Abstract
Background & Aims
Hepatitis D virus (HDV) superinfection in patients with hepatitis B virus (HBV) is associated with rapid progression to liver cirrhosis and hepatocellular carcinoma. Treatment options are limited, and no vaccine is available. Although HDV-specific CD8+ T cells are thought to control the virus, little is known about which HDV epitopes are targeted by virus-specific CD8+ T cells or why these cells ultimately fail to control the infection. We aimed to define how HDV escapes the CD8+ T-cell–mediated response.
Methods
We collected plasma and DNA samples from 104 patients with chronic HDV and HBV infection at medical centers in Europe and the Middle East, sequenced HDV, typed human leukocyte antigen (HLA) class I alleles from patients, and searched for polymorphisms in HDV RNA associated with specific HLA class I alleles. We predicted epitopes in HDV that would be recognized by CD8+ T cells and corresponded with the identified virus polymorphisms in patients with resolved (n = 12) or chronic (n = 13) HDV infection.
Results
We identified 21 polymorphisms in HDV that were significantly associated with specific HLA class I alleles (P < .005). Five of these polymorphisms were found to correspond to epitopes in HDV that are recognized by CD8+ T cells; we confirmed that CD8+ T cells in culture targeted these HDV epitopes. HDV variant peptides were only partially cross-recognized by CD8+ T cells isolated from patients, indicating that the virus had escaped detection by these cells. These newly identified HDV epitopes were restricted by relatively infrequent HLA class I alleles, and they bound most frequently to HLA-B. In contrast, frequent HLA class I alleles were not associated with HDV sequence polymorphisms.
Conclusions
We analyzed sequences of HDV RNA and HLA class I alleles that present epitope peptides to CD8+ T cells in patients with persistent HDV infection. We identified polymorphisms in the HDV proteome that associate with HLA class I alleles. Some variant peptides in epitopes from HDV were only partially recognized by CD8+ T cells isolated from patients; these could be mutations that allow HDV to escape the immune response, resulting in persistent infection. HDV escape from the immune response was associated with uncommon HLA class I alleles, indicating that HDV evolves, at the population level, to evade recognition by common HLA class I alleles.
Keywords: Cytotoxic T Cell, MHC Class I, TCR, Antigen Presentation
Abbreviations used in this paper: HDAg, hepatitis delta antigen; HBV, hepatitis B virus; HCV, hepatitis C virus; HDV, hepatitis D virus; HIV, human immunodeficiency virus; HLA, human leukocyte antigen; IC50, half maximal inhibitory concentration; IFN, interferon; IL-2, interleukin 2; L-HDAg, large variant of the hepatitis delta antigen; PBMC, peripheral blood mononuclear cell; PBS, phosphate-buffered saline; PCR, polymerase chain reaction; PD-1, programmed cell death protein 1; S-HDAg, small variant of the hepatitis delta antigen; TCF1, T-cell factor 1
Graphical abstract
What You Need to Know.
Background and Context
HDV infection is associated with severe liver disease, but little is known about the corresponding virus-specific CD8+ T cells, which are thought to mediate immune protection against HDV.
New Findings
In persistent infection, HDV mutates to escape from the virus-specific CD8+ T cell response. At the population-level, HDV has evolved to evade from recognition in the context of common HLA class I alleles.
Limitations
The observational nature of this study precludes formal identification of a causal association between viral escape and CD8+ T-cell failure in the context of infection with HDV.
Impact
These results suggest that viral escape should be considered as a key parameter in the design of immune-based interventions to prevent and treat liver disease associated with HDV.
Hepatitis D virus (HDV) has infected approximately 10% of hepatitis B virus (HBV)–seropositive individuals, affecting 15–20 million people worldwide.1 Patients with simultaneous HDV/HBV infection often experience severe acute hepatitis, with an enhanced risk of fulminant disease, but frequently clear both viruses. In contrast, HBV-seropositive patients who become superinfected with HDV generally develop chronic HDV/HBV infection. These patients have a high risk of rapid progression to liver cirrhosis and hepatocellular carcinoma. At present, the sole treatment option for chronic HDV/HBV infection is pegylated interferon (IFN) alfa, which is usually administered for 48 weeks. However, only approximately 30% of patients respond to treatment, and many subsequently develop viral relapse.2
Virus-specific CD8+ T cells are thought to play a key role in the outcome of HDV/HBV infection. This contention has been supported by vaccination studies in mice and woodchucks.3 However, little is known about naturally occurring HDV-specific CD8+ T-cell responses, despite the urgent need for novel prophylactic and therapeutic interventions. The HDV genome encodes a single viral protein, hepatitis delta antigen (HDAg), translated as large and small variants (L-HDAg and S-HDAg, respectively). The only difference between these 2 proteins is a 19–amino acid extension at the C-terminus of L-HDAg. An initial report described 2 HDV-specific CD8+ T-cell epitopes in human leukocyte antigen (HLA)-A*02 transgenic mice and patients with resolved HDV infection, but these findings were not confirmed in patients with persistent HDV infection.4 A subsequent report suggested that immune exhaustion may impair the efficacy of HDV-specific CD8+ T cells in vivo.5 In a recent study, we identified 2 HLA-B*27–restricted HDV-specific CD8+ T-cell epitopes, both of which were subject to viral escape by mutation.6 Of note, HLA-B*27 is associated with spontaneous clearance of hepatitis C virus (HCV) and elite control of human immunodeficiency virus (HIV), and HLA-B*27–restricted CD8+ T-cell responses drive viral evolution in patients infected with HCV or HIV.7 It nonetheless remains unclear if viral escape is a generalizable phenomenon that underlies immune failure in patients infected with HDV.
In the present study, we analyzed HLA class I–associated viral sequence polymorphisms across the entire HDV genome as surrogate markers of CD8+ T-cell–driven viral escape. These “HLA footprints” were used to identify CD8+ T-cell epitopes, confirm predicted escape mutations, and explore features of the host–virus interactome, providing mechanistic insights into adaptive immunity against HDV.
Materials and Methods
Patients and Samples
Patients with chronic HDV/HBV infection (n = 104) were recruited from 8 medical centers located in Germany (Bonn, Düsseldorf, Essen, Hannover, and Munich), Spain, Italy, and Iran.6 Viral sequences and HLA class I genotypes were determined according to standard protocols.6 CD8+ T-cell assays were performed with additional samples obtained from 12 patients with resolved HDV infection and 13 patients with chronic HDV/HBV infection, recruited from Freiburg, Hamburg, and Munich (Germany). All patients were infected with HDV genotype 1. Patient characteristics are summarized in Supplementary Tables 1 and 2. Ethical approval was granted by the Ethik-Kommission der Albert-Ludwigs-Universität Freiburg (no. 369/15). Written informed consent was obtained in all cases according to federal guidelines and the Declaration of Helsinki. Venous blood samples (50 mL per draw) were collected in EDTA-anticoagulated tubes. Peripheral blood mononuclear cells (PBMCs) were isolated with lymphocyte separation medium density gradients (Pancoll separation medium, PAN Biotech GmbH; Aidenbach, Germany) and resuspended in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% penicillin/streptomycin, and 1.5% HEPES buffer 1 mol/L (complete medium; all additives from Thermo Scientific (Waltham, MA).
