Abstract
HepG2 is one of the most widely used human cancer cell lines in biomedical research and one of the main cell lines of ENCODE. Although the functional genomic and epigenomic characteristics of HepG2 are extensively studied, its genome sequence has never been comprehensively analyzed and higher order genomic structural features are largely unknown. The high degree of aneuploidy in HepG2 renders traditional genome variant analysis methods challenging and partially ineffective. Correct and complete interpretation of the extensive functional genomics data from HepG2 requires an understanding of the cell line’s genome sequence and genome structure. Using a variety of sequencing and analysis methods, we identified a wide spectrum of genome characteristics in HepG2: copy numbers of chromosomal segments at high resolution, SNVs and Indels (corrected for aneuploidy), regions with loss of heterozygosity, phased haplotypes extending to entire chromosome arms, retrotransposon insertions and structural variants (SVs) including complex and somatic genomic rearrangements. A large number of SVs were phased, sequence assembled and experimentally validated. We re-analyzed published HepG2 datasets for allele-specific expression and DNA methylation and assembled an allele-specific CRISPR/Cas9 targeting map. We demonstrate how deeper insights into genomic regulatory complexity are gained by adopting a genome-integrated framework.
INTRODUCTION
Genomic instability is a hallmark of cancer where critical genomic changes create gene fusions, the disruption of tumor-suppressor and the amplification of oncogenes (1–3). A comprehensive knowledge of the mutations and larger structural changes that underlie a cancer genome is critical not only for a deeper understanding of the biological processes that drive tumor progression and evolution, but also for the development of targeted cancer therapies. The HepG2 cell line is one of the most widely used cancer cell lines used in many areas of biomedical research due to its extreme versatility, contributing to over 23 000 publications to date, even more than K562. It is a hepatoblastoma cell line derived from a 15-year-old male of European ancestry (4,5). Representing the human endodermal lineage, HepG2 cells are widely used as models for human toxicology studies (6–10), including toxicogenomic screens using CRISPR-Cas9 (11), in addition to studies on drug metabolism (12), cancer (13), liver disease (14), gene regulatory mechanisms (15) and biomarker discovery (16). As one of the main cell lines of the ENCyclopedia Of DNA Elements Project (ENCODE), HepG2 has been used to generate close to 1000 datasets for ENCODE (17).
The functional genomic and epigenomics aspects of HepG2 cells have been extensively studied with approximately 325 ChIP-Seq, 300 RNA-Seq and 180 eCLIP datasets available through ENCODE in addition to recent single-cell methylome and transcriptome datasets (18). However, the genome sequence and higher order genomic structural features of HepG2 have never been characterized in a comprehensive manner, even though the HepG2 cell line has been known to contain multiple chromosomal abnormalities (19,20). As a result, the extensive HepG2 functional genomics and epigenomics studies conducted to date were done without reliable genomic contexts for accurate interpretation.
Here, we report the first global, integrated and haplotype-resolved whole-genome characterization of the HepG2 cancer genome that includes copy numbers (CN) of large chromosomal regions at high-resolution, single-nucleotide variants (SNVs, also including single-nucleotide polymorphisms, i.e. SNPs) and small insertions and deletions (indels) with allele-frequencies corrected by CN in aneuploid regions, loss of heterozygosity, mega-base-scale phased haplotypes and structural variants (SVs), many of which are haplotype-resolved (Figure 1 and Supplementary Figure S1). The datasets generated in this study form an integrated, phased, high-fidelity genomic resource that can provide the proper contexts for future experiments that rely on HepG2’s unique characteristics. We show how knowledge about HepG2’s genomic sequence and structural variations can enhance the interpretation of functional genomics and epigenomics data. For example, we integrated HepG2 RNA-Seq data and whole-genome bisulfite sequencing data with ploidy and phasing information and identified many cases of allele-specific gene expression and allele-specific DNA methylation. We also compiled a phased CRISPR map of loci suitable for allele specific-targeted genome editing or screening. Finally, we demonstrate the power of this resource by providing compelling insights into the mutational history of HepG2 and oncogene regulatory complexity derived from our datasets. The technical framework demonstrated in this study is also suitable for the study of other cancer cell lines and primary tumor samples.
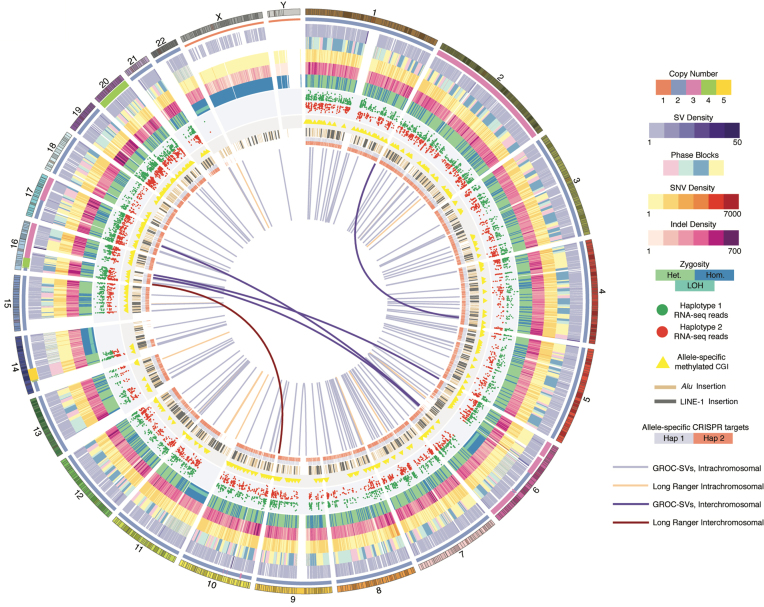
Figure 1.
Comprehensive Overview of the HepG2 Genome. Circos visualization of HepG2 genome variants with the following tracks in concentric order starting with outermost ‘ring’: human genome reference track (hg19); large CN changes (colors correspond to different CN, see legend panel); in 1.5 Mb windows, merged SV density (deletions, duplications, inversions) called using BreakDancer, BreakSeq, PINDEL, LUMPY and Long Ranger; phased haplotype blocks (demarcated with four colors for clearer visualization); SNV density in 1 Mb windows; Indel density in 1 Mb windows; dominant zygosity (heterozygous or homozygous > 50%) in 1 Mb windows; regions with loss of heterozygosity; allele-specific expression; CpG islands exhibiting allele-specific DNA methylation; non-reference LINE1 and Alu insertions; allele-specific CRISPR target sites; large-scale SVs resolved by using Long Ranger (peach: intrachromosomal: dark maroon: interchromosomal); by using GROC-SVs (light-purple: intrachromsomal; dark-purple: interchromosomal).
MATERIALS AND METHODS
HepG2 karyotyping and DNA extraction
HepG2 cells were acquired from the Stanford ENCODE Product Center for Mapping of Regulatory Regions (NHGRI Project 1U54HG006996-01). Karyotyping of HepG2 cells was conducted in the Cytogenetics Laboratory (cytogenetics.stanford.edu) at Stanford University Medical Center (Palo Alto, CA, USA), where 20 metaphase cells were analyzed by GTW banding. DNA extraction was performed using the Qiagen DNeasy Blood & Tissue Kit (Cat No. 69504), and concentration was measured using the Qubit dsDNA BR Assay Kit (Invitrogen, Waltham, MA, USA). Purity of DNA (OD260/280 > 1.8; OD260/230 > 1.5) was verified using NanoDrop (Thermo Scientific, Waltham, MA, USA). Using field-inversion gel electrophoresis on the Pippin Pulse System (Sage Science, Beverly, MA, USA), the extracted DNA was verified to be high molecular weight (mean > 35 kb).
CN of chromosome segments and allele frequencies of SNVs and Indels
Standard short-insert WGS (Supplementary Methods) coverage was calculated in 10-kb bins across the genome and plotted against the %GC content of each bin to verify the existence of discrete clusters corresponding to discrete CNs (Supplementary Figure S2). CN was assigned to a cluster based on the ratio of its mean coverage to that of the lowest cluster. For an example, the cluster with the lowest mean coverage was assigned CN1, and the cluster with twice as much mean coverage was assigned CN2 and so forth. The ratios for the five discrete clusters observed corresponded almost perfectly to CN1, CN2, CN3 and CN4. WGS coverage across the genome and across each chromosome was examined visually to assign CN for different chromosome segments or entire chromosomes based on the cluster analysis where adjacent chromosomal segments with different CNs could be identified by the clearly visible sharp and steep changes in sequencing coverage (Supplementary Figure S3 and Supplementary Data). For each chromosome segment, SNVs and Indels were called using by GATK Haplotypecaller (version 3.7) (21) by specifying the CN or ploidy of that chromosome segment (stand_emit_conf = 0.1, variant_index_type = LINEAR, variant_index_parameter = 128000, ploidy = {CN}). The resulting Haplotypecaller outputs from all chromosome segments were then concatenated, and variant quality scores were recalibrated using GATK VQSR with training datasets (dbSNP 138, HapMap 3.3, Omni 2.5 genotypes, 1000 Genomes Phase 1) as recommended by GATK Best Practices (22,23) and filtered with the setting tranche = 99.0. SNVs and Indels were annotated using dbSNP138 (24) followed by SnpEff (version 4.3; canonical transcripts) (25) and then filtered for protein altering variants using SnpSift (version 4.3; ‘HIGH’ and ‘MODERATE’ putative impact) (26). Protein-altering variants were intersected with the variants from the 1000 Genomes Project (27) and the Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) where overlapping variants were removed using Bedtools (version 2.26) (28). The resulting PPA variant calls were overlapped against the Catalogue of Somatic Mutations in Cancer (29) and Sanger Cancer Gene Census (30).
Identification of LOH
A Hidden Markov Model (HMM) was used to identify genomic regions exhibiting LOH. The HMM is designed with two states: LOH present and LOH absent. We used SNVs that were recalibrated and ‘PASS’-filtered from GATK VQSR as well as overlapped 1000 Genomes Project variants (31). The genome was split into 40-kb bins; heterozygous and homozygous SNVs were tallied for each bin, and bins with <12 SNVs were removed. A bin was classified as heterozygous if ≥50% of the SNVs within the bin are heterozygous; otherwise it was classified as homozygous. This classification was used as the HMM emission sequence. The HMM was initialized with the same initiation and transition probabilities (Prob = 10E-8) (3), and the Viterbi algorithm was used to estimate a best path. Adjacent LOH intervals were merged.
