Abstract
Ataxia with oculomotor apraxia type 1 (AOA1) is an early onset progressive spinocerebellar ataxia caused by mutation in aprataxin (APTX). APTX removes 5′-AMP groups from DNA, a product of abortive ligation during DNA repair and replication. APTX deficiency has been suggested to compromise mitochondrial function; however, a detailed characterization of mitochondrial homeostasis in APTX-deficient cells is not available. Here, we show that cells lacking APTX undergo mitochondrial stress and display significant changes in the expression of the mitochondrial inner membrane fusion protein optic atrophy type 1, and components of the oxidative phosphorylation complexes. At the cellular level, APTX deficiency impairs mitochondrial morphology and network formation, and autophagic removal of damaged mitochondria by mitophagy. Thus, our results show that aberrant mitochondrial function is a key component of AOA1 pathology. This work corroborates the emerging evidence that impaired mitochondrial function is a characteristic of an increasing number of genetically diverse neurodegenerative disorders.
INTRODUCTION
Ligation of DNA ends is the final step in almost all DNA repair pathways and is a critical step during DNA replication. Human cells have three DNA ligases named DNA ligase I, III and IV, and all use adenosine triphosphate (ATP) as a cofactor (1). During ligation, DNA becomes temporarily adenylated at the 5′-end (5′-AMP-DNA) by DNA ligase (1). Occasionally, DNA ligase dissociates from DNA after 5′-adenylation of DNA resulting in a 5′-AMP group that must be removed for DNA to be ligated later. In human cells, aprataxin (APTX) is the only protein that removes 5′-AMP from DNA (2). Human patients carrying mutation in APTX develop the progressive neurodegenerative disease ataxia with oculomotor apraxia type 1 (AOA1) (3,4).
APTX localizes to the nucleus and mitochondria (5). Evidence suggests that AOA1 pathology is related to mitochondrial dysfunction. Biochemical and cell biological analysis showed that APTX is more critical in mitochondrial DNA (mtDNA) repair than in the nuclear DNA repair (5–7). AOA1 patients display ataxia, neuropathy, cerebellar atrophy and coenzyme Q deficiency, traits seen in mitochondrial diseases (8,9). A mitochondrial disease database has been developed as a diagnostic tool to identify mitochondrial pathology in human diseases (10), which has been proven useful in previous studies (11). This database predicts that AOA1 is a disease with significant mitochondrial involvement (6,10).
Mitochondria are called the powerhouse of the cells because of their central role in cellular ATP production. Mitochondria also play other important biological roles including amino acids and lipid metabolism, Ca2+ signaling, cell-cycle regulation and apoptosis (12). Muscle and brain tissues are particularly vulnerable to mitochondrial abnormalities, probably because of their high ATP consumption and reliance on other mitochondrial functions. Accordingly, mitochondrial dysfunction has been identified in a number of ataxias and other types of neurodegenerative diseases (11,13–16).
Mitochondria are structurally highly dynamic organelles and their morphology is determined by the type of their host cell. Mitochondria undergo division (fission) and merge together (fusion). The ratio of fusion and fission determines the formation of the filamentous tubular network or punctate mitochondria (17). The processes of fusion and fission involve a group of dynamin-like and GTPase proteins. The major players in fusion include the outer mitochondrial membrane proteins mitofusion 1 (MFN1) and mitofusin 2 (MFN2), and the inner mitochondrial membrane protein optic atrophy type 1 (OPA1). The key fission proteins are the cytosolic dynamin-related protein 1 (DRP1), and several mitochondrial outer membrane proteins; mitochondrial fission factor (MFF), mitochondrial fission 1 protein (Fis1) and mitochondrial dynamic proteins MiD49, and MiD51 (18,19). The function, recruitment and assembly of these proteins are largely regulated by post-translational modifications (20).
Mitochondrial morphology is integral to mitochondrial quality control through a selective autophagic removal of dysfunctional mitochondria known as mitophagy (18). The processes of fusion, fission and mitophagy are collectively known as mitochondrial dynamics. Increasing evidence has identified a close interplay between mitochondrial dynamics, mitochondrial bioenergetics, cellular metabolism status and energy demand (21,22). Adding to the importance of the mitochondrial homeostasis network, recent research has identified a novel link between persistent nuclear DNA damage, activation of poly ADP-ribose polymerases (PARPs) and nicotinamide adenine dinucleotide (NAD+) consumption and mitochondrial dysfunction (23). The disruption of this axis has been identified as a central cause in many neurodegenerative diseases (14,24).
Previous studies suggested that APTX deficiency associates with mitochondrial abnormalities including mitochondrial morphology and network (5–7). However, a detailed investigation of the mitochondrial status in APTX-deficient cells is not available. The aim of this project is to elucidate the molecular mechanisms of mitochondrial dysfunction in APTX deficient cells by analyzing key players in mitochondrial maintenance and function in CRISPR mediated APTX−/− U2OS cells and in AOA1 patient-derived cells. We found significant changes in key mitochondrial parameters including disruption of mitochondrial morphology, network, decreased mitochondrial membrane potential (MMP), increased mitochondria reactive oxygen species (ROS) and impaired mitophagy response. Our results suggest that mitochondrial dysfunction is a key feature of AOA1 pathology.
MATERIALS AND METHODS
Synthetic oligonucleotides were from TAG Copenhagen. [γ-32P]ATP was from Perkin Elmer. 5′- DNA adenylation kit was from BioNordika (E2610S). MitoTracker Red CMXRos (M-7512), Mitosox red (M36008) and tetramethylrhodamine (TMRM) (T-668) were from Thermo Fisher Scientific- Life Technology. Saponin was from Sigma (74036). N-acetyl-l-cysteine (NAC) was from Sigma.
Cell lines and preparation of whole cell protein extracts (WCE)
U2OS cells were cultured in Dulbecco's modified Eagle's medium (DMEM)-Glutamax (Gibco). C2ABR and C3ABR (APTX proficient) and L938 (P206L/P206L) and L939 (P206L/V263G) (APTX deficient) patient-derived Epstein-Barr virus-transformed lymphoblast cell lines (25) were grown in RPMI medium 1640- Glutamax (Gibco). Both DMEM and Roswell Park Memorial Institute (RPMI) medium1640 were supplemented with 10% Fetal Bovine Serum (FBS) and 1% penicillin-streptomycin.
For whole cell extract (WCE) preparation, pelleted cells were suspended in lysis buffer (20 mM, 4-(2-hydroxyethyl)-1-piperazineethanesulfonicacid ( HEPES)-KOH, pH 7.5, 200 mM KCl, 10% glycerol, 1% Triton X-100, 1% non-ionic detergent, IGEPAL CA-630 (octylphenoxypolyethoxyethanol), 1 mM ethylenedinitrilo tetraaceticacid (EDTA), 1 mM Dithiothreitol (DTT), EDTA-free Complete protease inhibitor cocktail (Sigma) and PhosphoSTOP (Sigma)), and left on ice for 60 min. Cell debris was pelleted at 15 000 g for 15 min, and the supernatant ( WCE) was collected.
Preparation of mitochondria-enriched extracts
Cells were collected at 500 g, washed once with phosphate buffered saline (PBS) and suspended in isotonic buffer (20 mM HEPES-KOH pH 7.4, 5 mM KCl, 1 mM DTT, protease inhibitor cocktail) and left on ice to swell. The cells were broken in a Dounce tight-fit homogenizer in ice and equal volume of 2× mannitol-sucrose-HEPES (MSH) buffer (420 mM mannitol, 140 mM sucrose, 20 mM HEPES-KOH pH 7.4, 4 mM EDTA, 2 mM EGTA, 5 mM DTT) was added to the homogenate and centrifuged at 1000 g for 5 min (twice). The supernatant was centrifuged at 10 000 g for 30 min and the pellet containing mitochondria were suspended in lysis buffer (20 mM HEPES-KOH, pH 7.5, 200 mM KCl, 10% glycerol, 1% Triton X-100, 1% IGEPAL, 1 mM EDTA, 1 mM DTT, EDTA-free Complete protease inhibitor cocktail and PhosphoSTOP) and left on ice for 60 min followed with a mild sonication. The lysates were centrifuged at 15 000 g and the supernatants were collected and used as mitochondrial enriched extracts.
APTX knockdown
APTX-specific TRC shRNA-pLKO vector (clone ID TRCN0000083642; Sigma) and a negative control scramble shRNA-pLKO.1 construct (Addgene) were described previously (5). The plasmid (1 μg) was co-transfected with the packaging plasmid (pCMV-dr8.2DVPR, Addgene, 0.7 μg) and envelope plasmid (pCMV-VSV-G, Addgene, 0.3 μg) into human embryonic kidney 293T cells using PolyJet transfection reagent (SignaGen Laboratories). Lentivirus containing media were collected 48 h later and filtered through a 0.45 μM filter to remove the cell debris and used to infect U2OS cells. Puromycin-resistant U2OS cell colonies were propagated and tested by western blot analysis for aprataxin (ab31841; abcam).
CRISPR mediated APTX knockout
U2OS cells were seeded on a 6-well dish and transfected with aprataxin double Nickase plasmid (sc-417083-NIC, Santa Cruz Biotechnology) using PolyJet transfection reagent (SignaGen Laboratories). Transfected cells were selected in medium with 2 μg/ml puromycin for one week, then harvested and reseeded on 150 mm dishes at 20 cells per dish. Single cell colonies were propagated. Disruption of APTX was verified by polymerase chainreaction (PCR) amplification of the flanking target region and western blot analysis.
mtDNA integrity analysis
MtDNA integrity was analyzed using a PCR-based method (26). The PCR reactions were carried out as described previously (6). PCR products were separated in agarose gel and the intensity of the amplicons was measured using Image J. The resulting values were then converted to relative lesion frequencies per 10 kb DNA by application of the Poisson distribution (lesions/amplicon = −ln (At/A0)), where At represents the amplification of APTX−/− cells and A0 is the amplification of control cells (27). Lesions per 10 kb DNA = (−ln (At/A0)) × 10 000 [bp]/size of long amplicon [bp].
RNA-seq and Q-PCR analysis of gene expression
Total RNA was purified from APTX-KO U2OS cells, APTX-KO cells stably expressing APTX (APTX-Pos), two AOA1 patient cell lines (L938 and L939) and the corresponding control cell lines (C2ABR and C3ABR), using RNeasy Mini Kit (Qiagen). There were four biological replicates for the U2OS, APTX-KO and APTX-Pos cell lines, and two replicates for the patient control and AOA1 cell lines. The cells were in culture for 2–3 weeks before RNA extraction. The samples were sent to the Beijing Genomics Institute Genomics Services for analysis. Briefly, the Agilent 2100 Bio analyzer (Agilent RNA 6000 Nano Kit) was used for RNA sample quality control, RNA concentration, RIN value, 28S/18S and fragment length distribution analysis. For library construction, polyA-containing messenger RNA (mRNA) molecules were isolated with magnetic beads. Following purification, the mRNA was fragmented into small pieces. The cleaved RNA fragments were copied into first strand complementary DNA (cDNA) using reverse transcriptase and random primers. This is followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. These cDNA fragments then have the addition of a single ‘A' base and subsequent ligation of the adapter. The products are then purified and enriched with PCR amplification. We then quantified the PCR yield by Qubit and pooled samples together to make a single stranded DNA circle (ssDNA circle), which gave the final library. DNA nanoballs (DNBs) were generated with the ssDNA circle by rolling circle replication to enlarge the fluorescent signals at the sequencing process. The DNBs were loaded into the patterned nanoarrays and pair-end reads of 100 bp were read through on the BGISEQ-500 platform for the following data analysis study. For this step, the BGISEQ-500 platform combines the DNA nanoball-based nanoarrays and stepwise sequencing using Combinational Probe-Anchor Synthesis Sequencing Method. Low-quality reads were filtered out (more than 20% of the bases qualities are lower than 10), reads with adaptors and reads with unknown bases (N bases more than 5%) to get the clean reads (SOAPnuke v1.5.2). Then the clean reads were mapped onto reference genome, followed by novel gene prediction, SNP & INDEL calling and gene-splicing detection. Clean reads were mapped to reference using Bowtie2 v2.2.5 (28), then gene expression levels were calculated with RSEM v1.2.12 (29). Finally, DEGs were detected using DEseq2, performed as described in Michael et al. (30), adjusted P-value ≤ 0.05 and log2 fold change ≥ 1 or ≤ −1. Differentially expressed genes (DEGs) were subjected to GO functional enrichment analysis using Enrichr (31). Patient and control cells were treated similarly, however since there were only two of each, for each gene, the reads were averaged then log2 fold change was calculated between patient and control samples. Genes with a probability of ≥ 0.75 and log2 fold change ≥ 1 or ≤ -1 were considered a DEG. The RNA-seq data has been deposited to the GEO database.
