Machine learning reveals order in crumpled sheets using simulated flat-folding patterns as data surrogate in a data-limited regime.
Abstract
Machine learning has gained widespread attention as a powerful tool to identify structure in complex, high-dimensional data. However, these techniques are ostensibly inapplicable for experimental systems where data are scarce or expensive to obtain. Here, we introduce a strategy to resolve this impasse by augmenting the experimental dataset with synthetically generated data of a much simpler sister system. Specifically, we study spontaneously emerging local order in crease networks of crumpled thin sheets, a paradigmatic example of spatial complexity, and show that machine learning techniques can be effective even in a data-limited regime. This is achieved by augmenting the scarce experimental dataset with inexhaustible amounts of simulated data of rigid flat-folded sheets, which are simple to simulate and share common statistical properties. This considerably improves the predictive power in a test problem of pattern completion and demonstrates the usefulness of machine learning in bench-top experiments where data are good but scarce.
INTRODUCTION
Machine learning is a versatile tool for data analysis that has permeated applications in a wide range of domains (1). It has been particularly well suited to the task of mining large datasets to uncover underlying trends and structure, enabling breakthroughs in areas as diverse as speech and character recognition (2–5), medicine (6), games (7, 8), finance (9), and even romantic attraction (10). The prospect of applying machine learning to research in the physical sciences has likewise gained attention and excitement. Data-driven approaches have been successfully applied to data-rich systems such as classifying particle collisions in the Large Hadron Collider (LHC) (11, 12), classifying galaxies (13), segmenting large microscopy datasets (14, 15), or identifying states of matter (16, 17). Machine learning has also enhanced our understanding of soft matter systems: In a recent series of works, Cubuk, Liu, and collaborators (18–20) have used data-driven techniques to define and analyze a novel “softness” parameter governing the mechanical response of disordered, jammed systems.
All examples cited above address experimentally, computationally, or analytically well-developed scientific fields supplied by effectively unlimited data. By contrast, many systems of interest are characterized by scarce or poor-quality data, a lack of established tools, and a limited data acquisition rate that falls short of the demands of effective machine learning. As a result, the applicability of machine learning to these systems is problematic and would require additional tools. This would potentially be of high value to the experimental physics community and would require novel ways of circumventing the data limitations, either experimentally or computationally. Here, we study crumpling and the evolution of damage networks in thin sheets as a test case for machine learning–aided science in complex, data-limited systems that lack a well-established theoretical, or even a phenomenological, model.
Crumpling is a complicated and poorly understood process: As a thin sheet is confined to a small region of space, stresses spontaneously localize into one-dimensional regions of high curvature (21–23), forming a damage network of sharp creases (Fig. 1B) that can be classified according to the sign of the mean curvature: Creases with positive and negative curvature are commonly referred to as valleys and ridges, respectively. Previous works on crumpled sheets have established clear and robust statistical properties of these damage networks. For example, it has been shown that the number of creases at a given length follows a predictable distribution (24), and the cumulative amount of damage over repeated crumpling is described by an equation of state (25). However, these works do not account for spatial correlations, which is the structure we are trying to unravel. The goal of this work was to learn the statistical properties of these networks by solving a problem of network completion: Separating the ridges from valleys, can a neural net be trained to accurately recover the location of the ridges, presented only with the valleys? For later use, we call this problem partial network reconstruction. The predominant challenge we are addressing here is a severe data limitation. As detailed below, we were unable to perform this task using experimental data alone. However, by augmenting experimental data with computer-generated examples of a simple sister system that is well understood, namely, rigid flat folding, we trained an appropriate neural network with significant predictive power.
Fig. 1. Examples of crease networks.

(A) A 10 cm by 10 cm sheet of Mylar that has undergone a succession of rigid flat folds. (B) A sheet of Mylar that has been crumpled. (C) A simulated rigid flat-folded sheet. The sheet has been folded 13 times. Ridges are colored red, and valleys are blue.
The primary dataset used in this work was collected in a previous crumpling study (25), where the experimental procedures are detailed and are only reviewed here for completeness. Mylar sheets (10 cm by 10 cm) are crumpled by rolling them into a 3-cm-diameter cylinder and compressing them uniaxially to a specified depth within the cylindrical container, creating a permanent damage network of creasing scars embedded into the sheet. To extract the crease network, the sheet is carefully opened up and scanned using a custom-made laser profilometer, resulting in a topographic height map from which the mean curvature is calculated. The sheet is then successively recrumpled and scanned between 4 and 24 times, following the same procedure. The curvature map is preprocessed with a custom algorithm based on the Radon transform (for details, see section S1) to separate creases from the flat facets and fine texture in the data (Fig. 2A). The complete dataset consists of a total of 506 scans corresponding to 31 different sheets.
