Abstract
Filamentous bacterial belonged to Streptomyces species were novel drug source for medical and industrial applications. However, the detailed identification of Streptomyces species from Saudi Arabian extreme environment for the identification novel drug source for medical and industrial applications were rarely studied. The Streptomyces strain Al-Dhabi-2 obtained from the thermophilic region kingdom of Saudi Arabia, exhibited antimicrobial potentials against the pathogenic microorganism were characterized. Biochemical and phylogenetic analysis confirmed that the strain was closely associated to the Streptomyces species. The chromatogram of GC-MS analysis of this ethyl acetate extract (EA) had diverse of chemical compounds namely benzene acetic acid (7.81%), acetic acid, methoxy-, 2-phenylethyl ester (6.01%) were the major compounds. EA of Al-Dhabi-2 showed inhibition zone ranged from 14 to 25 mm at 5 mg/well concentration against the tested microbial pathogens. Results revealed that the significant MIC values were observed against B. cereus, and E. faecalis by (less than 39 μg/ml) and against S. agalactiae with (78 μg/ml). Minimum inhibitory concentrations (MIC) for fungi: were also reported against Cryptococcus neoformans and Trichophyton mentagrophytes by (156 μg/ml), whilst Candida albicans and Aspergillus niger by (312 μg/ml). Results of this study showed that thermophilic actinobacteria could be promise source in the context of searching for unique antimicrobial agents with novel properties.
Keywords: 16S rRNA gene, Thermophilic Streptomyces, Hot spring, Antimicrobial activity, GC-MS
1. Introduction
Saudi Arabia has a good source of clean and sustainable energy (Aljuhani, 2012), nearly ten geothermal hot springs available in Saudi Arabia (Khiyami et al., 2012). Thermophilic bacteria and archaea are populated and diverse in geothermal environments (Kaur et al., 2008). Microbes that thrive in harsh ecosystems are called extremophiles. During the last four decades, these microorganisms have been isolated form extreme sites (Khalil, 2011). Thermal systems scattered on different locations on the planet harbour several of thermophilic archaeal genera (Arab et al., 2000). Recently, thermophilic actinobacteria have gained notable interest by the researchers owing to the promising results they showed to produce variety of substances with biological activities and the economic potential for that. Theremophilic actinobacteria can be used in biodegradation and other biological processes (Kleeberg et al., 1998), produce bioactive molecules (Takeuchi et al., 1991) or enzymes (Uzel and Hames-Kocabas, 2007, Hames-Kocabas and Uzel, 2007). The genus belonged Streptomyces were known for the production of bioactive compounds such as streptothricin, streptomycin, actinomycin, chloramphenicol, tetracycline, erythromycin, leucomycin, vancomycin, gentamicin, teicoplanin, fortimicin, rosamicin, nocardicin and salinosporamide A respectively (Waksman et al., 2010, Arasu et al., 2013b, Al-Dhabi et al., 2014, Arasu et al., 2015, Al-Dhabi et al., 2019).
Actinobacteria, Gram positive bacteria, are the pioneers among all microbes in term of producing antimicrobial agents in drug discovery programs (Ellaiah et al., 2004, Stackebrandt et al., 1997, Arasu et al., 2013a, Balachandran et al., 2015, Al-Dhabi et al., 2018a, Al-Dhabi et al., 2018b). In nature, the distribution of actinobacteria are very diverse and represent a main source for commercially active molecules (Andrew and Cook Paul, 2003, Valli et al., 2012). Actinobacteria represent prolific source of several active compounds (Demain, 1995), with great impact in clinical treatment and other applications of human medicine (Watve et al., 2001). Thermophilic actinobacteria are potential as producer of such bioactive compounds due to they can grow rapidly, thus rapid mycelium autolysis in compared to mesophilic actinobacteria (Moreira et al., 1981). The genus Streptomyces produces the vast majority of the bioactive compounds more than any other genera within the actianobacteria phylum, therefore, Streptomyces has been extensively investigated and reported as a prolific mine for unique and new antibiotics (Watve et al., 2001). The current project aim to isolate thermophilic actianobacteria from the sediment sample collected from thermal spring located in kingdom of Saudi Arabia. Streptomyces Al-Dhabi-2 identified based on cultural, physiological, molecular level, biological parameters and sensitivity to different antibiotics. The secondary metabolites of the strain have been extracted to investigate their antimicrobial activities against pathogenic bacteria and fungi.
2. Materials and methods
2.1. Chemicals and medium
Cultivation media such as starch casein agar, nutrient agar, muller hinton agar and muller hinton broth were purchased from Himedia, India. Solvents such as hexane, chloroform, ethyl acetate, methanol, acetone and DMSO were obtained from Somato, Riyadh, Saudi Arabia. Commercial antibiotics such as, nalidixcic acid, actidione, streptomycin was procured from Himedia, India. Experiments were performed in mill Q water for the routine experiments.
2.2. Source of actinomycetes
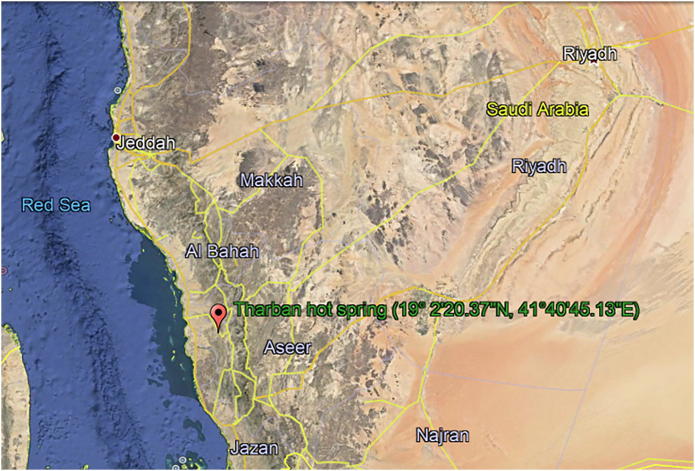
Sediments from hot spring of Tharban (19°2′20.37″N, 41°40′45.13″E) located in the southern west of Al-Mjardah province, in Aseer region, Saudi Arabia was collected aseptically (Fig. 1). The sediments samples were collected with sterilized scoop into the place. Each sample was taken in depth of 6–15 cm from ground’s surface. Sediments samples were put into sterile polythene bags and tightly covered. Immediately, the samples transferred to the our research laboratory, the samples were kept until to be dried and finally the dried sediments stored in a refrigerator until further use.
Fig. 1.
Hot spring of Saudi Arabia, taken from Google earth.
