Abstract
RNA polymerase (RNAP), the transcription machinery, shows dynamic binding across the genomic DNA under different growth conditions. The genomic features that selectively redistribute the limited RNAP molecules to dictate genome-wide transcription in response to environmental cues remain largely unknown. We chose the bacterial osmotic stress response model to determine genomic features that direct genome-wide redistribution of RNAP during the stress. Genomic mapping of RNAP and transcriptome profiles corresponding to the different temporal states after salt shock were determined. We found rapid redistribution of RNAP across the genome, primarily at σ70 promoters. Three subsets of genes exhibiting differential salt sensitivities were identified. Sequence analysis using an information-theory based σ70 model indicates that the intergenic regions of salt-responsive genes are enriched with a higher density of σ70 promoter-like sites than those of salt-sensitive genes. In addition, the density of promoter-like sites has a positive linear correlation with RNAP binding at different salt concentrations. The RNAP binding contributed by the non-initiating promoter-like sites is important for gene transcription at high salt concentration. Our study demonstrates that hyperdensity of σ70 promoter-like sites in the intergenic regions of salt-responsive genes drives the RNAP redistribution for reprograming the transcriptome to counter osmotic stress.
INTRODUCTION
RNA polymerase (RNAP) is the transcription machinery in bacteria, and its interaction with genomic DNA orchestrates gene expression in response to environmental cues. In contrast to eukaryotes, there is only one type of core RNAP, consisting of five subunits (α2ββ’ω), responsible for the synthesis of all RNA species in Escherichia coli. It is estimated that there are about 3600–6000 core RNAP molecules in a typical E. coli cell (1,2) to transcribe the genome (∼4.64 Mb) containing 4453 genes (3). There are multiple ways to direct the finite RNAP pool to different locations in the genome to regulate gene expression. One mechanism is mediated by sigma factors. E. coli core RNAP (E) needs to bind a σ factor to form the holoenzyme (Eσ) that recognizes promoters for transcription initiation. The major sigma factor σ70 is responsible for transcription of house-keeping genes, and the six alternative sigma factors are required for transcription of different gene sets (4,5). For example, σ38 is needed for transcription of stress genes under some unfavorable growth conditions, including stationary growth (6,7). In addition, hundreds of transcription factors and some small ligands also direct RNAP distribution across the genome (8–11).
RNAP redistribution to appropriate genes is necessary, especially when fast-growing cells are stressed by the environment. In fast-growing cells, most RNAPs transcribe the rRNA genes (12,13), which are involved in the synthesis of the translational machinery and encompass only a small percentage of the genome. RNAP forms foci at clusters of rRNA operons, resembling a eukaryotic nucleolus, which is compartmentalized to the edge of the relatively compact nucleoid (14–17). In stressed cells, RNAP distributes homogeneously in the expanded nucleoid (18). As a result, the amount of RNAP is limited for simultaneous genome-wide transcription of growth-related genes and stress genes (19). Despite many studies (20–23), the genome features that influence the redistribution of RNAP as the growth conditions change are incompletely understood. The osmotic stress response is an attractive model to address this issue.
The osmotic stress response in E. coli, which involves sequential changes in physiology and global gene expression, has been studied extensively (24–26). Notably, there are sequential changes in cytoplasmic K+, glutamate, compatible organic osmotic solutes (e.g. proline and glycine betaine), ubiquinone-8 and other molecules during the response (27–30). The rapid accumulation of cytoplasmic K+in vivo destabilizes RNAP–DNA interactions. Although molecular crowding, glutamate and other factors have been reported to stabilize the RNAP–DNA interaction under osmotic stress (29,31,32), the mechanism by which RNAP dynamically dissociates from some genes and re-associates with others is still unclear.
Co-imaging RNAP and DNA in fast-growing and salt shocked cells shows four phases of RNAP distribution (33). Prior to stress, RNAP formed distinct foci accompanying the relatively compact nucleoid. At 10 min post-NaCl treatment, significant amounts of RNAP dissociated from the nucleoid into the cytoplasm, and the nucleoid became hyper-condensed. RNAP then re-associated with the periphery of the nucleoid to form a ‘ring’ in 2-dimension. After 45 min, RNAP distribution and nucleoid compaction gradually returned to the starting state. Throughout the osmotic stress response, cells were well synchronized into the distinct states.
For this report, we determined the maps of genome-wide RNAP location in time-course experiments after salt shock and compared these data to global gene expression. Our study not only confirmed a massive dissociation of RNAP from the genomic DNA at the early phase of the osmotic stress response, but also identified a hyperdensity of σ70 promoter-like sites at the intergenic regions of salt shock response genes, contributing to RNAP redistribution and transcription of salt-responsive genes during osmotic stress response.
MATERIALS AND METHODS
Growth conditions and osmotic stress induction
The E. coli strain CC72 (MG1655 with the fluorescent protein venus fused to the 3′-terminal of rpoC) was grown in LB medium (tryptone 10 g/l, yeast extract 5 g/l, NaCl 5 g/l) overnight at 37°C. The overnight culture was diluted to an optical density (OD600) of 0.02 in fresh LB and incubated in a water bath at 37°C. The OD600 of the cell culture were measured at the indicated time points to construct the growth curve. When the OD600 of the cell culture reached 0.2, solid NaCl was added and dissolved to a final concentration of 0.5 M to induce osmotic stress as described (33).
Total RNA synthesis
The protocol to measure total RNA synthesis was essentially as described (34). When cells were grown to OD600 of 0.2 in LB, 4 μCi [5,6-3H] uridine (PerkinElmer) and 20 μg uridine were added to the cell culture (11 ml). Then osmotic stress was induced by adding 0.5 M NaCl. Immediately afterward, 0.5 ml of cell culture without and with osmotic stress was taken at indicated time points and precipitated by 3 ml of ice-cold 10% TCA (trichloroacetic acid). The precipitates were filtered through GF/C glass microfiber filters (Whatman) and washed with 3 ml of ice-cold 2% TCA. The dried filters were placed into scintillation vials with 5 ml scintillation fluid and counted by a scintillation counter.
Microscopy imaging
The samples taken before NaCl addition and 10, 20 and 45 min after inducing osmotic stress were fixed instantly by 3.7% formaldehyde at room temperature for 30 min. Then, the samples were washed twice by M9 medium and stained by Hoechst 33342 (1 μg/μl) for 15 min before being mounted on a slide for imaging. Super-resolution structured illumination microscopy was performed on a N-SIM (Nikon Inc.) with a back-illuminated 16-mm-pixel high-resolution EMCCD camera (Andor, DU897) and Apo TIRF 100× NA 1.49 Plan Apo oil objective. The SIM images were calibrated using fluorescence beads, reconstructed in the Analyze software (Nikon), further processed and analyzed by ImageJ.
