Abstract
Objective
To explore the expression differences of miRNA-21, miRNA-31 and miRNA-let7 between lung cancer patient and healthy people, thereby providing reference for early diagnosis of lung cancer.
Method
Real-time PCR was employed to determine the expression difference between lung cancer patients (50 cases) and healthy people (24 cases). The clinical data of lung cancer patients were analyzed to explore the correlation between clinicopathological characteristics and expression level of miRNA-21, miRNA-31, miRNA-let7.
Results
The relative expression levels of miRNA-21 and miRNA-31 in lung cancer group were obviously higher than those in healthy control group, and the relative expression level of miRNA-let7 in lung cancer group was slightly higher than that in healthy control group. Lung cancer patients with lymph node metastasis had higher expression level than those without lymph node metastasis. The ROC curve showed that the three miRNAs had clinical diagnosis efficiency for lung cancer, and the combined detection of the three miRNAs were more efficient in diagnosing lung cancer. Survival curve analysis suggested that the median survival times of patients in the miRNA-21 and miRNA-31 high expression groups were shorter than those in the low expression groups, and the median survival time of patients in miRNA-let7 high expression group was longer than that in the low expression group.
Conclusion
Plasma miRNA-21, miRNA-31 and miRNA-let7 may be diagnostic marker for lung cancer.
Keywords: Lung cancer, Plasma, miRNA-21, miRNA-31, miRNA-let7, Diagnosis
1. Introduction
Lung cancer is one of the malignancies with the highest incidence in the world, each year over 200,000 people being attacked (Takamizawa et al., 2004). Generally, without typical clinical manifestations in the early stage, patients with lung cancer have already been in advanced stages when diagnosed and have poor prognosis with a five-year survival rate less than 15% (Lopez-Camarillo et al., 2012). Provided that lung cancer can be detected and intervened in the early stage, the survival time of patients can be significantly prolonged (Sobin, 1982). Therefore, it is significant to seek an effective early diagnosis marker. Recent studies on MicroRNAs (miRNAs) found that this new type of oncogene or anti-oncogene is stable in mammalian plasma, and its unbalanced expression is closely linked to growth and development of different tumors (Kent and Mendell, 2006). It is indicated in a research that the expression level of miRNA-21 obviously changes in the process of tumor growth, which takes place in the early stage and is associated with tumor differentiation and metastasis (Frezzetti et al., 2011). In 2004, Japanese scholars first found that the expression of miRNA-let7 was significantly down-regulated in lung cancer cells and human lung cancer tissues (Takamizawa et al., 2004). In a study on the expression of miRNA-31 and LATS2 in tumor tissue and paracancerous normal tissue of 22 patients with non-small cell lung cancer, Liang et al. (2016) found that the expression of miRNA-31 was up-regulated in tumor issues and influenced the onset and development of tumor by negatively regulating the level of LATS2. In this study, three miRNAs that were significantly reported were selected, including miRNA-21, miRNA-31, miRNA-let7. Three miRNAs in plasma of lung cancer patients were detected so as to evaluate their diagnostic value for lung cancer.
2. Material and method
2.1. General data
Seventy three patients with middle-advanced lung cancer admitting to The Affiliated Cancer Hospital of Zhengzhou University from June 2008 to July 2009 were collected in this study, and 50 among them were selected through inclusion and exclusion criteria as lung cancer group, including 41 males and nine females. The cohort, aged between 32 and 79 years old, has a median age of 60 years old, and covers 22 cases of squamous cell carcinoma, 10 cases of adenocarcinoma, 8 cases of small cell carcinoma, 5 cases of adenosscale squamous cell carcinoma and 5 cases of other cancers. Inclusion criteria: (1) the patient was diagnosed as lung cancer by pathological diagnosis. (2) The patient had complete clinical diagnosis and treatment data. (3) The patient was not treated by radiotherapy, chemotherapy or biologics before obtaining the specimens. Exclusion criteria: (1) the patient has symptoms that may lead to anoxia, such as dyspnea; (2) the patient suffers from serious heart, liver, kidney or metabolic diseases. Twenty four healthy people who came to the hospital for physical examination at the same time period were selected as healthy control group. There is no significant difference in age and gander between two groups (P > 0.05). This study was approved by the hospital ethics committee. All the patients were aware of the purpose and significance of the study. Following obtaining informed consent from patients, 5 ml venous blood was taken and placed at room temperature for 15–30 min before being centrifuged for 10 min at 4 °C and 3000r/min. The upper layer of transparent yellowish plasma was drawn into the new RNA enzyme EP tube and stored at −80 °C for backup.
