Abstract
The mesenchymal (MES) subtype of glioblastoma (GBM) stem cells (GSCs) represents a subpopulation of cancer cells that are notorious for their highly aggressive nature and resistance to conventional therapy. Aldehyde dehydrogenase 1A3 (ALDH1A3) has been recently suggested as a key determinant for the maintenance of MES features of GSCs. However, the mechanisms underpinning aberrant ALDH1A3 expression remain elusive. Here, we identified ubiquitin-specific protease 9X (USP9X) as a bona fide deubiquitinase of ALDH1A3 in MES GSCs. USP9X interacted with, depolyubiquitylated, and stabilized ALDH1A3. Moreover, we showed that FACS-sorted USP9Xhi cells were enriched for MES GSCs with high ALDH1A3 activity and potent tumorigenic capacity. Depletion of USP9X markedly downregulated ALDH1A3, resulting in a loss of self-renewal and tumorigenic capacity of MES GSCs, which could be largely rescued by ectopic expression of ALDH1A3. Furthermore, we demonstrated that the USP9X inhibitor WP1130 induced ALDH1A3 degradation and showed marked therapeutic efficacy in MES GSC–derived orthotopic xenograft models. Additionally, USP9X strongly correlated with ALDH1A3 expression in primary human GBM samples and had a prognostic value for patients with the MES subgroup. Collectively, our findings unveil USP9X as a key deubiquitinase for ALDH1A3 protein stabilization and a potential target for GSC-directed therapy.
Keywords: Cell Biology, Oncology
Keywords: Brain cancer
Introduction
Glioblastoma (GBM) is the most common and devastating human brain malignancy characterized by extensive intratumoral heterogeneity (1, 2). Despite recent advances in multimodal treatments, the outcomes for GBM patients remain dismal, with a median survival of 12 to 15 months (3). GBM possesses a subpopulation of cells, termed GBM stem cells (GSCs) (4–6), closely resembling neural precursor cells and having self-renewal activity and multilineage differentiation potential as well as tumor-initiating capacity, which are thought to be responsible for disease progression, recurrence, and therapeutic resistance (7–10). Thus, there is an urgent need for the exploration of new treatment strategies against GSCs.
The aldehyde dehydrogenase (ALDH) superfamily comprises 19 enzymes that metabolize endogenous and exogenous aldehydes to their corresponding carboxylic acids (11). ALDH1A3 is the predominant ALDH isozyme that participates in various physiological processes in human cells. Growing evidence has shown that ALDH1A3 exhibits high activity in cancer stem cells (CSCs) and influences a diverse range of biological characteristics within these cells (12–15). In GBMs, ALDH1A3 has been found to function as a key molecule for the maintenance of self-renewal and tumorigenicity of mesenchymal (MES) GSCs (16–18). Nevertheless, the mechanisms underlying ALDH1A3 dysregulation in MES GSCs remain enigmatic.
Deubiquitinating enzymes (DUBs) are a large group of proteases that counteract the action of protein ubiquitination through removing ubiquitin moieties from target proteins. Thus far, there are approximately 115 recognized human DUBs consisting of 6 different families: ubiquitin-specific proteases (USPs), ubiquitin carboxy-terminal hydrolases (UCHs), ovarian tumor proteases (OTUs), Machado-Joseph disease proteases (MJDs), JAB1/MPN/Mov34 metalloenzymes (JAMMs), and motif interacting with Ub-containing novel DUB family (MINDY) (14, 19, 20). Accumulating evidence has suggested that DUBs are often deregulated in a variety of cancers and play a pivotal role in the homoeostasis of cellular proteins and the regulation of cellular processes that are involved in cancer initiation and progression (21, 22). USP9X is an important member of the USP subfamily that regulates a myriad of cellular processes, including cell growth and migration (23, 24), apoptosis and autophagy (25–27), immune response (28, 29), and neural development (30). USP9X has been shown to be deregulated in various types of malignancies and to display a unique ability to either promote or suppress tumorigenesis in a context-dependent manner (31–36). Notably, it has been recently demonstrated that USP9X is highly expressed in GBMs and that inhibition of USP9X could sensitize GBM cells to intrinsic and extrinsic apoptotic stimuli (37, 38). However, whether USP9X plays a role in GSC biology remains unclear.
In this study, we identify USP9X as a bona fide DUB that regulates ALDH1A3 polyubiquitylation and stabilization. We find that USP9X serves as a key molecule for predicting the enrichment of ALDH1A3hi MES GSCs and is required for the self-renewal and tumorigenicity of MES GSCs. We also assess the potential therapeutic effects of USP9X inhibition on patient-derived MES GSC xenograft models.
Results
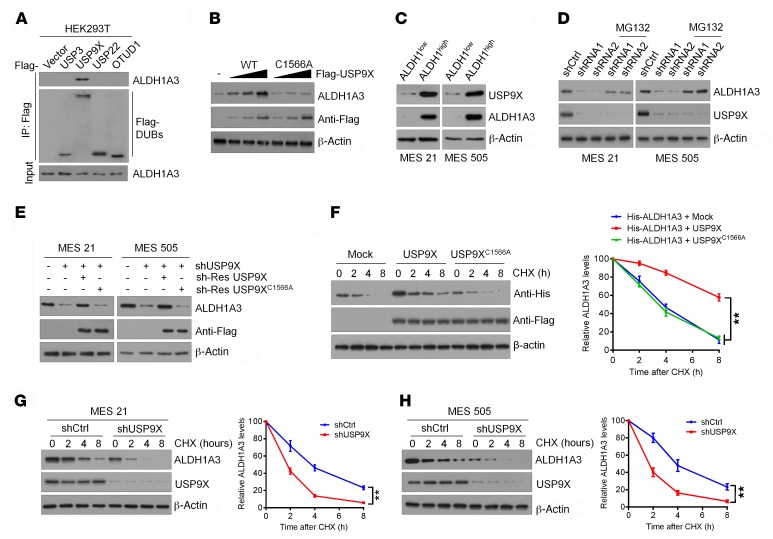
USP9X maintains ALDH1A3 stability.
To determine whether the ALDH1A3 protein is posttranslationally controlled by the ubiquitin-proteasome system (UPS), we first blocked de novo protein synthesis using cycloheximide (CHX) and chased the ALDH1A3 protein levels in HEK293T cells and normal human astrocytes (NHAs). Indeed, ALDH1A3 was gradually degraded and became almost undetectable within 8 hours of CHX treatment (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI126414DS1). Moreover, treatment of cells with the proteosomal inhibitor MG-132 resulted in a significant increase in ALDH1A3 protein levels (Supplemental Figure 1A). These data suggest that ALDH1A3 is degraded through the UPS. Next, we sought to identify the potential DUBs responsible for ALDH1A3 stabilization and performed an RNAi screening using the siGENOME RTF Library targeting 98 DUBs in HEK293T cells. This initial screen identified 4 DUBs (USP3, USP9X, USP22, and OTUD1) that are associated with ALDH1A3 expression (Supplemental Figure 1B). When these DUBs were coexpressed with ALDH1A3 in HEK293 cells, we found that only USP9X was able to interact directly with the endogenous ALDH1A3 (Figure 1A), thus implicating USP9X as a prominent candidate that controls ALDH1A3 stability. To test this, we transfected Flag-tagged WT USP9X or a catalytically inactive mutant C1566A USP9X (carrying a point mutation in one of the key cysteines of the catalytic domain) into HEK293T cells. Expression of WT USP9X, but not the C1566A mutant, increased the ALDH1A3 protein levels in a dose-dependent manner (Figure 1B), suggesting that USP9X modulates ALDH1A3 in a manner that depends on its DUB activity. In contrast, depletion of endogenous USP9X led to a drastic decrease in ALDH1A3 protein levels in 2 GBM cell lines (Supplemental Figure 1C). Neither overexpression nor knockdown of USP9X altered ALDH1A3 mRNA levels (Supplemental Figure 1, D and E).
Figure 1. USP9X maintains ALDH1A3 stability.