ChIP-chip and data analysis
The ChIP experiment was performed essentially as described previously (21). Briefly, the cell cultures at the above four phases were fixed by a 1% formaldehyde and 10 mM sodium phosphate (pH 7.6) solution for 20 min at room temperature. After cell lysis, RNase A treatment and sonication, the chromosome was fragmented to 100–1200 bp. RNAP binding DNA (IP DNA) was immunoprecipitated with RNAP using an antibody against the RNAP β’ subunit and pan mouse magnetic beads. The same sample without the β’ antibody was used as mock immunoprecipitation DNA (mock IP DNA). After washing and de-crosslinking, DNA was purified using the PCR purification kit (Qiagen). Then, the DNA samples were labeled and hybridized to a two-color tiling microarray (Roche-Nimblegen), consisting 389307 oligonucleotides (386486 from E. coli MG1655 genome and 2821 random oligonucleotides). The oligonucleotides were 50 nt long and spaced every 24 nt on both forward and reverse strands. Two to three biological replicates were performed per time point and the average data were used for the following analysis. The correlation between different replicates were measured by corPlot (Ringo) for LB (r = 0.748; n = 2), Na10 (r = 0.580; n = 2), Na20 (r = 0.635; n = 2) and Na45 (0.860, 0.647, 0.803; n = 3).
The Bioconductor package ‘Ringo’ in R was used to analyze ChIP-chip readouts (35). Briefly, raw data were normalized using the variance-stabilizing normalization method. Then, the signal was smoothed using a 400 bp moving median. The threshold intensity was calculated for each condition and a 0.999 quantile was used as upper bound for the null distribution. Each enriched region was detected by algorithms in the package directly. The RNAP-binding intensity across the genome was also extracted from R and mapped to the E. coli chromosome. Then, the RNAP-binding genes and enriched regions were determined and manually inspected, depending on the binding profiles. The coverage of RNAP under different condition was calculated by a custom script in Matlab (mathworks).
RNA extraction and gene expression microarray analysis
Each of the 800 μl cell culture at the above four phases was mixed with an equal volume of hot (65°C) acid phenol:chloroform:IAA (PCI; 125:24:1; pH 4.5) and incubated at 65°C for 5 min. After centrifugation, the upper aqueous phase was retained and extracted by acid PCI for two more times. After precipitation by the same volume of isopropanol at −20°C for 30 min, the RNA was treated by DNase I to remove the contaminant DNA. The RNA was extracted and precipitated again as the above steps, and finally dissolved in DEPC-treated water. The cDNA libraries were constructed using the Double-Stranded cDNA Synthesis Kit (Invitrogen) from the RNA. Labeling and microarray hybridization were performed by Roche-Nimblegen. The raw data were normalized by quantile normalization and Multichip Average (RMA) algorithm (36,37). A total of two technical replicates were performed per condition and each time point was a pool of two biological replicates. Complete linkage clustering was performed with the normalized, log-transformed data using Cluster 3.0 and visualized in TreeView.
Promoter analysis using the σ70 model
All 3736 sequences of E. coli intergenic regions with coordinates were collected from EcoGene 3.0 (38) and Genebank NC_000913.2. The neighboring 30 bases were added to each end of the reported intergenic regions, so that the σ70 promoter-like sites at the edges of the regions would not be cut off. Each extended intergenic region was analyzed by a previously built σ70 model (39), consisting of −35 and −10 individual information-theory based weight matrices that have a varying penalty in bits for each distance between these parts. The sequences were extracted by Pascal programs of the Delila system and then scanned to predict the possible σ70 promoter-like sites for both orientations using the multiscan program (39). The multiscan was performed with filtering to identify the strongest -10/-35 combination for each -10. Without this filter, the density increases linearly and is about 1.6–1.7 times higher. Either computation would give the same results. The number of predicted σ70 promoter-like sites was divided by the length of the region to obtain the density of sites in number of sites per base pair (#/bp). The individual information (bits) of each site in an intergenic region was plotted on the y-axis and the density of sites on the x-axis. The approximately Gaussianly distributed density promoter density and information content were converted to ellipses for simplicity.
Electrophoretic mobility shift assay (EMSA)
To make sure that all promoter-like sites spanning the open reading frames and intergenic regions are included, the selected intergenic regions with neighboring 30 bp extended at each end were amplified by polymerase chain reaction (PCR) using the listed primers containing a universal sequence (5′-TAATCAAATATGCCTT-3′) at the 5′-end (Supplementary Table S1). Then, a second round of PCR was performed to label the purified DNA fragments with Cy5 at both 5′-ends using the Cy5-labeled primer 5′-TAATCAAATATGCCTT-3′. About 1 nM labeled DNA and 100 nM Eσ70 were used in each EMSA experiment. Final KCl concentrations were used as indicated. The labeled probes were incubated with Eσ70 and KCl at 30°C for 30 min in the binding buffer (40 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.9, 4 mM dithiothreitol, 40 mM KCl, 10 mM MgCl2, 0.5 mg/ml calf bovine serum albumin (BSA) and 5% glycerol). After addition of heparin to 55 ng/μl, the reaction mixture was loaded onto a 1.5% agarose gel and electrophoresed in 1.0× TBE running buffer at 4°C. The gel was visualized by Typhoon Trio (GE Healthcare).
Atomic force microscopy imaging
The RNAP–DNA complexes were obtained by mixing 4 nM DNA and 12 nM Eσ70 in binding buffer (20 mM HEPES, pH 7.9, 10 mM MgCl2, 10 μM ZnCl2, 1 mM β-mercaptoethanol) with 50, 100 or 150 mM KCl. After incubating at 30°C for 30 min, the samples were diluted 2- to 4-fold by the binding buffer with the same KCl concentration. The samples were deposited onto 3-aminopropyl silatrane (APS) modified mica (Ted Pella) and incubated at room temperature for 2 min. Then, the surface was rinsed by Milli-Q water and dried by a stream of nitrogen. The samples were imaged with a Cypher VRS Video-Rate Atomic Force Microscope (Asylum Research) in air using a commercial silicon cantilever (OTESPA-R3, Bruker). Images were collected with a scan size of 1 μm in tapping mode at room temperature. Images were exported and analyzed by Gwyddion 2.5.1 software.
