Abstract
Eosinophilic esophagitis (EoE) is a recently described disease in which exposure to specific foods and allergens leads to type 2 inflammation, epithelial barrier dysfunction, and difficulty in swallowing. In the current issue of the JCI, Wen and colleagues investigate tissue T cell heterogeneity in patients with EoE using single-cell RNA sequencing (scRNA-seq). Esophageal epithelium from individuals with EoE convtained a prominent population of Th2 cells not seen in controls. The short-chain fatty acid (SCFA) receptor FFAR3 was found to be highly expressed in EoE Th2 cells. Experiments presented here provide evidence that SCFAs may promote type 2 inflammation in allergic diseases such as EoE and asthma. This study provides an early example of scRNA-seq for identifying relevant cell populations and mechanisms underlying allergic diseases.
EoE is a type 2 cytokine–driven disease
Eosinophilic esophagitis (EoE) was first recognized as a distinct entity in the 1990s (1). The subsequent increase in EoE prevalence reflects both increased awareness of this disease and a true increase in disease incidence (2). EoE presents both in children, where it causes difficulty in feeding, nausea and vomiting, heartburn, and failure to thrive, and in adults, where it commonly presents with difficulty swallowing and food impaction. The pathognomonic histologic finding is the presence of large numbers of eosinophils within the esophageal epithelium (3), and an increase in T cells and mast cells (4).
The evidence that EoE is driven by type 2 immune responses in the esophagus is now overwhelming. EoE is strongly associated with sensitization to food allergens and aeroallergens, and with other allergic disorders including allergic rhinitis, asthma, and atopic dermatitis. Genetic association studies revealed linkage to loci for the type 2 cytokine gene IL4, the epithelial-derived type 2–promoting cytokine gene TSLP, and two genes that are induced by the type 2 cytokine IL-13: CAPN14, which encodes a calpain protease that disrupts the epithelial barrier, and CCL26, which encodes the eosinophil chemoattractant eotaxin 3 (5, 6). Antibodies against the type 2 cytokines IL-5 and IL-13 reduce eosinophil levels in the esophagus but to date have not been shown to cause significant clinical improvement (7). Further research is therefore required to understand how type 2 responses are initiated and maintained in the esophagus. While many aspects of type 2 responses at other sites are undoubtedly at play in EoE, it is important to understand the unique features of immune responses in the esophageal environment that drive this disease in a growing subset of individuals with an allergic disease.
Single-cell RNA sequencing identifies tissue-resident EoE-associated T cell populations
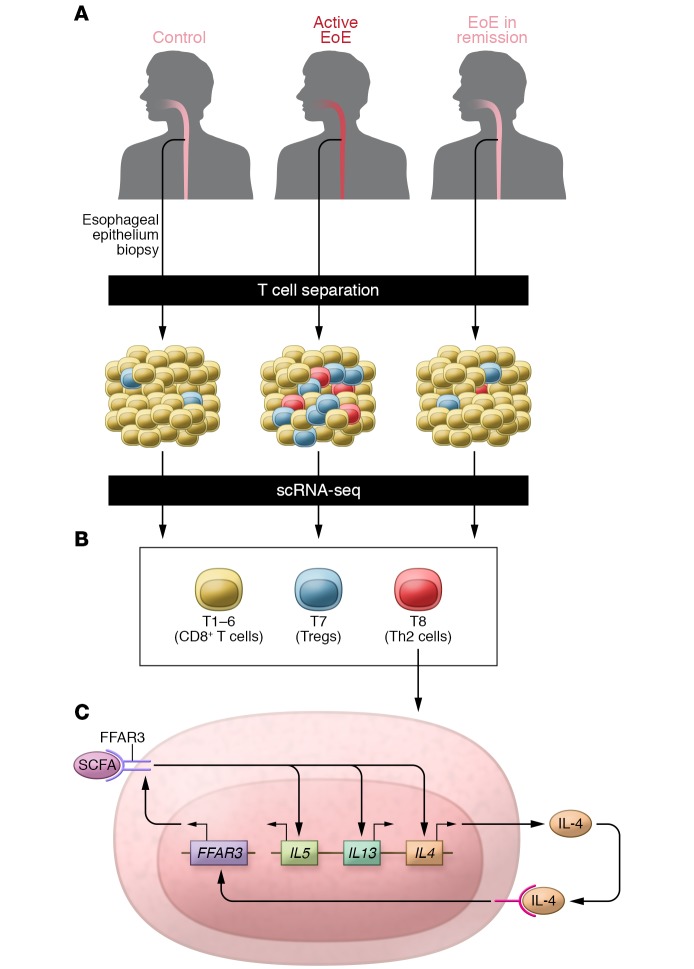
In this issue, Wen and colleagues (8) report the use of single-cell RNA sequencing (scRNA-seq) to study T cells from individuals with and without EoE (Figure 1A). Since becoming widely available over the last several years, scRNA-seq has been used by immunologists to study immune cell heterogeneity, infer lineage relationships and developmental trajectories, and study antigen receptor diversity (9, 10). This approach is just beginning to be applied to the study of allergic diseases (11, 12), and its application to EoE provides valuable lessons about the potential of scRNA-seq for identifying novel T cell populations in human disease. Two aspects of the approach used here are noteworthy. First, the authors concentrated on analyses of T cells from the affected tissue, the esophageal epithelium, rather than circulating T cells. Although obtaining T cells from biopsies performed during endoscopic procedures presents a challenge, it is clear that many essential characteristics of the local T cell response cannot be inferred from studies of circulating cells. Second, the authors generated a large amount of valuable information from a modest number of tissue T cells (239 cells from controls, 584 cells from individuals with active EoE, and 265 cells from individuals with EoE in complete remission). The microfluidic cell sorting scRNA-seq method used in this study is relatively costly on a per cell basis, but gives better coverage of each cell’s transcriptome compared with current droplet-based methods. Adequate sequencing depth allowed for detection of transcripts from gene families such as cytokines and transcription factors that can play critical roles even when expressed at a low copy number, and also permitted assessment of TCR diversity.