Haplotype phasing and variant analysis using linked-reads
Paired-end linked-reads (median insert size 396 bp, duplication rate 7.68%, Q30 Read1 78.7%, Q30 Read2 63.1%) were aligned to hg19 (alignment rate 90.4%, mean coverage 67.0x, zero coverage 0.117%) and analyzed using the Long Ranger Software (version 2.1.5) from 10x Genomics (32,33) (Pleasanton, CA, USA). Segmental duplications, reference gaps, unplaced contigs, regions with assembly issues and highly polymorphic sites (http://cf.10xgenomics.com/supp/genome/hg19/sv_blacklist.bed, http://cf.10xgenomics.com/supp/genome/hg19/segdups.bedpe) were excluded from the analysis. ENSEMBL annotations (http://cf.10xgenomics.com/supp/genome/gene_annotations.gtf.gz) were used for genes and exons. Phasing was performed by specifying the set of pre-called and filtered HepG2 heterozygous SNVs and Indels from GATK (see above) and formatted using mkvcf from Long Ranger (version 2.1.5). Heterozygous SNVs and Indels with more than two types of alleles in ploidy>2 regions were excluded from analysis. Large (>30 kb) SVs and large-scale complex rearrangements were identified using both the Long Ranger wgs module with the ‘–somatic’ option, GROC-SVs (default settings with breakpoint assembly) (34) and gemtools (35). The ‘–somatic’ option increases the sensitivity of the large-scale SV caller for somatic SVs by allowing the detection of sub-haplotype events and does not affect small-scale variant calling. Variants from Long Ranger analysis indicated as ‘PASS’ were retained. SV breakpoints identified using GROC-SVs were also analyzed for supporting evidence from mate-pair reads (see below). By using the method described in (36), the HepG2 haplotype-blocks identified from Long Ranger were ‘stitched’ to mega-haplotype blocks by leveraging the SNV haplotype imbalance in aneuploid regions where NA12878 linked-read sequencing data (https://support.10xgenomics.com/genome-exome/datasets/2.2.1/NA12878_WGS_v2) were used as the matching control. Only SNVs present in both genomes (HepG2 and NA12878) were included in the mega-haplotype blocks (Supplementary Data and Table 1). For details on linked-read library construction and allele-specific RNA expression, DNA methylation and CRISPR analysis, see Supplementary Methods.
Table 1.
Summary of HepG2 small variant calls and phasing results
| SNPs | INDELs | Phased WGS | |||
|---|---|---|---|---|---|
| All | 3 337 361 | 892 019 | % phased heterozygous SNPs | 99 | |
| Heterozygous/homozygous | 1 898 493/1 438 868 | 598 882/293 137 | % phased INDELs | 78 | |
| Protein altering | 11 460 (0.3%) | 1347 (0.2%) | Longest phase block | 31 106 135 | |
| dbSNP138 | 3 279 135 (98%) | 693 348 (78%) | Number of phase blocks | 1628 | |
| Heterozygous/homozygous | 1 845 345/1 433 790 | 439 143/254 205 | N50 phase block | 6 792 324 | |
| Novel | 58 226 (2%) | 198 671 (22%) | N50 Linked-reads per molecule | 61 | |
| Heterozygous/homozygous | 53 148/5078 | 159 739/38 932 | Barcodes detected | 1 532 287 | |
| 1000 Genomes Project & Exome Sequencing Project Overlap (with protein altering variants) | 11 083 (97%) | 1092 (81%) | Mean DNA per barcode (bp) | 633 889 | |
| Novel Protein Altering | 377 | 255 | |||
| COSMIC Overlap | 148 (39%) | 42 (16%) | |||
| Mega-haplotypes | |||||
| Chromosome | Start | End | Chromosome Arm | % of arm covered | P-value |
| 2 | 21 888 | 89 128 628 | 2p | 98% | 2.20E-16 |
| 2 | 98 803 025 | 243 046 591 | 2q | 98% | 2.20E-16 |
| 6 | 2 69 211 | 56 501 036 | 6p | 96% | 8.70E-07 |
| 6 | 62 383 957 | 170 631 019 | 6q | 99% | 3.92E-13 |
| 16 | 46 511 762 | 90 230 343 | 16q | 99% | 3.87E-05 |
| 17 | 34 819 191 | 80 982 386 | 17q | 83% | 4.25E-06 |
SV identification from short-insert and mate-pair WGS
For short-insert WGS, we identified structural variants using BreakDancer (version 1.4.5) (37), Pindel (version 0.2.4t) (38) and BreakSeq (version 2.0) (39) with default settings to obtain per-filtered calls. All SV calls were required to be >50 bp. We filtered out BreakDancer calls with <2 supporting paired-end reads and with confidence scores <90. Pindel calls were filtered for SVs with quality scores >400. No further filtering was performed for BreakSeq calls. From the mate-pair sequencing (Supplementary Methods), SV calls were made using LUMPY (version 0.6.11) (40). Split-reads and discordantly mapped reads were first extracted and sorted from the processed alignment file as described in github.com/arq5x/lumpy-sv (40). The lumpyexpress command was issued to obtain pre-filtered SV calls. Segmental duplications and reference gaps (hg19) downloaded from the UCSC Genome Browser (41,42) were excluded from the analysis through the ‘-x’ option. SV calls <50 bp were filtered out. To select for high-confidence calls, only SVs that have ≥5 supporting reads as well as both discordant and split-read support were retained. For details regarding experimental validation of SVs, see Supplementary Methods.
RESULTS
Karyotyping
We obtained HepG2 cells from the Stanford ENCODE Production Center. The cells exhibit a hyperdiploid karyotype of 49 to 52 chromosomes (Figure 2A). All 20 metaphase HepG2 cells analyzed using GTW banding were abnormal; 15 cells were very complex, characterized by multiple structural and numerical abnormalities, and the other 5 show a doubling (or tetraploid expansion) of this abnormal cell line, as typical of tumors both in vitro and in vivo. These include translocation between the chromosome 1p and 21p, trisomies of chromosomes 2, 16 and 17, tetrasomy of chromosome 20, uncharacterized arrangements of chromosomes 16 and 17, and a variable number of marker chromosomes. Five cells demonstrated >100 chromosomes and represent a tetraploid expansion of the stemline described. This tetraploid expansion is consistent with previously published results (19) but also absent from other published cytogenetic analyses of HepG2 (20), suggesting the clonal evolution arose during tumorigenesis or early in the establishment of the HepG2 cell line. Although the ploidies of all chromosomes in our HepG2 cell line were supported by previous published karyotypes (19,20,43), variations do exist and also among the various published analyses especially for chromosomes 16 and 17, suggesting that karyotypic differences exist between different HepG2 cell lines (Supplementary Table S1).
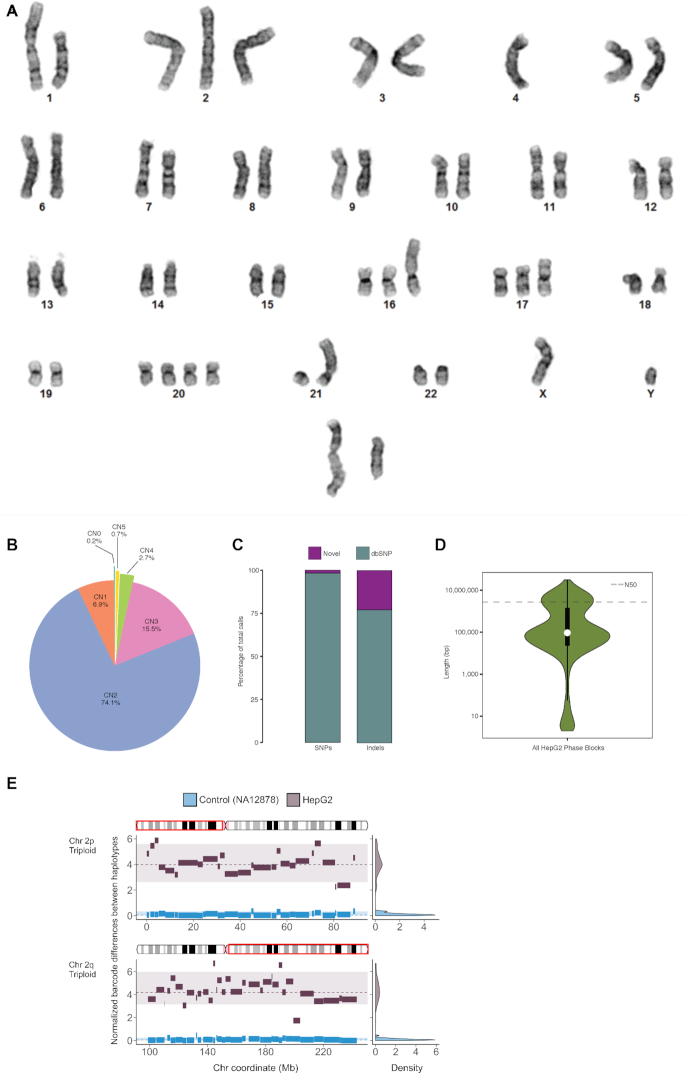
Figure 2.
