Abstract
It remains unknown what causes inflammatory bowel disease (IBD), including signaling networks perpetuating chronic gastrointestinal inflammation in Crohn’s disease (CD) and ulcerative colitis (UC), in humans. According to an analysis of up to 500 patients with IBD and 100 controls, we report that key transcripts of the IL-7 receptor (IL-7R) pathway are accumulated in inflamed colon tissues of severe CD and UC patients not responding to either immunosuppressive/corticosteroid, anti-TNF, or anti-α4β7 therapies. High expression of both IL7R and IL-7R signaling signature in the colon before treatment is strongly associated with nonresponsiveness to anti-TNF therapy. While in mice IL-7 is known to play a role in systemic inflammation, we found that in humans IL-7 also controlled α4β7 integrin expression and imprinted gut-homing specificity on T cells. IL-7R blockade reduced human T cell homing to the gut and colonic inflammation in vivo in humanized mouse models, and altered effector T cells in colon explants from UC patients grown ex vivo. Our findings show that failure of current treatments for CD and UC is strongly associated with an overexpressed IL-7R signaling pathway and point to IL-7R as a relevant therapeutic target and potential biomarker to fill an unmet need in clinical IBD detection and treatment.
Keywords: Gastroenterology, Immunology
Keywords: Cytokines, Inflammatory bowel disease, T cells
Introduction
Inflammatory bowel disease (IBD) consists of 2 major forms, ulcerative colitis (UC) and Crohn’s disease (CD), which are chronic relapsing gastrointestinal disorders characterized by chronic intestinal inflammation, dysregulated immune responses to intestinal microbiota, and dysfunction of the epithelial barrier (1, 2). The incidence and prevalence of IBD are increasing worldwide, and these diseases are associated with marked morbidity and have a major impact on quality of life and ability to work (3, 4). Current conventional treatments are aimed at dampening inflammation with the gradual use of antiinflammatory agents, steroids, immunosuppressive drugs, and biological agents targeting inflammatory cytokines (mainly TNF) for refractory and severe forms of IBD (5, 6). However, approximately one-third of patients receiving anti-TNF agents do not respond to treatment (primary failure), and a significant proportion (up to 50%) become refractory over time (secondary failure) (7). A key feature of IBD is also the rapid recruitment and prolonged persistence of leukocytes at the site of inflammation, which is facilitated by integrin interaction with cognate receptors expressed by endothelial cells allowing cell adhesion and transmigration (8, 9). Emerging therapies are targeting this entry point to the gut with antiadhesion molecules, specially targeting the gut-specific α4β7 integrin (10, 11). However, these therapies do not induce remission in more than half of patients, and relapse also occurs over time in primary responders. Thus, one major goal is to identify novel mechanisms and signaling networks that underlie the chronicity and relapse of IBD.
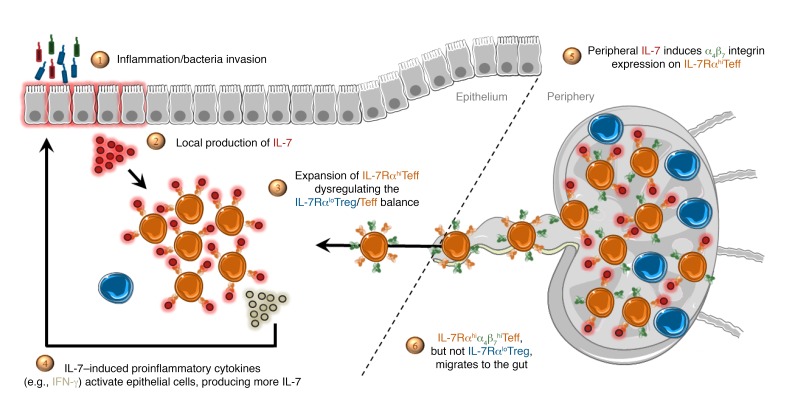
Interleukin-7 (IL-7) is a limiting and nonredundant cytokine produced mainly by epithelial and stromal cells that regulates T lymphocyte homeostasis (12, 13). Almost all conventional mature T lymphocytes express high IL-7 receptor (IL-7R) levels, with the specific exception of naturally occurring Tregs, constituting a rare opportunity to selectively target pathogenic effectors while preserving natural regulators (14–16). IL-7 signals through the cell surface IL-7R that consists of a specific IL-7R α chain (IL-7Rα, CD127) that dimerizes with the common cytokine receptor γ chain (γc, CD132, IL2RG) (17). IL-7Rα also interacts with the thymic stromal lymphopoietin receptor (TSLPR) chain to form the heterodimeric receptor of the TSLP cytokine (18). Upon activation, IL-7R delivers proliferative and antiapoptotic signals by activating PI3K and JAK/STAT pathways as well as regulating expression of anti- and proapoptotic BCL-2 family members (19). More than 2 decades of research has reproducibly highlighted the crucial systemic influence of the IL-7 pathway and depicted IL-7 as a fuel for chronic autoimmune and inflammatory diseases as well as transplant rejection in diverse rodent models (20–22). Remarkably, the colon is the major source of IL-7 outside of the lymphoid tissues in mice, and commensal microflora promotes steady-state IL-7 production by intestinal epithelial cells (23, 24). IL-7–transgenic mice spontaneously developed chronic colitis with IL-7Rαhi T cells infiltrating the gut mucosa, which, once isolated, transferred colitis to immunodeficient recipient mice, thereby demonstrating their pathogenicity (25–29). More recently, IL-7Rαhi innate lymphoid cells (ILCs) were described as new key effectors of IBD (30–32), and IL-7Rα antagonists suppressed both adaptive and innate inflammatory responses in different experimental models of colitis (26, 27, 33, 34).
In humans, however, research on IBD has mainly focused on a proinflammatory and antiinflammatory cytokine balance (35–37), while the IL-7/IL-7Rα axis remains poorly studied. IL-7 production by intestinal epithelial cells and IL7R expression by mucosal T lymphocytes were described in colon biopsies from healthy donors (38). One study reported genetic loci variation of the IL7R gene being associated with susceptibility to UC (39). Lastly, elevated IL-7 signaling pathway gene expression in blood CD8+ T cells at diagnosis was significantly associated with relapsing and a more aggressive IBD disease course (40). Whether IL-7 activities are implicated in the pathogenesis of IBD in humans and determine the fate of treatment is uncertain.
Results
Colon-specific increased mucosal IL7R signaling pathway signature in refractory IBD.
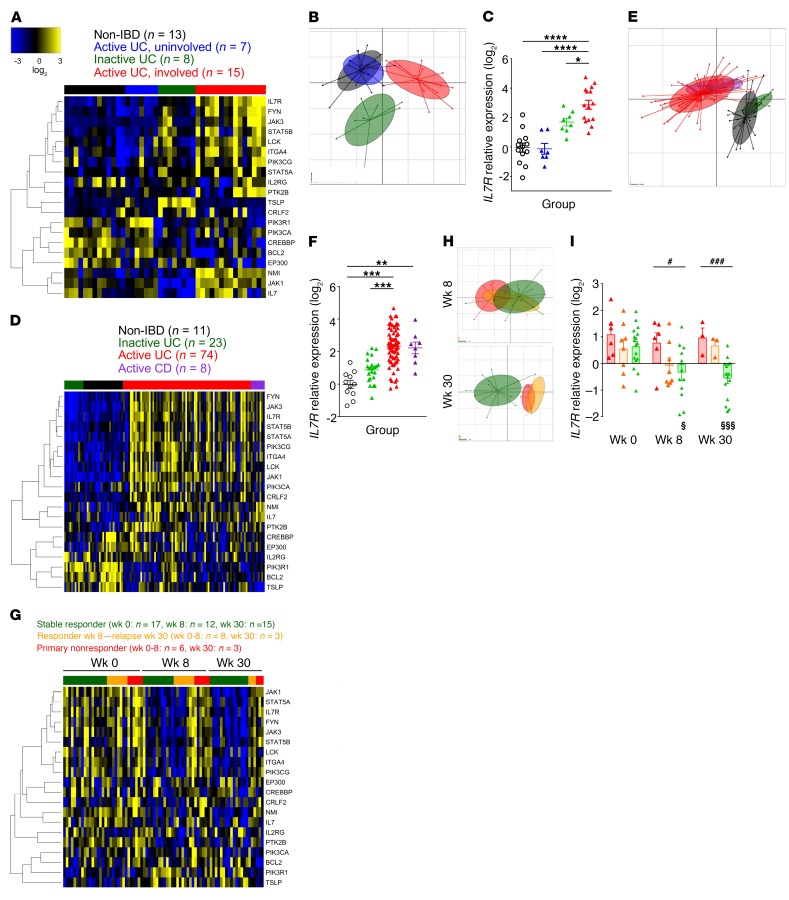
Higher gene expression of the IL-7 signaling pathway members in peripheral blood CD8+ T cells has been previously correlated with a relapsing and aggressive disease course of IBD (40). We therefore sought to address whether the local/mucosal IL-7/IL-7R pathway could be enriched in severe IBD and associated with the failure of conventional therapies. For this purpose, we focused our hypothesis-driven analysis on only the transcriptional IL-7/IL-7R signaling signature of 20 genes (curated from BioCarta by the Molecular Signatures Database [MSigDB]; ref. 40) in colon mucosal biopsies from previously published cohorts of IBD patients with clinically active or inactive disease after immunosuppressive therapy. We examined 2 independent cohorts (UC [n = 23] compared with controls without IBD [n = 13], ref. 41; and UC [n = 97] or CD [n = 8] compared with controls without IBD [n = 11]; ref. 42). We found that this 20-mRNA expression signature was altered according to the clinical status (Figure 1, A and D), with a particularly significant accumulation of IL7R mRNA transcripts in inflamed mucosa from refractory IBD patients relative to those in uninvolved control mucosa from the same patients (P < 0.0001), mucosa from responder patients (P < 0.001), or non-IBD controls (Figure 1, C and F). Principal component analysis (PCA) of the mucosal IL-7R signaling signature after immunosuppressive therapy displayed in both cohorts a clear and distinct separation between active versus inactive disease status and the group of uninvolved mucosa that gathered from non-IBD controls (Figure 1, B and E).
Figure 1. IL-7R signaling pathway expression is highly expressed in colonic mucosa of refractory IBD.
(A) Heatmap of the expression of the 20 selected genes of the IL-7R signaling pathway (40) in colon biopsies from non-IBD control patients (black; n = 13), inactive UC patients (green; n = 8), and active UC patients with involved (red; n = 15) or uninvolved proximal segments (blue; n = 7) after corticosteroids and/or immunosuppression therapy (data set GSE38713; ref. 41). The heatmap represents median-centered and colorized expression values. (B) PCA of the IL-7R signaling signature and (C) relative IL7R expression (log2 data were normalized to the control median; same patient and color groups as in A). (D) Heatmap of the IL-7R signaling signature in colon biopsies from non-IBD controls (black; n = 11), inactive (green; n = 23) or active (red; n = 74) UC patients, and active CD patients (purple; n = 8) after corticosteroids and/or immunosuppression therapy. (E) PCA of the IL-7R signaling signature and (F) relative IL7R expression (log2 data were normalized to the control median; same patient and color groups as in D). (G) Heatmap of the IL-7R signaling signature in colon biopsies from UC patients at weeks 0, 8, and 30 following anti-TNF therapy (green, stable responders, n = 12–17; orange, relapsing patients, n = 3–8; red, primary nonresponders, n = 3–6). (H) PCA after 8 (top) and 30 (bottom) weeks of anti-TNF treatment of the IL-7R signaling signature and (I) IL7R expression (log2 data were normalized to the median of the stable responder group; same patient and color groups as in G). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 between the indicated groups. #P < 0.05; ###P < 0.001 between responders and nonresponders at a single time point. §P < 0.05; §§§P < 0.001, Kruskal-Wallis with post hoc Dunn’s multiple comparisons test within a single response group across all 3 time points.
