Abstract
Multiple pathways counteract DNA replication stress to prevent genomic instability and tumorigenesis. The recently identified human SDE2 is a genome surveillance protein regulated by PCNA, a DNA clamp and processivity factor at replication forks. Here, we show that SDE2 cleavage after its ubiquitin-like domain generates Lys-SDE2Ct, the C-terminal SDE2 fragment bearing an N-terminal Lys residue. Lys-SDE2Ct constitutes a short-lived physiological substrate of the Arg/N-end rule proteolytic pathway, in which UBR1 and UBR2 ubiquitin ligases mediate the degradation. The Arg/N-end rule and VCP/p97UFD1-NPL4 segregase cooperate to promote phosphorylation-dependent, chromatin-associated Lys-SDE2Ct degradation upon UVC damage. Conversely, cells expressing the degradation-refractory K78V mutant, Val-SDE2Ct, fail to induce RPA phosphorylation and single-stranded DNA formation, leading to defects in PCNA-dependent DNA damage bypass and stalled fork recovery. Together, our study elucidates a previously unappreciated axis connecting the Arg/N-end rule and the p97-mediated proteolysis with the replication stress response, working together to preserve replication fork integrity.
INTRODUCTION
Achieving accurate genome duplication is a challenging task for the DNA replication machinery. Failure to overcome various replication-blocking obstacles causes perturbations of DNA synthesis, collectively referred to as replication stress (1). Increased formation of aberrant replication fork structures due to the loss of replication stress response pathways leads to deleterious mutational events that are highly associated with genome instability and tumorigenesis (2). Proliferating cell nuclear antigen (PCNA) is a processivity factor that plays a critical role in coordinating DNA replication by guiding replicative DNA polymerases at replication forks (3). Persistent replication stress, for instance, due to ultraviolet C (UVC)-induced replication-blocking lesions, results in the uncoupling of PCNA-associated polymerase and helicase activities that leads to an accumulation of single-stranded DNA (ssDNA) near replication forks (4). In turn, RPA-coated ssDNA activates the ATR-CHK1 pathway to resolve replication stress and preserve replication fork integrity (5). An extensive RPA-ssDNA platform recruits the RAD6/RAD18 ubiquitin (Ub) E2/E3 ligase complex to monoubiquitinate PCNA (PCNA-Ub) at stalled forks and further engages specialized polymerases that enable the bypass of DNA lesions via translesion DNA synthesis (TLS), a form of DNA damage tolerance mechanism regulated by the posttranslational modification of PCNA (6). In conjunction with the remodeling of stalled forks, such as replication fork reversal, these aforementioned pathways collectively facilitate stalled fork recovery and ensure efficient S phase progression against replication stress (7).
Regulated protein degradation mediated by the ubiquitin-proteasome system (UPS) keeps protein homeostasis in check and thus regulates multiple cellular processes. The N-end rule operates in the UPS through the recognition of proteins containing N-terminal (Nt) degradation signals, called N-degrons, by N-recognins. N-recognins are specialized ubiquitin E3 ligases that polyubiquitinate and degrade substrates with N-degrons via the proteasome (8). Thus, this defines the half-life of the N-end rule substrates, which is dependent upon the identity of a unique Nt residue. The Arg/N-end rule branch targets specific unacetylated and preferably positively charged Nt residues, such as Lys and Arg, that are exposed after proteolytic cleavage or generated through successive enzymatic modifications like deamination and arginylation (9) (Supplementary Figure S1A). Although the current understanding of the N-end rule has expanded over the past three decades and revealed its role in a broad range of cellular processes, only a few substrates have been identified in higher eukaryotes, including human. Hence, the identity of regulatory signaling pathways of the N-end rule-mediated proteolysis remains poorly understood, especially in the context of the DNA damage response (DDR).
Given the necessity of genome surveillance factors to function in close contact with DNA, fine-tuning their cellular levels and activity by proteolysis requires an additional regulatory element to extract substrates from the chromatin or macromolecular complexes. A growing body of evidence suggests that valosin-containing protein (VCP)/p97 segregase contributes to such process by utilizing ATP to extract and unfold chromatin-bound ubiquitinated substrates, thereby coordinating regulated turnover of DNA replication and repair factors by the proteasome (10). Importantly, disruption of the p97-driven chromatin-associated degradation (CAD) and persistent accumulation of its substrates lead to the so-called protein-induced chromatin stress (PICHROS) and impair genomic integrity, thus highlighting the importance of CAD in exerting spatiotemporal regulation of protein turnover and cellular function for genome maintenance (11).
We recently identified a new genome surveillance protein in human, SDE2, that regulates replication stress response via CAD at replication forks (12). The activity of SDE2 is regulated by the PCNA-dependent endolytic cleavage of its Nt Ub-like domain (UBL), which generates a C-terminal (Ct) SDE2 polypeptide, SDE2Ct, required for counteracting replication stress. We have demonstrated that timely degradation of SDE2Ct at chromatin is necessary for preserving replication fork integrity, thereby ensuring S phase progression and cellular survival. However, it remains unclear how dynamics of SDE2 proteolysis contributes to its function in modulating replication stress response and fork integrity. In this study, we identify the Arg/N-end rule-p97 proteolytic axis as a new regulator of SDE2 degradation and function. We demonstrate that timely degradation of SDE2Ct at replication forks is necessary for generating the RPA-ssDNA platform required for PCNA-dependent DNA damage bypass against UVC-induced replication stress, thereby promoting stalled fork recovery and S phase progression. We show that ATR-dependent SDE2Ct phosphorylation enhances its interaction with UFD1, the adaptor of the p97 complex, promoting SDE2Ct extraction from chromatin. Our study establishes a previously unappreciated connection between replication stress signaling and the Arg/N-end rule, whose coupling controls the CAD of genome surveillance factors to protect stalled forks and alleviate replication stress. We propose that regulated SDE2 degradation could be a key determinant for the dynamics of the replisome and for the remodeling of DDR factors at replication forks in order to optimize responses to replication stress.
MATERIALS AND METHODS
Cell culture and plasmid constructions
HeLa, U2OS, and 293T cell lines were acquired from the American Tissue Culture Collection (ATCC). UBR1−/− and UBR2−/− mouse embryonic cells were a kind gift from Alexander Varshavsky (Caltech). U2OS Flp-In T-REx cells were a kind gift from Daniel Durocher (The Lunenfeld-Tanenbaum Research Institute). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin, following standard culture conditions and procedures. Human SDE2 cDNA was acquired from Open Biosystems/Dharmacon (MHS1010–97228092), while full-length or deleted cDNA were PCR-amplified and subcloned into modified pcDNA3-Flag, pcDNA3-HA (Invitrogen), or pEGFP-C1 vectors (Clontech). Point mutations were introduced using the QuikChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) and confirmed by DNA sequencing. siRNA resistant SDE2 cDNA was generated by mutating four nucleotides of the target sequence for SDE2–1 siRNA oligo (acgCcaGtggccGacCaaa). Stable cell lines were generated by retroviral transduction of pMSCV-SDE2-Flag constructs using 8 μg/ml polybrene (Sigma-Aldrich), followed by selection with 2 μg/ml puromycin. Viruses were generated from 293T cells that were co-transfected with pMSCV-SDE2-Flag, pCMV-Gag/Pol and pCMV-VSV-G. Human UBR1 full-length cDNA was constructed by gene synthesis and cloned into pcDNA3-GFP KpnI-BamHI (Genscript). To stabilize the insert, plasmids were propagated and maintained at low copy in CopyCutter EPI400 strain (Lucigen), and plasmid replication was induced before DNA preparation. Human UFD1 cDNA was acquired from Open Biosystems (MHS6278–202828389) and cloned into pcDNA3-HA.
Plasmids and siRNA transfection
Plasmid transfection was performed using GeneJuice (Millipore) according to the manufacturer's protocols. siRNA duplexes were transfected at 25 nM using Lipofectamine RNAiMAX (Invitrogen). siRNA sequence information can be found on Supplementary Table S1.
Antibodies and chemicals
Antibodies used in this study are listed in Supplementary Table S2. Chemicals used in this study are listed in Supplementary Table S3.
Western blotting and fractionation
Cells were lysed in NETN300 buffer (1% NP40, 300 mM NaCl, 0.1 mM EDTA, and 50 mM Tris [pH 7.5]) supplemented with protease inhibitor cocktail (Roche), and halt phosphatase inhibitor cocktail (Thermo Fisher). Lysates were resolved by SDS-PAGE, transferred onto PVDF membranes (Millipore), and antibodies detected by an enhanced chemiluminescence method. Some immunoblot images were acquired by iBright CL1000 imaging system (Thermo Fisher). Subcellular fractionation was performed as previously described (13). Briefly, cells were lysed using cytoskeleton (CSK) buffer (10 mM Tris [pH 6.8], 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 1 mM EGTA, 1 mM EDTA, and 0.1% Triton X-100) for 5 min on ice. After centrifugation at 1500 g for 5 min, the supernatant (S) was separated from the pellet (P), and pellets were sequentially lysed in PBS and 2× boiling lysis buffer (50 mM Tris–Cl [pH 6.8], 2% SDS, and 850 mM β-mercaptoethanol).