(A) Four Flag-tagged DUBs (USP3, USP9X, USP22, and OTUD1) were expressed in HEK293T cells, and cell lysates were analyzed by IP with Flag beads followed by IB with antibodies against ALDH1A3 and Flag. (B) Increasing amounts of Flag-tagged USP9X (WT or C1566A mutant) were transfected into HEK293T cells, and cell lysates were analyzed by IB with antibody against ALDH1A3. (C) IB analysis of USP9X protein expression in ALDH1hi and ALDH1lo subpopulations isolated from MES 21 and 505 GSCs. (D) MES 21 and 505 GSCs transfected with 2 independent USP9X shRNA were treated with or without the proteasome inhibitor MG132 (20 μM, 8 hours), and then USP9X and ALDH1A3 were analyzed. (E) IB analysis of ALDH1A3 levels in MES 21 and 505 GSCs transduced with USP9X shRNA, together with either shRNA-resistant (sh-res) Flag-tagged USP9X WT or USP9X C1566A. (F) HEK293T cells were cotransfected with His-tagged ALDH1A3 and Flag-tagged USP9X WT or USP9X C1566A, treated with 100 μg/ml CHX, collected at the indicated times, and then subjected to IB with antibodies against His and Flag. Quantification of ALDH1A3 levels relative to β-actin is shown. (G and H) MES 21 (G) and 505 (H) GSCs stably expressing control shRNA or USP9X shRNA were treated with 100 μg/ml CHX, harvested at the indicated times, and then subjected to IB with antibodies against ALDH1A3 and USP9X. Quantification of ALDH1A3 levels relative to β-actin is shown. Data are represented as mean ± SD of 3 independent experiments. **P < 0.01, 1-way ANOVA with Dunnett’s post test (F); 2-tailed Student’s t test (G and H).
Given the importance of ALDH1A3 in the maintenance of GSCs (16–18), we investigated whether USP9X could affect ALDH1A3 expression levels in different subtypes of GSCs. To that end, we used cell-surface markers CD44 or CD133 to isolate 2 MES GSCs (MES 21 and 505) and 2 proneural (PN) GSCs (PN 35 and 182) from patient-derived GBM cells maintained through serial passages in immunocompromised mice as subcutaneous xenografts (Supplemental Figure 1, F–N). We observed that MES GSCs exhibited high levels of expression of USP9X and CD44, but minimal levels of the PN marker OLIG2. In contrast, PN GSCs expressed OLIG2 at high levels, but had minimal levels of USP9X and CD44 (Supplemental Figure 1O). It has been shown before that CD44+ MES GSCs include both ALDH1A3hi and ALDH1A3lo cells (16). Accordingly, we separated MES 21 and 505 GSCs into ALDH1hi and ALDH1lo subpopulations using FACS analysis after staining with ALDEFLUOR. Interestingly, we found that USP9X is highly expressed in the ALDH1hi subpopulation, but not the ALDH1lo subpopulation (Figure 1C). Next, we knocked down USP9X in MES 21 and 505 GSCs using 2 nonoverlapping lentiviral shRNAs. We noted that USP9X depletion dramatically reduced the expression of ALDH1A3 protein, which could be almost completely reversed by addition of the proteasomal inhibitor MG132 or overexpression of an shRNA-resistant WT, but not C1566A, mutant, USP9X (Figure 1, D and E). Conversely, expression of ALDH1A3 was dramatically increased when USP9X was ectopically expressed in PN 35 and 182 GSCs (Supplemental Figure 1P). However, USP9X depletion or overexpression had no significant effect on ALDH1A3 mRNA levels (Supplemental Figure 1, Q and R). To prove that USP9X could affect the stability of ALDH1A3 per se, we used CHX to cease protein synthesis and detected the ALDH1A3 protein levels after manipulation of USP9X. Enforced expression of WT USP9X, but not the C1566A mutant USP9X, resulted in a prominent increase in the stability of ectopically expressed ALDH1A3 protein in HEK293T cells (Figure 1F), whereas knocking down USP9X expression in MES 21 and 505 GSCs led to destabilization of the ALDH1A3 protein (Figure 1, G and H). Collectively, these results indicate that USP9X specifically regulates ALDH1A3 stability.
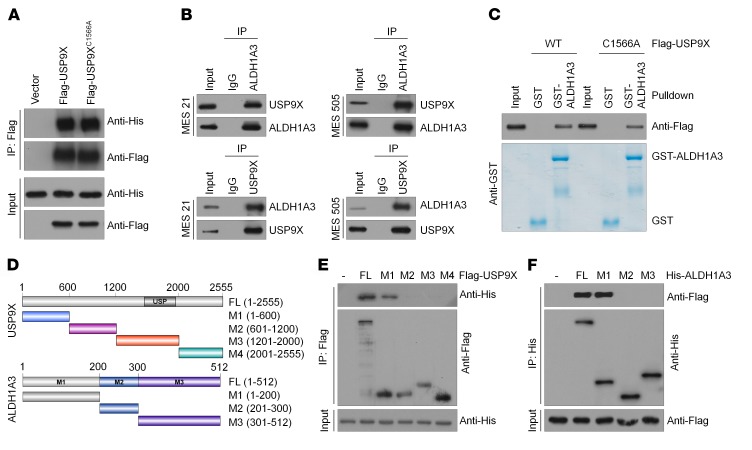
USP9X interacts with ALDH1A3.
We next sought to determine whether USP9X directly interacts with ALDH1A3. Co-IP assays revealed that His-tagged ALDH1A3 could be readily detected in either Flag-USP9X WT or Flag-USP9X C1566A immunoprecipitates in HEK293T cells and NHAs (Figure 2A and Supplemental Figure 2A), indicating that the DUB activity of USP9X is not required for such interaction. Similarly, a physical association between endogenous USP9X and ALDH1A3 proteins was validated in MES 21 and 505 GSCs as well as 2 established GBM cell lines (U87MG and T98G) (Figure 2B and Supplemental Figure 2B). Furthermore, we performed an in vitro GST pull-down assay by mixing purified GST-ALDH1A3 with purified recombinant protein Flag-USP9X WT or Flag-USP9X C1566A. As shown in Figure 2C, either USP9X WT or its C1566A mutant was able to bind to immobilized GST-ALDH1A3, but not to GST alone, thus confirming that the interaction between USP9X and ALDH1A3 is direct. To map the minimal essential regions required for their interaction, we generated various truncated mutants of Flag-USP9X and His-ALDH1A3 to narrow down the binding site (Figure 2D). Truncated mutation analysis showed that the N-terminal sequences (amino acids 1–600) of USP9X and the N-terminal sequences (amino acids 1–200) of ALDH1A3 are both required and sufficient for direct interaction with each other (Figure 2, E and F).
Figure 2. USP9X interacts with ALDH1A3.
(A) HEK293T cells were transfected with His-ALDH1A3 alone or in combination with Flag-tagged USP9X WT or USP9X C1566A, and cell lysates were analyzed by IP with Flag beads followed by IB with antibodies against His and Flag. (B) Cell lysates from MES 21 and 505 GSCs were analyzed by IP using antibodies against USP9X and ALDH1A3, then subjected to IB analysis. IgG was used as the isotype control. (C) Purified Flag-tagged USP9X WT or USP9X C1566A was incubated with GST or GST-ALDH1A3 coupled to glutathione-Sepharose beads. Proteins retained on Sepharose were then subjected to IB with indicated antibodies. Recombinant GST-ALDH1A3 was purified from bacteria and analyzed by SDS-PAGE and Coomassie blue staining. (D) Schematic representation of Flag-tagged full-length (FL) ZEB1, His-tagged FL ALDH1A3, and their various deletion mutants. (E) HEK293T cells were cotransfected with His-ALDH1A3 and Flag-tagged FL USP9X or its deletion mutants, and cell lysates were analyzed by IP with Flag beads followed by IB with antibodies against His and Flag. (F) HEK293T cells were cotransfected with Flag-USP9X and His-tagged FL ALDH1A3 or its deletion mutants, and cell lysates were analyzed by IP with His beads followed by IB with antibodies against Flag and His.
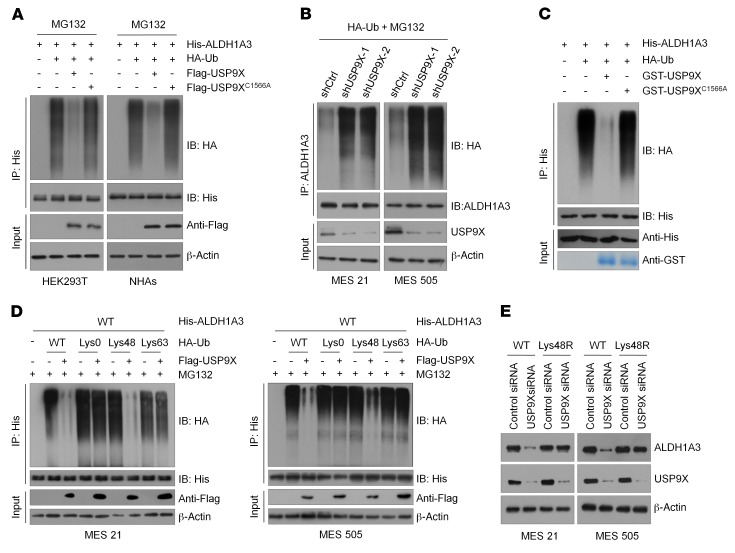
USP9X deubiquitinates ALDH1A3.