Figure 1. Single-cell RNA-seq–based identification of a role for FFAR3 in Th2 cells.
(A) T cells were isolated from esophageal biopsies obtained from subjects with EoE and healthy controls and analyzed by single-cell RNA-seq (scRNA-seq) on the Fluidigm C1 platform. (B) Two clusters of CD4+ T cells, T7 and T8, were enriched in subjects with EoE. (C) Cluster T8 included effector memory Th2 cells expressing IL4, IL5, and IL13, as well as FFAR3. Stimulation with FFAR ligands (SCFAs) increased production of Th2 cytokines in cell culture and augmented type 2 responses in an allergic airway disease model. In addition, IL-4 stimulated expression of FFAR3, suggesting the existence of a positive feedback loop that might amplify type 2 responses in EoE and other allergic diseases.
The scRNA-seq analysis of esophageal T cells identified eight populations of T cells, designated T1–T8 (Figure 1B). Clusters T1–T6 contained primarily or exclusively CD8+ T cells. Analysis of known marker genes determined that T7 was enriched for CD4+ Treg cells and T8 was enriched for CD4+ T cells producing type 2 cytokines (Th2 cells). In comparison to healthy controls, samples from active EoE tissue had more T7 and T8 cells. The differences were particularly dramatic for the T8 Th2 cell cluster, which was completely absent from healthy controls, represented a very small subset (0.4%) of T cells from subjects with EoE remission, and represented a much larger subset (6.4%) in subjects with active EoE. Analyses of TCR-β V-J usage and expression of cytokine, transcription factors, and other genes in the T7 and T8 CD4+ T cell populations revealed many findings that are consistent with prior studies and others that generated new hypotheses about how T cell responses develop in EoE and evolve during disease remission. One such hypothesis centered on the SCFA receptor FFAR3, which was differentially expressed in Th2 cells (T8) versus Treg cells (T7). Expression of FFAR3 was highly correlated with tissue eosinophilia and expression of other type 2 response genes. Of note, FFAR3 was not among the 150 genes identified as differentially expressed in the bulk RNA-seq comparison of T cells from EoE versus healthy control tissue, highlighting the value of scRNA-seq for measuring gene expression in cell subsets.
A potential role for the SCFA receptor FFAR3 in Th2 cells
FFAR2 (GPR43) and FFAR3 (GRP41) are G protein–coupled receptors activated by SCFAs, including acetate (C2), propionate (C3), and butyrate (C4), that are produced by gut bacteria (13). Both receptors are expressed in a variety of immune and nonimmune cell types, but FFAR2 and FFAR3 differ in their expression patterns, affinity for specific SCFAs, and ability to couple to different G proteins. SCFAs and their receptors have been implicated in the regulation of the gut epithelium, the endocrine pancreas, adipose tissue, and the sympathetic nervous system. Experiments with SCFA ligands and receptor knockout mice have demonstrated roles for these molecules in various inflammatory and immune responses. Although proinflammatory effects have been described in some systems (14), SCFAs often have antiinflammatory effects. For example, in a murine model of asthma, diet-induced increases in SCFAs were protective against type 2 airway inflammation, an effect that required FFAR3 but not FFAR2, and was attributed to an impairment in dendritic cell activation (15).
The finding of FFAR3 expression in EoE Th2 cells led the investigators to consider the regulation of this receptor and the effects of SCFAs on Th2 cell differentiation and function (Figure 1C). In studies with Jurkat T cells, which display FFAR3 on the cell surface, SCFAs induced expression of the type 2 cytokine IL-5. In addition, IL-4 stimulation of Jurkat cells increased expression of FFAR3, suggesting that a positive feedback loop involving this pathway may contribute to the development or persistence of Th2 inflammation in EoE. In vitro Th2 differentiation of primary mouse and human cells was augmented by the presence of the SCFA C4. Finally, in a murine allergic airway disease model, intranasal coadministration of C4 together with allergens increased IL-4, IL-5, and IL-13 production in lung T cells and augmented eosinophilia. Taken together, these findings are consistent with the hypothesis that SCFAs may act directly on CD4+ cells to promote type 2 responses. Understanding how this pathway may contribute to the pathogenesis of allergic diseases such as EoE and asthma will require a greater understanding of the nature of SCFAs present in the relevant tissues (esophagus and airway) and would benefit greatly from experimental systems such as T cell–specific SCFA receptor knockout mice to tease out the contributions of specific receptors on T cells. As the authors point out, this issue may be of clinical significance given the presence of SCFAs in the diet and in dietary supplements that have been touted for their antiinflammatory properties.