Generation of U2OS Flp-InTM T-RExTM cells
The host U2OS osteosarcoma cell line stably expresses the Tet-repressor (T-REx) and carries a single FRT locus (Flp-In). The SDE2 construct was cloned using the Gateway™ system and co-transfected with the pOG44 plasmid encoding the Flp recombinase (Invitrogen) to generate the U2OS Flp-In T-REx cell lines. SDE2 cDNA, first cloned into pENTR™ 3C double selection vector (Invitrogen), was previously mutated on four nucleotides to drive its resistance to the SDE2–1 siRNA oligo. Site-directed mutagenesis was used to generate the K78V mutation. The LR Clonase TM II was then used to transfer SDE2 cDNA into our custom destination plasmid, kindly provided by Dr. Jiricny (University of Zurich, Switzerland), and built upon pcDNA™ 5/FRT/TO vector (Invitrogen). SDE2 expressed from this vector has one Strep and one HA tag at its C-terminus. Forty-eight hours after transfection using X-tremeGENE™ 9 (Roche), hygromycin selection was started and stably transfected cells recovered after 4 weeks. Doxycycline was then used at 10 ng/ml to drive the expression of the transgene, usually after knocking down the endogenous SDE2 by transfecting SDE2 siRNA oligo-1 using Lipofectamine RNAiMAX, as indicated.
BrdU staining for ssDNA detection
Cells were grown on coverslips and incubated with 10 μM BrdU for 48 h before UVC irradiation. Cells were permeabilized with PBS/0.3% Triton X-100 for 3 min at 4 °C followed by fixation with 4% paraformaldehyde for 10 min at RT. After blocking with 5% BSA, cells were first incubated with 1:300 anti-BrdU (BU-1) for 2 h and, after washing with PBS, incubated with 1:1000 Alexa Fluor 488 goat anti-mouse IgG for 45 min in 1% BSA. After washing again with PBS, coverslips were mounted using DAPI-containing mounting medium (Vector Lab) and analyzed with Leica TCS SP8 X confocal microscope. Corrected total cell fluorescence (CTCF) intensity was quantified and calculated by Fiji, and analyzed by Prism (GraphPad).
DNA fiber combing
Exponentially growing cells were first labeled with 50 μM CldU for 20 min at 37 °C. After three washes with PBS, cells were treated or not with 30 J/m2 UV-C (UV Stratalinker 1800, Stratagene), and replenished with media containing 250 μM IdU for 30 min at 37 °C. DNA fibers were then prepared using the FiberPrep DNA extraction kit and the FiberComb Molecular Combing System (Genomic Vision, France), as recommended by the supplier. In brief, cells were harvested by trypsinization, counted, and 400,000 cells were subsequently washed in PBS before being embedded in low-melting point agarose, and cast in a plug mold. After full solidification, plugs were digested overnight with proteinase K. Over the next day, plugs were extensively washed before being shortly melted and digested with agarase overnight. The obtained DNA fibers were then combed onto a silanized coverslips (Genomic Vision, France). Combed coverslips were dehydrated, and DNA was denatured for 8 min using 0.5 M NaOH in 1 M NaCl. During the subsequent staining process, all incubations were done in humidified conditions at 37 °C. In short, coverslips were first blocked with 1% BSA for 30 min, then both primary antibodies were diluted together in 1% BSA (rat monoclonal anti-BrdU for CldU, 1:25, and mouse monoclonal anti-BrdU for IdU, 1:5) and incubated for 1 h. After washing the coverslips with PBS-Tween 0.05% (PBS-T), secondary antibodies were both diluted together in 1% BSA (Alexa Fluor 594 goat anti-rat and Alexa Fluor 488 goat anti-mouse, 1:100) and incubated for 45 min. After washing the coverslips with PBS-T and dehydrating them, they were mounted onto microscopic glass slides using ProLong™ Gold Antifade and cured overnight. DNA fibers were then imaged using the Leica TCS SP8 X confocal microscope and analyzed using the Fiji software.
Statistical analysis
Student's t-test was used to assess the statistical significance of our results, using Prism (GraphPad). Unpaired t-tests were performed with a 95% confidence interval, using two-tailed P-values, unless stated otherwise. For the DNA combing assay, the Mann–Whitney U-test was performed, with a 95% confidence interval, using Prism (GraphPad).
RESULTS
The natural fragment of SDE2 as a substrate of the Arg/N-end rule pathway
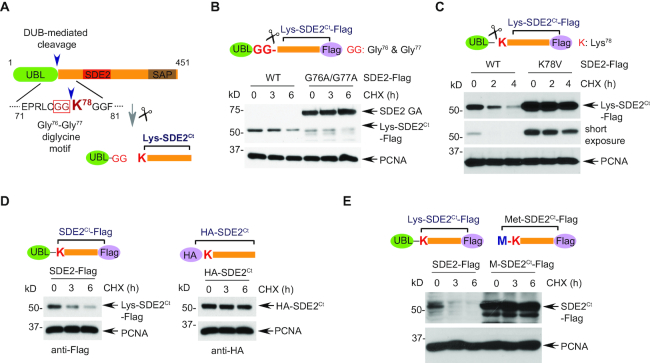
Our previous work showed that the first 77 residues of the human SDE2 comprises a UBL with conserved Ct-sequence of Gly76–Gly77 that is identical to the last two residues of Ub, and that cellular deubiquitinating enzymes (DUBs) could cleave SDE2 immediately after Gly77 (12) (Figure 1A). The resulting Ct-fragment of SDE2, termed SDE2Ct, was metabolically unstable in a cycloheximide (CHX)-based chase, whereas the full-length SDE2 mutant, made uncleavable through the G76A/G77A (GA) mutation, was long-lived (Figure 1B and Supplementary Figure S1B). Thus, the cleavage of SDE2 is required for the degradation of SDE2Ct. Since the latter was Lys-SDE2Ct, bearing Nt-Lys, a destabilizing residue in the Arg/N-end rule pathway, we asked whether human Lys-SDE2Ct might be a previously undescribed physiological substrate of this pathway. Indeed, mutation of Lys78 to a small hydrophobic residue such as Val or Ala within the C-terminally Flag-tagged full-length SDE2 resulted in a strong stabilization of the mutant, in addition to its increased level at the beginning of CHX chase. (Figure 1C and Supplementary Figure S1C and S1D). The K78V mutant was fully cleaved to generate Val-SDE2Ct, and its cellular levels were comparable to those of the GA mutant, indicating that degradation was impaired upon cleavage (Supplementary Figure S1E). Moreover, the degradation of SDE2Ct was inhibited by masking Lys78 of SDE2Ct either by HA-tag or Met (Figure 1D and E and Supplementary Figure S1F).
Figure 1.
SDE2 cleavage exposes an N-degron required for the Arg/N-end rule. (A) Schematic of SDE2 cleavage at a diglycine motif generating SDE2Ct with its Nt Lys78 exposed. (B) U2OS cells expressing wild-type (WT) or G76A/G77A full-length SDE2-Flag were treated with 50 μg/ml cycloheximide (CHX) or DMSO (vehicle) for the indicated times and analyzed by Western blotting (WB). (C–E) CHX chase and WB analysis of U2OS cells expressing either (C) WT or K78V full-length SDE2-Flag, or (D) WT full-length SDE2-Flag versus HA-SDE2Ct-Flag, or (E) WT full-length SDE2-Flag versus Met-SDE2Ct-Flag. Representative images are shown, and quantification graphs can be found in Supplementary Figure S1.
Similarly to other degradation signals recognized by the Ub system, N-degrons are at least bipartite. Their second determinant is a substrate's Lys residue, the site of a substrate-linked poly-Ub chain that is produced by an N-recognin Ub ligase following its binding to a substrate's destabilizing Nt-residue (8,14). We found that the disruption of Lys94, present closest to the N-terminus of Lys-SDE2Ct, stabilized the Lys-SDE2Ct, indicating that this Lys residue is one of the main (though not necessarily the sole) polyubiquitination sites (Supplementary Figure S1G).
Together, these results suggest that the cleavage of SDE2 downstream of its UBL exposes Lys, a destabilizing residue, and that Lys-SDE2Ct (hereinafter SDE2Ct in short) is a physiological substrate of the Arg/N-end rule pathway.
