Abstract
The human metapneumovirus (HMPV) fusion protein (F) mediates fusion of the viral envelope and cellular membranes to establish infection. HMPV F from some, but not all, viral strains promotes fusion only after exposure to low pH. Previous studies have identified several key residues involved in low pH triggering, including H435 and a proposed requirement for glycine at position 294. We analyzed the different levels of fusion activity, protein expression and cleavage of three HMPV F proteins not previously examined. Interestingly, low pH-triggered fusion in the absence of G294 was identified in one F protein, while a novel histidine residue (H434) was identified that enhanced low pH promoted fusion in another. The third F protein failed to promote cell-to-cell fusion, suggesting other requirements for F protein triggering. Our results demonstrate HMPV F triggering is more complex than previously described and suggest a more intricate mechanism for fusion protein function and activation.
Keywords: human metapneumovirus, HMPV, fusion, syncytia, strains, clade, low pH
Introduction
Human metapneumovirus (HMPV) is a recently discovered enveloped, negative-sense, single-stranded RNA virus. Although first identified in 2001, HMPV has now been shown to be a cause for respiratory tract infections in humans worldwide since at least 1958 (1–3). Nearly everyone is initially infected by five years of age and reinfection is common throughout life (4). Infection leads to a variety of symptoms ranging from coughing and wheezing to pneumonia and bronchiolitis, potentially requiring to hospitalization in severe cases. In addition, infants, immunocompromised, and elderly patients most likely to develop severe infections (1, 5–11). While HMPV is ubiquitous and responsible for severe upper and lower respiratory tract infections, there is still no FDA approved antiviral treatment or vaccination available. Therefore, a more thorough understanding of the viral lifecycle and molecular mechanisms required for infection are needed to discover novel antiviral targets.
HMPV is phylogenetically classified into two genetic lineages (A and B) and further characterized into sub-lineages (A1, A2, B1 and B2) based on the sequences of two surface glycoproteins: the fusion protein (F) and the attachment protein (G) (12). To infect cells, enveloped viruses fuse their membrane with host cell membranes, a process mediated by one or more viral surface glycoproteins. In the instance of HMPV, this process is mediated by F alone in vitro and in vivo, whereas closely related paramyxoviruses require both F and G (13–15). F is a homo-trimeric class I fusion protein present within viral membranes as well as membranes of infected host cells. To become activated, F is proteolytically cleaved from the precursor form (F0), into the metastable, disulfide-linked heterodimer (F1+F2) (16, 17). Cleavage can be accomplished by the addition of exogenous trypsin in vitro (15), although in vivo it is thought that F is cleaved by secreted or cell surface proteases present in the host. Once cleaved, HMPV F can be triggered to undergo an essentially irreversible and energetically favorable conformational change from the pre-fusion form to the post-fusion state with released potential energy driving membrane fusion (15, 18–20).
HMPV particles have been shown to be internalized via clathrin mediated endocytosis in human bronchial epithelial cells through a dynamin dependent mechanism (20, 21). Further evidence demonstrated that for HMPV, viral and host membrane fusion takes place within the endosomes (21). Some strains of HMPV, mainly within clade A, utilize low pH generated through endosomal acidification as a mechanism to trigger the fusion protein, similar to HA from influenza. However, this is proposed to not be true for all strains of HMPV (15, 18–21). For fusion proteins that are triggered by low pH, it is hypothesized that repulsive electrostatic forces between critical residues lead to global protein destabilization, initiating the conformational transition from the pre-fusion to post-fusion state (22–24). It has been proposed that specific histidine (H) residues become protonated at low pH and subsequently interact with neighboring basic residues to destabilize the pre-fusion state and initiate membrane fusion. In HMPV F, H435 within the globular head is thought to serve as a pH sensor (19, 20). Recently, a high resolution structure of a stabilized pre-fusion HMPV F [NL/1/00(A1)] was solved [PDB: 5WB0] (25). This structure revealed that lysine (K) 20 and glutamic acid (E) 433 interact to form a potential salt bridge. Under low pH conditions, protonation of the neighboring H435 may lead to cation electrostatic repulsion driving conformational changes and promotes membrane fusion. Studies with recombinant HMPV containing mutations in this region have confirmed its importance for viral infectivity (19, 26). Additional residues have been identified as playing a role in low pH triggered fusion, including K296, W396, and N404. Furthermore, studies using F proteins from prototype strains from each clade have suggested that fusion induced by low pH is restricted to clade A virus fusion proteins, and glycine (G) 294 is critical for low pH triggered fusion (18, 19). However, few HMPV F proteins have been studied in each clade and therefore additional analysis is needed to further understand this mechanism.
