Abstract
The four B-family DNA polymerases α, δ, ϵ and ζ cooperate to accurately replicate the eukaryotic nuclear genome. Here, we report that a Saccharomyces cerevisiae strain encoding the pol2-16 mutation that lacks Pol ϵ’s polymerase and exonuclease activities has increased dNTP concentrations and an increased mutation rate at the CAN1 locus compared to wild type yeast. About half of this mutagenesis disappears upon deleting the REV3 gene encoding the catalytic subunit of Pol ζ. The remaining, still strong, mutator phenotype is synergistically elevated in an msh6Δ strain and has a mutation spectrum characteristic of mistakes made by Pol δ. The results support a model wherein slow-moving replication forks caused by the lack of Pol ϵ’s catalytic domains result in greater involvement of mutagenic DNA synthesis by Pol ζ as well as diminished proofreading by Pol δ during replication.
INTRODUCTION
Eukaryotic nuclear DNA replication is largely conducted by the four B-family DNA polymerases (Pols), Pols α, δ, ϵ and ζ. Pol α initiates replication by synthesizing short RNA-DNA primers that are then used by Pols δ and ϵ to synthesize the majority of the lagging and leading DNA strands, respectively (1–4). The fourth B-family member, Pol ζ, is more specialized and contributes to DNA synthesis when more difficult-to-replicate sequences are encountered (5,6). Pols α and ζ lack intrinsic exonuclease activity, while Pols δ and ϵ have 3′-exonucleases that can proofread mismatches. Pols α and ζ lack intrinsic exonuclease activity, such that the accuracy with which they synthesize DNA depends primarily on their nucleotide selectivity. Pols δ and ϵ have high nucleotide selectivity and they also have 3′-exonucleases that can proofread mismatches to further improve accuracy. Thus, Pols ϵ and δ synthesize DNA with very high fidelity, with average base substitution error rates of <2.0 × 10−5 for Pol ϵ and less than 1.3 × 10−5 for Pol δ, and average single nucleotide deletions error rates of less than 5.0 × 10−7 for Pol ϵ and <1.3 × 10−5 for Pol δ (7,8). Thus, the high fidelity of nuclear DNA replication in unstressed eukaryotic cells is thought to reflect the ability of these four DNA polymerases to select and incorporate correct nucleotides, proofreading by Pols δ and ϵ during replication, and DNA mismatch repair (MMR) that corrects mismatches that escape proofreading (9–11).
This general understanding of how replication fidelity is achieved has been supported by many studies (see below), including those that attempt to more precisely understand where and when each of the four B-family DNA polymerases functions during replication of large and complex eukaryotic genomes (1). Studies published in the last few years suggest two different models for replication of the unstressed nuclear genome, one in which Pol δ is the major replicase for both DNA strands (12) and the other proposing that Pol ϵ has a major role in leading strand replication (2,13–21). The latter model is supported by a study published earlier this year of the yeast pol2-16 mutant (22), which lacks the catalytic domains for polymerization and proofreading by Pol ϵ. This strain survives by replicating the nuclear genome using Pol δ as the primary replicase for both the leading and lagging DNA strands. However, cell growth in the pol2-16 mutant is aberrant, as indicated by elongated S-phase an increased doubling time, larger than normal cells that contain aberrant nuclei, and rapid acquisition of suppressors. In the present study, we add another endpoint, a mutator phenotype indicating that replication fidelity is strongly reduced when the catalytic domains of Pol ϵ are missing. The new data suggest that this mutator effect is partly due to reduced proofreading by Pol δ and partly due to errors generated by Pol ζ.
MATERIALS AND METHODS
Yeast strains construction
Saccharomyces cerevisiae strains used in this study are listed in Supplemental Materials. All yeast strains were isogenic derivatives of AC402 and AC403, representing the W303 background. Wild type diploids of W303 background and the pol2-16 mutants were generated as described earlier (22). Strains bearing the pol3L612M polymerase variant were constructed via an integration-excision method using plasmid p170-pol3L612M (23). Strains with deletion of REV3 (rev3Δ) and MSH6 (msh6Δ) were constructed using one-step gene disruption as follows. PCR product containing the rev3Δ::KanMX4 cassette was amplified from genomic DNA of YPL167C using as primers 5_REV3_F and 3_REV3_R. The presence of the rev3Δ::KanMX4 in transformants that were G-418r was confirmed by PCR using primers up_REV3_f and pTEF. PCR product containing msh6Δ::Kl-LEU2 - cassette was amplified from pUG73 using primers MSH6-LEU2-5 and MSH6-LEU2-3′. The presence of msh6Δ::Kl-LEU2 in transformants that were LEU2+ was confirmed by PCR using primers up_msh6_5′_f and Kl-LEU2_5′_r. Primer sequences are provided in the Supplementary Data 1.
Mutation rate measurements
To determine spontaneous mutation rates, at least 24 independent cultures of each yeast strain (two independent isolates) were inoculated with a single yeast colony or a spore colony in 5 ml of liquid YPDA supplemented with adenine to a final concentration of 100 mg/l. Cultures were grown at 23°C to the stationary phase (for 5 days in case of the pol2-16 mutant or 3 days for POL2) and plated on selective and nonselective media. Plates were incubated at 23°C for 8 days and colonies were scored. The mean mutation rates as well as 95% confidence intervals were calculated as described in (24). To determine P-values for significance of differences of the mutation rates between strains the Mann–Whitney U non-parametric test and GraphPad Prism 7 software was used.
CAN1 mutation spectra analysis
The Canr colonies for mutational spectrum analysis at the CAN1 locus were collected as described previously (25). Primers Can1-AF and Can1-BR were used for CAN1 locus amplification and primers Can1_BR, Can1_AR, Can1_9R and Can1_10R were used for sequencing. Sequences of all oligonucleotides are listed in the Supplementary Data 1. Mutations in CAN1 were called using SeqMan DNASTAR Navigator sequence assembly software. Graphical representation of analyzed mutation spectra are presented in Supplementary Data 2. Statistical significance of differences between two spectra were determined using a Monte Carlo method as described in (24,26). Likewise for determining the significance of differences between ratios of reciprocal mutation rate between two data sets, i.e. a ratio of ratios. Significance cutoffs were selected with Šidák correction for multiple hypothesis testing based on a familywise error rate of 5% (27).
dNTP pools measurement
dNTP pools were measured in three independent spore colonies of each genotype, from a freshly dissected heterozygous diploid pol2-16/POL2. Cells were inoculated in YPD medium supplemented with 100 mg/l adenine and grown at 23°C to OD600 between 0.35 and 0.4. Cells equivalent to 30 OD units were harvested by filtration, immediately suspended in an ice-cold trichloroacetic acid-MgCl2 mixture, flash frozen in liquid nitrogen. Samples were further proceeded as described previously (28).
Flow cytometry
Cells from an asynchronously growing culture were processed and analyzed for cell cycle progression by Becton Dickinson FC500 flow cytometer as described previously (29).
Immunoblotting for Sml1 expression
5 OD units of yeast cells collected at log phase (OD600 between 0.3 and 0.7) were resuspended in TCA buffer (20 mM Tris, pH 8, 50 mM ammonium acetate, 2 mM EDTA) supplemented with protease inhibitors (cOmplete EDTA-free protease inhibitors, Roche) and vortexed with glass beads at 4°C. Sml1 was detected using anti-Sml1 antibody (AS10 847, Agrisera) at 1:1000 dilution. Pstair was used as loading control, and detected with an antibody against pstair (Sigma, P7962) at 1:5000 dilution. Western Blots were developed using chemiluminescent substrates for HRP (WesternBright Sirius, advansta), and images were taken using G:BOX (SYNGENE).
