Abstract
A key mechanism of tumor resistance to immune cells is mediated by expression of peptide-loaded HLA class I molecule (HLA-E) in tumor cells, which suppresses NK cell activity via ligation of the NK inhibitory receptor CD94/NK group 2 member A (NKG2A). Gene expression data from approximately 10,000 tumor samples showed widespread HLAE expression, with levels correlating with those of KLRC1 (NKG2A) and KLRD1 (CD94). To bypass HLA-E inhibition, we developed a way to generate highly functional NK cells lacking NKG2A. Constructs containing a single-chain variable fragment derived from an anti-NKG2A antibody were linked to endoplasmic reticulum–retention domains. After retroviral transduction in human peripheral blood NK cells, these NKG2A protein expression blockers (PEBLs) abrogated NKG2A expression. The resulting NKG2Anull NK cells had higher cytotoxicity against HLA-E–expressing tumor cells. Transduction of anti-NKG2A PEBL produced more potent cytotoxicity than interference with an anti-NKG2A antibody and prevented de novo NKG2A expression without affecting NK cell proliferation. In immunodeficient mice, NKG2Anull NK cells were substantially more powerful than NKG2A+ NK cells against HLA-E–expressing tumors. Thus, NKG2A downregulation evades the HLA-E cancer immune checkpoint and increases the antitumor activity of NK cell infusions. Because this strategy is easily adaptable to current protocols for clinical-grade immune cell processing, its clinical testing is feasible and warranted.
Keywords: Immunology, Oncology
Keywords: Immunotherapy, Innate immunity, NK cells
Introduction
Immunotherapy has become a mainstay of contemporary oncology. Major clinical responses achieved with T cell immune-checkpoint inhibitors and chimeric antigen receptor–directed (CAR-directed) T lymphocytes (1, 2) encourage the evaluation of other immunotherapeutic approaches. To this end, the anticancer potential of NK cell infusions has not yet been fully explored. In patients with acute myeloid leukemia (AML), responses associated with NK cells have been reported after allogeneic hematopoietic stem cell transplantation (3–5) and after infusion of NK cells activated ex vivo by cytokines (4, 6). Clinical activity, however, remains difficult to predict, responses are often not durable, and therapeutic potential against tumors beyond AML is unclear (7, 8).
NK cells have the inherent capacity to identify, and specifically eliminate, virally infected or transformed cells (7–9). NK activating and inhibitory receptors, ligated to varying degrees by molecules expressed on candidate target cells, regulate these functions (8, 9). The CD94/NK group 2 member A (NKG2A) heterodimeric receptor is one of the most prominent NK inhibitory receptors (10). It binds to a nonclassical minimally polymorphic HLA class I molecule (HLA-E), which presents peptides derived from leader peptide sequences of other HLA class I molecules, such as HLA-G (11–15). Upon ligation by peptide-loaded HLA-E, NKG2A transduces inhibitory signaling through 2 inhibitory immune-receptor tyrosine-based inhibition motifs, thus suppressing NK cytokine secretion and cytotoxicity (16–18).
Although HLA-E transcripts are ubiquitously expressed (19), surface expression of HLA-E is often weak or undetectable in tumor cell lines (20). During antitumor immune responses, however, IFN-γ secreted by immune cells upregulates HLA-E expression in tumor cells (21–23). Stabilized by peptide loading (24), the HLA-E/peptide complex dampens initial NK cell responses by binding CD94/NKG2A. HLA-E is indeed overexpressed in several tumors (25–33), and a correlation between higher HLA-E and poorer prognosis has been observed (25,27). Moreover, NKG2A+ NK cells are reportedly predominant in the tumor microenvironment. Thus, intratumoral NK cells in patients with non–small cell lung cancer have higher expression of NKG2A than those obtained from nontumoral distant sites (34, 35). Likewise, NKG2A expression is higher in NK cells infiltrating breast cancer (BRCA) tumors than in those isolated from symmetric normal breast tissue (36) and in peripheral blood NK cells of patients with AML compared with NK cells of age-matched controls (37). Because of the strong evidence for the suppressive effect of NKG2A on NK cell activity, this immune checkpoint has been targeted with antibodies, currently in clinical development, that block its interaction with HLA-E, currently in clinical development (38–41).
In this study, we determined HLAE expression in approximately 10,000 tumor samples and assessed its relation with that of KLRC1 (NKG2A) and KLRD1 (CD94). Then we sought to develop NK cells that were impervious to the inhibitory effects of HLA-E. To this end, we designed constructs, named protein expression blockers (PEBLs), that prevent the transport of NKG2A to the cell-surface membrane of NK cells. Finally, we tested to determine whether downregulation of NKG2A could overcome the HLA-E–mediated mechanism of tumor resistance to NK cell killing.
Results
HLAE expression in tumor samples, relation with immune cell infiltration, and KLRC1/KLRD1 expression.
We performed an unbiased analysis of HLAE expression in human tumors using data from 10,375 tumor samples, representing 33 tumor types, made available by The Cancer Genome Atlas (TCGA) Research Network. We found considerable inter- and intratumor heterogeneity in HLAE expression (Figure 1A). Tumor types with more than 100 samples whose median HLAE expression exceeded the median value (log10 transcripts per kilobase million of 3.92) included kidney renal clear cell carcinoma (KIRC), head and neck squamous cell carcinoma (HNSC), lung adenocarcinoma (LUAD), skin cutaneous melanoma (SKCM), prostate adenocarcinoma (PRAD), stomach adenocarcinoma (STAD), colon adenocarcinoma (COAD), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), lung squamous cell carcinoma (LUSC), and rectum adenocarcinoma (READ).
Figure 1. Expression of HLAE in tumors and its relation with KLRC1 (NKG2A) expression.
(A) Expression of HLAE in 10,375 specimens from 33 tumor types; box plots with data from every sample superimposed show log10 HLAE gene transcripts per kilobase million (TPM) in adrenocortical carcinoma (ACC), BLCA, BRCA, CESC, cholangiocarcinoma (CHOL), COAD, diffuse large B cell lymphoma (DLBC), esophageal carcinoma (ESCA), GBM, HNSC, kidney chromophobe (KICH), KIRC, kidney renal papillary cell carcinoma (KIRP), AML, LGG, liver hepatocellular carcinoma (LIHC), LUAD, LUSC, mesothelioma (MESO), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), PCPG, PRAD, READ, SARC, SKCM, STAD, testicular germ cell tumors (TGCT), THCA, THYM, uterine corpus endometrial carcinoma (UCEC), uterine carcinosarcoma (UCS), uveal melanoma (UVM). Box boundaries, first and third quartile range; whisker, interquartile range (first quartile to third quartile range) ×1.5. (B) Shown is log2 normalized expression of HLAE with KLRC1, KLRD1 (CD94), or KLRC2 (NKG2C) in 9520 tumors analyzed (tumors lacking KLRC1 expression were excluded). Pearson’s correlation coefficient and linear regression line are shown. (C) Relation between log2 normalized expression of HLAE and KLRC1. The 9520 tumor specimens were ordered by expression of HLAE or KLRC1; the corresponding expression of KLRC1 and HLAE is shown. (D) Relation between HLAE and KLRC1 expression in tumors with high Pearson’s correlation coefficient.
When all tumors were analyzed collectively, there was a direct relation between levels of HLAE expression and those of KLRC1 (NKG2A) (Pearson’s correlation coefficient r = 0.48) and KLRD1 (CD94) (r = 0.57), while there was no relation with the activating member of the NKG2 family KLRC2 (NKG2C) (r = 0.07) (Figure 1, B and C). The correlation between HLAE and KLRC1 was particularly evident in some tumors, such as bladder urothelial carcinoma (BLCA), sarcoma (SARC), CESC, HNSC, and PRAD (Figure 1D).
We further assessed the correlation between HLAE and KLRC1 using the Tumor IMmune Estimation Resource (TIMER; https://cistrome.shinyapps.io/timer/), which allows estimates of immune-cell infiltration (42). Among tumors with more than 100 samples, the highest correlation values were observed in BLCA, thyroid carcinoma (THCA), thymoma (THYM), HNSC, and BRCA; the lowest were in low-grade glioma (LGG), glioblastoma multiforme (GBM), KIRC, and pheochromocytoma and paraganglioma (PCPG) (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI123955DS1). Expression of HLAE correlated with that of genes encoding IFN-γ, markers associated with cytolytic immune cells (perforin, granzyme A), NK cells (NKG2D, NCR3, CD16), and T cells (CD3, CD8, CD4) (Supplemental Figure 1). The correlations with KLRC1 were maintained after conditioning the analysis for tumor purity (Supplemental Table 1 and Supplemental Figure 2), suggesting a relation between HLAE and KLRC1 regardless of the degree of immune cell infiltration. KLRC1 expression in tumor tissues was also strongly associated with that of genes encoding IFN-γ, perforin, and granzymes as well as with genes expressed in NK and T cells (Supplemental Figure 3).
Downregulation of NKG2A in NK cells.