To investigate whether USP9X actually catalyzes the deubiquitination of ALDH1A3, we coexpressed His-ALDH1A3 and HA-ubiquitin with either WT or the C1566A mutant of USP9X in HEK293 cells and NHAs. After IP ALDH1A3 from cells treated with MG132, we observed that ALDH1A3 was heavily ubiquitinated. However, coexpression of WT USP9X almost completely abolished ALDH1A3 ubiquitination, while the C1566A mutant USP9X did not have this effect (Figure 3A). Conversely, downregulation of USP9X by 2 independent shRNAs dramatically increased ALDH1A3 polyubiquitylation in MES 21 and 505 GSCs (Figure 3B). To prove that ALDH1A3 is a direct deubiquitinated substrate of USP9X, we incubated polyubiquitinated ALDH1A3 with purified GST-USP9X WT or GST-USP9X C1566A under cell-free conditions. We found that purified GST-USP9X WT, but not GST-USP9X C1566A, which is still able to interact with ALDH1A3, specifically disassembled ALDH1A3 ubiquitin moieties in vitro (Figure 3C), suggesting that USP9X stabilizes ALDH1A3 by removing its ubiquitination directly. To date, 2 major forms of polyubiquitin chains are known to be formed through distinct types of linkages (Lys48- or Lys63-linked chains). Lys48-linked ubiquitin chains serve as the main targeting signals for protein degradation by the proteasome, whereas Lys63-linked ubiquitin chains are involved in multiple cellular events that do not rely on the proteasome-mediated degradative pathway. Accordingly, we wondered which type of polyubiquitin modifications on ALDH1A3 protein was affected by USP9X. As shown in Figure 3D, USP9X effectively disassembled Lys48-linked polyubiquitylation of ALDH1A3, but had no significant effect on the nondegradative Lys63-linked polyubiquitylation of ALDH1A3. Furthermore, we expressed a Lys48-resistant (Lys48R) form of ubiquitin in USP9X-depleted MES 21 and 505 GSCs and found that enforced expression of Lys48R ubiquitin attenuated USP9X depletion–induced ALDH1A3 downregulation (Figure 3E), confirming that Lys48-linked polyubiquitination is critical for USP9X-mediated ALDH1A3 turnover. Together, these results demonstrate that USP9X is a bona fide DUB targeting ALDH1A3 protein for deubiquitination.
Figure 3. USP9X deubiquitinates ALDH1A3.
(A) HEK293T cells or NHAs were cotransfected with His-ALDH1A3, HA-ubiquitin (HA-Ub), and Flag-tagged USP9X WT or USP9X C1566A, and cell lysates were subjected to IP with His beads followed by IB with antibodies against HA and His. Cells were treated with 20 μM MG132 for 8 hours before harvesting. (B) MES 21 and 505 GSCs were cotransfected with the indicated siRNA and HA-Ub, and cell lysates were subjected to IP with ALDH1A3 antibody, followed by IB with antibodies against HA and ALDH1A3. Cells were treated with 20 μM MG132 for 8 hours before harvesting. (C) Unubiquitylated or ubiquitylated His-ALDH1A3 was incubated with GST-USP9X WT or GST-USP9X C1566A coupled to glutathione-Sepharose beads. His-ALDH1A3 was subjected to IP with His beads followed by IB with antibodies against HA and His. Recombinant GST-USP9X or GST-USP9X C1566A was analyzed by SDS-PAGE and Coomassie blue staining. (D) MES 21 and 505 GSCs were cotransfected with His-ALDH1A3, Flag-USP9X, and the indicated HA-Ub Lys0, Lys48-only, or Lys63-only plasmids, and then the ALDH1A3 ubiquitylation linkage was analyzed. (E) MES 21 and 505 GSCs transfected with Ub WT or Ub Lys48R were cultured for 72 hours in the presence of control siRNA or USP9X siRNA. Cell lysates were analyzed by IB using antibodies against ALDH1A3 and USP9X.
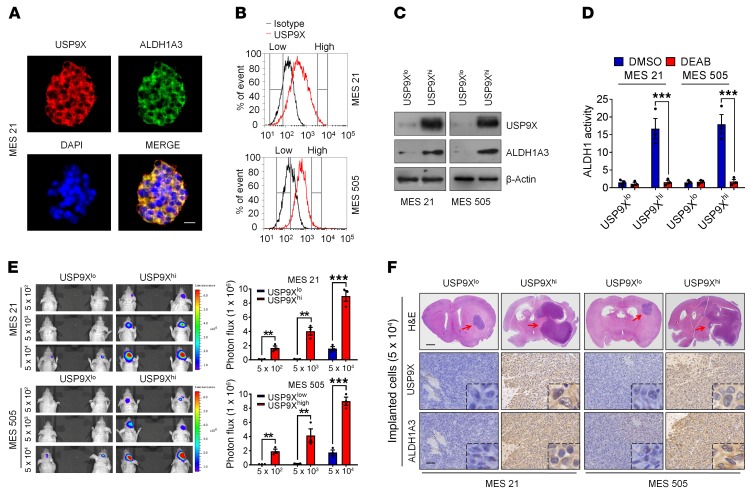
High USP9X expression predicts enrichment of ALDH1A3hi MES GSCs with potent tumorigenic capability.
Given the findings that both USP9X and ALDH1A3 are preferentially expressed in MES GSCs (Figure 1C and Figure 4A), coupled with the role of USP9X in regulating ALDH1A3 stability, we wondered whether high expression of USP9X might predict an enrichment of MES GSCs with high ALDH1A3 activity. To that end, we separated MES 21 and 505 GSCs into 2 distinct pools, expressing either high (USP9Xhi) or low (USP9Xlo) USP9X levels (Figure 4B). As expected, ALDH1A3 was highly expressed in the USP9Xhi subpopulation compared with the USP9Xlo subpopulation (Figure 4C). Furthermore, the results of ALDEFLUOR assay showed that the USP9Xhi subpopulation exhibited significantly higher ALDH1 activity than the USP9Xlo subpopulation (Figure 4D).
Figure 4. High USP9X expression predicts enrichment of ALDH1A3hi MES GSCs with potent tumorigenic capability.
(A) Confocal images showing colocalization of USP9X (red) and ALDH1A3 (green) in MES 21 GSCs. Nuclei were counterstained with DAPI (blue). (B) FACS sorting of USP9Xhi or USP9Xlo fractions isolated from MES 21 and 505 GSCs. (C) IB analysis of USP9X and ALDH1A3 levels in FACS-sorted USP9Xhi and USP9Xlo subpopulations. (D) Quantification of FACS analysis for ALDH1 activity in USP9Xhi or USP9Xlo subpopulations. DEAB was used to inhibit ALDH1 activity, serving as a negative control. (E) Representative bioluminescent images of intracranial GBM xenografts derived from FACS-sorted USP9Xhi or USP9Xlo subpopulations. Quantification of bioluminescent images is shown on the right. Colored scale bars represent photons/s/cm2/steradian. (F) H&E staining images and IHC images of USP9X and ALDH1A3 are shown in consecutive brain sections from mice implanted with 5 × 104 USP9Xhi or USP9Xlo subpopulations. Red arrows indicate tumors. Scale bars: 25 μm (A); 1 mm (H&E staining) and 100 μm (IHC staining) (F). Data are represented as mean ± SD of 3 independent experiments. **P < 0.01; ***P < 0.001, 2-tailed Student’s t test.
Next, we performed extreme limiting dilution intracranial implantation using USP9Xhi and USP9Xlo cells genetically engineered to stably express firefly luciferase. In vivo bioluminescence imaging demonstrated that as few as 5 × 102 USP9Xhi cells were sufficient to establish tumors in 60 days (Figure 4E). In stark contrast, a minimum of 5 × 104 USP9Xlo cells were required for tumor initiation (Figure 4, E and F). Below this level, the tumorigenic potential of the USP9Xlo cells was negligible. Immunohistochemical analysis demonstrated that mice bearing USP9Xhi cell–derived tumors exhibited high expression levels of ALDH1A3, whereas those bearing USP9Xlo cell–derived tumors were negative or only faintly positive for this molecule (Figure 4F). As a consequence, mice implanted with USP9Xhi cells had significantly shortened survival relative to those implanted with USP9Xlo cells (Supplemental Figure 3, A and B). Collectively, these results suggest that high expression of USP9X might signify great enrichment of ALDH1A3hi MES GSCs with potent tumorigenic capacity.
Ablation of USP9X expression impairs the self-renewal, tumorigenicity, and radio/chemoresistance of MES GSCs.
