Abstract
De novo loss-of-function (LoF) variants in the KMT2A gene are associated with Wiedemann−Steiner Syndrome (WSS). Recently, de novo KMT2A variants have been identified in sequencing studies of cohorts of individuals with neurodevelopmental disorders (NDDs). However, most of these studies lack the detailed clinical information required to determine whether those individuals have isolated NDDs or WSS (i.e. syndromic NDDs). We performed thorough clinical and neurodevelopmental phenotyping on six individuals with de novo KMT2A variants. From these data, we found that all six patients met clinical criteria for WSS and we further define the neurodevelopmental phenotypes associated with KMT2A variants and WSS. In particular, we identified a subtype of Autism Spectrum Disorder (ASD) in five individuals, characterized by marked rigid, repetitive and inflexible behaviours, emotional dysregulation, externalizing behaviours, but relative social motivation. To further explore the clinical spectrum associated with KMT2A variants, we also conducted a meta-analysis of individuals with KMT2A variants reported in the published literature. We found that de novo LoF or missense variants in KMT2A were significantly more prevalent than predicted by a previously established statistical model of de novo mutation rate for KMT2A. Our genotype−phenotype findings better define the clinical spectrum associated with KMT2A variants and suggest that individuals with de novo LoF and missense variants likely have a clinically unrecognized diagnosis of WSS, rather than isolated NDD or ASD alone. This highlights the importance of a clinical genetic and neurodevelopmental assessment for individuals with such variants in KMT2A.
Subject terms: Autism spectrum disorders, Disease genetics
Introduction
The KMT2A gene, also known as MLL, is on chr11q23 and encodes an H3K4 methyltransferase enzyme that regulates the expression of other genes including neuronal and homeobox-containing developmental genes.1 KMT2A is essential for embryonic development, hematopoiesis,2 and neural development3 and dysregulation of KMT2A function is associated with various human disorders. Recurrent somatic translocations between KMT2A and 70 different fusion partners result in leukaemia,4 whereas germline de novo loss-of-function (LoF) variants in KMT2A are associated with Wiedemann−Steiner Syndrome (WSS). WSS is characterized by hypertrichosis cubiti (hairy elbows), dysmorphic facial features, short stature, developmental delay and intellectual disability (ID).5
Since the initial association between WSS and KMT2A,5 additional patients with WSS and de novo LoF or missense variants in KMT2A have been reported5–7 (refer to Supplementary Table 1 for additional references). Although neurodevelopmental features and behavioural abnormalities were frequent, the lack of detailed neuropsychological assessments suggests the presence of these features may be underestimated and warrant further investigation.7 Whole-exome sequencing (WES) and whole-genome sequencing (WGS) studies in individuals with neurodevelopmental disorders (NDDs), including Autism Spectrum Disorder (ASD), have also revealed de novo KMT2A variants in 0.04−2.17% of individuals with NDDs8–12 (refer to Supplementary Table 1 for additional references). However, these studies that identified KMT2A variants in WSS and NDD cohorts were performed independently, and a comprehensive analysis of NDD and WSS phenotypes together in individuals with KMT2A variants has not been conducted. Here, we present the detailed clinical and genomic characterization of six unrelated individuals with de novo LoF or missense variants in KMT2A, and contextualize the findings with published literature.
Results
Clinical reports
Patient 1
A 10-year-old Caucasian male with a history of ASD, ID, attention deficit hyperactivity disorder (ADHD), hypotonia, growth and developmental delay was born to non-consanguineous parents of Ashkenazi Jewish ancestry, as a product of an in vitro fertilization twin pregnancy. He was delivered by Caesarian section at 34 weeks gestation, weighing 3 lb, 2 oz. He had poor growth with height and weight below the third percentile. A magnetic resonance imaging (MRI) of the brain at 2 years of age identified hypoplastic olfactory nerves and unusual configuration of the corpus callosum, showing a short dimension in anterior-posterior diameter and thinning of its body. His medical history is remarkable for delayed motor and speech milestones, hypotonia, bilateral cryptorchidism (surgically repaired), bilateral strabismus (surgically repaired) and constipation. He was diagnosed with ASD (age 5), ADHD (age 7), and ID (age 8). He takes Clonidine for ADHD and melatonin for trouble initiating sleep and frequent night awakenings.
G-banded karyotype, fragile X testing and chromosome microarray (CMA) were normal. WGS identified a de novo KMT2A frameshift variant, c.10324delG (p.Ala3442Profs*17; Supplementary Fig. 1). He was then clinically assessed at age 10 and diagnosed with WSS on the basis of characteristic facial features (Fig. 1a), short stature, microcephaly, generalized hypertrichosis, and aforementioned history of growth and developmental delay, hypotonia, constipation, and strabismus (details in Supplementary Table 2).
Fig. 1.
Facial features of six novel patients with de novo LoF or missense variants in KMT2A. a Patient 1 at 10 years; b patient 2 at 9 years; c patient 3 at 13 years; d Patient 4 at 25 years; e patient 5 at 5 years; f patient 6 at 1 year of age. Characteristic facial features noted in these patients include: thick and/or high arched eyebrows, long eyelashes, hypertelorism, downslanting palpebral fissures and a broad nasal tip. Written consent was obtained from all patients for open access publication of the photographs. LoF: loss-of-function
As a part of this study, neurodevelopmental testing at 10 years, 3 months of age (Table 1) was consistent with his previous diagnoses of ID, ASD, and ADHD. This assessment also identified emotional dysregulation and extremely low language and adaptive skills, but relative strength in vocabulary skills. We observed many repetitive and restrictive behaviours (e.g. fixation with technology, buses, music). Of note, he struggles with transitions between settings and activities, becoming easily upset and oppositional. Despite overall difficulties with social-communication, he demonstrates some appropriate skills, such as making good eye contact, sustaining short conversations, and asking/offering information, particularly when the topic revolved around his area of interest.
Table 1.