HepG2 Karyogram and Callset Overview. (A) Representative karyogram of HepG2 cells by GTW banding that shows multiple numerical and structural abnormalities including a translocation between the short arms of chromosomes 1 and 21, trisomies of chromosomes 12, 16 and 17, tetrasomy of chromosome 20, uncharacterized rearrangements of chromosomes 16 and 17 and a two marker chromosomes. ISCN 2013 description: 49∼52,XY,t(1:21)(p22;p11),+2,+16,add(16)(p13),?+17,?add(17)(p11.2),+20,+20,+1∼3mar[cp15]/101∼106,idemx2[cp5]. (B) CNs (by percentage) across the HepG2 genome. (C) Percentage of HepG2 SNVs and Indels that are novel and known (in dbSNP). (D) Violin plot with overlaid boxplot of phased haplotype block sizes, with N50 represented as a dashed line (N50 = 6 792 324 bp) with log-scaled Y-axis. (E) X-axis: chromosome coordinate (Mb). Y-axis: difference in unique linked-read barcode counts between major and minor haplotypes, normalized by SNV density. Haplotype blocks from of normal control sample (NA12878) in blue and from HepG2 in dark gray. Density plots on the right reflects the distribution of the differences in haplotype-specific barcode counts for control sample HepG2. Significant difference (one-sided t-test, P < 0.001) in haplotype-specific barcode counts indicates aneuploidy and haplotype imbalance. Haplotype blocks (with ≥100 phased SNVs) generated from Long Ranger (Dataset 2) for the major and minor haplotypes were then ‘stitched’ to mega-haplotypes encompassing the entire triploid chromosome arms of 2p and 2q.
High-resolution ploidy changes in HepG2
To obtain a high-resolution aneuploid map i.e. large CN changes by chromosomal region in HepG2, WGS coverage across the genome was first calculated in 10-kb bins and plotted against percent GC content where four distinct clusters were clearly observed (44) (Supplementary Figure S2). CNs were assigned to each cluster based on the ratio between its mean coverage and that of the lowest cluster (CN = 1). These assigned large CN changes by chromosomal region confirm the hyperdiploid state of the HepG2 genome as identified by karyotyping (Figure 2A; Supplementary Figure S2 and Supplementary Table S2). We see that 74.1% of the HepG2 genome has a baseline copy number of two (consistent with karyotype), 15.5% copy number of three, 2.7% copy number of four, 0.7% has a copy number of five and 6.9% in a haploid state (Figure 2B and Supplementary Table S2). Furthermore, these high-resolution CN changes across the HepG2 genome were also confirmed by two independent replicates of Illumina Infinium Multi-Ethnic Global-8 arrays (MEGA) array data (Supplementary Figure S3A and Supplementary Data). We found increased CN (CN = 3) over the oncogene VEGFA (6p21.1), which was found to be recurrently duplicated in cases of hepatocellular carcinoma (45).
SNVs and indels
We identified SNVs and indels in HepG2 by taking into account the CN of the chromosomal regions in which they reside so that heterozygous allele frequencies can be assigned accordingly (e.g. 0.33 and 0.67 in triploid regions; 0.25, 0.50 and 0.75 in tetraploid regions). Using GATK Haplotypecaller (21), we identified a total of ∼3.34M SNVs (1.90M heterozygous, 1.44M homozygous) and 0.90M indels (0.60M heterozygous, 0.29M homozygous) (Table 1, Dataset 1). Interestingly, there are 12 375 heterozygous SNVs and indels that have more than two haplotypes in chromosomal regions with CN > 2 (Dataset 1). In addition, chromosome 22 and large continuous stretches of chromosomes 6, 11 and 14 show striking loss of heterozygosity (LOH) (Figure 1 and Supplementary Table S3). Since genomic data from healthy tissue that correspond to HepG2 cells is not available, we intersected these SNVs and indels with dbsnp138 (24) and found the overlap to be ∼3.28M (98%) and ∼0.69M (78%), respectively (Figure 2C and Table 1). We found that 377 SNVs and 255 indels are private protein-altering (PPA) after filtering out those that overlapped with The 1000 Genomes Project (27) or the Exome Sequencing Project (46) (Table 1 and Supplementary Table S4). Moreover, the intersection between the filtered PPA variants and the Catalogue of Somatic Mutations in Cancer (COSMIC) is 39% and 16% for SNVs and indels, respectively (Supplementary Table S5). The gene overlap between HepG2 PPA and the Sanger Cancer Gene Census is 19 (Supplementary Table S6). HepG2 PPA variants include oncogenes and tumor suppressors such as NRAS (47), STK11/LKB1 (48) and PREX2 (49,50) as well as other genes recently found to play critical roles in driving cancer such as CDK12 (51) and IKBKB (52,53). RP1L1, which was recently found to be significantly mutated in hepatocellular carcinoma (45), is also present among the PPA variants.
Resolving haplotypes
We phased the heterozygous SNVs and indels in the HepG2 genome by performing 10X Genomics Chromium linked-read library preparation and sequencing (32,33). Post sequencing quality control analysis shows that 1.49 ng or approximately 447 genomic equivalents of high molecular weight (HMW) genomic DNA fragments (mean = 68 kb, 96.1% >20 kb, 22.0% >100 kb) were partitioned into 1.53 million oil droplets and uniquely barcoded (16 bp). This library was sequenced (2 × 151 bp) to 67x genome coverage with half of all reads coming from HMW DNA molecules with at least 61 linked reads (N50 Linked-Reads per Molecule) (Table 1). We estimate the actual physical coverage (CF) to be 247×. Coverage of the mean insert by sequencing (CR) is 18 176 bp (284 bp × 64) or 30.8%, thus the overall sequencing coverage C = CR × CF = 67×. Distributed over 1628 haplotype blocks (Table 1, Dataset 2), 1.87M (98.7%) of heterozygous SNVs and 0.67M (77.9%) of indels in HepG2 were successfully phased. The longest phased haplotype block is 31.1 Mbp (N50 = 6.80 Mbp) (Figure 2D and Table 1, Dataset 2); however, haplotype block lengths vary widely across different chromosomes (Figure 1 and Supplementary Figure S4). Poorly phased regions correspond to regions exhibiting LOH (Supplementary Table S3 and Figure 1, Dataset 2).
Construction of mega-haplotypes of entire chromosome arms
We constructed mega-haplotypes of entire chromosome arms by leveraging the haplotype imbalance in aneuploid regions in the HepG2 genome where phased haplotype blocks derived from linked-reads were ‘stitched’ together (Table 1 and Figure 2E; Supplementary Data). Briefly, by using a recently developed method (36) specifically for cancer genomes, we counted linked-read barcodes for each phased heterozygous SNVs in haplotype blocks with ≥100 phased SNVs (Dataset 2). Because each barcode is specific for an HMW DNA molecule, the total number of unique barcodes is directly correlated with the number of individual HMW DNA molecules that were sequenced. The fractional representation of a particular genomic sequence (or locus) can be obtained by counting the total number of unique barcodes associated with that particular genomic sequence. Consequently, for each phased haplotype with CN > 2, major and minor haplotypes can be assigned according to the number of barcodes associated with each haplotype (Figure 2E), where the major haplotype is the haplotype with more associated unique barcodes. In genomic regions where CN = 2, the two haplotypes are expected to have similar numbers of unique barcodes. In this method (36), a matched control for comparison is required to confidently discriminate between the major and minor haplotypes. Here, we used NA12878 as normal control because no matching normal tissue sample is available for HepG2 (Figure 2E). After performing the normalization procedures and statistical tests described in (36) to verify haplotype imbalance or aneuploidy genomic regions in HepG2, we then ‘stitched’ together contiguous blocks of phased major and minor haplotypes, respectively. Using this approach, a total of six autosomal mega-haplotypes were constructed (Table 1 and Supplementary Data); four of which encompass entire (or >96%) chromosome arms: 2p, 2q, 6p and 16q (Figure 3). The largest mega-haplotype is approximately 144 Mb long (2q).
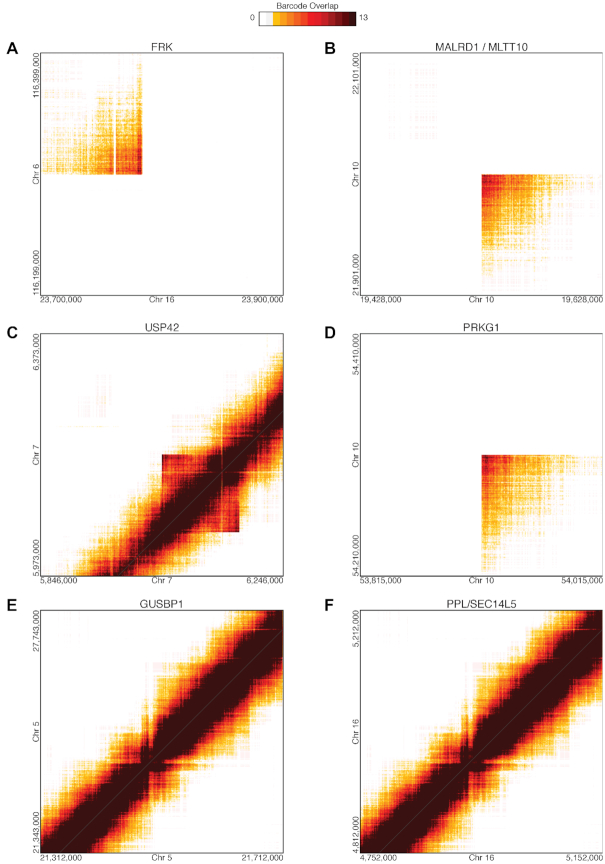
Figure 3.
Large SVs in HepG2 Resolved from Linked-Read Sequencing using Long Ranger. HepG2 SVs resolved by identifying identical linked-read barcodes in distant genomic regions with non-expected barcode overlap for identified using Long Ranger (32,33). (A) Disruption of FRK by translocation between chromosomes 6 and 16. (B) 2.47 Mb intra-chromosomal rearrangement between MALRD1 and MLLT10 on chromosome 10. (C) 127 kb duplication on chromosome 7 resulting in partial duplications of USP42 and PMS2. (D) 395 kb duplication within PRKG1 on chromosome 10. (E) 31.3 kb inversion within GUSBP1 on chromosome 5. (F) 60.4 kb inversion that disrupts PPL and SEC14L5.