Peptides, Antibodies, and Tetramers
Peptides were synthesized with a free amine NH2 terminus and a free acid COOH terminus with standard Fmoc chemistry (Genaxxon Bioscience, Ulm, Germany). Anti-CD8 (RPA-T8, 1:100; SK1, 1:300), anti-CD38 (Hb7, 1:200), anti-CD45RA (HI100, 1:50), anti-IFN-γ (4S.B3, 1:50), and anti–programmed cell death protein 1 (PD-1) (EH12.1, 1:33) were purchased from BD Biosciences (Germany). Anti-Bcl-2 (Bcl-2/100, 1:200), anti-CCR7 (G043H7, 1:33), anti-CD8 (RPA-T8, 1:400), and anti-CD127 (A019D5, 1:33) were purchased from BioLegend (San Diego, CA). Anti-CD14 (61D3, 1:100), anti-CD19 (HIB19, 1:100), anti-Eomes (WD1928, 1:50), anti-killer cell lectin-like receptor G1 (KLRG1) (13F12F2, 1:33), and anti-T-bet (4B10, 1:200) were purchased from Thermo Scientific. Anti-T-cell factor (1TCF1) (C63D9, 1:100) was purchased from Cell Signaling Technology (Danvers, MA). Tetrameric complexes of HLA-B*15:01/L-HDAg170–179 SMQGVPESPF were generated as described previously.8
Analysis of HDV Sequences and HLA Class I-Associated Viral Polymorphisms
Total RNA was extracted from serum samples and reverse transcribed into cDNA with Moloney Murine Leukemia Virus Reverse Transcriptase (Promega, Madison, WI) and the HDV-specific primer 771R (5′-CGGTCCCCTCGGAATGTTG-3′). The L-HDAg-encoding region was amplified from cDNA with Pfu DNA Polymerase (Promega) in a 2-step nested polymerase chain reaction (PCR) incorporating the HDV-specific primers 891-F (5′-AGGTCGGACCGCGAGGAGGT-3′), 339-R (5′-GCTGAAGGGGTCCTCTGGAGGTG-3′), 912-F (5′-GAGATGCCATGCCGACCCGAAGAG-3′), and 1674-R (5′-AGAAAAGAGTAAGAGYACTGAGG-3′). The following thermal profile was used for all PCRs: (i) 94°C for 10 minutes; (ii) 94°C for 30 seconds, 54°C for 45 seconds, and 72°C for 90 seconds (35 cycles); and (iii) 72°C for 7 minutes. PCR products were purified with a QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) and sequenced on an ABI 3730xl DNA Analyzer (Eurofins Genomics Germany GmbH; Ebersberg, Germany) with the internal primers 912-F and 1674-R. All sequences were submitted to GenBank (accession nos. MF175257–MF175360). HDV sequences were aligned with ClustalX2 software.9 HLA class I associations with amino acid residues at each alignment position were tested using the R package SeqFeatR (https://seqfeatr.zmb.uni-due.de).10 The Fisher exact test was used to determine significant associations in a 2 × 2 contingency table format with counts for observed combinations of amino acids and HLA class I allotypes at each alignment position. Further analyses were based on a cutoff P value of <.005.
Prediction of HDV-Specific CD8+ T-Cell Epitopes
Viral amino acid sequences, 15 residues N-terminal and 15 residues C-terminal, of the identified HLA class I–associated viral sequence polymorphisms were analyzed for the corresponding binding motifs with 4 online prediction tools: ANN 3.4 and netMHCpan 2.8 on the Immune Epitope Database website,11, 12 SYFPEITHI,13 and BIMAS.14 The 8mer, 9mer, and 10mer peptides were tested if available via the respective prediction tool. A half maximal inhibitory concentration (IC50) of ≤1000 nmol/L, a SYFPEITHI score of ≥20, and a BIMAS score of ≥20 were used as cutoffs. Candidate epitopes were ranked against epitopes predicted across the entire sequence of L-HDAg.
Peptide-Specific CD8+ T-Cell Lines
PBMCs were activated with peptides, as described previously.15 Briefly, 4 × 106 PBMCs were stimulated once with 10 μg/mL peptide and 0.5 μg/mL anti-CD28 (BD Biosciences) and fed every 3 days with complete medium containing 20 U/mL recombinant interleukin (IL)-2 (Miltenyi Biotec, Bergisch Gladbach, Germany). Peptide-specific CD8+ T-cell lines were used for experimental purposes after 14 days.
Intracellular IFN Gamma Staining
Procedures were carried out as described previously.16 Briefly, expanded CD8+ T cells or peptide-specific CD8+ T-cell lines (0.2 × 106 cells per well in a 96-well plate) were stimulated with peptides (10 μg/mL) in the presence of 50 U/mL recombinant IL-2 and 1 μL/mL brefeldin A (BD Biosciences). After 5 hours, cells were stained with 7-aminoactinomycin D and anti-CD8, fixed/permeabilized with Cytofix/Cytoperm, and stained with anti-IFN gamma (all reagents from BD Biosciences). Stained cells were fixed in phosphate-buffered saline (PBS) containing 2% paraformaldehyde. Data were acquired with an BD FACSCanto II flow cytometer (BD Biosciences) and analyzed with FlowJo software, version 10 (FlowJo, Ashland, OR).
HLA Class I Tetramer-Based Analysis and Cell Enrichment
Tetramer staining procedures were carried out as described previously.17 Briefly, 1 × 106 PBMCs per well were incubated in a 96-well plate with the relevant HLA class I tetramer for 15 minutes at 37°C. Cells were then washed 3 times with PBS containing 1% fetal calf serum and stained with the indicated surface and/or intracellular antibodies. Dead cells were excluded from the analysis with the fixable viability dye eFluor780 (1:5000, eBioscience, Germany). Cytoplasmic and nuclear molecules were shown with a FoxP3/Transcription Factor Staining Buffer Set (eBioscience). Stained cells were fixed in PBS containing 2% paraformaldehyde. Tetramer-based enrichment was performed as described by Alanio et al.18 Briefly, 10–15 × 106 PBMCs were labeled for 30 minutes at room temperature with the HLA-B*15:01/L-HDAg170–179 SMQGVPESPF tetramer coupled to phycoerythrin and enriched using anti-PE beads with MACS technology (Miltenyi Biotec, Germany). Frequencies of virus-specific CD8+ T cells were calculated as described previously.18 Data were acquired using an LSRFortessa flow cytometer and analyzed with FlowJo software version 10 (FlowJo, Ashland, OR).
Results
Identification of HLA Class I–Associated Viral Sequence Polymorphisms in L-HDAg
To assess the impact of HDV-specific CD8+ T-cell–mediated selection pressure in vivo, we analyzed a cohort of 104 patients with chronic HDV/HBV infection for HLA class I–associated viral sequence polymorphisms (HLA footprints) in L-HDAg. Using a highly significant cutoff value (P < .005) based on studies of other persistent viral infections,19, 20 we identified 21 viral sequence polymorphisms that were associated with specific HLA class I alleles (Table 1).
Table 1.