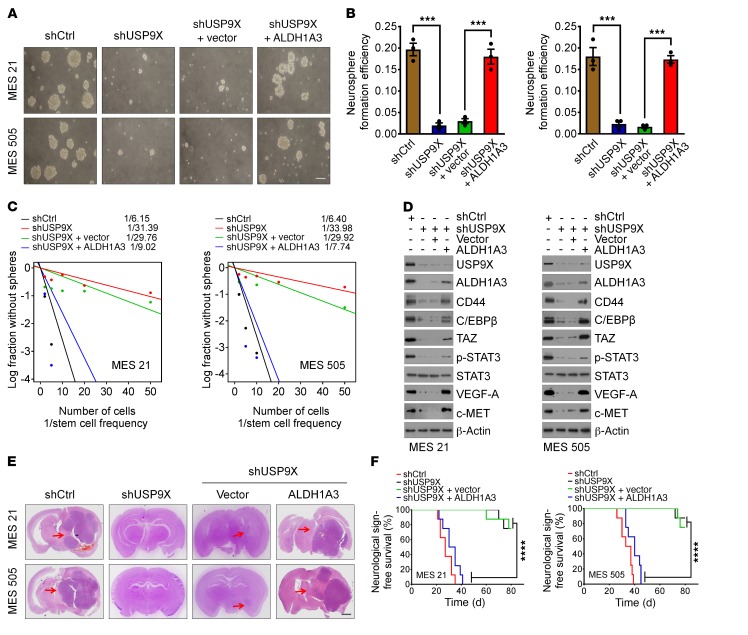
Given our results above, we were motivated to investigate whether USP9X is required for the self-renewal and tumorigenicity of MES GSCs. As expected, silencing of USP9X considerably attenuated cell growth (Figure 5, A and B) and decreased DNA replication, as revealed by EdU incorporation assay (Supplemental Figure 4, A and B). Moreover, in vitro limiting dilution assay demonstrated that knocking down USP9X expression remarkably reduced the tumorsphere formation frequency of MES21 and 505 GSCs (Figure 5C). Furthermore, reduced expression of ALDH1A3 and CD44 as well as master MES-specific markers, including C/EBPβ, TAZ, phosphorylated STAT3 (p-STAT3), VEGF-A, and c-MET, were observed in USP9X-depleted MES GSCs compared with controls (Figure 5D).
Figure 5. Ablation of USP9X expression impairs the self-renewal, tumorigenicity, and radio/chemoresistance of MES GSCs.
(A and B) Primary neurosphere formation was assessed in MES 21 and 505 GSCs transduced with shCtrl or shUSP9X, reconstituted with vector control or ALDH1A3. Representative images are shown (A). Neurosphere formation efficiency (spheres/cells plated) was quantified. (B) Data are represented as means ± SD of 3 independent experiments. ***P < 0.001, 2-tailed Student’s t test. (C) In vitro limiting dilution sphere-forming frequency of MES 21 and 505 GSCs transduced with shCtrl or shUSP9X, reconstituted with vector control or ALDH1A3. Stem cell frequencies were estimated as the ratio 1/x with the upper and lower 95% confidence intervals, where 1 = stem cell and x = all cells. (D) IB analysis of USP9X, ALDH1A3, CD44, C/EBPβ, TAZ, p-STAT3, STAT3, VEGF-A, and c-MET levels in MES 21 and 505 GSCs expressing shCtrl or shUSP9X, reconstituted with vector control or ALDH1A3. (E) H&E-stained brain sections from mice intracranially implanted with MES 21 or 505 GSCs with indicated modifications. Red arrows indicate tumors. (F) Kaplan-Meier survival curves of mice intracranially injected with MES 21 or 505 GSCs with indicated modifications (n = 8). ****P < 0.0001, log-rank (Mantel-Cox) test. Scale bars: 500 μm (A); 1 mm (E).
We next examined the impact of USP9X depletion on the tumorigenic potential of MES GSCs in vivo. Equal numbers of luciferase-labeled MES GSC 21 or 505 transduced with control shRNA (shCtrl) or shUSP9X were intracranially injected into NOD/SCID mice. Compared with the mice implanted with shCtrl-transduced MES GSCs, those implanted with shUSP9X-transduced MES GSCs displayed extended survival with a lower rate of tumor formation (Figure 5, E and F, and Supplemental Figure 4, C and D). To evaluate the role of ALDH1A3 in USP9X-mediated GSC stemness and tumorigenicity, we ectopically expressed ALDH1A3 in MES21 and 505 GSCs, in which endogenous USP9X had been depleted. We found that the inhibitory effect of USP9X depletion on cell proliferation, self-renewal, and tumorigenic potential could be largely rescued by ALDH1A3 (Figure 5, A–F, and Supplemental Figure 4, A–D). These results suggest the functional importance of the USP9X/ALDH1A3 axis in the maintenance of MES traits of GSCs.
Recent studies have shown that USP9X contributes to the radioresistance and/or chemoresistance of several types of cancer cells (31, 32, 34, 39). Thus, we wondered whether USP9X is involved in radio/chemoresistance of GSCs. To address this, PN 35 and 182 GSCs expressing low basal levels of USP9X were transduced with either empty or USP9X lentiviral vectors and stably transduced cells were treated with ionizing radiation (IR) (5 Gy) or temozolomide (TMZ) (100 μM). We found that enforced expression of USP9X in PN GSCs conferred resistance to both IR and TMZ (Supplemental Figure 4, E–G), whereas knockdown of USP9X enhanced the sensitivity of USP9X-overexpressing PN GSCs to IR and TMZ (Supplemental Figure 4, H–J). These results indicate that USP9X might contribute to the acquisition of radio/chemoresistance in PN GSCs.
Pharmacological inhibition of USP9X attenuates the tumor-initiating ability of MES GSCs with high ALDH1A3 activity.
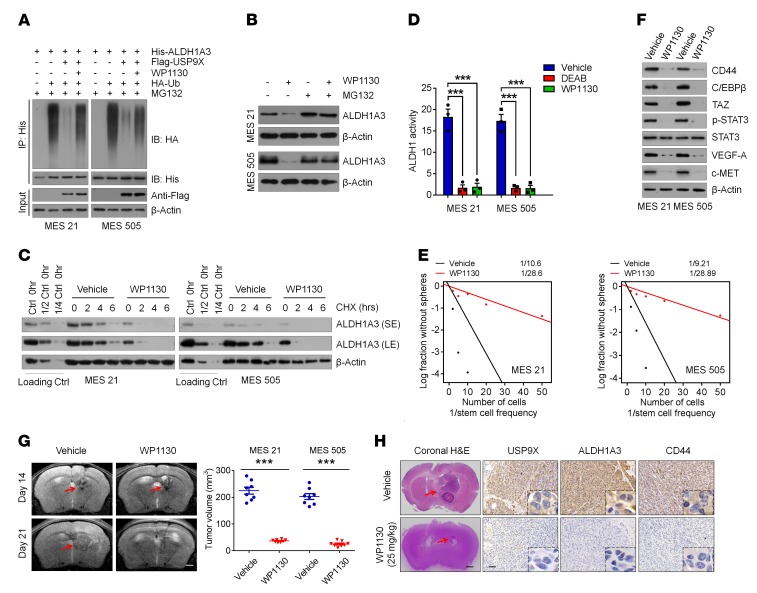
To leverage our findings for clinical application, we investigated the effect of USP9X pharmacologic inhibition on MES GSC–derived GBM models. We used a recently reported small-molecule inhibitor of USP9X, WP1130, and first examined whether the deubiquitinating activity of USP9X on ALDH1A3 is suppressed by WP1130 under our experimental conditions. Indeed, the ability of USP9X to remove ubiquitin moieties from polyubiquitinated ALDH1A3 was almost completely abrogated by 1 μmol/l WP1130 (Figure 6A). Correspondingly, WP1130 treatment reduced ALDH1A3 protein levels in MES 21 and 505 GSCs without affecting its mRNA levels (Figure 6B and Supplemental Figure 5A). This effect was reversed by MG132, suggesting that WP1130, like USP9X knockdown, promotes ALDH1A3 ubiquitination and degradation (Figure 6B). Indeed, cotreatment with WP1130 and CHX induced a marked decrease in the half-life of ALDH1A3 protein to less than 2 hours (Figure 6C). Furthermore, WP1130 substantially inhibited ALDH1 activity in MES 21 and 505 GSCs, as demonstrated by FACS analysis using the ALDEFLUOR assay (Figure 6D).
Figure 6. Pharmacological inhibition of USP9X attenuates the tumor-initiating ability of MES GSCs with high ALDH1A3 activity.
