Abstract
The serine/threonine kinases BRAF and CRAF are critical components of the MAPK signaling pathway that is activated in many cancer types. In approximately 1% of melanomas, BRAF or CRAF is activated through structural arrangements. We describe a metastatic melanoma with a GOLGA4-RAF1 fusion and pathogenic variants in catenin β 1 (CTNNB1) and cyclin-dependent kinase inhibitor 2A (CDKN2A). Anti–cytotoxic T-lymphocyte–associated protein 4/anti–programmed cell death 1 (anti-CTLA4/anti–PD-1) combination immunotherapy failed to control tumor progression. In the absence of other actionable variants, the patient was administered MEK inhibitor therapy on the basis of its potential action against RAF1 fusions. This resulted in a profound and clinically significant response. We demonstrated that GOLGA4-RAF1 expression was associated with ERK activation, elevated expression of the RAS/RAF downstream coeffector ETV5, and a high Ki67 index. These findings provide a rationale for the dramatic response to targeted therapy. This study shows that molecular characterization of treatment-resistant cancers can identify therapeutic targets and personalize therapy management, leading to improved patient outcomes.
Keywords: Genetics, Oncology
Keywords: Cancer, Melanoma, Molecular pathology
Introduction
MAPK pathway activation is a hallmark of many cancers. Many genes in this pathway, including RAS, RAF (encoding the serine/threonine kinases BRAF and CRAF), and MEK can contain activating mutations. BRAF mutations are found in 37% to 50% of melanomas (1, 2). CRAF, encoded by RAF1, forms homodimers as well as heterodimers with WT BRAF, thereby functioning as an alternative mitogenic signaling mechanism (3).
RAF1 structural variants have been reported in several malignancies, including melanoma, pilocytic glioma, and acinar pancreatic cancers (1, 4–11). These variants fuse the RAF1 kinase domain to diverse 5′ partners, but despite potential clinical actionability, patients in the majority of cases have not received targeted treatment. In vitro studies have demonstrated that RAF1 fusions are sensitive to the multi-kinase inhibitor sorafenib and are partially responsive to the MEK inhibitors (MEKi) selumetinib and trametinib (5, 11). A clinical response to trametinib was also reported in a patient with metastatic melanoma with an ANO10-RAF1 fusion (10).
We describe the case of a patient who carried WT BRAF and NRAS alleles and failed to respond to metastatic melanoma, who failed to respond to immune checkpoint therapy. Molecular analysis identified a GOLGA4-RAF1 fusion. The patient subsequently showed a profound, albeit transient, clinically significant response to MEKi therapy.
Results and Discussion
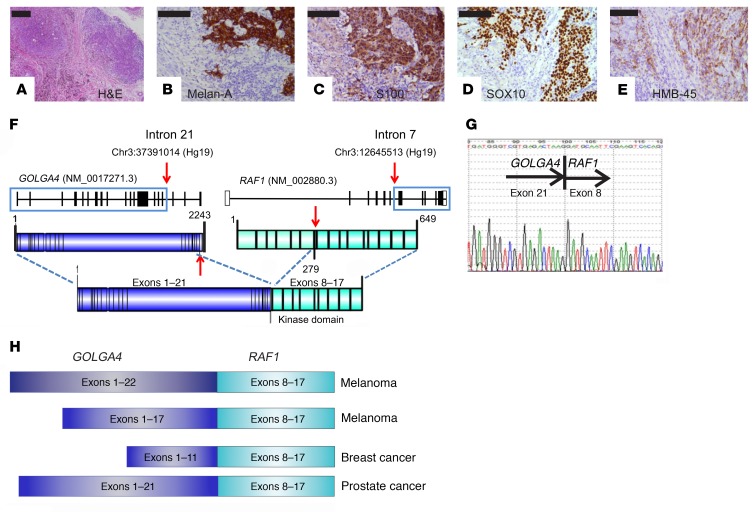
A 70-year-old man presented with bulky right inguinal lymph nodes, with the biopsy indicating metastatic melanoma. A fluorodeoxyglucose-PET CT (FDG-PET/CT) and MRI brain scan did not locate a primary malignancy or additional metastases. We performed right inguinal lymphadenectomy and retrieved 8 lymph nodes from the patient. Histological analysis showed near-complete replacement of 3 of these nodes by metastatic melanoma, up to 45 mm in maximal dimension, with 10 mm of extranodal extension. The tumor cells had mixed spindle and epithelioid morphology. IHC showed strong expression of S100 calcium-binding protein (S100), Melan-A, SRY-box 10 (SOX10), and human melanoma black (HMB-45) (Figure 1, A–E). Sequence analysis of BRAF (exons 11 and 15), KRAS (exons 2, 3 and 4), and NRAS (exons 2, 3, and 4) was performed using Fluidigm Access Array. All sequencing was confirmed as WT.