Summary of genetic results and neurodevelopmental profiles for six novel patients
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
|---|---|---|---|---|---|---|
| Sex | M | F | M | M | M | M |
| Technology used to identify KMT2A variant | WGSa | WESb | WGSa | WGSa | WESb | Targeted sequencing |
| KMT2A variantc | p.(Ala3442Profs*17) | p.(Ser2363Leufs*12) | p.(Val2057Tyrfs*18) | p.(Glu2566Lysfs*14) | p.(Leu2848Pro) | p.(Arg2699*) |
| Inheritance | De novo | De novo | De novo | De novo | De novo | De novo |
| Other variants | None | Maternally inherited 295 kb deletion chr4q31.3d (chr4:151,378,576-151,673,967) | Maternally inherited PAX1 p.(G166A)d, e | None | Maternally inherited CHRNA4 c.(274-1 G > A)d, f | None |
| Age at Neurodevelopmental Assessment | 10 y 3 m | 12 y 2 m | 13 y 1 m | 25 y 3 m | 5 y1m | 5y 9 m |
| Intellectual Disability Diagnosis | + | + | + | − | + | + |
| IQ (percentile) | <1st | 3rd | <1st | 4th | <1st | <1st |
| Overall Adaptive Skills (percentile) | 4th | 1st | <1st | 9th | <1st | <1st |
| Language Skills (OWLS-IIg percentile) | <1st | <1st | <1st | 32nd | <1st | 1st |
| Receptive Vocabulary (PPVT-4h percentile) | 16th | 9th | 1st | 70thi | <1st | 7th |
| Expressive Vocabulary (EVT-2j percentile) | 7th | 5th | <1st | N/Ak | <1st | 1st |
| ASD Diagnosis | + | − | + | + | + | + |
| Rigidity/ Poor flexibility | + | + | + | + | + | + |
| Anxiety Symptoms | − | +l | + | − | − | + |
| Attention Concerns | +m | +m | + | − | + | + |
| Emotional Regulation Concerns (Dysregulation) | + | + | + | −n | + | + |
| Externalizing Behaviours | + | − | + | − | + | + |
aWhole-genome sequencing
bWhole-exome sequencing
cKMT2A transcript used is NM_0011904.1
dVariant of unknown significance, not clinically significant
ePAX1 transcript used is NM_006192
fCHRNA4 transcript used is NM_000744.6
gOral and Written Language Scales, Second Edition
hPeabody Picture and Vocabulary Test, Fourth Edition
iBased on previous assessment at 19 y
jExpressive Vocabulary Test, Second Edition
kEVT-2 not administered. Expressive language skills from OWLS-II
lHas diagnosis of Generalized Anxiety Disorder
mHas diagnosis of Attention Deficit Hyperactivity Disorder
nBehavior Rating Inventory of Executive Function not administered due to age. Information based on Antisocial Personality Problem Scale on the Adult Behavior Checklist as well as parent report
Patient 2
A 12-year-old Caucasian female with a history of ADHD, ID, growth and developmental delay, and hypotonia was born to non-consanguineous Caucasian parents at 36 weeks gestation with a birth weight of 5 lb, 9 oz. She had poor growth in infancy, with height and weight below the third percentile. Her medical history is remarkable for delayed motor milestones, a ventricular septal defect (which closed spontaneously), strabismus, hypotonia, constipation, recurrent upper respiratory tract infections, and Klippel−Feil anomaly. An MRI of the brain at 10 months of age identified mildly prominent cerebral spinal fluid spaces with age appropriate myelination. At 12 years of age she presented with episodes of rigidity and flexion of the arms with tremulous movements. An electroencephalography (EEG) was normal and the neurology team suspected the movements could represent self-stimulating behaviours. She was diagnosed with ADHD and ID (age 9) and generalized anxiety disorder (age 10). She also has obsessive compulsive traits (compulsive hand washing) and has received behavioural therapy throughout childhood to present.
Clinical genetic assessments at 1 year 8 months of age included clinical CMA, fragile X testing, and metabolic screening. The latter two tests were normal; the microarray analysis identified a maternally inherited 295 kb deletion at chromosome 4q31.3 (chr4:151,378,576−151,673,967) that was not suspected to be clinically significant. After we identified a de novo KMT2A frameshift variant, c.7087_7090del (p.Ser2363Leufs*12; Supplementary Fig. 1) via WES, she was then clinically re-assessed at age 12 and diagnosed with WSS on the basis of characteristic facial features (Fig. 1b), generalized hypertrichosis (Fig. 2), and the history of growth and developmental delay, hypotonia, constipation, and strabismus (Supplementary Table 2).
Fig. 2.
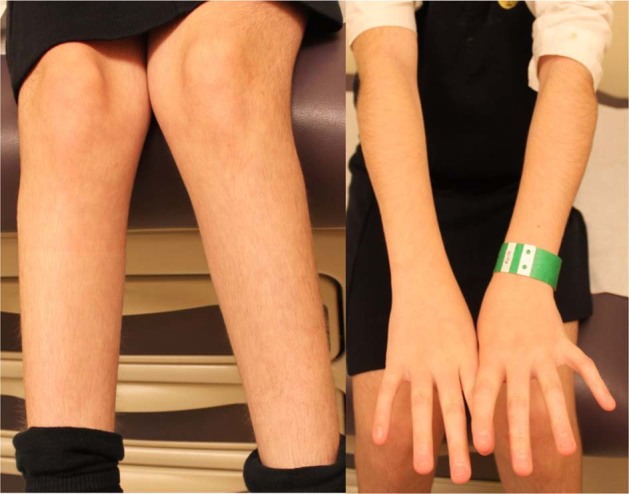
Hypertrichosis of the arms and legs of patient 2 at 9 years of age. Written consent was obtained from patient 2 for open access publication of the photographs
As a part of this study, neurodevelopmental testing at 12 years, 2 months of age (Table 1) was consistent with the previous diagnosis of ID, ADHD and anxiety disorder. The assessment also identified emotional dysregulation and extremely low language and adaptive skills, but relative strength in vocabulary skills. Standardized testing for ASD revealed that she met criteria on the Autism Diagnostic Observation Schedule, Second Edition (ADOS-2), but did not meet criteria on the Autism Diagnostic Interview—Revised (ADI-R). Based on overall clinical impression, she did not receive a diagnosis of ASD. Nevertheless, a pattern of restricted and repetitive behaviours was identified on the ADOS-2, ADI-R and parent report. She struggles with changes to routine and becomes easily frustrated. Of note, clinical impressions were of a socially motivated girl, whose relative strength in vocabulary masks her areas of difficulty and she presents as having a higher level of comprehension.
