applicable, ND: no data available, TMPS: targeted massively parallel sequencing, T13: trisomy 13, T18: trisomy 18, T21: trisomy 21.
aUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered.
cMissed cases per 100,000 tested. FN: false negatives.
dFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman.
eTraditional screening tests are first‐trimester combined test, second‐trimester quadruple test, second‐trimester fully integrated test, second‐trimester sequential test or second‐trimester triple test.
fSummary sensitivity and specificity were not obtained for traditional screening tests because the four studies used different cut‐offs to determine test positivity. Three of the four studies compared TMPS and traditional screening tests in the same population (direct comparison).
Summary of findings 7. Performance of gNIPT for detection of sex chromosome aneuploidies (45,X, 47,XXX, 47,XXY and 47,XYY combined)a.
| Performance of gNIPT for detection of sex chromosome aneuploidies (45,X, 47,XXX, 47,XXY and 47,XYY combined) | |||||||
| Test strategy |
Number of studies |
Number of affected pregnancies (Number of unaffected pregnancies)b |
Sensitivity % (95% CI) |
Specificity % (95% CI) |
Median prevalencec % (range) |
Missed cases (FN)d |
False positives (FP)e |
| Selected high‐risk pregnant women | |||||||
| MPSS | 12 | 151 (7452) | 91.9 (73.8 to 97.9) | 99.5 (98.8 to 99.8) | 1.53 (0.45 to 18.58) |
124 | 492 |
| TMPS | 4 | 96 (968) | 93.8 (86.8 to 97.2) | 99.6 (98.1 to 99.9) | 95 | 394 | |
| Difference between MPSS and TMPS | ‐1.85 (‐13.3 to 9.60) | ‐0.06 (‐0.82 to 0.71) | NA | ||||
| Implications |
|
||||||
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, MPSS: massively parallel shotgun sequencing, NA: not applicable, ND: no data available, TMPS: targeted massively parallel sequencing
aWe did not assess the accuracy of gNIPT individually for 47,XXX, 47,XXY and 47,XYY due to paucity data.
bUnaffected pregnancies: we included patients with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
cThe median prevalence and range were calculated by using all prospective or retrospective studies for each category considered.
dMissed cases per 100,000 tested. FN: false negatives.
eFalse positives per 100,000 tested. A false positive result may lead to unnecessary invasive tests depending on choices by the pregnant woman.
Background
Aneuploidies[1] are chromosomal abnormalities characterised by a different (additional or missing) number of chromosomes than the 23 pairs normally present in humans. These chromosomal anomalies are among the most common types of genetic disorders and they represent a significant cause of both childhood and adulthood morbidity or death. In addition, they may lead to perinatal complications (Wellesley 2012; Wu 2013a). The severity of associated symptoms is often variable and typically less severe in mosaic cases (not all cells affected) (Fishler 1991; Modi 2003; Zhu 2013). Although offering prenatal screening for fetal aneuploidies such as Down syndrome is now considered standard of care in routine antenatal care in most upper‐middle and high‐income countries, prenatal screening methods and strategies are evolving. Prenatal screening consists of blood‐based biochemical testing or ultrasound measurements or a combination of both, in addition to maternal age (Alldred 2012). Because of the serious health consequences of various aneuploidies and given their incurable nature, prenatal screening is an option available to pregnant women. An invasive diagnostic test (e.g. amniocentesis) is offered to pregnant women found to be at high risk of fetal aneuploidy after prenatal screening, but there is a procedure‐related risk of miscarriage. The discovery of circulating cell‐free DNA (ccfDNA) in maternal blood has enabled the development of genomics‐based non‐invasive prenatal testing (gNIPT) to analyse the fetal genome. Prenatal screening, and ultimately prenatal diagnosis, provides couples with the information necessary for taking informed decisions (the optimisation of medical intervention and psychological counselling for managing the identified condition or pregnancy termination). The decision to terminate pregnancy among women who received a positive diagnosis of fetal aneuploidy during the prenatal period varies between 86% and 97% (Choi 2012; Irving 2011). Many factors, such as religion, maternal age, gestational age at the time of diagnosis, number of existing children, past history of induced abortion and psychosocial factors (perceived parenting burden/reward, quality of life of a child with a chromosomal abnormality, attitudes toward, and comfort with individuals with disabilities, and support from others) influence women’s decision making following prenatal anomaly detection (Choi 2012).
In this systematic review, we assessed the accuracy of gNIPT for the detection of common fetal aneuploidies in pregnant women according to their prior risk of fetal aneuploidy. More specifically, we evaluated and compared the diagnostic performance of two new next‐generation sequencing approaches (i.e. massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS)) that have recently been proposed as methods of choice to detect fetal aneuploidies by analysing ccfDNA in maternal plasma. We also made comparisons between MPSS and TMPS or between gNIPT and their combination with other first‐tier screening approaches. gNIPT could be used as a first‐tier test in pregnant women without prior risk (i.e. in unselected pregnant women or the general population) or as a second‐tier test after a positive result for traditional first‐tier screening tests such as biochemical, ultrasound or both markers (with maternal age included in risk assessment) and previous maternal history when possible.
[1] For a glossary of terms, see Appendix 1. For a list of acronyms and abbreviations, see Appendix 2.
Target condition being diagnosed
The target conditions are fetal chromosomal abnormalities diagnosed in pregnant women. The seven target conditions assessed were Down syndrome (trisomy 21 or T21), Edward syndrome (trisomy 18 or T18), Patau syndrome (trisomy 13 or T13), Turner syndrome (45,X), Klinefelter syndrome (47,XXY), Triple X syndrome (47,XXX) and 47,XYY syndrome (47,XYY) (Table 8). The majority of aneuploidies are associated with an extra copy (trisomy) of one chromosome (e.g. three copies of chromosome 21 for T21 instead of two) or a loss of one chromosome (e.g. female 45,X). Chromosomal abnormality is usually caused by a chromosome division failure or a chromosomal translocation. For example, most cases (76.2%) of 45,X karyotype (all cells affected) are caused by paternal chromosome division failure (Uematsu 2002). The most common chromosomal abnormalities are T21 and 45,X, respectively. For T21, the prevalences reported for pregnant women are 0.11% and 0.44% at 25 and 35 years old, respectively at diagnosis procedure (Snijders 1999).
1. Characteristics of target conditions.
|
Target condition |
Affected birthsa /100,000 |
Clinical features | Prognosis |
| T21 | 140 to 230b,c | Intellectual disability (mild to moderate), neurodevelopmental problems, characteristic dysmorphic features, congenital defects (cardiac (44% to 58%) and gastrointestinal system (4% to 10%)), vision or hearing impairment (38% to 80%) and obstructive sleep apnoea syndrome (57%)d,e | Mean and median life expectancies are estimated to be 51 and 58 years oldf |
| T18 | 59c | Severe intellectual disability and a wide range of significant malformations (cardiac defects, gastrointestinal system defects, renal anomalies, central nervous system defects (apnoea and seizures))d,g | Most affected fetuses die in utero. Median survival has been estimated at 14 days (95% confidence interval (CI) 10 to 20) and 8% (95% CI 4 to 14) reach 1 year of ageh |
| T13 | 23c | Severe intellectual disability, seizures and several dysmorphic features, malformations of the extremities, cardiac defects, renal anomalies, and abdominal wall defectsd,i | Most affected fetuses die in utero. Median survival time has been estimated at 10 days (95% CI 7 to 19) and 8% (95% CI 4 to 14) reach 1 year of ageh |
| 45,X | 30 to 50c,j |
Learning disabilities (70%), short stature, congenital heart diseases (30%) and gonadal dysgenesis (90% with amenorrhoea and infertility due to early ovarian failure)k,l | Mortality in 45,X women is 3‐fold higher than in the general population with an average life span of 69 yearsm |
| 47,XXY | 12c | Learning disabilities (> 75%), small testes (> 95%), azoospermia (> 95%), male infertility (91% to 99%), decreased testosterone level (63% to 85%) and gynaecomastia (38% to 75%)l,n | Life expectancy is slightly shorter (approximately 2 years) than euploid menn |
| 47,XXX | 6c | Developmental delays (motor and speech), learning or intellectual disability, attention deficits (25% to 35%), mood disorders (anxiety and depression), tall stature (80% to 89%), clinodactyly (42% to 65%), hypotonia in infancy (55% to 71%), genitourinary malformations and congenital heart defectso | Mortality significantly increased with a median survival age of 70.9 years compare to 81.7 years for euploid femalesp |
| 47,XYY | 3c | Developmental delays (speech, language and motor), attention deficit disorder (52%), tall stature (78%), central adiposity, macrocephaly (33%), hypotonia (63%), clinodactyly (52%), hypertelorism (59%) and testicular enlargement for age (50%) but no increase in genital anomaliesq | Mortality increased with a reduction of life span of 10.3 years compared to euploid menr |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13.
aIncluding live births, fetal deaths and terminations of pregnancy.
b(Christianson 2006; Parker 2010)
e(Irving 2012; Weijerman 2010)
f(Wu 2013b)
g(Cereda 2012)
h(Wu 2013a)
i(Chen 2009)
k(Karnis 2012; Mazzanti 1998; Sybert 2004)
l(Tyler 2004)
m(Saenger 1996; Schoemaker 2008)
n(Groth 2013)
q(Bardsley 2013; Leggett 2010)
r(Stochholm 2010a).
Clinical characteristics and spectrum of severity are variable among aneuploidies. It has been reported that 50% of 45,X cases are mosaic (Sybert 2004). During the past few decades, caring for children with T21 or sex chromosomal abnormalities and provision of counselling to their family has changed fundamentally. These changes, including medical and surgical advances, specific interventions in the classroom for those with learning disabilities, interventions and support for parents and family members, have helped individuals with T21 live longer and enjoy an improved quality of life (Van Riper 2001). Many health problems associated with T21, 45,X, 47,XXY, 47,XXX and 47,XYY aneuploidies can be treated but fetuses with T18 and T13 are most affected and usually die in utero. The age at diagnosis varies widely depending on the condition. T21, T18 and T13 are generally detected during the perinatal period, while detection of 45,X, 47,XXX and 47,XYY is often delayed, sometimes up to 60 years old (Stochholm 2006; Stochholm 2010a; Tartaglia 2010). Around 10% of fetuses with 47,XXY are diagnosed prenatally and the mean age at diagnosis is in the mid‐30s. Most 47,XXY cases are never diagnosed (Groth 2013; Tyler 2004). The incidence, clinical features and prognosis of the target conditions are summarised in Table 8.
Index test(s)
Genomics‐based non‐invasive prenatal tests are based on the finding that placental cells continuously release detectable amounts of fetal ccfDNA into maternal blood. This fetal ccfDNA originates from normal placental cell death and consists mainly of relatively short fragments of < 300 base pairs (Bianchi 2004; Fan 2010). Proof‐of‐concept studies showed the feasibility of such tests to detect fetal aneuploidy in 2008 (Chiu 2008; Fan 2008).
We assessed these two gNIPT approaches (Figure 1):
1.
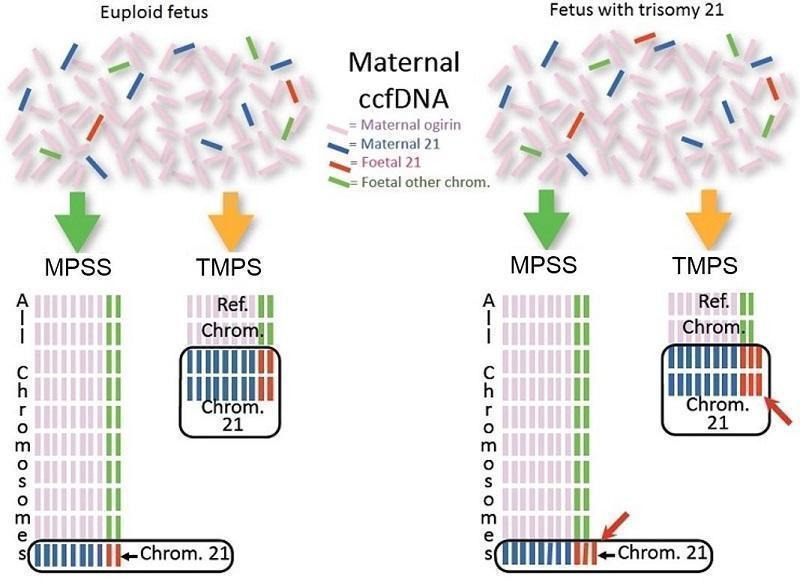
Difference between massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS). Genomics‐based non‐invasive prenatal testing (gNIPT) aims to count the number of copies of DNA fragments from the chromosomes of interest (chromosome 21 (Chrom. 21) in this example) present in circulating cell‐free DNA (ccfDNA) from a pregnant woman, relative to a reference set of chromosomes (Ref. Chrom.). DNA fragments circulating in maternal blood in the case of a euploid (left) and aneuploid (right) pregnancy are illustrated (top). MPSS produces a large number of sequence reads from all chromosomes while TMPS generates a larger proportion of reads from the chromosomes of interest (bottom). In both methods, sequence reads can be used to detect a slight excess of fetal genomic material coming from the chromosome of interest. Figure was created by FR.
massively parallel shotgun sequencing (MPSS) which randomly analyses all DNA fragments of a sample; and
targeted massively parallel sequencing (TMPS) which targets specific DNA fragments from the chromosomal regions of interest.
The fraction of the total ccfDNA in maternal circulation that is of fetal origin (the fetal fraction) is an important parameter for correctly identifying an aneuploid fetus by gNIPT (Canick 2013). Although the fetal ccfDNA fraction is a relatively small fraction (about 2% to 20%) of all ccfDNA in maternal blood, it can be detected from five weeks of gestation (Birch 2005; Canick 2013; Lo 1997; Lun 2008). Invasive procedures such as amniocentesis, may (Samura 2003) or may not be (Bussani 2011; Vora 2010) associated with a statistically significant increase of ccfDNA in maternal blood, which could affect fetal DNA concentration and affect gNIPT results. Therefore, in the context of clinical studies, maternal blood for gNIPT is usually collected either before or after waiting for a minimum of 24 hours following an invasive test. Indeed, the half‐life of ccfDNA has been estimated to be less than one day (Lo 1999; Yu 2013). On average, euploid multifetal pregnancies have a higher fetal ccfDNA fraction than euploid singleton pregnancies (Attilakos 2011; Canick 2012). There is no reported difference in ccfDNA concentration between monochorionic and dichorionic multifetal pregnancies (Attilakos 2011). However, dichorionic pregnancies complicate gNIPT analysis by the presence of an additional genome (or more in the presence of more than two fetuses) as opposed to the two genomes of mother and fetus present in singleton or monochorionic twin pregnancies.
Next generation sequencing (NGS) applied on DNA extracted from the plasma of pregnant women generates millions of DNA sequences from both maternal and fetal genomes in relative proportion to their original abundance (for technical details see Appendix 3). The data thus produced can be used to detect a slight excess (or loss) of fetal genomic material associated with cases of fetal aneuploidy (Papageorgiou 2012). These NGS technologies have paved the way for the development of gNIPT by alleviating the need for fetal‐specific genetic markers and with potentially better test accuracy than current fetal aneuploidy screening methods.
Currently, gNIPT for the detection of common aneuploidies has been developed by companies in America, Asia and Europe and are commercially available. As part of their marketing material, these companies have published the diagnostic performance of their respective tests on their websites (Table 9). In addition, several research and clinical laboratories have developed in‐house gNIPT.
2. Reported accuracy of commercially available genomics‐based non‐invasive prenatal testsa.
|
Test name (Company, country) |
Method | Aneuploidy |
Reported sensitivity % (95% CI) |
Reported specificity % (95% CI) |
Reported false positive rate % |
| Bambni™ Test (Berry Genomics Co. Ltd, China) |
MPSS | T21 | 100.0 (ND) | > 99.9 (ND) | < 0.1 |
| T18 | 100.0 (ND) | > 99.9 (ND) | < 0.1 | ||
| T13 | 100.0 (ND) | > 99.9 (ND) | < 0.1 | ||
| 45,X | 100.0 (ND) | 99.8 (ND) | 0.0 | ||
| 47,XXX | 100.0 (ND) | 100.0 (ND) | 0.1 | ||
| 47,XXY | 100.0 (ND) | 100.0 (ND) | 0.0 | ||
| 47,XYY | 100.0 (ND) | 100.0 (ND) | 0.0 | ||
| GENOMOM (Genome Care, Korea) |
MPSS | T21, T18 and T13 |
99.0 (ND) | ND | ND |
| SCA | 95.0 (ND) | ND | ND | ||
| Harmony™ prenatal test (Ariosa Diagnostics, Inc., USA) |
Oligo TMPS | T21 | > 99.0 (ND) | > 99.9 (ND) | < 0.1 |
| T18 | 97.4 (ND) | > 99.9 (ND) | < 0.1 | ||
| T13 | 93.8 (ND) | > 99.9 (ND) | < 0.1 | ||
| 45,Xb | 96.3 (81.7 to 99.8) | 99.5 (98.1 to 99.9) | 0.5 | ||
| 47,XXXb | 100.0 (ND) | 99.5 (98.1 to 99.9) | 0.5 | ||
| 47,XXYb | 100.0 (61.0 to 100.0) | 100.0 (99.0 to 100.0) | 0.0 | ||
| IONA® test (Premaitha Health plc, UK) |
MPSS | T21 | > 99.0 (ND) | > 99.0 (ND) | < 1.0 |
| T18 | > 99.0 (ND) | > 99.0 (ND) | < 1.0 | ||
| T13 | > 99.0 (ND) | > 99.0 (ND) | < 1.0 | ||
| (Laboratoire CERBA, France) |
MPSS | T21, T18 and T13 |
> 99.8 (ND) | > 99.8 (ND) | < 0.2 |
| MaterniT21™ Plus test (Sequenom Inc., USA) |
MPSS | T21 | 99.1 (96.6 to 99.9) | 99.9 (99.7 to 99.9) | 0.1 |
| T18 | > 99.9 (93.9 to 100.0) | 99.6 (99.3 to 99.7) | 0.4 | ||
| T13 | 91.7 (61.0 to 99.0) | 99.7 (98.5 to 99.5) | 0.3 | ||
| combined sex aneuploidies |
96.2 (ND) | 99.7 (ND) | 0.3 | ||
| MomGuard™ (LabGenomics, Korea) |
MPSS | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
> 99.0 (ND) | ND | ND |
| NIFTY™ test (Bejing Genomics Institute (BGI), China) |
MPSS | T21 | 99.2 (ND) | 100 (ND) | 0 |
| T18 | 98.2 (ND) | 100 (ND) | 0 | ||
| T13 | 100 (ND) | 100 (ND) | 0 | ||
| 45,X | > 99.9 (ND) | > 99.9 (ND) | < 0.1 | ||
| Panorama™ prenatal testc (Natera, Inc., USA) |
SNP TMPS | T21 | > 99.9 (ND) | 100 (ND) | 0 |
| T18 | > 96.4 (ND) | > 99.9 (ND) | < 0.1 | ||
| T13 | > 99.9 (ND) | 100 (ND) | 0 | ||
| 45,X | > 92.9 (ND) | > 99.9 (ND) | < 0.1 | ||
| PrenaTest® (LifeCodexx AG, Germany) |
MPSS | T21 | 98.7 (ND) | 99.9 (ND) | 0.1 |
| T18 | 100 (ND) | ||||
| T13 | 100 (ND) | ||||
| 45,X | 90.9 (ND) | 98.8 (ND) | 1.2 | ||
| 47,XYY | 100 (ND) | ||||
| Prendia (Genesupport, Switzerland) |
MPSS | T21 | 100.0 (88.8 to 100.0) | 100.0 (98.0 to 100.0) | 0.0 |
| T18 | 95.8 (76.8 to 99.7) | 100.0 (97.0 to 100.0) | 0.0 | ||
| T13 | 100.0 (74.6 to 100.0) | 100.0 (98.1 to 100.0) | 0.0 | ||
| 45,X | 100.0 (74.6 to 100.0) | 100.0 (98.1 to 100.0) | 0.0 | ||
| 47,XXX | 100.0 (46.2 to 100.0) | 100.0 (98.2 to 100.0) | 0.0 | ||
| Tranquility (Genoma, Switzerland) |
MPSS | T21 | 99.9 (ND) | 99.8 (ND) | 0.2 |
| T18 | 99.9 (ND) | 99.9 (ND) | 0.1 | ||
| T13 | 99.9 (ND) | 99.7 (ND) | 0.3 | ||
| verifi® prenatal test (Illumina, Inc., USA) |
MPSS | T21 | 99.5 (98.7 to 99.5) | 99.8 (98.9 to 99.9) | 0.2 |
| T18 | 97.3 (94.2 to 98.2) | 99.7 (99.5 to 99.9) | 0.3 | ||
| T13 | 98.0 (95.6 to 98.9) | 99.8 (99.8 to 99.9) | 0.2 | ||
| 45,X | 95.0 (75.1 to 99.9) | 99.0 (97.6 to 99.7) | 1.0 | ||
| VisibiliT™ (Sequenom Inc., USA) |
MPSS | T21 | > 99.0 (80.8 to 100) | > 99.9 (99.5 to 100) | < 0.1 |
| T18 | > 99.0 (65.5 to 100) | > 99.9 (99.5 to 100) | < 0.1 |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13 CI: confidence interval, MPSS: massively parallel shotgun sequencing, ND: no data available, TMPS: targeted massively parallel sequencing and SNP: single nucleotide polymorphism.
a(Ariosa Diagnostics 2016; BGI 2014; BGI 2016; Berry Genomics 2016; Genoma 2016; Genome Care 2016; Illumina 2014; Illumina 2016; LabGenomics 2016; LifeCodexx 2016; Natera 2016; Genesupport 2016; Premaitha Health plc 2016; Sequenom 2016).
b(Hooks 2014).
cDNA of maternal and paternal origin are needed.
Before taking a personal decision to accept or decline gNIPT, pregnant women should be given information on the screening process, which must include a discussion with a health professional (Gagnon 2010; Legare 2010; Legare 2011; St‐Jacques 2008). Following screening, the results should be explained in the context of the harms and benefits of definitive diagnosis through non directive counselling (Benn 2013b). In their recent guideline, the American College of Obstetricians and Gynecologists (ACOG) recommends that gNIPT should not be used to replace diagnostic testing and that all pregnant women with a positive gNIPT result should have a diagnostic procedure before undertaking any irreversible action such as pregnancy termination. Guidelines also recommend that pregnant women with an unreported, indeterminate or uninterpretable gNIPT result should receive further genetic counselling and be offered comprehensive ultrasound evaluation and diagnostic testing (ACOG #163 2016).
Clinical pathway
Prior test(s)
Prenatal screening for fetal aneuploidy (mostly T21) is part of public health programs in most upper‐middle and high‐income countries and is typically offered to all pregnant women (Benn 2013b; Chitayat 2011). Up to now, screening tests for aneuploidies have relied on blood‐based biochemical testing of placental markers with or without ultrasound imaging to assess for nuchal translucency thickness and other markers of fetal aneuploidy in the first trimester. The age of the pregnant woman is combined with levels of biomarkers and nuchal translucency as predictive markers for T21 in the first or second trimester (Benn 2011; Chitayat 2011; Summers 2007). Table 10 presents the various testing combinations (e.g. sequential, integrated or contingent algorithms) that have been described and are currently in use in prenatal clinics (Alldred 2017b). The screening performance of these algorithms is mostly related to the detection rates of different marker combinations and the accepted level of false positive rates. A large prospective Canadian study of 32,227 pregnant women showed that the detection rate of existing screening strategies for T21 can reach about 88.4%, with a screen‐positive rate of 3.3% when applying the integrated prenatal screening procedure (Okun 2008).
3. Traditional screening tests (mostly for T21)a.
| Screening tests |
First trimester (before 14 weeks’ gestation) |
Second trimester (14 to 20 weeks’ gestation) |
| Ultrasonography |
|
|
| Combined test |
|
NA |
| Triple test | NA |
|
| Quadruple test | NA |
|
| Sequential testb |
|
|
| Contingent testb |
|
|
| Serum integrated testc |
|
|
| Integrated testc |
|
|
Maternal age is often included in the algorithm for prenatal screening tests. AFP: alpha‐fetoprotein, hCG: human chorionic gonadotropin, NA: not applicable, NT: nuchal translucency, PAPP‐A: pregnancy associated plasma protein A and uE3: unconjugated estriol.
a(Gekas 2009; Okun 2008; Wald 2005). bA test result was available after first‐trimester screening test. cSingle test result available after second‐trimester screening test.
A woman is classified as screen‐positive if her risk is equal to or exceeds a predetermined threshold following prenatal screening result or due to some other factors such as personal or familial history of aneuploidies or translocations. Although these factors are considered to significantly increase the risk of fetal aneuploidy, the indications for invasive testing may vary between countries. To confirm the presence or absence of fetal aneuploidy in these high‐risk pregnant women, a diagnostic test involving karyotyping by an invasive procedure such as amniocentesis or chorionic villi sampling (CVS) is offered (ACOG #88 2007; Benn 2011; Chitayat 2011). Karyotyping by traditional banding techniques of fetal cells obtained from amniotic fluid or placental tissue has been considered the standard of care for prenatal diagnosis of aneuploidies (ACOG #545 2012; Benn 2013a; ICFMM 2013). Fluorescence in situ hybridisation (FISH) and quantitative fluorescence polymerase chain reaction (QF‐PCR) are appropriate standards of care for pregnant women at increased risk of common fetal aneuploidies based on screening results (Duncan 2011; Langlois 2011; South 2013). Microarray analysis by array comparative genomic hybridisation (aCGH) is recommended in pregnancies with fetal anomalies and it is increasingly replacing karyotyping (ACOG #682 2016).
Five reviews published in the Cochrane Library examined serum, urine, ultrasound or a combination of these tests for T21 screening. For first‐trimester serum tests (Alldred 2015a), the authors concluded that two markers in combination with maternal age, specifically pregnancy associated plasma protein A (PAPP‐A) and free human chorionic gonadotropin (hCG) are significantly better than those involving single markers combined with or without maternal age. For second‐trimester serum tests (Alldred 2012), the authors concluded that two or more markers, with or without inhibin A, in combination with maternal age are significantly more sensitive than one marker alone. Their review also showed that no test combination was superior to the others and therefore it was not possible to recommend a specific test combination. For first‐trimester ultrasound tests alone of in combination with first‐trimester serum tests (Alldred 2017a), the authors concluded that test strategies that combine ultrasound markers with serum markers, especially PAPP‐A and free ßhCG, and maternal age were significantly better than those involving only ultrasound markers (with or without maternal age) except nasal bone. For first‐ and second‐trimester serum tests with and without first‐trimester ultrasound tests (Alldred 2017b), the authors concluded that tests involving first‐trimester ultrasound with first‐ and second‐trimester serum markers in combination with maternal age are significantly better than those without ultrasound, or those evaluating first‐trimester ultrasound in combination with second‐trimester serum markers, without first‐trimester serum markers. For first‐ and second‐trimester urine tests (Alldred 2015b), the authors concluded that second‐trimester ß‐core fragment and oestriol with maternal age are significantly more sensitive than the single marker second‐trimester ß‐core fragment and maternal age. However, there were few studies and the evidence does not support the use of urine tests for T21 screening for the first 24 weeks of pregnancy.
Role of index test(s)
Genomics‐based non‐invasive prenatal testing such as MPSS or TMPS could be offered to pregnant women after a first‐tier screening and before a diagnostic test in order to better identify which pregnant women at increased risk of fetal aneuploidy should be offered further testing (triage) (Figure 2). The use of such NGS‐based approaches has also been suggested as a replacement for current first‐tier screening tests (biochemical, ultrasound or both) or as potential diagnostic tests to replace current diagnostic test (karyotyping of fetal cells from amniocentesis or CVS) (Bianchi 2012).
2.
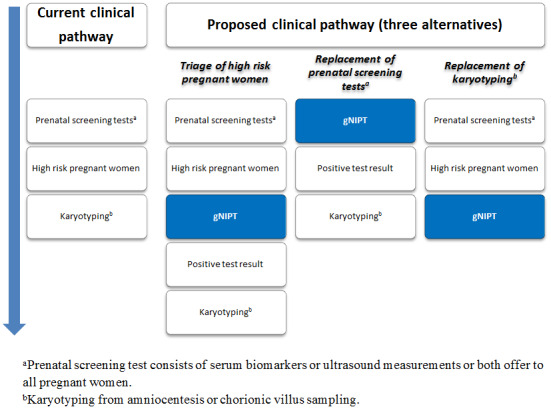
Current clinical pathway and three proposed uses of genomics‐based non‐invasive prenatal testing (gNIPT). Currently (on the left), pregnant women can have a prenatal screening test consisting of biomarkers or ultrasound, or both. For high‐risk pregnant women, an invasive diagnostic test (karyotyping) is offered. In the present review, we propose 3 different clinical pathways. First, gNIPT could be used as a triage test, to decide which pregnant women should receive further testing. Second, gNIPT could be used to replace current prenatal screening tests. Finally, gNIPT could be used to replace current invasive diagnostic tests (if diagnostic performance permits). At any point in a clinical pathway, a pregnant woman may decide not to proceed with other tests (not shown in the figure). Figure was designed by CL, JB, MB and YT.
Rationale
Current screening tests (biochemical, ultrasound or both) have relatively high false positive rates, which may result in undue anxiety for many pregnant women who will be offered an invasive diagnostic procedure. For example, at a prenatal screening risk cut‐off of 1:300, fetal aneuploidy is confirmed by karyotyping in only about 1/34 to 1/14 (3% to 7%) screen‐positive cases (Renshaw 2013; Wald 2005). As a result, many more women will undergo invasive diagnostic testing following positive screening tests than the number carrying a fetus with aneuploidy. In France, each year, about 800,000 pregnant women opt for prenatal T21 biochemical screening, ultrasound measurements or both, and about 24,000 of them (3%) will have karyotype testing (Basset 2013). Invasive testing methods for prenatal diagnosis of aneuploidy identify pregnancies with fetal chromosomal abnormalities, but contribute to an additional procedure‐related fetal loss rate (Wilson 2007). A recent meta‐analysis showed that weighted pooled procedure‐related risks of miscarriage of invasive testing methods before 24 weeks' gestation were 0.11% for amniocentesis and 0.22% for CVS (Akolekar 2015). The risk of miscarriage of normal fetuses associated with such invasive procedures has fostered the development of alternative screening and diagnostic approaches.
The discovery of fetal circulating cells and fetal ccfDNA in maternal blood during pregnancy has enabled the development of non‐invasive methods to analyse the fetal genome (Birch 2005; Lo 1997; Wright 2009). Fetal DNA offers advantages over circulating fetal cells because it is more easily extracted from maternal plasma samples and it disappears within hours after birth (undetectable about one to two days postpartum), as compared to the paucity and persistence of fetal cells in maternal blood over several consecutive pregnancies (up to 27 years) (Wright 2009; Yu 2013). At present, the analysis of ccfDNA by NGS technologies seems to be the most promising alternative gNIPT approach for the detection of fetal aneuploidies from maternal blood. This allows sequencing of tens of millions of these DNA fragments simultaneously, paving the way for the development of a non‐invasive, less psychologically stressful method potentially able of detecting fetal aneuploidies earlier and with better accuracy than current screening programs. As such, NGS technologies have the potential to radically change prenatal screening for fetal aneuploidy. Indeed, a study exploring the impact of gNIPT on prenatal care showed that more pregnant women with positive first‐trimester screening opt for further testing (from 47.2% to 78.8%) than before the introduction of gNIPT, while the rate of invasive diagnostic testing has decreased significantly (from 47.2% to 39.2%). Additionally, fewer pregnant women declined follow‐up testing when gNIPT was an option (from 52.8% to 21.2%) (Chetty 2013). Another study suggested that gNIPT could reduce procedure‐related fetal losses in high‐risk women by up to 88% (O'Leary 2013).
For instance, the new gNIPT approach is reported to detect aneuploidy with high sensitivity to select a subset of pregnant women for an invasive diagnostic procedure and could be performed in high‐risk pregnant women (as a second‐tier test) following a positive screening result (Benn 2013a). The major expected advantage of gNIPT by NGS over current (biochemical, ultrasound or both) screening tests is the significant decrease in false positive results and thus the reduction of invasive procedures and their associated normal fetus losses. Also, it was reported that a reduction of invasive prenatal procedures with the introduction gNIPT has indeed been documented (Chetty 2013; Larion 2014; Tiller 2014). Assessment of how NGS should be used in clinical practice for aneuploidy detection is currently being studied. NGS approaches could also be performed in general obstetrical population (as first‐tier test), in place of current screening algorithms (biochemical, ultrasound or both) (Figure 2). However, the field is moving rapidly. From January to July 2014, around 60 NIPT studies were published in PubMed compared to 70 studies in 2013 and 40 studies in 2012.
Up to now, no comprehensive systematic review including meta‐analyses has analysed and compared the diagnostic accuracy of MPSS and TMPS methods for the detection of fetal aneuploidies, either as a second‐tier test (i.e. in women at increased risk of fetal aneuploidy after current screening procedures) or as a first‐tier test (i.e. in all pregnant women). Benn 2013b published a review on gNIPT focused on providing the information needed by clinicians and public health providers before implementation of this technology in routine clinical practice. However, their review included only T21 and T18. Mersy 2013 published a systematic review on quality and outcome of diagnostic test accuracy studies on non‐invasive detection of fetal T21 only. One updated meta‐analysis (Gil 2015a) pooled all gNIPT methods but did not assess the relative performance of MPSS and TMPS technologies separately. More recently, Taylor‐Phillips 2016 published a meta‐analysis on gNIPT accuracy for major autosomal anomalies (T21, T18 and T13) without sex chromosome aneuploidies (SCAs) assessment and using restrictive inclusion criteria for included publications (e.g. limited to the English language, cohorts of more than 50 pregnant women) and including studies with incomplete follow‐up (pregnant women without reference standard). In the meta‐analysis of Mackie 2017, multifetal pregnancies and case‐control study design were excluded. In the meta‐analysis published by the Haute Autorité de Santé in France (HAS 2015), the accuracy of gNIPT was evaluated for T21 only and included studies with pregnant women selected at high risk of fetal aneuploidy as well as studies with pregnant women unselected for their risk (general population). Only studies published in English were included. The review of Agarwal 2013 described the properties of commercial tests available (e.g. type of gNIPT method, costs, turnaround times and reimbursement), intellectual property, commercialisation, patenting, patenting litigation and licensing landscape of technologies underlying these tests.
Genomics‐based non‐invasive prenatal tests are already advertised and marketed to North‐American, European and Asian healthcare providers. Leading companies are summarised in Table 9. Other entities are trying to make their way into the market (Birmingham Women's NHS; Counsyl; GENDIA; Genesis Genetics; Integrated Genetics; NIPD Genetics; Progenity; Quest Diagnostics; RAVGEN; Xcelom). Some of these assays have yet to be approved by the US Food and Drug Administration. There is significant pressure for increasing their use in clinical practice, but comparative effectiveness and cost‐effectiveness studies, as well as studies of the ethical, legal and social issues are scarce. Furthermore, tools needed for their patient value‐based implementation are not available or have not been validated.
Objectives
To evaluate and compare the diagnostic accuracy of massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS) using circulating cell‐free DNA (ccfDNA) in maternal blood for the detection of common fetal aneuploidies (T21, T18, T13, 45,X, 47,XXY, 47,XXX and 47,XYY) according to their prior risk of fetal aneuploidy. The genomics‐based non‐invasive prenatal testing (gNIPT) results were confirmed by a reference standard such as fetal karyotype or neonatal clinical examination.
To evaluate the screening performance of MPSS and TMPS as triage tests (a second‐tier screening test) for identifying which pregnant women at increased risk of fetal aneuploidy should be offered further testing, that is, after a first‐tier screening, but before a diagnostic test.
To assess the screening performance of MPSS and TMPS as a first‐tier test in pregnant women without prior risk (i.e. in unselected pregnant women or general population) as a replacement for current offered first‐tier tests (biochemical, ultrasound or both).
To assess the diagnostic performance of MPSS and TMPS as a second‐tier test as potential diagnostic tests to replace current invasive diagnostic tests.
Secondary objectives
To investigate potential sources of heterogeneity that may influence the diagnostic accuracy of MPSS and TMPS such as gestational age at the time of blood collection and type of reference standard used.
Methods
Criteria for considering studies for this review
Types of studies
We included studies that met the following inclusion criteria:
randomised studies where pregnant women were randomised to receive one gNIPT (MPSS or TMPS) as well as the reference standard;
retrospective and prospective cohort studies where all pregnant women were tested with one or more gNIPT methods and the reference standard (including head‐to‐head studies); and
retrospective and prospective case‐control studies comparing one or more of the gNIPT methods with the reference standard.
Although studies with a retrospective or case‐control design are prone to biases, we included such studies because we anticipated a paucity of other study designs. When data were sufficient, we explored the effect of excluding case‐control studies in sensitivity analyses.
We excluded studies for which it was not possible to extract or derive the number of true positives, false positives, false negatives and true negatives.
Participants
We included women of any age, ethnicity and gestational age with a singleton or multifetal (monochorionic and dichorionic) pregnancy.
Index tests
Genomics‐based non‐invasive prenatal tests based on plasma ccfDNA in maternal blood, analysis by either MPSS or TMPS methods.
Target conditions
We considered seven fetal aneuploidies, namely T21, T18, T13, 45,X, 47,XXY, 47,XXX and 47,XYY.
Reference standards
We considered the following test as reference standard: fetal karyotyping performed on cells obtained from chorionic villi sampling (CVS), amniotic fluid, placental tissue, a fetus lost by miscarriage or other equivalent and recognised methods on the same materials. By "fetal karyotyping" we mean traditional banding techniques, spectral karyotyping, fluorescence in situ hybridisation (FISH), array comparative genomic hybridisation (aCGH) or quantitative fluorescence polymerase chain reaction (QF‐PCR). If fetal karyotyping was not performed, we used neonatal clinical examination or medical records from birth as a secondary reference standard for T21, T18 or T13. For sex chromosome aneuploidies (SCA), only fetal karyotype was an appropriate reference standard because newborns usually have a normal phenotype.
Search methods for identification of studies
Electronic searches
We used a sensitive search strategy that included the following three sets of search terms and synonyms:
index test (e.g. cell‐free DNA, sequencing, non‐invasive and genetic diagnosis);
participants' description (e.g. pregnant women, fetus and prenatal); and
target condition (e.g. aneuploidy and chromosome anomalies).
We combined free‐text words and subject headings used within each set with the Boolean operator OR and then combined the three sets using AND. We reviewed publications from 1st January 2007 because MPSS and TMPS were introduced in the literature in 2008 (Chiu 2008; Fan 2008). We did not limit our search by language, search filter or publication type (e.g. journal article, clinical trial, validation study, review and comment).
We applied a comparable search strategy (Appendix 4) with adaptations for each of the following databases:
MEDLINE (Ovid) (January 2007 to July 2016);
Embase (January 2007 to July 2016);
Web of Science (ISI) (January 2007 to July 2016);
Cochrane Register of Diagnostic Test Accuracy Studies, Cochrane Library (January 2007 to October 2016);
ClinicalTrials.gov (January 2007 to September 2016);
European Clinical Trials Register (January 2007 to September 2016);
WHO ICTRP (January 2007 to September 2016);
The National Technical Information Service (NTIS) (January 2007 to September 2016);
OpenGrey (January 2007 to October 2016); and
National Guideline Clearing House (January 2007 to September 2016).
Searching other resources
We examined references cited in potentially relevant full‐text papers and those cited in previous reviews by cross‐checking bibliographies. We examined grey literature by searching data available on the websites of private prenatal diagnosis companies (Ariosa Diagnostics 2016; BGI 2016; Berry Genomics 2016; Genoma 2016; Genome Care 2016; Illumina 2016; LabGenomics 2016; LifeCodexx 2016; Natera 2016; Genesupport 2016; Premaitha Health plc 2016; Sequenom 2016) using gNIPT technologies (January 2007 to December 2016). We also searched for conference abstracts and theses in appropriate sources (e.g. TheseNet, Theses Canada Portal) (January 2007 to October 2016).
Data collection and analysis
We used the methods suggested by the Cochrane Diagnostic Test Accuracy Working Group (Deeks 2013). For selection of studies, data extraction and assessment of methodological quality, we conducted a pilot using 20 randomly selected articles to trial our forms in order to ensure criteria were applied consistently.
None of the review authors involved in conducting a gNIPT primary study (FL, FR, SL and YG) took part in the selection of studies, nor in any decisions/analyses related to their own studies. Furthermore, by the final date of data collection, these authors had not published a primary gNIPT study.
Selection of studies
Two review authors (MB and CL) independently identified relevant studies by screening the titles and abstracts of all studies identified by the search strategy. We obtained the full‐text version of all potentially relevant studies and assessed them for inclusion by using a study eligibility table based on prespecified inclusion criteria. The data collection form (Excel® format) for classifying studies during the full‐text assessment is presented in Appendix 5. We considered all comments, statements or errata related to included studies. We excluded studies that did not match the inclusion criteria and we recorded the reason(s) for exclusion. If results from the same study cohort were reported in multiple publications, we considered all the publications and included results from the most relevant and comprehensive publications. We excluded papers with preliminary results whose full published results were available. We resolved any disagreement between assessors (MB and CL) by iteration, discussion and consensus. If required, we consulted a third review author (JB or LN).
Data extraction and management
Two review authors (MB and CL, JB or LN) independently extracted information and data from each included study by using a data extraction form that we developed in Excel® format. We included the following items:
study characteristics (e.g. reference details allowing identification of the publication, language and study design);
population characteristics (e.g. gestational age, maternal age, ethnicity, total number of pregnant women, number of aneuploid cases, number of euploid cases, recruitment location (country, geographic locations or regions), recruitment period and other relevant tests carried out prior to index test (e.g. ultrasonography, biochemical screening));
features of the reference standard (e.g. fetal karyotyping, chromosome analysis or clinical examination);
features of the index test (e.g. technical details, commercial or in‐house gNIPT, cutpoint, failure rate, blood sample collection time (before or after reference standard) and first‐tier test or second‐tier test); and
data for constructing two‐by‐two tables (number of true positives, false positives, false negatives and true negatives) or summary statistics from which the data were derived. In the two‐by‐two tables, the true negative cases were patients with any other aneuploidy than the one under analysis and all euploid cases were considered unaffected. When data were presented in three‐by‐two tables due to unclassified index test results (defined as grey zone between positive and negative test results), we constructed two‐by‐two tables by considering all unclassified gNIPT results as test positives. This is because in practice such results will lead to further testing and investigation to ensure a case of fetal aneuploidy is not missed.
We cross‐checked all extracted and recorded data and we resolved any disagreement by iteration, discussion and consensus between two review authors (MB and CL, JB or LN). If required, we consulted a third author (JB, LN or CL). We wrote to the study contact author if information was missing or unclear or to clarify potential overlap between publications based on the same dataset to avoid including the same women more than once. If an article presented results including other aneuploidies than the ones under review, we considered only the subset of the cohort with the aneuploidies of interest.
Assessment of methodological quality
We used the revised QUality Assessment of Diagnostic Accuracy Studies (QUADAS‐2) tool for assessment of methodological quality of included studies (Whiting 2011). We tailored the tool to this review question using the operational criteria detailed in Appendix 6 to answer signalling questions and make the overall judgment of risk of bias and applicability concerns for each domain of the tool. We answered each signalling question with a ‘yes’, ‘no’ or ‘unclear’ response for each included study and we recorded the reason for the judgment made. If a study was recorded as ‘yes’ on all signalling questions related to risk of bias, then it was deemed appropriate to have an overall judgment of ‘low risk of bias’. If a study is recorded ‘no’ or ‘unclear’ on one or more signalling questions in a domain, then it was judged as having ‘high or unclear risk of bias’. Judgments about applicability concern were rated as ‘low’, ‘high’ or ‘unclear’ in relation to our review question. ‘Unclear concern’ was used only if insufficient information was available. Two review authors (MB and CL, JB or LN) independently applied the QUADAS‐2 tool to each included study and we resolved any disagreement by iteration, discussion and consensus. If required, we consulted a third review author (JB, LN or CL).
Statistical analysis and data synthesis
The unit of analysis was the pregnant woman irrespective of the type of pregnancy (multifetal or singleton pregnancy). We evaluated the performance of MPSS and TMPS for the detection of each type of aneuploidy under study both individually and globally for any type of aneuploidy (all autosomal aneuploidies combined and all sex chromosomal aneuploidies combined). We distinguished between each of the following groups of pregnant women and performed separate analyses for each subgroup:
unselected pregnant women undergoing aneuploidy screening (first‐tier gNIPT, i.e. offered to all pregnant women) and women selected at high risk of fetal aneuploidy (second‐tier gNIPT);
women with singleton and multifetal pregnancy because ccfDNA's fetal fraction in multifetal pregnancy is higher than in singleton pregnancy (Attilakos 2011; Canick 2012); and
pregnant women who underwent gNIPT during the first trimester (15 weeks or less), the first or second trimester (29 weeks or less) or at any time during pregnancy (42 weeks or less).
For each gNIPT method, we used Review Manager® to produce coupled forest plots of sensitivity and specificity, together with their 95% confidence intervals (CIs). We also plotted study‐specific estimates of sensitivity and specificity in receiver operating characteristic (ROC) space. All gNIPTs are laboratory‐developed tests based on differently calibrated assays with specific cutpoints to classify samples as euploid or aneuploid. There is no consensus on the cutpoints to use in practice. For this reason, we had planned to use a modelling strategy that focuses on the estimation of summary ROC curves (Macaskill 2010; Rutter 2001) and to estimate summary points (summary sensitivity and specificity) if a sufficient number of studies reported common cutpoints. However, given the qualitative nature of the cutpoints, which is highly dependent on each laboratory's developed gNIPT and study populations, it was not possible to identify a common cutpoint. Therefore, we reasoned that this was a special case where we can assume gNIPT results were binary (positive or negative). The rationale was further strengthened by the lack of apparent threshold effect when we examined the studies in ROC space. If a study reported more than one cutpoint, we considered all cutpoints and chose one cutpoint, the most commonly reported across all studies, such that only one pair of sensitivity and specificity from a study was included in meta‐analysis.
Due to limited or absence of threshold effect, there was no requirement to account for correlation between sensitivity and specificity across studies in meta‐analysis. Therefore, we removed the correlation parameter from the bivariate model (Chu 2006), thus simplifying the model to two univariate random‐effects logistic regression models for separate meta‐analyses of sensitivities and specificities (Takwoingi 2015). In cases where there were few studies in the meta‐analysis or a random‐effects analysis failed to converge, we used fixed‐effect logistic regression models. Where all studies in the meta‐analysis reported 100% sensitivity or 100% specificity, these fixed‐effect models fail as the prediction is perfect. Therefore, in such situations we used simple pooling by summing up the numbers of true positives and total cases to compute sensitivity, and the numbers of true negatives and unaffected pregnancies to compute specificity. CIs were obtained using the Wilson method (Newcombe 1998).
We compared the diagnostic accuracy of MPSS and TMPS by first using all available data (indirect comparison). If studies that compared MPSS and TMPS in the same population (head‐to‐head or direct comparison) were available, we had planned a second set of analyses restricted to direct comparisons. Comparative meta‐analyses were done by adding a covariate for test type to random‐effects or fixed‐effect models. We used likelihood ratio tests to assess the statistical significance of differences between tests by comparing models that included covariate terms for test type with models that did not include the terms. If data were available, comparisons between gNIPTs and traditional screening approaches were planned using a similar strategy to that described above. Meta‐analyses were performed using the xtmelogit and blogit functions in the Stata software package (version 13; StataCorp, College Station, Texas 77845, USA). When meta‐analyses of direct comparisons were not possible, we examined individual study results. For each comparative study, we computed differences in sensitivity and specificity, and 95% CIs were calculated for the differences using the Newcombe‐Wilson method without continuity correction (Newcombe 1998).
Investigations of heterogeneity
We examined forest plots of sensitivity and specificity and summary ROC plots for each gNIPT method to visually assess heterogeneity. If sufficient data were available for meta‐regression (by adding a covariate to a logistic regression model to explore its effect on sensitivity and specificity), we had planned to investigate the effect of the following:
study population (e.g. ethnicity, gestational age at blood collection); and
type of reference standard (i.e. karyotype or mixed reference standard).
However, formal investigations using meta‐regression were not possible due to limited data and little or no heterogeneity in test accuracy.
Sensitivity analyses
We performed sensitivity analyses to assess the effect of excluding case‐control studies and studies with a small number of cases of aneuploidy (less than 10 cases) on the summary estimates of test accuracy.
We had planned to also assess the effect of:
studies where pregnant women received an invasive diagnostic test less than one day before blood collection for gNIPT;
third trimester gestational age at the moment of blood collection for gNIPT;
studies available only as abstracts; and
studies at ‘high or unclear risk of bias’ according to the QUADAS‐2 assessment tool.
However, due to lack of data or lack of variability in estimates of sensitivity and specificity, only assessments of the impact of study design and number of cases were performed.
Results
Results of the search
We found a total of 11,912 articles through our electronic searches from January 2007 to October 2016 (see PRISMA study flow diagram in Figure 3). A total of 11,700 articles were identified through databases (941 through MEDLINE, 8381 through Embase, 1986 through Web of Science, 18 through Cochrane Diagnostic Test Accuracy register of studies, 245 through ClinicalTrial.gov, 43 through European Clinical Trials Register, 21 through WHO ICTRP, 34 through NTIS, 19 through OpenGrey and 12 through the National Guideline Clearing House). We found 212 publications through other sources (two articles received from the author, 175 from gNIPT company’s website, 27 from TheseNet and eight from These Canada Portal). After removing 2354 duplicates, two review authors independently screened the titles and abstracts of 9558 publications. Of the 9558 publications, 9209 were deemed irrelevant to our review question. We retrieved the full texts of the remaining 349 articles to assess their eligibility. After resolving disagreement between two or three review authors, 261 articles were excluded (see Characteristics of excluded studies for details) and 63 articles fulfilled our inclusion criteria (see Characteristics of included studies for details). Among these 63 articles, 62 were journal articles and one was a letter to the editor with sufficient information to be included (Jackson 2014). From the 63 articles, two articles presented two studies (two different cohort, two 2x2 tables). At all, we included 65 studies of 86,139 pregnant women (3141 aneuploids and 82,998 euploids). No studies are awaiting classification. We identified 25 ongoing trials through clinical trials databases (see Characteristics of ongoing studies for details). We will consider these trials in future updates.
3.