UBR1 and UBR2 mediate degradation of the SDE2Ct fragment
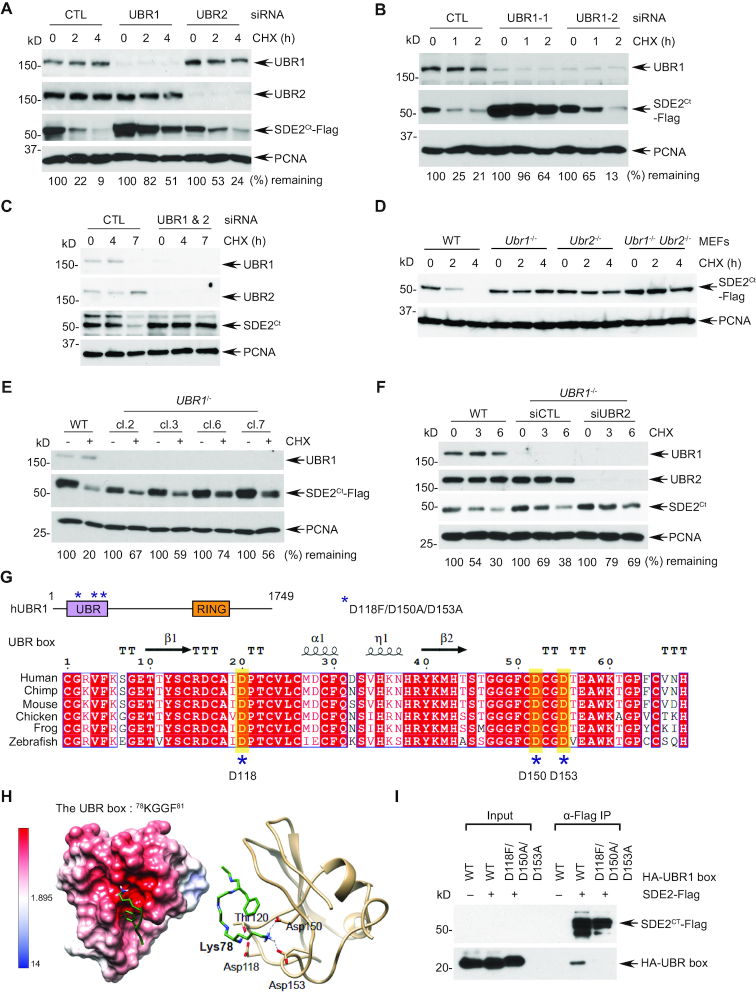
UBR1 and UBR2, the two 47% identical N-recognins, underlie the bulk of degradation of Arg/N-end rule substrates in mammalian cells (8). siRNA-mediated depletion of either UBR1 (using two independent siRNA oligonucleotides), or UBR2 significantly stabilized SDE2Ct in U2OS human osteosarcoma cells, although UBR1 deficiency results in a stronger effect on SDE2Ct degradation (Figure 2A and B). Additionally, co-depletion of both UBR1 and UBR2 led to a virtually complete cessation of degradation of endogenous and exogenous SDE2Ct (Figures 2C and Supplementary Figure S2A and B). Similar results were obtained using previously produced Ubr1−/− and Ubr2−/− mouse embryonic fibroblast (MEF) cell lines (15,16) (Figure 2D and Supplementary Figure S2C). To further substantiate our results, we generated UBR1 knockout U2OS cells using CRISPR/Cas9 (Supplementary Figure S2D). Multiple clones of UBR1−/− cells exhibited extended stability of SDE2Ct-Flag (Figure 2E), and knockdown of UBR2 in UBR1−/− cells prevented the degradation of both endogenous and exogenous SDE2Ct almost completely (Figure 2F and Supplementary Figure S2E). Variable potency of UBR1 deficiency on different experimental settings could be due to acute knockdown versus compensation during clonal selection of knockout cells. Together, these results from multiple cell lines support the idea that both UBR1 and UBR2 act as the primary ubiquitin E3 ligases required for SDE2Ct degradation in the Arg/N-end rule pathway.
Figure 2.
UBR1 and UBR2 function as an N-recognin for SDE2Ct degradation (A, B) U2OS cells sequentially transfected with the indicated siRNA oligos (versus control: CTL) and with full-length SDE2-Flag were treated with 50 μg/ml CHX for the indicated times and SDE2Ct-Flag levels were analyzed by WB. SDE2Ct levels were normalized by loading control and quantitated by ImageJ (0 h set as 100%). (C) U2OS cells were transfected with indicated siRNA oligos and treated with 100 μg/ml CHX for the indicated times and endogenous SDE2Ct levels were analyzed WB. Representative images are shown, and quantification graphs can be found in Supplementary Figure S2B. (D) Indicated MEFs overexpressing SDE2-Flag were subjected to CHX chase to analyze SDE2Ct-Flag degradation. (E) SDE2-Flag was transfected into independent U2OS UBR1 CRISPR knockout clones, and SDE2Ct-Flag levels were analyzed by WB after 4 h CHX. WT indicates an empty vector-transfected clone. (F) U2OS UBR1−/- clone 6 was transfected with UBR2 siRNA oligo (versus control in WT or UBR1−/- background), and the decay of endogenous SDE2Ct levels was monitored. (G) A schematic of human UBR1 structure, depicting the UBR box and the RING domain. Mutations introduced in this study are marked as asterisks. The sequence alignment was generated by ESPript 3. (H) (Left) Electrostatic potential surface representation of the UBR box adapted from the PDB structure 3NY3, showing a negatively charged pocket (red) with the SDE2Ct N-degron peptide, 78KGGF81. (Right) UBR-box recognition elements forming hydrogen bonds to the N-degron Lys78. (I) Anti-Flag co-IP of 293T cells co-expressing SDE2-Flag and HA-tagged UBR box (residue 1–176) from hUBR1 WT or mutant.
It is well established that the UBR family of N-recognins is characterized by an atypical zinc-finger domain named the UBR box at its N-terminus, which constitutes a negatively charged pocket that recognizes the positively charged N-degron of a substrate (17,18) (Figure 2G). We performed structure modeling based on the previously reported UBR box structure in complex with N-degron and found out that the Lys of the SDE2Ct peptide is predicted to form hydrogen bonds with negatively charged side chains from Asp118, Asp150 and Asp153 of the UBR box (19,20) (Figure 2H). We observed that GFP-tagged full-length UBR1 binds to SDE2Ct in the absence and presence of DNA damage (Supplementary Figure S2F), and mutations of the negatively charged residues in the UBR box in UBR1 abolished its interaction with SDE2Ct, confirming that the N-degron of SDE2Ct is recognized by the UBR box of UBR1 via electrostatic interactions (Figure 2I). Together, these results further confirm that the E3 ligase UBR1 recognizes the N-degron of SDE2Ct to trigger degradation of SDE2Ct.
The Arg/N-end rule regulates SDE2Ct degradation in response to UVC irradiation
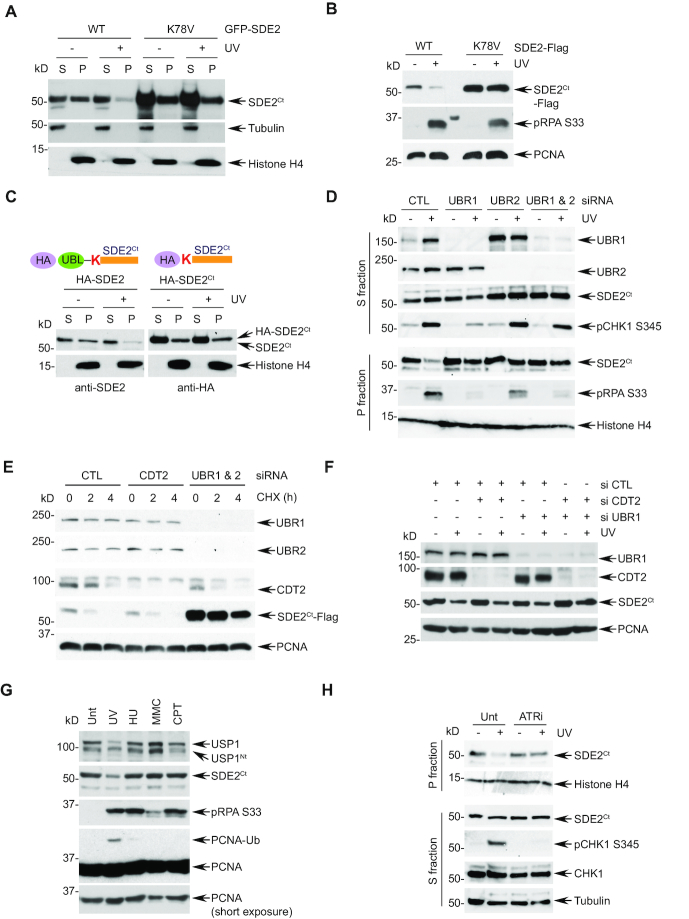
We previously showed that SDE2Ct undergoes degradation in response to UVC damage (12). This occurs to both endogenous and exogenously expressed SDE2Ct, specifically in the chromatin-enriched fraction, suggesting that SDE2Ct degradation occurs in the context of DNA interaction (Supplementary Figure S3A). Deletion of the putative DNA binding SAF-A/B, Acinus and PIAS (SAP) domain of SDE2 prevents its association with chromatin and abolishes its degradation, further supporting this idea (Supplementary Figure S3B) (12). Thus, we determined whether N-end rule signaling also regulates SDE2Ct degradation at chromatin during replication stress. Indeed, while wild-type (WT) SDE2Ct underwent degradation specifically in the chromatin-enriched fractions following UVC damage, the K78V mutant failed to do so (Figure 3A and Supplementary Figure S3C). We also observed similar results of defective degradation with the C-terminally Flag-tagged K78V or N-terminally HA-tagged SDE2Ct mutants (Figure 3B and C, and Supplementary Figure S3D and E). Additionally, co-depletion of UBR1 and UBR2 significantly abrogated the degradation of both endogenous and exogenous SDE2Ct in the chromatin, confirming that the N-end rule pathway is required for the CAD of SDE2Ct following DNA damage (Figure 3D and Supplementary Figure S3F and G). We have previously shown that CRL4CDT2 ubiquitin E3 ligase is required for damage-dependent degradation of SDE2Ct in the chromatin. Intriguingly, knockdown of UBR1 and 2, but not CDT2, prevented degradation of SDE2Ct during CHX chase, indicating the N-end rule primarily governs the turnover of SDE2Ct in unstressed cells (Figure 3E). In contrast, when we lowered the amount of siRNA oligo concentrations for transfection, we observed that knockdown of both CDT2 and UBR1 or UBR2 impeded the degradation of SDE2Ct following UVC damage in an additive manner (Figure 3F and Supplementary Figure S3H). These results indicate that the CAD of SDE2Ct under replication stress may be initiated by a bimodal activation of the N-end rule and CRL4CDT2 ubiquitin signaling. Of note, we have previously shown that SDE2Ct lacking a classic PCNA-interaction protein (PIP) degron was able to interact with CDT2 via the SAP domain (12), suggesting that damage-dependent SDE2Ct degradation should occur in the context of chromatin, where replication stress signaling may regulate localization and/or activity of both UBR1/2 and CRL4CDT2.