In vitro transcription
Supercoiled plasmids with the corresponding intergenic regions were used as templates in the in vitro transcription assays. E. coli rrnB T1 and T2 terminators were located downstream of the cloned intergenic regions in the plasmids so that discrete transcripts could be followed. In a 10 μl reaction mixture, 10 nM supercoiled plasmid, 30 nM Eσ70, 2 μl 5× transcription buffer (200 mM Tris–HCl at pH 8.0, 5 mM dithiothreitol, 0.5 mg/ml purified calf BSA, 50 mM MgCl2 and 50 mM KCl), water and corresponding KCl were mixed to a total volume of 8 μl and preincubated at 37°C for 15 min. Then, 2 μl of 5× NTP mixture (1 mM for ATP, CTP and GTP, 0.1 mM for UTP) containing 2 μCi of [α-32P] UTP (PerkinElmer) was added to the above mixture to initiate the transcription reaction. After incubation at 37°C for 20 min, the transcription reactions were terminated by the addition of 10 μl of stop solution (250 mM ethylenediaminetetraacetic acid, pH 8.0, 10 M urea, 0.05% xylene cyanol and bromphenol blue). Aliquots of the terminated reactions were analyzed on 8% sequencing gel (National Diagnostics). The RNA transcripts were visualized and quantified by autoradiography.
RESULTS
Mapping the temporal location of RNAP-binding sites reveals dynamics of genome wide RNAP–DNA interactions during osmotic stress response
The osmotic stress response is a dynamic process consisting of multiple steps. As shown in Figure 1A, cell growth was immediately arrested by salt shock, and shocked cells resumed growth only at or after 45 min. Most RNA synthesis was also shut down immediately after the addition of salt, and new RNA synthesis appeared largely only at 20 min after salt shock and continued to increase at 45 min after the stress (Figure 1B). Co-imaging of RNAP and DNA with super-resolution structured illumination microscopy (SR-SIM) captured dynamic interactions between RNAP and the nucleoid at the four defined phases during osmotic shock: early-exponential phase (LB, before 0.5 M NaCl treatment), which serves as a reference state for fast-growing cells prior to osmotic stress; shock phase (Na10, 10 min after 0.5 M NaCl treatment to the fast-growing bacterial culture in LB), osmoadaptation phase (Na20, 20 min after 0.5 M NaCl treatment) and recovery phase (Na45, 45 min after 0.5 M NaCl treatment) (Figure 1C). We hypothesize that the dynamic changes in the location of genome-wide RNAP-binding sites would reflect the salt sensitivity (or resistance) of the RNAP–DNA interaction, and/or of gene expression during the osmotic stress response. Thus, we performed ChIP-chip assays to identify the genome-wide location of RNAP-binding sites in the above four typical phases.
Figure 1.
Dynamic dissociation, re-association and redistribution of RNAP with the genome during osmotic stress. (A) Growth curve of CC72 in LB without (close circle) and with (open circle) osmotic stress in early-exponential phase. Red arrow indicates the time that osmotic stress was induced. Red and blue arrows indicate the time that samples were collected for imaging, ChIP-chip and transcriptome assays. Vertical bars represent standard deviations of three biological replicates. (B) The amount of RNA synthesized in LB without (close circle) and with (open circle) osmotic stress relative to time 0. The starting time of osmotic stress is designated as time 0. Vertical bars indicate the standard deviation of the mean for three independent experiments. (C) SR-SIM co-imaging of representative RNAP (green) and nucleoid (red) in the four typical phases. The scale bar represents 1 μm. (D) Genome browser representations of dynamic change in RNAP binding from LB to Na10, Na20 and Na45 with a moving average of 200 kb. Lower bar represents the E. coli genome with the locations of ori (light grey ellipse), ter (dark gray ellipse) and rrn regions (red circle). (E) Number of oligonucleotides with RNAP binding in each of the 10 kb bins. The coverage represents the proportion of oligonucleotides enriched in RNAP out of the number of total oligonucleotides. (F) Scatter plot of mean RNAP-binding signal intensity at Na10, Na20 and Na45 compared with LB. After normalizing the raw data, the relative intensity of RNAP binding was smoothed by a moving median of 400 bp. Then, the corresponding binding intensities of all probes in each 10 kb bin were averaged as the ‘mean RNAP binding’. Each dot represents a 10 kb bin. y = x (black diagonal) is shown to represent the same RNAP binding compared with LB. The smooth colored curves represent loess fit with a span of 0.75. The P values by two-sided t-test between LB and Na10, Na20 or Na45 are shown.
The results of ChIP-chip assays recapitulated the dissociation and re-association of RNAP to DNA at different phases during osmotic stress response, as revealed by image analysis (Figure 1C), but with detailed information on the changes in the number and location of the genome-wide RNAP-binding sites. Figure 1D shows a global view of dynamic RNAP-binding profiles across the E. coli genome (200 kb moving average window) for the four phases. In LB, most peaks of RNAP-binding sites appeared to surround the regions of the seven rRNA operons (rrn) and the origin of replication (ori), but it showed minimal binding around the terminus of replication (ter). In Na10, the loss in RNAP binding was massive across the genome, and only small fractions of RNAP-binding sites remained. At later time points (Na20 and Na45), although more RNAP-binding sites were recovered, the profiles were different from each other and that of LB.
To further study the differences in RNAP-binding profiles with a higher resolution, both RNAP-binding sites and mean RNAP-binding intensities in each 10 kb bin across the genome were calculated. In LB, only about 36% of the genome is ‘covered’ with RNAP; thus, vast regions of the genome appear to be devoid of RNAP binding (Figure 1E). This finding is consistent with electron microscope visualization of RNAP on chromatin spreads (40) and co-imaging of RNAP and the nucleoid in fast-growing cells (15). The coverage of RNAP-binding sites decreased 3-fold from 36% (LB) to 12% in Na10, and the mean length in RNAP binding was also significantly reduced (Supplementary Figure S1). Although the coverages of genome-wide RNAP-binding sites were raised to 25% in both Na20 and Na45, great differences on the RNAP-binding profiles (Figure 1E) and RNAP-binding intensities (t-test, P < 1 × 10−15) were still found across the genome, revealing the phase transition from osmoadaptation phase to recovery phase and further RNA synthesis at Na45 (Figure 1B). Analysis of the mean RNAP-binding signal intensities in each 10 kb bin in Na10, Na20 and Na45 compared with LB (Figure 1F) showed significantly decreased RNAP-binding intensities in 74% and 85% of the 465 bins in Na10 and Na45 (t-test, P < 1 × 10−13), respectively. In contrast, 49% of the bins were found with increased RNAP binding in Na20 (t-test, P = 0.25), which indicates that many RNAP molecules have re-associated with the nucleoid. But most regions with increased RNAP binding at Na20 have weak RNAP binding at LB. This result suggests that the osmotic response genes are likely masked to avoid RNAP binding before osmotic stress. It also shows that the RNAP is not strictly redistributed to some specific regions in the genome.