PRISMA flow diagram for selection of studies from January 2007 to October 2016.
#: number, DTA: diagnostic test accuracy, NTIS: The National Technical Information Service and WHO ICTRP: World Health Organization International Clinical Trials Registry Platform.
Basic features of the included studies
The clinical characteristics of pregnant women and sequencing method were generally well described or referenced. Some studies did not clearly report how patient selection was done and which inclusion and exclusion criteria were applied. Patients’ enrolment flow‐charts, pregnancy outcome flow‐chart and 2 x 2 tables were unclear in many studies. We therefore contacted study authors to clarify unclear information, to obtain missing data or to clarify potential overlap of patients between publications.
We described the characteristics of included studies in Characteristics of included studies table and provided a summary in Table 11. Forty‐two studies (65%) enrolled pregnant women selected at high risk of fetal aneuploidy (Alberti 2015; Ashoor 2012; Benachi 2015; Bianchi 2012; Bianchi 2013; Bijok 2014; Canick 2012; Chen 2011; Ehrich 2011; Hall 2014; Hooks 2014; Hou 2012; Huang 2014; Jeon 2014; Jiang 2012; Johansen 2016; Ke 2015; Kim 2016; Lee 2015; Lefkowitz 2016; Liang 2013; Liu 2012; Mazloom 2013; Nicolaides 2013; Nicolaides 2014a; Norton 2012; Palomaki 2012; Papageorghiou 2016a; Papageorghiou 2016b; Persico 2016; Poon 2016; Porreco 2014; Sehnert 2011; Song 2015; Sparks 2012a; Stumm 2014; Sukhikh 2015; Sung‐Hee 2015; Verweij 2013; Wang 2014; Wang 2015a; Zhang 2016); five studies (8%) enrolled pregnant women without prior risk of fetal aneuploidy (del Mar Gil 2014; Nicolaides 2012; Norton 2015; Quezada 2015; Song 2013); and 18 studies (28%) enrolled pregnant women from a mixed risk cohort of fetal aneuploidy (Ashoor 2013; Bevilacqua 2015; Bianchi 2014a; Chiu 2011; Comas 2015; Fiorentino 2016; Gil 2016; Jackson 2014; Korostelev 2014; Lau 2012; Ma 2016; Pergament 2014; Samango‐Sprouse 2013; Shaw 2014; Tynan 2016; Yao 2014; Zhou 2014a; Zhou 2014b). Mixed‐risk samples included a mixture of selected pregnant women with low, high or no prior risk of fetal aneuploidy. Such samples do not represent the real‐life situation (i.e. using gNIPT as a first‐tier screening test or as a second‐tier test) and so such studies were not used for addressing our research objectives. Nevertheless, as we did not pre‐specify exclusion of such studies, we analysed the data and the results are presented in Appendix 7.
4. Characteristics of included studies by type of gNIPT.
| Study ID | Target condition(s) |
Study design and participants |
Prior risk | Index test details | Cutpoint | Reference standard | Comparator |
| MPSS | |||||||
| Alberti 2015 | T21 |
|
High risk |
|
Z score of 3 | Fetal karyotypea | |
| Benachi 2015 | T21, T18, T13 |
|
High risk |
|
Z score of 3 for T21; 3.95 for T18 and T13 | Fetal karyotype or neonatal clinical examination | |
| Bianchi 2012 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype | |
| Bianchi 2013 | T21, T18, T13, 45,X |
|
High risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype | |
| Bianchi 2014a | T21, T18, T13 |
|
High, low and without prior risk |
|
NCV of 4; resequenced if NCV is between 3 and 4 | Fetal or postnatal karyotype, neonatal clinical examination or medical record from birth | Standard screening (T21 only with mixed cutpoints) which include first‐trimester combined test or a second‐trimester result (quadruple, serum integrated, fully integrated, or sequential). |
| Bijok 2014 | T21, T18, T13 |
|
High risk |
|
NR | Fetal karyotype | |
| Canick 2012 | T21, T18, T13 |
|
High risk |
|
Z score of 3 | Fetal karyotype | |
| Chen 2011 | T18, T13 |
|
High risk |
|
Z score of 3 | Fetal karyotype | |
| Chiu 2011 | T21 |
|
Mostly high (> 1/300) and some intermediate risk (between 1/300 and 1/1000) |
|
Z score of 3 | Fetal karyotype | |
| Ehrich 2011 | T21 |
|
High risk |
|
Z score of 2.5 | Fetal karyotype | |
| Fiorentino 2016 | T21, T18, T13 |
|
Mostly high risk and without prior risk |
|
NCV of 4; aneuploidy suspected if NCV is between 3 and 4 | Fetal karyotype or neonatal clinical examination | |
| Hou 2012 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
NR | Fetal karyotype | |
| Huang 2014 | T21, T18 |
|
High risk |
|
L score of 1 and t score of 2.5 including warning zone | Fetal karyotype | |
| Jeon 2014 | T21, T18 |
|
High risk |
|
Z score of 2.566 for T21; 2.459 for T18. | Fetal karyotype | |
| Jiang 2012 | T21, T18, T13, 45,X, 47,XXY, 47, XYY |
|
High risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype | |
| Johansen 2016 | T21, T18, T13 |
|
High risk |
|
Z score of 4 (unclassified if Z score is between 3 and 4) and WISECONDOR of 1% | Fetal karyotype | |
| Ke 2015 | T21, T18, T13 |
|
High risk |
|
T score of 3 | Fetal karyotype or newborn outcome | |
| Kim 2016 | T21 |
|
High risk |
|
Z score of 2.10 for Ion Proton™ | Fetal karyotype | |
| Lau 2012 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
Mostly high risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype | |
| Lee 2015 | T21, T18, T13 and SCA (no case found) |
|
High risk |
|
Z score of 4 (intermediate risk if Z score is between 2.5 and 4) for T21 and T18; 2.8 for T13 (intermediate risk if Z score is between 1.9 and 2.8) | Fetal or neonatal karyotype | |
| Lefkowitz 2016 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype | |
| Liang 2013 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype | |
| Liu 2012 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Z score of 3 | Fetal karyotype | |
| Ma 2016 | T21, T18, T13 |
|
High and low risk |
|
Z score of 3 | Fetal karyotype or postnatal follow‐up | |
| Mazloom 2013 | 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Different cutpoints used for the four SCAb | Fetal karyotype | |
| Palomaki 2012 | T21, T18, T13 |
|
High risk |
|
Z score of 3 for T21; 3.88 for T18; 7.17 for T13 | Fetal karyotype | |
| Papageorghiou 2016a | T21, T18, T13 |
|
High risk |
|
Likelihood ratio of 1 and maternal age‐adjusted probability risk score | Fetal karyotype or medical record from birth | |
| Papageorghiou 2016b | T21, T18, T13 |
|
High risk |
|
Likelihood ratio of 1 and maternal age‐adjusted probability risk score | Fetal karyotype or medical record from birth | |
| Poon 2016 | T21, T18, T13 |
|
High risk |
|
NR (authors used the same gNIPT than Papageorghiou 2016a) | Fetal karyotype | |
| Porreco 2014 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype or medical record from birth | |
| Sehnert 2011 | T21, T18, T13, 45,X |
|
High risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype | |
| Shaw 2014 | T21, T18, T13, 45,X, 47, XXX, 47,XXY, 47,XYY |
|
High and low risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype or medical record from birth | |
| Song 2013 | T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY (SCA data not shown in this review) |
|
Without prior risk |
|
Z score of 3 | Fetal or postnatal karyotype or medical record from birth | Triple test for T21 and T18 (cutpoint of 1 in 270). |
| Song 2015 | T21, T18, T13, 45,X, 47,XXX, 47,XYY |
|
High risk |
|
Z score of 3 | Fetal karyotype or neonatal clinical examination or both | |
| Stumm 2014 | T21, T18, T13 |
|
High risk |
|
MAD‐based Z score of 3 for T21; 3.2 for T18; 3.9 for T13 | Fetal karyotype | |
| Sukhikh 2015 | T21, T18, T13, 45,X |
|
High risk |
|
T score of 5 for T21 and T18; 4 for T13; 0.04 Chrom. X and 0.04 Chrom. Y for 45,X | Fetal karyotype | |
| Sung‐Hee 2015 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
L score of 1 and t score of 2.5 | Fetal karyotype or medical record from birth | |
| Tynan 2016 | T21, T18, T13 |
|
High and without prior risk |
|
risk score of 1% | Fetal karyotype or medical record from birth | |
| Wang 2014 | T21, T18, T13, 45,X |
|
High risk |
|
NR | Fetal or neonatal karyotype or clinical examination at 42 days after birth or both | |
| Wang 2015a | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Z score of 3 for T21, T18 and T13; ‐3 for Chrom. X and 3 for Chrom. Y for sex Chrom. classification. | Fetal karyotype or clinical follow‐up to 6 months from birth | |
| Yao 2014 | T21, T18, T13 and SCA (SCA data not shown in this review) |
|
High, low and without prior risk |
|
Different cutpoints used for autosomes and SCAb | Fetal karyotype or clinical follow‐up | |
| Zhang 2016 | T21, T18, 45,X, 47,XXX (SCA data not shown in this review) |
|
High risk |
|
Z score of 3 for T21 (no other cutpoint reported) | Fetal or neonatal karyotype or neonatal clinical examination | |
| Zhou 2014a | T21, T18, T13 |
|
High, low and without prior risk |
|
L score of 1 and t score of 2.5 | Fetal or neonatal karyotype or birth outcome | |
| Zhou 2014b | T21, T18, T13 |
|
High, low and without prior risk |
|
L score of 1 and t score of 2.5 | Fetal or neonatal karyotype or birth outcome | |
| TMPS | |||||||
| Ashoor 2012 | T21, T18 |
|
High risk |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype | |
| Ashoor 2013 | T13 |
|
High and low risk |
|
FORTE risk score of 1% | Fetal karyotype or neonatal clinical examination | |
| Bevilacqua 2015 | T21, T18, T13 |
|
High and without prior risk |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or neonatal karyotype | |
| Comas 2015 | T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY (SCA data not shown in this review) |
|
High and without prior risk |
|
Harmony™ prenatal test: NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) Panorama™ test: NR | Fetal karyotype or neonatal clinical examination | |
| del Mar Gil 2014 | T21, T18, T13 |
|
Without prior risk |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype | |
| Gil 2016 | T21, T18, T13 |
|
High and intermediate riskc |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or postnatal karyotype or neonatal clinical examination | |
| Hall 2014 | T13 |
|
High risk |
|
NR | Fetal karyotype or genetic testing of cord blood, buccal, saliva or products of conception | |
| Hooks 2014 | 45,X, 47,XXX, 47, XXY, 47,XYY |
|
High risk |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype | |
| Jackson 2014 | T21, T18, T13 |
|
High and low risk |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal karyotype or medical record from birth | |
| Korostelev 2014 | T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY |
|
High and without prior risk |
|
NR | Fetal karyotype or medical record from birth | |
| Nicolaides 2012 | T21, T18 |
|
Without prior risk |
|
Risk score of 1% | Fetal karyotype or neonatal clinical examination | First‐trimester combined test (cutpoint of 1 in 150). |
| Nicolaides 2013 | T21, T18, T13, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
NR | Fetal karyotype | |
| Nicolaides 2014a | 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
FORTE risk score of 1% | Fetal karyotype | |
| Norton 2012 | T21, T18 |
|
High risk |
|
FORTE risk score of 1% | Fetal karyotype | |
| Norton 2015 | T21, T18, T13 |
|
Without prior risk |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or postnatal karyotype, neonatal clinical examination or medical record from birth | First‐trimester combined test (cutpoint of 1 in 270 for T21 and 1 in 150 for T18 and T13). |
| Pergament 2014 | T21, T18, T13, 45,X |
|
High and low risk |
|
NR | Fetal karyotype or genetic testing of cord blood, buccal, saliva or products of conception or birth outcome | |
| Persico 2016 | T21, T18, 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High risk |
|
Risk score of 1% | Fetal karyotype | |
| Quezada 2015 | T21, T18, T13 |
|
Without prior risk |
|
NR (usually Harmony™ prenatal test uses FORTE risk score of 1%) | Fetal or postnatal karyotype, neonatal clinical examination or medical record from birth | First‐trimester combined test (cutpoint of 1 in 100 for T21). |
| Samango‐Sprouse 2013 | 45,X, 47,XXX, 47,XXY, 47,XYY |
|
High and low risk |
|
NR | Fetal karyotype or genetic testing of cord blood, buccal, saliva or products of conception | |
| Sparks 2012a | T21, T18 |
|
High risk |
|
NR | Fetal karyotype | |
| Verweij 2013 | T21 |
|
High risk |
|
FORTE risk score of 1% | Fetal karyotype | |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, DANSR: digital analysis of selected regions, FF: fetal fraction DNA, FORTE: fetal‐fraction optimised risk of trisomy evaluation, MAD: Median absolute deviation, MPSS: massively parallel shotgun sequencing, NATUS: Next‐generation Aneuploidy Test Using SNPs, NCV: normalised chromosome value, SCA: sex chromosome aneuploidy, SNP: single‐nucleotide polymorphism,TMPS: targeted massively parallel sequencing, T21: trisomy 21, T18: trisomy 18 and T13: trisomy 13.
aFetal karyotype include traditional banding techniques, spectral karyotype, fluorescence in situ hybridisation, array comparative genomic hybridisation or quantitative fluorescence polymerase chain reaction.
bDifferent cutpoints used for autosomes or SCA as follows:
Bianchi 2012: NCV of 4 (aneuploidy suspected if NCV is between 2.5 and 4) for T21, T18, and T13; NCV for Chrom. X of ‐4 and NCV for Chrom. Y of 2.5 for 45,X; NCV for Chrom. X of 4 and NCV for Chrom. Y of 2.5 for 47,XXX; NCV for Chrom. X between ‐2.5 and 2.5 and NCV for Chrom. Y > 33 for 47,XXY; NCV for Chrom. X of ‐4 and NCV for Chrom. Y of 4 for 47,XYY with NCV for Chrom. Y is two times greater than expected NCV Chrom. X.
Bianchi 2013: NCV of 4 (aneuploidy suspected if NCV is between 3 and 4) for T21, T18, and T13; NCV for Chrom. X of ‐3 and NCV for Chrom. Y of 3 for 45,X.
Jiang 2012: t score of 3 and logarithmic LR of 1 for T21, T18 and T13; if female fetus, t score of ‐2.5 for 45,X and 47,XXX; t score of 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X and Y independently for 47,XXY and 47,XYY.
Lau 2012: Z score of 3 for T21, T18 and T13; if female fetus, Z score for Chrom. X of ‐3 for 45,X; if female fetus, Z score for Chrom. X of 3 for 47,XXX; if male fetus, Z score for Chrom. Y of 3 for 47,XXY.
Lefkowitz 2016: Z score of 3 for T21; Z score of 3.95 for T18 and T13; Z scores for SCA see Mazloom 2013.
Liang 2013: Z score of 3 for T21; 5.91 for T18; 5.72 for T13; ± 2.91 for Chrom. X and ± 3 for Chrom. Y for sex chromosome classification.
Mazloom 2013: Z score of 3.5 for 47,XXX (non‐reportable regions between 2.5 and 3.5); Z score of ‐3.5 for 45,X (non‐reportable regions between ‐2.5 and ‐3.5); Z score of ‐3.5 for 47,XYY with Chrom. Y representation; between ‐3.5 and 3.5 for 47,XXY with Chrom. Y representation.
Porreco 2014: Z score of 3 for T21; Z score of 3,95 for T18 and T13; Z score of 3.5 for 47,XXX (non‐reportable regions between 2.5 and 3.5); Z score of ‐3.5 for 45,X (non‐reportable regions between ‐2.5 and ‐3.5); Z score of ‐3.5 for 47,XYY with Chrom. Y representation; Z score between ‐3.5 and 3.5 for 47,XXY with Chrom. Y representation.
Sehnert 2011: NCV of 4 (unclassified if NCV is between 2.5 and 4) for T21, T18, and T13; NCV for Chrom. Y of ‐2.0 SDs from the mean of male samples and NCV for Chrom. X of ‐3.0 SDs from the mean of female samples for sex chromosome classification.
Shaw 2014: Z score of 3 for T21, T18, and T13; Z score of ‐3 for Chrom. X and 3 for Chrom. Y for sex chromosome classification.
Yao 2014: T score of 2.5 for T21, T18 and T13; if female fetus, T score for Chrom. X of ‐2.5 for 45,X and 2.5 for 47,XXX; if male fetus, T score for Chrom. X of 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X (expected value of zero) for 47,XXY; if male fetus, T score for Chrom. X of 2.5 and R‐value (the ratio of the fetal DNA fraction estimated by chromosome Y to that estimated by chromosome X) between 1.8 and 2.2 for 47,XYY.
cPregnant women with a first‐trimester combined test selected for their risk of fetal aneuploidy (cutpoint of 1 in 100 for high risk and 1 in 101 to 1 in 2500 for intermediate risk).
The studies assessed MPSS and TMPS using various algorithms and cutpoints. Table 11 describes the specific gNIPT assay used in the included studies. Each assay was developed and validated by the testing laboratory. Among the 65 studies, 44 studies (68%) used a whole genome sequencing method (MPSS) (Alberti 2015; Benachi 2015; Bianchi 2012; Bianchi 2013; Bianchi 2014a; Bijok 2014; Canick 2012; Chen 2011; Chiu 2011; Ehrich 2011; Fiorentino 2016; Hou 2012; Huang 2014; Jeon 2014; Jiang 2012; Johansen 2016; Ke 2015; Kim 2016; Lau 2012; Lee 2015; Lefkowitz 2016; Liang 2013; Liu 2012; Ma 2016; Mazloom 2013; Palomaki 2012; Papageorghiou 2016a; Papageorghiou 2016b; Poon 2016; Porreco 2014; Sehnert 2011; Shaw 2014; Song 2013; Song 2015; Stumm 2014; Sukhikh 2015; Sung‐Hee 2015; Tynan 2016; Wang 2014; Wang 2015a; Yao 2014; Zhang 2016; Zhou 2014a; Zhou 2014b), and 21 (32%) used a targeted method (TMPS) (Ashoor 2012; Ashoor 2013; Bevilacqua 2015; Comas 2015; del Mar Gil 2014; Gil 2016; Hall 2014; Hooks 2014; Jackson 2014; Korostelev 2014; Nicolaides 2012; Nicolaides 2013; Nicolaides 2014a; Norton 2012; Norton 2015; Pergament 2014; Persico 2016; Quezada 2015; Samango‐Sprouse 2013; Sparks 2012a; Verweij 2013). Of the 65 studies, five studies compared gNIPT with traditional screening tests (Bianchi 2014a; Nicolaides 2012; Norton 2015; Quezada 2015; Song 2013). MPSS studies involved 50,864 pregnant women, TMPS studies involved 35,275 pregnant women and traditional screening tests involved 24,279 pregnant women. The most commonly (15 studies) used cutpoint for gNIPT assays was a chromosomal ratio Z score of 3. Thirteen studies used the FORTE risk score, eight studies used a normalised chromosome value (NCV) and 13 studies did not report their cutpoint. The remaining studies used other cutpoints (Table 11). Timing of blood sampling for gNIPT was before invasive procedure in 55 studies, before or more than 24 hours after invasive sampling in four studies (Ashoor 2013; Lefkowitz 2016; Pergament 2014; Samango‐Sprouse 2013), and was not reported in six studies (Bevilacqua 2015; Jiang 2012; Song 2013; Sparks 2012a; Wang 2014; Zhang 2016).
Among all aneuploidies considered, 36 studies (55%) reported analyses only for autosomes, four (6%) for only sex chromosome aneuploidies (SCA) and 25 studies (39%) for both autosomes and SCA. Fifty‐seven studies (82,620 pregnant women) evaluated T21, 50 studies (79,322 pregnant women) evaluated T18, 39 studies (68,958 pregnant women) evaluated T13, 20 studies (10,081 pregnant women) evaluated 45,X, seven studies (6035 pregnant women) evaluated 47,XXX, 12 studies (7609 pregnant women) evaluated 47,XXY and 10 studies (6987 pregnant women) evaluated 47,XYY (Table 11). Among all 65 included studies, there are a total of 2004 T21 cases, 634 T18 cases, 215 T13 cases, 232 45,X cases, 14 47,XXX cases, 25 47,XXY cases and 16 47,XYY cases. All 65 studies used an appropriate reference standard such as fetal or neonatal karyotype, genetic testing, neonatal clinical examination or medical records from birth. In 36 studies (55%), only one reference standard was used while 29 studies (45%) used more than one reference standard (Table 11).
Among the 65 studies, 40 (62%) studies were prospective cohort studies (Ashoor 2013; Bevilacqua 2015; Bianchi 2014a; Bijok 2014; Comas 2015; Fiorentino 2016; Gil 2016; Hou 2012; Huang 2014; Jackson 2014; Jeon 2014; Jiang 2012; Johansen 2016; Ke 2015; Kim 2016; Korostelev 2014; Lau 2012; Lee 2015; Liang 2013; Liu 2012; Mazloom 2013; Nicolaides 2013; Norton 2012; Norton 2015; Pergament 2014; Persico 2016; Porreco 2014; Quezada 2015; Samango‐Sprouse 2013; Shaw 2014; Song 2013; Song 2015; Stumm 2014; Sukhikh 2015; Verweij 2013; Wang 2014; Wang 2015a; Zhang 2016; Zhou 2014a; Zhou 2014b), eight (12%) studies were retrospective cohort studies (Benachi 2015; Bianchi 2013; del Mar Gil 2014; Nicolaides 2012; Sehnert 2011; Sung‐Hee 2015; Tynan 2016; Yao 2014), one (1%) study was a prospective and retrospective cohort study (Ma 2016) and 16 (25%) studies used a case‐control design (Alberti 2015; Ashoor 2012; Bianchi 2012; Canick 2012; Chen 2011; Chiu 2011; Ehrich 2011; Hall 2014; Hooks 2014; Lefkowitz 2016; Nicolaides 2014a; Palomaki 2012; Papageorghiou 2016a; Papageorghiou 2016b; Poon 2016; Sparks 2012a) (Table 11).
Forty‐eight (74%) studies included only singleton pregnancies, while five (8%) studies included only multifetal pregnancies. Four (6%) studies included women with either type of pregnancy and eight (12%) studies did not report the type of pregnancy. Ten (15%) studies included only pregnant women in the first trimester (15 weeks or less), 21 (33%) studies included pregnant women in the first two trimesters (29 weeks or less), 24 studies (37%) included pregnant women in the three trimesters (42 weeks or less) and 10 studies (15%) did not report gestational age. Eighteen studies (28%) had more than 50% Caucasian women in their cohort, 21 studies (32%) had more than 50% Asian women and 26 studies (40%) did not report ethnicity.
Thirty‐seven studies (57%) were industry‐funded or were written by one or more author affiliated with a company who sells gNIPT (Benachi 2015; Bianchi 2012; Bianchi 2013; Bianchi 2014a; Canick 2012; Chen 2011; Chiu 2011; Ehrich 2011; Hall 2014; Hooks 2014; Huang 2014; Jackson 2014; Jiang 2012; Kim 2016; Lau 2012; Lee 2015; Lefkowitz 2016; Ma 2016; Mazloom 2013; Nicolaides 2012; Nicolaides 2013; Norton 2012; Norton 2015; Palomaki 2012; Papageorghiou 2016a; Papageorghiou 2016b; Pergament 2014; Persico 2016; Porreco 2014; Samango‐Sprouse 2013; Sehnert 2011; Shaw 2014; Sparks 2012a; Stumm 2014; Tynan 2016; Verweij 2013; Yao 2014); 22 studies (34%) were not reported to be funded by industry but samples were sequenced and analysed by a commercial laboratory (Ashoor 2012; Ashoor 2013; Bevilacqua 2015; Bijok 2014; Comas 2015; del Mar Gil 2014; Fiorentino 2016; Gil 2016; Hou 2012; Jeon 2014; Ke 2015; Korostelev 2014; Liang 2013; Poon 2016; Quezada 2015; Song 2013; Song 2015; Sung‐Hee 2015; Wang 2014; Wang 2015a; Zhou 2014a; Zhou 2014b); three studies (4.5%) had no link with industry (Alberti 2015; Johansen 2016; Sukhikh 2015); and the funding source was not reported for three studies (4.5%) (Liu 2012; Nicolaides 2014a; Zhang 2016). Table 12 describes the specific gNIPT assay used in the included studies. Of the 65 studies, 61 (94%) used a commercial gNIPT (15 from Ariosa Diagnostics, Inc., 12 from Bejing Genomics Institute, four from Illumina (or Verinata Health), six from Natera, nine from Sequenom and 15 from other companies) (Table 12). It appears that, for three of the commercially available assays, there are nine studies or more adding up to a large number of cases and unaffected cases analysed. Further, only two assays (one TMPS and one MPSS) were used in one of the five studies involving unselected pregnant women and one assay (Ariosa’s Harmony™ test) was used in four of them. Twelve studies (19%) included their entire cohort in the analyses, 36 studies (55%) included between 80% to 99.9%, and 17 studies (26%) included less than 80%. We found 54 (83%) studies where patient exclusions and failed samples were reported (Table 13; Table 14).
5. Manufacturers of gNIPT used in the included studies by prior risk of fetal aneuploidy.
| Company |
Number of studies |
Number of affected/unaffected pregnanciesa |
Number of studies with pregnant women without prior risk of fetal aneuploidy |
Number of studies with high‐risk pregnant women |
Number of studies with mixed riskb cohort |
| Ariosa Diagnostics, Inc. |
15 | 594/32,302 | 4 | 6 | 5 |
| Bejing Genomics Institute (BGI) |
12 | 427/24,724 | 0 | 7 | 5 |
| Sequenom, Inc. | 9 | 904/8486 | 0 | 7 | 2 |
| Berry Genomics Co. Ltd |
6 | 147/3414 | 1 | 4 | 1 |
| Natera, Inc. | 6 | 276/2103 | 0 | 3 | 3 |
| Illumina, Inc. | 4 | 273/2342 | 0 | 3 | 1 |
| In‐house | 3 | 114/442 | 0 | 3 | 0 |
| Premaitha Health plc |
3 | 99/579 | 0 | 3 | 0 |
| Genome Care | 2 | 21/235 | 0 | 2 | 0 |
| CERBA | 1 | 113/745 | 0 | 1 | 0 |
| Genoma | 1 | 105/6977 | 0 | 0 | 1 |
| LabGenomics | 1 | 8/84 | 0 | 1 | 0 |
| LifeCodexx AG | 1 | 55/417 | 0 | 1 | 0 |
| Not reported | 1 | 5/148 | 0 | 1 | 0 |
| Total | 65 | 3141/82,998 | 5 | 42 | 18 |
aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bMixed‐risk cohort included a mix of pregnant women without prior risk, low risk or high risk of fetal aneuploidy.
6. Reasons for patient exclusion.
| Study ID | Number of pregnant women enrolled | Reasons for exclusion | Number of women with results for 2 x 2 table analysis |
| Alberti 2015 | 976 |
Total: 793 |
183 |
| Ashoor 2012 | 400 |
|
397 |
| Ashoor 2013 | 2167 |
Total: 218 |
1949 |
| Benachi 2015 | 900 |
Total: 14 |
886 |
| Bevilacqua 2015 | 2362 |
Total: 2022 |
340 |
| Bianchi 2012 | 2882 |
Total: 2366 In addition, other samples excluded from 2 x 2 tables for censored complex karyotype:
|
503 (T21) 502 (T18) 501 (T13) 489 (45,X) |
| Bianchi 2013 | 2882 |
|
113 |
| Bianchi 2014a | 2052 |
Total for T21 and T18: 100 Total for T13: 128 |
1952 (T21 and T18) 1914 (T13) |
| Bijok 2014 | 10 |
|
9 |
| Canick 2012 | 4664 |
|
27 |
| Chen 2011 | 392 |
|
289 |
| Chiu 2011 | 824 |
Total: 71 (8‐plex) |
753 (8‐plex) |
| Comas 2015 | 333 |
Total: 21 |
312 |
| del Mar Gil 2014 | 207 |
Total: 15 |
192 |
| Ehrich 2011 | 480 |
Total: 31 |
449 |
| Fiorentino 2016 | 7103 |
|
7082 |
| Gil 2016 | 11,692 |
Total: 8059 |
3633 |
| Hall 2014 | > 1000 |
Total: 936 |
64 |
| Hooks 2014 | 432 |
|
414 |
| Hou 2012 | 308 |
|
205 |
| Huang 2014 | 189 | NR | 189 |
| Jackson 2014 | 1228 |
Total: 67 |
1161 |
| Jeon 2014 | 155 | NR | 155 |
| Jiang 2012 | 903 | NR | 903 |
| Johansen 2016 | 375 |
Total: 202 |
173 |
| Ke 2015 | 2340 | NR | 2340 |
| Kim 2016 | 101 | NR | 101 |
| Korostelev 2014 | 1968 |
Total: 1283 |
685 |
| Lau 2012 | 108 | NR | 108 |
| Lee 2015 | 93 |
|
92 |
| Lefkowitz 2016 | 5321 |
Total: 4155 (autosomes) In addition:
Total: 4177 (SCA) |
1166 (autosomes) 1144 (SCA) |
| Liang 2013 | 435 |
Total: 23 |
412 |
| Liu 2012 | 153 | NR | 153 |
| Ma 2016 | 10,598 |
Total: 19 |
10,579 |
| Mazloom 2013 | 1975 |
|
411 |
| Nicolaides 2012 | 2230 |
Total: 281 |
1949 |
| Nicolaides 2013 | 242 |
|
229 |
| Nicolaides 2014a | 177 |
Total: 5 |
172 |
| Norton 2012 | 4002 |
Total: 922 |
3080 |
| Norton 2015 | 18,955 |
Total: 3114 |
15,841 |
| Palomaki 2012 | 4876 |
Total: 2905 |
1971 |
| Papageorghiou 2016a | 442 |
Total: 16 |
426 |
| Papageorghiou 2016b | 442 |
Total: 431 |
11 |
| Pergament 2014 | 1064 |
Total: 98 In addition,
|
963 (T21) 964 (T18 and 45,X) 965 (T13) |
| Persico 2016 | 259 |
Total: 10 |
249 |
| Poon 2016 | 242 |
|
241 |
| Porreco 2014 | 4170 |
In addition,
|
3322 (T21, T18, T13)
3278 (45,X, 47,XXX) 3201 (47,XXY, 47,XYY) |
| Quezada 2015 | 2905 |
Total: 120 |
2785 |
| Samango‐Sprouse 2013 | 201 |
Total: 15 |
186 |
| Sehnert 2011 | 1014 |
Total: 967 |
47 |
| Shaw 2014 | 201 |
|
200 |
| Song 2013 | 1916 |
Total: 175 |
1741 |
| Song 2015 | 213 |
Total: 9 |
204 |
| Sparks 2012a | 338 |
|
167 |
| Stumm 2014 | 522 |
Total: 50 |
472 |
| Sukhikh 2015 | 200 | NR | 200 |
| Sung‐Hee 2015 | 918 |
Total: 17 |
901 |
| Tynan 2016 | 1100 |
Total: 52 |
1048 |
| Verweij 2013 | 595 |
Total: 91 |
504 |
| Wang 2014 | 136 | NR | 136 |
| Wang 2015a | 917 | NR | 917 |
| Yao 2014 | 5950 |
|
5530 |
| Zhang 2016 | 87 | NR | 87 |
| Zhou 2014a | 306 |
|
301 |
| Zhou 2014b | 7705 |
Total: 3755 |
3950 |
ccfDNA: circulating cell‐free DNA, CVS: chorionic villi sampling, GA: gestational age, gNIPT: genomics‐based non‐invasive prenatal testing, NR: not reported by authors.
aSecond time: sample failed the second gNIPT assay.
bFirst time: sample failed the initial gNIPT assay. cComplex autosome karyotypes are mosaic, triploidies, unbalanced rearrangements with missing or duplicated genetic material. dOther are copy number variation of the X chromosome is confounded by maternal component and cannot be determined.
7. Proportion of pregnant women with a reference standard and assay failure during gNIPT process.
| Study ID |
Failure rate at first attempt (%) |
Repeated testsa (%) |
Failure rate of repeated tests (%) |
Final failure rate total (%) |
Aneuploidb samples (%) |
Euploidb samples (%) |
| MPSS | ||||||
| Alberti 2015 | 61/244 (25%) | 0 | NA | 61/244 (25%) | NR | NR |
| Benachi 2015 | 42/892 (4.7%) | 42 (100%) with second aliquot |
6/42 (14.3%) | 6/892 (0.7%) | 2.7% | 0.4% |
| Bianchi 2012 | 16/519 (3.1%) | 0 | NA | 16/356 (3.1%) | NR | NR |
| Bianchi 2014a | 18/1970 (0.9%) | 0c | NA | T21 and T18: 18/1970 (0.9%) T13: 18/1932 (0.9%) |
NR | NR |
| Bijok 2014 | 1/10 (10.0%) | 0 | NA | 1/10 (10.0%) | 50% | 0% |
| Chiu 2011 | 11/764 (1.4%) | 0 | NA | 11/764 (1.4%) | NR | NR |
| Ehrich 2011 | 20/467 (4.3%) | 20 (100%) resequenced | 18/20 (90%) | 18/467 (3.9%) | NR | NR |
| Fiorentino 2016 | 100/7103 (1.4%) | 79 (79%) with new sampling |
0 (0%) | 21/7103 (0.3%) | 0% | 0.3% |
| Johansen 2016 | NR | 2 with second aliquot or resequenced were in the grey zone (between affected and unaffected) |
NR | 11/184 (6%)d | 5.8% | 6.1% |
| Lee 2015 | 1/93 (1.1%) | 0 | NA | 1/93 (1.1%) | NR | NR |
| Lefkowitz 2016 | Autosomes: 42/1208 (3.5%) SCA: 64/1208 (5.3%) |
0 | NA | Autosomes: 42/1208 (3.5%) SCA: 64/1208 (5.3%) |
Autosomes: 3.8% SCA: 29.7% |
Autosomes: 3.4% SCA: 4.5% |
| Liang 2013 | 12/424 (2.8%) | 0 | NA | 12/424 (2.8%) | NR | NR |
| Ma 2016 | 5/10,584 (0.05%) | 0 | NA | 5/10,584 (0.05%) | NR | NR |
| Mazloom 2013 | 21/432 (4.9%) | 0 | NA | 21/432 (4.9%) | 11.8% | 4.3% |
| Palomaki 2012 | 110/1988 (5.5%) | 105 (95.5%) with second aliquot and 5 (4.5%) resequenced |
17/110 (15.5%) | 17/1988 (0.9%) | 1.0% | 0.8% |
|
Papageorghiou 2016a Papageorghiou 2016b |
5/431 (1.2%) | 0 | NA | 5/431 (1.2%) | NR | NR |
| Poon 2016 | 1/242 (0.4%) | 0 | NA | 1/242 (0.4%) | 0% | 0.5% |
| Porreco 2014 | Autosomes: 108/3430 (3.1%) 45,X and 47,XXX: 152/3430 (4.4%) 47,XXY and 47,XYY: 229/3430 (6.7%) |
0 | NA | Autosomes: 108/3430 (3.1%) 45,X and 47,XXX: 152/3430 (4.4%) 47,XXY and 47,XYY: 229/3430 (6.7%) |
NR | NR |
| Song 2013 | 73/1814 (4.0%) | 0 | NA | 73/1814 (4.0%) | 0% | 4.0% |
| Song 2015 | 1/205 (0.5%) | 0 | NA | 1/205 (0.5%) | NR | NR |
| Stumm 2014 | 32/504 (6.3%) | 0 | NA | 32/504 (6.3%) | 3.5% | 6.7% |
| Sung‐Hee 2015 | 21/908 (2.3%) | 16 (76.2%) with new sampling |
2/16 (12.5%) | 7/908 (0.8%) | NR | NR |
| Tynan 2016 | 52/1100 (4.7%) | 0 | NA | 52/1100 (4.7%) | 0% | 4.9% |
| Yao 2014 | 0 | 0 | NA | 0 | NA | NA |
| Zhou 2014a | 0 | 0 | NA | 0 | NA | NA |
| Zhou 2014b | 141/3954 (3.6%) | 141 (100%) with new sampling |
4/141 (2.8%) | 4/3954 (0.1%) | NR | NR |
| Overall range of final assay failure for MPSS | 0% to 25% | 0% to 50% | 0% to 6.7% | |||
| TMPS | ||||||
| Ashoor 2012 | 3/400 (0.8%) | 0 | NA | 3/400 (0.8%) | 0% | 1% |
| Ashoor 2013 | 53/2002 (2.6%) | 0 | NA | 53/2002 (2.6%) | 0% | 2.7% |
| Bevilacqua 2015 | 29/356 (8.1%) | 26 (90%) with 2nd aliquot |
13/26 (50%) | 16/356 (4.5%) | NR | NR |
| Comas 2015 | 9/316 (2.8%) | 6 (67%) with new sampling |
1/6 (16.7%) | 4/316 (1.3%) | NR | NR |
| del Mar Gil 2014 | 15/207 (7.2%) | 0 | NA | 15/207 (7.2%) | 23% | 6% |
| Gil 2016 | 99/3698 (2.8%) | 54 (54,5%) with new sampling |
20/54 (37%) | 65/3698 (1.8%) | NR | NR |
| Hall 2014 | 4/68 (5.9%) | 0 | NA | 4/68 (5.9%) | 11.8% | 3.9% |
| Hooks 2014 | 18/432 (4.2%) | 0 | NA | 18/432 (4.2%) | NR | NR |
| Jackson 2014 | NR | NR | 14 (NR) | 14/1175 (1.2%) | NR | NR |
| Nicolaides 2012 | 100/2049 (4.9%) | 0 | NA | 100/2049 (4.9%) | 9.1% | 4.9% |
| Nicolaides 2013 | 13/242 (5.4%) | 0 | NA | 13/242 (5.4%) | 6.3% | 5.2% |
| Nicolaides 2014a | 5/177 (2.8%) | 0 | NA | 5/177 (2.8%) | 5.1% | 1.7% |
| Norton 2012 | 148/3228 (4.6%) | 0 | NA | 148/3228 (4.6%) | NR | NR |
| Norton 2015 | 488/16,329 (3.0%) | 0 | NA | 488/16,329 (3.0%) | 20.6% | 2.9% |
| Pergament 2014 | T21: 88/1051 (8.4%) T18, 45,X: 87/1052 (8.3%) T13: 86/1053 (8.2%) |
0 | NA | T21: 88/1051 (8.4%) T18, 45,X: 87/1052 (8.3%) T13: 86/1053 (8.2%) |
All five chromosomes (n = 85): 15.2% |
All five chromosomes (n = 85): 7.1% |
| Persico 2016 | 10/259 (3.9%) | 0 | NA | 10/259 (3.9%) | 8.4% | 2.1% |
| Quezada 2015 | 122e/2838 (4.2%) | 110 (90.1%) with new sampling |
41/110 (37.3%) | 53/2838 (1.9%) | 4.1% | 1.8% |
| Samango‐Sprouse 2013 | 15/201 (7.5%) | 0 | NA | 15/201 (7.5%) | 6.3% | 7.6% |
| Verweij 2013 | 51/520 (9.8%) | 51 (100%) with 2nd aliquot |
16/51 (31.4%) | 16/520 (3.1%) NR |
NR | NR |
| Overall range of final assay failure for TMPS | 0.8% to 7.5% | 0% to 23% | 1% to 7.63% | |||
CVS: chorionic villi sampling, FF: fetal fraction DNA, GA: gestational age, NA: not applicable, NR: not reported by authors, QC: quality control. aRepeated tests included second aliquot (aliquot from first sampling), resequenced (same library) or new sampling.
baneuploid: proportion of failed samples of aneuploid cases out of all aneuploid tested with reference standard and gNIPT result. euploid: proportion of failed samples of euploid cases out of all euploid tested with reference standard and gNIPT result.
cAuthors decided to resequence 12 samples with gNIPT results. They were in the grey zone (between affected and unaffected) and were resequenced in uniplex. All repeated tests were in affected or unaffected zone.
dOnly the final failure rate was reported.The failure rate at first attempt was not reported nor the failure rate of repeated tests.
eAuthor reported 123 failed tests but this number included one sample lost in the mail and so did not undergo the sequencing process.
Summary of excluded studies
We described the excluded studies in the PRISMA flow diagram (Figure 3) as well as in Characteristics of excluded studies. After full‐text assessment, we excluded 261 articles.
Of these 261:
93 (36%) studies were not diagnostic test accuracy studies (e.g. implementation study, simulation model, method development, proof‐of‐concept, method without sequencing approach);
55 (21%) studies had overlapping samples and were excluded to avoid double counting;
54 (21%) studies had incomplete 2 X 2 data or insufficient information to derive a 2 X 2 table;
22 (8%) studies had either an inappropriate or no reference standard;
8 (3%) studies were identified as reviews or Health Technology Assessment reports;
11 (4%) studies had target conditions, methods or sampling schemes other than those specified in our review; and
18 (7%) studies were news, letters, comments, notes, replies or editorials without new data.
The 25 ongoing studies are described in Characteristics of ongoing studies.
Methodological quality of included studies
Figure 4 and Figure 5 show the risk of bias and applicability concerns for each included study for MPSS and TMPS, respectively. In Figure 6, the quality assessment results are summarised across all studies.
4.
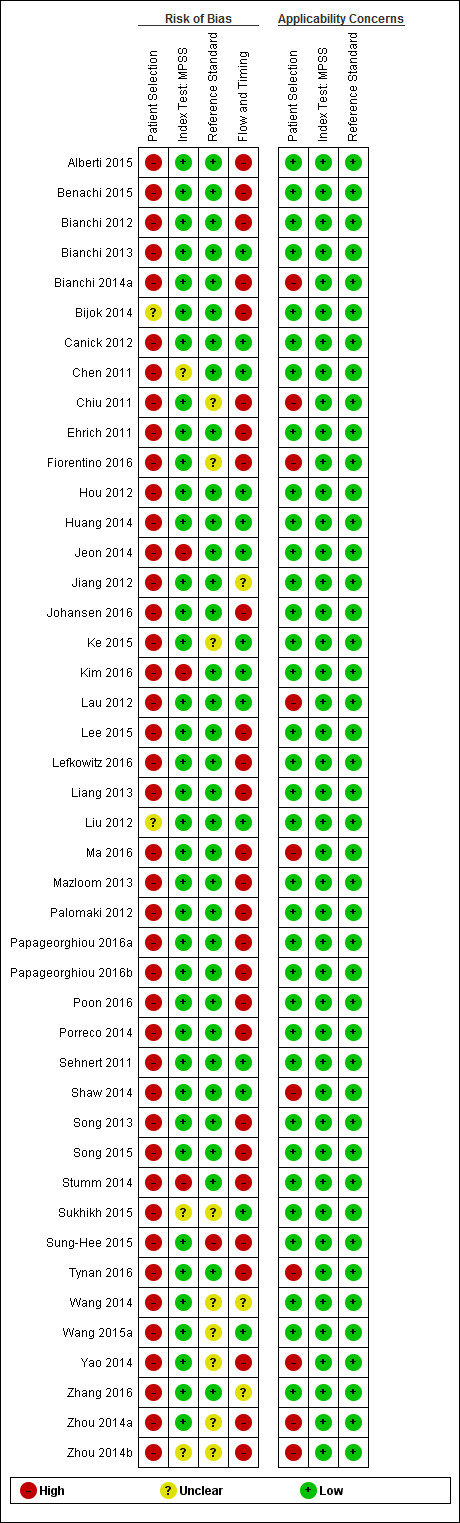
Risk of bias and applicability concerns summary: review authors' judgements about each domain for each of the studies included for massively parallel shotgun sequencing (MPSS).
5.

Risk of bias and applicability concerns summary: review authors' judgements about each domain for each study included for targeted massively parallel sequencing (TMPS).
6.
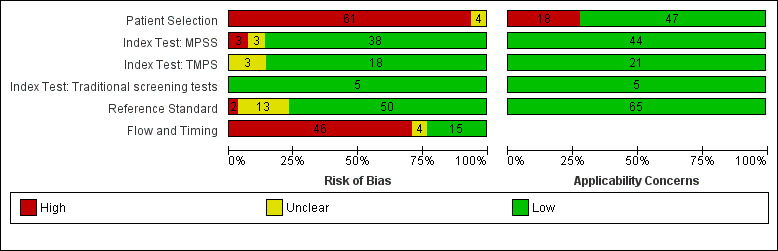
Risk of bias and applicability concerns (all tests included): review authors' judgements about each domains presented as percentages across included studies. MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing.
Risk of bias
No study was assessed as being at low risk of bias across all domains (Figure 4). For the patient selection domain, the 'Risk of bias' judgement was influenced mainly by inappropriate exclusions than the other signalling questions in this domain. Of the 61 studies judged to be at high risk of bias, 57 (93%) had inappropriate exclusions. The exclusions were mainly due to multifetal pregnancy, gestational age limits, and the prior risk of fetal aneuploidy. The remaining four (7%) studies were judged to be at unclear risk of bias (Figure 6).
In the index test domain, the risk of bias was considered to be low in 38 (58%) of the 44 MPSS studies and unclear in three (5%) studies. The remaining three (5%) MPSS studies were judged to be at high risk of bias because the index test was performed knowing the results of the reference standard or the threshold was not pre‐specified. The risk of bias was low in 18 (27%) of the 21 TMPS studies. The remaining three (5%) TMPS studies were judged to be at unclear risk of bias. All five studies that assessed traditional screening approaches were judged to be at low risk of bias for the index test domain (Figure 6).
In the reference standard domain, all studies used a reference standard likely to correctly classify the target condition. We considered 50 (77%) studies to be at low risk of bias because the studies stated that the reference standard results were interpreted without knowledge of the results of the index test. Of the remaining 15 studies, two (3%) studies were at high risk of bias because the reference standard was performed knowing the results of the index test while it was unclear what was done in the other 13 (20%) studies (Figure 6).
For the flow and timing domain, 46 (71%) studies were considered to be at high risk of bias because some pregnant women were excluded from 2 x 2 tables because gNIPT failed during the sequencing process. Fifteen (23%) studies were judged to be at low risk of bias. For the remaining four (6%) studies, information about the appropriate interval between the index test and reference standard was not provided (Figure 6).
Applicability concerns
We judged all studies to be of low applicability concern in the index test and reference standard domains because the studies matched the review question (Figure 4; Figure 6). All studies used a gNIPT method with ccfDNA in maternal blood and appropriate reference standard for the detection of common fetal aneuploidies. In the patient selection domain, 47 (72%) studies included cohort of pregnant women selected at high risk of fetal aneuploidy or cohort of unselected pregnant women and were judged to be of low applicability concern. In the other 18 (28%) studies, the cohorts comprised pregnant women with different prior risk of fetal aneuploidy (mixed risk cohorts). This population did not represent the real‐life situation and those cohorts were judged to be of high applicability concern.
Findings
The characteristics of the studies are summarised in Table 11 and Table 1. Results are presented separately for each of the main fetal aneuploidies (T21, T18, T13 and 45,X) and globally for all autosomes or all sex chromosome aneuploidies (SCA) combined (Table 2; Table 3; Table 4; Table 5; Table 6; Table 7). For each aneuploidy, results are presented according to the prior risk of chromosomal abnormality as high risk or unselected population and according to MPSS and TMPS methods. Results from mixed‐risk populations are summarised in Appendix 7. No study directly compared the accuracy of MPSS and TMPS. There were insufficient data to separately consider monochorionic and dichorionic pregnancies and four of the nine studies did not report chorionicity.
1. Trisomy 21 (T21 or Down syndrome)
A total of 57 studies assessed gNIPT for T21 in 2004 affected and 80,616 non T21 pregnancies. Five studies enrolled an unselected population of pregnant women undergoing aneuploidy screening, 36 studies enrolled pregnant women selected at high risk of fetal aneuploidy and 16 studies enrolled pregnant women with various prior risk and no a priori risk of fetal aneuploidy (mixed risk). Of the 57 studies, 41 assessed MPSS and 16 assessed TMPS. The results are summarised in Table 2.
a. Unselected population of pregnant women undergoing aneuploidy screening
Five cohort studies evaluated gNIPT in an unselected population of pregnant women undergoing aneuploidy screening. The studies included 22,412 non T21 pregnancies and 96 (0.43%) T21 cases. MPSS was assessed in one study and TMPS was assessed in four studies (Figure 7).
7.
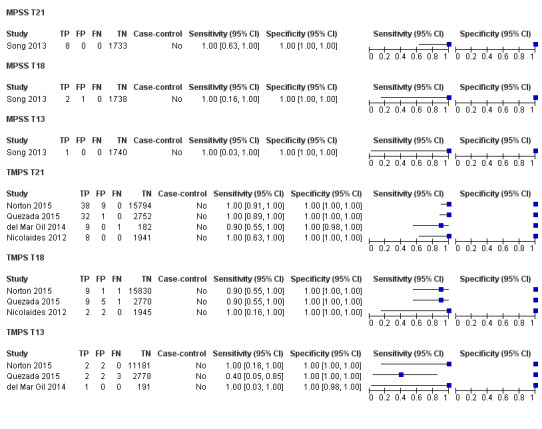
Forest plot of MPSS and TMPS for T21 in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
One prospective cohort study included eight T21 cases and 1733 non T21 pregnancies (Song 2013). The sensitivity (95% confidence interval (CI)) of MPSS was 100% (67.6% to 100%) and the specificity (95% CI) was 100% (99.8% to 100%).
ii. TMPS
TMPS was evaluated in four studies comprising 20,679 non T21 pregnancies and 88 T21 cases (del Mar Gil 2014; Nicolaides 2012; Norton 2015; Quezada 2015). The summary sensitivity (95% CI) was 99.2% (78.2% to 100%) and the summary specificity (95% CI) was 100% (> 99.9% to 100%).
iii. Comparative accuracy of MPSS and TMPS
It was not possible to compare the accuracy of MPSS and TMPS in a meta‐analysis because of limited data.
b. Selected population of pregnant women at high risk of fetal aneuploidy
Overall, 36 studies included pregnant women selected at high risk of fetal aneuploidy involving 20,317 non T21 pregnancies and 1294 (6.37%) T21 cases. MPSS was assessed in 30 studies and TMPS in six studies (Figure 8).
8.
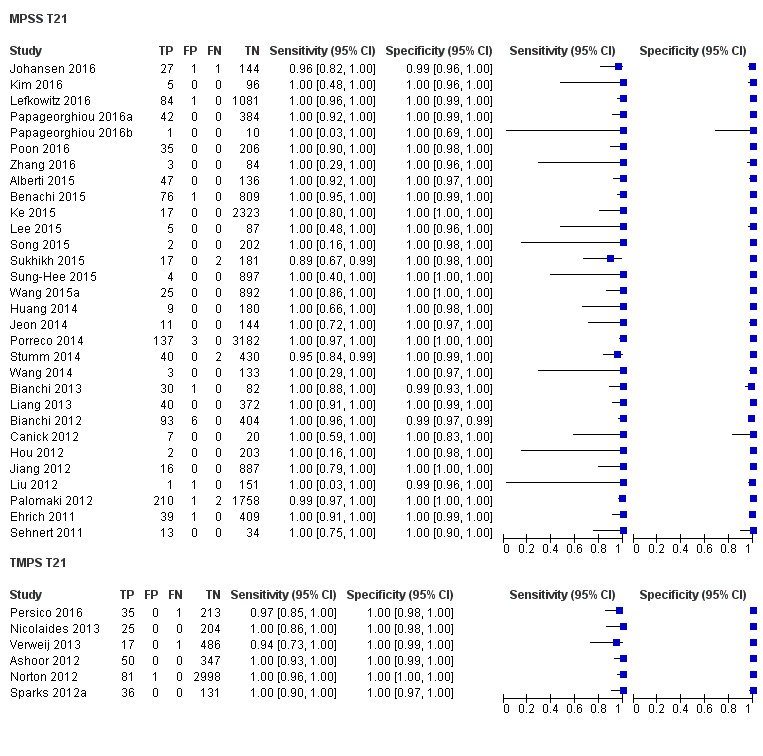
Forest plot of MPSS and TMPS for T21 in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
The 30 MPSS studies included 15,937 non T21 pregnancies and 1048 T21 cases (Alberti 2015; Benachi 2015; Bianchi 2012; Bianchi 2013; Canick 2012; Ehrich 2011; Hou 2012; Huang 2014; Jeon 2014; Jiang 2012; Johansen 2016; Ke 2015; Kim 2016; Lee 2015; Lefkowitz 2016; Liang 2013; Liu 2012; Palomaki 2012; Papageorghiou 2016a; Papageorghiou 2016b; Poon 2016; Porreco 2014; Sehnert 2011; Song 2015; Stumm 2014; Sukhikh 2015; Sung‐Hee 2015; Wang 2014; Wang 2015a; Zhang 2016). The summary sensitivity (95% CI) was 99.7% (98.0% to 100%) and the summary specificity (95% CI) was 99.9% (99.8% to 100%).
ii. TMPS
Six studies evaluated TMPS in 4380 non T21 pregnancies and 246 T21 cases (Ashoor 2012; Nicolaides 2013; Norton 2012; Persico 2016; Sparks 2012a; Verweij 2013). The summary sensitivity (95% CI) was 99.2% (96.8% to 99.8%) and the summary specificity (95% CI) was 100% (99.8% to 100%).
iii. Comparative accuracy of MPSS and TMPS
An indirect comparison of the 30 MPSS and six TMPS studies showed no statistical evidence of a difference in sensitivity or specificity or both (P value = 0.52). The differences in sensitivity and specificity were negligible (Table 2).
2. Trisomy 18 (T18)
Fifty studies assessed T18 in 634 cases and 78,688 non T18 pregnancies. Four studies enrolled unselected population of pregnant women undergoing aneuploidy screening, 33 studies enrolled pregnant women selected at high risk of fetal aneuploidy and 13 studies enrolled a cohort with mixed prior risk. Of the 50 studies, 38 evaluated MPSS and 12 evaluated TMPS. The results are summarised in Table 3.
a. Unselected population of pregnant women undergoing aneuploidy screening
Four studies, comprising 22,292 non T18 pregnancies and 24 (0.11%) T18 cases, assessed gNIPT for fetal aneuploidy in unselected pregnant women. One study assessed MPSS and three studies assessed TMPS (Figure 7).
i. MPSS
One MPSS study evaluated two T18 cases and 1739 non T18 pregnancies (Song 2013). The sensitivity (95% CI) was 100% (34.3% to 100%) and the specificity (95% CI) was 99.9% (99.7% to 100%).
ii. TMPS
Three studies evaluated TMPS in 20,553 non T18 pregnancies and 22 T18 cases (Nicolaides 2012; Norton 2015; Quezada 2015). The summary sensitivity (95% CI) was 90.9% (70.0% to 97.7%) and the summary specificity (95% CI) was 100% (99.9% to 100%).
iii. Comparative accuracy of MPSS and TMPS
It was not possible to compare the accuracy of MPSS and TMPS in a meta‐analysis because data were sparse.
b. Selected population of pregnant women at high risk of fetal aneuploidy
A total of 33 studies included pregnant women selected at high risk of fetal aneuploidy involving 444 (2.20%) T18 cases and 20,190 non T18 pregnancies. Of these, 28 studies assessed MPSS and five studies assessed TMPS (Figure 9).
9.
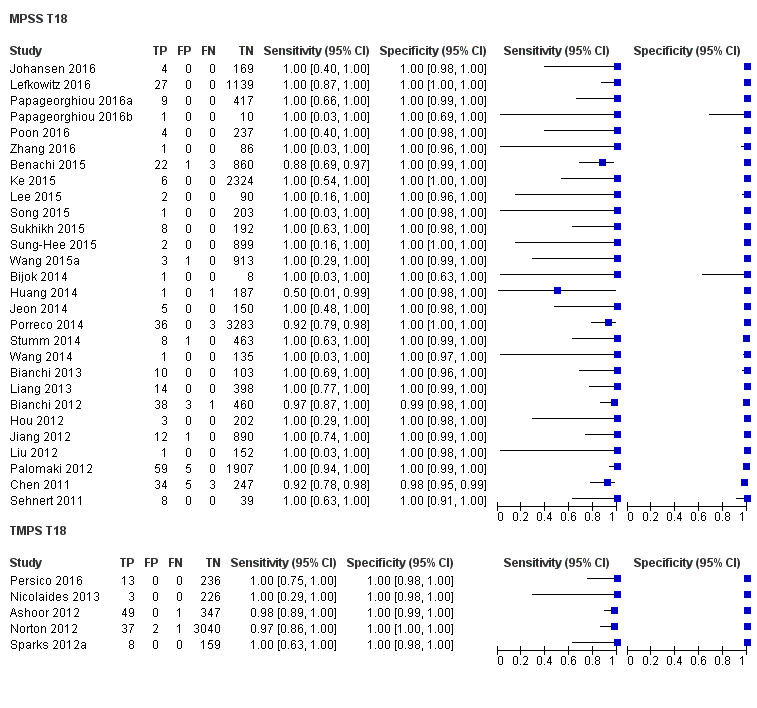
Forest plot of MPSS and TMPS for T18 in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
Twenty‐eight studies evaluated MPSS in 16,180 non T18 pregnancies and 332 T18 cases (Benachi 2015; Bianchi 2012; Bianchi 2013; Bijok 2014; Chen 2011; Hou 2012; Huang 2014; Jeon 2014; Jiang 2012; Johansen 2016; Ke 2015; Lee 2015; Lefkowitz 2016; Liang 2013; Liu 2012; Palomaki 2012; Papageorghiou 2016a; Papageorghiou 2016b; Poon 2016; Porreco 2014; Sehnert 2011; Song 2015; Stumm 2014; Sukhikh 2015; Sung‐Hee 2015; Wang 2014; Wang 2015a; Zhang 2016). The summary sensitivity (95% CI) was 97.8% (92.5% to 99.4%) and the summary specificity (95% CI) was 99.9% (99.8% to 100%).
ii. TMPS
Five studies evaluated TMPS in 4010 non T18 pregnancies and 112 T18 cases (Ashoor 2012; Nicolaides 2013; Norton 2012; Persico 2016; Sparks 2012a). The summary sensitivity (95% CI) was 98.2% (93.1% to 99.6%) and the summary specificity (95% CI) was 100% (99.8% to 100%).
iii. Comparative accuracy of MPSS and TMPS
An indirect comparison of the 28 MPSS and five TMPS studies showed no statistical evidence of a difference in sensitivity, specificity or both (P value = 0.47). The differences in sensitivity and specificity were negligible (Table 3).
3. Trisomy 13 (T13)
T13 was assessed in 39 studies comprising 215 affected and 68,743 non T13 pregnancies. Four studies evaluated unselected population of pregnant women undergoing fetal aneuploidy screening, while 22 studies evaluated women at high risk of fetal aneuploidy and 13 studies evaluated mixed prior risk cohorts. Of the 39 studies, 29 assessed MPSS and 10 assessed TMPS. The results are summarised in Table 4.
a. Unselected population of pregnant women undergoing aneuploidy screening
Four studies assessed gNIPT for T13 in unselected pregnant women. The studies included 15,894 non T13 pregnancies and nine (0.06%) T13 cases. Three studies evaluated TMPS and one study evaluated MPSS (Figure 7).
i. MPSS
One study evaluated MPSS in one T13 case and 1740 non T13 pregnancies (Song 2013). The sensitivity (95% CI) was 100% (20.7% to 100%) and the specificity (95% CI) was 100% (99.8% to 100%).
ii. TMPS
Three studies evaluated TMPS in 14,154 non T13 pregnancies and eight T13 cases (del Mar Gil 2014; Norton 2015; Quezada 2015). The summary sensitivity (95% CI) was 65.1% (9.2% to 97.2%) and the summary specificity (95% CI) was 100% (99.9% to 100%).
iii. Comparative accuracy of MPSS and TMPS
It was not possible to compare the accuracy of MPSS and TMPS in a meta‐analysis because data were sparse.
b. Selected population of pregnant women at high risk of fetal aneuploidy
A total of 22 studies evaluated pregnant women selected at high risk of fetal aneuploidy. The studies included 14,103 non T13 pregnancies and 148 (1.05%) T13 cases. Twenty studies assessed MPSS and two studies assessed TMPS (Figure 10).
10.
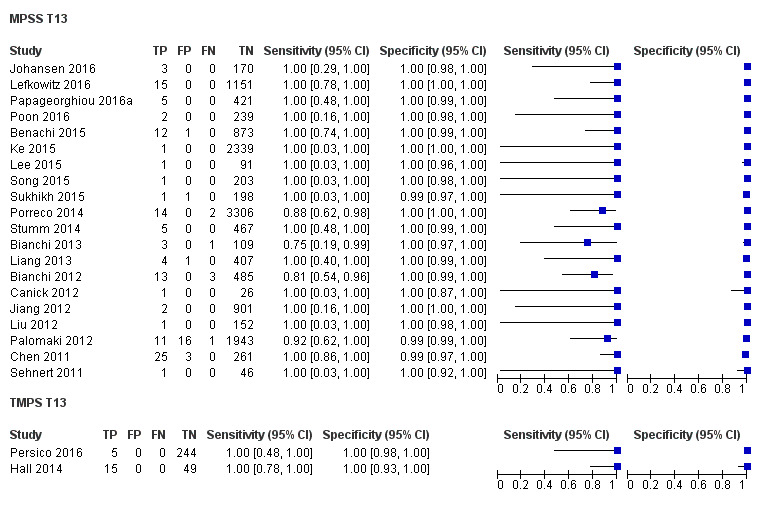
Forest plot of MPSS and TMPS for T13 in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
Twenty studies evaluated MPSS in 13,810 non T13 pregnancies and 128 T13 cases (Benachi 2015; Bianchi 2012; Bianchi 2013; Canick 2012; Chen 2011; Jiang 2012; Johansen 2016; Ke 2015; Lee 2015; Lefkowitz 2016; Liang 2013; Liu 2012; Palomaki 2012; Papageorghiou 2016a; Poon 2016; Porreco 2014; Sehnert 2011; Song 2015; Stumm 2014; Sukhikh 2015). The summary sensitivity (95% CI) was 95.8% (86.1% to 98.9%) and the summary specificity (95% CI) was 99.8% (99.8% to 99.9%).
ii. TMPS
Two studies evaluated TMPS in 293 non T13 pregnancies and 20 T13 cases (Hall 2014; Persico 2016). The summary sensitivity (95% CI) was 100% (83.9% to 100%) and the summary specificity (95% CI) was 100% (98.7% to 100%).
iii. Comparative accuracy of MPSS and TMPS
It was not possible to compare the accuracy of MPSS and TMPS in a meta‐analysis because data were sparse.
4. Turner syndrome (45,X)
Turner syndrome (45,X) was assessed in 20 studies, comprising 232 affected and 9849 non 45,X pregnancies. Among these studies, 16 enrolled pregnant women selected at high risk of fetal aneuploidy and four enrolled a cohort of pregnant women with mixed prior risk. Of the 20 studies, 14 evaluated MPSS and six evaluated TMPS. The results are summarised in Table 5.
a. Unselected population of pregnant women undergoing aneuploidy screening
No study assessed 45,X in this population.
b. Selected population of pregnant women at high risk of fetal aneuploidy
Sixteen studies included 198 (2.35%) affected and 8421 non 45,X pregnancies. MPSS and TMPS were assessed by 12 and four studies respectively (Figure 11).
11.
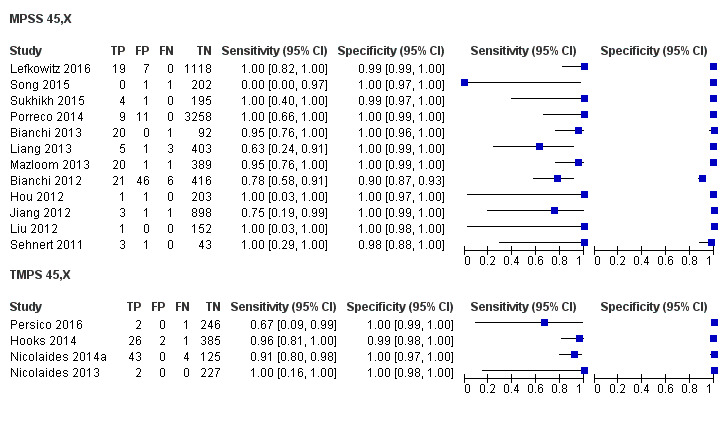
Forest plot of MPSS and TMPS for 45,X in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
Twelve studies evaluated MPSS in 119 affected and 7440 non 45,X pregnancies (Bianchi 2012; Bianchi 2013; Hou 2012; Jiang 2012; Lefkowitz 2016; Liang 2013; Liu 2012; Mazloom 2013; Porreco 2014; Sehnert 2011; Song 2015; Sukhikh 2015). The summary sensitivity (95% CI) was 91.7% (78.3% to 97.1%) and the summary specificity (95% CI) was 99.6% (98.9% to 99.8%).
ii. TMPS
Four studies evaluated TMPS in 79 affected and 985 non 45,X pregnancies (Hooks 2014; Nicolaides 2013; Nicolaides 2014a; Persico 2016). The summary sensitivity (95% CI) was 92.4% (84.1% to 96.5%) and the summary specificity (95% CI) was 99.8% (98.3% to 100%).
iii. Comparative accuracy of MPSS and TMPS
An indirect comparison of the 12 MPSS and four TMPS studies showed no statistical evidence of a difference in sensitivity, specificity or both (P value = 0.40). The differences in sensitivity and specificity were negligible (Table 5).
5. Triple X syndrome (47,XXX)
Seven studies assessed 47,XXX, comprising 14 (0.23%) affected and 6021 non 47,XXX pregnancies (Hooks 2014; Lefkowitz 2016; Liang 2013; Mazloom 2013; Nicolaides 2014a; Porreco 2014; Song 2015). The studies enrolled pregnant women selected at high risk of fetal aneuploidy. Five studies evaluated MPSS and two studies evaluated TMPS. (Figure 12; Table 15). We did not perform a separate meta‐analysis for 47,XXX due to sparse data (very few cases or studies, or one or more subgroups had no study).
12.
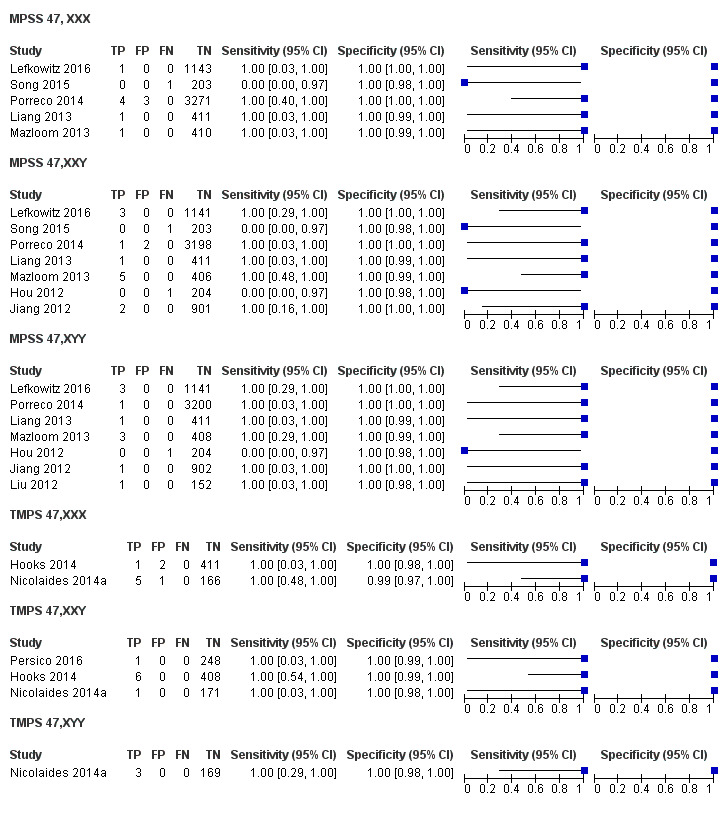
Forest plot of MPSS and TMPS for 47,XXX, 47,XXY and 47,XYY in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
8. Data for 47,XXX, 47,XXY and 47,XYY according to the prior risk of fetal aneuploidy and gNIPT approach.
| Test |
Number of studies |
Number of affected pregnancies |
Number of unaffected pregnanciesa |
|
| 47,XXX | ||||
| Selected high risk pregnant women |
MPSS | 5 | 8 | 5441 |
| TMPS | 2 | 6 | 580 | |
| 47,XXY | ||||
| Selected high risk pregnant women |
MPSS | 7 | 14 | 6466 |
| TMPS | 3 | 8 | 827 | |
| 47,XYY | ||||
| Selected high risk pregnant women |
MPSS | 7 | 11 | 6418 |
| TMPS | 1 | 3 | 169 | |
aUnaffected pregnancies: we included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unnon affected".
6. Klinefelter syndrome (47,XXY)
Twelve studies assessed 47,XXY in 25 (0.33%) affected and 7584 non 47,XXY pregnancies (Hooks 2014; Hou 2012; Jiang 2012; Lau 2012; Lefkowitz 2016; Liang 2013; Mazloom 2013; Nicolaides 2014a; Persico 2016; Porreco 2014; Samango‐Sprouse 2013; Song 2015). Ten studies enrolled pregnant women selected at high risk of fetal aneuploidy (Figure 12; Table 15) and two studies enrolled pregnant women with mixed risk (See Finding section 11). No study assessed 47,XXY in an unselected population of pregnant women undergoing aneuploidy screening. Eight studies assessed MPSS and four studies assessed TMPS. We did not perform a separate meta‐analysis for 47,XXY due to sparse data (very few cases or studies, or one or more subgroups had no study).
7. 47,XYY
Ten studies assessed 47,XYY in 16 (0.23%) affected and 6971 non 47,XYY pregnancies (Hou 2012; Jiang 2012; Lefkowitz 2016; Liang 2013; Liu 2012; Mazloom 2013; Nicolaides 2014a; Porreco 2014; Samango‐Sprouse 2013; Shaw 2014). Eight studies enrolled pregnant women selected at high risk of fetal aneuploidy (Figure 12; Table 15) and two studies enrolled pregnant women with mixed risk (See Finding section 11). Eight studies used MPSS and two studies used TMPS. We did not perform a separate meta‐analysis for 47,XXX due to sparse data (very few cases or studies, or one or more subgroups had no study).
8. All autosomes combined
Autosomal aneuploidies were assessed in 61 studies. The studies included 84,954 pregnant women of which 2853 were T21, T18 or T13 pregnancies and 82,073 were unaffected. Among these 61 studies, 43 assessed MPSS and 18 assessed TMPS. Of the 61 studies, five enrolled unselected pregnant women, 39 enrolled high‐risk pregnant women and 17 enrolled a cohort of mixed prior risk. The results are summarised in Table 6. The results for mixed risk cohorts are summarised in Appendix 7.
a. Unselected population of pregnant women undergoing aneuploidy screening
Five studies assessed 129 (0.58%) affected and 22,379 unaffected (non T21, T18 and T13) pregnancies. Of the five studies, one study assessed MPSS and four studies assessed TMPS (Figure 13).
13.
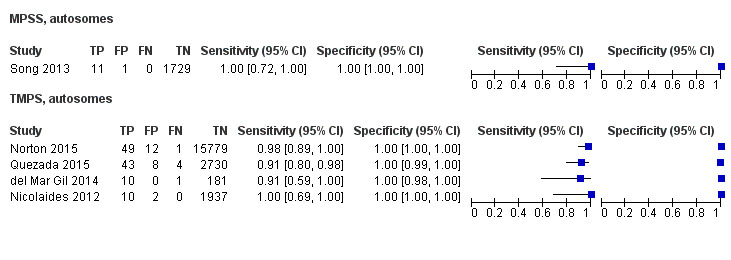
Forest plot of MPSS and TMPS for autosomes (T21, T18 and T13 combined) in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
Only one study assessed MPSS (Song 2013). The study evaluated 1730 unaffected (non T21, T18 and T13) pregnancies and 11 cases in women with singleton pregnancy. The sensitivity (95% CI) was 100% (74.1% to 100%) and the specificity (95% CI) was 99.9% (99.7% to 100%).
ii. TMPS
Four studies assessed TMPS in 20,649 unaffected (non T21, T18 and T13) pregnancies and 118 cases (del Mar Gil 2014; Nicolaides 2012; Norton 2015; Quezada 2015). Of the four studies, three studies included only women with singleton pregnancy and the remaining study included only women with multifetal pregnancy (Table 16). Based on the four studies, the summary sensitivity (95% CI) was 94.9% (89.1% to 97.7%) and the summary specificity (95% CI) was 99.9% (99.8% to 99.9%).
9. Subgroup analyses of MPSS and TMPS (type of pregnancy and gestational age).
| Test subgroups |
Number of studies |
Number of affected pregnancies |
Number of unaffected pregnanciesa |
Sensitivityb % (95% CI) |
Specificityb % (95% CI) |
|
| Pregnancy type | ||||||
| Autosomes (T21, T18 and T13 combined), unselected population | ||||||
| MPSS | singleton | 1 | 11 | 1730 | 100 (74.1 to 100) | 99.9 (99.7 to 100) |
| TMPS | singleton | 3 | 107 | 20,468 | 95.5 (87.4 to 98.4) | 99.9 (99.8 to 100) |
| multifetal | 1 | 11 | 181 | 90.9 (62.3 to 98.4) | 100 (97.9 to 100) | |
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | singleton | 19 | 1087 | 11,180 | 98.3 (97.3 to 98.9) | 99.6 (99.5 to 99.7) |
| multifetal | 3 | 21 | 206 | 95.2 (72.9 to 99.3) | 100 (98.2 to 100)c | |
| TMPS | singleton | 7 | 378 | 4282 | 98.9 (97.2 to 99.6) | 99.9 (99.8 to 100) |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | singleton | 7 | 101 | 4690 | 88.3 (52.9 to 98.1) | 99.3 (97.5 to 99.8) |
| TMPS | 4 | 96 | 968 | 93.8 (86.8 to 97.2) | 99.6 (98.1 to 99.9) | |
| Gestational age | ||||||
| Autosomes (T21, T18 and T13 combined), unselected population | ||||||
| MPSS | ≤29 weeks | 1 | 11 | 1730 | 100 (74.1 to 100) | 99.9 (99.7 to 100) |
| TMPS | ≤15 weeks | 4 | 118 | 20,649 | 94.9 (89.1 to 97.7) | 99.9 (99.8 to 99.9) |
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | ≤15 weeks | 3 | 49 | 532 | 100 (92.7 to 100)c | 100 (99.3 to 100)c |
| ≤29 weeks | 12 | 594 | 4605 | 98.3 (96.9 to 99.1) | 99.3 (99.0 to 99.5) | |
| ≤42 weeks | 13 | 729 | 7831 | 98.9 (95.0 to 99.8) | 99.9 (99.8 to 99.9) | |
| TMPS | ≤15 weeks | 2 | 128 | 498 | 99.2 (95.7 to 99.9)c | 100 (99.2 to 100)c |
| ≤29 weeks | 2 | 33 | 535 | 97.0 (84.7 to 99.5)c | 100 (99.3 to 100)c | |
| ≤42 weeks | 2 | 163 | 3084 | 99.4 (95.8 to 99.9) | 99.9 (99.7 to 100) | |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | ≤15 weeks | 1 | 2 | 202 | 0.00 (0.00 to 65.8) | 99.5 (97.2 to 99.9) |
| ≤29 weeks | 5 | 58 | 996 | 86.5 (63.1 to 96.0) | 95.1 (93.5 to 96.3) | |
| ≤42 weeks | 5 | 89 | 6103 | 95.8 (80.3 to 99.2) | 99.6 (99.4 to 99.7) | |
| TMPS | ≤15 weeks | 2 | 58 | 343 | 93.1 (83.0 to 97.4) | 99.7 (98.0 to 100) |
| ≤42 weeks | 1 | 34 | 380 | 97.1 (85.1 to 99.5) | 98.9 (97.3 to 99.6) | |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13 CI: confidence interval, MPSS: massively parallel shotgun sequencing, SCA: sex chromosome aneuploidies, TMPS: targeted massively parallel sequencing.
aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bFor two or more studies, the sensitivities and specificities are the summary estimates obtained from meta‐analysis.
cSimple pooling used to obtain summary estimates of sensitivity, specificity or both.
iii. Comparative accuracy of MPSS and TMPS
It was not possible to compare the accuracy of MPSS and TMPS in a meta‐analysis due to limited data.
b. Selected population of pregnant women at high risk of fetal aneuploidy
A total of 39 studies included 1886 (9.39%) affected and 20,079 unaffected (non T21, T18 and T13) pregnancies. Of the 39 studies, 32 assessed MPSS and seven assessed TMPS (Figure 14).
14.
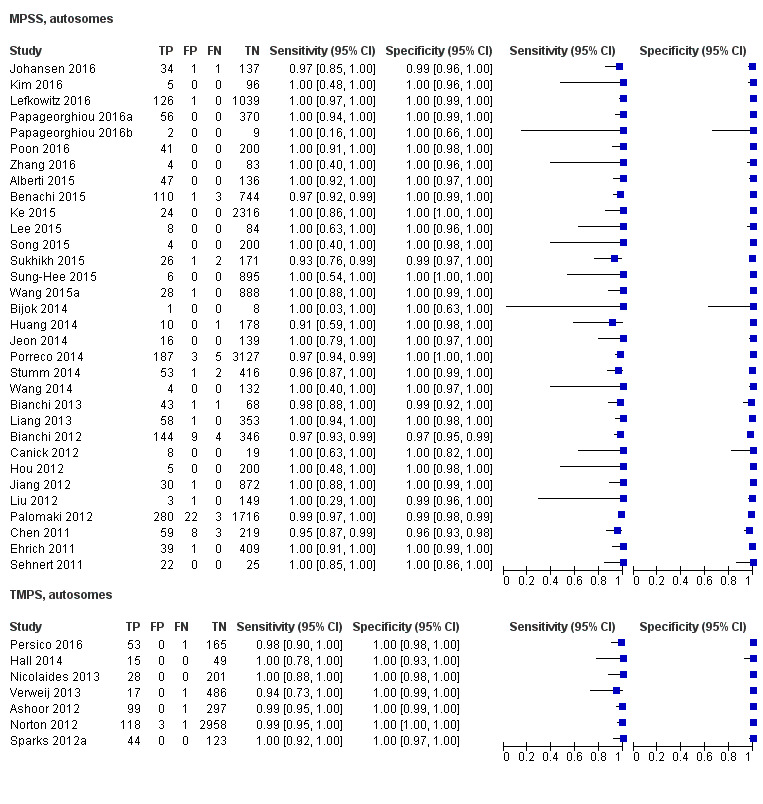
Forest plot of MPSS and TMPS for autosomes (T21, T18 and T13) in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
Thirty‐two MPSS studies evaluated 15,797 unaffected (non T21, T18 and T13) pregnancies and 1508 cases (Alberti 2015; Benachi 2015; Bianchi 2012; Bianchi 2013; Bijok 2014; Canick 2012; Chen 2011; Ehrich 2011; Hou 2012; Huang 2014; Jeon 2014; Jiang 2012; Johansen 2016; Ke 2015; Kim 2016; Lee 2015; Lefkowitz 2016; Liang 2013; Liu 2012; Palomaki 2012; Papageorghiou 2016a; Papageorghiou 2016b; Poon 2016; Porreco 2014; Sehnert 2011; Song 2015; Stumm 2014; Sukhikh 2015; Sung‐Hee 2015; Wang 2014; Wang 2015a; Zhang 2016). Of the 32 studies, 19 evaluated only singleton pregnancies, three evaluated only multifetal pregnancies, three evaluated singleton and multifetal pregnancies, and the remaining seven studies did not report type of pregnancy. Based on the 32 studies, the summary sensitivity (95% CI) was 98.8% (97.2% to 99.5%) and the summary specificity (95% CI) was 99.9% (99.7% to 100%). Results are presented separately for singleton and multifetal pregnancy studies in Table 16. The sensitivity tends to be lower in multifetal pregnancies but there are no enough studies in this subgroup to compare MPSS performance according to pregnancy type.
ii. TMPS
Seven TMPS studies evaluated 378 cases and 4282 unaffected (non T21, T18 and T13) pregnancies in women with singleton pregnancy (Ashoor 2012; Hall 2014; Nicolaides 2013; Norton 2012; Persico 2016; Sparks 2012a; Verweij 2013). The summary sensitivity (95% CI) was 98.9 (97.2% to 99.6%) and the summary specificity (95% CI) was 99.9% (99.8% to 100%) (Table 16).
iii. Comparative accuracy of MPSS and TMPS
An indirect comparison of the 32 MPSS and seven TMPS studies showed no statistical evidence of a difference in sensitivity, specificity or both (P value = 0.11). The differences in sensitivity and specificity were negligible (Table 6).
9. All sex chromosome aneuploidies (SCA) combined
The sex chromosome aneuploidies (45,X, 47,XXX, 47,XXY and 47,XYY) were considered together as one target condition. SCA was assessed in 20 studies, comprising 286 affected cases and 9839 non SCA pregnancies. MPSS and TMPS were assessed by 14 and six studies, respectively. Among the 20 studies, 16 enrolled pregnant women selected at high risk of fetal aneuploidy and four enrolled a cohort of pregnant women with mixed prior risk. The results are summarised in Table 7. The results for mixed risk cohorts are summarised in Appendix 7.
a. Unselected population of pregnant women undergoing aneuploidy screening
No study assessed SCA in an unselected population of pregnant women.
b. Selected population of pregnant women at high risk of fetal aneuploidy
Sixteen studies involving 247 (2.93%) affected and 8420 non SCA pregnancies were included. MPSS and TMPS were assessed by 12 and four studies respectively (Figure 15).
15.
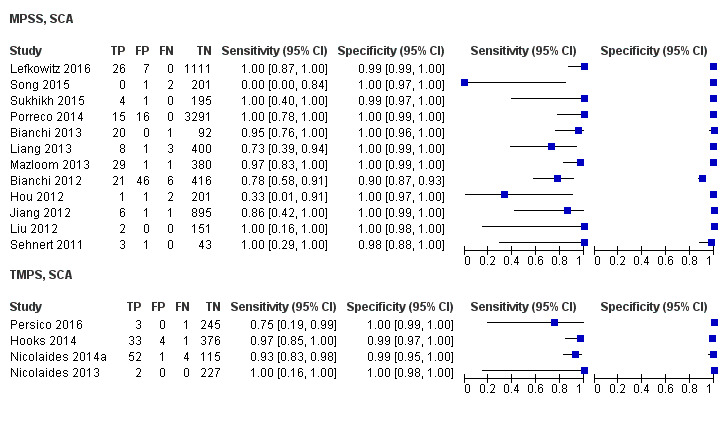
Forest plot of MPSS and TMPS for SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined) in pregnant women selected at high risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
i. MPSS
Twelve MPSS studies evaluated 151 affected and 7452 non SCA pregnancies (Bianchi 2012; Bianchi 2013; Hou 2012; Jiang 2012; Lefkowitz 2016; Liang 2013; Liu 2012; Mazloom 2013; Porreco 2014; Sehnert 2011; Song 2015; Sukhikh 2015). Of the 12 studies, seven included only women with singleton pregnancy, one evaluated singleton and multifetal pregnancies, and the remaining four studies did not report type of pregnancy. Results are presented separately for singleton and multifetal pregnancy studies in Table 16. Based on all 12 studies, the summary sensitivity (95% CI) was 91.9% (73.8% to 97.9%) and the summary specificity (95% CI) was 99.5% (98.8% to 99.8%).
ii. TMPS
Four TMPS studies evaluated 96 affected and 968 non SCA pregnancies in women with singleton pregnancy (Hooks 2014; Nicolaides 2013; Nicolaides 2014a; Persico 2016). The summary sensitivity (95% CI) was 93.8% (86.8% to 97.2%) and the summary specificity (95% CI) was 99.6% (98.1% to 99.9%).
iii. Comparative accuracy of MPSS and TMPS
An indirect comparison of the 12 MPSS and four TMPS studies showed no statistical evidence of a difference in sensitivity, specificity or both (P value = 0.41). The differences in sensitivity and specificity were negligible (Table 7).
10. gNIPT approach (MPSS or TMPS) against traditional screening tests
Five studies directly compared a gNIPT approach (MPSS or TMPS) and traditional screening tests for autosomal aneuploidies by using cohorts of pregnant women who were tested by both methods. Three studies compared TMPS and traditional screening tests, and two studies compared MPSS and traditional screening tests. The results are summarised in Table 2, Table 3, Table 4 and Table 6.
a. Unselected population of pregnant women undergoing aneuploidy screening
Only one study that compared TMPS and a traditional screening test evaluated T21, T18 and T13 individually in an unselected population of pregnant women undergoing aneuploidy screening (Norton 2015). This study evaluated 38, 10 and two cases of T21, T18 and T13, respectively and 15,803, 15,831 and 11,183 non T21, T18 and T13, respectively (Figure 16). Direct comparisons between gNIPT and traditional screening tests were not possible because there was only one study but authors observed eight, two and one cases of T21, T18 and T13 respectively missed by traditional screening test and only one T18 case missed by TMPS.
16.

