Abstract
We have characterized transcripts for three potassium channel homologs in the AKT/KAT subfamily (Shaker type) from the common ice plant (Mesembryanthemum crystallinum), with a focus on their expression during salt stress (up to 500 mm NaCl). Mkt1 and 2, Arabidopsis AKT homologs, and Kmt1, a KAT homolog, are members of small gene families with two to three isoforms each. Mkt1 is root specific; Mkt2 is found in leaves, flowers, and seed capsules; and Kmt1 is expressed in leaves and seed capsules. Mkt1 is present in all cells of the root, and in leaves a highly conserved isoform is detected present in all cells with highest abundance in the vasculature. MKT1 for which antibodies were made is localized to the plasma membrane. Following salt stress, MKT1 (transcripts and protein) is drastically down-regulated, Mkt2 transcripts do not change significantly, and Kmt1 is strongly and transiently (maximum at 6 h) up-regulated in leaves and stems. The detection and stress-dependent behavior of abundant transcripts representing subfamilies of potassium channels provides information about tissue specificity and the complex regulation of genes encoding potassium uptake systems in a halophytic plant.
Potassium, the most abundant cation in plant cells, plays essential roles in maintaining the membrane potential, ion homeostasis, in enzyme activation, signal transduction, and many other physiological processes. The molecular mechanism of potassium uptake by plant roots, loading, and transport within plants has been a focus of study during the last decade. Following the isolation of plant potassium channels by yeast (Saccharomyces cerevisiae) complementation (Anderson et al., 1992; Sentenac et al., 1992), several proteins capable of transporting potassium have been reported in Arabidopsis, potato (Solanum tuberosum), barley (Hordeum vulgare), wheat (Triticum aestivum), and a few other species. These membrane proteins include inwardly rectifying channels (IRC) and two types of carriers: those in the HKT and HAK/KUP families, respectively (Amtmann and Sanders, 1999; Chrispeels et al., 1999; Czempinski et al., 1999; Zimmermann and Sentenac, 1999). Here, we report the characterization of transcripts encoding potassium channel homologs in the Shaker-type subfamily from a halophytic plant, the common ice plant (Mesembryanthemum crystallinum). We focus on transcript behavior, comparing plants grown under control conditions with plants stressed by high sodium chloride.
As in animal systems, three families of plant potassium channels are known (Zimmermann and Sentenac, 1999). They contain a characteristic pore-forming (P) domain conferring ion selectivity, but they differ in the number of transmembrane (TM) and P domains. A nomenclature has become established by which two subfamilies of plant Shaker-type channels became known as members of either the AKT or KAT family (Zimmermann and Sentenac, 1999). All AKT- and KAT-type channels consist of six TM regions with one P region, but their electrophysiological features vary, as well as the regulation of channel activities (Marten et al., 1999; Hoth and Hedrich, 1999; for review, see Zimmermann and Sentenac, 1999). In contrast with the outward-rectifying animal Shaker channels (ORC), the functionally characterized KAT1 and AKT1 channels in Arabidopsis and their homologs in other species are IRC (Anderson et al., 1992; Sentenac et al., 1992). AKT differ from the KAT type by the presence of carboxy-terminal ankyrin repeat domains, possibly for anchoring to the cytoskeleton. So far, all potassium channels show high specificity for K+ over other alkali cations, making unlikely candidates for significant inadvertent sodium intrusion even at high Na+ to K+ ratios (Maathuis et al., 1997; Amtmann and Sanders, 1999).
Mainly expressed in guard cells, KAT1 constitutes a path for potassium influx during stomatal opening. KAT1 from Arabidopsis and KST1 from potato are activated by extracellular acidification (Mueller-Roeber et al., 1995; Very et al., 1995), initiated by increased activity of the plasma membrane H+-ATPase. Although earlier hypotheses, based on physiological observations, had assumed a distinction between channels as low-affinity transporters and carriers as high-affinity transporters, a more complex picture emerges at present. For example, KAT1, when expressed in Arabidopsis guard cells or yeast, mediates K+ uptake from media with as low as 10 μm of external K+ (Brüggemann et al., 1999). The expression of a second family member, KAT2, has been detected in Arabidopsis leaf mesophyll cells (Butt et al., 1997).
At high abundance, AKT1 is predominantly expressed in Arabidopsis roots (Cao et al., 1995; Lagarde et al., 1996). Mutant plants with a T-DNA insertion in Akt1 grow poorly on media with potassium concentrations in the micromolar range in comparison with wild type (Hirsch et al., 1998; Spalding et at., 1999), suggesting that AKT1-type channels can function in the high-affinity range. The expression of other family members, AKT2, has been located most strongly to the leaves (Cao et al., 1995), and AKT3 to leaf phloem (Marten et al., 1999). The latter seems to be responsible for phloem transport of potassium.
Here we report the characterization of three potassium channel transcripts, Mkt1, Mkt2, and Kmt1, from the halophyte common ice plant, which are homologs of the Arabidopsis IRC Akt1, Akt2/3, and Kat1. We analyzed tissue specificity of these genes at the transcript level, their regulated expression under salt stress, and protein amounts for MKT1.