2.3. Isolation, cultivation and maintenance thermophilic actinomycetes
A portion of sediment sample was dried at 100 °C by using hot air oven for one-hour support for the isolation of thermophilic actinobacteria (Uzel et al., 2011) dried sediment added to sterile DD water (10 g/100 ml) and thoroughly shaking for half hour (Kuster and Williams, 1981). Thermophilic actinobacteria were isolated via spreading dilutions of samples by adding 500 μl onto isolation media, nalidixic acid (25 µg/ml) and cycloheximide (50 µg/ml) antibiotics were supplemented to the both media to inhabit the grow of unwanted microes. The incubation of the plates were at 55 °C for 3–5 days with presence of humidity. The strains were repeatedly sub-cultured on SCA until obtain the pure culture. Pure isolates were stored in 50% (v/v) glycerol at freezer for long time storage.
2.4. Morphological characteristics of Streptomyces sp. Al-Dhabi-2
A loopful from the broth culture of strain Al-Dhabi-2 was spread on a clean glass slide to prepare the smear and then drying by heat. Crystal violet and iodine were added and washed with water and treated with alcohol at appropriate times. Finally, safranin stain was added onto the smear for 30 sec, the excessed of the stain was discard and the slide was drying. Slide was examined using phase-contrast microscope at 100×. Streptomyces sp. Al-Dhabi-2 was inoculated on 4 different ISP media (ISP2-ISP5) and MNGA medium, the incubation temperature was at 50 °C for 3–5 days. The morphological properties were observed under a magnifying lens, which include aerial, substrate mycelium, colour and the branching (Shirling and Gottlieb, 1966).
2.5. Characterizations of the strain by physiological and biochemical analysis
Standard method was followed for this analysis (Valan Arasu et al., 2008). Gelatin hydrolysis, starch hydrolysis, production of DNase, hydrogen sulphide production and ability to grow in different temperatures (29–55 °C), range of pH (5–9) and concentrations of NaCl (4% to 13%) on medium were tested using standard methods.
2.6. Molecular identification of Al-Dhabi-2
Thermophilic actinobacteria Al-Dhabi-2 inoculated in yeast peptone glucose (YPG) medium at 50 °C for 3–5 days. The extraction the DNA of strain Al-Dhabi-2 was performed by using the commercial kit and amplifications were done using the following primers [F 27 (5′-AGAGTTTGATCCTGGCTCAG-3) and R 1492 (5′-TACGGCTACCTTGTTACGACTT-3′)]. The PCR products were sequenced and analysed.
Clustal W software was used for aligning of the strain Al-Dhabi-2 16S rRNA sequences to those retrieved genes sequences form the NCBI. The resulted data was processed via neighbour-joining method used for analysed and analysing (Saitou and Nei, 1987). The evaluation of the topology of the resultant tree using bootstrapping assay by 1000 replications of the neighbour-joining tree.
2.7. Antimicrobial activity
2.7.1. Test microbes
The following pathogenic microorganisms were taken for preliminary screening and Minimum inhibitory concentration (MIC) studies. Escherichia coli, Bacillus cereus, Klebsiella pneumonia, Staphylococcus epidermidis, Enterococcus faecalis, Proteus vulgaris, Salmonella typhimurium, Staphylococcus aureus, Streptococcus agalactiae and Psudomonas aeruginosa. Fungi: Candida albicans and Cryptococcus neoformans were used. The tested microbes are ATCC except Cryptococcus neoformans (clinical isolate).
2.7.2. Inoculum preparation
Inoculums of the selected pathogenic bacteria inoculated into growth medium and incubated for 24 h at 35 °C. the inoculums were diluted via using sterile MHB medium to desired cell counts. Inoculums of fungi were prepared by grown in SDA slants for 10 days at 28 °C. Sterile D.D. water (5 ml) added to the fungal growth on slants and homogenized, Yeasts were inculcated into contain SDB media.
2.7.3. Preliminary antimicrobial activity
The inhibitory potential of strain Al-Dhabi-2 were screened and determined by cross-streak method (Duraipandiyan and Ignacimuthu, 2009). Secondary screening of fermented broth was tested against microbe by agar well diffusion method.
2.7.4. Optimization of media and antimicrobial metabolites production
The thermophilic strain Al-Dhabi-2 was inoculated into several broth media to evaluate the appropriate medium for bioactive secondary metabolite production (150 rpm at temperature 45 °C) in an orbital shaker. These media are; Antibiotic production medium, M6 medium, M3 medium-Micromonospora medium, yeast extract malt medium and MNG broth media. We have followed standard protocol which was published our previous paper (Al-Dhabi et al., 2016).
2.7.5. Extraction of antibacterial metabolites
Streptomyces Al-Dhabi-2 was grown in a shaker using modified nutrient glucose (MNG) broth medium, based on media optimization, for extraction of antimicrobial compounds for 13 days at 45 °C. Growth culture was filtered by using Whatman filter paper. About 5000 ml of culture filtrate was collected. The filtrate pH was reduced to 2 pH using HCl (0.1 N) and the organic solvent ethyl acetate was used as for extraction of antimicrobial metabololites by adding two different equal volume (v/v) of culture filtrate and organic solvent. Two layers were formed; the upper layer contains the organic solvent and the extracted secondary metabolites. Organic phase was separated by using separating funnel and the crude metabolites were collected after removing the organic solvent at 60 °C by using vacuum evaporator (IKA Rotary Evaporator model). The crude extract was stored in dark and cold place for further tests.
2.7.6. Preparation of stock crude extracts
Stock solution was prepared by dissolving 100 mg of EA into 0.5 ml of DMSO.
2.7.7. Agar-well diffusion method
Antibacterial and antifungal activities of the strain Al-Dhabi-2 were screened against selected human pathogenic microbes via using agar-well diffusion method. Wells with 5 mm diameter were made on the agar media using sterile cork borer followed by filling 25 µl of the prepared ethyl crude extract with concentration 5 mg/well. The standard positive control for bacteria and fungi are Streptomycin (25 µg) and caspofungin (200 µg) respectively, whilst the negative control is dimethyl sulfoxide (DMSO).
2.7.8. Determine MIC
Test concentrations were prepared via mixing Crude extract of strain Al-Dhabi-2 with DMSO:water (1:9). The evaluation of minimum inhibitory concentration (MIC) has conducted by previously published procedures (Duraipandiyan et al., 2010). The required concentrations (mg/ml) of the test extract (0.0781, 0.156, 0.312, 0.625, 1.25, 2.5 and 5 mg/ml) were placed into the 96 well plate which contain MHB medium. From each inoculum culture suspension 5 μl added into the wells and the 105 CFU/ml was the final inoculum size. The positive controls were caspofungin and streptomycin whilst DMSO was included as a negative control. The lowest concentration of the extract inhibited growth of microbe was used to determine the MIC values.