Forest plot of traditional screening tests for T21, T18 and T13 in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
Four studies compared a gNIPT approach with a traditional screening test for autosomal aneuploidies (T21, T18 and T13 combined) in 22,367 unselected pregnant women (Figure 17). Three studies (Nicolaides 2012; Norton 2015; Quezada 2015) compared TMPS and first‐trimester combined test (Figure 18), and one study (Song 2013) compared MPSS and a second‐trimester triple test. The three TMPS studies had similar characteristics. Meta‐analyses of direct comparisons between gNIPT and traditional screening tests were not possible because traditional screening tests used different cutpoints and there were very few studies to enable estimation of summary sensitivity and specificity at specific cutpoints. Individual study results are presented in Table 17. Overall, 16 aneuploid cases were missed by traditional screening test and only five cases were missed by gNIPT approach. While specificity was consistently higher for TMPS than traditional screening tests, sensitivity was not consistently higher as shown in Figure 18.
17.
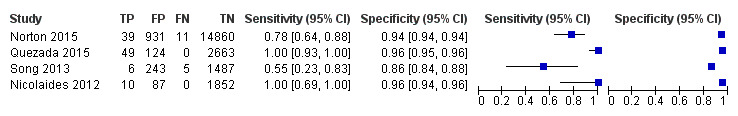
Forest plot of traditional screening tests for autosomes (T21, T18 and T13 combined) in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, TN: true negative and TP: true positive.
18.
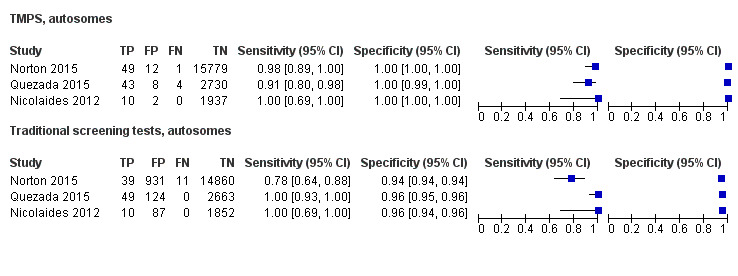
Forest plot of comparative studies of TMPS and traditional screening tests for autosomes (T21, T18 and T13 combined) in unselected pregnant women undergoing aneuploidy screening. FN: false negative, FP: false positive, TN: true negative and TP: true positive.
10. Direct comparisons of gNIPT and traditional screening tests for autosomes (T21, T18 and T13 combined) in unselected population of pregnant women undergoing aneuploidy screening.
| Study |
Sensitivity (true positives/cases) % |
Difference % (95% CI) |
Specificity (true negatives/unaffecteda) % |
Difference % (95% CI) |
||
| MPSS | Traditional screening tests | MPSS | Traditional screening tests | |||
| Song 2013 | 100 (11/11) | 54.6 (6/11) | 45.5 (10.0 to 72.0) | 99.9 (1729/1730) | 86.0 (1487/1730) | 14.0 (12.4 to 15.7) |
| TMPS | Traditional screening tests | TMPS | Traditional screening tests | |||
| Nicolaides 2012 | 100 (10/10) | 100 (10/10) | 0.00 (‐27.8 to 27.8) | 99.9 (1937/1939) | 95.5 (1852/1939) | 4.38 (3.51 to 5.40) |
| Norton 2015 | 98.0 (49/50) | 78.0 (39/50) | 20.0 (7.44 to 33.3) | 99.9 (15,779/15,791) | 94.1 (14,860/15,791) | 5.82 (5.46 to 6.20) |
| Quezada 2015 | 91.5 (43/47) | 100 (49/49) | ‐8.51 (‐19.9 to 0.40) | 99.7 (2730/2738) | 95.6 (2663/2787) | 4.16 (3.40 to 5.00) |
CI: confidence interval, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing.
aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
b. Mixed‐risk cohort of fetal aneuploidy
One study compared MPSS and traditional screening test for autosomal aneuploidies (T21, T18 and T13 combined) in a cohort with mixed prior risk of fetal aneuploidy including 1908 non T21, T18 and T31 pregnancies and four cases of autosomal aneuploidy (Bianchi 2014a). Traditional screening tests included first‐trimester combined test or a second‐trimester result (quadruple, serum integrated, fully integrated or sequential) (Figure 19). Overall, 80 unaffected pregnancies were detected as affected by traditional screening test against 12 for TMPS.
19.

Forest plot of traditional screening tests for autosomes (T21, T18 and T13 combined) in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
11. Pregnant women with mixed prior risk of fetal aneuploidy
Summary sensitivities and specificities for cohorts of pregnant women with mixed prior risk of fetal aneuploidy are presented in Appendix 7. For autosomal aneuploidies, 17 studies included 838 cases and 39,615 unaffected (non T21, T18 and T13) pregnancies. Of the 17 studies, 10 assessed MPSS and seven assessed TMPS (Figure 20). For T21, 16 studies included 614 cases (1.6%) and 37,887 non T21 pregnancies. Of the 16 studies, 10 assessed MPSS and six assessed TMPS. For T18, 13 studies included 166 cases (0.5%) and 36,206 non T18 pregnancies. Of the 13 studies, nine assessed MPSS and four assessed TMPS. For T13, 13 studies included 58 cases (0.1%) and 38,746 non T13 pregnancies. Eight of the 13 studies assessed MPSS and the other five assessed TMPS (Figure 21). For SCA, four studies included 39 cases and 1419 non SCA pregnancies; two of the studies assessed MPSS and the other two assessed TMPS (Figure 22). For 45,X, four studies included 34 cases (2.4%) and 1424 non 45,X pregnancies. Of the four studies, two studies assessed MPSS and two studies assessed TMPS. For 47,XXY, two studies (one of MPSS and one of TMPS) included three cases (1%) and 291 non 47,XXY pregnancies. For 47,XYY, two studies included two cases (0.5%) and 384 non 47,XYY pregnancies; one study assessed MPSS and the other study assessed TMPS. No study assessed gNIPT for 47,XXX in cohorts of pregnant women with mixed prior risk of fetal aneuploidy (Figure 23).
20.
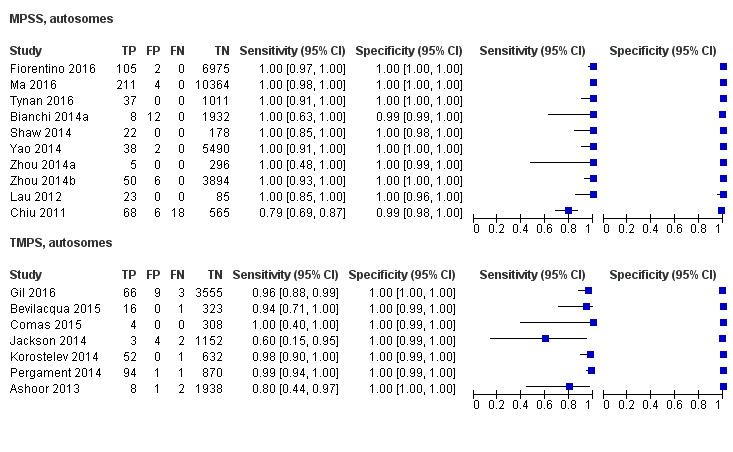
Forest plot of MPSS and TMPS for autosomes (T21, T18 and T13 combined) in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
21.
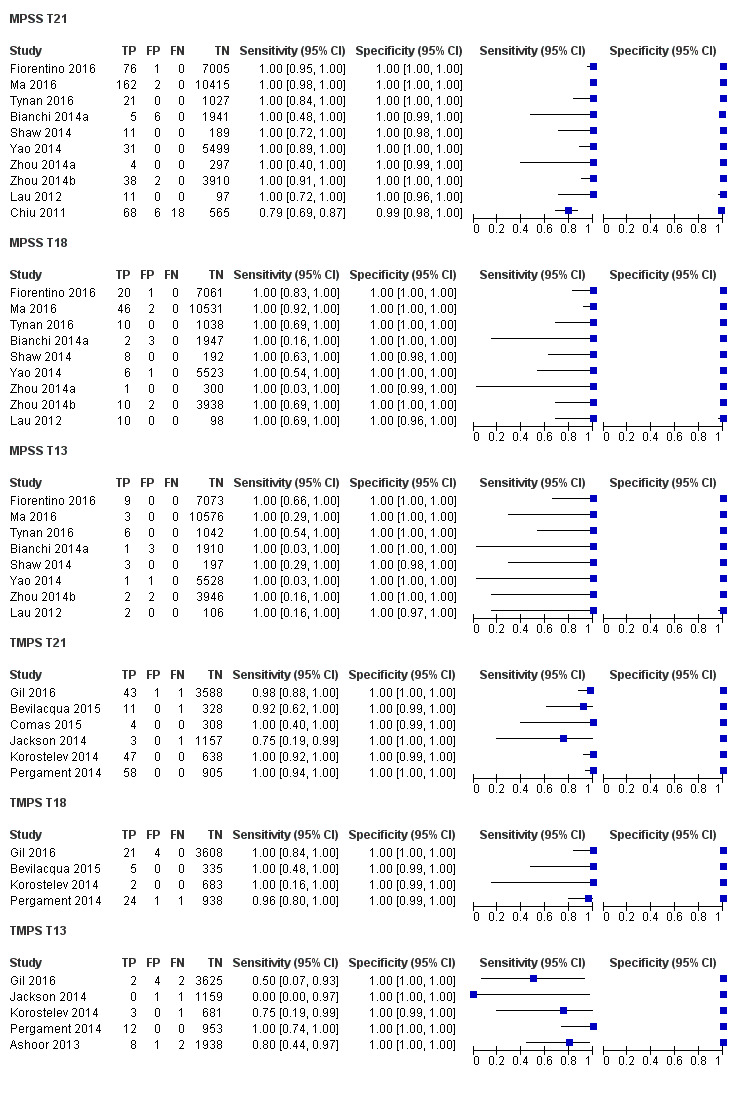
Forest plot of MPSS and TMPS for T21, T18 or T13 in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
22.
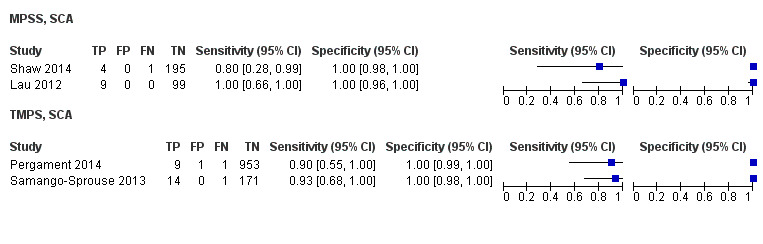
Forest plot of MPSS and TMPS for SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined) in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
23.
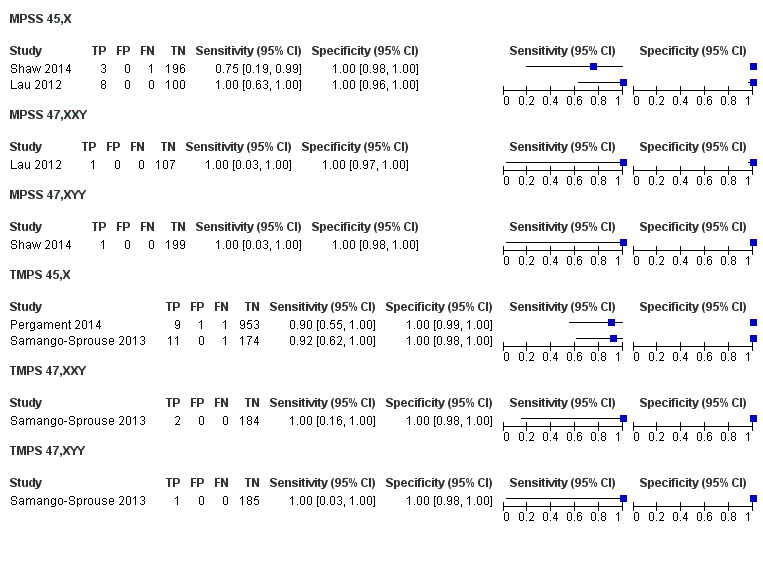
Forest plot of MPSS and TMPS for 45,X, 47,XXY or 47,XYY in pregnant women with mixed prior risk of fetal aneuploidy. FN: false negative, FP: false positive, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, TN: true negative and TP: true positive.
12. Failure rates
Table 14 shows the non‐negligible failure rate of gNIPT reported in the studies. gNIPT assay failure rate was reported in 46 out of 65 (71%) studies. The largest failure rate (25%) was observed in a study that used its own developed MPSS assay (Alberti 2015). The main reasons for assay failure included low amount of ccfDNA, low fetal fraction DNA and failure of sample to pass quality control. The failure rate ranged between 0% and 25% for MPSS and between 0.8% and 7.5% for TMPS. The number of aneuploid and euploid cases in failed samples was reported in 23 of 46 (50%) studies. Among these 23 studies, there were 1064 euploid cases and 79 aneuploid cases among 1143 failed samples. The failure rate among aneuploid cases, ranged between 0% and 50% for MPSS and between 0% and 23% for TMPS. The failure rate among euploid cases ranged between 0% and 6.7% for MPSS and between 1% and 7.6% for TMPS.
Investigation of heterogeneity
We planned to evaluate the effect of potential sources of heterogeneity such as type of reference standard and ethnicity. However, formal investigations using meta‐regression were not possible due to limited data and little or no heterogeneity in the sensitivities and specificities. Most studies (55%) used karyotyping while the remaining 29 studies (45%) used multiple reference standards. Ethnicity was not reported by 26 (40%) studies while the population in 21 (32%) studies was more than 50% Asian and in 18 (28%) studies the population was more than 50% Caucasian. In Appendix 8, the number of studies, affected and unaffected pregnancies are shown according to the gNIPT approach and prior risk of fetal aneuploidy. We also planned to assess gNIPT performance according to gestational age and gNIPT approach for autosomes and SCA aneuploidies. The accuracy of gNIPT appears to be high in all gestational age groups.
Sensitivity analyses
We did not perform sensitivity analyses to assess the effect of the interval between blood collection for gNIPT and fluid collection for reference standard because most studies had an acceptable interval between sample collection for index test and reference standard. Due to lack of data or lack of variability in estimates of sensitivity and specificity, analyses of the effect of high or unclear risk of bias according to the QUADAS‐2 domains were not done. We performed sensitivity analyses using data from all autosomes combined and all SCA combined in order to have enough studies to assess the impact of study design and number of cases. The results are presented in Table 18. Excluding case‐control studies or studies with less than 10 aneuploid cases had little or no impact on our findings.
11. Sensitivity analyses.
| Test |
Number of studies |
Number of affected pregnancies |
Number of unaffected pregnanciesa |
Summary sensitivity % (95% CI) |
Summary specificity % (95% CI) |
P valueb |
| Case‐control studies excluded | ||||||
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | 22 | 696 | 11,293 | 98.3 (95.1 to 99.4) | 99.9 (99.8 to 100) | 0.72 |
| TMPS | 4 | 219 | 3,813 | 98.6 (95.8 to 99.6) | 99.9 (99.8 to 100) | |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | 10 | 98 | 5,872 | 91.9 (73.8 to 97.9) | 99.5 (98.8 to 99.8) | 0.41 |
| TMPS | 2 | 6 | 472 | 93.8 (86.8 to 97.2) | 99.6 (98.1 to 99.9) | |
| Exclusion of studies with less than 10 pregnancies with aneuploidy | ||||||
| Autosomes (T21, T18 and T13 combined), selected high‐risk population | ||||||
| MPSS | 21 | 1458 | 13,921 | 98.7 (96.8 to 99.4) | 99.8 (99.5 to 100) | 0.07 |
| TMPS | 7 | 378 | 4,282 | 98.9 (97.2 to 99.6) | 99.9 (99.8 to 100) | |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), selected high‐risk population | ||||||
| MPSS | 6 | 130 | 5,761 | 94.5 (80.6 to 98.6) | 99.4 (97.6 to 99.8) | 0.28 |
| TMPS | 2 | 90 | 496 | 94.4 (87.3 to 97.7) | 99.0 (97.6 to 99.6) | |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13 CI: confidence interval, MPSS: massively parallel shotgun sequencing, SCA: sex chromosome aneuploidies, TMPS: targeted massively parallel sequencing.
aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bThe P value indicates the statistical significance of the difference in model fit and was obtained from likelihood ratio tests comparing models with and without a covariate for test type.
Discussion
Summary of main results
This review included data from 65 studies of 86,139 pregnant women (including 3141 aneuploids) tested by genomics‐based non‐invasive prenatal testing (gNIPT) and a reference standard. The gNIPT method used circulating cell‐free DNA (ccfDNA) in maternal blood for the detection of common fetal aneuploidies (T21, T18, T13, 45,X, 47,XXY, 47,XXX and 47,XYY). The number of gNIPT studies in unselected populations was limited (five studies), but 42 studies in high‐risk cohorts provided data for various meta‐analyses. Few (14%) studies included more than 100 aneuploid cases. Importantly, in almost all studies, the risk of bias was generally high with respect to patient selection as well as flow and timing. Some women can spontaneously lose their pregnancy after enrolment into a study. However, none of the studies reported such events. Since women with spontaneous abortions are likely to be lost to follow‐up, we believe that any risk of bias has been captured in the quality assessment of studies. Blood samples for gNIPT were mainly taken just before the invasive test (reference standard) and so pregnancies were unlikely to terminate naturally between the gNIPT and the reference standard. Across all studies, applicability concerns were low in the index test and reference standard domains.
These results show that massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS) perform similarly in terms of clinical sensitivity and specificity for the detection of fetal T21, T18, T13 and sex chromosome aneuploidy (SCA). However, no study compared the two approaches head‐to‐head in the same cohort of patients.
In high‐risk pregnancies, gNIPT methods (MPSS and TMPS) were highly accurate for detection of any of the three major trisomies (T21, T18 and T13) with sensitivities from 95.8% to 99.7% depending on specific trisomies and specificities above 99%. There were no statistically significant differences in accuracy between MPSS and TMPS.
In unselected cohorts of pregnant women, only one study evaluated MPSS. Based on meta‐analytic findings for each trisomy, TMPS appeared to be accurate for the detection of T21, with lower accuracy for T18 and T13. When compared to traditional prenatal screening tests, only four studies were identified (three for TMPS and one for MPSS). Genomics‐based non‐invasive prenatal testing showed greater specificity for T21 and T18 than traditional screening tests, while inconsistent results were observed for sensitivity. The inconsistency may be due to different cutpoints for traditional screening tests though one would expect that to also affect specificity. Given the small number of studies, the differences may be due to chance or there may be other differences between the studies that were not apparent.
With respect to the replacement of invasive tests, the performance of gNIPT observed in this review is not sufficient to replace current invasive diagnostic tests.
We also compared the diagnostic test accuracy of MPSS and TMPS for all three autosomes combined because gNIPT is being clinically proposed as one test during prenatal follow‐up to detect any of the three conditions. Under this scenario, in high‐risk pregnancies of fetal aneuploidy, there was no statistically significant difference in diagnostic accuracy between MPSS and TMPS. In unselected cohorts of pregnant women, a test comparison was not possible due to limited data.
There was paucity of data for each SCA. In high‐risk cohorts, all SCAs combined gave a pooled sensitivity (95% CI) and specificity (95% CI) of 91.9% (73.8% to 97.9%) and 99.5% (98.8% to 99.8%) from 12 MPSS studies. The pooled sensitivity (95% CI) and specificity (95% CI) were 93.8% (86.8% to 97.2%) and 99.6% (98.1% to 99.9%) from four TMPS studies. SCAs are considered “incidental” findings of current aneuploidy screening programs. It should be noted that SCAs are not of interest for prenatal screening since they do not lead to any intervention prior to birth.
The failure rate associated with gNIPT, which is higher than the current failure rate of traditional screening tests which is close to zero, is worrying and may be a source of bias. Futhermore, the large heterogeneity between laboratory‐developed assays in their protocol details and observed failure rates highlight the fact that each laboratory providing gNIPT services should determine its own failure rate and inform healthcare professionals ordering the test about this important test characteristic. Failed samples were excluded from the analyses in the studies. This systematic review found a slightly larger failure rate for TMPS than the MPSS approach. This was also reported by Yaron 2016. We also found that the proportion of failed samples for aneuploid samples was higher than the proportion of failed samples for euploid samples. If these failed samples were included in the summary statistics, the diagnostic performance of gNIPT would be lower.
Comparison with other systematic reviews with meta analysis
At the time of writing, there are four published systematic reviews with meta‐analyses of gNIPT (Gil 2015a; HAS 2015; Mackie 2017; Taylor‐Phillips 2016). Although these meta‐analyses had different criteria for including studies and analyses, they reported similar sensitivities and specificities to our findings.
As reported by Gil 2015a, the detection rate of gNIPT for autosomes was between 91.0% to 99.2% and specificity above 99.9% in singleton pregnancies. The detection rate for 45,X and SCA other than 45,X was 90.3% and 93.0%, respectively with specificity above 99.8% in singleton pregnancies. The results from HAS 2015 group for T21 were respectively 98.0% and 99.9% for sensitivity and specificity. Regarding Mackie 2017, the sensitivity was between 90.6% to 99.4% and specificity above 99.9% for autosomes. For 45,X, the sensitivity and specificity was 92.9% and 99.9%, respectively. They also pointed out that failed results were poorly reported across studies. Finally, Taylor‐Phillips 2016 reported sensitivity between 97.4% to 99.3% for autosomes and specificity of 99.9%.
This is the first Cochrane diagnostic test accuracy (DTA) review on gNIPT. There are five published Cochrane DTA reviews on prenatal screening tests (Alldred 2012; Alldred 2015; Alldred 2015a; Alldred 2017a; Alldred 2017b). The suite of reviews addressed traditional biochemical, ultrasound and urine markers for Down syndrome screening (Alldred 2010) and none of the other fetal aneuploidies considered in this review were evaluated in this suite. In the first of the three reviews, Alldred and colleagues evaluated second‐trimester serum markers and found that double and triple test combinations (involving alpha‐fetoprotein, human chorionic gonadotropin (hCG) (free and total) or unconjugated estriol) significantly outperformed individual markers, detecting six to seven out of every 10 Down syndrome pregnancies at a 5% false positive rate (Alldred 2012). The second review evaluated first‐trimester serum markers and found that a test strategy involving maternal age, PAPP‐A and free ßhCG significantly outperformed individual markers, detecting about seven out of every 10 Down’s syndrome pregnancies at a 5% false positive rate (Alldred 2015a). The third review evaluated urine markers and concluded there was a paucity of evidence to support the use of urine testing for Down syndrome screening (Alldred 2015b). The fourth review evaluated first‐trimester ultrasound tests alone or in combination with first‐trimester serum tests and found that a combination of ultrasound and serum markers (especially PAPP‐A and free ßhCG) and maternal age can detect about nine of 10 T21 affected pregnancies for a fixed 5% false positive rate (Alldred 2017a). The fifth review evaluated first‐ and second‐trimester serum tests with and without first‐trimester ultrasound tests and found that a combination of first‐trimester ultrasound with first‐ and second‐trimester serum markers with maternal age are significantly better than those without ultrasound or those evaluating first‐trimester ultrasound in combination with second‐trimester serum markers, without first‐trimester serum markers (the authors cannot make recommendations about a specific strategy) (Alldred 2017b).
Strengths and weaknesses of the review
Strengths
The review methodology was transparent with the full protocol published in the Cochrane Library (1 July 2015) and in PROSPERO (11 November 2015). The review evaluated the screening and diagnostic accuracy of gNIPT by MPSS and TMPS for seven common aneuploidies with no restriction imposed on population characteristics such as maternal age, gestational age, aneuploidy risk, number of fetuses and ethnicity. We performed a comprehensive search with no language restriction and we included studies in the languages used by various authors in the field, including Chinese, Bulgarian, Russian, Polish, Korean and Spanish. Study selection, data extraction and quality assessment were independently performed by two review authors. We contacted authors to clarify data and to avoid duplication of data as a result of overlapping populations.
We evaluated the performance of the two major gNIPT methods (MPSS and TMPS which included digital analysis of selected regions (DANSR) and single nucleotide polymorphism (SNP)‐based method) and included data on traditional screening tests when compared to gNIPT.
We collected and reported data on excluded and failed samples and presented the failure rate at first attempt, the number of repeated tests and the final failure rate for each study. When it was possible, we also reported separate failure rates among aneuploid and euploid cases. Where possible, we performed subgroups analyses to investigate heterogeneity, and also performed sensitivity analyses to assess the robustness of these findings.
Weaknesses
Fetal karyotyping is the reference standard for establishing a diagnosis of fetal aneuploidy. This is an invasive procedure with some risk for the fetus and the pregnant woman. Many pregnant women included in the studies, especially those involving unselected cohorts, were not tested by karyotyping. Rather, clinical examination of the newborn or medical records from birth were used as a secondary reference standard. We are aware that these secondary reference standards are not as accurate as fetal karyotype and some cases may have been missed.
Studies rarely reported the qualification of the person conducting the neonatal clinical examination at birth. Such examination is expected to be more reliable if it was made by a paediatrician or a geneticist. Ideally, this examination should be done a few months after birth because the phenotypic characteristics of aneuploidies are more apparent than at birth (Devlin 2004).
Genomics‐based non‐invasive prenatal testing assays are laboratory‐developed tests that are not standardised in their methods, sequencing platforms, sequencing data manipulation, measures used or cut‐offs for interpretation. Each assay was developed and validated by the testing laboratory and each laboratory has a different method. Usually detailed information about the assays were not available. As shown in Table 12, 15, different gNIPT assays were used in the studies included in this review. Thus, they may differ in various aspects and show different analytical and clinical validity. We have grouped them accordingly to the type of assay used (targeted versus shotgun), but there are also differences within each of these two subgroups that we were not able to account for, given the small number of studies published on most of these different assays. Thirteen of the assays were used only in studies of high‐risk pregnancies or mixed cohorts. Only a few gNIPT assays were used in a significant number of studies. Thus, caution should be used before generalising the diagnostic accuracy observed in this category of patients to all gNIPT assays. This limits the generalisability of these findings and we cannot infer that all gNIPT assays will show the same performance.
Applicability of findings to the review question
These findings suggest that gNIPT has high sensitivity and specificity for detection of fetal aneuploidies in high‐risk pregnancies. Performance varied depending on the type of aneuploidy. There was limited evidence of the performance of gNIPT in unselected cohorts of pregnant women. Most studies involved either high‐risk pregnancies or mixed populations where it was not possible to differentiate between high‐risk pregnancies and unselected pregnant women. Thus, more studies are needed in the general population of pregnant women before firm conclusions can be made about the sensitivity of gNIPT as a first‐tier screening test. The two major types of gNIPT method (MPSS and TMPS) appear to have comparable performance, but there are many different gNIPT assays for each approach. For many of these assays, very little data have been published about their diagnostic accuracy. Additionally, performance in the cohorts studied may not reflect performance in other populations owing to differences in fetal fraction distribution because of, for example, differences in mean body mass index or gestational age. Importantly, summary sensitivities and specificities derived from cohort data can be very different from the probability associated with any particular patient sample to be positive or negative depending on the sample’s specific fetal fraction. Thus, summary sensitivity, specificity and associated predictive values of an assay cannot be used as a straightforward measure of the probability of a specific patient’s sample to be affected given a positive or negative result. This underscores the importance, before clinically offering a laboratory developed gNIPT assay, that it is fully validated according to recognised best practice clinical laboratory molecular diagnostics guidelines. Finally, the methodological quality of studies was generally poor with high risk of bias, especially in terms of patient selection and flow and timing.
Authors' conclusions
Implications for practice.
Genomics‐based non‐invasive prenatal testing (gNIPT) appears to be an accurate prenatal screening test, its accuracy having been evaluated as a second‐tier screening test to identify pregnancies at very low risk of fetal aneuploidies (T21, T18 and T13) and thus to decrease the false positive rate of traditional screening approaches and avoid invasive procedures in those pregnant women. As a first‐tier aneuploidy screening test, based on limited data from comparative studies, gNIPT appears to have significantly better specificity than current screening approaches using maternal serum biochemical markers, ultrasound or both, but evidence about sensitivity is inconsistent. At current gNIPT pricing levels, gNIPT as a second‐tier screening test provides the best value for money, especially for publicly‐funded screening programs while gNIPT as a first‐tier screening test was found not to be cost‐effective (Nshimyumukiza 2017). The failure rate of gNIPT is a concern as it is substantially larger than the current failure rate of traditional prenatal screening approaches.
It is worth noting that gNIPT shows good performance for the detection of sex chromosome aneuploidies though data are sparse. The number of studies for sex chromosome aneuploidy (SCA) was small and confidence intervals on sensitivity and specificity estimates are therefore wide. Thus, sex chromosome aneuploidies appear to be more difficult to detect since performances of gNIPT are not as good as for detecting autosomal aneuploidies. SCAs are considered “incidental” findings of current aneuploidy screening programs and they do not lead to any intervention prior to birth.
Maternal serum screening, ultrasound fetal examination, gNIPT and invasive diagnostic tests are thus complementary approaches because in its current state, gNIPT cannot detect all chromosomal abnormalities or adverse obstetrical outcomes. About 44% to 64% of all chromosomal abnormalities found during prenatal diagnostic are common aneuploidies which gNIPT can detect (Kazerouni 2011; Shani 2016). Counselling expectant mothers and their partners is essential for explaining the advantages, limitations and risks of these procedures.
We conclude that given the current data on the performance of gNIPT, invasive fetal karyotyping is still the required diagnostic approach to confirm the presence of a chromosomal abnormality prior to making irreversible decisions relative to the pregnancy outcome.
Implications for research.
This systematic review has highlighted the fact that most published studies on gNIPT have high risk of bias in the patient selection and flow and timing domains. Many different gNIPT assays are in use and for the majority of them, there is insufficient published data to individually assess their clinical performance. Therefore, the results in this systematic review may not be generalisable to all gNIPT assays. Studies are needed that directly compare the accuracy of gNIPT with that of current traditional prenatal screening methods for fetal aneuploidy, especially in unselected populations of pregnant women. Such studies can provide valid evidence of the incremental accuracy of gNIPT if gNIPT is being considered as a first‐tier test. Particular attention should be paid to study design in order to minimise patient selection biases as well as biases in flow and timing domain. Further well‐designed, independent large‐scale studies on real life gNIPT’s implementation into prenatal care should be performed. Large scale randomised clinical trials of tests and patient outcomes are needed to validate the clinical utility of gNIPT in the various clinical settings. Given the rapid evolution of gNIPT and its capacity to detect other fetal chromosomal anomalies (Benn 2016), future systematic reviews may have to widen the scope of target conditions.
History
Protocol first published: Issue 7, 2015 Review first published: Issue 11, 2017
| Date | Event | Description |
|---|---|---|
| 2 July 2015 | Amended | Minor changes to author affiliations. |
Acknowledgements
We thank the Editorial Staff of Cochrane Pregnancy and Childbirth and the Editorial Team for Cochrane Diagnostic Test Accuracy Reviews for their support. We also thank Lynn Hampson for her help with development of the search strategy and members of "Cochrane Canada Francophone" for their support.
As part of the pre‐publication editorial process, this review has undergone dual peer review with Cochrane Pregnancy and Childbirth and the Cochrane Diagnostic Test Accuracy Reviews Unit and has been commented by peer reviewers.
This project was supported by the National Institute for Health Research, via Cochrane Infrastructure funding to Cochrane Pregnancy and Childbirth. The views and opinions expressed therein are those of the authors and do not necessarily reflect those of the Systematic Reviews Programme, NIIHR, NHS or the Department of Health.
We wish to thank our translators. Thanks to Dr Ze Zhang for his help with Chinese publications, Roumiana Gulemetova for her help with Bulgarian and Russian publications, Sylvie Giroux for her help with Spanish publications, Richard Szostak for his help with Polish publication and Hyun Jin Park for her help with Korean publication.
Appendices
Appendix 1. Screening tests and medical terms glossarya
| Terms | Definitions |
| Amniocentesis | Invasive procedure under continuous ultrasound guidance (performed between 15 to 19 weeks of gestational age). A sterile needle is passed through the mother’s abdomen, uterus and amniotic sac. A sample of fetal cells present in the amniotic fluid surrounding the fetus is aspirated with a syringe and sent for analysis to test for a range of chromosomal and inherited disorders. |
| Aneuploidy | The state of having a different (additional or missing) number of chromosomes than the 23 pairs normally present in humans. |
| Attention deficit disorder (ADD) | ADD is a neurodevelopmental disorder defined by impairing levels of inattention and disorganisation. Inattention manifests behaviourally in ADD as wandering off task, lacking persistence, having difficulty sustaining focus and being disorganised. |
| Case‐control study | In the context of diagnostic accuracy, existing records are used to identify a group of people known to have the target condition (cases) and another group (controls) without the target condition. The control group may consist of healthy individuals or those with other conditions similar to the target condition Cases and controls are then compared with respect to certain variables hypothesised to increase the risk of having the disease. |
| Chorionic villus sampling (CVS) | An abdominal or cervical procedure performed under continuous ultrasound guidance to obtain a sample of placental tissue for chromosomal or genetic analysis (between 12 to 19 weeks of gestational age). The range of chromosomal and genetic conditions that can be detected is similar to those for amniocentesis. |
| Clinodactyly | Permanent deflection of one or more fingers. |
| Cut‐off | Synonyms: cutpoint or threshold. |
| Cutpoint | A value for a test result measured on an ordinal or continuous scale which divides the group of people tested into a group at lower risk of the condition being screened for and a group at higher risk (for whom further investigations may be offered). Synonyms: cut‐off or threshold. |
| Detection rate | The proportion of affected individuals with a positive screening result. The detection rate is the same as the sensitivity of a test. |
| Developmental delay | An individual with this neurodevelopmental disorder fails to meet expected developmental milestones in several areas of intellectual functioning. |
| Diagnostic accuracy | The ratio of true positive and true negative results to the total number of test results (true positives, true negatives, false positives and false negatives). Represents the level of agreement between the information from the index test and the reference standard. |
| Diagnostic test | A test recognised as having best performances to provide sufficient information allowing a definite diagnosis (as opposed to screening test results that need to be confirmed before a final diagnosis can be reached). |
| Dysgenesis | Defective or abnormal formation of an organ or part, primarily during embryonic development. Gonodal dysgenesis is a defective development of the gonads, which may be accompanied by abnormalities of the sex chromosomes. |
| False negative | A negative test result in someone with the target condition. |
| False positive | A positive test result in someone without the target condition. |
| Fluorescence in situ Hybridisation |
Describes a type of DNA analysis by the hybridisation of fluorescently‐labelled probes complementary to certain genomic regions. In the context of fetal aneuploidy detection, describes a diagnostic test in which chromosome‐specific fluorescently‐labelled DNA probes are used on uncultured cells from chorionic villi or amniotic fluid to assess the number of homologous chromosome copies present. |
| High risk of fetal aneuploidy | A pregnancy is considered at high risk of fetal aneuploidy if the result of the prenatal screening test puts the fetus at increased risk for aneuploidy. |
| Hypertelorism | Abnormal distance between two paired organs. |
| Intellectual disability or intellectual developmental disorder |
This neurodevelopmental disorder is characterised by deficits in general mental abilities, such as reasoning, problem solving, planning, abstract thinking, judgment, academic learning and learning from experience. It is a condition diagnosed before age 18. In the past, the term mental retardation was used to describe this condition but this term is no longer used. |
| Invasive procedure | A method used to obtain a biological sample that involves significant disruption of the physical integrity of a patient. Examples include amniocentesis and chorionic villi sampling. |
| Karyotype | A photomicrograph of an individual’s chromosomes arranged in a standard format and visualised by various staining methods, showing the number, size, and shape of each chromosome; used to correlate chromosomal anomalies with specific diseases (karyotyping). In humans, there are a total of 23 pairs of homologous chromosomes (total of 46 chromosomes). |
| Learning disability | Learning disability refers to inadequate development of specific academic, language and speech skills such as reading disability, mathematics disability and writing disability. |
| Low risk of fetal aneuploidy | A pregnancy is considered at low risk of fetal aneuploidy if the result of the prenatal screening test puts the fetus at decreased risk for aneuploidy. |
| Meta‐analysis | The use of statistical techniques in a systematic review to integrate the results of included studies. Sometimes misused as a synonym for systematic reviews which may or may not include a meta‐analysis. |
| Mosaic | An individual who has some cells with an unusual genetic or chromosomal make‐up while the rest of the cells in the body have the typical genetic or chromosomal constitution. |
| Mixed risk population | Mixed risk population included a mixture of selected pregnant women with low, high or no prior risk of fetal aneuploidy. |
| Negative predictive value | A measure of test performance. Defined as the proportion of people with a negative test result who do not have the target condition. |
| Nuchal translucency scan | The thickness of fluid in the tissue space within the nape of the fetal neck typically measured by ultrasonography. An increased amount of fluid is associated with Down syndrome and other structural or genetic anomalies. |
| Positive predictive value | A measure of test performance. Defined as the proportion of people with a positive test result who do have the target condition. |
| Probability | The chance or risk of an event happening. |
| Prospective study | A study in which a group of individuals is followed through time in order to detect the occurrence of a disease or another outcome of interest. |
| Reference standard | The best available test to detect the presence or absence of the target condition. |
| Retrospective study | A study in which all or part of the data collection occurred before initiation of the study. |
| Screening | Testing asymptomatic people for the likelihood of the presence of a disease, either with the aim of reducing risk of an adverse outcome, or with the aim of giving information about risk. |
| Seizure | A sudden attack, spasm, or convulsion caused by abnormal electrical conduction in the brain. |
| Sensitivity | A measure of test performance. Defined as the proportion of individuals with the target condition who have a positive test result. Higher sensitivity values means that a higher proportion of affected individuals will be detected by the test (few false negatives).Sensitivity is the same as the detection rate. |
| Single nucleotide polymorphism |
Single nucleotide polymorphisms are the most common type of genetic variation among people. A difference in a single DNA nucleotide (A, T, C or G) in a DNA sequence. |
| Specificity | A measure of test performance. Defined as the proportion of individuals without the target condition who have a negative test result. Higher specificity values means that a smaller proportion of unaffected individuals will be wrongly classified as having the target condition (few false positives). |
| Threshold | Synonyms: cutpoint or cut‐off. |
| True negative | An individual with a negative test result who does not have the target condition. |
| True positive | An individual with a positive test result who has the target condition. |
| Trisomy | Three copies of a particular chromosome rather than the usual pair. |
| Unselected pregnant women | A pregnant women who did not undergo any prenatal screening test at the time of enrolment. |
aAdapted in part from the United Kingdom National Screening Committee Glossary, MedlinePlus Medical Encyclopedia, American Psychiatric Association and The Cochrane Collaboration’s Glossary of terms (APA 2013; Cochrane Glossary 2014; MedlinePlus 2014; UK Screening Glossary 2012).
Appendix 2. List of acronyms and abbreviations
| Acronyms or abbreviations | Terms |
| 45,X | monosomy X or Turner syndrome |
| 47,XXX | trisomy X or triple X syndrome |
| 47,XXY | Klinefelter syndrome |
| aCGH | array comparative genomic hybridisation |
| AFP | alpha‐fetoprotein |
| Bioch/US | biochemical or ultrasound or both screening test |
| ccfDNA | circulating cell‐free DNA |
| Chrom. 21 | chromosome 21 |
| CVS | chorionic villi sampling |
| FISH | fluorescence in situ hybridisation |
| gNIPT | genomics‐based non‐invasive prenatal testing |
| hCG | human chorionic gonadotropin |
| HSROC | hierarchical summary receiver operating characteristic |
| MPSS | massively parallel shotgun sequencing |
| NA | not applicable |
| ND | no data available |
| NGS | next generation sequencing |
| NR | not reported |
| NT | nuchal translucency |
| PAPP‐A | pregnancy associated plasma protein A |
| QF‐PCR | quantitative fluorescent polymerase chain reaction |
| QUADAS‐2 | QUality Assessment of Diagnostic Accuracy Studies |
| Ref. Chrom | reference chromosome |
| RS | reference standard |
| SCA | sex chromosome aneuploidy |
| SNP | single nucleotide polymorphism |
| T13 | trisomy 13 or Patau syndrome |
| T18 | trisomy 18 or Edward syndrome |
| T21 | trisomy 21 or Down syndrome |
| TMPS | targeted massively parallel sequencing |
| uE3 | unconjugated estriol |
Appendix 3. Index test technical details
Typically, blood samples from pregnant women are obtained by venous puncture in the first or second trimester. After two centrifugation steps, plasma is separated from maternal whole blood and ccfDNA is extracted from plasma with commercial kits. DNA is converted into a genomic library where each of the DNA fragments are ligated with platform specific adapters. For TMPS only, libraries are clonally amplified before being sequenced. Then the libraries of several pregnant women are loaded on a next generation sequencer. The produced sequencing reads are aligned on a reference human genome to their respective chromosomal location and the number of sequence reads from each chromosome is computed (Rothberg 2011). MPSS randomly sequences DNA fragments from across the whole genome while TMPS sequences DNA fragments from selected regions (Figure 1). Ultimately, all gNIPT for aneuploidies rely on assigning sequence reads of DNA fragments to their chromosome of origin and comparing total number or proportions of reads or single nucleotide polymorphisms (SNP) genotype between each chromosome of interest (e.g. 13, 18, 21, X and Y) and a reference set of chromosomes. A Z score (or other statistics) are computed and a patient‐specific risk can be assessed based on a risk threshold determined from read counts from a series of known euploid and aneuploid pregnancies. For MPSS, the counts from chromosomes of interest are normalised using the counts from all other chromosome sequences, while, for TMPS, the counts are normalised against a subset of selected sequences. Bioinformatic approaches vary according to the testing approach (MPSS or TMPS) and research team. Besides the use of normalised chromosome read counts, TMPS also allows for the use of additional allelic information when polymorphic loci such as SNP are targeted, such as an estimate of fetal DNA concentration (fetal DNA proportion) (Liao 2012). Thus, while MPSS produces a larger number of total sequence reads, TMPS will generates a larger number of reads from each targeted chromosomes.
Appendix 4. Search strategy
| MEDLINE (Ovid) | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | 'cell‐free dna'.mp | Index test |
| 2 | 'cell free dna'.mp | |
| 3 | cfdna.mp | |
| 4 | ffdna.mp | |
| 5 | cffdna.mp | |
| 6 | 'free foetal dna'.mp | |
| 7 | 'free fetal dna'.mp | |
| 8 | nipd.mp | |
| 9 | nipt.mp | |
| 10 | (non invasive or noninvasive or non‐invasive).mp | |
| 11 | (genetic adj2 (diagnos* or detect* or test* or screen*)).mp | |
| 12 | exp Genetic Testing/ | |
| 13 | exp Sequence Analysis, DNA/ | |
| 14 | ((antenatal or ante natal) adj2 (diagnos* or detect* or test* or screen*)).mp | |
| 15 | ((prenatal or pre natal) adj2 (diagnos* or detect* or test* or screen*)).mp | |
| 16 | exp Prenatal Diagnosis/ | |
| 17 | or/1‐16 | |
| 18 | maternal.mp | Patient description |
| 19 | exp Pregnancy/ | |
| 20 | exp Pregnancy Complications/ | |
| 21 | pregnant.mp | |
| 22 | pregnanc*.mp | |
| 23 | exp Fetus/ | |
| 24 | fetus.mp | |
| 25 | foetus.mp | |
| 26 | fetal.mp | |
| 27 | foetal.mp | |
| 28 | or/18‐27 | |
| 29 | trisom*.mp | Target condition |
| 30 | aneuploid*.mp | |
| 31 | (down* adj syndrome*).mp | |
| 32 | exp Aneuploidy/ | |
| 33 | exp Trisomy/ | |
| 34 | exp Down Syndrome/ | |
| 35 | chromosome disorders.mp | |
| 36 | or/29‐35 | |
| 37 | or/1‐5,11‐13 | Combined sets |
| 38 | 6 or 7 or 8 or 9 or 14 or 15 or 16 | |
| 39 | 36 and 37 and 28 and 10 | |
| 40 | 38 and 37 and 36 | |
| 41 | 39 or 40 | Final combined set |
| Embase (Embase.com) | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | 'cell‐free dna' | Index test |
| 2 | 'cell free dna' | |
| 3 | cfdna | |
| 4 | ffdna | |
| 5 | cffdna | |
| 6 | 'free foetal dna' | |
| 7 | 'free fetal dna' | |
| 8 | nipd | |
| 9 | nipt | |
| 10 | 'non invasive' | |
| 11 | noninvasive | |
| 12 | 'non‐invasive' | |
| 13 | genetic NEXT/1 (diagnos* or screen* or test* or detect*) | |
| 14 | 'genetic screening'/exp | |
| 15 | 'genetic testing'/exp | |
| 16 | 'sequence analysis dna'/exp | |
| 17 | antenatal NEXT/1 (diagnos* or screen* or test* or detect*) | |
| 18 | prenatal NEXT/1 (diagnos* or screen* or test* or detect*) | |
| 19 | pre?natal NEXT/1 (diagnos* or screen* or test* or detect*) | |
| 20 | 'prenatal diagnosis'/exp | |
| 21 | 1 OR 2 OR 3 OR 4 OR 5 OR 6 OR 7 OR 8 OR 9 OR 10 OR 11 OR 12 OR 13 OR 14 OR 15 OR 16 OR 17 OR 18 OR 18 OR 19 OR 20 |
|
| 22 | antenatal | Patient description |
| 23 | prenatal | |
| 24 | pre?natal | |
| 25 | maternal | |
| 26 | foetus | |
| 27 | fetus | |
| 28 | foetal | |
| 29 | fetal | |
| 30 | pregnanc* | |
| 31 | pregnant | |
| 32 | 'pregnancy'/exp | |
| 33 | 'pregnancy complications'/exp | |
| 34 | 'pregnant woman'/exp | |
| 35 | 'pregnant women'/exp | |
| 36 | 22 OR 23 OR 24 OR 25 OR 26 OR 27 OR 28 OR 29 OR 30 OR 31 OR 32 OR 33 OR 34 OR 35 |
|
| 37 | trisom* | Target condition |
| 38 | aneuploid* | |
| 39 | down* NEXT/1 syndrome | |
| 40 | 'aneuploid'/exp | |
| 41 | 'aneuploidy'/exp | |
| 42 | 'trisomy'/exp | |
| 43 | 'downs syndrome'/exp | |
| 44 | 'down syndrome'/exp | |
| 45 | 'chromosome disorders' | |
| 46 | 37 OR 38 OR 39 OR 40 OR 41 OR 42 OR 43 OR 44 OR 45 | |
| 47 | 21 and 36 and 46 | Final combined set |
| Web of Science (ISI) | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | TOPIC: (down* syndrome) ORTOPIC: (trisom*) ORTOPIC: (aneuploid*) | Target condition |
| 2 | TOPIC: (pregnan*) | Patient description |
| 3 | TOPIC: (dna) ORTOPIC: (blood) | Index test |
| 4 |
TOPIC: (pre?natal screen*) ORTOPIC: (prenatal screen*) ORTOPIC: (pre?natal test*) ORTOPIC: (prenatal test*) ORTOPIC: (genetic test*) ORTOPIC: (genetic screen*) ORTOPIC: (prenatal diagnos*) ORTOPIC: (pre?natal diagnos*) ORTOPIC: (detection) ORTOPIC: (genetic diagnos*) ORTOPIC: (non invasive) ORTOPIC: (non‐invasive) ORTOPIC: (noninvasive) |
Index test |
| 5 | #1 AND #2 AND #3 AND #4 | Final combined set |
| Cochrane Register of Diagnostic Test Accuracy Studies |
| Hand search: in diagnostic test accuracy database, there are 18 publications from the Cochrane Pregnancy and Childbirth group. |
| Clinicaltrials.gov | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | (down syndrome OR trisomy OR aneuploidy) | Target condition |
| 2 | (testing OR screening OR diagnosis OR detection) | Index test |
| 3 | #1 AND #2 | Final combined set |
| European Clinical Trials Register | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | pregnan* | Population |
| 2 | trisom* OR aneuploid* | Target condition |
| 3 | #1 OR #2 | Final combined set |
| Who ICTRP | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | screen* OR detect* OR diagnos* OR test* OR pregnan* | Index test and population |
| 2 | «down syndrome » OR trisom* OR aneuploid* | Target condition |
| 3 | #1 AND #2 | Final combined set |
| NTIS.gov | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | (down syndrome OR trisomy OR aneuploidy) | Target condition |
| 2 | (testing OR screening OR diagnosis OR detection) | Index test |
| 3 | #1 AND #2 | Final combined set |
| OpenGrey | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | "down syndrome" OR trisom* OR aneuploid* | Target condition |
| 2 | screen* OR detect* OR diagnos* OR test* OR pregnan* | Index test and population |
| 3 | #1 AND #2 | Final combined set |
| National Guideline Clearing House (NGCH) | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | diagnosis (guideline category) | Index test |
| 2 | screening (guideline category) | Index test |
| 3 | aneuploid OR trisomy OR « down syndrome » | Target condition |
| 4 | (#1 OR #2) AND #3 | Final combined set |
| TheseNet | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | trisomy | Target condition |
| 2 | screening | Index test |
| 3 | #1 AND #2 | Final combined set |
| These Canada Portal | ||
| Steps | Text words and subject headings | Sets of search |
| 1 | trisomy OR (down AND syndrome) | Target condition |
| 2 | screening | Index test |
| 3 | #1 AND #2 | Final combined set |
Appendix 5. Data collection form for study classification during full‐text assessment
| Heading | Detailed instructions | Data |
| Study ID | Last name of the first author and year of publication | Name: Year: |
| Reference details | Details allowing identification of the publication | Journal: Volume: Issue: Pages: Accession number (e.g. PMIDa): |
| Multiple reports of this study |
For example, duplicate publications or follow‐up studies. Provide the study ID linked to this classified study |
Study ID: |
| Type of report | Check the appropriate box | Journal article ❑ Conference/abstract ❑ Ongoing trial ❑ Others ❑ Specify: |
| Language | In which language was the study written | Language: Translation needed? Yes ❑ No ❑ |
| Eligibility | Provide reason for exclusion or awaiting classification (e.g. why authors should be contacted and what issues should be clarified) |
Study excluded? Yes ❑ No ❑
Reason: Awaiting classification? Yes ❑ No ❑ Reason: |
| Report author contact details for further information |
Date when the authors were contacted (dd/mm/yyyy) | No need for further contact ❑ Authors have been contacted on: Reply received on: |
| Review author ID | Who completed the form | Name: |
| Date of classification | (dd/mm/yyyy) | Date: |
| Notes, questions or reminders |
||
aPMID: PubMed identifier.
Appendix 6. QUADAS‐2 tool for assessing methodological quality of included studies
| Signalling question | Signalling question | Signalling question | Risk of bias |
Concerns about applicability |
|
| Domain 1: Patient selection | |||||
|
Patient selection |
Was a consecutive or random sample of patients enrolled? | Was a case‐control design avoided? | Did the study avoid inappropriate exclusions? | Could the selection of patients have introduced bias? | Are there concerns that the included patients and setting do not match the review question? |
|
Yes: if all consecutive or random samples or convenient samples or all eligible pregnant women were enrolled. No: if selected pregnant women were enrolled. Unclear: if this was not clear from the report. |
Yes: if a case‐control design was avoided. No: If a case‐control design was not avoided. Unclear: if this was not clear from the report. |
Yes: if the study avoided inappropriate exclusions. No: if pregnant woman was excluded based on family's situation, maternal age, ethnicity, maternal cancer history, type of pregnancy, gestational age, assisted reproductive technology or any other aneuploidies. Unclear: if this was not clear from the report. |
Low risk: if ‘yes’ for all signalling question. High or unclear risk: if ‘no’ or ‘unclear’ was reported for at least one signalling question. |
Low concern: if the selected pregnant women represent the women indicated by the review questiona. High concern: if selected pregnant women differ from those targeted by the review questiona. Unclear concern: if insufficient information was available. |
|
| Domain 2: Index test | |||||
|
Index testb |
Were the index test results interpreted without knowledge of the results of the reference standard? | If a threshold was used, was it prespecified? | Could the conduct or interpretation of the index test have introduced bias? | Are there concerns that the index test, its conduct, or interpretation differ from the review question? | |
|
Yes: if the gNIPT results were interpreted without knowledge of the results of the reference standardc. No: if the gNIPT results were interpreted with knowledge of the results of the reference standardc. Unclear: if this was not clear from the report. |
Yes: if criteria for a positive test were prespecified. No: if the criteria for a positive test were not prespecified. Unclear: if this was not clear from the report. |
Low risk: if ‘yes’ for all signalling question. High or unclear risk: if ‘no’ or ‘unclear’ was reported for at least one signalling question. |
Low concern: if the gNIPT was performed such as described in the review questiona. High concern: if gNIPT vary from those specified in the review question. Unclear concern: if insufficient information was available. |
||
| Domain 3: Reference standard | |||||
|
Reference Standardc |
Is the reference standardc likely to correctly classify the target conditiond? | Were the reference standard results interpreted without knowledge of the results of the index testb? | Could the reference standard, its conduct, or its interpretation have introduced bias? | Are there concerns that the target condition as defined by the reference standard does not match the review question? | |
|
Yes: if one appropriate reference standardc was used. No: if pregnant women did not undergo appropriate reference standardc. Unclear: if this was not clear from the report. |
Yes: if karyotype results were interpreted without knowledge of results of the index testb. No: if karyotype results were interpreted with the knowledge of results of the index testb. Unclear: if this was not clear from the report. |
Low risk: if ‘yes’ for all signalling questions. High or unclear risk: if ‘no’ or ‘unclear’ was reported for at least one signalling question. |
Low concern: if the reference standardsc were used as described in the review questiona. High concern: if the reference standardc vary from those specified in the review questiona. Unclear concern: if insufficient information was available. |
||
| Domain 4: Flow and timing | |||||
|
Flow and timing |
Was there an appropriate interval between gNIPT and reference standard? | Did all analysed patients receive the reference standard? | Were all patients included in the analysis? | Could the patient flow have introduced bias? | |
|
Yes: if the interval between blood collection for gNIPT and fluid collection for reference standardc was more than one day (only if blood collection occurred after fluid collection). If blood collection occurred before fluid collection, there is no time limite. No: if the interval between blood collection for gNIPT and the fluid collection for reference standardc was less than one day if the blood collection occurred after the fluid collection. Unclear: if this was not clear from the report. |
Yes: if all pregnant women analysed have appropriate reference standardc. No: if some pregnant women analysed do not have a karyotype result. Unclear: if this was not clear from the report. |
Yes: if all pregnant women recruited into the study were included in the analysis or if failed samples occurred before NGS process. No: if all pregnant women recruited into the study were not included in the analysis or if failed samples occurred during NGS process. Unclear: if this was not clear from the report. |
Low risk: if ‘yes’ for all signalling questions. High or unclear risk: if ‘no’ or ‘unclear’ was reported for at least one signalling question. |
||
aReview question: what is the diagnostic accuracy of massively parallel shotgun sequencing (MPSS) and targeted massively parallel sequencing (TMPS) using circulating cell‐free DNA (ccfDNA) in maternal blood for the detection of common fetal aneuploidies (T21, T18, T13, 45,X, 47,XXY, 47,XXX and 47,XYY) in pregnant women according to their prior risk of fetal aneuploidy?
bIndex test refers to genomics‐based non‐invasive prenatal testing (gNIPT) methods such as MPSS or TMPS.
cThe appropriate reference standard is karyotyping (traditional banding techniques or spectral karyotyping from invasive methods like chorionic villi sampling or amniocentesis), chromosome analysis (e.g. FISH, aCGH and QF‐PCR), clinical examination or medical record from birth (for T21, T18 or T13). For sex chromosome aneuploidies, only fetal karyotype was appropriate reference standard because they usually have a normal phenotype.
dTarget conditions (aneuploidies) are T21, T18, T13, 45,X, 47,XXY, 47,XXX and 47,XYY. eTarget conditions (aneuploidies) do not vary over time.
Appendix 7. gNIPT accuracy in mixed prior risk of fetal aneuploidy
| Test |
Number of studies |
Number of affected pregnancies |
Number of unaffected pregnanciesa |
Sensitivityb % (95% CI) |
Specificityb % (95% CI) |
P valuec |
| T21, mixed risk | ||||||
| MPSS | 10 | 445 | 30,962 | 96.0 (93.7 to 97.4) | 99.9 (99.9 to 100) | |
| TMPS | 6 | 169 | 6925 | 98.2 (94.6 to 99.4) | 100 (99.9 to 100) | |
| Difference between MPSS and TMPS | ‐2.27 (‐4.97 to 0.43) | ‐0.04 (‐0.08 to ‐0.002) | 0.10 | |||
| Traditional screening testsd | 1 | 3 | 1909 | 100 (43.9 to 100) | 96.4 (95.5 to 97.1) | |
| T18, mixed risk | ||||||
| MPSS | 9 | 113 | 30,637 | 100 (98.3 to 100)e | 99.9 (99.8 to 100) | |
| TMPS | 4 | 53 | 5569 | 98.1 (87.8 to 99.7) | 99.9 (99.8 to 100) | |
| Traditional screening tests | 1 | 1 | 1905 | 100 (20.7 to 100) | 99.4 (99.0 to 99.7) | |
| T13, mixed riskf | ||||||
| MPSS | 8 | 27 | 30,384 | 100 (87.5 to 100)e | 100 (> 99.9 to 100)e | |
| TMPS | 5 | 31 | 8362 | 78.6 (48.3 to 93.5) | 99.9 (99.8 to 100) | |
| 45X, mixed risk | ||||||
| MPSS | 2 | 12 | 296 | 91.7 (58.7 to 98.8) | 100 (98.7 to 100)e | |
| TMPS | 2 | 22 | 1128 | 90.9 (70.0 to 97.7) | 99.9 (99.4 to 100) | |
| 47,XXY, mixed risk | ||||||
| MPSS | 1 | 1 | 107 | ND | ||
| TMPS | 1 | 2 | 184 | ND | ||
| 47,XYY, mixed risk | ||||||
| MPSS | 1 | 1 | 199 | ND | ||
| TMPS | 1 | 1 | 185 | ND | ||
| Autosomes (T21, T18 and T13 combined), mixed risk | ||||||
| MPSS | 10 | 585 | 30,822 | 96.9 (95.2 to 98.1) | 99.9 (99.9 to 99.9) | |
| TMPS | 7 | 253 | 8793 | 96.0 (92.8 to 97.9) | 99.8 (99.7 to 99.9) | |
| Difference between MPSS and TMPS | 0.88 (‐1.90 to 3.65) | 0.07 (‐0.02 to 0.16) | 0.25 | |||
| Traditional screening tests | 1 | 4 | 1908 | 100 (51.0 to 100) | 95.8 (94.8 to 96.6) | |
| SCA (45,X, 47,XXX, 47,XXY and 47,XYY combined), mixed risk | ||||||
| MPSS | 2 | 14 | 294 | 92.9 (63.0 to 99.0) | 100 (98.7 to 100)e | |
| TMPS | 2 | 25 | 1125 | 92.0 (73.1 to 98.0) | 99.9 (99.4 to 100) | |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, ND: no data available, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13, CI: confidence interval, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, SCA: sex chromosome aneuploidies.
aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bFor two or more studies, the sensitivities and specificities are the summary estimates obtained from meta‐analysis. Sensitivity and specificity, and their 95% CIs are reported as percentages.
cThe P value indicates the statistical significance of the difference in model fit and was obtained from likelihood ratio tests comparing models with and without a covariate for test type.
dTraditional screening test are first trimester combined test, second trimester quadruple test, second trimester fully integrated test, second trimester sequential test or second trimester triple test.
eSimple pooling used to obtain summary estimates of sensitivity and/or specificity.
fTest comparison analysis did not converge.
Appendix 8. Investigation of heterogeneity
| Test subgroups |
Number of studies |
Number of affected pregnancies |
Number of unaffected pregnanciesa |
|
| Reference standard | ||||
| Autosomes, unselected population | ||||
| MPSS | mixed reference standardb | 1 | 11 | 1730 |
| TMPS | karyotypingc | 1 | 11 | 181 |
| mixed reference standard | 3 | 107 | 20,468 | |
| Autosomes, selected high‐risk population | ||||
| MPSS | karyotyping | 22 | 1075 | 7028 |
| mixed reference standard | 10 | 433 | 8769 | |
| TMPS | karyotyping | 7 | 378 | 4282 |
| SCA, selected high‐risk population | ||||
| MPSS | karyotyping | 10 | 134 | 3943 |
| mixed reference standard | 2 | 17 | 3509 | |
| TMPS | karyotyping | 4 | 96 | 968 |
| Ethnicity | ||||
| Autosomes, unselected population | ||||
| MPSS | more than 50% Asiand | 1 | 11 | 1730 |
| TMPS | more than 50% Caucasiane | 3 | 107 | 20,468 |
| not reported | 1 | 11 | 181 | |
| Autosomes, selected high‐risk population | ||||
| MPSS | more than 50% Asian | 14 | 206 | 6589 |
| more than 50% Caucasian | 7 | 843 | 6262 | |
| not reported | 11 | 459 | 2946 | |
| TMPS | more than 50% Caucasian | 3 | 237 | 3744 |
| not reported | 4 | 141 | 538 | |
| SCA, selected high‐risk population | ||||
| MPSS | more than 50% Asian | 5 | 25 | 1852 |
| more than 50% Caucasian | 5 | 96 | 4286 | |
| not reported | 2 | 30 | 1314 | |
| TMPS | more than 50% Caucasian | 1 | 56 | 116 |
| not reported | 3 | 40 | 852 | |
45,X: Turner syndrome, 47,XXX: triple X syndrome, 47,XXY: Klinefelter syndrome, T21: trisomy 21, T18: trisomy 18, T13: trisomy 13, MPSS: massively parallel shotgun sequencing, TMPS: targeted massively parallel sequencing, SCA: sex chromosome aneuploidies
aWe included pregnancies with any other aneuploidy than the one under analysis with all euploid cases as "unaffected" pregnancies.
bMixed RS: include karyotyping and neonatal clinical examination or medical records from birth.
cKaryotyping: include fetal karyotyping performed on cells obtained from chorionic villi sampling (CVS), amniotic fluid, placental tissue, a fetus lost by miscarriage or other equivalent and recognised methods on the same materials.
dMore than 50% Asian: in the cohort, more than 50% of all pregnant women were Asian ethnicity.
eMore than 50% Caucasian: in the cohort, more than 50% of all pregnant women were Caucasian ethnicity.
Data
Presented below are all the data for all of the tests entered into the review.
Tests. Data tables by test.
| Test | No. of studies | No. of participants |
|---|---|---|
| 1 MPSS T21 | 41 | 50133 |
| 2 MPSS T18 | 38 | 49003 |
| 3 MPSS T13 | 29 | 46090 |
| 4 MPSS 45,X | 14 | 7867 |
| 5 MPSS 47, XXX | 5 | 5449 |
| 6 MPSS 47,XXY | 8 | 6588 |
| 7 MPSS 47,XYY | 8 | 6629 |
| 8 MPSS all 7 aneuploidies | 44 | 50864 |
| 9 MPSS, autosomes | 43 | 50453 |
| 10 MPSS, SCA | 14 | 7911 |
| 11 TMPS T21 | 16 | 32487 |
| 12 TMPS T18 | 12 | 30319 |
| 13 TMPS T13 | 10 | 22868 |
| 14 TMPS 45,X | 6 | 2214 |
| 15 TMPS 47,XXX | 2 | 586 |
| 16 TMPS 47,XXY | 4 | 1021 |
| 17 TMPS 47,XYY | 2 | 358 |
| 18 TMPS all 7 aneuploidies | 21 | 35275 |
| 19 TMPS, autosomes | 18 | 34473 |
| 20 TMPS, SCA | 6 | 2214 |
| 21 Traditional screening tests, autosomes | 5 | 24279 |
| 22 Traditional screening tests T21 | 2 | 17753 |
| 23 Traditional screening tests T18 | 2 | 17747 |
| 24 Traditional screening tests T13 | 1 | 11185 |
1. Test.