Similarly, the IL-7R signaling signature after anti-TNF treatment allows distinguishing between biopsies according to disease status in a third cohort of UC patients involved in the phase III ACT-1 clinical trial of infliximab (including long-term responders [n = 17], short-term responders and then relapse [n = 8], and primary nonresponders [n = 6]; ref. 43) (Figure 1G). PCA of the mucosal IL-7R signature clearly separates responder from nonresponder patients 30 weeks after maintenance with anti-TNF treatment (Figure 1H). Furthermore, mucosal IL7R expression was significantly higher in clinically active versus inactive disease in anti-TNF–treated patients at both early (week 8, P < 0.05) and late (week 30, P < 0.001) time points (Figure 1I). IL7R expression significantly decreased after anti-TNF therapy in long-term responders compared with baseline pretreatment levels (P < 0.001); however, no significant difference was measurable before treatment between groups in this clinical trial cohort.
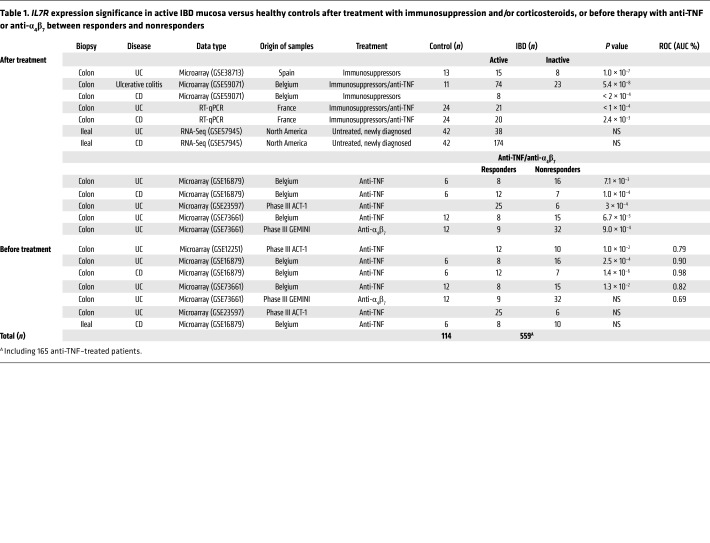
In contrast to colon mucosa, the IL-7R signature in ileal biopsies was not associated with clinical status in a fourth cohort (44) of untreated pediatric patients with newly diagnosed ileal (iCD; n = 143) or colon-only (cCD, n = 30) CD or UC (n = 38) as compared with pediatric healthy controls (n = 42) (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI121668DS1). IL7R expression was only higher in ileal biopsies with histological microinflammation versus uninflamed biopsies of cCD; however, no difference was observed in ulcerated or nonulcerated iCD relative to healthy controls (Supplemental Figure 1C). Furthermore, IL7R was not differentially expressed in colon mucosa of different non-IBD types of controls, including in inflammatory non-IBD pathology such as diverticulitis (Supplemental Figure 2). Altogether, the data indicate that mucosal expression of the IL-7R signaling pathway, and specifically IL7R, is strongly and selectively associated with colonic but not ileal IBD inflammation. Expression is related to disease activity in IBD patients (Table 1) and could be one mechanism associated with refractory and severe IBD.
Table 1. IL7R expression significance in active IBD mucosa versus healthy controls after treatment with immunosuppression and/or corticosteroids, or before therapy with anti-TNF or anti-α4β7 between responders and nonresponders.
Colonic IL-7Rα expression predicts resistance to anti-TNF therapy.
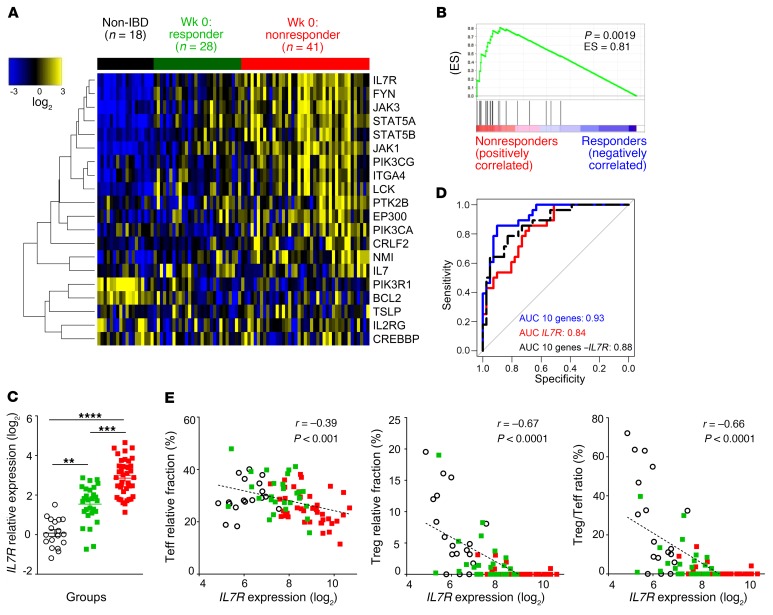
We then asked whether the mucosal IL-7R signaling signature is associated with responsiveness to anti-TNF before initiation of biologic therapy in patients refractory to corticosteroids and/or immunosuppression. We thus performed a meta-analysis of publicly available transcriptional data sets of 3 cohorts of UC patients with colon mucosa biopsies performed before anti-TNF treatment (within a week) (45–47). In these 3 cohorts, anti-TNF response was defined as histological healing analyzed 4–6 weeks after their first anti-TNF infusion (altogether: n = 18 non-IBD controls, n = 41 UC nonresponders, and n = 28 UC responders). Differential analysis of all expressed genes of the meta–data set (17,037 genes) identified only 85 genes significantly (adjusted P value < 0.05) and differentially (log2 FC > 1) expressed between responders and nonresponders, including IL7R (Supplemental Figure 3), which clustered with a large group of genes associated with adaptive immune response, T cell activation, and leukocyte migration/adhesion (Supplemental Figure 4). Analysis of the transcriptional IL-7R signaling signature (20 genes previously described) revealed that the IL-7R signaling signature, in particular IL7R, was already elevated in colon biopsies of primary UC nonresponder patients before initiation of anti-TNF therapy (Figure 2, A–C). A lasso (least absolute shrinkage and selection operator) regression analysis identified a combination of 10 genes (IL7R, IL2RG, JAK1, PIK3CA, LCK, PTK2B, EP300, NMI, CRLF2, and TSLP) within this IL-7R signaling signature able to discriminate anti-TNF nonresponders from responders. Pretreatment expression of IL7R or the 10-gene IL-7R–restricted signature strongly discriminated primary nonresponsiveness to anti-TNF therapy in this meta-analysis (IL7R receiver operating characteristic [ROC] AUC = 84%, 10-gene IL-7R signature ROC AUC = 93%) (Figure 2D). The absence of IL7R expression in the signature decreased correlation with anti-TNF response (AUC = 88%). These data suggest that the entire pathway is associated with the nonresponse to anti-TNF, from IL-7R to transcriptionally regulated proteins (i.e., ITGA4) to major key signaling components (JAK/STAT, PI3K) (Supplemental Figure 5). Intestinal oncostatin M (OSM) has been also recently reported to be associated with anti-TNF unresponsiveness in IBD (48). OSM was also identified as differentially expressed among 85 genes in our meta-analysis (Supplemental Figure 3). OSM expression correlated in these UC biopsies with IL7R expression but was not superior to IL7R in discriminating primary nonresponse to anti-TNF therapy (Supplemental Figure 6). While combining OSM or OSMR with IL7R expression slightly improved ROC analysis, the 10-gene IL-7R–restricted signature remained more potent. Finally, the abundance of immune cell populations in this meta–data set was estimated using the deconvolution CIBERSORT tool (49). IL7R expression was not correlated with relative abundance of myeloid cells, B cells, or NK cells (data not shown) or with any subsets of effector T lymphocytes (CD8+, CD4+, naive, or memory; or resting, activated, or follicular Th cells) (Figure 2E). In contrast, deconvolution of the meta–data set revealed that IL7R expression was significantly and inversely correlated with the relative fraction of Tregs (r = –0.67, P < 0.0001) in colon mucosa of UC patients before anti-TNF therapy and associated with an imbalanced local ratio of Tregs to effector T lymphocytes (Teffs) (r = –0.66, P < 0.0001) (Figure 2E). IL7R expression also correlated significantly and positively with both IFNG and Tbx21 (T-bet) gene expression, hallmarks of Th1 effector cells, and with expression of some Th17 hallmark genes (such as IL17A, IL22, and IL23, but, surprisingly, in an inverse correlation with the RORC gene), as well as the GATA3 hallmark of Th2 (though no correlation was observed with IL4, IL5, or IL13) (Supplemental Figure 7). Finally, using a recently published gene expression signature of human tonsil CD127+ ILCs characterized by single-cell RNA sequencing (50, 51), we found that signatures of all ILC subsets (ILC1, ILC2, and ILC3) were enriched in UC patients from the meta–data set as compared with non-IBD controls, and that ILC signatures were significantly overexpressed in anti-TNF nonresponder versus responder patients (Supplemental Figure 8). However, these signatures were built using isolated tonsil ILC subsets and not to estimate the relative abundance of ILCs in a bulk RNA data set. Furthermore, the low abundance of ILCs in human colon (52, 53) makes them unlikely to explain transcriptomic differences detected in bulk transcriptomic data.
Figure 2. Colonic mucosal IL7R and IL-7R signaling pathway expression at baseline is associated with nonresponse to anti-TNF therapy.
(A) Heatmap of the expression of the 20 selected genes previously reported as key members of the IL-7R signaling pathway (40) in colon biopsies of non-IBD controls (n = 18), responders (green; n = 28), and nonresponders (red; n = 41) before anti-TNF therapy. Meta-analysis of 3 UC cohorts with histological healing as the anti-TNF response criterion: data sets GSE16879 (46), GSE12251 (45), and GSE73661 (47). The heatmap represents median centered colorized expression values in which yellow values indicate overexpression and blue values indicate underexpression. (B) Gene set enrichment analysis from the meta–data set at week 0 of the IL-7R signaling signature (20 selected genes). ES, enrichment score. (C) Relative IL7R expression before anti-TNF treatment (log2 data normalized to the control median) in the same groups of patients and colors as in A. (D) ROC analysis of expression of IL7R, the IL7R 10-gene signature (IL7R, IL2RG, JAK1, PIK3CA, LCK, PTK2B, EP300, NMI, CRLF2, and TSLP), and the signature without IL7R (9 genes) distinguishing anti-TNF responders and nonresponders. (E) Correlation of IL7R expression with enrichment of leukocyte subsets as estimated by CIBERSORT in the same groups of patients and colors as in A. Left: Effector T lymphocytes (Teff); middle: regulatory T lymphocytes (Treg); right: Treg/Teff ratio. **P < 0.01; ***P < 0.001; ****P < 0.0001 between indicated groups, Kruskal-Wallis test with post hoc Dunn’s multiple comparisons test.