Figure 1. Metastatic melanoma with a GOLGA4-RAF1 fusion.
(A) H&E staining of a lymph node metastasis. (B–E) IHC for Melan-A (B), S100 (C), SOX10 (D), and HMB-45 (E). (F) Schematic illustration of the GOLGA4-RAF1 fusion. (G) Sanger sequence of the RT-PCR product. (H) Illustration of previously described GOLGA4-RAF1 fusions showing consistent retention of the RAF1 kinase domain (exons 8–17). Scale bars: 100 μm.
Immune checkpoint therapy is the standard of care for patients with metastatic melanoma who do not have an activating BRAF mutation (see the Supplemental Material; supplemental material available online with this article; doi:10.1172/JCI123089DS1). The patient was administered 3 cycles of treatment with the anti–PD-1 inhibitor pembrolizumab but experienced a rapid clinical decline. Imaging confirmed marked interval disease progression with FDG-avid nodal, soft tissue, pulmonary, hepatic, splenic, and cardiac involvement. He was then administered anti-CTLA4 (ipilimumanb) and anti–PD-1 (nivolumab) combination immunotherapy.
We performed extensive molecular testing to identify an actionable target, and detected a GOLGA4-RAF1 gene fusion (Figure 1F). Subsequent tests using both reverse transcription PCR (RT-PCR) with Sanger sequencing and the Archer FusionPlex Oncology Research Panel showed an in-frame chimeric mRNA of GOLGA4 exons 1–21 fused to RAF1 exons 8–17 (Figure 1G). The entire coding regions of other MAPK pathway genes (BRAF, NRAS, KIT, GNAS, GNAQ, KRAS, MAP2K1/2, MAPK1/2, and NF1) were WT.
RAF1 and BRAF gene fusions are rare but potentially clinically significant (1, 4–11). They involve fusion of a novel N-terminal sequence to the C-terminal kinase domain of CRAF. GOLGA4 encodes golgin-245 and is a recurrent 5′ partner in RAF1 fusions that have been previously described in melanoma (1), breast carcinoma (8), and prostate cancer (9) (Figure 1H). In the present case, the fusion included the entire CRAF kinase domain but lacked the CRAF N-terminal Ras-binding domain (RBD) and C1 autoinhibitory domain (Figure 1F). It also lacked the GRIP domain of Golgin-245 that is required for recruitment to the trans-Golgi network.
We also observed gains of chromosomal regions 5q, 8q, 13q, and 17q21.31-qter, focal amplification (6 copies) of 12p11.21-12.2 (which harbors KRAS), and losses of chromosome arms 6q, 9p (which harbors CDKN2A), 10p, and 10q. Mutational signatures of exposure to high UV and alkylating agents were present. We also detected 2 somatic pathogenic variants: a missense mutation in CTNNB1 (NCBI reference sequence NM_001098210.1) c.110C>G p.(Ser37Cys) and a truncating mutation in CDKN2A (NCBI reference sequence NM_000077.4) c.148C>T p.(Gln50*).