Fig. 2. A schematic of the processing pipeline.
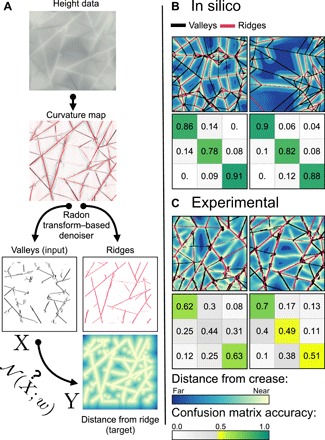
(A) From the height map, a mean curvature map is calculated and denoised with a Radon transform–based method. Valleys (black) and ridges (red) are separated. The binary image of the valleys (X) is the input to the neural network (). The distance transform of the binary image of the ridges is the target (Y). Brighter colors represent regions closer to ridges. These color conventions are consistent through all the figures in this paper. (B) Two samples of predictions on generated data. The true fold network is superimposed on the predicted distance map. It is seen that the true ridges (red) coincide perfectly with the bright colors, demonstrating strong predictive power. Below the predictions, we show confusion matrices, with the nearest third of pixels, the middle third, and the furthest third. (C) Two predictions, as well as their corresponding confusion matrices, using the network trained on generated data (without noise) and applied to experimental scans.
RESULTS
Failures with only experimental data
As stated above, the task we tried to achieve is partial network reconstruction: inferring the location of the ridges given only the valleys (Fig. 2A). Our first attempts were largely unsatisfactory and demonstrated little to no predictive power. Strategies for improving our results included subdividing the input data into small patches of different length scales, varying the network architecture, data representation, and loss function, and denoising the data in different ways. We approached variants of the original problem statement, trying to predict specific crease locations, distance from a crease, and changes in the crease network between successive frames. In all these cases, our network invariably learned specific features of the training set rather than general principles that hold for unseen test data, a common problem known as overfitting. The main culprit for this failure is insufficient data: The dataset of a few hundred scans available for this study is small compared with standard practices in machine learning tasks [for example, the problem of handwritten digit classification using the MNIST database, which is commonly given as an introductory exercise in machine learning, consists of 70,000 images (5)]. Moreover, as creases produce irreversible scars, images of successive crumples of the same sheet are highly correlated, rendering the effective size of our dataset considerably smaller.
Overfitting can be addressed by constraining the model complexity through insights from physical laws, geometric rules, symmetries, or other relevant constraints. Alternatively, it can be mediated by acquiring more data. Sadly, neither of these avenues is viable: Current theory of crumpling cannot offer significant constraints about the structure or evolution of crease networks. Furthermore, adding a significant amount of experimental data is prohibitively costly: Achieving a dataset of the size typically used in deep learning problems, say 104 scans, would require thousands of lab hours, given that a single scan takes about 10 min. Last, data cannot be efficiently simulated since, while preliminary work on simulating crumpling is promising (26, 27), generating a simulated crumpled sheet still takes longer than an actual experiment. A different approach is needed.
Turning to a sister system: Rigid flat folding
An alternative strategy is to consider a reference system free from data limitations alongside the target system, with the idea that similarities between the target and reference systems allow a machine learning model of one to inform that of the other. This is similar to transfer learning (28), but in this case, rather than repurpose a network, we supplement the training data with that of a reference system. In our case, a natural choice of such a system is a rigid flat-folded thin sheet, effectively a more constrained version of crumpling that is well understood. Rigid flat folding is the process of repeatedly folding a thin sheet along straight lines to create permanent creases, keeping all polygonal faces of the sheet flat during folding. For brevity, we will henceforth omit the word “rigid” and refer simply to flat folding.
Known rules constrain the structure of the flat-folded crease network: Creases cannot begin or terminate in the interior of a sheet—they must either reach the boundary or create closed loops; the number of ridge and valley creases that meet at each vertex differs by two (Maekawa’s theorem); last, alternating sector angles must sum to π (Kawasaki’s theorem) (29). Given these rigid geometric rules, we expect partial network reconstruction of rigid flat-folded sheets to be a much more constrained problem than that of crumpled ones.
However, while experimentally collecting flat-folding data is only marginally less costly than collecting crumpling data, simulating it on a computer is a straightforward task, which provides a dataset of a practically unlimited size. We wrote a custom code to do this using the Voro++ library (30) for rapid manipulation of the polygonal folded facets, as described in section S2. Typical examples are shown in Figs. 1C and 2B and fig. S1.