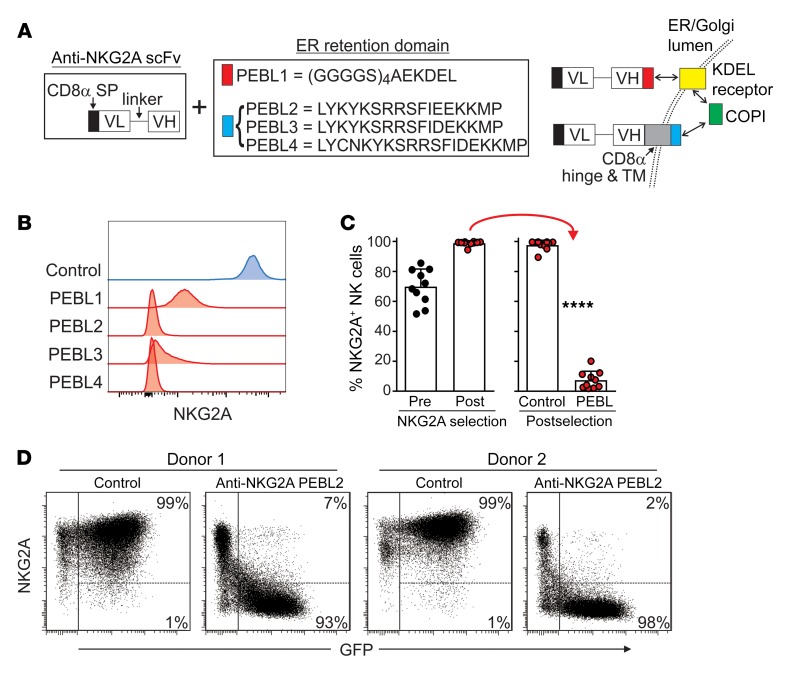
We reasoned that the antitumor capacity of NK cell infusions in an HLA-E–rich tumor microenvironment would increase if NK cells could bypass the HLA-E checkpoint. To this end, we designed PEBLs consisting of a single-chain variable fragment (scFv) derived from the sequence of an anti-NKG2A monoclonal antibody (Z199) (17, 43) linked to 4 different endoplasmic reticulum (ER) retention domains (Figure 2A). PEBL1 contains a (GGGGS)4AEKDEL domain; it binds the KDEL (lysine, aspartic acid, glutamic acid, leucine) peptide receptor, linked to the coat protein complex I (COPI) vesicular transport, which mediates protein traffic between ER and Golgi (44, 45). PEBLs 2–4 contain different peptides, including KKMP (lysine, lysine, methionine, proline) domains, together with CD8α hinge and transmembrane domains; these allow direct binding to COPI (45, 46).
Figure 2. Downregulation of NKG2A expression in NK cells with anti-NKG2A PEBLs.
(A) Schematic representation of the anti-NKG2A PEBL constructs and their mechanisms of action. The PEBL constructs consist of a CD8α signal peptide and an anti-NKG2A scFv followed, at the C terminus, by the sequences listed in the box, according to each PEBL. PEBL1 binds to the KDEL receptor, which joins the COPI. PEBLs 2–4 bind directly to COPI. VL, light chain variable domain; VH, heavy chain variable domain. (B) Downregulation of NKG2A expression in NK92 cells. Flow cytometric histograms show surface expression of NKG2A, as detected by anti-NKG2A APC (Miltenyi Biotech), after transduction with a vector containing GFP only (Control) or GFP plus PEBLs 1–4. (C) NKG2A+ expanded human NK cells were purified by magnetic bead–positive selection and transduced with anti–NKG2A-PEBL2 or with GFP only. Shown are the percentages of NKG2A+ cells before and after purification and after transduction (10 experiments with NK cells from 9 donors), as measured by flow cytometry. ****P < 0.0001, t test. (D) Representative flow cytometry dot plots of 2 of the experiments shown in C. The right area in each dot plot encloses GFP+ (i.e., transduced) NK cells; the percentages of NKG2A+ and NKG2A cells among these cells are shown.
To test the capacity of PEBLs to downregulate NKG2A, we inserted the constructs into a murine stem cell virus (MSCV) retroviral vector containing GFP and used it to express the anti-NKG2A PEBLs in the NKG2A+ cell line NK92. NKG2A expression was essentially abrogated in all GFP+ NK92 cells (Figure 2B). PEBLs were not detectable on the cell surface, whereas a surface membrane-bound anti-NKG2A scFv used as a control was highly expressed (Supplemental Figure 4), indicating that PEBLs were confined to the intracellular space.
To determine whether anti-NKG2A PEBLs could also downregulate NKG2A expression in primary human NK cells, we expanded peripheral blood NK cells (n = 10 from 9 donors) by 5- to 6-day stimulation with the K562-mb15-41BBL cell line (47, 48). We then purified NKG2A+ NK cells by magnetic bead–positive selection and transduced them with anti-NKG2A-PEBL2. The percentage of NKG2A+ cells before purification was 69.8% ± 11.8%, and this was increased to 98.8% ± 1.6% after purification. Transduction of anti-NKG2A PEBL markedly reduced NKG2A. Among GFP+ cells, representing 49.1 % to 82.0% (median, 68.2%) of all NK cells, NKG2A+ cells were 7.5% ± 5.9%, while in cells transduced with a vector containing only GFP, NKG2A expression remained high (97.6% ± 3.2%; P < 0.0001) (Figure 2, C and D).
Cell markers, gene expression, and functional features of NKG2Anull NK cells.
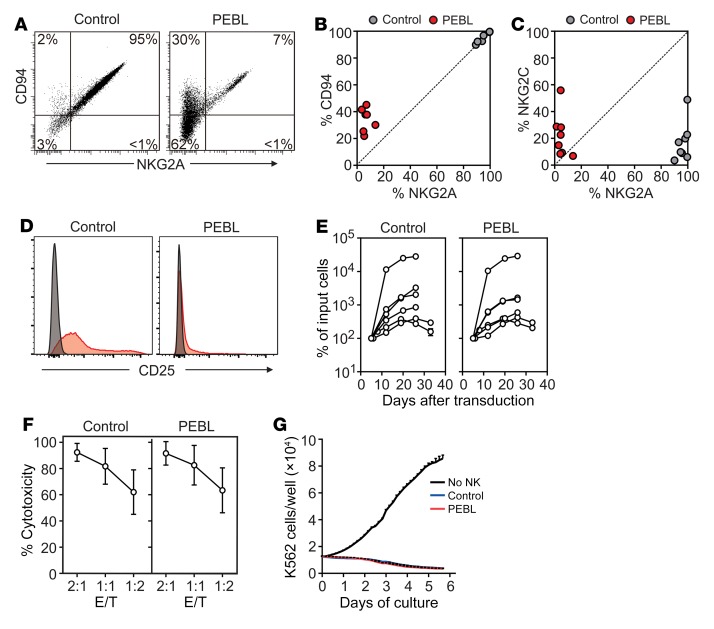
Anti-NKG2A PEBL transduction resulted in downregulation of CD94 surface expression (Figure 3, A and B). In the 7 donors tested, residual CD94+ cells represented 21.9%–45.1% (median, 37.8%) of NK cells transduced with PEBL versus 89.9%–99.7% (median, 96.8%) of NK cells transduced with GFP alone. After staining the cells with the anti-NKG2A antibody and testing for CD94 expression after cell permeabilization, we found that CD94 was expressed in PEBL cells at levels similar to those measured in control cells (Supplemental Figure 5), suggesting that CD94 was synthesized normally in PEBL cells, but was less expressed on the cell membrane owing to the lack of NKG2A expression.
Figure 3. Phenotypic and functional effects of NKG2A downregulation by PEBL.
(A) Downregulation of NKG2A is accompanied by decrease of surface CD94 expression. Flow cytometry dot plots show expression of NKG2A (CD159a PE) and of CD94 (anti-CD94 APC) in a representative sample of NK cells expanded by coculture with K562-mb15-41BBL and transduced with either anti-NKG2A PEBL-2 or GFP alone (Control). (B) Summary of CD94 versus NKG2A expression results obtained with NK cells from 7 donors. (C) NKG2C (CD159c APC) versus NKG2A expression obtained with NK cells from 8 donors. (D) Expression of CD25 in PEBL-transduced and control NK cells. Flow cytometry histograms show expression of CD25 (red, detected with anti–CD25 PE-Cy7); staining with isotype-matched nonreactive antibody (mouse IgG1 PE-Cy7) is shown in gray. (E) Survival and expansion of PEBL-transduced and control NK cells from 6 donors cultured with IL-2 (400 IU/ml). Data are shown as mean (± SD) of triplicate measurements at each time point. (F) Results of 4-hour cytotoxicity assays performed against luciferase-labeled K562 cells. BrightGlo was added after 4 hours of coculture, and luminescence was measured using a Flx 800 plate reader. Data are shown as mean (± SD) of triplicate measurements with NK cells from 8 donors. (G) Long-term cytotoxicity of PEBL-transduced and control NK cells against mCherry-transduced K562 cells at 1:8 E/T. K526 cell growth was measured with IncuCyte Zoom System. Data are shown as mean (± SD) of triplicate measurements with NK cells from 1 donor and of cultures without NK cells.
Anti-NKG2A PEBL transduction did not affect expression of other NK cell molecules, including CD56, CD16, CD69, CD57, CD335 (NKp46), CD336 (NKp44), CD337 (NKp30), CD158a, CD158b, CD158e, CD226 (DNAM-1), NKG2D and CD137 (4-1BB), granzyme A, granzyme B, and perforin (Supplemental Figure 6A). Expression of CD159c (NKG2C) was also similar in PEBL-transduced and control NK cells (Figure 3C). In the 8 donors tested, NKG2C+ cells represented 6.9%–55.9% (median, 18.8%) of NK cells transduced with PEBL versus 3.5%–48.9% (median, 13.5%) of NK cells transduced with GFP alone. We noted that expression of CD25 was lower in NK cells transduced with anti-NKG2A PEBL than in those transduced with GFP alone (Figure 3D). Nevertheless, PEBL-transduced NK cells could expand as well as control NK cells with IL-2 (Figure 3E). Likewise, the IL-2–dependent cell line NK92 continued to proliferate normally after downregulation of NKG2A (Supplemental Figure 6B).
We applied RNA-Seq to examine the expression of 44 genes encoding NK activating and inhibitory receptors, cytokines and cytokine receptors, and cytolytic molecules. There were no differences in mRNA expression for NKG2A, CD94, and NKG2C, and none of the other 41 genes was marked as a “discovery” after controlling for false discovery rate (Supplemental Table 2).
To further examine the functionality of NKG2Anull NK cells, we tested their capacity to lyse HLA-E–negative K562 leukemia cells. In both short- and long-term cultures, anti-NKG2A PEBL-transduced NK cells were as powerful as control NK cells (Figure 3, F and G). In line with these results, NKG2Anull cells and control NK cells had equal capacities to exocytose cytotoxic granules in the presence of target cells, as shown by CD107a staining (Supplemental Figure 6C). IFN-γ secretion after target stimulation was also maintained (Supplemental Figure 6D). Hence, downregulation of NKG2A by PEBL did not impair NK cell function.