Analysis within each cohort independently showed that pretreatment individual baseline IL7R expression was significantly increased in UC but also in cCD nonresponder colon mucosa and did not decrease after anti-TNF therapy (Supplemental Figure 9). Colon mucosal IL7R and the 10-gene IL-7R signature were strongly and repeatedly discriminative of primary responsiveness to anti-TNF therapy in 4 different cohorts of UC and cCD patients (10-gene IL-7R signature ROC AUC = 88.3%, 95.3%, 95.2%, 96.7%). In contrast, while the ileum was also inflamed in active iCD patients, the IL-7R signaling signature or IL7R alone in ileal mucosa was not differentially expressed before or after anti-TNF induction therapy and was not associated with anti-TNF responsiveness in iCD patients (Supplemental Figure 1, D–F). Finally, colon mucosa gene expression analysis of a single recent cohort of UC patients treated with anti-α4β7 (vedolizumab) therapy confirmed also that IL-7R signaling signature was altered and IL7R was differentially expressed between responders and nonresponders at early (week 6) or late (week 52) time points after treatment (Supplemental Figure 10). Altogether, our meta-analysis argues that the pretreatment mucosal IL-7R signaling signature and IL7R expression in the colon are strongly associated with nonresponsiveness to anti-TNF, and presumably also anti-α4β7 therapy, in UC and cCD patients (Table 1) and are associated with a local deficit in Tregs.
Characterization of mucosal IL7/IL7R expression.
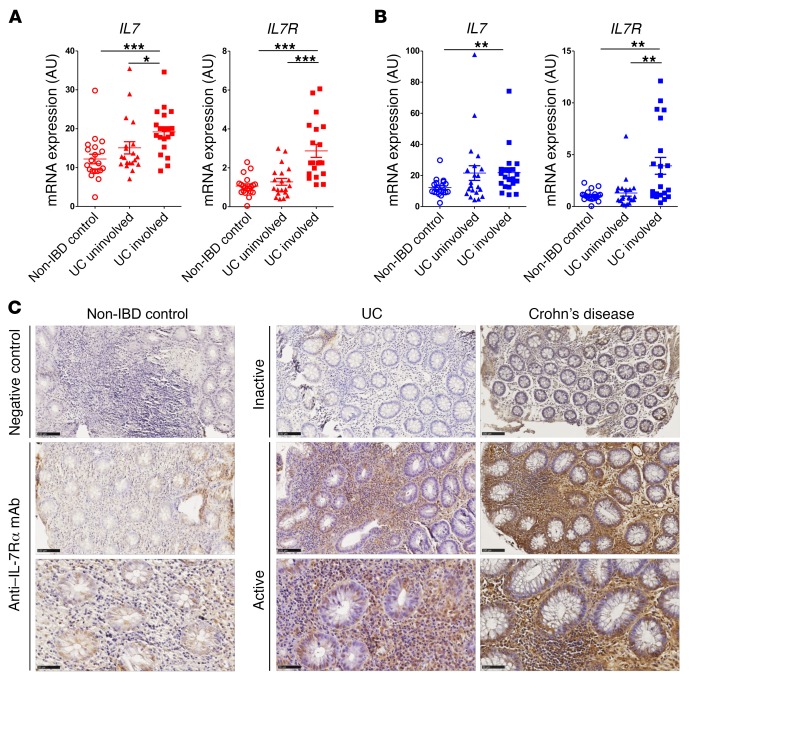
To validate the meta-analysis results, we first performed quantitative reverse transcription PCR (RT-qPCR) on freshly isolated mucosal colon biopsy specimens from UC (n = 21) or CD (n = 24) patients with active disease (based on clinical and endoscopic findings) and treated with immunosuppressive or anti-TNF therapies (Supplemental Table 1). Paired biopsies from both uninvolved (healthy) and involved (diseased) colonic areas of IBD patients were analyzed and compared with those from non-IBD control patients (n = 20) who underwent endoscopy at Nantes University Hospital in France. This analysis confirmed significantly higher expression of IL7, IL7R, and IL2RG (γ chain) in involved colon mucosa of patients with immunosuppressive- or anti-TNF–refractory UC and CD, compared with uninvolved paired tissue from the same patients or non-IBD controls (Figure 3, A and B, and Supplemental Figure 11). The TSLPR chain, which also forms a heterodimer with IL-7Rα (18), was also elevated in the inflamed colon of IBD patients (Supplemental Figure 11). In line with a previous report (54), full-length TSLP cytokine transcripts were not significantly different, while the short-form TSLP variant, which does not signal IL-7Rα/TSLPR but has characteristics of an antimicrobial peptide (55), exhibited reduced mRNA levels in both healthy and inflamed colonic areas of UC and CD patients (Supplemental Figure 11). In parallel, we found increased mRNA expression of ITGA4 (α4 integrin) and ITGB7 (β7 integrin) as well as TNF and TNF receptors (TNFR1 and TNFR2) in involved IBD samples, confirming the inflammatory profile of these biopsies. Both IL7 and IL7R mRNA expression significantly correlated with ITGA4/ITGB7 and TNF/TNFR expression (Supplemental Figure 12) with a marked correlation for α4β7 integrin. Several IL-7 and IL-7Rα splice variants have been described (56, 57), including a functional soluble IL-7Rα isoform (58). Expression of membrane-bound IL7R and full-length IL7 strongly correlated with IL7R and all IL7 variants by RT-qPCR analysis, suggesting that alternative splicing is not modified in inflammatory settings (Supplemental Figure 13).
Figure 3. Increased IL7R mRNA and IL-7Rα protein expression in UC and CD colon mucosa.
(A and B) Relative IL7 and IL7R expression, as measured by RT-PCR, of colon biopsies from inflamed and healthy areas of UC (n = 21) (A) and CD (n = 24) (B) patients compared with non-IBD control patients (n = 20). Each symbol represents 1 patient, bars represent means, and error bars show SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 between indicated groups. (C) Representative IL-7Rα immunohistochemical staining of colonic biopsy specimens from non-IBD control (infectious acute colitis) and inactive and active UC or CD (scale bars: top and middle, 100 μm; bottom, higher-magnification images of the panels above, 50 μm).
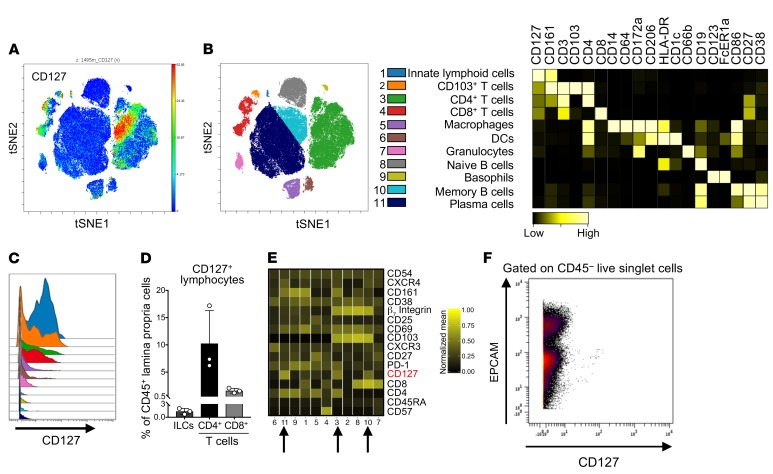
We then confirmed increased IL-7Rα expression at the protein level first by immunohistochemistry in colon biopsy specimens of active UC and CD patients as compared with inactive IBD or non-IBD controls (Figure 3C). IL-7Rα staining was mainly present on inflammatory infiltrates in these biopsies as well as by some epithelial cells in the inflammatory microenvironment. To decipher which immune cell types express IL-7Rα in inflamed colon, mass cytometry (CyTOF) analyses, using a panel of 30 detection antibodies, were performed on human lamina propria cell suspensions prepared from the colon of UC patients (n = 3) before treatment with immunotherapy. IL-7Rα expression was observed only on mucosal CD4+ or CD8+ T lymphocytes as well as ILCs but not on other leukocyte cell types (Figure 4, A–C). Remnant epithelial (EPCAM+CD45–) or stromal cells (EPCAM–CD45–) in these lamina propria cell suspensions did not express IL-7Rα (Figure 4F). As anticipated, ILCs expressed the highest level of IL-7Rα. However, the low abundance of ILCs within mucosal leukocytes (Figure 4D) is unlikely to explain overexpression in the bulk transcriptomic data set. Phenograph analysis identified 3 clusters of IL-7Rα+ T lymphocytes, 2 of them corresponding to mucosal CD4+ or CD8+ T cells coexpressing mucosal integrins αE and β7 (Figure 4E). In contrast, the mucosal CD4+ T cell subset expressing the highest level of CD25 (IL-2R), which might be enriched in regulatory T cells, did not express a significant level of IL-7Rα, confirming that Tregs in inflamed mucosal tissues do not express IL-7R. Altogether, our biocollection transcriptomic and proteomic analyses confirmed that IL-7Rα is overexpressed in inflamed colon mucosa from IBD patients and that IL-7Rα is mainly expressed by mucosal ILCs and T lymphocytes coexpressing mucosal integrins such as β7 integrin.
Figure 4. Characterization of CD127 expression among UC lamina propria cells.
(A) Representative t-distributed stochastic neighbor embedding (tSNE) analysis showing single-cell intensity of CD127 among CD45+ lamina propria leukocytes from UC inflamed colon (n = 3 UC patients). (B) Left: Major immune populations were manually gated and identified on the tSNE map on the basis of canonical marker expression patterns. Right: Heatmap showing the corresponding color-coded normalized expression of canonical markers in gated immune populations. (C) Histograms showing medians of CD127 signal intensity within each of the immune populations defined in B. (D) Frequencies of CD127+ lymphocyte populations among lamina propria leukocytes. Bars correspond to the mean ± SEM (n = 3 UC patients). (E) Phenograph analysis identifies 3 clusters of CD127+ T cells in UC inflamed colon. Each column corresponds to a cluster. Rows indicate the color-coded normalized mean expression intensity of indicated markers in each cluster. CD127+ T cell clusters are indicated with arrows. (F) CD127 expression on lamina propria remnant EPCAM+ epithelial cells or EPCAM– stromal cells.
IL-7 signaling controls α4β7 gut-homing integrin expression on effector human T cells.
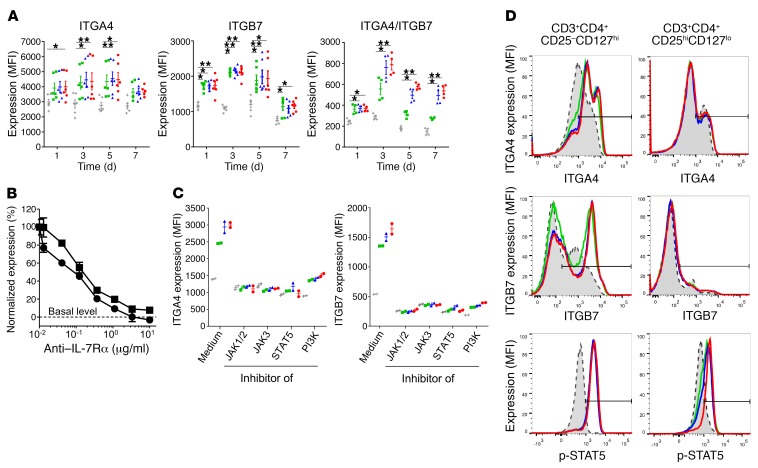
Our validation transcriptional analysis of colon biopsies and CyTOF analysis of mucosal leukocytes revealed a marked correlation between IL-7/IL-7Rα and α4β7 integrin expression in humans. Injection of recombinant IL-7 has recently been shown to provoke rapid and significant T cell accumulation in the gut of macaques and chronically HIV-infected patients (59, 60). Another study described that in vivo IL-7 treatment increased α4β7 expression on circulating T cells from HIV-infected individuals (61). Based on these findings, we aimed to further decipher the relationship between IL-7/IL-7Rα and α4β7 integrin expression in the human setting. Indeed, the link between IL-7 and α4β7 expression could not be easily deduced from previous experimental models, since we found that mouse IL-7 did not regulate mouse T lymphocyte α4 and β7 expression at the mRNA or protein level (Supplemental Figure 14). We first confirmed that human and mouse IL-7 dose-dependently and selectively induced in vitro α4, β7, and α4β7 heterodimer expression on the surface of human T lymphocytes from healthy donors (Figure 5A and Supplemental Figure 15, A and B). As expected, IL-7Rα expression was downregulated. However, no modification was observed for other major gut-homing receptors (αE, CCR6, CCR9, or GPR15), other homing receptors (CCR5, CXCR3), nor β1 integrin, which associates with α4 (Supplemental Figure 16). IL-7–induced α4β7 overexpression was long-lasting and required continuous stimulation, since removal of IL-7 after 24 hours dramatically prevented maintenance of α4β7 overexpression (Supplemental Figure 17A). Similarly, an anti–human IL-7Rα antagonist mAb dose-dependently prevented IL-7–induced α4β7 overexpression (Figure 5B).