Patient 3
A 13-year-old Caucasian male with a history of ASD, growth and developmental delay and hypotonia was born to non-consanguineous Caucasian parents at term by caesarean section with a birth weight of 6 lb, 14 oz. His mother was on citalopram during the pregnancy for treatment of depression. He was diagnosed with grade five vesicoureteral reflux in infancy with a dysplastic kidney on the left. He experienced absence seizures at 3 months of age and again at 9 years. An EEG at 9 months of age was normal. A computed tomography scan of the brain at three months showed delayed myelination. A brain MRI at two and a half years showed hypoplastic olfactory nerves, a Klippel−Feil anomaly, and incomplete myelin maturation in the inferior frontal lobes and temporal tips. Growth parameters were at or below the third percentile throughout childhood. He was hypotonic and there was a history of severe constipation. All of his developmental milestones were delayed. At about two and a half years, he was diagnosed with ASD. At about 11 years of age, he had symptoms of anxiety and Oppositional Defiant Disorder (ODD) although no official diagnosis was given. He was trialled on several anti-anxiety medications with no effect and is currently on resperidone. He has received extensive behavioural therapy from the time of ASD diagnosis to the present.
Clinical genetic assessments at 3 and 5 years of age did not identify a specific genetic diagnosis. Clinical CMA, fragile X testing, and metabolic testing were reported to be normal. We identified a de novo KMT2A frameshift variant, c.6169del (p.Val2057Tyrfs*18; Supplementary Fig. 1) via WGS. He was then clinically re-assessed at age 13 and diagnosed with WSS on the basis of characteristic facial features (Fig. 1c), microcephaly, hypertrichosis and history of growth and developmental delay, hypotonia, constipation, and ASD (details in Supplementary Table 2).
As a part of this study, neurodevelopmental testing at 13 years and 1 month of age (Table 1) confirmed previous diagnoses of ASD and ID. The assessment also identified emotional dysregulation and extremely low language/vocabulary and adaptive skills and symptoms of anxiety and ODD as reported previously. Concerns with attention were also endorsed by parents. Of note, he has significant difficulty with restricted and repetitive behaviours as identified by scores on the ADOS-2, as well as observations during the assessment. He struggled with transitions between settings and activities, becoming easily upset and requiring frequent breaks from work. Despite overall difficulties with social-communication, he demonstrates emerging skills, such as interest and engagement in social interactions as observed clinically and on multiple items on the ADOS-2.
Patient 4
A 25-year-old Caucasian male who was born to non-consanguineous Caucasian parents with a birth weight of 6 lb, 11 oz. has a history of short stature with growth consistently at the third percentile, hypotonia and delayed motor milestones. He was diagnosed with ASD (Asperger syndrome) at 10 years of age and had hyperlexia and symptoms of Tourette syndrome, specifically verbal tics.
Chromosomal microarray was performed on a research basis and the results were normal. After we identified a de novo KMT2A frameshift variant, c.7695_7696del (p.Glu2566Lysfs*14; Supplementary Fig. 1) via WGS, he was clinically re-assessed at age 25 and diagnosed with WSS on the basis of characteristic facial features (Fig. 1d), history of generalized hypertrichosis, and history of growth and developmental delay, cryptorchidism, hypotonia, and ASD (Supplementary Table 2).
Neurodevelopmental testing, as part of this study, at 25 years, 3 months of age (Table 1) confirmed previous diagnoses of ASD. He has developed good social-communication skills over the years, which contributed to an ADOS-2 score below clinical cut-off. However, based on overall clinical impressions he continues to meet criteria for ASD with notable difficulties being flexible in conversations, providing insight into others’ emotions, and understanding subtle nuances in social situations. Abnormality in speech rhythm, intonation and volume were also noted. This assessment identified borderline cognitive skills, low average adaptive skills, and average language and vocabulary skills. No concerns around anxiety, attention, aggression and emotional regulation were endorsed by parents or through clinical observation.
Patient 5
A 5-year-old Caucasian male with a history of ASD, growth and developmental delay, microcephaly, hypotonia, and esotropia was born at term to non-consanguineous Caucasian parents and had a birth weight of 6 lb, 7 oz. At 2 months of age he was hospitalized for investigation of nonepileptic paroxysmal events, with recurrent agitation, fist clenching, movement of arms and legs and screaming. Investigations including EEG and barium swallow were reported to be normal. At 3 months of age he began experiencing feeding difficulties with poor growth (weight below the third percentile). Due to ongoing feeding difficulties, a G-tube was inserted at 11 months. A brain MRI at 11 months identified cystic lesions in the pineal region and the pituitary fossa. Repeat MRI at 3 years also noted a dysplastic corpus callosum, hypoplastic optic nerves and a Klippel-Feil anomaly. His medical history is also remarkable for microcephaly, hypotonia, esotropia, constipation, bilateral orchidopexy and surgery for a tongue-tie release. All of his developmental milestones were delayed. He was subsequently diagnosed with ASD at 5 years of age and is on the waitlist for behavioural therapy.
Initial clinical genetics assessment at 8 months of age included clinical CMA, metabolic investigations and molecular testing for Prader−Willi syndrome and spinal muscular atrophy, which were all negative. At 19 months of age, a gene panel of 392 ID genes (University of Chicago) identified a maternally inherited variant in CHRNA4 and not suspected to be clinically significant. To date, he does not have evidence of seizures. At three and a half years of age WES was clinically requested and identified a de novo missense variant in KMT2A, c.8543 T > C (p.Leu2848Pro). He was clinically re-assessed at 5 years of age and noted to have facial features characteristic for WSS (Fig. 1e), generalized hypertrichosis and the history described above (Supplementary Table 2).
Neurodevelopmental testing, as part of this study, was conducted at 5 years, 1 month of age (Table 1) and confirmed the previous diagnosis of ASD. Based on the ADOS-2, ADI-R, and clinical observations, he had most difficulty with flexibility, following another person’s lead, and sensory-seeking behaviour. Despite difficulties with areas of social-communication, he demonstrated motivation for social interaction and appropriate use of facial expressions. This assessment also identified extremely low cognitive, language and adaptive skills, leading to a diagnosis of ID. Of note, when his demands are not met, he exhibits aggressive and self-injurious behaviours as reported on the Child Behavior Checklist (CBCL) and ADI-R. We observed concerns around attention and scores on the CBCL were significantly elevated. Assessment of emotional regulation showed significantly elevated scores, indicating dysregulation.