In this study, we examined three previously uncharacterized HMPV F proteins for their fusion activity, protein expression, and cleavage activation levels. The first F protein, cloned from TN83–1211, contained a unique H434 residue, adjacent to a previously characterized histidine at 435 demonstrated to be critical for low pH fusion. Protein mutagenesis in our reference strain supports its contribution to increased fusion at low pH and more efficient cleavage by trypsin. The second F protein, cloned from TN94–49, is able to promote low pH mediated fusion without G294, although this residue was previously identified as critical for this mechanism of membrane fusion. The third HMPV F protein, cloned from TN96–12, contains E at position 294. Interestingly, TN96–12 was unable to mediate fusion at either neutral or low pH conditions, or in the presence of the attachment protein, G. This finding suggests additional factors are necessary to trigger the F protein. Taken together, these results further demonstrate the complexity of HMPV F mediated membrane fusion and the significant phenotypic differences observed with only a few amino acid changes.
Materials and Methods
Cell lines
Vero cells(ATCC) and BSR cells (provided by Karl-Klaus Conzelmann, Max Pettenkofer Institut) were grown in Dulbecco’s modified Eagle’s medium (DMEM; Gibco Invitrogen, Carlsbad, CA), supplemented with 10% fetal bovine serum (FBS, Sigma). BSR cell media was supplemented with 0.5 mg/ml G-418 sulfate (Gibco Invitrogen, Carlsbad, CA) every third passage to select for T7 polymerase-expressing cells.
Plasmids and antibodies
HMPV virus strains were kindly provided by the designated sources: CAN97–83 (Peter Collins and Ursula J. Buchholz, NIAID), TN94–49, TN96–12 and TN83–1211 (BEI Resources). Viral RNA (vRNA) was isolated from viruses propagated in Vero cells by phenol-chloroform extraction and resuspended in 50μL DEPC treated water.
The F genes were amplified from vRNA by RT-PCR using the SuperscriptIII One Step RT-PCR system (Invitrogen) according to the manufacturer’s protocol with the following settings: 1 cycle at 55°C for 30 min, 1 cycle at 94°C for 2 min, 40 cycles of the following three steps: 94°C for 15 sec, 60°C for 2 min, 68°C for 1 min, then 1 cycle at 68°C for 5 min. The amplified product was cloned into the pCAGGS-MCS eukaryotic expression plasmid. The CAN97–83 434H F mutant was created using the gene cloned in pGEM-3Zf(+) by site-directed mutagenesis using QuikChange (Stratagene) and subsequently sub-cloned into pCAGGS-MCS. All F expression plasmids were sequenced in their entirety, and BioEdit was used for sequence analysis. Monoclonal antibody 54G10, which maps to the antigenic site 5/6 of HMPV F, was used for immunoprecipitation and expression assays. The epitope is conserved within the F proteins used in this study. (27, 28).
Syncytia assay
Subconfluent monolayers of Vero cells were transiently transfected with a total of 2 μg of DNA consisting of pCAGGS-HMPV F derived from each isolate or empty pCAGGS-MCS vector using Lipofectamine and Plus Reagents (Invitrogen) according to the manufacturer’s instructions, and a syncytia assay performed as described (15). Syncytia images were taken with a Nikon Coolpix995 camera mounted on a Nikon TS100 inverted phase-contrast microscope using a 5× objective. Quantification of syncytia formation is reported as a fusion index, as previously described (29). Briefly, the fusion index was calculated using the equation f = [1−(C/N)] where C is the number of cells in a field after fusion and N, the number of nuclei. Six fields were scored per condition representative of 3 independent experiments.
Reporter gene fusion assay
Vero cells were transfected using Lipofectamine and Plus Reagents with 1.5 μg pCAGGS-HMPV F derived from each F isolate or empty pCAGGS-MCS vector, and 1.5 μg of plasmid containing luciferase cDNA under the control of the T7 promoter (Promega). Three independent experiments were conducted for the luciferase reporter gene assay was performed as described (15).
Surface expression of proteins, metabolic labeling, and immunoprecipitation
Vero cells were transiently transfected with 2 μg pCAGGS expression vectors using Lipofectamine and Plus Reagents. At 18 to 24 hrs post-transfection, cells were starved in methionine- and cysteine-deficient DMEM for 45 min and then metabolically labeled with Tran35S-label (100 μCi/ml; MP Biomedicals) for 3 hr in the presence of 3.0 μg/mL TPCK-trypsin. Surface biotinylation and immunoprecipition using monoclonal antibody 54G10 and protein A-conjugated Sepharose beads (Amersham, Piscataway, N.J.) were performed as previously described (30). The immunoprecipitated F proteins were analyzed via SDS-15% polyacrylamide gel electrophoresis (SDS-PAGE) and visualized using the Typhoon imaging system. Four independent experiments were quantified.
Statistical analysis
One-way ANOVA analysis with post hoc Bonferroni multiple comparisons test was used to analyze the data using GraphPad Prism 7. Statistical significance is noted for P< 0.05 (*), P<0.005 (**), P<0.0005 (***) and P<0.0001 (****). Error bars indicate standard deviation.