For real-time quantitative-PCR (Q-PCR) analysis, cDNA was prepared using Maxima Reverse Transcriptase and Oligo dT (12–18) (Thermo Fisher Scientific- Life tech). Q-PCR was carried out using a real-time PCR kit (Bio-Rad1725271) following the manufacturer’s protocol.
For analysis of alternative splicing of OPA1 mRNA, cDNA was prepared from the cells and used as template to PCR amplify exons three to nine using primers; forward, F1- 5′-GGATTGTGCCTGACATTGTG-3, and reverse, R1-5′-TCTGATACTTCAACTGAGTGTGC. PCR amplification of exons three to seven was carried out using primers; forward, F2-5′- GTGTGGGAAATTGATGAGTATATCG, and reverse R2- 5′-GCACTCTGATCTCCAACCAC. The PCR products were separated in 8% non-denaturing polyacrylamide gel (1 × Tris-Borate-EDTA (TBE) buffer, 37.5:1 acrylamide: bis).
FACS analysis for mitochondrial membrane potential and mitochondrial ROS
Fluorescence-activated cell sorting (FACS) analysis was performed at least three times independently with CytoFLEX flow cytometry platform (Beckman Coulter). All analyses were done in ice-cold sorting buffer (PBS, 0.1% bovine serum albumin (BSA)), and each experiment included ∼100 000 events per group. MMP and ROS levels were evaluated with TMRM and Mitosox Red, respectively. TMRM (40 nM) or Mitosox Red (5 uM) were added to the cells, and incubated for 15 min in 5% CO2, 37°C. U2OS cells were trypsinized and collected. After washing, cells were suspended in ice-cold sorting buffer for sorting. AOA1 cells were spun down to remove free dyes, suspended in sorting buffer and subjected to FACS. Both TMRM and Mitosox Red stained cells were analyzed at 561 nm lasers with 586 nm emission filters. All FACS data were analyzed using CytExpert Software (Beckman Coulter).
Live cell confocal microscopy
Cells were seeded on 30 mm diameter coverslips (0787, Thermo Scientific) placed in 6 cm culture dishes with an optimal density (200 000 cells per dish). After 24 h, the cells were stained with different reagents (100 nM Mitotracker red, 40 nM TMRM or Mitosox) for 15 min. After washing, coverslips were mounted on chambers for live imaging. Microscope images were acquired using LSM780 confocal system with a 63× oil immersion objective. Quantification of mitochondria parameters was carried out in Image J software as described previously (32).
Mitophagy assay
Mitophagy in live cells was monitored by Mitophagy detection kit (Dojindo Molecular Technologies). Urolithin A (UA, 5 μM) was used to trigger mitophagy as described previously (33). The level of mitophagy was defined by the area of Mtphagy dye per cell. At least 50 cells were qualified in each group. The level of colocalization of Mtphagy dye and lysosome dye was also analyzed. Quantification analysis was carried out using Image J.
Oxygen consumption rate (OCR)
Oxygen consumption was measured using the Seahorse XF24 instrument, according to the manufacturer’s instruction (Seahorse Biosciences, North Billerica, MA). Cells were seeded into a Seahorse tissue culture plate at a density of 50 000 cells per well in DMEM with 1 mM sodium pyruvate, 2 mM glutamine and 10% FBS. After 24 h, the medium was replaced with un-buffered XF assay medium (Seahorse Biosciences), pH 7.4, supplemented with 25 mM glucose, 1 mM sodium pyruvate and 2 mM glutamine. One hour later, the oxygen consumption rate (OCR) was measured in the Seahorse XF24 analyzer in four blocks of three 3-min periods. The first block measured the basal respiration rate. Next, 1 μM oligomycin was added to inhibit complex 5 and the second block was measured. Then, 0.3 μM carbonyl cyanide 4-trifluoromethoxy-phenylhydrazone (FCCP) was added to uncouple respiration, and the third block was measured. Finally, 2 μM antimycin A was added to inhibit complex 3, and the last measurements were acquired. The cells were counted after the experiment and the results were normalized to cell number in each well. Three biological experiments were done with technical replicates of 2–5 for each genotype.
Immunocytochemistry
Cells were seeded at 20 000 cells per well on 12 mm diameter coverslips in 24-well dishes. After 24 h, the cells were fixed in 4% paraformaldehyde in PBS for 10 min at room temperature and washed three times with PBS. Cells were quenched in 50 mM NH4Cl in PBS for 10 min and permeabilized and blocked at the same time in blocking buffer (3% bovine serum albumin (BSA), 0.1% saponin, PBS, pH 7.4) for 60 min. Primary antibodies (diluted in blocking buffer) were added and incubated at 4°C overnight. The samples were washed three times in 0.1% saponin in PBS and incubated with Alexa Fluor-conjugated secondary antibodies diluted in blocking buffer for 1 h at room temperature protected from light. For nuclei standing, 5 μg/ml 4,6-Diamidino-2-phenylindoledihydrochloride (DAPI) solution (D1306, Thermo Fischer Scientific) was applied to the samples for 5 min at room temperature following incubation with secondary antibody. Cells were washed three times with PBS and mounted on SuperFrost glass slides (3302775, Thermo Scientific) with mounting medium (S3023, Dako).
Transmission electron microscopy (TEM) analysis
Cells were grown to confluence on Thermanox coverslips and fixed with 2% v/v glutaraldehyde in 0.05 M sodium phosphate buffer (pH 7.2). The samples were rinsed three times in 0.15 M phosphate buffer (pH 7.2) and post-fixed in 1% w/v OsO4 in 0.12 M sodium phosphate buffer (pH 7.2) for 2 h. The samples were dehydrated in graded series of ethanol, transferred to propylene oxide and embedded in Epon according to standard procedures. Following polymerization, the Thermanox coverslip was peeled off. Sections, ∼60 nm thick, were cut with a Leica UC7 microtome (Leica Microsystems, Wienna, Austria) and collected on copper grids with Formvar supporting membranes, stained with uranyl acetate and lead citrate and subsequently examined with a Philips CM 100 Transmission EM (Philips, Eindhoven, The Netherlands), operated at an accelerating voltage of 80 kV and equipped. Digital images were recorded with an OSIS Veleta digital slow scan 2k × 2k CCD camera and the ITEM software package. Quantification of cristae length was done using Image J. Twenty cells were quantified in each group.
ATP measurement
Cells were plated in 6-well dishes at 6 × 105 cells per dish. Intracellular ATP was measured using a luciferase-based assay (ATPlite Luminescence Assay Kit, PerkinElmer) following manufacturer’s guidelines. A standard curve was generated and used to calculate samples ATP concentration. Protein concentration was determined using Bradford protein assay reagents (Bio-Rad). The content of ATP was normalized for protein content and presented as percentage of control.
Expression of APTX in the APTX−/− U2OS cells
Total RNA was purified from U2OS cells using RNeasy Kit (Qiagen). cDNA was prepared using SuperScript III RT (Invitrogen) and used as a template to PCR amplifying the APTX isoform variant 6 containing a putative mitochondrial localization signal (5) (Supplementary Figure S1). Primers used in PCR were; forward 5′- AACTAGATCTATGAGTAACGTGAATTTGTCCGTCTCC, and reverse 5′- AATC GGATCCTCACTGTGTCCAGTGCTTCCTGAG. The PCR products were digested with BglII and BamHI restriction enzymes and cloned into pAcGFP1-Hyg-N1 vector (Clontech). APTX−/− cells were transfected with the construct using PolyJet transfection reagent (SignaGen Laboratories) and stable APTX-GFP expressing cells were selected in medium containing hygromycine and enriched in FACS.
DNA substrates
Duplex 22-mer DNA containing a 5′-AMP was prepared as follows: oligonucleotide 5′-GATCCTCTAGAGTCGACCTGCA-3′ was first end-labeled at the 5′-end by [γ-32P]ATP and T4 polynucleotide kinase, and then adenylated to generate an oligonucleotide with 5′-AMP-32p-DNA. Or 3′-end fluorophore TAMRA labeled oligo was 5′-end phosphorylated followed with 5′-adenylation of DNA to generate 5′-AMP-DNA—3′-TAMRA oligo. The adenylated oligo was annealed to a complementary oligo at 1:1.25 ratio in 20 mM HEPES-KOH, pH 7.5 and 100 mM NaCl, heated at 90°C for 5 min and slowly cooled down to room temperature to prepare 5′-AMP-containing duplex DNA.
5′-AMP-DNA repair assay
Repair reactions were carried out using 15 μg WCEs in 40 mM HEPES-KOH, pH 7.8, 1 mM DTT, 0.36 mg/ml BSA, 100 mM KCl, 2 mM EDTA and 5 pmol DNA substrate at 30°C for the indicated times. The reactions were stopped by adding loading buffer (10 mM EDTA, 90% formamide, xylene cyanol, bromphenol blue) and heating the samples at 85°C for 5 min. DNA was separated in 20% denaturing acrylamide gel (19:1 acrylamide:bis, 1 × TBE and 7.5 M urea).
Western blot analysis
WCEs were separated in Tris-glycine sodium dodecyl sulphate (SDS) gels and transferred onto Polyvinylidenedifluoride (PVDF) membrane. Each experiment was done three to five times. The images are shown as three technical replicates of one biological replicate. The primary antibodies used were: APTX (sc-374108), Fis1 (sc-376469), proliferating cell nuclear antigen (PCNA) (sc-56), MFN1 (sc-166644), MFN2 (sc-515647) from Santa Cruz. OPA1 (67589S), superoxide dismutase 2 (SOD2) (13194S), AMP-activated protein kinase (AMPK) (2535S), Phospho-AMPK (Thr172), DRP1 (8570S), Phospho-DRP1 (Ser616) (3455S), anti-acetyl lysine (9681S) and Sirtuin (SIRT)1 (8469) were from CellSignaling. TOMM20 (WH0009804M1), and actin (A5441) were from Sigma-Aldrich. SIRT3 (10099-1-AP), and PARP1 (LS-B3432) were from Nordic Biosite. VDAC1 (ab14734) and APTX (ab31841) were from Abcam, PGC-1α (NBP1-04676) and light chain 3 (LC3) (NB600-1384) were from Novus Biologicals, TFAM (H00007019-B01P) was from Abnova.
To detect oxidative phosphorylation (OXPHOS) complex assembly, we used an assembly-dependent total OXPHOS rodent antibody cocktail (ab110413, Abcam). The antibodies in the cocktail are against a subunit that is labile when its complex is not assembled. The samples were prepared following the manufacturer’s protocol.
Colony forming assay
Cells were seeded on 6-well dishes at 200 cells per well. Next day, the cells were treated with 0.8 mM methyl methanesulfonate (MMS) or 0.020 mM menadione in serum-free medium for 60 min. The drugs were removed, and cells were gently washed once with PBS and normal medium was added to each well. Cells were left to recover for 10 days. Cells were stained in 0.5% crystal violet, dissolved in 20% ethanol for 1 h and washed in water, and colonies were counted.
Caenorhabditis elegans strains and methods
Caenorhabditis elegans eat-3 (ad426) and N2 control strains were obtained from Caenorhabditis Genetics Center (CGC, University of Minnesota). Strains were maintained at 20°C. Nematode Growth medium (NGM) agar plates seeded with OPA50 bacteria. Synchronous worm cultures were obtained by allowing gravid adults lay eggs onto NGM/OP50 plates for 2–4 h. Eggs were then collected and seeded onto NGM agar plates.
For mitochondrial network imaging, Day 1 (D1) worms were incubated for 24 h at 20°C on NGM OP50 plates containing 1 μM tetramethylrhodamine methyl ester (TMRM). Worms were subsequently transferred to fresh NGM plates containing only OP50 for 1 h to clear their intestinal tract of residual dye, after which they were transferred to cover slides and paralyzed. Photographs were immediately taken with LSM780 (Zeiss) confocal microscope.