2.8. Chemical profile of crude extract
The profile of substances contained bioactive compounds in the crude extract of the strain were investigated via using gas chromatography (GC-MS) as described by Al-Dhabi et al., 2016, Valsalam et al., 2019).
3. Results
Actinobacteria have been extensively studied, earlier researchers have been published several reports regarding to the morphology, physiology, biochemical and biological properties of the actinobacteria (Al-Dhabi et al., 2016). However, there is not much report on thermophilic actinobacteria with antibacterial and antifungal properties from the extreme environmental regions of Saudi Arabia (Fig. 1).
3.1. Cultural properties
Strain Al-Dhabi-2 was Gram positive and filamentous. Cultural properties results revealed that the strain can grow well on various ISP and MNGA media. Arial mycelium of Al-Dhabi-2 was grey and brown color. Diffusible pigment was not produced in the agar plate (Table 1).
Table 1.
Morphological traits of the strain Al-Dhabi-2.
| Media | Growth | Colour of aerial mycelium | Reverse side colour | Pigment |
|---|---|---|---|---|
| MNGA | +++ | Grey | Yellow-brown | Absent |
| ISP-2 | ++ | Grey | Yellow-brown | Absent |
| ISP-3 | ++ | Grey | Brown | Absent |
| ISP-4 | ++ | Grey | White | Absent |
| ISP-5 | +++ | Whitesh-grey | White | Absent |
+++: prolific growth and ++: good growth.
3.2. Biochemical and physiological characteristics
Physiological and biochemical traits of Al-Dhabi-2 were studied. The results showed that the strain could be cultured at a salt concentrations ranging from 4% to 7%. The isolated strain Al-Dhabi-2 grew normally at the pH ranging between 5 and 9 and the optimal pH was 7.0. The strain grew in a range of the different temperatures (29 °C to 55 °C) on all tested media. The optimal growth temperature was 50 °C. Based on these results the isolated actinomycete Al-Dhabi-2 was classified as thermophilic actinobacteria. The tolerance of temperature, pH, variation of NaCl concentrations, utilization of different carbon source, production of amylase, DNase, gelatinase and sensitivity to the different antibiotics indicated that the strain belongs to actinobacteria (Table 2).
Table 2.
Biochemical and physiological traits of the strain Al-Dhabi-2.
| Characteristics | Results |
|---|---|
| Gram stain | Positive |
| Growth | Filamentous aerial growth |
| diffusible pigment production | – |
| Melanoid production | – |
| Growth temperature range | 29 °C to 55 °C |
| Growth optimal temperature | 50 °C |
| Growth pH range | 5–9 |
| Optimal pH for growth | 7.0 |
| Production of H2S | − |
| Amylase | + |
| Protease | − |
| Gelatinase | + |
| deoxyribonuclease (DNase) production | + |
| NaCl tolerant | 4% to 7% |
| Carbon sources utilization | |
| Negative control; none carbon source | − |
| Positive control; glucose | ++ |
| Sucrose | − |
| D-fructose | + |
| D-rhamnose | ++ |
| D-Xylose | ++ |
| D-arabinose | ++ |
| D-manitol | ++ |
| Sensitivity to antibiotics | |
| Ciprofloxacin (5 µg) | S |
| Gentamicin (10 µg) | S |
| Penicillin (10 µg) Ampicillin (25 µg) |
R R |
| Chloramphenicol (30 µg) | S |
| Vancomycin (30 µg) | S |
| Tetracycline (30 µg) | S |
| Nystatin (100 µg) | R |
+: presence; −: absence; S: Sensitive; R: Resistance.
3.3. DNA extraction, sequencing, and phylogenetic analysis
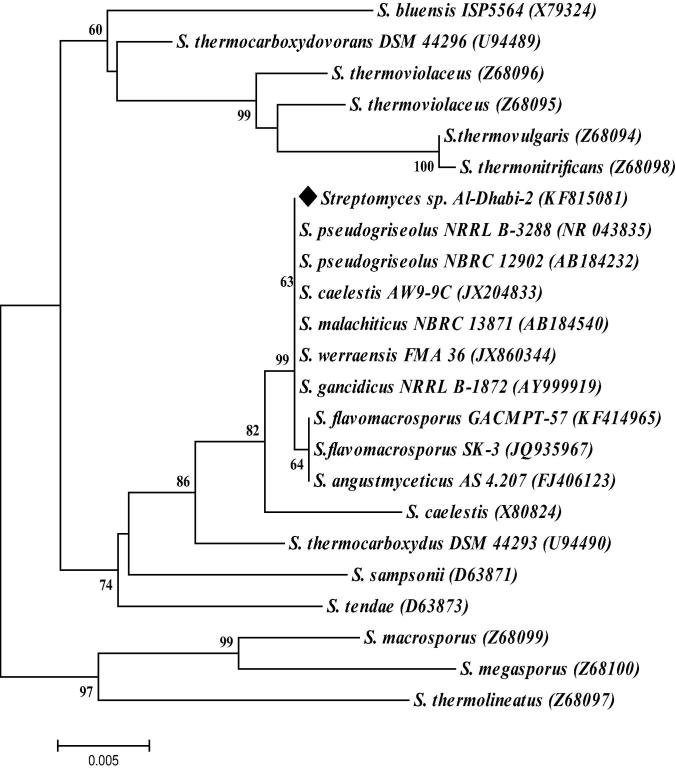
Based on phenotypic characteristics and according to the Bergey’s Manual, Al-Dhabi-2 was identified as Streptomyces sp. 1238 bp was resulted from the sequencing of 16S rRNA gene was and resulted, the sequences were deposited to NCBI and got the accession number (KF815081). 16S rRNA gene sequences showed high similarity to those sequences of 16S rRNA gene deposited in NCBI, sequences of Streptomyces pseudogriseolus was the closest to Al-Dhabi-2 sequences with 100% (E Value: 0.0) identity. Phylogenetic analysis confirmed these results as shown in phylogenetic tree of strain Al-Dhabi-2 (Fig. 2). Eventually the molecular results are consistent and supported the phenotyping identification of the strain Al-Dhabi-2 as Streptomyces sp.
Fig. 2.
Neighbour-joining tree based on partial 16S rRNA gene sequences showing the relationship between strain Al-Dhabi-2 and 22 species of the genus Streptomyces. Values at nodes point to bootstrap support (%) levels based on analysis of 1000 resampled datasets the values >50% are given only. Numbers between parentheses are NCBI accession numbers for each sequence.