The relative changes of gene expression and RNAP binding after salt shock were also found to be positively correlated across the whole genome at all time points (Supplementary Figure S2). Taken together, the above results demonstrated the dynamic nature of RNAP redistribution, including RNAP dissociation and re-association across the genome during the time course of osmotic stress, and they indicate that the redistributed RNAP across the genome could orchestrate transcription of stress-responsive genes.
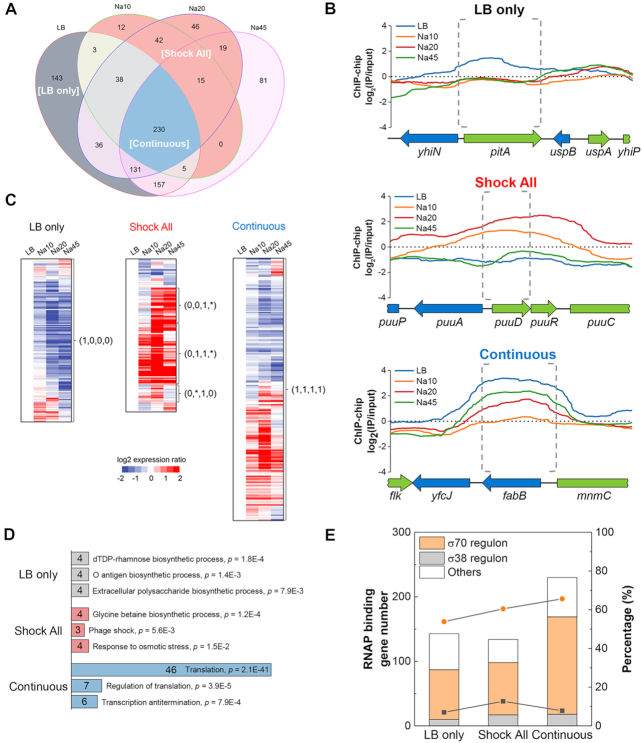
‘LB only’, ‘Shock All’ and ‘Continuous’ genes show different salt sensitivities to RNAP binding
We next used the temporal maps of RNAP localization in the whole genome to determine the RNAP-binding genes in each phase (Supplementary Table S2). The relationship of detected genes with RNAP binding at different time points during the stress response is depicted in Figure 2A. In total, there were 743, 345, 557 and 638 genes with detected RNAP binding in LB, Na10, Na20 and Na45, respectively. These detected genes can be classified into several categories, and we focused on three subsets of genes that exhibited differential salt sensitivities during the osmotic stress response: ‘LB only’, ‘Shock All’ and ‘Continuous’. The ‘LB only’ category contains 143 salt-sensitive genes in which RNAP binding is evident only in LB (Figure 2A). The RNAP binding to these genes was particularly sensitive to elevated cytoplasmic salt concentration after the stress, as indicated by the RNAP-binding profile of a typical ‘LB only’ gene (Figure 2B, top). The temporal transcriptome profiles indicate that the expressions of most ‘LB only’ genes are down-regulated during osmotic stress compared to LB (Figure 2C, left). These results demonstrate that, in general, the lack of RNAP binding corresponds to reduced gene expression for this class of genes.
Figure 2.
‘LB only’, ‘Shock All’ and ‘Continuous’ genes with differential salt sensitivities to RNAP binding. (A) Venn diagram depicting the overlap in the number of RNAP-binding genes among LB, Na10, Na20 and Na45. LB only, gray background; Shock All, red background; Continuous, blue background. (B) Representative RNAP-binding profiles of the genes in ‘LB only’, ‘Shock All’ and ‘Continuous’. The upstream intergenic and coding regions of the target genes in each subset are indicated by gray dashed brackets. The binding profiles below ‘0’ means the RNAP-binding intensity is lower than the negative control. (C) Different patterns of gene expression in ‘LB only’, ‘Shock All’ and ‘Continuous’. The Log2 ratio of gene expression at Na10, Na20 and Na45 relative to LB is shown. The four numbers in parentheses from left to right represent the RNAP-binding state at LB, Na10, Na20 and Na45, respectively. ‘0’, no RNAP binding; ‘1’, RNAP binding. ‘*’ represents ‘0’ or ‘1’. (D) Enriched Go biological processes in ‘LB only’, ‘Shock All’ and ‘Continuous’. Only the top three biological processes are listed. Numbers in the bar indicate the number of genes enriched in each corresponding biological process. (E) Gene number (columns) and percentage (lines) of σ70 and σ38 regulons in RNAP-binding genes of each group. Orange circle, percentage of σ70 regulon; black square, percentage of σ38 regulon. ‘Others’ refers to genes controlled by unassigned sigma factor(s) and/or by sigma factors other than σ70 and σ38.
The ‘Shock All’ subset includes 134 salt-responsive genes (Figure 2A) and RNAP binding at these genes was apparent only in Na10, and/or Na20, but not in LB. The temporal RNAP-binding profile of a typical ‘Shock All’ gene (Figure 2B, middle) showed robust RNAP binding after osmotic stress, which means RNAP binding to these genes was resistant to high salt in vivo and represented osmotic stress-responsive genes. This is further confirmed by DAVID analysis (41), which showed that the ‘Shock All’ genes were enriched in the biosynthesis of glycine betaine important for osmoprotection (28) and other osmotic response processes (Figure 2D, middle). Some genes related to phage shock and iron ion transport were also enriched (Supplementary Table S3), but their function during osmotic response is still unknown. Transcription of most ‘Shock All’ genes were up-regulated during osmotic stress (Figure 2C, middle), they thus showed a strong positive correlation with enriched RNAP bindings. Because there were no apparent RNAP bindings in these genes in LB, it is apparent that expression of the ‘Shock All’ genes is silent in fast-growing cells.
The third subset referred to as ‘Continuous’ contains 230 genes bound by RNAP in all four time-points (Figure 2A). The RNAP-binding profile of a typical ‘Continuous’ gene shows that RNAP binding is maximum in LB prior to the stress, but is significantly reduced in Na10, followed by different extents of recovery at later time points after salt shock (Figure 2B, bottom). Temporal transcriptional profiles of the ‘Continuous’ genes reveal that about half of these genes have increased RNA levels, while the other half have decreased levels (Figure 2C, right). The ‘Continuous’ genes are predominantly involved in translation processes (Figure 2D, bottom). Like rrn genes, translation genes are involved in the production of ribosomes, and expression of these growth genes is robust in fast-growing cells, but they are sensitive to growth rate (19). A significant decrease in RNAP binding in the ‘Continuous’ genes in Na10 indicates that they are partially sensitive to high salt in vivo.