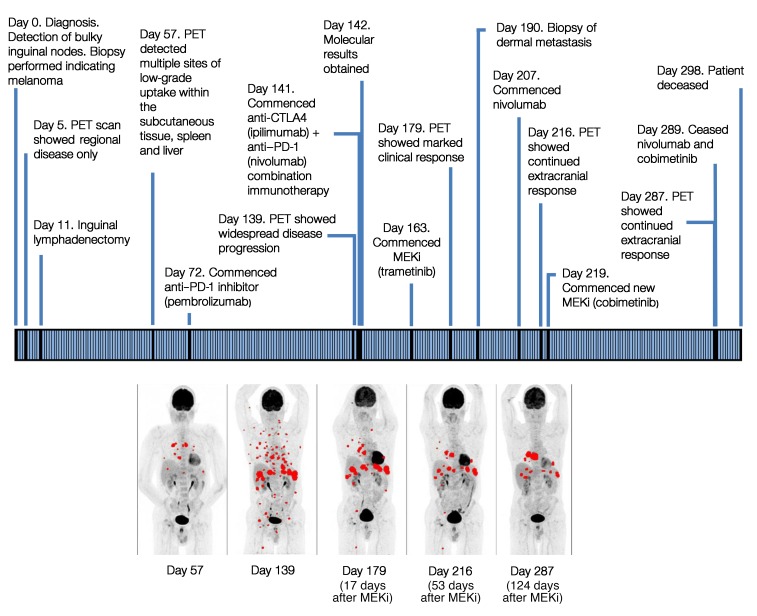
At the time of returning the genetic results, the patient had developed further disease progression with new onset of brain metastatic disease despite the use of combination immunotherapy (Figure 2). His case was reviewed by our institute’s molecular tumor board. In view of the experimental evidence that tumor cells with BRAF-CRAF fusion proteins are potentially sensitive to multikinase inhibitors (5–7, 12) or MEKi (10), the decision was made to proceed with the single agent trametinib, a reversible, highly selective allosteric inhibitor of MEK1/-2 (see the Supplemental Material). An FDG-PET performed 17 days after treatment commencement showed a marked response, with both metabolic and structural responses seen on extracranial imaging (Figure 2). The patient’s Eastern Cooperative Oncology Group (ECOG) performance status also improved from 3 to 1. However, we observed progressive intracranial disease. This is a well-documented phenomenon in melanoma treatment, with both immunotherapy and targeted agents demonstrating lower rates of response in the brain (13, 14). The patient was subsequently treated with whole-brain radiotherapy, and nivolumab treatment was added. After 6 weeks of therapy, the treatment was changed to another potent and highly selective MEK1/-2 inhibitor, cobimetinib, because of problems with trametinib access (see Supplemental Material). Treatment continued for 5 months, during which the patient continued to have good extracranial control of the disease (Figure 2). Imaging showed improvement in subcutaneous and visceral disease, with a rapid decline in blood LDH levels. Unfortunately, the tumor progressed within the brain, and the patient ceased MEKi and nivolumab therapy 10 days prior to his death.
Figure 2. Clinical timeline for the patient, with maximum-intensity projection FDG-PET imaging times noted.
Maximum-intensity projection (MIP) FDG-PET (MIP PET) images show a profound clinical response after MEKi treatment. Day 57: image shows baseline metastatic disease before MEKi treatment. Day 139: disease progression seen on follow-up study. Day 179 (17 days after MEKi): significant metabolic response. Day 216 (53 days after MEKi) and day 287 (124 days after MEKi): continued metabolic response. The maximum standardized uptake value (SUVmax) of a representative target lesion declined from 9.0 on day 139 (before MEKi) to 6.0 on day 179, to 4.2 on day 216, and to 1.8 on day 287. All FDG-avid metastatic sites are shown in red.