3.4. Antibacterial and antifungal activities
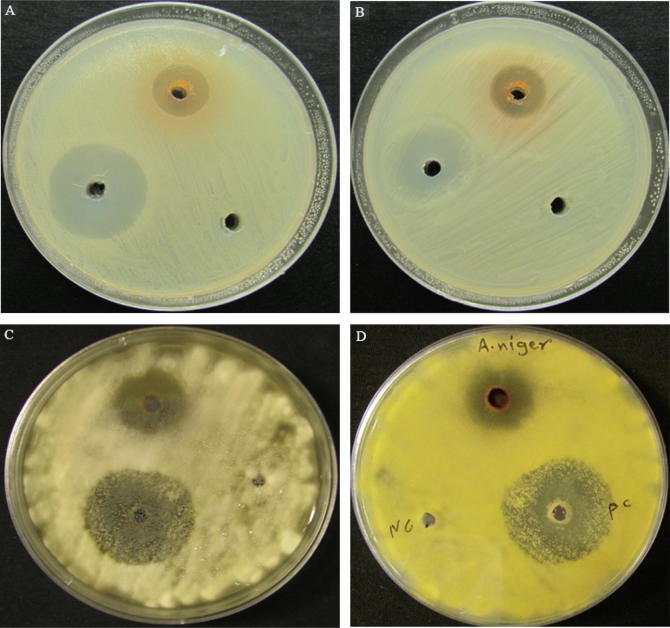
Preliminary antimicrobial screening showed that Streptomyces Al-Dhabi-2 inhibited the growth of tested microbes in streak method. Significant antimicrobial activities observed against Escherichia coli, Bacillus cereus, Enterococcus faecalis, Klebsiella pneumonia, Staphylococcus epidermidis, Proteus vulgaris, Streptococcus agalactiae, Salmonella typhimurium, Psudomonas aeruginosa, Staphylococcus aureus, C. albicans and Cryptococcus neoformans (Table 3). Based on the results of preliminary screening, further studies have been conducted in the secondary screening. The levels of bioactive compounds production in different growth media were determined. Results showed that the medium MNG was the best one for antibiotic production. Thus, strain Al-Dhabi-2 was grown on MNG broth medium. Ethyl acetate was used to extract bioactive compounds. Crude extract yielded 800 mg. Agar well-diffusion assay was used for screening the crude extract of Al-Dhabi-2 against bacterial and fungal pathogens. Al-Dhabi-2 exhibited good antimicrobial activity against the determined ATCC microbes (Table 4). The highest antimicrobial activities were reported against E. Faecalis (16 mm) followed by, S. epidermidis, B. cereus and S. agalactiae by 15 mm zone of inhibition. However, the activity against filaments fungi was interested by 19 mm for A. niger followed by 16 mm zone of inhibition for T. mentagrophytes (Fig. 3).
Table 3.
Antimicrobial activity of Al-Dhabi-2 in preliminary screening.
| Tested microbes | ATCC* strain No. | Inhibition activity |
|---|---|---|
| B. cereus | 11778 | ++ |
| S. epidermidis | 12228 | ++ |
| S. aureus | 6538P | + |
| E. faecalis | 49532 | +++ |
| S. agalactiae | 27956 | ++ |
| E. coli | 10536 | ++ |
| P. vulgaris | 33420 | + |
| P. aeruginosa | 27853 | ++ |
| S. typhimurium | 13311 | ++ |
| K. pneumonia | 13882 | ++ |
| C. albicans | 2091 | ++ |
| C. neoformans | Clinical strain | ++ |
ATCC: American type culture collection, +: moderate; ++: good; +++: Significant.
Table 4.
The activity of Al-Dhabi-2 ethyl acetate extract against list of human pathogens.
| Microorganisms | ATCC No. | Zone of inhibition in mm |
|
|---|---|---|---|
| Bacteria | Al-Dhabi-2 (5 mg/well) | Streptomycin 10 µg | |
| B. cereus | 11778 | 15 | 22 |
| S. epidermidis | 12228 | 15 | – |
| S. aureus | 6538P | 11 | 12 |
| E. faecalis | 49532 | 16 | 24 |
| S. agalactiae | 27956 | 15 | 20 |
| E. coli | 10536 | 14 | 15 |
| P. vulgaris | 33420 | 10 | 15 |
| P. aeruginosa | 27853 | 14 | 16 |
| S. typhimurium | 13311 | 10 | 17 |
| K. pneumoniae | 13882 | 11 | 17 |
| Fungi Caspofungin 200 µg | |||
| A. niger | 16888 | 19 | 27 |
| T. mentagophytes | 9533 | 16 | – |
| C. albicans | 2091 | 10 | 22 |
| C. neoformans | Clinical strain | 10 | – |
–: no activity.
Fig. 3.
Antimicrobial activity of Al-Dhabi-1 ethyl acetate extract against human pathogenic microbes. (A) E. faecalis; (B) S. agalactiae; (C) A. niger and (D) A. niger (reverse side of the plate). PC; positive control and NC; negative control.
Based on the well diffusion assay results the extract was also studied for minimum inhibitory concentrations for tested bacteria and fungi. The MIC evaluation the extract was conducted using standard method. The values of MIC of ethyl acetate extract have been ranged between less than 0.039 to 0.625 mg/ml as shown in Table 5. The highest values were observed against B. cereus, E. faecalis with <0.039 mg/ml and S. agalactiae by 0.078, while in fungi the highest value is 0.156 mg/ml against C. neoformans and T. mentagrophytes.
Table 5.
MIC values of Al-Dhabi-2 ethyl acetate extract using micro-broth dilution method.