2.2. Detection of expression level of miRNA-21, miRNA-31 and miRNA-let7 in plasma
Extraction of total miRNA: The total miRNA was extracted in accordance with instructions of miRNA extraction kit. Purity detection was conducted by spectrophotometer, and the A260/A280 value between 1.8 and 2.1 indicates a qualified purity.
2.3. Detection of expression of miRNA-21
The Tapman miRNA reverse tanscription kit (purchased from ABI company in the United States) was used to synthesize cDNA. Taking cDNA as a template, the Applied Biosystems 7500 PCR instrument (purchased from ABI company in the United States) was employed to perform quantitative detection for miRNA-21 using qRT-PCR method, and miRNA-16 was selected as the reference. According to instructions of the kit, the reaction conditions are: initial denaturation was carried out at 95 °C for 10 min; denaturation was carried out at 95 °C for 15 s and at 60 °C for 1 min, and 40 cycles were carried out to obtain the CT value. The expression (F) of miRNA-21 was calculated using relative quantitative method by Schmittgen and Livak (2008): F = 2−△CT; △CT = CTmiRNA-21-CTmiRNA-16, in which CT refers to the number of cycles corresponding to the setting threshold of the fluorescent signal in each reaction tube.
The expression of miRNA-31 and miRNA-let7 was detected by the same method as above.
2.4. Statistical method
SPSS19.0 was used to carry out statistical analysis of the above data. The normality test of measurement data was performed followed by independent-sample T-test for normally-distributed data and Mann-Whitney test for non-normally distributed. With α = 0.05 as the test level, the results are expressed as [median (Interquartile range)]. Receiver operating characteristic (ROC) curve was used to analyze the efficacy of plasma miRNA in the diagnosis of lung cancer, and Kaplan-Meier method was adopted to carry out the survival analysis of lung cancer patients.
3. Results
3.1. Comparison of expression of miRNA-21, miRNA-31 and miRNA-let7 in lung cancer group and healthy control group
The relative expression level of plasma miRNA-21 in lung cancer group was 0.1227(0.0396,0.2944) and in healthy control group was 0.0511(0.0068,0.1253), indicating that lung cancer group had a obviously higher level than healthy control group, with a statistically significant difference (P < 0.05). Similarly, the relative expression level of plasma miRNA-31 in lung cancer group was 0.0089(0.0002,0.0621) and in healthy control group was 0.0004(0,0.0025), showing that lung cancer group had a obviously higher level than healthy control group, with a statistically significant difference (P < 0.05). The relative expression level of miRNA-let7 in lung cancer group was 0.0016(0.0004,0.1606) and in healthy control group was 0.0010(0,0.0038), suggesting that lung cancer group had a slightly higher level than healthy control group, with a statistically significant difference (P < 0.05). Details are shown in Table 1.
Table 1.
Comparison of expression (2−△CT) of miRNA-21, miRNA-31 and miRNA-let7 in lung cancer group and healthy control group [median (quartile)].
| miRNA-21 | miRNA-31 | miRNA-let7 | |
|---|---|---|---|
| Lung cancer group | 0.1227* (0.0396,0.2944) | 0.0089** (0.0002,0.0621) | 0.0016* (0.0004,0.1606) |
| Healthy control group | 0.0511 (0.0068,0.1253) | 0.0004 (0,0.0025) | 0.0010 (0,0.0038) |
| P-value | 0.035 | 0.001 | 0.019 |
Note: * indicates a statistical difference; ** indicates a statistically significant different.
3.2. Relationship between clinical characteristics and plasma miRNA expression of lung cancer patients
Fifty cases of lung cancer were classified according to the clinicopathological factors (age, gender, smoking history, mass size, lymph node metastasis) so as to analyze the relationship between the relative expression level of miRNA-21, miRNA-31, miRNA-let7 and the clinicopathological factors. Results indicated that lung cancer patients with lymph node metastasis had higher expression level than those without lymph node metastasis (P < 0.05), but other clinicopathological factors were not correlated to the relative expression level of miRNA-21, miRNA-31, miRNA-let7 (P > 0.05). Details are shown in Table 2.
Table 2.