Results
To examine HMPV fusion, F protein genes were cloned from three available HMPV strains: TN94–49, TN96–12 and TN83–1211, all isolated in the Williams laboratory and propagated and stored at BEI. These F isolates were sequenced and compared to a low pH prototype strain, CAN97–83, used as a positive control in these studies. HMPV F is highly conserved, and therefore few amino acid changes were detected between strains. The TN94–49 F clone differed from CAN97–83 F by only 6 amino acids (Fig. 1D, Fig. S1). One notable difference was residue 294, which contained K294 instead of G294, a glycine residue previously suggested as essential for low pH triggered fusion for clade A HMPV F proteins (18). When comparing TN96–12 F clone to CAN97–83 F, 9 amino acid changes were present, including E294 instead of G294. Lastly, the F isolate from TN83–1211 only differed from CAN97–83 by two amino acids, S175 and H434, and did not match the published isolate sequence of TN83–1211. However, this mutation was present in several clones analyzed, suggesting the viral stock may have had a heterogeneous population of virus present. For clarity, we refer to this protein as S175H434 F. The presence of a unique amino acid in S175H434, H434, instead of the highly conserved Q434 in all other published strains, led us to further study this F isolate. The neighboring residue H435 is hypothesized to play a critical role in electrostatic repulsion with neighboring residues after protonation at low pH, acting as a physiologic timing sensor to promote fusion. Based on these findings, we hypothesized that the second histidine at position 434 could potentially be involved in fusion of S175H434 after exposure to low pH, similar to the function observed for H435. In addition, TN94–49 containing K294 and TN96–12 containing E294 were of interest to assess the role of these amino acids in low pH mediated fusion compared to G294.
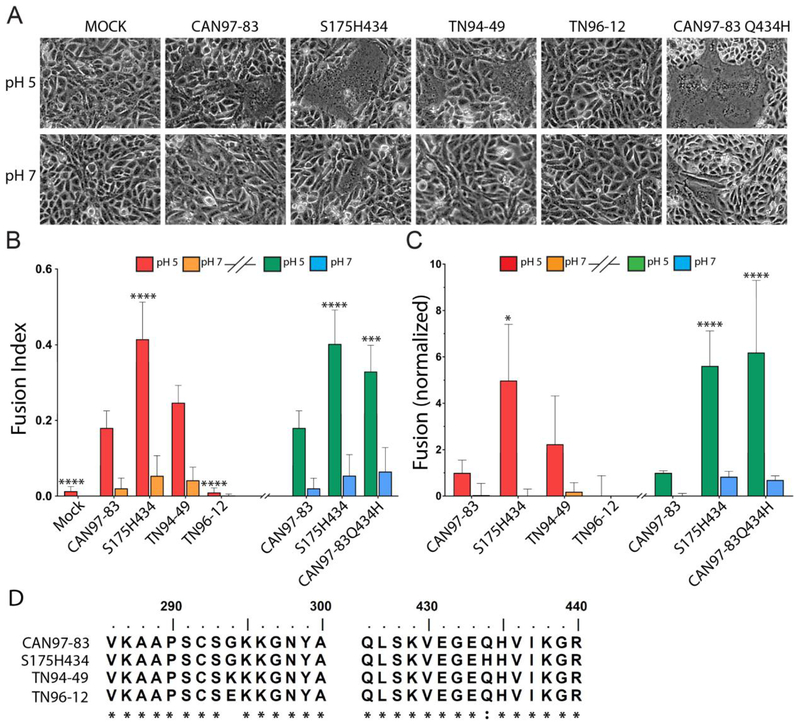
Fig 1. HMPV F proteins from different strains exhibit variable fusion activity promoted by low pH.
(A) Representative images of syncytia formation of cells expressing the HMPV F proteins after pulses at pH 5 or pH 7 (n = 3). (B) The fusion index was calculated using the equation f = [1 − (C/N)] where C is the number of cells in a field after fusion and N, the number of nuclei. Six fields were scored per condition representative of 3 independent experiments. “*”s indicate statistical significance compared to fusion for CAN97–83 (A2) F after pH 5 pulses (n=3) [* p<0.05, ** p<0.005, *** p<0.0005 and **** p<0.0001]. Graph was broken into two experiments (denoted by the graph break and colors: red/orange bars represent independent experiment from green/blue). Statistical significance within these assays is compared to CAN97–83 within each independent experiment. (C) Luciferase reporter gene assay of Vero cells transfected with HMPV F upon which BSR cells were overlaid and subjected to two pH pulses. Data are presented and normalized to CAN97–83 (A2) F luminescence (fusion) at pH 5 (n = 3) +/− standard deviation. * Indicates statistical significance compared to CAN97–83 F after pH 5 pulses. Graphical representation and statistics were conducted as described in B. (D) Partial protein sequence analysis of F from 4 strains of HMPV surrounding key residues at positions 294 and 435. Sequence alignment was generated using ClustalW. The asterisk “*” indicates identical residues and the colon “:” indicates conserved substitutions.