Having flat folding as a reference system provides foremost a convenient setting for comparing the performance of different network architectures. The vast parameter space of neural networks requires testing different hyperparameters, loss functions, optimizers, and data representations with no standard method for finding the optimal combination. This problem is exacerbated when it is not at all clear where the failure lies: Is the task at all feasible? If so, is the network architecture appropriate? If so, is the dataset sufficiently large? Answering these questions with our limited amount of experimental data is very difficult. In contrast, for flat-folded sheets, we are certain that the task is feasible and our dataset is comprehensive, so experimentation with different networks is easier. After testing many architectures, we identified a network capable of learning the desired properties of our data, reproducing linear features and maintaining even nonlocal angle relationships between features.
Network structure
The chosen network is a modified version of the fully connected SegNet (31) deep convolutional neural net. As outlined in Fig. 2A, each crease network is separated into its valleys and ridges. The neural net, , is given as an input binary image of the valleys, denoted X (“input” in Fig. 2). The output of the network, , is the predicted distance transform of the ridges, Y. That is, for each pixel, Y is the distance to the nearest ridge pixel (“target” in Fig. 2). Training is performed by minimizing the L2 distance (the “loss”) between the predicted distance transform, , and the real one
| (1) |
where the summation index i represents image pixels. The motivation for this choice of representation is that creases are sharp and narrow features, and therefore, if we require to predict the precise location of a crease, even slight inaccuracies would lead to vanishing gradients of L, making training harder. See Materials and Methods below for full details of the implementation.
In silico flat folding
For exclusively in silico–generated flat-folding data, the trained network performs partial network reconstruction with nearly perfect accuracy, as demonstrated in Fig. 2B: The agreement between the true location of the valleys (red lines) and their predicted location (bright colors) is visibly flawless. As a means of quantifying accuracy, we present the confusion matrices of the predicted and true output (Fig. 2B).
Confusion matrices are a common way to quantify classification errors, and since we are predicting the distance from a crease, the problem can be thought of as a classification problem: Choosing some thresholds according to typical values of the distances, we can ask for each point in space whether it is close to a crease, far from it, or at an intermediate distance. The confusion matrix measures what percentage of each class is correctly classified and, if not, what class it is wrongly classified as. We define three equal bins, based on the relative distance from the predicted ridges. The upper row in the matrix corresponds to pixels that are closest to the ridges, and the lower row corresponds to the farthest pixels. Similarly, the first and last columns correspond to the closest and farthest predicted distances. Thus, the top left entry in the matrix contains the probability of correctly predicting regions closest to a ridge, which is approximately 90%.
Partial network reconstruction of in silico flat-folded sheets is itself a nontrivial task requiring the knowledge of a complicated set of geometrical rules. Tasked to a human, inferring these rules from the data would require non-negligible effort in writing an explicit algorithm. The neural network, however, solves this problem with relative ease.
Experimental flat folding
As an intermediate step between in silico flat-folding and experimental crumpling data, we next examine the performance of the neural network on experimental flat-folding scans. Figure 2C reveals that the resulting prediction weakens by comparison, a consequence of noise present in experimental data that is absent from the in silico samples. Noise occurs in the form of varying crease widths, fine texture, and missing creases that are undetected in image processing. In some cases, even the true creases that are missed during processing are correctly predicted, which also introduces error to our accuracy metric (see, for example, the center of the second panel of Fig. 2C). While sufficient data of experimental flat folding would likely allow the network to distinguish signal from noise, in our data-limited regime, noise must be added to the generated in silico data to help the network learn to accurately predict experimental scans and avoid overfitting.
We examine the effect of adding several types of noise on the prediction accuracy on experimental input (Fig. 3, A to E). We observe considerable improvement and find that adding experimentally realistic noise (Fig. 3E) is more effective than toggling individual pixels randomly (Fig. 3, B and D). We found that the noise type that leads to optimal training is to randomly add and remove patches of input that are approximately the same length scale as the noise in the experimental scans. We also find that it is important to provide input data with lines of variable width to prevent the network from expecting only creases of a particular width. For complete details of the different noise properties, see Materials and Methods.
Fig. 3. Effect of noise type on prediction.
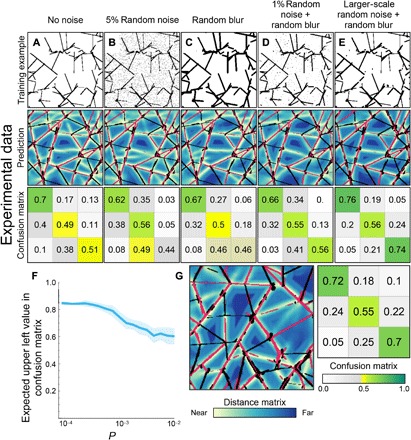
(A to E) An example noised image (top), an example prediction (middle), and the corresponding confusion matrix (bottom) for different types of artificial noise. Noise types are described concisely in the title of each panel, and complete specifications are given in Materials and Methods. (F) The upper left value of the confusion matrix when each pixel of the near-perfect prediction from Fig. 2B was randomly toggled with probability P. (G) The network from (E) applied on an additional experimental scan (from left panel of Fig. 2C). The average confusion matrix on all experimental scans is shown.