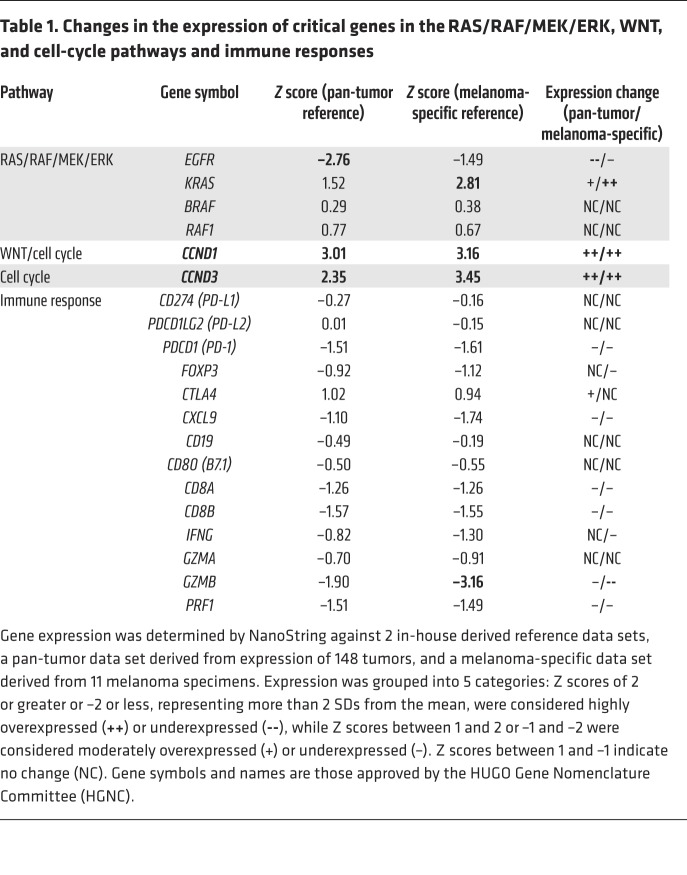
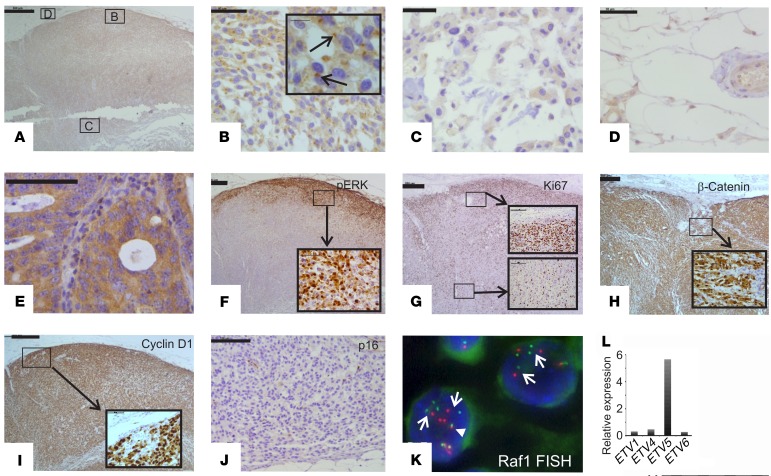
We investigated the tumor biology in order to better understand the consequences of the RAF1 fusion and other genomic alterations. Expression levels of RAF1 and BRAF RNA were not significantly elevated (Table 1). However, IHC with an anti-CRAF antibody showed intense paranuclear and moderate diffuse cytoplasmic staining in the peripheral tumor cells (Figure 3B), but only weak paranuclear and cytoplasmic staining in the center of the tumor (Figure 3C). This pattern resembles that seen with WT GOLGA4, which localizes to the Golgi and cytosol in human tissue (15). Benign tissue either adjacent to or within the tumor (vasculature and adipose stroma) showed weak cytoplasmic or no staining (Figure 3D). Diffuse cytoplasmic staining without paranuclear expression was seen in our on-slide positive control (a colorectal adenocarcinoma) for the CRAF antibody (Figure 3E). Strong nuclear phosphorylated ERK (p-ERK) staining was restricted to the same peripheral regions that showed intense CRAF expression (Figure 3F). The E twenty-six (ETS) family of transcription factors (ETV proteins) are critical downstream coeffectors of MAPK signaling and modulate MEK and RAF inhibitor responses (16). Expression of ETV5 in particular has been found to correlate with BRAF V600E activation in melanoma (17). RNA-Seq using Archer FusionPlex showed markedly elevated expression of ETV5, but not ETV1, -4, or -6 (Figure 3L), consistent with activated MAPK signaling. We predicted that activation of MAPK signaling would lead to increased cell proliferation. Ki67 labeling for actively proliferating tumor cells showed marked regional variation (Figure 3G). It was highest within the same peripheral foci that also showed strong expression of CRAF and p-ERK (Figure 3, A–C and F, performed on adjacent serial sections). The central parts of the tumor and the peripheral regions with low CRAF and p-ERK expression had a low Ki67-labeling index (Figure 3G).