Single-cell–based approaches to allergic and other immune-mediated diseases
This study gives an early view of the challenges and rewards associated with using scRNA-seq and related methods for studying human allergic diseases. Obtaining sufficient numbers of immune cells from relevant tissues obtained from clinically well-characterized individuals will often be challenging, but is critical to the success of this approach. Deployment of methods that allow for efficient, in-depth analysis of much larger numbers of cells will be necessary for understanding disease heterogeneity in human populations, fully characterizing antigen receptor repertoires, and studying critical but rare immune cell types. Equally important will be the development of tools and resources for sharing and analyzing these complex data sets. While transcriptome-wide profiling of isolated cells is already a powerful tool, emerging methods will add additional layers of data about the epigenetic state (16) or protein levels, and allow spatial analysis of gene expression in tissues (17). While studies in mouse and other model systems remain indispensable, these “deep phenotyping” methods are an important step toward advancing the future of humans as model organisms (18).
Acknowledgments
WLE is supported by NIH grant K99HL135403. DJE is supported by NIH grants U19AI077439, R01HL138424, and R35HL1452345.
Version 1. 04/08/2019
Electronic publication
Version 2. 05/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(5):1830–1832. https://doi.org/10.1172/JCI128479.
See the related article at Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis.
References
- 1.Attwood SE, Smyrk TC, Demeester TR, Jones JB. Esophageal eosinophilia with dysphagia. A distinct clinicopathologic syndrome. Dig Dis Sci. 1993;38(1):109–116. doi: 10.1007/BF01296781. [DOI] [PubMed] [Google Scholar]
- 2.Furuta GT, Katzka DA. Eosinophilic esophagitis. N Engl J Med. 2015;373(17):1640–1648. doi: 10.1056/NEJMra1502863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dellon ES, et al. Distribution and variability of esophageal eosinophilia in patients undergoing upper endoscopy. Mod Pathol. 2015;28(3):383–390. doi: 10.1038/modpathol.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108(6):954–961. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 5.Martin LJ, et al. Eosinophilic esophagitis (EoE) genetic susceptibility is mediated by synergistic interactions between EoE-specific and general atopic disease loci. J Allergy Clin Immunol. 2018;141(5):1690–1698. doi: 10.1016/j.jaci.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard C, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest. 2006;116(2):536–547. doi: 10.1172/JCI26679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ko E, Chehade M. Biological therapies for eosinophilic esophagitis: where do we stand? Clin Rev Allergy Immunol. 2018;55(2):205–216. doi: 10.1007/s12016-018-8674-3. [DOI] [PubMed] [Google Scholar]
- 8.Wen T, et al. Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis. J Clin Invest. 2019;129(5):2014–2028. doi: 10.1172/JCI125917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017;358(6359):58–63. doi: 10.1126/science.aan6828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vegh P, Haniffa M. The impact of single-cell RNA sequencing on understanding the functional organization of the immune system. Brief Funct Genomics. 2018;17(4):265–272. doi: 10.1093/bfgp/ely003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ordovas-Montanes J, et al. Allergic inflammatory memory in human respiratory epithelial progenitor cells. Nature. 2018;560(7720):649–654. doi: 10.1038/s41586-018-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang D, et al. Single-cell profiling of peanut-responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. J Allergy Clin Immunol. 2018;141(6):2107–2120. doi: 10.1016/j.jaci.2017.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolognini D, Tobin AB, Milligan G, Moss CE. The pharmacology and function of receptors for short-chain fatty acids. Mol Pharmacol. 2016;89(3):388–398. doi: 10.1124/mol.115.102301. [DOI] [PubMed] [Google Scholar]
- 14.Sina C, et al. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol. 2009;183(11):7514–7522. doi: 10.4049/jimmunol.0900063. [DOI] [PubMed] [Google Scholar]
- 15.Trompette A, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20(2):159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 16.Cao J, et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science. 2018;361(6409):1380–1385. doi: 10.1126/science.aau0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salmén F, et al. Barcoded solid-phase RNA capture for Spatial Transcriptomics profiling in mammalian tissue sections. Nat Protoc. 2018;13(11):2501–2534. doi: 10.1038/s41596-018-0045-2. [DOI] [PubMed] [Google Scholar]
- 18.FitzGerald G, et al. The future of humans as model organisms. Science. 2018;361(6402):552–553. doi: 10.1126/science.aau7779. [DOI] [PubMed] [Google Scholar]