For western blot analysis, D1 worms were collected and lysed in RIPA buffer (Life technologies) with complete proteases cocktail (Roche). Worms were disrupted by sonication using a Branson sonifier (Thomas Scientific) with the following settings: 10% amplitude; 10 s on; 30 s off, total on time 5 min. Samples were kept on ice during sonication to avoid excessive heating. Afterward, the debris was cleared by centrifugation at 12 000 × g for 10 min at 4°C. The supernatant was resolved in laemmli sample buffer and used for western blot (WB).
ATP was measured with ATPlite Luminescence Assay (Perkin Elmer). D1 worms were collected and washed three times with M9 buffer. Worms were pelted and resuspended in cell lysis solution from the ATP assay kit. Assays were performed according to the manufacturer’s protocol.
Chemotaxis to isoamyl alcohol was performed at 20°C, on 9 cm agar plates as described earlier (34). The chemotaxis index was calculated by subtracting the number of animals found at the trap from the number of animals at the source of the chemical, divided by the total number of animals entered into the assay (34). The resulting values were expressed and graphed as percentiles. About 200 adult animals for each strain were assayed in each experiment.
For swimming movement, D1 worms were randomly selected and transferred to a 6 cm petri dish containing 1 ml of M9 buffer. The worms were allowed to acclimate for ∼10 s, and then movements were scored for 1 min. Thirty worms were scored in each group.
Statistical analysis
Error bars represent SE or range (for experiments with less than three replicates) as indicated in the figure legends. Data were processed in Excel and statistical analyses were performed using GraphPad Prism 7 (GraphPad Software). Statistical analysis of differences between two groups was performed using a two-tailed, unpaired t-test and between more than two groups using a one-way ANOVA analysis of variance test followed by a Tukey’s post-hoc comparison; two-way ANOVA was used for comparison between control and AOA1 patient cells; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
RESULTS
CRISPR-mediated APTX−/− U2OS cells display hallmarks of mitochondrial dysfunction
We elected U2OS cells for this work for several reasons. U2OS cells have active and functional mitochondria and rely on OXPHOS for ATP production (35). They grow in monolayer, are amenable to transfection and have an overall structure that makes them particularly suitable for mitochondrial morphology and network analysis (35). We used a lentiviral delivery system and knocked down (KD) APTX in U2OS cells. Western blot analysis showed 90–95% reduction in APTX concentration in the APTX-KD cells compared with control cells (Figure 1A, KD 20 days), in line with a previous report (5). Surprisingly, extracts from APTX-KD cells displayed a robust 5′-AMP removal activity (Figure 1B), indicating that even at a very low concentration, APTX efficiently repairs 5′-adenylated DNA. This demonstrates that in studies where APTX is not completely depleted, it may mask biological consequences of APTX deficiency. Thus, we completely depleted APTX in the cells using CRISPR technology (APTX-KO). Western blot analysis of protein extracts from APTX-KO and lymphoblastoid cells derived from AOA1 patients completely lacked APTX (Figure 1C). DNA repair analysis of the extracts showed absence of 5′-AMP-DNA removal activity in APTX-KO and patient cell extracts indicating that APTX is the sole enzyme for removal of 5′-adenylated DNA in human cells, and also confirms the specificity of our DNA substrate (Figure 1D).
Figure 1.
Knockdown and knockout of APTX in U2OS cells. (A) Western blot analysis to determine APTX knockdown (KD) efficiency in U2OS cells using a lentiviral-delivered shRNA system. (B) Repair analysis of 5′-AMP DNA in extracts from APTX KD cells and control cells. (C) Western blot analysis showing complete absence of APTX in CRISPR-mediated APTX knockout (KO) U2OS cells (lane 2) and in cells prepared from AOA1 patients. (D) APTX deficient cells are devoid of 5′-AMP removal activity confirming the specificity of our DNA substrate for APTX activity.
The MMP is generated by OXPHOS through the electron transport chain (ETC). MMP measurement has frequently been used to evaluate mitochondrial health and function. Mitochondrial ROS production is highly regulated by MMP and mitochondrial dysfunction is often linked to increased ROS production (36). APTX-knock out cells (APTX−/−) showed significantly lower MMP (Figure 2A and C confocal microscopy image), and higher mitochondrial ROS production (Figure 2B and C) compared with the control cells. Superoxide dismutases (SODs) are cellular antioxidants that catalyze the conversion of superoxide anions to oxygen and hydrogen peroxide. Mitochondrial SOD2 is a primary defense enzyme against mitochondrial superoxide. The SOD2 level was markedly higher in APTX−/− cells compared with control cells (Figure 2D), probably in response to increased mitochondrial ROS production. Mitochondrial SIRT3 is a NAD+-dependent lysine deacetylase that regulates the function of mitochondrial proteins (37), including SOD2 (38). APTX−/− cells displayed a significantly higher level of SIRT3 compared with the control cells (Figure 2D). Thus, impaired MMP and enhanced ROS production associate with APTX deficiency.
Figure 2.
Knockout of APTX causes mitochondrial dysfunction in U2OS cells. (A) FACS analysis and quantifications of MMP using TMRM. Bars display TMRM fluorescence intensity as mean ± S.E.M., N = 3 (* P < 0.05, *** P < 0.001). (B) FACS analysis and quantifications of mitochondrial ROS levels using MitoSOX, Bars display MitoSOX fluorescence intensity as mean ± S.E.M., N = 3 (*P < 0.05). (C) Confocal images showing TMRM and MitoSOX staining in live APTX−/- and control U2OS cells. The scale bar represents 10 μm. (D) A representative western blot image of SOD2 and SIRT3 in the indicated WCEs. Graphs on the right show quantifications of SOD2 and SIRT3 protein levels normalized to actin. Values are mean ± S.E.M., N = 3 (**P < 0.01 and ***P < 0.001).
Impaired mitochondrial network and cristae structure in APTX−/− cells
Previously, knockdown of APTX in human neuroblastoma SH-SY5Y cells was suggested to disrupt the mitochondrial network (5), which may be related to the changes in the level of mitochondrial fission and fusion proteins in APTX−/− cells observed here. However, the molecular mechanism of mitochondrial network impairment and the possibility of mitochondrial fission and fusion links have not been investigated. Thus, we investigated the status of the mitochondrial network and morphology in these cells. Live cell confocal microscopy analysis showed elongated and highly branched tubular mitochondria in control cells (Figure 3A, Control). APTX−/− cells, however, contained more punctate and less connected mitochondria, which mostly clustered in perinuclear regions (Figure 3A, APTX−/−). APTX−/− cells had more mitochondria but they were smaller and shorter than in control cells (Figure 3A andgraphs). Immunocytochemistry analysis of fixed cells using an antibody targeting the translocase of the outer mitochondrial membrane, TOMM20, also showed punctate staining of mitochondria in APTX−/− cells (Figure 3B). Furthermore, transmission electron microscopy (TEM) confirmed the confocal microscopy results showing more abundant, but smaller and fragmented mitochondria in APTX−/− cells (Figure 3C). Notably, altered mitochondrial morphology did not significantly change mitochondrial content in APTX−/− cells, as judged by western blot analysis of the outer mitochondrial membrane proteins VDAC1 and TOMM20 (Figure 3D). Collectively, these results show that APTX deficiency may alter mitochondrial morphology without significant effect on the overall mitochondrial content.
Figure 3.
Knockout of APTX alters mitochondrial morphology and network. (A) Representative images of live APTX−/− and control cells pre-incubated with MitoTracker Red showing mitochondrial network formation; scale bar represents 10 μm. Graphs on the right show quantifications of mitochondrial morphology (number, length, size and shape) from more than 200 cells, values are mean ± S.E.M. (**P < 0.01, ****P < 0.0001). (B) Immunocytochemistry analysis of fixed cells against the outer mitochondrial membrane protein TOMM20. Nuclei are visualized by DAPI staining; scale bar represents 10 μm. (C) TEM images showing detailed mitochondrial morphology and cristae structures; scale bar represents 1 μm. The graph shows the quantification analysis of cristae length using Image J. Twenty cells were quantified in each group (*** P < 0.001). (D) Western blot analysis of mitochondrial outer membrane proteins VDAC1 and TOMM20 to determine relative mitochondrial abundance in APTX−/− and control cells.
The mitochondrial inner membrane folds into the matrix and forms distinct structures called cristae. Emerging data suggests that the morphology and shape of cristae modulate the organization and function of OXPHOS complexes and thus directly influences cell metabolism (39). Electron microscopy (EM) data showed that control cells contained long and fine cristae stretched out from the membrane. APTX−/− cells, however, contained shorter and reduced cristae density (Figure 3C and the graph), indicating a marked alteration in cristae shape and density in APTX−/− cells.
Diminished OPA1 expression in APTX−/− cells
Mitochondria are highly dynamic organelles. Size and morphology of mitochondria are determined by the rate of mitochondrial fission and fusion (40). Emerging findings are unraveling an intricate connection between mitochondrial morphology and network organization and a number of key cellular processes including the clearance of dysfunctional mitochondria by mitophagy, mtDNA maintenance and integrity, mitochondrial stress signaling, and cellular energy demand and metabolism (22,39–45).
Western blot analysis of some commonly accepted key regulators of mitochondrial fission and fusion showed that the concentration of OPA1, a central regulator of mitochondrial inner membrane fusion and a key protein in mitochondrial dynamics and cristae structure formation (18,46,47), and also of MFN1, another regulator of mitochondrial fusion, were considerably lower in APTX−/− cells (Figure 4A and the graphs). Phosphorylation of DRP1 is thought to regulate the translocation of DRP1 from cytosol to the outer mitochondrial membrane, a key step in the initiation of fission. The concentration of DRP1 but not the phosphorylated DRP1 (p-DRP1) was lower in APTX−/−cells (Figure 4A). These results show that APTX deficiency may affect the expression and the stability of some mitochondrial fusion and fission proteins in U2OS cells. Q-PCR analysis showed significantly lower OPA1 mRNA expression in APTX−/− cells (Figure 4B).
Figure 4.
APTX depletion decreases OPA1 protein and mRNA levels in U2OS cells. (A) A representative western blot image for key mitochondrial morphology proteins. Graphs on the right show quantifications of different protein levels normalized to actin. Both OPA1 bands were included in the quantification analysis. Values are mean ± S.E.M., N = 3 (*P < 0.05, **P < 0.01,****P < 0.0001). (B) Q-PCR analysis of OPA1 gene expression. OPA1 gene expression was normalized to the expression of the house-keeping gene actin. Bars show relative OPA1 mRNA level as mean ± S.E.M., N = 3 (****P < 0.0001).