Figure 5. IL-7 controls α4β7 expression on human effector, but not regulatory, T lymphocytes.
(A) Surface expression of α4, β7, and α4β7 integrin heterodimers on human T lymphocytes from the blood of healthy volunteers cultured for the indicated periods of time in medium alone (white bars) or supplemented with recombinant human IL-7 at 1 ng/ml (green), 5 ng/ml (blue), or 25 ng/ml (red). Data are expressed as median fluorescent intensity ± SEM measured by flow cytometry (n = 6). (B) Human T cells were cultured for 3 days with 5 ng/ml of recombinant human IL-7 and a blocking anti–human IL-7Rα mAb at different concentrations. ITGA4 (squares) and ITGB7 (circles) overexpression was normalized to the condition without anti–IL-7Rα mAb and expressed as mean ± SEM. Dotted line represents basal level expression at t0. (C) Surface expression of α4 and β7 integrin on human T lymphocytes cultured for 48 hours in medium alone (gray) or supplemented with recombinant human IL-7 at 1 ng/ml (green), 5 ng/ml (blue), or 25 ng/ml (red) and, when indicated, with 10 μg/ml JAK1/2 inhibitor (ruxolitinib), 5 μM JAK3 inhibitor (CAS 202475-60-3), 5 μM STAT5 inhibitor (CAS 285986-31-4), or 12.5 μM PI3K inhibitor (LY294002). Data are expressed as median fluorescent intensity ± SEM measured by flow cytometry. (D) Human effector (CD25–CD127hi) and regulatory (CD25hiCD127lo) CD4+ T cells, purified by flow cytometry using a nonantagonistic anti–human CD127 (IL-7Rα) mAb, were cultured with medium alone (gray histogram) or supplemented with 1 ng/ml (green), 5 ng/ml (blue), or 25 ng/ml (red) of human IL-7. IL-7 signaling (STAT5 phosphorylation) was measured after 15 minutes of culture, while ITGA4 and ITGB7 expression was evaluated at 48 hours. Data show 1 representative of 5 experiments. *P < 0.05; **P < 0.01 between indicated groups, Kruskal-Wallis test with post hoc Dunn’s multiple comparisons test.
IL-7 induced not only upregulation but also functional activation of the human α4β7 heterodimer, since it increased the binding to mucosal vascular addressin cell adhesion molecule-1 (MADCAM1), the specific ligand of α4β7 (Supplemental Figure 18A). Expression of α4 and β7 protein levels did not differ after cell permeabilization, suggesting the absence of a preformed intracellular pool that might be mobilized by IL-7 (Supplemental Figure 18, B–D). In contrast, we found that ITGA4 mRNA expression increased in human T cells within 2 hours after IL-7 exposure while ITGB7 mRNA levels remained unmodified (Supplemental Figure 19A). Using chemical inhibitors for each step of protein synthesis, we confirmed that IL-7–induced α4 expression required DNA transcription, RNA translation, and endoplasmic reticulum and post-Golgi protein export (Supplemental Figure 19B). While IL-7 differentially regulated α4 and β7 transcription, similar results were obtained with protein synthesis inhibitors for α4 and β7 expression, suggesting that IL-7 only indirectly controlled β7 RNA translation (Supplemental Figure 19C).
Because IL-7 did not regulate ITGA4 expression on mouse T cells (Supplemental Figure 14), we explored species-specific transcription factors. The transcription factor CREB is predicted to bind to the human but not the mouse ITGA4 gene promoter (Gene Transcription Regulation Database [GTRD]; http://gtrd.biouml.org). Previous genome-wide analysis of CREB occupancy and phosphorylation showed ITGA4 gene activation in human cells and tissues (62) but not in mouse (63). We found that a CREB/CBP association inhibitor prevented α4β7 overexpression in humans (Supplemental Figure 17B), while a CBP/P300 histone acetyltransferase inhibitor had no effect (Supplemental Figure 17C), implicating CREB in α4 expression. In addition, blocking the JAK/STAT pathway using JAK1/2, JAK3, or STAT5 inhibitors or the PI3K pathway with specific inhibitors prevented IL-7–induced α4β7 overexpression, demonstrating that it requires complete IL-7R signaling (Figure 5C).
Finally, one potential advantage of IL-7Rα–targeted therapy is that it specifically spares Tregs over effector T cells (21), based on a high differential expression level (14, 15). We found that IL-7 was able to dose-dependently induce STAT5 phosphorylation in Tregs, in spite of the low expression level of its receptor on these cells, but did not induce α4β7 overexpression in contrast to its effect on effector T cells (Figure 5D).
A pathogenic role of the IL-7Rα pathway in colon inflammation of humanized mice.
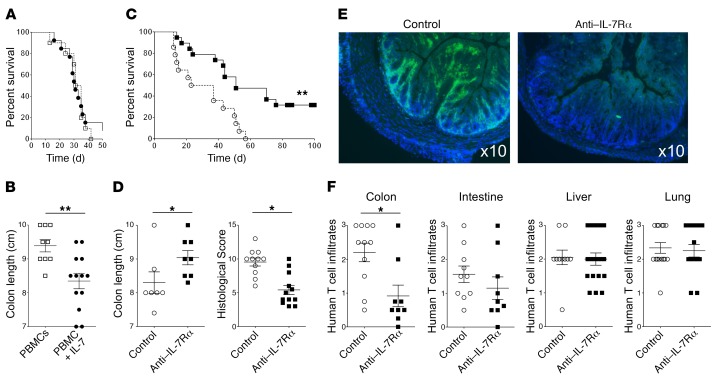
Specific control of human effector T cell upregulation of α4β7 integrin expression via the IL-7/IL-7Rα axis suggests that a possible pathogenic role for the IL-7R pathway in human IBD was not previously appreciated in conventional rodent colitis models. To further explore the possible link between IL-7/IL-7Rα, α4β7 integrin expression, and IBD, we reconstituted NOD-scid IL-2Rγ–/– (NSG) immunodeficient mice with human peripheral blood mononuclear cells (PBMCs) from healthy donors. In this model, human T cells led to xenogenic graft-versus-host disease (GVHD) with significant infiltration of colon, small intestine, liver, and lung as previously described (64). We found in a suboptimal GVHD model induced by 10 × 106 human PBMCs that pretreatment of human PBMCs with human IL-7 upregulated α4β7 expression and led to a reduced colon length in recipient mice, a hallmark of colonic inflammation (Figure 6, A and B). This, however, did not accelerate the lethality of GVHD. Conversely, in accordance with the observation that mouse IL-7 could also induce α4β7 expression on human T cells (Supplemental Figure 15A) and the confirmation in the literature (65) and experimentally that human α4β7 integrin could interact with mouse MADCAM1 (Supplemental Figure 15C), in the severe GVHD model induced by 50 × 106 human PBMCs, administration of an antagonist anti–human IL-7Rα mAb significantly delayed GVHD development in NSG mice reconstituted with unmanipulated human PBMCs (Figure 6C). It also prevented colon retraction and histological lesions of colon GVHD (Figure 6D) as well as specifically reduced human T lymphocyte infiltration in the colon but not in other target tissues of GVHD (Figure 6, E and F). To determine whether anti–IL-7Rα therapy in this model also acts systemically, we performed a back-to-back comparison with the anti-α4β7 mAb vedolizumab (Entyvio, Millennium Pharmaceuticals). Surprisingly, we found that unlike the anti–IL-7Rα mAb, vedolizumab had no effect on GVHD and colon inflammation (Supplemental Figure 20), suggesting also a systemic action of IL-7 besides α4β7 upregulation in this humanized GVHD model.
Figure 6. IL-7/IL-7Rα pathway controls human T cell–mediated colon inflammation.
(A and B) Survival (A) and colon length (B) of immunodeficient NOD-scid IL-2Rγ–/– (NSG) mice reconstituted with 10 × 106 freshly purified human PBMCs from healthy donors (white squares; n = 11) or the same cells cultured 3 days with 5 ng/ml of recombinant human IL-7 (black circles; n = 13). Colon length was measured when mice had lost 20% of body weight. (C) Survival of NSG mice reconstituted with 50 × 106 freshly purified human PBMCs from healthy donors and treated 3 times per week for 4 weeks from day 0 with 5 mg/kg of a blocking anti–human IL-7Rα mAb (black squares; n = 19) or equivalent volume of excipient (vehicle; white circles; n = 14). (D) Colon length and colon histological GVHD score of the same NSG mice as in C that were sacrificed when they had lost 20% of their body weight or after 3 months without clinical signs of GVHD. (E) Representative images of human CD3+ T cell infiltration (green) in the colon of NSG mice treated with excipient or anti–IL-7Rα mAb. Blue staining represents nuclei. (F) Histological human T cell infiltration score in colon, intestine, liver, and lung tissues of NSG mice as in C. Each symbol represents 1 mouse; horizontal bars indicate the mean ± SEM. *P < 0.05; **P < 0.01.
IL-7Rα blockade protects from colitis in humanized mice.
We next sought to address whether the local and systemic effects of IL-7Rα blockade would be effective in a more relevant colitis model in humanized mice containing both innate and adaptive immune cells. We therefore used NSG mice reconstituted with hematopoietic CD34+ cord blood stem cells and induced colitis by intrarectal injection of the haptenating substance TNBS in ethanol. Ethanol is required to break the mucosal barrier, whereas TNBS is believed to haptenize colonic autologous and/or microbiota proteins, rendering them immunogenic to the host immune system (66). In this humanized mouse model, human T cell subsets (CD4+ and CD8+ naive, central memory, and effector memory T cells) as well as human ILC subsets (ILC1, ILC2, ILC3 NCR+, and ILC3 NCR–) located in the spleen expressed membrane IL-7Rα, except CD4+CD25+ T cells, since, as anticipated, this last subset is enriched in Tregs expressing low levels of IL-7Rα (Supplemental Figure 21). While ILCs were undetectable in colon mucosa from humanized mice, we found that mucosal human T cells expressed a high level of IL-7Rα as compared with peripheral human T cells from the same humanized mice (Supplemental Figure 22).