NKG2Anull NK cells avert suppression by HLA-E–expressing tumor cells.
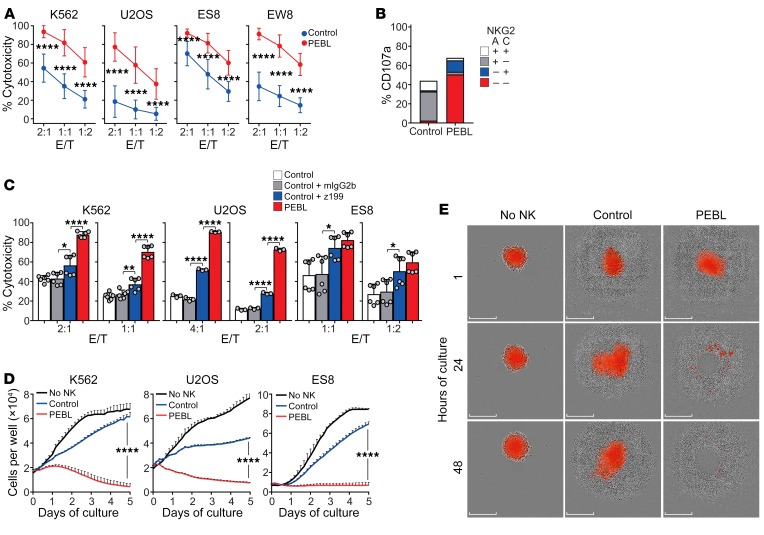
To determine whether NKG2A downregulation released NK cells from HLA-E–mediated inhibition, we generated tumor cells with strong NKG2A-binding potential. For this purpose, we transduced the ligand of NKG2A, i.e., HLA-E plus HLA-G signal peptide (GpHLA-E) (49, 50), in tumor cell lines derived from AML (K562), osteosarcoma (U2OS), and Ewing’s sarcoma (ES8 and EW8) (Supplemental Figure 7A). These cell lines express a variable profile of inhibitory and activating NK ligands: the inhibitory HLA-class I is absent or dim in K562 and ES8, but highly expressed in U2OS and EW8; the activating NKG2D ligands MICA/B and ULBP1-3 and the DNAM-1 ligands CD112 and CD155 are expressed in all 4 lines, but at generally low levels in ES8 and EW8 (51). Regardless, GpHLA-E expression markedly inhibited CD107a secretion by NKG2A+ NK cells and NK cytotoxicity overall (Supplemental Figure 7, B and C).
We used these targets to test the function of NKG2Anull NK cells. In 4-hour cytotoxicity assays, NKG2Anull NK cells had a markedly higher killing capacity than control NK cells (P < 0.0001 for all comparisons; Figure 4A). Exocytosis of lytic granules marked by CD107a expression was not confined to NKG2C+ cells; in fact, most CD107a+ cells lacked both NKG2A and NKG2C expression (Figure 4B). Importantly, NKG2A downregulation did not increase cytotoxicity against autologous activated CD4+ T cells (Supplemental Figure 8A), an effect that NKG2A had been previously reported to suppress (52, 53). Cytotoxicity against bone marrow–derived mesenchymal cells (54) also remained low, regardless of IFN-γ stimulation (Supplemental Figure 8B). Thus, gains in antitumor activity by NKG2A downregulation were not accompanied by increased toxicity against nontransformed cells.
Figure 4. Downregulation of NKG2A increases NK cell cytotoxicity against tumor cells expressing HLA-E with HLA-G signal peptide (GpHLA-E).
(A) Four-hour cytotoxicity assays with NKG2A+ NK cells transduced with anti-NKG2A PEBL or GFP only (Control). Target cell lines were transduced with GpHLA-E (see Supplemental Figure 7) and luciferase. BrightGlo was added after 4 hours of coculture, and luminescence was measured using a Flx 800 plate reader. Data are shown as mean (± SD) of cell killing using target cells cultured without NK cells as reference. NK cells from 11 donors were tested with K562, 6 with U2OS, 8 with ES8, and 5 with EW8, all in triplicate. (B) Expression of CD107a among NK cell subsets after 4-hour coculture with K562-GpHLA-E cells. Percentages are shown as mean of triplicate measurements with NK cells from 1 donor. (C) Four-hour cytotoxicity (measured as in A) of GFP-transduced NK cells against GpHLA-E–transduced target cells in the presence of the anti-NKG2A antibody Z199 compared with that of anti-NKG2A PEBL-transduced NK cells. An isotype-matched (mIgG2b) nonreactive immunoglobulin served as a control. Data are shown as mean (± SD) of triplicate measurements with NK cells from 2 donors (K562, ES8) or 1 donor (U2OS). (D) Long-term cytotoxicity of PEBL-transduced and control NK cells against GpHLA-E+ target cells. Experiments were performed at E/T 1:8 for K562, 1:2 for U2OS, and 1:4 for ES8. Tumor cell growth was measured with IncuCyte Zoom System. Data are shown as mean (± SD) of triplicate measurements with NK cells from 1 representative donor (out of 3 tested) compared with growth of the cell line without NK cells. (E) Spheroid tumors formed with U2OS-GpHLA-E cells transduced with mCherry were cocultured with PEBL-transduced or control NK cells at a 2:1 E/T. Images were collected with the IncuCyte Zoom System. Scale bars: 300 μm. Numerical data are shown in Supplemental Figure 9. *P < 0.05; **P < 0.01; ****P < 0.0001, t test.
Antibody binding to NKG2A may reduce its NK inhibitory activity (38, 39, 41, 55). To compare the effect of anti-NKG2A PEBL to that of antibody interference with NKG2A, we preincubated NKG2A+ cells with either the anti-NKG2A antibody Z199 or an isotype-matched nonreactive antibody. We then tested their capacity to kill GpHLA-E–transduced K562, U2OS, and ES8 cells. Although Z199 significantly improved the cytotoxicity of control NK cells against all 3 targets, the same NK cells transduced with anti-NKG2A PEBL were consistently more cytotoxic (Figure 4C).
To further test the gains in antitumor activity afforded by NKG2A downregulation, we performed long-term cytotoxicity assays at low (1:2–1:8) effector/target cell (E/T) ratios against GpHLA-E–transduced cell lines. As shown in Figure 4D, tumor cell killing was again markedly superior with the anti-NKG2A PEBL cells. These results were corroborated in experiments in which U2OS-GpHLA-E was grown as a spheroid. As shown in Figure 4E and Supplemental Figure 9, NKG2Anull cells were clearly more powerful.
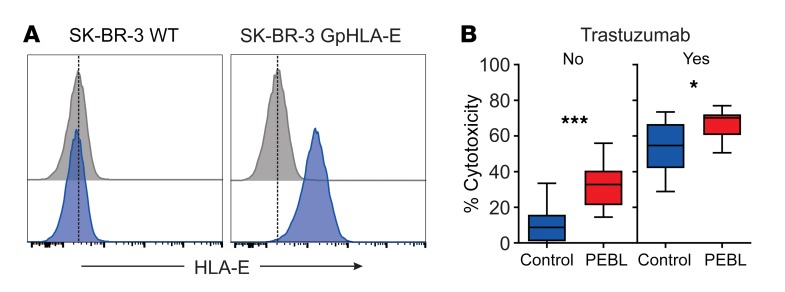
Finally, we determined whether antibody-dependent cell cytotoxicity (ADCC) was affected by the downregulation of NKG2A with anti-NKG2A PEBL. For this purpose, we transduced the HER2+ BRCA cell line SK-BR-3 with GpHLA-E (Figure 5A) and used these cells as targets for the anti-Her2 antibody trastuzumab. As shown in Figure 5B, NKG2Anull cells were more powerful against GpHLA-E–SK-BR-3 cells in the absence of antibody (P = 0.0001) and maintained superior cytotoxicity in the presence of trastuzumab (P = 0.018).
Figure 5. Downregulation of NKG2A increases ADCC activity.
(A) Expression of HLA-E in the SK-BR-3 cell line transduced with HLA-E plus HLA-G signal peptide (GpHLA-E) or nontransduced (WT). Cells were labeled with APC-conjugated anti–HLA-E APC (blue) or isotype-matched nonreactive antibody (gray). (B) Four-hour cytotoxicity with NK cells transduced with anti-NKG2A PEBL or GFP alone (Control) against SK-BR-3-GpHLA-E cells expressing luciferase. BrightGlo was added after 4 hours of coculture, and luminescence was measured using a Flx 800 plate reader. Box (25th–75th percentile, median) and whiskers (minimum-maximum) graphs indicate the collective results of triplicate measurements obtained with NK cells from 2 donors, at a 1:1 or 1:2 E/T. Trastuzumab was added at 10 μg/ml. Horizontal bars correspond to median value. ***P = 0.0001; *P = 0.018, t test.
NKG2A downregulation increases NK cell killing of tumor cells exposed to IFN-γ and suppresses NKG2A upregulation by IL-12.