Figure 3.
The N-end rule promotes damage-dependent SDE2Ct degradation. (A) U2OS cells expressing Nt GFP-tagged full-length SDE2 WT or K78V were treated with 40 J/m2 UVC and 4 h later, fractionated into cytosolic/nucleoplasmic (S) versus chromatin-enriched (P) fractions, followed by WB analysis. (B) The P fraction of U2OS cells expressing Ct Flag-tagged full-length SDE2 WT or K78V and treated or not with UVC were analyzed by WB. (C) U2OS cells expressing Nt HA-tagged full-length SDE2 or SDE2Ct were UVC irradiated, fractionated, and analyzed by anti-SDE2 and anti-HA WB, respectively. (D) U2OS cells transfected with the indicated siRNA were UVC irradiated, fractionated, and analyzed by WB. The SDE2Ct levels in the P fraction were quantified by ImageJ, and remaining SDE2Ct following UVC was determined compared to untreated. (E) CHX chase and WB analysis of U2OS cells transfected with the indicated siRNA oligos and SDE2-Flag. (F) U2OS cells were transfected with the indicated combination of siRNA oligos and GFP-SDE2, irradiated with 40 J/m2 UVC, and P fractions were analyzed by WB (UBR1 blot from S fraction). (G) U2OS cells were left untreated (Unt) or treated with 40 J/m2 UVC (4 h recovery), 2 mM hydroxyurea (HU) for 16 h, 100 ng/ml mitomycin C (MMC) for 16 h, or 100 ng/ml camptothecin (CPT) for 16 h, and P fractions were analyzed by WB. (H) U2OS cells were pretreated with 10 μM ATR inhibitor (VE-821) for 1 h, UVC irradiated and let recover in the presence of the inhibitor. Endogenous SDE2Ct was analyzed by WB in the P fractions.
Interestingly, SDE2Ct degradation appears to be the most pronounced following UVC irradiation in comparison to other sources of replication stress, and this coincides with the induction of PCNA monoubiquitination (Figure 3G). Depletion of RAD18, a ubiquitin E3 ligase for PCNA, did not prevent SDE2Ct degradation, indicating that SDE2Ct degradation is upstream of PCNA monoubiquitination (Supplementary Figure S3I). As previously reported, another Arg/N-end rule substrate USP1, a DUB for PCNA-Ub, exhibited specific degradation upon UVC irradiation (21) (Figure 3G). These results point to the importance of the N-end rule in regulating both USP1 and SDE2Ct in replication-associated DNA repair, to adjust the levels of PCNA-Ub. Thus, we reasoned that ATR checkpoint signaling might be coupled to the N-end rule-mediated SDE2Ct degradation in response to the replication stress caused by UVC lesions. Notably, treatment of cells with the ATR inhibitor VE-821 prevented endogenous SDE2Ct degradation following UVC irradiation (Figure 3H). These results establish a new link between replication stress-induced ATR signaling and the proteolytic pathway regulated by the N-end rule.
The p97UFD1-NPL4 segregase complex promotes chromatin-associated degradation of SDE2Ct
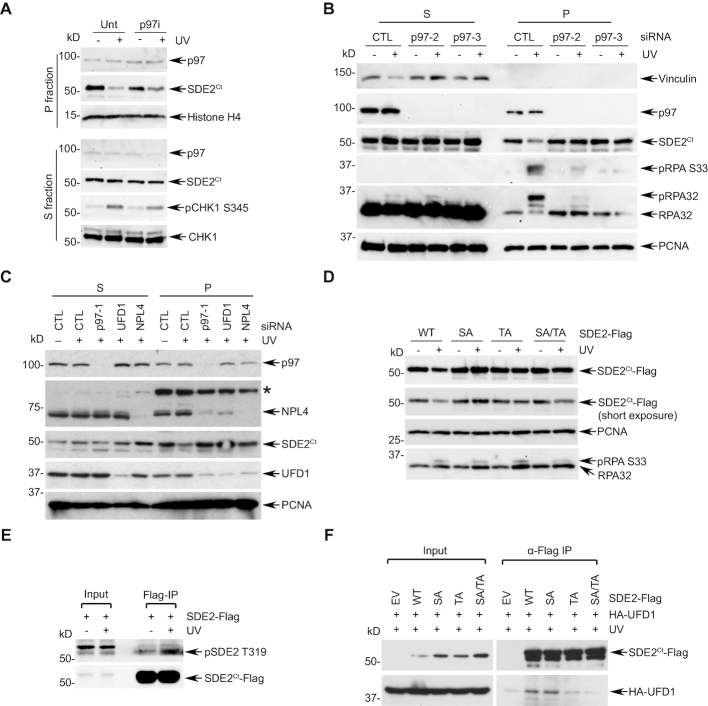
Specific SDE2Ct degradation in the chromatin upon UVC damage prompted us to investigate the element that couples replication stress and N-end rule signaling in the context of DNA interaction. The VCP/p97 segregase could be a reasonable candidate as it has emerged as a critical factor for the CAD of multiple DNA replication and damage checkpoint proteins (10). Indeed, inhibition of p97 activity via its specific inhibitor NMS-873 attenuated the degradation of endogenous SDE2Ct degradation upon UVC irradiation (Figure 4A and Supplementary Figure S4A). We observed a similar antagonistic effect of p97 inhibition to the degradation of both non-tagged and Flag-tagged exogenous SDE2Ct (Supplementary Figure S4B). p97 depletion using independent siRNA oligos also prevented the degradation of SDE2Ct specifically in the chromatin-enriched fraction following UVC irradiation (Figure 4B). Substrate specificity of p97 is determined by its adaptor proteins that contain p97-interating motifs and ubiquitin-binding domains. Notably, depletion of ubiquitin fusion degradation 1 (UFD1) or nuclear protein localization protein 4 (NPL4), two components of the heterodimeric p97 adaptor complex, protected SDE2Ct from UVC-induced degradation specifically at the chromatin, indicating that the p97UFD1-NPL4 complex mediates CAD of SDE2Ct (Figure 4C). In contrast, depletion of another adaptor, UBXN7, required for targeting the NER factors DDB2 and XPC, did not affect SDE2Ct degradation (22) (Supplementary Figure S4C). Together, these data indicate that the N-end rule-p97UFD1-NPL4 axis promotes the CAD of SDE2Ct upon UVC-induced replication stress.
Figure 4.
p97UFD1-NPL4 mediates the CAD of SDE2Ct. (A) U2OS cells were pretreated with 10 μM p97 inhibitor (NMS-873) for 1 h, UVC irradiated and let recover in the presence of the inhibitor. Endogenous SDE2Ct was analyzed by WB. (B) U2OS cells transfected with two independent p97 siRNA oligos (versus CTL) were irradiated with 40 J/m2 UVC, and S/P fractions were analyzed by WB. (C) U2OS cells transfected with indicated siRNA oligos were irradiated with 40 J/m2 UVC, fractionated, and analyzed by WB. * chromatin-bound nonspecific protein. (D) U2OS cells expressing WT, S266A, T319A, or S266A/T319A SDE2-Flag were irradiated with 40 J/m2 UVC, and P fractions were analyzed by WB. (E) U2OS cells expressing SDE2-Flag were irradiated with 40 J/m2 UVC, recovered in the presence of 20 μM MG132 for 2 h. Chromatin fraction was subjected to anti-Flag IP and WB with anti-pSDE2 T319 and Flag antibodies. (F) 293T cells transfected with the indicated plasmids were subjected to anti-Flag IP and WB.
Damage-inducible SDE2Ct phosphorylation promotes association with the p97UFD1-NPL4 complex for its degradation
We next sought to determine the mechanisms by which SDE2Ct is degraded in a damage-dependent manner. SDE2 contains two putative ATM/ATR-mediated S/TQ phosphorylation sites at Ser266 and Thr319, located in the linker region between the conserved SDE2 and SAP domains. Phosphorylation at Thr319 upon UVC damage was previously reported in a phosphoproteomics analysis (23). Since ATR inhibition reduces SDE2Ct degradation, we hypothesized that damage-inducible SDE2Ct phosphorylation may contribute to its degradation. Notably, mutations of either Ser266 or Thr319, or of both, elevated steady-state SDE2Ct levels and significantly attenuated SDE2Ct degradation following UVC irradiation, indicating that SDE2Ct phosphorylation contributes to its own degradation (Figure 4D and Supplementary Figure S4D). The phospho-specific antibody that we generated was able to recognize damage-inducible pThr319 of exogenous SDE2 enriched by anti-Flag immunoprecipitation (Figure 4E), which is abolished in the T319A mutant or ATR inhibition, and removed by phosphatase treatment (Supplementary Figure S4E–G). To further determine how phosphorylation affects SDE2Ct degradation, we examined whether phosphorylation promotes interactions with players in the N-end rule-p97 signaling axis. Consistent with our identification of the UFD1–NPL4 heterodimer as an adaptor for p97-mediated SDE2Ct degradation, we observed the interaction between SDE2Ct-Flag and HA-UFD1 (Figure 4F). Importantly, mutations of the phosphorylation sites significantly abolished the co-immunoprecipitation of UFD1 by SDE2Ct, indicating that SDE2Ct phosphorylation is required for UFD1 to efficiently recognize SDE2Ct. Consistent with this, ATR inhibition abolished this interaction (Supplementary Figure S4H). In contrast, interactions of the non-phosphorylatable SDE2 mutants with UBR1 upon UVC damage were largely unaffected (Supplementary Figure S4I). Together, these data suggest that ATR-dependent SDE2Ct phosphorylation promotes N-end rule-dependent SDE2Ct degradation by enhancing the SDE2Ct interaction with the p97UFD1-NPL4 complex to facilitate the extraction of ubiquitinated SDE2Ct from chromatin.