HLA Class I–Associated Viral Sequence Polymorphisms and HDV-Specific CD8+ T Cell Epitopes
| No. | HLA | AA position | Mutation | P value | AA sequence epitope candidate | AA range | Positive responses/number of patients tested |
|---|---|---|---|---|---|---|---|
| 1 | B*15 | 170 | S170N | 2.9 × 10–8 | SMQGVPESPF | 170–179 | 10/14 |
| 2 | B*13 | 33 | D33E | .0001 | DLRKVKKKI | 33–41 | 0/5 |
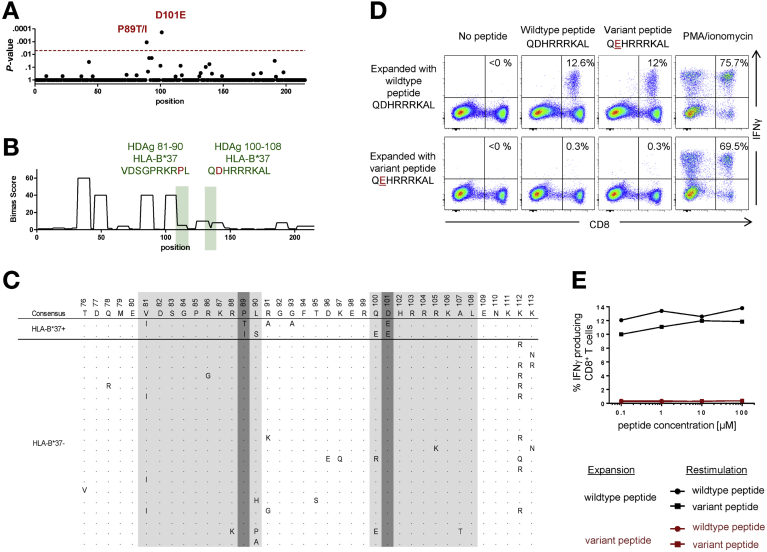
| 3 | B*37 | 101 | D101E | .0002 | QDHRRRKAL | 100–108 | 1/1 |
| 4 | A*29 | 63 | K63R | .0002 | — | — | — |
| 5 | A*30 | 47 | D47E | .0010 | KVKKKIKK | 36–43 | 0/3 |
| 6 | B*37 | 89 | P89T/I | .0011 | VDSGPRKRPL | 81–90 | 1/1 |
| 7 | B*49 | 37 | V37A/T | .0015 | EELERDLRKV | 28–37 | ND |
| 8 | B*13 | 100 | Q100K | .0018 | RQDHRRRKAL | 99–108 | 0/5 |
| 9 | B*51 | 81 | V81I | .0019 | — | — | — |
| 10 | B*13 | 43 | K43R | .0021 | DLRKVKKKI | 33–41 | 0/5 |
| 11 | B*41 | 158 | G158A/D/M | .0023 | — | — | — |
| 12 | B*18 | 47 | E47D | .0027 | DENPWLGNI | 46–54 | 1/5 |
| 13 | A*33 | 37 | V37T/A | .0028 | EELERDLR | 28–35 | ND |
| 14 | B*14 | 107 | A107T | .0028 | DHRRRKAL | 101–108 | 0/1 |
| 15 | A*30 | 49 | P49L/S | .0031 | KVKKKIKK | 36–43 | 0/3 |
| 16 | B*41 | 139 | R139K | .0034 | RERRVAGPPV | 140–149 | 1/1 |
| 17 | B*13 | 96 | D96E | .0035 | RQDHRRRKAL | 99–108 | 0/5 |
| 18 | B*27 | 105 | R105K | .0039 | RRKALENKK | 104–112 | Previously described6 |
| RRDHRRRKAL | 99–108 | Previously described6 | |||||
| 19 | B*38 | 131 | K131G | .0039 | — | — | — |
| 20 | B*13 | 113 | K113R | .0043 | RQDHRRRKAL | 99–108 | 0/5 |
| KQLSAGGKNL | 113–122 | 0/5 | |||||
| 21 | A*68 | 134 | T134A | .0045 | LTEEDERR | 133–140 | 0/5 |
NOTE. Confirmed epitopes are displayed in bold.
AA, amino acid; ND, not done.
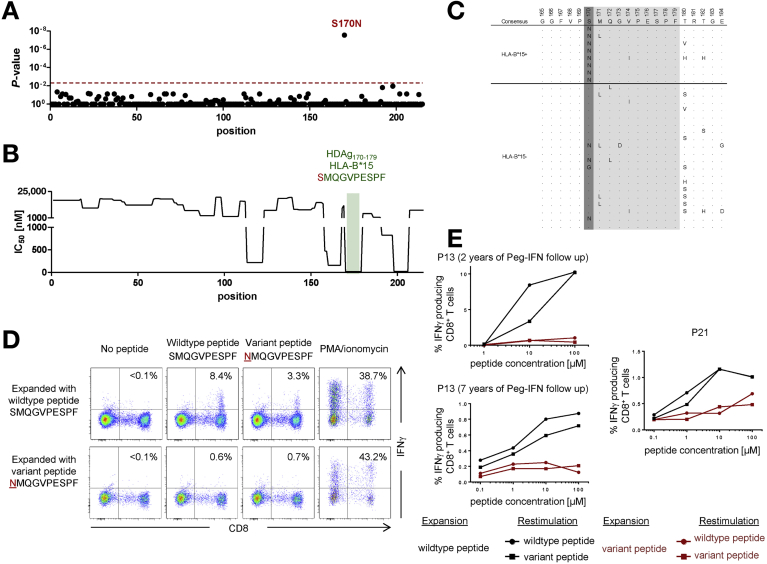
The strongest association linked residue 170 in L-HDAg with HLA-B*15. All 8 HLA-B*15+ patients harbored viruses with a serine (S) to asparagine (N) substitution at this position (S170N) compared with only 16 of 96 HLA-B*15– patients (P = 2.9 × 10–8) (Figure 1A and C).
Figure 1.
HLA-B*15–restricted CD8+ T-cell responses specific for L-HDAg170–179 drive viral escape. (A) P values for the associations between HDV sequence polymorphisms and the presence of HLA-B*15, plotted for each amino acid residue in the L-HDAg protein. The cutoff for significance was set at P = .005 (dotted red line). (B) Predicted HLA-B*15 binding affinity of candidate peptide epitopes in L-HDAg. IC50 values were predicted for 8mers, 9mers, and 10mers with the ANN 3.4 method (www.iedb.org). The best hit corresponding to the confirmed HLA-B*15–restricted epitope L-HDAg170–179 is highlighted in green. (C) Viral sequences from individual HLA-B*15+ and HLA-B*15– patients compared with the consensus sequence derived from all 104 patients (all HLA-B*15+ patients are shown above the line, and the first 20 HLA-B*15– patients are shown below the line). Dots indicate agreement with consensus, and single-letter amino acid codes indicate variation from consensus. The HLA-B*15–associated polymorphism S170N is highlighted in dark grey, and the confirmed HLA-B*15–restricted epitope L-HDAg170–179 is highlighted in grey. (D) PBMCs from HLA-B*15+ patient 13 (2 years after IFN treatment) were expanded in the presence of the wild-type or variant peptide corresponding to the L-HDAg170–179 epitope. After 14 days, cells were restimulated in parallel with each peptide and tested for IFN gamma production. Negative controls without peptide restimulation and positive controls stimulated with phorbol myristate acetate/ionomycin are also shown. (E) Equivalent results after restimulation with each peptide in serial dilution assays. PBMCs from patient 13 (2 and 7 years after IFN treatment) and HLA-B*15+ patient 21 were processed and tested as in D. M, mol/L; P, patient; PMA, phorbol myristate acetate.
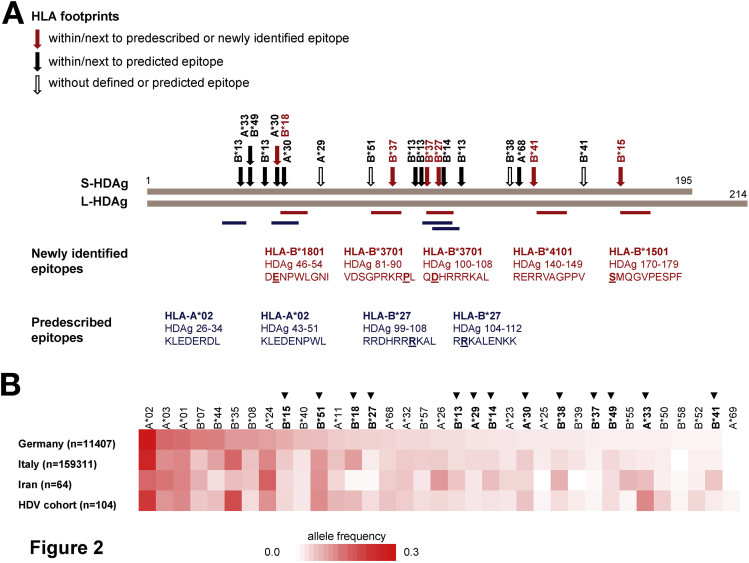
Only 5 of 21 significant polymorphisms (24%) were associated with HLA-A alleles, whereas 16 of 21 significant polymorphisms (76%) were associated with HLA-B alleles (Table 1). Moreover, these HLA footprints were distributed quite evenly across the central region of HDAg, sparing the N-terminal 30 amino acids of S/L-HDAg, the C-terminal 25 amino acids of S-HDAg, and the entire C-terminal extension unique to L-HDAg (Figure 2A).