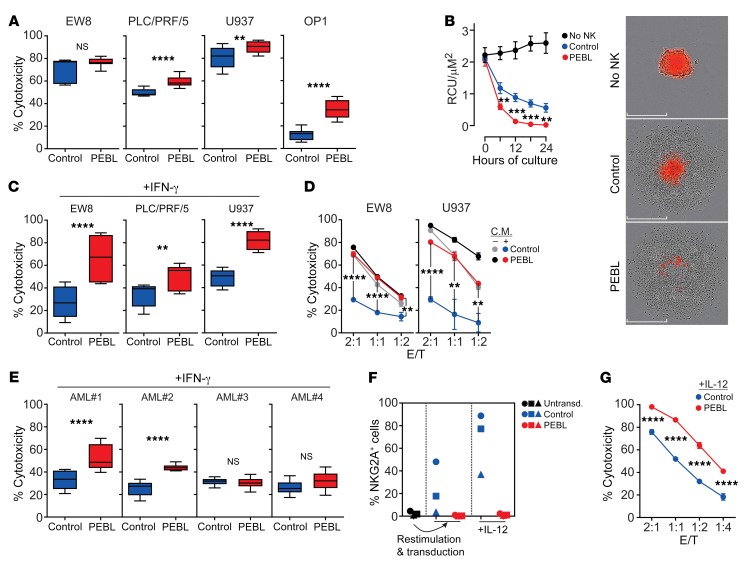
The above experiments showed that downregulation of NKG2A dramatically reduced the inhibition exerted by transduced GpHLA-E. We identified cell lines with endogenous surface HLA-E expression (Supplemental Figure 10) and tested their relative susceptibility to cell killing by NK cells with or without NKG2A downregulation. As shown in Figure 6A, even after only 4 hours of coculture, NKG2Anull NK cells exerted significantly higher cytotoxicity in 3 of the 4 cell lines (P < 0.01 for the AML cell line U937, and P < 0.0001 for the hepatocellular carcinoma cell line PLC/PRF/5 and the acute lymphoblastic leukemia cell line OP-1); in the remaining cell line, the Ewing’s sarcoma cell line EW8, 4-hour cell killing was higher with NKG2Anull cells (76.5% ± 1.3% vs. 66.4 ± 6.2; n = 9), but the difference was not statistically significant. When tested against a spheroid model of the U2OS cell line not transduced with GpHLA-E (this cell line expresses low surface HLA-E; Supplemental Figure 7A), NKG2Anull cells were significantly more powerful than their NKG2A+ counterparts (P < 0.01 at 6 and 24 hours; P < 0.001 at 12 and 18 hours; Figure 6B).
Figure 6. Cytotoxicity of NKG2Anull NK cells against tumor cells with endogenous HLA-E expression.
(A) Four-hour cytotoxicity of NK cells transduced with anti-NKG2A PEBL or GFP only (Control) against cell lines expressing endogenous HLA-E (see Supplemental Figure 10). EW8 and PLC/PRF/5 were transduced with luciferase. BrightGlo was added after 4 hours of coculture, and luminescence was measured using a Flx 800 plate reader. Cytotoxicity of U937 and OP-1 was measured by flow cytometry. Box (25th–75th percentile, median) and whiskers (minimum-maximum) plots from 3 experiments with cells from 3 donors (EW8, PLC/PRF/5), and 6 experiments with cells from 2 donors (U937, OP-1) in triplicate, at 2:1, 1:1, or 1:2 E/T. (B) Spheroid tumors of U2OS-mCherry were cocultured with NK cells at 1:2 E/T in triplicate and analyzed with IncuCyte Zoom System. Data are shown as mean (± SD) red calibrated unit (RCU)/μM2. Representative images at end of culture are shown. Scale bars: 300 μm. (C) Four-hour cytotoxicity against cell lines exposed to IFN-γ (300 ng/ml; 12 hours). Plots are from 3 experiments with NK cells from 3 donors (EW8, PLC/PRF/5), and 6 with NK cells from 2 donors (U937) in triplicate at 2:1, 1:1, or 1:2 E/T. (D) Similar experiments targeting cells exposed for 12 hours to conditioned medium (C.M.) from 24-hour cocultures of NK cells with the respective cell lines. Four-hour cytotoxicity was compared with that against cells not exposed to conditioned medium. (E) Four-hour cytotoxicity against primary AML cells from 4 patients, exposed to IFN-γ (300 ng/ml; 12 hours). Data are from 4 experiments with NK cells from 2 donors in triplicate at 2:1 and 1:1 E/T. (F) NKG2A-negative NK cells from 3 donors were stimulated with K562-mb15-41BBL for 7 days, transduced with anti-NKG2A PEBL or GFP alone, and then exposed to IL-12 (20 ng/ml) for 5 days. Percentage of NKG2A+ cells at each stage is shown. (G) PEBL-transduced and control NK cells were exposed to IL-12 and tested in 4-hour cytotoxicity assays against K562-GpHLA-E cells. Data are shown as mean (± SD) of triplicate measurements at each E/T. **P < 0.01; ***P < 0.001; ****P < 0.0001, t test.
It is known that IFN-γ secreted by immune cells reactive to tumors increases expression of HLA-E, which, in turn, promotes resistance to NK cells via NKG2A ligation (23). We determined whether improvements in tumor cell killing brought about by NKG2A downregulation with PEBL extended to tumor cells exposed to IFN-γ. After 12 hours of exposure to IFN-γ (300 ng/ml), EW8, PLC/PRF/5, and U937 overexpressed HLA-E (Supplemental Figure 10). These cells were consistently more susceptible to killing by NKG2Anull NK cells than by control NK cells (P < 0.01 for PLC/PRF/5; P < 0.0001 for both EW8 and U937) (Figure 6C).
Because NK cells secrete IFN-γ during activation (Supplemental Figure 6), we postulated that exposure to the supernatant from cocultures of NK cells and target cells would also induce NK cell resistance in tumor cells. To test this notion, we cultured NK cells with EW8 or U937 cells for 24 hours, collected and filtered the supernatant, and added it to fresh EW8 and U937. After 12 hours, we compared NK cells with and without NKG2A downregulation in 4-hour cytotoxicity assays. As shown in Figure 6D, exposure to the NK conditioned medium induced considerable resistance to NKG2A+ cells, but NKG2Anull cytotoxicity remained high (for EW8, P < 0.0001 at 2:1 and 1:1 and P < 0.01 at 1:2; for U937, P < 0.0001 at 2:1 and P < 0.01 at 1:1 and 1:2).
Previous studies have shown that AML cells from patients overexpress HLA-E after exposure to IFN-γ, suggesting that this might favor the growth of NK-resistant leukemic cells (33). We obtained 4 bone marrow specimens from patients with AML collected at diagnosis (n = 3) or relapse (n = 1) and containing over 90% AML blasts. We exposed the cells to IFN-γ (300 ng/ml for 12 hours) and used them as targets of NKG2A+ NK cells that had been transduced with either anti-NKG2D PEBL or GFP alone. As shown in Figure 6E, NKG2Anull cells exerted a significantly higher cytotoxicity (P < 0.0001) in 2 of the 4 samples. Interestingly, the 2 sensitive samples were also those expressing the highest levels of HLA-E (MFI: 17171 and 12267 versus 8595 and 6390).
IL-12, secreted by macrophages and dendritic cells, is an important activator of immune responses against tumor cells, including the generation of NK cells with memory-like properties (56, 57). However, exposure of NK cells to IL-12 also increases expression of CD94/NKG2A (58–60). In the context of enhanced HLA-E expression in tumor cells caused by IFN-γ secretion, this is likely to further dampen NK cell responses. We purified NKG2A-negative cells from peripheral blood NK cells from 3 donors and transduced them with either GFP alone or anti-NKG2A PEBL. We noticed that coculture with K562-mb15-41BBL cells during the transduction procedure induced an increase in NKG2A expression in control NK cells, while NKG2A remained undetectable in PEBL-transduced cells (Figure 6F). Exposure to IL-12 (20 ng/ml for 5 days) further increased NKG2A expression in control NK cells, but not in PEBL-transduced NK cells (Figure 6F). Thus, the PEBL strategy not only downregulated NKG2A expression in NKG2A-positive NK cells, but also suppressed NKG2A expression after stimulation and IL-12 exposure. When the cytotoxicity of PEBL-transduced NK cells exposed to IL-12 was tested using GpHLA-E K562 cells, there was a clear improvement in cytotoxicity over that of control NK cells (P < 0.0001 at all E/T ratios tested) (Figure 6G).
Antitumor capacity of NKG2Anull cells in vivo.
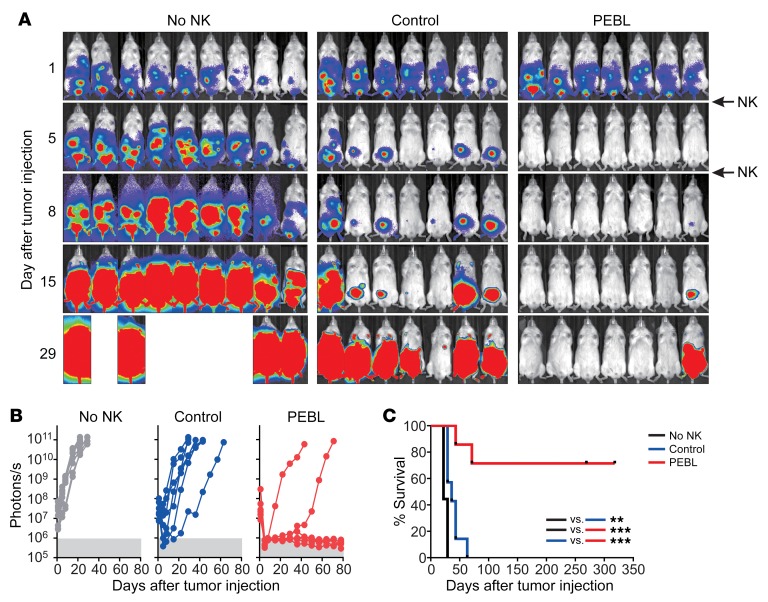
The above results indicated that NKG2A downregulation markedly enhanced the antitumor capacity of NK cells. To further challenge this finding, we engrafted the Ewing’s sarcoma cell line ES8 transduced with GpHLA-E in immunodeficient mice. As shown in Figure 7, treatment of mice with 2 infusions of control NK cells (transduced with GFP alone and expressing NKG2A) on days 1 and 5 after tumor injection only delayed tumor development. Median survival for the 2 groups was 22 and 36 days from tumor infusion. In contrast, the same NK cells with downregulated NKG2A by PEBL induced profound responses, which resulted in long-term survival for most mice (median survival was not reached after more than 269 days follow-up; P < 0.001, log rank test compared with control NK cells). In a second model, we engrafted the osteosarcoma line U2OS expressing GpHLA-E and treated mice with 2 infusions of NK cells 3 and 10 days after tumor injection (Supplemental Figure 11). Again, control NK cells only slightly delayed relapse (median survival, 25 and 32 days, respectively). Also, in this model NKG2Anull cells had clearly superior antitumor activity to that of their NKG2A+ counterparts (median survival, not reached after 60 days follow-up; P < 0.01). Of note, no evidence of xenoreactivity (weight loss, ruffled fur, abnormal posture, or decreased activity) was observed.