Patient 6
A 5-year-old Caucasian male who was born to non-consanguineous Caucasian parents at term by Caesarian section with a birth weight of 7 lb, 11 oz. had poor growth in infancy, with height and weight below the third percentile. His medical history is remarkable for feeding difficulties with gastroesophageal reflux, a ventricular septal defect (which closed spontaneously), hypotonia, severe constipation (requiring hospitalization for bowel cleanout on several occasions) and recurrent urinary tract and upper respiratory infections. At 22 months of age he experienced seizures during the process of bowel cleanout. These were suspected to be related to hypoglycemia secondary to fasting. Investigations did not identify a metabolic aetiology for the hypoglycemia. An EEG identified abnormal epileptiform discharges. MRI identified a pineal cyst, craniocervical junction stenosis and a Klippel−Feil anomaly. He subsequently experienced several seizure-like episodes with eye-rolling and arm extension in association with intercurrent illnesses and stress related to medical procedures. A repeat EEG was reported to be normal. It was suspected that these episodes may be due to atypical vasovagal syncope. All of his motor milestones were delayed. A school behavioural assessment at 4 years of age noted concerns around non-compliance, physical aggression and tantrums, disruptive behaviours, and touching/taking other’s possessions. No diagnosis was given at this time, but extensive accommodations were implemented at school.
Initial clinical genetics assessment at 23 months of age included clinical CMA, and molecular testing for Russell−Silver syndrome and Smith−Lemli Opitz syndrome, which were all negative. He was re-assessed at 31 months of age and based on the observation of hypertrichosis (arms and back), dysmorphic facial features, failure to thrive and constipation, targeted testing of KMT2A was requested clinically, which identified a de novo nonsense variant, c.8095 C > T (p.Arg2699*). He was clinically re-assessed at 5 years of age as part of this study and noted to have facial features characteristic for WSS (Fig. 1f), generalized hypertrichosis and the history described above (see Supplementary Table 2).
His first neurodevelopmental assessment was conducted as part of this study at the age of 5 years, 9 months (Table 1), at which time he received a diagnosis of ASD and ID. He met criteria for ASD based on the ADOS-2, ADI-R and clinical judgement. He had most difficulty with repetitive interests, fixations with items, and transitions and changes in routine. He demonstrated shared enjoyment during social interactions, appropriate eye contact and facial expressions, and was able to point to objects of interest, indicating a relative strength in social-communication. This assessment also identified extremely low language and adaptive skills, but relative strength in receptive vocabulary skills. Of note, he demonstrates rigidity and poor flexibility in his behaviour and difficulty regulating his emotions, becoming easily upset and anxious when things are not done his way, which can lead to aggressive behaviours. Concerns around attention were significantly elevated based on the CBCL and clinical observations. He is currently on a waitlist for behavioural therapy.
Nuclear family assessments
In all patients, assessment of cognitive functioning of parents (N = 11) and all siblings (N = 4) identified an IQ ranging from average to superior. Symptoms of anxiety were commonly endorsed by parents, although we did not perform formal anxiety diagnostic assessments. In both patients where the proband had a diagnosis of ADHD, one parent reported having attention difficulties in themselves. In one of these families a sibling also had a diagnosis of ADHD, although did not report current symptoms. In five of the six patients (except for patient 4), ASD traits were present in at least one parent, although we did not perform formal ASD diagnostic assessments. There were no ASD traits identified in any of the siblings and diagnostic testing for ASD was not warranted. No other obvious familial patterns impacting neurodevelopment were identified within or across the six patients.
Meta-analysis of KMT2A variants
We conducted a meta-analysis to identify other KMT2A variants in individuals with WSS and/or NDDs. Including our six patients, we identified 127 individuals with WSS or NDD and de novo KMT2A LoF and missense variants (Fig. 3 and Supplementary Data). Using KMT2A variants from WES and WGS studies (Supplementary Table 1 and Supplementary Data), we found that individuals ascertained for NDDs were significantly enriched for de novo LoF and missense variants (p = 6.15 × 10−4) and de novo LoF variants alone (p = 1.92 × 10−3) when compared with controls, but not for de novo missense variants alone (p = 0.321) (Table 2). The lack of statistical significance for de novo missense variants might have been due to the decreased power from the limited number of control samples used to detect the smaller effect sizes of missense variants. Therefore, we also determined whether our observed numbers of de novo LoF and missense variants among individuals with NDD were greater than those expected based on a statistical model of de novo LoF and missense mutation rates for this gene, as determined previously.13 The observed numbers of de novo LoF variants (p = 5.96 × 10−69), de novo missense variants (p = 0.016), and both combined (p = 8.92 × 10−41) were all significantly increased in the NDD cohort, compared with the expected number of de novo LoF and/or missense variants derived from the mutation rate of KMT2A13 (Table 2).
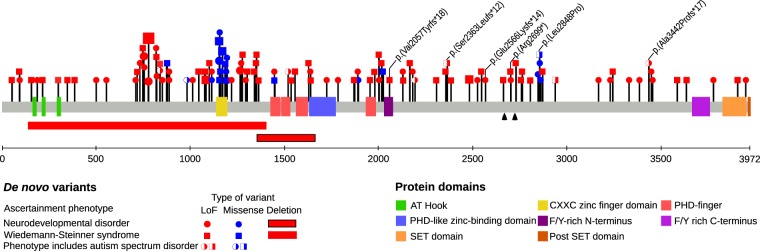
Fig. 3.
Distribution of de novo LoF, missense, and deletion variants in KMT2A from novel cases and individuals from meta-analysis. One hundred and three de novo LoF, 24 de novo missense, and two de novo deletion variants are plotted on the KMT2A protein (NM_001197104.1) with the protein coordinates below. The six new patients are shown with Human Genome Variation Society nomenclature. The two black arrows indicate the two taspase 1 cleavage sites that generate the N-terminal and C-terminal fragment. Size of the circle or square is proportional to the number of individuals with that specific KMT2A variant. LoF: loss-of-function
Table 2.