RESULTS
Potassium Channel Transcript Isolation
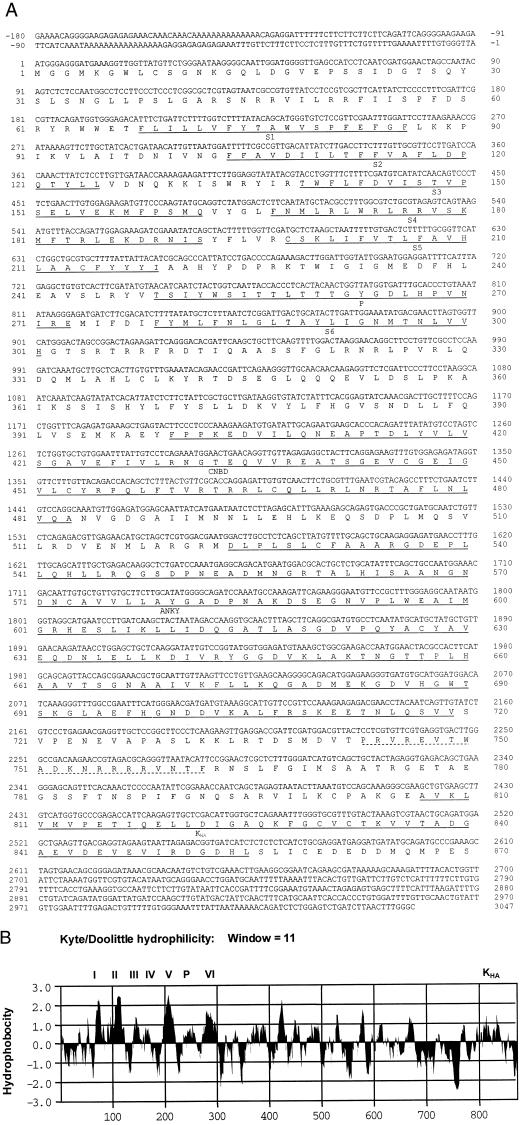
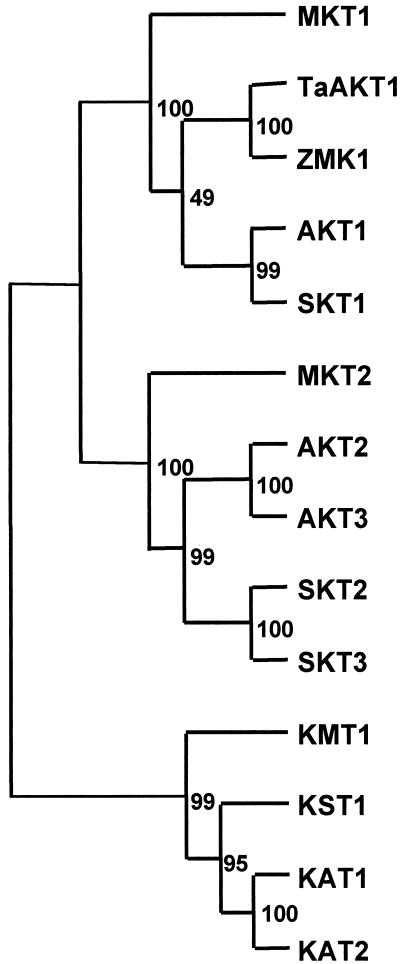
Three members of the AKT/KAT subfamily of inward-rectifying potassium channels were isolated from the common ice plant by a combination of reverse transcription (RT)-PCR amplification with degenerate primers and cDNA library screening (Figs. 1 and 2). Partial cDNAs of Mkt1 were obtained by RT-PCR from degenerate primers and 5′-RACE amplifications, and full-length cDNAs were isolated from cDNA libraries. The deduced amino acid sequence of the ice plant Akt1 homolog, Mkt1 (accession no. AF267753), specifies 870 residues (Fig. 1A). Partial cDNAs for Mkt2 and Kmt1 were isolated from total RNA by RT-PCR amplification with degenerate primers (Mkt2, accession no. AF267755; and Kmt1, accession no. AF267754). All RT-PCR products and cDNAs were sequenced on both strands. Hydropathy plots of MKT1 show the putative membrane topology of this channel, and additional domains are indicated (Fig. 1B). The three ice plant sequences show similar hydrophobicity profiles with respect to TM regions. The membrane-embedded P region between TM5 and TM6 and the voltage-sensing region in TM4 are conserved characteristics of these channels (Fig. 1). The deduced proteins of the three sequences aligned with other AKT/KAT-type proteins as members of the subfamily characterized by six TM domains. The phylogenetic analysis (Fig. 2) places the ice plant sequences among the Arabidopsis, potato, and grass Shaker-type IRC.
Figure 1.
Characterization of the ice plant Mkt1 potassium channel transcript. A, DNA and deduced protein sequence of the MKT1 potassium channels from the common ice plant (accession no. AF267753). Putative functional domains are underlined. They are labeled S1 through S6: TM regions (S4 is also the voltage sensor); P, the pore-forming domain; CNBD, cyclic nucleotide-binding domain; ANKY, ankyrin repeats (underlined); and KHA, a conserved domain rich in hydrophobic and acidic residues is located at the carboxy terminal end (underlined). A region used for the generation of oligopeptide-directed antibodies is indicated by a dotted line. B, Hydrophobicity plot of MKT1 protein according to Kyte and Doolittle (1982; MacVector 6.5, Oxford Molecular Ltd., Oxford). Putative TM domains (I–VI), the pore-forming domain (P), and the KHA domain are indicated.
Figure 2.
Phylogenetic comparison of ice plant MKT1, MKT2, and KMT1 deduced amino acid sequences with sequences of other plant potassium channel proteins. The phylogenetic tree, including bootstrap values, was constructed using Clustal W AKT1,2,3 and KAT1,2—Arabidopsis; SKT1,2,3 and KST1—potato; MKT1,2 and KMT1—common ice plant (Mkt1, accession no. AF267753; Mkt2, accession no. AF267755; and Kmt1, accession no. AF267754). TaAKT1—wheat; ZMK1—Zea mays. KAT2, SKT3, MKT2, and KMT1 are partial sequences.
Table I compares the deduced amino acid sequences of the three ice plant genes with their counterparts in Arabidopsis (BestFit in Genetics Computer Group; Wisconsin Package Version 10.0, Genetics Computer Group, Madison, WI). The comparable regions of MKT1, MKT2, and KMT1, respectively, share 52% to 62% amino acid sequence identity, and 60% to 64% identity in nucleotide sequence (also based on best fit). The phylogenetic tree of the channels, including a number of sequences from other species (Fig. 2), can rely only on few sequences at present, and is complicated by low sequence homology between the AKT and KAT subfamilies. The alignments separate the AKT1 and AKT2/3 subfamilies and place the KAT1 subfamily separate with high bootstrap values. The inclusion of the ice plant sequences places them separate from the other angiosperm orders, most likely reflecting the evolutionary separation of the ice plant (order Caryophyllales). The inclusion of AKT1 homologs from grasses (T. aestivum and Z. mays, respectively) similarly introduces low bootstrap values separating monocot and dicot AKT1 subfamily members.
Table I.