Figure 7. Antitumor capacity of anti-NKG2A PEBL-transduced NK cells in immunodeficient mice.
(A) ES8 cells (2 × 105) transduced with GpHLA-E and luciferase were injected i.p. in 23 NOD/SCID IL2RGnull mice. One day later, mice were treated with 1 × 107 expanded NKG2A+ NK cells transduced with either GFP alone (Control) or with anti-NKG2A PEBL (n = 7 for each group); another group of mice received tissue culture medium instead (no NK; n = 9). One additional injection of NK cells or medium was given 4 days later. All mice received i.p. injections of IL-2 (20,000 IU each) 3 times per week. Bioluminescence was measured with a Xenogen IVIS Spectrum System, with imaging beginning 5 minutes after i.p. injection of d-luciferin (150 μg/g body weight) and analyzed with Living Image 3.0 software. Collated ventral images of mice in 2 independent experiments are shown, 1 with 9 mice (3 in each group) and the other with 14 mice (6 injected with tumor alone, 4 treated with control NK cells, and 4 with anti-NKG2A PEBL NK cells). (B) Luminescence measurements of tumor cell growth. Each symbol corresponds to the sum of bioluminescence measurements by ventral and dorsal imaging in each mouse. (C) Kaplan-Meier curves and log-rank test for overall survival. Mice were euthanized when the sum of ventral and dorsal bioluminescence signal reached 5 × 1010 photons per second. **P = 0.0034; ***P < 0.001.
Discussion
The unique capacity of NK cells to recognize and kill tumor cells (8, 9) has been corroborated by reports of major clinical activity in some patients (6, 7), suggesting that NK cell infusions could have considerable potential for the treatment of cancer. Tumors, however, develop mechanisms of resistance that can nullify the NK cell effect. In this study, we developed a way to overcome the potent NK resistance mechanism mediated by HLA-E ligation of the inhibitory CD94/NKG2A receptor. We sought to abrogate its impact by removing NKG2A from the surface of NK cells and achieved this goal with an approach based on constructs designed to prevent protein surface expression. Anti-NKG2A PEBLs stopped NKG2A surface expression together with that of its coreceptor, CD94. The resulting NK cells not only retained their general function, but they were substantially more cytotoxic than control NK cells against target cells expressing HLA-E molecules loaded with the HLA-G signal peptide. NKG2A downregulation by PEBL also produced higher antitumor activity than interference with an anti-NKG2A antibody, while cell killing of nontransformed cells did not increase. The gains in cytotoxicity observed in short- and long-term in vitro cultures were confirmed in xenograft models, where NKG2Anull cells induced durable tumor remission. These results indicate that NKG2A downregulation is an effective way to increase the antitumor activity of NK cell therapies.
In tumor cells, downregulation of classical HLA class I molecules decreases susceptibility to T cell recognition and cytotoxicity (61). At the same time, overexpression of nonclassical HLA class I molecules, such as HLA-E and HLA-G, generates resistance to NK cells through binding of CD94/NKG2A (40). CD94 has no signaling capacity, and as we showed in our study, its expression on the cell surface of NKG2A+ NK cells is largely dependent on the presence of NKG2A. CD94 can also form heterodimers with NKG2C, which activates NK cells via DAP12 (10, 62). Although HLA-E can bind both inhibitory CD94/NKG2A and activating CD94/NKG2C, it has a lower affinity for the latter (50, 63). Expression of HLA-E, and hence resistance to NK cells, can be further enhanced by exposure to IFN-γ secreted by tumor-reactive immune cells (21–23). In our experiments, we recapitulated this scenario by exposing tumor cells to IFN-γ or to supernatant collected from cocultures of NK cells and tumor cells or by enforcing expression of GpHLA-E (49, 50, 64). In all these settings, resistance to NK cells increased, but downregulation of NKG2A on NK cells overcame resistance and markedly increased cytotoxicity. Another potential mechanism of resistance during immune response to tumors is expression of NKG2A generated by exposure of NK cells (including cells originally lacking surface NKG2A expression) to higher levels of IL-12 in the tumor microenvironment (58–60). Transduction of NK cells with anti-NKG2A PEBL blocked such upregulation and restored cytotoxicity.
The importance of the CD94/NKG2A-HLA-E immune checkpoint in regulating NK cell activity against tumors has led to the ongoing clinical testing of an anti-NKG2A inhibitory antibody (39, 40). We used the sequence of the heavy and light chain regions of an anti-NKG2D antibody as a basis for developing an anti-NKG2A scFv. We found that when the scFv was coupled to selected ER/Golgi retention domains, downregulation of NKG2A was rapid and nearly absolute. Importantly, NKG2A+ NK cells with NKG2A downregulation had more powerful cytotoxicity than those exposed to the anti-NKG2A antibody. This could be explained by the fact that CD94/NKG2A recycles between cell surface and cytoplasm independently of HLA-E ligation (65), yielding a continuous supply of new receptors on the cell surface that may eventually escape antibody blockade. NKG2A is a major NK inhibitory receptor, but there are other receptors that also regulate NK cell function. To this end, we have used PEBLs to successfully downregulate the NK inhibitory receptors KIR2DL1 and KIR2DL2/3 in primary NK cells (T. Kamiya and D. Campana, unpublished results). The relative antitumor capacity of these cells, NKG2Anull cells, and those lacking multiple inhibitory receptors remains to be investigated.
Downregulation of NKG2A might have also been achieved by using modern gene-editing methodologies based on meganucleases, TALEN, or CRISPR/Cas9, which have already been applied to genetically modify T lymphocytes (66–69). As shown in this study, PEBL-mediated downregulation of NKG2A is highly efficient and specific. It also has several potential practical advantages. First, anti-NKG2A PEBL expression can be accomplished with viral vectors identical to those currently used for expression of CAR in T lymphocytes. Therefore, the PEBL method can be readily adapted to current protocols for clinical-grade processing of genetically modified immune cells. Second, PEBL composition is similar to that of currently used CARs and should not pose the risk of nonspecific gene editing. Therefore, the introduction of PEBLs in clinical cell processing should not elicit additional regulatory concerns. Finally, anti-NKG2A PEBL can be combined with activating receptors to further enhance NK cell cytotoxicity. PEBL constructs can be assembled in multicistronic vectors that allow simultaneous expression of PEBLs and activating receptors. We have used this approach to effectively downregulate surface molecules in T cells while simultaneously expressing CARs (70, 71).
Expression of PEBL in NK cells relied on a culture system that has already been translated into a GMP-grade protocol or infusion of autologous or allogeneic NK cells in patients with cancer (8). Several approaches are being tested clinically to improve the anticancer potential of NK cell infusions, such as ex vivo expansion (8, 72, 73), activation with cytokines (4, 6), and infusion of antibodies against inhibitory receptors (39, 40, 74). Other approaches, such as CAR expression (47, 75), expression of activating receptors (76), and expression of membrane-bound cytokines (77), suggest possibilities to further enhance NK cell potency that warrant clinical exploration. The NKG2Anull cells generated in this study can evade a central mechanism of tumor resistance to NK cells. Importantly, NKG2A is also expressed in a subset of activated CD3+ T lymphocytes and its ligation can inhibit their function (23, 78, 79). In mice, loss of function of Qa-1b (equivalent to HLA-E in humans) resulting from in vivo CRISPR screening increased sensitivity of tumor cells to immune cells (80). Correlative studies in ovarian and cervical carcinomas have shown a predominant infiltration of CD8+ lymphocytes expressing NKG2A (26, 81), and recently published studies indicate that an anti-NKG2A antibody can augment T cell antitumor activity (41, 82). Thus, the approach described here could potentially be extended to protocols of adoptive T cell therapy of cancer.
Methods
Gene expression studies.
RNAseqv2 expression data from 10,375 individual tumor annotated samples were downloaded from the TCGA web portal. Thirty-three tumor types, as defined by TCGA codes, were included (Figure 1). Pairwise gene expression correlation values and associated P values were calculated in R. Gene expression box plots were generated using R graphics package GGPLOT2 (83). Ordered column plots, scatter plots, and linear regression lines were plotted in Microsoft Excel or GraphPad. Analyses were also performed using TIMER (42).
Cells.
The human cell lines NK92, K562, SK-BR-3, PLC/PRF/5, U2OS, and U937 were obtained from ATCC. ES8 and EW8 were from the tissue repository of St. Jude Children’s Research Hospital (Memphis, Tennessee, USA). OP-1 was developed in our laboratory (84). We transduced K562, SK-BR-3, PLC/PRF/5, U2OS, ES8, EW8, and U937 cells with a MSCV-IRES-GFP (where IRES indicates internal ribosome entry site) retroviral vector containing the firefly luciferase gene (48). K562, U2OS, and ES8 cells were also transduced with MSCV-IRES-mCherry vector. Transduced cells were selected for their expression of GFP or mCherry with a MoFlo (Beckman Coulter) or a FACSAria (BD Biosciences) cell sorter. Bone marrow–derived mesenchymal stromal cells immortalized by expression of human telomerase reverse transcriptase were previously developed in our laboratory (54). NK92 cells were cultured in RPMI 1640 (Thermo Fisher Scientific) with 12.5% FBS, 12.5% horse serum, 200 IU/ml IL-2, and antibiotics; other cell lines were maintained in RPMI 1640 or in DMEM (GE Healthcare; U2OS and PLC/PRF/5) with 10% FBS (GE Healthcare) and antibiotics. Tumor cell lines were periodically validated by DNA fingerprinting analysis (DSMZ).