While the values in the confusion matrices in Fig. 3E might seem low, it is noteworthy that the metric used here is not trivial to interpret: It compares the L2 distance from a distance map, which is particularly sensitive to noise since a localized noise speckle in a region remote from valleys perturbs a large region of space (essentially, of the size of its Voronoi cell). To gauge the effect of noise on the accuracy metric, we randomly toggle a fraction P of pixels in an otherwise perfect flat-folding example and recompute the entries of its confusion matrix, as presented in Fig. 3F. With realistic noise levels, i.e., P ~ 10−3, we can expect accuracy values between 0.75 and 0.80 in the upper left and lower right entries of the confusion matrix, comparable to the values reported in Fig. 3E. That is, for experimental flat folding, we achieve accuracy levels that are comparable to what is expected for a perfect prediction with noisy preprocessing.
Experimental crumpling
For crumpling, we train the neural network using a combination of 30% experimental crumpling and 70% in silico flat-folding data, which was noised as described above. We also tried pretraining on in silico data prior to training on crumpling data but observed no improvement. Training on this combined dataset, the resulting predictions accurately reconstruct key features of the crease networks in crumpled sheets, which were not achieved in prior attempts. In Fig. 4, we present predictions on entire sheets (Fig. 4A) and a few closeups on selected regions (Fig. 4B). The confusion matrices suggest that the network is often relatively accurate in predicting regions that are directly near a crease (upper left entry) and large open spaces (lower right entry), classifying these regions with 50 to 60% accuracy. In addition, fig. S3 shows the prediction on each of the 16 successive crumples of the same sheet held out from training.
Fig. 4. Predictions on crumpling.

(A) One sheet that was successively crumpled, shown after four and seven crumpling iterations. Color code follows Fig. 2. (B) Closeups on selected smaller patches from the same image, broken down to prediction, prediction and target, and prediction and input.
The ratio of 70% in silico data was chosen since it provides optimal predictions, as shown in Fig. 5A. We present three different metrics to quantify the predictive power: the L2 loss of Eq. 1, the Pearson correlation between the prediction and the target, and the average of the upper left and lower right of the confusion matrix (classification accuracy). We find that all accuracy metrics are optimized for training on 50 to 70% in silico data. It is also interesting to see in what way this affects the prediction: In Fig. 5D, we show that when trained solely on experimental data, the neural network produces a blurred and indecisive prediction, while for 100% flat-folding data, the network predicts only unrealistic straight and long creases.
Fig. 5. Effect of fraction generated data.
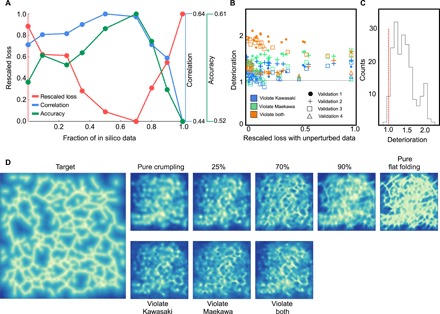
(A) Three quantifications of the predictive power of the model when trained on varying amounts of generated data and a constant amount of crumpling data. Strong predictive power corresponds to low loss (red) and large Pearson correlation and classification accuracy (blue and green, respectively). (B) Deterioration (see main text) for each sheet in the validation set, as a function of the rescaled loss. Colors correspond to different perturbations and marker styles to cross-validation sample. It is seen that all tested perturbations lead to worse predictive power (above the gray reference line). The few points below the reference line occur at high crumple number and low absolute loss. (C) Histogram of all points in (B). Values to the right of the red line correspond to deterioration when using unphysical data. (D) Example target and predictions for the various models considered in previous panels.
In addition to these metrics, one can compare the network’s output to “random” network completion, i.e., to a network that construes a pattern having the statistical properties of a crease network but is only weakly correlated with the input image. Although a generative model for crease networks is not available, we can sample crease patterns from the experimental data and compare the predicted distance maps to those measured from these randomly selected samples. This is discussed in section S5, where it is seen that our prediction for a given crease pattern is overwhelmingly closer to the truth than any sampled patch from other experiments (fig. S5).
The similarity of flat folding and crumpling
These results demonstrate that augmenting the dataset with in silico–generated flat-folding data allows the network to discern some underlying geometric order in crease networks of experimental crumpling data. This suggests that the two systems share some common statistical properties, and it is interesting to ask how robust this similarity is. One may suspect that the main contribution of the in silico data is merely having a multitude of intersecting straight lines, which are the main geometric feature that is analyzed, but that the specific statistics of these lines is not crucial.