Figure 2.
Viral sequence polymorphisms are associated with infrequent HLA class I alleles and spare the N- and C-termini of L-HDAg. (A) Distribution of identified HLA footprints across L-HDAg. Red arrows indicate HLA footprints corresponding to confirmed (predescribed or newly identified) epitopes. Filled black arrows indicate HLA footprints corresponding to predicted epitopes. Empty black arrows indicate HLA footprints without defined or predicted epitopes. Newly identified epitopes are shown as red bars, and pre-described epitopes are shown as blue bars. (B) HLA-A and HLA-B allele frequencies in the study cohort compared with reference populations from Germany, Italy, and Iran. HLA class I alleles identified in at least 2 patients in the study cohort are shown (HLA class I alleles present in 1 patient cannot reach statistical power for the identification of HLA class I–associated viral sequence polymorphisms). Footprint-linked HLA class I alleles are shown in bold and marked with an arrowhead.
Validation of CD8+ T-Cell Epitopes and Viral Escape Mutations in L-HDAg
To confirm the relevance of these associations, we used in silico prediction tools to analyze viral amino acid sequences for HLA class I binding motifs, guided by the corresponding footprint data (see Materials and Methods). In addition to 8mer, 9mer, and 10mer peptides, an extended window incorporating 15 residues either side of the HLA footprint was tested to allow the identification of CD8+ T-cell epitopes associated with flanking mutations, which can disrupt antigen processing and facilitate viral escape.21, 22, 23, 24, 25 Using this approach with defined cutoffs (an IC50 ≤ 1000 nmol/L, a SYFPEITHI score ≥ 20, or a BIMAS score ≥ 20), we identified 18 candidate CD8+ T-cell epitopes spanning 17 of the 21 HLA footprints (Figures 1B, 3B, and 4B; Table 1; and Supplementary Table 3). As expected, these candidates included 2 HLA-B*27-restricted epitopes identified previously with the same sequence data set, validating the overall strategy.6 Of note, 12 of 18 HLA footprints were located within the respective candidate epitopes, whereas 6 of 18 HLA footprints were located in the regions flanking the respective candidate epitopes. These latter footprints did not affect antigen processing in silico, as predicted with an online algorithm (www.iedb.org),26 but negative results were also obtained in similar analyses of flanking mutations known to impair the generation of epitopes derived from HCV and HIV (Supplementary Table 4).
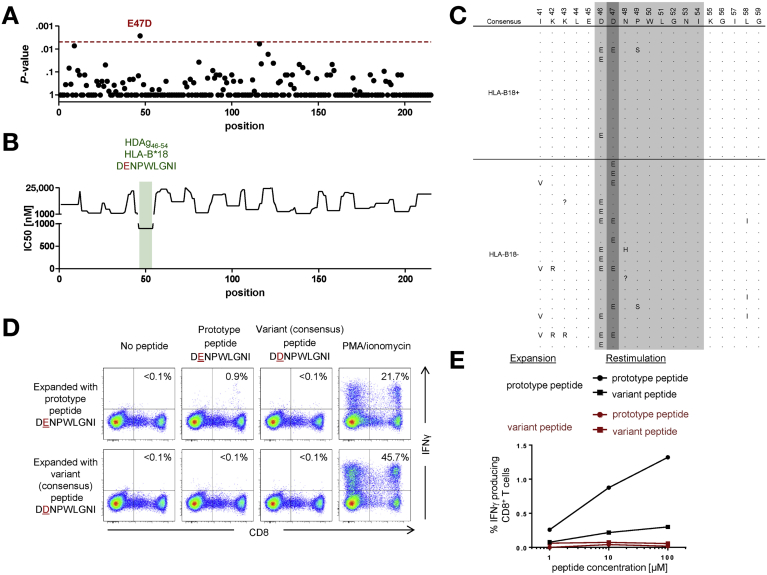
Figure 3.
HLA-B*18–restricted CD8+ T-cell responses specific for L-HDAg46–54 drive viral escape and population-level evolution of HDV. (A–E) Details as per the corresponding panels in Figure 1. In this case, the ancestral peptide is denoted as the prototype, and the variant peptide is denoted as the consensus, reflecting a negative association between HLA-B*18 and the viral polymorphism D47E. Representative data from patient 3 are shown in (D) and (E). M, mol/L; PMA, phorbol myristate acetate.
Figure 4.
The rare allotype HLA-B*37 selects for viral escape in the CD8+ T-cell epitopes L-HDAg81–90 and L-HDAg100–108. (A–E) Details as per the corresponding panels in Figure 1. (B) the BIMAS score (higher scores indicate higher binding affinities) was used in place of the ANN 3.4 method, which does not include a prediction algorithm for HLA-B*37 binders. (D, E) Representative data from patient 1 (epitope L-HDAg100–108). PMA, phorbol myristate acetate.
We then evaluated the immunogenicity of candidate epitopes in functional assays using peptide-stimulated CD8+ T-cell lines generated from HLA class I–matched patients with resolved HDV infection (n = 12) or chronic HDV/HBV infection (n = 13) (Supplementary Table 2). Five novel HDV-specific CD8+ T-cell epitopes were identified with this approach (Table 1), including 1 restricted by HLA-B*15 (Figure 1), 1 restricted by HLA-B*18 (Figure 3), 2 restricted by HLA-B*37 (Figure 4), and 1 restricted by HLA-B*41 (data not shown). In further experiments, we generated peptide-specific CD8+ T-cell lines targeting either the prototype or variant epitopes and tested the impact of viral sequence variation using IFN gamma production assays. As shown in Figures 1D, 3D, and 4D, and more clearly in serial dilution assays (Figures 1E, 3E, and 4E), the variant peptides were recognized suboptimally by some, but not all, prototype-specific CD8+ T cells. Conversely, variant-specific CD8+ T cells responded poorly to the corresponding prototype and variant peptides without exception, consistent with population-level viral escape (Figures 1D and E, 3D and E, 4D and E).
An association of special interest was detected at residue 47 (Figure 3A and C). Here, the consensus aspartate (D) was conserved in 13 of 14 HLA-B*18+ patients, whereas the variant glutamate (E) was found in 42 of 90 HLA-B*18– patients (P = .0027). This observation is compatible with the notion that HLA-B*18 drives population-level evolution from glutamate, which likely represents the ancestral residue, to aspartate and further suggests that the E47D substitution does not markedly impair viral fitness, given the relative paucity of sequence revertants in the absence of HLA-B*18. In line with these interpretations, the variant peptide (47E) induced a small but reproducible IFN gamma response, whereas the consensus peptide (47D) was not recognized in parallel assays (Figure 3D and E).