Using linked-reads to identify and reconstruct large and complex SVs
From the linked reads, breakpoints of large-scale SVs can be identified by searching for distant genomic regions with linked-reads that have large numbers of overlapping barcodes. SVs can also be assigned to specific haplotypes if the breakpoint-supporting reads contain phased SNVs or indels (32,33). Using this approach (implemented by the Long Ranger software from 10X Genomics), we identified 97 large SVs >30 kb (99% phased) (Dataset 3) and 3473 deletions between 50 bp and 30 kb (78% phased) (Dataset 4). The large SVs include inter- and intra-chromosomal rearrangements (54) (Figure 3A and B), duplications (Figure 3C and D) and inversions (Figure 3E and F). A remarkable example is the haplotype-resolved translocation between chromosomes 16 and 6 (Figure 3A) resulting in the disruption of the non-receptor Fyn-related tyrosine kinase gene FRK, which has been identified as a tumor suppressor (55,56). Another example is the 127 kb tandem duplication on chromosome 7 (Figure 3C) that results in the partial duplication of genes PMS2, encoding a mismatch repair endonuclease, and USP42, encoding the ubiquitin-specific protease 42. An interesting large SV is the 395 kb duplication within PRKG1 (Figure 3D), which encodes the soluble l-α and l-β isoforms of cyclic GMP-dependent protein kinase. We also identified a 193 kb homozygous deletion in PDE4D for HepG2 using linked-read sequencing where six internal exons within the gene are deleted (Figure 3D).
Furthermore, we also used the long-range information from the deep linked-reads sequencing dataset to identify, assemble and reconstruct the breakpoints of SVs in the HepG2 genome using a recently developed method called Genome-wide Reconstruction of Complex Structural Variants (GROC-SVs) (34). Here, HMW DNA fragments that span breakpoints are statistically inferred and refined by quantifying the barcode similarity of linked-reads between pairs of genomic regions similar to Long Ranger (32). Sequence reconstruction is then achieved by assembling the relevant linked reads around the identified breakpoints from which SVs are automatically reconstructed. Breakpoints that also have supporting evidence from the 3 kb-mate pair dataset (see ‘Materials and methods’ section) are indicated as high-confidence events. GROC-SVs called a total of 140 high-confidence breakpoints including four inter-chromosomal events (Figure 1, Dataset 5 and Figure 4A–D); 138 of the breakpoints were successfully sequence-assembled with nucleotide-level resolution of breakpoints as well the exact sequence in the cases where nucleotides have been added or deleted. We identified striking examples of inter-chromosomal rearrangements or translocations in HepG2 between chromosomes 1 and 4 (Figure 4A) and between chromosomes 6 and 17 (Figure 4B) as well as breakpoint-assembled large genomic deletions (Figure 4C, Dataset 5). This break-point assembled 335 kb heterozygous deletion is within the NEDD4L on chromosome 18. Finally, we identified a large (1.3 mb) intra-chromosomal rearrangement that deletes large portions of RBFOX1 and RP11420N32 in one haplotype on chromosome 16 using deep linked-read sequencing (Figure 4D, Dataset 3, Dataset 5).
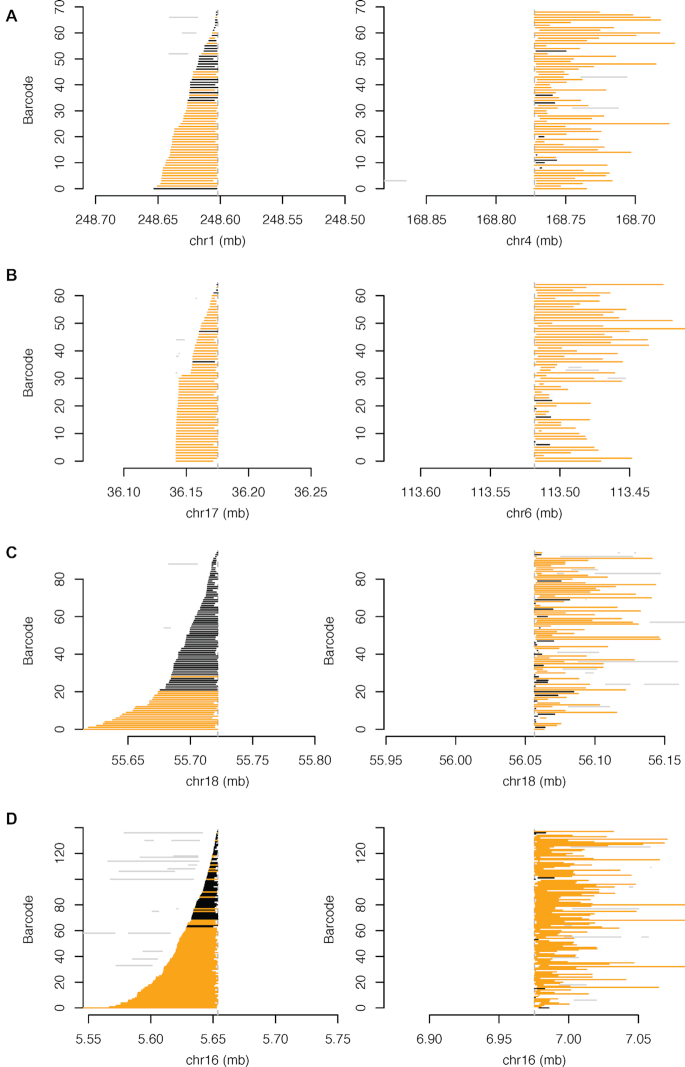
Figure 4.
HepG2 SVs Reconstructed and Assembled Using GROC-SVs in HepG2. (A–D) Each line depicts a fragment inferred from 10X-Genomics data based on clustering of reads with identical barcodes (Y-axis) identified from GROC-SVs (34). Abrupt ending (dashed vertical line) of fragments indicates location of SV breakpoint. All breakpoints depicted are validated by 3 kb-mate-pair sequencing data. Fragments are phased locally with respect to surrounding SNVs (haplotype-specific) are in orange, and black when no informative SNVs are found nearby. Gray lines indicate portions of fragments that do not support the current breakpoint. (A) Translocation between chromosomes 1 and 4. Linked-read fragments containing overlapping barcodes that map to chromosome 1 end abruptly near 248.60 mb indicating a breakpoint, and then continues simultaneously near 168.75 mb on chromosome 4. (B) Translocation between chromosomes 6 and 17. Linked-read fragments containing overlapping barcodes that map to chromosome 17 end abruptly near 36.17 mb indicating a breakpoint and then continues simultaneously near 113.52 mb on chromosome 6. (C) Large (335 kb) heterozygous deletion within NEDD4L on chromosome 18. (D) Large (1.3 mb) intra-chromosomal rearrangement that deletes large portions of RBFOX1 and RP11420N32 on chromosome 16.
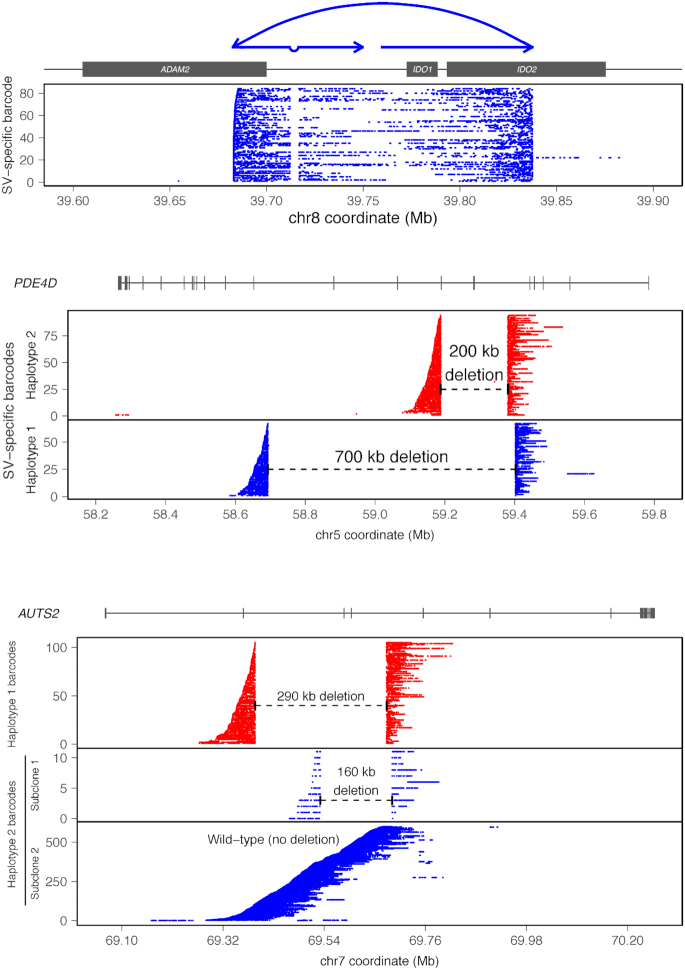
We then employed ‘gemtools’ (35) to resolve and phase large and complex SVs in the HepG2 genome. We identified a complex SV on chromosome 8 that involves a small deletion downstream of ADAM2 that is also within a larger tandem duplication leading to the amplification of the oncogene IDO1 (57) and the first half of IDO2 (Figure 5). Two allele-specific deletions 700 and 200 kb, respectively, were identified in the PDE4D on chromosome 5 (Figure 5). Since chromosome 5 is triploid in HepG2 (Figures 1 and 2A; Supplementary Data), we see approximately twice as much linked-reads barcode representation for the allele harboring the 200 kb deletion, suggesting that this allele of PDE4D has two copies and the allele harboring the 700 kb deletion has one copy (Figure 5). Similarly, we also identified two allele-specific deletions, 290 and 160 kb respectively within AUTS2 on chromosome 7 (Figure 5). Interestingly, for the allele harboring the 160 kb deletion, the non-deleted reference allele is also present at much larger frequency as indicted by the total number of linked-read barcodes, suggesting that the allele harboring the 160 kb deletion within AUTS2 occurs in a fraction of HepG2 cells or sub-clonally (Figure 5). From the total number of linked-read barcodes associated with this 160 kb allele-specific deletion in AUTS2, we estimate that this deletion occurs in 10% of HepG2 cells. All breakpoints identified using ‘gemtools’ were individual polymerase chain reaction (PCR) and Sanger sequencing verified (Supplementary Table S7).
Figure 5.