RESULTS
Aberrant cell cycle progression, S-phase checkpoint activation and increased dNTP pools in pol2-16
Because the pol2-16 mutant quickly accumulates suppressors (22), here and throughout this study we use freshly isolated haploid pol2-16 colonies obtained from spores germinated from meiotic progeny of heterozygous diploid pol2-16/POL2 strains. Compared to wild type colonies, three independent pol2-16 mutant colonies exhibited aberrant progression through the cell cycle when analyzed by flow cytometry (Figure 1A). These results are consistent with our earlier study (22) demonstrating that, as compared to wild type, pol2-16 mutant cells are larger, grow more slowly and have aberrant nuclei (22). Moreover, pol2-16 cells also have dNTP concentrations that are elevated from 3-fold (for dGTP) to 5.5-fold (for dCTP) (Figure 1B). Consistent with such stress-related phenotypes, the pol2-16 mutant also has an activated S-phase checkpoint. The level of Sml1, an indicator of S-phase checkpoint activation, is significantly decreased to the level observed in wild type yeast treated with 4-NQO (Figure 1C).
Figure 1.
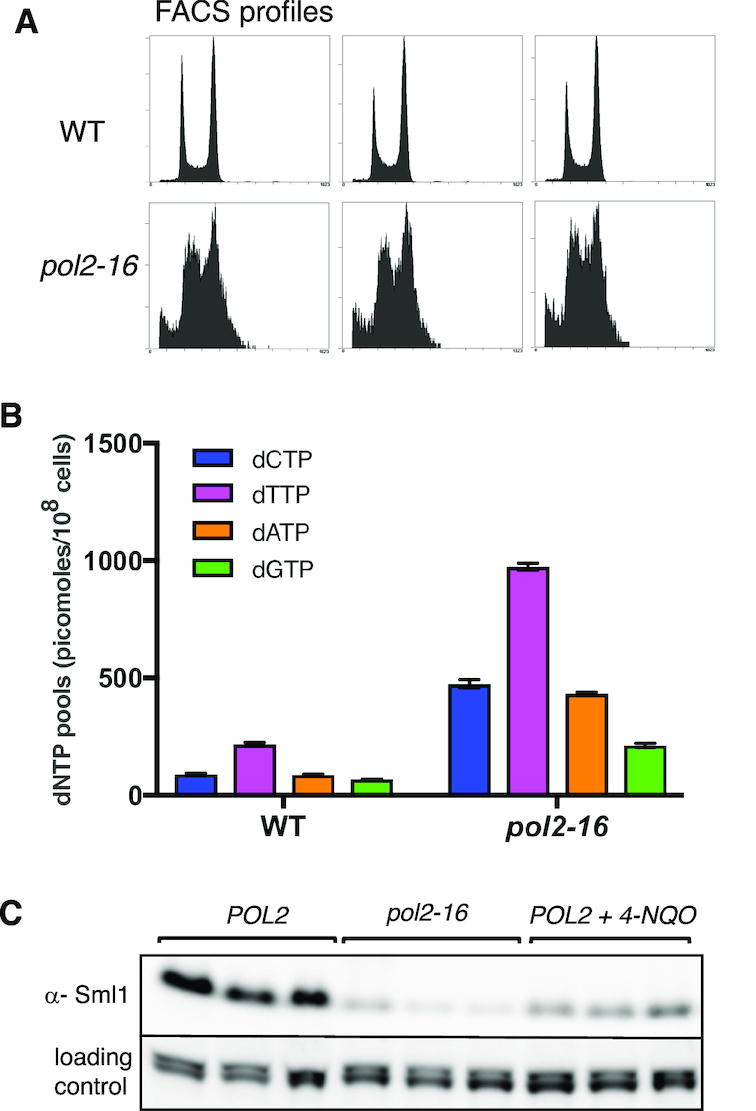
Lack of Pol ϵ catalytic domains (pol2-16) leads to replication stress. (A) Flow cytometry profiles of log phase yeast cultures of wild type and pol2-16 strains used for dNTP pool measurements; (B) intracellular dNTP levels; presented as mean values ± SD (n = 3); (C) western blot detection of Sml1 levels in whole cell extracts of the wild type, pol2-16 and wild type strains treated with the 4-nitroquinoline 1-oxide (4-NQO) at 0.2 μg/ml for 4 h, representative of two independent measurements is presented.
An increased mutation rate in pol2-16 mutant
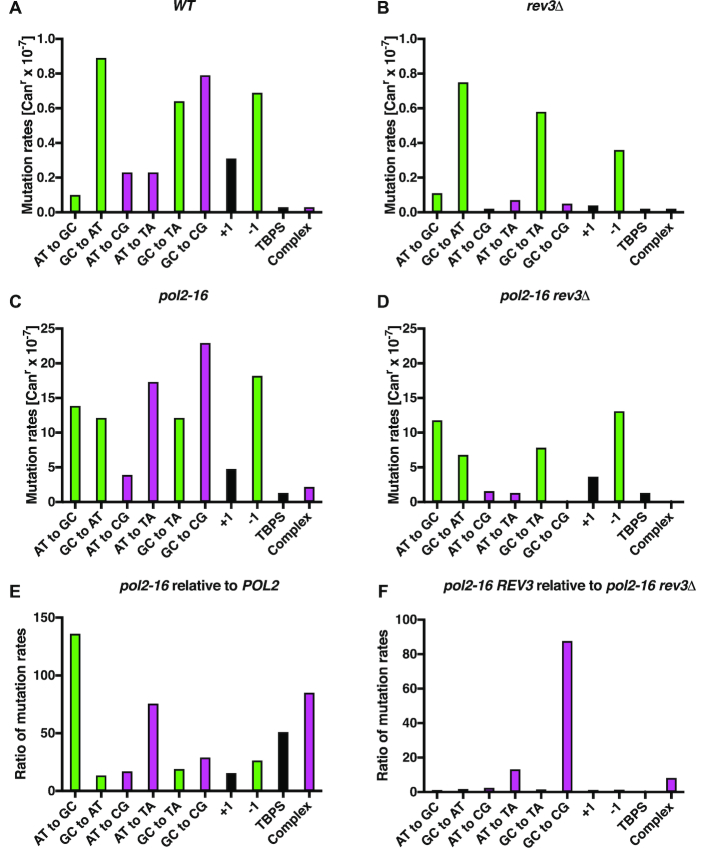
Next we measured the spontaneous mutation rates in the pol2-16 and wild type strains using the CAN1 reporter gene. Compared to a mutation rate of 4.1 × 10−7 in wild type cells, the pol2-16 mutant has mutation rate of 110 × 10−7 (Table 1). This 27-fold increase is substantial (P < 0.0001, Supplementary Data 1), being six times larger than that observed in a pol2-4 strain that lacks Pol ϵ proofreading and about three times larger than the rate in an msh6Δ strain that is partially defective in MMR of replication errors (Table 1). To understand the source of the mutations that spontaneously occur in the pol2-16 mutant, we sequenced Canr colonies (Table 2 and Supplementary Table S1), calculated mutation rates for various substitutions and insertions/deletions (indels) (Table 2 and Supplementary Table S1), and then compared the rates in wild type yeast (Figure 2A) to those in the pol2-16 mutant (Figure 2C). The mutation rates are increased in the pol2-16 mutant by factors ranging from about 10- to 130-fold (Figure 2E, Table 2 and Supplementary Table S1).