Sequence comparisons between Arabidopsis and common ice plant potassium channel proteins in the AKT and KAT family
| Gene | AKT1
(Arabidopsis)
|
AKT2/3 (Arabidopsis)
|
KAT1
(Arabidopsis)
|
KAT2 (Arabidopsis)
|
||||
|---|---|---|---|---|---|---|---|---|
| I | S | I | S | I | S | I | S | |
| MKT1 | 63.8 | 72.2 | 46.0 | 55.7 | – | – | – | – |
| MKT2 | 55.8 | 66.3 | 77.9 | 84.2 | – | – | – | – |
| KMT1 | – | – | – | – | 55.4 | 64.6 | 55.7 | 64.2 |
Gene Complexity
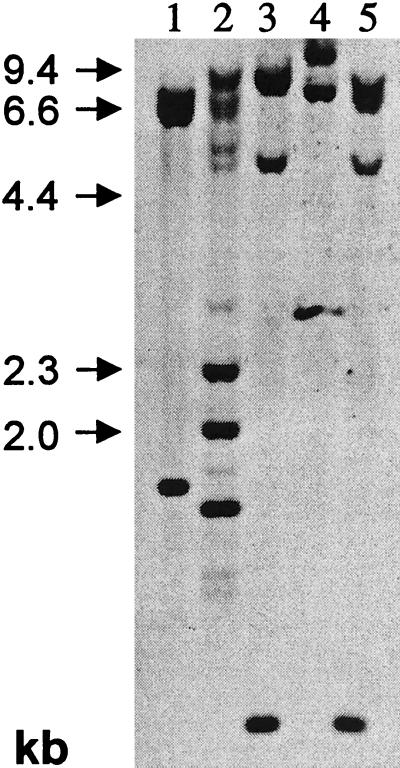
Southern-type hybridizations were performed with the ice plant Mkt1 cDNA. The results of copy number reconstructions (not included), based on an nDNA content of 390 Mb (DeRocher et al., 1990), indicated Mkt1 as one copy of a small gene family with two to three members (Fig. 3). For example, when probed with a full-length Mkt1 coding sequence, genomic DNA digested with XmnI with two restriction sites in the cDNA revealed strong signals at 8.5, 2.3, 2.0, and 1.3 kb. Weaker bands at 7.3, 6.7, 5.5, 5.0, 2.9, 1.5, 0.9, and 0.8 kb (Fig. 3, lane 2) likely identify a different Mkt1 isoform or isoforms.
Figure 3.
DNA-blot analysis of Mkt1. The entire coding region was used as the probe. Restriction endonucleases were: 1, EcoRI; 2, Xmn1; 3, HindIII; 4, XhoI; and 5, HindIII and XhoI.
Expression Patterns
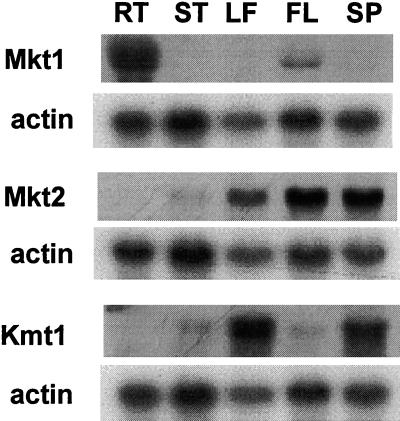
RNA was isolated from roots, stems, leaves, flowers, and seed capsules to obtain information on tissue-specific expression by RNA-blot hybridization. The three channel transcripts are expressed at different abundance, and each channel shows distinct tissue-specific patterns (Fig. 4). For Mkt1 and Kmt1, 3′-UTR regions, or sequences close to the 3′ end, were used as probes, and the entire RT-PCR product was used for Mkt2. Mkt1 is mainly expressed in root tissues, similar to its counterpart Akt1 in Arabidopsis (Cao et al.,, 1995). Mkt2 is expressed in all aerial tissues, including leaves, stems (at low abundance), flowers, and seed capsules. Kmt1 transcripts are most abundant in leaves and seed capsules. No signal was detected in leaves for Mkt1with a 3′-UTR-specific probe, whereas Mkt2 and Kmt1 are absent from roots. Tissue specificity may suggest distinct physiological roles for these potassium channel proteins.
Figure 4.
Tissue-specific expression of Mkt1, Mkt2, and Kmt1. 3′-untranslated (UTR) regions or sequence-divergent carboxy terminal regions of transcripts were used as probes. Actin was included as a loading standard. Twenty micrograms of total RNA from roots (RT), stems (ST), leaves (LF), flowers (FL), and seed pods (SP) was loaded in each lane.
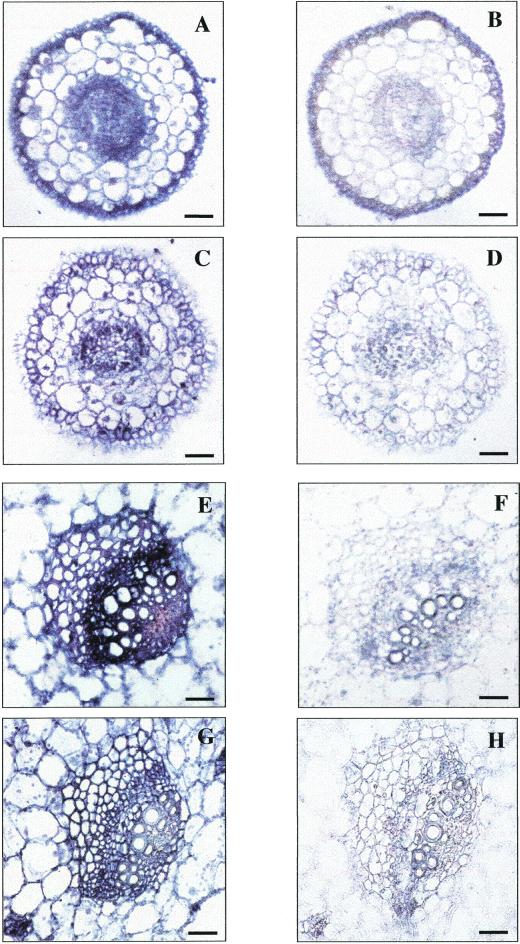
To describe the tissue- or cell-type-specific expression patterns in more detail, in situ hybridizations were performed for Mkt1, using RNA probes corresponding to a fragment downstream of the sixth TM region (amino acid 327–470). Hybridizations with antisense (Fig. 5, A, C, E, and G) and sense (Fig. 5, B, D, F, and H) probes are compared. In root tissue, Mkt1 signals were detected in cells of the epidermis, cortical cells, and in the stele (Fig. 5A). Salt stress reduced the absolute signal intensity but did not affect cell specificity (Fig. 5C). Signals were also detected in leaf phloem-related cells and in phloem and xylem parenchyma regions of the leaf with lower intensity signals in mesophyll cells (Fig. 5E). As in the roots, the intensity of the signals declined under salt stress conditions (Fig. 5F). We consider the signals in leaves to be caused by a leaf-specific homolog of the root-specific Mkt1. It is unlikely that this form is Mkt2 or Kmt1 because of low sequence identity between Mkt1 and either Mkt2 (60.5%) or Kmt1 (63.7%). Quantitative PCR analysis of root and leaf RNA with the different coding region and 3′-end-specific probes used in RNA-blot and in situ experiments showed that northern hybridization reported the presence of Mkt1 precisely, whereas the in situ data reveal another Mkt-like transcript (data not shown).