Because these three subsets of genes show different RNAP-binding patterns before and after osmotic stress, they provide a proper entry point to investigate why RNAP dissociates with some genes but still binds to others. Since high salt concentration could generally destabilize the protein–DNA complexes, there should be some specific protein-binding sites existing in different subsets of the intergenic regions or some specific features of these promoters that could help RNAP redistribution.
Hyperdensity of σ70 promoter-like sites is identified at the intergenic regions of salt-responsive genes
It has been reported that there are abundant putative σ70 promoters in the intergenic regions and true promoters occur mostly within dense regions of overlapping promoter-like sites (42); however, their functions are still unknown. Due to the primary function of sigma factors in recognizing a set of gene promoters for RNAP, we propose that some features of the σ70 promoter-like sites may contribute to differential RNAP binding to different classes of genes during the osmotic stress response. We assigned the ‘LB only’, ‘Shock All’ and ‘Continuous’ genes to the known sigma factor regulon(s) based on the RegulonDB (43) and Ecocyc (44) databases. Consistent with the transcription analysis (Supplementary Figure S3 and Table S4), the predominant genes in all three categories belong to the σ70 regulon, and only small fractions of these genes belong to the regulon of global stress regulator σ38 (Figure 2E). A possible reason is that σ38 is rarely expressed in the early-exponential phase cells grown in rich medium (LB) and here only early osmotic response (10, 20 and 45 min) after salt shock are followed. These results indicate that the interactions of RNAP and σ70 promoters are mostly relevant and may be responsible for RNAP redistribution during the osmotic stress response.
To test this hypothesis, a flexible information-theory based σ70 model was used to predict the number of potential σ70 promoter-like sites (39). The σ70 model consists of -10 and -35 weight matrices with a flexible distance of 15 to 20 bases between these two parts. Each promoter-like site has a total ‘strength’ or conservation in bits (Ri, bits/site), which is computed from the sum of the information including the −10 and −35 hexamers and subtracting the entropic cost of the variable distance between them, regardless of whether the site is functional or not in initiating transcription of the downstream gene. To correct for the difference in the length of the intergenic regions of different genes, which range from 1 to 1455 bp, the density of promoter-like sites (sites per base pair, #/bp) is computed and compared.
The sequence conservation (y-axis) and density (x-axis) of predicted σ70 promoters in all E. coli intergenic regions were first analyzed and shown as the colored background (Figure 3A–D), which served as a reference data set. The mean information content of all promoter-like sites was 3.15 bits. The average density of promoter-like sites in all E. coli intergenic regions was 0.042 #/bp, the equivalent to having 4.2 promoter-like sites for a 100-bp intergenic region. Next, all promoter-like sites at each intergenic region of ‘LB only’, ‘Shock All’ and ‘Continuous’ genes were predicted and presented as dots over the colored reference (Figure 3B–D). Since each intergenic region has a particular density and the predicted σ70 promoter-like sites within the region usually have different information, the dots often appear in vertical lines. It is evident that the cloud of dots from ‘Shock All’ is further to the right than those from ‘LB only’ and ‘Continuous’, which means ‘Shock All’ genes had an overall higher density of σ70 promoter-like sites at the intergenic regions.
Figure 3.
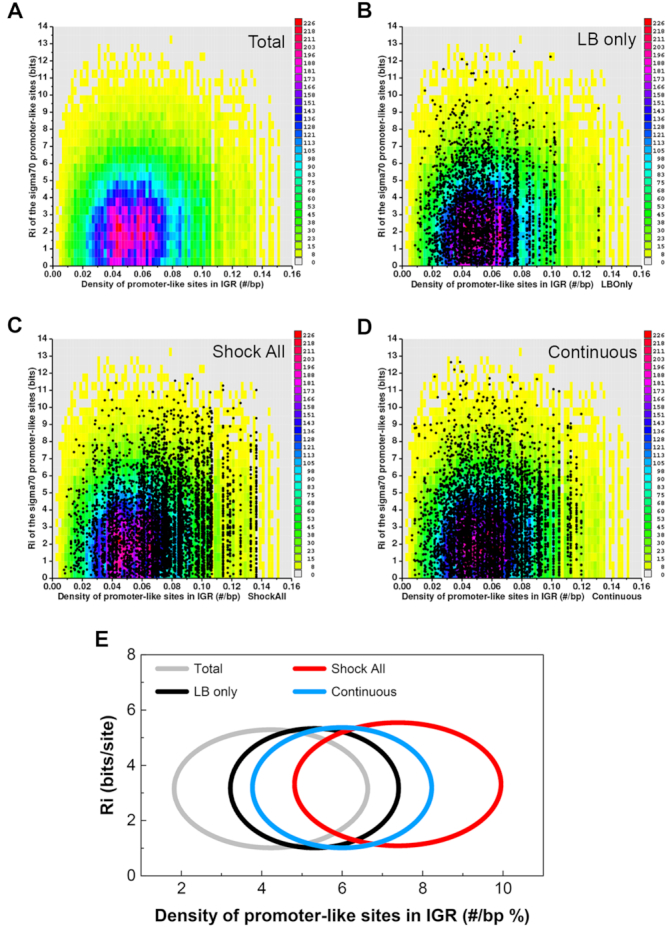
Hyperdensity of promoter-like sites at the intergenic regions of ‘Shock All’ genes. (A) Conservation and density of the σ70 promoter-like sites for all E. coli intergenic regions. Zero bits were used as the cutoff for finding the sites. Each colored rectangle represents the corresponding number of sites in the indicated conservation (bits) and density (#/bp). Scatter plots of all σ70 promoter-like sites at the intergenic regions of ‘LB only’ (B), ‘Shock All’ (C) and ‘Continuous’ (D) genes. The same colored background (as shown in panel A) represents a baseline. Each dot represents a predicted σ70 promoter site and is already represented in the colored reference background. (E) Conservation and density of the σ70 promoter-like sites in each subset are shown by ellipses. Total, the σ70 promoter-like sites of all intergenic regions in the E. coli genome are used as a control. The data shown above are summarized using ellipses. The center of each ellipse represents the average information content (y-axis) and density of the predicted promoters (x-axis) of the corresponding subset. Standard deviation of the information content and promoter density is used as the semi-minor axis and semi-major axis to plot the ellipse, respectively.