Large and complex haplotype-resolved SVs using gemtools. Each SV is identified from linked-reads clustered by identical barcodes (i.e. SV-specific barcodes, Y-axis) indicative of single HMW DNA molecules (depicted by each row) that span SV breakpoints. Haplotype-specific SVs are represented in blue and red. X-axis: hg19 genomic coordinate. (Top) Complex SV on chromosome 8 involving a 4585 bp deletion downstream of ADAM2. This deletion is within a tandem duplication leading to the amplification of the IDO1 and the first half of IDO2. The presence of HMW molecules sharing the same linked-read barcodes spanning both breakpoints indicates a cis orientation and occurrence on only one allele of this locus. Schematic diagram of the rearranged structures drawn above the plot. (Middle) Two haplotype-resolved deletions 700 kb (blue) and 200 kb (red), respectively, occurring on two separate alleles within of PDE4D on chromosome 5—the spanning HMW molecules for each deletion do not share SV-specific barcodes, indicating that these deletions are in trans. Two haplotype-resolved deletions, 290 kb (red) and 160 kb (blue) respectively, within AUTS2 on chromosome 7. The reference allele of AUTS2 without the deletion (Haplotype 2) is also detected and resolved by linked-reads (blue, bottom panel). The 160 kb deletion on Haplotype 2 occurs sub-clonally.
SVs from mate-pair sequencing
To obtain increase sensitivity in the detection medium-sized (1–100 kb) SVs in HepG2, we prepared a 3 kb-mate pair library and sequenced (2 × 151 bp) it to a genome coverage of 7.9x after duplicate removal. The sequencing coverage of each 3 kb insert (CR) is 302 bp (or 10% of the insert size) that translates to a physical coverage (CF) of 79×. Deletions, inversions and tandem duplications from the mate-pair library were identified from analysis of discordant read pairs and split reads using LUMPY (40). Only SVs that are supported by both discordant read-pair and split-read supports were retained. Using this approach, we identified 122 deletions, 41 inversions and 133 tandem duplications (Dataset 6). Approximately 76% of these SVs are between 1 and 10 kb, 86% are between 1 and 100 kb (9% between 10 and 100 kb) and 3% are >100 kb (Dataset 6). Twenty SVs (16 deletions and 4 duplications) were randomly selected for experimental validation using PCR and Sanger sequencing in which 15/16 were successfully validated (93.8%) (Supplementary Table S7).
SVs identified from deep short-insert WGS
Deletions, inversions, insertions and tandem duplications were identified from the HepG2 WGS dataset using Pindel (38), BreakDancer (37) and BreakSeq (39). Since similar categories of SVs were also identified using mate-pair and linked-read sequencing, these SVs were combined with the SVs identified previously using Long Ranger and LUMPY where variations with support from multiple methods and with >50% reciprocal overlap were merged. In total, 6405 SVs were obtained from all methods that include 5226 deletions, 245 duplications, 428 inversions and 494 insertions (only BreakDancer (37) was designed to call insertions) (Supplementary Data). A set of deletion (n = 27) and tandem duplication calls (n = 4) was randomly selected to confirm by PCR and Sanger sequencing, and 30/32 (94%) events were successfully validated (Supplementary Table S7). Consistent with previous analysis (58), deletions show the highest concordance among the various methods of detection compared to duplication and inversion calls (Supplementary Figure S5). As expected, we detected a 520 bp deletion in exon 3 of the β-catenin (CTNNB1) gene (Dataset 4, Supplementary Data), which was previously documented to exist in HepG2 (59). Interestingly, we found no SVs or PPA mutations in the Wnt-pathway gene CAPRIN2 (60), which had been previously reported for hepatoblastoma (61).
Identification of non-reference Alu and LINE1 insertions
From our deep-coverage short-insert WGS data, we also analyzed the HepG2 genome for non-reference LINE1 and Alu retrotransposon insertions using RetroSeq (62) with some modifications. These insertions were identified from paired-end reads that have one of the pair mapping to hg19 uniquely and other mapping to an Alu or LINE1 consensus sequence in full or split fashion (see ‘Materials and methods’ section). Retrotransposon insertion events with greater than five supporting reads were categorized as high confidence and retained (Supplementary Table S8). We identified 1899 and 351 non-reference Alu and LINE1 insertions in the HepG2 genome, respectively (Figure 1). We randomly chose 8 Alu and 10 LINE1 insertions with split-read support for confirmation using PCR and Sanger sequencing where 87.5% and 100% were successfully validated, respectively (Supplementary Table S8).
Allele-specific gene expression
Due to the abundance of aneuploidy in the HepG2 genome, CN changes of genomic regions should be taken into account when analyzing for allele-specific gene expression in order to reduce false positives and false negatives. Using the heterozygous SNV allele frequencies in HepG2 (Dataset 1), we re-analyzed two replicates of HepG2 ENCODE RNA-Seq data. We identified 3189 and 3022 genes that show allele-specific expression (P < 0.05) in replicates one and two, respectively (Figure 1 and Supplementary Table S9). Furthermore, we also identified 862 and 911 genes that would have been falsely identified to have allele-specific expression (false positives), if the copy numbers of SNV allele frequencies were not taken into consideration as well as 446 and 407 genes that would not have been identified (false negatives) in replicates one and two, respectively (Supplementary Table S10).
Allele-specific DNA methylation
Using the phasing information for HepG2 SNVs (Dataset 2), we also identified 384 CpG islands (CGIs) that exhibit allele-specific DNA methylation (Figure 1 and Supplementary Table S11). We obtained two independent replicates of HepG2 whole-genome bisulfite sequencing data (2 × 125 bp, experiment ENCSR881XOU) from the ENCODE Portal (17). Read alignment to hg19 was performed using Bismark (63); 70.0% of reads were uniquely aligned, 44.7% of cytosines were methylated in a CpG context. We then phased methylated and unmethylated CpGs to their respective haplotypes by identifying reads that overlap both CpGs and phased heterozygous SNVs (Dataset 2). We grouped the phased individual CpGs into CGIs and totaled the number of reads that contain methylated and unmethylated cytosines for each CGI allele, normalizing by CN in cases of aneuploidy. Fisher’s exact test was used to evaluate allele-specific methylation, and significant results were selected using a target false discovery rate of 10% (64) (see ‘Materials and methods’ section). Ninety-eight CGIs reside within promoter regions (defined as 1 kb upstream of a gene); 277 are intragenic and 96 lie within 1 kb downstream of 348 different genes (Supplementary Table S11). The following 11 genes are within 1 kb of a differentially methylated CGI and also overlap with the Sanger Cancer Gene Census: FOXA1, GNAS, HOXD13, PDE4DIP, PRDM16, PRRX1, SALL4, STIL, TAL1 andZNF331. Twenty-seven unique CGIs with allele-specific methylation overlap with allele-specific RNA expression (Supplementary Table S9).
Allele-specific CRISPR targets
We identified 38 551 targets in the HepG2 genome suitable for allele-specific CRSIPR targeting (Figure 1 and Supplementary Table S12). Phased variant sequences (including reverse complement) that differ by >1 bp between the alleles were extracted to identify all possible CRISPR targets by pattern searching for [G, C or A]N20GG (see ‘Materials and methods’ section). Only conserved high-quality targets were retained by using a selection method previously described and validated (65). We took the high-quality target filtering process further by taking the gRNA function and structure into account. Targets with multiple exact matches, extreme GC fractions and those with TTTT sequence (which might disrupt the secondary structure of gRNA) were removed. Furthermore, we used the Vienna RNA-fold package (66) to identify gRNA secondary structure and eliminated targets for which the stem–loop structure for Cas9 recognition is not able to form (67). Finally, we calculated the off-target risk score using the tool as described for this purpose (68). A very strict off-target threshold score was chosen in which candidates with a score below 75 were rejected to ensure that all targets are as reliable and as specific as possible.
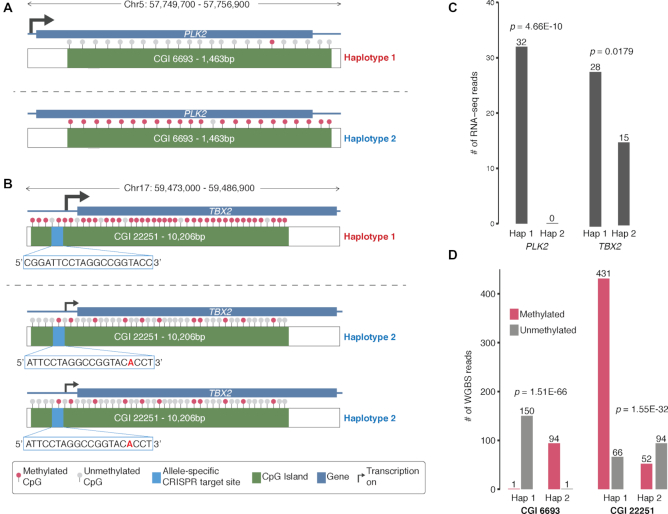
Genomic sequence and structural context provides insight into regulatory complexity
We show examples of how deeper insights into gene regulation and regulatory complexity can be obtained by integrating genomic sequence and structural contexts with functional genomics and epigenomics data (Figure 6A–D). One example is the allele-specific RNA expression and allele-specific DNA methylation in HepG2 at the PLK2 locus on chromosome 5 (Figure 6A). By incorporating the genomic context in which PLK2 is expressed in HepG2 cells, we see that PLK2 RNA is only expressed from Haplotype 1 (P = 4.66E-10) in which the CGI within the gene is completely unmethylated (P = 1.51E-66) in the expressed allele and completely methylated in the non-expressed allele (Figure 6A, C and D). The second example is allele-specific RNA expression and allele-specific DNA methylation of the TBX2 gene in HepG2 (Figure 6B). The TBX2 locus on chromosome 17 is triploid, and we see that TBX2 is preferentially expressed from Haplotype 1 that has one copy and lower expression is observed from the two copies of Haplotype 2 (P = 0.0179) (Figure 6B and C). We also observed highly preferential DNA methylation of the CGI in Haplotype 1 (P = 1.55E-32) (Figure 6B and D). In addition, there is also an allele-specific CRISPR targeting site for both haplotypes in the promoter region of TBX2 and inside CGI 22251 (1937 bp upstream of TBX2 gene and 2259 bp downstream of the 5′ end of CGI 22251) (Figure 6B).