To determine whether these F isolates were able to promote neutral or low pH-mediated membrane fusion, we first conducted a syncytia assay. As previously reported, CAN97–83 F promoted fusion when exposed to low pH and was therefore used as a positive control for low pH triggered fusion (Fig. 1A). TN94–49 F generated syncytia following low pH treatment, similar to CAN97–83 F, but no syncytia formation was observed at neutral pH. Interestingly, TN96–12 F was unable to generate cell-to-cell fusion at both neutral and low pH, whereas S175H434 F generated minimal background syncytia at neutral pH and robust syncytia formation after exposure to low pH that was significantly higher than the syncytia observed with CAN97–83 F (Fig. 1A, quantified in 1B). To confirm our findings in the syncytia assay, we utilized a luciferase reporter assay as a second cell-to-cell fusion assay metric. Consistent with the syncytia assay, TN94–49 F exhibited no fusion above background at neutral pH, and low pH induced activity similar to that of CAN97–83 F, while S175H434 F exhibited significantly higher (nearly 500%) low pH-induced fusion activity compared to CAN97–83 F. Again, TN96–12 F-mediated fusion was undetectable above background levels (Fig. 1C).
F S175H434 differs from CAN97–83 F by only two amino acids yet demonstrates significantly increased low pH-induced membrane fusion (Fig.1 A–C). H435 has been implicated in low pH-mediated triggering, so it appeared that H434 might serve a similar role. To examine the contribution of H434 in fusion, we generated a point mutation in CAN97–83 WT F using site-directed mutagenesis, changing Q at position 434 to H (CAN97–83 Q434H). We then conducted both syncytia and luciferase reporter assays to examine fusion activity of this mutant. Interestingly, syncytia assays demonstrated that CAN97–83 Q434H was able to recapitulate the high level of fusion observed for S175H434 (Fig. 1A), and quantification of the fusion activity demonstrated a similar fusion profile to that of S175H434 (Fig. 1B). The luciferase reporter assay again confirmed that CAN97–83 Q434H demonstrated fusion activity similar to S175H434 and significantly higher than WT CAN97–83 F (Fig. 1C). Together, these results demonstrate that the hyperfusogenic phenotype observed in S175H434 is primarily mediated by a single amino acid change, Q434H.
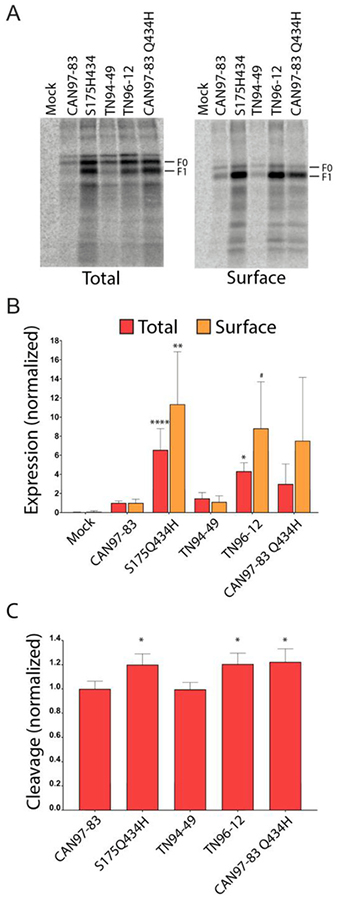
Fusion mediated by F requires that the protein is synthesized, trafficked to the surface and proteolytically processed. As fusion is correlated with cell surface expression and cleavage activation, we utilized radioactive metabolic labelling coupled with surface biotinylation to examine both total and surface protein expression and cleavage activation profiles for each F isolate. Interestingly, we identified significant differences in the total and surface protein expression levels between the F proteins. Compared to CAN97–83 WT F, TN94–49 F had similar levels of total (F0+F1) and surface expression, (Fig. 2A and quantified in B), which correlated with the similar fusion activities found for these proteins (Fig. 1C). The two F proteins shown to promote high levels of fusion, S175H434 and CAN97–83 Q434H, both displayed higher average total protein expression (approximately 6-fold and 3-fold, respectively, compared to CAN97–83 F; Figure 2A and B), though this difference was only statistically significant for S175H434 F. The two highly fusing F proteins also displayed higher average surface protein expression (approximately 10-fold and 7-fold, compared to CAN97–83), though again statistical significance was only reached for S175H434 F. These results suggest that higher surface expression levels may be at least partially responsible for the higher levels of fusion observed for S175H434. However, TN96–12 F failed to promote fusion, despite similarly high surface expression, demonstrating that surface expression is only one factor contributing to overall fusion (Fig. 1B). These findings demonstrate that a single amino acid change was able to increase the fusion protein expression compared to WT, suggesting that this area of the protein is involved in protein stability or turnover and offering a potential explanation for the phenotypes observed.
Fig 2. HMPV F protein expression and cleavage by exogenous trypsin.