Defective mitophagy response in APTX−/− cells
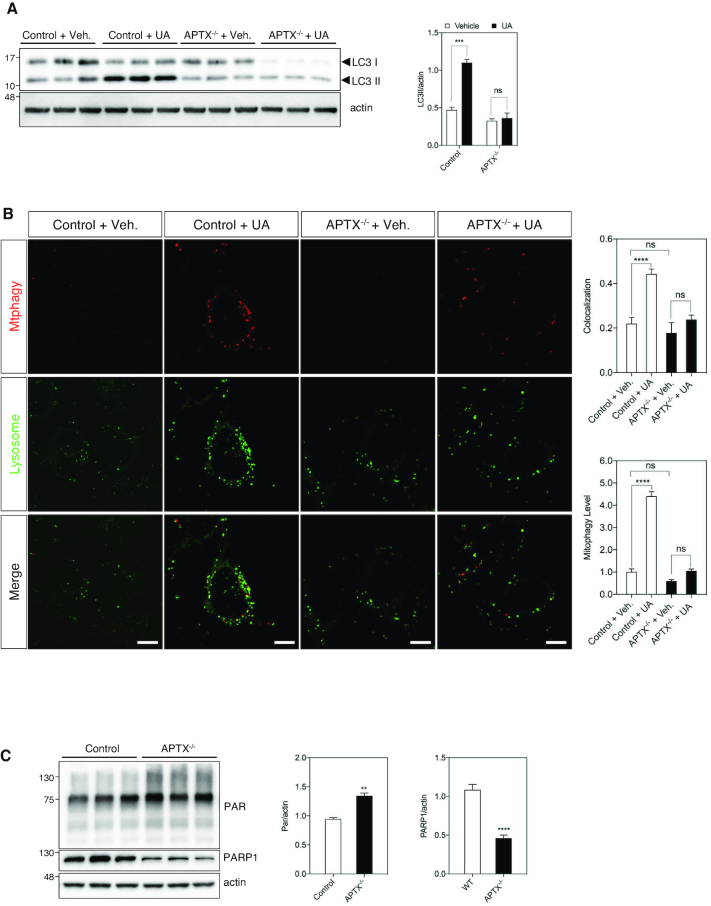
Autophagy is a process whereby intracellular components are engulfed with membrane bound autophagic vesicles (autophagosome), then fuse with lysosomes, and the contents of the cargo is degraded (48). Western blot analysis of microtubule-associated protein 1 LC3 is frequently used as a marker to measure autophagic flux. LC3-I is conjugated to phosphatidylethanolamine to form LC3-II, which is localized to autophagosomes (49). There were no detectable differences in the levels of LC3-I, LC3-II between the control and APTX−/− cells (Figure 5A, control + vehicle and APTX−/− + vehicle, respectively), suggesting comparable basal autophagic activity in these cells. The selective autophagic elimination of damaged mitochondria is called mitophagy (48). Urolithin A (UA) is a metabolite of natural compounds known as ellagitannins (33), and it was recently shown to induce mitophagy (33). Following UA treatment, the level of LC3-II in control cells, but not in APTX−/− cells increased significantly. This suggests robust autophagosome formation in response to UA treatment in control cells but not in APTX−/− cells. It could also reflect slower autophagosome-lysosome fusion rate, or impaired lysosomal degradation of the cargo in control cells compared with APTX−/− cells (Figure 5A). To clarify these alternatives and to specifically measure mitophagy, we used a commercially available dye kit. The dye (Mtphagy), accumulates in mitochondria following the induction of mitophagy, the damaged mitochondria then fuse to lysosomes resulting in a higher fluorescence signal. Pre-treatment of the cells with UA induced a strong mitophagy signal in control cells that colocalized with the lysosomal signal (Figure 5B, control + UA, merged, yellow spots and the graphs), suggesting a robust mitophagy response in control cells. The intensity of the dye signal was considerably less in APTX−/− cells than control cells following UA treatment (Figure 5B, APTX−/− + UA, and the graphs). Collectively, these results suggest that the ability to induce mitophagy by UA is compromised in APTX−/− cells.
Figure 5.
Mitophagy induction is compromised in APTX deficient cells. (A) Western blot analysis of autophagy marker LC3 in different cells. The ratio of LC3II (lower band) to actin was calculated to demonstrate autophagic response to urolithin (UA). Data are shown as mean ± S.E.M., N = 3, (***P < 0.001). (B) Assessment of mitophagy in UA treated cells. The merged images show colocalization between the mitophagy dye and the lysosome dye. The upper graph on the right shows the quantification analysis of colocalization of lysosome and mitopahgy dyes. The lower graph shows the quantification of mitophagy signal. Data is presented as mean ± S.E.M., (****P < 0.0001). scale bar represents 10 μm. (C) Western blot analysis of PARylation (PAR) and PARP1 levels in different WCEs as indicated. Quantifications are from three independent experiments. Data are shown as mean ± S.E.M., (**P < 0.01, ****P < 0.0001).
APTX−/− cells show elevated PARylation but unchanged NAD+ content
DNA damage response (DDR) proteins poly (ADP-ribose) polymerases 1 and 2 (PARP1/2) signal the location of DNA damage on the genome to DNA repair proteins by adding ADP-ribosepolymers (PAR) to themselves and to nearby proteins consuming NAD+ in the process (50). PARP1 is responsible for ∼90–95% of the total cellular PARP activity (51). The overall level of PAR was higher in APTX−/− cells (Figure 5C and the graph). Surprisingly, the level of PARP1 was markedly lower in APTX−/− cells (Figure 5C and the graph). The elevated level of PARylation, however, did not seem to be associated with detectable effect on the cellular NAD+ content (Supplementary Figure S2), suggesting that the NAD+-dependent reactions were not significantly affected in APTX−/− cells.
Re-introduction of APTX into the APTX−/− U2OS cells reverses some phenotypes back to the APTX positive control cells
To further confirm the role of APTX in the observed changes in mitochondrial parameters, we tested whether re-introduction of APTX into APTX−/− cells could rescue key features that were significantly altered by APTX depletion. APTX cDNA was prepared from U2OS cells and used as template to construct an APTX-GFP expressing vector. APTX−/− cells were transfected with the construct and stable APTX expressing cells were selected and propagated (hereafter, referred to APTX-positive). By RNAseq analysis, APTX is expressed about eight times more than in control cells. Approximately 10% of the cells showed a mitochondrial APTX-GFP signal as estimated by confocal microscopy inspection of the cells (Supplementary Figure S3). Expression of APTX restored 5′-AMP removal activity in APTX−/− cells, indicating that the cells were expressing a catalytically functional protein (Figure 6A).
Figure 6.
Expression of human APTX variant 6 containing an N-terminal putative mitochondrial localization signal in APTX−/− cells. (A) Repair analysis of 5′-AMP DNA in the indicated WCEs. Lower band corresponds to repaired DNA substrate. (B) WB analysis of SOD2 and APTX in the indicated cell lines. (C) Measurement of ATP level in the indicated cells. Values are mean ± S.E.M., N = 3, (***P < 0.001). (D) A representative western blot image of AMPK and activated AMPK (pAMPK). Quantifications are shown as mean ± S.E.M., N = 3, (**P < 0.01). (E) Western blot image of the subunits of OXPHOS complexes I-V. Quantifications are shown as mean ± S.E.M., N = 3, (***P < 0.001). (F) Seahorse measurements of OCR in APTX−/−, APTX-Pos and control cells. Statistical significant was calculated using paired-t-test, N = 3 (*P < 0.05). (G) PARylation (PAR) analysis by western blot. Quantifications are shown as mean ± S.E.M., N = 3, (***P < 0.001). (H) PCR-based mtDNA damage analysis. Quantifications are shown as mean ± S.E.M., N = 3, (**P < 0.01). (I) Colony forming analysis to determine the ability of the cells to recover following treatment with the genotoxic drugs MMS and menadione. Quantifications are shown as mean ± S.E.M., N = 3, (***P < 0.001).
Western blot analysis of APTX-positive extracts identified two major bands and a few weaker bands that migrated faster in the gel (Figure 6B). These may be truncated APTX-GFP protein, or the product of alternative transcription or translation of the APTX-GFP construct. The upper band corresponds to the long isoform of APTX-GFP. The lower bands may represent truncated APTX-GFP or the product of alternative transcription or translation of the cloned APTX. The level of APTX-GFP was ∼5.5-times higher than the level of APTX in control cells.
SOD2 was highly abundant in APTX−/− cells (Figures 2D and 6B). The expression of APTX restored it to the level of control cells (Figure 6B, APTX-Pos). Mitochondria are the major site of ATP production and the level of cellular ATP may reflect the status of mitochondrial function. ATP levels were significantly lower in APTX−/− cells (Figure 6C). Expression of APTX restored ATP to the level of control cells (Figure 6C). The lower ATP levels in APTX−/− cells may be caused by a higher rate of ATP consumption, or low MMP (Figure 2A) that can result in suboptimal mitochondrial ATP production.
The serine/threonine AMPK complex, is a key sensor of cellular energy status by ADP- and AMP:ATP ratio and possibly also of glucose availability (52). In general, activated AMPK phosphorylated at Thr172 (pAMPK) increases catabolic processes while decreasing anabolic reactions through the phosphorylation of key proteins in several pathways including mTOR, glycolysis and mitochondrial homeostasis. Activated AMPK promotes mitophagy through phosphorylation of ULK1 (53). The level of pAMPK but not of total AMPK was considerably lower in APTX−/− cells. Expression of APTX increased the level of pAMPK in APTX−/− cells nearly to the level of control cells that was statistically significant (Figure 6D and the graphs). Low level of activated AMPK may in part account for the delayed mitophagy response in APTX−/− cells (Figure 5B). 5-aminoimidazole-4-carboxamide ribonucleoside (AICAR) becomes converted in the cells to an AMP analog and has been used as an AMPK activating compound (54). APTX−/− cells showed ∼40% lower level of activated AMPK (p-AMPK) compared to control cells following AICAR treatment suggesting a somewhat less efficient AMPK response to increased AMP:ATP ratio (Supplementary Figure S4). This may partially explain low basal level of activated AMPK in APTX−/− cells (Figure 6D) despite lower cellular ATP content (Figure 6C).
The OXPHOS system consists of five multipeptide complexes (CI-CV) composed of over 90 different structural proteins, which are encoded by nuclear and mtDNA. They require assembly protein factors for proper assembly and function. The stability of the complexes is interdependent (55,56). Diminished MMP and increased mitochondrial ROS production may be caused by OXPHOS system dysfunctions and changes in the protein levels of OXPHOS subunits may reflect altered ETC function. We examined the abundance of selected OXPHOS subunits using an assembly specific antibody cocktail. We found that the mitochondrial OXPHOS complexes I, II, III and IV were more abundant in APTX−/− cells compared with control cells (Figure 6E). Expression of APTX corrected them back to the level of control cells (Figure 6E and the graphs).
The effects of APTX deficiency on mitochondrial respiration was tested by measuring OCR, using the Seahorse XF24 analyzer. APTX−/− and APTX-Pos cells showed lower ATP-linked respiration and higher leak relative to control cells (Figure 6F and the graph). In contrast, APTX-Pos cells showed a higher reserve capacity (Figure 6F and the graph). Together, these measurements show that the presence or absence of APTX can alter mitochondrial respiration which together with the low MMP (Figure 2A), and high ROS (Figure 2B) in APTX−/− cells suggests mitochondrial dysfunction.
The level of PARylation was higher in APTX−/− cell extracts compared with control cells (Figures 6G and 5C, PAR). APTX expression corrected it back to the level of control cells (Figure 6G and the graph).
MtDNA in APTX-deficient cells is more susceptible to damage than nuclear DNA (5,6). Using a PCR-base method, we detected more damage in mtDNA from APTX−/− cells than in mtDNA from control cells (Figure 6H). Expression of APTX in APTX−/− cells corrected it back to the level of control cells (Figure 6H). These results support a role for APTX in mtDNA repair and integrity, in line with and extension of previous reports (5–7).
The number of mtDNA molecules, also known as mtDNA copy number, varies significantly depending on the cell type and is thought to correlate with the cellular need for energy and ATP production. Previously, lentiviral-mediated knockdown of APTX resulted in reduced mtDNA copy number (5). However, we did not find any differences in mtDNA copy number between APTX−/− and AOA1 patient cells and their corresponding control cells (Supplementary Figure S5). These differences might be related to the cell type differences used in these studies; here, we used U2OS cells and AOA1 patient-derived lymphoblast cell lines, whereas in the previous study (5), SH-SY5Y and primary human skeletal muscle cells were used.
The colony forming assay is used for monitoring the ability of cells to recover from genotoxic treatment. Next, we determined the recovery capacity of the cells following treatment with the genotoxic compounds MMS and menadione. MMS modifies guanine to 7-methylguanine, and adenine to 3-methyladenine, which can result in base mispairing and replication blocks, respectively (57). Menadione exposure results in the accumulation of ROS in mitochondria that attack and damage macromolecules including mtDNA (58). Menadione and MMS treatments showed that APTX−/− cells were more sensitive to these agents than control cells (Figure 6I). APTX re-expression reduced the sensitivity of APTX−/− cells to these genotoxic agents back to the level of control cells (Figure 6I).