Table 1.
Spontaneous mutation rates in the pol2-16, rev3Δ, msh6Δ, pol3L612M mutant alone and in combinations
| Strain | Mutation Rate [Canr x 10−7] | Relative rate (mutants vs. wt) |
|---|---|---|
| Wild type | 4.1a (3.7–4.4) b | 1.0c |
| pol2-16 | 110 (96–120) | 26.8 |
| pol2-4 | 18.0 d | 4.4 |
| msh6Δ | 40 (35–46) | 9.8 |
| pol2-16 msh6Δ | 570 (450–730) | 139.0 |
| rev3Δ | 2.4 (2.1–2.6) | 0.6 |
| pol2-16 rev3Δ | 50 (42–59) | 12.2 |
| rev3Δ msh6Δ | 49 (43–57) | 12.0 |
| pol2-16 rev3Δ msh6Δ | 320 (260–410) | 78.1 |
| pol3L612M | 29 (22–37) | 7.1 |
| pol2-16 pol3L612M | 670 (490–910) | 163.4 |
aMean value of the mutation rates are presented as Canr x 10 −7;
b95% CL range of mutation rates are presented in parentheses;
cRelative rate is the mutation rate in a given strain divided by the mutation rate in the wild type strain;
dMutation rate taken from (25).
Table 2.
Mutation rates of specific mutation types detected in Canr yeast colonies
| Type of mutation/ Strain | WT | pol2-16 | rev3Δ | pol2-16 rev3Δ | pol2-16 rev3Δ msh6Δ | pol2-16 pol3L612M | pol3L612M |
|---|---|---|---|---|---|---|---|
| Base substitutions | 2.88 a (113) b | 82.28 (190) | 1.58 (87) | 29.58 (113) | 285.22 (164) | 368.66 (115) | 18.7 (118) |
| Transitions | 0.99 (39) | 25.98 (60) | 0.85 (47) | 18.59 (71) | 205.22 (118) | 182.73 (57) | 12.68 (80) |
| AT→GC | 0.1 (4) | 13.86 (32) | 0.11 (6) | 11.78 (45) | 76.52 (44) | 57.7 (18) | 5.55 (35) |
| GC→AT | 0.89 (35) | 12.13 (28) | 0.75 (41) | 6.81 (26) | 128.7 (74) | 125.02 (39) | 7.13 (45) |
| Transversions | 1.88 (74) | 56.3 (130) | 0.73 (40) | 10.99 (42) | 80 (46) | 185.93 (58) | 6.02 (38) |
| AT→CG | 0.23 (9) | 3.9 (9) | 0.02 (1) | 1.57 (6) | 3.48 (2) | 32.06 (10) | 1.9 (12) |
| AT→TA | 0.23 (9) | 17.32 (40) | 0.07 (4) | 1.31 (5) | 1.74 (1) | 32.06 (10) | 1.27 (8) |
| GC→TA | 0.64 (25) | 12.13 (28) | 0.58 (32) | 7.85 (30) | 74.78 (43) | 102.58 (32) | 2.22 (14) |
| GC→CG | 0.79 (31) | 22.95 (53) | 0.05 (3) | 0.26 (1) | <1.74 (<1) | 19.23 (6) | 0.63 (4) |
| InDels c | 0.99 (39) | 22.95 (53) | 0.4 (22) | 16.75 (64) | 31.3 (18) | 298.13 (93) | 9.83 (62) |
| + 1 | 0.31 (12) | 4.76 (11) | 0.04 (2) | 3.66 (14) | 12.17 (7) | 48.09 (15) | 1.27 (8) |
| − 1 | 0.69 (27) | 18.19 (42) | 0.36 (20) | 13.09 (50) | 19.13 (11) | 250.05 (78) | 8.56 (54) |
| Insertions ≥2 | 0.08 (3) | 0.43 (1) | 0.07 (4) | 0.79 (3) | 1.74 (1) | <3.21 (<1) | 0.16 (1) |
| Deletions ≥2 | 0.15 (6) | 0.87 (2) | 0.35 (19) | 1.57 (6) | 1.74 (1) | <3.21 (<1) | 0.16 (1) |
| TBPS d | <0.03 (<1) | 1.3 (3) | 0.02 (<1) | 1.31 (5) | <1.74 (<1) | <3.21 (<1) | 0.16 (1) |
| Complex c | <0.03 (<1) | 2.17 (5) | <0.02 (<1) | <0.26 (<1) | <1.74 (<1) | 3.21 (1) | <0.16 (<1) |
| Total | 4.1 (161) | 110 (254) | 2.4 (132) | 50 (191) | 320 (184) | 670 (209) | 29 (183) |
aRates [Canr x 10−7] for particular types of mutations were calculated as described previously (24);
bNumber of events for specific classes of mutations are shown in brackets;
cIndels include minus and plus one nucleotide mutations;
dTBPS are tandem base pair substitutions;
eComplex mutations are defined as multiple changes within short DNA stretches (separated by up to 10 nt).
5% or less of sequenced Canr yeast colonies were wild type and they were not included in the analysis.
Figure 2.
Pol ζ is responsible for a fraction of mutations in the pol2-16 mutant. Diagrams A–D show the mutation rates of specific mutation classes measured for wild type, pol2-16, rev3Δ and pol2-16 rev3Δ yeast. Data in panels A–D are from Table 2. Panels E and F present ratios of mutation rates of specific mutation classes. Pink bars indicate mutation types characteristic for Pol ζ, green bars indicate mutation types characteristic for Pol δ (see text). Black bars represent other mutations (not Pol δ or ζ-dependent). TBPS are tandem base pair substitutions.
Partial suppression of pol2-16 mutator effect by deletion of REV3
The results in Figure 2C suggest that the loss of Pol ϵ catalytic activities in the pol2-16 mutant may promote two different sets of replication errors. One set includes A•T to G•C, G•C to A•T and G•C to T•A substitutions and single-base deletion mutations (colored green in Figure 2). This is interesting because, although there are many types of base-base substitution and indel mismatches that theoretically can be made during DNA replication, it is these specific mutations that are preferentially made by Pol δ, through T•dGMP, G•dTMP, C•dTMP and single-base deletion mistakes (30). Moreover, Pol δ is the polymerase implicated by HydEn-seq analysis of ribonucleotide incorporation to perform the bulk of replication of both DNA strands in the pol2-16 mutant (22).