(A) Representative gels of total and surface protein expression in metabolically labeled Vero cells expressing empty vector pCAGGS-MCS (Mock), CAN97–83 F, S175H434 F, TN94–49 F, TN96–12 F, and mutant CAN97–83 Q434H F in the presence of 3.0 μg/ml of TPCK-trypsin. (B) Quantification of the total and surface protein populations for F (F0 and F1 forms) in metabolically labeled Vero cells. Data are presented as normalization to CAN97–83 (A2) F expression, which was set to 1 (n = 4). “*”s Indicate statistical significance compared to F for CAN97–83 F (n= 4) [# P<0.07, * p<0.05, ** p<0.005, *** p<0.0005 and **** p<0.0001]. (C) Quantification of the relative amount of fusion protein cleavage within the surface population of F calculated using F1/(F1 + F0) and normalized to CAN97–83, set as 1.
HMPV F is synthesized as an inactive, monomeric protein that must be proteolytically processed into the heterodimeric, disulfide linked F1 and F2, in order to mediate fusion. In vitro, exogenous trypsin cleaves the protein within the cleavage motif to generate the fusogenically active form. Due to the requirement of cleavage for fusion, we examined the relative amount of trypsin cleavage for each F variant. To identify a potential role for proteolytic activation by trypsin on fusion activity, percent cleavage was quantified for the surface population of F (Fig. 2C). Interestingly, F variants that were more highly expressed, S175H434, TN96–12, and CAN97–83 Q434H were cleaved at significantly higher levels than CAN97–83 or TN94–49, potentially contributing to the observed hyperfusogenic phenotypes (Fig. 2C). Though the higher levels of CAN97–83 Q434H protein expression did not reach statistical significance, this protein displayed significantly higher levels of protein cleavage compared to CAN97–83 WT F.
These findings suggest the hyperfusogenic phenotypes observed for S175H434 F were due in part to the presence of higher levels of F at the surface as well as increased cleavage activation. The finding that CAN97–83 Q434H yielded significantly increased membrane fusion and cleavage suggests H434 is important for this phenotype. Conversely, TN96–12, which failed to mediate cell-to-cell fusion in both syncytia and luciferase reporter assays, was significantly more abundant in total, and approaching significantly higher surface populations (p=0.0685). In addition, it wascleaved significantly higher when compared to CAN97–83 F. Although cleaved significantly higher when compared to CAN97–83 F, TN96–12 F was unable to mediate fusion in cell-to-cell fusion assays suggesting there may be other factors necessary for triggering of this F isolate which are not present on the cell surface.
Discussion
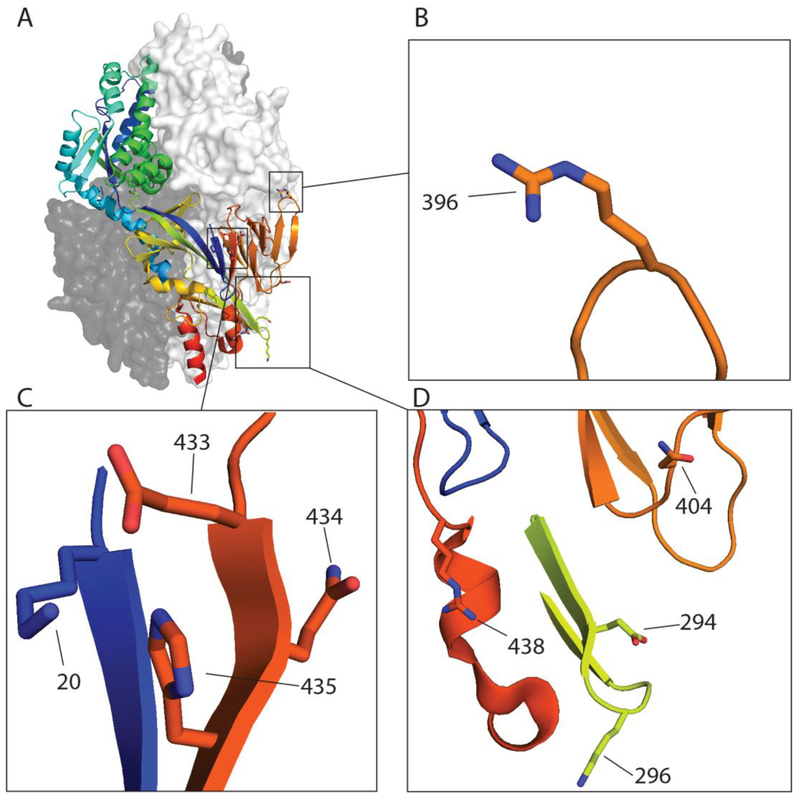
In this study we examined the fusion activity, expression and cleavage of three previously uncharacterized F proteins. Previously, we have reported low pH-promoted membrane fusion for CAN97–83, and others have reported this phenomenon only within clade A strains of HMPV (15, 18–20), with a glycine residue at position 294 (G294) described as a requirement for low pH-promoted fusion (18). However, TN94–49 F contains a lysine (K) at this position and promotes fusion after exposure to low pH, indicating that either of these residues can be present at position 294 in an HMPV F protein which promotes low pH-induced fusion (Fig. 3). Genetic variability analysis of HMPV F proteins demonstrated that position 294 is one of two positively selected sites with relaxed selective constraints for the amino acids G, K or E, indicating that when one of these are present at this position, viral fitness is unaffected (31). Analyses from the Melero group (18, 19) indicated that E294 was present with neutral pH fusing F proteins, while previous results from several groups showed low pH fusion with F proteins containing G294. Our findings demonstrate that K294 can also be present in a low pH induced F protein. Additional amino acids at positions 296, 396 and 404 (Fig. 3) have been described to modulate low pH fusion sensitivity (19). However, these amino acids are completely conserved in the strains of HMPV examined in this study (Fig. S1).