Viral Evolution During Superinfection With HDV
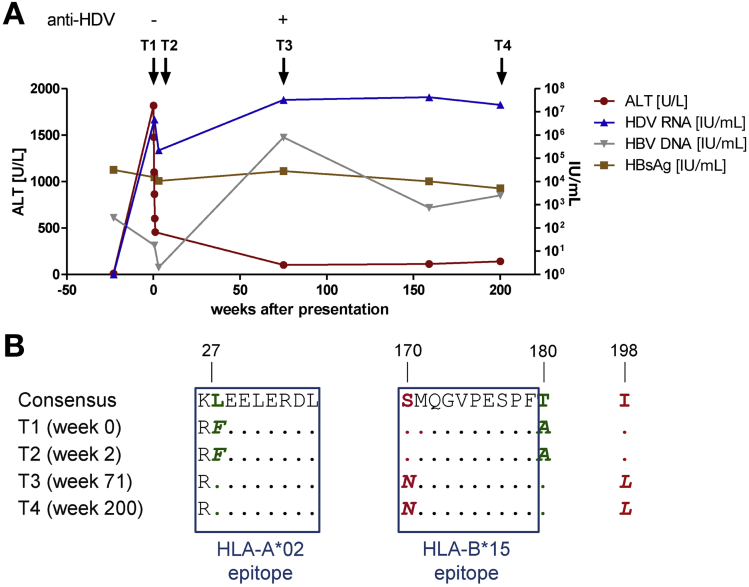
To probe the biological implications of these findings, we characterized the emergence of viral escape mutations in longitudinal samples from an HLA-B*15+ patient with acute HDV/HBV superinfection (P18 in Supplementary Table 2). This patient presented with alanine aminotransferase levels approaching 2000 U/L, which declined to <500 U/L over a period of 1 week, and serum HDV RNA levels approaching 107 IU/mL, which declined by approximately 2 orders of magnitude in the first week and subsequently relapsed to plateau at >107 IU/mL (Figure 5A). Over a period of >200 weeks before initiation of antiviral therapy with pegylated IFN alfa, viral sequence analysis showed a total of 4 amino acid substitutions in L-HDAg. Two of these mutations were reversions toward the consensus sequence, and 2 mutations encoded de novo substitutions away from the consensus sequence (Figure 5B). One of the reversion mutations occurred at an HLA-A*02 anchor residue, suggesting primary acquisition of an escape virus from an HLA-A*02+ individual (patient P18 lacked HLA-A*02). More importantly, 1 of the de novo mutations (S170N) was a bone fide escape variant located in L-HDAg170–179 SMQGVPESPF, matching the HLA-B*15 footprint identified in our cohort of patients with chronic HDV/HBV infection (Figure 1 and Table 1). These observations clearly show the longitudinal accumulation of viral escape mutations in L-HDAg and tentatively link this phenomenon with CD8+ T-cell failure in the context of superinfection with HDV.
Figure 5.
Viral escape in the HLA-B*15–restricted epitope L-HDAg170–179 after superinfection with HDV. (A) Clinical course of HDV/HBV superinfection in patient 18 (HLA-A*24:02+, -B*15:01+, -B*35:02+). (B) Viral sequences in patient 18 at early (week 0 and week 2) and late time points (week 71 and week 200) after superinfection with HDV. All longitudinal sequence variants are shown (n = 4). Dots indicate agreement with consensus across the study cohort. Residues with de novo mutations are shown in red, and residues that revert to wild type are shown in green. The de novo mutation S170N is located in the confirmed HLA-B*15–restricted epitope L-HDAg170–179 and corresponds to the linked footprint (Table 1). Also, the F27L reversion colocalizes with an anchor residue for the pre-described HLA-A*02–restricted epitope L-HDAg26–34. ALT, alanine aminotransferase; T, time.
HDV Sequence Polymorphisms Are Associated With Infrequent HLA Class I Alleles
As noted, most of the HLA class I footprints identified in our cohort were associated with HLA-B alleles (Table 1). In further analyses, we realized that common HLA class I alleles, such as HLA-A*01, -A*02, -A*03, -B*07, -B*08, -B*35, and -B*44, all of which occurred at frequencies >10% in the German reference population, were not associated with viral sequence polymorphisms (Figure 2B). Conversely, significant footprints in the HDV genome were almost exclusively associated with relatively uncommon HLA class I alleles, namely HLA-A*29, -A*30, -B*13, -B*14, -B*15, -B*18, -B*27, -B*37, -B*38, -B*41, -B*49, and -B*51 (Figure 2B and Table 1).
To extend these observations, we compared HLA class I allele frequencies in our cohort of patients with chronic HDV/HBV infection with the corresponding HLA class I allele frequencies in Germany, Italy, and Iran, where the primary medical centers were located for recruitment purposes (Figure 2B). Additional patients were recruited from a medical center in Barcelona, but HLA class I allele frequency data were not available for the general population in Spain. The overall distribution of HLA class I allele frequencies in our cohort of patients with chronic HDV/HBV infection was similar to the overall distribution of HLA class I allele frequencies in Germany, Italy, and Iran (Figure 2B). However, the footprint-linked allele HLA-B*51 was relatively common in our study cohort and in the general populations of Italy and Iran (10%–11%), and we were unable to identify an HLA-B*51–restricted candidate epitope in HDV (Table 1 and Supplementary Table 3). Other immune selection pressures, such as interactions between the Bw4 motif and killer cell immunoglobulin-like receptors, may therefore drive viral mutation in the context of HLA-B*51. Similarly, the footprint-linked allele HLA-B*18 was relatively common in our study cohort (6.7%) and in the general population of Italy (9.7%). This observation is compatible with the negative association between HLA-B*18 and sequence variation at residue 47 in L-HDAg.
These data can be explained by the concept of viral evolution at the population level. Accordingly, relatively infrequent HLA class I allotypes continue to drive the accumulation of escape variants in vivo, whereas more common HLA class I allotypes shape the circulating quasi-species and therefore no longer select for de novo mutations in HDV.
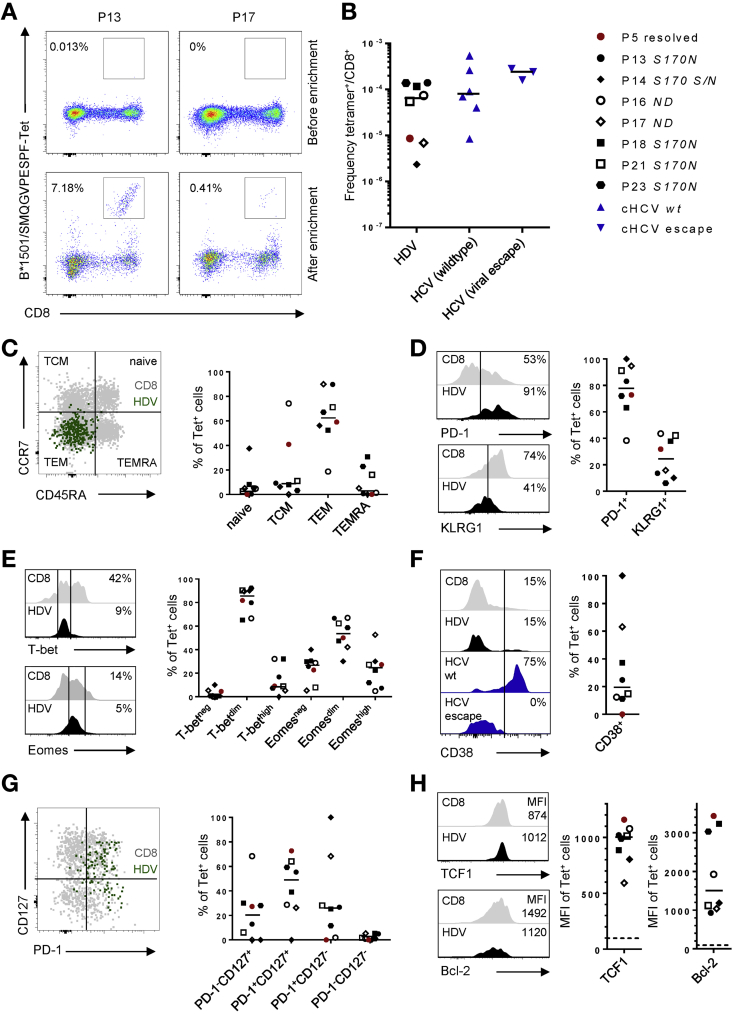
HDV-Specific CD8+ T Cells Are Maintained at Very Low Frequencies After Viral Escape
In line with previous studies,4, 6 we were unable to detect HDV-specific CD8+ T cells directly ex vivo, even with the aid of fluorochrome-labeled tetramers corresponding to the HLA-B*15-restricted epitope L-HDAg170–179 SMQGVPESPF (Figure 6A, upper panels). We therefore characterized these cells using a tetramer-based enrichment strategy.27, 28 Distinct populations of tetramer+ CD8+ T cells were detected after enrichment in all 7 HLA-B*15+ patients with chronic HDV/HBV infection (patients 13, 14, 16, 17, 18, 21, and 23) (Figure 6A, lower panels, and B). The frequencies of these HLA-B*15–restricted HDV-specific CD8+ T cells were low, however, subordinate even to rarely detectable virus-specific CD8+ T cells in patients with chronic HCV infection (Figure 6B). In patients 13, 14, 17, 18, 21, and 23, SMQGVPESPF-specific CD8+ T cells displayed a predominant effector-memory phenotype, whereas in patient 16, SMQGVPESPF-specific CD8+ T cells displayed a predominant central-memory phenotype (Figure 6C). These HDV-specific CD8+ T cells expressed intermediate to high levels of PD-1 (Figure 6D), intermediate levels of the inhibitory receptor KLRG1 (Figure 6D), and intermediate levels of the transcription factors T-bet and Eomes (Figure 6E). In line with these characteristics, which contrast with the typical phenotype of terminally exhausted cells (PD-1hiKLRG1+T-betdimEomeshi), SMQGVPESPF-specific CD8+ T cells also expressed low levels of the activation marker CD38, akin to HCV-specific CD8+ T cells targeting escape variants (Figure 6F). Moreover, SMQGVPESPF-specific CD8+ T cells preferentially displayed a PD-1+CD127+ phenotype (Figure 6G) and expressed relatively high levels of TCF1 (Figure 6H), a transcription factor that defines memory-like cells with proliferative capacity.29 Similar levels of TCF1 were expressed by SMQGVPESPF-specific CD8+ T cells in a patient (patient 5) with resolved HDV infection (Figure 6H). Expression of the pro-survival factor BCL2 was more variable, however, with the highest levels expressed by SMQGVPESPF-specific CD8+ T cells in patient 5 (Figure 6H). These phenotypic characteristics broadly mirror those reported previously for HCV-specific CD8+ T cells in the absence of antigenic stimulation, reflecting viral escape or viral clearance.17, 29 Although further studies are required to establish such parallels across different viral infections, it is notable that SMQGVPESPF-specific CD8+ T cells in patient 14, who retained a subpopulation of prototype viruses, expressed relatively low levels of TCF1 and BCL2 in conjunction with an atypical CD38+PD-1+CD127– phenotype (Figure 6F and G).