Then, these data were simplified using ellipses for better visualization and comparison (Figure 3E). Little variation of the average sequence conservation (Ri, bits/site) of the predicted σ70 promoter-like sites was found. However, it is apparent that the intergenic regions of ‘Shock All’ genes had the highest mean density of promoter-like sites (7.4 #/100 bp), followed by that of ‘Continuous’ genes (6.0 #/100 bp) and of ‘LB only’ genes (5.4 #/100 bp), respectively. This analysis thus reveals a possible positive correlation between the density of promoter-like sites and RNAP-binding ratio in different classes of genes in vivo. For this reason, we propose that the hyperdensity of the σ70 promoter-like sites in intergenic regions of salt-responsive genes, represented by ‘Shock All’ genes, could provide more ways to effectively capture and retain Eσ70 released from those salt-sensitive genes after an osmotic shock.
The intergenic regions with higher promoter density have higher RNAP-binding ratio with increasing salt concentrations
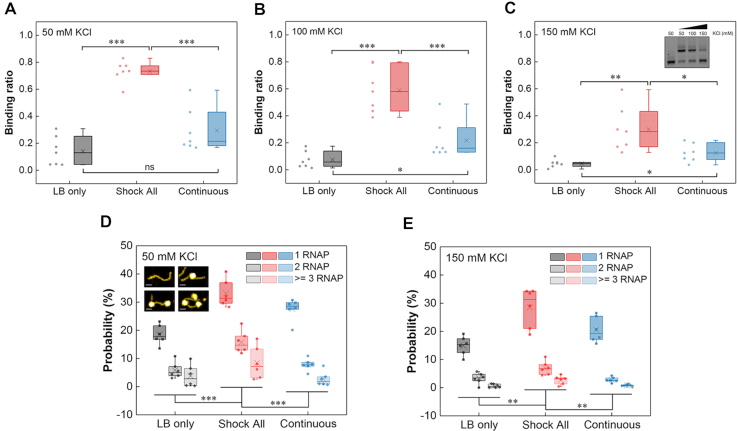
To test the above hypothesis, we synthesized a series of ‘LB only’, ‘Shock All’ and ‘Continuous’ intergenic DNA regions, whose density of promoter-like sites is close to the mean value of each subset (LB only, 5.4 #/100 bp; Shock All, 7.4 #/100 bp; Continuous, 6.0 #/100 bp). Then, we determined their RNAP-binding ratio (the ratio of shifted DNA band with RNAP binding to total DNA) at low (50 mM), medium (100 mM) and high (150 mM) KCl concentrations using electrophoretic mobility shift assays (EMSA). The EMSA results generally showed a stronger RNAP binding to the ‘Shock All’ intergenic regions compared with ‘LB only’ and ‘Continuous’ (Supplementary Figure S4). After quantifying the binding ratio, the data indicated that RNAP always has significantly higher binding to ‘Shock All’ intergenic regions than ‘LB only’ and ‘Continuous’ at each KCl concentration (Figure 4A–C). But ‘Continuous’ intergenic regions did not show significantly higher RNAP binding compared with ‘LB only’ at low salt concentration until 100 and 150 mM KCl (Figure 4A–C). These results indicate a gradient increase in RNAP binding to the intergenic regions of ‘LB only’, ‘Continuous’ and ‘Shock All’ genes. Furthermore, as the salt concentration increases, RNAP prefers binding to the ‘Shock All’ and ‘Continuous’ intergenic regions.
Figure 4.
RNAP binding to the intergenic regions of ‘LB only’, ‘Shock All’ and ‘Continuous’ genes. Boxplots show the RNAP binding to the intergenic regions of ‘LB only’, ‘Shock All’ and ‘Continuous’ at 50 mM KCl (A), 100 mM KCl (B) and 150 mM KCl (C). Each boxplot is based on the dots on the left side. Inset shows the representative RNAP binding to a ‘Shock All’ intergenic region (dhaK-dhaR) at different KCl concentrations. AFM analysis showing the RNAP binding number per DNA molecule in the intergenic regions of ‘LB only’, ‘Shock All’ and ‘Continuous’ at 50 mM KCl (D) and 150 mM KCl (E). Generally, 150–850 DNAs of each intergenic region from two repeats in every viewing field were examined for analysis. Insets show the DNA without and with RNAP (1, 2, and 3) binding by AFM; scale bar, 25 nm. Each boxplot shows the probability of RNAP binding. Each dot represents an intergenic region with the mean promoter density of the corresponding subset. The boxes indicate the lower (25%) and upper (75%) quartiles; the solid lines and crosses represent the median and mean, respectively. The data were compared by two-sided t-test (***P ≤ 0.001; **P ≤ 0.01; *P ≤ 0.05; ns, P > 0.05).
As the EMSA results only represented the overall RNAP–DNA interaction in the population, atomic force microscopy (AFM) was used to obtain snapshots of RNAP–DNA interaction per molecule at low (50 mM) and high salt (150 mM) concentrations. Six intergenic regions with the mean promoter density of each subset (LB only: yceQ-rne, speA-speB, yciM-pyrF, yjhZ-yjhX, yhhQ-tusA and djlA-yabP; Shock All: fepD-entS, astE-spy, nadK-recN, yjdA-yjdM, yciT-osmB and nrdF-proV; Continuous: rlmN-rodZ, ybjL-ybjM, lspA-fkpB, nusG-rplK, yjiS-yjiR and bamD-raiA) were used for AFM imaging. The AFM images showed that 1, 2 or more RNAPs could bind to a single DNA (Figure 4D and Supplementary Figure S5). To quantify and compare the RNAP-binding probability between different groups, we calculated the RNAP binding number per DNA in >150 DNA molecules. It is evident that the percentage of ‘Shock All’ DNA with RNAP binding at both low and high salt concentrations (57%, 41%) was significantly higher compared to the ‘Continuous’ (39%, 23%) and ‘LB only’ (28%, 19%) intergenic regions (Figure 4D and E). These results elucidate that RNAP binding number gradually increases from the intergenic regions of ‘LB only’ to ‘Continuous’ to ‘Shock All’.