(A) MES 21 and 505 GSCs were cotransfected with His-ALDH1A3, HA-Ub, and Flag-USP9X in the absence or presence of 1 μM WP1130, and cell lysates were subjected to IP with His beads followed by IB with antibodies against HA and His. Cells were treated with 20 μM MG132 for 8 hours before harvesting. (B) IB analysis of ALDH1A3 in MES 21 and 505 GSCs treated with 1 μM WP1130 or vehicle with or without MG132. (C) MES 21 and 505 GSCs were treated with 1 μM WP1130 or vehicle for 24 hours, followed by 100 μg/ml CHX, harvested at the indicated times, and then subjected to IB with antibodies against ALDH1A3. SE, short exposure; LE, long exposure. (D) Quantification of FACS analysis for ALDH1 activity in MES 21 and 505 GSCs following treatment with 1 μM WP1130, 150 μM DEAB, or vehicle. (E) In vitro limiting dilution sphere-forming frequency of MES 21 and 505 GSCs after treatment with 1 μM WP1130 or vehicle. (F) IB analysis of the indicated proteins in MES 21 and 505 GSCs after treatment with 1 μM WP1130 or vehicle. (G) T2-weighted MRI images (left) and quantification of tumor volume (right) in mice bearing xenografts derived from MES 21 or 505 GSCs following treatment with 25 mg/kg WP1130 or vehicle. Red arrows indicate tumors. (H) H&E- and IHC-stained images of USP9X, ALDH1A3, and CD44 in mice intracranially implanted with MES 21 or 505 GSCs after treatment with WP1130 (25 mg/kg) or vehicle. Red arrows indicate tumors. Scale bars: 1 mm (G); 1 mm (H&E) and 100 μm (IHC) (H). Data are represented as mean ± SD of 3 independent experiments. ***P < 0.001, 1-way ANOVA with Dunnett’s post test (D); 2-tailed Student’s t test (G).
Next, we examined the sensitivity of MES 21 and 505 GSCs to WP1130. We found that treatment of 2 MES GSCs with WP1130 resulted in a significant inhibition of cell proliferation and tumorsphere-forming ability accompanied by a pronounced loss of MES features (Figure 6, E and F, and Supplemental Figure 5, B and C). We then assessed the therapeutic effects of WP1130 on mice bearing intracranial tumors derived from MES 21 and 505 GSCs. WP1130 or vehicle was administered locally by convection-enhanced delivery (CED). Consistent with our in vitro findings, tumor-bearing mice receiving WP1130 treatment by 7-day continuous CED infusion showed retarded tumor growth and improved survival compared with vehicle-treated mice (Figure 6G and Supplemental Figure 5D). In these tumors, expression levels of USP9X, ALDH1A3, and CD44 were strongly attenuated by WP1130, as demonstrated by immunohistochemical analysis (Figure 6H). Together, these results suggest that pharmacological inhibition of USP9X might effectively eliminate MES GSCs via promoting ALDH1A3 destabilization.
USP9X shows a positive correlation with ALDH1A3 protein levels and is associated with poor survival of ALDH1A3hi MES GBMs.
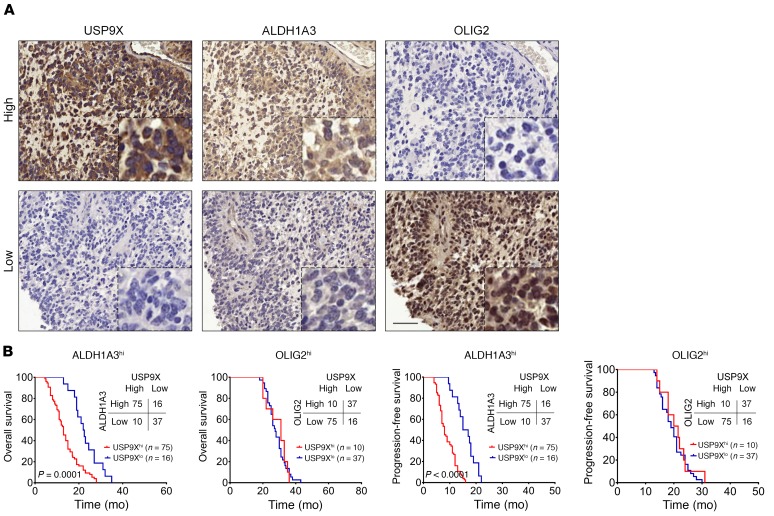
Finally, we performed immunohistochemical staining of USP9X, ALDH1A3 (MES GBM marker), and OLIG2 (PN GBM marker) in a tissue microarray, including 138 primary GBM specimens. As shown in Figure 7A and Supplemental Figure 6A, USP9X is strongly correlated with ALDH1A3 expression and mutually exclusive with OLIG2. Specifically, about 88.2% of the high USP9X samples exhibited high ALDH1A3 expression, whereas 69.8% of samples with low USP9X displayed low ALDH1A3 expression. Furthermore, Kaplan-Meier survival analysis demonstrated that USP9X expression levels had no predictive value for patient survival in the OLIG2hi GBM patients. In contrast, USP9Xhi GBM patients displayed significantly shorter overall survival and progression-free survival in the ALDH1A3hi patient group (Figure 7B), suggesting that USP9X has a prognostic value for patients within the MES subgroup. Collectively, these human GBM data strongly align with our experimental findings of USP9X-mediated ALDH1A3 stabilization and associated MES features in GSCs.
Figure 7. USP9X shows a positive correlation with ALDH1A3 protein levels and is associated with poor survival of ALDH1A3hi MES GBMs.
(A) IHC staining of 138 human GBM specimens for USP9X and ALDH1A3. Representative consecutive sections from 2 specimens are shown. Scale bars: 50 μm. (B) Kaplan-Meier curves showing overall survival (upper panels) and progression-free survival (lower panels) of GBM patients divided based on USP9X expression in ALDH1A3hi MES tumors or OLIG2hi PN tumors (P values by log-rank test).
Discussion
One of the major hindrances for cancer treatment and therapeutic design is intratumor heterogeneity, shaped by phenotypically distinct tumor cell subpopulations. GSCs are at the apex of the cellular hierarchy in GBMs, which are thought to be responsible for the aggressive phenotype and therapeutic resistance (40, 41). Emerging evidence has shown that high levels of ALDH1A3 activity are critical for maintaining MES features of GSCs (16–18). Therefore, identification of regulatory mechanisms that govern ALDH1A3 expression is critical for developing GSC-targeted therapy. In this study, using unbiased DUB-focused siRNA screening, we identified USP9X as a bona fide DUB that interacts with ALDH1A3, reverses ALDH1A3 polyubiquitylation, and protects ALDH1A3 from proteasomal degradation, thereby leading to the stabilization and accumulation of ALDH1A3 in MES GSCs. We provide evidence that this event requires the catalytic activity of USP9X because a catalytically inactive mutant C1566A could not stabilize ALDH1A3. We also mapped and identified the N-terminal region (amino acids 1–600) of USP9X and the N-terminal region (amino acids 1–200) of ALDH1A3 essential for their mutual interactions. Moreover, the results from human GBM specimens further underscored the existence of a strong association between USP9X and ALDH1A3.
It has been recently demonstrated that most CD44+ MES GSCs express high levels of ALDH1A3, whereas a small subset of CD44+ MES GSCs express relatively low levels of ALDH1A3 (16), suggesting that both CD44+ALDH1A3hi and CD44+ALDH1A3lo exist in individual tumors. In this study, we fractionated CD44+ MES GSCs into USP9Xhi and USP9Xlo subpopulations and observed that the USP9Xhi fraction exhibited substantially higher ALDH1A3 expression and activity than the USP9Xlo fraction. When USP9Xhi and USP9Xlo subpopulations were assayed for intracranial tumorigenicity by the extreme limiting dilution analysis, we found that mice implanted with the USP9Xhi subpopulation showed markedly higher tumorigenic potential compared with those implanted with the USP9Xlo subpopulation. Our findings indicate that CD44+ MES GSCs include both USP9X/ALDH1A3hi and USP9X/ALDH1A3lo cells, that CD44+ USP9X/ALDH1A3hi cells represent a particular subpopulation responsible for more aggressive behavior of MES GSCs, and that these cells might be potential targets for treatment of GBMs. Indeed, silencing of USP9X resulted in ALDH1A3 destabilization and reduced expression of CD44 and master MES-associated transcription factors (TFs), including C/EBPβ, TAZ, and p-STAT3, leading to the inhibition of self-renewal and tumor-initiating capacities of MES GSCs. This effect could be largely rescued by ectopic expression of ALDH1A3. Thus, our work expands our understanding of how GSCs possess high ALDH1A3 expression and activity and also identifies the USP9X/ALDH1A3 axis as an important target for GSC-based therapy.
In the current neurooncology clinic, adjuvant IR combined with TMZ is a mainstay of postsurgical GBM treatment. Our current study showed that enforced expression of USP9X in PN GSCs conferred resistance to both IR and TMZ. This effect could be substantially abrogated by knocking down USP9X. Thus, our findings reveal USP9X as a critical factor for the acquisition of radio/chemoresistance in PN GSCs and suggest that targeting USP9X could potentially attenuate radio/chemoresistance of PN GSCs following IR or TMZ treatment.