MPSS T21.
2. Test.

MPSS T18.
3. Test.

MPSS T13.
4. Test.

MPSS 45,X.
5. Test.

MPSS 47, XXX.
6. Test.

MPSS 47,XXY.
7. Test.

MPSS 47,XYY.
8. Test.

MPSS all 7 aneuploidies.
9. Test.

MPSS, autosomes.
10. Test.

MPSS, SCA.
11. Test.

TMPS T21.
12. Test.

TMPS T18.
13. Test.

TMPS T13.
14. Test.

TMPS 45,X.
15. Test.

TMPS 47,XXX.
16. Test.

TMPS 47,XXY.
17. Test.

TMPS 47,XYY.
18. Test.

TMPS all 7 aneuploidies.
19. Test.

TMPS, autosomes.
20. Test.

TMPS, SCA.
21. Test.

Traditional screening tests, autosomes.
22. Test.

Traditional screening tests T21.
23. Test.

Traditional screening tests T18.
24. Test.

Traditional screening tests T13.
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Alberti 2015.
| Study characteristics | |||
| Patient sampling | Study design: case‐control study (1:2) from a prospective cohort. Participants: pregnant women selected from a high‐risk population of fetal aneuploidy. Inclusion criteria: pregnant women who had a risk of fetal trisomy 21 (> 1 in 250), based on the combination of maternal age with ultrasound and maternal serum markers during the first or second trimester and prior invasive testing. Exclusion criteria: multifetal pregnancies, absence of medical coverage by the National Health System and women declining an invasive procedure. | ||
| Patient characteristics and setting | Number enrolled: 976 pregnant women. Number available for 2 x 2 table: 183 pregnant women (subgroup of 19%). 23 euploid samples were used as reference set and 8 samples randomly chosen for pretesting phase. Setting: 3 centres in France. Recruitment period: March 2010 to April 2013. Ethnicity: not reported. Mean gestational age (± SD): 14 (± 2) weeks. Mean maternal age (± SD): 35.2 (± 6.7) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 without multiplexing. Each library was sequenced using 50 bases‐length reads chemistry in a single end‐flow cell. Mean fetal fraction DNA: (male only) euploid: 20.11% and T21: 16.86%. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 3. In‐house gNIPT. |
||
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 701/976 samples were not selected for the case‐control study. 50/275 samples were excluded during DNA extraction (47 for low amount of DNA and 3 for haemolysis) (no gNIPT results). 31/225 samples were excluded from analysis (8 for pretesting phase and 23 for reference set). 11/194 samples were excluded from analysis for insufficient fetal fraction DNA (no gNIPT results). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate the implementation of gNIPT for trisomy 21 into a cytogenetics laboratory in a university teaching hospital as well as validate gNIPT’s clinical use on samples collected prospectively. | ||
| Funding source or sponsor of the study | Study not funded by industry. | ||
| Informations about the authors contacted | Authors were contacted on: 23 March and 4 May 2016. Last reply received on: 16 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Ashoor 2012.
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control study (1:3) from a prospective cohort.
Participants: pregnant women selected from a high‐risk population (archived maternal plasma samples) of fetal aneuploidy. Inclusion criteria: singleton pregnancies between 11 to 13 weeks' gestation. Exclusion criteria: pregnancies that were conceived by in vitro fertilization. |
||
| Patient characteristics and setting | Number enrolled: 400 pregnant women. Number available for 2 x 2 table: 397 archived plasma samples (subgroup of 99%). Setting: 1 centre. Tertiary Referral Centre, King’s College Hospital, London, United Kingdom. Recruitment period: March 2006 to August 2011. Ethnicity: Caucasian (88.5%), Afro‐Caribbean (5%), South Asian (4%), East Asian (2%) and multiracial (0.5%). Mean gestational age (range): 13.3 (12.1 to 13.7) weeks. Mean maternal age (range): 36.2 (29.9 to 41.2) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ prenatal test by Ariosa Diagnostics, Inc. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standard: fetal karyotype of chorionic villi. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 3/400 samples failed amplification and sequencing (no gNIPT result). 25 samples did not meet Ariosa Diagnostics, Inc acceptance criteria but they were replaced with the next available cases. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To assess the prenatal detection rate of T21 and T18 and the false‐positive rate by chromosome‐selective sequencing of maternal plasma ccfDNA. | ||
| Funding source or sponsor of the study | Study not funded by industry but samples were analysed at Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Ashoor 2013.
| Study characteristics | |||
| Patient sampling | Study design: blinded prospective cohort (second phase). First phase (case‐control study) not shown in the present review. Participants: euploid pregnancies underwent routine first‐trimester combined screening and confirmed T13 cases were selected. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 2167 pregnant women. Number available for 2 x 2 table: 1949 pregnant women (subgroup of 90%). Setting: several centres. Euploid pregnancies were from King’s College Hospital, London, UK and T13 cases were from the USA. Recruitment period: October 2010 to January 2011 for euploid pregnancies. Not reported for T13 cases. Ethnicity: Caucasian (70.8%), African (20%), Asian (6.8%), mixed (2.6%). Mean gestational age (± SD; range): 12.7 (± 0.62; 13 to 26) weeks. Mean maternal age (± SD): 31.8 (± 5.6) years. Relevant tests carried out prior to index test: not reported. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Median fetal fraction DNA (range): euploids: 10.0% (4.1% to 31.0%) and T21: 14.0% (6.1% to 24.0%). Blood samples for gNIPT were collected before reference standard for euploid pregnancies. T13 samples were collected post‐confirmation of trisomy by karyotyping (reference standard). Cutpoint: positive if FORTE algorithm risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test by Ariosa Diagnostics, Inc. |
||
| Target condition and reference standard(s) | Target condition: T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or neonatal clinical examination. | ||
| Flow and timing | Blood samples were obtained at the time of screening for euploid pregnancies (before reference standard). Blood samples were obtained after T13 confirmation following invasive procedure (reference standard). gNIPT was a first‐ or a second‐tier test. 165/2167 samples were excluded because they were used in the first phase. 53/2002 samples failed during amplification or sequencing (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To assess the performance of chromosome‐selective sequencing of maternal plasma cell‐free DNA (cfDNA) in non‐invasive prenatal testing for trisomy 13. | ||
| Funding source or sponsor of the study | Study not funded by industry but samples were analysed at Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Benachi 2015.
| Study characteristics | |||
| Patient sampling | Study design: blinded, retrospective analysis from a prospective cohort. Participants: all pregnant women considered at high risk of fetal aneuploidies who were willing to undergo invasive procedure. Inclusion criteria: at least 18 years old, more than 10 weeks of gestation and singleton or twin pregnancies. Exclusion criteria: vanishing twin or < 18 years old. | ||
| Patient characteristics and setting | Number enrolled: 900 pregnant women. Number available for 2 x 2 table: 886 pregnant women (subgroup of 98%). Setting: 29 centres. French Fetal Medicine Centres in France. Recruitment period: December 2012 to October 2013. Ethnicity: Caucasian (84.2%), Black or Caribbean (4.6%), Asian (2.0%), mixed (5.7%) and unknown (3.5%). Median gestational age (range): 15.1 (10.2 to 34.6) weeks. Median maternal age (range): 35 (30 to 39) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS with Illumina v3 flow‐cell on a HiSeq 1500 sequencer in 12‐plex. Mean fetal fraction DNA: group 1 (patients without abnormal fetal ultrasound findings, but at high risk of fetal aneuploidy): 10.9% and group 2 (high risk of fetal aneuploidy after ultrasound finding): 11.2%. Blood samples for gNIPT were collected just before reference standard. Cutpoint: positive if Z score > 3 (T21) or > 3.95 (T18 and T13). Commercial test: Laboratoire CERBA's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or neonatal clinical examination. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 8/900 samples without karyotype result were excluded. 42 samples failed the initial MPSS testing for technical issues. 42/42 repeated tests using a second aliquot and 36/42 samples obtained gNIPT results. 6/892 samples failed during gNIPT process (low fetal fraction DNA or result appeared atypical) (no gNIPT result). |
||
| Comparative | |||
| Aim to study | To evaluate the performance of the gNIPT (using fetal ccfDNA) for detection of the 3 main autosomal fetal trisomies in a very high‐risk population of patients whose fetuses display ultrasonographically identified anomalies by comparing the results with those obtained by conventional fetal karyotyping. | ||
| Funding source or sponsor of the study | Funding source not reported. 1 author is an employee of Laboratoire CERBA and also a shareholder. | ||
| Informations about the authors contacted | Authors were contacted on: 25 May 2016. Reply received on: 26 May 2016. | ||
| Notes | Authors are from de Collaborative SEquençage a Haut Debit et Aneuploidies (SEHDA) Study Group. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Bevilacqua 2015.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women between 10 to 28 weeks’ gestation selected at high risk of fetal trisomy or women who wanted to have the new test as a primary method of screening (unselected population). Inclusion criteria: singleton (not reported in the present review) or twin pregnancies between 10 to 28 weeks’ gestation. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 2362 pregnant women including 1847 singleton pregnancies (data not reported in the present review) and 515 twin pregnancies.
Number available for 2 x 2 table: 340 twin pregnancies (subgroup of 66%).
Setting: multicentre.
Recruitment period: May 2013 to September 2014 (twin).
Ethnicity: not reported.
Median gestational age (range): 13.0 (10 to 28) weeks.
Median maternal age (range): 36.8 (19 to 50.3) years. Chorionicity (368/515): 13% monochorionic and 58.4% dichorionic. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening for some women. Language of the study: English. |
||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Mean fetal fraction DNA (range): twins: 8.7% (4.1% to 30.0%) and singleton: 11.7% (4.0% to 38.9%). Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test by Ariosa Diagnostics, Inc. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or neonatal karyotype. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 164/515 samples without follow‐up were excluded including138 lost to follow‐up, 19 still pregnant and 7 miscarriages or stillbirths without karyotype of fetal tissue. 29/515 samples failed the initial TMPS testing. 26/29 samples resequenced with a second aliquot of the first sampling and 13/26 samples obtained a gNIPT result. 16/515 samples failed during sequencing process (no gNIPT result). |
||
| Comparative | |||
| Aim to study | To report the clinical implementation of cfDNA analysis of maternal blood in screening for T21, T18 and T13 in a large series of twin pregnancies and examine variables that could influence the failure rate of the test. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc made blinded sequencing and analysis. | ||
| Informations about the authors contacted | Author was contacted on: 1 June and 27 September 2016. No replies received from the author. |
||
| Notes | gNIPT results from singleton pregnancies were not reported in the present review for incomplete 2 x 2 tables. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Bianchi 2012.
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control (1:4) study from the MELISSA prospective cohort. Participants: pregnant women randomly selected from a high‐risk population (archived maternal plasma samples). Inclusion criteria: singleton pregnancies at high risk of fetal aneuploidy between 8 and 22 weeks of gestation. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 2882 pregnant women. Number available for 2 x 2 table: 503 pregnant women for T21, 502 for T18, 501 for T13 and 489 for 45,X (subgroup of 17%). Setting: 60 centres. Medical centre in 25 states in USA. Samples from 53 centres were analysed. Recruitment period: June 2010 to August 2011. Ethnicity: Caucasian (72.7%), Afro American (10.9%), Asian (9.9%), Native American or Alaska Native (0.9%) and multiracial (5.6%). Mean gestational age (± SD; range): 15.1 (± 3.16; 10 to 23) weeks. Mean maternal age (± SD; range): 35.2 (± 6.40; 18 to 46) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 6‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) for T21, T18, and T13: positive if NCV > 4 (aneuploidy suspected if NCV is between 2.5 and 4). 2) for 45,X: positive if NCV for Chrom. X < ‐4 and NCV for Chrom. Y < 2.5. 3) for 47,XXX: positive if NCV for Chrom. X > 4 and NCV for Chrom. Y < 2.5. 4) for 47,XXY: positive if NCV for Chrom. X between ‐2.5 and 2.5 and NCV for Chrom. Y > 33. 5) for 47,XYY: positive if NCV for Chrom. X < ‐4 and NCV for Chrom. Y > 4 with NCV for Chrom. Y is 2 times greater than expected NCV Chrom. X. Commercial test: Verinata's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXX, 47,XXY and 47,XYY. Reference standard: fetal karyotype of chorionic villi (42.7%), amniotic fluid (56.4%) or products of conception (0.9%). | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 257/2882 samples were excluded (127 were ineligibles, 45 without karyotype and 85 for multifetal pregnancies). 2091/2625 samples were not selected for this case‐control study. 2/534 samples were excluded for tracking issue. 16/532 samples without fetal DNA detected were excluded during process (no gNIPT result). 13/516 samples were excluded of T21 2 x 2 table for censored complex karyotype. 14/516 samples were excluded of T18 2 x 2 table for censored complex karyotype. 15/516 samples were excluded of T13 2 x 2 table for censored complex karyotype. 27/516 samples were excluded of 45,X 2 x 2 table for censored complex karyotype. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To prospectively determine the diagnostic accuracy of massively parallel sequencing to detect whole chromosome fetal aneuploidy from maternal plasma. | ||
| Funding source or sponsor of the study | Study funded by Verinata Health, Inc. (a wholly owned subsidiary of Illumina, Inc.). | ||
| Informations about the authors contacted | Authors were contacted on: 1 March and 30 November 2016. Replies received on: 1 March and 8 December 2016. | ||
| Notes | This study is a clinical trial. MELISSA study. Clinicaltrials.gov NCT01122524. Data for 47,XXY, 47,XYY and 47,XXX were incomplete in the publication (data not shown in the present review). |
||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Bianchi 2013.
| Study characteristics | |||
| Patient sampling | Study design: retrospective study (archived maternal plasma samples) from a prospective cohort. Participants: pregnant women selected from a high‐risk population (archived maternal plasma samples). Inclusion criteria: eligible blood samples, singleton pregnancies with karyotype result and nuchal cystic hygroma on fetal ultrasound. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 2882 pregnant women.
Number available for 2 x 2 table: 113 pregnant women (subgroup of 4%).
Setting: 60 centres in USA.
Recruitment period: June 2010 to August 2011.
Ethnicity: Caucasian (73%), Afro‐American (10%), Asian (9%) and multiracial (8%).
Mean gestational age (± SD): 13.2 (± 2.0) weeks. Median gestational age (range): 12.6 (10 to 21) weeks. Mean maternal age (± SD): 32.2 (± 5.8) years. Median maternal age (range): 32.9 (18 to 44) years. Relevant test carried out prior to index test: ultrasonography (nuchal translucency measurement). Language of the study: English. |
||
| Index tests | gNIPT by MPSS with the sequencing chemistry Illumina TrueSeq 3.0. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) for T21, T18 and T13: positive if NCV > 4 (aneuploidy suspected zone between 3 and 4). 2) for 45,X: positive if NCV Chrom. X < ‐3 and NCV Chrom. Y < 3. Commercial test: Verinata's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. Reference standard: fetal karyotype of chorionic villi (78%), amniotic fluid (20%) or products of conception (2%). | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 2769/2882 samples were not selected for this study. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To estimate the accuracy and potential clinical effect of using massively parallel sequencing of maternal plasma DNA to detect fetal aneuploidy in a population of pregnant women carrying fetuses with nuchal cystic hygroma. | ||
| Funding source or sponsor of the study | Study funded by Verinata Health, Inc. (a wholly owned subsidiary of Illumina, Inc.). | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | 74/113 samples were previously sequenced during the MELISSA trial. In this study, all 113 samples were newly resequenced (no overlap) with MELISSA study. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Bianchi 2014a.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women who planned to undergo (without prior risk) or had completed (high‐ or low‐risk) standard prenatal serum screening for fetal aneuploidy. Inclusion criteria: pregnant women of 18 years or older, gestational age ≥ 8 weeks, able to provide consent and pregnancy records accessible and available for data collection. Exclusion criteria: invasive procedure (amniocentesis or CVS) performed within 2 weeks prior enrolment or prenatal screening determination by nuchal translucency measurement only. | ||
| Patient characteristics and setting | Number enrolled: 2052 pregnant women.
Number available for 2 x 2 table: 1952 for T21 and T18 (subgroup of 95%) and 1914 for T13 (subgroup of 93%).
Setting: 21 centres. In 14 states (USA).
Recruitment period: 2 July 2012 to 4 January 2013.
Ethnicity: Caucasian (65.4%), Afro‐American (22.3%), Asian (7.3%) and other (5%).
Mean gestational age (± SD; range): 20.3 (± 8.6; 8 to 39.4) weeks. Mean maternal age (± SD; range): 29.6 (± 5.54; 18 to 48.6) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 8‐plex. Mean fetal fraction DNA: more than 35 years old: 11.3%, less than 35 years old: 11.6%, and at third trimester only: 24.6%. Blood samples for gNIPT were collected before or after reference standard. Cutpoint: positive if NCV ≥ 4. Resequenced if NCV is between 3 and 4. Commercial test: verifi® prenatal test by Verinata Health. The traditional screening tests (first‐trimester combined test or a second‐trimester result (quadruple, serum integrated, fully integrated or sequential)) were also assessed. Mixed cutpoints used. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi, amniotic fluid or products of conception, neonatal clinical examination or medical record from birth. | ||
| Flow and timing | Blood samples were obtained prior or after the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 10/2052 samples failed blood quality control before sequencing process. 72/2042 samples without clinical outcome. 38/2042 samples without standard screening result. 17/2042 samples without gNIPT result. 1/2042 samples without standard screening result and without gNIPT result. 12 resequenced samples were in the grey zone (between affected and unaffected) and were successfully resequenced in uniplex. |
||
| Comparative | |||
| Aim to study | To compare the results of gNIPT with ccfDNA for fetal autosomal aneuploidy with the results of conventional screening for T21 and T18 in a general obstetrical population. To compare false positive rates with the use of each method. To compare false positive rates for T13 in a subset of pregnant women in whom standard screening results included a risk assessment for trisomy 13. To compare fetal ccfDNA fractions in low‐risk patients and those in high‐risk patients in the CARE study population to assess the potential effects of demographic differences on test performance. | ||
| Funding source or sponsor of the study | Study funded by Illumina, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 10 February, 1 June and 28 June 2016. No replies received from the author. | ||
| Notes | This study is a clinical trial (Comparison of Aneuploidy Risk Evaluations; CARE study). ClinicalTrials.gov number: NCT0166335. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Bijok 2014.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy. Inclusion criteria: pregnant women at high risk of fetal aneuploidy with invasive test result. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 10 pregnant women. Number available for 2 x 2 table: 9 pregnant women (subgroup of 90%). Setting: obstetric and gynaecology clinic in Warsaw, Poland. Recruitment period: not reported. Ethnicity: not reported. Median gestational age (range): 16 (13 to 23) weeks. Median maternal age (range): 31 (26 to 36) years. Relevant test carried out prior to index test: ultrasonography (nuchal translucency measurement). Language of the study: Polish. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in multiplex with BGI's algorithm. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Commercial test: NIFTY™ test by Bejing Genomics Institute. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, and T13. Reference standard: fetal karyotype of chorionic villi (30%) or amniotic fluid (70%). | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 1/10 sample failed during sequencing process (no gNIPT result) for low fetal fraction DNA. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To present initial results of non‐invasive prenatal diagnosis of common aneuploidies (T21, T18, and T13) based on ccfDNA in maternal plasma in high‐risk pregnant women, and to compare the results with routine karyotyping. | ||
| Funding source or sponsor of the study | Study not funded by industry but NIFTY™ tests were provided by Beijing Genomics Institute, Shenzen, China. | ||
| Informations about the authors contacted | Authors were contacted on: 2 May and 4 July 2016. Replies received on: 4 and 16 May 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Canick 2012.
| Study characteristics | |||
| Patient sampling | Study design: case‐control study. Participants: all multifetal pregnant women with T21, T18 or T13 fetus were selected along with all euploid triplet pregnancies and a random selection of euploid twin pregnancies. Inclusion criteria: multifetal pregnant women, at least 18 years old, between about 10 weeks and 21 weeks 6 days of gestation, at high risk of aneuploidies and who undergo an invasive procedure. Exclusion criteria: singleton pregnancies or low risk of aneuploidy. | ||
| Patient characteristics and setting | Number enrolled: 4664 pregnant women. Number available for 2 x 2 table: 27 multifetal pregnancies (25 twin and 2 triplet pregnancies) (subgroup of 0.6%). Setting: 27 centres. Prenatal diagnostic centres (Canada, Italy, Spain, Czech Republic, Argentina, Ireland, Hungary, USA, Israel and Australia). Recruitment period: April 2009 to February 2011. Ethnicity: not reported. Mean gestational age (range): 15.0 (10.9 to 19) years. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 4‐plex. Fetal fraction DNA range: 7% to 55%. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score ≥ 3. Commercial test: Sequenom's test. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T13. T18 was also assessed but no case was found. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples were obtained immediately prior the invasive procedure (reference standard). gNIPT was a second‐tier test. 4637/4664 samples were not selected for this case‐control study. No failed sample was reported in multifetal pregnancies. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To study prenatal testing for T21, T18, and T13 by MPSS of fetal ccfDNA in high‐risk multifetal pregnant women. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. Some authors are employees and shareholders of Sequenom, Inc. or of Sequenom Center for Molecular Medicine. | ||
| Informations about the authors contacted | Author was contacted on: 10 March 2016. Reply received on: 16 March 2016. |
||
| Notes | This study is a clinical trial "A New Prenatal Blood Test for Down Syndrome" ClinicalTrials.gov number: NCT00877292. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Chen 2011.
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control study from a prospective cohort and archived plasma. Participants: pregnant women with clinical indications of fetal aneuploidy (high risk of fetal aneuploidy) for invasive procedure. Inclusion criteria: singleton pregnancies with and without trisomy 13, 18 or 21, matched for gestational ages. Exclusion criteria: twin pregnancies. |
||
| Patient characteristics and setting | Number enrolled: 392 pregnant women (252 from the prospective cohort and 140 were archived plasma). Number available for 2 x 2 table: 289 pregnant women (subgroup of 74%). Setting: 10 centres in Hong Kong, the Netherlands, and UK. Recruitment period for the prospective cohort: October 2008 to May 2009. Recruitment period for the archived plasma samples collection: October 2003 to September 2008. Ethnicity: not reported. Gestational age: not reported. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx in 2‐plex. Feta fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 3. Commercial test: Sequenom's test. |
||
| Target condition and reference standard(s) | Target conditions: T18 and T13. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. |
||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 103/392 samples were selected as reference control. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To assess the prenatal diagnostic performance by MPSS of maternal plasma DNA on a cohort of pregnant women with T13 and T18 fetuses. | ||
| Funding source or sponsor of the study | Study co‐sponsored by Sequenom, Inc and Life Technologies. Some authors have filed patent on gNIPT (part of this patent has been licensed to Sequenom, Inc). | ||
| Informations about the authors contacted | Author was contacted on: 14 December 2015 and 10 May 2016. Reply received on: 12 May 2016. |
||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Chiu 2011.
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:5) from a prospective cohort and archived plasma. Participants: pregnant women with clinical indications for invasive procedure, mixed risk (mostly high risk (> 1/300 at traditional screening test), intermediate risk (between 1/300 and 1/1000) or other risk factors). T21 and non T21 pregnancies matched for gestational ages. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. |
||
| Patient characteristics and setting | Number enrolled: 824 pregnant women. Number available for 2 x 2 table: 753 (8‐plex) (subgroup of 91%). Setting: 10 centres in Hong Kong, the Netherlands, and UK. Recruitment period for the prospective cohort: October 2008 to May 2009. Recruitment period for the archived plasma samples collection: October 2003 to September 2008. Ethnicity: not reported. Median gestational age: 13.1 weeks. Median maternal age: 35.4 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer II in 8‐plex and 2‐plex (not reported in the present review). Median fetal fraction DNA (interquartile 1 and 3): male euploid: 15.2% (10.6% and 19.1%), archived samples: 14.7%, and prospective samples: 15.4%. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 3. Commercial test: Sequenom's test. |
||
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. |
||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 60/824 samples were excluded before sequencing process (2 twin pregnancies, 12 without karyotype and 46 failed quality control for blood sampling). 11/764 samples failed quality control during sequencing process (no gNIPT result). 96/753 samples were also used for reference controls (8‐plex). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To validate the diagnostic performance and practical feasibility of massively parallel genomic sequencing for the non‐invasive prenatal assessment of trisomy 21 in pregnant women who had undergone conventional screening and were clinically indicated for definitive testing. | ||
| Funding source or sponsor of the study | Study sponsored by Sequenom, Inc. Some authors have filed patent applications on gNIPT (part of this patent has been licensed to Sequenom, Inc). | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | Data from 2‐plex sequencing were excluded from the present review to avoid double counting. We kept data from 8‐plex because it is the method most likely to be used for routine testing. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Comas 2015.
| Study characteristics | |||
| Patient sampling | Study design: blinded, observational prospective cohort study. Participants: all pregnant women who underwent conventional first‐trimester combined screening for fetal aneuploidies (without prior risk of fetal aneuploidy). Some pregnant women were referred after their combined test (high risk of fetal aneuploidy). Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies, cases of ultrasound anomalies, nuchal translucency > 99 centile, combined risk at first‐trimester screening > 1/10, or women at high risk of other genetic conditions. | ||
| Patient characteristics and setting | Number enrolled: 333 pregnant women (85.5% without prior risk and 16.5% were at high risk of fetal aneuploidy). Number available for 2 x 2 table: 312 pregnant women (subgroup of 95%). Setting: 1 private prenatal diagnostics centre in Barcelona, Spain (Hospital Universitari Quiron Dexeus). Recruitment period: January to December 2013. Ethnicity: not reported. Mean gestational age (range): 14.6 (9.5 to 23.5) weeks. Mean maternal age (range): 37 (21 to 46) years. Relevant test carried out prior to index test: biochemical screening for a part of the cohort. Language of the study: English | ||
| Index tests | gNIPT by TMPS (DANSR assay or SNP‐based method). Mean fetal fraction DNA (range): 12.7% (4.2% to 27.9%), Harmony™ prenatal test: 13.1%, and Panorama™ prenatal test: 12.7%. Blood samples for gNIPT were collected before reference standard. DANSR assay cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. SNP‐based method cutpoint: not reported. Commercial test: Panorama™ prenatal test by Natera, Inc. or Harmony™ prenatal test by Ariosa Diagnostics, Inc. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXX, 47, XXY, 47,XYY. SCA data were not reported in the present review. T18 and T13 were also assessed but no case was found. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or neonatal clinical examination. |
||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 17/333 samples excluded because still pregnant at the time of publication (no follow‐up). 9/333 samples failed the initial TMPS testing. 6/9 repeated sampling was performed and results were obtained in 5/6. 3/333 samples without gNIPT result were excluded (unrepeated samples). 1/333 samples without gNIPT result and follow‐up were excluded (still pregnant). |
||
| Comparative | |||
| Aim to study | To evaluate gNIPT of ccfDNA as a screening method for major chromosomal anomalies in a clinical setting. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc and Natera, Inc. made sequencing and analysis. | ||
| Informations about the authors contacted | Author was contacted on: 27 May 2016 and 31 May 2016. Reply received on: 31 May 2016. | ||
| Notes | gNIPT is offered to pregnant women at their own cost. 45,X, 47,XXY, 47,XYY and 47,XXX were also screened but inappropriate reference standard for the present review was used. gNIPT data from SCA were not shown in this review. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
del Mar Gil 2014.
| Study characteristics | |||
| Patient sampling | Study design: retrospective cohort study. Data from prospective cohort were not shown in the present review. Participants: pregnant women without a priori risk who undergo first‐trimester screening for trisomies (archived maternal plasma samples). Inclusion criteria: multifetal pregnancies between 11 to 13 weeks' gestation. Exclusion criteria: singleton pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 207 pregnant women from the retrospective cohort.
Number available for 2 x 2 table: 192 pregnant women (subgroup of 93%). Setting: 1 centre at Kings’ College Hospital in London, UK. Recruitment period: not reported. Ethnicity: not reported. Median gestational age (range): 13.0 (12.4 to 13.9) weeks. Median maternal age (range): 33.7 (26.7 to 37.9) years. Chorionicity: 41% of pregnancies were monochorionic (85/207) and 59% of pregnancies were dichorionic (122/207). Relevant tests carried out prior to index test: none. Language of the study: English. |
||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Mean fetal fraction DNA (range): euploids: 9.8% (7.4% to 12.1%), T21: 10.8% (6.8% to 12.1%), and T13: 7%. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ prenatal test by Ariosa Diagnostics, Inc. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T13. T18 was also assessed but no case was found. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐tier test. 15/207 samples failed during sequencing process (11 for low fetal fraction DNA and 4 for laboratory processing failures) (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To examine the clinical implementation of TMPS of ccfDNA in maternal blood and an algorithm that relies on the lower fetal fraction DNA contribution of the 2 fetuses in the assessment of risk for trisomies in twin pregnancies. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc have performed gNIPT at their own expense. Study funded by a grant from The Fetal Medicine Foundation, UK. | ||
| Informations about the authors contacted | Author was contacted on: 27 May and 27 September 2016. No reply received from the author. |
||
| Notes | Data from prospective cohort study were not shown in the present review because patients with gNIPT negative result were without follow‐up to confirm gNIPT result. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Ehrich 2011.
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:11) from a prospective cohort. Participants: pregnant women selected from a high‐risk population. Inclusion criteria: not reported. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 480 pregnant women. Number available for 2 x 2 table: 449 pregnant women (subgroup of 94%). Setting: in clinical practice and pregnancy termination centres. Recruitment period: May 2009 to unknown date. Ethnicity: not reported. Median gestational age (range): 16 (8 to 36) weeks. Mean maternal age (range): 37 (18 to 47) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx in 4‐plex. Minimum fetal fraction DNA as estimated with the fetal quantifier assay: 3.9%. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 2.5. Commercial test: Sequenom's test. |
||
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype of chorionic villi (19%) or amniotic fluid (81%). | ||
| Flow and timing | Blood samples were obtained prior or after the invasive procedure (reference standard). gNIPT was a second‐tier test. 13/480 samples excluded before sequencing process (9 for plasma volume < 3.5 mL and 4 for processing errors). 20/467 samples failed the initial MPSS testing. 20/20 samples were resequenced using the same library (10 samples in 4‐plex and 10 in monoplex) and 2/20 samples obtained a gNIPT results. 18/467 samples failed quality control during sequencing process, including 7 samples for low fetal fraction DNA (no gNIPT result). |
||
| Comparative | |||
| Aim to study | To evaluate a multiplexed massively parallel shotgun sequencing assay for noninvasive trisomy 21 detection using circulating cell‐free fetal DNA. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | Author was been contacted on: 5 May and 28 September 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Fiorentino 2016.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Retrospective cohort (training set) not reported in the present review. Participants: mostly pregnant women selected from a high‐risk population and pregnant women without prior risk (14%). Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 7103 pregnant women. Number available for 2 x 2 table: 7082 pregnant women (subgroup of 99.7%). Setting: in Italy. Recruitment period: September to December 2014. Ethnicity: not reported. Mean gestational age (± SD; range): 12.8 (± 2.3; 10 to 30) weeks. Mean maternal age (± SD; range): 36.4 (± 4.7; 24 to 54) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2500 sequencer in 15‐plex with SAFeR™ algorithm. Fetal fraction DNA: the limit of detection (the lowest fetal fraction DNA with a detectable aneuploidy) for T21 was determined at 2% fetal fraction level. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if NCV > 4 (aneuploidy suspected if NCV was between 3 and 4). Commercial test: Genoma's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or neonatal clinical examination. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a first‐ or a second‐tier test.
100/7103 samples failed the initial MPSS testing. 79/100 repeated samples with a second blood draw and all obtained a gNIPT result. 21/100 unrepeated samples failed quality control metrics (no gNIPT result). |
||
| Comparative | |||
| Aim to study | To determine the limit of detection of a gNIPT method, in order to define the actual lower fetal fraction DNA required to detect common fetal autosomal trisomies, using a model system to simulate samples at different proportions of fetal ccfDNA. Secondly, to assess the impact of low fetal fraction DNA on the performance of ccfDNA‐based maternal plasma testing for aneuploidies. | ||
| Funding source or sponsor of the study | Study not funded by industry but the samples were analysed in the GENOMA laboratory (Rome, Italy). | ||
| Informations about the authors contacted | Authors were contacted on: 30 August and 6 September 2016. Reply received on: 6 September. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Gil 2016.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women with a first‐trimester combined test selected for their risk of fetal aneuploidy (cut‐off of 1 in 100 for high risk and 1 in 101 to 1 in 2500 for intermediate risk). Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies, terminations of pregnancy, miscarriages or stillbirths without follow‐up. | ||
| Patient characteristics and setting | Number enrolled: 11,692 pregnant women.
Number available for 2 x 2 table: 3633 pregnant women (subgroup of 31%).
Setting: 2 centres. King’s College Hospital, London, and Medway Maritime Hospital, Gillingham, Kent in UK.
Recruitment period: October 2013 to February 2015.
Ethnicity: Caucasian (70%), Afro‐Carabbean (20%), Asian (7%) and mixed (3%). Gestational age: not reported. Median maternal age (range): 31.6 (25.8 to 39.5) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. |
||
| Index tests | gNIPT by TMPS (DANSR assay). Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ prenatal test by Ariosa Diagnostics, Inc. Traditional screening test was also assessed but 2 x 2 tables were incomplete. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi, postnatal karyotype or neonatal clinical examination. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test. 7994/11,692 samples did not undergo a gNIPT (no gNIPT result). 99/3698 samples failed the initial TMPS testing. 54/99 repeated sampling were processed and 34/54 gNIPT results were obtained. 65/3698 samples without gNIPT result. |
||
| Comparative | |||
| Aim to study | To report the feasibility of implementing gNIPT. To examine the factors affecting patient decisions concerning their options for screening and decisions on the management of affected pregnancies. To report the prenatal diagnosis of fetal trisomies and outcome of affected pregnancies following the introduction of contingent screening. | ||
| Funding source or sponsor of the study | Study not funded by industry but the cost of collection and analysis of the blood samples for the cell‐free DNA test was covered by Ariosa Diagnostics, Inc. These organisations had no role in study design, data collection, data analysis, data interpretation or writing of the report. Study was funded by a grant from The Fetal Medicine Foundation, UK. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| High | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Hall 2014.
| Study characteristics | |||
| Patient sampling | Study design: case‐control study (1:3), age‐matched randomly selected from a larger cohort.
Participants: pregnant women with an affected fetus or considered to be at high risk of fetal aneuploidy were recruited. Inclusion criteria: pregnant women at least 18 years of age who had signed an informed consent, and with singleton pregnancy. Exclusion criteria: fetal mosaicism. |
||
| Patient characteristics and setting | Number enrolled: more than 1000 pregnant women. Number available for 2 x 2 table: 64 pregnant women (subgroup of 6%). Setting: 6 centres. Western Institutional (WA, USA), Einstein Institutional (CA and MO, USA), Polish Mother’s Memorial Hospital Institutional (Polish), Bio Medical Research Institute of America (CA, USA), and the Mt. Sinai School of Medicine (NY, USA). Recruitment period: March to December of 2012. Ethnicity: not reported. Median gestational age (range): 16.0 (12.1 to 22.7) weeks. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencer. Samples were amplified using 11,000‐plex or 19,488‐plex targeted polymerase chain reaction (targets included SNPs from chromosomes 13, 18, 21, X, and Y). Mean fetal fraction DNA (median; range): 12.1% (11.1%; 2.2% to 30.4%). Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Commercial test: Natera's prenatal test. |
||
| Target condition and reference standard(s) | Target condition: T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or genetic testing of the cord blood, buccal, saliva or products of conception. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. About 932 samples were not selected for this case‐control study. 4/68 samples failed DNA quality threshold for low fetal fraction DNA (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To determine how a single nucleotide polymorphism (SNP)‐ and informatics‐based non‐invasive prenatal aneuploidy test performs in detecting trisomy 13. | ||
| Funding source or sponsor of the study | Study funded by Natera, Inc. (involved in study design, data collection and analysis, decision to publish, and preparation of the manuscript). | ||
| Informations about the authors contacted | Authors were contacted on: 21 April 2016, and 27 May 2016. No reply received from the authors. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Hooks 2014.
| Study characteristics | |||
| Patient sampling | Study design: case‐control study from archived plasma samples from a prospective cohort. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: pregnant women 18 years and older, with a singleton pregnancy at gestational age 10 weeks or greater, and who were planning to undergo invasive prenatal diagnosis. Exclusion criteria: multifetal pregnancies, pregnant women with a known maternal aneuploidy, active malignancy or a history of metastatic cancer, or those who had already undergone chorionic villus sampling or amniocentesis during the current pregnancy. | ||
| Patient characteristics and setting | Number enrolled: not reported. 432 maternal plasma samples were retrieved from the prospective cohort. Number available for 2 x 2 table: 414 samples (subgroup of 96%). Setting: 16 centres. Selected prenatal care centres in the USA, the Netherlands and Sweden. Recruitment period: not reported. Ethnicity: not reported. Mean gestational age (± SD; range): 15.4 (± 3.7; 10 to 34.1) weeks. Mean maternal age (± SD; range): 35.6 (± 5.7; 18.5 to 45.5) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test by Ariosa Diagnostics, Inc. |
||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXY and 47,XXX. 47,XYY was also assessed but no case was found. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples were obtained prior the invasive procedure (reference standard). gNIPT was a second‐tier test. 18/432 samples failed during sequencing process (no gNIPT result) for low fetal fraction DNA, unusually high variation in ccfDNA counts or failure to pass the quality control measures of the DANSR assay. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To assess the performance of a directed chromosomal analysis approach in the prenatal evaluation of fetal sex chromosome aneuploidy. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | BGI‐Shenzhen were contacted on: 19 May 2016. Author was contacted on: 16 June 2016. No reply received from the author. |
||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Hou 2012.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 308 pregnant women. Number available for 2 x 2 table: 205 pregnant women (subgroup of 67%). Setting: 1 centre. Henan Province People's Hospital in China. Recruitment period: October 2010 to January 2012. Ethnicity: Asian. Gestational age range: 14 to 24 weeks. Mean maternal age (range): 31 (21 to 44) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: Chinese. | ||
| Index tests | gNIPT by MPSS on IIIumina HiSeq 2000 sequencer with BGI's algorithm. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected just before reference standard. Cutpoint: not reported. Commercial test: BGI‐Shenzhen's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, 45,X, 47,XXY and 47,XYY. T13 and 47,XXX were also assessed but no cases were found. Reference standard: fetal karyotype of amniotic fluid. | ||
| Flow and timing | Blood samples were obtained just prior the invasive procedure (reference standard). gNIPT was a second‐tier test. 103/308 patients did not undergo gNIPT (no gNIPT result). No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigate the clinical value of gNIPT using ccfDNA in maternal blood. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI‐Shenzhen provided the test. | ||
| Informations about the authors contacted | Author was contacted on: 11 April 2016 (author) and 19 May 2016 (BGI's contact). No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Huang 2014.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy (as real clinical samples). Inclusion criteria: twin pregnancies with live fetuses and karyotype result. Exclusion criteria: singleton pregnancies, twins with intrauterine fetal demise at the time of sampling or without fetal karyotype result. | ||
| Patient characteristics and setting | Number enrolled: 189 pregnant women.
Number available for 2 x 2 table: 189 pregnant women (whole cohort included in analyses).
Setting: 7 centres. Hospitals in China.
Recruitment period: April 2012 to April 2013.
Ethnicity: most Asian.
Median gestational age (range): 19 (11 to 39) weeks.
Median maternal age (range): 31 (22 to 44) years.
Chorionicity: 17% monochorionics (33/189), 80% dichorionics (152/189) and 2% unknown (4/189). Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 platform. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected 30 minutes before reference standard. Cutpoint: positive if t score > 2.5 and L score risk > 1 (warning zone if t score > 2.5 or L score > 1). Commercial test: BGI's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standard: fetal karyotype of chorionic villi (2.1%), amniotic fluid (94.2%) or cord blood (3.7%). | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To assess the performance of noninvasive prenatal testing for trisomies 21 and 18 on the basis of MPSS of ccfDNA from maternal plasma in twin pregnancies. | ||
| Funding source or sponsor of the study | Funded by the Shenzhen Engineering Laboratory for Clinical Molecular Diagnostic, the China National GeneBank‐Shenzhen, the Medical Centre for Critical Pregnant Women in Guangzhou and Prenatal monitoring, In utero therapy and Follow‐up after birth in the complexity of Twin Pregnancy. Some authors worked for BGI‐Shenzhen. | ||
| Informations about the authors contacted | Author was contacted on: 10 February 2016. BGI‐Shenzhen were contacted on: 19 May 2016. No reply received from author. |
||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Jackson 2014.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected at high risk and low risk of fetal aneuploidy presenting for screening. Inclusion criteria: not reported. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 1228 pregnant women screened at first‐trimester, including 1184 pregnant women with normal first‐trimester ultrasound and 44 with abnormal ultrasound.
Number available for 2 x 2 table: 1161 pregnant women (subgroup of 95%).
Setting: 1 centre. South Shore Hospital in USA. Recruitment period: June 2012 to January 2013. Ethnicity: not reported. Gestational age: not reported. Median maternal age: 31.5 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. |
||
| Index tests | gNIPT by TMPS (DANSR assay). Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid, or medical record from birth. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 67/1228 samples excluded of 2 x 2 tables, including 7 women with other abnormal ultrasound, 14 women opted for CVS only without gNIPT, 32 women declined all testing and 14 samples failed after 2 attempts during sequencing process (no gNIPT result). |
||
| Comparative | |||
| Aim to study | To assess the performance of nuchal translucency measurement followed by gNIPT in the first‐trimester to screen for aneuploidy in a community‐based average‐risk population. | ||
| Funding source or sponsor of the study | Funding source not reported but 1 author is employed by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 22 February 2016 and 15 March 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Unclear | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Jeon 2014.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: women who gave written informed consent participated in the study if they were ≥ 19 years old and had a singleton pregnancy with a gestational age of at least 12 weeks. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 155 pregnant women. Number available for 2 x 2 table: 155 pregnant women (whole cohort included in analyses). Setting: 1 centre. Xiamen Maternal & Child Health Care Hospital, Xiamen, Fujian, China. Recruitment period: March 2012 to October 2013. Ethnicity: Asian. Gestational age ranges: 12 to 16 weeks (18.1%), 17 to 21 weeks (55.5%), ≥ 22 weeks (26.5%). All between 12 to 24 weeks. Mean maternal age (± SD; range): 30.73 (± 4.99; 19 to 43) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Ion Torrent PGM sequencer with 10 samples per chip. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected just before reference standard. Cutpoint: positive if Z score > 2.566 (T21) or > 2.459 (T18). Commercial test: Genome Care's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standard: fetal karyotype of amniotic fluid. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigated whether fetal T18 and T21 were sensitively and specifically detectable by semiconductor sequencer: Ion Proton™. | ||
| Funding source or sponsor of the study | Study funded by the Industrial Strategic Technology Development Program, "Bioinformatics platform development for next generation bioinformation analysis" funded by the Ministry of Knowledge Economy (MKE, Korea). | ||
| Informations about the authors contacted | Author was contacted on: 6 and 11 April 2016. Reply received on: 11 April 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| High | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Jiang 2012.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women at high risk of fetal aneuploidy presenting for invasive testing selected from the cohort. Inclusion criteria: not reported. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 903 pregnant women. Number available for 2 x 2 table: 903 pregnant women (whole cohort included in analyses). Setting: 3 centres in Shenzen, China. Recruitment period: June 2009 to August 2010. Ethnicity: Asian. Gestational age range: 10 to 39 weeks. Maternal age range: 20 to 45 years. Relevant tests carried out prior to index test: not reported. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on platforms Illumina Genome Analyzer IIx or Illumina HiSeq 2000 by multiplex sequencing. Fetal fraction DNA (range): quality control criteria > 3.5% (1% to 33%). It is not reported if the blood samples for gNIPT were collected before or after reference standard. Cutpoint: 1) Positive if binary hypothesis t score (first hypothesis) > 3 and t score (second hypothesis) < 3 and if logarithmic LR > 1 (autosomal aneuploidy). 2) Positive if t score < ‐2.5 (45,X and 47,XXX) without Chrom. Y representation. 3) Positive if t score > 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X and Y independently (47,XXY and 47,XYY) for male fetus. Commercial test: NIFTY™ prenatal test by Bejing Genomics Institute (BGI). |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Reference standard: fetal karyotype of amniotic fluid. | ||
| Flow and timing | It is not reported if the blood samples were obtained prior or after the invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To develop an advanced gNIPT method based on MPSS. | ||
| Funding source or sponsor of the study | Study funded by industry. BGI was involved in the study design, conduct of the study, analysis and interpretation of results. | ||
| Informations about the authors contacted | Author was contacted on: 19 May 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Johansen 2016.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 375 pregnant women (184 for the validation set). Number available for 2 x 2 table: 173 pregnant women (subgroup of 94%). Setting: Danish public health setting. Recruitment period: not reported. Ethnicity: not reported. Median gestational age (range): 13.4 (10.6 to 31) weeks. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer in 5‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected just before reference standard. Cutpoint: positive if Z score ≥ 4 and WISECONDOR ≥ 1% (unclassified if Z score between 3 and 4). In‐house test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior the invasive procedure (reference standard).
gNIPT was a second‐tier test. 191/375 not selected, samples for the validation set were excluded. 11/184 samples failed during sequencing process for low fetal fraction DNA (no gNIPT result). 2/173 samples were resequenced because gNIPT results were in the inconclusive zone and 2 results were obtained. |
||
| Comparative | |||
| Aim to study | To introduce gNIPT for fetal autosomal trisomies and gender in a Danish public health setting, using semi‐conductor sequencing and published open source scripts for analysis. | ||
| Funding source or sponsor of the study | No funding source was reported. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Ke 2015.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women considered at high risk of fetal aneuploidy. Inclusion criteria: singleton pregnancies. Pregnant women at high risk of fetal aneuploidy describe as follows: over age 35, the histories of abnormal pregnancy including children with T21 and repeated spontaneous abortion, stillbirth in pregnancy periods, abnormal serological screening for T21 at early and mid pregnancy, abnormal screening for fetal nuchal translucency using colour duplex ultrasonography between 11‐14 weeks of gestation. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 2340 pregnant women. Number available for 2 x 2 table: 2340 pregnant women (whole cohort included in analyses). Setting: 1 centre. Clinical setting at Shenzhen Second People’s Hospital in China. Recruitment period: March 2012 to May 2013. Ethnicity: Asian. Gestational age: positive cases were between 16 to 24 weeks. All cohort: 95% were between 15 to 20 weeks, 3% were between 12 to 14 weeks and 0.9% were ≥ 24 weeks. Maternal age: 88% were less than 35 years old and 12% were 35 years old or more. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if t score > 3. Commercial test: BGI‐Shenzhen's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, and T13. Reference standards: fetal karyotype or newborn outcome. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To validate the efficacy of detection of fetal cell‐free DNA in maternal plasma of trisomy 21, 18 and 13 in a clinical setting. | ||
| Funding source or sponsor of the study | Study not funded by industry but patients had obtained insurance plans on behalf of Shenzhen Huada Genomics Institute. | ||
| Informations about the authors contacted | Author was contacted on: 22 April 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Kim 2016.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: not reported. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 101 pregnant women. Number available for 2 x 2 table: 101 pregnant women (whole cohort included in analyses). Setting: 3 centres (Mirae & Heemang, Namujungwon and GN hospitals) in Korea. Recruitment period: December 2014 to April 2015. Ethnicity: Asian. Gestational age range: 11 to 18 weeks. Mean maternal age (± SD; range): 35.45 (± 3.64; 25 to 42) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening (quadruple test screening). Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Ion Torrent PGM (data not shown in the present review) and Ion Proton™ sequencer in multiplex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 2.10 for Ion Proton™. Commercial test: Genome Care's prenatal test. |
||
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype of amniotic fluid. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To compare the Ion Torrent PGM and Ion Proton™ platforms for gNIPT for fetal T21 directly using PGM and Ion Proton™ simultaneously for the same set of samples. | ||
| Funding source or sponsor of the study | Study funded by Genome Care internal research funding. The first author is employee of Genome Care. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | Data from PGM sequencer are not shown in the present review to avoid patients overlap. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | No | ||
| High | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Korostelev 2014.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected from a population at high risk or without prior risk of fetal aneuploidy. Inclusion criteria: women who had a singleton pregnancy and more than 9 weeks of gestation. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 1968 pregnant women. Number available for 2 x 2 table: 685 pregnant women (subgroup of 35%). Setting: private clinics in Moscow, Russia. Recruitment period: 2012 to 2014. Ethnicity: not reported. Median gestational age (range): 14 (9 to 33) weeks. Mean maternal age (range): 34.4 (26 to 45) years. Relevant tests carried out prior to index test: biochemical screening or ultrasonography (nuchal translucency measurement) or both. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers with NATUS algorithm. Fetal fraction DNA: not reported (usually NATERA used quality control criteria > 4%). Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Commercial test: Natera's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. 45,X, 47,XXY, 47,XYY and 47,XXX were also screened but inappropriate reference standard for the present review was used (data not shown in this review). Reference standards: fetal karyotype of chorionic villi or amniotic fluid or medical record from birth. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test. 240/1968 samples did not undergo gNIPT (no gNIPT result). 1043/1728 samples without follow‐up were excluded. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To examine possibility to use combination of gNIPT and chromosomal microarray analysis for prenatal diagnostics and their advantages between combined first‐trimester screen with confirmation by karyotyping of CVS or amniocytes. | ||
| Funding source or sponsor of the study | Study not funded by industry but gNIPT was carried out by Natera, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 21 June 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Lau 2012.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women mostly at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: not reported. Exclusion criteria: not reported. |
||
| Patient characteristics and setting | Number enrolled: 108 pregnant women. Number available for 2 x 2 table: 108 pregnant women (whole cohort included in analyses). Setting: 1 centre in Japan. Recruitment period: not reported. Ethnicity: Asian. Median gestational age (range): 12.7 (11.6 to 28) weeks, 89.8% < 14 weeks. Mean maternal age (± SD): 37 (± 4.3) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected immediately before reference standard. Cutpoint: 1) positive if Z score ≥ 3 (T21, T18 and T13). 2) for female fetus, positive if Chrom. X Z score ≤ ‐3 (45,X). 3) for female fetus, positive if Chrom. X Z score ≥ 3 (47,XXX). 4) for male fetus, positive if Chrom. Y Z score ≥ 3 (47,XXY). Commercial test: NIFTY™ prenatal test by BGI‐Shenzhen. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X and 47,XXY. 47,XYY and 47,XXX were also assessed but no case was found. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. |
||
| Flow and timing | Blood samples for gNIPT were collected immediately before invasive procedure (reference standard). gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigate the inclusion of an internal reference in the noninvasive prenatal identification of common fetal aneuploidies using massively parallel sequencing on maternal plasma. | ||
| Funding source or sponsor of the study | Study funded by BGI‐Shenzhen. | ||
| Informations about the authors contacted | BGI‐Shenzhen contacted on: 19 May 2016. No reply received from the author. |
||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Lee 2015.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: pregnant women who were > 18 years old and gestational age > 8 weeks, multifetal and singleton pregnancies. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 93 pregnant women. Number available for 2 x 2 table: 92 pregnant women (subgroup of 99%). Setting: 1 centre at Asan Medical Centre, Seoul, Korea. Recruitment period: August 2014 to February 2015. Ethnicity: Asian. Median gestational age (range): 21.1 (8.2 to 31.1) weeks. Median maternal age (range): 32 (21 to 43) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina MiSeq sequencer in 12‐plex or on NextSeq 500 sequencer in 96‐plex. Median fetal fraction DNA (range): male fetus only: 10.2% (3.85% to 25.0%). Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) positive if Z score > 4 (intermediate risk if Z score between 2.5 and 4) for T21 and T18. 2) positive if Z score > 2.8 (intermediate risk if Z score between 1.9 and 2.8) for T13. Commercial test: MomGuard™ by LabGenomics. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. SCA were also assessed but no case was found. Reference standards: fetal karyotype of chorionic villi, amniotic fluid, cord blood or products of conception or neonatal karyotype from peripheral blood. | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test.
1/93 samples failed during sequencing process for low fetal fraction DNA (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate the performance of MomGuard™, a gNIPT, for detecting T21, T18, T13, and SCA abnormalities recently developed in Korea. | ||
| Funding source or sponsor of the study | Study funded by a grant from the LabGenomics Clinical Research Institute. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Lefkowitz 2016.
| Study characteristics | |||
| Patient sampling | Study design: Retrospective cohort, blinded case‐control study. Participants: pregnant women selected at high risk of fetal aneuploidy from 4 cohorts (archived maternal plasma samples). Inclusion criteria: not reported. Exclusion criteria: cases of fetal mosaicism or incomplete karyotype or microarray information. | ||
| Patient characteristics and setting | Number enrolled: 5321 pregnant women in all 4 cohorts. 1222 pregnant women selected for this study. Number available for 2 x 2 table: 1166 pregnant women (subgroup of 95%) for autosomes and 1144 pregnant women (subgroup of 94%) for SCA. Setting: multicentre. Recruitment period: not reported. Ethnicity: not reported. Median gestational age (range): 17 (8 to 38) weeks. Median maternal age (range): 36.0 (17.8 to 47) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 6‐plex or uniplex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before (for 1189 pregnant women) or after (for 24 pregnant women) reference standard. Cutpoint: 1) positive for T21 if Z score ≥ 3. 2) positive for T18 or T13 if Z score ≥ 3.95. 3) positive for 45,X if Z score < ‐3.5 (non‐reportable regions between ‐2.5 and ‐3.5). 4) positive for 47,XXX if Z score > 3.5 (non‐reportable regions between 2.5 and 3.5). 5) positive for 47,XYY if Z score < ‐3.5 with Chrom. Y representation. 6) positive for 47,XXY if Z score is between ‐3.5 and 3.5 with Chrom. Y representation. Commercial test: Sequenom's test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. copy number variants ≥ 7 Mb were also assessed but data not shown in the present review. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior or after the invasive procedure (reference standard).