Figure 3: HMPV residues identified thus far involved in low pH mediated fusion.
A) The pre-fusion homo-trimeric structure of HMPV F of NL/1/00 (pdb: 5WB0) with residue positions identified for low pH fusion highlighted including (B) 396 (C) 20, 433, 434, 435(D) 294, 296, 396, 404 and 438. The HRB region was predicted using LearnCoil score for viral membrane fusion proteins as described (39, 40) and stretches from position 453–487 of the fusion protein.
HMPV F S175H434 displayed a hyperfusogenic phenotype and significantly increased protein expression of both total and surface populations compared to CAN97–83 F, but only two amino acid changes were present between the two proteins. One notable amino acid difference was H434, which is in close proximity to a previously identified amino acid, H435, shown to be important for low pH triggered fusion. We generated a CAN97–83 mutant, containing Q434H, and demonstrated this single amino acid mutation could recapitulate the hyperfusogenic phenotype and expression patterns observed for S175H434 F. Due to its close proximity, H434 may contribute to low pH mediated fusion through a similar mechanism to H435, which has been hypothesized to interact with surrounding residues and, upon protonation, destabilize the pre-fusion form to initiate refolding to the post-fusion form. Recently, a high resolution pre-fusion structure of HMPV F was published. Examination of the structure suggests the need for destabilization in the heptad repeat B region to trigger the refolding event to the post-fusion conformation (25). This structure also suggests a model for the role of H435 in fusion, as a potential electrostatic disruption between E433 and K20 by protonated H435 upon acidification could provide this destabilization (Fig. 3A and C). Additionally, K438 could play a role given the close proximity of the residue to the H434 and H435 amino acid positions, potentially enhancing this destabilization and increasing fusion activity (Fig. 3A and D) (26). Lastly, there are significant differences in the overall protein expression of F from the examined strains, suggesting a potential role in protein folding, overall stability, or turnover rates for H434. When examining known sequences of HMPV F proteins, H434 was not present, which suggests that the hyperfusogenicity conferred by this amino acid may not be beneficial to HMPV infectivity. For parainfluenza virus 3 (PIV3), a closely related paramyxovirus, enhancement of receptor binding and fusion within the monolayer was detrimental for replication in human airway epithelium and in vivo during infection in cotton rats (32). However, when examining a hyperfusogenic F protein mutant from a more closely related pneumovirus, respiratory syncytial virus, there were increased viral loads, severe lung pathology and weight loss in mice compared to controls (33), so whether hyperfusogenicity is preferentially selected by HMPV during infection is not well understood. Interestingly, when HMPV was incubated with the neutralizing monoclonal antibody 54G10, one of the detected escape mutations was Q434H. No changes in binding affinity of 54G10 for the Q434H mutant were detected, suggesting that this mutation may introduce structural changes into the F protein which provide some potential benefit (34).
Our results show that TN96–12 was also highly expressed in both total and surface populations but does not mediate fusion in cell-to-cell fusion assays. Co-expression of TN96–12 F with G at neutral or low pH was not sufficient to induce syncytia formation in cell culture (data not shown), indicating that lack of the G protein is not the reason for the absence of fusion. TN96–12 HMPV is a clinical isolate, able to initiate infection in patients as well as propagate in cell culture, and thus must have a functional fusion protein. Therefore, our findings point toward the need for other host factors required for fusion of some HMPV strains in order to escape the endosome and initiate infection, and the lack of fusion in our assays suggests these factors may not be present on the cell surface. Currently, heparan sulfate proteoglycans and RGD binding integrins are proposed cellular factors for association and entry of HMPV through endocytosis (21, 35–37). It is possible that there are other critical factor(s) within the endosome that interact with HMPV F and trigger the fusion protein, similar to the use of the endosomal receptor NPC-1 by Ebola virus GP (38).