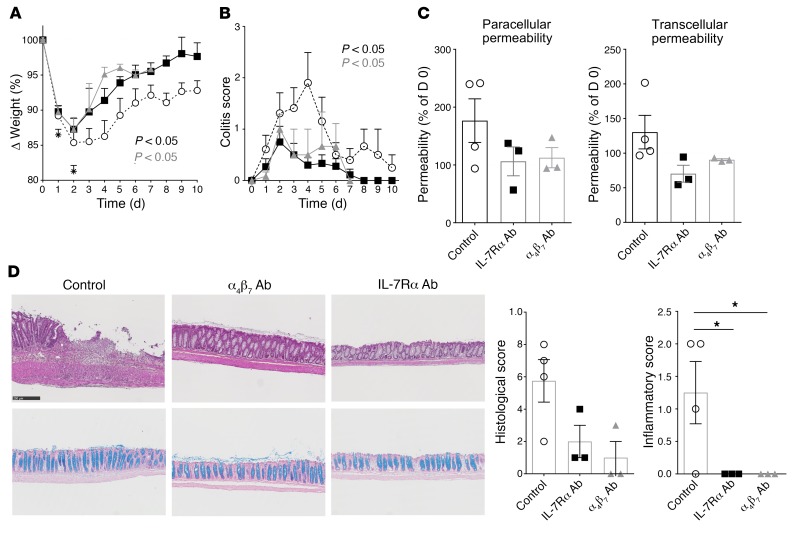
While the immunodeficient NSG mice dramatically succumbed to TNBS/ethanol injection, the humanized mice were partially protected, but developed acute signs of colitis including diarrhea and delayed weight recovery (Figure 7, A and B), in accordance with a dual protective and deleterious role for immune cells in gut homeostasis and inflammation (67). Similarly to vedolizumab, administration of anti–human IL-7Rα mAbs from day 0 was already effective from day 3 after injection in this acute humanized colitis model, since mice started to recover weight and presented significantly less diarrhea (Figure 7, A and B). Although it did not reach significance because of the limited number of humanized mice analyzed and interindividual variability, we observed that intestinal epithelial barrier homeostasis returned to basal levels with either therapy, as measured by the paracellular and transcellular intestinal permeability (Figure 7C) and the significant reduction of histological inflammatory score (Figure 7D) 1 week after treatment.
Figure 7. IL-7Rα blockade efficiency in vivo in humanized acute TNBS colitis.
(A and B) Weight variation (mean ± SEM) (A) and colitis score (mean ± SEM) (B) of NSG mice previously reconstituted with 50 × 103 human cord blood CD34+ hematopoietic stem cells and challenged several months later with intrarectal administration of 5 mg of 2,4,6-trinitrobenzenesulfonic acid (TNBS) reconstituted in 50% ethanol. Mice were treated intraperitoneally 3 times a week from day 0 with control vehicle (white circles; n = 14), 5 mg/kg of blocking anti–human IL-7Rα mAb (black squares; n = 16), or blocking anti–human α4β7 (vedolizumab) mAb (gray triangles; n = 6). (C) Paracellular and transcellular intestinal permeability (mean ± SEM) measured in vivo in NSG mice as in A after oral administration of sulfonic acid–FITC (SAF) and horseradish peroxidase (HRP) performed at day 0 and 1 week after TNBS/ethanol injection, and measured in sera 2 hours after oral administration. Results were normalized for each mouse to day 0. (D) Left: Representative colon hematoxylin-eosin-safranin staining (top) and cyan blue coloration (bottom) for each group 1 week after TNBS injection. Right: Histological and inflammatory mucosal score quantification. Scale bar: 250 μm. *P < 0.05 between indicated groups, Kruskal-Wallis test with post hoc Dunn’s multiple comparisons test.
Finally, we explored the use of blocking anti–human IL-7Rα mAbs in vivo in a treatment model using immunodeficient NSG mice previously reconstituted with a low number (2.5 × 106) of human PBMCs and receiving a chronic cycle of dextran sulfate sodium (DSS; 1% in drinking water). Humanized mice received vehicle or 5 mg/kg anti–IL-7Rα mAbs at the initiation of the second cycle of DSS and until the end of the protocol. No colitis stool score difference was observed between vehicle and anti–IL-7Rα–treated mice before treatment, whereas after treatment initiation mice treated with anti–IL-7Rα mAbs displayed significantly lower signs of diarrhea and mucosal histological lesions as compared with control mice in response to the iterative chronic cycle of DSS (Supplemental Figure 23).
IL-7Rα mAb alters mucosal effector T cells in UC biopsies cultured ex vivo.
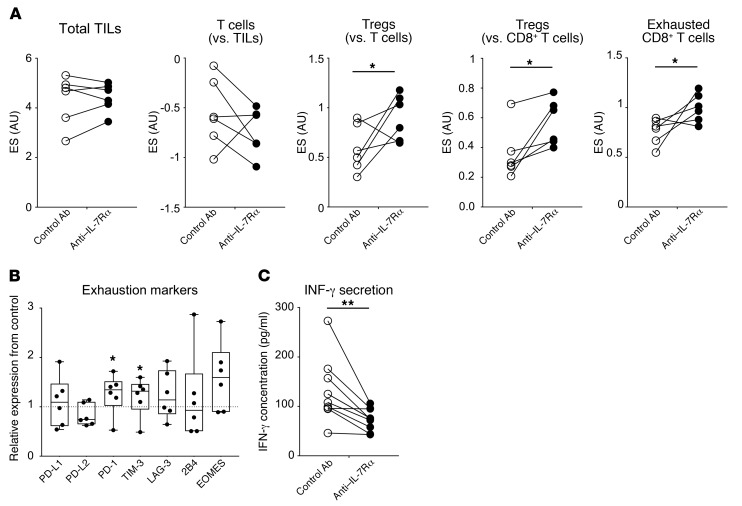
To evaluate the local protective effect of the anti–IL-7Rα antagonist, we used fresh mucosal biopsies from UC patients as an ex vivo inflammatory model of human IBD as previously described (68). Anti–human IL-7Rα mAb (10 μg/ml) or isotype control (10 μg/ml) was added to organ-cultured samples from 10 patients with UC (Supplemental Table 2). After 24 hours of culture, supernatants were analyzed by ELISA, and 6 pairs of biopsies were processed for NanoString transcriptional technology analysis using a panel of 255 human genes related to inflammation. Transcriptomic analysis first showed that samples clustered by donor origin and not by treatment conditions, confirming high interindividual variability in these biopsies (Supplemental Figure 24). Nevertheless, using the NanoString immune cell signature, the analysis revealed that the relative fraction of T cells tended to decrease in the anti–IL-7R condition (not significantly) without impacting Tregs (Figure 8A). Hence, the relative frequency of Tregs among total T cells or effector CD8+ T cells significantly increased in the anti–IL-7R condition. We also observed a significant increase of the exhaustion signature within CD8+ T cells, in particular due to the overexpression of PD-1 and TIM-3 as compared with isotype control (Figure 8B). Finally, in accordance with the result on effector T cell exhaustion, we found that anti–IL-7R significantly inhibited IFN-γ secretion in the supernatant (Figure 8C). Altogether, this analysis showed that anti–IL-7Rα mAb acts locally and rapidly in UC colon samples grown ex vivo and alters effector T cells while tipping the immune balance toward regulatory T cells.
Figure 8. IL-7Rα blockade efficiency ex vivo in human UC organ cultures.
(A) Immune cell signature enrichment scores and CD8+ T cell exhaustion signature using NanoString transcriptional analysis of inflamed colon fragments from UC patients (n = 6). Samples from each patient were cultured ex vivo at 37°C for 24 hours in medium with 10 μg/ml of control IgG mAb or blocking anti–human IL-7Rα mAb. (B) Individual expression of exhaustion molecules in anti–IL-7Rα–treated conditions normalized to isotype control. (C) IFN-γ cytokine concentration measured by ELISA in the supernatant of UC (n = 10) inflamed colon fragments in the same assay. Each pair of symbols represents samples from the same patient. *P < 0.05; **P < 0.01, Mann-Whitney U test. TILs, tumor-infiltrating lymphocytes.
Discussion
Since IBD is characterized by chronic inflammation, most targeted therapies are directed to dampen downstream effectors of dysregulated immune responses (i.e., inflammatory cytokines). However, these strategies are not particularly effective in a substantial proportion of patients, and significant rates of acquired resistance are observed. The identification of alternative therapeutic approaches targeting more upstream mechanisms is desired to prevent relapse and maintain patients under long-term remission. Our study elucidates a previously unknown dysregulated pathway in human IBD, whereby IL-7R signaling specifically regulates human effector but not regulatory T cell homing to the gut through a species-specific control of α4 and β7 integrin expression. Importantly, while an association of IL-7 and colitis has been consistently described in rodents, in humans this pathway has not been studied in IBD tissues, and its impact on anti-TNF responsiveness has not previously been evaluated.
Excepting a meta-analysis reporting 29 additional UC risk loci including the IL7R gene (39), only one study has reported an association between the IL-7 pathway and severe IBD, whereby the transcriptional profile of circulating T cells correlated with the clinical course in UC and CD (40).
Our analysis of 7 transcriptional data sets of mucosal biopsies from UC and CD patients, and validation with local cohorts by RT-qPCR quantification and protein detection by immunohistochemistry, revealed that the IL-7R signaling signature is reproducibly altered in colon mucosa of active diseases and clearly discriminates the disease status from inactive states or non-IBD controls. This is not the case for mucosal ileal biopsies, suggesting a specific association of the IL-7/IL-7R pathway with colonic IBD inflammation. Furthermore, several components of the entire pathway were found to be reproducibly associated and overexpressed, suggesting a potential biologically active pathway specifically located in diseased tissues. Our study revealed that key players of the IL-7R signaling signature and, notably, individual expression of IL7R at baseline are reproducibly and strongly associated with a considerable risk of resistance to anti-TNF in 3 different cohorts of UC and 1 cohort of CD. Association of the IL-7R pathway with resistance to anti-α4β7 therapy has also been observed, although it remains to be confirmed since only 1 cohort has been available so far. In an additional UC cohort (43), the IL-7R signature was elevated in nonresponder patients after anti-TNF therapy, and IL7R was significantly associated with nonresponse to anti-TNF treatment, but with substantially elevated IL7R seen only in primary nonresponders at baseline (Figure 1, G–I). It should be noted that for previous cohorts, an anti-TNF or anti-α4β7 response was defined by complete endoscopic and histological mucosal healing, while in this different cohort the definition of responsiveness was less robust since it was based only on reduction of the clinical Mayo score and rectal bleeding subscore (43). Our data also highlight potentially useful biomarkers of colon inflammation capable of discriminating patients who will achieve full remission from those who will be completely nonresponsive following therapy. Colon fibrosis has also been associated with stenosis and high need of surgery in CD patients treated with anti-TNF (69), and nonresponse to anti-TNF has recently been associated with higher fibronectin and collagen deposition in comparison with responder patients (70). Even if it is impossible to formally rule out the presence of significant fibrosis based on the available data concerning the patients included in the meta-analysis and the meta-cohort, the predominance of UC patients makes it unlikely since fibrosis in UC is much less common and benign stenosis is rarely detected. A recently published observation concerning the oncostatin M (OSM) pathway, where 64 candidate cytokines have been profiled, also includes IL7, which was not discriminant among the top cytokines differentially expressed in IBD biopsies (48). While we found significantly increased IL7 cytokine expression by RT-qPCR in our local cohort, IL7 cytokine expression assessed in previous studies was found to be significantly increased in only 2 of 7 data sets. In response to commensal microflora invasion, gut epithelial cells were shown to produce IL-7 (23, 71, 72) and to regulate the proliferation of intestinal mucosal lymphocytes (38) as well as production of inflammatory cytokines by human ILC3s also expressing IL-7R (73). Thus, it is conceivable that high IL7 expression in the colon is more related to bacterial tissue invasion, while our results reproducibly show an IL7R and IL-7R signaling signature accumulation that is associated with clinical IBD status and nonresponsiveness to anti-TNF therapy.