Evaluation of different variant classes in KMT2A in individuals with neurodevelopmental disorders (NDDs)
| Count (%) | p value | |||
|---|---|---|---|---|
| Individuals with NDDs | Control | vs. controla | vs. expected number of variantsb | |
| SNV/indel | (n = 15,070) | (n = 2299) | ||
| De novo LoF | 44 (0.29%) | 0 (0%) | 1.92 × 10−3 | 5.96 × 10−69 |
| De novo missense | 8 (0.05%) | 0 (0%) | 0.321 | 0.016 |
| Combined | 52 (0.35%) | 0 (0%) | 6.15 × 10−4 | 8.92 × 10−41 |
| CNV | (n = 5815 c) | (n = 46,652) | ||
| Deletions | 2 | 2 | 0.063 | NA |
| Duplications | 2 | 7 | 0.19 | NA |
aFisher’s exact test, one-sided
bBinomial test. Expected number of variants for 15,070 individuals was determined using the de novo mutation rate model in Samocha et al.
cFor deletions, n = 5815 and for duplications, n = 4681
To determine whether the de novo LoF and missense variants assort to certain regions of the KMT2A protein according to associated phenotypes, we examined the distribution of these variants. We found de novo LoF and missense variants to be dispersed throughout KMT2A and no obvious genotype−phenotype correlation (with respect to WSS, NDDs and ASD) (Fig. 3). There is a cluster of 13 de novo missense variants in the CXXC zinc finger binding domain, of which p.(Arg1154Trp), p.(Cys1155Tyr), and p.(Gly1168Asp) are recurrent variants found at similar frequencies in individuals with WSS or NDDs (Fig. 3 and Supplementary Table 3). With regards to copy number variants (CNVs), deletions and duplications (p = 0.063 and 0.19, respectively) in KMT2A were not significantly enriched compared with controls (Table 2, Supplementary Fig. 2).
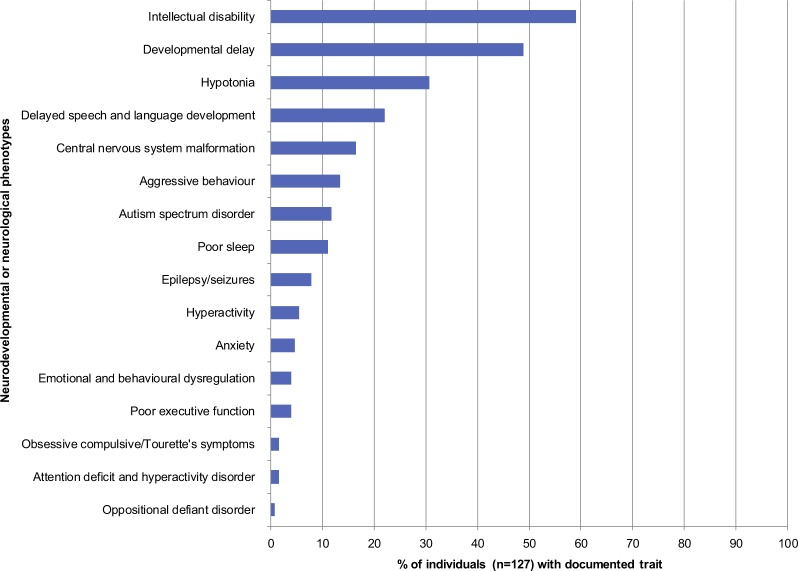
To dissect the neurodevelopmental spectrum of de novo KMT2A variants, we examined the reported phenotypes of the 127 individuals with de novo LoF or missense variants in KMT2A. The top five reported phenotypes were ID (59.1%), developmental delay (48.8%), hypotonia (30.7%), delayed speech and language development (22.0%), and malformations in central nervous system (16.5%) (Fig. 4). ASD was described in 11.8% of individuals (Fig. 4). Of note, detailed phenotyping was not available for the majority of individuals. ADHD, ODD, Tourette Syndrome, and obsessive compulsive symptoms were observed in our patients and have not been previously reported in individuals with de novo KMT2A variants.
Fig. 4.
Spectrum of reported neurodevelopmental and neurological phenotypes of 127 individuals with de novo LoF or missense variants in KMT2A. Frequencies of neurodevelopmental and neurological phenotypes reported in publications are shown. Note that absence of phenotype from a publication either means the phenotype was not assessed or there was a negative finding, which we could not distinguish in most publications. LoF: loss-of-function
Discussion
We describe six individuals with WSS and a characteristic spectrum of NDDs with de novo LoF or missense variants in KMT2A. We did not identify other clinically significant variants that contributed to the observed NDD phenotypes, suggesting that heterozygous mutations in KMT2A alone is sufficient to explain the observed NDD phenotypes.14 In line with recent reports that identified KMT2A as a candidate gene for ASD11,15 and NDDs,8,12 our findings support KMT2A as a high confidence NDD susceptibility gene similar to ARID1B and KMT2C. De novo LoF and missense variants in KMT2A are highly penetrant and can explain the neurodevelopmental and phenotypic features of WSS. This is supported by a high constraint for LoF and missense mutations in KMT2A (the probability of LoF intolerance score is 1.00 and the z-score for missense mutations is 6.64 16). Collectively, our findings can contribute to increased molecular diagnostic yields for NDDs and ASD.17,18
Prior to identifying the KMT2A variants, patients 1−5 had a diagnosis of NDD (three were ascertained for ASD), but no identified syndrome. Only patient 6 had a clinical diagnosis of WSS, which was confirmed molecularly. After identification of KMT2A variants and clinical reassessment, patients 1−5 met the criteria for WSS, suggesting that WSS is often under-recognized. With the increased utilization of WES/WGS in the clinical and research settings, we anticipate that the diagnostic rate of WSS will increase.7,19
Prior to this study, the NDD phenotype in individuals with KMT2A variants had not been systematically assessed using specific neuropsychological tests. A recent paper by Baer et al. highlighted the need for more in-depth NDD phenotyping.7 Our deep phenotyping analysis delineated a particular ASD subtype in association with KMT2A variants. Poor language skills/delayed speech has been previously reported in individuals with KMT2A variants. The six individuals in our study were able to speak in short phrases or full sentences, giving the impression of higher abilities than were found on direct assessments. Specifically, vocabulary, higher-order language skills and cognitive functioning of five of our patients were delayed. The emerging ASD subtype included social motivation to interact with others (despite social-communication difficulties), paired with rigid, repetitive and inflexible behaviours (e.g. difficulty switching from preferred topics), emotional dysregulation and externalizing behaviours that limited their ability to sustain social interactions. Although some ASD features were present in at least one parent of the four out of five patients with ASD, autistic traits have been shown to exist in a continuous distribution in the general population with family members of individuals with ASD having a greater likelihood of exhibiting subclinical autistic traits.20–22 We did not find clinically significant inherited rare variants that contributed to ASD risk in our patients. This does not rule out additional genetic factors that may be contributing to ASD risk in these families,14 such as common variants, rare variants with smaller effect sizes, noncoding variants, other structural variants, or variants whose contribution is still unknown. Nevertheless, our findings suggest that de novo LoF and missense variants in KMT2A are associated with WSS, a syndromic form of NDD with increased risk of a particular ASD subtype, providing further delineation of the neurodevelopmental phenotype associated with WSS.