Figure 5.
Cell-specific expression of Mkt1. In situ hybridization of Mkt1 with a probe derived from a conserved region (nucleotides 980–1,410, corresponding with amino acids 327–470; see Fig. 1). A, Unstressed root tip; C, stressed root tip (500 mm NaCl, 3 d); E, unstressed plant, vascular tissue in the primary leaf; G, stressed plant, vascular tissue in the primary leaf. B, D, F, and H are tissue sections from the same plants as those in the other panels probed with sense RNA. Bars represent 40 μm.
Transcript Amounts during Salt Stress
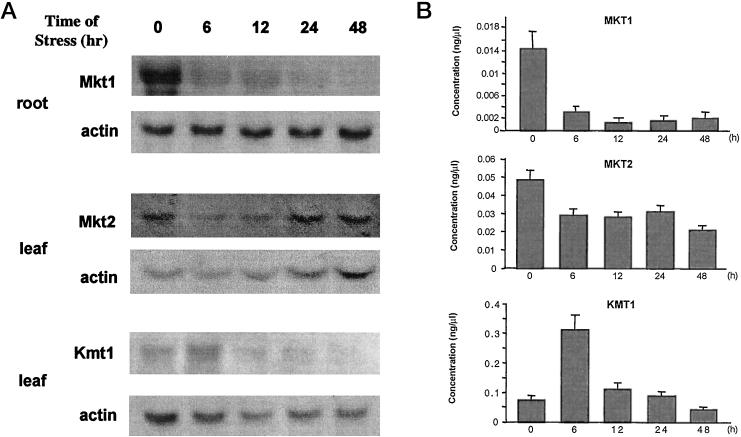
We were interested in transcript behavior during salt stress. Figure 6 shows results from RNA-blot hybridizations. Mature ice plants were used, which had been exposed to 400 mm NaCl for various time periods. RNA was probed in those tissues in which a particular transcript was most abundant (see Fig. 4), i.e. Mkt1 in root and Mkt2 and Kmt1 in leaf tissue. Mkt1 amounts decreased drastically within 6 h following stress, confirming in situ hybridization data (Fig. 5), and suggesting a switch in potassium uptake from Mkt1 to other systems in the roots (Fig. 6A; see “Discussion”). Root potassium content, based on earlier results (Adams et al., 1992; Nelson et al., 1999), declines long term under salt stress conditions to approximately 60% of the amount found in controls. Potassium content similarly decreased by about 50% in juvenile leaves in plants stressed for 3 d, compared with unstressed plants (Adams et al., 1998). In contrast with whole leaf or root content, the concentration of potassium in the xylem, measured by pressure bomb extrusion of xylem sap in side shoots of the ice plant (Table II), shows no decline under stress conditions.
Figure 6.
Mkt1, Mkt2, and Kmt1 transcript expression during NaCl stress. A, Northern blots of Mkt1, Mkt2, and Kmt1. Five- to 6-week-old plants grown in hydroponic tanks were stressed with 400 mm NaCl for the times (hours) indicated. Ten micrograms of total RNA was loaded per lane and actin served as the loading control. B, Transcript levels (concentration based on cDNA amounts generated from cloned transcripts that produced the same signal intensity as transcripts in total RNA) of Mkt1, Mkt2, and Kmt1 in stressed plants detected by semiquantitative RT-PCR.
Table II.
Changes in ion and polyol content in xylem sap of stressed ice plants
| Controla | NaCl Stressa | |
|---|---|---|
| Sodium | 0.9 ± 0.2 | 59.5 ± 9.7 |
| Potassium | 48.7 ± 4.2 | 58.1 ± 6.3 |
| Myo-inositol | <0.1 | 0.6 ± 0.3 |
| d-ononitol | <0.1 | 1.5 ± 0.6 |
In micromoles/injection volume (50 μL of undiluted xylem sap in each experiment from three plants; n = 6). Plants, grown hydroponically in 0.25 Hoagland nutrient solution and 6 weeks of age, were stressed for 24 h at 400 mm in the medium. Xylem sap was collected from side-shoot segments by pressure bomb extrusion (Nelson et al., 1998). Ions and polyols were determined by HPLC analysis (Adams et al., 1992). Suc was absent or below the limit of detection in the samples.
Amounts of Mkt2 transcripts in leaves declined to approximately 50% within 6 h, but did not change significantly later during salt stress (Fig. 6A). In contrast, expression of Kmt1 showed a different behavior following salt stress. Kmt1 amounts, which were measured in leaf and stem tissue (only leaf shown), increased dramatically but transiently, approaching a peak at 6 h and decreasing rapidly afterward (Fig. 6A). This behavior may indicate an important role of KMT1 in ion homeostasis in leaves during the onset and early periods of salt stress in the ice plant. Very similar patterns were indicated for the three transcripts when RT-PCR was used (Fig. 6B). As an average of three repeat experiments using semiquantitative measurements, the decline of root Mkt1 was by a factor of approximately seven, leaf Mkt2 declined to approximately 50% of the prestress value, and the increase of leaf Kmt1 amounted to an approximately 4- to 6-fold transient increase.