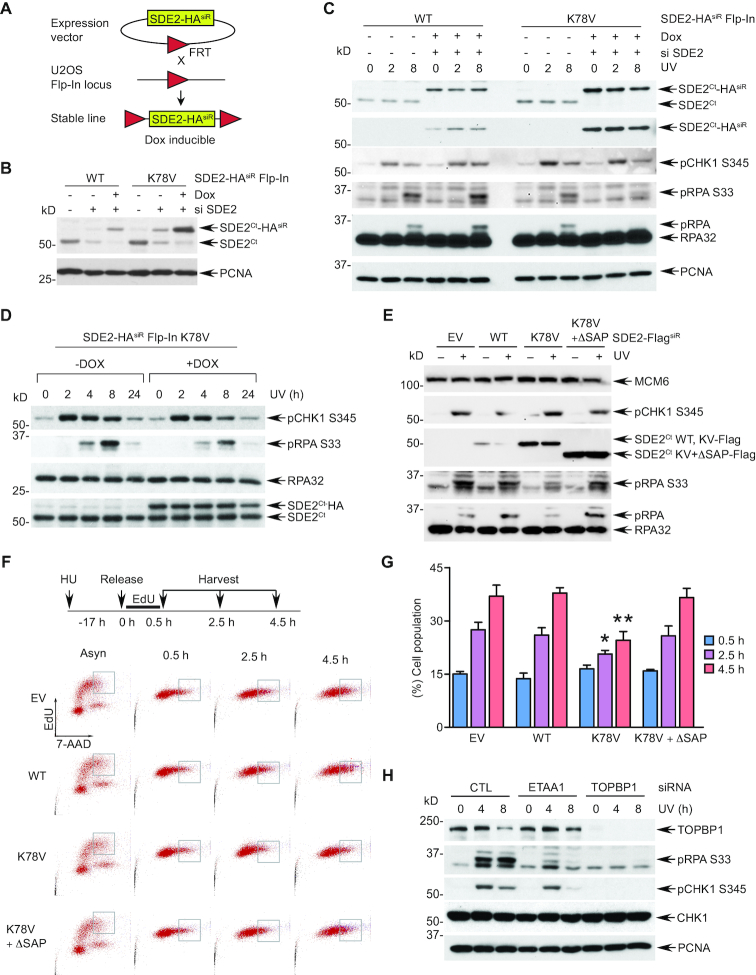
N-end rule-dependent degradation of SDE2Ct promotes RPA phosphorylation and recovery from stalled forks
To explore the specific role of SDE2Ct degradation regulated by the N-end rule in counteracting replication stress, we employed an inducible Flp-In™ T–REx™ system to replace endogenous SDE2 with exogenous SDE2 WT or K78V. In this system, siRNA-resistant SDE2-HA WT or K78V cDNA was integrated into a specific locus in U2OS cells via Flp recombinase-mediated DNA recombination at the FRT site, where SDE2-HA expression is induced by doxycycline (dox) while endogenous SDE2 is simultaneously eliminated by siRNA to ‘flip’ the expression (24) (Figure 5A). This system allows generation of isogenic stable cell lines where the protein expression is controlled to an endogenous level. Upon knockdown of the endogenous SDE2 and dox induction of the exogenous SDE2Ct-HA WT or K78V, we observed that the SDE2Ct K78V proteins are maintained at higher levels than WT ones, indicating that SDE2-HA is under the control of the N-end rule (Figure 5B). In this setting, exogenous SDE2Ct-HA WT was able to fully complement endogenous SDE2Ct for RPA phosphorylation and CHK1 activation following UVC irradiation (Figure 5C). In contrast, cells expressing the K78V mutant exhibited a marked reduction of RPA phosphorylation in comparison to the WT ‘flipped’ cells, indicating that defective SDE2Ct degradation compromises RPA loading and modification at stalled replication forks (Figure 5C). Of note, the kinetics of CHK1 phosphorylation were not affected, suggesting that the observed RPA phenotype is not due to a global defect in checkpoint signaling. Furthermore, induction of the K78V mutant expression on top of endogenous SDE2 was sufficient to attenuate RPA phosphorylation, suggesting that aberrant SDE2Ct accumulation interferes with RPA activation and recruitment to stalled forks (Figure 5D). We observed similar results of RPA phosphorylation impairment from the ‘flipped’ cells reconstituted with the K78V mutant (Supplementary Figure S5A) and attenuation of RPA phosphorylation after induction of K78V on top of endogenous SDE2 in the K78V Flp-In cells (Supplementary Figure S5B).
Figure 5.
N-end rule-dependent SDE2Ct degradation is necessary for optimal RPA phosphorylation and loading at stalled replication forks. (A) A schematic of generation of the Flp-In U2OS cells. (B) Validation of the Flp-In system by induction of expression of the WT or K78V siRNA-resistant (siR) SDE2-HA WT or K78V using 10 ng/ml doxycycline (dox) for 72 h and by siRNA-mediated knockdown for 84 h. (C) SDE2 WT or K78V expression was induced in Flp-In cells by siRNA transfection and dox induction, cells were irradiated by 10 J/m2 UVC, and lysates harvested after the indicated periods of recovery were analyzed by WB. (D) SDE2 K78V Flp-In cells were induced or not with 10 ng/ml dox for 64 h, followed by UVC irradiation and WB. (E) U2OS cells stably expressing MSCV-based SDE2-FlagsiR WT, K78V, or K78V + ΔSAP (versus EV) were irradiated with 30 J/m2 followed by 3 h recovery, and cell lysates were analyzed by WB. (F) U2OS cells in (E) were incubated with 2 mM HU for 17 h, pulsed with 10 μM EdU for 30 min, and released into fresh medium. Cells were harvested at indicated times, and progression of EdU+ cells was monitored by flow cytometry. A representative gating of EdU+ cells at late S phase is shown. (G) The percentage of gated cells tracked by EdU staining is represented. Mean ± SD (n = 3 independent experiments), * P < 0.05 and ** P < 0.01 compared to EV, WT, or K78V + ΔSAP, unpaired two-tailed Student's t-test. (H) siRNA-transfected U2OS cells were irradiated with 10 J/m2 UVC, harvested at the indicated times, and cell lysates were analyzed by WB.
To complement our findings, we generated an independent system to replace endogenous SDE2 with siRNA-resistant Flag-tagged SDE2 variants by MSCV retroviral vector-mediated transduction (Supplementary Figure S5C). Interestingly, deletion of the DNA-binding SAP domain (ΔSAP) within the K78V mutant (K78V ΔSAP) further stabilized the protein in stable cell lines, indicating that the CAD of SDE2Ct is a key downstream step for its degradation, and that other proteolytic pathways may converge to the p97 signaling to promote SDE2Ct extraction from the chromatin. Using this system, we again showed that cells reconstituted with the K78V mutant following SDE2 knockdown fail to induce robust RPA phosphorylation upon UVC damage, without compromising CHK1 phosphorylation (Supplementary Figure S5D). Following UVC irradiation, phosphorylated RPA was exclusively present in the chromatin-enriched fraction, which was abrogated in the K78V-expressing cells, confirming that RPA loading and phosphorylation at stalled forks were inhibited due to defective SDE2Ct degradation (Supplementary Figure S5E). Moreover, stable overexpression of the K78V mutant in endogenous SDE2 background was sufficient to reduce pRPA levels induced by UVC damage in comparison to vector control or WT (Figure 5E). Importantly, deletion of the SAP domain in the K78V mutant failed to suppress pRPA levels, indicating that the harmful effect of SDE2Ct accumulation manifests itself in the context of DNA association (Figure 5E). Similarly, transient overexpression of the K78A mutant, but not the ΔSAP mutant, decreased UVC-induced pRPA levels compared to control despite their similar expression levels (Supplementary Figure S5F). Together, these data suggest that the N-end rule-mediated SDE2Ct-degradation facilitates RPA recruitment and phosphorylation at stalled forks.
Then, we explored the impact of SDE2 degradation by the N-end rule in cell cycle progression and stalled fork recovery using the above-mentioned MSCV stable cells. Overexpression of the SDE2Ct wild-type or mutant did not significantly impact cell proliferation, as measured by cells traversing S phase via EdU labeling and flow cytometry (Supplementary Figure S5G). In contrast, when assessed following replication stress, enforced expression of the K78V mutant, but not of the WT or K78V ΔSAP, significantly impaired S phase progression (Figure 5F and G). These results indicate that disruption of SDE2Ct degradation prevents efficient DNA replication and stalled fork recovery under replication stress, highlighting the physiological role of the N-end rule in genome maintenance.