As explained above, flat-folding networks are characterized by two theorems: Maekawa’s theorem, which constrains the curvatures (ridge/valley) of creases joined in each vertex, and Kawasaki’s theorem, which constrains the relative angles at vertices. We tested the sensitivity of our prediction to replacing the in silico data used in training with crease networks that violate these rules: We obtained crease networks that violate Maekawa’s theorem by taking flat-folding networks and randomly reassigning curvatures to each crease, and crease networks that violate Kawasaki’s theorem by perturbing all vertex positions. Last, we obtained crease networks that violate both rules by performing both perturbations simultaneously. Examples of perturbed networks are shown in fig. S6, with additional details about the perturbation process.
The effect is quantified in Fig. 5 (B and C). We define, for a given sheet, the “deterioration” as the ratio between the loss of a network trained on 70% experimental data and 30% perturbed flat-folding data to that of a network trained on the same ratio of experimental and unperturbed data. It is seen that breaking the flat-folding rules leads to consistently worse performance for all types of perturbations.
We cross-validated with four different experiments covering a total of 198 sheets. Although for some small fraction (<5%) of the sheets training on perturbed data has led to marginally better performance, this happened mostly in sheets with low loss, and the improvement is negligible. On average, the network trained on perturbed data has a loss approximately 35% higher than that of the network trained on unperturbed data.
These results, namely, that training on perturbed flat-folding networks led to inferior performance, again suggest a similarity between crumpled crease networks and flat-folded networks. We did not quantitatively study the detailed effect of the different kinds of perturbations—i.e., whether violating Kawasaki’s rule, Maekawa’s rule, or both results in more or less accurate predictions. Instead, equipped with this physical insight, we propose to directly probe the statistical similarity with traditional methods by measuring vertex properties in crease networks, a study that will be reported elsewhere.
DISCUSSION
Experimental data are paramount to our understanding of the physical world. However, prohibitive data acquisition rates in many experimental settings require augmenting experimental data to draw meaningful conclusions. In particular, computer simulations now play a significant role in exploratory science; many experimental conditions can be accurately simulated to corroborate our understanding of empirical results.
Despite these advances, the simulation of certain phenomena is inhibited by insufficient theoretical knowledge of the system or by demanding computational resources and development time. For crumpling, without a deeper understanding that would allow the use of simplified/reduced models, simulations require prohibitively small time steps, small domain discretization, or both (26). Here, we show that even with a small experimental training set, augmenting the dataset by computer-generated, artificially noised data of flat folding, salient features of the ridge network can be predicted from the surrounding valleys: The network successfully predicts the presence of certain creases, as well as their pronounced absence in certain locations (see Fig. 4B). Moreover, our results demonstrate a statistical similarity between flat folding and crumpling, evidenced by the fact that when flat-folding data are replaced with data of similar geometry but different statistics, the algorithm does not succeed in learning the underlying distribution to the same extent (Fig. 5B).
Our results demonstrate the capacity of a neural network to learn, at least partially, the structural relationship of ridges and valleys in a crease pattern of crumpled sheets. The next step is to understand the network’s decision process, with the aim of uncovering the physical principles responsible for the observed structure. However, while interpretation of trained weights is currently a heavily researched topic [see (32–34), among many others], there is not yet a standard method to do so. Our ongoing work seeks to probe the network’s inner workings by perturbing the input data. For example, we can individually alter input pixels and quantify the effect of perturbation on the prediction relative to the original target. Alternatively, we can examine the effect of adding or removing creases or test the prediction on inputs that do not occur naturally in crumpled sheets. Some preliminary results are discussed in section S4.
Improving the experimental dataset by performing dedicated experiments or replacing the simulated flat folding with simulated crumpling data is also a promising future direction. While we have only demonstrated the advantages of data augmentation for one problem, it is tempting to imagine how it may apply to other systems in experimental physics. In addition to providing insights into the structure of crease patterns, a quantitative predictive model (i.e., an oracle) could serve as an important experimental tool that allows for targeted experiments, especially when experiments are costly or difficult. As shown above, a trained neural network is able to shed light on where order exists, even if the source of the order is not apparent.
Replacing the scientific discovery process with an automated procedure is risky. Frequently, hypotheses that were initially proposed are not the focal points of the final works they germinated, as observations and insights along the way sculpt the research toward its final state. This serendipitous aspect of discovery has been of immense importance to the sciences and is difficult to include in automated data exploration methods, which is an area of ongoing research (35–37). By showing that data-driven techniques are able to make nontrivial predictions on complicated systems, even in a severely data-limited regime, we hope to demonstrate that these tools should become a valuable tool for experimentalists in many different fields.