Functional studies in conditional knockout mice deficient in Kmt2a in various brain regions also support a role for KMT2A in neurodevelopment. Kmt2a is known to regulate genes related to neurogenesis23 and is required for neurogenesis in neural stem cells in the subventricular zone.3 Moreover, neurons from Camk-Cre+ Kmt2a2lox/2lox mice exhibit significant changes in H3K4me3 and gene expression levels in orthologues of neuropsychiatric genes,24 and significant changes in expression of genes related to the synapse, axon, membrane,24 cognition and mood.25 These findings are reflected in the behavioural phenotypes in the mutant mice: ablation of Kmt2a in the forebrain resulted in increased anxiety, hyperactivity, spatial working memory deficits, and impaired social behaviours,24,25 whereas ablation of Kmt2a in the striatum, it resulted in increased anxiety.25 Some of these phenotypes (i.e. impaired social behaviours, anxiety, and hyperactivity) are also observed in individuals carrying de novo KMT2A variants.
Skeletal anomalies are a known feature of WSS with a recent study documenting Klippel−Feil anomaly in five patients with WSS and de novo KMT2A variants.7 Among our six patients, four have Klippel−Feil anomaly. Skeletal phenotypes have also been observed in Kmt2a+/− mice, including fusion of cervical vertebrae.26 In mouse embryonic fibroblasts, Kmt2a regulates the expression and H3K4 trimethylation of Gdf6 (a gene involved in bone morphogenesis that causes Klippel–Feil Syndrome (KFS)), Pax1 (KFS candidate gene27), and Pax9 (a functionally relevant gene in vertebrate segmentation28). WES or WGS in patients 2, 3, and 5 did not identify variants in genes known to be associated with KFS (GDF6, MEOX1, GDF3, MYO18B), except for a maternally inherited missense variant in PAX1 in patient 3 (Table 1). These findings suggest that skeletal anomalies may be a more common feature of WSS than previously reported, due to dysregulation of genes involved in bone morphogenesis, as a result of KMT2A dysregulation.
In conclusion, we have demonstrated that individuals with de novo LoF or missense variants in KMT2A likely have a clinically unrecognized diagnosis of WSS, rather than non-syndromic NDD. Our findings further characterize the neurodevelopmental phenotype associated with WSS, revealing a characteristic subtype of ASD. We recommend that any individual with a de novo KMT2A LoF or missense variant undergo both clinical genetics and neurodevelopmental assessments.
Methods
Study participants
Participants for the clinical report series were recruited at the Hospital for Sick Children. Four of the six patients were identified as part of an ongoing research study investigating the genetic aetiology of ASD and neurodevelopmental conditions. The remaining two patients were identified through the Division of Clinical and Metabolic Genetics at the Hospital for Sick Children. Written informed consent approved by the Research Ethics Board at The Hospital for Sick Children was obtained for the study. The authors affirm that human research participants provided informed consent for open access publication of the images in figures. All six families underwent a clinical assessment by a geneticist (R.W.) and a neurodevelopmental assessment supervised by a psychologist (I.D. and A.R.). The clinical genetics assessment involved a review of the proband’s medical and family histories and a physical dysmorphology examination. The neurodevelopmental assessment involved psychological assessment of the proband and available first-degree relatives as described below.
Neurodevelopmental phenotype
Probands were assessed using validated, standardized multi-method approaches and assessments. Where available, and still valid (i.e. completed within past 2 years), the scores from assessments completed at other clinics were used to decrease test burden. Direct (individualized assessments, interviews) and indirect (questionnaires) assessments used to evaluate ASD, cognitive functioning, language skills (including receptive and expressive), vocabulary (receptive and expressive), executive functioning skills, behavioural and emotional concerns (specifically anxiety/depressive symptoms, attention concerns, obsessive compulsive tendencies, emotional dysregulation, externalizing behaviours), and adaptive functioning are described in supplementary methods. The available nuclear family members of each patient were assessed using validated, standardized multi-method approaches to evaluate any familial patterns or impacts on neurodevelopment. Areas evaluated included cognitive functioning, vocabulary (receptive and expressive), ASD traits, and behavioural and emotional concerns.
WGS analysis for patients 1, 3, and 4
Through the Autism Speaks MSSNG portal,11 we screened WGS data of 2414 ASD parent−child trios for rare (<1% in population control databases) de novo LoF and damaging missense (predicted damaging in five of seven prediction algorithms as previously described29) variants in KMT2A (NM_0011904.1). We selected those with a genotype quality score ≥99 and with 30–70% of reads supporting the alternative allele, for validation using Sanger sequencing. Rare SNVs and indels were downloaded from MSSNG and annotated as previously described.11 We used a previously recommended workflow30 to detect CNVs larger than 1 kb from WGS data, using read depth-based algorithms: ERDS and CNV-nator. We calculated an internal frequency filter using the ERDS and CNVnator calls of the 4828 unaffected parents in MSSNG and prioritized rare (<1% allele frequency in unaffected parents) CNVs.
WES analysis for patient 2
We sequenced DNA from whole blood of patient 2 using Illumina HiSeq2500, after exome enrichment with SureSelect V5 Capture Kit (Aligent). We used Burrows−Wheeler Aligner v.0.5.9 31 to align WES reads, Genome Analysis Toolkit Haplotype caller v.1.1-28 32 to call variants. We selected variants relevant to NDDs (discussed below) for Sanger sequencing confirmation using DNA extracted from the blood of patient 2 and her parents.