Correlation between the expression (2−△CT) of plasma miRNA and clinical characteristics of lung cancer patients [median (quartile)] * 10−2.
| Patients | N | miRNA-21 | miRNA-31 | miRNA-Let7 |
|---|---|---|---|---|
| Age | ||||
| ≥60 | 28 | 17.45(5.31,45.29) | 0.99(0.03,7.44) | 0.15(0.04,19.11) |
| <60 | 22 | 5.89(2.45,15.74) | 0.62(0.02,3.85) | 0.20(0.02,9.71) |
| Z-value, P-value | −2.248,0.25 | −0.645,0.519 | −0.489,0.625 | |
| Sex | ||||
| Male | 41 | 9.48(2.88,24.09) | 0.84(0.03,5.69) | 0.23(0.04,16.44) |
| Female | 9 | 20.08(5.10,37.45) | 1.85(0.01,17.80) | 0.15(0.03,19.51) |
| Z-value, P-value | −1.557,0.119 | −0.048,0.962 | −0.564,0.573 | |
| Smoking | ||||
| Ex smokers/smokers | 36 | 10.41(3.66,29.77) | 0.59(0.03,6.46) | 0.13(0.04,13.90) |
| Never | 14 | 4.66(1.43,14.75) | 0.23(0.02,2.82) | 0.15(0.02,10.10) |
| Z-value, P-value | −0.281,0.779 | −0.259,0.795 | −0.454,0.650 | |
| Lump size | ||||
| Max. Diameter ≥ 5 mm | 33 | 8.03(1.68,21.86) | 0.79(0.04,6.55) | 2.21(0.08,17.72) |
| Max. Diameter < 5 mm | 17 | 5.16(1.10,28.74) | 0.22(0.01,2.03) | 0.11(0.02,8.32) |
| Z-value, P-value | −0.891,0.373 | −1.116,0.264 | −1.014,0.311 | |
| Lymph node metastasis | ||||
| Metastasis | 14 | 13.28(5.89,31.43) | 1.31(0.07,8.21) | 2.93(0.05,28.70) |
| Never | 36 | 10.41(3.19,28.57) | 0.03(0,1.55) | 0.06(0.03,0.16) |
| Z-value, P-value | −0.821,0.412 | −2.269,0.023 | −2.182,0.029 | |
3.3. Diagnostic efficacy of combined detection of miRNAs for lung cancer
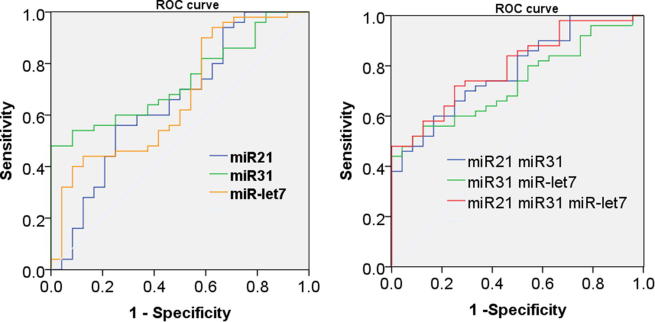
As indicated in ROC curve, the area under the curve (AUC) of plasma miRNA-21 was 0.653, and the sensitivity and specificity were 60% and 66.7%, respectively, with a statistical difference (P = 0.035). The AUC, sensitivity and specificity of miRNA-31 were 0.733, 60%, 75%, respectively, with a statistical difference (P = 0.001), and those of miRNA-let7 were 669, 54% and 58.3%, respectively, with a statistical difference (P = 0.019). The combined detection of miRNA-21 and miRNA-31 showed the AUC, sensitivity and specificity of 0.781, 70% and 70.8%, respectively. For the combined detection of miRNA-31 and miRNA-let7, the AUC, sensitivity and specificity were 0.732, 60% and 75%, respectively, but the combined detection of miRNA-21 and miRNA-let7 showed no statistical difference. The combined detection of the three markers showed the AUC, sensitivity and specificity of 0.798, 72% and 75%, respectively, indicating the diagnostic efficiency of these three markers, and that their combination is more efficient for diagnosis. Details are sown in Fig. 1.
Fig. 1.
ROC curve of miRNA for lung cancer diagnosis.