To summarize, putting back APTX into APTX-KO cells corrected the protein levels of SOD2, pAMPK, OXPHOS complexes I, II, III and IV back to the level of control cells. It also corrected cellular ATP levels, DDR via parylation signal, mtDNA integrity and sensitivity to MMS and menadione treatment back to the level of control cells, and improved mitochondrial respiration reserve capacity, altogether suggesting the role of APTX in the observed changes in APTX−/− cells. However, the expression of OPA1, SIRT3 and PARP1 were not corrected by re-introducing APTX into the APTX−/− cells (Figure 9; Supplementary Figure S6 and Supplementary Tables S1–S3), even though, the levels of these proteins were consistently altered in APTX−/− cells and in AOA1 patient-derived cells (Figures 2D, 4, 5C, 7C and E, 8A and B, 9; Supplementary Tables S1–S4).
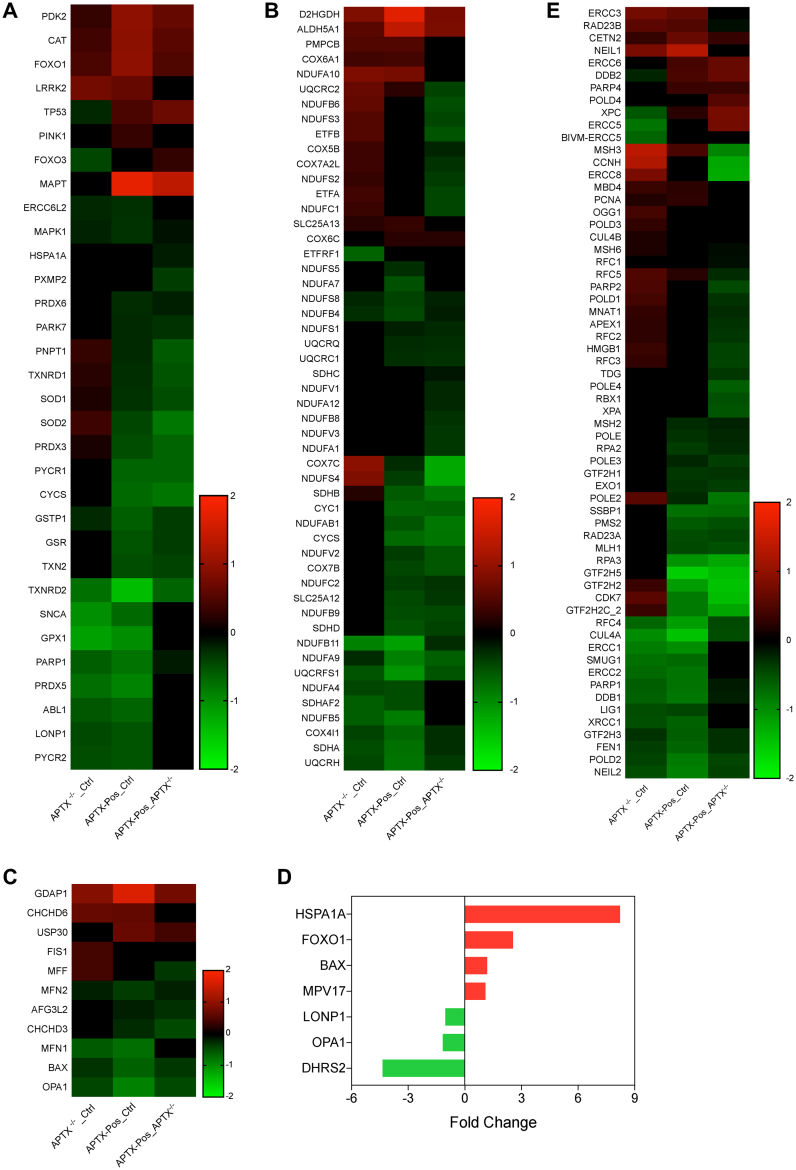
Figure 9.
RNA Seq analysis. After comparison of the shared GO terms from GO cellular compartment, three terms were of interest GO:0034599 cellular response to oxidative stress, GO:0022904 respiratory ETC and GO:0008053 mitochondrial fusion. The genes in those three terms are shown in (A–C), respectively. (D) Panel shows genes from the cellular response to oxidative stress (GO:0034599) and mitochondrial fusion (GO:0008053) from AOA1 patient cells versus controls. No mitochondrial ETC genes were found in the AOA1 patient-derived lymphoblastoid cell lines. (E) Genes from the DNA repair pathways base excision, mismatch and NER were also collected and displayed. Note: a few genes were obtained from various lists, were expressed wildly differently and were removed. From the stress response gene list, the gene DHRS2 (KO_Ctrl -11.1; Pos_Ctrl -9, Pos_KO 2.2), from the BER list, APTX (KO_Ctrl -0.52; Pos_Ctrl 2.6; Pos_KO 3.1) and from the NER gene list, the genes GTF2H2C (KO_Ctrl 0.97; Pos_Ctrl -6.9; Pos_KO -7.1) were each removed from the heat maps. All heat maps were centered on zero. Heat maps were made using GraphPad Prism 7.
Figure 7.
AOA1 patient cell lines also display mitochondrial dysfunctions. (A) FACS analysis and quantifications of MMP using TMRM. Bars display TMRM fluorescence intensity as mean ± S.E.M., N = 3 (*P < 0.05). (B) FACS analysis and quantifications of mitochondrial ROS level using MitoSOX. Bars display MitoSOX fluorescence intensity as mean ± S.E.M., N = 3 (*P < 0.05, **P < 0.01). (C) A representative western blot image of SOD2 and SIRT3 in the indicated WCEs. Graphs show the quantifications of APTX, SOD2, SIRT3, VDAC1 and PGC1α protein levels normalized to PCNA. Values are mean ± S.E.M., N = 3 (*P < 0.05, **P < 0.01). (D) Western blot analysis of protein acetylation in mitochondria enriched extracts from control and AOA1 cells. (E) Western blot analysis of PARylation (Par) and PARP1 levels in different WCEs as indicated, quantifications are from three independent experiments. Data are shown as mean ± S.E.M., (**P < 0.01, ***P < 0.001). (F) Western blot analysis of the indicated cells treated with 1 mM NAC for 48 h for PAR.
Figure 8.
OPA1 protein and mRNA levels are diminished in AOA1 cells. (A) A representative western blot image of mitochondrial fusion and fission proteins in extracts from the APTX-deficient patient-derived cells L938 and L939 and the control cells C2ABR and C3ABR. Quantifications are shown as mean ± S.E.M., N = 3, (*P < 0.05, **P < 0.01, ***P < 0.001). (B) Q-PCR analysis of OPA1 gene expression in the patients and control cells. Quantifications are shown as mean ± S.E.M., N = 3, (*P < 0.05, **P < 0.01). (C) Western blot analysis of the subunits of OXPHOS, using PCNA as loading control. Quantifications are shown as mean ± S.E.M., N = 3, (**P < 0.01).
AOA1 patient-derived cells recapitulate the abnormal mitochondrial features seen in APTX−/− U2OS cells
Next, we used AOA1 patient-derived cells (L938 and L939) and control cells (C3ABR and C2ABR) that are Epstein–Barr virus-transformed lymphoblastoid cell lines (5,25). MMP analysis showed a significant reduction in MMP (Figure 7A), and a significantly higher level of mitochondrial ROS production (Figure 7B), and higher SOD2 (Figure 7C) in AOA1 cells compared with the control cells. Together with the results from the APTX−/− cells (Figure 2A, B and D), impaired MMP and increased oxidative stress appear to associate strongly with APTX deficiency.
Increased acetylation of mitochondrial proteins in AOA1 cells
The key function of SIRT3 deacetylase in mitochondrial homeostasis is probably the repair of non-enzymatic acetylation of mitochondrial proteins by acetyl-CoA (59). Like in APTX−/− cells (Figure 2D), the level of SIRT3 was considerably higher in AOA1 patient cells compared with the control cells (Figure 7C). Western blot analysis of mitochondria enriched extracts showed an overall increased level of mitochondrial protein acetylation in AOA1 cells (Figure 7D). Together, these results show that AOA1 patient cells undergo mitochondrial stress in the form of higher ROS production and acetylation of mitochondrial proteins.
Significantly reduced level of PARP1 but enhanced PARylation in AOA1 cells
The PARP superfamily consists of 16 members (60). PARP1 is responsible for most of the cellular PARP activity (51). The level of total poly(AD-ribose) (PAR) was markedly higher in AOA1 cells (Figure 7E), suggesting elevated DDR signaling. The level of PARP1, however, was significantly reduced in AOA1 cells compared with control cells (Figure 7E). Together, both AOA1 and APTX−/− U2OS (Figure 5C) cells showed elevated PAR that was inversely correlated with PARP1 concentration.
ROS can chemically modify DNA bases and cause DNA stranded breaks (61). To test whether elevated PAR signaling in AOA1 cells was a result of ROS generated DNA damage, we treated the cells with the ROS scavenger NAC. Western blot analysis showed a significant reduction in PAR signaling (Figure 7F). Collectively, these results suggest that loss of APTX resulted in a higher ROS production that together with reduced DNA repair capacity results, at least in part, in elevated DNA damage level and DDR in the form of PARylation.
Diminished overall OPA1 level is a common feature of APTX deficient cells
Western blot analysis of key regulators of mitochondrial fission and fusion showed that the level of the mitochondrial inner membrane fusion protein OPA1, and DRP1, a cytosolic protein that is recruited to the outer membrane surface to catalyze the fission, were significantly reduced in AOA1 cells (Figure 8A). We found no detectable changes in the amounts of Fis1, an outer mitochondrial membrane protein that mediates fission by acting as DRP1 receptor (20,62), or the mitochondrial fusion protein MFN1, in AOA1 cells compared to control cells (Figure 8A). Phosphorylation of DRP-1 at Serine 616 is required for the recruitment of DRP1 to the surface of mitochondria (63). The ratio of p-DRP1 to total DRP1 was unchanged (Figure 8A). The reduced level of DRP1 in patient cells may be a protective response to OPA1 reduction in order to maintain a balance between fusion and fission (64).
Like in APTX−/− cells (Figure 4B), the expression of OPA1 was significantly reduced in AOA1 cells compared with the control cells (Figure 8B). Thus, APTX-deficiency results in down-regulation of OPA1 gene expression. To test whether loss of OPA1 is a characteristic of APTX-deficient cells, we knocked out APTX in HEK cells. APTX−/− HEK cells also showed a significantly lower OPA1 level compared with control HEK cells (Supplementary Figure S7). Collectively, these results demonstrate that diminished OPA1 expression and protein level is a common feature of APTX-deficient cells.
OPA1 mRNA undergoes alternative splicing (65) and some isoforms seem to be specifically important for mitochondrial network formation (66). To test the possible presence of OPA1 isoforms specific to APTX-deficient cells, we carried out reverse transcription-PCR amplification of the region flanking exons three to nine (Supplementary Figure S8A). Two putative mRNA isoforms were differently expressed between AOA1 and control cells (primers F1/R1, Supplementary Figure S8B, arrows). These were further verified by a second primer set to amplify a shorter region between exons three to seven (primers F2/R2). Furthermore, better separation of OPA1 protein in 6% polyacrylamide SDS gel revealed that of the five apparent isoforms, three were differentially expressed between APTX-deficient and control cells (Supplementary Figure S8C, arrows). In conclusion, both the overall level of OPA1 expression and the expression of some putative OPA1 isoforms are altered in APTX-deficient cells compared to control cells.
Specific up-regulation of OXPHOS complexes III and V in AOA1 cells
We examined the abundance of selected OXPHOS subunits in AOA1 cells and found that complexes CIII and V were more abundant in AOA1 cells than in control cells (Figure 8C). Taken together, the results show that APTX deficiency consistently leads to the up-regulation of OXPHOS protein subunits probably as a part of a compensatory transcriptional response to mitochondrial dysfunction (67,68). Such response seems to be tissue specific (69), which may explain why complexes I and II were not affected in AOA1 cells but were up-regulated in APTX−/− cells (Figure 6E). Moreover, it is known that the type and the level of mitochondrial proteins can vary between tissues (70). We speculate that the differences in OXPHOS between AOA1 and APTX-KO cells may be a result of differential expression of OXPHOS subunits and assembly factors in these cells.
Collectively, AOA1 and APTX−/− share key features of mitochondrial dysfunctions supporting the involvement of APTX in these phenotypes in particular, and in mitochondrial homeostasis as a whole.