Protein Expression and Localization
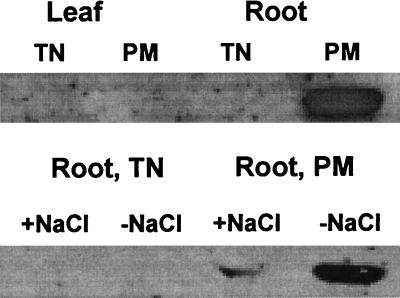
Antibodies, which had been generated against an MKT1 oligopeptide sequence in a region that was variable in all alignments (underlined in Fig. 1), were used to probe for the presence of MKT1 (Fig. 7). Plasma membrane and tonoplast fractions were isolated by discontinuous Suc gradient centrifugation of membrane vesicles. The presence of MKT1 protein was detected in the plasma membrane in roots (Fig. 7A), consistent with the information obtained for AKT1 (Lagarde et al., 1996), which is expressed in the root epidermis and functions as the potassium uptake machinery in roots from the soil. Like the transcript, MKT1 protein amount decreases under salt stress (Fig. 7B).
Figure 7.
MKT1 protein expression in the ice plant. Tissue and membrane localization of MKT1 protein in roots and leaves. TN, Tonoplast; PM, plasma membrane. MKT1 expression in root tissues in different membrane fractions. +NaCl, Plants stressed with 200 mm NaCl for 2 weeks; −NaCl, membranes from plants without NaCl.
DISCUSSION
We are interested in plant sodium uptake, particularly the effects of an excess of sodium on potassium homeostasis. Little is known about the entry of sodium into plants, but possible pathways for sodium entry and transport across plant cell membranes are uptake systems for monovalent cations, including the nonselective cation channels (Schachtman and Liu, 1999). Significant among those are potassium import proteins, which satisfy the demand for K+ as the most abundant cation in the cytosol. The available information seems to indicate that the high selectivity of the characterized potassium channels does not lead to significant sodium influx (Amtmann and Sanders, 1999; Santa-Maria et al., 2000). The ubiquitously high K+ selectivity of potassium channels is exemplified by Arabidopsis KAT1, which exhibits a discrimination ratio of 20 for K+ over Na+ when expressed in yeast and analyzed by patch clamping (Bertl et al., 1995). High selectivity also has been elegantly demonstrated by calculations based on models of potassium channel structures (Doyle et al., 1998; Åqvist and Luzhkov, 2000). The recent detection of genes for sodium/proton antiporters highlighted one of the systems for sodium mobility between cytosol and vacuole (Apse et al., 1999; Gaxiola et al., 1999), but how sodium enters cells in the first instance and how this might interfere with potassium uptake is not explained by the activity of these antiporters.
The characterization of three potassium channel transcripts from the halophytic ice plant reinforces reports about the presence, known only from few species other than Arabidopsis, of a multitude of potassium transport proteins. DNA analysis (Fig. 3) in the common ice plant indicated that the Mkt type alone should be represented by two or three isoforms with Mkt1 the root-specific member of the family. An additional highly conserved form, an interpretation that we deduce from the in situ hybridizations (Fig. 5), is expressed in the leaves. It is likely that these forms perform identical functions, distinguished by promoters that determine organ-, tissue-, or cell-specific expression patterns. The structures of these channels place Mkt1 and Mkt2 in the AKT group, and Kmt1 is a homolog of the KAT group of IRC, all proteins with the domain structure of the Shaker family of K+ channels. Also, the three orthologous ice plant transcripts show similar expression patterns in different tissues from Arabidopsis. Their coincident isolation in evolutionarily distant species indicates further that these channels represent abundant members of the potassium channel gene complement in these species. The relative concentrations of the potassium channel transcripts in total RNA under normal conditions, as detected by RT-PCR, is approximately 1:3:5 (Mkt1:Mkt2:Kmt1).
The expression patterns, distinct for each of the three transcripts, reveal a surprisingly complex salt stress response. The repression of Mkt1 expression in roots, to approximately 15% of the prestress amount within 6 h (Fig. 6), is interesting because potassium export from the roots is not inhibited (Table II). The decline in root potassium is significant only during long-term stress periods (Adams et al., 1992). If we equalize transcript and protein amounts (documented only for MKT1; Figs. 6 and 7) and assume constant activity, decreased root K+ channel amounts could indicate that other channels or transporters are active or activated during stress episodes in the ice plant. Studies with the ice plant have indicated accelerated protein turnover under stress but transcript amounts are typically good indicators for the amount of the corresponding proteins (Vernon et al., 1988; Adams et al., 1998; Cushman and Bohnert, 1999).
Transcripts for Mkt2 in leaves are little affected by salt stress with an approximately 50% decline within 6 h, which persists for several days. We assume that Mkt2 amounts increase again as suggested by strong signals in flowers, where it may act in opening of the flowers, and seed capsules. Although no comparison is possible with flowers in unstressed plants because salinity is a prerequisite for the ice plant to enter reproductive development (Adams et al., 1998), it seems possible that Mkt2 is responsible for potassium homeostasis in reproductive organs because a flower-specific Mkt1 homolog is expressed at low levels (Fig. 4).
Kmt1 expression in leaves and seed capsules represents a surprisingly strong signal, if we assume that Kmt1 is expressed, as is its Arabidopsis ortholog, in the guard cells of leaves, stems, and seed capsules of the ice plant. The signal in leaves (and stems, not shown) increases transiently early during stress with a maximum approximately 6 h after salt stress. This coincides with extreme stress-induced wilting of leaves, although recovery occurs within 1 to 2 d (Adams et al., 1998). During that time, sodium concentrations increase in the leaves, compensated by the accumulation of compatible solutes (Adams et al., 1992; Adams et al., 1998). The transient increase of transcripts for Kmt1 might support continued opening of the stomata during the early stress period, and this may foster the drastic increase of sodium based on sustained transpiration. The transient wilting may then initiate further defense mechanisms and the ice plant's switch to Crassulacean acid metabolism. This view of a function for KMT1 would indicate that the channels, although playing no role or only a minor role in sodium uptake, could also have a function in supporting osmotic adjustment.
Whether this response is general or specific for the ice plant is unknown because few studies have concentrated on potassium channel transcripts and none have targeted their function in halophytes. Whether the regulation of transcript amounts for each channel is by transcriptional, posttranscriptional, or (post) translational processes is not known. So far, studies have been targeted toward aspects of activity in heterologous systems, and those that addressed the control of expression have been conducted with stress-sensitive species. The detection of regulated transcript amounts for some potassium channels seemingly in response to salt stress is novel. We could not detect any changes in transcript amounts for the ice plant Mkt1, Mkt2, and Kmt1 when the plants were starved for potassium (data not shown). This is consistent with observations of an AKT1 ortholog in Brassica napus (Lagarde et al., 1996), but in contrast with a recent report documenting up-regulation of wheat AKT1 in response to potassium starvation (Buschmann et al., 2000). Induction of transcripts by potassium starvation seems more common among potassium transporters: HKT1 (wheat), HvHAK1 (barley), and AtKUP3 (Arabidopsis) respond to K+ starvation (Santa-Maria et al., 1997; Fu and Luan, 1998; Kim et al., 1998; Wang et al., 1998).