MATERIALS AND METHODS
Experiments
Experimental flat-folding and crumpling data were performed on 10 cm by 10 cm sheets of 0.05-mm-thick Mylar. Flat folds were performed successively at random, without allowing the paper to unfold between successive creases. Crumpled sheets were obtained by first rolling the sheet into a 3-cm-diameter cylinder and then applying axial compression to a specified depth between 7.5 and 55 mm. Sheets were successively crumpled between 4 and 24 times.
To image the experimental crease network, crumpled/flat-folded sheets were opened up, and their height profile was scanned using a home-built laser profilometer. The mean curvature map was calculated by differentiating the height profile and then denoised using a custom Radon-based denoiser (the implementation details of which are given in section S1). A total of 506 scans were collected from 31 different experiments.
Network architecture and training
Data were fed into a fully convolutional network, based on the SegNet architecture (31) with the final soft-max layer removed, as we did not perform a classification problem. The depth of the network allows for long-range interactions to be incorporated without fully connected layers. The network was implemented in Mathematica, and optimization was performed using the Adam optimizer (38) on a Tesla 40c graphics processing unit (GPU) with 256 GB of random-access memory (RAM) and a computer with a Titan V GPU and 128 GB of RAM. Code is freely available. See “materials availability” below.
For training, the in silico–generated input data were augmented with standard data augmentation methods: Symmetric copies of each original were generated by reflection and rotation. All images were down sampled to have dimensions of 224 by 224 pixels. For crumpling data, creases were also linearized to look more similar to the experimental input. An example of the effect of linearizing is shown in fig. S2.
Noise
Noise was added to the input in a few different ways (presented in Fig. 3B). The noise of each panel was generated as follows:
A. No noise.
B. “White” noise: Each pixel was randomly toggled with 5% probability.
C. Random blur: Input was convolved with a Gaussian with a width drawn uniformly between 0 and 3. The array was then thresholded at 0.1. Here and below, “thresholded at z” means a pointwise threshold was imposed on the array, such that values smaller than z were set to 0 and otherwise set to 1.
D. Each pixel was randomly toggled with 1% probability and then passed through random blur (C).
E. Input was random blurred [as in (C)] but thresholded at 0.55. We denote the blurred-and-thresholded input as . Then, was noised using both additive and multiplicative noise, as follows: Y and Z are two random fields drawn from a pointwise uniform distribution between 0 and 1 and convolved with a Gaussian of width seven (pixels) and thresholded at 0.55. Last, the “noised” input is
Supplementary Material
Acknowledgments
We thank an anonymous referee for the suggestion to perturb the in silico input data. Funding: This work was supported by the National Science Foundation through the Harvard Materials Research Science and Engineering Center (DMR-1420570). S.M.R. acknowledges support from the Alfred P. Sloan Research Foundation. The GPU computing unit was obtained through the NVIDIA GPU grant program. J.H. was supported by a Computational Science Graduate Fellowship (DOE CSGF). Y.B.-S. was supported by the JSMF postdoctoral fellowship for the study of complex systems. C.H.R. was partially supported by the Applied Mathematics Program of the U.S. DOE Office of Advanced Scientific Computing Research under contract no. DE-AC02-05CH11231. Author contributions: S.M.R. and C.H.R. conceived the concepts of the research. Y.B.-S., J.H., S.M.R., and C.H.R. designed the research. L.M.L. performed new experiments. J.A. and J.H. performed the image preprocessing. J.A. wrote the Radon transform methods code. Y.B.-S., J.H., and S.M. contributed to the machine learning. C.H.R. and J.H. wrote the flat-folding code. All authors analyzed the data and wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: Source code is available at https://github.com/hoffmannjordan/Crumpling. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaau6792/DC1
Section S1. Radon transform–based detection method
Section S2. In silico generation of flat-folding data
Section S3. Prediction on 16 sheets
Section S4. Probing the network: Ongoing work
Section S5. Another approach to error quantification
Section S6. Perturbing the in silico data
Fig. S1. In silico–generated flat-folded crease networks.
Fig. S2. Comparison between the preprocessed curvature map and the linearized version.
Fig. S3. Prediction on a sheet that was crumpled 16 times.
Fig. S4. Additional test results.
Fig. S5. Prediction accuracy.
Fig. S6. Examples of perturbed in silico data.