Table 1. Changes in the expression of critical genes in the RAS/RAF/MEK/ERK, WNT, and cell-cycle pathways and immune responses.
Figure 3. Expression of key MAPK, WNT, and cell-cycle proteins.
(A–E) IHC for CRAF in (A–D) a lymph node metastasis and in (E) an on-slide positive control (colorectal adenocarcinoma). The locations of the images in B–D (boxes) are shown in A. Arrows in the enlarged inset image in B point to strong paranuclear staining in peripheral tumor cells in the lymph node metastasis. Weak cytoplasmic staining is evident in the central tumor cells in the lymph node metastasis (C) and in the perinodal stromal cells (D), and strong diffuse cytoplasmic staining is seen, without paranuclear accentuation, in the colorectal adenocarcinoma (E). (F) IHC for p-ERK shows strong nuclear staining and weak cytoplasmic staining in peripheral tumor cells in the lymph node metastasis (enlarged inset image). (G) IHC for Ki67 shows a high percentage of labeled tumor cells in the periphery but not the center of the lymph node metastasis. Note the expression pattern, which is similar to that of p-ERK. (H and I) IHC for β-catenin (H) and cyclin D1 (I) shows strong nuclear and moderate cytoplasmic staining in both peripheral and central tumor cells (enlarged inset images). (J) IHC for p16 shows no staining in tumor cells (the occasional positive staining is seen in stromal cells). (K) Break-apart RAF1 FISH in tumor cells in the lymph node metastasis. Note the multiple widely separated 5′ (green, centromeric) and 3′ (red, telomeric) signals in tumor cells (arrows). A fused 5′ and 3′ signal is shown for comparison (arrowhead). (L) Archer Dx RNA analysis of relative expression of ETS family members shows elevated expression of the RAS/RAF downstream coeffector ETV5. Scale bars: 500 μm (A and F–I), 100 μm (E and J), and 50 μm (B–D); enlarged insets: 10 μm (B), 100 μm (G), 50 μm (F, H, and I).
CTNNB1 activation and CDKN2A inactivation are known drivers of melanoma (18, 19). CTNNB1 Ser37Cys impairs a critical GSK-3β phosphorylation site, leading to aberrant β-catenin accumulation in the nucleus (20), where it functions as a coactivator of transcription factor 4 (TCF4) and lymphoid enhancer binding factor 1 (LEF1), resulting in the transcription of genes that include cyclin D1 (CCND1) and thereby promoting cell proliferation (21). NanoString analysis revealed markedly upregulated CCND1 (Table 1). IHC showed strong nuclear and moderate cytoplasmic expression of β-catenin and CCND1 throughout the nodal metastasis (Figure 3, H and I). NanoString analysis also showed elevated expression of cyclin D3 (CCND3) and KRAS (Table 1), the latter being consistent with the KRAS amplification. Expression of KIT was moderately elevated, as shown by NanoString analysis, and anti-KIT IHC showed moderately intense cytoplasmic staining throughout the nodal tumor (data not shown). Loss of functional p16 (also known as INK4A), primarily through deletion, occurs in approximately 80% to 90% of metastatic melanomas (22). p16 is an inhibitor of cyclin-dependent kinases 4 and 6 ((CDK4) and CDK6), which regulate progression through the G1 phase of the cell cycle (23). IHC showed no expression of p16 in tumor cells, consistent with a “double hit” of CDKN2A, i.e., chromosome 9p loss and an inactivating c.148C>T p.(Gln50*) mutation (Figure 3J).