Figure 6.
Characterization of HLA-B*15–restricted HDV-specific CD8+ T cells in patients with resolved or persistent HDV infection. (A) HLA-B*15–restricted CD8+ T cells specific for L-HDAg170–179 were tetramer-enriched from PBMCs isolated from patients with chronic HDV/HBV infection. Representative tetramer stainings from patient 13 (relatively large tetramer+ population) and patient 17 (relatively small tetramer+ population) are shown before (upper panels) and after enrichment (lower panels). (B) Frequency of tetramer+ cells among total CD8+ T cells. HDV-specific CD8+ T cells from 7 patients with chronic HDV/HBV infection and 1 patient with resolved HDV infection (patient 5, highlighted in red) were compared with HCV-specific CD8+ T cells targeting epitopes with conserved or escaped viral sequences in chronic HCV infection. Autologous viral sequences corresponding to the L-HDAg170–179 epitope are displayed next to the patient codes. S170N indicates the presence of the viral escape sequence, and S170S/N indicates presence of the prototype and the viral escape sequence, respectively. ND indicates that sequences were not obtained from patients with low levels of HDV RNA. (C–H) Enriched HDV-specific CD8+ T cells were characterized by flow cytometry. Gray histograms indicate non-naive bulk CD8+ T cells. Representative plots are derived from patient 21. (C) Distribution among naive (CD45RA+CD27+CCR7+), central memory (CD45RO+CCR7+), effector memory (CD45RO+CCR7–), and terminally differentiated effector memory subsets (CD45RO–CCR7–). (D) Expression levels of the exhaustion markers PD-1 and KLRG1. (E) Expression levels of the transcriptions factors T-bet and Eomes. (F) Expression of the activation marker CD38 compared with HCV-specific CD8+ T cells targeting epitopes with conserved (wildtype) or escaped viral sequences in chronic HCV infection. (G) Distribution among subsets defined by expression levels of PD-1 and CD127. (H) Expression of the transcription factors TCF1 and BCL2. Dotted lines indicates fluorescence-minus-1 controls. MFI, median fluorescence intensity; ND, not done; P, patient; TCM, central memory; TEM, effector memory; TEMRA, terminally differentiated effector-memory subset; wt, wild type.
Discussion
HDV/HBV infection is one of the few remaining difficult-to-treat conditions in viral hepatitis, leading to liver cirrhosis, liver failure, and hepatocellular carcinoma in many patients.1, 2 Despite the urgent need for novel prophylactic and therapeutic interventions, remarkably little is known about the natural repertoire of virus-specific CD8+ T cells, which are thought to play a key role in immune protection against HDV. In this study, we analyzed HLA class I–associated viral sequence polymorphisms in L-HDAg, giving new insights into the immunobiology of HDV infection. First, we extended the repertoire of defined HDV-specific CD8+ T-cell epitopes from 4, encompassing 2 restricted by HLA-A*024 and 2 restricted by HLA-B*27,6 to 9, now also encompassing 1 restricted by HLA-B*15, 1 restricted by HLA-B*18, 2 restricted by HLA-B*37, and 1 restricted by HLA-B*41. Second, we showed that viral escape occurs during persistent HDV infection. Third, we uncovered a link between viral escape and infrequent HLA class I alleles, indicating population-level adaptation of HDV. Finally, we showed that HDV-specific CD8+ T cells in patients with resolved or persistent HDV infection display similar non–terminally exhausted phenotypes, consistent with viral escape in the absence of clearance. In line with this interpretation, HLA-B*15–restricted HDV-specific CD8+ T cells that reproducibly select for viral escape could be expanded from patients with chronic HDV/HBV infection, whereas HDV-specific CD8+ T cells restricted by HLA class I allotypes that less commonly select for viral escape could not be expanded from patients with chronic HDV/HBV infection.
The phenomenon of viral adaptation at the population level may lead to a relative paucity of CD8+ T-cell epitopes restricted by common HLA class I allotypes, which in turn may impede the development of an effective vaccine against HDV. In line with this prediction, viral sequence polymorphisms were not associated with the common HLA alleles A*01, A*02, A*03, A*24, B*07, B*08, B*35, and B*44, and suitable binding motifs for many of the corresponding allotypes were not present in the HDV proteome (L-HDAg).6 However, these findings need to be confirmed with direct immunogenicity assays, because prediction algorithms are potentially fallible, and certain epitopes may not be subject to viral escape, particularly if they are located in biologically constrained regions of L-HDAg. In this context, our experimental strategy was not designed to identify CD8+ T-cell epitopes that are not subject to viral escape. Moreover, our study cohort did not include patients with acute HDV infection, potentially limiting the detection of novel CD8+ T-cell specificities. An unbiased approach will therefore be required to define the full range of immunogenic epitopes derived from HDV.30 The association between immunogenicity and infrequent allotypes reported here may also be confounded by HLA class I polymorphisms. For example, the viral mutation S170N was found in all patients with HLA-B*15:01 but was not found in patients with HLA-B*15:10 or HLA-B*15:18 (data not shown). Similarly, common polymorphisms in HLA-B*27 can lead to subtype-specific patterns of epitope targeting in the context of other persistent viral infections, including HCV and HIV.31, 32
Viral escape has direct implications for immune efficacy and the development of immunotherapeutic strategies in the setting of chronic HDV/HBV infection. Recent vaccine studies in chimpanzees and humans with chronic HCV infection showed good immunogenicity against nonconserved viral epitopes, but the corresponding vaccine-boosted CD8+ T cells were ineffective, because they failed to recognize the circulating virus.33, 34 Effective therapeutic vaccines will therefore likely need to prime and/or boost virus-specific CD8+ T cells that target conserved epitopes.