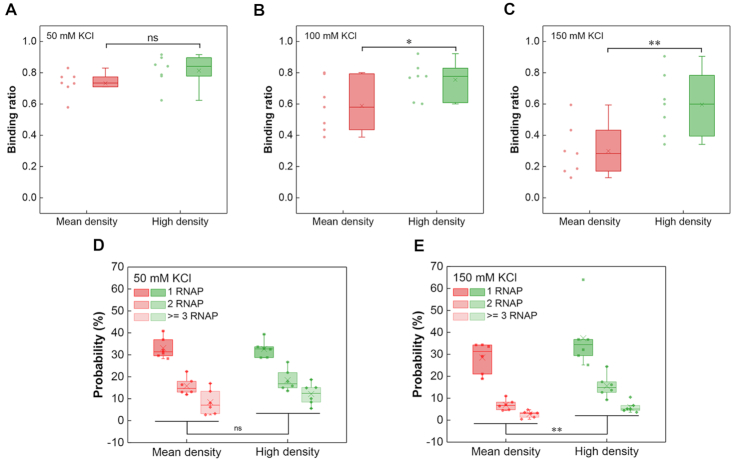
To eliminate the effects of possible conserved sequences or other elements responsible for osmotic response in the ‘Shock All’ intergenic regions, the intergenic regions in the same subset with different promoter densities should be compared. Thus, another set of intergenic regions with a higher density of σ70 promoter-like sites (10.5 #/100 bp) than the mean density (7.4 #/100 bp) in ‘Shock All’ was chosen for EMSA analysis (Supplementary Figure S6). We found that at 50 mM KCl, both sets of intergenic regions (mean, high promoter density) had high RNAP-binding ratio (73%, 81%) without significant difference between them (Figure 5A). However, the RNAP-binding ratio of intergenic regions containing high promoter density dropped slower than those containing mean promoter density with increasing salt concentrations, resulting in significant differences between these two groups at 100 and 150 mM KCl (Figure 5B and C). Consistent with the EMSA assays, the AFM analyses showed similar results at low and high salt conditions (Figure 5D and E). All these results demonstrate that the intergenic regions with higher density of σ70 promoter-like sites have more RNAP binding when the salt concentration increases.
Figure 5.
High promoter density of ‘Shock All’ genes corresponds to high RNAP binding. Boxplots of RNAP binding to the ‘Shock All’ intergenic regions with mean or high promoter density at 50 mM KCl (A), 100 mM KCl (B) and 150 mM KCl (C). RNAP binding number per DNA molecule at 50 mM KCl (D) and 150 mM KCl (E) in the ‘Shock All’ intergenic regions measured by AFM. The ‘Shock All’ intergenic regions with high promoter density (puuA-puuD, gspA-gspC, leuL-leuO, ycgX-iraM, bluF-ycgZ and dcuC-pagP) were used for AFM imaging. The analysis was based on 150–850 DNAs examined for the above intergenic regions in each viewing field. Each dot represents an intergenic region with the mean or high promoter density of the corresponding group. The two-sided t-test was applied for statistical analysis (**P ≤ 0.01; *P ≤ 0.05; ns, P > 0.05).
Density of σ70 promoter-like sites has a direct and positive linear correlation with RNAP binding
The average RNAP-binding ratio of ‘LB only’, ‘Shock All’ and ‘Continuous’ intergenic regions decreased 67% (14.1% to 4.7%), 59% (73.3% to 29.9%) and 57% (29.3% to 12.5%) from 50 mM KCl to 150 mM KCl (Figure 4A–C), respectively. The similar magnitude of RNAP binding decrease suggests that the salt sensitivity of RNAP binding was similar among these three subsets. But the intergenic regions with higher promoter density do have more RNAP binding at high salt. To further determine whether the high density of σ70 promoter-like sites directly contributes to RNAP binding, we manually changed the promoter density of the intergenic regions and tested whether the RNAP-binding ratio was modified accordingly. Thus, a set of chimeras was systematically constructed based on two completely different intergenic regions with similar lengths. The first intergenic region mipA-yeaG from ‘LB only’ has a relatively low density of σ70 promoter-like sites (0.057 #/bp), while the other intergenic region bdm-osmC from ‘Shock All’ has higher promoter density (0.087 #/bp). To make different chimeras, the first quarter, first half and first three-quarters of mipA-yeaG and bdm-osmC were swapped with each other to generate six constructs with increasing promoter densities (0.059 to 0.080 #/bp, Figure 6A). These six chimeras and the two parental intergenic regions were then used to determine the RNAP-binding ratio at low and high salt concentrations (Supplementary Figure S7).
Figure 6.
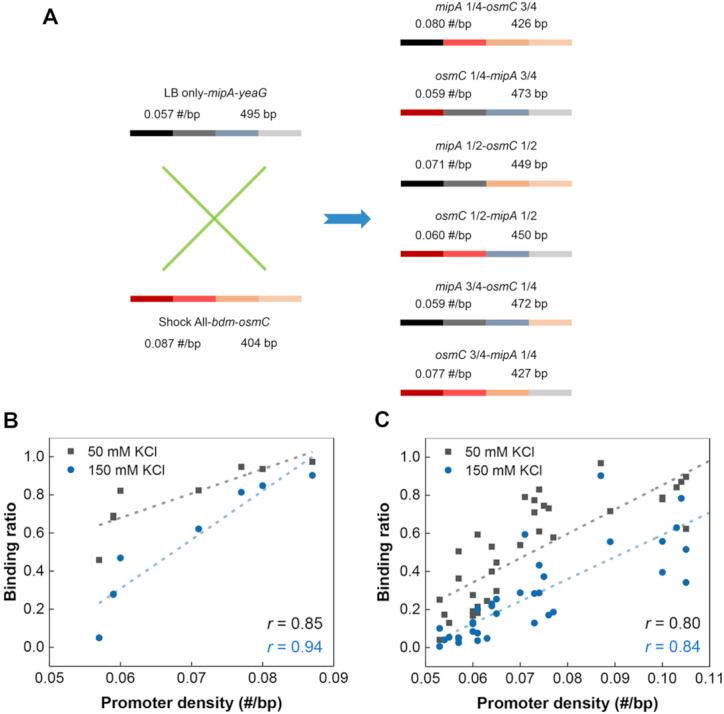
Positive correlation between RNAP binding and the density of promoter-like sites. (A) An overview of the DNA components of six chimeras derived from mipA-yeaG and bdm-osmC. The numbers above the bar indicate the corresponding density of promoter-like sites and length of DNA. Plots of binding ratio with RNAP measured by EMSA against promoter density based on mipA-yeaG, bdm-osmC and six chimeras (B) or the natural intergenic regions (C). Each filled square or circle denotes the data collected from a chimera or natural intergenic region. The linear regression lines are drawn across the plots (dashed line). The Pearson's correlation coefficient (r) is indicated.
From the scatter plots, we observed a strong positive linear correlation (r = 0.85) between RNAP-binding ratio and promoter density of the eight DNA regions at 50 mM KCl (Figure 6B). Although the binding ratio decreased, a similar strong linear correlation (r = 0.94) was also uncovered at 150 mM KCl (Figure 6B). Consistently, similar positive linear correlations (r = 0.80, 0.84) were maintained when more natural intergenic regions were tested (Figure 6C and Supplementary Table S5). These findings clearly demonstrate that hyperdensity of σ70 promoter-like sites in the intergenic regions directly determine one important property—binding to RNAP with high probability at different salt concentrations. Furthermore, the almost identical slopes of the linear regressions (12.8 at 50 mM KCl; 11.6 at 150 mM KCl; Figure 6C) show that RNAP binding to the intergenic regions with varying promoter densities have similar salt sensitivity. It means that as the salt concentration increases, RNAP may completely dissociate with the intergenic regions of low promoter density, but not with those containing high promoter density. Because of the higher promoter density in ‘Shock All’ intergenic regions, in general, this property could lead to the redistribution of RNAP to the ‘Shock All’ intergenic regions to transcribe stress-responsive genes during osmotic stress.