| Microorganisms | ATCC No. | MIC |
|
|---|---|---|---|
| Bacteria | Al-Dhabi-2 (mg/ml) | Streptomycin µg/ml | |
| B. cereus | 11778 | <0.039 | 0.156 |
| S. epidermidis | 12228 | 0.156 | >10 |
| S. aureus | 6538P | 0.625 | 2.5 |
| E. faecalis | 49532 | <0.039 | 1.25 |
| S. agalactiae | 27956 | 0.078 | 1.25 |
| E. coli | 10536 | 0.312 | 0.625 |
| P. vulgaris | 33420 | 0.625 | 5 |
| P. aeruginosa | 27853 | 0.625 | 1.25 |
| S. typhimurium | 13311 | 0.312 | 5 |
| K. pneumoniae | 13882 | 0.156 | 0.625 |
| Fungi Caspofungin µg/ml | |||
| A. niger | 16888 | 0.312 | 62.5 |
| T. mentagophytes | 9533 | 0.156 | >500 |
| C. albicans | 2091 | 0.312 | <4 |
| C. neoformans | Clinical strain | 0.156 | 250 |
3.5. GC-MS profile of crude extract
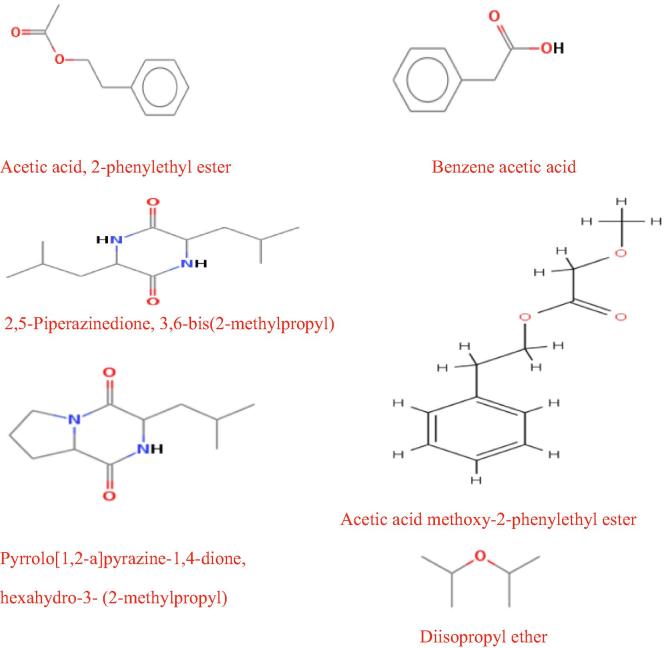
GC-MS chromatograph has been used for analysis the diversity of the compounds contained within the extract of Sterptomyces sp. Al-Dhabi-2. Results showed that crude extract consists of many compounds (Table 6). The acetic acid, 2-phenylethyl ester (10.45%), benzene acetic acid (7.81%), acetic acid, 3,6-bis(2-methylpropyl)- (6.62%), acetic acid methoxy-2-phenylethyl ester (6.01%) and diisopropyl ether (5.25%) were the major compounds (Fig. 4).
Table 6.
Chemical profile of ethyl acetate extract.
| Sl. no | RT | Compound | Quality | Molecular weight | Peak area % |
|---|---|---|---|---|---|
| 1 | 4.059 | Diisopropyl ether | 45 | 102.104 | 5.25 |
| 2 | 5.700 | Hexylene glycol | 59 | 118.099 | 3.06 |
| 3 | 8.082 | Phenylethyl Alcohol | 94 | 122.073 | 2.59 |
| 4 | 9.084 | Benzoic acid | 53 | 122.037 | 3.00 |
| 5 | 10.463 | Acetic acid, 2-phenylethyl ester | 90 | 164.084 | 10.45 |
| 6 | 10.594 | Benzene acetic acid | 94 | 136.052 | 7.81 |
| 7 | 12.526 | 1-Tetradecene | 99 | 196.219 | 0.85 |
| 8 | 13.136 | Trans-Cinnamic acid | 96 | 148.052 | 1.95 |
| 9 | 13.557 | Acetic acid, methoxy-, 2-phenylethyl ester | 83 | 194.094 | 6.01 |
| 10 | 14.878 | N-Acetyltyramine | 50 | 179.095 | 0.95 |
| 11 | 15.241 | Cetene | 99 | 224.25 | 2.41 |
| 12 | 16.853 | 7-Acetyl-1,7-diazabicyclo[2.2.0]he ptane | 22 | 140.095 | 1.26 |
| 13 | 17.652 | 1-Octadecene | 99 | 252.282 | 2.26 |
| 14 | 17.783 | Coumarin | 58 | 146.037 | 0.88 |
| 15 | 18.044 | Phenol, 3,5-dimethoxy- | 49 | 154.063 | 2.94 |
| 16 | 18.262 | N-Acetyltyramine | 91 | 179.095 | 1.85 |
| 17 | 19.105 | l-Proline, N-allyloxycarbonyl-, heptyl ester | 43 | 297.194 | 3.32 |
| 18 | 19.293 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(2-methylpropyl)- | 95 | 210.137 | 5.45 |
| 19 | 19.831 | E-15-Heptadecenal | 99 | 252.245 | 1.46 |
| 20 | 21.573 | 2,5-Piperazinedione, 3-methyl-6-(p henylmethyl)- | 95 | 218.106 | 1.01 |
| 21 | 21.733 | 2,5-Piperazinedione, 3,6-bis(2-met hylpropyl)- | 43 | 266.168 | 6.62 |
| 22 | 21.820 | 1-Docosene | 97 | 308.344 | 1.11 |
| 23 | 22.227 | 2-(3-Amino-1,2,4-triazol-2-yl)(4,5H)imidazoline | 35 | 152.081 | 0.64 |
| 24 | 23.331 | Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro-3-(phenylmethyl)- | 91 | 244.121 | 2.64 |
| 25 | 23.389 | 9-Octadecenamide, (Z)- | 99 | 281.272 | 2.44 |
| 26 | 25.422 | Fumaric acid, propyl 2,3,6-trichlo rophenyl ester | 35 | 335.972 | 1.63 |
| 27 | 25.800 | Formamide, N-(2,4-diamino-1,6-dihy dro-6-oxo-5-pyrimidinyl)- | 37 | 169.06 | 3.13 |
| 28 | 26.264 | Bicyclo[3.1.1]heptan-2-one, 6,6-di methyl-, (1R)- | 55 | 138.104 | 1.49 |
| 29 | 26.511 | Tartaric acid, dimenthyl ester | 64 | 426.298 | 2.60 |
| 30 | 27.150 | Squalene | 99 | 410.39 | 0.63 |
Fig. 4.
Chemical structure of major compounds of Al-Dhabi-2 crude extract.
4. Discussion
Since the beginning of the revolutionary era for antibiotic production in the last century, actinobacteria come to the forefront as a prolific source for bioactive compounds production. Thousands of bioactive compounds have been extracted from microorganisms, actinobacteria are produced around 50% of these compounds as secondary metabolites (Berdy, 2005). In present study, the thermophilic Streptomyces strain was isolated from thermal spring in Saudi Arabia and identified based on to standard methods via using phenotypic and molecular identification techniques. The phenotypic and molecular characteristics of Al-Dhabi-2 are consistent with those of the genus Streptomyces. Al-Dhabi-2 exhibited moderate antibacterial and antifungal activities in the streak method. Sequences analysis of the gene 16S rRNA was used for molecular characterization of Streptomyces Al-Dhabi-2. The sequencing of 16S rRNA illustrated that Streptomyces sp. Al-Dhabi-2 (accession No. KF815081) had close family relationship with sequences of different species belong to Streptomyces when compared to Gen Bank data also.