Specific downregulation of pRPA, but not pCHK1, by the K78V mutant following replication stress suggests that a subset of ATR signaling is impaired by defective SDE2Ct degradation. Similarly, knockdown of ETAA1, a recently identified ATR activator that is recruited to stalled forks via direct RPA interaction (25,26), specifically abrogated RPA phosphorylation upon UVC irradiation and camptothecin (CPT) treatment, while pCHK1 activation was largely unaffected (Figure 5H and Supplementary Figure S5H). In contrast, knockdown of TOPBP1, another ATR activator at the 5’ ssDNA-dsDNA junction of collapsed forks that works in parallel to ETAA1, was sufficient to abolish CHK1 phosphorylation. These results indicate that aberrant accumulation of SDE2Ct K78V may selectively impair ATR activation cascade regulated by ETAA1, whose activity directly relies on the generation of a ssDNA-RPA platform in the vicinity of stalled forks (27).
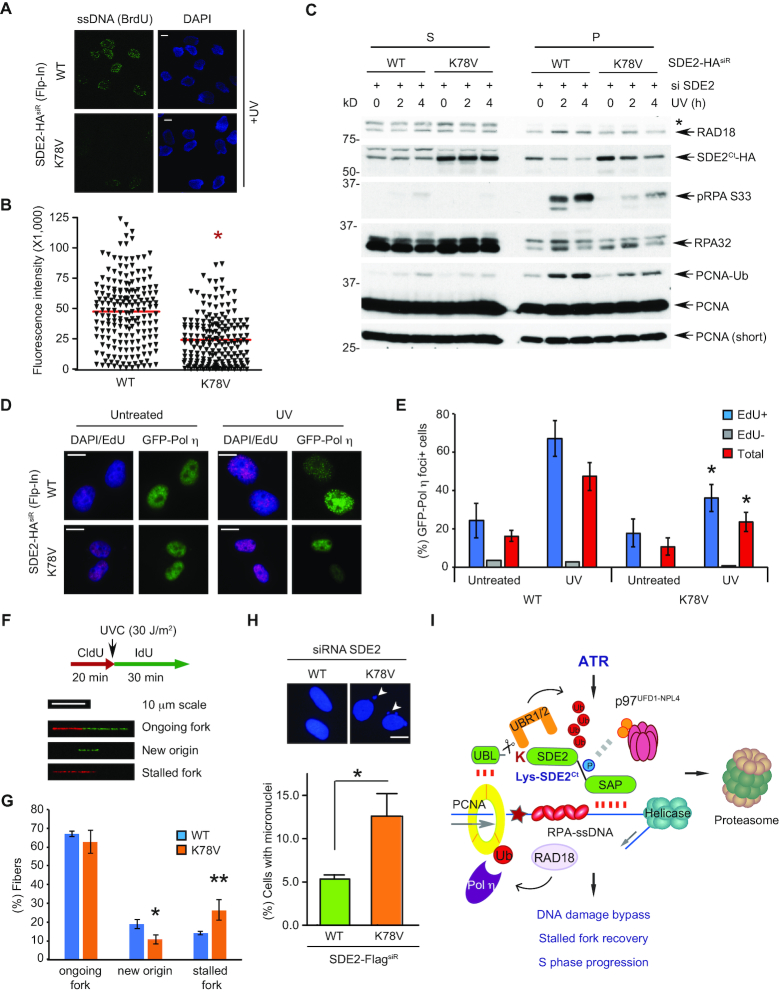
SDE2Ct degradation facilitates ssDNA formation and signaling for DNA damage bypass at stalled replication forks
We reasoned that the RPA-loading defect seen in the K78V mutant cells could result from reduced ssDNA formation at stalled forks due to the improper clearance of SDE2Ct in response to replication stress. Indeed, native BrdU staining revealed that the flipped cells expressing the K78V mutant displayed reduced generation of ssDNA in response to UVC irradiation, compared to those expressing WT (Figure 6A and B). We observed similar results in the MSCV-based stable cell lines expressing WT versus K78V following knockdown of endogenous SDE2 (Supplementary Figure S6A and B). A stretch of RPA-coated ssDNA at stalled replication forks provides a platform for recruitment of the RAD18 ubiquitin E3 ligase, which in turn monoubiquitinates PCNA and triggers polymerase switching required for TLS. Subcellular fractionation demonstrated that the flipped cells expressing K78V mutant have impaired RPA phosphorylation and RAD18 association to chromatin, which coincided with reduced UVC-inducible PCNA monoubiquitination in comparison to the WT cells (Figure 6C and Supplementary Figure S6C). Similarly, reduced levels of RPA phosphorylation, RAD18 recruitment, and PCNA monoubiquitination were observed in MSCV-based stable cell lines reconstituted with the K78V mutant (Supplementary Figure S6D). Overexpression of the K78A mutant was also sufficient to antagonize PCNA monoubiquitination after UVC irradiation (Supplementary Figure S6E). Subsequently, reconstitution of SDE2-depleted cells with the K78V mutant decreased the foci formation of TLS polymerase η to UVC-induced DNA lesions in both Flp-In and MSCV systems, indicating that the downregulation of PCNA-Ub compromises the switching to the TLS polymerases due to a defect in SDE2 degradation after UVC-induced damage (Figure 6D and E, and Supplementary Figure S6F and G). Damage-dependent Pol η foci formation was predominantly observed in EdU-labeled S-phase cells, indicating that the foci were mostly induced against replication-associated DNA lesions (Figure 6E). Moreover, knockdown of UBR1 and UBR2 increased cellular SDE2 levels and compromised UVC-induced RPA phosphorylation, which recapitulates the effect of K78V mutant, although UBR1 and UBR2 may have other unknown N-end rule substrates contributing replication stress signaling (Supplementary Figure S6H).
Figure 6.
UVC-induced SDE2Ct degradation promotes DNA damage bypass at stalled replication forks. (A) SDE2 WT or K78V flipped cells were incubated with 10 μM BrdU for 48 h, and ssDNA presence was analyzed by anti-BrdU immunostaining in non-denaturing conditions. Representative images are shown. Scale bar, 10 μm. (B) Quantification of BrdU intensity using Fiji. A representative result from three independent experiments is shown. * P < 0.0001, unpaired Student's t-test. (C) SDE2 WT or K78V flipped cells were irradiated with 40 J/m2 UVC and harvested 4 h later, then fractionated and analyzed by WB. * ubiquitinated RAD18 at S. (D) WT or K78V flipped cells were reverse transfected with GFP-Pol η on coverslips, pretreated with 10 μM EdU for 30 min to mark S-phase cells, irradiated with 40 J/m2 UVC, and after 2 h, GFP-Pol η foci were visualized by anti-GFP immunofluorescence. Representative images of GFP-Pol η from EdU+ cells (Alexa Fluor 647; violet) are shown. Scale bars, 10 μm. (E) Quantification of GFP-Pol η foci from EdU+, EdU–, or total (both EdU+ and EdU-) cells. Foci+ indicates >20 defined GFP-Pol η foci. Mean ± SD (n = 3), >100 cells per condition. * P < 0.01 compared to WT, unpaired Student's t-test. (F) Experimental conditions for DNA combing and representative images of DNA fiber tracks as classified. (G) Quantification of ongoing forks, IdU-labeled only (new origin firing), and CldU-labeled only (stalled fork) tracks. Mean ± SD (n = 3), >100 tracks per condition. * P = 0.0154 ** P = 0.0210, unpaired Student's t-test. (H) SDE2-FlagsiR WT or K78V-expressing U2OS cells were DAPI-stained, and the percentage of nuclei exhibiting micronuclei (arrowhead) were quantified. Mean ± SEM (n = 3), * P = 0.0102, unpaired Student's t-test. Scale bars, 10 μm. (I) A model depicting the ATR-N-end rule-p97 signaling axis that regulates SDE2Ct degradation after UVC damage.
One consequence of the defective TLS due to the disruption of SDE2 turnover would be cell's inability to recover from stalled replication forks upon UVC damage. Analysis of replication fork dynamics using a DNA combing assay revealed that cells expressing the K78V mutant have a significant increase in the frequency of fork stalling after UVC irradiation (Figure 6F and G). Cells expressing the K78V mutant exhibited increased numbers of shorter replication tracks in comparison to WT, although overall track length distribution was similar, indicating slower fork recovery (Supplementary Figure S6I and J). Subsequently, cells expressing the K78V mutant displayed elevated levels of micronuclei, which represents increased chromosome missegregation, likely caused by underreplicated DNA and chromosomal aberrations (Figure 6H). Taken together, these results support the idea that timely degradation of SDE2Ct in response to UVC-induced replication stress promotes the signaling for PCNA-dependent DNA damage bypass, thereby ensuring continuous fork progression at DNA lesions and chromosome stability.