Taken together, the results in this study highlight the diversity of HMPV F activity and the complexity associated with fusion. Our results indicate that the contribution of a single amino acid can be responsible for observed phenotypes, demonstrating that minor evolutionary changes can lead to significant phenotypic differences that alter HMPV infection and tropism. Further studies are necessary to better understand and elucidate key contributing factors of fusion protein stability, cleavage and host factor interaction required for HMPV infection, as well as which key residues and regions of the F protein are vital for fusion and entry of the virus.
Supplementary Material
Sequence alignment was generated using ClustalW. The asterisk “*”s indicates a site that is not conserved.
Highlights.
The fusion protein from several analyzed strains of human metapneumovirus displays significant differences in fusion activity.
Changes in one or a few amino acids can result in alterations in protein stability or function.
An F protein with a lysine at 294 efficiently promoted low-pH triggered fusion, in contrast to previous reports that a glycine residue at this position was essential for low-pH triggering.
Acknowledgments
We thank members of the Dutch lab for discussion and evaluation of experiments and the manuscript.
Funding Information
This work was supported by CCTS TL1 training program (TL1TR000115) and individual NRSA (F30AI114194) to EK, A Great Rivers Affiliate predoctoral fellowship (55710PRE4230022) to AC, and NIAID grant R01AI051517, NIGMS grant P30GM110787, and UK University Research Professor funds to RED.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van den Hoogen BG, Bestebroer TM, Osterhaus AD, Fouchier RA. 2002. Analysis of the genomic sequence of a human metapneumovirus. Virology 295:119–132. [DOI] [PubMed] [Google Scholar]
- 2.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, Osterhaus AD. 2001. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 7:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van den Hoogen BG, Osterhaus DM, Fouchier RA. 2004. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J 23:S25–32. [DOI] [PubMed] [Google Scholar]
- 4.Don M, Korppi M, Valent F, Vainionpaa R, Canciani M. 2008. Human metapneumovirus pneumonia in children: results of an Italian study and mini-review. Scand J Infect Dis 40:821–826. [DOI] [PubMed] [Google Scholar]
- 5.Feuillet F, Lina B, Rosa-Calatrava M, Boivin G. 2012. Ten years of human metapneumovirus research. J Clin Virol 53:97–105. [DOI] [PubMed] [Google Scholar]
- 6.Boivin G, De Serres G, Cote S, Gilca R, Abed Y, Rochette L, Bergeron MG, Dery P. 2003. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis 9:634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deffrasnes C, Hamelin ME, Boivin G. 2007. Human metapneumovirus. Semin Respir Crit Care Med 28:213–221. [DOI] [PubMed] [Google Scholar]
- 8.Esper F, Boucher D, Weibel C, Martinello RA, Kahn JS. 2003. Human metapneumovirus infection in the United States: clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 111:1407–1410. [DOI] [PubMed] [Google Scholar]
- 9.Esper F, Martinello RA, Boucher D, Weibel C, Ferguson D, Landry ML, Kahn JS. 2004. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis 189:1388–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahn JS. 2006. Epidemiology of human metapneumovirus. Clin Microbiol Rev 19:546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van den Hoogen BG, van Doornum GJ, Fockens JC, Cornelissen JJ, Beyer WE, de Groot R, Osterhaus AD, Fouchier RA. 2003. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis 188:1571–1577. [DOI] [PubMed] [Google Scholar]
- 12.van den Hoogen BG, Herfst S, Sprong L, Cane PA, Forleo-Neto E, de Swart RL, Osterhaus AD, Fouchier RA. 2004. Antigenic and genetic variability of human metapneumoviruses. Emerg Infect Dis 10:658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4:613–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naylor CJ, Brown PA, Edworthy N, Ling R, Jones RC, Savage CE, Easton AJ. 2004. Development of a reverse-genetics system for Avian pneumovirus demonstrates that the small hydrophobic (SH) and attachment (G) genes are not essential for virus viability. Journal of General Virology 85:3219–3227. [DOI] [PubMed] [Google Scholar]
- 15.Schowalter RM, Smith SE, Dutch RE. 2006. Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J Virol 80:10931–10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamb RA, Parks GD. 2007. Paramyxoviridae: The viruses and their replication, p 1449–1496. In Knipe DM, Howley PM (ed), Fields Virology, Fifth ed. Lippincott, Williams and Wilkins, Philadelphia. [Google Scholar]