Thermophilic actinobacteria can be isolated form the hot environments. They were isolated from various hot springs in several countries over the world and and identified by different techniques from the numerical taxonomy (Şahin et al., 2002), analysis of the composition of fatty acids (McNabb et al.,1997), through the use analysis of ribosomal protein (Ochi, 1992) and to phylogentic analysis the sequences of 16S rRNA (Song et al., 2001, Yoon and Park, 2000). It can be said that, identification of bacteria, includes actinobacteria, via sequencing of 16S rRNA represent a robust and accurate technique among the previous methods for identification (Songara and Swarnjeet, 2013, Woese, 1987). Morphological characteristics still represent one of the leading traits for identifying strains of the genus Streptomyces at species level, such as the colour of the aerial mycelium (Pridham and Tresner, 1974).
Singh and Kapoor (2013) used the biochemical characterization to identify thermophilic Streptomyces sp. MSC702 strain was isolated from mushroom compost samples. The strain Al-Dhabi-2 was grown at different temperatures from 29 °C to 55 °C. The optimum temperature was 50 °C. Production of antimicrobial metabolites was observed at 45 °C for Al-Dhabi-2. James and Edwards, 1988, James and Edwards, 1989 reported that S. thermoviolaceus utilized a broad spectrum of substances as sole source for carbon. Several of these compounds enhanced the production of antibiotics at temperature range from 30 to 55 °C, However the optimal production was at 50 °C. The strain Al-Dhabi-2 produced good enzyme activity. Amylase, gelatinase and deoxyribonuclease (DNase) enzymes were observed in the agar plate using appropriate media. Stress molecules can be produced by the microorganisms that thrive in extreme environments by which these microbes are surviving in such systems, therefore these unique molecules can be extracted as novel compounds and can be investigated in many aspects.
Hence, thermophilic actinobacteria isolates were screened against pathogenic microbes for their antimicrobial activities (Uzel et al., 2011). Thermophilic actinobacteria Al-Dhabi-2 showed inhibitory activity against tested microbes. Extract of Al-Dhabi-2 inhibited the growth of tested microbes at 5 mg/ml concentration. The appropriate carbon source was glucose for antimicrobial agent production by Al-Dhabi-2. Temperature and pH are important factors for the growth of actinobacteria and production of antimicrobial metabolites. Al-Dhabi-2 grew optimally at 45–50 °C; optimum pH was 7.0. El–Abyad et al. (1996) reported that media composition is very specific for the antimicrobial potentials of the Streptomyces species.
Thermomycin and granaticin are antibiotics produced by thermophilic strains belonging to Streptomyces thermophiles and Streptomyces thermoviolaceus respectively (Edwards, 1993, Arokiyaraj et al., 2015). Songara and Swarnjeet (2013) reported that thermophilic actinobacteria identified as Streptomyces sp. from Rajasthan exhibited antibacterial activity to Klebsiella sp and S. aureus. The extracts of thermophilic Thermoactinomyces sp. isolated suppressed the growth of methicillin resistant S. aureus (MRSA) (Uzel, 2011, Korkmaz et al., 2007). The crude extract of the strain Al-Dhabi-2 exhibited antimicrobial activities which inhibited the growth of almost all tested microbes including K. pneumonia, B. cereus, S. typhi S. aureus, E. coli at 5 mg level. The chemical profile of the extract of Al-Dhabi-2 was analysed by GC-MS. The results showed that 30 compounds were present in the extract. Benzene acetic acid was one of those compounds found. A previous study has reported that benzene acetic acid exhibited antifungal and antibacterial properties (Tayade and Jadhao, 2012, Arasu et al., 2017, Arasu et al., 2019). Therefore, further chemical analyses will be conducted on crude extract of Al-Dhabi-2 to determine the compound/compounds responsible for the antimicrobial activity.
5. Conclusion
This study was conducted in attempts searching for new antimicrobial compounds that produced via microorganisms such as Streptomyces from unexplored environments in particular the extreme habitats. The antimicrobial activities results of crude extract antagonistic to bacteria and fungi were promising. GC-MS techniques guided to detect acetic acid, 2-phenylethyl ester (10.45%), benzene acetic acid (7.81%), acetic acid, 3,6-bis(2-methylpropyl)-(6.62%), methoxy- 2-phenylethyl ester (6.01%) and diisopropyl ether (5.25%) from the crude extract. The future work is aiming to isolate the bioactive compound, followed by testing its antimicrobial activity against antibiotic resistant bacteria.
Acknowledgments
This project was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdulaziz City for Science and Technology, Kingdom of Saudi Arabia, Award Number (11-BIO1873-02).
Footnotes
Peer review under responsibility of King Saud University.
References
- Al-Dhabi N.A., Ghilan A.K.M., Arasu M.V., Duraipandiyan V. Green biosynthesis of silver nanoparticles produced from marine Streptomyces sp. Al-Dhabi-89 and their potential applications against wound infection and drug resistant clinical pathogens. J. Photochem. Photobiol., B. 2018;189:176–184. doi: 10.1016/j.jphotobiol.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Al-Dhabi N.A., Ghilan A.K.M., Arasu M.V., Duraipandiyan V. Green biosynthesis of silver nanoparticles produced from marine Streptomyces sp. Al-Dhabi-89 and their potential applications against wound infection and drug resistant clinical pathogens. J. Photochem. Photobiol., B. 2018;189:176–184. doi: 10.1016/j.jphotobiol.2018.09.012. [DOI] [PubMed] [Google Scholar]
- Al-Dhabi N.A., Ghilan A.K.M., Esmail G.A., Arasu M.V., Duraipandiyan V., Ponmurugan K. Bioactivity assessment of the Saudi Arabian Marine Streptomyces sp. Al-Dhabi-90, metabolic profiling and its in vitro inhibitory property against multidrug resistant and extended-spectrum beta-lactamase clinical bacterial pathogens. J. Infect. Public Health. 2019 doi: 10.1016/j.jiph.2019.01.065. [DOI] [PubMed] [Google Scholar]