Overall, the strong spatial correlation between Ki67 labeling and elevated expression of CRAF and p-ERK as well as the significant extracranial response to the MEKi indicated that GOLGA4/CRAF signaling through p-ERK was the critical oncogenic driver. The heterogeneity of GOLGA4-CRAF expression was intriguing and prompted us to perform RAF1 break-apart FISH to determine whether all tumor cells carried GOLGA4-RAF1. We detected split RAF1 5′ and 3′ signals in all tumor cells, i.e., in both the periphery and center of the tumor (Figure 3K). We speculate that the differences in CRAF expression are due to differences in GOLGA4-CRAF protein stability between the periphery and center, possibly because peripheral tumor cells may be able to more readily access nutrients and oxygen than central tumor cells can. Alternatively, since the periphery only represented a minority of the total tumor input in the NanoString analysis, GOLGA4-RAF1 mRNA expression may be elevated at the periphery of the tumor. The RAF1 FISH analysis also showed that GOLGA4-RAF1 was amplified in all tumor cells. There were 5–10 rearranged RAF1 copies per tumor nucleus (Figure 3K), with a RAF1/chromosome 3 ratio of approximately 4. GOLGA4-RAF1 amplification further indicates that activated CRAF is a critical driver. We excised a dermal metastasis from the patient on day 190, which was 28 days after the commencement of MEKi treatment. The excised metastasis had low tumor cell proliferative activity and morphological features of tumor regression, both likely due to the MEKi treatment (see the Supplemental Material and Supplemental Figure 1).
The GOLGA4-CRAF fusion is likely to have high CRAF kinase activity, since the RBD and C1 domains, which suppress CRAF activity (24), are absent, and Golgin proteins can spontaneously multimerize the kinase domains of their fusion partners, leading to increased kinase activity (25). The likely Golgi expression of GOLGA4-CRAF is intriguing, since the GRIP domain of GOLGA4, which is sufficient for Golgi localization, was absent. Nevertheless, it is possible that GOLGA4-CRAF promotes tumor cell proliferation by aiding Golgi complex fragmentation at cell division, because CRAF and MEK1 are both implicated in this process (26).
Biomarker analysis may partly explain the lack of response to immune checkpoint therapy. We found no increase in the expression of programmed cell death 1 (PD-1), programmed death–ligand 1 (PD-L1), or PD-L2 compared with pan-cancer and melanoma-specific reference data sets (Table 1). Expression levels of cytotoxic T cell markers, including granzyme B and perforin, which are typically associated with cytotoxic T cell killing (27), were low compared with levels indicated in both reference data sets. The lack of tumor-infiltrating lymphocytes in the tumor may partly account for the low expression of these T cell markers.
In summary, we describe a metastatic melanoma with a GOLGA4-RAF1 fusion that showed a profound extracranial response to MEKi. This adds to a growing body of evidence demonstrating success in the therapeutic targeting of RAF structural variants. The correlation of activated RAS/RAF/MEK/ERK signaling and cell proliferation provided a rationale for the therapeutic response. We also identified aberrant WNT activation and probable CDK4 and CDK6 activation mediated through both increased cyclin D1 and cyclin D3 expression and p16 loss. While inhibitors of WNT signaling remain at an early stage of development, CDK4 and CDK6 inhibitors targeting cell-cycle dysregulation have been approved by the FDA for specific breast cancer indications. Therefore, combined anti-MEK and anti-CDK4 and anti-CDK6 therapy may be a potential therapeutic option in RAF fusion–driven tumors.
Methods
A complete description of the methods is provided in the Supplemental Material.
Study approval.
All experiments were performed under protocols reviewed and approved by iPREDICT (approval no. HREC/13/MH/326) and SUPER (Solving Unknown Primary cancER) (approval no. 11/117) studies and the Peter MacCallum Cancer Centre (Melbourne, Australia).