- 17.Smith EC, Popa A, Chang A, Masante C, Dutch RE. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. Febs j 276:7217–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herfst S, Mas V, Ver LS, Wierda RJ, Osterhaus AD, Fouchier RA, Melero JA. 2008. Low-pH-induced membrane fusion mediated by human metapneumovirus F protein is a rare, strain-dependent phenomenon. J Virol 82:8891–8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mas V, Herfst S, Osterhaus AD, Fouchier RA, Melero JA. 2011. Residues of the human metapneumovirus fusion (F) protein critical for its strain-related fusion phenotype: implications for the virus replication cycle. J Virol 85:12650–12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schowalter RM, Chang A, Robach JG, Buchholz UJ, Dutch RE. 2009. Low-pH Triggering of Human Metapneumovirus Fusion: Essential Residues and Importance in Entry. J Virol 83:1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox RG, Mainou BA, Johnson M, Hastings AK, Schuster JE, Dermody TS, Williams JV. 2015. Human Metapneumovirus Is Capable of Entering Cells by Fusion with Endosomal Membranes. PLoS Pathog 11:e1005303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison SC. 2008. Viral membrane fusion. Nat Struct Mol Biol 15:690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang Q, Sivaramakrishna RP, Ludwig K, Korte T, Bottcher C, Herrmann A. 2003. Early steps of the conformational change of influenza virus hemagglutinin to a fusion active state: stability and energetics of the hemagglutinin. Biochim Biophys Acta 1614:3–13. [DOI] [PubMed] [Google Scholar]
- 24.Kampmann T, Mueller DS, Mark AE, Young PR, Kobe B. 2006. The Role of histidine residues in low-pH-mediated viral membrane fusion. Structure 14:1481–1487. [DOI] [PubMed] [Google Scholar]
- 25.Battles MB, Mas V, Olmedillas E, Cano O, Vazquez M, Rodriguez L, Melero JA, McLellan JS. 2017. Structure and immunogenicity of pre-fusion-stabilized human metapneumovirus F glycoprotein. Nat Commun 8:1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang A, Hackett BA, Winter CC, Buchholz UJ, Dutch RE. 2012. Potential electrostatic interactions in multiple regions affect human metapneumovirus F-mediated membrane fusion. J Virol 86:9843–9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulbrandt ND, Ji H, Patel NK, Barnes AS, Wilson S, Kiener PA, Suzich J, McCarthy MP. 2008. Identification of antibody neutralization epitopes on the fusion protein of human metapneumovirus. J Gen Virol 89:3113–3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster JE, Cox RG, Hastings AK, Boyd KL, Wadia J, Chen Z, Burton DR, Williamson RA, Williams JV. 2015. A Broadly Neutralizing Human Monoclonal Antibody Exhibits In Vivo Efficacy Against Both Human Metapneumovirus and Respiratory Syncytial Virus. The Journal of Infectious Diseases 211:216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White J, Matlin K, Helenius A. 1981. Cell fusion by Semliki Forest, influenza, and vesicular stomatitis viruses. J Cell Biol 89:674–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson RG, Lamb RA. 1993. The molecular biology of influenza viruses and paramyxoviruses, p 35–73. In Davidson A, Elliott RM (ed), Molecular Virology: A Practical Approach. IRL Oxford University Press, Oxford. [Google Scholar]
- 31.Lo Presti A, Cammarota R, Apostoli P, Cella E, Fiorentini S, Babakir-Mina M, Ciotti M, Ciccozzi M. 2011. Genetic variability and circulation pattern of human metapneumovirus isolated in Italy over five epidemic seasons. New Microbiol 34:337–344. [PubMed] [Google Scholar]
- 32.Palmer SG, Porotto M, Palermo LM, Cunha LF, Greengard O, Moscona A. 2012. Adaptation of human parainfluenza virus to airway epithelium reveals fusion properties required for growth in host tissue. MBio 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hotard AL, Lee S, Currier MG, Crowe JE Jr., Sakamoto K, Newcomb DC, Peebles RS Jr., Plemper RK, Moore ML. 2015. Identification of residues in the human respiratory syncytial virus fusion protein that modulate fusion activity and pathogenesis. J Virol 89:512–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schuster JE, Cox RG, Hastings AK, Boyd KL, Wadia J, Chen Z, Burton DR, Williamson RA, Williams JV. 2015. A broadly neutralizing human monoclonal antibody exhibits in vivo efficacy against both human metapneumovirus and respiratory syncytial virus. J Infect Dis 211:216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang A, Masante C, Buchholz UJ, Dutch RE. 2012. Human metapneumovirus (HMPV) binding and infection are mediated by interactions between the HMPV fusion protein and heparan sulfate. J Virol 86:3230–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cox RG, Livesay SB, Johnson M, Ohi MD, Williams JV. 2012. The human metapneumovirus fusion protein mediates entry via an interaction with RGD-binding integrins. J Virol 86:12148–12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei Y, Zhang Y, Cai H, Mirza AM, Iorio RM, Peeples ME, Niewiesk S, Li J. 2014. Roles of the putative integrin-binding motif of the human metapneumovirus fusion (f) protein in cell-cell fusion, viral infectivity, and pathogenesis. J Virol 88:4338–4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, Dal Cin P, Dye JM, Whelan SP, Chandran K, Brummelkamp TR. 2011. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477:340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh M, Berger B, Kim PS. 1999. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J Mol Biol 290:1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger B, Singh M. 1997. An iterative method for improved protein structural motif recognition. J Comput Biol 4:261–273. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment was generated using ClustalW. The asterisk “*”s indicates a site that is not conserved.