Systemic IL-7 is known for its deleterious role in chronic colitis in rodents (74, 75). In this study we have unraveled an important species difference, whereby IL-7 imprints gut-homing specificity on T cells in humans by upregulating α4β7 integrin expression, a phenomenon not observed in rodents. We show that this is due to the recruitment of CREB to the ITGA4 promoter in humans but not in mice. Other major integrin or chemokine receptors implicated in T cell homing to tissues were not modified, suggesting a specific control of T cell entry in the gut in humans in response to IL-7 consumption. Confirming this IL-7 mechanism of action, human CD127+ T lymphocytes analyzed by CyTOF in the inflamed lamina propria of colon UC patients predominantly expressed also the mucosal integrin β7. Besides IL-7, to our knowledge only retinoic acid has been previously described to control α4β7 integrin expression (76). Interestingly, similar to our results for IL-7, retinoic acids control both α4 and β7 protein expression by directly controlling α4, but not β7, gene transcription (77). Retinoic acids control both effector and Treg α4β7 integrin expression (78). We found that in spite of the low expression of IL-7Rα, IL-7 still induced signaling in Tregs, however without modulating α4 and β7 expression. This suggested that IL-7 selectively imprints effector, but not regulatory, T lymphocyte migration to the gut. These results confirm data by Fischer et al., who studied integrin and chemokine receptor expression on effector and regulatory T cells from UC patients (65). In particular, this study demonstrated that an anti-α4β7 mAb (vedolizumab) impairs gut homing of both human effector and regulatory T cells in humanized mice, and confirmed recent observations that colitis is exacerbated in β7-knockout mice as a result of deletion of the colonic Treg population (79).
Finally, we confirmed a deleterious role for IL-7 in gut inflammation mediated by human immune cells using different in vivo humanized mouse models and ex vivo punch colon biopsy cultures from UC patients. Human T lymphocytes overexpressing α4β7 after incubation with IL-7 induced colonic inflammation when transferred into immunodeficient mice. Blocking IL-7Rα with an antagonistic mAb delayed human T lymphocyte–mediated GVHD, with a prolonged action specifically in the colon. In contrast, blocking α4β7 using vedolizumab, which presumably acts locally but not systemically, did not protect from chronic xenogeneic GVHD. Otherwise, anti–IL-7Rα therapy demonstrated in vivo efficacy in restoring gut homeostasis in an acute and chronic colitis humanized mouse model reconstituted with hematopoietic stem cells or peripheral human leukocytes and challenged by intrarectal ethanol/TNBS to induce acute colonic inflammation or receiving a chronic cycle of DSS to induce chronic colon inflammation. Although IL-7R blockade shares some mechanism of action with anti-α4β7 on colon mucosa human T cell migration, these humanized inflammatory models also showed that anti–IL-7R has a broader impact, potentially through systemic mechanisms on chronic inflammation.
While targeting a cytokine pathway known for its activity on T cell homeostasis, we were surprised by the rapid action of IL-7Rα blockade in the acute colitis humanized model. We therefore applied anti–IL-7Rα mAb on surgical specimens of mucosal intestine from UC patients cultured ex vivo. We found that anti–IL-7Rα mAb significantly inhibited IFN-γ production in the culture supernatant within 24 hours, which definitively demonstrated that the anti–IL-7Rα therapy is efficient locally and rapidly on gut mucosal inflammation. Transcriptional analysis revealed that anti–IL-7Rα mAb alters mucosal effector T lymphocytes while respecting regulatory T cells and hence tips the balance toward immune regulation. It showed also that IL-7Rα blockade seems to induce a transcriptional program of T cell exhaustion (e.g., PD-1 and TIM-3 expression), which could also explain the inhibition of IFN-γ secretion. Close linkage between IL-7 and IFN-γ secretion has been previously proposed, since IL-7 production by human intestinal epithelial cells in response to Citrobacter rodentium was described to be dependent on IFN-γ (71) and others demonstrated that IFN-γ directly enhanced IL7 mRNA expression in intestinal epithelial cells (80). Finally, Shalapour et al. (23) suggested a feedback loop in which commensal bacteria drive IFN-γ production by lymphocytes, which in turn promotes epithelial cell IL-7 production and IL-7–dependent local survival of lymphocytes. IL7R expression correlated also with OSM/OSMR gene expression in colon biopsies of UC patients before anti-TNF therapy. It remains to be explored whether blocking IL-7R or OSM pathways would be active or induce similar clinical responses in the same subgroup of patients. It was previously described that OSM and other members of the IL-6 family could induce IL-7 secretion by fibroblasts (81) and hence could increase proliferation of T lymphocytes. Altogether, local production of IL-7 by stromal, myeloid, and/or epithelial cells in response to danger or stress signals might amplify the dysregulated Teff/Treg balance by increasing the expansion of effector T lymphocytes, which in turn secrete more IFN-γ, initiating an inflammatory feedback loop.
Human infants with IL-7Rα mutations have severe T cell lymphopenia necessitating bone marrow transplantation (82). In contrast to most studies performed in rodent models where IL-7R blockade also induces broad lymphodepletion (21), administration of high doses of anti–IL-7Rα antagonistic mAb in baboons (83), cynomolgus monkey (84), or marmosets (85) did not induce lymphopenia or a significant decrease in peripheral T lymphocyte numbers. Similarly, early clinical results from phase I trials with 2 different antagonist anti–IL-7Rα mAbs, in healthy volunteers (GSK2618960; NCT02293161, ClinicalTrials.gov) or in type 1 diabetic adult patients (PF-06342674; NCT02038764), did not demonstrate induction of lymphopenia (86). No information in the IBD population is available so far. Altogether these observations confirm that IL-7 biology in mice significantly differs from that in humans, but the long-term impact of IL-7R blockade on the human peripheral T cell pool will have to be closely examined.
Our results indicate that the IL-7R pathway is locally dysregulated in the colon of severe IBD patients and may contribute to the maintenance of chronic inflammation. More importantly, our study identified that a high colonic IL-7R signaling gene signature, particularly IL7R, is reproducibly and strongly associated with the absence of response to anti-TNF and presumably also to anti-α4β7 therapies both in UC and CD patients before initiation of the therapy, constituting potentially a new predictive biomarker to identify these refractory patients. Finally, we have unraveled human-specific IL-7 control of gut α4β7 integrin expression and confirmed in preclinical in vivo animal and ex vivo human models the therapeutic potential of IL-7Rα blockade. Altogether, our findings suggest that anti–IL-7Rα therapy may offer a novel therapeutic opportunity to prevent relapse and reinstate immune tolerance in IBD patients.
Methods
Data sets and anti-TNF or anti-α4β7 response criteria.
Gene expression data sets were obtained from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/): GSE38713 (41) and GSE59071 (42) for colon biopsies from UC patients after treatment with immunosuppressants and/or corticosteroids; GSE57945 (44) for ileal biopsies from pediatric healthy controls and patients with newly diagnosed and untreated ileal CD, colonic CD, or UC; GSE16879 (46) for colon biopsies from UC patients as well as colon and ileal biopsies from CD patients before and 4–6 weeks after treatment with anti-TNF (infliximab); GSE12251 (45) for colon biopsies of UC patients before anti-TNF (infliximab) therapy; GSE23597 (43) for colon biopsies from UC patients before and 8 weeks and 30 weeks after anti-TNF (infliximab) therapy; and GSE73661 (47) for colon biopsies from UC patients before and 4–6 weeks after treatment with anti-TNF (infliximab) as well as UC patients before and 6 weeks and 52 weeks after treatment with anti-α4β7 (vedolizumab). Patient characteristics as well as criteria and definition for determining primary responsiveness to anti-TNF or anti-α4β7 therapy are described in Supplemental Methods. Briefly, patients with active IBD refractory to corticosteroids and/or immunosuppression underwent biopsy collection within a week before anti-TNF or anti-α4β7 therapy and at early (week 4–8) or late (week 30 or 52) time points after the first infusion. In all cohorts except GSE23597 (43), anti-TNF or anti-α4β7 response was defined by complete histological and endoscopic healing. In GSE23597 (43), anti-TNF response was defined by a decrease from baseline of the total Mayo score of at least 3 points and 30% with an accompanying decrease of the bleeding subscore of at least 1 point and an absolute bleeding subscore of 0 or 1. This cohort of UC patients treated with anti-TNF was not included in our meta-analysis.
Ex vivo human colon culture model.
Human colon tissues were collected during endoscopy or during routine surgery. Biopsies or small mucosal fragments were cut (3-mm2 pieces) from resected intestinal tissue collected from macroscopically and microscopically inflamed areas of intestine from patients with either CD or UC. For UC patients, 6 patients gave biopsies and 4 gave resected bowel. For CD, 3 patients gave biopsies and 7 gave resected bowel. When mucosal resection tissue was used, small biopsy-size fragments were cut using scissors. Biopsies or biopsy-sized fragments were placed (1 biopsy per well) in 24-well plates in 300 μl serum-free HL-1 medium (Lonza Bioscience) supplemented with l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μg/ml gentamycin (Sigma-Aldrich). Mucosal explants were incubated for 24 hours at 37°C and 5% CO2. The anti–human CD127 mAb (clone N13B2, OSE Immunotherapeutics) or IgG control was added at 10 μg/ml into the medium at the beginning of incubation time. Supernatants were collected after 24 hours of culture and stored at –70°C. Cytokine production in biopsy supernatants was measured by ELISA. Human recombinant IL-8 (DY208) DuoSet ELISA kit (R&D Systems) and human IFN-γ (31673539, ImmunoTools) were used according to the manufacturer’s instructions.
Animals.
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NOD-scid IL2Rγ-null [NSG]) immunodeficient mice were initially obtained from The Jackson Laboratory and then bred in the Humanized Rodent Platform of LabEx IGO (University of Nantes). All animals were housed under specific pathogen–free conditions, according to institutional guidelines.
Humanized GVHD model.
Seven- to ten-week old NSG mice were sublethally irradiated (1.5 Gy) and infused i.p. with 50 × 106 freshly isolated human PBMCs from healthy donors (EFS [Etablissement Français du Sang]) as previously described (64). Mice were treated i.p. 3 times per week, from day 0 until day 28, with 5 mg/kg of anti–human IL-7Rα mAb (clone N13B2, OSE Immunotherapeutics), anti–human α4β7 mAb (vedolizumab, Entyvio), or equivalent volume of excipient (vehicle). In some experiments, NSG mice were infused i.p. with 10 × 106 human PBMCs, pretreated or not overnight in vitro with 5 ng/ml of recombinant human IL-7 (Bio-Rad). Animals were maintained in aseptic conditions and were monitored 3 times a week for weight and clinical evaluation. The endpoint for the survival study was set when mice lost more than 20% of body weight.
Humanized TNBS-induced colitis model.
Four-week-old NSG mice were sublethally irradiated (1.5 Gy) and infused i.v. with 50 × 103 human CD34+ mononuclear cells from umbilical cord blood (University Hospital, Nantes). Humanized mice were included in the colitis protocol after achieving sufficient immune blood reconstitution defined by a human T lymphocyte engraftment superior to 10%. Colitis was induced via intrarectal injection of 200 μl of a solution containing 5 mg of 2,4,6-trinitrobenzenesulfonic acid (TNBS) (Sigma-Aldrich, France) in 50% ethanol (VWR International, France). Mice were also treated i.p. from day –1 three times a week with 5 mg/kg of anti–human IL-7Rα mAb (clone N13B2, OSE Immunotherapeutics), anti–human α4β7 mAb (vedolizumab, Entyvio, Takeda, Osaka, Japan), or equivalent volume of excipient (vehicle). Intestinal permeability was determined after oral gavage at day 0 and day 7 with 150 μl of carboxymethyl cellulose solution at 0.5% with sulfonic acid coupled to FITC (SAF) at 10 mg/ml (478.32 D, Thermo Fisher Scientific, Saint Aubin, France) for paracellular permeability measurement and horseradish peroxidase (HRP) at 10 mg/ml for transcellular permeability evaluation. The concentration of SAF and HRP in plasma was evaluated 2 hours after gavage using a Varioskan microplate reader (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Animals were also monitored daily for body weight and clinical evaluation. Mice were scored using a colitis score that sums a clinical score and a stool score (clinical score: 0, normal behavior; 1, asthenia/low movement; 2, absence of movement; 3, tottering/lethargic; stool score: 0, normal consistency; 1, soft consistency but still formed; 2, diarrhea; 3, diarrhea with rectal bleeding). Mice were sacrificed if they achieved up to 25% of body weight loss.