DISCUSSION
The ATR-N-end rule-p97 proteolytic axis modulates replication stress via SDE2 degradation
In this study, we have shed light on a new ATR-N-end rule-p97 proteolytic axis, which governs the degradation of SDE2Ct at replication forks to counteract replication stress and preserve fork integrity. Regulated SDE2Ct degradation at stalled forks is necessary for generating an optimal ssDNA-RPA platform required for PCNA-dependent DNA damage bypass and stalled fork recovery (Figure 6I). Endolytic cleavage of SDE2 exposes an N-degron recognized by the UBR1/2 E3 ligases to polyubiquitinate SDE2Ct, which in turn undergoes chromatin extraction and degradation mediated by the p97UFD1-NPL4 segregase complex. This exclusively occurs in the context of DNA interaction, highlighting the cell's ability to specifically recognize and eliminate the protein pools directly influencing DNA replication and repair. p97 plays a regulatory role in this process, and p97-dependent recognition of polyubiquitinated substrates and extraction from chromatin may be a key rate-liming step for SDE2Ct degradation mediated by the N-end rule. Importantly, the N-end rule-mediated SDE2Ct degradation is coupled to ATR checkpoint signaling to facilitate SDE2Ct degradation in response to replication stress, by enhancing the interaction of SDE2Ct with p97UFD1-NPL4. In this regard, our study provides an important mechanistic insight into how proteolysis is regulated by phosphorylation-dependent ubiquitin signaling in the context of the DDR.
Growing evidence supports the idea that p97-driven CAD constitutes a critical regulatory mechanism for successful DNA replication and repair processes. Its multifaceted roles have been revealed by the identification of a wide range of substrates in multiple genome maintenance pathways, including initiation of DNA replication and S phase progression (licensing factor CDT1 and histone methyltransferase SET8), chromosome segregation (Aurora B), TLS (polymerase η), DNA interstrand cross-link repair (FANCD2), termination of DNA replication (MCM7 in the CMG helicase), nucleotide excision repair (XPC and DDB2) and double-strand breaks repair (Ku70/80) (22,28–34). Interestingly, p97 activity is targeted to specific situations by distinct cofactors, including UFD1-NPL4, SPRTN, UBXN7 and FAF1, emphasizing the versatility of CAD and its multiple layers of regulation (35,36). A recent study also identified RTF2 in the replisome, which needs to be removed by the proteasome shuttles DDI1/2 to suppress excessive ssDNA accumulation and fork collapse, thereby preventing replication stress (37). Although somewhat counterintuitive, these examples highlight the notion that proper elimination of key genome surveillance factors is essential for fine-tuning their activity in order to promote both DNA damage checkpoint signaling and recovery from necessary repairs. In the case of SDE2Ct, its persistent association with DNA via its SAP domain appears to be detrimental to cells, indicating that timely removal of SDE2Ct from stalled forks may be necessary for paving the way to optimal checkpoint signaling, mediated by the formation of RPA-coated ssDNA at relevant levels and positions within the replication fork structure, and/or active remodeling of the replisome components to regulate their activity at stalled forks. Identification of the interacting protein of SDE2 at replication forks would be necessary to better understand the role of regulated SDE2Ct degradation, thereby preventing replication stress.
SDE2 damage-specific mechanisms of action for promoting the replication stress response
Currently, the exact mechanisms by which SDE2 specifically responds to UVC-induced DNA lesions are not clear. Replication stress caused by distinct genotoxic insults may initiate different checkpoint sub-pathways to counteract specific damage most effectively. Indeed, a recent study demonstrated that hydroxyurea and aphidicolin, which both stall replication and induce ssDNA accumulation, relay distinct signaling pathways to remodel the replisome at replication forks (38). Additionally, the PCNA-interacting E3 ligase TRAIP is also specifically required for ssDNA formation and replication fork restart against UVC-induced lesions (39). To the best of our knowledge, what appears so far to be unique to UVC-induced lesions when encountered during DNA replication is that they are largely subject to TLS. As TLS is known to be regulated by the upstream processes of PCNA-monoubiquitination and DNA polymerase switching stemming from RPA-coated ssDNA, we propose that acute but dynamic changes of the uncoupling between the helicase and the polymerase may be uniquely promoted by SDE2Ct degradation to facilitate downstream TLS polymerase switch. We envision that the structure of DNA itself, distorted by a UVC lesion in close vicinity of a stalled replicative DNA polymerase, may also contribute to the propagation of specific signaling. SDE2 was previously identified at active replication forks by iPOND (40). SDE2 may be a part of the replisome complex, whose regulated turnover may determine the dissociation and degradation of other factors at stalled forks, which has been similarly proposed in the removal of RTF2 from the replisome upon replication stress to achieve optimal checkpoint signaling (37). Notably, USP1, a deubiquitinating enzyme for PCNA-Ub is exclusively degraded upon UVC damage-dependent autocleavage to promote PCNA monoubiquitination and TLS, arguing for the presence of specific DDR signaling that is utilized to promote stalled fork recovery from UVC-induced DNA lesions (41). More importantly, the cleaved Ct USP1 is another known substrate of the Arg/N-end rule (21), indicating that the N-end rule modulates repair of UVC-damage in at least two ways – by removing SDE2 to generate an optimal ssDNA-RPA platform for RAD18 recruitment and by degrading USP1 to inhibit its DUB activity, both of which promote PCNA monoubiquitination.
SDE2: a new substrate of the mammalian Arg/N-end rule for the DDR
In line with our study, Mishra and colleagues have recently reported that the cleaved C-terminal product of Sde2 in Schizosaccharomyces pombe modulates pre-mRNA splicing as a regulatory component of the spliceosome and is degraded via the Arg/N-end rule (42). It is remarkable to observe a highly conserved mechanism of proteolysis that regulates two biologically distinct processes in remotely related organisms. This report in conjunction with our current study highlights the role of regulated proteolysis by the Arg/N-end rule in nucleic acid metabolism and genome maintenance. Intriguingly though, Sde2 in S. pombe lacks both the DNA binding domain and the S/TQ motifs, which we have shown to fine-tune SDE2 turnover and function in mammalian cells, indicating that SDE2 in higher eukaryotes may have evolved to acquire additional functions in DNA damage signaling and repair. Since proper mRNA splicing is essential for counteracting aberrant accumulation of the DNA:RNA hybrids, R-loops, which are implicated as a major source of replication stress, SDE2 may have a broader role of genome surveillance, encompassing processes such as DNA replication and transcription as well as telomere maintenance (43).
The existence of the N-end rule, which suggests that a DUB could mediate the cleavage of an Nt ubiquitin moiety of a substrate, has been proposed for 30 years; however, it has only been demonstrated by an engineered Nt-fused ubiquitin model substrate so far (44–46). On the other hand, USP1, another validated mammalian N-end rule substrate, undergoes auto-cleavage via the UBL of its C-terminus (21). In this regard, our study demonstrates that SDE2 is the first physiological substrate in mammalian cells that undergoes the exposure of an N-degron via DUB-mediated cleavage ‘at its N-terminus’, as it was originally proposed.
We show in this study that while bulk degradation of SDE2Ct in unstressed cells is primarily dependent on the N-end rule, both the N-end rule and CRL4CDT2 appear to work in parallel to promote CAD of SDE2Ct under replication stress. Different ubiquitin signaling pathways may have evolved to participate in the ubiquitination of SDE2Ct, which converges on p97-dependent extraction of proteins from stalled forks. Whether the N-end rule or CRL4CDT2 is regulated by replication stress signaling or p97UFD1-NPL4-dependent chromatin removal is a rate-limiting step of SDE2Ct degradation is currently not clear. A proteomics study indicates that UBR1 is phosphorylated upon UVC damage, thus its activity could be regulated by the DDR as well (23). We anticipate that the N-end rule-p97 proteolytic axis may regulate the turnover of other unknown factors required for DNA replication and fork stability. Interestingly, the replisome is associated with various players involved in proteolysis including TRAIP (E3), RNF4 (E3), USP7 (DUB), and SPRTN (metalloprotease of DNA-protein cross-links) (47–51). It would be interesting to examine whether the N-end rule coordinates with these proteolytic factors to preserve replication fork integrity.