gNIPT was a second‐tier test. 4099/5321 samples not selected for this study. 14/1222 samples were excluded before sequencing process (11/14 samples excluded for incomplete diagnostic information and 3/14 samples excluded for confirmed mosaicism). 42/1208 samples failed during autosome sequencing process (no gNIPT result) including 11/42 failed samples for low fetal fraction DNA, 29/42 failed samples for technical reasons and 2/42 failed samples for other biological reasons (maternal event). 22/1166 samples failed SCA sequencing process (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To provide a clinical validation of the sensitivity and specificity of a novel NIPT for detection of genome wide abnormalities. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Liang 2013.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women considered at high risk for fetal T21. Inclusion criteria: singleton and twin pregnancies underwent conventional serum screening and ultrasound scanning, and who invasive prenatal diagnostics were offered. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 435 pregnant women. Number available for 2 x 2 table: 412 pregnant women (subgroup of 94.7%). Setting: 3 hospitals in China. Recruitment period: March 2009 to June 2011. Ethnicity: Asian. Median gestational age (range): 21.4 (11.4 to 39.4) weeks. Most pregnant women (60%) are between 21 to 40 weeks. Only 1 case is in the first trimester (0.23%). Mean maternal age (± SD): 31 (± 5.9) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or multiple screening tests. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 8‐plex or 12‐plex. Fetal fraction DNA: for a Z score cutoff value of 3 for chromosome 21, fetal DNA was estimated to 5.52%. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) positive if Z score > 3 (T21). 2) positive if Z score > 5.91 (T18). 3) positive if Z score > 5.72 (T13). 4) positive if Z score Chrom. X < ‐2.91 and Z score Chrom. Y < 3 (45,X). 5) positive if Z score Chrom. X range from ‐2.91 to +2.91 and Z score Chrom. Y > 3 (47,XXY). 6) positive if Z score Chrom. X > 2.91 and Z score Chrom. Y < 3 (47,XXX). 7) positive if Z score Chrom. X < ‐2.91 and Z score Chrom. Y > 3 (47,XYY). Commercial test: Berry Genomics's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY, and 47,XXX. Reference standard: fetal karyotype of chorionic villi (1%) or amniotic fluid (77%) or cord blood (22%). | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 12/435 samples failed sequencing process quality control (no gNIPT result). 11/423 samples without karyotype were excluded (no reference standard result). |
||
| Comparative | |||
| Aim to study | To determine whether gNIPT by maternal plasma DNA sequencing can uncover all fetal chromosome aneuploidies in 1 simple sequencing event. | ||
| Funding source or sponsor of the study | Study not funded by industry but Berry Genomics Co. Ltd performed the sequencing analysis for free. This study was supported by the grants from the National High Technology Research and Development Program of China (863 Program) (No.2011AA02A112), the National Key Basic Research Program of China (2012CB944600) and the National Key Technology R&D Program of China (2012BAI09B05). | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Yes | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Liu 2012.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: women who planned an invasive testing for 1 or more of the following reasons: abnormality in plasma test, older than 35 years old, infant deformity (ultrasound), taken drugs (teratogen) during early pregnancy or history of malformation caused by virus infection, history of birth defect caused by abnormal chromosome, history of fetus stopping growth or repeated spontaneous abortion or dead fetus or dead birth for unknown reason, history of chromosome abnormality in family or either of the couple, too much or little amniotic fluid. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 153 pregnant women.
Number available for 2 x 2 table: 153 pregnant women (whole cohort included in analyses). Setting: Henan Province People Hospital Medical. Recruitment period: October to November 2011. Ethnicity: Asian. Gestational age: more than 14 weeks. Mean maternal age (± SD; range): 32.3 (± 1.2; 20 to 44) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: Chinese. |
||
| Index tests | gNIPT by MPSS on Illumina HiSeq sequencer in multiplex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected 30 minutes before reference standard. Cutpoint: positive if Z score ≥ 3. It is not reported if gNIPT was a commercial or an in‐house test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X and 47,XYY. 47,XXY and 47,XXX were also assess but no case were found. Reference standard: fetal karyotype of amniotic fluid. | ||
| Flow and timing | Blood samples for gNIPT were obtained 30 minutes prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To determine the feasibility and accuracy of detecting numerical chromosomal abnormalities by high‐flux sequencing analysis of ccfDNA from maternal plasma. | ||
| Funding source or sponsor of the study | Study funded by by the Nalional Natural Science Foundation of China and a Medical Science and Technology Research Project of Henan Province. | ||
| Informations about the authors contacted | Author was contacted on 11 April 2016 but contact author's email is no longer valid. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Ma 2016.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective and retrospective cohort study. Participants: pregnant women selected from a high‐risk population presenting for invasive testing (prospective cohort) and archived maternal plasma from mixed‐risk (high and low risk of fetal aneuploidy) pregnant women (retrospective cohort). Inclusion criteria: singleton pregnancies with gestational age of 12 weeks or above at the time of sampling. Exclusion criteria: women with twin pregnancy or organ donation history or maternal chromosome abnormality. | ||
| Patient characteristics and setting | Number enrolled: 10,598 pregnant women. 2439 from prospective cohort and 8159 from retrospective cohort. Number available for 2 x 2 table: 10,579 pregnant women (subgroup of 99.8%). Setting: 20 centres. Prenatal diagnosis clinics in China. Recruitment period: January 2012 to January 2014 (retrospective) and February to May 2014 (prospective). Ethnicity: Asian. Median gestational age: 19 weeks. Median maternal age (range): 32 (16 to 53) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on BGISEQ‐1000 platform in 16 or 24‐plex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 3. Commercial test: BGI‐Shenzhen's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi, amniotic fluid or cord blood, or postnatal follow‐up. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐ or a first‐tier test. 19/10,598 samples were excluded from the analysis including 5 from retrospective cohort (4 samples had incomplete clinical information and 1 sample failed quality control during sequencing) and 14 from prospective cohort (10 samples had incomplete clinical information and 4 samples failed quality control during sequencing). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To report the established gNIPT screening system and the clinical performance of a new ultrahigh throughout gNIPT method based on combinatorial probe‐anchor ligation sequencing (cPAL) of ccfDNA in detecting T21, T18 and T13 in the multicentre network using a centralised testing mode. | ||
| Funding source or sponsor of the study | Some authors are employees of BGI‐Shenzhen, BGI‐Manufacture or BGI‐DX. Study funded by Shenzhen Birth Defect Screening Project Lab, Key Laboratory of Cooperation Project in Guangdong Province, Shenzhen Municipal Government of China, Pilot projects of regional strategic emerging industry cluster development by Hubei provincial development and Reform Commission and Action plan for the development of high‐tech industry in biotechnology and new medicine in 2012 by Wuhan Science and Technology Bureau. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Mazloom 2013.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study (validation set). Participants: pregnant women selected at high risk of fetal aneuploidy. Inclusion criteria: ≥ 18 years old and singleton pregnancies between 10.5 and 20 weeks of gestation. Exclusion criteria: multifetal pregnancies, mosaic cases for sex chromosomes, or samples without documented karyotype report available. | ||
| Patient characteristics and setting | Number enrolled: 1975 pregnant women including 1564 in the training set (data not shown in the present review) and 411 in the validation set. Number available for 2 x 2 table: 411 pregnant women (subgroup of 95% of validation set). Setting: not reported. Recruitment period: not reported. Ethnicity: Caucasian (58.4%), Asian (18.5%), Afro‐American (7.5%), other and not specified (15.6%). Median gestational age (range): 17 (8 to 29) weeks. Median maternal age (range): 36 (19 to 47) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina v3 flow cells on HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) positive for 45,X if Z score < ‐3.5 (non‐reportable regions between ‐2.5 and ‐3.5). 2) positive for 47,XXX if Z score > 3.5 (non‐reportable regions between 2.5 and 3.5). 3) positive for 47,XYY if Z score < ‐3.5 with Chrom. Y representation. 4) positive for 47,XXY if Z score is between ‐3.5 and 3.5 with Chrom. Y representation. Commercial test: Sequenom's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXY, 47,XYY and 47,XXX. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 1564/1975 excluded samples were used for the training set. 21/411 failed samples were in the non reportable region and were considered positive gNIPT result by authors. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To extend the detection of autosomal aneuploidies by MPSS of ccfDNA from maternal plasma to include common sex chromosome aneuploidies. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. and Sequenom Center for Molecular Medicine (SCMM). | ||
| Informations about the authors contacted | Author was contacted on: 26 May 2016. No reply received from the author. | ||
| Notes | Data from the training set were not shown in the present review. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Nicolaides 2012.
| Study characteristics | |||
| Patient sampling | Study design: retrospective study from a prospective cohort. Participants: selected archived plasma samples from pregnant women without prior risk of fetal aneuploidy (general population) attending for their routine first‐trimester combined screening for aneuploidies. Inclusion criteria: singleton pregnancies between 11 to 13.9 weeks' gestation. Archived samples of at least 2 mL. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 2230 pregnant women. Number available for 2 x 2 table: 1949 pregnant women (subgroup of 87%). Setting: not reported. Recruitment period: October 2010 to January 2011. Ethnicity: Caucasian (69.8%), African (20.6%), South Asian (4%), East Asian (2.8%) and mixed (2.8%). Gestational age range: 11 to 13.9 weeks. Median maternal age (range): 31.8 (27.7 to 35.4) years. Relevant tests carried out prior to index test: none. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay). Median fetal fraction DNA (interquartile range): euploids: 10.0% (7.8% to 13.0%), T21: 12.5% (9.2% to 21.3%), and T18: 9.3% (5.6% to 13.0%). Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if FORTE algorithm risk score ≥ 1%. Commercial test: Harmony™ Prenatal test by Ariosa Diagnostics, Inc. The traditional screening test (combined test at the first trimester) was also assessed. Cutpoint of combined test: 1 in 150. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or neonatal clinical examination. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐tier test. 181/2230 samples were ineligible (no fetal karyotype or follow‐up, miscarriage, stillbirth, termination of pregnancy or other abnormalities). 100/2049 samples failed during sequencing process including 46 for low fetal DNA and 54 had assay failures (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To assess performance of noninvasive prenatal testing for fetal trisomy in a routinely screened first‐trimester pregnancy population. | ||
| Funding source or sponsor of the study | The study was supported by a grant from the Fetal Medicine Foundation (UK). The cost of collection and analysis of the samples was covered by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Nicolaides 2013.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: singleton pregnancies at high risk of fetal aneuploidy between 11 to 13 weeks' gestation. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 242 pregnant women. Number available for 2 x 2 table: 229 pregnant women (subgroup of 95%). Setting: 1 centre. Fetal Medicine Centre, in UK. Recruitment period: not reported. Ethnicity: not reported. Median gestational age (range): 13.1 (11.3 to 13.9) weeks. Median maternal age (range): 35.7 (18.5 to 46.5) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers with NATUS algorithm. Fetal fraction DNA: the lowest fetal fraction DNA on a case that returned a result was 3.95%. Blood samples for gNIPT were collected immediately before reference standard. Cutpoint: not reported. Commercial test: Natera's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, 45,X. 47,XXY, 47,XYY and 47,XXX were also assessed but no case was found. T13 was also assessed but the only 1 case presented in this publication was published thereafter in Hall 2014. T13 case was excluded to avoid double counting. Reference standard: fetal karyotype of chorionic villi. | ||
| Flow and timing | Blood samples were obtained just before the invasive procedure (reference standard). gNIPT was a second‐tier test. 13/242 samples failed sequencing process quality control (no gNIPT result). No repeated test reported. 1 T13 cases was excluded to avoid double counting because it was published thereafter in Hall 2014. |
||
| Comparative | |||
| Aim to study | To assess the performance of ccfDNA testing in maternal blood for detection of fetal aneuploidy of chromosomes 13, 18, 21, X, and Y using TMPS of single‐nucleotide polymorphisms. | ||
| Funding source or sponsor of the study | Study funded by a grant from the Fetal Medicine Foundation (UK Charity No: 1037116). Analysis of samples was performed at their own expense by Natera, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | T13 cases data are not shown in the present review. They were excluded to avoid double counting because they are also published in Hall 2014. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Nicolaides 2014a.
| Study characteristics | |||
| Patient sampling | Study design: case‐control study. Participants: pregnant women selected from a high‐risk population (archived maternal plasma samples). Inclusion criteria: singleton pregnancies. Exclusion criteria: cases of fetal mosaicism and multifetal pregnancies. |
||
| Patient characteristics and setting | Recruited participants: 177 archived maternal plasma. Number available for 2 x 2 table: 172 samples (subgroup of 97%). Setting: recruitment in London, UK. Ethnicity: Caucasian (90%), Afro‐Caribbean (4%), Asian (5%) and other (1%). Gestational age range: 11.2 to 14.1 weeks. Maternal age range: 17.3 to 47.8 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. |
||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Median fetal fraction DNA (range): euploids: 13.0% (4.8% to 32.0%), 45,X: 10.0% (6.3% to 18.0%), and 47,XXX, 47,XXY, and 47,XYY: 12.0% (6.4% to 16.0%). Blood samples for gNIPT were collected just before reference standard. Cutpoint: positive if FORTE algorithm risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test by Ariosa Diagnostics, Inc. |
||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXX, 47,XXY, and 47,XYY. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. |
||
| Flow and timing | Blood samples for gNIPT were collected just before invasive procedure (reference standard). gNIPT was a second‐tier test. 5/177 samples failed during sequencing process (no gNIPT result), including 1 sample failed laboratory quality control metrics and 4 samples failed for an insufficient fetal ccfDNA fraction. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To report the clinical performance of chromosome‐selective sequencing of cfDNA in maternal blood and the FORTE algorithm for the assessment of fetal sex chromosome aneuploidies. | ||
| Funding source or sponsor of the study | No funding source was reported. | ||
| Informations about the authors contacted | Author was contacted on: 10 February 2016. No reply received from the author. |
||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Norton 2012.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: pregnant women aged ≥ 18 years, at gestational age ≥ 10 weeks, with a singleton pregnancy, who were planning to undergo invasive prenatal diagnosis for any indication. Exclusion criteria: multifetal pregnancies, women with know aneuploidy, had active malignancy or a history of metastatic cancer, or had already undergone CVS or amniocentesis during the current pregnancy. | ||
| Patient characteristics and setting | Number enrolled: 4002 pregnant women. Number available for 2 x 2 table: 3080 pregnant women (subgroup of 77%). Setting: 48 centres. Selected prenatal care Centres in USA, the Netherlands and Sweden. Recruitment period: not reported. Ethnicity: Caucasian (49.6%), Afro‐American (6.4%), Asian (13.4%), Hispanic (22.7%) and other (7.9%). Mean gestational age (± SD; range): 16.9 (± 4.1; 10 to 38.7) weeks. Mean maternal age (± SD; range): 34.3 (± 6.4; 18 to 50) years. Relevant test carried out prior to index test: not reported. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex. Mean fetal fraction DNA (± SD; range): euploids: 11% (± 4.5%; 4.2% to 51.3%), T21: 11.6% (± 4.2%; 5.1% to 23.3%), and T18: 10% (± 3.8%; 4.9% to 20.8%). Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if FORTE algorithm risk score ≥ 1%. Commercial test: Ariosa Diagnostics, Inc's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standard: fetal karyotype of chorionic villi (74.7%) or amniotic fluid (25.3%). | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test.
774/4002 samples excluded for ineligible criteria. 148/3228 samples failed during sequencing process (no gNIPT result), including 57 samples failed for low fetal fraction DNA and 91 samples failed sequencing process. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate performance of a gNIPT of fetal T21 and T18. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Norton 2015.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: unselected population of pregnant women undergoing aneuploidy screening (without prior risk of fetal aneuploidy). Inclusion criteria: singleton pregnancies, at least 18 years of age, and between 10 to 14 weeks of gestation. Exclusion criteria: women who had a miscarriage, chose to terminate the pregnancy or had a stillbirth without confirmatory genetic testing. |
||
| Patient characteristics and setting | Number enrolled: 18,955 pregnant women. Number available for 2 x 2 table: 15,841 pregnant women (subgroup of 84%). Setting: 35 centres in USA States, Canada, Sweden, the Netherlands, Belgium, and Italy. Recruitment period: March 2012 to April 2013. Ethnicity: Caucasian (70.9%), Afro‐American (8.2%), Asian (10.5%), Native American (0.6%), multiracial (2.7%), other (6.7%) and missing data (0.5%). Mean gestational age (range): 12.5 (10.0 to 14.3) weeks. Mean maternal age (range): 31 (18 to 48) years whose 76% of pregnant women analysed had < 35 years old. Relevant tests carried out prior to index test: none. Language of the study: English. |
||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 sequencer in 96‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ Prenatal Test by Ariosa Diagnostics, Inc. The traditional screening tests (combined test at the first trimester) were also assessed. Cutpoint of combined test: 1 in 270 for T21 or 1 in 150 for T18 and T13. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi, amniotic fluid or products of conception or neonatal karyotype, neonatal clinical examination or medical record from birth. |
||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐tier test. 3114/18,955 samples excluded of analysis including 229 samples did not meet inclusion criteria or meet exclusion criteria, 31 had twins, 121 had unknown ovum‐donor status, 64 withdrew or were withdrawn by investigator, 384 had sample‐handling errors, 308 without standard screening test result, 488 failed sequencing and have no gNIPT result (192 for low fetal fraction DNA, 83 for non fetal fraction DNA and 213 for high assay variance or assay failures) and 1489 were lost to follow‐up. |
||
| Comparative | |||
| Aim to study | To test the hypothesis that gNIPT has better performance than standard first‐trimester screening (with measurement of nuchal translucency and biochemical analytes) in risk assessment for trisomy 21 in a large, unselected population of women presenting for aneuploidy screening. To also evaluate the performance of gNIPT and standard screening in the assessment of risk for trisomies 18 and 13. |
||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc and Perinatal Quality Foundation. | ||
| Informations about the authors contacted | Author was contacted on: 10 February 2016. Reply received on: 11 February 2016. |
||
| Notes | This study is a clinical trial (Noninvasive Examination of Trisomy (NEXT) ClinicalTrials.gov number, NCT01511458). | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Palomaki 2012.
| Study characteristics | |||
| Patient sampling | Study design: nested case‐control (1:3) study. Participants: pregnant women at high risk of fetal aneuploidy presenting for invasive testing were selected. Inclusion criteria: ≥ 18 years old, between about 10 weeks and 21 weeks 6 days of gestation, at high risk of aneuploidies and who underwent a diagnostic procedure. Exclusion criteria: multifetal pregnancies or low risk of fetal aneuploidy. | ||
| Patient characteristics and setting | Number enrolled: 4664 pregnant women. 1776 pregnant women selected for this study and 212 reanalysed samples from Palomaki 2011. Number available for 2 x 2 table: 1971 pregnant women (1759 from this study + 212 from Palomaki 2011) (subgroup of 42%). Setting: 27 centres. Prenatal diagnostic centres (Canada, Italy, Spain, Czech Republic, Argentina, Ireland, Hungary, USA, Israel and Australia). Recruitment period: April 2009 to February 2011. Ethnicity (only for 293 pregnant women): Caucasian (84.9%), Afro‐american (4.1%), Asian (5.5%) and unknown (5.5%). Mean gestational age (range): 14.7 (9 to 22) weeks. Mean maternal age (± SD): 37.2 (± 5) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 4‐plex. Mean (geometric) fetal fraction DNA (range): 13.4% (4% to 50%). Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score > 3 (T21), > 3.88 (T18) or > 7.17 (T13). Commercial test: Sequenom's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype of chorionic villi, amniotic fluid or products of conception. | ||
| Flow and timing | Blood samples were obtained immediately prior the invasive procedure (reference standard). gNIPT was a second‐tier test. 2888/4664 samples were not selected for this study. 110/1776 samples failed the initial MPSS testing. 105/110 samples required repeat testing using a second aliquot and 5/110 samples were resequenced with the same library. 93/110 samples obtained a gNIPT results. 17/1776 samples failed during sequencing process, most for low fetal fraction DNA (no gNIPT result). |
||
| Comparative | |||
| Aim to study | To determine whether maternal plasma ccfDNA sequencing can identify T18 and T13 as well as T21. | ||
| Funding source or sponsor of the study | Study fully funded by Sequenom, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | This study is a clinical trial "A New Prenatal Blood Test for Down Syndrome" ClinicalTrials.gov number: NCT00877292. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Papageorghiou 2016a.
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:9). Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: at least 18 years of age, singleton or twin pregnancies of at least 10 weeks’ gestation and a clinical indication for an invasive procedure. Exclusion criteria: higher‐order multiple pregnancies (triplets or more), known mosaicism, partial trisomy or translocations, fetal demise, disappearing twin, malignancy or known aneuploidy in the pregnancy. | ||
| Patient characteristics and setting | Number enrolled: 442 pregnant women. Number available for 2 x 2 table: 426 singleton pregnancies (subgroup of 96%). Setting: 6 hospital centres in England, UK. Recruitment period: April 2008 to November 2014. Ethnicity: not reported. Median gestational age (range): 15.4 (11 to 36.6) weeks. Median maternal age (range): 35 (18 to 55) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer in 8‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if likelihood ratio > 1 and maternal age‐adjusted probability risk score. Commercial test: IONA® test by Premaitha Health (public limited company). |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or medical record from birth. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test.
5/442 samples failed during sequencing process including 3 samples for low fetal fraction DNA and 2 samples did not have sufficient DNA fragment counts (no gNIPT result). 11/437 twin pregnancies were not selected. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigate the accuracy of the IONA® test in the discrimination between euploid pregnancies and those affected by fetal trisomies 21, 18 and 13. | ||
| Funding source or sponsor of the study | Study funded by Premaitha Health (public limited company). Some authors are employees of Premaitha Health plc. | ||
| Informations about the authors contacted | Author was contacted on: 19 September 2016. Reply received on: 20 September 2016. | ||
| Notes | Data from singleton pregnancies only reported here. See Papageorghiou 2016b for data on twin pregnancies. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Papageorghiou 2016b.
| Study characteristics | |||
| Patient sampling | Study design: blinded, case‐control study (1:9). Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: at least 18 years of age, a singleton or twin pregnancies of at least 10 weeks’ gestation and a clinical indication for an invasive procedure. Exclusion criteria: higher‐order multiple pregnancies (triplets or more), known mosaicism, partial trisomy or translocations, fetal demise, disappearing twin, malignancy or known aneuploidy in the pregnancy. | ||
| Patient characteristics and setting | Number enrolled: 442 pregnant women. Number available for 2 x 2 table: 11 twin pregnancies (subgroup of 2%). Setting: 6 hospital centres in England, UK. Recruitment period: April 2008 to November 2014. Ethnicity: not reported. Median gestational age (range): 15.4 (11 to 36.6) weeks. Median maternal age (range): 35 (18 to 55) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer in 8‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if likelihood ratio > 1 and maternal age‐adjusted probability risk score. Commercial test: IONA® test by Premaitha Health (public limited company). |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or medical record from birth. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test. 5/442 samples failed during sequencing process including 3 samples for low fetal fraction DNA and 2 samples did not have sufficient DNA fragment counts (no gNIPT result). 426/437 singleton pregnancies were not selected. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigate the accuracy of the IONA® test in the discrimination between euploid pregnancies and those affected by fetal trisomies 21, 18 and 13. | ||
| Funding source or sponsor of the study | Study funded by Premaitha Health (public limited company). Some authors are employees of Premaitha Health plc. | ||
| Informations about the authors contacted | Author was contacted on: 19 September 2016. Reply received on: 20 September 2016. | ||
| Notes | Data from twin pregnancies only reported here. Data from singleton pregnancies reported in Papageorghiou 2016a. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Pergament 2014.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women from a population with mixed risk of fetal aneuploidy presenting for aneuploidy screening (51% high risk and 49% low risk). Inclusion criteria: women were 18 years of age or older with a singleton pregnancy of at least 7 weeks of gestation and signed an informed consent. Exclusion criteria: women with confirmed sex chromosome abnormality (47,XXX, XXY, XYY), confirmed triploidy, confirmed fetal mosaicism or multifetal pregnancy or egg donor. | ||
| Patient characteristics and setting | Number enrolled: 1064 pregnant women. Number available for 2 x 2 table: 963 pregnant women for T21, 964 for T18 and 45,X and 965 for T13 (subgroup of 91%). Setting: 35 centres. Prenatal care centres worldwide in Czech Republic, Japan, USA, Ireland and Spain. Recruitment period: not reported. Ethnicity: not reported. Mean gestational age (± SD; range): 17.0 (± 4.1; 7.6 to 40.6) weeks. Median gestational age: 14.3 weeks. Mean maternal age (± SD; range): 30.3 (± 7.4; 18 to 47) years. Median maternal age: 30.0 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers, 19,488‐plex targeted PCR with NATUS algorithm. Range fetal fraction DNA: 2% to 50%. Blood samples for gNIPT were collected before (93%) or 4 days or later after (7%) reference standard. Cutpoint: not reported. Commercial test: Natera's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. Reference standards: fetal karyotype with confirmatory fluorescence in situ hybridisation or cytogenetic karyotype analysis or by genetic testing of cord blood, buccal sample, saliva, or products of conception, post‐natal or post‐live birth follow‐up. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior (93%) or after (7%) to the invasive procedure (reference standard).
gNIPT was a second‐tier test. 13/1064 samples excluded for other aneuploidies, including 6 cases with triploidy, 3 fetal mosaics, 2 cases with 47,XXY, 1 case with 47,XXX and 1 case with 47,XYY. 85/1051 samples failed quality control (no gNIPT result) including 64 low fetal fraction DNA, 12 low DNA, 6 contaminations, 2 loss of heterozygosity and 1 poor model fit. Between 1 to 3 samples did not passed quality control for all 5 chromosomes. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To estimate performance of a single nucleotide polymorphism–based gNIPT (TMPS) for fetal aneuploidy in high‐risk and low‐risk populations on single venipuncture. | ||
| Funding source or sponsor of the study | Study funded by Natera, Inc. and a grant from the National Institute of Health, National Institute of Child Health and Human Development (4R44HD062114‐02). The majority of the authors are employees of Natera, Inc. and hold stock or options to hold stock in the company. | ||
| Informations about the authors contacted | Author was contacted on: 22 June 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Persico 2016.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected from a high‐risk population. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 259 pregnant women. Number available for 2 x 2 table: 249 pregnant women (subgroup of 96%). Setting: 4 fetal medicine centres in Italy. Recruitment period: March to December 2014. Ethnicity: not reported. Gestational age: not reported. Median maternal age (range): 36 (20 to 46) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina Genome Analyzer IIx or HiSeq sequencers, 19,488‐plex targeted PCR with NATUS algorithm. Fetal fraction DNA: amount measured but not reported (usually NATERA used quality control criteria > 4%). Blood samples for gNIPT were collected just before reference standard. Cut‐off value: positive if risk score > 1%. Commercial test: Natera's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY and 47,XXX. 47,XYY was also assessed but no case was found. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test.
10/259 samples failed during sequencing process (no gNIPT result) including 2 samples failed internal quality control and 8 samples had low fetal fraction DNA. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigate a strategy for clinical implementation of ccfDNA testing in high‐risk pregnancies after first‐trimester combined screening. | ||
| Funding source or sponsor of the study | Study not funded by industry but the cost of ccfDNA testing were covered by Natera, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Poon 2016.
| Study characteristics | |||
| Patient sampling | Study design: retrospective cohort, blinded nested case‐control study. Participants: archived maternal plasma from pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing (CVS). Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 242 pregnant women. Number available for 2 x 2 table: 241 pregnant women (subgroup of 99.6%). Setting: 1 centre at King’s College Hospital, London, UK. Recruitment period: April 2007 to June 2012. Ethnicity: Caucasian (75%), Afro‐Caribbean (17%), Asian (5%) and mixed (3%). Median gestational age (range): 12.7 (11.4 to 13.6) weeks. Median maternal age (range): 33.7 (29.2 to 40.5) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected just before reference standard. Cutpoint: not reported but authors used the same prenatal test than Papageorghiou 2016a (cutpoint: positive if likelihood ratio > 1 and maternal age‐adjusted probability risk score). Commercial test: IONA® test by Premaitha Health (public limited company). |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype of chorionic villi. | ||
| Flow and timing | Blood samples for gNIPT were obtained just before the invasive procedure (reference standard).
gNIPT was a second‐tier test.
1/242 samples failed for low fetal fraction DNA (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To assess the potential performance of screening for fetal T21, T18 and T13 by ccfDNA analysis of maternal blood using the IONA® test. | ||
| Funding source or sponsor of the study | Study not funded by industry but the IONA® test was provided by Premaitha Health plc, Manchester, UK. Study supported by a grant from The Fetal Medicine Foundation. | ||
| Informations about the authors contacted | Author was contacted on: 19 September 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Porreco 2014.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort, observational study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing when research personnel have been available. Inclusion criteria: singleton pregnancy in a patient 18 years of age or older who had provided written informed consent and who had made the decision to pursue invasive prenatal diagnosis by CVS or amniocentesis. Exclusion criteria: inability to give written informed consent, multifetal pregnancies, or fetal demise of an additional embryo during the current pregnancy at 8 weeks or more of gestation. | ||
| Patient characteristics and setting | Number enrolled: 4170 pregnant women. Number available for 2 x 2 table: 3322 for autosomes (subgroup of 80%), 3278 for 45,X and 47,XXX (subgroup of 79%) and 3201 for 47,XXY and 47,XYY (subgroup of 77%). Setting: 31 centres in USA. Recruitment period: September 2009 to April 2011. Ethnicity: Caucasian (60,1%), Asian (18,7%), Afro‐American (4,5%) and other (16.7%). Mean gestational age (± SD; range): 16.3 (± 3.5; 9.0 to 37.0). Mean maternal age (± SD; range): 35.1 (± 5.6; 18.0 to 50.0). Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 in 12‐plex. Range fetal fraction DNA: 4% to 50%. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) for T21, positive if Z score ≥ 3. 2) for T18 and T13, positive if Z score ≥ 3.95. 3) positive for 45,X if Z score < ‐3.5 (non‐reportable regions between ‐2.5 and ‐3.5). 4) positive for 47,XXX if Z score > 3.5 (non‐reportable regions between 2.5 and 3.5). 5) positive for 47,XYY if Z score risk < ‐3.5 with Chrom. Y representation. 6) positive for 47,XXY if Z score risk is between ‐3.5 and 3.5 with Chrom. Y representation. Commercial test: Sequenom's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXX, 47,XXY and 47,XYY. Reference standards: fetal karyotype of chorionic villi or amniotic fluid, or medical record from birth. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a second‐tier test. 740/4170 samples excluded before sequencing process including 320 samples for insufficient sample volume,120 samples processed outside of the 6 hours laboratory process window, 270 failed laboratory quality control set, 24 for incomplete case report form and 6 without invasive procedure performed). For autosomes: 54/3430 autosomes samples excluded for quality control deviation (low fetal DNA fraction, library concentration, total counts, and amplification bias). For autosomes: 54/3376 samples excluded for complex autosome karyotypes (mosaic, triploidies, unbalanced rearrangements with missing or duplicated genetic material). For 45,X and 47,XXX: 102/3430 samples excluded for low fetal fraction DNA or copy number variation of the Chrom. X is confounded by maternal component and cannot be determined. For 45,X and 47,XXX: 50/3328 samples excluded for complex SCA karyotype. For 47,XXY and 47,XYY: 182/3430 samples excluded for low fetal fraction DNA or copy number variation of the Chrom. X is confounded by maternal component and cannot be determined. For 47,XXY and 47,XYY: 47/3248 samples excluded for complex SCA karyotype. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To validate the clinical performance of MPSS of ccfDNA contained in specimens from pregnant women at high risk of fetal aneuploidy to test fetuses for T21, T18, T13, 45,X, 47,XXX, 47,XXY and 47,XYY. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | Author was contacted on: 30 May 2016. Reply received on: 31 May 2016. | ||
| Notes | This study is a clinical trial (Non‐Invasive Screening for Fetal Aneuploidy) ClinicalTrials.gov number, NCT00847990. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Quezada 2015.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: self‐selected pregnant women from the general population presenting for aneuploidy screening (without prior risk of fetal aneuploidy). Inclusion criteria: pregnant women between 10 to 11 weeks’ gestation with singleton pregnancy who underwent the combined test. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 2905 pregnant women. Number available for 2 x 2 table: 2785 pregnant women (subgroup of 96%). Setting: 1 centre. Fetal Medicine Centre in London, UK. Recruitment period: October 2012 to January 2014. Ethnicity: Caucasian (88.5%), South Asian (6.0%), East Asian (3.3%), Afro‐Caribbean (0.7%) and mixed (1.5%). Median gestational age (range): 10.6 (10 to 11.9) weeks. Median maternal age (range): 36.9 (20.4 to 51.9) years. Relevant tests carried out prior to index test: none. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay). Median fetal fraction DNA (range): 11% (4% to 40%). Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Usually, Harmony™ prenatal test uses FORTE algorithm; positive if FORTE risk score ≥ 1%. Commercial test: Harmony™ Prenatal test by Ariosa Diagnostics, Inc. The traditional screening tests (combined test at the first trimester) was also assessed. Cutpoint of combined test: 1 in 100 for T21. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi, amniotic fluid or products of conception, neonatal karyotype, neonatal clinical examination or medical record from birth. | ||
| Flow and timing | Blood samples were obtained prior to invasive procedure (reference standard). gNIPT was a first‐tier test. 122/2905 failed the initial TMPS testing (122 = 123 ‐ 1 sample lost in mail). 66/2851 samples without follow‐up were excluded. 110/122 required repeat testing using a second blood sample and results were obtained in 69/110 samples. 53/2905 samples failed during sequencing process (41 samples failed second sequencing and 12 unrepeated tests) (no gNIPT result). |
||
| Comparative | |||
| Aim to study | To examine, in a general population (pregnant women without prior risk of fetal aneuploidy), the performance of ccfDNA testing for T21, T18 and T13 at 10 to 11 weeks’ gestation and compare it to that of the combined test at 11 to 13 weeks' gestation. | ||
| Funding source or sponsor of the study | Study not funded by industry but Ariosa Diagnostics, Inc made sequencing and analyses. | ||
| Informations about the authors contacted | Author was contacted on: 21 April 2016 and 30 May 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Samango‐Sprouse 2013.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women at high or low risk of fetal aneuploidy (known sex chromosome aneuploidy and euploid pregnancies). Inclusion criteria: women were at least 18 years of age, had singleton pregnancy, or with known sex chromosome aneuploidy. Exclusion criteria: pregnant women with known mosaicism, autosomal trisomy, or triploidy. | ||
| Patient characteristics and setting | Number enrolled: 201 pregnant women.
Number available for 2 x 2 table: 186 pregnant women (subgroup of 93%).
Setting: 8 prenatal care centres in UK, USA, Poland, and Czech Republic.
Recruitment period: not reported.
Ethnicity: not reported.
Mean gestational age: euploid pregnancies 13.2 weeks, and aneuploid pregnancies 15.3 weeks. Gestational age range: overall 9.4 to 36.4 weeks. Maternal age: not reported. Relevant test carried out prior to index test: not reported. Language of the study: English. |
||
| Index tests | gNIPT by TMPS (SNP‐based method) on Illumina HiSeq 2000 sequencer with NATUS algorithm. Mean fetal fraction DNA: euploids: 10.9% and aneuploids: 12.1%. Overall range: 2.9% to 37.7%. Blood samples for gNIPT were collected just before or at least 4 days after reference standard. Cutpoint: not reported. Commercial test: Natera's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: 45,X, 47,XXX, 47,XXY, and 47,XYY. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or genetic testing of cord blood, buccal sample, saliva, or products of conception. | ||
| Flow and timing | Blood samples were collected just before or at least 4 days after invasive procedure (reference standard).
gNIPT was a first‐ or second‐tier test.
14/201 samples failed sequencing process quality control (no gNIPT result) including 12 for low fetal fraction or poor DNA quality and 2 samples did not return a result for SCA. 1/187 sample excluded for conflicting algorithm metrics (no meaningful gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To develop a SNP‐based and informatics‐based gNIPT that detects sex chromosome aneuploidies early in pregnancy. | ||
| Funding source or sponsor of the study | It is unclear if the study was funded by industry but all authors are employees of Natera, Inc. except the first author (Carole Samango‐Sprouse). This study was supported in part by a grant from the National Institute of Health, National Institute of Child Health and Human Development. | ||
| Informations about the authors contacted | Author was contacted on: 22 April, 4 July and 29 September 2016. Replies received on: 29 and 30 September 2016. |
||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Sehnert 2011.
| Study characteristics | |||
| Patient sampling | Study design: blinded retrospective study (archived maternal plasma samples). Participants: pregnant women selected from a high risk of fetal aneuploidy population. Inclusion criteria: pregnant women age 18 years or older with singleton or multifetal pregnancy. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: overall: 1014 pregnant women including 71 women selected on 435 for the training set (not shown in the present review) and 48 women selected on 575 for the test set. Number available for 2 x 2 table: 47 (subgroup of 8%). Setting: 13 centres in USA. Recruitment period: January 2010 to June 2010. Ethnicity: Caucasian (62.7%), Hispanic (16.5%), Asian (6.2%), multiethnic (5.2%), Afro‐American (4.0%), Native American (0.9 %) and other or not specified (1.8%). Mean gestational age (range): 15.4 (10.6 to 28.4) weeks. Mean maternal age (± SD; range): 34.2 (± 8.22; 18 to 46) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx sequencer in uniplex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint prespecified with the training set: 1) positive if NCV > 4 for autosomes. There is a "no call zone" between 2.5 and 4 considering as gNIPT positive result for the present review. 2) positive if NCV for Chrom. Y < ‐2.0 SDs from the mean of male samples and if NCV for Chrom. X < ‐3.0 SDs from the mean of female samples for 45,X. Commercial test: Verinata's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. Reference standard: fetal karyotype of chorionic villi (58,3%) or amniotic fluid (41.7%). | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test. 895/1014 samples were not selected for sequencing. 71/119 samples were selected for the training set (not shown in the present review). 1/48 sample from twin gestation in the test set was removed from the final analysis. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To develop and test an optimised algorithm from MPSS data and demonstrated the potential universality of the sequence tag mapping and chromosome quantification method for the detection of multiple chromosomal abnormalities. | ||
| Funding source or sponsor of the study | Study funded by Illumina (formerly Verinata Health). The funding organizations played a direct role in the design of the study, the choice of enrolled patients, the review and interpretation of data, and the preparation and final approval of the manuscript. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Shaw 2014.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: consecutive pregnant women were selected from a mixed‐risk population. They were classified in extremely high‐risk group for T21 with a screening T21 risk > 1:30 or nuchal translucency > 3.0 mm and low‐risk group with a screening T21 risk < 1:1500. Inclusion criteria: pregnant women at > 12 weeks’ gestation, singleton or multifetal pregnancies. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 201 pregnant women.
Number available for 2 x 2 table: 200 pregnant women (subgroup of 99.5%).
Setting: 11 medical centres in Taiwan.
Recruitment period: June to December 2012.
Ethnicity: Asian.
Mean gestational age (± SD): high‐risk pregnant women 17.3 (± 2.1) weeks, and low‐risk pregnant women 16.1 (± 3.0) weeks. Gestional age range: overall 12 to 20 weeks. Mean maternal age (± SD): high‐risk pregnant women 35.1 (± 3.2) years, and low‐risk pregnant women 34.6 (± 2.6) years. Chorionicity: all dichorionic (4/4). Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) positive if Z score > 3 (T21, T18, and T13). 2) positive if Z score Chrom. X < ‐3 and Z score Chrom. Y < 3 (45,X). 3) positive if Z score Chrom. X < ‐3 and Z score Chrom. Y > 3 (47,XYY). Commercial test: Berry Genomics' prenatal test. The traditional screening test (combined test at the first trimester) was also assessed but complete data for 2 x 2 tables were unavailable. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, and 47,XYY. 47,XXX and 47,XXY were also screened but no case was found. Reference standards: fetal karyotype of amniotic fluid or medical record from birth. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test.
1 sample excluded for early gestational age (< 12 weeks). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate the performance of gNIPT for all fetal chromosomal aneuploidies in an extremely high‐risk group undergoing first‐trimester combined T21 screening. | ||
| Funding source or sponsor of the study | Funding sources were not reported but 2 authors are affiliated to Berry Genomics Co. Ltd., Beijing, PR China. | ||
| Informations about the authors contacted | Author was contacted on: 10 February and 23 June 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Song 2013.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women without a priori risk of fetal aneuploidy who undergo routine prenatal screening. Inclusion criteria: singleton pregnancies and pregnant women younger than 35 years old. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 1916 pregnant women. Number available for 2 x 2 table: 1741 pregnant women (subgroup of 91%). Setting: 2 clinical centres in Beijing, China. Recruitment period: April 2011 to December 2011. Ethnicity: Asian. Mean gestational age (± SD; range): 16.57 (± 1.56; 11 to 21.9) weeks. Mean maternal age (± SD; range): 29.03 (± 2.70; 20 to 34) years. Relevant tests carried out prior to index test: none. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score ≥ 3. Commercial test: Berry Genomics' prenatal test. The traditional screening test (second‐trimester triple test) was also assessed. Cutpoint of triple test: 1 in 270 for T21 and T18. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. 45,X, 47,XXX, 47, XXY, 47,XYY were also screened but inappropriate reference standard for the present review was used. Reference standards: fetal karyotype of chorionic villi, amniotic fluid or cord blood or medical record from birth. | ||
| Flow and timing | It is not reported if the blood samples were collected before or after invasive procedure (reference standard).
It is not reported if the gNIPT was a first‐ or second‐tier test.
64/1916 samples failed sequencing process (failed DNA quality control criteria or sequencing quality control) (no gNIPT result). 102/1916 samples without follow‐up were excluded. 9/1916 samples were without follow‐up and failed sequencing process (no gNIPT result). No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate the performance of gNIPT for detection of fetal aneuploidies in a Chinese cohort of women younger than 35 years old in a prospective clinical setting. Also, to compare the performance of gNIPT with the routine prenatal screening (second‐trimester combined test). | ||
| Funding source or sponsor of the study | Study not funded by industry. This study was supported by a grant (2006BAI05A10) from the National Key Technology Research and Development Program of China during the ‘11th Five‐Year Plan’. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | SCA were also screened but inappropriate reference standard for the present review was used. gNIPT data from SCA were not shown in this review. | ||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 2: Index Test Traditional screening tests | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | No | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Song 2015.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected arbitrarily at high risk of fetal aneuploid presenting for aneuploidy screening by gNIPT. Inclusion criteria: advanced maternal age (≥ 35 years) and singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 213 pregnant women. Number available for 2 x 2 table: 204 pregnant women (subgroup of 96%). Setting: 1 centre. Peking Union Medical College Hospital (PUMCH), Beijing, China. Recruitment period: May 2012 to August 2013. Ethnicity: Asian. Median gestational age (range): 9.9 (8 to 12.9) weeks. Mean maternal age (range): 37.25 (35 to 45) years. Relevant tests carried out prior to index test: none. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 sequencer in 12‐plex. Median fetal fraction DNA (range): only male fetus: 8.54% (2.69% to 18.75%). Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if Z score ≥ 3. Commercial test: Berry Genomics' prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY and 47,XXX. 47,XYY were also assessed but no case was found. Reference standards: fetal karyotype of chorionic villi or amniotic fluid for all analysed women (178/178) and neonatal clinical examination (198/212). | ||
| Flow and timing | Blood samples for gNIPT were obtained prior the invasive procedure (reference standard).
gNIPT was a first‐tier test.
1/213 sample failed quality control (haemolysis). 8/212 samples without reference standard were excluded including 5 miscarriages, 2 intrauterine fetal deaths and 1 termination of pregnancy. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate the feasibility of gNIPT of maternal plasma samples collected from pregnant Chinese women in early gestation, between 8 and 12.9 weeks’ gestation. | ||
| Funding source or sponsor of the study | Study not funded by industry but gNIPTs were done and analysed at Berry Genomics Co. Ltd. Study funded by a grant from the National Natural Science Foundation of China. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Sparks 2012a.
| Study characteristics | |||
| Patient sampling | Study design: case‐control study from a prospective cohort. Participants: pregnant women selected from a high risk of fetal aneuploidy population. Inclusion criteria: women at least 18 years of age, at least 10 weeks’ gestation and have singleton pregnancy. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: not reported. A subset of 338 pregnant women including 171 women in the training set (data not shown in the present review) and 167 women in the validation set were selected for this study. Number available for 2 x 2 table: 167 pregnant women (subgroup of 49%). Setting: not reported. Recruitment period: not reported. Ethnicity: not reported. Mean gestational age (± SD; range): 18.6 (± 4.0; 11 to 36.1) weeks. Mean maternal age (± SD; range): 33.5 (± 7.1; 18 to 51) years. Relevant tests carried out prior to index test: not reported. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 sequencer in multiplex with FORTE algorithm. Range fetal fraction DNA: 3% to 33%. It is not reported if the blood samples for gNIPT were collected before or after reference standard. Cutpoint: not reported. Commercial test: Ariosa Diagnostics, Inc's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standards: fetal karyotype or chromosome analysis by FISH or both. | ||
| Flow and timing | It is not reported if the blood samples were collected before or after invasive procedure (reference standard).
gNIPT was a second‐tier test.
171/338 samples were excluded for the training set. No failed sample reported in the validation set. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To develop a novel biochemical assay and algorithm for the prenatal evaluation of risk for fetal T21 and T18 using ccfDNA obtained from maternal blood. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. All authors are employees of Aria Dx Inc. (now Ariosa Diagnostics). K Sparks is a member of the board of the company. | ||
| Informations about the authors contacted | Author was contacted on: 23 June 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | No | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Stumm 2014.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Blinded for T21 and unblinded for T18 and T13. Participants: all consecutively enrolled pregnant women selected at high risk of fetal aneuploidy. Inclusion criteria: pregnant women at least 18 years old, at high risk for chromosomal aberrations, signed informed consent, planned a conventional karyotyping procedure (invasive diagnostic), had singleton pregnancy and blood drawn before the invasive procedure. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 522 pregnant women. Number available for 2 x 2 table: 472 pregnant women (subgroup of 90%). Setting: 5 clinical centres in Germany and Switzerland. Recruitment period: not reported. Ethnicity: not reported. Mean gestational age (range): 15.6 (11.0 to 32.1) weeks. Mean maternal age (range): 36.0 (19 to 47) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer in 12‐plex with DAP.21 algorithm without CG correction. Mean fetal fraction DNA (range): male fetus only: 12.3% (3.7% to 36.8%). Blood samples for gNIPT were collected just before reference standard. Cutpoint: 1) positive if MAD‐based Z ‐score ≥ 3 for T21. 2) positive if MAD‐based Z score ≥ 3.2 for T18. 3) positive if MAD‐based Z score ≥ 3.9 for T13. Commercial test: LifeCodexx's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype of chorionic villi (30.3%), amniotic fluid (69.1%) or cord blood (0.6%). | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior the invasive procedure (reference standard).
gNIPT was a second‐tier test.
18/522 samples excluded, including 8 without reference standard result, 9 without consent and 1 was previously analysed. 32/504 samples failed during sequencing process (no gNIPT result), including 14 samples failed sequencing quality criteria and 18 samples failed libraries. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To validate the diagnostic accuracy of a gNIPT for detecting T21, T18 and T13 for a population in Germany and Switzerland. | ||
| Funding source or sponsor of the study | Study funded by LifeCodexx AG and GATC Biotech AG. | ||
| Informations about the authors contacted | Author was contacted on: 22 February 2016, 24 February and 19 May 2016. Reply received on: 24 February 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | No | ||
| If a threshold was used, was it pre‐specified? | No | ||
| High | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Sukhikh 2015.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: not reported. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 200 pregnant women. Number available for 2 x 2 table: 200 pregnant women (whole cohort included in analyses). Setting: not reported. Recruitment period: not reported. Ethnicity: not reported. Median gestational age (range): 14 (10 to 20) weeks. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: Russian. | ||
| Index tests | gNIPT by MPSS on Ion Proton™ sequencer. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) Positive for T21 and T18 if T score > 5. 2) Positive for T13 if T score > 4. 3) Positive for 45,X if T score for chrom. X > 0.04 and for chrom. Y < 0.04. In‐house test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. Reference standard: fetal karyotype of chorionic villi, amniotic fluid or placenta. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test.
No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To estimate the feasibility of using a next‐generation sequencing technique for the noninvasive prenatal diagnosis of fetal aneuploidies. | ||
| Funding source or sponsor of the study | Funding source not reported. | ||
| Informations about the authors contacted | Author was contacted on: 9 September and 4 October 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Sung‐Hee 2015.
| Study characteristics | |||
| Patient sampling | Study design: retrospective study from a prospective cohort. Participants: pregnant women selected from a high risk of fetal aneuploidy population. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 918 pregnant women. Number available for 2 x 2 table: 901 pregnant women (subgroup of 99%). Setting: various medical sites in Korea. Recruitment period: May 2012 to December 2013. Ethnicity: Asian. Mean gestational age (± SD; range): 16.6 (± 2.2; 11 to 25) weeks. Mean maternal age (± SD; range): 35.3 (± 4.1; 22 to 46) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening (59%) or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if L score > 1 and t score > 2.5 (warning zone if t score risk > 2.5 or L score risk > 1). Commercial test: BGI‐Shenzhen's test. Biochemical serum‐screening results were reported in the study but 2 x 2 tables could not be derived. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. T13 were also assessed but the only case found was without follow‐up. SCA were also screened but inappropriate reference standard for the present review was used. Reference standards: fetal karyotype for gNIPT positive cases and medical record from birth for gNIPT negative cases. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a first‐ or a second‐tier test.
8/918 samples were ineligible. 9/910 samples without follow‐up were excluded (all samples had positive gNIPT result). 2/9 women had abortion and 7/9 women declined invasive testing. 21/910 samples failed to give a risk score for gNIPT of the first blood samples including 1 haemolysed sample, 8 samples thawing due to transport delay, 3 due to cell‐free DNA extraction failures and 9 samples had low fetal fraction. 16/21 samples were repeated with new sampling. 14/16 samples obtained a gNIPT results and 2/16 samples failed to provide informative results and were classified as test failures. 7/910 samples failed during sequencing process (no gNIPT result). 5/7 samples failed the initial MPSS testing and were not resequenced and 2/7 samples failed the second MPSS testing. |
||
| Comparative | |||
| Aim to study | To report the initial clinical performance of gNIPT in detecting fetal chromosomal aneuploidies, especially T21, T18 and T13, in singleton pregnancies in Korea. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI performed sequencing and analysis. Study funded by Seoul Clinical Laboratories Research Grant (2015, President: Kyoung‐Ryul Lee). | ||
| Informations about the authors contacted | Author was contacted on: 13, 19 and 26 September 2016. Replies received on: 19 and 25 September and 11 October 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | No | ||
| High | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Tynan 2016.
| Study characteristics | |||
| Patient sampling | Study design: blinded, retrospective clinical evaluation study. Participants: pregnant women selected from 3 internal clinical studies (archived maternal plasma samples). 84.5% without prior risk and 15.8% had high risk of fetal aneuploidy. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 1100 pregnant women. Number available for 2 x 2 table: 1048 pregnant women (subgroup of 95%). Setting: multicentre. Recruitment period: beginning in November 2009. Ethnicity: not reported. Gestational age (range): 9 to 38.1 weeks. Maternal age (range): 18 to 45 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 or HiSeq 2500 sequencer in multiplex. Mean fetal fraction DNA (± SD): low‐risk group: 10% (± 3.1%), high‐risk group (< 35 years): 11.9% (± 4.8%), and high‐risk group (≥ 35 years): 11.1% (± 3.4%). Median fetal fraction DNA (range): low‐risk group: 10.7% (3.1% to 22.9%), high‐risk group (< 35 years): 10.7% (4.9% to 28.3%), and high‐risk group (≥ 35 years): 11% (3.1% to 25.5%). Blood samples for gNIPTwere collected before reference standard. Cutpoint: positive if risk score ≥ 1%. Commercial test: VisibiliT™ test by Sequenom, Inc. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or medical record from birth. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a first‐ or second‐tier test.
52/1100 samples failed during sequencing process (no gNIPT result) including 28 for technical failures (library preparation or low aligned reads counts) and 24 for discretionary non‐reporting because of factors such as sequencing bias. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To demonstrate the clinical performance of a simplified, low coverage, low cost MPSS assay (VisibiliT™) that combines a maternal age‐based risk for T21, T18, and T13, the fractional concentration of fetal DNA, and the representation of chromosomes 21, 18, and 13 in the sample to provide a risk score for T21, T18 and T13, with classification of fetal sex result. | ||
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Verweij 2013.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: consecutive pregnant women selected at high risk of fetal aneuploidy presenting for invasive testing. Inclusion criteria: women who sign informed consent, ≥ 18 years old and carrying a singleton pregnancy with a gestational age of at least 10 weeks. Exclusion criteria: multifetal pregnancies, an invasive procedure performed prior to the blood sampling, history or active significant malignancy requiring major surgery or systemic chemotherapy, or language restriction with failure to understand the study information. | ||
| Patient characteristics and setting | Number enrolled: 595 pregnant women. Number available for 2 x 2 table: 504 pregnant women (subgroup of 85%). Setting: multicentres in the Netherlands and Sweden. Recruitment period: May 2011 to March 2012. Ethnicity: Caucasian (84.8%), Mediterranean (6%), Asian (3.3%), Afro‐European (1.3%), and other (4.6%). Mean gestational age (± SD; range): 14.0 (± 2.1; 10 to 28) weeks. Mean maternal age (± SD; range): 36.4 (± 4.6; 20 to 47) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by TMPS (DANSR assay) on Illumina HiSeq 2000 in 96‐plex with FORTE algorithm. Mean fetal fraction DNA (± SD; range): 11.1% (± 4.1%; 4% to 30%). Blood samples for gNIPT were collected just before reference standard. Cutpoint: positive if FORTE risk score ≥ 1%. Commercial test: Ariosa Diagnostics, Inc's test. |
||
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype of chorionic villi (54%) or amniotic fluid (46%). | ||
| Flow and timing | Blood samples for gNIPT were obtained just prior the invasive procedure (reference standard).
gNIPT was a second‐tier test.
75/595 samples were ineligible. 51/520 samples failed the initial TMPS testing. 51/51 samples were repeated with a second aliquot of the first sampling and 35/51 samples obtained a gNIPT results. 16/520 samples failed during sequencing process (no gNIPT result), including 7 samples with low fetal DNA fraction and 9 samples failed laboratory processing or specimen issues. |
||
| Comparative | |||
| Aim to study | To evaluate the performance of a directed gNIPT method of ccfDNA analysis for fetal T21 by shipping the whole blood samples from Europe to a laboratory in the USA. | ||
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. 2 authors are paid employees of Ariosa Daignostics. 1 author is a board member of Ariosa Diagnostics. | ||