- Al-Dhabi N.A., Esmail G.A., Duraipandiyan V., Valan Arasu M., Salem-Bekhit M.M. Isolation, identification and screening of antimicrobial thermophilic Streptomyces sp. Al-Dhabi-1 isolated from Tharban hot spring, Saudi Arabia. Extremophiles. 2016;20:79–90. doi: 10.1007/s00792-015-0799-1. [DOI] [PubMed] [Google Scholar]
- Al-Dhabi N.A., Duraipandiyan V., Arasu M.V., Ponmurugan K., Ignacimuthu S. Antifungal metabolites from sponge associated marine Streptomyces sp. strain (ERIMA-01). Journal of Pure and Applied. Microbiology. 2014;8(2):115–128. [Google Scholar]
- Aljuhani S.G. Long Beach; California Article: 2012. The Potential of Enhanced Geothermal Energy Systems in Saudi Arabia 2012 Adapted from oral presentation at AAPG Annual Convention and Exhibition; p. 80249. [Google Scholar]
- Andrew E., Cook Paul R.M. Rapid identification of filamentous actinomycetes to the genus level using genus-specific 16S rRNA gene restriction fragment patterns. Int. J. Syst. Evol. Microbiol. 2003;53:1907–1915. doi: 10.1099/ijs.0.02680-0. [DOI] [PubMed] [Google Scholar]
- Arab H., Volker H., Thomm M. Thermococcus aegaeicus sp. nov. and Staphylothermus hellenicus sp. nov., two novel hyperthermophilic archaea isolated from geothermally heated vents off Palaeochori Bay, Milos. Greece. Int. J. Syst. Evol. Microbiol. 2000;50:2101–2108. doi: 10.1099/00207713-50-6-2101. [DOI] [PubMed] [Google Scholar]
- Arasu M.V., Thirumamagal R., Srinivasan M.P., Al-Dhabi N.A., Ayeshamariam A., Saravana Kumar D., Punithavel N., Jayachandran M. Green chemical approach towards the synthesis of CeO2 doped with seashell and its bacterial applications intermediated with fruit extracts. J. Photochem. Photobiol., B. 2017;172:50–60. doi: 10.1016/j.jphotobiol.2017.05.032. [DOI] [PubMed] [Google Scholar]
- Arasu M.V., Al-Dhabi N.A., Saritha V., Duraipandiyan V., Muthukumar C., Kim S.-J. Antifeedant, larvicidal and growth inhibitory bioactivities of novel polyketide metabolite isolated from Streptomyces sp. AP-123 against Helicoverpa armigera and Spodoptera litura. BMC Microbiol. 2013;13(1):105. doi: 10.1186/1471-2180-13-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasu M.V., Arokiyaraj S., Viayaraghavan P., Kumar T.S.J., Duraipandiyan V., Al-Dhabi N.A., Kaviyarasu K. One step green synthesis of larvicidal, and azo dye degrading antibacterial nanoparticles by response surface methodology. J. Photochem. Photobiol., B. 2019;190:154–162. doi: 10.1016/j.jphotobiol.2018.11.020. [DOI] [PubMed] [Google Scholar]
- Arasu M.V., Duraipandiyan V., Ignacimuthu S. Antibacterial and antifungal activities of polyketide metabolite from marine Streptomyces sp. AP-123 and its cytotoxic effect. Chemosphere. 2013;90(2):479–487. doi: 10.1016/j.chemosphere.2012.08.006. [DOI] [PubMed] [Google Scholar]
- Arasu M.V., Al-Dhabi N.A., Choi K.I. Identification of novel quinine metabolite from marine actinomycetes with antifungal and anticancer bio-prospective. Fresenius Environ. Bull. 2015;24(10a):3281–3287. [Google Scholar]
- Arokiyaraj S., Saravanan M., Badathala V. Green synthesis of Silver nanoparticles using aqueous extract of Taraxacum officinale and its antimicrobial activity. South Indian J. Bioogical Sci. 2015;2:115–118. [Google Scholar]
- Balachandran C., Duraipandiyan V., Emi N., Ignacimuthu S. Antimicrobial and cytotoxic properties of Streptomyces sp. (ERINLG-51) isolated from Southern Western Ghats. South Indian J. Biol. Sci. 2015;1:7–14. [Google Scholar]
- Berdy J. Bioactive microbial metabolites; a personal view. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Demain A. Cambridge University Press; 1995. Why do Microorganisms Produce Antimicrobial? Proceeding of Symposium on Society of General Microbiology; pp. 205–228. [Google Scholar]
- Duraipandiyan V., Ignacimuthu S. Antibacterial and antifungal activity of Flindersine isolated from the traditional medicinal plant, Toddalia asiatica (L.) Lam. J. Ethnopharmacol. 2009;123:494–498. doi: 10.1016/j.jep.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Duraipandiyan V., Sasi A.H., Islam V.I.H., Valanarasu M., Ignacimuthu S. Antimicrobial properties of actinomycetes from the soils of Himalaya. J. de Mycol. Médicale. 2010;20:15–20. [Google Scholar]
- Edwards C. Isolation properties and potential applications of thermophilic actinomycetes. Appl. Biochem. Biotechnol. 1993;42:161–179. [Google Scholar]
- El–Abyad M., El–Sayed M.A., El–Shanshoury A.R., El–Sabbagh S.M. Antimicrobial activities of Streptomyces pilcher, Streptomyces canescens and Streptomyces citrofluorescens against fungal and bacterial pathogens of tomato in vitro. Folia. Micobiol. 1996;41:321–328. doi: 10.1007/BF02814708. [DOI] [PubMed] [Google Scholar]
- Ellaiah P., Ramana T., Bapiraju K.V.V.S., Sujatha P., Uma Sankar A. Investigation on marine actinomycetes from Bay of Bengal near Kakinada coast of Andhra Pradesh. Asian. J. Microbiol. Biotech. Env. Sci. 2004;6:53–56. [Google Scholar]
- Hames-Kocabas E.E., Uzel A. Alkaline protease production by an actinomycete MA 1–1 isolated from marine sediments. Ann. Microbiol. 2007;57:71–75. [Google Scholar]
- James P.D.A., Edwards C. The effects of cultural conditions on growth and secondary metabolism in Streptomyces thermoviolecus. FEMS Microbiol. Lett. 1988;52:1–6. [Google Scholar]
- James P.D.A., Edwards C. The effects of temperature on growth and production of the antibiotic granaticin by a thermotolerant Streptomycetes. J. Gen. Microbiol. 1989;135:1997–2003. doi: 10.1099/00221287-135-7-1997. [DOI] [PubMed] [Google Scholar]
- Kaur G., Mountain B., Pancost R. Microbial membrane lipids in active and inactive sinters from Champagne Pool, New Zealand: Elucidating past geothermal chemistry and microbiology. Org. Geochem. 2008;39:1024–1028. [Google Scholar]
- Khalil A. Isolation and characterization of three thermophilic bacterial strains (lipase, cellulose and amylase producers) from hot springs in Saudi Arabia. Afr. J. Biotechno. 2011;10:8834–8839. [Google Scholar]
- Khiyami M.A., Serour E.A., Shehata M.M., Bahkli A.H. Thermo-aerobic bacteria from geothermal springs in Saudi Arabia. Afr. J. Biotechnol. 2012;11:4053–4062. [Google Scholar]