Mononucleated cells from peripheral blood of healthy donors, and from peripheral blood or bone marrow of patients with AML were separated by centrifugation on a Lymphoprep Density Step (Axis-Shield) and washed twice in RPMI 1640.
NK cell expansion and NKG2A selection.
To expand NK cells, peripheral blood mononucleated cells were cocultured with the genetically modified K562-mb15-41BBL cell line in SCGM medium (CellGenix) containing 10% FBS and 40 IU/ml IL-2 (Proleukin, Novartis) (8, 47, 48, 51, 76, 77, 85). After 5 to 7 days of culture, residual T cells were removed using Dynabeads CD3 (Thermo Fisher). NK cells were labeled with allophycocyanin-conjugated (APC-conjugated) anti-CD159a (NKG2A) antibody, followed by anti-APC MicroBeads (Miltenyi Biotec); the LS and LD columns (Miltenyi Biotec) were used to select or remove NKG2A+ cells, respectively. Selected NK cells were restimulated with K562-mb15-41BBL cells and 400 IU/ml IL-2. Recombinant human IL-12 (R&D Systems; 20 ng/ml) was used in some experiments.
PEBL constructs.
We designed a scFv containing the variable regions of the heavy and light chains of the murine anti-human NKG2A monoclonal antibody Z199 (17, 43) and a 20–amino acid linker. To construct the anti-NKG2A PEBLs, the nucleotide sequence was joined to the CD8α signal peptide and sequences encoding ER/Golgi retention peptides (GGGGS)4AEKDEL (PEBL1), or CD8α hinge and transmembrane domain followed by LYKYKSRRSFIEEKKMP (PEBL2), LYKYKSRRSFIDEKKMP (PEBL3), or LYCNKYCKSRRSFIEEKKMP (PEBL4; Figure 1A). Constructs were subcloned into the MSCV-derived retroviral vector containing IRES and GFP. Preparation of retroviral supernatant and transduction were performed as previously described (85). Briefly, MSCV retroviral vector–conditioned medium was added to polypropylene tubes coated with RetroNectin (Takara); after centrifugation and removal of the supernatant, NKG2A-selected and restimulated NK cells were added to the tubes and left at 37°C for 12 hours; fresh viral supernatant was added on 2 other successive days. Transduced NK cells were maintained in SCGM medium with FBS, antibiotics, and 400 IU/ml of IL-2 until the time of experiments.
Cell marker and mRNA expression.
Expression of PEBLs was detected using a biotin-conjugated goat anti-mouse F(ab′)2 antibody (Jackson ImmunoResearch), followed by streptavidin conjugated to APC (Jackson ImmunoResearch). Other antibodies used to determine cell-marker expression are listed in Supplemental Table 3. Intracellular staining for CD94 was performed after surface staining with anti-NKG2A and permeabilization with 8E, a reagent developed in our laboratory. Cell staining was analyzed using Accuri C6 or Fortessa flow cytometers (BD Bioscience), with Diva (BD Biosciences) or FlowJo software.
Whole-transcriptome mRNA-Seq library preparation of NK cells with or without NKG2A downregulation was performed using the TruSeq Stranded mRNA Library Prep Kit (Illumina) and sequenced on a NextSeq 500 platform with a read length of 2 × 151 bp. The reads were mapped to the human genome reference hg19 using STAR (86). Reads aligned to each gene were counted using featureCounts (87), and the expression levels were calculated as fragments per kilobase of transcript per million mapped reads (FPKM). Expression levels were log transformed, followed by batch effect correction using ComBat of the sva package for downstream analysis (88). All original mRNA-Seq data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE127081).
Cytokine production, CD107a staining, and cytotoxicity assays.
To measure IFN-γ production, NK cells were cocultured at 37°C 5% CO2 with target cells for 8 hours in the presence of 0.1% Brefeldin A (GolgiPlug, BD Biosciences); cells were then labeled with phycoerythrin-conjugated (PE-conjugated) anti–IFN-γ (clone 25723.11, BD Biosciences) after cell membrane permeabilization with 8E (a permeabilization reagent developed in our laboratory). To measure exocytosis of lytic granules, target and effector cells were cocultured in the presence of PE-Cyanin 7–conjugated (Cy7-conjugated) anti-human CD107a antibody (H4A3; BioLegend). After 1 hour, monensin (GolgiStop, BD Biosciences) was added to the plate, and cultures were prolonged for another 3 hours.
To test cytotoxicity, NK cells were cocultured with target cells labeled with calcein red–orange AM (Thermo Fisher Scientific). After 4 hours, the number of viable target cells was counted by flow cytometry (76). To test cytotoxicity against luciferase-labeled target cells, BrightGlo (Promega) was added after 4 hours of coculture, and luminescence was measured using a Flx 800 plate reader (BioTek). To test ADCC, trastuzumab (10 μg/ml; Herceptin, Roche) was added to SK-BR-3 cells expressing GpHLA-E prior to coculture with NK cells. For long-term cytotoxicity assay, NK cells were cocultured with mCherry-transduced target cells, and mCherry-expressing cells were counted with the IncuCyte Zoom System (Essen BioScience). To generate tumor spheroids, U2OS cells expressing mCherry cells were seeded into ultra-low attachment 96-well round bottom plates (Corning) at a concentration of 500 cells per 100 μl of RPMI 1640. After centrifugation, cells were cultured for 7 days. Plates were then transferred into the IncuCyte Zoom System. On day 8, NK cells were added. Images were captured with the IncuCyte Zoom System and a ×10 objective lens at 6-hour intervals. Tumor cell growth was quantified based on the fluorescence intensity of mCherry.
To interfere with NKG2A function, we used a purified nonconjugated anti-NKG2A antibody (Z199, Beckman Coulter); an isotype-matched nonreactive immunoglobulin (R&D) was used as a control. In some experiments, we used as targets cell lines (K562, ES8, EW8, U2OS, and SK-BR-3) transduced with a construct encoding HLA-E with its signal peptide (MVDGTLLLLLSEALALTQTWA) replaced with the HLA-G signal peptide (MVVMAPRTLFLLLSGALTLTETWA) (49, 50). The nucleotide sequence was cloned into the MSCV vector containing IRES and GFP and transduced in cells that were previously transduced with firefly luciferase and GFP. To detect HLA-E expression, transduced cells were labeled with APC-conjugated anti–HLA-E antibody (Thermo Fisher Scientific) and sorted with the MoFlo cell sorter. In some experiments, target cells were cultured for 12 hours with either IFN-γ (300 ng/ml; Sigma-Aldrich) or culture supernatant from cocultures of NK cells and tumor cells before the cytotoxicity assays. The culture supernatant was collected from 24-hour cocultures, centrifuged at high speed, and filtered before adding it to fresh tumor cells.
Xenograft models.
ES8 cells expressing luciferase and GpHLA-E were injected i.p. into NOD.Cg-Prkdcscid IL2rgtm1Wjl/SzJ (NOD/SCID IL2RGnull) mice (Jackson Laboratory) at 2 × 105 per mouse. NK cells were expanded for 5 days. After CD3 depletion and NKG2A+ selection, NK cells were restimulated with K562-mb15-41BBL cells and transduced with anti-NKG2A PEBL or GFP only. One and five days after ES8 cell injection, PEBL-transduced and control NK cells were administered at 1 × 107 cells per mouse (7 mice per group). Another group of 9 mice received RPMI 1640 with 10% FBS instead of NK cells. All mice received i.p. injections of IL-2 (20,000 IU each) 3 times per week. Similar experiments were performed with U2OS-GpHLA-E cells injected i.p. in 15 mice, with the difference being that NK cells were administered on days 3 and 10 after tumor injection. Bioluminescence was measured with a Xenogen IVIS Spectrum system (Caliper Life Sciences) after i.p. injection of d-luciferin potassium salt (PerkinElmer; 150 μg/g body weight). Luminescence emitted was analyzed with Living Image 3.0 software. Mice were euthanized when the sum of ventral and dorsal bioluminescence signal reached 2 to 5 × 1010 photons per second or earlier if physical signs warranting euthanasia appeared. Mice were distributed in each group to achieve an even representation of tumor engraftment based on the luminescence signal recorded before NK cell administration. The only inclusion criteria were tumor engraftment by luminescence before NK cell infusion.
Statistics.
For RNA-Seq analysis of NK cells, paired t test was used to compare expression levels, with statistical analysis performed using R. Controlling for false discovery rate was done using the 2-stage linear step-up procedure of Benjamini, Krieger, and Yekutieli, with a Q of 1% using GraphPad. Comparisons of NK cell function were analyzed using t test; P < 0.05 was considered significant. Mouse survival was analyzed by log-rank test.
Study approval.
Peripheral blood samples were obtained from discarded anonymized by-products of platelet donations from healthy adult donors at the National University Hospital or the Health Science Authority Blood Banks, Singapore. Samples from patients with AML were obtained from banked surplus material. Studies were performed with approval from the Institutional Review Board, National University of Singapore. All mouse procedures were approved by the Institutional Animal Care and Use Committee and conducted in accordance with current guidelines.
Author contributions
TK developed PEBLs, performed experiments, and analyzed data. SVS and DW performed experiments and analyzed data. MR analyzed gene expression data. DC designed the study, analyzed data, and wrote the manuscript with TK, SVS, DW, and MR.
Supplementary Material
Acknowledgments
We thank Rick Nicoletti for help with TCGA data analysis, Jiang Nan and Li Zhenhua for performing RNA-Seq analysis, and Amit Jain for assistance with the spheroid model. This work was supported by grant NMRC/STaR/0025/2015a from the National Medical Research Council of Singapore.