Identifying clinically significant variants
To identify clinically significant variants in patients 1−4, we prioritized rare (<1% allele frequency in population controls) LoF or missense variants predicted to be damaging by at least five of seven criteria, as previously described,29 and variants reported in ClinVar33 and the Human Gene Variant Database.34 We also prioritized rare CNVs (<1% allele frequency in population controls and <1% allele frequency in parents from the MSSNG database), including those overlapping syndromic regions in ClinGen Genome Dosage Sensitivity Map35 databases, which contains expert-curated CNVs involved in developmental disorders. Genes affected by such variants were compared with ASD candidate genes,11,12,36–38 candidate genes for NDDs,12 and genes implicated in neurodevelopmental or behavioural phenotypes according to the Human and Mouse Phenotype Onotologies.39,40 Additionally, we considered the mode of inheritance from the Online Mendelian Inheritance in Man and Clinical Genomics Database41 and the segregation in the family. Then we classified the variants as clinically significant (pathogenic or likely pathogenic) or of unknown significance, based on the American College of Medical Genetics and Genomics (ACMG) guidelines.42,43
Meta-analysis of KMT2A variants
We conducted a literature search on the PubMed database for articles published between January 2011 and August 2018, to gather additional data from individuals with WSS or NDDs and variants in KMT2A. Search terms were: KMT2A, MLL, WSS, Wiedemann−Steiner Syndrome, ID, autism spectrum disorder, developmental delay, epilepsy, ADHD, OCD, schizophrenia, bipolar disorder, whole exome sequencing, and whole genome sequencing. We found 37 publications that identified KMT2A variants in individuals with WSS or NDDs (Supplementary Table 1).
To determine whether there was an enrichment of de novo KMT2A variants in NDD patients, we used WGS data from 250 Dutch parent−offspring trios44 and WES data from 138 parent−offspring trios45–48 and 1911 unaffected siblings46 (Supplementary Table 1) as controls. The total number of control trios was 2299.
We also screened a microarray database of 4681 ASD37 and 15,812 control individuals (unpublished data; and see Zarrei et al.49 for details about published controls) for CNVs that overlapped KMT2A. We considered rare CNVs (≤0.1% allele frequency in controls) that overlapped KMT2A and were called by two algorithms within the same microarray platform and validated these with Taq-Man Copy Number Assays. We also searched the Database of Genomic Variants50 for additional CNVs impacting KMT2A in controls.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The authors thank The Centre for Applied Genomics at the Hospital for Sick Children for technical assistance and helpful discussions. Supported by Autism Speaks, Genome Canada, Canada Foundation for Innovation, Canadian Institute for Advanced Research (CIFAR), Ontario Brain Institute, Government of Ontario, Canadian Institutes of Health Research, The Hospital for Sick Children and University of Toronto McLaughlin Centre. The Autism Speaks MSSNG-genome sequencing and open science platform is supported by Autism Speaks and partners. S.W.S. holds the GlaxoSmithKline-CIHR Chair in Genome Sciences at The Hospital for Sick Children and University of Toronto. Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome‐wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The authors acknowledge the contribution of data from Genetic Architecture of Smoking and Smoking Cessation accessed through dbGaP. Funding support for genotyping, which was performed at the Center for Inherited Disease Research (CIDR), was provided by 1X01 HG005274‐01. CIDR is fully funded through a federal contract from the National Institutes of Health to The Johns Hopkins University, contract number HHSN268200782096C. Assistance with genotype cleaning, as well as with general study coordination, was provided by the Gene Environment.
Author contributions
A.J.S.C., S.W.S., M.U., and R.K.C.Y. conceived the clinical reports. A.J.S.C. conducted the meta-analysis, statistical analysis, and wrote first draft of the manuscript. C.C., N.H., P.M.A., R.W., I.D., A.R., and R.S. performed the clinical genetics, neurodevelopmental assessments, and interpretation of clinical phenotype data. A.J.S.C., S.W., M.U., and M.Z. were involved in analyzing genetics data. A.J.S.C., M.Z., and R.K.C.Y. were involved in data interpretation. A.J.S.C., M.Z., R.K.C.Y., C.C., N.H., R.W., I.D., R.S. and S.W.S. contributed to the preparation of this manuscript. All authors read and approved the final manuscript.
Data availability
WGS data can be accessed through the Autism Speaks MSSNG database (for access, see http://www.mss.ng/researchers). WGS data for patients 1, 3, and 4 and WES data for patient 2 are deposited in the European Genome-phenome Archive (www.ebi.ac.uk/ega/) under accession number EGSA00001003521. CNV control datasets used for the analyses described in this manuscript were obtained from dbGaP at https://www-ncbi-nlm-nih-gov.myaccess.library.utoronto.ca/projects/gap/cgi517bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p1. Datasets supporting the conclusions of this article are included within the article, supplementary tables, and supplementary data. Other data that support the findings of this study are available from the corresponding author on reasonable request.
Competing interests
S.W.S. is on the Scientific Advisory committees of Population Bio and Deep Genomics. Athena Diagnostics and Lineagen license intellectual property from the Hospital for Sick Children based on research from his laboratory. These relationships did not influence data interpretation or presentation during this study, but are still being disclosed for potential future considerations. The other authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies the paper on the npj Genomic Medicine website (10.1038/s41525-019-0083-x).