| Informations about the authors contacted | Author was contacted on: 22 April 2016. Reply received on: 25 April 2016. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Yes | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test TMPS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Wang 2014.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women in the first trimester of pregnancy with advanced maternal ages or ultrasound abnormality (high risk of fetal aneuploidy). Inclusion criteria: singleton pregnancies between 11 to 14 weeks' gestation. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 136 pregnant women. Number available for 2 x 2 table: 136 pregnant women (whole cohort included in analyses). Setting: 1 centre. General Hospital of PLA, Beijing, China. Recruitment period: March 2011 to August 2013. Ethnicity: Asian. Gestational age range: 11 to 13.9 weeks. Maternal age range: 35 to 44 years. Relevant test carried out prior to index test: ultrasonography for some women. Language of the study: Chinese. | ||
| Index tests | gNIPT by MPSS on Illumina HiSeq 2000 sequencer with NIFTY™ algorithm. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: not reported. Commercial test: BGI‐Shenzhen's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. T13 was also assessed but no case was found. 45,X was also screened but inappropriate reference standard for the present review was used for pregnant women with gNIPT negative result. gNIPT data from 45,X were not shown in this review. Reference standards: fetal karyotype of amniotic fluid or cord blood or neonatal clinical examination at 42 days after birth or both. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a first‐ or second‐tier test.
No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigate the value of maternal plasma ccfDNA examination in detection of fetal chromosomal aneuploidy in pregnant women at advanced maternal age during the first trimester of pregnancy. | ||
| Funding source or sponsor of the study | Study not funded by industry. Study funded by National Science & Technology Pillar Program during the Twelfth Five‐year Plan Period (2012BA131B06). | ||
| Informations about the authors contacted | Author was contacted on: 19 May and 27 June 2016. No replies received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | Unclear | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Wang 2015a.
| Study characteristics | |||
| Patient sampling | Study design: prospective cohort study. Participants: pregnant women selected from a high risk of fetal aneuploidy population. Inclusion criteria: pregnant women at high risk of fetal aneuploidy between 14 and 26 weeks of gestation. Exclusion criteria: not reported. | ||
| Patient characteristics and setting | Number enrolled: 917 pregnant women. Number available for 2 x 2 table: 917 pregnant women (whole cohort included in analyses). Setting: 1 centre at prenatal clinic, Lianyungang Maternal and Child Health Hospital, Lianyungang, Jiangsu 222001, China. Recruitment period: January 2012 to December 2013. Ethnicity: Asian. Gestational age range: 14 to 26 weeks. Maternal age range: 18 to 46 years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina v2 HiSeq 2000 flow cell on a HiSeq sequencer. Fetal fraction DNA: not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) for T21, T18 and T13, positive if Z score > 3. 2) for 47,XXY and 47,XYY, positive if Z score Chrom. X > ‐3 and Z score Chrom. Y < 3. 3) for 45,X and 47,XXX, positive if Z score Chrom. X between ‐3 and 3 without Chrom. Y representation. Commercial test: Berry Genomics' prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21 and T18. T13 was also assessed but no case was found. SCA was also assessed but inappropriate reference standard for the present review was used. Reference standards: fetal karyotype of amniotic fluid or clinical follow‐up (once per month) from birth to 6 months. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a second‐tier test. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To investigate the clinical efficiency of gNIPT identifying fetal chromosomal aneuploidies. | ||
| Funding source or sponsor of the study | Study not funded by industry but Berry Genomics Co. Ltd give technical support. Study funded by the Community Development Fund, granted by the Department of Family Planning and Healthcare, Jiangsu Province, China. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Low | |||
Yao 2014.
| Study characteristics | |||
| Patient sampling | Study design: retrospective study. Participants: pregnant women presenting with low‐, high‐ or without prior risk factors of fetal aneuploidy (gNIPT was offered routinely as a prenatal screening test). Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 5950 pregnant women. Number available for 2 x 2 table: 5530 pregnant women (subgroup of 93%). Setting: 1 centre. The Prenatal Diagnosis Centre, Southwest Hospital, Chongqing, China. Recruitment period: June 2011 to December 2012. Ethnicity: Asian. Mean gestational age (range): 19.6 weeks (65% of the cohort were between 16 to 20.9 weeks). Mean maternal age (± SD): 30 (± 5) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both for some women. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex with NIFTY™ algorithm. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: 1) positive if t score ≥ 2.5 for autosomes. 2) positive if t score for Chrom. X < ‐2.5 for female fetuses for 45,X. 3) positive if t score for Chrom. X > 2.5 for female fetuses for 47,XXX. 4) positive if t score for Chrom. X > 2.5 combined with estimation of fetal ccfDNA concentration by Chrom. X (expected value of zero) for 47,XXY. 5) positive if t score for Chrom. X > 2.5 and R‐value (the ratio of the fetal DNA fraction estimated by chromosome Y to that estimated by chromosome X) between 1.8 and 2.2 for 47,XYY. Commercial test: BGI‐Shenzhen's prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18, T13. 45,X, 47,XXY, 47,XYY and 47,XXX were also screened but inappropriate reference standard for the present review was used. gNIPT data from SCA were not shown in this review. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or follow‐up by telephone interview with the clinician after the expected delivery date. | ||
| Flow and timing | Blood samples for gNIPT were obtained prior to the invasive procedure (reference standard).
gNIPT was a first‐ or second‐tier test.
420/5950 samples without follow‐up were excluded. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate the performance of a MPSS in detecting fetal sex chromosome aneuploidy (SCA) and to present a comprehensive clinical counselling protocol for SCA‐positive patients. Author also assessed autosomes aneuploidies. | ||
| Funding source or sponsor of the study | Funding source not reported but many authors are employees of the Clinical Laboratory of BGI Health, BGI‐Shenzen or of the Shenzen Birth Defect Screening Projet Lab. | ||
| Informations about the authors contacted | No need for further contact. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Zhang 2016.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected from a high risk of fetal aneuploidy population. Inclusion criteria: women aged ≥ 35 years at the time of delivery, single birth, high risk of T21 or single abnormal multiple of the median, elevated fetal nuchal translucency in the early pregnancy, a soft marker in the genetic scan, or cardiac structural abnormalities in the second‑trimester genetic sonography, not suitable for invasive prenatal diagnosis, such as those with human immunodeficiency virus infection, placenta previa, low‑set placenta, oligohydramnios, Rh‐negative blood type, a history of abortion, threatened abortion or precious pregnancy. Exclusion criteria: multifetal pregnancies, maternal with chromosomal diseases, or received allogeneic blood transfusion, organ transplantation, stem cell therapy, or with a gestational age of < 12 weeks. | ||
| Patient characteristics and setting | Number enrolled: 87 pregnant women. Number available for 2 x 2 table: 87 pregnant women (whole cohort included in analyses). Setting: 1 centre at the Obstetrics and Gynecology Hospital of Fudan University (Shanghai, China). Recruitment period: January 2012 to December 2013. Ethnicity: Asian. Median gestational age (range): 19 (12.4 to 32.5) weeks. Mean maternal age (± SD): 37.48 (± 2.17) years. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) or biochemical screening or both. Language of the study: English. | ||
| Index tests | gNIPT by MPSS on Illummina Hiseq 2000 sequencer in 12‐plex. Fetal fraction DNA: not reported. It is not reported if the blood samples for gNIPT were collected before or after reference standard. Cutpoint for T21: positive if Z score ≥ 3. No other cutpoint reported. Commercial test: Berry Genomics' prenatal test. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18. 45,X and 47,XXX were also screened but inappropriate reference standard for the present review was used. Reference standards: fetal karyotype of amniotic fluid or cord blood or neonatal clinical examination by neonatologists. | ||
| Flow and timing | It is not reported if the blood samples were collected before or after invasive procedure (reference standard).
gNIPT was a second‐tier test.
No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To evaluate the efficacy of using gNIPT technology in screening T21 among women of advanced maternal age and to provide evidence for prenatal screening of T21. | ||
| Funding source or sponsor of the study | Funding source not reported. | ||
| Informations about the authors contacted | Author was contacted on: 7 September 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | Low | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Yes | ||
| Low | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Unclear | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | Yes | ||
| Unclear | |||
Zhou 2014a.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected at high risk for T21 (51.3%), low risk for T21 (2.6%) or without a priori risk (46.1%). gNIPT was integrated in clinical workflow. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 306 pregnant women. Number available for 2 x 2 tables: 301 pregnant women in the pilot validation set (subgroup of 98%). See Zhou 2014b for the integration set. Setting: 1 centre. Women’s Hospital, Zhejiang University School of Medicine, Hangzhouin, China. Recruitment period: September 2011 to October 2011. Ethnicity: Asian. Gestational age range: 12 to 24 weeks. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening for a part of this cohort. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if T score > 2.5 and L score > 1 (warning zone if t score > 2.5 or L score > 1). Commercial test: NIFTY™ prenatal test by BGI‐Shenzhen. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of amniotic fluid or neonatal karyotype or birth outcome. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. For the pilot validation set: 5/306 samples without follow‐up were excluded. No failed sample reported. No repeated test reported. |
||
| Comparative | |||
| Aim to study | To report the clinical application of gNIPT to detect chromosomal aneuploidies, especially T21, T18 and T13 in Chinese singleton pregnancies. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI‐Shenzhen made sequencing and analysis. Some authors are employees of BGI‐Shenzhen. | ||
| Informations about the authors contacted | Author was contacted on: 31 May 2016. No reply received from author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Yes | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Low | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
Zhou 2014b.
| Study characteristics | |||
| Patient sampling | Study design: blinded, prospective cohort study. Participants: pregnant women selected at high risk, low risk for T21 or without a priori risk. gNIPT was integrated in clinical workflow. Inclusion criteria: singleton pregnancies. Exclusion criteria: multifetal pregnancies. | ||
| Patient characteristics and setting | Number enrolled: 7705 pregnant women. Number available for 2 x 2 tables: 3950 pregnant women in the integration set (subgroup of 51%). Setting: 1 centre. Women’s Hospital, Zhejiang University School of Medicine, Hangzhouin, China. Recruitment period: September 2011 to July 2013. Ethnicity: Asian. Gestational age range: 12 to 24 weeks. Maternal age: not reported. Relevant tests carried out prior to index test: ultrasonography (nuchal translucency measurement) and biochemical screening for a part of this cohort. Language of the study: English. |
||
| Index tests | gNIPT by MPSS on Illumina Genome Analyzer IIx or HiSeq 2000 sequencer in 12‐plex. Fetal fraction DNA: amount measured but not reported. Blood samples for gNIPT were collected before reference standard. Cutpoint: positive if T score > 2.5 and L score > 1 (warning zone if t score > 2.5 or L score > 1). Commercial test: NIFTY™ prenatal test by BGI‐Shenzhen. |
||
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of amniotic fluid or neonatal karyotype or birth outcome. | ||
| Flow and timing | Blood samples were obtained prior to the invasive procedure (reference standard). gNIPT was a first‐ or second‐tier test. 141/7705 samples failed the initial MPSS testing. 141/141 samples were repeated with a new sampling and 137/141 samples obtained a gNIPT results. 4/7705 samples failed the second MPSS testing for low fetal fraction DNA (no gNIPT result). 3751/7701 samples without birth outcome were excluded (no reference standard). |
||
| Comparative | |||
| Aim to study | To report the clinical application of gNIPT to detect chromosomal aneuploidies, especially T21, T18 and T13 in Chinese singleton pregnancies. | ||
| Funding source or sponsor of the study | Study not funded by industry but BGI‐Shenzhen made sequencing and analysis. Some authors are employees of BGI‐Shenzhen. | ||
| Informations about the authors contacted | Author was contacted on: 31 May 2016. No reply received from the author. | ||
| Notes | |||
| Methodological quality | |||
| Item | Authors' judgement | Risk of bias | Applicability concerns |
| DOMAIN 1: Patient Selection | |||
| Was a consecutive or random sample of patients enrolled? | No | ||
| Was a case‐control design avoided? | Yes | ||
| Did the study avoid inappropriate exclusions? | No | ||
| High | High | ||
| DOMAIN 2: Index Test MPSS | |||
| Were the index test results interpreted without knowledge of the results of the reference standard? | Unclear | ||
| If a threshold was used, was it pre‐specified? | Yes | ||
| Unclear | Low | ||
| DOMAIN 3: Reference Standard | |||
| Is the reference standards likely to correctly classify the target condition? | Yes | ||
| Were the reference standard results interpreted without knowledge of the results of the index tests? | Unclear | ||
| Unclear | Low | ||
| DOMAIN 4: Flow and Timing | |||
| Was there an appropriate interval between index test and reference standard? | Yes | ||
| Did all analysed patients receive the reference standard? | Yes | ||
| Were all patients included in the analysis? | No | ||
| High | |||
CVS: chorionic villi sampling DANSR: digital analysis of selected regions FISH: fluorescence in situ hybridisation gNIPT: genomics‐based non‐invasive prenatal testing MAD: Median absolute deviation MPSS: massively parallel shotgun sequencing NCV: normalised chromosome value SD: standard deviation SNP: single nucleotide polymorphism TMPS: targeted massively parallel sequencing
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Anderson 2015 | Not a diagnostic test accuracy study. Poster abstract. |
| Anselem 2016 | Decision making study. Observational study. Not a diagnostic test accuracy study. |
| Bayindir 2015 | Samples overlap with Brady 2016. Most gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. |
| Beamon 2013 | Poster abstract of the 33rd Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Observational study with incomplete follow‐up. Samples overlap with Beamon 2014. |
| Beamon 2014 | Observational study with incomplete follow‐up. Not a diagnostic test accuracy study. Some gNIPT results unconfirmed by a reference standard test. |
| Belloin 2016 | Most women (95%) completed a questionnaire to report their birth outcome (inappropriate reference standard for this review). |
| Benachi 2015b | Tribune. Not a diagnostic test accuracy study. |
| Benachi 2016 | All samples overlap with Benachi 2015. |
| Benn 2015 | Letter to the editor on Zhang 2015 without data. Not a diagnostic test accuracy study. |
| Bhatt 2014 | Poster abstract of the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014. Patients with gNIPT negative result were without follow‐up (no reference standard). Incomplete 2 x 2 tables. |
| Bianchi 2012a | Samples overlap with Bianchi 2012. Data excluded to avoid double counting. |
| Bianchi 2014b | Editorial. Not a diagnostic test accuracy study. |
| Bianchi 2014c | Data excluded to avoid double counting. Samples overlap with Bianchi 2015b. |
| Bianchi 2015a | Not a diagnostic test accuracy study. Author presented some false positive cases in women with malignancy. |
| Bianchi 2015b | Incomplete 2 x 2 table. In this observational study, most women (98.9%) had no follow‐up (no reference standard). |
| Bianchi 2015c | Not a diagnostic test accuracy study. Author presented some false positive cases in women with malignancy. Samples overlap with Bianchi 2015a. |
| Bianchi 2015d | Poster abstract. Not a diagnostic test accuracy study. Author presented some false positive cases in women with malignancy. Samples overlap with Bianchi 2015a. |
| BlueCross BlueShield Asssociation 2014 | Technology Evaluation Center Assessment. Review. |
| Brady 2016 | Review with new data but most gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. |
| Chen 2013 | Poster abstract of the ISPD 17th International Conference on Prenatal Diagnosis and Therapy. Samples overlap with Huang 2014. |
| Chen 2014 | Poster abstract. Samples overlap with Yeang 2014. |
| Cherry 2014 | Poster abstract. Samples overlap with Meck 2015. |
| Cheung 2015 | Incomplete 2 x 2 table. This letter presented women who had positive results after screening and were referred for invasive procedure to confirm the presence of fetal aneuploidy. Only, the true positive and false positive gNIPT results were reported. |
| Chiu 2008 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Chiu 2010 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Christina 2012 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Cinnioglu 2012 | Poster abstract. Samples overlap with Rabinowitz 2012a. |
| Cirigliano 2013 | Full poster from the 17th International Conference on Prenatal Diagnosis and Therapy, ISPD 2013 received. Samples overlap with Cirigliano 2014 and Ordoñez 2015. Insufficient information to derive 2 x 2 tables. |
| Cirigliano 2014 | Full poster from the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014 received. Samples overlap with Cirigliano 2013 and Ordoñez 2015. Insufficient information to derive 2 x 2 tables. |
| Cuckle 2015 | Review with simulation model for gNIPT. |
| Curnow 2014 | Poster abstract of the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014. Samples overlap with Dar 2014. |
| Dan 2012 | Incomplete 2 x 2 table. Women with gNIPT negative results completed a questionnaire to report their birth outcome (inappropriate reference standard for this review). |
| Dar 2014 | Implementation study. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). Some women had follow‐up by telephone (inappropriate reference standard for this review). |
| De Ligt 2013 | Case report (deletion). |
| Denona 2016 | Poster abstract. Retrospective observational study. Insufficient information to derive 2 x 2 tables. |
| Discenza 2015 | Poster abstract. Some gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. |
| Dobson 2015 | Poster abstract. Insufficient information to derive 2 x 2 tables (gNIPT positive results only). Decision making. Samples overlap with Dobson 2016. |
| Dobson 2016 | Insufficient information to derive 2 x 2 tables (gNIPT positive results only). Decision making. |
| Dong 2016 | Sequencing not based on maternal plasma ccfDNA. |
| Duenwald 2016 | Method development. Analytical accuracy. Not a diagnostic test accuracy study. |
| Ehrich 2011a | Editorial comment without new data. |
| Eiben 2014 | Review. Not a diagnostic test accuracy study. |
| Ellison 2015 | Poster abstract. All gNIPT results (TMPS) were confirmed with a previous gNIPT result (MPSS) (inappropriate reference standard for this review). Insufficient information to derive 2 x 2 tables. |
| Faas 2011 | Poster abstract of the 8th European Cytogenetics Conference. Samples overlap with Faas 2012. |
| Faas 2012 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Fairbrother 2013a | Observational study. Incomplete 2 x 2 data. Most patients were without follow‐up (no reference standard). |
| Fairbrother 2013b | Conference abstract of the 17th International Conference on Prenatal Diagnosis and Therapy, ISPD 2013. Samples overlap with Fairbrother 2013a. |
| Fan 2008 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Fang 2015 | Insufficient information to derive 2 x 2 tables. |
| Ferres 2013 | Not a diagnostic test accuracy study (implementation study). |
| Fiorentino 2015 | Poster abstract. All samples overlap with Fiorentino 2016. |
| Fosler 2015 | Poster abstract of the 35th Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Observational study. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). |
| Futch 2013 | Observational study with incomplete follow‐up. Incomplete 2 x 2 table. Many gNIPT results unconfirmed by a reference standard test. |
| Gabriel 2014 | Conference abstract. Proof‐of‐concept. |
| Galea 2014 | Full poster from the 18th International Conference on Prenatal Diagnosis and Therapy, ISPD 2014. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). |
| Gao 2014 | News, comment on Liao 2014 and Yuan 2013 without new data. |
| Gao 2015 | Poster abstract. Insufficient information to derive 2 x 2 tables. |
| Geifman‐Holtzman 2013 | Poster abstract of the 33rd Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Samples overlap with Xiong 2015. |
| Geifman‐Holtzman 2014 | Poster abstract of the 34th Annual Meeting of the Society for Maternal‐Fetal Medicine: The Pregnancy Meeting. Samples overlap with Xiong 2015. |
| Gerundino 2017 | Women were asked to complete a questionnaire to report their birth outcome (inappropriate reference standard for this review). Insufficient information to derive 2 x 2 tables. |
| Gil 2013 | Most patients with gNITP negative result were without follow‐up (no reference standard) because 962 women had not yet delivered at the time of writing the publication. Insufficient information to derive 2 x 2 tables. Some patients overlap with del Mar Gil 2014. |
| Gil 2015 | Decision making including gNIPT accuracy data. All samples overlap with Gil 2016. |
| Gnetetskaya 2015 | Poster abstract. Samples overlap with Kurtser 2015. |
| Grati 2014 | Not a diagnostic test accuracy study. No sequencing data. |
| Gray 2013 | Observational study. Not a diagnostic test accuracy study. Full poster received from the authors. Poster of the 17th International Conference on Prenatal Diagnosis and Therapy, ISPD 2013. |
| Gromminger 2014 | Data excluded to avoid double counting. Blinded DNA sequencing libraries were provided by Sequenom from their clinical trial cohort (NCT00877292) and were resequenced by LifeCodexx. |
| Guex 2013 | Research letter. Samples overlap with Pescia 2017. |
| Halks‐Miller 2015 | In reply to Bianchi 2015a. Not a diagnostic test accuracy study. |
| Harasim 2016 | Poster abstract. Insufficient information to derive 2 x 2 tables. |
| Hernandez‐Gomez 2015 | Implementation study. Not a diagnostic test accuracy study. |
| Hofmann 2013 | Poster abstract. Samples overlap with Stumm 2014. |
| Hofmann 2014 | Conference abstract. Insufficient information to derive 2 x 2 tables. |
| Hofmann 2015 | Method development. Data were reanalysed by a new algorithmic approach of PraenaTest®. Not a diagnostic test accuracy study. |
| Hu 2014 | Not a next generation sequencing publication. NIPT was ultrasound and serum biomarkers. |
| Hu 2015 | Incomplete 2 x 2 tables. Only gNIPT positive results presented. |
| Hui 2015a | Poster abstract. All samples overlap with Hui 2015b. |
| Hui 2015b | Implementation study. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without follow‐up (no reference standard). |
| Jackson 2013 | Poster abstract. All samples overlap with Jackson 2014. |
| Jensen 2013 | Proof‐of‐concept. Not a diagnostic test accuracy study. Samples overlap with Palomaki 2012. |
| Jensen 2015 | Proof‐of‐concept study with unblinded samples. Not a diagnostic test accuracy study. |
| Jin 2014 | Incomplete 2 x 2 table. Women with gNIPT negative results were followed‐up by telephone (inappropriate reference standard for this review). |
| Johnson 2013 | Not a next generation sequencing method. |
| Juneau 2014 | Method development. Incomplete 2 x 2 table. Most patients were without follow‐up (no reference standard). |
| Kagan 2015 | Not a diagnostic test accuracy study. Simulation model. |
| Kalantar 2014 | Not next generation sequencing method. |
| Karlsson 2015 | Methodological publication. Not a diagnostic test accuracy study. |
| Kershberg 2015 | Poster abstract. Some gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. |
| Kinde 2012 | Methodological publication. Not a diagnostic test accuracy study. |
| Korabecna 2012 | Bioinformatic simulation with Palomaki 2011 data. |
| Koumbaris 2016 | Method development (proof‐of‐concept study). Development of an advanced fetal fraction estimation method and aneuploidy determination algorithm. Not a diagnostic test accuracy study. |
| Kurtser 2015 | Most patients with gNIPT negative result were without follow‐up (no reference standard). Incomplete 2 x 2 tables. |
| Lambert‐Messerlian 2014 | Samples overlap with Palomaki 2011 and Palomaki 2012. Data excluded to avoid double counting. |
| Larion 2015 | Poster abstract. Implementation study. |
| Lau 2012a | Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone or by email (inappropriate reference standard for this review). All samples overlap with Lau 2014. |
| Lau 2013 | Sample overlap with Lau 2014. |
| Lau 2014 | Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone or by email (inappropriate reference standard for this review). |
| Lebo 2015 | Incomplete 2 x 2 table. |
| Leung 2013 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Levandoski 2015 | Poster abstract. Observational study about discordant gNIPT results. Insufficient information to derive 2 x 2 tables. |
| Levy 2013 | Poster abstract. Incomplete 2 x 2 table. |
| Levy 2013a | Poster abstract. Incomplete 2 x 2 table. |
| Levy 2013b | Proof‐of‐concept. Not a diagnostic test accuracy study. Samples overlap with Zimmermann 2013. |
| Li 2012 | Methodological publication about relation between fetal fraction and multiple clinical factors. |
| Li 2015 | Observational study. Unavailable information about gNIPT approach used. It is unclear if patients with gNIPT negative result were followed up (no reference standard). |
| Liao 2011 | Not a diagnostic test accuracy study. No aneuploid case. |
| Liao 2012 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Liao 2013 | This is a poster abstract. The full publication was also excluded. See Liao 2014 for reasons of exclusion. |
| Liao 2014 | Incomplete 2 x 2 table for the retrospective and the prospective cohort. In prospective cohort, most patients were without follow‐up (no reference standard). For the retrospective cohort, number of gNIPT results was not reported. Sensitivity and specificity were presented for the retrospective cohort but 2 x 2 tables could not be derived. |
| Liao 2014a | Letter to the editor about Bianchi 2014b without new data. |
| Liu 2015 | Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone (inappropriate reference standard for this review). |
| Lo 2014 | Bioinformatic development. Comparison of sensitivity and specificity using 3 different count normalisation methods. |
| Lo 2014a | Poster abstract. Bioinformatic development. Comparison of sensitivity and specificity using 3 different count normalisation methods. Samples overlap with Lo 2014. |
| Loucký 2013 | Samples overlap with Palomaki 2012. Data excluded to avoid double counting. |
| Louis‐Jacques 2014 | Full poster. Observational study. Not a diagnostic test accuracy study. |
| Ma 2015 | Samples overlap with Ma 2016. |
| Ma 2015a | Poster abstract. All samples overlap with Ma 2016. |
| Manotaya 2016 | Insufficient information to derive 2 x 2 tables. Women without invasive testing results were encouraged to report birth outcomes through the insurance policy reimbursed (inappropriate reference standard for this review). |
| Marchili 2015 | Poster abstract. Implementation study. Not a diagnostic test accuracy study. Insufficient information to derive 2 x 2 tables. |
| Mayen 2015 | Observational study. Not a diagnostic test accuracy study. |
| Mazloom 2013a | Poster abstract. Samples overlap with Mazloom 2013. |
| McCullough 2014 | Incomplete 2 x 2 table. The clinician of women who passed gNIPT was encouraged to send ad hoc feedback to the lab (inappropriate reference standard for this review). |
| McCullough 2014a | Poster abstract. Incomplete 2 x 2 table. Samples overlap with McCullough 2014. |
| McCullough 2015 | Full poster of the 19th International Conference on Prenatal Diagnosis and Therapy, ISPD 2015. Most gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 × 2 tables. Some patients overlap with McCullough 2014. |
| McLennan 2016 | Most patients with gNIPT negative result were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. |
| Meck 2014 | Poster abstract. Samples overlap with Meck 2015. |
| Meck 2015 | Not a diagnostic test accuracy study. Observational study. |
| Meck 2015a | Poster abstract. Samples overlap with Meck 2015. |
| Mennuti 2015 | Review without original data. |
| Minarik 2015 | gNIPT negative results unconfirmed by a reference standard test. Not a diagnostic test accuracy study. |
| Miron 2011 | Not a diagnostic test accuracy study. This these explore traditional screening tests. |
| Mundy 2008 | Health Technology Assessment. Not a diagnostic test accuracy study. |
| Mundy 2009 | Health Technology Assessment. Not a diagnostic test accuracy study. |
| Musci 2014 | Poster abstract. Samples overlap with Norton 2015. |
| Musci 2014a | Poster abstract. Samples overlap with Hooks 2014 and Nicolaides 2014a. |
| NCT00770458 | Not a gNIPT method (other method). |
| NCT00877292 | Not with ccfDNA (other sampling). |
| NCT00891852 | Not a gNIPT method (other method). |
| NCT00971334 | Completed clinical trial but no published data. |
| NCT01052688 | Incomplete 2 x 2 data (ongoing study with cases only). |
| NCT01256606 | Not a gNIPT method (other method). |
| NCT01451671 | Incomplete 2 x 2 data (ongoing study with cases only). |
| NCT01451684 | Observational study on gNIPT without fetal karyotype. |
| NCT01555346 | Completed clinical trial but no published data. |
| NCT01574781 | Completed clinical trial but no published data. |
| NCT01597063 | Completed clinical trial but no published data. |
| NCT01661010 | Not a diagnostic test accuracy study. |
| NCT01663675 | Adult with T21. Not with pregnant women (other population). |
| NCT01668251 | Not a diagnostic test accuracy study. |
| NCT01725438 | Not with ccfDNA (other sampling). |
| NCT01837979 | Incomplete 2 x 2 data. |
| NCT01966991 | Completed clinical trial but no published data. |
| NCT02127515 | Not a diagnostic test accuracy study. Pregnant women with gNIPT have not a reference standard. |
| NCT02226315 | Inappropriate reference standard for this review (pregnancy outcome data obtained from the patient). |
| NCT02872948 | Not a gNIPT method (other method). |
| Neufeld‐Kaiser 2015 | Observational study. Not a diagnostic test accuracy study. Incomplete 2 x 2 tables. Most gNIPT results unconfirmed by a reference standard test. |
| Neveling 2015 | Method validation for the NextSeq 500 platform. Not a diagnostic test accuracy study. |
| Nickolich 2016 | Not a diagnostic test accuracy study. |
| Nicolaides 2013a | Poster abstract. All samples overlap with Nicolaides 2012. |
| Nicolaides 2014 | Simulation model on gNIPT implantation in first‐ or second‐tier test. |
| Nicolaides 2014b | Note on Nicolaides 2014a without new data. |
| Nicolaides 2014c | Target condition presented in this publication is not the focus of this review. Publication of next generation sequencing with ccfDNA for fetal triploidy. |
| Norem 2015 | Full poster received from authors. Most patients were without follow‐up (no reference standard). Incomplete 2 x 2 tables. |
| Norton 2014 | Bioinformatic simulation. |
| Norton 2014a | Poster abstract. Samples overlap with Norton 2015. |
| Norton 2015a | Bioinformatic simulation. |
| Norton 2015b | Editorial on Norton 2015 without new data. |
| Norton 2015c | Author reply to comments from Sentilhes 2015 and Smith‐Bindman 2015 about Norton 2015 without new data. |
| Norton 2016 | Simulation model to compare sequential and ccfDNA screening with data published in the literature. Not a diagnostic test accuracy study. |
| O'Leary 2014 | Bioinformatic simulation. |
| Oepkes 2015 | Most patients were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. |
| Oneda 2016 | Poster abstract. Insufficient information to derive 2 x 2 tables. |
| Ordoñez 2015 | Full poster received. Some gNIPT results unconfirmed by a reference standard test. Insufficient information to derive 2 x 2 tables. |
| Palomaki 2011 | Samples overlap with Palomaki 2012 (samples in Palomaki 2011 have been reanalysed in Palomaki 2012). Study excluded to avoid double counting. |
| Palomaki 2012a | Samples overlap with Palomaki 2012. Conference abstract about Palomaki 2012 data. |
| Palomaki 2012b | Editorial on Palomaki 2011 without new data. |
| Palomaki 2015 | Not a diagnostic test accuracy study. |
| Palomaki 2015a | Note about Palomaki 2015. Not a diagnostic test accuracy study. |
| Perez‐Pedregosa 2015 | Incomplete 2 x 2 tables. Some women with gNIPT negative results were followed up by telephone (inappropriate reference standard for this review). |
| Pescia 2017 | Follow‐up for gNIPT negative results was ensured by an inquiry of two sets of randomly selected samples (inappropriate reference standard for this review). |
| Petersen 2014 | Not a next generation sequencing publication. NIPT was ultrasound measurement and serum biomarkers. |
| Pettit 2014 | Most patients with gNIPT negative result were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. |
| Porreco 2014a | Reply to Grati 2014 without sequencing data. |
| Rabinowitz 2012 | Poster abstract. Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Rabinowitz 2012a | Poster abstract with incomplete 2 x 2 tables. |
| Rabinowitz 2012b | Poster abstract. Samples overlap with Rabinowitz 2012a. |
| Rabinowitz 2013 | Poster abstract. Samples overlap with Pergament 2014. |
| Rabinowitz 2014 | Poster abstract. Sample overlap with Pergament 2014. |
| Rad 2014 | Implementation study without sequencing data presented. |
| Radoi 2015 | Incomplete 2 x 2 tables. Most patients were without follow‐up (no reference standard). |
| Rava 2012 | Poster abstract. Samples overlap with Bianchi 2012. |
| Rava 2014 | Methodological publication about fetal DNA fraction with MELISSA samples. |
| Reiff 2015 | Insufficient information to derive 2 x 2 tables. |
| Reiff 2016 | Insufficient information to derive 2 x 2 tables. |
| Reimers 2015 | Conference abstract from the 19th International Conference on Prenatal Diagnosis and Theraphy, ISPD 2015. Simulation model. Not a diagnostic test accuracy study. |
| Revello 2016 | Not a diagnostic test accuracy study. All samples overlap with Gil 2016 and Quezada 2015. |
| Ryan 2016 | Method development of version 2 to SNP‐based gNIPT. Not a diagnostic test accuracy study. |
| Sachse 2015 | Proof‐of‐concept of fetal fraction quantification by qPCR. |
| Samura 2015 | Most patients were without follow‐up (no reference standard). Insufficient information to derive 2 x 2 tables. Samples overlap with Sago 2015. |
| Sarno 2016 | Some women reported their birth outcome (inappropriate reference standard for this review). Information about false positive results were insufficient to derive all 2 x 2 tables. |
| Schöck 2015 | Poster abstract. Bioinformatics development with unblinded samples. |
| Sehnert 2013 | Poster abstract. Incomplete 2 x 2 table. |
| Sehnert 2014 | Poster abstract. Samples overlap with Bianchi 2014b. |
| Sentilhes 2015 | Comment about Norton 2015 without new data. |
| Seo 2015 | Women with gNIPT result were without follow‐up at birth. |
| Settler 2015 | Full poster received. Insufficient information to derive 2 x 2 tables. Some gNIPT results unconfirmed by a reference standard test. |
| Shani 2016 | Simulation model. Not a diagnostic test accuracy study. |
| Shaohua 2012 | Poster abstract. Full poster not received. Incomplete 2 x 2 table. |
| Sharma 2015 | Poster abstract about patient perceptions of gNIPT from the multi‐centered Canadian PEGASUS trial. gNIPT results compared with first trimester combined test (inappropriate reference standard for this review). |
| Shaw 2013 | Poster abstract. Samples overlap with Shaw 2014. |
| Shen 2016 | Method development. Not a diagnostic test accuracy study. |
| Shi 2015 | Incomplete 2 x 2 table. gNIPT negative result unconfirmed by a reference standard test. |
| Shulman 2014 | Poster abstract. Incomplete 2 x 2 table. Most patients with gNIPT negative result were without a reference standard test. |
| Sistermans 2015a | Letter to the editor on Bianchi 2015a without data. |
| Smith‐Bindman 2015 | Comment about Norton 2015 without new data. |
| Song 2012 | Poster abstract. Some samples overlap with Sparks 2012a. |
| Sparks 2012 | Method development (all unblinded samples). Incomplete 2 x 2 table. Most patients with gNIPT negative result were unconfirmed by a reference standard test. |
| Srinivasan 2013 | Poster abstract. Samples from MELISSA study (potentially overlap). |
| Stokowski 2015 | Not a next generation sequencing method. |
| Strah 2015 | Women were followed up by telephone interview to find out their birth outcome (inappropriate reference standard for this review). |
| Straver 2014 | Proof‐of‐concept. |
| Strom 2015 | Incomplete 2 x 2 table. Only women with gNIPT positive result were reported. |
| Stumm 2011 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Stumm 2012 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Stumm 2012a | Poster abstract. Samples overlap with Stumm 2014. |
| Stumm 2013 | Poster abstract. Samples overlap with Stumm 2014. |
| Stumm 2016 | Not a diagnostic test accuracy study. |
| Swanson 2012 | Publication about Bianchi 2012 without new data. |
| Syngelaki 2014 | Not a diagnostic test accuracy study. Simulation model. |
| Tan 2016 | Women with gNIPT negative results were followed up by telephone interview (inappropriate reference standard for this review). Insufficient information to derive 2 x 2 tables. |
| Taneja 2016 | Incomplete follow‐up. Incomplete 2 x 2 table. Many gNIPT results unconfirmed by a reference standard test. |
| Taneja 2017 | Most patients with gNIPT negative result were without reference standard test. Providers were encouraged to report discordant clinical outcomes. Insufficient information to derive 2 x 2 tables. |
| Tarrier 2015 | gNIPT results unconfirmed by a reference standard test. Their reference method is verifi® results (inappropriate reference standard for this review). |
| Taylor 2014 | Not a diagnostic test accuracy study. Observational study and decision making about gNIPT uptake in their center. |
| Togneri 2016 | Full poster received. Internal verification set and implantation in their centre. Not a diagnostic test accuracy study. |
| Tong 2016 | Not a next‐generation sequencing method with ccfDNA. |
| Valderramos 2016a | Poster abstract. Insufficient information to derive 2 x 2 tables. |
| Valderramos 2016b | Poster abstract. Samples overlap with Valderramos 2016c. |
| Valderramos 2016c | Insufficient information to derive 2 x 2 tables. Retrospective cohort of patients with gNIPT positive results. |
| van den Oever 2012a | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| van den Oever 2012b | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| van den Oever 2013 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Van Opstal 2016 | Simulation model. Not a diagnostic test accuracy study. |
| Verweij 2013a | Poster abstract. All samples overlap with Verweij 2013. |
| Wald 2015a | Not a diagnostic test accuracy study. Prenatal screening workflow proposed. |
| Wald 2015b | Not a diagnostic test accuracy study. Prenatal screening workflow proposed. |
| Wang 2012 | Incomplete 2 x 2 table. Women with gNIPT negative results were followed up by telephone (inappropriate reference standard for this review). |
| Wang 2015b | Not a diagnostic test accuracy study. |
| Wang 2015c | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Wang 2015d | Editorial on Wang 2015b without new data. |
| Wang 2015e | Not a diagnostic test accuracy study. |
| Xiong 2015 | Full poster received. Observational study and incomplete follow‐up. |
| Yankova 2015 | Simulation model for gNIPT implantation. Not a diagnostic test accuracy study. |
| Yaron 2015 | Commentary about gNIPT for microdeletion syndromes and rare autosomal trisomies. Not a diagnostic test accuracy study. |
| Yeang 2014 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Yu 2014 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Yuan 2013 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Zhang 2015 | Incomplete 2 x 2 table. Women with gNIPT negative results were followed‐up by telephone (inappropriate reference standard for this review). |
| Zhou 2013 | Poster abstract. Incomplete 2 x 2 table. |
| Zimmermann 2012 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Zimmermann 2013 | Proof‐of‐concept. Not a diagnostic test accuracy study. |
| Zwiefelhofer 2013 | Implementation assessment of 2 sequencing platforms for gNIPT in a routine clinical environment. Not a diagnostic test accuracy study. |
ccfDNA: circulating cell‐free DNA gNIPT: genomics‐based non‐invasive prenatal testing MPSS: massively parallel shotgun sequencing TMPS: targeted massively parallel sequencing
Characteristics of ongoing studies [author‐defined order]
Basaran 2015.
| Trial name or title | Publication's title: False positive and false negative results of cell free DNA testing. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. |
| Index and comparator tests | gNIPT by TMPS or MPSS by commercial company providing gNIPT in Turkey (Ariosa Diagnostics, Inc., BGI‐Shenzhen, Illumina, Inc, Natera, Inc. and Sequenom, Inc). Blood samples for gNIPT were collected before reference standard. |
| Starting date | Not reported. |
| Contact information | Dr Seher Basaran Department of Medical Genetics Istanbul University, Istanbul Medical Faculty TURKEY 90 (212) 4142000 basarabs@istanbul.edu.tr |
| Aim to study | To demonstrate the importance of confirmation of fetus genotype by invasive testing after gNIPT. |
| Funding source or sponsor of the study | The genetic centre is not affiliated with any commercial company providing gNIPT. |
| Information about the authors contacted | Author was contacted on: 12, 14 and 18 January 2016. Last reply received on: 19 January 2016. |
| Notes | At the time of this writing, the authors plan to publish a full publication soon. |
Buresch 2016.
| Trial name or title | Poster's title: Actual rates of recommended diagnostic testing after first‐trimester screening vs same‐day screening by cell free DNA. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Reference standard: not reported. |
| Index and comparator tests | MPS. |
| Starting date | January to June 2015. |
| Contact information | Susan Klugman Department of Obstetrics and Gynecology and Women's Health, Albert Einstein College of Medicine, Montefiore Medical Center 1695 Eastchester Road, Bronx, NY 10461, United States. sklugman@montefiore.org |
| Aim to study | To compare actual patient referrals for post‐screen diagnostic tests following first‐trimester screening vs same day ccfDNA. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 1 and 23 September 2016. Reply received on: 23 September 2016. |
| Notes | Authors are working on data at the time of writing and they plan to submit for publication. |
Chen 2011a.
| Trial name or title | Oral presentation's title: Noninvasive prenatal diagnosis of fetal aneuploidy by massively parallel sequencing of maternal plasma DNA. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and SCA. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by MPSS on Illumina GAIIx/HiSeq 2000 sequencer. Cutpoint: positive if t score < ‐4. Commercial test: BGI's test. |
| Starting date | Not reported. |
| Contact information | Fang Chen, Beijing Genomics Institute, Shenzhen, China |
| Aim to study | To assess gNIPT with ccfDNA performance on fetal aneuploidies. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | BGI was contacted on: 19 May 2016. No reply received from the author. |
| Notes | Cohort of 5268 pregnant women. They successfully identified 62 cases of T21, 40 cases of T18, 3 cases of T13, 13 cases of SCA. In a cohort of karyotyping cases, the sensitivity and specificity of the aneuploidy fetus detection was 100% and 100%, respectively. |
Da Fonseca 2015.
| Trial name or title | Abstract's title: Non‐Invasive prenatal testing for the most common aneuploidies (trisomies 21, 18, and 13) using a semiconductor‐sequencing platform: a French multicenter pilot study. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT on semiconductor sequencing platform (MPSS). Blood sample collection not reported. Cutpoint: not reported. |
| Starting date | Not reported. |
| Contact information | J.P. Da Fonseca, Inserm U1016 Plateforme Génomique, Paris, France. |
| Aim to study | To validate a common protocol and to evaluate the efficiency and reliability of gNIPT of the most common chromosomal aneuploidies using a semiconductor sequencing platform. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 19 January 2016 and 23 March 2016. Reply received on: 16 February 2016. |
| Notes | Conference Abstract of the 10th European Cytogenetics Conference of the European Cytogenetics Association, ECA 2015. Prospective study of 500 pregnant women at high risk of fetal aneuploidy who undergo fetal karyotyping. The NIPT results matched the fetal karyotyping results in all of the cases: all trisomies were detected. |
ISRCTN11174071.
| Trial name or title | Comparison of false positive rates in prenatal combined screening and cell free DNA screening for trisomy 21 (ReFaPo study). |
| Target condition and reference standard(s) | Target condition: T21. Reference standard: prenatal or postnatal karyotype. |
| Index and comparator tests | gNIPT. |
| Starting date | July 2016. |
| Contact information | Karl Oliver Kagan University of Tuebingen Department of Obstetrics and Gynaecology Calwerstrasse 7 Tuebingen 72076 Germany |
| Aim to study | To compare the false positive rate of cell‐free DNA and traditional screening methods in a randomised controlled trial in a cohort without prior risk of fetal aneuploidy. |
| Funding source or sponsor of the study | Study funded by CENATA GmbH who does the analysis. |
| Information about the authors contacted | No need for further contact. |
| Notes | Target number of participants: 1400. Recruitment end date: March 2017. Intention to publish date: October 2018. DOI 10.1186/ISRCTN11174071 |
Lin 2014.
| Trial name or title | Clinical implementation of noninvasive prenatal testing in twin pregnancies with assisted reproductive technique treatment. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype or clinical outcomes. |
| Index and comparator tests | gNIPT by MPSS. Commercial test: BGI Shenzhen’s prenatal test. |
| Starting date | Not reported. |
| Contact information | BGI‐Shenzhen Shenzhen, China |
| Aim to study | To assess the clinical implementation of MPS‐based NIPT in twin pregnancies with assisted reproductive technique treatment. |
| Funding source or sponsor of the study | Not reported but BGI‐Shenzhen made sequencing and analyses. |
| Information about the authors contacted | No need for further contact. |
| Notes | Some women were still pregnant at the time of writing this poster abstract. |
Mu 2014.
| Trial name or title | Maternal non‐invasive fetal DNA test used in prenatal diagnosis. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. Reference standard: fetal karyotype of amniotic fluid. |
| Index and comparator tests | gNIPT by MPSS. NIFTY™ prenatal test by BGI‐Shenzhen. |
| Starting date | In 2012. |
| Contact information | Mu Y. Beijing United Family Hospital Beijing, China. |
| Aim to study | Not reported. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 19 April and 19 May 2016. No reply received from the author. |
| Notes | Poster abstract. Some women were still pregnant at the time of writing this poster abstract. |
NCT01429389.
| Trial name or title | Specimen collection from pregnant women at increased risk for fetal aneuploidy. |
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT. |
| Starting date | May 2011. |
| Contact information | Sequenom, Inc. |
| Aim to study | To develop a prenatal aneuploidy test using ccfDNA from blood samples from pregnant women who have an increased risk indicator/s for fetal chromosomal aneuploidy detection (T21). |
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
NCT01472523.
| Trial name or title | A safer pre‐natal diagnosis using free DNA in maternal blood (IONA®). |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and other chromosomal abnormalities yet to be determined. Reference standards: prenatal karyotype and follow‐up for 1 year. |
| Index and comparator tests | gNIPT by TMPS (selective amplification of fetal DNA) by Premaitha Health. |
| Starting date | April 2007. |
| Contact information | Brenda Kelly National Health Service, United Kingdom |
| Aim to study | To validate a novel gNIPT method that could increase the titre of fetal DNA within a given sample. |
| Funding source or sponsor of the study | Study funded by Premaitha Health. |
| Information about the authors contacted | No need for further contact. |
| Notes |
NCT01545674.
| Trial name or title | Prenatal Non‐invasive Aneuploidy Test Utilizing SNPs trial (PreNATUS). |
| Target condition and reference standard(s) | Target conditions: aneuploidy in a fetus at chromosomes 13, 18, 21, X and Y. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by TMPS (SNP based technology by Natera, Inc.). |
| Starting date | January 2012. |
| Contact information | Ronald Wapner, MD, Columbia University |
| Aim to study | To assess the diagnostic capability of an informatics enhanced SNP based technology (Parental Support) to identify pregnant women who are carrying a fetus with an aneuploidy from free floating DNA in the maternal blood. |
| Funding source or sponsor of the study | Study funded by Natera, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
NCT01925742.
| Trial name or title | Study of the efficacy of new non‐invasive prenatal tests for screening for fetal trisomies using maternal blood (PEGASUS). |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: prenatal or neonatal karyotype or medical record from birth. |
| Index and comparator tests | gNIPT by Semiconductor MPSS (Ion Torrent Proton™) or optical‐based MPSS (Illumina) or by TMPS with Harmony™ prenatal test by Ariosa Diagnostics, Inc. |
| Starting date | August 2013. |
| Contact information | François Rousseau CHU de Québec Québec, Canada |
| Aim to study | To perform a pan‐Canadian large‐scale validation study comparing the relative effectiveness and clinical performances of 2 index gNIPT methods using fetal ccfDNA in maternal blood in Canadian clinical laboratories between themselves and with that of fetal karyotype for detecting fetal aneuploidy of chromosomes 13, 18 and 21 and to compare the accuracy of this new gNIPT method with traditional prenatal screening methods. |
| Funding source or sponsor of the study | Study funded by Centre Hospitalier Universitaire de Québec, Laval University, Genome Canada, Genome Quebec, Genome British Columbia and Canadian Institutes of Health Research (CIHR). |
| Information about the authors contacted | No need for further contact. |
| Notes | Recruitment of patients completed (near 5000 pregnant women enrolled). at the time of writing, they are sequencing 3600 pregnant women with the 2 gNIPT MPSS platforms. A subsample of about 2300 blood samples was analysed by Ariosa Diagnostics, Inc (TMPS). Estimated study completion date: June 2017. |
NCT02201862.
| Trial name or title | Non‐invasive Chromosomal Evaluation of Trisomy study (NICHE). |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by TMPS by Ariosa Diagnostics, Inc. |
| Starting date | April 2014. |
| Contact information | Romielle Aquino 408‐209‐9098 raquino@ariosadx.com Or Thomas Musci 408‐229‐7500 tmusci@ariosadx.com |
| Aim to study | To provide clinically annotated samples to support continued improvements in the Ariosa Diagnostics, Inc Test content, methodology, specimen processing and quality control. |
| Funding source or sponsor of the study | Study funded by Ariosa Diagnostics, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
NCT02278536.
| Trial name or title | Multiple gestation study. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and SCA. Reference standards: fetal karyotype (amniocentesis or CVS) or genetic testing from cheek swab or saliva from live‐born children. |
| Index and comparator tests | gNIPT by TMPS by Natera, Inc. |
| Starting date | March 2013. |
| Contact information | Brian Kirshon Houston Perinatal Associates Or Zach Demko Natera, Inc. |
| Aim to study | To demonstrate the accuracy of our new NATUS diagnostic method to determine the genetic health of the developing fetuses in a multiple gestation pregnancy from a maternal blood sample. |
| Funding source or sponsor of the study | Study funded by Natera, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
NCT02278874.
| Trial name or title | High risk multiple gestation study. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and SCA. Reference standards: fetal karyotype (amniocentesis or CVS) or genetic testing from cheek swab or saliva from live‐born children. |
| Index and comparator tests | gNIPT by TMPS by Natera, Inc. |
| Starting date | August 2014. |
| Contact information | Joanne Stone Mt. Sinai Hospital, New York |
| Aim to study | To demonstrate the accuracy of our proprietary algorithm method to determine the genetic health of the developing fetuses in a multiple gestation pregnancy from a maternal blood sample. |
| Funding source or sponsor of the study | Study funded by Natera, Inc., Mount Sinai Hospital New York, Montefiore Medical Center, Long Island Jewish Medical Center and Tufts Medical Center. |
| Information about the authors contacted | No need for further contact. |
| Notes |
NCT02317965.
| Trial name or title | Non‐invasive screening for fetal aneuploidy. |
| Target condition and reference standard(s) | Target conditions: T21 and T18. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by MPSS by Progenity, Inc. |
| Starting date | March 2015. |
| Contact information | Richard Porreco Obstetrix Medical Group of Colorado |
| Aim to study | To detect whole chromosome abnormalities on all chromosomes 13, 16, 18, 21, X and Y, in the fetus through analysis of ccfDNA and compound sample DNA in maternal blood. |
| Funding source or sponsor of the study | Study funded by Progenity, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
NCT02424474.
| Trial name or title | T21,18 and 13 screening by cell free fetal DNA in low risk patients (DEPOSA). |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by MPSS. |
| Starting date | June 2015. |
| Contact information | Alexandra Benachi Antoine Béclère Hospital |
| Aim to study | To evaluate the performance of gNIPT in a population of pregnant women with and without in vitro fertilisation (IVF) concomitantly to regular first‐trimester trisomy 21 (T21) screening using maternal age, nuchal fold measurement and serum screening. |
| Funding source or sponsor of the study | Study funded by Assistance Publique ‐ Hôpitaux de Paris. |
| Information about the authors contacted | No need for further contact. |
| Notes | Recruitment of patients completed (933 pregnant women enrolled). |
NCT02787486.
| Trial name or title | Expanded Noninvasive Genomic Medical Assessment: the Enigma study. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, microdeletion syndromes, sex chromosome abnormalities, infectious and other diseases, and blood group typing. Reference standard: fetal karyotype or medical records. |
| Index and comparator tests | gNIPT by MPSS provided by Progenity, Inc. |
| Starting date | October 2015. |
| Contact information | Paul Bien 760‐494‐1743 paul.bien@progenity.com |
| Aim to study | To evaluate the relative clinical sensitivity, specificity, and performance of the laboratory‐developed test as a screening test for fetal chromosomal aneuploidy, infectious and other diseases, and RhD genotyping in the general population of pregnant women. |
| Funding source or sponsor of the study | Study funded by Progenity, Inc. |
| Information about the authors contacted | No need for further contact. |
| Notes |
Sago 2015.
| Trial name or title | Nationwide demonstration project of next‐generation sequencing of cell‐free DNA in maternal plasma in Japan: 1‐year experience |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or tissues of the miscarriage or medical record from birth. |
| Index and comparator tests | gNIPT by MPSS. Blood samples were collected before invasive procedure. Commercial test: MaterniT21™ prenatal test from Sequenom, Inc. |
| Starting date | 15 November 2012. Recruitment period between April 2013 to March 2014. |
| Contact information | Haruhiko Sago National center for Child‐health and development Perinatal Center 2‐10‐1Ookura, Setagaya‐ku, Tokyo 03‐3416‐0181 sagou‐h@ncchd.go.jp |
| Aim to study | To evaluate the quality of the genetic counselling in Japan. Sago 2015 reported the 1‐year experience of a nationwide demonstration project to introduce gNIPT of fetal aneuploidy from maternal plasma and discuss how to implement this program in Japan. |
| Funding source or sponsor of the study | Study supported by the Grant of the National Center for Child Health and Development 24‐3, Japan. Sequenom, Inc made gNIPT. |
| Information about the authors contacted | Author was contacted on: 6 April and 14 June 2016. No reply received from the author. |
| Notes | Authors continue collecting follow‐up data in the study population. |
Sanchez‐Usabiaga 2015.
| Trial name or title | Clinical implementation of non‐invasive prenatal study for detecting aneuploidies by fetal DNA based on single nucleotide polymorphisms: 2 years in Mexico. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or medical record from birth. |
| Index and comparator tests | gNIPT by TPMS. Commercial test: Natera’s prenatal test. |
| Starting date | Recruitment period: March 2013 to February 2015. |
| Contact information | Dr. Rafael Sánchez Usabiaga rsanchez@medicafertil.com.mx |
| Aim to study | To describe our experience of 2 years integrating gNIPT by ccfDNA in its variant of single nucleotide polymorphism (SNPs) as a screening method for the detection of common aneuploidies, since 9 weeks of gestation. |
| Funding source or sponsor of the study | Not reported but Natera, Inc. made gNIPT sequencing and analyses. |
| Information about the authors contacted | No need for further contact. |
| Notes | There are 270 pregnant women included in this study. |
Sistermans 2015.
| Trial name or title | TRIDENT: or monitored NIPT implementation in the Netherlands. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype of chorionic villi or amniotic fluid is recommended in case of abnormal gNIPT test results. Neonatal clinical examination not mentioned. |
| Index and comparator tests | gNIPT by MPSS. |
| Starting date | 01 April 2014. |
| Contact information | Dr. Erik Sistermans. VU University Medical Center Dept. of Clinical and Human Genetics Van der Boechorststraat 7 1081 BT Amsterdam NETHERLANDS +31‐20‐020‐4448346 Email: e.sistermans@vumc.nl |
| Aim to study | To investigate and evaluate all relevant aspects of the introduction of NIPT in the Dutch prenatal screening program. |
| Funding source or sponsor of the study | The TRIDENT study was designed and proposed by the national multidisciplinary NIPT consortium. |
| Information about the authors contacted | Author have been contacted on: 9 December 2015 and 15 March 2016. Reply received on: 16 March 2016. |
| Notes | Conference abstract presented at the Annual conference of the European Society of Human Genetics at Glasgow, Scotland, UK. http://www.emgo.nl/research/quality‐of‐care/research‐projects/1451/trident‐study‐trial‐by‐dutch‐laboratories‐for‐evaluation‐of‐non‐invasive‐prenatal‐testing‐nipt/background/ The authors plan to publish a full publication soon. |
Torres 2015.
| Trial name or title | Genetic non invasive prenatal testing: A clinical and technical experience of 3000 cases with follow‐up. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Reference standard: fetal karyotype of amniotic fluid. |
| Index and comparator tests | gNIPT by MPSS. Commercial test: TrisoNIM® prenatal test by NIMGenetics Genomics. |
| Starting date | Not reported. |
| Contact information | Juan C Cigudosa NIMGenetics Genomics Madrid, Spain. |
| Aim to study | To show a NIPT protocol, called TrisoNIM®, which has been partially performed in our laboratory, based in massive parallel sequencing. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author were contacted on: 29 February, 22 March, 15 and 27 June 2016. Reply received on: 20 June 2016. |
| Notes | Full poster received from the authors. |
Van Wymersch 2015.
| Trial name or title | Introduction of noninvasive prenatal testing for fetal trisomies: preliminary results and consequences on invasive samplings. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13, 45,X, 47,XXY, 47,XYY and 47,XXX. Microdeletion syndromes can also be detected. Reference standards: fetal karyotype of chorionic villi or amniotic fluid or medical record from birth. |
| Index and comparator tests | gNIPT by MPSS. Blood samples were collected before invasive procedure. Cutpoint: not reported. Commercial test: NIFTY™ test (Bejing Genomics Institute, Hong‐Kong, China). |
| Starting date | December 2013. |
| Contact information | Dr Van Wymersch Didier, Service de Gynécologie Obstétrique, Centre Hospitalier de Luxembourg, 4 Rue Barblé, L‐1210 Luxembourg E‐mail: vanwymersch.didier@chl.lu |
| Aim to study | To analyse a year of gNIPT implantation in our institute and to analyse gNIPT implication in chromosomal abnormalities screening politic. |
| Funding source or sponsor of the study | No reported. Samples analysed at BGI. |
| Information about the authors contacted | Author was contacted on: 12 September 2016. Reply received on: 29 September 2016. |
| Notes | This publication showed the first 683 samples. At the time of writing, authors have a much larger population of 2132 pregnant women. No false negative results have been observed to date for all the pregnancies that have already come to term. |
Willems 2014.
| Trial name or title | The first 3000 Non‐Invasive Prenatal Tests (NIPT) with the Harmony test in Belgium and the Netherlands. |
| Target condition and reference standard(s) | Target conditions: T21, T18 and T13. Reference standard: fetal karyotype of chorionic villi or amniotic fluid. |
| Index and comparator tests | gNIPT by TMPS. Commercial test: Harmony™ prenatal test by Ariosa Diagnostics, Inc. |
| Starting date | Recruitment period: March 2013 to December 2013. |
| Contact information | Patrick Willems patrick.willems@genetic‐diagnostic.net |
| Aim to study | To report the results of the first 3000 consecutive gNIPT tests performed in pregnant women from Belgium and the Netherlands. |
| Funding source or sponsor of the study | Not reported. Ariosa Diagnostics, Inc made sequencing and analysis. |
| Information about the authors contacted | No need for further contact. |
| Notes |
Yu 2014a.
| Trial name or title | Maternal non‐invasive fetal DNA test used in prenatal diagnosis. |
| Target condition and reference standard(s) | Target conditions: T21, T18, T13 and 45,X. Reference standards: cytogenetic tests and postnatal follow‐up. |
| Index and comparator tests | gNIPT by MPSS by BGI‐Shenzhen. |
| Starting date | Patients recruited in 2012. |
| Contact information | Yu M or Fei S. Beijing United Family Hospital. |
| Aim to study | To determine gNIPT accuracy in Chinese population. |
| Funding source or sponsor of the study | Not reported. |
| Information about the authors contacted | Author was contacted on: 15 May 2016. BGI was contacted on: 19 May 2016. No reply received from the author or BGI. |
| Notes | Conference abstract. Some women were still pregnant at the time of writing their conference abstract. |
Zwiefelhofer 2014.
| Trial name or title | Prenatal detection of fetal aneuploidy on the Ion Torrent Proton™ platform. |
| Target condition and reference standard(s) | Target condition: T21. Reference standard: fetal karyotype. |
| Index and comparator tests | gNIPT by MPSS on the Proton™ platform. |
| Starting date | Not reported. |
| Contact information | Sequenom, Inc. |
| Aim to study | To examine the performance of a gNIPT for fetal aneuploidy on the Ion Torrent Proton™ platform. |
| Funding source or sponsor of the study | Study funded by Sequenom, Inc. |
| Information about the authors contacted | Author was contacted on: 19 April and 15 June 2016. Reply received on: 22 June 2016. |
| Notes | Full poster received from authors. This study includes 156 samples including 16 women carrying a T21 fetus. All patient samples were correctly identified according to their karyotype results. |
CVS: chorionic villi sampling gNIPT: genomics‐based non‐invasive prenatal testing MPSS: massively parallel shotgun sequencing TMPS: targeted massively parallel sequencing