The most significant footprint-linked allele in the present study was HLA-B*15, which has also been shown to select for viral escape mutations during persistent HCV infection.35 In addition, genetic association studies have linked HLA-B*15 and/or its serological equivalent (B62) with better outcomes after infection with HCV.36, 37, 38 However, it remains unclear if HLA-B*15 is associated with spontaneous clearance of HDV, and, moreover, any such associations will be extremely difficult to identify, given the clinical heterogeneity of HDV-infected patients and the requirement for co-infection with HBV. In our study cohort, the presence of HDV-specific CD8+ T-cell responses did not correlate with the outcome of infection (clearance vs persistence). However, the presence of HDV amino acid variations corresponding to polymorphisms associated with the patient’s HLA alleles (Supplementary Table 1) displayed a trend for association with both lower HDV RNA titers (median, 20,638 IU/mL [range, 6 –20,098,000,000 IU/mL] in patients with ≥1 variation vs median, 61,930 IU/mL [range, 10–9,804,000 IU/mL] in patients without any variation; P = .054 [Mann–Whitney test]) and lower alanine aminotransferase levels (median, 73 U/L [range, 19–281 U/L] in patients with ≥1 variation vs median 86 U/L [range, 31–507 U/L] in patients without any variation; P = .070 [Mann–Whitney test]), respectively. These results suggest that HLA class I footprint mutations indicate efficient CD8+ T cell pressure, leading to partial control of viremia and liver inflammation. This interpretation is based on the assumption, however, that partial cross-recognition of escape variants occurs and is sufficient to exert pressure on the virus.
It is particularly notable that our findings considerably overlap with those reported previously in the setting of persistent infection with HCV or HIV. For example, in all 3 infections, virus-specific CD8+ T-cell pressure can select escape variants to fixation (HDV: HLA-B*15–associated polymorphism S170N; HCV: HLA-A*01–associated polymorphism Y1444F)39 and drive viral evolution at the population level to extinguish commonly restricted epitopes (HDV: HLA-B*18–restricted epitope L-HDAg47–54; HCV: HLA-A*01–restricted epitope NS31436–1444;39 HIV: HLA-B*51–-restricted epitope RT128–135).40 Similarly, in all 3 infections, virus-specific CD8+ T cells are preferentially restricted by HLA-B,41, 42, 43, 44 rare allotypes confer a particular advantage,45 and viral escape mutations undergo reversion in the absence of selection pressure after transmission.46, 47 Finally, in all 3 infections, CD8+ T cells targeting escape variants display a memory-like phenotype, indicating a loss of antigenic drive.17, 25, 29
Despite these striking parallels across distinct viruses with respect to adaptive host–pathogen interactions, the immunobiology of HDV infection is complicated by the mandatory requirement for co-infection with HBV. Much work will therefore be required to unravel the parameters that govern clinical outcome in the midst of constantly shifting immune responses that simultaneously target rapidly evolving antigens derived from 2 different viruses in a genetically diverse host population. The findings reported here are nonetheless informative with respect to the underlying cellular and molecular processes and can be considered as an early step on a challenging journey.
Acknowledgments
We thank the individuals who donated blood for this study, the physicians and nurses involved in patient recruitment and sample acquisition, and the technicians who isolated PBMCs. We are especially grateful to Christin Ackermann, Thomas Schirdewahn, and Ulrich Spengler for providing information from patients.
Author contributions: Hadi Karimzadeh, Muthamia M. Kiraithe, Michael Roggendorf, and Christoph Neumann-Haefelin designed the study and interpreted the data; Hadi Karimzadeh performed viral sequence and footprint analyses; Muthamia M. Kiraithe, Valerie Oberhardt, and Elahe Salimi Alizei performed immunological experiments; Jan Bockmann, Julian Schulze zur Wiesch, Heiner Wedemeyer, Markus Cornberg, Adalbert Krawczyk, Jassin Rashidi-Alavijeh, Francisco Rodríguez-Frías, Rosario Casillas, Maria Buti, Antonina Smedile, Seyed Moayed Alavian, Marcus Panning, Bijan Raziorrouh, and Christoph Neumann-Haefelin enrolled patients and performed clinical evaluations; Bettina Budeus and Daniel Hoffmann assisted with statistical analyses; Andreas Heinold, and Florian Emmerich performed HLA class I genotyping; Emma Gostick, and David A. Price generated HLA class I tetramers; Jörg Timm, Maike Hofmann, Robert Thimme, and Ulrike Protzer helped design and interpret the experiments; Hadi Karimzadeh, Muthamia M. Kiraithe, Valerie Oberhardt, David A. Price, Michael Roggendorf, and Christoph Neumann-Haefelin wrote the manuscript; all authors read and approved the final manuscript.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported in part by the Deutsche Forschungsgemeinschaft via grants to Bijan Raziorrouh (SFB-TR179/TP3 project number 272983813), Julian Schulze zur Wiesch (SFB841, SFB1328), Daniel Hoffmann (TRR60/B1), Jörg Timm (TI323/4-1), Maike Hofmann, Robert Thimme (SFB-TRR-179/TP1), Ulrike Protzer (SFB-TR179/TP14 and TRR36/B13), and Christoph Neumann-Haefelin (SFB-TRR-179/TP2). Additional funding was provided by the Medical Faculty of the University of Duisburg-Essen/Technical University of Munich and by a registry grant from the European Association for the Study of the Liver. Hadi Karimzadeh was supported by a grant from the German Academic Exchange Service. David A. Price was supported by a Wellcome Trust Senior Investigator Award (100326Z/12/Z). Julian Schulze zur Wiesch, Heiner Wedemeyer, Markus Cornberg, Ulrike Protzer, and Michael Roggendorf were supported by the German Center for Infection Research.
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2019.02.003.
Contributor Information
Michael Roggendorf, Email: michael.roggendorf@tum.de.
Christoph Neumann-Haefelin, Email: christoph.neumann-haefelin@uniklinik-freiburg.de.