Differential gene expression analysis
We performed RNA-seq analyses on APTX-KO, and APTX-KO cells stably expressing APTX variant 6 (APTX-Pos), and two AOA1 patient-derived cell lines (L939, and L939) and the corresponding control cells (Ctrl) (Supplementary Tables S1–S4). There were similar numbers of DEGs, APTX versus Ctrl, 1532 (57% down); APTX-Pos versus Ctrl, 2590 (59.2% up); APTX-Pos versus APTX-KO, 1384 (72.8% up). For the ATPX-KO cell lines, the significant differentially expressed genes were subjected to GO cellular compartment analysis via Enrichr (31) to uncover potential mitochondrial terms. However, the term mitochondrion (GO 0005739) was not among the top terms for any pairwise comparison. However, since we were interested in defining the mitochondrial terms that were changed after APTX KO and APTX add back, we proceeded and acquired the DEG gene lists from the mitochondrion GO term and wanted to subject that list to GO biological function analysis but each pairwise comparison had too few genes (APTX KO_Ctrl, 42; APTX POS_Ctrl, 107, APTX POS_Ctrl) for this to be meaningful. So, we collected all the genes with an adjusted P-values ≤ 0.05, dropped the log2 fold change cutoff requirement, and collected the mitochondrion term (GO 0005739) gene lists from each pairwise comparison. Not imposing a cutoff change is routinely done in Gene Set Enrichment Analysis (71). Mitochondrion was a top scoring term in each pairwise comparison using this input gene list (Supplementary Table S5). After GO biological process analysis, the GO terms to cellular response to oxidative stress (Figure 9A), respiratory ETC (Figure 9B), mitochondrial fusion (Figure 9C), and several terms related to translation were of interest to us (see Supplementary Table S6 for the entire top 20 list of terms). SOD2 was among the up-regulated genes in ATPX KO cells that was down-regulated after adding back APTX (Figure 9A). Among the ETC genes, there is one clear block of genes (*, Figure 9D) whose expression seems to be up in the APTX KO and normalized by APTX rescue. Likewise, the mitochondrial FIS1 and MFF genes are up-regulated in APTX KO but normalized after APTX re-expression. In contrast, MFN1, OPA1 and BAX genes were down-regulated in the KO cells and remained that way after APTX rescue and mitochondrial network changes in APTX−/− cells (Figure 9C). We should note here that the expression change of OPA1 was very modest, −0.45 log2 fold change in the APTX KO_Ctrl comparison, so OPA1 does not appear to be strongly regulated at the level of transcription in these cells.
We conducted a similar analysis on the AOA1 patient cells versus controls; roughly half were up- or down-regulated (1180 genes, 57% down) (Supplementary Table S4). From the AOA1 set, the genes from the term mitochondrion were extracted and the expression of OPA1 (down), LONP (down) and FOXO1 (up) were changed similar as in the APTX KO cells (Figure 9D). There was only one mitochondrial electron transport gene in the AOA1 set, ATPAF1, the assembly factor for the F1 complex, which was down-regulated.
Since APTX is involved in DNA base excision repair (BER), we also sought to determine if DNA repair genes were responsive to either the loss or rescue of APTX expression. BER, nucleotide excision repair (NER) and mismatch repair (MMR) terms were found in KEGG pathways, and interestingly, NER was statistically significant (adj. P-value ≤ 0.05) in all three comparisons (when using the set of genes that have adjusted P-values ≤ 0.05 and dropping the log2 fold change requirement). MMR was significantly changed in APTX-Pos versus APTX-KO comparison (adj. P-value 0.0005), while BER was only trending (adj. P-value 0.06). A heat map of the combined DNA repair genes list is shown in Figure 9E. Notably, PARP1 is down-regulated in all pairwise comparisons, while the expression of PARP2 is up-regulated in APTX KO and normalized after APTX rescue.
The Caenorhabditis elegans model of OPA-1 deficiency recapitulates the stress phonotypes in APTX-deficient cells
To date, there is no known C. elegans homolog of APTX (72). Therefore, to isolate the functional consequences of OPA1 loss on an organismal level, the C. elegans eat-3 strain was examined. Caenorhabditis elegans eat-3 (ad426) strain (73) has a mutation in the D2013.5 gene, which encodes the ortholog of yeast Mgm1 and mammalian Opa1 (74). The ad426 mutation leads to fragmented mitochondria similar to those cause by mutations in Opa1 and Mgm1 (74). As previously reported, the eat-3 (ad426) strain show disrupted mitochondrial network by TMRM live staining (Figure 10A). Similar to AOA1 patient cells, the level of PARylation and acetylated proteins is also increased in eat-3 (ad426) worms (Figure 10B and C). We also measured ATP levels and found that eat-3 (ad426) worms have a much lower level of ATP compared with N2 worms (Figure 10D), suggesting mitochondrial function is impaired. To further explore the biological importance of the integrity of mitochondrial network, we performed chemotaxis assay by using isoamyl alcohol, which is an attractive odor to the worms. In N2 worms, ∼75% were attached by isoamyl alcohol after 1 h. In eat-3 (ad426) worms, only 22% were attracted by isoamyl alcohol (Figure 10E). To exclude the possibility that the difference in chemotaxis index is caused by motor dysfunction, we performed a swimming assay. The result showed no significant differences between N2 and eat-3 (ad426) worms (Figure 10F). Thus, decreased chemotaxis index, together with normal motor function, suggest that eat-3 (ad426) worms have deficiencies in sensing isoamyl alcohol. In worms, two pairs of amphid sensory neurons, AWC and AWA, are required for chemotaxis to isoamyl alcohol (75). Taken together, we conclude that in nematode, eat-3 (Opa1) deficiency leads to the disruption of mitochondrial network, and impairment of sensory neurons.
Figure 10.
The Caenorhabditis elegans Opa1 homolog eat-3 recapitulates some of the stress phenotypes and responses seen in APTX-defect cells. (A) Visualization of mitochondrial network by treating the worms with TMRM. Scale bar represents 10 μm. (B) Western blot analysis of C. elegans extracts for (B) parylation (Par) and (C) total protein acetylation, (D) ATP measurements, (E) chemotaxis assay and (F) swimming assays. Data is presented as mean ± S.E.M., (** P < 0.01, *** P < 0.001).
DISCUSSION
In this study, we show that APTX-deficient cells displayed hallmarks of mitochondrial dysfunction and stress, i.e. low MMP, increased mitochondrial ROS production, susceptibility to mtDNA damage, elevated acetylation of mitochondrial proteins, changed OXPHOS protein abundance, altered mitochondrial morphology and impaired mitophagy.
A significant reduction in the level of a protein does not necessarily recapitulate the phenotypes caused by a complete loss of that protein. Germline deletion of DNA repair protein Xrcc1 in mouse is embryonic lethal; however, Xrcc1 at levels nearly 10% of normal cells support embryonic viability (76). We also found that extracts from cells with APTX as low as 5% of the normal cells displayed robust 5′-AMP removal activity compared with the control cells (Figure 1A and B). This result may partially explain the diverse clinical symptoms and complex genotype-phenotype correlations in AOA1 patients given different rates of degradation and instability of APTX mutants (9,77,78). This also demonstrates that it is of great importance to achieve a complete depletion in order to characterize the functions of aprataxin. Such a strategy might be necessary in the study of most DNA repair proteins.
Mitochondria are structurally highly dynamic organelles. They constantly change shape, size, and form interconnected networks in response to environmental cues. Mitochondrial size and morphology are determined by the rate of fission and fusion, which together with mitophagy and biogenesis, regulate the characteristic mitochondrial network. Mitochondrial morphology varies across cell types and tissues and is highly adaptive to metabolic signaling and stress (22,40). Defects or changes in the expression of components of mitochondrial fission and fusion cause disease including neurodegeneration and are associated with the normal aging process (18,24). The expression and protein level of the inner membrane fusion protein OPA1 was consistently lower in APTX-deficient cells (Figures 4A and B. 8A and B, 9Supplementary Tables S1–S3). OPA1 is a dynamin-like GTPase localized in the mitochondrial inner membrane and is commonly thought to play key roles in mitochondrial structure and dynamics by mediating inner mitochondrial membrane fusion and by controlling the cristae shape (46,47). OPA1 mutations were first identified in dominant optic atrophy (DOA), a disease specifically affecting retinal ganglion cells (79,80). OPA1 deficiency has also been identified in patients with clinically diverse symptoms and with significant variations of age of onset and severity of the disease (81). The extra-ocular symptoms of OPA1 deficiency include deafness, ataxia, myopathy and spinocerebellar degeneration (82–84). Thus, AOA1 and OPA1-related neurodegeneration share phenotypes suggesting that mitochondrial dysfunction is a common pathway in these diseases. Moreover, a mitochondrial disease database (10) predicts AOA1 and DOA diseases to have significant mitochondrial involvement (Supplementary Figure S9). Fibroblasts from patients with OPA1 mutation display mitochondrial network fragmentation (85,86). Homozygous Opa1 mutant mice die in utero during embryogenesis, but heterozygous Opa1 mutants display the main features of human DOA including abnormal mitochondrial morphology, disorganized cristae structure, mitochondrial dysfunction and mtDNA instability (87–89). Eat-3 is the C. elegans homolog of human OPA1. Mitochondria in eat-3 mutant C. elegans strain are fragmented and show phenotypes consistent with defects in OXPHOS system (74). Thus, ample evidence shows a key role of OPA1 in mitochondrial function and network formation across species. As such, OPA1 reduction may, at least in part, explain the observed loss of mitochondrial network (Figure 3A and B), and impaired cristae in APTX-deficient cells (Figure 3C).
In humans, OPA1 is present in multiple isoforms generated by alternative splicing of mRNA at exons 4, 4b and 5b, which further undergo proteolysis by the mitochondrial inner-membrane peptidases YME1L and OMA1 (65,90,91). PCR amplification of the OPA1 cDNA, and WB analysis showed specific changes in the expression of OPA1 isoforms between AOA1 and control cells (Supplementary Figure S8). Furthermore, the alternative splicing analysis of OPA1 in RNA-seq data showed the preferential expression of isoforms 1, 7 and 8 in U2OS and AOA1-patient cells (Supplementary Figure S10). Interestingly, isoform 7 was down-regulated in both AOA1-patient cells and in APTX−/− cells. The OPA1 isoform 1 was, in addition, down-regulated in AOA1-patient cells. Thus, APTX-deficiency is linked to altered expression of OPA1 isoforms, which may contribute to the mitochondrial morphology and network changes in APTX-deficient cells. To determine the biological significance of these differences is technically challenging, because both loss and overexpression of OPA1 causes fragmentation of the mitochondrial network and disorganization of cristae (66,92), indicating that balanced expression of OPA1 is important for proper mitochondrial function and network formation.
One limitation of this study is that re-introducing APTX into APTX−/− cells did not correct the expression of OPA1 and mitochondrial network in APTX−/− cells. Moreover, the specific function of APTX in the nucleus and mitochondria in the observed mitochondrial phenotypes was not addressed here.
It has been shown that APTX regulates the expression and the stability of PARP1 (93). RNA-seq results showed that the expression of PARP1 was somewhat lower in AOA1 patient cells and in APTX−/− cells compared to control cells (Supplementary Tables S1–S4 and Figure 9). PARP2 is another PARP family member with PAR synthesis activity and is expressed at ∼15% of PARP1 (60). The level of the expression of PARP2 was somewhat higher in APTX−/− cells compared to control cells (Figure 9 and Supplementary Table S1–S3). In AOA1 patient cells, however, there were no significant changes in PARP2 expression compared with control cells (Supplementary Table S4). In APTX−/− cells, PARP2 also showed increased expression. Several of the PARP members were differentially expressed in APTX-deficient cells (Figure 9 and Supplementary Tables S1–S4). Poly-(ADP-ribose)glycohydrolyase (PARG) is responsible for the degradation of poly-(ADP-ribose). There was no significant difference in the level of expression of PARG in AOA1 or APTX−/− cells compared to their corresponding control cells (Supplementary Tables S1–S3) suggesting that the rate of PAR turnover did not account for the observed elevated PAR levels in APTX-deficient cells. Taken together, defect in APTX results in an elevated level of PARylation.