Differences between protocol and review
In the “reference standards” section in the methods, the protocol did not specify that neonatal clinical examination or medical records from birth are not appropriate reference standards for SCA diagnosis. These aneuploidies (SCA) usually have a normal phenotype and can not be detected with neonatal clinical examination or medical records from birth.
In the “sensitivity analyses” section, of the protocol, the authors stated they intended to investigate “studies where not all pregnant women received neither reference standard (no karyotyping confirmation nor birth follow‐up); authors who have taken for granted that the baby is normal". We decided to remove this analysis because this goes against one of our criteria for considering studies for this review (pregnant women with MPSS or TMPS and a reference standard). These type of studies were excluded.
We decide to remove Google scholar from our electronic searches databases list. Google scholar found more than 100,000 publications about our topic but only the first 1000 are retrievable. This database is not reproducible and search fields did not allow us to specify the search strategy. All first most relevant publications were already found with other databases.
We changed OpenSIGLE to Opengrey because the first has been replaced by the second.
Although studies used different cutpoints, there was little or no variation in threshold and no requirement to estimate the correlation between sensitivity and specificity across studies in a meta‐analysis. Therefore, we did not estimate summary ROC curves using the HSROC model. As the cutpoints were regarded as qualitative, we estimated summary sensitivities and specificities using random‐effects and fixed‐effect logistic regression models, and simple pooling as appropriate. Further details are available in the statistical analysis and data synthesis section. We used the Stata software package for the analyses instead of SAS.
We made changes in the QUADAS‐2 tool. In domain 1, at third signalling question, we added "maternal cancer history, type of pregnancy, gestational age, assisted reproductive technology" in "No" answer. In concerns of applicability in domain 3, concerns about applicability, we modified conditions for low and high concern according to the review question. In domain 4, at second signalling question, we added "analysed" for clarify and we changed "yes" answer by removing "karyotype result" for "appropriate reference standard".
Contributions of authors
FR initiated this review project and recruited all authors. MB co‐ordinated the production of the review.
MB, CL, JB, YT and FR drafted the protocol and all authors critically commented on the protocol. The final protocol was read and approved by all authors.
MB, CL and WW developed and applied the literature search. MB, CL, JB and LN applied eligibility criteria, collected, filled the data form and assessed methodological quality of the included studies. MB contacted authors of the included studies. YT checked data collection, undertook statistical analysis and data synthesis. MB, CL and FR wrote the draft of the review. MB, CL, JB, LN, YT, SL, YG and FR contributed with redrafting. SL, FL, YG and FR provide senior clinical input. FR oversaw the review process. The final review was read and approved by all authors.
Sources of support
Internal sources
No sources of support supplied
External sources
-
Research Chair in Health Technology Assessment in laboratory medicine to FR, Canada.
From Ministère de la Santé et des Services Sociaux, Fonds de la Recherche en Santé du Québec, Centre Hospitalier Universitaire de Québec.
-
Canadian Institutes of Health Research (CIHR), Canada.
Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) (AFT, FR, FL, SL)
-
Canadian Institutes of Health Research (CIHR), Canada, Canada.
Canada Research Chair in Critical Care Neurology and Trauma (to AFT).
Declarations of interest
Mylene Badeau: none known.
Jonatan Blais: none known.
Yves Giguère (and some other members of the review team ‐ FR, SL, FL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors may eventually publish results from clinical trials that could be considered in this review. None of the four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest
Sylvie Langlois (and some other members of the review team ‐ FR, YG, FL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors (FR, SL, YG and FL) may eventually publish results from clinical trials that could be considered in this review. None of these four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest. This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
France Légaré (and some other members of the review team ‐ FR, YG, SL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors (FR, SL, YG and FL) may eventually publish results from clinical trials that could be considered in this review. None of these four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest. This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
Carmen Lindsay: none known.
Leon Nshimyumukiza: none known.
François Rousseau (and some other members of the review team ‐ SL, YG, FL) are investigators in a Research Project funded under the auspices of Genome Canada and the Canadian Institutes for Health Research (both non‐for‐profit organisations funded by the Canadian government) but that call for some mandatory in‐kind contributions from other partners. This Research Project thus receives in‐kind funding from private corporations which either offer commercial NIPT tests (Ariosa Diagnostics Inc, San Jose, CA) or offer reagents and/or equipment that can be used to perform NIPT assays (Life Technologies Inc, NY, USA; Illumina, San Diego, CA, USA; QIAGEN, Hilden, GER; Perkin Elmer, Waltham, MASS, USA). This funding is at arms length from the scientific components of the research project. The present review is not funded by this Project. Some of the authors (FR, SL, YG and FL) may eventually publish results from clinical trials that could be considered in this review. None of these four authors involved in gNIPT studies will take part in the selection of studies, nor in any decisions/analyses related to their own studies. All authors declared no other conflict of interest. This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
Yemisi Takwoingi: The University of Birmingham received consultancy fees from Universite Laval for statistical analysis.
Alexis F Turgeon: This review was supported by a Canadian Institutes of Health Research (CIHR) Knowledge Synthesis Grant: Fall 2014 Competition (2014‐11‐17) awarded to AFT, FR, FL, and SL. CIHR in no way influenced the results or conclusions of this review.
William Witteman: none known.
New
References
References to studies included in this review
Alberti 2015 {published and unpublished data}
- Alberti A, Salomon LJ, Lorc'h M, Couloux A, Bussieres L, Goupil S, et al. Non‐invasive prenatal testing for trisomy 21 based on analysis of cell‐free fetal DNA circulating in the maternal plasma. Prenatal Diagnosis 2015;35(5):471‐6. [PUBMED: 25643828] [DOI] [PubMed] [Google Scholar]
- NCT01118507. Trisomy of chromosome 21 diagnosis by high output sequencing. clinicaltrials.gov/show/NCT01118507 Date first received: 28 April 2010.
Ashoor 2012 {published data only}
- Ashoor G, Syngelaki A, Wagner M, Birdir C, Nicolaides KH. Chromosome‐selective sequencing of maternal plasma cell‐free DNA for first‐trimester detection of trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology 2012;206(4):322.e1‐5. [PUBMED: 22464073] [DOI] [PubMed] [Google Scholar]
Ashoor 2013 {published data only}
- Ashoor G, Syngelaki A, Wang E, Struble C, Oliphant A, Song K, et al. Trisomy 13 detection in the first trimester of pregnancy using a chromosome‐selective cell‐free DNA analysis method. Ultrasound in Obstetrics & Gynecology 2013;41(1):21‐5. [PUBMED: 22996646] [DOI] [PubMed] [Google Scholar]
Benachi 2015 {published data only}
- Benachi A, Letourneau A, Kleinfinger P, Senat MV, Gautier E, Favre R, et al. Cell‐free DNA analysis in maternal plasma in cases of fetal abnormalities detected on ultrasound examination. Obstetrics and Gynecology 2015;125(6):1330‐7. [PUBMED: 26000504] [DOI] [PubMed] [Google Scholar]
Bevilacqua 2015 {published data only}
- Bevilacqua E, Gil MM, Nicolaides KH, Ordonez E, Cirigliano V, Dierickx H, et al. Performance of screening for aneuploidies by cell‐free DNA analysis of maternal blood in twin pregnancies. Ultrasound in Obstetrics & Gynecology 2015;45(1):61‐6. [PUBMED: 25297464] [DOI] [PubMed] [Google Scholar]
Bianchi 2012 {published data only}
- Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP, et al. Genome‐wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstetrics and Gynecology 2012;119(5):890‐901. [PUBMED: 22362253] [DOI] [PubMed] [Google Scholar]
Bianchi 2013 {published and unpublished data}
- Bianchi DW, Prosen T, Platt LD, Goldberg JD, Abuhamad AZ, Rava RP, et al. Massively parallel sequencing of maternal plasma DNA in 113 cases of fetal nuchal cystichygroma. Obstetrics and Gynecology 2013;121(5):1057‐62. [DOI] [PubMed] [Google Scholar]
- NCT01122524. MatErnal bLood IS Source to accurately diagnose fetal Aneuploidy (MELISSA). clinicaltrials.gov/show/NCT01122524 Date first received: 11 May 2010.
Bianchi 2014a {published and unpublished data}
- Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, et al. DNA sequencing versus standard prenatal aneuploidy screening. New England Journal of Medicine 2014;370(9):799‐808. [PUBMED: 24571752] [DOI] [PubMed] [Google Scholar]
- NCT01663350. Comparison of aneuploidy risk evaluations. clinicaltrials.gov/show/NCT01663350 Date first received: 31 July 2012.
Bijok 2014 {published data only}
- Bijok J, Gorzelnik K, Massalska D, Ilnicka A, Pawlowska B, Zimowski JG, et al. Non‐invasive prenatal diagnosis of the most common aneuploidies with cell‐free fetal DNA in maternal serum‐‐preliminary results [Nieinwazyjna diagnostyka prenatalna najczestszych aneuploidii na podstawie plodowego DNA we krwi matki ‐‐doniesienie wstepne]. Ginekologia Polska 2014;85(3):208‐13. [PUBMED: 24783433] [DOI] [PubMed] [Google Scholar]
Canick 2012 {published data only}
- Canick JA, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Ehrich M, Boom D, et al. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenatal Diagnosis 2012;32(8):730‐4. [PUBMED: 22585317] [DOI] [PubMed] [Google Scholar]
Chen 2011 {published data only}
- Chen EZ, Chiu RW, Sun H, Akolekar R, Chan KC, Leung TY, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLOS One 2011;6(7):e21791. [PUBMED: 21755002] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chiu 2011 {published data only}
- Chiu RW, Akolekar R, Zheng YW, Leung TY, Sun H, Chan KC, et al. Non‐invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. BMJ (Clinical Research Ed.) 2011;342:c7401. [PUBMED: 21224326] [DOI] [PMC free article] [PubMed] [Google Scholar]
Comas 2015 {published data only}
- Comas C, Echevarria M, Rodriguez MA, Prats P, Rodriguez I, Serra B. Initial experience with non‐invasive prenatal testing of cell‐free DNA for major chromosomal anomalies in a clinical setting. Journal of Maternal‐fetal & Neonatal Medicine 2015;28(10):1196‐201. [PUBMED: 25048745] [DOI] [PubMed] [Google Scholar]
del Mar Gil 2014 {published data only}
- Mar Gil M, Quezada MS, Bregant B, Syngelaki A, Nicolaides KH. Cell‐free DNA analysis for trisomy risk assessment in first‐trimester twin pregnancies. Fetal Diagnosis and Therapy 2014;35(3):204‐11. [PUBMED: 24247435] [DOI] [PubMed] [Google Scholar]
Ehrich 2011 {published data only}
- Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. American Journal of Obstetrics and Gynecology 2011;204(3):205.e1‐11. [PUBMED: 21310373] [DOI] [PubMed] [Google Scholar]
Fiorentino 2016 {published data only}
- Fiorentino F, Bono S, Pizzuti F, Mariano M, Polverari A, Duca S, et al. The importance of determining the limit of detection of non‐invasive prenatal testing methods. Prenatal Diagnosis 2016;36(4):304‐11. [PUBMED: 26815144] [DOI] [PubMed] [Google Scholar]
Gil 2016 {published data only}
- Gil MM, Revello R, Poon LC, Akolekar R, Nicolaides KH. Clinical implementation of routine screening for fetal trisomies in the UK NHS: cell‐free DNA test contingent on results from first‐trimester combined test. Ultrasound in Obstetrics & Gynecology 2016;47(1):45‐52. [PUBMED: 26498918] [DOI] [PubMed] [Google Scholar]
Hall 2014 {published data only}
- Hall MP, Hill M, Zimmermann B, Sigurjonsson S, Westemeyer M, Saucier J, et al. Non‐invasive prenatal detection of trisomy 13 using a single nucleotide polymorphism‐ and informatics‐based approach. PLOS One 2014;9(5):e96677. [PUBMED: 24805989] [DOI] [PMC free article] [PubMed] [Google Scholar]
Hooks 2014 {published data only}
- Hooks J, Wolfberg AJ, Wang ET, Struble CA, Zahn J, Juneau K, et al. Non‐invasive risk assessment of fetal sex chromosome aneuploidy through directed analysis and incorporation of fetal fraction. Prenatal Diagnosis 2014;34(5):496‐9. [PUBMED: 24510887] [DOI] [PubMed] [Google Scholar]
Hou 2012 {published data only}
- Hou QF, Wu D, Chu Y, Kang B, Liao SX, Yang YL, et al. Clinical application of noninvasive prenatal diagnosis using cell free fetal DNA in maternal plasma. Zhonghua Fu Chan Ke za Zhi 2012;47(11):813‐7. [PUBMED: 23302120] [PubMed] [Google Scholar]
Huang 2014 {published data only}
- Huang X, Zheng J, Chen M, Zhao Y, Zhang C, Liu L, et al. Noninvasive prenatal testing of trisomies 21 and 18 by massively parallel sequencing of maternal plasma DNA in twin pregnancies. Prenatal Diagnosis 2014;34(4):335‐40. [PUBMED: 24357023] [DOI] [PubMed] [Google Scholar]
Jackson 2014 {published data only}
- Jackson J, Hamar B, Lazar E, Lim K, Rodriguez D, Stock K, et al. Nuchal translucency measurement plus non‐invasive prenatal testing to screen for aneuploidy in a community‐based average‐risk population. Ultrasound in Obstetrics & Gynecology 2014; Vol. 44, issue 4:491. [PUBMED: 24890031] [DOI] [PubMed]
Jeon 2014 {published data only}
- Jeon YJ, Zhou Y, Li Y, Guo Q, Chen J, Quan S, et al. The feasibility study of non‐invasive fetal trisomy 18 and 21 detection with semiconductor sequencing platform. PLOS One 2014;9(10):e110240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Jiang 2012 {published data only}
- Jiang F, Ren J, Chen F, Zhou Y, Xie J, Dan S, et al. Noninvasive Fetal Trisomy (NIFTY™) test: an advanced noninvasive prenatal diagnosis methodology for fetal autosomal and sex chromosomal aneuploidies. BMC Medical Genomics 2012;5:57. [PUBMED: 23198897] [DOI] [PMC free article] [PubMed] [Google Scholar]
Johansen 2016 {published data only}
- Johansen P, Richter SR, Balslev‐Harder M, Miltoft CB, Tabor A, Duno M, et al. Open source non‐invasive prenatal testing platform and its performance in a public health laboratory. Prenatal Diagnosis 2016;36(6):530‐6. [PUBMED: 27027563] [DOI] [PubMed] [Google Scholar]
Ke 2015 {published data only}
- Ke WL, Zhao WH, Wang XY. Detection of fetal cell‐free DNA in maternal plasma for Down syndrome, Edward syndrome and Patau syndrome of high risk fetus. International Journal of Clinical and Experimental Medicine 2015;8(6):9525‐30. [PUBMED: 26309618] [PMC free article] [PubMed] [Google Scholar]
Kim 2016 {published data only}
- Kim S, Jung H, Han SH, Lee S, Kwon J, Kim MG, et al. Comparison of two high‐throughput semiconductor chip sequencing platforms in noninvasive prenatal testing for Down syndrome in early pregnancy. BMC Medical Genomics 2016;9(1):22. [PUBMED: 27129388] [DOI] [PMC free article] [PubMed] [Google Scholar]
Korostelev 2014 {published data only}
- Korostelev S, Totchiev G, Kanivets I, Gnetetskaya V. Association of non‐invasive prenatal testing and chromosomal microarray analysis for prenatal diagnostics. Gynecological Endocrinology 2014;30:13‐6. [DOI] [PubMed] [Google Scholar]
Lau 2012 {published data only}
- Lau TK, Chen F, Pan X, Pooh RK, Jiang F, Li Y, et al. Noninvasive prenatal diagnosis of common fetal chromosomal aneuploidies by maternal plasma DNA sequencing. Journal of Maternal‐Fetal & Neonatal Medicine 2012;25(8):1370‐4. [PUBMED: 22070770] [DOI] [PubMed] [Google Scholar]
Lee 2015 {published data only}
- Lee MY, Cho DY, Won HS, Hwang AR, Jeong B, Kim J, et al. Performance of Momguard, a new non‐invasive prenatal testing protocol developed in Korea. Obstetrics & Gynecology Science 2015;58(5):340‐5. [PUBMED: 26430657] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lefkowitz 2016 {published data only}
- Lefkowitz RB, Tynan JA, Liu T, Wu Y, Mazloom AR, Almasri E, et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. American Journal of Obstetrics and Gynecology 2016;215(2):227.e1‐227.e16. [PUBMED: 26899906] [DOI] [PubMed] [Google Scholar]
Liang 2013 {published data only}
- Liang D, Lv W, Wang H, Xu L, Liu J, Li H, et al. Non‐invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing. Prenatal Diagnosis 2013;33(5):409‐15. [PUBMED: 23299662] [DOI] [PubMed] [Google Scholar]
Liu 2012 {published data only}
- Liu HY, Wu D, Li H, Guo SK, Zhang CY, Liao SX, et al. Significance of detecting free DNA from maternal plasma for the diagnosis of fetal chromosomal aneuploidies. Zhonghua yi xue yi chuan xue za zhi = Zhonghua yixue yichuanxue zazhi [Chinese journal of medical genetics] 2012;29(4):435‐8. [PUBMED: 22875501] [DOI] [PubMed] [Google Scholar]
Ma 2016 {published data only}
- Ma J, Wang Y, Wang W, Dong Y, Xu C, Zhou A, et al. Validation of combinatorial probe‐anchor ligation (cPAL) based sequencing method for non‐invasive prenatal testing in trisomy detection by a central laboratory. Ultrasound in Obstetrics & Gynecology 2016;50(1):49‐57. [PUBMED: 27363706] [DOI] [PubMed] [Google Scholar]
Mazloom 2013 {published data only}
- Mazloom AR, Dzakula Z, Oeth P, Wang H, Jensen T, Tynan J, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell‐free DNA from maternal plasma. Prenatal Diagnosis 2013;33(6):591‐7. [PUBMED: 23592550] [DOI] [PubMed] [Google Scholar]
Nicolaides 2012 {published data only}
- Nicolaides KH, Syngelaki A, Ashoor G, Birdir C, Touzet G. Noninvasive prenatal testing for fetal trisomies in a routinely screened first‐trimester population. American Journal of Obstetrics and Gynecology 2012;207(5):374.e1‐6. [PUBMED: 23107079] [DOI] [PubMed] [Google Scholar]
Nicolaides 2013 {published data only}
- Nicolaides KH, Syngelaki A, Gil M, Atanasova V, Markova D. Validation of targeted sequencing of single‐nucleotide polymorphisms for non‐invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y. Prenatal Diagnosis 2013;33(6):575‐9. [DOI] [PubMed] [Google Scholar]
Nicolaides 2014a {published data only}
- Nicolaides KH, Musci TJ, Struble CA, Syngelaki A, Gil MM. Assessment of fetal sex chromosome aneuploidy using directed cell‐free DNA analysis. Fetal Diagnosis and Therapy 2014;35(1):1‐6. [PUBMED: 24335155] [DOI] [PubMed] [Google Scholar]
Norton 2012 {published data only}
- Norton ME, Brar H, Weiss J, Karimi A, Laurent LC, Caughey AB, et al. Non‐Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology 2012;207(2):137.e1‐8. [PUBMED: 22742782] [DOI] [PubMed] [Google Scholar]
Norton 2015 {published and unpublished data}
- NCT01511458. Non‐invasive chromosomal examination of Trisomy Study. clinicaltrials.gov/show/NCT01511458 Date first received: 13 January 2012.
- Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, et al. Cell‐free DNA analysis for noninvasive examination of trisomy. New England Journal of Medicine 2015;372(17):1589‐97. [PUBMED: 25830321] [DOI] [PubMed] [Google Scholar]
Palomaki 2012 {published data only}
- Palomaki GE, Deciu C, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genetics in Medicine 2012;14(3):296‐305. [PUBMED: 22281937] [DOI] [PMC free article] [PubMed] [Google Scholar]
Papageorghiou 2016a {published data only}
- Papageorghiou AT, Khalil A, Forman M, Hulme R, Mazey R, Mousa HA, et al. Clinical evaluation of the IONA® test: a non‐invasive prenatal screening test for trisomies 21, 18 and 13. Ultrasound in Obstetrics & Gynecology 2016;47(2):188‐93. [PUBMED: 26493543] [DOI] [PMC free article] [PubMed] [Google Scholar]
Papageorghiou 2016b {published data only}
- Papageorghiou AT, Khalil A, Forman M, Hulme R, Mazey R, Mousa HA, et al. Clinical evaluation of the IONA® test: a non‐invasive prenatal screening test for trisomies 21, 18 and 13. Ultrasound in Obstetrics & Gynecology 2016;47(2):188‐93. [PUBMED: 26493543] [DOI] [PMC free article] [PubMed] [Google Scholar]
Pergament 2014 {published data only}
- Pergament E, Cuckle H, Zimmermann B, Banjevic M, Sigurjonsson S, Ryan A, et al. Single‐nucleotide polymorphism‐based noninvasive prenatal screening in a high‐risk and low‐risk cohort. Obstetrics and Gynecology 2014;124(2 Pt 1):210‐8. [PUBMED: 25004354] [DOI] [PMC free article] [PubMed] [Google Scholar]
Persico 2016 {published data only}
- Persico N, Boito S, Ischia B, Cordisco A, Robertis V, Fabietti I, et al. Cell‐free DNA testing in the maternal blood in high‐risk pregnancies after first‐trimester combined screening. Prenatal Diagnosis 2016;36(3):232‐6. [PUBMED: 26749576] [DOI] [PubMed] [Google Scholar]
Poon 2016 {published data only}
- Poon LC, Dumidrascu‐Diris D, Francisco C, Fantasia I, Nicolaides KH. IONA® test for first‐trimester detection of trisomies 21, 18 and 13. Ultrasound in Obstetrics & Gynecology 2016;47(2):184‐7. [PUBMED: 26387684] [DOI] [PMC free article] [PubMed] [Google Scholar]
Porreco 2014 {published and unpublished data}
- NCT00847990. Non‐Invasive screening for fetal aneuploidy. clinicaltrials.gov/show/NCT00847990 Date first received: 18 February 2009.
- Porreco RP, Garite TJ, Maurel K, Marusiak B, Ehrich M, Boom D, et al. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. American Journal of Obstetrics and Gynecology 2014;211(4):365.e1‐12. [PUBMED: 24657131] [DOI] [PubMed] [Google Scholar]
Quezada 2015 {published data only}
- Quezada MS, Gil MM, Francisco C, Orosz G, Nicolaides KH. Screening for trisomies 21, 18 and 13 by cell‐free DNA analysis of maternal blood at 10‐11 weeks' gestation and the combined test at 11‐13 weeks. Ultrasound in Obstetrics & Ggynecology 2015;45(1):36‐41. [PUBMED: 25251385] [DOI] [PubMed] [Google Scholar]
Samango‐Sprouse 2013 {published data only}
- Samango‐Sprouse C, Banjevic M, Ryan A, Sigurjonsson S, Zimmermann B, Hill M, et al. SNP‐based non‐invasive prenatal testing detects sex chromosome aneuploidies with high accuracy. Prenatal Diagnosis 2013;33(7):643‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sehnert 2011 {published data only}
- Sehnert AJ, Rhees B, Comstock D, Feo E, Heilek G, Burke J, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell‐free fetal DNA from maternal blood. Clinical Chemistry 2011;57(7):1042‐9. [PUBMED: 21519036] [DOI] [PubMed] [Google Scholar]
Shaw 2014 {published data only}
- Shaw SW, Hsiao CH, Chen CY, Ren Y, Tian F, Tsai C, et al. Noninvasive prenatal testing for whole fetal chromosomal aneuploidies: a multicenter prospective cohort trial in Taiwan. Fetal Diagnosis and Therapy 2014;35(1):13‐7. [PUBMED: 24296685] [DOI] [PubMed] [Google Scholar]
Song 2013 {published data only}
- Song Y, Liu C, Qi H, Zhang Y, Bian X, Liu J. Noninvasive prenatal testing of fetal aneuploidies by massively parallel sequencing in a prospective Chinese population. Prenatal Diagnosis 2013;33(7):700‐6. [DOI] [PubMed] [Google Scholar]
Song 2015 {published data only}
- Song Y, Huang S, Zhou X, Jiang Y, Qi Q, Bian X, et al. Non‐invasive prenatal testing for fetal aneuploidies in the first trimester of pregnancy. Ultrasound in Obstetrics & Gynecology 2015;45(1):55‐60. [DOI] [PubMed] [Google Scholar]
Sparks 2012a {published data only}
- Sparks AB, Struble CA, Wang ET, Song K, Oliphant A. Noninvasive prenatal detection and selective analysis of cell‐free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. American Journal of Obstetrics and Gynecology 2012;206(4):319.e1‐9. [PUBMED: 22464072] [DOI] [PubMed] [Google Scholar]
Stumm 2014 {published data only}
- Stumm M, Entezami M, Haug K, Blank C, Wustemann M, Schulze B, et al. Diagnostic accuracy of random massively parallel sequencing for non‐invasive prenatal detection of common autosomal aneuploidies: a collaborative study in Europe. Prenatal Diagnosis 2014;34(2):185‐91. [PUBMED: 24222400] [DOI] [PubMed] [Google Scholar]
Sukhikh 2015 {published data only}
- Sukhikh GT, Karetnikova NA, Shubina ES, Baranova EE, Korostin DO, Ekimov AN, et al. Noninvasive prenatal diagnosis of aneuploidies by next‐generation sequencing (NGS) in a group of high‐risk women. Akusherstvo i Ginekologiia 2015;4:5‐10. [Google Scholar]
Sung‐Hee 2015 {published data only}
- Sung‐Hee H, Young‐Ho Y, Jae‐Song R, Myung‐Soo K, Young‐Jin K, Kyoung‐Ryul L. Noninvasive prenatal test for fetal chromosomal aneuploidies by massively parallel sequencing of cell‐free fetal DNA in maternal plasma: The first clinical experience in Korea. Journal of Genetic Medicine 2015;12(2):85‐91. [Google Scholar]
Tynan 2016 {published data only}
- Tynan JA, Kim SK, Mazloom AR, Zhao C, McLennan G, Tim R, et al. Application of risk score analysis to low‐coverage whole genome sequencing data for the noninvasive detection of trisomy 21, trisomy 18, and trisomy 13. Prenatal Diagnosis 2016;36(1):56‐62. [PUBMED: 26505614] [DOI] [PubMed] [Google Scholar]
Verweij 2013 {published data only}
- Verweij EJ, Jacobsson B, Scheltema PA, Boer MA, Hoffer MJ, Hollemon D, et al. European non‐invasive trisomy evaluation (EU‐NITE) study: a multicenter prospective cohort study for non‐invasive fetal trisomy 21 testing. Prenatal Diagnosis 2013;33(10):996‐1001. [PUBMED: 23794121] [DOI] [PubMed] [Google Scholar]
Wang 2014 {published data only}
- Wang S, Gao Z, Lu Y, Li Y, Jiang S, Wang L, et al. Detection of fetal chromosomal aneuploidy in pregnant women at advanced maternal age during the first trimester. Nan Fang Yi Ke da Xue Xue Bao [Journal of Southern Medical University] 2014;34(5):655‐8. [PUBMED: 24849430] [PubMed] [Google Scholar]
Wang 2015a {published data only}
- Wang L, Meng Q, Tang X, Yin T, Zhang J, Yang S, et al. Maternal mosaicism of sex chromosome causes discordant sex chromosomal aneuploidies associated with noninvasive prenatal testing. Taiwanese Journal of Obstetrics & Gynecology 2015;54(5):527‐31. [PUBMED: 26522104] [DOI] [PubMed] [Google Scholar]
Yao 2014 {published data only}
- Yao H, Jiang F, Hu H, Gao Y, Zhu Z, Zhang H, et al. Detection of fetal sex chromosome aneuploidy by massively parallel sequencing of maternal plasma DNA: initial experience in a Chinese hospital. Ultrasound in Obstetrics & Gynecology 2014;44(1):17‐24. [PUBMED: 24616044] [DOI] [PubMed] [Google Scholar]
Zhang 2016 {published data only}
- Zhang J, Zhang B. Second‐generation non‐invasive high‐throughput DNA sequencing technology in the screening of Down's syndrome in advanced maternal age women. Biomedical Reports 2016;4(6):715‐8. [PUBMED: 27313855] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zhou 2014a {published data only}
- Zhou Q, Pan L, Chen S, Chen F, Hwang R, Yang X, et al. Clinical application of noninvasive prenatal testing for the detection of trisomies 21, 18, and 13: a hospital experience. Prenatal Diagnosis 2014;34(11):1061‐5. [PUBMED: 24899146] [DOI] [PubMed] [Google Scholar]
Zhou 2014b {published data only}
- Zhou Q, Pan L, Chen S, Chen F, Hwang R, Yang X, et al. Clinical application of noninvasive prenatal testing for the detection of trisomies 21, 18, and 13: a hospital experience. Prenatal Diagnosis 2014;34(11):1061‐5. [PUBMED: 24899146] [DOI] [PubMed] [Google Scholar]
References to studies excluded from this review
Anderson 2015 {published data only}
- Anderson B, Zhang K, Nguyen Q, Tsao D, Liu Y, Livingston K, et al. An automated, non‐invasive prenatal screening assay (NIPS) for trisomy 21,18,13 in singleton and twin gestations. International Journal of Gynecology and Obstetrics 2015;131:E264. [Google Scholar]
Anselem 2016 {published data only}
- Anselem O, Keroui S, Deput‐Rampon C, Chartier M, Costa JM, Goffinet F, et al. Analysis of cell‐free DNA in maternal blood for detection of fetal trisomy 21 in high‐risk population: Couples acceptance and grounds for refusal [Étude de l'ADN foetal dans le sang maternel pour la détection de la trisomie 21 en population à risque accru : adhésion des couples et motifs de refus]. Journal de Gynecologie, Obstetrique et Biologie de la Reproduction 2016;45(8):918‐23. [PUBMED: 26780844] [DOI] [PubMed] [Google Scholar]
Bayindir 2015 {published data only}
- Bayindir B, Dehaspe L, Brison N, Brady P, Ardui S, Kammoun M, et al. Noninvasive prenatal testing using a novel analysis pipeline to screen for all autosomal fetal aneuploidies improves pregnancy management. European Journal of Human Genetics 2015;23(10):1286‐93. [PUBMED: 25585704] [DOI] [PMC free article] [PubMed] [Google Scholar]
Beamon 2013 {published data only}
- Beamon C, Hardisty E, Harris S, Vora N. Promises and pitfalls of a new technology: a single center experience with noninvasive prenatal testing (NIPT). American Journal of Obstetrics and Gynecology 2013;208(1):S244‐S5. [Google Scholar]
Beamon 2014 {published data only}
- Beamon CJ, Hardisty EE, Harris SC, Vora NL. A single center's experience with noninvasive prenatal testing. Genetics in Medicine 2014;16(9):681‐7. [PUBMED: 24675675] [DOI] [PubMed] [Google Scholar]
Belloin 2016 {published data only}
- Belloin C, Jacquemard F, Bernabe‐Dupont C, Viot G, Lohmann L, Grange G. The noninvasive prenatal testing for Down's Syndrome. Retrospective study of 8821 patients [Le dépistage prénatal non invasif de la trisomie 21. Étude rétrospective à propos de 8821 patientes]. Journal de Gynecologie, Obstetrique et Biologie de la Reproduction 2016;45(9):1127‐32. [PUBMED: 27091545] [DOI] [PubMed] [Google Scholar]
Benachi 2015b {published data only}
- Benachi A, Costa JM. Non invasive prenatal diagnosis of trisomy 21. La Revue du Praticien 2015;65(2):160‐2. [PUBMED: 25939212] [PubMed] [Google Scholar]
Benachi 2016 {published data only}
- Benachi A, Letourneau A, Kleinfinger P, Senat MV, Gautier E, Favre R, et al. Performance and indications of noninvasive prenatal testing using cell free circulating fetal DNA (cffDNA) for the detection of fetal trisomy 21, 18 and 13 in France [Performance et indication du dépistage des trisomies 21, 18 et 13 en France par l'analyse de l'ADN foetal dans le sang maternel.]. Journal de Gynécologie, Obstétrique et Biologie de la Reproduction 2016;45(6):633‐40. [PUBMED: 26518155] [DOI] [PubMed] [Google Scholar]
Benn 2015 {published data only}
- Benn P. Re: Non‐invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146 958 pregnancies. Zhang H, Gao Y, Jiang F, Fu M, Yuan Y, Guo Y, et al. Ultrasound in Obstetrics & Gynecology 2015; 45(5): 530‐8.. Ultrasound in Obstetrics & Gynecology 2015;45(5):512‐3. [PUBMED: 25914392] [DOI] [PubMed] [Google Scholar]
Bhatt 2014 {published data only}
- Bhatt S, Parsa S, Snyder H, Taneja P, Halks‐Miller M, Seltzer W, et al. Clinical laboratory experience with noninvasive prenatal testing: Update on clinically relevant metrics. Prenatal Diagnosis 2014;34(Suppl 1):48. [Google Scholar]
Bianchi 2012a {published data only}
- Bianchi D, Platt L, Goldberg J, Abuhamad A, Sehnert A, Rava R. Whole genome maternal plasma DNA sequencing accurately detects autosomal and sex chromosome aneuploidies. Prenatal Diagnosis 2012;32((Bianchi D.) Tufts Medical Center, Mother Infant Research Institute, Boston, United States):3‐4. [Google Scholar]
Bianchi 2014b {published data only}
- Bianchi DW, Lamar Parker R, Wentworth J, Madankumar R, Saffer C, Das AF, et al. DNA sequencing versus standard prenatal aneuploidy screening. Obstetrical and Gynecological Survey 2014;69(6):319‐21. [EMBASE: 2014389701] [Google Scholar]
Bianchi 2014c {published data only}
- Bianchi D, Swanson A, Parsa S, Bhatt S, Halks‐Miller M, Sehnert A, et al. NIPT for sex chromosome aneuploidy: Initial clinical laboratory experience and biologic reasons for discordant results. American Journal of Obstetrics and Gynecology 2014;210(1):S87‐S8. [Google Scholar]
Bianchi 2015a {published data only}
- Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, et al. Noninvasive prenatal testing and incidental detection of occult maternal malignancies. JAMA 2015;314(2):162‐9. [PUBMED: 26168314] [DOI] [PubMed] [Google Scholar]
Bianchi 2015b {published data only}
- Bianchi DW, Parsa S, Bhatt S, Halks‐Miller M, Kurtzman K, Sehnert AJ, et al. Fetal sex chromosome testing by maternal plasma DNA sequencing: clinical laboratory experience and biology. Obstetrics and Gynecology 2015;125(2):375‐82. [DOI] [PubMed] [Google Scholar]
Bianchi 2015c {published data only}
- Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, et al. Noninvasive prenatal yesting and incidental detection of occult maternal malignancies. Obstetrical and Gynecological Survey 2015;70(12):744‐6. [DOI] [PubMed] [Google Scholar]
Bianchi 2015d {published data only}
- Bianchi D, Chudova D, Sehnert A, Bhatt S, Murray K, Prosen T, et al. Incidental detection of occult maternal malignancies by noninvasive prenatal testing. Prenatal Diagnosis 2015;35:10‐1. [DOI] [PubMed] [Google Scholar]
BlueCross BlueShield Asssociation 2014 {published data only}
- BlueCross BlueShield Asssociation. Noninvasive prenatal cell‐free fetal DNA‐based screening for aneuploidies other than trisomy 21. Technology Evaluation Center Assessment Program Executive summary 2014;29(7):1‐7. [PUBMED: 25577816] [PubMed] [Google Scholar]
Brady 2016 {published data only}
- Brady P, Brison N, Bogaert K, Ravel T, Peeters H, Esch H, et al. Clinical implementation of NIPT ‐ technical and biological challenges. Clinical Genetics 2016;89(5):523‐30. [PUBMED: 25867715] [DOI] [PubMed] [Google Scholar]
Chen 2013 {published data only}
- Chen F, Zheng J, Chen M, Fang Q, Wei Y, Zhang C, et al. Noninvasive prenatal testing of Trisomy 21 and 18 by maternal plasma sequencing in twin pregnancies. Prenatal Diagnosis 2013;33:14. [Google Scholar]
Chen 2014 {published data only}
- Chen M, Ma GC, Yeang CH. Genome‐wide normalized score: A novel counting algorithm to detect fetal trisomies in non‐invasive prenatal testing. Prenatal Diagnosis 2014;34:49. [DOI] [PubMed] [Google Scholar]
Cherry 2014 {published data only}
- Cherry AM, Williams JM Jr, Manning MA. Accuracy of non‐invasive prenatal screening for trisomies 13, 18 and 21 and sex chromosome aneuploidy. Cytogenetic and Genome Research 2014;142(3):240‐1. [Google Scholar]
Cheung 2015 {published data only}
Chiu 2008 {published data only}
- Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W, Leung TY, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proceedings of the National Academy of Sciences of the United States of America 2008;105(51):20458‐63. [PUBMED: 19073917] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chiu 2010 {published data only}
- Chiu RW, Sun H, Akolekar R, Clouser C, Lee C, McKernan K, et al. Maternal plasma DNA analysis with massively parallel sequencing by ligation for noninvasive prenatal diagnosis of trisomy 21. Clinical Chemistry 2010;56(3):459‐63. [PUBMED: 20026875] [DOI] [PubMed] [Google Scholar]
Christina 2012 {published data only}
- Christina Fan H, Gu W, Wang J, Blumenfeld YJ, El‐Sayed YY, Quake SR. Non‐invasive prenatal measurement of the fetal genome. Nature 2012;487(7407):320‐4. [EMBASE: 2012416711] [DOI] [PMC free article] [PubMed] [Google Scholar]
Cinnioglu 2012 {published data only}
- Cinnioglu C. The application of bioinformatics to genetic testing for the detection of human aneuploidy and genotyping. Reproductive BioMedicine Online 2012;24:S37. [Google Scholar]
Cirigliano 2013 {published data only}
- Cirigliano V, Ordonez E, Rueda L, Moreno M, Palao B, Paz Canadas M. Introduction of cfDNA based screening for common trisomies in Spain. Prenatal Diagnosis 2013;33:85‐6. [Google Scholar]
Cirigliano 2014 {published data only}
- Cirigliano V, Ordoñez E, Rueda L, Cañadas P, Moreno M, Palao B. cfDNA based aneuploidy screening in Spain: Results of one year clinical application. Prenatal Diagnosis 2014;34:51. [Google Scholar]
Cuckle 2015 {published data only}
- Cuckle H, Benn P, Pergament E. Cell‐free DNA screening for fetal aneuploidy as a clinical service. Clinical Biochemistry 2015;48(15):932‐41. [PUBMED: 25732593] [DOI] [PubMed] [Google Scholar]
Curnow 2014 {published data only}
- Curnow K, Ryan A, Banjevic M, Shchegrova S, Babiarz J, Constantin T, et al. Observations following clinical implementation of a cfDNA and single‐nucleotide polymorphism‐based non‐invasive prenatal aneuploidy test (NIPT). Prenatal Diagnosis 2014;34:51‐2. [Google Scholar]
Dan 2012 {published data only}
- Dan S, Wang W, Ren J, Li Y, Hu H, Xu Z, et al. Clinical application of massively parallel sequencing‐based prenatal noninvasive fetal trisomy test for trisomies 21 and 18 in 11,105 pregnancies with mixed risk factors. Prenatal Diagnosis 2012;32(13):1225‐32. [PUBMED: 23138752] [DOI] [PubMed] [Google Scholar]
Dar 2014 {published data only}
- Dar P, Curnow KJ, Gross SJ, Hall MP, Stosic M, Demko Z, et al. Clinical experience and follow‐up with large scale single‐nucleotide polymorphism‐based noninvasive prenatal aneuploidy testing. American Journal of Obstetrics and Gynecology 2014;211(5):527.e1‐527.e17. [PUBMED: 25111587] [DOI] [PubMed] [Google Scholar]
De Ligt 2013 {published data only}
- Ligt J, Janssen I, Bon B, Buysse K, Gomes I, Eggink A, et al. Detecting partial fetal aneuploidies by MPS: An unexpected discrepancy between amniotic fluid and ccffDNA. Prenatal Diagnosis 2013;33(Suppl 1):72‐3. [ISSN 01973851] [Google Scholar]
Denona 2016 {published data only}
- Denona B, Mone F, Cathcart B, Mahony R, Carroll S, Higgins S, et al. Changing rates of noninvasive prenatal testing and prenatal invasive testing in a tertiary centre. BJOG: an international journal of obstetrics and gynaecology 2016;123:75‐6. [Google Scholar]
Discenza 2015 {published data only}
- Discenza M, Ahern D, Coles K, Dobson L, Reiss R. Sex chromosome aneuploidy detection by NIPT: helpful or hazardous?. Prenatal Diagnosis 2015;35:105‐6. [DOI] [PubMed] [Google Scholar]
Dobson 2015 {published data only}
- Dobson L, Reiff E, Little S, Bromley B, Wilkins‐Haug L. Clinical course and patient outcomes following positive noninvasive prenatal testing (NIPT). Prenatal Diagnosis 2015;35:3. [DOI] [PubMed] [Google Scholar]
Dobson 2016 {published data only}
- Dobson LJ, Reiff ES, Little SE, Wilkins‐Haug L, Bromley B. Patient choice and clinical outcomes following positive noninvasive prenatal screening for aneuploidy with cell‐free DNA (cfDNA). Prenatal Diagnosis 2016;36(5):456‐62. [PUBMED: 26938930] [DOI] [PubMed] [Google Scholar]
Dong 2016 {published data only}
- Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian Q, et al. Low‐pass whole‐genome sequencing in clinical cytogenetics: a validated approach. Genetics in Medicine 2016;18(9):940‐8. [PUBMED: 26820068] [DOI] [PubMed] [Google Scholar]
Duenwald 2016 {published data only}
- Duenwald S, Barbacioru C, Deciu C, Chen G, Comstock D, Skvortsov D, et al. Development of a novel paired‐end sequencing‐based noninvasive prenatal test. American Journal of Obstetrics and Gynecology 2016;214(1):S254‐5. [Google Scholar]
Ehrich 2011a {published data only}
- Ehrich M, Deciu C, Zwiefelhofer T, Tynan JA, Cagasan L, Tim R, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Obstetrical & Gynecological Survey 2011;66(6):342‐4. [DOI] [PubMed] [Google Scholar]
Eiben 2014 {published data only}
- Eiben B, Glaubitz R, Kagan KO. Non‐invasive prenatal diagnostics: ETS and NGS‐based tests. Medizinische Genetik 2014;26(4):382‐90. [EMBASE: 2014948590] [Google Scholar]
Ellison 2015 {published data only}
- Ellison C, Sun Y, Hogg G, Fox J, Tao H, McCarthy E, et al. Leveraging targeted sequencing of paired homologs for noninvasive detection of fetal aneuploidies. Prenatal Diagnosis 2015;35:54. [Google Scholar]
Faas 2011 {published data only}
- Faas B, Vissers L, Janssen I, Ligt J, Eggink A, Veltman J, et al. Multiplex massively parallel sequencing for noninvasive prenatal diagnosis. Chromosome Research 2011;19:S35‐S6. [Google Scholar]
Faas 2012 {published data only}
- Faas BH, Ligt J, Janssen I, Eggink AJ, Wijnberger LD, Vugt JM, et al. Non‐invasive prenatal diagnosis of fetal aneuploidies using massively parallel sequencing‐by‐ligation and evidence that cell‐free fetal DNA in the maternal plasma originates from cytotrophoblastic cells. Expert Opinion on Biological Therapy 2012;12 Suppl 1:S19‐26. [PUBMED: 22500971] [DOI] [PubMed] [Google Scholar]
Fairbrother 2013a {published data only}
- Fairbrother G, Johnson S, Musci TJ, Song K. Clinical experience of noninvasive prenatal testing with cell‐free DNA for fetal trisomies 21, 18, and 13, in a general screening population. Prenatal Diagnosis 2013;33(6):580‐3. [PUBMED: 23494956] [DOI] [PubMed] [Google Scholar]
Fairbrother 2013b {published data only}
- Fairbrother G, Johnson S, Musci T, Song K. Clinical experience of Harmony™ Prenatal Test, a noninvasive prenatal test, in a general screening population. Prenatal Diagnosis 2013;33:78. [DOI] [PubMed] [Google Scholar]
Fan 2008 {published data only}
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proceedings of the National Academy of Sciences of the United States of America 2008;105(42):16266‐71. [PUBMED: 18838674] [DOI] [PMC free article] [PubMed] [Google Scholar]
Fang 2015 {published data only}
- Fang Y, Wang G, Wang C, Suo F, Gu M, Xia Y. The diagnosis pattern of mid‐trimester fetal chromosomal aneuploidy in Xuzhou and the clinical applications. Cell Biochemistry and Biophysics 2015;73(2):267‐70. [PUBMED: 25733332] [DOI] [PubMed] [Google Scholar]
Ferres 2013 {published data only}
- Ferres MA, Lichten L, Sachs A, Lau KM, Bianchi DW. Early experience with noninvasive DNA testing for aneuploidy in prenatal care. Prenatal Diagnosis 2013;33:73. [Google Scholar]
Fiorentino 2015 {published data only}
- Fiorentino F, Spinella F, Bono S, Pizzuti F, Mariano M, Polverari A, et al. Feasibility of noninvasive prenatal testing for common fetal aneuploidies in maternal serum with low levels circulating fetal cell‐free DNA fraction. Prenatal Diagnosis 2015;35:1. [Google Scholar]
Fosler 2015 {published data only}
- Fosler L, Winters P, Jones K, Platt L. Clinical laboratory experience with noninvasive prenatal testing in twin gestations. American Journal of Obstetrics and Gynecology 2015;212(1):S330‐S1. [Google Scholar]
Futch 2013 {published data only}
- Futch T, Spinosa J, Bhatt S, Feo E, Rava RP, Sehnert AJ. Initial clinical laboratory experience in noninvasive prenatal testing for fetal aneuploidy from maternal plasma DNA samples. Prenatal Diagnosis 2013;33(6):569‐74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Gabriel 2014 {published data only}
- Gabriel, H.Biskup, S. Parallel prenatal testing for trisomies (‐13,‐18 and‐21), turner syndrome, 22q11.2 del/dup‐syndromes (e.g. DiGeorge), Smith‐Lemli‐Opitz syndrome and Noonan syndrome in fetuses with increased nuchal translucency using the AmpliSeq™ technology (Life Tech). Medizinische Genetik 2014;26(1):192‐3. [ISSN 09365931] [Google Scholar]
Galea 2014 {published data only}
- Galea M, Grehan S, Ives D, Evans K, Harraway J. A one‐year experience of non‐invasive prenatal testing in an Australian private pathology setting. Prenatal Diagnosis 2014;34:54. [Google Scholar]
Gao 2014 {published data only}
- Gao Y, Xie B, Liu R. Delivering noninvasive prenatal testing in a clinical setting using semiconductor sequencing platform. Science China Life Sciences 2014;57(7):737‐8. [PUBMED: 24969704] [DOI] [PubMed] [Google Scholar]
Gao 2015 {published data only}
- Gao Y, Zeng F, Fu M, Zhang H, Jiang F, Chen F, et al. Comparison of NIPT clinical performance in 72,382 high risk pregnant women and 40,287 low‐risk pregnant women. Prenatal Diagnosis 2015;35:2. [Google Scholar]
Geifman‐Holtzman 2013 {published data only}
- Geifman‐Holtzman O, Berman J, Ness AL, Cohen A, Fischer R, Carre A, et al. NIPDT registry‐a critical utilization of non‐invasive prenatal diagnosis test recently introduced to clinical practice. American Journal of Obstetrics and Gynecology 2013;208(1):S254. [Google Scholar]
Geifman‐Holtzman 2014 {published data only}
- Geifman‐Holtzman O, Berman J, Xiong Y, Carre A, Ness A, Fang YMV, et al. Noninvasive prenatal testing (NIPT) registry patients' results and providers' perspective. American Journal of Obstetrics and Gynecology 2014;210(1):S96. [Google Scholar]
Gerundino 2017 {published data only}
- Gerundino F, Giachini C, Contini E, Benelli M, Marseglia G, Giuliani C, et al. Validation of a method for noninvasive prenatal testing for fetal aneuploidies risk and considerations for its introduction in the Public Health System. Journal of Maternal‐Fetal & Neonatal Medicine 2017;30(6):710‐6. [PUBMED: 27226231] [DOI] [PubMed] [Google Scholar]
Gil 2013 {published data only}
- Gil MM, Quezada MS, Bregant B, Ferraro M, Nicolaides KH. Implementation of maternal blood cell‐free DNA testing in early screening for aneuploidies. Ultrasound in Obstetrics & Gynecology 2013;42(1):34‐40. [PUBMED: 23744609] [DOI] [PubMed] [Google Scholar]
Gil 2015 {published data only}
- Gil MM, Giunta G, Macalli EA, Poon LC, Nicolaides KH. UK NHS pilot study on cell‐free DNA testing in screening for fetal trisomies: factors affecting uptake. Ultrasound in Obstetrics & Gynecology 2015;45(1):67‐73. [PUBMED: 25302655] [DOI] [PubMed] [Google Scholar]
Gnetetskaya 2015 {published data only}
- Gnetetskaya V, Kurtser M. Diagnosis of chromosomal aneuploidies using non‐invasive prenatal test in Moscow. Journal of Perinatal Medicine 2015;43(s1):1300‐64. [Google Scholar]
Grati 2014 {published data only}
- Grati FR, Malvestiti F, Ferreira JC, Bajaj K, Gaetani E, Agrati C, et al. Fetoplacental mosaicism: potential implications for false‐positive and false‐negative noninvasive prenatal screening results. Genetics in Medicine 2014;16(8):620‐4. [PUBMED: 24525917] [DOI] [PubMed] [Google Scholar]
Gray 2013 {published data only}
- Gray KJ, Wilkins‐Haug L, Gerrol P, Reiss R. Introduction of ffDNA testing for aneuploidy screening: Indications, changes in invasive testing rates, and caveats. Prenatal Diagnosis 2013;33:74. [Google Scholar]
Gromminger 2014 {published data only}
- Gromminger S, Yagmur E, Erkan S, Nagy S, Schock U, Bonnet J, et al. Fetal aneuploidy detection by cell‐free DNA sequencing for multiple pregnancies and quality issues with vanishing twins. Journal of Clinical Medicine 2014;3(3):679‐92. [PUBMED: 26237471] [DOI] [PMC free article] [PubMed] [Google Scholar]
Guex 2013 {published data only}
- Guex N, Iseli C, Syngelaki A, Deluen C, Pescia G, Nicolaides KH, et al. A robust second‐generation genome‐wide test for fetal aneuploidy based on shotgun sequencing cell‐free DNA in maternal blood. Prenatal Diagnosis 2013;33(7):707‐10. [DOI] [PubMed] [Google Scholar]
Halks‐Miller 2015 {published data only}
Harasim 2016 {published data only}
- Harasim T, Wagner A, Rath S, Behaqi P, Rost I, Klein H‐G. Prenatalis® NIPT: Accredited high resolution non‐invasive prenatal testing by using massive parallel ultra‐deep sequencing. Medizinische Genetik 2016;28(1):152‐3. [Google Scholar]
Hernandez‐Gomez 2015 {published data only}
- Hernandez‐Gomez M, Ramirez‐Arroyo E, Melendez‐Hernandez R, Garduno‐Zaraza LM, Mayen‐Molina DG. Non invasive prenatal test (NIPT) in maternal blood by parallel massive sequencing. Initial experience in Mexican women and literature review [Prueba prenatal no invasiva (NIPT) en sangre materna a traves de secuenciacion masiva paralela (MPS). Experiencia inicial en mujeres mexicanas y revision de la bibliografia]. Ginecologia y Obstetricia de Mexico 2015;83(5):277‐88. [PUBMED: 26233973] [PubMed] [Google Scholar]
Hofmann 2013 {published data only}
- Hofmann W, Entezami M, Haug K, Blank C, Wustemann M, Schulze B, et al. Diagnostic accuracy for the noninvasive prenatal detection of common autosomal aneuploidies. Prenatal Diagnosis 2013;33:75. [DOI] [PubMed] [Google Scholar]
Hofmann 2014 {published data only}
- Hofmann W, Grömminger S, Schöck U, Bonnet J, Smerdka P. Non‐invasive prenatal testing (NIPT): Laboratory experiences of PrenaTest®. Medizinische Genetik 2014;26(1):194. [Google Scholar]
Hofmann 2015 {published data only}
- Hofmann W, Grömminger S, Sachse M, Bonnet J, Schöck U. Recent bioinformatic advances of non‐invasive prenatal detection to enhance diagnostic accuracy and aneuploidy discovery. Prenatal Diagnosis 2015;35:57. [Google Scholar]
Hu 2014 {published data only}
- Hu Y, Duan H, Li J, Zhu R. Study of clinical indications for non‐invasive prenatal testing based on distributions of fetal chromosomal abnormalities. Prenatal Diagnosis 2014;34:59. [Google Scholar]
Hu 2015 {published data only}
- Hu HJ, Kwon YJ, Oh M, Kim J, Cho DY, Seo DH. Evaluating the results of the Momguard™ noninvasive prenatal test. Journal of Genetic Medicine 2015;12(2):96‐9. [DOI: 10.5734/JGM.2015.12.2.96] [DOI] [Google Scholar]
Hui 2015a {published data only}
- Hui L, Teoh M, Silva Costa F, Ramsay P, Palma‐Dias R, Richmond Z, et al. Clinical implementation of noninvasive prenatal testing by Australian sonologists. BJOG: a international journal of obstetrics and gynaecology 2015;122:52. [Google Scholar]
Hui 2015b {published and unpublished data}
- Hui L, Teoh M, Silva C, Ramsay P, Palma‐Dias R, Richmond Z, et al. Clinical implementation of cell‐free DNA‐based aneuploidy screening: perspectives from a national audit. Ultrasound in Obstetrics & Gynecology 2015;45(1):10‐5. [DOI] [PubMed] [Google Scholar]
Jackson 2013 {published data only}
- Jackson J, Dever K, Hamar B, Lazar E, Lim KH, Rodriguez D, et al. Nuchal translucency plus noninvasive prenatal testing screening for aneuploidy in a population of low‐and high‐risk patients. Prenatal Diagnosis 2013;33:22. [Google Scholar]
Jensen 2013 {published data only}
- Jensen TJ, Zwiefelhofer T, Tim RC, Dzakula Z, Kim SK, Mazloom AR, et al. High‐throughput massively parallel sequencing for fetal aneuploidy detection from maternal plasma. PLOS One 2013;8(3):e57381. [PUBMED: 23483908] [DOI] [PMC free article] [PubMed] [Google Scholar]
Jensen 2015 {published data only}
- Jensen TJ, Deciu C, Ehrich M, Geis J, Kim SK, Tao H. Selective enrichment of genomic loci for the noninvasive detection of fetal aneuploidies. Obstetrics and Gynecology 2015;125:92S. [Google Scholar]
Jin 2014 {published data only}
- Jin Y, Miao Z, Ge J, Zhang W, Li S, Liu X. Prenatal diagnosis of fetal chromosome aneuploidy by massively parallel genomic sequencing. National Medical Journal of China 2014;94(23):1788‐90. [PubMed] [Google Scholar]
Johnson 2013 {published data only}
- Johnson J, Pastuck M, Metcalfe A, Connors G, Krause R, Wilson D, et al. First‐trimester Down syndrome screening using additional serum markers with and without nuchal translucency and cell‐free DNA. Prenatal Diagnosis 2013;33(11):1044‐9. [DOI] [PubMed] [Google Scholar]
Juneau 2014 {published data only}
- Juneau K, Bogard PE, Huang S, Mohseni M, Wang ET, Ryvkin P, et al. Microarray‐based cell‐free DNA analysis improves noninvasive prenatal testing. Fetal Diagnosis and Therapy 2014;36(4):282‐6. [DOI] [PubMed] [Google Scholar]
Kagan 2015 {published data only}
- Kagan KO, Schmid M, Hoopmann M, Wagner P, Abele H. Screening performance and costs of different strategies in prenatal screening for trisomy 21. Geburtshilfe und Frauenheilkunde 2015;75(3):244‐50. [PUBMED: 25914417] [DOI] [PMC free article] [PubMed] [Google Scholar]
Kalantar 2014 {published data only}
- Kalantar SM, Sitar G, Carabresy J. Is time for using non‐invasive pre‐natal diagnosis for genetic disorders?. Iranian Journal of Reproductive Medicine 2014;12(6):15. [Google Scholar]
Karlsson 2015 {published data only}
- Karlsson K, Sahlin E, Iwarsson E, Westgren M, Nordenskjold M, Linnarsson S. Amplification‐free sequencing of cell‐free DNA for prenatal non‐invasive diagnosis of chromosomal aberrations. Genomics 2015;105(3):150‐8. [DOI] [PubMed] [Google Scholar]
Kershberg 2015 {published data only}
- Kershberg H, Greenberg J, Liou A, Logg A, Alvarado M, Natoli J, et al. Positive and negative predictive values of NIPT in an integrated health care system: summary of NIPT outcomes in Kaiser Permanente Southern California. Prenatal Diagnosis 2015;35:58. [Google Scholar]
Kinde 2012 {published data only}
- Kinde I, Papadopoulos N, Kinzler KW, Vogelstein B. FAST‐SeqS: a simple and efficient method for the detection of aneuploidy by massively parallel sequencing. PLOS One 2012;7(7):e41162. [PUBMED: 22815955] [DOI] [PMC free article] [PubMed] [Google Scholar]
Korabecna 2012 {published data only}
- Korabecna M. Non invasive prenatal diagnostics of most frequent chromosomal aneuploidies in the language of numbers. Aktualni Gynekologie a Porodnictvi 2012;4(1):114‐5. [Google Scholar]
Koumbaris 2016 {published data only}
- Koumbaris G, Kypri E, Tsangaras K, Achilleos A, Mina P, Neofytou M, et al. Cell‐free DNA analysis of targeted genomic regions in maternal plasma for non‐invasive prenatal testing of trisomy 21, trisomy 18, trisomy 13, and fetal sex. Clinical Chemistry 2016;62(6):848‐55. [PUBMED: 27117469] [DOI] [PubMed] [Google Scholar]
Kurtser 2015 {published data only}
- Kurtser MA, Gnetetskaya VA. A noninvasive prenatal test in the diagnosis of chromosome aneuploidies. Akushersivo i Ginekologiya 2015;8:65‐9. [Google Scholar]
Lambert‐Messerlian 2014 {published data only}
- Lambert‐Messerlian G, Kloza EM, Williams J 3rd, Loucky J, O'Brien B, Wilkins‐Haug L, et al. Maternal plasma DNA testing for aneuploidy in pregnancies achieved by assisted reproductive technologies. Genetics in Medicine 2014;16(5):419‐22. [PUBMED: 24091801] [DOI] [PubMed] [Google Scholar]
Larion 2015 {published data only}
- Larion S, Warsof S, Romary L, Mlynarczyk M, Abuhamad A. Three year clinical experience with noninvasive prenatal testing in 3000 high risk cases in the United States. Prenatal Diagnosis 2015;35:59. [Google Scholar]
Lau 2012a {published data only}
- Lau TK, Chan MK, Lo PS, Chan HY, Chan WS, Koo TY, et al. Clinical utility of noninvasive fetal trisomy (NIFTY™) test‐‐early experience. Journal of Maternal‐fetal & Neonatal Medicine 2012;25(10):1856‐9. [PUBMED: 22471583] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lau 2013 {published data only}
- Lau TK, Jiang F, Chan MK, Zhang H, Lo PS, Wang W. Non‐invasive prenatal screening of fetal Down syndrome by maternal plasma DNA sequencing in twin pregnancies. Journal of Maternal‐Fetal & Neonatal Medicine 2013;26(4):434‐7. [PUBMED: 23035860] [DOI] [PubMed] [Google Scholar]
Lau 2014 {published data only}
- Lau TK, Cheung SW, Lo PS, Pursley AN, Chan MK, Jiang F, et al. Non‐invasive prenatal testing for fetal chromosomal abnormalities by low‐coverage whole‐genome sequencing of maternal plasma DNA: review of 1982 consecutive cases in a single center. Ultrasound in Obstetrics & Gynecology 2014;43(3):254‐64. [PUBMED: 24339153] [DOI] [PubMed] [Google Scholar]
Lebo 2015 {published data only}
- Lebo RV, Novak RW, Wolfe K, Michelson M, Robinson H, Mancuso MS. Discordant circulating fetal DNA and subsequent cytogenetics reveal false negative, placental mosaic, and fetal mosaic cfDNA genotypes. Journal of Translational Medicine 2015;13:260. [PUBMED: 26260800] [DOI] [PMC free article] [PubMed] [Google Scholar]
Leung 2013 {published data only}
- Leung TY, Qu JZZ, Liao GJW, Jiang P, Cheng YKY, Chan KCA, et al. Noninvasive twin zygosity assessment and aneuploidy detection by maternal plasma DNA sequencing. Prenatal Diagnosis 2013;33(7):675‐81. [DOI] [PubMed] [Google Scholar]
Levandoski 2015 {published data only}
- Levandoski K, Marchand K, Briere L, Ferres M. One clinic's experience with discordant NIPT results for trisomies 18 and 13: Practical and psychosocial implications of non‐invasive prenatal testing. Prenatal Diagnosis 2015;35:106‐7. [Google Scholar]
Levy 2013 {published data only}
- Levy B, Banjevic M, Hill M, Zimmermann B, Ryan A, Sigurjonsson S, et al. Targeted sequencing of SNPs results in highly accurate noninvasive detection of fetal aneuploidy of chromosomes 13, 18, 21, X, and Y: A validation study. Human Reproduction 2013;28:i26‐i7. [Google Scholar]
Levy 2013a {published data only}
- Levy B, Banjevic M, Hill M, Zimmermann B, Ryan A, Sigurjonsson S, et al. Targeted sequencing of single‐nucleotide polymorphisms for accurate, noninvasive fetal aneuploidy detection at autosomal and sex chromosomes. Prenatal Diagnosis 2013;33:76. [Google Scholar]
Levy 2013b {published data only}
- Levy, Ryan B, Banjevic A, Hill M, Zimmermann M, Sigurjonsson B, et al. Highly multiplexed single‐nucleotide polymorphism amplification and sequencing to identify fetal aneuploidy from maternal cell‐free DNA. Journal of Perinatal Medicine 2013;41(Suppl 1):34‐218. [ISSN 03005577] [Google Scholar]
Li 2012 {published data only}
- Li X, Sehnert A, Rava R. Percentage of fetal cell‐free DNA in maternal plasma: Dependence on multiple clinical factors. Prenatal Diagnosis 2012;32:6. [Google Scholar]
Li 2015 {published data only}
- Li WH, Wang PH, Chuang CM, Chang YW, Yang MJ, Chen CY, et al. Noninvasive prenatal testing for fetal trisomy in a mixed risk factors pregnancy population. Taiwanese Journal of Obstetrics & Gynecology 2015;54(2):122‐5. [PUBMED: 25951714] [DOI] [PubMed] [Google Scholar]
Liao 2011 {published data only}
- Liao GJ, Lun FM, Zheng YW, Chan KC, Leung TY, Lau TK, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clinical Chemistry 2011;57(1):92‐101. [PUBMED: 21078840] [DOI] [PubMed] [Google Scholar]
Liao 2012 {published data only}
- Liao GJ, Chan KC, Jiang P, Sun H, Leung TY, Chiu RW, et al. Noninvasive prenatal diagnosis of fetal trisomy 21 by allelic ratio analysis using targeted massively parallel sequencing of maternal plasma DNA. PLOS One 2012;7(5):e38154. [PUBMED: 22666469] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liao 2013 {published data only}
- Liao C, Fu YG, Huang SY, Fu F, Xie GE. Rapid noninvasive prenatal diagnosis of Down syndrome with Ion Proton™. Prenatal Diagnosis 2013;33:76‐7. [Google Scholar]
Liao 2014 {published data only}
- Liao C, Yin AH, Peng CF, Fu F, Yang JX, Li R, et al. Noninvasive prenatal diagnosis of common aneuploidies by semiconductor sequencing. Proceedings of the National Academy of Sciences of the United States of America 2014;111(20):7415‐20. [PUBMED: 24799683] [DOI] [PMC free article] [PubMed] [Google Scholar]
Liao 2014a {published data only}
Liu 2015 {published data only}
- Liu J, Wang H, Xi H, Jia Z, Zhou Y, Wu L. Application of next‐generation DNA sequencing for prenatal testing of fetal chromosomal aneuploidies. Zhonghua Yi Xue Yi Chuan Xue za Zhi [Chinese Journal of Medical Genetics] 2015;32(4):533‐7. [PUBMED: 26252102] [DOI] [PubMed] [Google Scholar]
Lo 2014 {published data only}
- Lo KK, Boustred C, Chitty LS, Plagnol V. RAPIDR: an analysis package for non‐invasive prenatal testing of aneuploidy. Bioinformatics (Oxford, England) 2014;30(20):2965‐7. [PUBMED: 24990604] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lo 2014a {published data only}
- Lo K, Boustred C, McKay F, Fielding S, Plagnol V, Chitty L. A comparison of statistical methods for NIPT for aneuploidies and development of the RAPID‐R package. Prenatal Diagnosis 2014;34:59‐60. [Google Scholar]
Loucký 2013 {published data only}
- Loucký J, Zemánek M. Non invasive prenatal testing of most frequent chromosomal aneuploidies ‐ Some other aspects [Neinvazivní prenatální testóvání nejčastějsích chromozomálních aneuploidií ‐ Některé dalši aspekty]. Actual Gynecology and Obstetrics 2013;5(1):6‐7. [Google Scholar]
Louis‐Jacques 2014 {published data only}
- Louis‐Jacques A, Burans C, Robinson S, Schofield E, Smulian J, Rochon M. Use of commercial tests for aneuploidy screening using cell‐free fetal DNA in clinical practice. Obstetrics and Gynecology 2014;123(Suppl 1):154S. [Google Scholar]
Ma 2015 {published data only}
- Ma J, Pan H, Fu J, Yu L, Yang H. Perspective study of non‐invasive prenatal testing using cell‐free fetal DNA in high‐risk population. Zhonghua Yi Xue za Zhi 2015;95(11):849‐52. [PUBMED: 26080919] [PubMed] [Google Scholar]
Ma 2015a {published data only}
- Ma J, Yang H, Chen F, Wang Y. Nationwide evaluation on noninvasive prenatal testing through maternal plasma DNA sequencing under governmental regulation in China. Prenatal Diagnosis 2015;35:67. [Google Scholar]
Manotaya 2016 {published data only}
- Manotaya S, Xu H, Uerpairojkit B, Chen F, Charoenvidhya D, Liu H, et al. Clinical experience from Thailand: noninvasive prenatal testing as screening tests for trisomies 21, 18 and 13 in 4736 pregnancies. Prenatal Diagnosis 2016;36(3):224‐31. [PUBMED: 26748603] [DOI] [PubMed] [Google Scholar]
Marchili 2015 {published data only}
- Marchili JP, Chaulet A, Guetmonovitch O, Margulies ND, Solarz V. One clinic's experience with cell free fetal DNA testing in Argentina. Prenatal Diagnosis 2015;35:60. [Google Scholar]
Mayen 2015 {published data only}
- Mayen DG, Hernandez‐Gomez M, Ramirez E, Meléndez R, Garduño LM. Noninvasive prenatal test, one year of experience in the genetics clinic at the Hospital Angeles Lomas in Mexico City. Prenatal Diagnosis 2015;35:60‐1. [Google Scholar]
Mazloom 2013a {published data only}
- Mazloom A, Dzakula Z, Wang H, Oeth P, Jensen T, Tynan J, et al. Detection of fetal sex chromosomal aneuploidies by sequencing circulating cell‐free DNA from maternal plasma. Prenatal Diagnosis 2013;33:77. [DOI] [PubMed] [Google Scholar]
McCullough 2014 {published data only}
- McCullough RM, Almasri EA, Guan X, Geis JA, Hicks SC, Mazloom AR, et al. Non‐invasive prenatal chromosomal aneuploidy testing‐‐clinical experience: 100,000 clinical samples. PLOS One 2014;9(10):e109173. [DOI] [PMC free article] [PubMed] [Google Scholar]
McCullough 2014a {published data only}
- McCullough R, Almasri E, Guan X, Oeth P, Bombard A, Saldivar JS. Noninvasive prenatal testing: 100,000 patients clinical impact. American Journal of Obstetrics and Gynecology 2014;210(1):S58. [Google Scholar]
McCullough 2015 {published data only}
- McCullough R, Almasri E, Boomer T, Wardrop J, Oeth P, Paxton W, et al. High volume clinical laboratory noninvasive prenatal testing: A synopsis (or summary) of results from >375,000 patients. Prenatal Diagnosis 2015;35:51‐2. [Google Scholar]
McLennan 2016 {published data only}
- McLennan A, Palma‐Dias R, Silva Costa F, Meagher S, Nisbet DL, Scott F. Noninvasive prenatal testing in routine clinical practice‐‐an audit of NIPT and combined first‐trimester screening in an unselected Australian population. Australian & New Zealand Journal of Obstetrics & Gynaecology 2016;56(1):22‐8. [PUBMED: 26817523] [DOI] [PubMed] [Google Scholar]
Meck 2014 {published data only}
- Meck JM, Kramer DE, Aviram A, Trunca C, Riethmaier D, Pineda AD, et al. Non‐invasive prenatal screening from viewpoint of the cytogenetics laboratory. Cytogenetic and Genome Research 2014;142(3):241. [Google Scholar]
Meck 2015 {published data only}
- Meck JM, Kramer Dugan E, Matyakhina L, Aviram A, Trunca C, Pineda‐Alvarez D, et al. Noninvasive prenatal screening for aneuploidy: positive predictive values based on cytogenetic findings. American Journal of Obstetrics and Gynecology 2015;213(2):214.e1‐5. [PUBMED: 25843063] [DOI] [PubMed] [Google Scholar]
Meck 2015a {published data only}
- Meck J, Dugan EK, Pineda‐Alvarez D, Wray A, Richard G, Matyakhina L. Comparison of results from noninvasive prenatal screening and diagnostic cytogenetic testing. Prenatal Diagnosis 2015;35:61. [Google Scholar]
Mennuti 2015 {published data only}
- Mennuti MT, Chandrasekaran S, Khalek N, Dugoff L. Cell‐free DNA screening and sex chromosome aneuploidies. Prenatal Diagnosis 2015;35(10):980‐5. [PUBMED: 26088741] [DOI] [PubMed] [Google Scholar]
Minarik 2015 {published data only}
- Minarik G, Repiska G, Hyblova M, Nagyova E, Soltys K, Budis J, et al. Utilization of Benchtop Next Generation Sequencing Platforms Ion Torrent PGM and MiSeq in noninvasive prenatal testing for chromosome 21 trisomy and testing of impact of in silico and physical size selection on its analytical performance. PLOS One 2015;10(12):e0144811. [PUBMED: 26669558] [DOI] [PMC free article] [PubMed] [Google Scholar]
Miron 2011 {published data only}
- Miron P. Prenatal screening for trisomy 21 and other aneuploidies at first trimester [Dépistage prénatal de la trisomie 21 et autres aneuploïdies au premier trimestre]. Université de Montréal (Faculté de Médecine) 2011:244 pages. [No AMICUS: 39273224]
Mundy 2008 {published data only}
- Mundy L, Hiller JE. Non‐invasive prenatal diagnostic test for Down's Syndrome (Structured abstract). Health Technology Assessment Database. Adelaide; Adelaide Health Technology Assessment (AHTA), 2008, issue 2. [HTA‐32012000229]
Mundy 2009 {published data only}
- Mundy L, Hiller JE. Non‐invasive prenatal diagnostic test for trisomy‐21 (Down's Syndrome) (Structured abstract). Health Technology Assessment Database. Adelaide Health Technology Assessment (AHTA) on behalf of National Horizon Scanning Unit (HealthPACT and MSAC), 2009, issue 2. [HTA‐32010000766]
Musci 2014 {published data only}
- Musci T. The largest study on NIPT thus far: The next study. Journal of Maternal‐fetal & Neonatal Medicine 2014;27:16. [Google Scholar]
Musci 2014a {published data only}
- Musci T, Struble C, Wang E, Hooks J, Syngelaki A, Mar Gil M, et al. Risk assessment for fetal sex chromosome aneuploidies using digital analysis of selected regions (DANSRTM) assays. Prenatal Diagnosis 2014;34:1‐2. [Google Scholar]
NCT00770458 {unpublished data only}
- NCT00770458. Non‐invasive screening for fetal aneuploidy: a new maternal plasma marker. ClinicalTrials.gov/show/NCT00770458 Date first received: 8 October 2008.
NCT00877292 {unpublished data only}
- NCT00877292. A new prenatal blood test for Down syndrome. ClinicalTrials.gov/show/NCT00877292 Date first received: 6 April 2009.
NCT00891852 {unpublished data only}
- NCT00891852. Non‐invasive determination of fetal chromosome abnormalities. ClinicalTrials.gov/show/NCT00891852 Date first received: 29 April 2009.
NCT00971334 {unpublished data only}
- NCT00971334. Noninvasive screening for fetal aneuploidy: a new maternal plasma marker. ClinicalTrials.gov/show/NCT00971334 Date first received: 1 September 2009.
NCT01052688 {unpublished data only}
- NCT01052688. Noninvasive screening for affected pregnancies: assay development and optimization in affected pregnancies. ClinicalTrials.gov/show/NCT01052688 Date first received: 15 January 2010.
NCT01256606 {unpublished data only}