Figure 6.
Genomic Sequence and Structural Context Provides Insight into Regulatory Complexity in HepG2. (A) Chr5:57,755,334-57,756,803 locus containing the serine/threonine-protein kinase gene PLK2 and CGI 6693 (1463 bp) where phased Haplotype 1 and Haplotype 2. Allele-specific transcription of PLK2 from Haplotype 2 only. CpGs in CGI 6693 are mostly unmethylated in Haplotype 2 (expressed) and highly methylated in Haplotype 1 (repressed). (B) Chr17:59,473,060-59,483,266 locus (triploid in HepG2) containing T-box transcription factor gene TBX2 and CpG Island (CGI) 22251 (10 206 bp) where phased Haplotype 2 has two copies and Haplotype 1 has one copy. Allele-specific transcription of TBX2 from Haplotype 2 only. CpGs in CGI 22251 are unmethylated in Haplotype 1 (repressed) and methylated in Haplotype 2 (expressed). Allele-specific CRISPR targeting site 1937 bp inside the 5′ region of TBX2 for both Haplotypes. (C) Number of allele-specific RNA-Seq reads in Haplotypes 1 and 2 for PLK2 and TBX2 where both genes exhibit allele-specific RNA expression (P = 0.4.66E-10 and P = 0.0179, respectively). (D) Number of methylated and unmethylated phased whole-genome bisulfite-sequencing reads for Haplotypes 1 and 2 in CGI 6693 and CGI 22251 where both CGIs exhibit allele-specific DNA methylation (P = 1.51E-66 and P = 1.55E-32, respectively).
DISCUSSION
As one of the most widely used cell lines in biomedical research, HepG2’s genomic sequence and structural features have never been characterized in a comprehensive manner beyond its karyotype (19,20) and SNVs identified from ChIP-Seq data and 10× coverage WGS that do not take aneuploidy or CN into consideration (69,70). Here, in summary, we performed a comprehensive analysis of genomic structural features (Figure 1) for the HepG2 cell line that includes SNVs (Dataset 1), Indels (Dataset 1), large CN or ploidy changes across chromosomal regions at 10 kb resolution (Supplementary Table S2), phased haplotype blocks (Dataset 2), phased CRISPR targets (Supplementary Table S12), novel retrotransposon insertions (Supplementary Table S8) and SVs (Datasets 3–6) including deletions, duplications, inversions, translocations, and those that are the result of complex genomic rearrangements. Many of the HepG2 SVs are also phased, assembled and experimentally verified (Dataset 5, Supplementary Tables S7 and S8).
We illustrate, using PLK2 and TBX2 (Figure 6A and B), examples where genomic context can enhance the interpretation of function genomics and epigenomics data to derive novel insights into the complexity of oncogene regulation. The Polo-like kinase gene PLK2 (SNK) is a transcriptional target of p53 and also a cancer biomarker (71,72). It has been studied in the contexts of many human cancers (71,73–75). Disruption of PLK2 has also been proposed to have therapeutic value in sensitizing chemo-resistant tumors. Its roles in Burkitt’s lymphoma (76), hepatocellular carcinoma (73) and epithelial ovarian cancer (74) are consistent with that of tumor suppressors while its role in colorectal cancer is consistent with that of an oncogene (75). Interestingly, promoter methylation and/or LOH were linked to the down-regulation of PLK2 in human hepatocellular carcinoma (73). Chemotherapy resistance of epithelial ovarian cancer can be conferred by the down-regulation of PLK2 at the transcriptional level via DNA methylation of the CpG island in the PLK2 promoter (74). Here we show that the down-regulation of PLK2 in HepG2 cancer cells could be achieved through what appears to be allele-specific transcriptional silencing via allele-specific DNA methylation of a large CGI within the gene body (Figure 6A).
The T-box transcription factor TBX2 is a critical regulator of cell fate decisions, cell migration and morphogenesis in the development of many organs (77–80). It regulates cell cycle progression (81), and its overexpression has been demonstrated in promoting or maintaining the proliferation of many cancers including melanomas (82), nasopharyngeal cancer (83), breast cancer (84,85), prostate cancer (86) and gastric cancer (87). Here, we show that three copies of the TBX2 gene exist in HepG2 cancer cells as a result of duplication in Haplotype 2. However, it is preferentially expressed in Haplotype 1 possibly due to the highly allele-specific DNA methylation in the CGI that spans its promoter region and most of the gene body (Figure 6B). It is plausible that overexpression of TBX2 in other cancer types are caused by similar genomic rearrangements and/or epigenetic mechanisms where duplication of TBX2 may result in the overexpression and DNA methylation (possibly allele-specific) may contribute an additive effect to TBX2 overexpression or act as the sole contributor where TBX2 is not duplicated (see Supplementary Discussion for detailed discussion of other oncogenes, tumor-suppressors and other genes associated with cancer that are disrupted as a consequence of genomic variation in HepG2).
Combining orthogonal methods and signals greatly improves SV-calling sensitivity and accuracy (40,88). We compared SVs identified from various methods. For deletion SVs, linked reads show the highest sensitivity. The linked-reads analysis software Long Ranger detects SVs (deletions, duplications and inversions) larger than 30 kb (Dataset 3) and deletions smaller than 30 kb (Dataset 4)—a wider size spectrum for deletions. Out of the total 4771 unique deletion calls in HepG2, 3364 (71%) can be detected using linked-reads alone; the lower-coverage mate-pair dataset (analyzed using LUMPY (40)) added another 31 calls, and the deep-coverage WGS dataset added the rest (Supplementary Figure S5A). However, for duplications and inversions, we see that many more calls were added by bringing in the mate-pair and deep short-insert whole-genome datasets and incorporating analyses of other mapping signals such as discordant-read-pair and split-reads (Supplementary Figure S5B and C). Overall, we see considerable overlap as well as variant calls specific to each method for deletions (Supplementary Figure S5A), and much less overlap for duplications and inversions (Supplementary Figure S5B and C). This is consistent with what has been shown previously (58) as inversions and duplications are more difficult in principle to accurately resolve. Experimental validation of specific SVs of interest should be conducted by individual laboratories prior to functional follow-up studies. This study is primarily focused on the utilization of Illumina sequencing to resolve SVs in HepG2. In the relatively near future, long-read technologies such as Oxford Nanopore or Pacific Biosciences can be expected to become of considerable utility in the analysis of complex cancer genomes. The eventual incorporation of long-reads can be expected to improve the ability to resolve challenging variants for example inside or flanked by repetitive regions i.e. segmental duplications, and also the sensitivity to detect nested SVs (89,90). Long-reads are accompanied with much higher error-rates compared to short Illumina sequencing, but it is foreseeable that the continuing development of computational tools for and SV detection will at least partially offset this challenge (89–92).
All data and results generated from this global whole-genome analysis of HepG2 is publicly available through ENCODE (encodeproject.org) (17). This analysis serves as a valuable reference for further understanding the vast amount of existing HepG2 ENCODE data. Our results also guide future study designs that utilize HepG2 cells including CRISPR/Cas9 experiments where knowledge of the phased genomic variants can extend or modify the number of editing targets including those that are haplotype-specific (Supplementary Table S12) while knowledge of aberrant chromosomal CN changes will allow for more accurate interpretation of functional data in non-diploid regions. This study may serve as a technical archetype for advanced, integrated and global analysis of genomic sequence and structural variants for other widely cell lines with complex genomes.
Since HepG2 has been passaged for decades and across many different laboratories, additional genome variation may be present in HepG2 cells that had been long separated from the ENCODE HepG2 production line. Many of the results we discuss here are supported by previous studies, for instance, karyotyping and mutation in CTNNB1 (59), but there are minor differences such as the lack of a mutation in CAPRIN2 (60). We expect that the vast majority of genomic variants that we describe here to be shared across the different versions of HepG2 cells, thus when taking into account for future global studies, substantial insights are expected to be gained. However, if specific loci is of interest for follow-up studies, a first step should always be to experimentally confirm the presence the particular working line of HepG2 as distinct lines may harbor slight variations. While the complexity of the HepG2 genome renders the design and interpretation of functional genomic and epigenomic studies more challenging, the results of this study enables researchers to continue to use HepG2 to investigate the effects of different types of genomic variations on the multiple layers of functionality and regulation for which ENCODE data are already available and continues to be produced.
DATA AVAILABILITY
All raw and processed data files are publicly released on the ENCODE portal (encodeproject.org) via accession ENCBS760ISV. Datasets 1–6 can individually be accessed via ENCODE accession numbers ENCFF336CFC, ENCFF853HHD, ENCFF467ETN, ENCFF717TPE, ENCFF330UFT, ENCFF241CEK, respectively.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Arineh Khechaduri for performing genomic DNA preparation. We thank Aditi Narayanan, Dr. Idan Gabdank, Nathaniel Watson, Zachary A. Myers, Khine Zin Lin, and Dr. Cricket Sloan for data organization and upload to ENCODE. Dr. Athena Cherry and the Stanford Cytogenetics Laboratory for karyotype analysis. A.E.U. is a Tashia and John Morgridge Faculty Scholar of the Stanford Child Health Research Institute.
Authors’ Contributions: B.Z. and A.E.U conceived and designed the study. B.Z., R.P., I.S., and Y.H. performed experiments. B.Z., S.S.H., S.U.G., N.S., J.M.B., X.L.Z., X.W.Z., J.G.A., S.B., G.S., D.P., and A.A. performed analysis. H.P.J, W.H.W., and A.E.U. contributed resources and supervised the study. B.Z., S.S.H., and A.E.U. wrote the manuscript with inputs from S.U.G., J.M.B., and H.P.J.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NIH/NHGRI [P50-HG007735 to A.E.U.]; Stanford Medicine Faculty Innovation Program (to A.E.U); NIH/NHLBI [T32-HL110952 to B.Z.]; NIH/NHGRI [R01-HG007834, P50-HG007735 to W.H.W.]; NSF Graduate Research Fellowship [DGE-114747 to J.G.A]; NIH/NIGMS [T32-GM096982 to J.G.A]; NIH/NCI [U24CA220242 to A.A.]; Intermountain Healthcare Research Award (to H.P.J.); NIH/NHGRI [R01-HG006137, P01-HG00205 to H.P.J.]. Funding for open access charge: NHGRI [P50 HG007735].