The fork-head associated (FHA) domain is a phosphopeptide interacting domain associated with proteins involved in a number of processes including intracellular signal transduction, transcription, protein transport, DNA repair and protein degradation (94). APTX seems to interact with several proteins through its FHA domain. In the nucleus, APTX was reported to interact with DNA repair proteins XRCC1, PARP1 and also with the transcription factor p53 (25,95–97). A recently developed concept connects nuclear DNA damage signaling to mitochondrial function and maintenance. According to this model, persistent DNA damage, e.g. because of a defect in DNA repair or diminished DNA repair capacity, activates prolonged DDR by PARP1/2 in the form of PAR being added to proteins in the vicinity of the DNA damage. This process consumes NAD+, an essential cofactor involved in a number of key processes including mitochondrial bioenergetics and mitophagy. Thus, persistent DDR leads to a competition between these processes for the available NAD+ leading to mitochondrial dysfunction (23). Moreover, the cellular bioavailability of NAD+ declines during normal aging (98,99). In support of this model, NAD+ replenishment improves health and lifespan in animal models of human disease (11). APTX-deficient cells consistently contained reduced levels of PARP1 but elevated PARylation compared with control cells (Figures 5C, 6G and 7E and F). Treatment of the cells with the ROS scavenger NAC significantly reduced PARylation in APTX-deficient cells (Figure 7F). Thus, we speculate that the lack of APTX leads to higher ROS production and reduced PARP1 level, which in turn results in diminished basal DDR and DNA repair, persistence of PARylation, which over time may contribute to the impairment of mitochondrial homeostasis in APTX-deficient cells.
In conclusion, our data links impaired mitochondrial homeostasis to APTX deficiency and AOA1 pathology. In recent years, mitochondria have become targets for pharmacological interventions for treatment of neurodegenerative diseases with promising results (23,100). In the future, perhaps such strategies should be tested on AOA1 patients.
DATA AVAILABILITY
The RNA-seq data has been deposited to the GEO database under series number GSE124412.
Supplementary Material
ACKNOWLEDGEMENTS
We like to thank Jane Tian for bioenergetic experiments and for help with the microarray analysis. We would like to thank Dr Elin Lehmann, Dr Yongqing Zhang and Dr Kevin G. Becker of the Gene Expression and Genomics Unit, NIA Intramural Program, NIH. We would like to thank Kavya Achanta for her help with C. elegans study. We would like to thank Dr Daniel R. McNeill for C2/3ABR and L938/9 cell lines. We would like to thank Dr Beimeng Yang and Dr Anthony Moore for critically reading the manuscript. Confocal and electron microscopy experiments were carried out at the Core Facility of Integrated Microscopy (CFIM), University of Copenhagen.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NORDEA Foundation, Denmark (02-2013-0220); EU Joint Programme-Neurodegenerative Disease Research (JPND); Innovation Fund Denmark (5188-00001); Olav Thon foundation Norway (531811-710131); Novo Nordisk foundation Denmark (NNF17OC0027812); Intramural Research Program of the NIH, National Institute on Aging (AG000733). Funding for open access charge: University of Copenhagen.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ellenberger T., Tomkinson A.E.. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008; 77:313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ahel I., Rass U., El-Khamisy S.F., Katyal S., Clements P.M., McKinnon P.J., Caldecott K.W., West S.C.. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006; 443:713–716. [DOI] [PubMed] [Google Scholar]
- 3. Date H., Onodera O., Tanaka H., Iwabuchi K., Uekawa K., Igarashi S., Koike R., Hiroi T., Yuasa T., Awaya Y. et al.. Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene. Nat. Genet. 2001; 29:184–188. [DOI] [PubMed] [Google Scholar]
- 4. Moreira M.C., Barbot C., Tachi N., Kozuka N., Uchida E., Gibson T., Mendonca P., Costa M., Barros J., Yanagisawa T. et al.. The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin. Nat. Genet. 2001; 29:189–193. [DOI] [PubMed] [Google Scholar]
- 5. Sykora P., Croteau D.L., Bohr V.A., Wilson D.M. 3rd.. Aprataxin localizes to mitochondria and preserves mitochondrial function. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:7437–7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akbari M., Sykora P., Bohr V.A.. Slow mitochondrial repair of 5′-AMP renders mtDNA susceptible to damage in APTX deficient cells. Sci. Rep. 2015; 5:12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Caglayan M., Prasad R., Krasich R., Longley M.J., Kadoda K., Tsuda M., Sasanuma H., Takeda S., Tano K., Copeland W.C. et al.. Complementation of aprataxin deficiency by base excision repair enzymes in mitochondrial extracts. Nucleic Acids Res. 2017; 45:10079–10088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quinzii C.M., Kattah A.G., Naini A., Akman H.O., Mootha V.K., DiMauro S., Hirano M.. Coenzyme Q deficiency and cerebellar ataxia associated with an aprataxin mutation. Neurology. 2005; 64:539–541. [DOI] [PubMed] [Google Scholar]
- 9. Renaud M., Moreira M.C., Ben Monga B., Rodriguez D., Debs R., Charles P., Chaouch M., Ferrat F., Laurencin C., Vercueil L. et al.. Clinical, biomarker, and molecular delineations and Genotype-Phenotype correlations of ataxia with oculomotor apraxia type 1. JAMA Neurol. 2018; 75:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scheibye-Knudsen M., Scheibye-Alsing K., Canugovi C., Croteau D.L., Bohr V.A.. A novel diagnostic tool reveals mitochondrial pathology in human diseases and aging. Aging. 2013; 5:192–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fang E.F., Kassahun H., Croteau D.L., Scheibye-Knudsen M., Marosi K., Lu H., Shamanna R.A., Kalyanasundaram S., Bollineni R.C., Wilson M.A. et al.. NAD(+) replenishment improves lifespan and healthspan in ataxia telangiectasia models via mitophagy and DNA repair. Cell Metab. 2016; 24:566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brand M.D., Nicholls D.G.. Assessing mitochondrial dysfunction in cells. Biochem. J. 2011; 435:297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cornelius N., Wardman J.H., Hargreaves I.P., Neergheen V., Bie A.S., Tumer Z., Nielsen J.E., Nielsen T.T.. Evidence of oxidative stress and mitochondrial dysfunction in spinocerebellar ataxia type 2 (SCA2) patient fibroblasts: effect of coenzyme Q10 supplementation on these parameters. Mitochondrion. 2017; 34:103–114. [DOI] [PubMed] [Google Scholar]
- 14. Fang E.F., Scheibye-Knudsen M., Brace L.E., Kassahun H., SenGupta T., Nilsen H., Mitchell J.R., Croteau D.L., Bohr V.A.. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014; 157:882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bose A., Beal M.F.. Mitochondrial dysfunction in Parkinson's disease. J. Neurochem. 2016; 139:216–231. [DOI] [PubMed] [Google Scholar]
- 16. Costa V., Scorrano L.. Shaping the role of mitochondria in the pathogenesis of Huntington's disease. EMBO J. 2012; 31:1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Scheibye-Knudsen M., Fang E.F., Croteau D.L., Wilson D.M. 3rd, Bohr V.A.. Protecting the mitochondrial powerhouse. Trends Cell Biol. 2015; 25:158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sebastian D., Palacin M., Zorzano A.. Mitochondrial dynamics: coupling mitochondrial fitness with healthy aging. Trends Mol. Med. 2017; 23:201–215. [DOI] [PubMed] [Google Scholar]
- 19. Loson O.C., Song Z., Chen H., Chan D.C.. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013; 24:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otera H., Wang C., Cleland M.M., Setoguchi K., Yokota S., Youle R.J., Mihara K.. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J. Cell Biol. 2010; 191:1141–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liesa M., Shirihai O.S.. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013; 17:491–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wai T., Langer T.. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 2016; 27:105–117. [DOI] [PubMed] [Google Scholar]
- 23. Fang E.F., Scheibye-Knudsen M., Chua K.F., Mattson M.P., Croteau D.L., Bohr V.A.. Nuclear DNA damage signalling to mitochondria in ageing. Nat. Rev. Mol. Cell Biol. 2016; 17:308–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Archer S.L. Mitochondrial dynamics. mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 2013; 369:2236–2251. [DOI] [PubMed] [Google Scholar]
- 25. Gueven N., Becherel O.J., Kijas A.W., Chen P., Howe O., Rudolph J.H., Gatti R., Date H., Onodera O., Taucher-Scholz G. et al.. Aprataxin, a novel protein that protects against genotoxic stress. Hum. Mol. Genet. 2004; 13:1081–1093. [DOI] [PubMed] [Google Scholar]
- 26. Santos J.H., Mandavilli B.S., Van Houten B.. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol. Biol. 2002; 197:159–176. [DOI] [PubMed] [Google Scholar]
- 27. Hunter S.E., Jung D., Di Giulio R.T., Meyer J.N.. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods. 2010; 51:444–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Merrill Ronald A. Strack S, Usachev Y. Techniques to investigate mitochondrial function in neurons. 2017; 123:NY: Humana Press; 31–48. [Google Scholar]
- 33. Ryu D., Mouchiroud L., Andreux P.A., Katsyuba E., Moullan N., Nicolet-Dit-Felix A.A., Williams E.G., Jha P., Lo Sasso G., Huzard D. et al.. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat. Med. 2016; 22:879–888. [DOI] [PubMed] [Google Scholar]
- 34. Bargmann C.I., Horvitz H.R.. Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron. 1991; 7:729–742. [DOI] [PubMed] [Google Scholar]
- 35. Akbari M., Keijzers G., Maynard S., Scheibye-Knudsen M., Desler C., Hickson I.D., Bohr V.A.. Overexpression of DNA ligase III in mitochondria protects cells against oxidative stress and improves mitochondrial DNA base excision repair. DNA Repair. 2014; 16:44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Starkov A.A., Fiskum G.. Regulation of brain mitochondrial H2O2 production by membrane potential and NAD(P)H redox state. J. Neurochem. 2003; 86:1101–1107. [DOI] [PubMed] [Google Scholar]
- 37. Lombard D.B., Alt F.W., Cheng H.L., Bunkenborg J., Streeper R.S., Mostoslavsky R., Kim J., Yancopoulos G., Valenzuela D., Murphy A. et al.. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol. Cell. Biol. 2007; 27:8807–8814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qiu X., Brown K., Hirschey M.D., Verdin E., Chen D.. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010; 12:662–667. [DOI] [PubMed] [Google Scholar]
- 39. Cogliati S., Frezza C., Soriano M.E., Varanita T., Quintana-Cabrera R., Corrado M., Cipolat S., Costa V., Casarin A., Gomes L.C. et al.. Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency. Cell. 2013; 155:160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta. 2012; 1817:1833–1838. [DOI] [PubMed] [Google Scholar]
- 41. Walczak J., Partyka M., Duszynski J., Szczepanowska J.. Implications of mitochondrial network organization in mitochondrial stress signalling in NARP cybrid and Rho0 cells. Sci. Rep. 2017; 7:14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Al-Mehdi A.B., Pastukh V.M., Swiger B.M., Reed D.J., Patel M.R., Bardwell G.C., Pastukh V.V., Alexeyev M.F., Gillespie M.N.. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci. Signal. 2012; 5:ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Lower B., Wunderlich F.T., von Kleist-Retzow J.C., Waisman A., Westermann B., Langer T.. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria. Genes Dev. 2008; 22:476–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yamaguchi R., Lartigue L., Perkins G., Scott R.T., Dixit A., Kushnareva Y., Kuwana T., Ellisman M.H., Newmeyer D.D.. Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization. Mol. Cell. 2008; 31:557–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Elachouri G., Vidoni S., Zanna C., Pattyn A., Boukhaddaoui H., Gaget K., Yu-Wai-Man P., Gasparre G., Sarzi E., Delettre C. et al.. OPA1 links human mitochondrial genome maintenance to mtDNA replication and distribution. Genome Res. 2011; 21:12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Frezza C., Cipolat S., Martins de Brito O., Micaroni M., Beznoussenko G.V., Rudka T., Bartoli D., Polishuck R.S., Danial N.N., De Strooper B. et al.. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006; 126:177–189. [DOI] [PubMed] [Google Scholar]