The emergence of ALDH1A3 as a potentially therapeutic target for GSCs serves as an impetus for the need for the development of selective inhibitors. However, no clinically active drugs that specifically target ALDH1A3hi subpopulation are currently available (11). Accumulating evidence suggests that selective inhibition of DUBs has been proposed as a promising treatment option for cancer therapy, and a number of DUB inhibitors have been identified and are currently being tested in preclinical studies and clinical trials (42, 43). In the current study, we used the USP9X inhibitor WP1130 to assess the impact of pharmacological inhibition of USP9X on ALDH1A3 protein expression and activity as well as MES GSC–derived tumor growth. We found that WP1130 is able to promote robust polyubiquitylation of ALDH1A3, which results in a marked reduction in ALDH1A3 protein levels and functional activity, leading to attenuation of the tumor-initiating ability of MES GSCs both in vitro and in vivo. Therefore, our findings suggest that USP9X-specific inhibitors might hold promise for future therapy of patients with ALDH1A3hi MES GSCs, and it would be interesting to explore whether these results might be generalizable to the treatment of other CSCs as well.
In summary, this study identifies USP9X as a critical deubiquitinase for stabilizing ALDH1A3 and maintaining its high activity, which leads to the maintenance of MES properties, self-renewal, and tumorigenic capability of GSCs (Supplemental Figure 6B). Targeting ALDH1A3 stabilization through pharmacological inhibition of USP9X may thus open an avenue for therapeutic intervention in GBMs.
Methods
Human subjects.
Four freshly resected GBM specimens were obtained from the Department of Neurosurgery, First Affiliated Hospital of Nanjing Medical University (Supplemental Table 1). Tissue microarray consisting of 138 formalin-fixed, paraffin-embedded GBM tissues was obtained from the Department of Neurosurgery, Second and Fourth Affiliated Hospitals of Harbin Medical University (44).
Cell lines, primary cell cultures, and GSCs.
Human GBM U87MG and T98G cell lines and human embryonic kidney HEK293T cell lines were obtained from ATCC. These cell lines were cultured in DMEM with 10% FBS. NHAs were obtained from ScienCell Research Laboratories and cultured in astrocyte growth media supplemented with rhEGF, insulin, ascorbic acid, GA-1000, l-glutamine, and 5% FBS. MES or PN GSCs were isolated from primary GBM tumors or patient-derived GBM xenografts, as previously described (45, 46). Briefly, primary GBM cells were dissociated from the freshly resected GBM specimens using the Papain Dissociation System (Worthington Biochemical). The isolated GBM cells were recovered in DMEM/F12 medium supplemented with B27 (Invitrogen), bFGF, and EGF (20 ng/ml each). Cells were then labeled with anti-CD44 (Miltenyi Biotec, catalog 130-113-334) or anti-CD133 (Miltenyi Biotec, catalog 130-110-962) antibody followed by FACS to isolate MES or PN GSCs. The sorted GSCs were validated by GSC enrichment markers and were functionally characterized by self-renewal potential, multilineage differentiation potency, and tumorigenic capacity. GSCs were constantly maintained as GBM xenografts and were only dissociated, sorted, and cultured in the neurobasal medium for the functional experiments. Only early passage GSC cells were used for the study. The unique identities of GBM cell lines and patient-derived GSC cells were authenticated by short tandem-repeat analysis, as described in Supplemental Table 2. All the cells were routinely tested for mycoplasma contamination bimonthly using MycoAlert PLUS Kits (Lonza).
Cell sorting.
Alexa Fluor 488–conjugated USP9X (Abcam, ab203270) was incubated with the Protein Transfection Reagent (Thermo Scientific, Pierce Protein Biology) for 5 minutes in serum-free medium. The protein/reagent mixture was then added to GSCs. After 4 hours of incubation in a 5% CO2 incubator at 37°C, GSCs were washed 3 times with PBS and analyzed by a BD FACSAria II SORP.
ALDEFLUOR assay and separation of the ALDH1hi cell population by FACS analysis.
ALDH1 enzyme activity was determined using a fluorogenic dye–based ALDEFLUOR assay kit (Stem Cell Technologies) according to the manufacturer’s instructions. Cells were suspended in ALDEFLUOR assay buffer containing ALDH1 substrate (BODIPY-aminoacetaldehyde) and incubated for 45 minutes at 37 °C. ALDEFLUOR-stained cells treated with the specific ALDH1 enzyme inhibitor diethylaminobenzaldehyde (DEAB) (150 μM) were used as negative controls. ALDH1hi and ALDH1lo cells were isolated using a BD FACSAria II SORP based on fluorescence signals of these cells.
siRNA library screening.
The Dharmacon siGENOME RTF SMARTpool siRNA library for human DUBs was used to screen for human deubiquitylases. Briefly, HEK293T cells were added to the rehydrated Dharmacon RTF siRNA library plates. After 48 hours, cell lysates were extracted and the expression of endogenous ALDH1A3 was examined by IB.
Plasmids, siRNA, shRNA, and cell transfections.
Lentiviral constructs expressing Flag-tagged USP9X, Flag-tagged USP9X C1566A, and His-tagged ALDH1A3 were generated by cloning their ORF with the N-terminal Flag or His sequence into the pCDH-CMV-MCS-EF1α-Puro vector. Plasmids coding for HA-tagged ubiquitin-Lys63 (pRK5-HA-ubiquitin-Lys63) and HA-tagged ubiquitin-Lys48 (pRK5-HA-ubiquitin-Lys48) were obtained from Addgene. Site-directed mutagenesis in USP9X was performed with a QuikChange Mutagenesis Kit (Agilent Technologies), according to the manufacturer’s instructions. The authenticity of all the constructs was confirmed by DNA sequencing. Lentiviral shRNA plasmids for targeting USP9X and nonspecific control shRNA were purchased from Dharmacon (Supplemental Table 3). All transfections were performed using Lipofectamine 3000 (Invitrogen) following the manufacturer’s instructions.
RNA isolation and quantitative real-time PCR.
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN) and then reverse transcribed with an iScript cDNA Synthesis Kit (Bio-Rad). Expression levels of ALDH1A3 were determined using the 2–ΔΔCt method and normalized to the housekeeping gene GAPDH. The primer sequences of ALDH1A3 are shown in Supplemental Table 3.
IB, immunofluorescence, and immunohistochemistry.
The following primary antibodies were used in this study: anti–CD44-FITC (catalog 130-113-334) and anti–CD133-PE (catalog 130-110-962) from Miltenyi Biotec; anti-USP9X (catalog ab19879), anti-TAZ (catalog ab84927), anti-C/EBPβ (catalog ab32358), anti-VEGFA (catalog ab51745), and anti-c-MET (catalog ab74217) from Abcam; anti-CD44 (catalog 3570), anti–p-STAT3 (catalog 9145), anti-STAT3 (catalog 9139), anti–His-Tag (catalog 2366), anti–HA-Tag (catalog 3724), anti-mouse IgG (catalog 5415), anti-rabbit IgG (catalog 3900), anti-GST (catalog 2624), and anti–β-actin (catalog 4970) from Cell Signaling Technology; anti-ALDH1A3 (catalog ab129815) from Abcam; and anti-Flag-Tag (catalog F3165) from Sigma-Aldrich.
For IB, cells were harvested and lysed in NETN buffer containing protease and phosphatase inhibitors (Sigma-Aldrich). Cell lysates were subjected to SDS-PAGE and then electrotransferred onto polyvinylidene difluoride membranes that were incubated with the indicated primary antibodies, washed, and probed with HRP-conjugated secondary antibodies.
For immunofluorescence staining, patient-derived GSCs were fixed with 4% formaldehyde, permeabilized with 0.25% Triton X-100, and then blocked with 1% BSA for 1 hour at room temperature. Cells were probed with the indicated primary antibodies. Following wash with PBS-T, cells were incubated with appropriate Alexa Fluor 488– or Alexa Fluor 594–labeled secondary antibodies (Thermo Fisher Scientific) and DAPI-containing Vectashield mounting medium (Vector Laboratories), then visualized by confocal laser-scanning microscopy (Zeiss LSM 5).
For immunohistochemical staining, human GBM tissue microarray or GBM xenografts were deparaffinized and rehydrated through a descending alcohol series, which was followed by antigen retrieval with sodium citrate buffer. Tumor sections were blocked with 1% BSA with 0.25% Triton X-100 and 3% H2O2 in PBS for 1 hour at room temperature, then incubated with the indicated primary antibodies overnight at 4°C. Thereafter, sections were incubated with HRP conjugates using diaminobenzidine detection. The staining signal was scored according to the proportion of positive cells and staining intensity, as described previously (45). Briefly, the quantification of IHC staining was done according to the proportion of positively stained tumor cells and the intensity of staining. The proportion of positively stained tumor cells was graded as follows: 0, no positive tumor cells; 1, 0.01%–25% positive tumor cells; 2, 25.01%–50% positive tumor cells; 3, 50.01%–75% positive tumor cells; and 4, 75% or greater positive tumor cells. The cells at each intensity of staining were recorded on a scale of 0 (no signal), 1 (weak), 2 (moderate), and 3 (strong). The IHC score for each section was computed by the following formula: IHC score = staining intensity × proportion of positively stained tumor cells. A total score of 0–12 was calculated and graded as negative (–, score: 0), weak (+, score: 1–4), moderate (++, score: 5–8), or strong (+++, score: 9–12). Cutoff values to define the high and low expression of USP9X and ALDH1A3 were chosen on the basis of a measurement of heterogeneity with the log-rank test statistic with respect to overall survival. Because the optimal cutoff thresholds were identified from the current study as 4, tumors with IHC scores of 4 or more were considered as high expression, and tumors with IHC scores of less than 4 were considered as low expression.