The second set of errors in the pol2-16 mutant are A•T to C•G, A•T to T•A and G•C to C•G transversions and complex errors involving multiple clustered changes (all colored pink in Figure 2C). This second set of errors has previously been observed to disappear in yeast strains defective in Pol ζ (rev3Δ) (31–35), suggesting that these errors may be generated by Pol ζ. We therefore deleted the REV3 gene encoding the catalytic subunit of Pol ζ and then compared the mutation rate and specificity in the double mutant pol2-16 rev3Δ strain to that in the single mutants. Consistent with a role for Pol ζ in spontaneous mutagenesis, and as expected based on previous results (25,31–33,36–38) (and see discussion below), the rev3Δ strain has an approximately 2-fold lower mutation rate than the wild type yeast (Table 1 and Figure 2B; P < 0.0001, Supplementary Table S1). Importantly for the present study, this is also the case for the pol2-16 mutant, where the mutation rate dropped from 110 × 10−7 in pol2-16 to 50 × 10−7 in the pol2-16 rev3Δ double mutant (Table 1; P < 0.0001, Supplementary Data 1). Moreover, the analysis of mutational specificity reveals that in the pol2-16 rev3Δ double mutant strain, the rates for the second set of mutations (pink in Figure 2C and D) are diminished by 88-, 13- and 8- fold, respectively, for G•C to C•G, A•T to T•A, and complex mutations (Figure 2F; for all three mutation types P ≤ 0.0001, Supplementary Data 1), while the first set of mutations that includes A•T to G•C, G•C to A•T, G•C to T•A and one nucleotide deletions (in green) is only marginally affected by the rev3Δ, being decreased by 1.2-, 1.8-, 1.5- and 1.4-fold, respectively (for all four mutation types, p is >0.00029, the Šidák cutoff for a familywise error rate of 0.05).
Suppression of pol2-16 rev3Δ mutator effect by mismatch repair
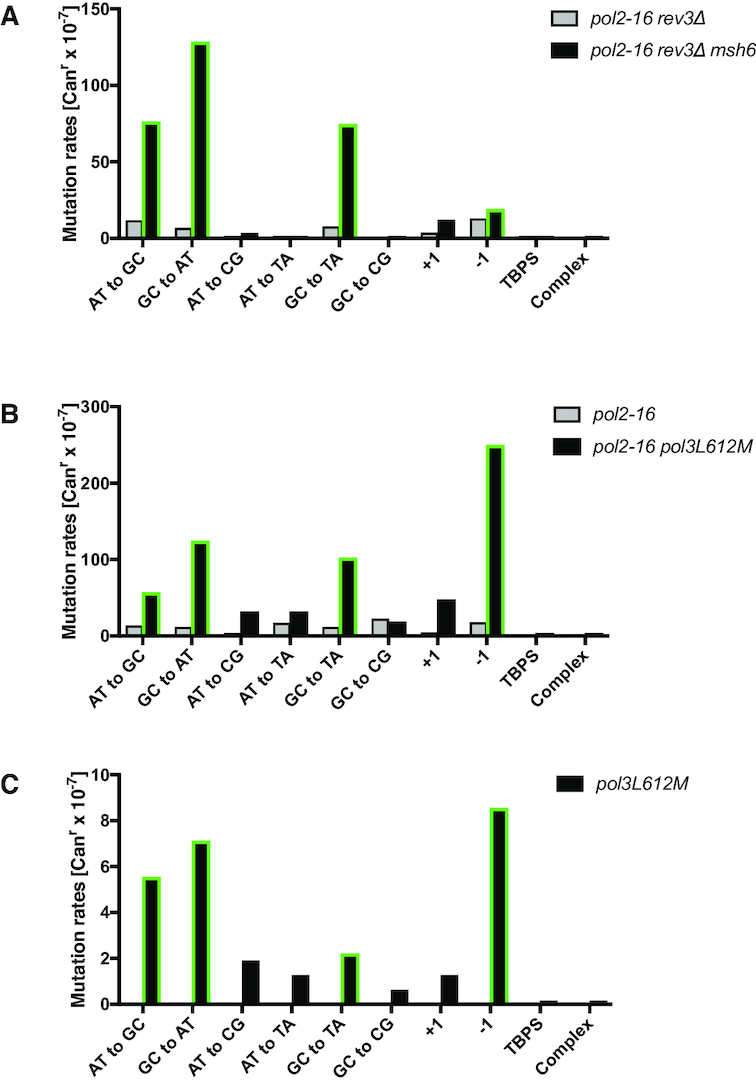
To determine whether the mutator effects in the pol2-16 rev3Δ strain depend on Pol δ, we performed two types of experiments. First, we partially inactivated MMR by deleting the MSH6 gene from the pol2-16 rev3Δ strain. MSH6 is a component of MutSα, the heterodimer that initiates the correction of base•base mismatches and small indel loops made by Pols α, δ and ϵ (14,39), but not mismatches made by Pol ζ (40,41). Consistent with previous reports (42), the mutation rate in the msh6Δ strain was 40 × 10−7 (Table 1), representing a 10-fold increase over the rate in wild type cells (P < 0.0001, Supplementary Data 1). Also consistent with other studies indicating that Pol ζ-dependent mutagenesis is independent of MMR (40,43), the mutation rate was largely unaffected by also deleting REV3 (msh6Δ rev3Δ, 49 × 10−7, Table 1). However, the mutation rate in the pol2-16 rev3Δ msh6Δ triple mutant strain is 320 × 10−7 (Table 1). This large increase in rate when Msh6 is missing is consistent with robust MMR of replication errors created in the pol2-16 rev3Δ mutant (P < 0.0001, Supplementary Data 1).
Evidence that mutations in pol2-16 rev3Δ are generated by Pol δ
Sequence analysis of Canr yeast colonies from pol2-16 rev3Δ versus pol2-16 rev3Δ msh6Δ strains (Table 2, Figure 3A and Supplementary Table S1) reveals that loss of Msh6-dependent MMR largely increases rates for the three base substitutions that are most commonly created by Pol δ (30), namely A•T to G•C transitions via template T•dGMP mismatches, G•C to A•T transitions via template G•dTMP mismatches, and G•C to T•A transversions via template C•dTMP mismatches (Figure 3A; for all three mutation types P ≤ 0.0001, Supplementary Data 1). These data are consistent with our previous interpretation based on HydEn-seq analysis that Pol δ is the primary replicase for both DNA strands in the pol2-16 mutant (22).
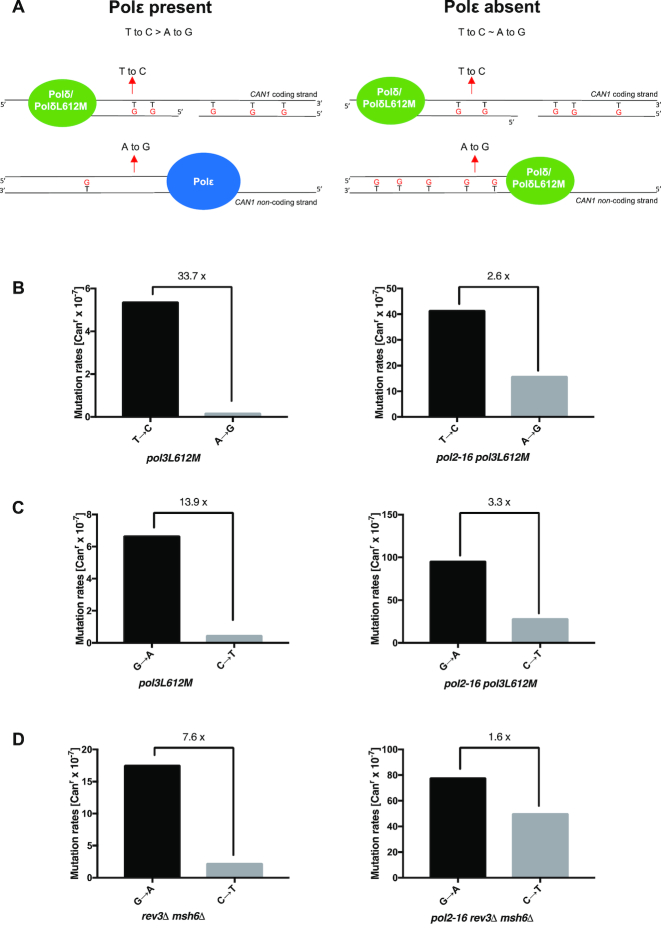
Figure 3.