In summary, our studies provide new insights into how the N-end rule coordinates with replication stress signaling to exert proteolytic control over genome surveillance factors at replication forks. As cancer cells exhibit high levels of replication stress, this knowledge would be helpful to develop therapeutic strategies that exploit the N-end rule pathway of SDE2 in order to exacerbate the elevated replication stress of cancer cells and trigger a catastrophic failure of cancer cell viability.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Alexander Varshavsky (Caltech) for his insightful discussion and reagents. We thank Dr. Daniel Durocher (The Lunenfeld-Tanenbaum Research Institute) for a cell line and Dr. Orlando Schärer (UNIST, Korea) for critically reading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [CA218132]; American Cancer Society [RSG-18-037-01-DMC]; Concern Foundation; Carol M. Baldwin Breast Cancer Research Award; and Startup fund from the Research Foundation and the Cancer Center at Stony Brook University (to H.K.). Funding for open access charge: American Cancer Society.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zeman M.K., Cimprich K.A.. Causes and consequences of replication stress. Nat. Cell Biol. 2014; 16:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaillard H., Garcia-Muse T., Aguilera A.. Replication stress and cancer. Nat. Rev. Cancer. 2015; 15:276–289. [DOI] [PubMed] [Google Scholar]
- 3. Moldovan G.L., Pfander B., Jentsch S.. PCNA, the maestro of the replication fork. Cell. 2007; 129:665–679. [DOI] [PubMed] [Google Scholar]
- 4. Saldivar J.C., Cortez D., Cimprich K.A.. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017; 18:622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marechal A., Zou L.. RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res. 2015; 25:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lehmann A.R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J.F., Kannouche P.L., Green C.M.. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair. 2007; 6:891–899. [DOI] [PubMed] [Google Scholar]
- 7. Quinet A., Lemacon D., Vindigni A.. Replication fork Reversal: Players and guardians. Mol. Cell. 2017; 68:830–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011; 20:1298–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sriram S.M., Kim B.Y., Kwon Y.T.. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 2011; 12:735–747. [DOI] [PubMed] [Google Scholar]
- 10. Ramadan K., Halder S., Wiseman K., Vaz B.. Strategic role of the ubiquitin-dependent segregase p97 (VCP or Cdc48) in DNA replication. Chromosoma. 2016; 126:17–32. [DOI] [PubMed] [Google Scholar]
- 11. Vaz B., Halder S., Ramadan K.. Role of p97/VCP (Cdc48) in genome stability. Front. Genet. 2013; 4:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jo U., Cai W., Wang J., Kwon Y., D’Andrea A.D., Kim H.. PCNA-Dependent cleavage and degradation of SDE2 regulates response to replication stress. PLoS Genet. 2016; 12:e1006465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J., Jo U., Joo S.Y., Kim H.. FBW7 regulates DNA interstrand cross-link repair by modulating FAAP20 degradation. Oncotarget. 2016; 7:35724–35740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ravid T., Hochstrasser M.. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 2008; 9:679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kwon Y.T., Xia Z., An J.Y., Tasaki T., Davydov I.V., Seo J.W., Sheng J., Xie Y., Varshavsky A.. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 2003; 23:8255–8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kwon Y.T., Xia Z., Davydov I.V., Lecker S.H., Varshavsky A.. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol. Cell. Biol. 2001; 21:8007–8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tasaki T., Mulder L.C., Iwamatsu A., Lee M.J., Davydov I.V., Varshavsky A., Muesing M., Kwon Y.T.. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005; 25:7120–7136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tasaki T., Zakrzewska A., Dudgeon D.D., Jiang Y., Lazo J.S., Kwon Y.T.. The substrate recognition domains of the N-end rule pathway. J. Biol. Chem. 2009; 284:1884–1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Choi W.S., Jeong B.C., Joo Y.J., Lee M.R., Kim J., Eck M.J., Song H.K.. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat. Struct. Mol. Biol. 2010; 17:1175–1181. [DOI] [PubMed] [Google Scholar]
- 20. Matta-Camacho E., Kozlov G., Li F.F., Gehring K.. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat. Struct. Mol. Biol. 2010; 17:1182–1187. [DOI] [PubMed] [Google Scholar]
- 21. Piatkov K.I., Colnaghi L., Bekes M., Varshavsky A., Huang T.T.. The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol. Cell. 2012; 48:926–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Puumalainen M.R., Lessel D., Ruthemann P., Kaczmarek N., Bachmann K., Ramadan K., Naegeli H.. Chromatin retention of DNA damage sensors DDB2 and XPC through loss of p97 segregase causes genotoxicity. Nat. Commun. 2014; 5:3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R. 3rd, Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y. et al.. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007; 316:1160–1166. [DOI] [PubMed] [Google Scholar]
- 24. O’Gorman S., Fox D.T., Wahl G.M.. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991; 251:1351–1355. [DOI] [PubMed] [Google Scholar]
- 25. Bass T.E., Luzwick J.W., Kavanaugh G., Carroll C., Dungrawala H., Glick G.G., Feldkamp M.D., Putney R., Chazin W.J., Cortez D.. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016; 18:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haahr P., Hoffmann S., Tollenaere M.A., Ho T., Toledo L.I., Mann M., Bekker-Jensen S., Raschle M., Mailand N.. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 2016; 18:1196–1207. [DOI] [PubMed] [Google Scholar]
- 27. Feng S., Zhao Y., Xu Y., Ning S., Huo W., Hou M., Gao G., Ji J., Guo R., Xu D.. Ewing Tumor-associated antigen 1 interacts with replication protein A to promote restart of stalled replication forks. J. Biol. Chem. 2016; 291:21956–21962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Raman M., Havens C.G., Walter J.C., Harper J.W.. A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Mol. Cell. 2011; 44:72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ramadan K., Bruderer R., Spiga F.M., Popp O., Baur T., Gotta M., Meyer H.H.. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007; 450:1258–1262. [DOI] [PubMed] [Google Scholar]
- 30. Davis E.J., Lachaud C., Appleton P., Macartney T.J., Nathke I., Rouse J.. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nat. Struct. Mol. Biol. 2012; 19:1093–1100. [DOI] [PubMed] [Google Scholar]
- 31. Mosbech A., Gibbs-Seymour I., Kagias K., Thorslund T., Beli P., Povlsen L., Nielsen S.V., Smedegaard S., Sedgwick G., Lukas C. et al.. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nat. Struct. Mol. Biol. 2012; 19:1084–1092. [DOI] [PubMed] [Google Scholar]
- 32. Maric M., Maculins T., De Piccoli G., Labib K.. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014; 346:1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gibbs-Seymour I., Oka Y., Rajendra E., Weinert B.T., Passmore L.A., Patel K.J., Olsen J.V., Choudhary C., Bekker-Jensen S., Mailand N.. Ubiquitin-SUMO circuitry controls activated fanconi anemia ID complex dosage in response to DNA damage. Mol. Cell. 2015; 57:150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van den Boom J., Wolf M., Weimann L., Schulze N., Li F., Kaschani F., Riemer A., Zierhut C., Kaiser M., Iliakis G. et al.. VCP/p97 extracts sterically trapped Ku70/80 rings from DNA in Double-Strand break repair. Mol. Cell. 2016; 64:189–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Franz A., Orth M., Pirson P.A., Sonneville R., Blow J.J., Gartner A., Stemmann O., Hoppe T.. CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Mol. Cell. 2011; 44:85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van den Boom J., Meyer H.. VCP/p97-Mediated unfolding as a principle in protein homeostasis and signaling. Mol. Cell. 2018; 69:182–194. [DOI] [PubMed] [Google Scholar]
- 37. Kottemann M.C., Conti B.A., Lach F.P., Smogorzewska A.. Removal of RTF2 from stalled replisomes promotes maintenance of genome integrity. Mol. Cell. 2018; 69:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Somyajit K., Gupta R., Sedlackova H., Neelsen K.J., Ochs F., Rask M.B., Choudhary C., Lukas J.. Redox-sensitive alteration of replisome architecture safeguards genome integrity. Science. 2017; 358:797–802. [DOI] [PubMed] [Google Scholar]
- 39. Harley M.E., Murina O., Leitch A., Higgs M.R., Bicknell L.S., Yigit G., Blackford A.N., Zlatanou A., Mackenzie K.J., Reddy K. et al.. TRAIP promotes DNA damage response during genome replication and is mutated in primordial dwarfism. Nat. Genet. 2016; 48:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dungrawala H., Rose K.L., Bhat K.P., Mohni K.N., Glick G.G., Couch F.B., Cortez D.. The replication checkpoint prevents two types of fork collapse without regulating replisome stability. Mol. Cell. 2015; 59:998–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D.. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006; 8:339–347. [DOI] [PubMed] [Google Scholar]
- 42. Thakran P., Pandit P.A., Datta S., Kolathur K.K., Pleiss J.A., Mishra S.K.. Sde2 is an intron-specific pre-mRNA splicing regulator activated by ubiquitin-like processing. EMBO J. 2018; 37:89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sugioka-Sugiyama R., Sugiyama T.. Sde2: a novel nuclear protein essential for telomeric silencing and genomic stability in Schizosaccharomyces pombe. Biochem. Biophys. Res. Commun. 2011; 406:444–448. [DOI] [PubMed] [Google Scholar]
- 44. Bachmair A., Varshavsky A.. The degradation signal in a short-lived protein. Cell. 1989; 56:1019–1032. [DOI] [PubMed] [Google Scholar]
- 45. Varshavsky A. Ubiquitin fusion technique and related methods. Methods Enzymol. 2005; 399:777–799. [DOI] [PubMed] [Google Scholar]
- 46. Bachmair A., Finley D., Varshavsky A.. In vivo half-life of a protein is a function of its amino-terminal residue. Science. 1986; 234:179–186. [DOI] [PubMed] [Google Scholar]
- 47. Hoffmann S., Smedegaard S., Nakamura K., Mortuza G.B., Raschle M., Ibanez de Opakua A., Oka Y., Feng Y., Blanco F.J., Mann M. et al.. TRAIP is a PCNA-binding ubiquitin ligase that protects genome stability after replication stress. J. Cell Biol. 2016; 212:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lecona E., Rodriguez-Acebes S., Specks J., Lopez-Contreras A.J., Ruppen I., Murga M., Munoz J., Mendez J., Fernandez-Capetillo O.. USP7 is a SUMO deubiquitinase essential for DNA replication. Nat. Struct. Mol. Biol. 2016; 23:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ragland R.L., Patel S., Rivard R.S., Smith K., Peters A.A., Bielinsky A.K., Brown E.J.. RNF4 and PLK1 are required for replication fork collapse in ATR-deficient cells. Genes Dev. 2013; 27:2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stingele J., Bellelli R., Alte F., Hewitt G., Sarek G., Maslen S.L., Tsutakawa S.E., Borg A., Kjaer S., Tainer J.A. et al.. Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease SPRTN. Mol. Cell. 2016; 64:688–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vaz B., Popovic M., Newman J.A., Fielden J., Aitkenhead H., Halder S., Singh A.N., Vendrell I., Fischer R., Torrecilla I. et al.. Metalloprotease SPRTN/DVC1 orchestrates replication-coupled DNA-Protein crosslink repair. Mol. Cell. 2016; 64:704–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.