Conflict of interest statement. None declared.
REFERENCES
- 1. Negrini S., Gorgoulis V.G., Halazonetis T.D.. Genomic instability–an evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010; 11:220–228. [DOI] [PubMed] [Google Scholar]
- 2. Hanahan D., Weinberg R.A.. Hallmarks of Cancer: the next generation. Cell. 2011; 144:646–674. [DOI] [PubMed] [Google Scholar]
- 3. Adey A., Burton J.N., Kitzman J.O., Hiatt J.B., Lewis A.P., Martin B.K., Qiu R., Lee C., Shendure J.. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013; 500:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aden D.P., Fogel A., Plotkin S., Damjanov I., Knowles B.B.. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979; 282:615–616. [DOI] [PubMed] [Google Scholar]
- 5. López-Terrada D., Cheung S.W., Finegold M.J., Knowles B.B.. Hep G2 is a hepatoblastoma-derived cell line. Hum. Pathol. 2009; 40:1512–1515. [DOI] [PubMed] [Google Scholar]
- 6. Schoonen W.G.E.J., Westerink W.M.A., de Roos J.A.D.M., Débiton E.. Cytotoxic effects of 100 reference compounds on Hep G2 and HeLa cells and of 60 compounds on ECC-1 and CHO cells. I mechanistic assays on ROS, glutathione depletion and calcein uptake. Toxicol. In Vitro. 2005; 19:505–516. [DOI] [PubMed] [Google Scholar]
- 7. Sahu S.C., O’Donnell M.W., Sprando R.L.. Interactive toxicity of usnic acid and lipopolysaccharides in human liver HepG2 cells. J. Appl. Toxicol. 2012; 32:739–749. [DOI] [PubMed] [Google Scholar]
- 8. Dias da Silva D., Carmo H., Lynch A., Silva E.. An insight into the hepatocellular death induced by amphetamines, individually and in combination: the involvement of necrosis and apoptosis. Arch. Toxicol. 2013; 87:2165–2185. [DOI] [PubMed] [Google Scholar]
- 9. Menezes C., Alverca E., Dias E., Sam-Bento F., Pereira P.. Involvement of endoplasmic reticulum and autophagy in microcystin-LR toxicity in Vero-E6 and HepG2 cell lines. Toxicol. In Vitro. 2013; 27:138–148. [DOI] [PubMed] [Google Scholar]
- 10. Kamalian L., Chadwick A.E., Bayliss M., French N.S., Monshouwer M., Snoeys J., Park B.K.. The utility of HepG2 cells to identify direct mitochondrial dysfunction in the absence of cell death. Toxicol. In Vitro. 2015; 29:732–740. [DOI] [PubMed] [Google Scholar]
- 11. Xia P., Zhang X., Xie Y., Guan M., Villeneuve D.L., Yu H.. Functional toxicogenomic assessment of triclosan in human HepG2 cells using Genome-Wide CRISPR-Cas9 screening. Environ. Sci. Technol. 2016; 50:10682–10692. [DOI] [PubMed] [Google Scholar]
- 12. Alzeer S., Ellis E.M.. Metabolism of gamma hydroxybutyrate in human hepatoma HepG2 cells by the aldo-keto reductase AKR1A1. Biochem. Pharmacol. 2014; 92:499–505. [DOI] [PubMed] [Google Scholar]
- 13. Xu D., He X., Chang Y., Xu C., Jiang X., Sun S., Lin J.. Inhibition of miR-96 expression reduces cell proliferation and clonogenicity of HepG2 hepatoma cells. Oncol. Rep. 2013; 29:653–661. [DOI] [PubMed] [Google Scholar]
- 14. Hao L., Ito K., Huang K.-H., Sae-tan S., Lambert J.D., Ross A.C.. Shifts in dietary carbohydrate-lipid exposure regulate expression of the non-alcoholic fatty liver disease-associated gene PNPLA3/adiponutrin in mouse liver and HepG2 human liver cells. Metabolism. 2014; 63:1352–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huan L.C., Wu J.-C., Chiou B.-H., Chen C.-H., Ma N., Chang C.Y., Tsen Y.-K., Chen S.C.. MicroRNA regulation of DNA repair gene expression in 4-aminobiphenyl-treated HepG2 cells. Toxicology. 2014; 322:69–77. [DOI] [PubMed] [Google Scholar]
- 16. Mangrum J.B., Martin E.J., Brophy D.F., Hawkridge A.M.. Intact stable isotope labeled plasma proteins from the SILAC-labeled HepG2 secretome. Proteomics. 2015; 15:3104–3115. [DOI] [PubMed] [Google Scholar]
- 17. Sloan C.A., Chan E.T., Davidson J.M., Malladi V.S., Strattan J.S., Hitz B.C., Gabdank I., Narayanan A.K., Ho M., Lee B.T. et al.. ENCODE data at the ENCODE portal. Nucleic Acids Res. 2016; 44:D726–D732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou Y., Guo H., Cao C., Li X., Hu B., Zhu P., Wu X., Wen L., Tang F., Huang Y. et al.. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016; 26:304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simon D., Aden D.P., Knowles B.B.. Chromosomes of human hepatoma cell lines. Int. J. Cancer. 1982; 30:27–33. [DOI] [PubMed] [Google Scholar]
- 20. Chen H.-L., Chiu T.-S., Chen P.-J., Chen D.-S.. Cytogenetic studies on human liver cancer cell lines. Cancer Genet. Cytogenet. 1993; 65:161–166. [DOI] [PubMed] [Google Scholar]
- 21. McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al.. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010; 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. et al.. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinforma. 2013; 43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. DePristo M.a., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.a., del Angel G., Rivas M.a., Hanna M. et al.. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011; 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M.. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cingolani P., Patel V.M., Coon M., Nguyen T., Land S.J., Ruden D.M., Lu X.. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012; 3:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. The 1000 Genomes Project Consortium Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A. et al.. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S. et al.. COSMIC: Exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015; 43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R.. A census of human cancer genes. Nat. Rev. Cancer. 2004; 4:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Hsi-Yang Fritz M. et al.. An integrated map of structural variation in 2,504 human genomes. Nature. 2015; 526:75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng G.X.Y., Lau B.T., Schnall-Levin M., Jarosz M., Bell J.M., Hindson C.M., Kyriazopoulou-Panagiotopoulou S., Masquelier D.A., Merrill L., Terry J.M. et al.. Haplotyping germline and cancer genomes with high-throughput linked-read sequencing. Nat. Biotechnol. 2016; 34:303–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Marks P., Garcia S., Barrio A.M., Belhocine K., Bernate J., Bharadwaj R., Bjornson K., Catalanotti C., Delaney J., Fehr A. et al.. Resolving the Full Spectrum of Human Genome Variation using Linked-Reads. 2018; (12 May 2018, date last accessed)https://www.biorxiv.org/content/10.1101/230946v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spies N., Weng Z., Bishara A., McDaniel J., Catoe D., Zook J.M., Salit M., West R.B., Batzoglou S., Sidow A.. Genome-wide reconstruction of complex structural variants using read clouds. Nat. Methods. 2017; 14:915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Greer S.U., Nadauld L.D., Lau B.T., Chen J., Wood-Bouwens C., Ford J.M., Kuo C.J., Ji H.P.. Linked read sequencing resolves complex genomic rearrangements in gastric cancer metastases. Genome Med. 2017; 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bell J.M., Lau B.T., Greer S.U., Wood-Bouwens C., Xia L.C., Connolly I.D., Gephart M.H., Ji H.P.. Chromosome-scale mega-haplotypes enable digital karyotyping of cancer aneuploidy. Nucleic Acids Res. 2017; 45:e162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen K., Wallis J.W., McLellan M.D., Larson D.E., Kalicki J.M., Pohl C.S., McGrath S.D., Wendl M.C., Zhang Q., Locke D.P. et al.. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009; 6:677–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z.. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009; 25:2865–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lam H.Y.K., Mu X.J., Stütz A.M., Tanzer A., Cayting P.D., Snyder M., Kim P.M., Korbel J.O., Gerstein M.B.. Nucleotide-resolution analysis of structural variants using BreakSeq and a breakpoint library. Nat. Biotechnol. 2010; 28:47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Layer R.M., Chiang C., Quinlan A.R., Hall I.M.. LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014; 15:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J.. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004; 32:D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wong N., Lai P., Pang E., Leung T.W., Lau J.W., Johnson P.J.. A comprehensive karyotypic study on human hepatocellular carcinoma by spectral karyotyping. Hepatology. 2000; 32:1060–1068. [DOI] [PubMed] [Google Scholar]
- 44. Abyzov A., Urban A.E., Snyder M., Gerstein M.. CNVnator: An approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011; 21:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cancer Genome Atlas Research Network Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell. 2017; 169:1327–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Fu W., O’Connor T.D., Jun G., Kang H.M., Abecasis G., Leal S.M., Gabriel S., Altshuler D., Shendure J., Nickerson D.A. et al.. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2012; 493:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pylayeva-Gupta Y., Grabocka E., Bar-Sagi D.. RAS oncogenes: weaving a tumorigenic web. Nat. Rev. Cancer. 2011; 11:761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou W., Zhang J., Marcus A.I.. LKB1 tumor suppressor: Therapeutic opportunities knock when LKB1 is inactivated. Genes Dis. 2014; 1:64–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Berger M.F., Hodis E., Heffernan T.P., Deribe Y.L., Lawrence M.S., Protopopov A., Ivanova E., Watson I.R., Nickerson E., Ghosh P. et al.. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012; 485:502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang J., Gong X., Ouyang L., He W., Xiao R., Tan L.. PREX2 promotes the proliferation, invasion and migration of pancreatic cancer cells by modulating the PI3K signaling pathway. Oncol. Lett. 2016; 12:1139–1143. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51. Paculová H., Kohoutek J.. The emerging roles of CDK12 in tumorigenesis. Cell Div. 2017; 12:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xia Y., Yeddula N., Leblanc M., Ke E., Zhang Y., Oldfield E., Shaw R.J., Verma I.M.. Reduced cell proliferation by IKK2 depletion in a mouse lung-cancer model. Nat. Cell Biol. 2012; 14:257–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kai X., Chellappa V., Donado C., Reyon D., Sekigami Y., Ataca D., Louissaint A., Mattoo H., Joung J.K., Pillai S.. IκB Kinase β (IKBKB) mutations in lymphomas that constitutively activate canonical nuclear factor κB (NFκB) signaling. J. Biol. Chem. 2014; 289:26960–26972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fernandez-Banet J., Lee N.P., Chan K.T., Gao H., Liu X., Sung W.-K., Tan W., Fan S.T., Poon R.T., Li S. et al.. Decoding complex patterns of genomic rearrangement in hepatocellular carcinoma. Genomics. 2014; 103:189–203. [DOI] [PubMed] [Google Scholar]