Pol δ is responsible for a fraction of mutations in the pol2-16 mutant. Data in diagrams A–C are from Table 2 and show the mutation rates of specific mutation classes measured for pol2-16 rev3Δ, pol2-16 rev3Δ msh6Δ (panel A), pol2-16, pol2-16 pol3L612M (panel B) and pol3L612M yeast (panel C). Grey and black bars represent different genotypes. Green borderline of bars indicates mutation classes characteristic for L612M Pol δ.
As a further test of this hypothesis, we compared mutation rates and specificity in strains encoding a mutator allele of Pol δ, pol3L612M. Strains harboring this mutator allele generate Pol δ-dependent replication errors at an increased rate (14,30), as exemplified here by the 7-fold increase in mutation rate of the pol3L612M mutant compared to the wild type strain (Table 1). In this strain background, the addition of the pol2-16 mutation increases the mutation rate to 670 × 10−7 (P < 0.0001, Supplementary Data 1), representing a 160-fold increase over the rate in the wild type strain. Moreover, the CAN1 mutation spectrum (Table 2 and Supplementary Table S1) reveals that this strong increase is largely due to increases in the same three base-base mismatches mentioned immediately above (for all three mutation types P < 0.0001, Supplementary Data 1), plus single-base deletions (P < 0.0001, Supplementary Data 1) that are also characteristic of replication errors generated by L612M Pol δ (Figure 3C and (14,30)).
To further examine the role of Pol δ in replication of both DNA strands in the pol2-16 mutant, we next performed a pairwise comparisons of the CAN1 mutation spectra quantified in Figure 4 and Supplementary Table S1. Both wild type and L612M Pol δ have established preferences for T•dGMP over complementary A•dCMP mismatches (30). Given that CAN1 is replicated by forks originating from ARS507 roughly 98% of the time (15), we can infer the coding strand of CAN1 is usually the template for lagging strand synthesis. If Pol δ replicates only one strand (Figure 4A-left panel), then we expect a high ratio of T to C versus A to G substitutions at CAN1 (Figure 4A, left panel). If Pol δ replicates both strands we would expect a ratio closer to 1 (how close depends on details of sequence specificity and whether the Pol δ mutation rate would be the same on both strands; Figure 4A, right panel). In the pol3L612M strain, the ratio of T to C versus A to G substitutions is 33.7 (Figure 4B-left panel). However, the ratio drops to only 2.6 in the pol2-16 pol3L612M double mutant that lacks Pol ϵ catalytic activities (P = 0.002, Supplementary Data 1). A similar result is seen for G to A versus C to T substitutions in the latter two strains, where the ratio drops from 13.9 to 3.3 (Figure 4C; P = 0.02, Supplementary Data 1). In the rev3Δ msh6Δ strain, the ratio of G to A versus C to T substitutions is 7.6 (Figure 4D-left panel). However, the ratio drops to only 1.6 in the pol2-16 rev3Δ msh6Δ triple mutant that lacks Pol ϵ catalytic activities (P ≤ 0.001, Supplementary Data 1). These data are consistent with the interpretation that Pol δ is the major replicase for one DNA strand in wild type cells and both strands when the catalytic activities of Pol ϵ are missing.
Figure 4.
Mutation rates of specific events (reciprocal mistakes). Panel A represent a model of what is expected if Pol δ does only lagging strand synthesis (left) as opposed to the synthesis of both leading and lagging strands (right). Panels B–D represent the mutation rate [Canr x 10−7] for a specific event types. See text for explanations. A full list of all detected events for pol3L612M, pol2-16 pol3L612M, rev3Δ msh6Δ and pol2-16 rev3Δ msh6Δ strains is presented in Supplementary Table S1.
In wild type yeast at the CAN1 locus Pol δ synthesizes the majority of the lagging strand while Pol ϵ works predominantly on the leading strand. Replacement of the wild type Pol δ with the pol3L612M variant results in a seven-fold increase in mutation rate as compared to wild type yeast (29 × 10−7 in the po3L612M vs 4 × 10−7 for wild type; Table 1). When we remove the Pol ϵ catalytic domains in the pol2-16pol3L612M double mutant, we observe a multiplicative increase in the mutation rates as compared to single mutants (from 29 × 10−7 for pol3L612M and 110 × 10−7 for the pol2-16 to 670 × 10−7 in pol2-16pol3L612M; Table 1). These results suggest that there are additional factors affecting the fidelity of DNA synthesis due to lack of Pol ϵ’s catalytic domains (pol2-16). The effect could reflect the dNTP pool increases observed in the pol2-16 yeast, which is known to be a very important determinant of the fidelity of DNA polymerases (28,44–48).
DISCUSSION
The results presented here are consistent with the following interpretations regarding the fidelity of nuclear DNA replication when the catalytic activities of Pol ϵ are missing.
As we and others have shown earlier, the lack of Pol ϵ’s catalytic domains causes impaired cell cycle progression and an elongated S-phase, and it significantly enlarges the size of the cell and the nucleus (22,49,50). Here we show that the pol2-16 mutant also has a significantly decreased level of Sml1, a ribonucleotide reductase (RNR) inhibitor, and increased dNTP levels (Figure 1). All of these phenotypes suggest that lack of Pol ϵ’s catalytic domains leads to a replication stress and S-phase checkpoint activation. Such replication stress and accompanying increased dNTPs pool are associated with genome instability and increased spontaneous mutation rates (51–56).
Previous studies by Sugino et al. (49) indicated that the pol2-16 mutant has mutation rates that are only slightly greater than wild type, e.g., by only 1.6-fold at the URA3 locus. This contrasts with our results demonstrating that pol2-16 is a much more robust mutator, with a mutation rate at CAN1 that is 27-fold higher than for wild type cells and even higher than that of a MMR defective strain (Table 1). The difference between our measurements and those reported earlier is potentially explained by the rapid accumulation of suppressors acquired by the pol2-16 mutant (22). The nature of the suppression is under investigation. To minimize selection of suppressors that affect growth (22) and may affect mutagenesis, in all experiments we used pol2-16 spore colonies from a freshly dissected heterozygous diploid pol2-16/POL2. The strong increase in mutation rate in the pol2-16 mutant cannot be explained simply by loss of Pol ϵ proofreading activity, because inactivation of this activity in the pol2-4 mutant leads only to a moderate increase in mutation rate ((4,25,41) and Table 1), and does so without increasing dNTP pools (55). In addition, while Pol ϵ proofreads its own mismatches, it may not proofread mismatches made by Pol α (57) and possibly Pol δ (58).
In agreement with results published earlier (32,33), REV3 deletion in otherwise wild type yeast causes an ∼2-fold decrease in the spontaneous mutation rate (Table 1). A similar decrease is conferred in the pol2-16 mutant upon REV3 deletion. The mutations that disappear (colored pink in Figure 2) are those reported to be generated by Pol ζ, namely complex mutations and transversions. We previously proposed that complex mutations, which contain two or more single base substitutions and indels within about 10 base pairs of each other, are generated by Pol ζ during short stretches of processive DNA synthesis (59). The substitutions include A•T to C•G, A•T to T•A and G•C to C•G transversions (Figure 2C and D). All four classes of mutations are hallmarks of error-prone DNA synthesis by Pol ζ (25,33–35,37,38,41,60–62). In particular, the complex mutations and GC to CG transversions have been suggested to occur when replication stalls due to presence of atypical DNA structures, such as hairpins or non-B-form DNA structures (6). Under normal conditions Pol ϵ is physically connected to the moving fork via the CMG helicase (63,64), but when Pol ϵ catalytic domains are absent (pol2-16), Pol δ, which is excluded from the CMG (65), becomes the leading strand replicase. This physical uncoupling of unwinding (2) and leading strand synthesis may allow ssDNA secondary structure formation and thus an increase in mutagenic synthesis by Pol ζ. By extrapolation, this implies that under circumstances when Pol ϵ is fully active, Pol ζ will contribute less to mutagenic synthesis of genomic DNA.