Author contributions
CRM, HX, and OWJP coordinated the project and wrote the manuscript. KS analyzed and prepared the clinical data. CRM and DYC performed molecular analysis. HX, HSL, and APF analyzed the sequencing results. DE performed the NanoString experiments and analysis. DJB and JB performed IHC. VN and BMB performed FISH. OWJP analyzed tumor morphology, IHC results, and FISH. AI analyzed the patient’s imaging results. CK was the treating clinician. SB, JD, DDB, and LM coordinated patient recruitment and ensured human ethics compliance. RWT, AHB, SBF, and all other authors provided intellectual input.
Supplementary Material
Acknowledgments
We thank Melbourne Genomic Health Alliances (MGHA) and the Australian Genomic Health Alliance (NHMRC grant 1113531) for funding the I-PREDICT study. We also thank the following individuals at the Peter MacCallum Cancer Centre: Richard Lupat for assistance with GRIDSS (Genome Rearrangement IDentification Software Suite) analysis; Jason Li for setting up the analysis pipeline for the nCounter data set; Jenna Stewart and David Yoannidis for technical assistance; Kelly Waldek for advice on IHC; Chung-Yan Ma for discussions on immunomarkers; and Michael McKay and Glen Gurra for critical discussions of the manuscript.
Version 1. 03/05/2019
In-Press Preview
Version 2. 04/02/2019
Electronic publication
Version 3. 05/01/2019
Print issue publication
Funding Statement
No specific Grant number available
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, The American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(5):1940–1945.https://doi.org/10.1172/JCI123089.
Contributor Information
Christopher R. McEvoy, Email: christopher.mcevoy@petermac.org.
Huiling Xu, Email: huiling.xu@petermac.org.
Kortnye Smith, Email: kortnye.smith@petermac.org.
Dariush Etemadmoghadam, Email: detemad@gmail.com.
Huei San Leong, Email: HueiSan.Leong@petermac.org.
David Y. Choong, Email: david.choong@petermac.org.
David J. Byrne, Email: david.byrne@petermac.org.
Amir Iravani, Email: Amir.Iravani@petermac.org.
Sophie Beck, Email: sophie.beck@petermac.org.
Linda Mileshkin, Email: linda.mileshkin@petermac.org.
Richard W. Tothill, Email: rtothill@unimelb.edu.au.
David D. Bowtell, Email: david.bowtell@petermac.org.
Bindi M. Bates, Email: bindi.bates@petermac.org.
Violeta Nastevski, Email: Vicky.Nastevski@petermac.org.
Judy Browning, Email: Judy.Browning@petermac.org.
Anthony H. Bell, Email: anthony.bell@petermac.org.
Chloe Khoo, Email: chloe.khoo@hotmail.com.
Jayesh Desai, Email: Jayesh.Desai@mh.org.au.
Andrew P. Fellowes, Email: andrew.fellowes@petermac.org.
Stephen B. Fox, Email: Stephen.Fox@petermac.org.
Owen W.J. Prall, Email: owen.prall@petermac.org.
References
- 1.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network Genomic Classification of Cutaneous Melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lavoie H, Therrien M. Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol. 2015;16(5):281–298. doi: 10.1038/nrm3979. [DOI] [PubMed] [Google Scholar]
- 4.Chmielecki J, et al. Comprehensive genomic profiling of pancreatic acinar cell carcinomas identifies recurrent RAF fusions and frequent inactivation of DNA repair genes. Cancer Discov. 2014;4(12):1398–1405. doi: 10.1158/2159-8290.CD-14-0617. [DOI] [PubMed] [Google Scholar]
- 5.Palanisamy N, et al. Rearrangements of the RAF kinase pathway in prostate cancer, gastric cancer and melanoma. Nat Med. 2010;16(7):793–798. doi: 10.1038/nm.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yde CW, et al. A new NFIA:RAF1 fusion activating the MAPK pathway in pilocytic astrocytoma. Cancer Genet. 2016;209(10):440–444. doi: 10.1016/j.cancergen.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Jones DT, Kocialkowski S, Liu L, Pearson DM, Ichimura K, Collins VP. Oncogenic RAF1 rearrangement and a novel BRAF mutation as alternatives to KIAA1549:BRAF fusion in activating the MAPK pathway in pilocytic astrocytoma. Oncogene. 2009;28(20):2119–2123. doi: 10.1038/onc.2009.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmaier RJ, et al. High-Throughput Genomic Profiling of Adult Solid Tumors Reveals Novel Insights into Cancer Pathogenesis. Cancer Res. 2017;77(9):2464–2475. doi: 10.1158/0008-5472.CAN-16-2479. [DOI] [PubMed] [Google Scholar]