Sustained potassium supply is essential for plant performance under excess sodium. A survey of the literature seems to support a hypothesis suggesting HKT-type transporters, originally viewed as high-affinity potassium transporters (Schachtman and Schroeder, 1994; Rubio et al., 1995), contributing significantly to sodium uptake. An Arabidopsis HKT transporter has been identified as a protein with high specificity for sodium and low affinity toward potassium (Uozumi et al., 2000). Similar uptake characteristics with high affinity for sodium are also indicated for the HKT homologs of rice (Oryza sativa) and the common ice plant (Golldack et al., 1997; D. Golldack, H. Su, F. Quigley, U.R. Kawasawi, C. Muñoz-Garay, J. Bennett, H.J. Bolwert, and O. Pantoja, unpublished data; H. Su and H.J. Bohnert, unpublished data), but other possible avenues for sodium uptake still exist. Such uptake systems might also be found among nonselective cation channels. Several of these ORC have been detected and their action, following the sodium-induced depolarization of root plasma membranes, could lead to sodium influx (see Schachtman and Liu, 1999; Blumwald et al., 2000). Voltage-independent cation channels alternatively or additionally exhibit properties that could make them a major route for sodium influx at a high Na+ to K+ ratio (Amtmann and Sanders, 1999; Blumwald et al., 2000). That such proteins should exist can be deduced. For example, in the Arabidopsis akt1 mutant, K+ uptake by systems other than AKT1 was inhibited by NH4+ but stimulated by Na+ (Spalding et al., 1999), suggesting an influence of Na+ on the regulation of at least some high-flux K+-uptake proteins. This is consistent with our results (Table II) and data on ice plant HAK- and HKT-type potassium transport systems, both of which include isoforms that are up-regulated by the presence of sodium (Su and Bohnert, unpublished data). A switch to high-affinity K+ transporters other than the AKT/KAT-type at high salinity and low external NH4+ (0.5 mm in our experiments) may be due to an altered K+ to Na+ ratio (Spalding et al., 1999). In Arabidopsis salt-overly-sensitive (sos) mutants, sensitivity is more closely related to potassium availability than to sodium concentrations in the plants (Wu et al., 1996; Zhu et al., 1998). The complexity of proteins that take up monovalent cations is further exemplified by the detection, in the form of genes, expressed sequence tags, cDNAs, or activities, of a total of 15 ORC or IRC in Arabidopsis. Although isoforms in these gene families may function in a tissue- or cell-specific manner, changes in combination or isoform abundance along the plant axis may play a crucial role in K+ homeostasis, impossible to gauge by measuring the activity of individual proteins. The regulation of expression or activity of potassium channels and transporters illustrates the important roles of K+-uptake systems in reestablishing homeostasis to support plant salinity tolerance.
MATERIALS AND METHODS
Plant Material
Seedlings of the common ice plant (Mesembryanthemum crystallinum) were transferred to aerated hydroponic tanks about 2 weeks after germination. Plants were grown in 0.5× Hoagland nutrition solution with the amount of iron doubled and 3 mm potassium. For salt stress treatments, the plants were watered with one-half-strength Hoagland solution including 400 mm NaCl at the age of 4 to 5 weeks for the time periods indicated. Unstressed control plants were grown in parallel and harvested at the same time.
Nucleic Acid Isolation and Hybridizations
For RNA isolations, each 4 g of frozen, ground tissues was suspended in 16 mL of extraction medium containing 100 mm Tris-HCl (pH 8.0), 150 mm NaCl, 10 mm EDTA, 2 mm aurin tricarboxylic acid, 1% (w/v) SDS, and 1% (w/v) sarcosyl. After adding an equal volume of phenol, the extracts were incubated on ice for 1 h with shaking. The aqueous phases, collected by centrifugation at 5,000g for 10 min at 4°C, was incubated with an equal volume of isopropyl alcohol overnight at −20°C. Total precipitated nucleic acids were collected by centrifugation at 6,000g for 20 min at 4°C and dissolved in diethyl pyrocarbonate-treated water. RNA was precipitated by incubation with an equal volume of 4 m LiCl twice at 4°C for 4 h and collected by centrifugation. Concentrations of nucleic acids were measured at 260 nm. Genomic DNA was isolated from 2- to 3-week-old plants as described (Sambrook, et al., 1989). Poly(A+) mRNA was selected from total RNA by using the PolyATract mRNA Isolation System IV (Promega, Madison, WI).
For Southern-blot analyses, 10 μg of genomic DNA was completely digested with different restriction enzymes and DNA fragments were separated on 0.7% (w/v) agarose gels in 0.5× TBE buffer. For RNA blot analysis, 10 to 20 μg of total RNA was separated on 1% (w/v) agarose gel containing 5.5% (w/v) formaldehyde. Gel treatment, transfer of nucleic acid, and the hybridizations were performed according to the instruction manual of membranes (Stratagene, La Jolla, CA). All blot hybridizations were conducted with Duralon-UVTM membranes (Stratagene) and probed with 32P-labeled DNA fragments (ICN Biomedicals, Inc., Irvine, CA).
Semiquantitative RT-PCR
One to 5 μg of total RNA or 50 to 500 ng of poly(A+)-RNA was used for reverse transcription. RNA and 50 ng of oligo(dT)12-18 in a volume of 12 μL were incubated at 70°C for 10 min. After quenching on ice, 4 μL of 5× first-strand buffer, 2 μL of 0.1 m dithiothreitol, and 1 μL of 10 mm dNTP mix were added and tubes were incubated at 42°C for 2 min. One microliter of Supercript II (Life Technologies, Inc., Rockville, MD) was added and the incubation was continued for 50 min. The reaction was stopped by heating (70°C, 15 min) and 1 μL of ribonuclease A (2 units) was added to remove RNA complementary to the cDNA. Amplification reactions were carried out with 1 μL of the first-strand cDNA, 1× PCR buffer, 1.5 mm MgCl2, 0.2 mm dNTPs, 0.4 mm primers, and 2.5 units Taq DNA polymerase in a volume of 50 μL (Life Technologies, Inc.). After denaturation (94°C, 3 min), samples were subjected to 32 cycles of 1.5 min at 94°C, 1.5 min (55°C or temperature gradient), and 2 min at 72°C. PCR products were separated on 0.8% (w/v) agarose gels (0.5× TBE).