The synergistic increase in mutation rates in the pol2-16 rev3Δ msh6Δ mutant as compared to those in the pol2-16 rev3Δ and rev3Δ msh6Δ mutants reveals that the errors made in the pol2-16 rev3Δ mutant strain are substrate for MMR, indicating that they are generated during DNA replication. Moreover, the mutational specificity (green bars in Figure 3A) is consistent with mutations made by Pol δ. These include the two transitions, A•T to G•C and G•C to A•T, the G•C to T•A transversions and also indel mutations, as seen in previous studies (14,30) and as observed here in the pol2-16 pol3-L612M strain (Figure 3B). The high rates at which all these mutations are generated are also consistent with the high dNTP concentrations observed in the pol2-16 background.
The L686M variant of Pol δ extends mismatches more efficiently as compared to wild type Pol δ and therefore proofreads mismatches less efficiently, despite retaining normal 3′ exonuclease activity (30). In the pol2-16 mutant studied here, dNTP pools are increased, and the same classes of base substitution mutations are observed as those in the pol3L686M mutant (Figure 3B and C). These facts suggest that the mutator effect in the pol2-16 mutant partly reflects impaired proofreading by Pol δ that is caused by the higher concentration of dNTPs. This interpretation is consistent with data in vitro demonstrating that proofreading is suppressed as the concentration of the next correct dNTP is increased (28,66,67).
Analysis of reciprocal mutation classes using polymerase variants have been used to assign DNA polymerases to specific DNA strands (13,14,23) and (Figure 4A). Here, we analyzed the specificity of mutations in strains with and without Pol ϵ’s catalytic subunit (POL2 and pol2-16, respectively), as well as bearing either the wild type or a mutator variant of Pol δ (POL3 and pol3L612M, respectively). Studies of the pol3L612M mutant in vitro have previously demonstrated that this variant of Pol δ incorporates dG opposite T in the template about 28-fold more frequently than it incorporates the complementary dC opposite A in the template (30). This fact, coupled with the fact that the coding strand of CAN1 is the template for lagging strand synthesis, predicts more T to C mutations than A to G mutations in the pol3L612M yeast, where Pol δ predominantly synthesizes the lagging strand (Figure 4). By analyzing the ratio of T to C versus A to G, we can determine whether Pol δ works on one (Figure 4A, left panel) or both DNA strands (Figure 4A, right panel). The ratio of T to C versus A to G mutations was 33.7 in the pol3L612M (Figure 4B, left panel) and decreased to only 2.6 in the pol2-16 pol3L612M (Figure 4B, right panel), indicating that both strands are synthesized by Pol δ. Similarly, the ratio of G to A vs C to T was 13.9 in the pol3L612M strain (Figure 4C, left panel) and decreased to only 3.3 in the pol2-16 pol3L612M (Figure 4C, right panel). Moreover, whole genome mutation accumulation experiments in yeast strains bearing the pol3L612M variant of Pol δ reveal that the rates of A to G vs T to C and of G to A vs C to T mutations are also biased when MMR is active (14). In the present study, the ratio of G to A vs C to T dropped from 7.6 in the rev3Δ msh6Δ strain to 1.6 (Figure 4D, left panel) in the pol2-16 rev3Δ msh6Δ triple mutant (Figure 4D, right panel). The stronger mutational biases in the strain with Pol ϵ catalytic activity than in the pol2-16 mutant strain without Pol ϵ catalytic domains are also consistent with previous HydEn-seq results showing Pol δ synthesis of both leading and lagging DNA strands when Pol ϵ catalytic domains are not present (22).
Here we present an extreme case wherein Pol δ is the major replicase for both the leading strand and the lagging strand across the entire genome due to lack of catalytic domains of Pol ϵ. This situation is likely to be relevant even in cells with wild type Pol ϵ. This is because in some circumstances, Pol δ has been shown to synthesize both the leading and lagging strands in a more local manner, for example during break-induced replication (68,69) and during homologous recombination-dependent replication fork restart (70). Both of these processes are mutagenic.
The catalytic subunit of Pol ϵ is composed of an amino terminal region possessing its two catalytic activities, and a carboxy-terminal region involved in checkpoint control (71–73). Both regions of Pol ϵ are involved in a network of interactions with other components of the replisome. For example, crosslinking mass spectrometry analysis identified Pol2p interactors that include Cdc45, Psf1, Mcm2, Mcm 5 and Mcm6 (74,75). Lack of the N-terminal lobe of Pol2p in the pol2-16 mutant may disturb interactions with other components of the replisome affecting both the replication initiation as well as DNA replication progression, thereby allowing other DNA polymerases to have access to the primer terminus more frequently.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kasia Bebenek and Roel Schaaper for critical reading of and thoughtful comments on the manuscript. We thank all members of the DNA replication fidelity group for helpful discussions throughout the work. We are grateful to Dmitry Gordenin for providing the source of rev3Δ::KanMX4 cassette.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Division of Intramural Research of the NIH [Z01 ES065070 to T.A.K.]; NIEHS; Swedish Cancer Society and the Swedish Research Council (to A.C.). Funding for open access charge: Division of Intramural Research of the NIH, NIEHS [Z01 ES065070 to T.A.K.].
Conflict of interest statement. None declared.