3.4. Survival curve analysis
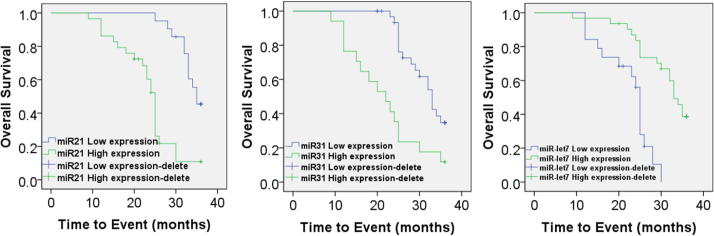
According to the maximum Youden index of ROC curve, we obtained the optimal diagnostic critical values 2−△CT of miRNA-21, miRNA-31 and miRNA-let7, which were 0.065, 0.0025 and 0.0015. Based on that, patients were divided into high expression groups and low expression groups. Results showed that the median survival time in miRNA-21 high expression group was 24 months, and that in miRNA-21 low expression group was 35 months, and the difference was statistically significant (P < 0.05). The median survival time in miRNA-31 high expression group was 22 months, and that in miRNA-31 low expression group was 30 months, and the difference was statistically significant (P < 0.05). The median survival time in miRNA-let7 high expression group was 33 months, and that in miRNA-let7 low expression group was 24 months, and the difference was statistically significant (P < 0.05). Details are shown in Fig. 2.
Fig. 2.
Kaplan-Meier curves for OS (a) and PFS (b) according to lung cancer patient.
4. Discussion
Micro RNA, about 19 to 25 nucleotides in length, is a non coding small RNA found in eucaryote cells that can regulate the development process, and it is involved in the important post transcriptional regulation mechanism of eukaryotic cells (Bartel, 2004). Studies have shown that miRNA, which accounts for less than 3% of the human genome, regulates up to 30% of the genes. What’s more, over 50% miRNA is located in the regulatory region of tumor related genes, so the imbalance of miRNA is probably one of the important mechanisms for tumor (Wang, 2015). Given the complex pathogenesis of lung cancer, miRNA may influence its occurrence and development by a variety of ways; for instance, studies showed the low expression of miRNA-145 in lung cancer patients, indicating that miRNA may act as an anti-oncogene (Melkamu et al., 2010), while studies also proved the high expression of miRNA-141 in lung cancer patients, suggesting that miRNA may act as an oncogene (Barba et al., 2011). In recent years, more and more studies have shown that miRNA can be used as a potential diagnostic marker for cancer (Guo et al., 2015). In a lung cancer research, Wang et al. (2013) studied the expression level of miRNA-21 in 25 patients with lung squamous cell carcinoma, and results found that the expression level of miRNA-21 in lung squamous cell carcinoma was 8.1 times higher than that in adjacent normal tissues. And studies also found that high expression level of miRNA-21 suggested a poor prognosis (Starkey Lewis et al., 2011). A study by Calin et al. (2002) found that the expression level of miRNA-let7 showed obvious down-regulation in lung cancer tissues compared with that in adjacent normal tissues but no obvious down-regulation in breast cancer and colorectal cancer tissues. In addition, lung cancer patients with low expression of miRNA-let7 had shorter survival time than those with high expression (Wang, 2015), and the high expression of miRNA-let7 inhibited the growth of lung cancer A549 cell line (Bowman, 2006). Cui et al. (2013) confirmed in the serum of patients with non-small cell lung cancer that miR-125b can be used as an indicator for diagnosis and prediction of chemotherapy efficacy, thus providing a new idea for solving the drug resistance of lung cancer. However, the application is restricted to a large extent because of the invasive tissue detection and the demand of accurately locating lesion in the process of specimen collection (He and Tu, 2017). A rich, stable expression of miRNA can be detected in the human blood, and it is acknowledged that tumor cells in the blood system are the key to tumor metastasis (Lu, 2010). Currently, most researchers believe that serum miRNA originates from necrotic cells or apoptotic cells, namely, tissue-specific cells are released from necrotic cells and enter into blood, or endogenous miRNA are released from tumor cells and enter into blood (Chen et al., 2012). Serum miRNA detection began in 1948, Mandel and Metais (1948) showed the presence of RNA (circulating RNA) in plasma and serum in healthy people and patients. Compared with traditional serum tumor markers, its endogenous structure endows it with ability to fight ribonuclease and maintain its expression after repeated freezing thawing and pH changes (Chen et al., 2008), which lays a foundation for plasma miRNA being a diagnostic marker. Previous studies are mostly based on unbalanced expression of two kinds of miRNA in lung cancer, and the combined detection of three kinds of miRNA in lung cancer is rarely reported. Therefore, we selected three miRNAs that were significantly reported, including miRNA-21, miRNA-31, miRNA-let7. Three miRNAs in plasma of lung cancer patients were detected so as to evaluate their diagnostic value for lung cancer.