Statistics.
Continuous variables are expressed as mean ± SEM, unless otherwise indicated, and compared with the nonparametric Mann-Whitney 2-sided U test or Kruskal-Wallis tests with Dunn’s ad hoc pairwise comparisons for more than 2 groups. Graft survival was calculated using the Kaplan-Meier method. The log-rank test was used to compare survival times between different groups. The Spearman test was used for correlation analyses. P values of less than 0.05 were considered statistically significant. All statistical analyses were performed on GraphPad Prism (GraphPad Software).
Study approval.
Animal studies were approved by the French National Ethics Committee (CEEA-2012-155, CEEA-2013-779, CEEA-2013-1466, and CEEA-2015-2926). Research ethics committee approval (10/H0704/73) for studies using human tissue was obtained from the NRES Committee London – City and East. The study was also approved by the Queen Mary University of London (QMUL) Joint Research and Development office of Barts and The London School of Medicine and Dentistry. All aspects of the work described were performed following Good Clinical Practice and Good Clinical Laboratory Practice guidelines. All patients provided informed written consent before participating in the study. Colonic biopsies were obtained and stored in the biocollection of the IMAD. All patients whose samples were included in the biocollection gave written and informed consent before the endoscopic work-up. The clinical and demographic data were collected at the time of colonoscopy and recorded in a computerized database securely coupled to the biological collection. This biocollection was started in 2008 and was approved and registered by the French Ministry of Science and Research (DC-2008-402).
Author contributions
NP, BV, PN, and JPS conceived the study. LB, NP, BV, and PN designed and supervised the experiments. LB, RD, LK, VND, SP, TD, CM, VG, AK, VT, GD, CT, BM, NG, and SLB performed the experiments. NP, LB, RD, EK, FM, JM, CC, AR, MM, JFM, IN, MN, EC, TTM, PD, HLM, SB, AB, GB, SB, NSLG, CB, AA, RJ, and JPS analyzed data and/or provided human samples. NP, LB, and BV wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the French Public Bank of Investment (Effimab grant I 1302011W), the Fondation pour la Recherche Médicale (grant LBS20130627235), and the Région of Pays de la Loire. We thank the humanized rodent platform and MicroPICell cellular and tissular imaging core facility of Nantes University, and the NIH AIDS Research & Reference Reagent program for its gift of anti–human ITGA4-ITGB7 (Act-1 clone) mAb. This work was realized in the context of the IHU-Cesti project, the DHU Oncogreffe, and LabEx IGO thanks to French government financial support managed by the National Research Agency via the Investment into the Future program (ANR-10-IBHU-005 and ANR-11- LABX-0016-01). Richard Danger is supported by a Marie Skłodowska-Curie fellowship (IF-EF) from the European Union’s Horizon 2020 Research and Innovation Programme (grant agreement 706296).
Version 1. 04/02/2019
Electronic publication
Version 2. 05/01/2019
Print issue publication
Footnotes
Conflict of interest: CM, JPS, SB, BV, and NP are shareholders of OSE Immunotherapeutics, a company owning IL-7 receptor antagonists.
Copyright: © 2019, The American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(5):1910-1925.https://doi.org/10.1172/JCI121668.
Contributor Information
Lyssia Belarif, Email: lyssia.belarif@ose-immuno.com.
Richard Danger, Email: richard.danger@univ-nantes.fr.
Laetitia Kermarrec, Email: Laetitia.Kermarrec@umanitoba.ca.
Sabrina Pengam, Email: sabrina.pengam@ose-immuno.com.
Tony Durand, Email: tony.durand@univ-nantes.fr.
Caroline Mary, Email: caroline.mary@ose-immuno.com.
Elise Kerdreux, Email: Elise.KERDREUX@chu-nantes.fr.
Vanessa Gauttier, Email: vanessa.gauttier@ose-immuno.com.
Aneta Kucik, Email: a.k.kucik@qmul.ac.uk.
Virginie Thepenier, Email: virginie.thepenier@ose-immuno.com.
Christie Chang, Email: Christie.Chang@mssm.edu.
Nina Salabert-Le Guen, Email: nina.salabert@chu-nantes.fr.
Ahmed Abidi, Email: Ahmed.abidi@univ-nantes.fr.
Grégoire David, Email: gregoire_david@hotmail.fr.
Florent Malard, Email: florent.malard@inserm.fr.
Celine Takoudju, Email: celinetakoudju@hotmail.fr.
Bernard Martinet, Email: bernard.martinet@univ-nantes.fr.
Isabelle Neveu, Email: Isabelle.Neveu@univ-nantes.fr.
Michel Neunlist, Email: michel.neunlist@univ-nantes.fr.
Emmanuel Coron, Email: emmanuel.coron@chu-nantes.fr.
Pierre Desreumaux, Email: pdesreumaux@hotmail.com.
Hoa-Le Mai, Email: le.hoa-mai@univ-nantes.fr.
Jean-François Mosnier, Email: jeanfrancois.mosnier@chu-nantes.fr.
Miriam Merad, Email: miriam.merad@mssm.edu.
Sophie Brouard, Email: Sophie.Brouard@univ-nantes.fr.
Jean-Paul Soulillou, Email: soulillou@yahoo.fr.
Gilles Blancho, Email: gblancho@chu-nantes.fr.
Arnaud Bourreille, Email: arnaud.bourreille@chu-nantes.fr.
Philippe Naveilhan, Email: philippe.naveilhan@univ-nantes.fr.
Bernard Vanhove, Email: bernard.vanhove@ose-immuno.com.
Nicolas Poirier, Email: nicolas.poirier@univ-nantes.fr.
References
- 1.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474(7351):307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham C, Cho JH. Inflammatory bowel disease. N Engl J Med. 2009;361(21):2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Danese S, Fiocchi C. Ulcerative colitis. N Engl J Med. 2011;365(18):1713–1725. doi: 10.1056/NEJMra1102942. [DOI] [PubMed] [Google Scholar]
- 4.Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet. 2012;380(9853):1590–1605. doi: 10.1016/S0140-6736(12)60026-9. [DOI] [PubMed] [Google Scholar]
- 5.Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet. 2017;389(10080):1756–1770. doi: 10.1016/S0140-6736(16)32126-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torres J, Mehandru S, Colombel JF, Peyrin-Biroulet L. Crohn’s disease. Lancet. 2017;389(10080):1741–1755. doi: 10.1016/S0140-6736(16)31711-1. [DOI] [PubMed] [Google Scholar]
- 7.Allez M, et al. Report of the ECCO pathogenesis workshop on anti-TNF therapy failures in inflammatory bowel diseases: definitions, frequency and pharmacological aspects. J Crohns Colitis. 2010;4(4):355–366. doi: 10.1016/j.crohns.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Adams DH, Eksteen B. Aberrant homing of mucosal T cells and extra-intestinal manifestations of inflammatory bowel disease. Nat Rev Immunol. 2006;6(3):244–251. doi: 10.1038/nri1784. [DOI] [PubMed] [Google Scholar]
- 9.Agace WW. Tissue-tropic effector T cells: generation and targeting opportunities. Nat Rev Immunol. 2006;6(9):682–692. doi: 10.1038/nri1869. [DOI] [PubMed] [Google Scholar]
- 10.Feagan BG, et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med. 2013;369(8):699–710. doi: 10.1056/NEJMoa1215734. [DOI] [PubMed] [Google Scholar]
- 11.Sandborn WJ, et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med. 2013;369(8):711–721. doi: 10.1056/NEJMoa1215739. [DOI] [PubMed] [Google Scholar]
- 12.Mackall CL, Fry TJ, Gress RE. Harnessing the biology of IL-7 for therapeutic application. Nat Rev Immunol. 2011;11(5):330–342. doi: 10.1038/nri2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7(2):144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med. 2006;203(7):1701–1711. doi: 10.1084/jem.20060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seddiki N, et al. Expression of interleukin (IL)-2 and IL-7 receptors discriminates between human regulatory and activated T cells. J Exp Med. 2006;203(7):1693–1700. doi: 10.1084/jem.20060468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel L, et al. Patients with relapsing-remitting multiple sclerosis have normal Treg function when cells expressing IL-7 receptor alpha-chain are excluded from the analysis. J Clin Invest. 2008;118(10):3411–3419. doi: 10.1172/JCI35365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9(7):480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandey A, et al. Cloning of a receptor subunit required for signaling by thymic stromal lymphopoietin. Nat Immunol. 2000;1(1):59–64. doi: 10.1038/76923. [DOI] [PubMed] [Google Scholar]
- 19.Carrette F, Surh CD. IL-7 signaling and CD127 receptor regulation in the control of T cell homeostasis. Semin Immunol. 2012;24(3):209–217. doi: 10.1016/j.smim.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dooms H. Interleukin-7: fuel for the autoimmune attack. J Autoimmun. 2013;45:40–48. doi: 10.1016/j.jaut.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 21.Mai HL, et al. IL-7 receptor blockade following T cell depletion promotes long-term allograft survival. J Clin Invest. 2014;124(4):1723–1733. doi: 10.1172/JCI66287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piotti G, Ma J, Adams E, Cobbold S, Waldmann H. Guiding postablative lymphocyte reconstitution as a route toward transplantation tolerance. Am J Transplant. 2014;14(7):1678–1689. doi: 10.1111/ajt.12756. [DOI] [PubMed] [Google Scholar]
- 23.Shalapour S, et al. Commensal microflora and interferon-gamma promote steady-state interleukin-7 production in vivo. Eur J Immunol. 2010;40(9):2391–2400. doi: 10.1002/eji.201040441. [DOI] [PubMed] [Google Scholar]
- 24.Shalapour S, et al. Interleukin-7 links T lymphocyte and intestinal epithelial cell homeostasis. PLoS One. 2012;7(2):e31939. doi: 10.1371/journal.pone.0031939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe M, et al. Interleukin 7 transgenic mice develop chronic colitis with decreased interleukin 7 protein accumulation in the colonic mucosa. J Exp Med. 1998;187(3):389–402. doi: 10.1084/jem.187.3.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamazaki M, et al. Mucosal T cells expressing high levels of IL-7 receptor are potential targets for treatment of chronic colitis. J Immunol. 2003;171(3):1556–1563. doi: 10.4049/jimmunol.171.3.1556. [DOI] [PubMed] [Google Scholar]
- 27.Okada E, et al. IL-7 exacerbates chronic colitis with expansion of memory IL-7Rhigh CD4+ mucosal T cells in mice. Am J Physiol Gastrointest Liver Physiol. 2005;288(4):G745–G754. doi: 10.1152/ajpgi.00276.2004. [DOI] [PubMed] [Google Scholar]
- 28.Totsuka T, et al. IL-7 is essential for the development and the persistence of chronic colitis. J Immunol. 2007;178(8):4737–4748. doi: 10.4049/jimmunol.178.8.4737. [DOI] [PubMed] [Google Scholar]
- 29.Shinohara T, et al. Upregulated IL-7 receptor α expression on colitogenic memory CD4+ T cells may participate in the development and persistence of chronic colitis. J Immunol. 2011;186(4):2623–2632. doi: 10.4049/jimmunol.1000057. [DOI] [PubMed] [Google Scholar]
- 30.Buonocore S, et al. Innate lymphoid cells drive interleukin-23-dependent innate intestinal pathology. Nature. 2010;464(7293):1371–1375. doi: 10.1038/nature08949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernink JH, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14(3):221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- 32.Goldberg R, Prescott N, Lord GM, MacDonald TT, Powell N. The unusual suspects — innate lymphoid cells as novel therapeutic targets in IBD. Nat Rev Gastroenterol Hepatol. 2015;12(5):271–283. doi: 10.1038/nrgastro.2015.52. [DOI] [PubMed] [Google Scholar]