In Situ Hybridizations
Tissues from the root tip and from the second leaf pair of the plants were fixed in formaldehyde, dehydrated, and embedded as described by McKhann and Hirsch (1993). The tissues were embedded in Paraplast Plus (Fisher Scientific, Pittsburgh) and 10-μm sections were mounted on poly-L-Lys-coated slides. Sense and antisense RNA transcripts labeled with digoxigenin-UTP (Boehringer, Mannheim, Germany) were synthesized by T3 and T7 RNA polymerase from linearized pBluescript harboring the cDNA from position 980 to 1,410 in the MKT1 cDNA. Transcripts were hydrolyzed to an average length of 200 nucleotides by alkaline treatment (Cox and Goldberg, 1989). In situ hybridizations were performed as described by Yamada et al. (1995). Signal detection was done with antidigoxigenin alkaline phosphatase-conjugated Fab fragments (Boehringer) and 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium as a substrate.
Membrane Isolation and Immunological Detection
Antibodies for MKT1 were raised against an oligopeptide, PRVREVTWADKNRRRRVNTFCOOH (amino acids 743–762), close to the carboxyl terminus of MKT1 (HTI Bio-Products, Inc., Ramona, CA). Membrane fractions were isolated as described (Barkla, et al., 1995). Tonoplast membranes were collected at the 0% to 22% (w/v) Suc interface and plasma membranes at the 32% to 38% (w/v) interface. SDS-PAGE and protein blotting followed established procedures (Sambrook et al., 1989).
ACKNOWLEDGMENTS
The authors gratefully acknowledge support from the German Research Council (Bonn), the Japanese Society for the Promotion of Science (Tokyo), and the Ministry of Agriculture (China), respectively.
Footnotes
This work was supported by the Arizona Agricultural Experiment Station and in part by the U.S. Department of Agriculture-National Research Initiative Competitive Grants Program (Plant Responses to the Environment).
LITERATURE CITED
- Adams P, Nelson DE, Yamada S, Chmara W, Jensen RG, Bohnert HJ, Griffiths H. Growth and development of Mesembryanthemum crystallinum (Aizoaceae) New Phytol. 1998;138:171–190. doi: 10.1046/j.1469-8137.1998.00111.x. [DOI] [PubMed] [Google Scholar]
- Adams P, Thomas JC, Vernon DM, Bohnert HJ, Jensen RG. Distinct cellular and organismic responses to salt stress. Plant Cell Physiol. 1992;33:1215–1223. [Google Scholar]
- Amtmann A, Sanders D. Mechanisms of Na+ uptake by plant cells. Adv Bot Res. 1999;29:75–112. [Google Scholar]
- Anderson JA, Huprikar SS, Kochian LV, Lucas WJ, Gaber RF. Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:3736–3740. doi: 10.1073/pnas.89.9.3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiporter in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- Åqvist J, Luzhkov V. Ion permeation mechanism of the potassium channel. Nature. 2000;404:881–884. doi: 10.1038/35009114. [DOI] [PubMed] [Google Scholar]
- Barkla BJ, Zingarelli L, Blumwald E, Smith JAC. Tonoplast Na+/H+ antiport activity and its energization by the vacuolar H+-ATPase in the halophytic plant Mesembryanthemum crystallinum L. Plant Physiol. 1995;109:549–556. doi: 10.1104/pp.109.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertl A, Anderson JA, Slayman CL, Gaber RF. Use of Saccharomyces cerevisiae for patch-clamp analysis of heterologous membrane proteins: characterization of Kat1, an inward-rectifying K+ channel from Arabidopsis thaliana, and comparison with endogenous yeast channels and carriers. Proc Natl Acad Sci USA. 1995;92:2701–2705. doi: 10.1073/pnas.92.7.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta. 2000;1465:140–151. doi: 10.1016/s0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- Brüggemann L, Dietrich P, Becker D, Dreyer I, Palme K, Hedrich R. Channel-mediated high-affinity K+ uptake into guard cells from Arabidopsis. Proc Natl Acad Sci USA. 1999;96:3298–3302. doi: 10.1073/pnas.96.6.3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann PH, Vaidyanathan R, Gassmann W, Schroeder JI. Enhancement of Na+ uptake currents, time-dependent inward-rectifying K+ channel currents, and K+ channel transcripts by K+ starvation in wheat root cells. Plant Physiol. 2000;122:1387–1397. doi: 10.1104/pp.122.4.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AD, Blatt MR, Ainsworth CC. Expression, evolution and genomic complexity of potassium ion channel genes of Arabidopsis thaliana. J Plant Physiol. 1997;150:652–660. [Google Scholar]
- Cao Y, Ward JM, Kelly WB, Ichida AM, Gaber RF, Anderson JA, Uozumi N, Schroeder JI, Crawford NM. Multiple genes, tissue specificity, and expression-dependent modulation contribute to the functional diversity of potassium channels in Arabidopsis. Plant Physiol. 1995;109:1093–1106. doi: 10.1104/pp.109.3.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrispeels MJ, Crawford NM, Schroeder JI. Proteins for transport of water and mineral nutrients across the membranes of plant cells. Plant Cell. 1999;11:661–675. doi: 10.1105/tpc.11.4.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Goldberg RB. Analysis of plant gene expression. In: Shaw CH, editor. Plant Molecular Biology: A Practical Approach. Oxford: IRL Press; 1989. pp. 1–35. [Google Scholar]
- Cushman JC, Bohnert HJ. Crassulacean acid metabolism: molecular genetics. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:305–332. doi: 10.1146/annurev.arplant.50.1.305. [DOI] [PubMed] [Google Scholar]
- Czempinski K, Gaedeke N, Zimmermann S, Mueller-Roeber B. Molecular mechanisms and regulation of plant ion channels. J Exp Bot. 1999;50:955–966. [Google Scholar]
- DeRocher EJ, Harkins KR, Galbraith DW, Bohnert HJ. Systemic endopolyploidy in ice plant is developmentally regulated and may be common to succulents with small genomes. Science. 1990;250:99–101. doi: 10.1126/science.250.4977.99. [DOI] [PubMed] [Google Scholar]