Reference (39)
REFERENCES AND NOTES
- 1.LeCun Y., Bengio Y., Hinton G., Deep learning. Nature 521, 436–444 (2015). [DOI] [PubMed] [Google Scholar]
- 2.A.-R. Mohamed, G. Dahl, G. Hinton, Deep belief networks for phone recognition, in Proceedings of the NIPS Workshop on Deep Learning for Speech Recognition and Related Applications (Neural Information Processing Systems Foundation, 2009), p. 39. [Google Scholar]
- 3.G. Dahl, D. Yu, L. Deng, A. Acero, Large vocabulary continuous speech recognition with context-dependent DBN-HMMS, in Proceedings of the 2011 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (IEEE, 2011), pp. 4688–4691. [Google Scholar]
- 4.L. Deng, J. Li, J.-T. Huang, K. Yao, D. Yu, F. Seide, M. Seltzer, G. Zweig, X. He, J. Williams, Y. Gong, A. Acero, Recent advances in deep learning for speech research at Microsoft, in Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP) (IEEE, 2013), pp. 8604–8608. [Google Scholar]
- 5.LeCun Y., Bottou L., Bengio Y., Haffner P., Gradient-based learning applied to document recognition. Proc. IEEE 86, 2278–2324 (1998). [Google Scholar]
- 6.Shameer K., Johnson K. W., Glicksberg B. S., Dudley J. T., Sengupta P. P., Machine learning in cardiovascular medicine: Are we there yet? Heart 104, 1156–1164 (2018). [DOI] [PubMed] [Google Scholar]
- 7.V. Mnih, K. Kavukcuoglu, D. Silver, A. Graves, I. Antonoglou, D. Wierstra, M. Riedmiller, Playing atari with deep reinforcement learning. arXiv:1312.5602 [cs.LG] (19 December 2013).
- 8.Silver D., Schrittwieser J., Simonyan K., Antonoglou I., Huang A., Guez A., Hubert T., Baker L., Lai M., Bolton A., Chen Y., Lillicrap T., Hui F., Sifre L., van den Driessche G., Graepel T., Hassabis D., Mastering the game of go without human knowledge. Nature 550, 354–359 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Heaton J. B., Polson N. G., Witte J. H., Deep learning for finance: Deep portfolios. Appl. Stoch. Models Bus. Ind. 33, 3–12 (2017). [Google Scholar]
- 10.Joel S., Eastwick P. W., Finkel E. J., Is romantic desire predictable? Machine learning applied to initial romantic attraction. Psychol. Sci. 28, 1478–1489 (2017). [DOI] [PubMed] [Google Scholar]
- 11.W. Bhimji, S. A. Farrell, T. Kurth, M. Paganini, Prabhat, E. Racah, Deep neural networks for physics analysis on low-level whole-detector data at the LHC. arXiv:1711.03573 [hep-ex] (9 November 2017).
- 12.Baldi P., Sadowski P., Whiteson D., Searching for exotic particles in high-energy physics with deep learning. Nat. Commun. 5, 4308 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Banerji M., Lahav O., Lintott C. J., Abdalla F. B., Schawinski K., Bamford S. P., Andreescu D., Murray P., Raddick M. J., Slosar A., Szalay A., Thomas D., Vandenberg J., Galaxy zoo: Reproducing galaxy morphologies via machine learning. Mon. Not. R. Astron. Soc. 406, 342–353 (2010). [Google Scholar]
- 14.C. Sommer, C. Straehle, U. Koethe, F. A. Hamprecht, Ilastik: Interactive learning and segmentation toolkit, in Proceedings of the 2011 IEEE International Symposium on Biomedical Imaging: From Nano to Macro (IEEE, 2011), pp. 230–233. [Google Scholar]
- 15.Gulshan V., Peng L., Coram M., Stumpe M. C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P. C., Mega J. L., Webster D. R., Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316, 2402–2410 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Carrasquilla J., Melko R. G., Machine learning phases of matter. Nat. Phys. 13, 431–434 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spellings M., Glotzer S. C., Machine learning for crystal identification and discovery. AIChE J. 64, 2198–2206 (2018). [Google Scholar]
- 18.Cubuk E. D., Schoenholz S. S., Rieser J. M., Malone B. D., Rottler J., Durian D. J., Kaxiras E., Liu A. J., Identifying structural flow defects in disordered solids using machine-learning methods. Phys. Rev. Lett. 114, 108001 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Sussman D. M., Schoenholz S. S., Cubuk E. D., Liu A. J., Disconnecting structure and dynamics in glassy thin films. Proc. Natl. Acad. Sci. U.S.A. 114, 10601–10605 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenholz S. S., Cubuk E. D., Kaxiras E., Liu A. J., Relationship between local structure and relaxation in out-of-equilibrium glassy systems. Proc. Natl. Acad. Sci. U.S.A. 114, 263–267 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amar M. B., Pomeau Y., Crumpled paper. Proc. R. Soc. Lond. A 453, 729 (1997). [Google Scholar]