- 33.Powell N, et al. The transcription factor T-bet regulates intestinal inflammation mediated by interleukin-7 receptor+ innate lymphoid cells. Immunity. 2012;37(4):674–684. doi: 10.1016/j.immuni.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willis CR, et al. Interleukin-7 receptor blockade suppresses adaptive and innate inflammatory responses in experimental colitis. J Inflamm (Lond) 2012;9(1):39. doi: 10.1186/1476-9255-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neurath MF. Cytokines in inflammatory bowel disease. Nat Rev Immunol. 2014;14(5):329–342. doi: 10.1038/nri3661. [DOI] [PubMed] [Google Scholar]
- 36.MacDonald TT, Biancheri P, Sarra M, Monteleone G. What’s the next best cytokine target in IBD? Inflamm Bowel Dis. 2012;18(11):2180–2189. doi: 10.1002/ibd.22967. [DOI] [PubMed] [Google Scholar]
- 37.de Souza HS, Fiocchi C. Immunopathogenesis of IBD: current state of the art. Nat Rev Gastroenterol Hepatol. 2016;13(1):13–27. doi: 10.1038/nrgastro.2015.186. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe M, et al. Interleukin 7 is produced by human intestinal epithelial cells and regulates the proliferation of intestinal mucosal lymphocytes. J Clin Invest. 1995;95(6):2945–2953. doi: 10.1172/JCI118002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43(3):246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JC, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest. 2011;121(10):4170–4179. doi: 10.1172/JCI59255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Planell N, et al. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62(7):967–976. doi: 10.1136/gutjnl-2012-303333. [DOI] [PubMed] [Google Scholar]
- 42.Vanhove W, et al. Strong upregulation of AIM2 and IFI16 inflammasomes in the mucosa of patients with active inflammatory bowel disease. Inflamm Bowel Dis. 2015;21(11):2673–2682. doi: 10.1097/MIB.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 43.Toedter G, et al. Gene expression profiling and response signatures associated with differential responses to infliximab treatment in ulcerative colitis. Am J Gastroenterol. 2011;106(7):1272–1280. doi: 10.1038/ajg.2011.83. [DOI] [PubMed] [Google Scholar]
- 44.Haberman Y, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124(8):3617–3633. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arijs I, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut. 2009;58(12):1612–1619. doi: 10.1136/gut.2009.178665. [DOI] [PubMed] [Google Scholar]
- 46.Arijs I, et al. Mucosal gene expression of antimicrobial peptides in inflammatory bowel disease before and after first infliximab treatment. PLoS One. 2009;4(11):e7984. doi: 10.1371/journal.pone.0007984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arijs I, et al. Effect of vedolizumab (anti-α4β7-integrin) therapy on histological healing and mucosal gene expression in patients with UC. Gut. 2018;67(1):43–52. doi: 10.1136/gutjnl-2016-312293. [DOI] [PubMed] [Google Scholar]
- 48.West NR, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med. 2017;23(5):579–589. doi: 10.1038/nm.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Björklund ÅK, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol. 2016;17(4):451–460. doi: 10.1038/ni.3368. [DOI] [PubMed] [Google Scholar]
- 51.Koues OI, et al. Distinct gene regulatory pathways for human innate versus adaptive lymphoid cells. Cell. 2016;165(5):1134–1146. doi: 10.1016/j.cell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simoni Y, et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity. 2017;46(1):148–161. doi: 10.1016/j.immuni.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krämer B, et al. Compartment-specific distribution of human intestinal innate lymphoid cells is altered in HIV patients under effective therapy. PLoS Pathog. 2017;13(5):e1006373. doi: 10.1371/journal.ppat.1006373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fornasa G, et al. Dichotomy of short and long thymic stromal lymphopoietin isoforms in inflammatory disorders of the bowel and skin. J Allergy Clin Immunol. 2015;136(2):413–422. doi: 10.1016/j.jaci.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bjerkan L, et al. The short form of TSLP is constitutively translated in human keratinocytes and has characteristics of an antimicrobial peptide. Mucosal Immunol. 2015;8(1):49–56. doi: 10.1038/mi.2014.41. [DOI] [PubMed] [Google Scholar]
- 56.Vudattu NK, Magalhaes I, Hoehn H, Pan D, Maeurer MJ. Expression analysis and functional activity of interleukin-7 splice variants. Genes Immun. 2009;10(2):132–140. doi: 10.1038/gene.2008.90. [DOI] [PubMed] [Google Scholar]
- 57.Gregory SG, et al. Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet. 2007;39(9):1083–1091. doi: 10.1038/ng2103. [DOI] [PubMed] [Google Scholar]
- 58.Lundström W, et al. Soluble IL7Rα potentiates IL-7 bioactivity and promotes autoimmunity. Proc Natl Acad Sci U S A. 2013;110(19):E1761–E1770. doi: 10.1073/pnas.1222303110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beq S, et al. Injection of glycosylated recombinant simian IL-7 provokes rapid and massive T-cell homing in rhesus macaques. Blood. 2009;114(4):816–825. doi: 10.1182/blood-2008-11-191288. [DOI] [PubMed] [Google Scholar]
- 60.Sereti I, et al. Decreases in colonic and systemic inflammation in chronic HIV infection after IL-7 administration. PLoS Pathog. 2014;10(1):e1003890. doi: 10.1371/journal.ppat.1003890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cimbro R, et al. IL-7 induces expression and activation of integrin α4β7 promoting naive T-cell homing to the intestinal mucosa. Blood. 2012;120(13):2610–2619. doi: 10.1182/blood-2012-06-434779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang X, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102(12):4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lesiak A, et al. A genome-wide screen of CREB occupancy identifies the RhoA inhibitors Par6C and Rnd3 as regulators of BDNF-induced synaptogenesis. PLoS One. 2013;8(6):e64658. doi: 10.1371/journal.pone.0064658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poirier N, et al. Preclinical efficacy and immunological safety of FR104, an antagonist anti-CD28 monovalent Fab′ antibody. Am J Transplant. 2012;12(10):2630–2640. doi: 10.1111/j.1600-6143.2012.04164.x. [DOI] [PubMed] [Google Scholar]
- 65.Fischer A, et al. Differential effects of α4β7 and GPR15 on homing of effector and regulatory T cells from patients with UC to the inflamed gut in vivo. Gut. 2016;65(10):1642–1664. doi: 10.1136/gutjnl-2015-310022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wirtz S, Neufert C, Weigmann B, Neurath MF. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2(3):541–546. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 67.Pelczar P, et al. A pathogenic role for T cell-derived IL-22BP in inflammatory bowel disease. Science. 2016;354(6310):358–362. doi: 10.1126/science.aah5903. [DOI] [PubMed] [Google Scholar]
- 68.Rovedatti L, et al. Differential regulation of interleukin 17 and interferon gamma production in inflammatory bowel disease. Gut. 2009;58(12):1629–1636. doi: 10.1136/gut.2009.182170. [DOI] [PubMed] [Google Scholar]
- 69.Louis E, et al. Treatment of small bowel subocclusive Crohn’s disease with infliximab: an open pilot study. Acta Gastroenterol Belg. 2007;70(1):15–19. [PubMed] [Google Scholar]
- 70.de Bruyn JR, et al. Intestinal fibrosis is associated with lack of response to Infliximab therapy in Crohn’s disease. PLoS One. 2018;13(1):e0190999. doi: 10.1371/journal.pone.0190999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang W, Du JY, Yu Q, Jin JO. Interleukin-7 produced by intestinal epithelial cells in response to Citrobacter rodentium infection plays a major role in innate immunity against this pathogen. Infect Immun. 2015;83(8):3213–3223. doi: 10.1128/IAI.00320-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imanishi J. Expression of cytokines in bacterial and viral infections and their biochemical aspects. J Biochem. 2000;127(4):525–530. doi: 10.1093/oxfordjournals.jbchem.a022636. [DOI] [PubMed] [Google Scholar]
- 73.Glatzer T, et al. RORγt+ innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity. 2013;38(6):1223–1235. doi: 10.1016/j.immuni.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 74.Tomita T, et al. Systemic, but not intestinal, IL-7 is essential for the persistence of chronic colitis. J Immunol. 2008;180(1):383–390. doi: 10.4049/jimmunol.180.1.383. [DOI] [PubMed] [Google Scholar]
- 75.Nemoto Y, et al. Bone marrow-mesenchymal stem cells are a major source of interleukin-7 and sustain colitis by forming the niche for colitogenic CD4 memory T cells. Gut. 2013;62(8):1142–1152. doi: 10.1136/gutjnl-2012-302029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21(4):527–538. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 77.Kang SG, Park J, Cho JY, Ulrich B, Kim CH. Complementary roles of retinoic acid and TGF-β1 in coordinated expression of mucosal integrins by T cells. Mucosal Immunol. 2011;4(1):66–82. doi: 10.1038/mi.2010.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bakdash G, Vogelpoel LT, van Capel TM, Kapsenberg ML, de Jong EC. Retinoic acid primes human dendritic cells to induce gut-homing, IL-10-producing regulatory T cells. Mucosal Immunol. 2015;8(2):265–278. doi: 10.1038/mi.2014.64. [DOI] [PubMed] [Google Scholar]
- 79.Zhang HL, et al. Regulatory T-cell depletion in the gut caused by integrin β7 deficiency exacerbates DSS colitis by evoking aberrant innate immunity. Mucosal Immunol. 2016;9(2):391–400. doi: 10.1038/mi.2015.68. [DOI] [PubMed] [Google Scholar]
- 80.Thang PH, et al. The role of IL-1β in reduced IL-7 production by stromal and epithelial cells: a model for impaired T-cell numbers in the gut during HIV-1 infection. J Intern Med. 2010;268(2):181–193. doi: 10.1111/j.1365-2796.2010.02241.x. [DOI] [PubMed] [Google Scholar]
- 81.Sawa S, et al. Autoimmune arthritis associated with mutated interleukin (IL)-6 receptor gp130 is driven by STAT3/IL-7-dependent homeostatic proliferation of CD4+ T cells. J Exp Med. 2006;203(6):1459–1470. doi: 10.1084/jem.20052187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Giliani S, et al. Interleukin-7 receptor alpha (IL-7Ralpha) deficiency: cellular and molecular bases. Analysis of clinical, immunological, and molecular features in 16 novel patients. Immunol Rev. 2005;203:110–126. doi: 10.1111/j.0105-2896.2005.00234.x. [DOI] [PubMed] [Google Scholar]
- 83.Belarif L, et al. IL-7 receptor blockade blunts antigen-specific memory T cell responses and chronic inflammation in primates. Nat Commun. 2018;9(1):4483. doi: 10.1038/s41467-018-06804-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kern B, Li W, Bono C, Lee LF, Kraynov E. Receptor occupancy and blocking of STAT5 signaling by an anti-IL-7 receptor α antibody in cynomolgus monkeys. Cytometry B Clin Cytom. 2016;90(2):191–198. doi: 10.1002/cyto.b.21247. [DOI] [PubMed] [Google Scholar]
- 85.Dunham J, et al. Blockade of CD127 exerts a dichotomous clinical effect in marmoset experimental autoimmune encephalomyelitis. J Neuroimmune Pharmacol. 2016;11(1):73–83. doi: 10.1007/s11481-015-9629-6. [DOI] [PubMed] [Google Scholar]
- 86. Ganguly BJ. An IL-7 receptor blocking antibody increases the ratio of Tregs to effector memory T cells (TEM) and increases PD-1 expression on T cell subsets in the peripheral blood of patients with type 1 diabetes. American Diabetes Association 76th Scientific Sessions; June 10–14, 2016; New Orleans, Louisiana, USA. https://professional.diabetes.org/content/previous-scientific-sessions-abstracts-posters-and-webcasts Accessed March 1, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.