GST-tagged protein purification and GST pull-down assays.
GST-ALDH1A3, GST-USP9X-WT, and GST-USP9X-C1566A in the bacterial expression plasmid pGEX-4T-1 were expressed in the Escherichia coli strain BL21 by induction with 0.4 mM isopropyl β-d-1-thiogalactopyranoside at 16°C and purified with GST beads (Sigma-Aldrich) according to the manufacturer’s protocol.
For GST pull-down assay, bacterial-expressed GST, GST-ALDH1A3, GST-USP9X-WT, or GST-USP9X-C1566A bound to glutathione-Sepharose 4B beads (GE Healthcare) was incubated with Flag-USP9X WT, Flag-USP9X C1566A, or His-ALDH1A3 expressed in HEK293T cells for 2 hours at 4°C. After reaction, complexes were washed at least 4 times with GST-binding buffer, eluted by boiling in SDS-PAGE loading buffer, and subjected to IB with the indicated antibodies.
IP.
Cells transfected with the indicated constructs were collected and lysed in NETN buffer containing protease inhibitors (Sigma-Aldrich). Cell lysates were precleared with protein A/G agarose (Thermo Fisher Scientific) for 1 hour at 4 °C. Precleared lysates were immunoprecipitated with the indicated antibodies overnight at 4°C and then incubated with protein A/G agarose for an additional 2 hours at 4°C. The immunocomplexes were washed with NETN buffer 4 times, and the bound proteins were eluted by boiling and subjected to SDS-PAGE and IB.
In vivo and in vitro deubiquitylation of ALDH1A3.
For in vivo ALDH1A3 ubiquitylation assay, cells were transfected with the indicated plasmids, then treated with 20 μM MG132 for 8 hours. Cells were collected and lysed in NETN buffer plus 0.1% SDS, 20 μM MG132, and protease inhibitors. Lysates were incubated with anti-ALDH1A3 antibody for 3 hours and protein A/G agarose beads for a further 8 hours at 4°C. The precipitated proteins were then released from the beads by boiling for 10 minutes in SDS-PAGE loading buffer and were subjected to IB with the anti-HA antibody.
For preparation of ubiquitinated ALDH1A3 as the substrate for the in vitro deubiquitination assay, HEK293T cells were transfected with both His-ALDH1A3 and HA-ubiquitin and were treated with 20 μM MG132 for 8 hours. Ubiquitylated ALDH1A3 was purified from the cell extracts with anti-His affinity column (Thermo Fisher Scientific) and then incubated with the recombinant GST-USP9X-WT or GST-USP9X-C1566A protein in a deubiquitylation buffer for 2 hours at 37°C. Reactions were subjected to IB analysis.
Protein half-life assay.
Cells transfected with the indicated plasmids were treated with the protein synthesis inhibitor CHX (100 μg/ml; Sigma-Aldrich) for the indicated durations before collection.
EdU incorporation.
EdU staining was performed using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen). Briefly, dissociated GSCs were plated into wells of laminin-precoated 8-well chamber slides. EdU was added to the culture media at a final concentration of 10 μmol/l for 3 hours. Cells were then fixed with 4% paraformaldehyde in PBS for 15 minutes and penetrated with 0.5% Triton X-100 for 30 minutes. Nuclei were counterstained with DAPI. Five fields of view per slide were examined for EdU-positive cells.
Neurosphere formation assay and extreme limiting dilution assay.
For neurosphere formation assay, dissociated single cells were plated at a density of 1 cell/μl and the spheres that formed after 7 days were counted. For extreme limiting dilution assay, GSCs with indicated modification or treatment were dissociated to single cells and then plated in 96-well plates at densities of 1, 5, 10, 20, or 50 cells per well. After incubation for 7 days, each well was examined for formation of tumor spheres. Stem cell frequency was calculated using Extreme Limiting Dilution Analysis (ELDA) (http://bioinf.wehi.edu.au/software/elda/).
Intracranial xenograft tumor models and treatments.
For tumorigenicity studies, 1 × 105 luciferase-expressing GSCs with indicated modification were intracranially injected into the right caudate nucleus of immunocompromised mice using a stereotactic apparatus (coordinates: 2 mm anterior, 2 mm lateral, 3 mm depth from the dura). All mice were monitored daily for the development of neurological symptoms. The tumor growth was monitored by bioluminescence imaging with an IVIS Lumina Imaging System. The mice were humanely euthanized 2 to 10 weeks after implantation, and their brains were harvested, paraffin embedded, stained with H&E to confirm the presence of tumor, and subjected to immunohistochemical staining. For in vivo limiting dilution assays, mice were implanted with 5 × 102, 5 × 103, and 5 × 104 FACS-sorted USP9Xhi or USP9Xlo cells. For animal survival analysis, mice were maintained until manifestation of neurological symptoms (i.e., hunched back, loss of body weight, reduced food consumption, and inactivity) from tumor burden developed or until 80 days after injection.
For testing in vivo inhibition effect of USP9X inhibitor WP1130, 1 × 105 MES GSCs were implanted intracranially into individual mice. From day 10, MRI was performed to verify the formation of MES GSC-derived tumors. There were no significant differences in tumor size between groups at time of treatment allocation. ALZET micro-osmotic pumps (DURECT Corp.) and infusion apparatus were implanted into tumor-bearing mice, and CED of either WP1130 (25 mg/kg) or vehicle was initiated. Treatment was delivered at a rate of 0.5 μl/h for 7 days. To verify the efficacy of intracranial administration of WP1130, MRI was carried out after removal of the pump system. The investigators were blinded to the group allocation and study outcome assessments of all mice.
MRI of orthotopic xenograft tumors.
MRI studies were performed on a Bruker 7.0T scanner (Bruker BioSpin GmbH) with a 16 cm bore. T2-weighted coronal images were acquired by rapid acquisition with relaxation enhancement (RARE) sequence with the following parameters: TR, 3000 ms; TE, 60 ms; RARE factor, 12; average, 4; FOV, 40 × 30 mm; in-plane resolution, 156 × 156 μmol/l2; slice thickness, 0.75 mm; and slice gap, 0.25 mm. Tumor volume was assessed by contouring the lesions in the T2-weighted images using ImageJ (NIH).
Statistics.
Data in all graphs are represented as mean ± SD of biological triplicates. Significance was calculated by a 2-tailed Student’s t test or 1-way ANOVA with Dunnett’s post test for multiple comparisons using GraphPad Prism 7.0 software. Kendall’s τ-β was used to test for correlations in the immunohistochemical results. A log-rank (Mantel-Cox) analysis was used to determine statistical significance of Kaplan-Meier survival curves. For all statistical tests, P values of less than 0.05 were considered significant.
Study approval.
All tumor collection and analysis was approved by the IRBs and the ethics committees of Nanjing Medical University and Harbin Medical University. Informed consent was obtained from all individual participants. All animal experiments were conducted with the approval of IACUC of Nanjing Medical University and in conformity with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011).
Author contributions
ZC, HWW, SW, LF, and HW conceived the project and designed the study. ZC, HWW, SW, LF, SF, XC, CP, XW, and JL performed the experiments. DC, YC, WW, DL, NL, YY, and HW analyzed and interpreted the data. HW wrote the manuscript and provided study supervision.
Supplementary Material
Acknowledgments
We thank Chengyi Hu and He Wang for assistance with collecting fresh GBM patient specimens; the Nanjing Medical University Animal Core Facility for excellent mouse husbandry and care; the Nanjing Medical University Pathology Facility for assistance with paraffin processing, embedding, and histopathological analysis; and the Nanjing Medical University Flow Facility for expert cell sorting. This work was supported by grants from the National Natural Science Foundation of China (81772681 and 81201978 to HW; 81670153 and 81200362 to SW), the Excellent Youth Foundation of Jiangsu Province (BK20160098 to HW), the Natural Science Foundation of Jiangsu Province (BK2012483 to HW), the Program for Advanced Talents within Six Industries of Jiangsu Province (2012-WSN-019 to HW), the Natural Science Foundation of Heilongjiang Province (H2015095 to HWW), the National High Technology Research Development Program of China (863 Program) (2012AA02A508) and the National Key Research and Development Plan (2016YFC0902500), the Program for Development of Innovative Research Team in the First Affiliated Hospital of Nanjing Medical University, and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Version 1. 04/08/2019
Electronic publication
Version 2. 05/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(5):2043–2055.https://doi.org/10.1172/JCI126414.