Results showed that the relative expression levels of miRNA-21 and miRNA-31 were higher than those in healthy control group, and the relative expression level of miRNA-let7 was slightly higher than in healthy control group, with statistically significant differences. These results are consistent with studies of Wang Jiajia (Wang et al., 2015) and Liang Zhu (Liang et al., 2016), but the part related to miRNA-let7 doesn’t agree with the study by Calin (Barba et al., 2011), and the following causes are considered: (1) the reference genes were different; (2) the experiment reagent used in two studies were purchased from different companies; (3) there is individual difference in expression level of miRNA in patients; (4) there are racial, gender, geographical and other differences in patients. Through the analysis of correlation between clinical characteristics and plasma miRNA expression of miRNA-21, miRNA-31, miRNA-let7 in lung cancer patients, it was found that lung cancer patients with lymph node metastasis had higher expression level of miRNA-31, miRNA-let7 than those without lymph node metastasis (P < 0.05), indicating that miRNA-31 and miRNA-let7 may be linked to metastasis and invasion of tumor cells. The ROC curve showed that the three miRNA had clinical diagnosis efficiency for lung cancer, and the combined detection of the three miRNAs were more efficient in diagnosing lung cancer. Survival curve analysis suggested that the median survival times of patients in the miRNA-21 and miRNA-31 high expression groups were shorter than those in the low expression groups, and the median survival time of patients in miRNA-let7 high expression group was longer than that in the low expression group, indicating that the expression level of miRNA may be related to disease progression (Dey et al., 2017, Gao et al., 2017, Ge et al., 2018, Peng et al., 2017, Bokhari et al., 2018, Yildirim et al., 2017).
In summary, the miRNA-21 and miRNA-31 in plasma of lung cancer patients were highly expressed, which is correlated to clinical characteristics. Combined detection of miRNA-21, miRNA-31 and miRNA-let7 is valuable for diagnosis of lung cancer, so the application of plasma miRNA in diagnosis, treatment and prognosis is significant. However, there are several limitations. Considering the sample size in this study, there is a possibility of accidental error. Therefore, the sensitivity, the specificity and the selection of critical value remain to be verified in a large scale samples.
Acknowledgement
The work was financially supported by The Science and Technology Breakthrough Project of Henan Province (Grant No.: 172102310521) and The Medical Science and Technology Breakthrough Project of Henan Province (Grant No.: 201702253).
Footnotes
Peer review under responsibility of King Saud University.
References
- Barba M., Felsani A., Rinaldi M., Giunta S., Malorni W., Paggi M.G. Reducing the risk of overdiagnosis in lung cancer: a support from molecular biology. J. Cell. Physiol. 2011;226(9):2213–2214. doi: 10.1002/jcp.22558. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bokhari T.H., Roohi S., Hina S., Saeed S. Synthesis, optimization and biological evaluation of (99)Mtc-digoxin as possible cardiac imaging agent. Pak. J. Pharmac. Sci. 2018;31(1):19–24. [PubMed] [Google Scholar]
- Bowman E. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9(3):189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Frequentl deletions and down-regulation of microRNA genes miR15 and Mir-16 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Zhang Y., Wang J., Jiang X., Xiang Y., Xu C., Zheng P., Zhang J., Li R., Zhang H., Shang X., Gong T., Ning G., Wang J., Zen K., Zhang J., Zhang C.Y. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Chen F., Lu H.Q., Zang F.R., Zhou X.F. Correlation study on serum levels of three miRNA and early diagnosis of non-small cell lung cancer. Chin. Mod. Doctor. 2012;50(25):56–57. [Google Scholar]
- Cui E.H., Li H.J., Hua F., Wang B., Mao W., Feng X.R., Li J.Y., Wang X. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol. Sin. 2013;34(2):309–313. doi: 10.1038/aps.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S., Ghosh M., Rangra N.K., Kant K., Shah S.R., Pradhan P.K., Singh S. High-performance liquid chromatography determination of praziquantel in rat plasma; application to pharmacokinetic studies. Ind. J. Pharmac. Sci. 2017;79(6):885–892. [Google Scholar]