- 47. MacVicar T., Langer T.. OPA1 processing in cell death and disease - the long and short of it. J. Cell Sci. 2016; 129:2297–2306. [DOI] [PubMed] [Google Scholar]
- 48. Pickles S., Vigie P., Youle R.J.. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr. Biol. 2018; 28:R170–R185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T.. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000; 19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pascal J.M., Ellenberger T.. The rise and fall of poly(ADP-ribose): an enzymatic perspective. DNA Repair. 2015; 32:10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ame J.C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Hoger T., Menissier-de Murcia J., de Murcia G.. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 1999; 274:17860–17868. [DOI] [PubMed] [Google Scholar]
- 52. Lin S.C., Hardie D.G.. AMPK: sensing glucose as well as cellular energy status. Cell Metab. 2018; 27:299–313. [DOI] [PubMed] [Google Scholar]
- 53. Kim J., Kundu M., Viollet B., Guan K.L.. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011; 13:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hardie D.G., Schaffer B.E., Brunet A.. AMPK: an energy-sensing pathway with multiple inputs and outputs. Trends Cell Biol. 2016; 26:190–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Koopman W.J., Distelmaier F., Smeitink J.A., Willems P.H.. OXPHOS mutations and neurodegeneration. EMBO J. 2013; 32:9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Winge D.R. Sealing the mitochondrial respirasome. Mol. Cell Biol. 2012; 32:2647–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beranek D.T. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat. Res. 1990; 231:11–30. [DOI] [PubMed] [Google Scholar]
- 58. Fukui M., Choi H.J., Zhu B.T.. Rapid generation of mitochondrial superoxide induces mitochondrion-dependent but caspase-independent cell death in hippocampal neuronal cells that morphologically resembles necroptosis. Toxicol. Appl. Pharmacol. 2012; 262:156–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weinert B.T., Moustafa T., Iesmantavicius V., Zechner R., Choudhary C.. Analysis of acetylation stoichiometry suggests that SIRT3 repairs nonenzymatic acetylation lesions. EMBO J. 2015; 34:2620–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ame J.C., Spenlehauer C., de Murcia G.. The PARP superfamily. Bioessays. 2004; 26:882–893. [DOI] [PubMed] [Google Scholar]
- 61. Akbari M., Krokan H.E.. Cytotoxicity and mutagenicity of endogenous DNA base lesions as potential cause of human aging. Mech. Ageing Dev. 2008; 129:353–365. [DOI] [PubMed] [Google Scholar]
- 62. Mozdy A.D., McCaffery J.M., Shaw J.M.. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J. Cell Biol. 2000; 151:367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kashatus D.F., Lim K.H., Brady D.C., Pershing N.L., Cox A.D., Counter C.M.. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 2011; 13:1108–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Manczak M., Kandimalla R., Fry D., Sesaki H., Reddy P.H.. Protective effects of reduced dynamin-related protein 1 against amyloid beta-induced mitochondrial dysfunction and synaptic damage in Alzheimer's disease. Hum. Mol. Genet. 2016; 25:5148–5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Delettre C., Griffoin J.M., Kaplan J., Dollfus H., Lorenz B., Faivre L., Lenaers G., Belenguer P., Hamel C.P.. Mutation spectrum and splicing variants in the OPA1 gene. Hum. Genet. 2001; 109:584–591. [DOI] [PubMed] [Google Scholar]
- 66. Del Dotto V., Mishra P., Vidoni S., Fogazza M., Maresca A., Caporali L., McCaffery J.M., Cappelletti M., Baruffini E., Lenaers G. et al.. OPA1 isoforms in the hierarchical organization of mitochondrial functions. Cell Rep. 2017; 19:2557–2571. [DOI] [PubMed] [Google Scholar]
- 67. Heddi A., Stepien G., Benke P.J., Wallace D.C.. Coordinate induction of energy gene expression in tissues of mitochondrial disease patients. J. Biol. Chem. 1999; 274:22968–22976. [DOI] [PubMed] [Google Scholar]
- 68. Fakouri N.B., Durhuus J.A., Regnell C.E., Angleys M., Desler C., Hasan-Olive M.M., Martin-Pardillos A., Tsaalbi-Shtylik A., Thomsen K., Lauritzen M. et al.. Rev1 contributes to proper mitochondrial function via the PARP-NAD(+)-SIRT1-PGC1alpha axis. Sci. Rep. 2017; 7:12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van der Westhuizen F.H., van den Heuvel L.P., Smeets R., Veltman J.A., Pfundt R., van Kessel A.G., Ursing B.M., Smeitink J.A.. Human mitochondrial complex I deficiency: investigating transcriptional responses by microarray. Neuropediatrics. 2003; 34:14–22. [DOI] [PubMed] [Google Scholar]
- 70. Pagliarini D.J., Calvo S.E., Chang B., Sheth S.A., Vafai S.B., Ong S.E., Walford G.A., Sugiana C., Boneh A., Chen W.K. et al.. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008; 134:112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E. et al.. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003; 34:267–273. [DOI] [PubMed] [Google Scholar]
- 72. Rass U., Ahel I., West S.C.. Molecular mechanism of DNA deadenylation by the neurological disease protein aprataxin. J. Biol. Chem. 2008; 283:33994–34001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Avery L. The genetics of feeding in Caenorhabditis elegans. Genetics. 1993; 133:897–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kanazawa T., Zappaterra M.D., Hasegawa A., Wright A.P., Newman-Smith E.D., Buttle K.F., McDonald K., Mannella C.A., van der Bliek A.M.. The C. elegans Opa1 homologue EAT-3 is essential for resistance to free radicals. PLoS Genet. 2008; 4:e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bargmann C.I., Hartwieg E., Horvitz H.R.. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993; 74:515–527. [DOI] [PubMed] [Google Scholar]
- 76. Tebbs R.S., Thompson L.H., Cleaver J.E.. Rescue of Xrcc1 knockout mouse embryo lethality by transgene-complementation. DNA Repair. 2003; 2:1405–1417. [DOI] [PubMed] [Google Scholar]
- 77. Hirano M., Matsumura R., Nakamura Y., Saigoh K., Sakamoto H., Ueno S., Inoue H., Kusunoki S.. Unexpectedly mild phenotype in an ataxic family with a two-base deletion in the APTX gene. J. Neurol. Sci. 2017; 378:75–79. [DOI] [PubMed] [Google Scholar]
- 78. Yokoseki A., Ishihara T., Koyama A., Shiga A., Yamada M., Suzuki C., Sekijima Y., Maruta K., Tsuchiya M., Date H. et al.. Genotype-phenotype correlations in early onset ataxia with ocular motor apraxia and hypoalbuminaemia. Brain. 2011; 134:1387–1399. [DOI] [PubMed] [Google Scholar]
- 79. Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G. et al.. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000; 26:211–215. [DOI] [PubMed] [Google Scholar]
- 80. Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E. et al.. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 2000; 26:207–210. [DOI] [PubMed] [Google Scholar]
- 81. Chao de la Barca J.M., Prunier-Mirebeau D., Amati-Bonneau P., Ferre M., Sarzi E., Bris C., Leruez S., Chevrollier A., Desquiret-Dumas V., Gueguen N. et al.. OPA1-related disorders: diversity of clinical expression, modes of inheritance and pathophysiology. Neurobiol. Dis. 2016; 90:20–26. [DOI] [PubMed] [Google Scholar]
- 82. Schaaf C.P., Blazo M., Lewis R.A., Tonini R.E., Takei H., Wang J., Wong L.J., Scaglia F.. Early-onset severe neuromuscular phenotype associated with compound heterozygosity for OPA1 mutations. Mol. Genet. Metab. 2011; 103:383–387. [DOI] [PubMed] [Google Scholar]
- 83. Carelli V., Sabatelli M., Carrozzo R., Rizza T., Schimpf S., Wissinger B., Zanna C., Rugolo M., La Morgia C., Caporali L. et al.. ‘Behr syndrome' with OPA1 compound heterozygote mutations. Brain. 2015; 138:e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bonneau D., Colin E., Oca F., Ferre M., Chevrollier A., Gueguen N., Desquiret-Dumas V., N’Guyen S., Barth M., Zanlonghi X. et al.. Early-onset Behr syndrome due to compound heterozygous mutations in OPA1. Brain. 2014; 137:e301. [DOI] [PubMed] [Google Scholar]
- 85. Liao C., Ashley N., Diot A., Morten K., Phadwal K., Williams A., Fearnley I., Rosser L., Lowndes J., Fratter C. et al.. Dysregulated mitophagy and mitochondrial organization in optic atrophy due to OPA1 mutations. Neurology. 2017; 88:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Bonifert T., Karle K.N., Tonagel F., Batra M., Wilhelm C., Theurer Y., Schoenfeld C., Kluba T., Kamenisch Y., Carelli V. et al.. Pure and syndromic optic atrophy explained by deep intronic OPA1 mutations and an intralocus modifier. Brain. 2014; 137:2164–2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Alavi M.V., Bette S., Schimpf S., Schuettauf F., Schraermeyer U., Wehrl H.F., Ruttiger L., Beck S.C., Tonagel F., Pichler B.J. et al.. A splice site mutation in the murine Opa1 gene features pathology of autosomal dominant optic atrophy. Brain. 2007; 130:1029–1042. [DOI] [PubMed] [Google Scholar]
- 88. Chen L., Liu T., Tran A., Lu X., Tomilov A.A., Davies V., Cortopassi G., Chiamvimonvat N., Bers D.M., Votruba M. et al.. OPA1 mutation and late-onset cardiomyopathy: mitochondrial dysfunction and mtDNA instability. J. Am. Heart Assoc. 2012; 1:e003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Davies V.J., Hollins A.J., Piechota M.J., Yip W., Davies J.R., White K.E., Nicols P.P., Boulton M.E., Votruba M.. Opa1 deficiency in a mouse model of autosomal dominant optic atrophy impairs mitochondrial morphology, optic nerve structure and visual function. Hum. Mol. Genet. 2007; 16:1307–1318. [DOI] [PubMed] [Google Scholar]
- 90. Anand R., Wai T., Baker M.J., Kladt N., Schauss A.C., Rugarli E., Langer T.. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 2014; 204:919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Song Z., Chen H., Fiket M., Alexander C., Chan D.C.. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 2007; 178:749–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Cipolat S., Martins de Brito O., Dal Zilio B., Scorrano L.. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:15927–15932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Harris J.L., Jakob B., Taucher-Scholz G., Dianov G.L., Becherel O.J., Lavin M.F.. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum. Mol. Genet. 2009; 18:4102–4117. [DOI] [PubMed] [Google Scholar]
- 94. Durocher D., Jackson S.P.. The FHA domain. FEBS Lett. 2002; 513:58–66. [DOI] [PubMed] [Google Scholar]
- 95. Clements P.M., Breslin C., Deeks E.D., Byrd P.J., Ju L., Bieganowski P., Brenner C., Moreira M.C., Taylor A.M., Caldecott K.W.. The ataxia-oculomotor apraxia 1 gene product has a role distinct from ATM and interacts with the DNA strand break repair proteins XRCC1 and XRCC4. DNA Repair. 2004; 3:1493–1502. [DOI] [PubMed] [Google Scholar]
- 96. Date H., Igarashi S., Sano Y., Takahashi T., Takahashi T., Takano H., Tsuji S., Nishizawa M., Onodera O.. The FHA domain of aprataxin interacts with the C-terminal region of XRCC1. Biochem. Biophys. Res. Commun. 2004; 325:1279–1285. [DOI] [PubMed] [Google Scholar]
- 97. Horton J.K., Stefanick D.F., Caglayan M., Zhao M.L., Janoshazi A.K., Prasad R., Gassman N.R., Wilson S.H.. XRCC1 phosphorylation affects aprataxin recruitment and DNA deadenylation activity. DNA Repair. 2018; 64:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Camacho-Pereira J., Tarrago M.G., Chini C.C.S., Nin V., Escande C., Warner G.M., Puranik A.S., Schoon R.A., Reid J.M., Galina A. et al.. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Metab. 2016; 23:1127–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Fang E.F., Lautrup S., Hou Y., Demarest T.G., Croteau D.L., Mattson M.P., Bohr V.A.. NAD(+) in aging: molecular mechanisms and translational implications. Trends Mol. Med. 2017; 23:899–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sorrentino V., Menzies K.J., Auwerx J.. Repairing mitochondrial dysfunction in disease. Annu. Rev. Pharmacol. Toxicol. 2018; 58:353–389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data has been deposited to the GEO database under series number GSE124412.