Version 1. 03/12/2019
In-Press Preview
Version 2. 04/15/2019
Electronic publication
Version 3. 05/01/2019
Print issue publication
Footnotes
Conflict of interest: TK, SVS, and DC are coinventors in patents and patent applications (8,026,097; 9,511,092; 62/112,765; 62/477,311; 62/628,788) describing the technologies used or related technologies. DC is a scientific founder and stockholder of Nkarta Therapeutics and MediSix Therapeutics, which hold licenses for some of the technologies described. MR is an employee of MediSix Therapeutics.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(5):2094–2106.https://doi.org/10.1172/JCI123955.
See the related Commentary at Setting traps for NKG2A gives NK cell immunotherapy a fighting chance.
Contributor Information
Takahiro Kamiya, Email: kamiped@tmd.ac.jp.
See Voon Seow, Email: paessv@nus.edu.sg.
Desmond Wong, Email: paewmhd@nus.edu.sg.
Murray Robinson, Email: murray@medisixtx.com.
Dario Campana, Email: paedc@nus.edu.sg.
References
- 1.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27(4):450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sadelain M, Rivière I, Riddell S. Therapeutic T cell engineering. Nature. 2017;545(7655):423–431. doi: 10.1038/nature22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruggeri L, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 4.Miller JS, et al. Successful adoptive transfer and in vivo expansion of human haploidentical NK cells in patients with cancer. Blood. 2005;105(8):3051–3057. doi: 10.1182/blood-2004-07-2974. [DOI] [PubMed] [Google Scholar]
- 5.Cooley S, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116(14):2411–2419. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romee R, et al. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med. 2016;8(357):357ra123. doi: 10.1126/scitranslmed.aaf2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller JS. Therapeutic applications: natural killer cells in the clinic. Hematology Am Soc Hematol Educ Program. 2013;2013:247–253. doi: 10.1182/asheducation-2013.1.247. [DOI] [PubMed] [Google Scholar]
- 8.Shimasaki N, Coustan-Smith E, Kamiya T, Campana D. Expanded and armed natural killer cells for cancer treatment. Cytotherapy. 2016;18(11):1422–1434. doi: 10.1016/j.jcyt.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Morvan MG, Lanier LL. NK cells and cancer: you can teach innate cells new tricks. Nat Rev Cancer. 2016;16(1):7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 10.Lazetic S, Chang C, Houchins JP, Lanier LL, Phillips JH. Human natural killer cell receptors involved in MHC class I recognition are disulfide-linked heterodimers of CD94 and NKG2 subunits. J Immunol. 1996;157(11):4741–4745. [PubMed] [Google Scholar]
- 11.Braud VM, et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391(6669):795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 12.Lee N, et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc Natl Acad Sci U S A. 1998;95(9):5199–5204. doi: 10.1073/pnas.95.9.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187(5):813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller JD, Weber DA, Ibegbu C, Pohl J, Altman JD, Jensen PE. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J Immunol. 2003;171(3):1369–1375. doi: 10.4049/jimmunol.171.3.1369. [DOI] [PubMed] [Google Scholar]
- 15.Hoare HL, et al. Subtle changes in peptide conformation profoundly affect recognition of the non-classical MHC class I molecule HLA-E by the CD94-NKG2 natural killer cell receptors. J Mol Biol. 2008;377(5):1297–1303. doi: 10.1016/j.jmb.2008.01.098. [DOI] [PubMed] [Google Scholar]
- 16.Houchins JP, Lanier LL, Niemi EC, Phillips JH, Ryan JC. Natural killer cell cytolytic activity is inhibited by NKG2-A and activated by NKG2-C. J Immunol. 1997;158(8):3603–3609. [PubMed] [Google Scholar]
- 17.Carretero M, et al. The CD94 and NKG2-A C-type lectins covalently assemble to form a natural killer cell inhibitory receptor for HLA class I molecules. Eur J Immunol. 1997;27(2):563–567. doi: 10.1002/eji.1830270230. [DOI] [PubMed] [Google Scholar]
- 18.Gornalusse GG, et al. HLA-E–expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol. 2017;35(8):765–772. doi: 10.1038/nbt.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei XH, Orr HT. Differential expression of HLA-E, HLA-F, and HLA-G transcripts in human tissue. Hum Immunol. 1990;29(2):131–142. doi: 10.1016/0198-8859(90)90076-2. [DOI] [PubMed] [Google Scholar]
- 20.Marín R, et al. Analysis of HLA-E expression in human tumors. Immunogenetics. 2003;54(11):767–775. doi: 10.1007/s00251-002-0526-9. [DOI] [PubMed] [Google Scholar]
- 21.Gustafson KS, Ginder GD. Interferon-gamma induction of the human leukocyte antigen-E gene is mediated through binding of a complex containing STAT1alpha to a distinct interferon-gamma-responsive element. J Biol Chem. 1996;271(33):20035–20046. doi: 10.1074/jbc.271.33.20035. [DOI] [PubMed] [Google Scholar]
- 22.Cerboni C, Mousavi-Jazi M, Wakiguchi H, Carbone E, Kärre K, Söderström K. Synergistic effect of IFN-gamma and human cytomegalovirus protein UL40 in the HLA-E–dependent protection from NK cell-mediated cytotoxicity. Eur J Immunol. 2001;31(10):2926–2935. doi: 10.1002/1521-4141(2001010)31:10<2926::AID-IMMU2926>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 23.Malmberg KJ, et al. IFN-gamma protects short-term ovarian carcinoma cell lines from CTL lysis via a CD94/NKG2A-dependent mechanism. J Clin Invest. 2002;110(10):1515–1523. doi: 10.1172/JCI15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchesi M, et al. HLA-dependent tumour development: a role for tumour associate macrophages? J Transl Med. 2013;11:247. doi: 10.1186/1479-5876-11-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Kruijf EM, et al. HLA-E and HLA-G expression in classical HLA class I-negative tumors is of prognostic value for clinical outcome of early breast cancer patients. J Immunol. 2010;185(12):7452–7459. doi: 10.4049/jimmunol.1002629. [DOI] [PubMed] [Google Scholar]
- 26.Gooden M, et al. HLA-E expression by gynecological cancers restrains tumor-infiltrating CD8(+) T lymphocytes. Proc Natl Acad Sci U S A. 2011;108(26):10656–10661. doi: 10.1073/pnas.1100354108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andersson E, et al. Non-classical HLA-class I expression in serous ovarian carcinoma: Correlation with the HLA-genotype, tumor infiltrating immune cells and prognosis. Oncoimmunology. 2016;5(1):e1052213. doi: 10.1080/2162402X.2015.1052213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy EM, et al. Human leukocyte antigen-E protein is overexpressed in primary human colorectal cancer. Int J Oncol. 2008;32(3):633–641. [PubMed] [Google Scholar]
- 29.Guo ZY, et al. Predictive value of HLA-G and HLA-E in the prognosis of colorectal cancer patients. Cell Immunol. 2015;293(1):10–16. doi: 10.1016/j.cellimm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 30.Silva TG, et al. Expression of the nonclassical HLA-G and HLA-E molecules in laryngeal lesions as biomarkers of tumor invasiveness. Histol Histopathol. 2011;26(12):1487–1497. doi: 10.14670/HH-26.1487. [DOI] [PubMed] [Google Scholar]
- 31.Hirankarn N, Kimkong I, Mutirangura A. HLA-E polymorphism in patients with nasopharyngeal carcinoma. Tissue Antigens. 2004;64(5):588–592. doi: 10.1111/j.1399-0039.2004.00311.x. [DOI] [PubMed] [Google Scholar]
- 32.Derré L, et al. Expression and release of HLA-E by melanoma cells and melanocytes: potential impact on the response of cytotoxic effector cells. J Immunol. 2006;177(5):3100–3107. doi: 10.4049/jimmunol.177.5.3100. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen S, et al. HLA-E upregulation on IFN-gamma-activated AML blasts impairs CD94/NKG2A-dependent NK cytolysis after haplo-mismatched hematopoietic SCT. Bone Marrow Transplant. 2009;43(9):693–699. doi: 10.1038/bmt.2008.380. [DOI] [PubMed] [Google Scholar]
- 34.Platonova S, et al. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71(16):5412–5422. doi: 10.1158/0008-5472.CAN-10-4179. [DOI] [PubMed] [Google Scholar]
- 35.Gillard-Bocquet M, et al. Lung tumor microenvironment induces specific gene expression signature in intratumoral NK cells. Front Immunol. 2013;4:19. doi: 10.3389/fimmu.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mamessier E, et al. Human breast cancer cells enhance self tolerance by promoting evasion from NK cell antitumor immunity. J Clin Invest. 2011;121(9):3609–3622. doi: 10.1172/JCI45816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stringaris K, et al. Leukemia-induced phenotypic and functional defects in natural killer cells predict failure to achieve remission in acute myeloid leukemia. Haematologica. 2014;99(5):836–847. doi: 10.3324/haematol.2013.087536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spee P, Chen J, Padkjaer S, Jing S, Zhang J, Yu J, inventors. Humanized anti-human NKG2A monoclonal antibody. U.S. Patent 9422368B2. 2016. [Google Scholar]
- 39.Ruggeri L, et al. Effects of anti-NKG2A antibody administration on leukemia and normal hematopoietic cells. Haematologica. 2016;101(5):626–633. doi: 10.3324/haematol.2015.135301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muntasell A, et al. Targeting NK-cell checkpoints for cancer immunotherapy. Curr Opin Immunol. 2017;45:73–81. doi: 10.1016/j.coi.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 41.André P, et al. Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell. 2018;175(7):1731–1743.e13. doi: 10.1016/j.cell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li T, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sivori S, et al. CD94 functions as a natural killer cell inhibitory receptor for different HLA class I alleles: identification of the inhibitory form of CD94 by the use of novel monoclonal antibodies. Eur J Immunol. 1996;26(10):2487–2492. doi: 10.1002/eji.1830261032. [DOI] [PubMed] [Google Scholar]