References
- 1.Rao RC, Dou Y. Hijacked in cancer: the KMT2 (MLL) family of methyltransferases. Nat. Rev. Cancer. 2015;15:334–346. doi: 10.1038/nrc3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muntean AG, Hess JL. The pathogenesis of mixed-lineage leukemia. Annu. Rev. Pathol. 2012;7:283–301. doi: 10.1146/annurev-pathol-011811-132434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lim DA, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- 5.Jones WD, et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am. J. Hum. Genet. 2012;91:358–364. doi: 10.1016/j.ajhg.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyake N, et al. Delineation of clinical features in Wiedemann-Steiner syndrome caused by KMT2A mutations. Clin. Genet. 2016;89:115–119. doi: 10.1111/cge.12586. [DOI] [PubMed] [Google Scholar]
- 7.Baer S, et al. Wiedemann−Steiner syndrome as a major cause of syndromic intellectual disability: a study of 33 French cases. Clin. Genet. 2018;94:141–152. doi: 10.1111/cge.13254. [DOI] [PubMed] [Google Scholar]
- 8.Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–438. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Euro ERESC, Epilepsy Phenome/Genome P, Epi KC. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am. J. Hum. Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lelieveld SH, et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 11.Yuen R, et al. Whole genome sequencing resource identifies 18 new candidate genes for autism spectrum disorder. Nat. Neurosci. 2017;20:602–611. doi: 10.1038/nn.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Mantilla AJ, Moreno-De-Luca A, Ledbetter DH, Martin CL. A cross-disorder method to identify novel candidate genes for developmental brain disorders. JAMA Psychiatry. 2016;73:275–283. doi: 10.1001/jamapsychiatry.2015.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samocha KE, et al. A framework for the interpretation of de novo mutation in human disease. Nat. Genet. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang N, Cytrynbaum C, Scherer SW. Communicating complex genomic information: a counselling approach derived from research experience with Autism Spectrum Disorder. Patient Educ. Couns. 2018;101:352–361. doi: 10.1016/j.pec.2017.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Hoang N, Buchanan JA, Scherer SW. Heterogeneity in clinical sequencing tests marketed for autism spectrum disorders. NPJ Genom. Med. 2018;3:27. doi: 10.1038/s41525-018-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tammimies K, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903. doi: 10.1001/jama.2015.10078. [DOI] [PubMed] [Google Scholar]
- 18.Carter MT, Scherer SW. Autism spectrum disorder in the genetics clinic: a review. Clin. Genet. 2013;83:399–407. doi: 10.1111/cge.12101. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez BA, Scherer SW. Syndromic autism spectrum disorders: moving from a clinically defined to a molecularly defined approach. Dialogues Clin. Neurosci. 2017;19:353–371. doi: 10.31887/DCNS.2017.19.4/sscherer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Constantino JN, Todd RD. Autistic traits in the general population: a twin study. Arch. Gen. Psychiatry. 2003;60:524–530. doi: 10.1001/archpsyc.60.5.524. [DOI] [PubMed] [Google Scholar]
- 21.Whitehouse AJ, Hickey M, Ronald A. Are autistic traits in the general population stable across development? PLoS ONE. 2011;6:e23029. doi: 10.1371/journal.pone.0023029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piven J, Palmer P, Jacobi D, Childress D, Arndt S. Broader autism phenotype: evidence from a family history study of multiple-incidence autism families. Am. J. Psychiatry. 1997;154:185–190. doi: 10.1176/ajp.154.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Potts MB, et al. Analysis of Mll1 deficiency identifies neurogenic transcriptional modules and Brn4 as a factor for direct astrocyte-to-neuron reprogramming. Neurosurgery. 2014;75:472–482. doi: 10.1227/NEU.0000000000000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakovcevski M, et al. Neuronal Kmt2a/Mll1 histone methyltransferase is essential for prefrontal synaptic plasticity and working memory. J. Neurosci. 2015;35:5097–5108. doi: 10.1523/JNEUROSCI.3004-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen EY, et al. Neuronal deletion of Kmt2a/Mll1 histone methyltransferase in ventral striatum is associated with defective spike-timing-dependent striatal synaptic plasticity, altered response to dopaminergic drugs, and increased anxiety. Neuropsychopharmacology. 2016;41:3103–3113. doi: 10.1038/npp.2016.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayton P, et al. Truncation of the Mll gene in exon 5 by gene targeting leads to early preimplantation lethality of homozygous embryos. Genesis. 2001;30:201–212. doi: 10.1002/gene.1066. [DOI] [PubMed] [Google Scholar]
- 27.McGaughran JM, Oates A, Donnai D, Read AP, Tassabehji M. Mutations in PAX1 may be associated with Klippel−Feil syndrome. Eur. J. Hum. Genet. 2003;11:468–474. doi: 10.1038/sj.ejhg.5200987. [DOI] [PubMed] [Google Scholar]
- 28.Tracy MR, Dormans JP, Kusumi K. Klippel-Feil syndrome: clinical features and current understanding of etiology. Clin. Orthop. Relat. Res. 2004;424:183–190. doi: 10.1097/01.blo.0000130267.49895.20. [DOI] [PubMed] [Google Scholar]
- 29.Yuen RK, et al. Genome-wide characteristics of de novo mutations in autism. NPJ Genom. Med. 2016;1:160271–1602710. doi: 10.1038/npjgenmed.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trost B, et al. A comprehensive workflow for read depth-based identification of copy-number variation from whole-genome sequence data. Am. J. Hum. Genet. 2018;102:142–155. doi: 10.1016/j.ajhg.2017.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows−Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Landrum MJ, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stenson PD, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 35.Rehm HL, et al. ClinGen—the clinical genome resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanders SJ, et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pinto D, et al. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrahams BS, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol. Autism. 2013;4:36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blake JA, et al. The Mouse Genome Database: integration of and access to knowledge about the laboratory mouse. Nucleic Acids Res. 2014;42:D810–D817. doi: 10.1093/nar/gkt1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohler S, et al. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomon BD, Nguyen AD, Bear KA, Wolfsberg TG. Clinical genomic database. Proc. Natl. Acad. Sci. USA. 2013;110:9851–9855. doi: 10.1073/pnas.1302575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Richards S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearney HM, et al. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet. Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 44.Genome of the Netherlands C. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet. 2014;46:818–825. doi: 10.1038/ng.3021. [DOI] [PubMed] [Google Scholar]
- 45.Gulsuner S, et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell. 2013;154:518–529. doi: 10.1016/j.cell.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–1682. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 48.Xu B, et al. De novo gene mutations highlight patterns of genetic and neural complexity in schizophrenia. Nat. Genet. 2012;44:1365–1369. doi: 10.1038/ng.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zarrei M, et al. De novo and rare inherited copy-number variations in the hemiplegic form of cerebral palsy. Genet. Med. 2018;20:172–180. doi: 10.1038/gim.2017.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacDonald JR, Ziman R, Yuen RK, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WGS data can be accessed through the Autism Speaks MSSNG database (for access, see http://www.mss.ng/researchers). WGS data for patients 1, 3, and 4 and WES data for patient 2 are deposited in the European Genome-phenome Archive (www.ebi.ac.uk/ega/) under accession number EGSA00001003521. CNV control datasets used for the analyses described in this manuscript were obtained from dbGaP at https://www-ncbi-nlm-nih-gov.myaccess.library.utoronto.ca/projects/gap/cgi517bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p1. Datasets supporting the conclusions of this article are included within the article, supplementary tables, and supplementary data. Other data that support the findings of this study are available from the corresponding author on reasonable request.