- NCT01256606. Prenatal test for fetal aneuploidy detection. ClinicalTrials.gov/show/NCT01256606 Date first received: 22 November 2010.
NCT01451671 {unpublished data only}
- NCT01451671. Development of a prenatal test for fetal aneuploidy detection. ClinicalTrials.gov/show/NCT01451671 Date first received: 10 October 2011.
NCT01451684 {unpublished data only}
- NCT01451684. Development of a non‐invasive prenatal test. ClinicalTrials.gov/show/NCT01451684 Date first received: 10 October 2011.
NCT01555346 {unpublished data only}
- NCT01555346. Clinical evaluation of the SEQureDx T21 test In high risk pregnancies. ClinicalTrials.gov/show/NCT01555346 Date first received: 15 February 2012.
NCT01574781 {unpublished data only}
- NCT01574781. Non‐invasive prenatal diagnostic validation study. ClinicalTrials.gov/show/NCT01574781 Date first received: 2 April 2012.
NCT01597063 {unpublished data only}
- NCT01597063. Clinical evaluation of the SEQureDx Trisomy test in low risk pregnancies. ClinicalTrials.gov/show/NCT01597063 Date first received: 9 May 2012.
NCT01661010 {unpublished data only}
- NCT01661010. The clinical study of sex chromosome variants. ClinicalTrials.gov/show/NCT01661010 Date first received: 7 August 2012.
NCT01663675 {unpublished data only}
- NCT01663675. Trisomy 21 in adulthood. ClinicalTrials.gov/show/NCT01663675 Date first received: 8 August 2012.
NCT01668251 {unpublished data only}
- NCT01668251. Turner syndrome prenatal diagnosis study. ClinicalTrials.gov/show/NCT01668251 Date first received: 3 November 2011.
NCT01725438 {unpublished data only}
- NCT01725438. Non invasive prenatal diagnosis of trisomy 21 by genetic analysis of circulating fetal cells. ClinicalTrials.gov/show/NCT01725438 Date first received: 8 November 2012.
NCT01837979 {unpublished data only}
- NCT01837979. Down syndrome screening based on dried blood spots and cell‐free fetal DNA. ClinicalTrials.gov/show/NCT01837979 Date first received: 14 April 2013.
NCT01966991 {unpublished data only}
- NCT01966991. Prenatal screening for Down syndrome with DNAFirst. ClinicalTrials.gov/show/NCT01966991 Date first received: 17 October 2013.
NCT02127515 {unpublished data only}
- NCT02127515. Non invasive prenatal testing of Down syndrome. ClinicalTrials.gov/show/NCT02127515 Date first received: 2 April 2014.
NCT02226315 {unpublished data only}
- NCT02226315. Clinical performance of the MaterniT21™ PLUS LDT in multiple gestation pregnancies. ClinicalTrials.gov/show/NCT02226315 Date first received: 20 August 2014.
NCT02872948 {unpublished data only}
- NCT02872948. Diagnosis accuracy of noninvasive screening by PCR Digital for Down syndrome. ClinicalTrials.gov/show/NCT02872948 Date first received: 16 August 2016.
Neufeld‐Kaiser 2015 {published data only}
- Neufeld‐Kaiser WA, Cheng EY, Liu YJ. Positive predictive value of non‐invasive prenatal screening for fetal chromosome disorders using cell‐free DNA in maternal serum: independent clinical experience of a tertiary referral center. BMC Medicine 2015;13:129. [PUBMED: 26033224] [DOI] [PMC free article] [PubMed] [Google Scholar]
Neveling 2015 {published data only}
- Neveling K, Thung DT, Beulen L, Buijsman W, Gomes I, Heuvel S, et al. Clinical utility of NextSeq 500 sequencing for noninvasive prenatal testing. Prenatal Diagnosis 2015;35:54‐5. [Google Scholar]
Nickolich 2016 {published data only}
- Nickolich S, Farahi N, Jones K, Mounsey A. PURLs: Aneuploidy screening: Newer noninvasive test gains traction. Journal of Family Practice 2016;65(1):49‐52. [PUBMED: 26845165] [PMC free article] [PubMed] [Google Scholar]
Nicolaides 2013a {published data only}
- Nicolaides K, Syngelaki A, Ashoor G, Musci T, Wang E, Song K. Clinical performance comparison of Harmony™ Prenatal Test and first‐trimester combined screening in general pregnancy population. Prenatal Diagnosis 2013;33:3. [Google Scholar]
Nicolaides 2014 {published data only}
- Nicolaides KH, Syngelaki A, Poon LC, Gil MM, Wright D. First‐trimester contingent screening for trisomies 21, 18 and 13 by biomarkers and maternal blood cell‐free DNA testing. Fetal Ddiagnosis and Therapy 2014;35(3):185‐92. [PUBMED: 24192489] [DOI] [PubMed] [Google Scholar]
Nicolaides 2014b {published data only}
- Nicolaides KH, Musci TJ, Struble CA, Syngelaki A, Mar Gil M. Assessment of fetal sex chromosome aneuploidy using directed cell‐free DNA analysis. Obstetrical and Gynecological Survey 2014;69(5):249‐50. [DOI] [PubMed] [Google Scholar]
Nicolaides 2014c {published data only}
- Nicolaides KH, Syngelaki A, Mar Gil M, Quezada MS, Zinevich Y. Prenatal detection of fetal triploidy from cell‐free DNA testing in maternal blood. Fetal Diagnosis and Therapy 2014;35(3):212‐7. [PUBMED: 24135152] [DOI] [PubMed] [Google Scholar]
Norem 2015 {published data only}
- Norem C, Obolensky E, Bijesse E, Turocy J, Blumberg B, Fehlen‐Quizon P, et al. Non‐invasive prenatal screening for trisomies‐2 years experience in a large Health Maintenance Organization (HMO). Prenatal Diagnosis 2015;35:62. [Google Scholar]
Norton 2014 {published data only}
- Norton ME, Jelliffe‐Pawlowski LL, Currier RJ. Chromosome abnormalities detected by current prenatal screening and noninvasive prenatal testing. Obstetrics and Gynecology 2014;124(5):979‐86. [DOI] [PubMed] [Google Scholar]
Norton 2014a {published data only}
- Norton ME, Jacobsson B, Swamy G, Laurent L, Ranzini A, Brar H, et al. Non‐invasive EXamination of Trisomy using directed cell‐free DNA analysis: The NEXT study. Prenatal Diagnosis 2014;34:e2. [Google Scholar]
Norton 2015a {published data only}
- Norton M, Wapner R, Kuppermann M, Jelliffe‐Pawlowski L, Currier R. Cell free DNA analysis vs sequential screening as primary testing considering all fetal chromosomal abnormalities. American Journal of Obstetrics and Gynecology 2015;212(1):S2. [DOI] [PubMed] [Google Scholar]
Norton 2015b {published data only}
- Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, et al. Cell‐Free DNA analysis for noninvasive examination of trisomy. Obstetrical & Gynecological Survey 2015;70(8):483‐4. [DOI: 10.1097/01.ogx.0000470657.58577.f2] [DOI] [PubMed] [Google Scholar]
Norton 2015c {published data only}
Norton 2016 {published data only}
- Norton ME, Baer RJ, Wapner RJ, Kuppermann M, Jelliffe‐Pawlowski LL, Currier RJ. Cell‐free DNA vs sequential screening for the detection of fetal chromosomal abnormalities. American Journal of Obstetrics and Gynecology 2016;214(6):727.e1‐6. [PUBMED: 26709085] [DOI] [PubMed] [Google Scholar]
O'Leary 2014 {published data only}
- O'Leary P, Maxwell S, Murch A, Dickinson J. What could non‐invasive prenatal testing miss in highrisk pregnancies and would it change outcomes?. Prenatal Diagnosis 2014;34:52‐3. [Google Scholar]
Oepkes 2015 {published data only}
- Oepkes D, Schuring‐Blom H, Pajkrt E, Faas B, Bax C, Coumans A, et al. TRIDENT: Or monitored NIPT implementation in the Netherlands. Prenatal Diagnosis 2015;35:10. [Google Scholar]
Oneda 2016 {published data only}
- Oneda B, Steindl K, Masood R, Reshetnikova I, Krejci P, Baldinger R, et al. Noninvasive prenatal testing: More caution in counseling is needed in high risk pregnancies with ultrasound abnormalities. European Journal of Obstetrics Gynecology and Reproductive Biology 2016;200:72‐5. [DOI] [PubMed] [Google Scholar]
Ordoñez 2015 {published data only}
- Ordoñez E, Rueda LR, Cañadas MP, Moreno M, Palao B, Cirigliano V. cfDNA‐Based aneuploidy screening, the more the better?. Prenatal Diagnosis 2015;35:107. [Google Scholar]
Palomaki 2011 {published data only}
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genetics in Medicine 2011;13(11):913‐20. [PUBMED: 22005709] [DOI] [PubMed] [Google Scholar]
Palomaki 2012a {published data only}
- Palomaki GE, Kloza EM, Canick JA. Evidence that next‐generation sequencing of maternal plasma will identify aneuploidies in a low risk population. Prenatal Diagnosis 2012;32:4. [Google Scholar]
Palomaki 2012b {published data only}
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Haddow JE, Neveux LM, Ehrich M, et al. Editorial comment on DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Obstetrical and Gynecological Survey 2012;67(2):86‐8. [ISSN 0029‐7828] [DOI] [PubMed] [Google Scholar]
Palomaki 2015 {published data only}
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Boom D, Ehrich M, Deciu C, et al. Circulating cell free DNA testing: Are some test failures informative?. Prenatal Diagnosis 2015;35(3):289‐93. [DOI] [PubMed] [Google Scholar]
Palomaki 2015a {published data only}
- Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Boom D, Ehric M, Deciu C, et al. Circulating cell free DNA testing: are some test failures informative?. Obstetrical and Gynecological Survey 2015;70(8):492‐4. [DOI] [PubMed] [Google Scholar]
Perez‐Pedregosa 2015 {published data only}
- Pérez‐Pedregosa J, Paredes Ros B, Calles Hernandez LC, Izquierdo López L, Cabrillo Rodriguez E, Hurtado Caballero IV, et al. Non‐invasive prenatal screening for aneuploidy through analysis of cell‐free fetal DNA from maternal blood [Cribado prenatal no invasivo de aneuploidías mediante análisis de ADN fetal en sangre materna]. Progresos de Obstetricia y Ginecología 2015;58(3):113‐7. [Google Scholar]
Pescia 2017 {published data only}
- Pescia G, Guex N, Iseli C, Brennan L, Osteras M, Xenarios I, et al. Cell‐free DNA testing of an extended range of chromosomal anomalies: clinical experience with 6,388 consecutive cases. Genetics in Medicine 2017;19(2):169‐75. [PUBMED: 27362910] [DOI] [PMC free article] [PubMed] [Google Scholar]
Petersen 2014 {published data only}
- Petersen OB, Vogel I, Ekelund C, Hyett J, Tabor A. Potential diagnostic consequences of applying non‐invasive prenatal testing: population‐based study from a country with existing first‐trimester screening. Ultrasound in Obstetrics & Gynecology 2014;43(3):265‐71. [DOI] [PubMed] [Google Scholar]
Pettit 2014 {published data only}
- Pettit KE, Hull AD, Korty L, Jones MC, Pretorius DH. The utilization of circulating cell‐free fetal DNA testing and decrease in invasive diagnostic procedures: an institutional experience. Journal of Perinatology 2014;34(10):750‐3. [PUBMED: 24875410] [DOI] [PubMed] [Google Scholar]
Porreco 2014a {published data only}
- Porreco RP. Reply. American Journal of Obstetrics and Gynecology 2014;211(6):712. [DOI] [PubMed] [Google Scholar]
Rabinowitz 2012 {published data only}
- Rabinowitz M, Gemelos G, Hill M, Demko Z, McAdoo S, Levy B. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and y. Fertility and Sterility 2012;98(3):S28‐S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rabinowitz 2012a {published data only}
- Rabinowitz M, Gemelos G, Banjevic M, Zimmermann B, Baner J, Levy B, et al. Non‐invasive detection of fetal aneuploidy across five chromosomes in the first trimester by targeted sequencing. Prenatal Diagnosis 2012;32:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Rabinowitz 2012b {published data only}
- Rabinowitz M, Gemelos G, Banjevic M, Zimmermann B, Baner J, Levy B, et al. Highly accurate first trimester non‐invasive detection of fetal aneuploidy of 13, 18, 21 and sex chromosomes by targeted sequencing. Human Reproduction 2012;27(Suppl 2):ii106‐ii108. [ISSN 0268‐1161] [Google Scholar]
Rabinowitz 2013 {published data only}
- Rabinowitz M, Hill M, Demko Z, McAdoo S, Zimmermann B, Levy B. Using targeted sequencing of SNPs to achieve a highly accurate non‐invasive detection of fetal aneuploidy of 13,18, 21 and sex chromosomes. American Journal of Obstetrics and Gynecology 2013;208(1):S253. [Google Scholar]
Rabinowitz 2014 {published data only}
- Rabinowitz M, Valenti E, Pettersen B, Sigurjonsson S, Hill M, Zimmermann B. Noninvasive aneuploidy detection by multiplexed amplification and sequencing of polymorphic loci. Obstetrics and Gynecology 2014;123 Suppl 1(5):167S‐S. [Google Scholar]
Rad 2014 {published data only}
- Rad S, Turner AL, Beauchamp S, Aghajanian P, Williams J 3rd, Esakoff TF. Noninvasive prenatal testing compared with invasive diagnostic testing in the setting of an abnormal state aneuploidy screen. Obstetrics and Gynecology 2014;123 Suppl 1:100s. [Google Scholar]
Radoi 2015 {published data only}
- Radoi VE, Bohiltea CL, Bohiltea RE, Albu DN. Cell free fetal DNA testing in maternal blood of Romanian pregnant women. Iranian Journal of Reproductive Medicine 2015;13(10):623‐6. [PUBMED: 26644790] [PMC free article] [PubMed] [Google Scholar]
Rava 2012 {published data only}
- Rava R, Bianchi D, Platt L, Goldberg J, Abuhamad A, Sehnert A. Genome wide fetal aneuploidy detection by sequencing of maternal plasma DNA: diagnostic accuracy in a prospective, blinded, multicenter study. American Journal of Obstetrics and Gynecology 2012;206(1):S367‐S. [Google Scholar]
Rava 2014 {published data only}
- Rava RP, Srinivasan A, Sehnert AJ, Bianchi DW. Circulating fetal cell‐free DNA fractions differ in autosomal aneuploidies and monosomy X. Clinical Chemistry 2014;60(1):243‐50. [DOI] [PubMed] [Google Scholar]
Reiff 2015 {published data only}
- Reiff E, Little S, Dobson L, Agarwala V, Wilkins‐Haug L, Bromley B. The role of first trimester ultrasound in prenatal aneuploidy screening for women with a negative cell free fetal DNA. Prenatal Diagnosis 2015;35:16‐7. [Google Scholar]
Reiff 2016 {published data only}
- Reiff ES, Little SE, Dobson L, Wilkins‐Haug L, Bromley B. What is the role of the 11‐ to 14‐week ultrasound in women with negative cell‐free DNA screening for aneuploidy?. Prenatal Diagnosis 2016;36(3):260‐5. [PUBMED: 26748490] [DOI] [PubMed] [Google Scholar]
Reimers 2015 {published data only}
- Reimers R, Mason‐Suares H, Little S, Reiff E, Bromley B, Dobson L, et al. What will be missed if Noninvasive Prenatal Testing (NIPT) is the only assessment for “Ultrasound Indicated” genetic studies?. Prenatal Diagnosis 2015;35:63‐4. [Google Scholar]
Revello 2016 {published data only}
- Revello R, Sarno L, Ispas A, Akolekar R, Nicolaides KH. Screening for trisomies by cell‐free DNA testing of maternal blood: consequences of a failed result. Ultrasound in Obstetrics & Gynecology 2016;47(6):698‐704. [PUBMED: 26743020] [DOI] [PubMed] [Google Scholar]
Ryan 2016 {published data only}
- Ryan A, Hunkapiller N, Banjevic M, Vankayalapati N, Fong N, Jinnett KN, et al. Validation of an enhanced version of a single‐nucleotide polymorphism‐based noninvasive prenatal test for detection of fetal aneuploidies. Fetal Diagnosis and Therapy 2016;40:219‐23. [PUBMED: 27028530] [DOI] [PubMed] [Google Scholar]
Sachse 2015 {published data only}
- Sachse M, Grömminger S, Schöck U, Bonnet J, Hofmann W. Development of a non‐invasive prenatal test (NIPT) assay for trisomy 21 (T21) based on comparative quantification of chromosome 21 and a reference chromosome via quantitative real‐time PCR. Medizinische Genetik 2015;27(1):177. [Google Scholar]
Samura 2015 {published data only}
- Samura O, Sago H, Sekizawa A. Nationwide project using next‐generation sequencing of cellfree DNA in maternal plasma in Japan: One and a half years of experience. Prenatal Diagnosis 2015;35:64. [DOI] [PubMed] [Google Scholar]
Sarno 2016 {published data only}
- Sarno L, Revello R, Hanson E, Akolekar R, Nicolaides KH. Prospective first‐trimester screening for trisomies by cell‐free DNA testing of maternal blood in twin pregnancy. Ultrasound in Obstetrics & Gynecology 2016;47(6):705‐11. [PUBMED: 26970114] [DOI] [PubMed] [Google Scholar]
Schöck 2015 {published data only}
- Schöck U, Grömminger S, Bonnet J, Sachse M, Hofmann W. Recent bioinformatic advances of non‐invasive prenatal detection to enhance diagnostic accuracy and aneuploidy discovery. Medizinische Genetik 2015;27(1):177. [Google Scholar]
Sehnert 2013 {published data only}
- Sehnert A, Futch T, Seltzer W, Bianchi D, Rava R. Clinical laboratory experience using noninvasive prenatal testing for fetal aneuploidy in late gestation. Prenatal Diagnosis 2013;33:80‐1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Sehnert 2014 {published data only}
- Sehnert A, Bianchi D, Rava R. Maternal cell‐free DNA (cfDNA) sequencing versus standard prenatal aneuploidy screening in a general obstetrical population. Prenatal Diagnosis 2014;34:8. [Google Scholar]
Sentilhes 2015 {published data only}
Seo 2015 {published data only}
- Seo DH, Cho DY, Kim J, Kim SY, Cho SE, Oh M. Clinical study of non‐invasive prenatal testing using next‐generation sequencing. Journal of Laboratory Medicine and Quality Assurance 2015;37:214‐8. [DOI: 10.15263/jlmqa.2015.37.4.214] [DOI] [Google Scholar]
Settler 2015 {published data only}
- Settler C, Dohany L. Clinical laboratory experience in a general obstetrical population of noninvasive prenatal screening for fetal aneuploidy from cell free DNA. Prenatal Diagnosis 2015;35:64‐5. [Google Scholar]
Shani 2016 {published data only}
- Shani H, Goldwaser T, Keating J, Klugman S. Chromosomal abnormalities not currently detected by cell‐free fetal DNA: a retrospective analysis at a single center. American Journal of Obstetrics and Gynecology 2016;214(6):729.e1‐729.e11. [PUBMED: 26721783] [DOI] [PubMed] [Google Scholar]
Shaohua 2012 {published data only}
- Shaohua T, Huanzheng L, Erle Z, Xueqing X. Noninvasive prenatal diagnosis for aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Prenatal Diagnosis 2012;32:79. [Google Scholar]
Sharma 2015 {published data only}
- Sharma P, Metcalfe A, Pastuck M, Laberge A‐M, Haidar H, Ravitsky V, et al. Women's understanding of non‐invasive prenatal testing based on cell free DNA versus first trimester combined screening. Prenatal Diagnosis 2015;35:106. [Google Scholar]
Shaw 2013 {published data only}
- Shaw SWS, Chen CY, Hsiao CH, Ren Y, Tian F, Tsai C, et al. Non‐invasive prenatal testing for whole fetal chromosomal aneuploidies: A multi‐center prospective cohort trial in Taiwan. Prenatal Diagnosis 2013;33:81. [DOI] [PubMed] [Google Scholar]
Shen 2016 {published data only}
- Shen J, Wen Z, Qin X, Shi Y. Noninvasive fetal trisomy detection by multiplexed semiconductor sequencing: a barcoding analysis strategy. Journal of Human Genetics 2016;61(3):247‐52. [PUBMED: 26657936] [DOI] [PubMed] [Google Scholar]
Shi 2015 {published data only}
- Shi X, Zhang Z, Cram DS, Liu C. Feasibility of noninvasive prenatal testing for common fetal aneuploidies in an early gestational window. Clinica Chimica Acta 2015;439:24‐8. [DOI] [PubMed] [Google Scholar]
Shulman 2014 {published data only}
- Shulman L, Dungan J, Ginsberg N. The use of noninvasive prenatal screening (NIPS) in the assessment of an abnormal fetal ultrasound. Prenatal Diagnosis 2014;34:62. [Google Scholar]
Sistermans 2015a {published data only}
Smith‐Bindman 2015 {published data only}
Song 2012 {published data only}
- Song K, Ashoor G, Syngelaki A, Wagner M, Birdir C, Struble C, et al. Clinical evaluation of a directed cfDNA analysis method for non‐invasive prenatal fetal trisomy detection. Prenatal Diagnosis 2012;32:16‐7. [Google Scholar]
Sparks 2012 {published data only}
- Sparks AB, Wang ET, Struble CA, Barrett W, Stokowski R, McBride C, et al. Selective analysis of cell‐free DNA in maternal blood for evaluation of fetal trisomy. Prenatal Diagnosis 2012;32(1):3‐9. [PUBMED: 22223233] [DOI] [PMC free article] [PubMed] [Google Scholar]
Srinivasan 2013 {published data only}
- Srinivasan A, Bianchi D, Liao W, Sehnert A, Rava R. Maternal plasma DNA sequencing: Effects of multiple gestation on aneuploidy detection and the relative cell‐free fetal DNA (cffDNA) per fetus. American Journal of Obstetrics and Gynecology 2013;208(1):S31. [Google Scholar]
Stokowski 2015 {published data only}
- Stokowski R, Wang E, White K, Batey A, Jacobsson B, Brar H, et al. Clinical performance of non‐invasive prenatal testing (NIPT) using targeted cell‐free DNA analysis in maternal plasma with microarrays or next generation sequencing (NGS) is consistent across multiple controlled clinical studies. Prenatal Diagnosis 2015;35(12):1243‐6. [PUBMED: 26332378] [DOI] [PMC free article] [PubMed] [Google Scholar]
Strah 2015 {published data only}
- Strah D, Ovniček P, Bernik J. Non‐invasive prenatal cell‐free fetal DNA testing for down syndrome and other chromosomal abnormalities [Neinvazivno predrojstveno testiranje prostih plodov DNA za downov sindrom in ostale kromosomske nepravilnosti]. Zdravniški Vestnik 2015;84(11):727‐33. [Google Scholar]
Straver 2014 {published data only}
- Straver R, Sistermans EA, Holstege H, Visser A, Oudejans CBM, Reinders MJT. WISECONDOR: Detection of fetal aberrations from shallow sequencing maternal plasma based on a within‐sample comparison scheme. Nucleic Acids Research 2014;42(5):e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Strom 2015 {published data only}
- Strom C. The positive predictive value of noninvasive prenatal screening (NIPS). Chromosome Research 2015;23(1):S23. [Google Scholar]
Stumm 2011 {published data only}
- Stumm M, Trunk N, Beck M, Entezami M, Becker R, Locherbach J, et al. Non‐invasive prenatal detection of chromosome aneuploidies using next‐generation sequencing: First steps towards clinical application. Medizinische Genetik 2011;23(1):86. [Google Scholar]
Stumm 2012 {published data only}
- Stumm M, Entezami M, Trunk N, Beck M, Locherbach J, Wegner RD, et al. Noninvasive prenatal detection of chromosomal aneuploidies using different next generation sequencing strategies and algorithms. Prenatal Diagnosis 2012;32(6):569‐77. [PUBMED: 22573401] [DOI] [PubMed] [Google Scholar]
Stumm 2012a {published data only}
- Stumm M, Entezami M, Haug K, Blank C, Wustemann M, Schulze B, et al. Non‐invasive prenatal detection of trisomy 21 using massively parallel sequencing: A collaborative study in Europe. Prenatal Diagnosis 2012;32:63‐4. [DOI] [PubMed] [Google Scholar]
Stumm 2013 {published data only}
- Stumm M, Entezami M, Haug K, Blank C, Wustemann C, Schulze B, et al. Diagnostic accuracy of PrenaTest® for non‐invasive prenatal detection of common autosomal aneuploidies. Medizinische Genetik 2013;25(1):171‐2. [DOI] [PubMed] [Google Scholar]
Stumm 2016 {published data only}
- Stumm M. Noninvasive prenatal tests from the viewpoint of human geneticists: Possibilities and limits in daily routine [Nichtinvasive pränatale Tests aus Sicht des Humangenetikers: Möglichkeiten und Grenzen in der täglichen Routine]. Der Gynäkologe 2016;49(6):429‐36. [Google Scholar]
Swanson 2012 {published data only}
- Swanson A, Coffeen C, Sehnert AJ. Non‐invasive prenatal testing for fetal aneuploidy by massively parallel DNA sequencing of maternal plasma: The future has arrived today. Laboratoriums Medizin 2012;36(5):269‐75. [Google Scholar]
Syngelaki 2014 {published data only}
- Syngelaki A, Pergament E, Homfray T, Akolekar R, Nicolaides KH. Replacing the combined test by cell‐free DNA testing in screening for trisomies 21, 18 and 13: impact on the diagnosis of other chromosomal abnormalities. Fetal Diagnosis and therapy 2014;35(3):174‐84. [PUBMED: 24525399] [DOI] [PubMed] [Google Scholar]
Tan 2016 {published data only}
- Tan Y, Gao Y, Lin G, Fu M, Li X, Yin X, et al. Noninvasive prenatal testing (NIPT) in twin pregnancies with treatment of assisted reproductive techniques (ART) in a single center. Prenatal Diagnosis 2016;36(7):672‐9. [PUBMED: 27150972] [DOI] [PubMed] [Google Scholar]
Taneja 2016 {published data only}
- Taneja PA, Snyder HL, Feo E, Kruglyak KM, Halks‐Miller M, Curnow KJ, et al. Noninvasive prenatal testing in the general obstetric population: clinical performance and counseling considerations in over 85 000 cases. Prenatal Diagnosis 2016;36(3):237‐43. [PUBMED: 26715197] [DOI] [PMC free article] [PubMed] [Google Scholar]
Taneja 2017 {published data only}
- Taneja PA, Prosen TL, Feo E, Kruglyak KM, Halks‐Miller M, Curnow KJ, et al. Fetal aneuploidy screening with cell‐free DNA in late gestation. Journal of Maternal‐Fetal & Neonatal Medicine 2017;30(3):338‐42. [PUBMED: 27124739] [DOI] [PubMed] [Google Scholar]
Tarrier 2015 {published data only}
- Tarrier B, Sekedat M, Mann T, Stoerker J, Bombard A. Whole genome maternal plasma DNA sequencing for fetal aneuploidy accurately detects autosomal and sex chromosome abnormalities: A validation study with independent fetal fraction observations. Prenatal Diagnosis 2015;35:65‐6. [Google Scholar]
Taylor 2014 {published data only}
- Taylor JB, Chock VY, Hudgins L. NIPT in a clinical setting: an analysis of uptake in the first months of clinical availability. Journal of Genetic Counseling 2014;23(1):72‐8. [DOI] [PubMed] [Google Scholar]
Togneri 2016 {published data only}
- Togneri F, Court S, Parks M, Clokie S, Hamilton S, Bibb N, et al. Noninvasive prenatal testing for fetal aneuploidy: The experience of an NHS regional genetics laboratory. BJOG: an international journal of obstetrics and gynaecology 2016;123:70. [Google Scholar]
Tong 2016 {published data only}
- Tong H, Jin Y, Xu Y, Zou B, Ye H, Wu H, et al. Prenatal diagnosis of trisomy 21, 18 and 13 by quantitative pyrosequencing of segmental duplications. Clinical Genetics 2016;90(5):451‐5. [PUBMED: 26948280] [DOI] [PubMed] [Google Scholar]
Valderramos 2016a {published data only}
- Valderramos SG, Rao R, Scibetta EW, Clark K, Krakow D, Silverman NS, et al. Clinical accuracy of abnormal cell‐free fetal DNA (cfDNA) results for the sex chromosomes. American Journal of Obstetrics and Gynecology 2016;214(1):S402‐3. [Google Scholar]
Valderramos 2016b {published data only}
- Valderramos SG, Rao R, Scibetta EW, Clark K, Krakow D, Silverman NS, et al. Clinical accuracy of abnormal autosomal cell‐free fetal DNA (cfDNA) screening. American Journal of Obstetrics and Gynecology 2016;214(1):S330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Valderramos 2016c {published data only}
- Valderramos SG, Rao RR, Scibetta EW, Silverman NS, Han CS, Platt LD. Cell‐free DNA screening in clinical practice: abnormal autosomal aneuploidy and microdeletion results. American Journal of Obstetrics and Gynecology 2016;215(5):626.e1‐626.e10. [PUBMED: 27371353] [DOI] [PMC free article] [PubMed] [Google Scholar]
van den Oever 2012a {published data only}
- Oever JM, Balkassmi S, Verweij EJ, Iterson M, Adama van Scheltema PN, Oepkes D, et al. Single molecule sequencing of free DNA from maternal plasma for noninvasive trisomy 21 detection. Clinical Chemistry 2012;58(4):699‐706. [PUBMED: 22278607] [DOI] [PubMed] [Google Scholar]
van den Oever 2012b {published data only}
- Oever JME, Balkassmi S, Verweij EJ, Iterson M, Scheltema PNA, Oepkes D, et al. Single molecule sequencing of free DNA from maternal plasma for non‐invasive trisomy 21 detection. Prenatal Diagnosis 2012;32:60‐1. [Google Scholar]
van den Oever 2013 {published data only}
- Oever JM, Balkassmi S, Johansson LF, Adama van Scheltema PN, Suijkerbuijk RF, Hoffer MJ, et al. Successful noninvasive trisomy 18 detection using single molecule sequencing. Clinical Chemistry 2013;59(4):705‐9. [PUBMED: 23315481] [DOI] [PubMed] [Google Scholar]
Van Opstal 2016 {published data only}
- Opstal D, Srebniak MI, Polak J, Vries F, Govaerts LC, Joosten M, et al. False negative NIPT results: risk figures for chromosomes 13, 18 and 21 based on chorionic villi results in 5967 cases and literature review. PLOS One 2016;11(1):e0146794. [PUBMED: 26771677] [DOI] [PMC free article] [PubMed] [Google Scholar]
Verweij 2013a {published data only}
- Verweij EJ, Jacobsson B, Scheltema PNA, Boer MA, Hoffer MJV, Hollemon D, et al. European Noninvasive Trisomy Evaluation (EU‐NITE) study: Multicenter prospective study for noninvasive fetal trisomy 21 testing. Prenatal Diagnosis 2013;33:83‐4. [DOI] [PubMed] [Google Scholar]
Wald 2015a {published data only}
- Wald NJ, Bestwick JP. Performance of antenatal reflex DNA screening for Down's syndrome. Journal of Medical Screening 2015;22(4):168‐74. [PUBMED: 25882667] [DOI] [PubMed] [Google Scholar]
Wald 2015b {published data only}
Wang 2012 {published data only}
- Wang SJ, Gao ZY, Lu YP, Li YL, You YQ, Zhang LW, et al. Value of detection of cell‐free fetal DNA in maternal plasma in the prenatal diagnosis of chromosomal abnormalities. Zhonghua Fu Chan Ke za Zhi 2012;47(11):808‐12. [PUBMED: 23302119] [PubMed] [Google Scholar]
Wang 2015b {published data only}
- Wang JC, Sahoo T, Schonberg S, Kopita KA, Ross L, Patek K, et al. Discordant noninvasive prenatal testing and cytogenetic results: a study of 109 consecutive cases. Genetics in Medicine 2015;17(3):234‐6. [PUBMED: 25101914] [DOI] [PubMed] [Google Scholar]
Wang 2015c {published data only}
- Wang S, Huang S, Ma L, Liang L, Zhang J, Zhang J, et al. Maternal X chromosome copy number variations are associated with discordant fetal sex chromosome aneuploidies detected by noninvasive prenatal testing. Clinica Chimica Acta; International Journal of Clinical Chemistry 2015;444:113‐6. [PUBMED: 25689220] [DOI] [PubMed] [Google Scholar]
Wang 2015d {published data only}
- Wang JC, Sahoo T, Schonberg S, Kopita KA, Ross L, Patek K, et al. Discordant noninvasive prenatal testing and cytogenetic results: A study of 109 consecutive cases. Obstetrical and Gynecological Survey 2015;70(7):434‐6. [DOI] [PubMed] [Google Scholar]
Wang 2015e {published data only}
- Wang E, Struble C, Kingsley C, Steeke R, Batey A, Hollemon D, et al. Importance of fetal fraction analysis for CFDNA testing in the general pregnancy population. Reproductive Sciences 2015;22:176A. [Google Scholar]
Xiong 2015 {published data only}
- Xiong Y, Berman J, Weiner S, Carre A, Seligman NS, Steinbach G, et al. Noninvasive prenatal testing (NIPT) registry‐patients' results and providers' perspective. Reproductive Sciences 2015;22:176A‐7A. [Google Scholar]
Yankova 2015 {published data only}
- Yankova M, Chaveeva P, Stratieva V. Models of clinical implementation of cell free fetal DNA in the maternal serum screening test‐analysis. Akusherstvo i Ginekologiia 2015;54(7):15‐21. [PUBMED: 27025103] [PubMed] [Google Scholar]
Yaron 2015 {published data only}
- Yaron Y, Jani J, Schmid M, Oepkes D. Current status of testing for microdeletion syndromes and rare autosomal trisomies using cell‐free DNA technology. Obstetrics and Gynecology 2015;126(5):1095‐9. [PUBMED: 26444108] [DOI] [PubMed] [Google Scholar]
Yeang 2014 {published data only}
- Yeang CH, Ma GC, Hsu HW, Lin YS, Chang SM, Cheng PJ, et al. Genome‐wide normalized score: a novel algorithm to detect fetal trisomy 21 during non‐invasive prenatal testing. Ultrasound in Obstetrics & Gynecology 2014;44(1):25‐30. [PUBMED: 24700679] [DOI] [PubMed] [Google Scholar]
Yu 2014 {published data only}
- Yu SC, Chan KC, Zheng YW, Jiang P, Liao GJ, Sun H, et al. Size‐based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proceedings of the National Academy of Sciences of the United States of America 2014;111(23):8583‐8. [PUBMED: 24843150] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yuan 2013 {published data only}
- Yuan Y, Jiang F, Hua S, Du B, Hao Y, Ye L, et al. Feasibility study of semiconductor sequencing for noninvasive prenatal detection of fetal aneuploidy. Clinical Chemistry 2013;59(5):846‐9. [PUBMED: 23364181] [DOI] [PubMed] [Google Scholar]
Zhang 2015 {published data only}
- Zhang H, Gao Y, Jiang F, Fu M, Yuan Y, Guo Y, et al. Non‐invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound in Obstetrics & Gynecology 2015;45(5):530‐8. [PUBMED: 25598039] [DOI] [PubMed] [Google Scholar]
Zhou 2013 {published data only}
- Zhou D, Liang D, Lv W, Tian F, Song Z, Zhang J, et al. Noninvasive prenatal testing in China: Results and learning from a clinical study of more than 50,000 pregnancies. Prenatal Diagnosis 2013;33:1. [Google Scholar]
Zimmermann 2012 {published data only}
- Zimmermann B, Hill M, Gemelos G, Demko Z, Banjevic M, Baner J, et al. Noninvasive prenatal aneuploidy testing of chromosomes 13, 18, 21, X, and Y, using targeted sequencing of polymorphic loci. Prenatal Diagnosis 2012;32(13):1233‐41. [PUBMED: 23108718] [DOI] [PMC free article] [PubMed] [Google Scholar]
Zimmermann 2013 {published data only}
- Zimmermann B, Banjevic M, Hill M, Lacroute P, Dodd M, Sigurjonsson S, et al. Highly multiplexed single‐nucleotide polymorphism amplification and sequencing to identify fetal aneuploidy from maternal cell‐free DNA. Prenatal Diagnosis 2013;33:68. [Google Scholar]
Zwiefelhofer 2013 {published data only}
- Zwiefelhofer T, Whitley P, Roy K, Jean‐Jacques R, Ehrich M. Prenatal detection of fetal aneuploidy using bench top sequencing. Prenatal Diagnosis 2013;33:85. [Google Scholar]
References to ongoing studies
Basaran 2015 {published and unpublished data}
- Basaran S, Yuksel A, Has R, Kirgiz M, Dehgan T, Karaman B. False positive and false negative results of cell free DNA testing. Chromosome Research 2015;23(1):S124‐S5. [Google Scholar]
Buresch 2016 {published data only}
- Buresch A, Rosner M, Suskin B, Einstein F, Bircaj E, Lister R, et al. Actual rates of recommended diagnostic testing after first trimester screening vs. same‐day screening by cell free DNA. American Journal of Obstetrics and Gynecology 2016;214(1):S326. [Google Scholar]
Chen 2011a {published data only}
- Chen F, Wang W, Shan D, Ren J, Xie J, Huang Y, et al. Noninvasive prenatal diagnosis of fetal aneuploidy by massively parallel sequencing of maternal plasma DNA. Journal of Perinatal Medicine 2011;39:299. [Google Scholar]
Da Fonseca 2015 {published data only}
- Fonseca JP, Khattabi L, Brun S, Chatron N, Gueguen P, Nectoux J, et al. Non‐Invasive prenatal testing for the most common aneuploidies (trisomies 21, 18, and 13) using a semiconductor‐sequencing platform: a French multicenter pilot study. Chromosome Research 2015;23(1):S110‐S1. [Google Scholar]
ISRCTN11174071 {unpublished data only}
- ISRCTN11174071. Comparison of false positive rates in prenatal combined screening and cell free DNA screening for trisomy 21. isrctn.com/ISRCTN11174071 Date first received: 26 July 2016. [DOI: 10.1186/ISRCTN11174071] [DOI]
Lin 2014 {published data only}
- Lin G, Gao Y, Yin X, Tan Y, Zhang H, Lu G, et al. Clinical implementation of noninvasive prenatal testing in twin pregnancies with assisted reproductive technique treatment. Prenatal Diagnosis 2014;34:14. [Google Scholar]
Mu 2014 {published data only}
- Mu Y, Sun F. Maternal non‐invasive fetal DNA test used in prenatal diagnosis. International Journal of Laboratory Hematology 2014;36:106‐7. [Google Scholar]
NCT01429389 {unpublished data only}
- NCT01429389. Specimen collection from pregnant women at increased risk for fetal aneuploidy. ClinicalTrials.gov/show/NCT01429389 Date first received: 2 September 2011.
NCT01472523 {unpublished data only}
- NCT01472523. A safer pre‐natal diagnosis using free DNA in maternal blood. ClinicalTrials.gov/show/NCT01472523 Date first received: 11 October 2011.
NCT01545674 {unpublished data only}
- NCT01545674. Prenatal non‐invasive aneuploidy test utilizing SNPs trial. ClinicalTrials.gov/show/NCT01545674 Date first received: 1 March 2012.
NCT01925742 {unpublished data only}
- NCT01925742. Study of the efficacy of new non‐invasive prenatal tests for screening for fetal trisomies using maternal blood. ClinicalTrials.gov/show/NCT01925742 Date first received: 16 August 2013.
NCT02201862 {unpublished data only}
- NCT02201862. Non‐invasive chromosomal evaluation of trisomy study. ClinicalTrials.gov/show/NCT02201862 Date first received: 22 July 2014.
NCT02278536 {unpublished data only}
- NCT02278536. Multiple gestation study. ClinicalTrials.gov/show/NCT02278536 Date first received: 28 October 2014.
NCT02278874 {unpublished data only}
- NCT02278874. High risk multiple gestation study. ClinicalTrials.gov/show/NCT02278874 Date first received: 27 August 2014.
NCT02317965 {unpublished data only}
- NCT02317965. Non‐invasive screening for fetal aneuploidy. ClinicalTrials.gov/show/NCT02317965 Date first received: 9 December 2014.
NCT02424474 {unpublished data only}
- NCT02424474. T21,18 and 13 screening by cell free fetal DNA in low risk patients. ClinicalTrials.gov/show/NCT02424474 Date first received: 8 April 2015.
NCT02787486 {unpublished data only}
- NCT02787486. Expanded noninvasive genomic medical assessment: the Enigma Study. ClinicalTrials.gov/show/NCT02787486 Date first received: 26 May 2016.
Sago 2015 {published data only}
- Sago H, Sekizawa A, and Japan NIPT consortium. Nationwide demonstration project of next‐generation sequencing of cell‐free DNA in maternal plasma in Japan: 1‐year experience. Prenatal Diagnosis 2015;35(4):331‐6. [DOI] [PubMed] [Google Scholar]
Sanchez‐Usabiaga 2015 {published data only}
- Sanchez‐Usabiaga RA, Aguinaga‐Rios M, Batista‐Espinoza A, Hurtado‐Amador R, Romero‐Tovar S. [Clinical implementation of non‐invasive prenatal study for detecting aneuploidies by fetal DNA based on single nucleotide polymorphisms: two years in Mexico] [Implementacion clinica del estudio prenatal no invasivo para Ia deteccion de aneuplodias mediante ADN fetal con base en polimorfismos de nucleotido unico: dos anos en Mexico]. Ginecologia y Obstetricia de Mexico 2015;83(4):220‐31. [PUBMED: 26727755] [PubMed] [Google Scholar]
Sistermans 2015 {published data only}
- Sistermans EA, Schuring‐Blom GH, Faas BHW, Boon EMJ, Bax CJ, Coumans ABC, et al. TRIDENT: or monitored NIPT implementation in the Netherlands. European Journal of Human Genetics 2015;23(Suppl 1):C01.2. [Google Scholar]
Torres 2015 {published data only}
- Torres Y, Suela J, Cigudosa JC. Genetic non invasive prenatal testing: A clinical and technical experience of 3.000 cases with follow‐up. Chromosome Research 2015;23(1):S140‐S141. [ISSN 09673849] [Google Scholar]
Van Wymersch 2015 {published data only}
- Wymersch D, Gilson G. Introduction of noninvasive prenatal testing for fetal trisomies: preliminary results and consequences on invasive samplings [Introduction du depistage sanguin des trisomies foetales par recherche d'ADN foetal circulant: Resultats preliminaires d'une annee et evolution des prelevements invasifs]. Bulletin de la Societe des Sciences Medicales du Grand‐Duche de Luxembourg 2015;1:65‐72. [PUBMED: 26946853] [PubMed] [Google Scholar]
Willems 2014 {published data only}
- Willems PJ, Dierickx H, Vandenakker E, Bekedam D, Segers N, Deboulle K, et al. The first 3,000 Non‐Invasive Prenatal Tests (NIPT) with the Harmony™ test in Belgium and the Netherlands. Facts, Views & Vision in ObGyn 2014;6(1):7‐12. [PUBMED: 25009720] [PMC free article] [PubMed] [Google Scholar]
Yu 2014a {published data only}
- Yu M, Fei S. Maternal non‐invasive fetal DNA test used in prenatal diagnosis. Clinical Chemistry and Laboratory Medicine 2014;52:S1570. [Google Scholar]
Zwiefelhofer 2014 {published data only}
- Zwiefelhofer T, Whitley P, Roy K, Saha M, Burcham T, Boom D, et al. Prenatal detection of fetal aneuploidy on the Ion Torrent Proton™ platform. Prenatal Diagnosis 2014;34:64‐5. [Google Scholar]
Additional references
ACOG #163 2016
- American College of Obstetricians and Gynecologists. Practice Bulletin No. 163 Summary: Screening for fetal aneuploidy. Obstetrics and Gynecology 2016;127(5):979‐81. [PUBMED: 27101120] [DOI] [PubMed] [Google Scholar]
ACOG #545 2012
- American College of Obstetricians and Gynecologists Committee on Genetics. Committee Opinion No. 545: Noninvasive prenatal testing for fetal aneuploidy. Obstetrics and Gynecology 2012;120(6):1532‐4. [PUBMED: 23168792] [DOI] [PubMed] [Google Scholar]
ACOG #682 2016
- American College of Obstetricians and Gynecologists and Society for Maternal‐Fetal Medicine. Committee Opinion No.682: Microarrays and next‐generation sequencing technology: The use of advanced genetic diagnostic tools in obstetrics and gynecology. Obstetrics and Gynecology 2016;128(6):e262‐8. [PUBMED: 27875474] [DOI] [PubMed] [Google Scholar]
ACOG #88 2007
- American College of Obstetricians and Gynecologists. ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstetrics and Gynecology 2007;110(6):1459‐67. [PUBMED: 18055749] [DOI] [PubMed] [Google Scholar]
Agarwal 2013
- Agarwal A, Sayres LC, Cho MK, Cook‐Deegan R, Chandrasekharan S. Commercial landscape of noninvasive prenatal testing in the United States. Prenatal Diagnosis 2013;33(6):521‐31. [PUBMED: 23686656] [DOI] [PMC free article] [PubMed] [Google Scholar]
Akolekar 2015
- Akolekar R, Beta J, Picciarelli G, Ogilvie C, D'Antonio F. Procedure‐related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta‐analysis. Ultrasound in Obstetrics & Gynecology 2015;45(1):16‐26. [PUBMED: 25042845] [DOI] [PubMed] [Google Scholar]
Alldred 2010
- Alldred SK, Deeks JJ, Neilson JP, Alfirevic Z. Antenatal screening for Down's syndrome: generic protocol. Cochrane Database of Systematic Reviews 2010, Issue 4. [DOI: 10.1002/14651858.CD007384.pub2; CD007384] [DOI] [Google Scholar]
Alldred 2012
- Alldred SK, Deeks JJ, Guo B, Neilson JP, Alfirevic Z. Second trimester serum tests for Down's Syndrome screening. Cochrane Database of Systematic Reviews 2012, Issue 6. [DOI: 10.1002/14651858.CD009925] [DOI] [PMC free article] [PubMed] [Google Scholar]
Alldred 2015a
- Alldred SK, Takwoingi Y, Guo B, Pennant M, Deeks JJ, Neilson JP, et al. First trimester serum tests for Down's syndrome screening. Cochrane Database of Systematic Reviews 2015, Issue 11. [DOI: 10.1002/14651858.CD011975; PUBMED: 26617074] [DOI] [PMC free article] [PubMed] [Google Scholar]
Alldred 2015b
- Alldred SK, Guo B, Takwoingi Y, Pennant M, Wisniewski S, Deeks JJ, et al. Urine tests for Down's syndrome screening. Cochrane Database of Systematic Reviews 2015, Issue 12. [DOI: 10.1002/14651858.CD011984; PUBMED: 26662198] [DOI] [PMC free article] [PubMed] [Google Scholar]
Alldred 2017a
- Alldred SK, Takwoingi Y, Guo B, Pennant M, Deeks JJ, Neilson JP, et al. First trimester ultrasound tests alone or in combination with first trimester serum tests for Down's syndrome screening. Cochrane Database of Systematic Reviews 2017, Issue 3. [DOI: 10.1002/14651858.CD012600; PUBMED: 28295158] [DOI] [PMC free article] [PubMed] [Google Scholar]
Alldred 2017b
- Alldred SK, Takwoingi Y, Guo B, Pennant M, Deeks JJ, Neilson JP, et al. First and second trimester serum tests with and without first trimester ultrasound tests for Down's syndrome screening. Cochrane Database of Systematic Reviews 2017, Issue 3. [DOI: 10.1002/14651858.CD012599; PUBMED: 28295159] [DOI] [PMC free article] [PubMed] [Google Scholar]
APA 2013
- American Psychiatric Association. Neurodevelopmental disorders. Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DMS‐5®). Arlington: American Psychiatric Publishing, 2013. [DOI: 10.1176/appi.books.9780890425596.514988] [DOI] [Google Scholar]
Ariosa Diagnostics 2016
- Ariosa Diagnostics, Inc, USA. www.ariosadx.fr/healthcare‐professionals/ (accessed 25 October 2016).
Attilakos 2011
- Attilakos G, Maddocks DG, Davies T, Hunt LP, Avent ND, Soothill PW, et al. Quantification of free fetal DNA in multiple pregnancies and relationship with chorionicity. Prenatal Diagnosis 2011;31(10):967‐72. [PUBMED: 21769896] [DOI] [PubMed] [Google Scholar]
Bardsley 2013
- Bardsley MZ, Kowal K, Levy C, Gosek A, Ayari N, Tartaglia N, et al. 47,XYY syndrome: clinical phenotype and timing of ascertainment. Journal of Pediatrics 2013;163(4):1085‐94. [PUBMED: 23810129] [DOI] [PMC free article] [PubMed] [Google Scholar]
Basset 2013
- Basset C, Beaufils F, Dreifuss‐Netter F, Droit RP, Gaudray P, Matuchansky C, et al. Avis N° 120: Questions éthiques associées au développement des tests génétiques foetaux sur sang maternel. Comité Consultatif National d’Ethique (CCNE) pour les sciences de la vie et de la santé, 2013:50 pages. [Google Scholar]
Benn 2011
- Benn P, Borrell A, Crossley J, Cuckle H, Dugoff L, Gross S, et al. Aneuploidy screening: a position statement from a committee on behalf of the Board of the International Society for Prenatal Diagnosis, January 2011. Prenatal Diagnosis 2011;31(6):519‐22. [PUBMED: 21604286] [DOI] [PubMed] [Google Scholar]
Benn 2013a
- Benn P, Borell A, Chiu R, Cuckle H, Dugoff L, Faas B, et al. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenatal Diagnosis 2013;33(7):622‐9. [PUBMED: 23616385] [DOI] [PubMed] [Google Scholar]
Benn 2013b
- Benn P, Cuckle H, Pergament E. Non‐invasive prenatal testing for aneuploidy: current status and future prospects. Ultrasound in Obstetrics & Gynecology 2013;42(1):15‐33. [PUBMED: 23765643] [DOI] [PubMed] [Google Scholar]
Benn 2016
- Benn P. Expanding non‐invasive prenatal testing beyond chromosomes 21, 18, 13, X and Y. Clinical Genetics 2016;90(6):477‐85. [PUBMED: 27283893] [DOI] [PubMed] [Google Scholar]
Berry Genomics 2016
- Berry Genomics Co. Ltd, China. www.berrygenomics.com/bambnitest/ (accessed 5 December 2016).
BGI 2014
- Bejing Genomics Institute, China. www.niftytest.com/plus/list.php?tid=14 (accessed 20 June 2014).
BGI 2016
- Bejing Genomics Institute, China. www.niftytest.com/healthcare‐providers/clinical‐data/ (accessed 25 October 2016).
Bianchi 2004
- Bianchi DW. Circulating fetal DNA: its origin and diagnostic potential‐a review. Placenta 2004;25 Suppl A:S93‐S101. [PUBMED: 15033315] [DOI] [PubMed] [Google Scholar]
Birch 2005
- Birch L, English CA, O'Donoghue K, Barigye O, Fisk NM, Keer JT. Accurate and robust quantification of circulating fetal and total DNA in maternal plasma from 5 to 41 weeks of gestation. Clinical Chemistry 2005;51(2):312‐20. [PUBMED: 15608152] [DOI] [PubMed] [Google Scholar]
Bussani 2011
- Bussani C, Tommaso M, Cioni R, Pasquini L, Quitadamo L, Scarselli G. Quantitative variation analysis of fetal DNA in maternal plasma samples collected before and after amniocentesis. Journal of Obstetrics and Gynaecology Research 2011;37(6):571‐4. [PUBMED: 21375672] [DOI] [PubMed] [Google Scholar]
Canick 2013
- Canick JA, Palomaki GE, Kloza EM, Lambert‐Messerlian GM, Haddow JE. The impact of maternal plasma DNA fetal fraction on next generation sequencing tests for common fetal aneuploidies. Prenatal Diagnosis 2013;33(7):667‐74. [PUBMED: 23592541] [DOI] [PubMed] [Google Scholar]
Cereda 2012
- Cereda A, Carey JC. The trisomy 18 syndrome. Orphanet Journal of Rare Diseases 2012;7:81. [PUBMED: 23088440] [DOI] [PMC free article] [PubMed] [Google Scholar]
Chen 2009
- Chen CP. Prenatal sonographic features of fetuses in trisomy 13 pregnancies (I). Taiwanese Journal of Obstetrics & Gynecology 2009;48(3):210‐7. [PUBMED: 19797008] [DOI] [PubMed] [Google Scholar]
Chetty 2013
- Chetty S, Garabedian MJ, Norton ME. Uptake of noninvasive prenatal testing (NIPT) in women following positive aneuploidy screening. Prenatal Diagnosis 2013;33(6):542‐6. [PUBMED: 23592525] [DOI] [PubMed] [Google Scholar]
Chitayat 2011
- Chitayat D, Langlois S, Wilson RD, Genetics Committee of the Society of Obstetricians and Gynaecologists of Canada, Prenatal Diagnosis Committee of the Canadian College of Medical Geneticists. Prenatal screening for fetal aneuploidy in singleton pregnancies. Journal of Obstetrics and Gynaecology Canada : JOGC 2011;33(7):736‐50. [PUBMED: 21749752] [DOI] [PubMed] [Google Scholar]
Choi 2012
- Choi H, Riper M, Thoyre S. Decision making following a prenatal diagnosis of Down syndrome: an integrative review. Journal of Midwifery & Women's Health 2012;57(2):156‐64. [PUBMED: 22432488] [DOI] [PubMed] [Google Scholar]
Christianson 2006
- Christianson A, Howson CP, Modell B. March of dimes. Global report on birth defects. The hidden toll of dying and disabled children. March of Dimes Birth Defects Foundation 2006:1‐76. [http://www.marchofdimes.com/glue/files/global‐report‐on‐birth‐defects‐the‐hidden‐toll‐of‐dying‐and‐disabled‐children‐full‐report.pdf]
Chu 2006
Cochrane Glossary 2014
- The Cochrane Collaboration’s Glossary of terms. community‐archive.cochrane.org/glossary (accessed 17 July 2014).
Deeks 2013
- Deeks JJ, Wisniewski S, Davenport C. Chapter 4: Guide to the contents of a Cochrane Diagnostic Test Accuracy Protocol. In: Deeks JJ, Bossuyt PM, Gatsonis C (editors), Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0.0. The Cochrane Collaboration, 2013. Available from: methods.cochrane.org/sdt/handbook‐dta‐reviews.
Devlin 2004
- Devlin L, Morrison PJ. Accuracy of the clinical diagnosis of Down syndrome. Ulster Medical Journal 2004;73(1):4‐12. [PUBMED: 15244118] [PMC free article] [PubMed] [Google Scholar]
Driscoll 2009
- Driscoll DA, Gross S. Clinical practice. Prenatal screening for aneuploidy. New England Journal of Medicine 2009;360(24):2556‐62. [PUBMED: 19516035] [DOI] [PubMed] [Google Scholar]
Duncan 2011
- Duncan A, Langlois S. Use of array genomic hybridization technology in prenatal diagnosis in Canada. Journal of Obstetrics and Gynaecology Canada 2011;33(12):1256‐9. [PUBMED: 22166281] [PubMed] [Google Scholar]
Fan 2010
- Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Analysis of the size distributions of fetal and maternal cell‐free DNA by paired‐end sequencing. Clinical Chemistry 2010;56(8):1279‐86. [PUBMED: 20558635] [DOI] [PubMed] [Google Scholar]
Fishler 1991
- Fishler K, Koch R. Mental development in Down syndrome mosaicism. American Journal of Mental Retardation 1991;96(3):345‐51. [PUBMED: 1836734] [PubMed] [Google Scholar]
Gagnon 2010
- Gagnon S, Labrecque M, Njoya M, Rousseau F, St‐Jacques S, Legare F. How much do family physicians involve pregnant women in decisions about prenatal screening for Down syndrome?. Prenatal Diagnosis 2010;30(2):115‐21. [PUBMED: 20013876] [DOI] [PubMed] [Google Scholar]
Gekas 2009
- Gekas J, Gagne G, Bujold E, Douillard D, Forest JC, Reinharz D, et al. Comparison of different strategies in prenatal screening for Down's syndrome: cost effectiveness analysis of computer simulation. BMJ (Clinical Research Ed.) 2009;338:b138. [PUBMED: 19218323] [DOI] [PMC free article] [PubMed] [Google Scholar]
Genesupport 2016
- Genesupport, Switzerland. www.prendia.ch/fileadmin/user_upload/pdf/Tableau_Anomalies‐Numeriques.pdf (accessed 25 October 2016).
Genoma 2016
- Genoma, Switzerland. genoma.com/product_page.php?id=2 (accessed 10 November 2016).
Genome Care 2016
- Genome Care, Korea. http://genomecare.net/en/ (accessed 5 December 2016).
Gil 2015a
- Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell‐free DNA in maternal blood in screening for fetal aneuploidies: updated meta‐analysis. Ultrasound in Obstetrics & Gynecology 2015;45(3):249‐66. [PUBMED: 25639627] [DOI] [PubMed] [Google Scholar]
Groth 2013
- Groth KA, Skakkebaek A, Host C, Gravholt CH, Bojesen A. Clinical review: Klinefelter syndrome‐‐a clinical update. Journal of Clinical Endocrinology and Metabolism 2013;98(1):20‐30. [PUBMED: 23118429] [DOI] [PubMed] [Google Scholar]
HAS 2015
- Haute Autorité de Santé. Screening test accuracy for fetal trisomy 21 with circulating cell‐free DNA. [Les performances des tests de dépistage de la trisomie 21 foetale par analyse de l’ADN libre circulant]. Collège de la Haute Autorité de Santé 2015.
ICFMM 2013
- Anonymous. Position Statement from the Italian College of Fetal Maternal Medicine: Non‐invasive prenatal testing (NIPT) by maternal plasma DNA sequencing. Journal of Prenatal Medicine 2013;7(2):19‐20. [PUBMED: 23991274] [PMC free article] [PubMed] [Google Scholar]
Illumina 2014
- Verinata Health, Inc. www.verifitest.com/clinical‐data/ (accessed 3 March 2014).
Illumina 2016
- Illumina, Inc, USA. www.illumina.com/clinical/reproductive‐genetic‐health/nipt.html (accessed 25 October 2016).
Irving 2011
- Irving C, Richmond S, Wren C, Longster C, Embleton ND. Changes in fetal prevalence and outcome for trisomies 13 and 18: a population‐based study over 23 years. Journal of Maternal‐Fetal & Neonatal Medicine 2011;24(1):137‐41. [PUBMED: 20384468] [DOI] [PubMed] [Google Scholar]
Irving 2012
- Irving CA, Chaudhari MP. Cardiovascular abnormalities in Down's syndrome: spectrum, management and survival over 22 years. Archives of Disease in Childhood 2012;97(4):326‐30. [PUBMED: 21835834] [DOI] [PubMed] [Google Scholar]
Karnis 2012
- Karnis MF. Fertility, pregnancy, and medical management of Turner syndrome in the reproductive years. Fertility and Sterility 2012;98(4):787‐91. [PUBMED: 23020910] [DOI] [PubMed] [Google Scholar]
Kazerouni 2011
- Kazerouni NN, Currier RJ, Flessel M, Goldman S, Hennigan C, Hodgkinson C, et al. Detection rate of quadruple‐marker screening determined by clinical follow‐up and registry data in the statewide California program, July 2007 to February 2009. Prenatal Diagnosis 2011;31(9):901‐6. [PUBMED: 21706514] [DOI] [PubMed] [Google Scholar]
LabGenomics 2016
- LabGenomics Clinical Research Institute, Korea. www.momguard.com/eng/home.php?go=main (accessed 5 December 2016).
Langlois 2011
- Langlois S, Duncan A. Use of a DNA method, QF‐PCR, in the prenatal diagnosis of fetal aneuploidies. Journal of Obstetrics and Gynaecology Canada 2011;33(9):955‐60. [PUBMED: 21923994] [DOI] [PubMed] [Google Scholar]
Larion 2014
- Larion S, Warsof SL, Romary L, Mlynarczyk M, Peleg D, Abuhamad AZ. Association of combined first‐trimester screen and noninvasive prenatal testing on diagnostic procedures. Obstetrics and Gynecology 2014;123(6):1303‐10. [PUBMED: 24807333] [DOI] [PubMed] [Google Scholar]
Legare 2010
- Legare F, Kearing S, Clay K, Gagnon S, D'Amours D, Rousseau M, et al. Are you SURE?: Assessing patient decisional conflict with a 4‐item screening test. Canadian Family Physician Medecin de Famille Canadien 2010;56(8):e308‐14. [PUBMED: 20705870] [PMC free article] [PubMed] [Google Scholar]
Legare 2011
- Legare F, St‐Jacques S, Gagnon S, Njoya M, Brisson M, Fremont P, et al. Prenatal screening for Down syndrome: a survey of willingness in women and family physicians to engage in shared decision‐making. Prenatal Diagnosis 2011;31(4):319‐26. [PUBMED: 21268046] [DOI] [PubMed] [Google Scholar]
Leggett 2010
- Leggett V, Jacobs P, Nation K, Scerif G, Bishop DV. Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XYY, or XXY: a systematic review. Developmental Medicine and Child Neurology 2010;52(2):119‐29. [PUBMED: 20059514] [DOI] [PMC free article] [PubMed] [Google Scholar]
LifeCodexx 2016
- LifeCodexx AG, Germany. lifecodexx.com/en/for‐physicians/methods‐technology/clinical‐validation/ (accessed 10 November 2016).
Lo 1997
- Lo YM, Corbetta N, Chamberlain PF, Rai V, Sargent IL, Redman CW, et al. Presence of fetal DNA in maternal plasma and serum. Lancet 1997;350(9076):485‐7. [PUBMED: 9274585] [DOI] [PubMed] [Google Scholar]
Lo 1999
- Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. American Journal of Human Genetics 1999;64(1):218‐24. [PUBMED: 9915961] [DOI] [PMC free article] [PubMed] [Google Scholar]
Lun 2008
- Lun FM, Chiu RW, Allen Chan KC, Yeung Leung T, Kin Lau T, Dennis Lo YM. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clinical Chemistry 2008;54(10):1664‐72. [PUBMED: 18703764] [DOI] [PubMed] [Google Scholar]
Macaskill 2010
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: Analysing and presenting results. In: Deeks JJ, Bossuyt PM, Gatsonis C, (editors), Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0. The Cochrane Collaboration 2010. Available from: methods.cochrane.org/sdt/handbook‐dta‐reviews.
Mackie 2017
- Mackie FL, Hemming K, Allen S, Morris RK, Kilby MD. The accuracy of cell‐free fetal DNA‐based non‐invasive prenatal testing in singleton pregnancies: a systematic review and bivariate meta‐analysis. BJOG: an international journal of obstetrics and gynaecology 2017;124(1):32‐46. [PUBMED: 27245374] [DOI] [PubMed] [Google Scholar]
Mazzanti 1998
- Mazzanti L, Cacciari E. Congenital heart disease in patients with Turner's syndrome. Italian Study Group for Turner Syndrome (ISGTS). Journal of Pediatrics 1998;133(5):688‐92. [PUBMED: 9821430] [DOI] [PubMed] [Google Scholar]
MedlinePlus 2014
- Medical Encyclopedia, A service of the U.S. National Library of Medicine, National Institutes of Health. medlineplus.gov/mplusdictionary.html (updated daily, accessed 17 July 2014).
Mersy 2013
- Mersy E, Smits LJ, Winden LA, Die‐Smulders CE, Paulussen AD, Macville MV, et al. Noninvasive detection of fetal trisomy 21: systematic review and report of quality and outcomes of diagnostic accuracy studies performed between 1997 and 2012. Human Reproduction Update 2013;19(4):318‐29. [PUBMED: 23396607] [DOI] [PubMed] [Google Scholar]
Modi 2003
- Modi D, Berde P, Bhartiya D. Down syndrome: a study of chromosomal mosaicism. Reproductive Biomedicine Online 2003;6(4):499‐503. [PUBMED: 12831601] [DOI] [PubMed] [Google Scholar]
Natera 2016
- Natera, Inc, USA. www.natera.com/panorama‐test/clinical‐information (accessed 25 October 2016).
Newcombe 1998
- Newcombe RG. Interval estimation for the difference between independent proportions: comparison of eleven methods. Statistics in Medicine 1998;17(8):873‐90. [PUBMED: 9595617] [DOI] [PubMed] [Google Scholar]
Nshimyumukiza 2017
O'Leary 2013
- O'Leary P, Maxwell S, Murch A, Hendrie D. Prenatal screening for Down syndrome in Australia: costs and benefits of current and novel screening strategies. Australian & New Zealand Journal of Obstetrics & Gynaecology 2013;53(5):425‐33. [PUBMED: 24090461] [DOI] [PubMed] [Google Scholar]
Okun 2008
- Okun N, Summers AM, Hoffman B, Huang T, Winsor E, Chitayat D, et al. Prospective experience with integrated prenatal screening and first trimester combined screening for trisomy 21 in a large Canadian urban center. Prenatal Diagnosis 2008;28(11):987‐92. [PUBMED: 18925623] [DOI] [PubMed] [Google Scholar]
Papageorgiou 2012
- Papageorgiou EA, Patsalis PC. Non‐invasive prenatal diagnosis of aneuploidies: new technologies and clinical applications. Genome Medicine 2012;4(5):46. [PUBMED: 22640877] [DOI] [PMC free article] [PubMed] [Google Scholar]
Parker 2010
- Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, Meyer RE, et al. Updated National Birth Prevalence estimates for selected birth defects in the United States, 2004‐2006. Birth Defects Research. Part A, Clinical and Molecular Teratology 2010;88(12):1008‐16. [PUBMED: 20878909] [DOI] [PubMed] [Google Scholar]
Premaitha Health plc 2016
- Premaitha Health public limited company, UK. (accessed 15 November 2016).
Renshaw 2013
- Renshaw R, Ellis K, Jacobs P, Morris J. Antenatal screening for Down syndrome: a quantitative demonstration of the improvements over the past 20 years. Journal of Health Services Research & Policy 2013;18(4):195‐201. [PUBMED: 23864125] [DOI] [PubMed] [Google Scholar]
Rothberg 2011
- Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, et al. An integrated semiconductor device enabling non‐optical genome sequencing. Nature 2011;475(7356):348‐52. [PUBMED: 21776081] [DOI] [PubMed] [Google Scholar]
Rutter 2001
- Rutter CM, Gatsonis CA. A hierarchical regression approach to meta‐analysis of diagnostic test accuracy evaluations. Statistics in Medicine 2001;20(19):2865‐84. [PUBMED: 11568945] [DOI] [PubMed] [Google Scholar]
Saenger 1996
- Saenger P. Turner's syndrome. New England Journal of Medicine 1996;335(23):1749‐54. [PUBMED: 8929268] [DOI] [PubMed] [Google Scholar]
Samura 2003
- Samura O, Miharu N, Hyodo M, Honda H, Ohashi Y, Honda N, et al. Cell‐free fetal DNA in maternal circulation after amniocentesis. Clinical Chemistry 2003;49(7):1193‐5. [PUBMED: 12816922] [DOI] [PubMed] [Google Scholar]
Schoemaker 2008
- Schoemaker MJ, Swerdlow AJ, Higgins CD, Wright AF, Jacobs PA. Mortality in women with turner syndrome in Great Britain: a national cohort study. Journal of Clinical Endocrinology and Metabolism 2008;93(12):4735‐42. [PUBMED: 18812477] [DOI] [PubMed] [Google Scholar]
Sequenom 2016
- Sequenom, Inc. www.sequenom.com/tests/reproductive‐health/maternit21‐plus#provider‐references and www.sequenom.com/tests/reproductive‐health/visibilit#provider‐test‐details (accessed 25 October 2016).
Snijders 1999
- Snijders RJ, Sundberg K, Holzgreve W, Henry G, Nicolaides KH. Maternal age‐ and gestation‐specific risk for trisomy 21. Ultrasound in Obstetrics & Gynecology 1999;13(3):167‐70. [PUBMED: 10204206] [DOI] [PubMed] [Google Scholar]
South 2013
- South ST, Lee C, Lamb AN, Higgins AW, Kearney HM. ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013. Genetics in Medicine 2013;15(11):901‐9. [PUBMED: 24071793] [DOI] [PubMed] [Google Scholar]
St‐Jacques 2008
- St‐Jacques S, Grenier S, Charland M, Forest JC, Rousseau F, Legare F. Decisional needs assessment regarding Down syndrome prenatal testing: a systematic review of the perceptions of women, their partners and health professionals. Prenatal Diagnosis 2008;28(13):1183‐203. [PUBMED: 19097031] [DOI] [PubMed] [Google Scholar]
Stochholm 2006
- Stochholm K, Juul S, Juel K, Naeraa RW, Gravholt CH. Prevalence, incidence, diagnostic delay, and mortality in Turner syndrome. Journal of Clinical Endocrinology and Metabolism 2006;91(10):3897‐902. [PUBMED: 16849410] [DOI] [PubMed] [Google Scholar]
Stochholm 2010a
- Stochholm K, Juul S, Gravholt CH. Diagnosis and mortality in 47,XYY persons: a registry study. Orphanet Journal of Rare Diseases 2010;5:15. [PUBMED: 20509956] [DOI] [PMC free article] [PubMed] [Google Scholar]
Stochholm 2010b
- Stochholm K, Juul S, Gravholt CH. Mortality and incidence in women with 47,XXX and variants. American Journal of Medical Genetics. Part a 2010;152A(2):367‐72. [PUBMED: 20101696] [DOI] [PubMed] [Google Scholar]
Summers 2007
- Summers AM, Langlois S, Wyatt P, Wilson RD, Society of Obstetricians and Gynaecologists of Canada. Prenatal screening for fetal aneuploidy. Journal of Obstetrics and Gynaecology Canada : JOGC 2007;29(2):146‐79. [PUBMED: 17346485] [DOI] [PubMed] [Google Scholar]
Sybert 2004
- Sybert VP, McCauley E. Turner's syndrome. New England Journal of Medicine 2004;351(12):1227‐38. [PUBMED: 15371580] [DOI] [PubMed] [Google Scholar]
Takwoingi 2015
- Takwoingi Y, Guo B, Riley RD, Deeks JJ. Performance of methods for meta‐analysis of diagnostic test accuracy with few studies or sparse data. Statistical Methods in Medical Research 2015;26(4):1896‐911. [PUBMED: 26116616] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tartaglia 2010
- Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L. A review of trisomy X (47,XXX). Orphanet Journal of Rare Diseases 2010;5:8. [PUBMED: 20459843] [DOI] [PMC free article] [PubMed] [Google Scholar]
Taylor‐Phillips 2016
- Taylor‐Phillips S, Freeman K, Geppert J, Agbebiyi A, Uthman OA, Madan J, et al. Accuracy of non‐invasive prenatal testing using cell‐free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta‐analysis. BMJ Open 2016;6(1):e010002. [PUBMED: 26781507] [DOI] [PMC free article] [PubMed] [Google Scholar]
Tiller 2014
- Tiller GE, Kershberg HB, Goff J, Coffeen C, Liao W, Sehnert AJ. Women's views and the impact of noninvasive prenatal testing on procedures in a managed care setting. Prenatal Diagnosis 2014;34:1‐6. [PUBMED: 25201151] [DOI] [PubMed] [Google Scholar]
Tyler 2004
- Tyler C, Edman JC. Down syndrome, Turner syndrome, and Klinefelter syndrome: primary care throughout the life span. Primary Care 2004;31(3):627‐48, x‐xi. [PUBMED: 15331252] [DOI] [PubMed] [Google Scholar]
Uematsu 2002
- Uematsu A, Yorifuji T, Muroi J, Kawai M, Mamada M, Kaji M, et al. Parental origin of normal X chromosomes in Turner syndrome patients with various karyotypes: implications for the mechanism leading to generation of a 45,X karyotype. American Journal of Medical Genetics 2002;111(2):134‐9. [PUBMED: 12210339] [DOI] [PubMed] [Google Scholar]
UK Screening Glossary 2012
- United Kingdom National Screening Committee Glossary. webarchive.nationalarchives.gov.uk/20150408175925/http://www.screening.nhs.uk/glossary (Last reviewed June 2012, accessed 18 November 2013).
Van Riper 2001
- Riper M, Cohen WI. Caring for children with Down syndrome and their families. Journal of Pediatric Health Care 2001;15(3):123‐31. [PUBMED: 11353361] [DOI] [PubMed] [Google Scholar]
Vora 2010
- Vora NL, Johnson KL, Peter I, Tighiouart H, Ralston SJ, Craigo SD, et al. Circulating cell‐free DNA levels increase variably following chorionic villus sampling. Prenatal Diagnosis 2010;30(4):325‐8. [PUBMED: 20127897] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wald 2005
- Wald NJ, Rodeck C, Hackshaw AK, Rudnicka A. SURUSS in perspective. Seminars in Perinatology 2005;29(4):225‐35. [PUBMED: 16104673] [DOI] [PubMed] [Google Scholar]
Weijerman 2010
- Weijerman ME, Winter JP. Clinical practice. The care of children with Down syndrome. European Journal of Pediatrics 2010;169(12):1445‐52. [PUBMED: 20632187] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wellesley 2012
- Wellesley D, Dolk H, Boyd PA, Greenlees R, Haeusler M, Nelen V, et al. Rare chromosome abnormalities, prevalence and prenatal diagnosis rates from population‐based congenital anomaly registers in Europe. European Journal of Human Genetics 2012;20(5):521‐6. [PUBMED: 22234154] [DOI] [PMC free article] [PubMed] [Google Scholar]
Whiting 2011
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529‐36. [PUBMED: 22007046] [DOI] [PubMed] [Google Scholar]
Wilson 2007
- Wilson RD, Langlois S, Johnson JA. Mid‐trimester amniocentesis fetal loss rate. Journal of Obstetrics and Gynaecology Canada : JOGC 2007;29(7):586‐95. [PUBMED: 17623573] [DOI] [PubMed] [Google Scholar]
Wright 2009
- Wright C. Cell‐free fetal nucleic acids for non‐invasive prenatal diagnosis, Report of the UK expert working group. PHG foundation (2009).
Wu 2013a
- Wu J, Springett A, Morris JK. Survival of trisomy 18 (Edwards syndrome) and trisomy 13 (Patau Syndrome) in England and Wales: 2004‐2011. American Journal of Medical Genetics. Part a 2013;161(10):2512‐8. [PUBMED: 23949924] [DOI] [PubMed] [Google Scholar]
Wu 2013b
- Wu J, Morris JK. The population prevalence of Down's syndrome in England and Wales in 2011. European Journal of Human Genetics 2013;21(9):1016‐9. [PUBMED: 23321618] [DOI] [PMC free article] [PubMed] [Google Scholar]
Yaron 2016
- Yaron Y. The implications of non‐invasive prenatal testing failures: a review of an under‐discussed phenomenon. Prenatal Diagnosis 2016;36(5):391‐6. [PUBMED: 26941176] [DOI] [PubMed] [Google Scholar]
Yu 2013
- Yu SC, Lee SW, Jiang P, Leung TY, Chan KC, Chiu RW, et al. High‐resolution profiling of fetal DNA clearance from maternal plasma by massively parallel sequencing. Clinical Chemistry 2013;59(8):1228‐37. [PUBMED: 23603797] [DOI] [PubMed] [Google Scholar]
Zhu 2013
- Zhu JL, Hasle H, Correa A, Schendel D, Friedman JM, Olsen J, et al. Survival among people with Down syndrome: a nationwide population‐based study in Denmark. Genetics in Medicine 2013;15(1):64‐9. [PUBMED: 22878506] [DOI] [PMC free article] [PubMed] [Google Scholar]
References to other published versions of this review
Badeau 2015
- Badeau M, Lindsay C, Blais J, Takwoingi Y, Langlois S, Légaré F, et al. Genomics‐based non‐invasive prenatal testing for detection of fetal chromosomal aneuploidy in pregnant women. Cochrane Database of Systematic Reviews 2015, Issue 7. [DOI: 10.1002/14651858.CD011767] [DOI] [PMC free article] [PubMed] [Google Scholar]