- 9.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161(5):1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. doi: 10.1200/PO.17.00138. Kim KB, et al. Significant clinical response to a MEK inhibitor therapy in a patient with metastatic melanoma harboring an RAF1 fusion [published online January 17, 2018]. JCO Precision Oncology . [DOI] [PubMed]
- 11.Jain P, et al. CRAF gene fusions in pediatric low-grade gliomas define a distinct drug response based on dimerization profiles. Oncogene. 2017;36(45):6348–6358. doi: 10.1038/onc.2017.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Phillips JJ, et al. Activating NRF1-BRAF and ATG7-RAF1 fusions in anaplastic pleomorphic xanthoastrocytoma without BRAF p.V600E mutation. Acta Neuropathol. 2016;132(5):757–760. doi: 10.1007/s00401-016-1616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davies MA, et al. Dabrafenib plus trametinib in patients with BRAFV600-mutant melanoma brain metastases (COMBI-MB): a multicentre, multicohort, open-label, phase 2 trial. Lancet Oncol. 2017;18(7):863–873. doi: 10.1016/S1470-2045(17)30429-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long GV, et al. A randomized phase II study of nivolumab or nivolumab combined with ipilimumab in patients (pts) with melanoma brain metastases (mets): The Anti-PD1 Brain Collaboration (ABC) J Clin Oncol. 2017;35(no. 15_suppl):9508. doi: 10.1200/JCO.2017.35.15_suppl.9508. [DOI] [Google Scholar]
- 15.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 16.Wang B, et al. ATXN1L, CIC, and ETS Transcription Factors Modulate Sensitivity to MAPK Pathway Inhibition. Cell Rep. 2017;18(6):1543–1557. doi: 10.1016/j.celrep.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pratilas CA, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106(11):4519–4524. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275(5307):1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 19.Grafström E, Egyházi S, Ringborg U, Hansson J, Platz A. Biallelic deletions in INK4 in cutaneous melanoma are common and associated with decreased survival. Clin Cancer Res. 2005;11(8):2991–2997. doi: 10.1158/1078-0432.CCR-04-1731. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, et al. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108(6):837–847. doi: 10.1016/S0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- 21.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149(6):1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 22.Sheppard KE, McArthur GA. The cell-cycle regulator CDK4: an emerging therapeutic target in melanoma. Clin Cancer Res. 2013;19(19):5320–5328. doi: 10.1158/1078-0432.CCR-13-0259. [DOI] [PubMed] [Google Scholar]
- 23.O’Leary B, Finn RS, Turner NC. Treating cancer with selective CDK4/6 inhibitors. Nat Rev Clin Oncol. 2016;13(7):417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 24.Cutler RE, Stephens RM, Saracino MR, Morrison DK. Autoregulation of the Raf-1 serine/threonine kinase. Proc Natl Acad Sci U S A. 1998;95(16):9214–9219. doi: 10.1073/pnas.95.16.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Troadec E, et al. A novel t(3;13)(q13;q12) translocation fusing FLT3 with GOLGB1: toward myeloid/lymphoid neoplasms with eosinophilia and rearrangement of FLT3? Leukemia. 2017;31(2):514–517. doi: 10.1038/leu.2016.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colanzi A, Sutterlin C, Malhotra V. RAF1-activated MEK1 is found on the Golgi apparatus in late prophase and is required for Golgi complex fragmentation in mitosis. J Cell Biol. 2003;161(1):27–32. doi: 10.1083/jcb.200208099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voskoboinik I, Whisstock JC, Trapani JA. Perforin and granzymes: function, dysfunction and human pathology. Nat Rev Immunol. 2015;15(6):388–400. doi: 10.1038/nri3839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.