- Frezzetti D., De Menna M., Zoppoli P., Guerra C., Ferraro A., Bello A.M., De Luca P., Calabrese C., Fusco A., Ceccarelli M., Zollo M., Barbacid M., Di Lauro R., De Vita G. Upregulation of miRNA-21 by Ras in vivo and its role in tumor growth. Oncogene. 2011;30(3):275–286. doi: 10.1038/onc.2010.416. [DOI] [PubMed] [Google Scholar]
- Gao W., Baig A.Q., Ali H., Sajjad W., Farahani M.R. Margin based ontology sparse vector learning algorithm and applied in biology science. Saudi J. Biol. Sci. 2017;24(1):132–138. doi: 10.1016/j.sjbs.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge S., Wang L., Liu Z., Jiang S., Yang X., Yang W., Peng W. Properties of nonvolatile and antibacterial bioboard produced from bamboo macromolecules by hot pressing. Saudi J. Biol. Sci. 2018;25(3):474–478. doi: 10.1016/j.sjbs.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Zhang H., Zhang L., He Y., Weng S., Dong Z., Wang J., Zhang P., Nao R. MicroRNA-21 regulates non-small cell lung cancer cell proliferration by affecting cell affecting cell apoptosis via COX-19. Int. J. Clin. Exp. Med. 2015;8(6):8835–8841. [PMC free article] [PubMed] [Google Scholar]
- He Y.L., Tu H.Y. Progress of relativity between miRNA and the diagnosis and treatment of non-small cell cancer. Shandong Med. J. 2017;57(4):110–112. [Google Scholar]
- Kent O.A., Mendell J.T. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–6196. doi: 10.1038/sj.onc.1209913. [DOI] [PubMed] [Google Scholar]
- Liang Z., Chen J., He Z., Lin L.Y., Xie Z.Q., Xie J.L., Chen C.Y. The expression of miRNA-31 and LAST2 protein in the non-small cell lung cancer tissues. J. Guangdong Med. Coll. 2016;34(6):596–598. [Google Scholar]
- Lopez-Camarillo C., Marchat L.A., Arechaga-Ocampo E., Perez-Plasencia C., Del Moral-Hernandez O., Castaneda-Ortiz E.J., Rodriguez-Cuevas S. MetastamiRs: non-coding microRNAs driving cancer invasion and metastasis. Int. J. Molecul. Sci. 2012;13(2):1347–1379. doi: 10.3390/ijms13021347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, S.H., 2010. Discovery and validation of miRNA biomarkers in tissue and plasma of the patients with different types of lung cancer. Ph.D. Dissertation, Fudan University.
- Mandel P., Metais P. Les acides nucleiques du plasma sanguin cbez l’Homme. C R Seances Soc. Biol. Fil. 1948;142:241–242. [PubMed] [Google Scholar]
- Melkamu T., Zhang X., Tan J., Zeng Y., Kassie F. Alteration of microRNA expression in vinyl carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2010;31(2):252–258. doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- Peng W., Ge S., Ebadi A.G., Hisoriev H., Esfahani M.J. Syngas production by catalytic co-gasification of coal-biomass blends in a circulating fluidized bed gasifier. J. Clean. Prod. 2017;168:1513–1517. [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sobin L.H. The world health organization’s histological classification of lung tumors: a comparison of the first and second editions. Cancer Detect. Prevent. 1982;5:391–406. [PubMed] [Google Scholar]
- Starkey Lewis P.J., Dear J., Platt V., Simpson K.J., Craig D.G., Antoine D.J., French N.S., Dhaun N., Webb D.J., Costello E.M., Neoptolemos J.P., Moggs J., Goldring C.E., Park B.K. Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology. 2011;54(5):1767–1776. doi: 10.1002/hep.24538. [DOI] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Takamizawa J., Konishi H., Yanagisawa K., Tomida S., Osada H., Endoh H., Harano T., Yatabe Y., Nagino M., Nimura Y., Mitsudomi T., Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64(11):3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Wang Q. Advances of MicroRNA in diagnosis and treatment of lung cancer. J. Clin. Pulm. Med. 2015;9:1728–1730. [Google Scholar]
- Wang J.J., Lei W., Chen C., Mu C.Y., Shen X.C., Liu Z.Y., Huang J.A. Expression and significance of microRNA-205 in non-small cell lung cancer tissues and plasma. Jiangsu Med. J. 2015;41(10):1167–1169. [Google Scholar]
- Wang Y., Xi L.F., Yang T.T. Correlation of miRNA-21 expression with C-myc in lung squamous carcinoma. Chin. J. Anat. 2013;36(4):758–761. [Google Scholar]
- Yildirim N., Simsek M., Turkeli M., Bilici M., Ozmen H.K. Early onset of radiation induced sarcoma: a case report and review of the literature. Acta Medica Mediterranea. 2017;33(4):565–568. [Google Scholar]