Supplementary Material
References
- 1.Hughes S.A., Wedemeyer H., Harrison P.M. Hepatitis delta virus. Lancet. 2011;378:73–85. doi: 10.1016/S0140-6736(10)61931-9. [DOI] [PubMed] [Google Scholar]
- 2.Heidrich B., Yurdaydin C., Kabacam G. Late HDV RNA relapse after peginterferon alpha-based therapy of chronic hepatitis delta. Hepatology. 2014;60:87–97. doi: 10.1002/hep.27102. [DOI] [PubMed] [Google Scholar]
- 3.Roggendorf M. Perspectives for a vaccine against hepatitis delta virus. Semin Liver Dis. 2012;32:256–261. doi: 10.1055/s-0032-1323631. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y.H., Tao M.H., Hu C.P. Identification of novel HLA-A*0201-restricted CD8+ T-cell epitopes on hepatitis delta virus. J Gen Virol. 2004;85:3089–3098. doi: 10.1099/vir.0.80183-0. [DOI] [PubMed] [Google Scholar]
- 5.Schirdewahn T., Grabowski J., Owusu Sekyere S. The third signal cytokine interleukin 12 rather than immune checkpoint inhibitors contributes to the functional restoration of hepatitis D virus-specific T cells. J Infect Dis. 2017;215:139–149. doi: 10.1093/infdis/jiw514. [DOI] [PubMed] [Google Scholar]
- 6.Karimzadeh H., Kiraithe M.M., Kosinska A.D. Amino acid substitutions within HLA-B*27-restricted T cell epitopes prevent recognition by hepatitis delta virus-specific CD8+ T cells. J Virol. 2018;92 doi: 10.1128/JVI.01891-17. e01891–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neumann-Haefelin C. HLA-B27-mediated protection in HIV and hepatitis C virus infection and pathogenesis in spondyloarthritis: two sides of the same coin? Curr Opin Rheumatol. 2013;25:426–433. doi: 10.1097/BOR.0b013e328362018f. [DOI] [PubMed] [Google Scholar]
- 8.Price D.A., Brenchley J.M., Ruff L.E. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202:1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larkin M.A., Blackshields G., Brown N.P. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 10.Budeus B., Timm J., Hoffmann D. SeqFeatR for the discovery of feature-sequence associations. PLoS One. 2016;11:e0146409. doi: 10.1371/journal.pone.0146409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoof I., Peters B., Sidney J. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen M., Lundegaard C., Worning P. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rammensee H., Bachmann J., Emmerich N.P. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics. 1999;50:213–219. doi: 10.1007/s002510050595. [DOI] [PubMed] [Google Scholar]
- 14.Parker K.C., Bednarek M.A., Coligan J.E. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J Immunol. 1994;152:163–175. [PubMed] [Google Scholar]
- 15.Neumann-Haefelin C., McKiernan S., Ward S. Dominant influence of an HLA-B27 restricted CD8+ T cell response in mediating HCV clearance and evolution. Hepatology. 2006;43:563–572. doi: 10.1002/hep.21049. [DOI] [PubMed] [Google Scholar]
- 16.Thimme R., Oldach D., Chang K.M. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bengsch B., Seigel B., Ruhl M. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLoS Pathog. 2010;6:e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alanio C., Lemaitre F., Law H.K. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 2010;115:3718–3725. doi: 10.1182/blood-2009-10-251124. [DOI] [PubMed] [Google Scholar]
- 19.Carlson J.M., Brumme C.J., Martin E. Correlates of protective cellular immunity revealed by analysis of population-level immune escape pathways in HIV-1. J Virol. 2012;86:13202–13216. doi: 10.1128/JVI.01998-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kefalakes H., Budeus B., Walker A. Adaptation of the hepatitis B virus core protein to CD8+ T-cell selection pressure. Hepatology. 2015;62:47–56. doi: 10.1002/hep.27771. [DOI] [PubMed] [Google Scholar]
- 21.Allen T.M., Altfeld M., Yu X.G. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milicic A., Price D.A., Zimbwa P. CD8+ T cell epitope-flanking mutations disrupt proteasomal processing of HIV-1 Nef. J Immunol. 2005;175:4618–4626. doi: 10.4049/jimmunol.175.7.4618. [DOI] [PubMed] [Google Scholar]
- 23.Walker A., Skibbe K., Steinmann E. Distinct escape pathway by hepatitis C virus genotype 1a from a dominant CD8+ T cell response by selection of altered epitope processing. J Virol. 2016;90:33–42. doi: 10.1128/JVI.01993-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifert U., Liermann H., Racanelli V. Hepatitis C virus mutation affects proteasomal epitope processing. J Clin Invest. 2004;114:250–259. doi: 10.1172/JCI20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Timm J., Walker C.M. Mutational escape of CD8+ T cell epitopes: implications for prevention and therapy of persistent hepatitis virus infections. Med Microbiol Immunol. 2015;204:29–38. doi: 10.1007/s00430-014-0372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenzer S., Peters B., Bulik S. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci. 2005;62:1025–1037. doi: 10.1007/s00018-005-4528-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nitschke K., Flecken T., Schmidt J. Tetramer enrichment reveals the presence of phenotypically diverse hepatitis C virus-specific CD8+ T cells in chronic infection. J Virol. 2015;89:25–34. doi: 10.1128/JVI.02242-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schmidt J., Neumann-Haefelin C., Altay T. Immunodominance of HLA-A2-restricted hepatitis C virus-specific CD8+ T cell responses is linked to naive-precursor frequency. J Virol. 2011;85:5232–5236. doi: 10.1128/JVI.00093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wieland D., Kemming J., Schuch A. TCF1+ hepatitis C virus-specific CD8+ T cells are maintained after cessation of chronic antigen stimulation. Nat Commun. 2017;8:15050. doi: 10.1038/ncomms15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landahl J., Bockmann J.H., Scheurich C. Detection of a broad range of low-level major histocompatibility complex class II–restricted, hepatitis delta virus (HDV)–specific T-cell responses regardless of the clinical status. J Infect Dis. 2019;219:568–577. doi: 10.1093/infdis/jiy549. [DOI] [PubMed] [Google Scholar]
- 31.Adland E., Hill M., Lavandier N. Differential immunodominance hierarchy of CD8+ T-cell responses in HLA-B*27:05- and -B*27:02-mediated control of HIV-1 infection. J Virol. 2018;92 doi: 10.1128/JVI.01685-17. e01685–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nitschke K., Barriga A., Schmidt J. HLA-B*27 subtype specificity determines targeting and viral evolution of a hepatitis C virus-specific CD8+ T cell epitope. J Hepatol. 2014;60:22–29. doi: 10.1016/j.jhep.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Callendret B., Eccleston H.B., Satterfield W. Persistent hepatitis C viral replication despite priming of functional CD8+ T cells by combined therapy with a vaccine and a direct-acting antiviral. Hepatology. 2016;63:1442–1454. doi: 10.1002/hep.28309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly C., Swadling L., Capone S. Chronic hepatitis C viral infection subverts vaccine-induced T-cell immunity in humans. Hepatology. 2016;63:1455–1470. doi: 10.1002/hep.28294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruhl M., Knuschke T., Schewior K. CD8+ T-cell response promotes evolution of hepatitis C virus nonstructural proteins. Gastroenterology. 2011;140:2064–2073. doi: 10.1053/j.gastro.2011.02.060. [DOI] [PubMed] [Google Scholar]
- 36.Fitzmaurice K., Hurst J., Dring M. Additive effects of HLA alleles and innate immune genes determine viral outcome in HCV infection. Gut. 2015;64:813–819. doi: 10.1136/gutjnl-2013-306287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hraber P., Kuiken C., Yusim K. Evidence for human leukocyte antigen heterozygote advantage against hepatitis C virus infection. Hepatology. 2007;46:1713–1721. doi: 10.1002/hep.21889. [DOI] [PubMed] [Google Scholar]
- 38.Nitschke K., Luxenburger H., Kiraithe M.M. CD8+ T-cell responses in hepatitis B and C: the (HLA-) A, B, and C of hepatitis B and C. Dig Dis. 2016;34:396–409. doi: 10.1159/000444555. [DOI] [PubMed] [Google Scholar]
- 39.Neumann-Haefelin C., Frick D.N., Wang J.J. Analysis of the evolutionary forces in an immunodominant CD8 epitope in hepatitis C virus at a population level. J Virol. 2008;82:3438–3451. doi: 10.1128/JVI.01700-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawashima Y., Pfafferott K., Frater J., Matthews P. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gaudieri S., Rauch A., Park L.P. Evidence of viral adaptation to HLA class I-restricted immune pressure in chronic hepatitis C virus infection. J Virol. 2006;80:11094–11104. doi: 10.1128/JVI.00912-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kiepiela P., Leslie A.J., Honeyborne I. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 43.Neumann-Haefelin C., Timm J., Spangenberg H.C. Virological and immunological determinants of intrahepatic virus-specific CD8+ T-cell failure in chronic hepatitis C virus infection. Hepatology. 2008;47:1824–1836. doi: 10.1002/hep.22242. [DOI] [PubMed] [Google Scholar]
- 44.Timm J., Li B., Daniels M.G. Human leukocyte antigen-associated sequence polymorphisms in hepatitis C virus reveal reproducible immune responses and constraints on viral evolution. Hepatology. 2007;46:339–349. doi: 10.1002/hep.21702. [DOI] [PubMed] [Google Scholar]
- 45.Trachtenberg E., Korber B., Sollars C. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 46.Leslie A.J., Pfafferott K.J., Chetty P. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 47.Timm J., Lauer G.M., Kavanagh D.G. CD8 epitope escape and reversion in acute HCV infection. J Exp Med. 2004;200:1593–1604. doi: 10.1084/jem.20041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.