REFERENCES
- 1. Stillman B. Reconsidering DNA polymerases at the replication fork in eukaryotes. Mol. Cell. 2015; 59:139–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yeeles J.T., Janska A., Early A., Diffley J.F.. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell. 2017; 65:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schauer G.D., O’Donnell M.E.. Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Georgescu R., Yuan Z., Bai L., de Luna Almeida Santos R., Sun J., Zhang D., Yurieva O., Li H., O’Donnell M.E.. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E697–E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Makarova A.V., Burgers P.M.. Eukaryotic DNA polymerase zeta. DNA Repair (Amst.). 2015; 29:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Northam M.R., Moore E.A., Mertz T.M., Binz S.K., Stith C.M., Stepchenkova E.I., Wendt K.L., Burgers P.M., Shcherbakova P.V.. DNA polymerases zeta and Rev1 mediate error-prone bypass of non-B DNA structures. Nucleic Acids Res. 2014; 42:290–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shcherbakova P.V., Pavlov Y.I., Chilkova O., Rogozin I.B., Johansson E., Kunkel T.A.. Unique error signature of the four-subunit yeast DNA polymerase epsilon. J. Biol. Chem. 2003; 278:43770–43780. [DOI] [PubMed] [Google Scholar]
- 8. Fortune J.M., Pavlov Y.I., Welch C.M., Johansson E., Burgers P.M., Kunkel T.A.. Saccharomyces cerevisiae DNA polymerase delta: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J. Biol. Chem. 2005; 280:29980–29987. [DOI] [PubMed] [Google Scholar]
- 9. Burgers P.M.J., Kunkel T.A.. Eukaryotic DNA replication fork. Annu. Rev. Biochem. 2017; 86:417–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kunkel T.A., Burgers P.M.J.. Arranging eukaryotic nuclear DNA polymerases for replication: Specific interactions with accessory proteins arrange Pols alpha, delta, and in the replisome for leading-strand and lagging-strand DNA replication. Bioessays. 2017; 39:doi:10.1002/bies.201700070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lujan S.A., Williams J.S., Kunkel T.A.. DNA polymerases divide the labor of genome replication. Trends Cell Biol. 2016; 26:640–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson R.E., Klassen R., Prakash L., Prakash S.. A major role of DNA polymerase δ in replication of both the leading and lagging DNA strands. Mol. Cell. 2015; 59:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pursell Z.F., Isoz I., Lundstrom E.B., Johansson E., Kunkel T.A.. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007; 317:127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lujan S.A., Clausen A.R., Clark A.B., MacAlpine H.K., MacAlpine D.M., Malc E.P., Burkholder A.B., Fargo D.C., Gordenin D.A., Kunkel T.A.. Heterogeneous polymerase fidelity and mismatch repair bias genome variation and composition. Genome Res. 2014; 24:1751–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clausen A.R., Lujan S.A., Burkholder A.B., Orebaugh C.D., Williams J.S., Clausen M.F., Malc E.P., Mieczkowski P.A., Fargo D.C., Smith D.J. et al.. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 2015; 22:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Daigaku Y., Keszthelyi A., Muller C.A., Miyabe I., Brooks T., Retkute R., Hubank M., Nieduszynski C.A., Carr A.M.. A global profile of replicative polymerase usage. Nat Struct Mol Biol. 2015; 22:192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams J.S., Lujan S.A., Kunkel T.A.. Processing ribonucleotides incorporated during eukaryotic DNA replication. Nat. Rev. Mol. Cell Biol. 2016; 17:350–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Koh K.D., Balachander S., Hesselberth J.R., Storici F.. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat. Methods. 2015; 12:251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langston L.D., Zhang D., Yurieva O., Georgescu R.E., Finkelstein J., Yao N.Y., Indiani C., O’Donnell M.E.. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15390–15395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shinbrot E., Henninger E.E., Weinhold N., Covington K.R., Goksenin A.Y., Schultz N., Chao H., Doddapaneni H., Muzny D.M., Gibbs R.A. et al.. Exonuclease mutations in DNA polymerase epsilon reveal replication strand specific mutation patterns and human origins of replication. Genome Res. 2014; 24:1740–1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haradhvala N.J., Polak P., Stojanov P., Covington K.R., Shinbrot E., Hess J.M., Rheinbay E., Kim J., Maruvka Y.E., Braunstein L.Z. et al.. Mutational strand asymmetries in cancer genomes reveal mechanisms of DNA damage and repair. Cell. 2016; 164:538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garbacz M.A., Lujan S.A., Burkholder A.B., Cox P.B., Wu Q., Zhou Z.X., Haber J.E., Kunkel T.A.. Evidence that DNA polymerase delta contributes to initiating leading strand DNA replication in Saccharomyces cerevisiae. Nat. Commun. 2018; 9:858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nick McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M., Kunkel T.A.. Division of labor incorporation into DNA at the eukaryotic replication fork. Moll Cell. 2008; 30:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lujan S.A., Williams J.S., Pursell Z.F., Abdulovic-Cui A.A., Clark A.B., McElhinny S.A.N., Kunkel T.A.. Mismatch repair balances leading and lagging strand DNA replication fidelity. PLoS Genet. 2012; 8:e1003016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garbacz M., Araki H., Flis K., Bebenek A., Zawada A.E., Jonczyk P., Makiela-Dzbenska K., Fijalkowska I.J.. Fidelity consequences of the impaired interaction between DNA polymerase epsilon and the GINS complex. DNA Repair (Amst.). 2015; 29:23–35. [DOI] [PubMed] [Google Scholar]
- 26. Lujan S.A., Williams J.S., Clausen A.R., Clark A.B., Kunkel T.A.. Ribonucleotides are signals for mismatch repair of leading-strand replication errors. Mol. Cell. 2013; 50:437–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zbyněk Š. Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 1967; 62:626–633. [Google Scholar]
- 28. Watt D.L., Buckland R.J., Lujan S.A., Kunkel T.A., Chabes A.. Genome-wide analysis of the specificity and mechanisms of replication infidelity driven by imbalanced dNTP pools. Nucleic Acids Res. 2016; 44:1669–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sabouri N., Viberg J., Goyal D.K., Johansson E., Chabes A.. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 2008; 36:5660–5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nick McElhinny S.A., Stith C.M., Burgers P.M., Kunkel T.A.. Inefficient proofreading and biased error rates during inaccurate DNA synthesis by a mutant derivative of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 2007; 282:2324–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stone J.E., Lujan S.A., Kunkel T.A., Kunkel T.A.. DNA polymerase zeta generates clustered mutations during bypass of endogenous DNA lesions in Saccharomyces cerevisiae. Environ. Mol. Mutagen. 2012; 53:777–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Northam M.R., Garg P., Baitin D.M., Burgers P.M., Shcherbakova P.V.. A novel function of DNA polymerase zeta regulated by PCNA. EMBO J. 2006; 25:4316–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Northam M.R., Robinson H.A., Kochenova O.V., Shcherbakova P.V.. Participation of DNA polymerase zeta in replication of undamaged DNA in Saccharomyces cerevisiae. Genetics. 2010; 184:27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhong X., Garg P., Stith C.M., Nick McElhinny S.A., Kissling G.E., Burgers P.M., Kunkel T.A.. The fidelity of DNA synthesis by yeast DNA polymerase zeta alone and with accessory proteins. Nucleic Acids Res. 2006; 34:4731–4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kochenova O.V., Bezalel-Buch R., Tran P., Makarova A.V., Chabes A., Burgers P.M., Shcherbakova P.V.. Yeast DNA polymerase zeta maintains consistent activity and mutagenicity across a wide range of physiological dNTP concentrations. Nucleic Acids Res. 2017; 45:1200–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Roche H., Gietz R.D., Kunz B.A.. Specificity of the yeast Rev3-Delta antimutator and Rev3 dependency of the mutator resulting from a defect (Rad1-Delta) in nucleotide Excision-Repair. Genetics. 1994; 137:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kraszewska J., Garbacz M., Jonczyk P., Fijalkowska I.J., Jaszczur M.. Defect of Dpb2p, a noncatalytic subunit of DNA polymerase varepsilon, promotes error prone replication of undamaged chromosomal DNA in Saccharomyces cerevisiae. Mutat. Res. 2012; 737:34–42. [DOI] [PubMed] [Google Scholar]