- Doyle DA, Cabral JM, Pfuetzer RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- Fu HH, Luan S. AtKUP1: a dual-affinity K+ transporter from Arabidopsis. Plant Cell. 1998;10:63–73. doi: 10.1105/tpc.10.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SA, Fink GR. The Arabidopsis thaliana proton transporters AtNhx1 and Avp1 can function in cation detoxification. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golldack D, Kamasani UR, Quigley F, Bennett J, Bohnert HJ. Salt stress-dependent expression of a HKT-type high-affinity potassium transporter in rice. Plant Physiol Suppl. 1997;114:118. [Google Scholar]
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Science. 1998;280:918–921. doi: 10.1126/science.280.5365.918. [DOI] [PubMed] [Google Scholar]
- Hoth S, Hedrich R. Distinct molecular bases for pH sensitivity of the guard cell K+ channels KST1 and KAT1. J Biol Chem. 1999;274:11599–11603. doi: 10.1074/jbc.274.17.11599. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI. AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell. 1998;10:51–62. doi: 10.1105/tpc.10.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lagarde D, Basset M, Lepetit M, Conejero G, Gaymard F, Astruc S, Grignon C. Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 1996;9:195–203. doi: 10.1046/j.1365-313x.1996.09020195.x. [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Ichida AM, Sanders D, Schroeder JI. Roles of higher plant K+ channels. Plant Physiol. 1997;114:1141–1149. doi: 10.1104/pp.114.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marten I, Hoth S, Deeken R, Ache P, Ketchum KA, Hoshi T, Hedrich R. AKT3, a phloem-localized K+ channel, is blocked by protons. Proc Natl Acad Sci USA. 1999;96:7581–7586. doi: 10.1073/pnas.96.13.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann HI, Hirsch AM. In situ localization of specific mRNAs in plant tissue. In: Glick BR, Thompson JE, editors. Methods in Plant Molecular Biology and Biotechnology. Boca Raton, FL: CRC Press; 1993. pp. 179–205. [Google Scholar]
- Mueller-Roeber B, Ellenberg J, Provart N, Willmitzer L, Busch H, Becker D, Dietrich P, Hoth S, Hedrich R. Cloning and electrophysiological analysis of KST1, an inward rectifying K+ channel expressed in potato guard cells. EMBO J. 1995;14:2409–2416. doi: 10.1002/j.1460-2075.1995.tb07238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Koukoumanos M, Bohnert HJ. Myo-inositol amounts in roots control sodium uptake in a halophyte. Plant Physiol. 1999;119:165–172. doi: 10.1104/pp.119.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE, Rammesmayer G, Bohnert HJ. The regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell. 1998;10:753–764. doi: 10.1105/tpc.10.5.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio F, Gassmann W, Schroeder JI. Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science. 1995;270:1660–1663. doi: 10.1126/science.270.5242.1660. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santa-Maria GE, Danna CH, Czibener C. High-affinity potassium transport in barley roots: ammonium-sensitive and -insensitive pathways. Plant Physiol. 2000;123:297–306. doi: 10.1104/pp.123.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Maria GE, Rubio F, Dubcovsky J, Rodriguez-Navarro A. The HAK1 gene of barley is a member of a large gene family and encodes a high-affinity potassium transporter. Plant Cell. 1997;9:2281–2289. doi: 10.1105/tpc.9.12.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman DP, Liu W. Molecular pieces to the puzzle of the interaction between potassium and sodium uptake in plants. Trends Plant Sci. 1999;4:281–286. doi: 10.1016/s1360-1385(99)01428-4. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Schroeder JI. Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature. 1994;370:655–658. doi: 10.1038/370655a0. [DOI] [PubMed] [Google Scholar]
- Sentenac H, Bonneaud N, Minet M, Lacroute F, Salmon J-M, Gaymard F, Grignon C. Cloning and expression in yeast of a plant potassium ion transport system. Science. 1992;256:663–665. doi: 10.1126/science.1585180. [DOI] [PubMed] [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD. Potassium uptake supporting plant growth in the absence of AKT1 channel activity: inhibition by ammonium and stimulation by sodium. J Gen Physiol. 1999;113:909–918. doi: 10.1085/jgp.113.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uozumi N, Kim EJ, Rubio F, Yamaguchi T, Muto S, Tsuboi A, Bakker EP, Nakamura T, Schroeder JI. The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 2000;122:1–11. doi: 10.1104/pp.122.4.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon DM, Ostrem JA, Schmitt JM, Bohnert HJ. PEPCase transcript levels in Mesembryanthemum crystallinum decline rapidly upon relief from salt stress. Plant Physiol. 1988;86:1002–1004. doi: 10.1104/pp.86.4.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Very AA, Gaymard F, Bosseux C, Sentenac H, Thibaud JB. Expression of a cloned plant K+ channel in Xenopus oocytes: analysis of macroscopic currents. Plant J. 1995;7:321–332. doi: 10.1046/j.1365-313x.1995.7020321.x. [DOI] [PubMed] [Google Scholar]
- Wang T-B, Gassmann W, Rubio F, Schroeder JI, Glass ADM. Rapid up-regulation of HKT1, a high-affinity potassium transporter gene, in roots of barley and wheat following withdrawal of potassium. Plant Physiol. 1998;118:651–659. doi: 10.1104/pp.118.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-J, Ding L, Zhu J-K. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell. 1996;8:617–627. doi: 10.1105/tpc.8.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Katsuhara M, Kelly WB, Michalowski CB, Bohnert HJ. A family of transcripts encoding water channel proteins: tissue-specific expression in the common ice plant. Plant Cell. 1995;7:1129–1142. doi: 10.1105/tpc.7.8.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK, Liu L, Xiong L. Genetic analysis of salt tolerance in Arabidopsis: evidence for a critical role of potassium nutrition. Plant Cell. 1998;10:1181–1191. doi: 10.1105/tpc.10.7.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann S, Sentenac H. Plant ion channels: from molecular structures to physiological functions. Curr Opin Plant Biol. 1999;2:477–482. doi: 10.1016/s1369-5266(99)00020-5. [DOI] [PubMed] [Google Scholar]