- 55. Brauer P.M., Tyner A.. RAKing in AKT: a tumor suppressor function for the intracellular tyrosine kinase FRK. Cell Cycle. 2009; 8:2728–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yim E.-K., Siwko S., Lin S.-Y.. Exploring Rak tyrosine kinase function in breast cancer. Cell Cycle. 2009; 8:2360–2364. [DOI] [PubMed] [Google Scholar]
- 57. Platten M., von Knebel Doeberitz N., Oezen I., Wick W., Ochs K.. Cancer immunotherapy by targeting IDO1/TDO and their downstream effectors. Front. Immunol. 2015; 5:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lam H.Y.K., Pan C., Clark M.J., Lacroute P., Chen R., Haraksingh R., O’Huallachain M., Gerstein M.B., Kidd J.M., Bustamante C.D. et al.. Detecting and annotating genetic variations using the HugeSeq pipeline. Nat. Biotechnol. 2012; 30:226–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. López-Terrada D., Gunaratne P.H., Adesina A.M., Pulliam J., Hoang D.M., Nguyen Y., Mistretta T.-A., Margolin J., Finegold M.J.. Histologic subtypes of hepatoblastoma are characterized by differential canonical Wnt and Notch pathway activation in DLK+ precursors. Hum. Pathol. 2009; 40:783–794. [DOI] [PubMed] [Google Scholar]
- 60. Ding Y., Xi Y., Chen T., Wang J., Tao D., Wu Z.-L., Li Y., Li C., Zeng R., Li L.. Caprin-2 enhances canonical Wnt signaling through regulating LRP5/6 phosphorylation. J. Cell Biol. 2008; 182:865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jia D., Dong R., Jing Y., Xu D., Wang Q., Chen L., Li Q., Huang Y., Zhang Y., Zhang Z. et al.. Exome sequencing of hepatoblastoma reveals novel mutations and cancer genes in the Wnt pathway and ubiquitin ligase complex. Hepatology. 2014; 60:1686–1696. [DOI] [PubMed] [Google Scholar]
- 62. Keane T.M., Wong K., Adams D.J.. RetroSeq: Transposable element discovery from next-generation sequencing data. Bioinformatics. 2013; 29:389–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Krueger F., Andrews S.R.. Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011; 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Storey J.D., Tibshirani R.. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 2003; 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sunagawa G.A., Sumiyama K., Ukai-Tadenuma M., Perrin D., Fujishima H., Ukai H., Nishimura O., Shi S., Ohno R.-I., Narumi R. et al.. Mammalian reverse genetics without crossing reveals Nr3a as a Short-Sleeper gene. Cell Rep. 2016; 14:662–677. [DOI] [PubMed] [Google Scholar]
- 66. Lorenz R., Bernhart S.H., Höner Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O.. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014; 156:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Huang D., Ovcharenko I.. Identifying causal regulatory SNPs in ChIP-seq enhancers. Nucleic Acids Res. 2015; 43:225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cavalli M., Pan G., Nord H., Wallén Arzt E., Wallerman O., Wadelius C.. Allele-specific transcription factor binding in liver and cervix cells unveils many likely drivers of GWAS signals. Genomics. 2016; 107:248–254. [DOI] [PubMed] [Google Scholar]
- 71. Burns T.F., Fei P., Scata K.A., Dicker D.T., El-Deiry W.S.. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (Taxol)-Exposed cells. Mol. Cell. Biol. 2003; 23:5556–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Coley H.M., Hatzmichael E., Blagden S.P., McNeish I.A., Thompson A., Crook T., Syed N.. Polo Like Kinase 2 Tumour Suppressor and cancer biomarker: new perspectives on drug sensitivity/resistance in ovarian cancer. Oncotarget. 2012; 3:78–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pellegrino R., Calvisi D.F., Ladu S., Ehemann V., Staniscia T., Evert M., Dombrowski F., Schirmacher P., Longerich T.. Oncogenic and tumor suppressive roles of polo-like kinases in human hepatocellular carcinoma. Hepatology. 2010; 51:857–868. [DOI] [PubMed] [Google Scholar]
- 74. Syed N., Coley H.M., Sehouli J., Koensgen D., Mustea A., Szlosarek P., McNeish I., Blagden S.P., Schmid P., Lovell D.P. et al.. Polo-like kinase Plk2 is an epigenetic determinant of chemosensitivity and clinical outcomes in ovarian cancer. Cancer Res. 2011; 71:3317–3327. [DOI] [PubMed] [Google Scholar]
- 75. Ou B., Zhao J., Guan S., Wangpu X., Zhu C., Zong Y., Ma J., Sun J., Zheng M., Feng H. et al.. Plk2 promotes tumor growth and inhibits apoptosis by targeting Fbxw7/Cyclin E in colorectal cancer. Cancer Lett. 2016; 380:457–466. [DOI] [PubMed] [Google Scholar]
- 76. Syed N., Smith P., Sullivan A., Spender L.C., Dyer M., Karran L., O’Nions J., Allday M., Hoffmann I., Crawford D. et al.. Transcriptional silencing of Polo-like kinase 2 (SNK/PLK2) is a frequent event in B-cell malignancies. Blood. 2006; 107:250–256. [DOI] [PubMed] [Google Scholar]
- 77. Harrelson Z., Kelly R.G., Goldin S.N., Gibson-Brown J.J., Bollag R.J., Silver L.M., Papaioannou V.E.. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004; 131:5041–5052. [DOI] [PubMed] [Google Scholar]
- 78. Suzuki T., Takeuchi J., Koshiba-Takeuchi K., Ogura T.. Tbx genes specify posterior digit identity through Shh and BMP signaling. Dev. Cell. 2005; 8:971–972. [DOI] [PubMed] [Google Scholar]
- 79. Manning L., Ohyama K., Saeger B., Hatano O., Wilson S.A., Logan M., Placzek M.. Regional morphogenesis in the Hypothalamus: A BMP-Tbx2 pathway coordinates fate and proliferation through shh downregulation. Dev. Cell. 2006; 11:873–885. [DOI] [PubMed] [Google Scholar]
- 80. Cho G.-S., Choi S.-C., Park E.C., Han J.-K.. Role of Tbx2 in defining the territory of the pronephric nephron. Development. 2011; 138:465–474. [DOI] [PubMed] [Google Scholar]
- 81. Bilican B., Goding C.R.. Cell cycle regulation of the T-box transcription factor tbx2. Exp. Cell Res. 2006; 312:2358–2366. [DOI] [PubMed] [Google Scholar]
- 82. Vance K.W., Carreira S., Brosch G., Goding C.R.. Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 2005; 65:2260–2268. [DOI] [PubMed] [Google Scholar]
- 83. Lv Y., Si M., Chen N., Li Y., Ma X., Yang H., Zhang L., Zhu H., Xu G.-Y., Wu G.-P. et al.. TBX2 over-expression promotes nasopharyngeal cancer cell proliferation and invasion. Oncotarget. 2017; 8:52699–52707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wang B., Lindley L.E., Fernandez-Vega V., Rieger M.E., Sims A.H., Briegel K.J.. The T Box transcription factor TBX2 promotes Epithelial-Mesenchymal transition and invasion of normal and malignant breast epithelial cells. PLoS One. 2012; 7:e41355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. D’Costa Z.C., Higgins C., Ong C.W., Irwin G.W., Boyle D., McArt D.G., McCloskey K., Buckley N.E., Crawford N.T., Thiagarajan L. et al.. TBX2 represses CST6 resulting in uncontrolled legumain activity to sustain breast cancer proliferation: a novel cancer-selective target pathway with therapeutic opportunities. Oncotarget. 2014; 5:1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Du W.-L., Fang Q., Chen Y., Teng J.-W., Xiao Y.-S., Xie P., Jin B., Wang J.-Q.. Effect of silencing the T-Box transcription factor TBX2 in prostate cancer PC3 and LNCaP cells. Mol. Med. Rep. 2017; 16:6050–6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yu H., Liu B.O., Liu A., Li K., Zhao H.. T-box 2 expression predicts poor prognosis in gastric cancer. Oncol. Lett. 2015; 10:1689–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Mohiyuddin M., Mu J.C., Li J., Bani Asadi N., Gerstein M.B., Abyzov A., Wong W.H., Lam H.Y.K.. MetaSV: An accurate and integrative structural-variant caller for next generation sequencing. Bioinformatics. 2015; 31:2741–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Nattestad M., Goodwin S., Ng K., Baslan T., Sedlazeck F.J., Rescheneder P., Garvin T., Fang H., Gurtowski J., Hutton E. et al.. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 2018; 28:1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sedlazeck F.J., Rescheneder P., Smolka M., Fang H., Nattestad M., von Haeseler A., Schatz M.C.. Accurate detection of complex structural variations using single-molecule sequencing. Nat. Methods. 2018; 15:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gong L., Wong C.-H., Cheng W.-C., Tjong H., Menghi F., Ngan C.Y., Liu E.T., Wei C.-L.. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat. Methods. 2018; 15:455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018; 34:3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed data files are publicly released on the ENCODE portal (encodeproject.org) via accession ENCBS760ISV. Datasets 1–6 can individually be accessed via ENCODE accession numbers ENCFF336CFC, ENCFF853HHD, ENCFF467ETN, ENCFF717TPE, ENCFF330UFT, ENCFF241CEK, respectively.