- Kleeberg I., Hetz C., Kroppenstedt R.M. Biodegradation of aliphatic-aromatic copolyesters by Thermomonospora fusca and other thermophilic compost isolates. Appl. Environ. Microb. 1998;64:1731–1735. doi: 10.1128/aem.64.5.1731-1735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkmaz C.A., Hames-Kocabas E.E., Uzel A., Bedir E. Tryptamine derived amides with thiazole ring system from Thermoactinomyces strain TA66-2. Magn. Reson. Chem. 2007;46:80–83. doi: 10.1002/mrc.2101. [DOI] [PubMed] [Google Scholar]
- Kuster E., Williams S.T. Selection of media for isolation of streptomycetes. Nature. 1964;202:928–939. doi: 10.1038/202928a0. [DOI] [PubMed] [Google Scholar]
- McNabb A., Shuttleworth R., Behme R. Fatty acid characterization of rapidly growing pathogenic aerobic actinomycetes as a means of identification. J. Clin. Microbiol. 1997;35:1361–1368. doi: 10.1128/jcm.35.6.1361-1368.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A.R., Philips A., Humprey A.E. Production of cellulases by therniomonosporn species. Biotechnol. Bioeng. 1981;23:1339–1347. [Google Scholar]
- Ochi K. Polyacrylamide gel electrophoresis analysis of ribosomal protein: a new approach for actinomycete taxonomy. Gene. 1992;115:261–265. doi: 10.1016/0378-1119(92)90568-a. [DOI] [PubMed] [Google Scholar]
- Pridham T.C., Tresner H.D. Family VII Streptomycetaceae, Waksmand and Herici 1943. In: Buchanan R.E., Gibons N.E., editors. Bergey‘s Manual of Determinative Bacteriology. 8th ed. Williams and Wilkins; Baltimore: 1974. pp. 747–829. [Google Scholar]
- Şahin N., Öztürk E., Işık K. Selective isolation and numerical classification of novel thermophilic Streptomycetes. Turk. J. Biol. 2002;26:13–24. [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evo. 1987;l4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shirling E.B., Gottlieb D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966;16:313–340. [Google Scholar]
- Singh R., Kapoor V. Biochemical characterization of thermophilic actinomycete (Streptomyces sp. MSC702) from mushroom compost in India. Int. J. Curr. Biotechnol. 2013;1:1–7. [Google Scholar]
- Song J., Weon H.Y., Yoon S.H. Phylogenetic diversity of thermophilic actinomycetes and Thermoactinomyces spp. isolated from mushroom composts in Korea based on 16S rRNA gene sequence analysis. FEMS Microbiol. Lett. 2001;202:97–102. doi: 10.1111/j.1574-6968.2001.tb10786.x. [DOI] [PubMed] [Google Scholar]
- Songara D., Swarnjeet K. DNA based identification and characterization of thermophilic Streptomyces sp. from desert soil of Rajasthan. Int. J. Curr. Microbiol. App. Sci. 2013;2:418–427. [Google Scholar]
- Stackebrandt E., Rainey F., Ward-Rainey N.L. Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int. J. Syst. Bacteriol. 1997;47:479–481. [Google Scholar]
- Takeuchi M., Takahashi S., Inukai M. Helvecardins A and B, novel glycopeptide antibiotics. II. Structural elucidation. J. Antibiot. 1991;44:271–277. doi: 10.7164/antibiotics.44.271. [DOI] [PubMed] [Google Scholar]
- Tayade D.T., Jadhao N.G. Attempt in the synthesis of 2-[(2,6 disubstitutedthiocarbamidophenyl)amino] benzeneacetic acid and their antimicrobial study. J. Pure. Appl. Microbiol. 2012;6:2025–2028. [Google Scholar]
- Uzel A., Hameş kocabaş E.E., Bedir E. Prevalence of Thermoactinomyces thalpophilus and T. Sacchari strains with biotechnological potential at hot springs and soils from West Anatolia in Turkey. Turk. J. Biol. 2011;35:195–202. [Google Scholar]
- Uzel A., Hames-Kocabas E.E. Production of organic solventstable alkaline protease from a marine Streptomyces strain Marac 1–4. Fresen. Environ. Bull. 2007;16:1523–1525. [Google Scholar]
- Valan Arasu M., Duraipandiyan V., Agastian P., Ignacimuthu S. Antimicrobial activity of Streptomyces spp. ERI-26 recovered from Western Ghats of Tamil Nadu. J. Mycol. Me. 2008;18:147–153. [Google Scholar]
- Valli S., Suvathi S.S., Aysha O.S., Nirmala P., Vinoth K.P., Reena A. Antimicrobial potential of Actinomycetes species isolated from marine environment. Asian. Pac. J. Trop. Biomed. 2012;2:469–473. doi: 10.1016/S2221-1691(12)60078-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsalam S., Agastian P., Arasu M.V., Al-Dhabi N.A., Ghilan A.K.M., Kaviyarasu K., Ravindran B., Chang S.W., Arokiyaraj S. Rapid biosynthesis and characterization of silver nanoparticles from the leaf extract of Tropaeolum majus L. and its enhanced in-vitro antibacterial, antifungal, antioxidant and anticancer properties. J. Photochem. Photobiol., B. 2019;191:65–74. doi: 10.1016/j.jphotobiol.2018.12.010. [DOI] [PubMed] [Google Scholar]
- Waksman S.A., Schatz A., Reynolds D.M. Production of antibiotic substances by actinomycetes. Ann. NY. Acad. Sci. 2010;1213:112–124. doi: 10.1111/j.1749-6632.2010.05861.x. [DOI] [PubMed] [Google Scholar]
- Watve M.G., Tickoo R., Jog M.M., Bhole B.D. How many antibiotics are produced by genus Streptomyces? Arch. Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- Woese C.R. Bacterial evolution. Microbiol. Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J.H., Park Y.H. Phylogenetic analysis of the genus Thermoactinomyces based on 16S rDNA sequences. Int. J. Syst. Evol. Micr. 2000;50:1081–1086. doi: 10.1099/00207713-50-3-1081. [DOI] [PubMed] [Google Scholar]
Further reading
- Bull A.T., Stach J.E. Marine actinobacteria new opportunities for natural product search and discovery. Trends. Microbiol. 2007;15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsinomy methods. Mol. Bio. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgin D.G., Gibson T.J. CLUSTAL W. improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl. Acid Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vining L.C. Secondary metabolism, inventive evolution and biochemical diversity: A review. Gene. 1992;115:135–140. doi: 10.1016/0378-1119(92)90551-y. [DOI] [PubMed] [Google Scholar]
- Xu L.H., Jiang Y., Li W.J., Wen M.L., Li M.G., Jiang C.L. Streptomyces roseoalbus sp. nov., an Actinomycete isolated from soil in Yunnan, China. Antonie Van Leeuwenhoek. 2005;87:189–194. doi: 10.1007/s10482-004-3720-y. [DOI] [PubMed] [Google Scholar]