See the related Commentary at Deubiquitinating ALDH1A3 key to maintaining the culprit of aggressive brain cancer.
Contributor Information
Zhengxin Chen, Email: zhengxinchen1989@126.com.
Hong-Wei Wang, Email: hongweiwang1969@yeah.net.
Shuai Wang, Email: wangooshuai@126.com.
Ligang Fan, Email: ligangfan1983@126.com.
Shuang Feng, Email: shuangfeng9191@126.com.
Xiaomin Cai, Email: xiaomincai1993@163.com.
Chenghao Peng, Email: chenghaopeng1991@163.com.
Xiaoting Wu, Email: xiaotingwu1990@163.com.
Jiacheng Lu, Email: jiachenglu1994@163.com.
Dan Chen, Email: simpledanchen@126.com.
Yuanyuan Chen, Email: xoochen@126.com.
Wenting Wu, Email: wentingwu1983@163.com.
Daru Lu, Email: darulu@126.com.
Ning Liu, Email: ningliu1958@163.com.
Yongping You, Email: yongpingyou1967@yeah.net.
Huibo Wang, Email: hbwang@njmu.edu.cn.
References
- 1.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frattini V, et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet. 2013;45(10):1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Hemmati HD, et al. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci U S A. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh SK, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 6.Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 7.Hu B, et al. Epigenetic activation of WNT5A drives glioblastoma stem cell differentiation and invasive growth. Cell. 2016;167(5):1281–1295.e18. doi: 10.1016/j.cell.2016.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Z, et al. Targeting self-renewal in high-grade brain tumors leads to loss of brain tumor stem cells and prolonged survival. Cell Stem Cell. 2014;15(2):185–198. doi: 10.1016/j.stem.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 11.Koppaka V, et al. Aldehyde dehydrogenase inhibitors: a comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol Rev. 2012;64(3):520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao C, et al. Essential role of aldehyde dehydrogenase 1A3 for the maintenance of non-small cell lung cancer stem cells is associated with the STAT3 pathway. Clin Cancer Res. 2014;20(15):4154–4166. doi: 10.1158/1078-0432.CCR-13-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin TY, Lee CC, Chen KC, Lin CJ, Shih CM. Corrigendum to “Inhibition of RNA transportation induces glioma cell apoptosis via downregulation of RanGAP1 expression” [Chem. Biol. Interact. 232 (2015 May 5) 49-57] Chem Biol Interact. 2017;268:149. doi: 10.1016/j.cbi.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 14.Abdul Rehman SA, et al. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell. 2016;63(1):146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan JJ, Cai J, Guo YF, Bian XW, Yu SC. ALDH1A3, a metabolic target for cancer diagnosis and therapy. Int J Cancer. 2016;139(5):965–975. doi: 10.1002/ijc.30091. [DOI] [PubMed] [Google Scholar]
- 16.Mao P, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110(21):8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng P, et al. FOXD1-ALDH1A3 signaling is a determinant for the self-renewal and tumorigenicity of mesenchymal glioma stem cells. Cancer Res. 2016;76(24):7219–7230. doi: 10.1158/0008-5472.CAN-15-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sullivan KE, Rojas K, Cerione RA, Nakano I, Wilson KF. The stem cell/cancer stem cell marker ALDH1A3 regulates the expression of the survival factor tissue transglutaminase, in mesenchymal glioma stem cells. Oncotarget. 2017;8(14):22325–22343. doi: 10.18632/oncotarget.16479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weisberg EL, et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat Chem Biol. 2017;13(12):1207–1215. doi: 10.1038/nchembio.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbé S. Deubiquitylases from genes to organism. Physiol Rev. 2013;93(3):1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 21.Pal A, Young MA, Donato NJ. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 2014;74(18):4955–4966. doi: 10.1158/0008-5472.CAN-14-1211. [DOI] [PubMed] [Google Scholar]
- 22.Suresh B, Lee J, Kim H, Ramakrishna S. Regulation of pluripotency and differentiation by deubiquitinating enzymes. Cell Death Differ. 2016;23(8):1257–1264. doi: 10.1038/cdd.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homan CC, et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet. 2014;94(3):470–478. doi: 10.1016/j.ajhg.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dupont S, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136(1):123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 25.Nagai H, et al. Ubiquitin-like sequence in ASK1 plays critical roles in the recognition and stabilization by USP9X and oxidative stress-induced cell death. Mol Cell. 2009;36(5):805–818. doi: 10.1016/j.molcel.2009.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Grasso D, et al. Zymophagy, a novel selective autophagy pathway mediated by VMP1-USP9x-p62, prevents pancreatic cell death. J Biol Chem. 2011;286(10):8308–8324. doi: 10.1074/jbc.M110.197301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rott R, et al. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Natl Acad Sci U S A. 2011;108(46):18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naik E, Webster JD, DeVoss J, Liu J, Suriben R, Dixit VM. Regulation of proximal T cell receptor signaling and tolerance induction by deubiquitinase Usp9X. J Exp Med. 2014;211(10):1947–1955. doi: 10.1084/jem.20140860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park Y, Jin HS, Liu YC. Regulation of T cell function by the ubiquitin-specific protease USP9X via modulating the Carma1-Bcl10-Malt1 complex. Proc Natl Acad Sci USA. 2013;110(23):9433–9438. doi: 10.1073/pnas.1221925110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jolly LA, Taylor V, Wood SA. USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol Biol Cell. 2009;20(7):2015–2029. doi: 10.1091/mbc.e08-06-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engel K, et al. USP9X stabilizes XIAP to regulate mitotic cell death and chemoresistance in aggressive B-cell lymphoma. EMBO Mol Med. 2016;8(8):851–862. doi: 10.15252/emmm.201506047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfsperger F, et al. Deubiquitylating enzyme USP9x regulates radiosensitivity in glioblastoma cells by Mcl-1-dependent and -independent mechanisms. Cell Death Dis. 2016;7:e2039. doi: 10.1038/cddis.2015.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toloczko A, et al. Deubiquitinating enzyme USP9X suppresses tumor growth via LATS kinase and core components of the hippo pathway. Cancer Res. 2017;77(18):4921–4933. doi: 10.1158/0008-5472.CAN-16-3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li L, et al. The deubiquitinase USP9X promotes tumor cell survival and confers chemoresistance through YAP1 stabilization. Oncogene. 2018;37(18):2422–2431. doi: 10.1038/s41388-018-0134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khan OM, et al. The deubiquitinase USP9X regulates FBW7 stability and suppresses colorectal cancer. J Clin Invest. 2018;128(4):1326–1337. doi: 10.1172/JCI97325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murtaza M, Jolly LA, Gecz J, Wood SA. La FAM fatale: USP9X in development and disease. Cell Mol Life Sci. 2015;72(11):2075–2089. doi: 10.1007/s00018-015-1851-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karpel-Massler G, et al. Inhibition of deubiquitinases primes glioblastoma cells to apoptosis in vitro and in vivo. Oncotarget. 2016;7(11):12791–12805. doi: 10.18632/oncotarget.7302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang B, et al. Deubiquitinase USP9X deubiquitinates β-catenin and promotes high grade glioma cell growth. Oncotarget. 2016;7(48):79515–79525. doi: 10.18632/oncotarget.12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trivigno D, Essmann F, Huber SM, Rudner J. Deubiquitinase USP9x confers radioresistance through stabilization of Mcl-1. Neoplasia. 2012;14(10):893–904. doi: 10.1593/neo.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhat KPL, et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim SH, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-κB-dependent manner. Cancer Cell. 2016;29(2):201–213. doi: 10.1016/j.ccell.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Arcy P, Linder S. Molecular pathways: translational potential of deubiquitinases as drug targets. Clin Cancer Res. 2014;20(15):3908–3914. doi: 10.1158/1078-0432.CCR-14-0568. [DOI] [PubMed] [Google Scholar]
- 43.D’Arcy P, Wang X, Linder S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol Ther. 2015;147:32–54. doi: 10.1016/j.pharmthera.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 44.Peng C, et al. The error-prone DNA polymerase κ promotes temozolomide resistance in glioblastoma through Rad17-dependent activation of ATR-Chk1 signaling. Cancer Res. 2016;76(8):2340–2353. doi: 10.1158/0008-5472.CAN-15-1884. [DOI] [PubMed] [Google Scholar]
- 45.Hu J, et al. MiR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell. 2016;29(1):49–60. doi: 10.1016/j.ccell.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, et al. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J Clin Invest. 2014;124(10):4489–4502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.