- 22.Witten T. A., Stress focusing in elastic sheets. Rev. Mod. Phys. 79, 643–675 (2007). [Google Scholar]
- 23.Aharoni H., Sharon E., Direct observation of the temporal and spatial dynamics during crumpling. Nat. Mater. 9, 993–997 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Andresen C. A., Hansen A., Schmittbuhl J., Ridge network in crumpled paper. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 76, 026108 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Gottesman O., Andrejevic J., Rycroft C. H., Rubinstein S. M., A state variable for crumpled thin sheets. Commun. Phys. 1, 70 (2018). [Google Scholar]
- 26.Narain R., Pfaff T., O’Brien J. F., Folding and crumpling adaptive sheets. ACM Trans. Graph. 32, 1–8 (2013). [Google Scholar]
- 27.Guo Q., Han X., Fu C., Gast T., Tamstorf R., Teran J., A material point method for thin shells with frictional contact. ACM Trans. Graph. 37, 147 (2018). [Google Scholar]
- 28.Pan S. J., Yang Q., A survey on transfer learning. IEEE Trans. Knowl. Data Eng. 22, 1345–1359 (2010). [Google Scholar]
- 29.Turner N., Goodwine B., Sen M., A review of origami applications in mechanical engineering. Proc. Inst. Mech. Eng. C J. Mech. Eng. Sci. 230, 2345–2362 (2016). [Google Scholar]
- 30.Rycroft C. H., Voro++: A three-dimensional Voronoi cell library in C++. Chaos 19, 041111 (2009). [DOI] [PubMed] [Google Scholar]
- 31.Badrinarayanan V., Kendall A., Cipolla R., Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 39, 2481–2495 (2017). [DOI] [PubMed] [Google Scholar]
- 32.D. Smilkov, N. Thorat, C. Nicholson, E. Reif, F. B. Vi’egas, M. Wattenberg, Embedding projector: Interactive visualization and interpretation of embeddings. arXiv:1611.05469 [stat.ML] (16 November 2016).
- 33.N. Frosst, G. Hinton, Distilling a neural network into a soft decision tree. arXiv:1711.09784 [cs.LG] (27 November 2017).
- 34.M. Sundararajan, A. Taly, Q. Yan, Axiomatic attribution for deep networks. arXiv:1703.01365 [cs.LG] (4 March 2017).
- 35.Raccuglia P., Elbert K. C., Adler P. D. F., Falk C., Wenny M. B., Mollo A., Zeller M., Friedler S. A., Schrier J., Norquist A. J., Machine-learning-assisted materials discovery using failed experiments. Nature 533, 73–76 (2016). [DOI] [PubMed] [Google Scholar]
- 36.Baltz E. A., Trask E., Binderbauer M., Dikovsky M., Gota H., Mendoza R., Platt J. C., Riley P. F., Achievement of sustained net plasma heating in a fusion experiment with the optometrist algorithm. Sci. Rep. 7, 6425 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren F., Ward L., Williams T., Laws K. J., Wolverton C., Hattrick-Simpers J., Mehta A., Accelerated discovery of metallic glasses through iteration of machine learning and high-throughput experiments. Sci. Adv. 4, eaaq1566 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D. P. Kingma, J. Ba, Adam: A method for stochastic optimization. arXiv:1412.6980 [cs.LG] (22 December 2014).
- 39.J. Lehman, J. Clune, D. Misevic, C. Adami, L. Altenberg, J. Beaulieu, P. J. Bentley, S. Bernard, G. Beslon, D. M. Bryson, P. Chrabaszcz, N. Cheney, A. Cully, S. Doncieux, F. C. Dyer, K. O. Ellefsen, R. Feldt, S. Fischer, S. Forrest, A. Frénoy, C. Gagné, L. L. Goff, L. M. Grabowski, B. Hodjat, F. Hutter, L. Keller, C. Knibbe, P. Krcah, R. E. Lenski, H. Lipson, R. MacCurdy, C. Maestre, R. Miikkulainen, S. Mitri, D. E. Moriarty, J.-B. Mouret, A. Nguyen, C. Ofria, M. Parizeau, D. Parsons, R. T. Pennock, W. F. Punch, T. S. Ray, M. Schoenauer, E. Shulte, K. Sims, K. O. Stanley, F. Taddei, D. Tarapore, S. Thibault, W. Weimer, R. Watson, J. Yosinski, The surprising creativity of digital evolution: A collection of anecdotes from the evolutionary computation and artificial life research communities. arXiv:1803.03453 [cs.NE] (9 March 2018).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/4/eaau6792/DC1
Section S1. Radon transform–based detection method
Section S2. In silico generation of flat-folding data
Section S3. Prediction on 16 sheets
Section S4. Probing the network: Ongoing work
Section S5. Another approach to error quantification
Section S6. Perturbing the in silico data
Fig. S1. In silico–generated flat-folded crease networks.
Fig. S2. Comparison between the preprocessed curvature map and the linearized version.
Fig. S3. Prediction on a sheet that was crumpled 16 times.
Fig. S4. Additional test results.
Fig. S5. Prediction accuracy.
Fig. S6. Examples of perturbed in silico data.
Reference (39)