- 38. Szwajczak E., Fijalkowska I.J., Suski C.. The CysB motif of Rev3p involved in the formation of the four-subunit DNA polymerase zeta is required for defective-replisome-induced mutagenesis. Mol. Microbiol. 2017; 106:659–672. [DOI] [PubMed] [Google Scholar]
- 39. Marsischky G.T., Filosi N., Kane M.F., Kolodner R.. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996; 10:407–420. [DOI] [PubMed] [Google Scholar]
- 40. Huang M.E., Rio A.G., Galibert M.D., Galibert F.. Pol32, a subunit of Saccharomyces cerevisiae DNA polymerase delta, suppresses genomic deletions and is involved in the mutagenic bypass pathway. Genetics. 2002; 160:1409–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Aksenova A., Volkov K., Maceluch J., Pursell Z.F., Rogozin I.B., Kunkel T.A., Pavlov Y.I., Johansson E.. Mismatch repair-independent increase in spontaneous mutagenesis in yeast lacking non-essential subunits of DNA polymerase epsilon. PLoS Genet. 2010; 6:e1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Flores-Rozas H., Kolodner R.D.. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:12404–12409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lehner K., Jinks-Robertson S.. The mismatch repair system promotes DNA polymerase zeta-dependent translesion synthesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:5749–5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chabes A., Georgieva B., Domkin V., Zhao X., Rothstein R., Thelander L.. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003; 112:391–401. [DOI] [PubMed] [Google Scholar]
- 45. Kumar D., Viberg J., Nilsson A.K., Chabes A.. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010; 38:3975–3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kumar D., Abdulovic A.L., Viberg J., Nilsson A.K., Kunkel T.A., Chabes A.. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011; 39:1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mertz T.M., Sharma S., Chabes A., Shcherbakova P.V.. Colon cancer-associated mutator DNA polymerase delta variant causes expansion of dNTP pools increasing its own infidelity. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2467–E2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bebenek K., Roberts J.D., Kunkel T.A.. The effects of dNTP pool imbalances on frameshift fidelity during DNA replication. J. Biol. Chem. 1992; 267:3589–3596. [PubMed] [Google Scholar]
- 49. Ohya T., Kawasaki Y., Hiraga S., Kanbara S., Nakajo K., Nakashima N., Suzuki A., Sugino A.. The DNA polymerase domain of pol(epsilon) is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J. Biol. Chem. 2002; 277:28099–28108. [DOI] [PubMed] [Google Scholar]
- 50. Kesti T., Flick K., Keranen S., Syvaoja J.E., Wittenberg C.. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell. 1999; 3:679–685. [DOI] [PubMed] [Google Scholar]
- 51. Davidson M.B., Katou Y., Keszthelyi A., Sing T.L., Xia T., Ou J., Vaisica J.A., Thevakumaran N., Marjavaara L., Myers C.L. et al.. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012; 31:895–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Williams J.S., Clausen A.R., Nick McElhinny S.A., Watts B.E., Johansson E., Kunkel T.A.. Proofreading of ribonucleotides inserted into DNA by yeast DNA polymerase epsilon. DNA Repair (Amst.). 2012; 11:649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chabes A., Stillman B.. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:1183–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kumar D., Abdulovic A.L., Viberg J., Nilsson A.K., Kunkel T.A., Chabes A.. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011; 39:1360–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Williams L.N., Marjavaara L., Knowels G.M., Schultz E.M., Fox E.J., Chabes A., Herr A.J.. dNTP pool levels modulate mutator phenotypes of error-prone DNA polymerase epsilon variants. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2457–E2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mertz T.M., Sharma S., Chabes A., Shcherbakova P.V.. Colon cancer-associated mutator DNA polymerase delta variant causes expansion of dNTP pools increasing its own infidelity. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2467–E2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pavlov Y.I., Frahm C., Nick McElhinny S.A., Niimi A., Suzuki M., Kunkel T.A.. Evidence that errors made by DNA polymerase alpha are corrected by DNA polymerase delta. Curr. Biol. 2006; 16:202–207. [DOI] [PubMed] [Google Scholar]
- 58. Flood C.L., Rodriguez G.P., Bao G., Shockley A.H., Kow Y.W., Crouse G.F.. Replicative DNA polymerase delta but not epsilon proofreads errors in Cis and in Trans. PLoS Genet. 2015; 11:e1005049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stone J.E., Kissling G.E., Lujan S.A., Rogozin I.B., Stith C.M., Burgers P.M., Kunkel T.A.. Low-fidelity DNA synthesis by the L979F mutator derivative of Saccharomyces cerevisiae DNA polymerase zeta. Nucleic Acids Res. 2009; 37:3774–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pavlov Y.I., Shcherbakova P.V., Kunkel T.A.. In vivo consequences of putative active site mutations in yeast DNA polymerases alpha, epsilon, delta, and zeta. Genetics. 2001; 159:47–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shcherbakova P.V., Fijalkowska I.J.. Translesion synthesis DNA polymerases and control of genome stability. Front. Biosci. 2006; 11:2496–2517. [DOI] [PubMed] [Google Scholar]
- 62. Zheng D.Q., Zhang K., Wu X.C., Mieczkowski P.A., Petes T.D.. Global analysis of genomic instability caused by DNA replication stress in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E8114–E8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sengupta S., van Deursen F., de Piccoli G., Labib K.. Dpb2 integrates the leading-strand DNA polymerase into the eukaryotic replisome. Curr. Biol. 2013; 23:543–552. [DOI] [PubMed] [Google Scholar]
- 64. Muramatsu S., Hirai K., Tak Y.S., Kamimura Y., Araki H.. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010; 24:602–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Georgescu R.E., Schauer G.D., Yao N.Y., Langston L.D., Yurieva O., Zhang D., Finkelstein J., O’Donnell M.E.. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife. 2015; 4:e04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Creighton S., Goodman M.F.. Gel kinetic analysis of DNA polymerase fidelity in the presence of proofreading using bacteriophage T4 DNA polymerase. J. Biol. Chem. 1995; 270:4759–4774. [DOI] [PubMed] [Google Scholar]
- 67. Beckman R.A., Loeb L.A.. Multi-stage proofreading in DNA replication. Q. Rev .Biophys. 1993; 26:225–331. [DOI] [PubMed] [Google Scholar]
- 68. Deem A., Keszthelyi A., Blackgrove T., Vayl A., Coffey B., Mathur R., Chabes A., Malkova A.. Break-induced replication is highly inaccurate. PLoS Biol. 2011; 9:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lydeard J.R., Jain S., Yamaguchi M., Haber J.E.. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007; 448:820–823. [DOI] [PubMed] [Google Scholar]
- 70. Miyabe I., Mizuno K., Keszthelyi A., Daigaku Y., Skouteri M., Mohebi S., Kunkel T.A., Murray J.M., Carr A.M.. Polymerase delta replicates both strands after homologous recombination-dependent fork restart. Nat. Struct. Mol. Biol. 2015; 22:932–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dua R., Levy D.L., Campbell J.L.. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol e and its unexpected ability to support growth in the absence of the DNA polymerase domain. J. Biol. Chem. 1999; 274:22283–22288. [DOI] [PubMed] [Google Scholar]
- 72. Goswami P., Abid Ali F., Douglas M.E., Locke J., Purkiss A., Janska A., Eickhoff P., Early A., Nans A., Cheung A.M.C. et al.. Structure of DNA-CMG-Pol epsilon elucidates the roles of the non-catalytic polymerase modules in the eukaryotic replisome. Nat .Commun. 2018; 9:5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tahirov T.H., Makarova K.S., Rogozin I.B., Pavlov Y.I., Koonin E.V.. Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol. Direct. 2009; 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sun J., Shi Y., Georgescu R.E., Yuan Z., Chait B.T., Li H., O’Donnell M.E.. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol. 2015; 22:976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yuan Z., Bai L., Sun J., Georgescu R., Liu J., O’Donnell M.E., Li H.. Structure of the eukaryotic replicative CMG helicase suggests a pumpjack motion for translocation. Nat. Struct. Mol. Biol. 2016; 23:217–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.