- 44.Rabouille C, Klumperman J. Opinion: The maturing role of COPI vesicles in intra-Golgi transport. Nat Rev Mol Cell Biol. 2005;6(10):812–817. doi: 10.1038/nrm1735. [DOI] [PubMed] [Google Scholar]
- 45.Gao C, et al. Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci. 2014;19(8):508–515. doi: 10.1016/j.tplants.2014.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Nilsson T, Jackson M, Peterson PA. Short cytoplasmic sequences serve as retention signals for transmembrane proteins in the endoplasmic reticulum. Cell. 1989;58(4):707–718. doi: 10.1016/0092-8674(89)90105-0. [DOI] [PubMed] [Google Scholar]
- 47.Imai C, Iwamoto S, Campana D. Genetic modification of primary natural killer cells overcomes inhibitory signals and induces specific killing of leukemic cells. Blood. 2005;106(1):376–383. doi: 10.1182/blood-2004-12-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fujisaki H, et al. Expansion of highly cytotoxic human natural killer cells for cancer cell therapy. Cancer Res. 2009;69(9):4010–4017. doi: 10.1158/0008-5472.CAN-08-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Llano M, et al. HLA-E–bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: preferential response to an HLA-G-derived nonamer. Eur J Immunol. 1998;28(9):2854–2863. doi: 10.1002/(SICI)1521-4141(199809)28:09<2854::AID-IMMU2854>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 50.Valés-Gómez M, Reyburn HT, Erskine RA, López-Botet M, Strominger JL. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 1999;18(15):4250–4260. doi: 10.1093/emboj/18.15.4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cho D, Shook DR, Shimasaki N, Chang YH, Fujisaki H, Campana D. Cytotoxicity of activated natural killer cells against pediatric solid tumors. Clin Cancer Res. 2010;16(15):3901–3909. doi: 10.1158/1078-0432.CCR-10-0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takao S, Ishikawa T, Yamashita K, Uchiyama T. The rapid induction of HLA-E is essential for the survival of antigen-activated naive CD4 T cells from attack by NK cells. J Immunol. 2010;185(10):6031–6040. doi: 10.4049/jimmunol.1000176. [DOI] [PubMed] [Google Scholar]
- 53.Nielsen N, Ødum N, Ursø B, Lanier LL, Spee P. Cytotoxicity of CD56(bright) NK cells towards autologous activated CD4+ T cells is mediated through NKG2D, LFA-1 and TRAIL and dampened via CD94/NKG2A. PLoS ONE. 2012;7(2):e31959. doi: 10.1371/journal.pone.0031959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mihara K, et al. Development and functional characterization of human bone marrow mesenchymal cells immortalized by enforced expression of telomerase. Br J Haematol. 2003;120(5):846–849. doi: 10.1046/j.1365-2141.2003.04217.x. [DOI] [PubMed] [Google Scholar]
- 55.Pende D, et al. HLA-G recognition by human natural killer cells. Involvement of CD94 both as inhibitory and as activating receptor complex. Eur J Immunol. 1997;27(8):1875–1880. doi: 10.1002/eji.1830270809. [DOI] [PubMed] [Google Scholar]
- 56.Kerkar SP, Restifo NP. Cellular constituents of immune escape within the tumor microenvironment. Cancer Res. 2012;72(13):3125–3130. doi: 10.1158/0008-5472.CAN-11-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fehniger TA, Cooper MA. Harnessing NK cell memory for cancer immunotherapy. Trends Immunol. 2016;37(12):877–888. doi: 10.1016/j.it.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sáez-Borderías A, Romo N, Magri G, Gumá M, Angulo A, López-Botet M. IL-12-dependent inducible expression of the CD94/NKG2A inhibitory receptor regulates CD94/NKG2C+ NK cell function. J Immunol. 2009;182(2):829–836. doi: 10.4049/jimmunol.182.2.829. [DOI] [PubMed] [Google Scholar]
- 59.Béziat V, Descours B, Parizot C, Debré P, Vieillard V. NK cell terminal differentiation: correlated stepwise decrease of NKG2A and acquisition of KIRs. PLoS ONE. 2010;5(8):e11966. doi: 10.1371/journal.pone.0011966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Romee R, et al. Cytokine activation induces human memory-like NK cells. Blood. 2012;120(24):4751–4760. doi: 10.1182/blood-2012-04-419283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malmberg KJ, Sohlberg E, Goodridge JP, Ljunggren HG. Immune selection during tumor checkpoint inhibition therapy paves way for NK-cell “missing self” recognition. Immunogenetics. 2017;69(8-9):547–556. doi: 10.1007/s00251-017-1011-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lanier LL, Corliss B, Wu J, Phillips JH. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity. 1998;8(6):693–701. doi: 10.1016/S1074-7613(00)80574-9. [DOI] [PubMed] [Google Scholar]
- 63.Kaiser BK, Pizarro JC, Kerns J, Strong RK. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc Natl Acad Sci U S A. 2008;105(18):6696–6701. doi: 10.1073/pnas.0802736105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Das J, Khakoo SI. NK cells: tuned by peptide? Immunol Rev. 2015;267(1):214–227. doi: 10.1111/imr.12315. [DOI] [PubMed] [Google Scholar]
- 65.Borrego F, Kabat J, Sanni TB, Coligan JE. NK cell CD94/NKG2A inhibitory receptors are internalized and recycle independently of inhibitory signaling processes. J Immunol. 2002;169(11):6102–6111. doi: 10.4049/jimmunol.169.11.6102. [DOI] [PubMed] [Google Scholar]
- 66.Torikai H, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119(24):5697–5705. doi: 10.1182/blood-2012-01-405365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poirot L, et al. Multiplex genome-edited T-cell manufacturing platform for “off-the-shelf” adoptive T-cell immunotherapies. Cancer Res. 2015;75(18):3853–3864. doi: 10.1158/0008-5472.CAN-14-3321. [DOI] [PubMed] [Google Scholar]
- 68.Qasim W, et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017;9(374):eaaj2013. doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- 69.Eyquem J, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Png YT, Vinanica N, Kamiya T, Shimasaki N, Coustan-Smith E, Campana D. Blockade of CD7 expression in T cells for effective chimeric antigen receptor targeting of T-cell malignancies. Blood Adv. 2017;1(25):2348–2360. doi: 10.1182/bloodadvances.2017009928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kamiya T, Wong D, Png YT, Campana D. A novel method to generate T-cell receptor-deficient chimeric antigen receptor T cells. Blood Adv. 2018;2(5):517–528. doi: 10.1182/bloodadvances.2017012823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shah NN, et al. Acute GVHD in patients receiving IL-15/4-1BBL activated NK cells following T-cell-depleted stem cell transplantation. Blood. 2015;125(5):784–792. doi: 10.1182/blood-2014-07-592881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ciurea SO, et al. Phase 1 clinical trial using mbIL21 ex vivo-expanded donor-derived NK cells after haploidentical transplantation. Blood. 2017;130(16):1857–1868. doi: 10.1182/blood-2017-05-785659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Benson DM, et al. A phase 1 trial of the anti-KIR antibody IPH2101 in patients with relapsed/refractory multiple myeloma. Blood. 2012;120(22):4324–4333. doi: 10.1182/blood-2012-06-438028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu E, et al. Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia. 2018;32(2):520–531. doi: 10.1038/leu.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang YH, Connolly J, Shimasaki N, Mimura K, Kono K, Campana D. A chimeric receptor with NKG2D specificity enhances natural killer cell activation and killing of tumor cells. Cancer Res. 2013;73(6):1777–1786. doi: 10.1158/0008-5472.CAN-12-3558. [DOI] [PubMed] [Google Scholar]
- 77.Imamura M, et al. Autonomous growth and increased cytotoxicity of natural killer cells expressing membrane-bound interleukin-15. Blood. 2014;124(7):1081–1088. doi: 10.1182/blood-2014-02-556837. [DOI] [PubMed] [Google Scholar]
- 78.Mingari MC, et al. HLA class I-specific inhibitory receptors in human T lymphocytes: interleukin 15-induced expression of CD94/NKG2A in superantigen- or alloantigen-activated CD8+ T cells. Proc Natl Acad Sci U S A. 1998;95(3):1172–1177. doi: 10.1073/pnas.95.3.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J, Matsuoka M, Cantor H, Homer R, Enelow RI. Cutting edge: engagement of NKG2A on CD8+ effector T cells limits immunopathology in influenza pneumonia. J Immunol. 2008;180(1):25–29. doi: 10.4049/jimmunol.180.1.25. [DOI] [PubMed] [Google Scholar]
- 80.Manguso RT, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017;547(7664):413–418. doi: 10.1038/nature23270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheu BC, et al. Up-regulation of inhibitory natural killer receptors CD94/NKG2A with suppressed intracellular perforin expression of tumor-infiltrating CD8+ T lymphocytes in human cervical carcinoma. Cancer Res. 2005;65(7):2921–2929. doi: 10.1158/0008-5472.CAN-04-2108. [DOI] [PubMed] [Google Scholar]
- 82.van Montfoort N, et al. NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell. 2018;175(7):1744–1755.e15. doi: 10.1016/j.cell.2018.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wickham H. Elegant Graphics for Data Analysis. New York, NY: Springer-Verlag; 2009. [Google Scholar]
- 84.Manabe A, et al. Interleukin-4 induces programmed cell death (apoptosis) in cases of high-risk acute lymphoblastic leukemia. Blood. 1994;83(7):1731–1737. [PubMed] [Google Scholar]
- 85.Kamiya T, Chang YH, Campana D. Expanded and activated natural killer cells for immunotherapy of hepatocellular carcinoma. Cancer Immunol Res. 2016;4(7):574–581. doi: 10.1158/2326-6066.CIR-15-0229. [DOI] [PubMed] [Google Scholar]
- 86.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 88.Chakraborty S, Datta S, Datta S. Surrogate variable analysis using partial least squares (SVA-PLS) in gene expression studies. Bioinformatics. 2012;28(6):799–806. doi: 10.1093/bioinformatics/bts022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.