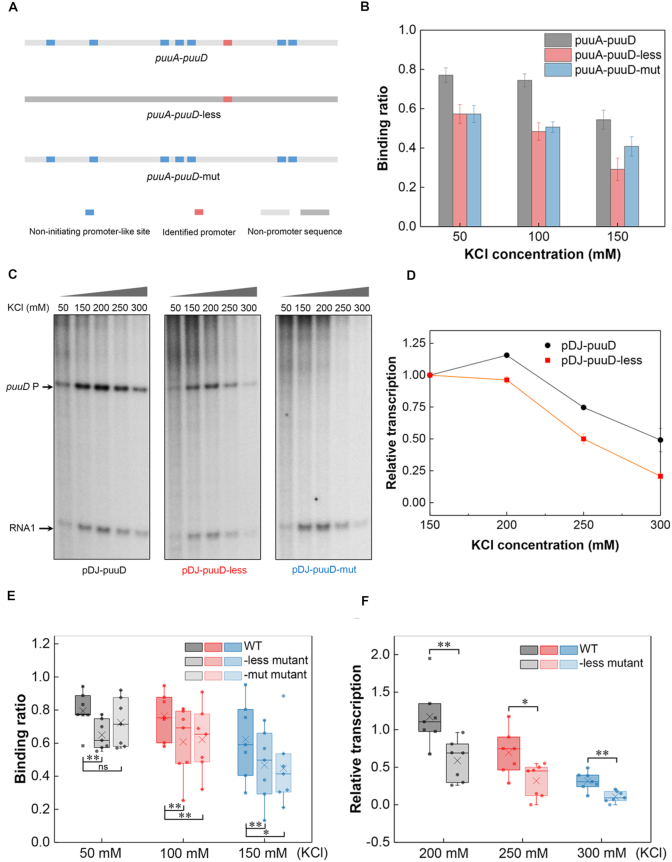
The non-initiating promoter-like sites contribute to more transcription from the actively transcribed promoter at high salt
Consistent with the above results, our in vitro transcription assays revealed relatively higher transcription levels at high salt concentrations when the intergenic region had a higher promoter density (Supplementary Figure S8). We next attempted to dissect whether the non-initiating promoter-like sites contributing to RNAP binding could lead to more transcription from the actively transcribed promoter at high salt. To this end, the ‘Shock All’ intergenic region, puuA-puuD (0.100 #/bp), which has a clearly identified promoter for puuD (45) was chosen for mutational analysis. One mutant construct (puuA-puuD-less) kept the real promoter intact, but all other non-initiating promoter-like sites were replaced with non-promoter sequences derived from gene yagH. The other construct (puuA-puuD-mut) kept the non-initiating promoter-like sites intact, but two mutations were introduced to inactivate the real promoter (Figure 7A and Supplementary Table S6). As expected, puuA-puuD-less showed reduced RNAP binding compared with wild-type at each KCl concentration (Figure 7B).
Figure 7.
The non-initiating promoter-like sites contribute to RNAP binding and transcription. (A) Schematic of the identified promoter and promoter-like sites in puuA-puuD and two derived constructs. (B) RNAP binding to puuA-puuD, puuA-puuD-less and puuA-puuD-mut at varying KCl concentrations. Three independent EMSA experiments were used for the analysis. (C) Effects of KCl concentration on puuD transcription from plasmids pDJ-puuD, pDJ-puuD-less and pDJ-puuD-mut. KCl concentration used in each lane is shown on the top. Arrows indicate the transcripts from puuD promoter and internal control RNA1. (D) Relative quantification of the transcripts from pDJ-puuD and pDJ-puuD-less with increasing KCl. The puuD signal at 150 mM KCl was set as 1. RNA1 was used to normalize the signal. The error bars indicate the standard deviation of three independent experiments. (E) RNAP binding to the wild-type intergenic regions, -less mutants and -mut mutants at 50 mM KCl, 100 mM KCl and 150 mM KCl. -less mutant, all the non-initiating promoter-like sites were removed; -mut mutant, only the actively initiating promoter was mutated. (F) Transcription from the wild-type intergenic regions and -less mutants with increasing KCl concentrations relative to 150 mM KCl. **P ≤ 0.01; *P ≤ 0.05; ns, P > 0.05; two-sided t-test.
Then, the effects of the two mutants on puuD transcription were determined by in vitro transcription assays. As anticipated, there was no puuD transcript (167 nt) made from the DNA template containing puuA-puuD-mut, except from wild-type and puuA-puuD-less DNA templates (Figure 7C). In addition, the relative transcription levels of puuD from wild-type were higher than those from puuA-puuD-less as the KCl concentration increased (Figure 7C and D), in agreement with the results from the EMSA assays. To further confirm the above results, we chose another seven ‘Shock All’ intergenic regions (fepD-entS, nrdF-proV, yjdA-yjdM, bdm-osmC, yiaT-yiaU, gspA-gspC and bluF-ycgZ) analyzed by the previous EMSA to construct the mutants, and then repeated the EMSA and in vitro transcription experiments (Supplementary Figures S9 and S10). With the exception of bdm-osmC, which did not show transcripts from the wild-type intergenic region, similar correlation was also obtained from the other promoter constructs. The RNAP-binding ratio and relative transcription levels from the wild-types were significantly higher than the mutants (Figure 7E and F). These results demonstrate that the non-initiating promoter-like sites in the ‘Shock All’ intergenic regions not only improve the effective RNAP binding, but also contribute to gene transcription during osmotic stress.
DISCUSSION
In this study, we attempted to determine how RNAP redistributes across the genome during osmotic stress response and to understand the mechanism underlying the redistribution. As previously reported, a conserved G-element (GCGG) close to the -35 region of σ38 promoters was found to mediate the transcription of osmo-regulated genes, and the -35 region elements of σ38 promoters were also proposed to explain the binding of RNAP to DNA during osmotic shock (46,47). These mechanisms may hold true for the cells grown in minimal media during osmotic stress response (48–50); however, in fast-growing cells of our study, the global stress regulator σ38 is rarely involved until Na45. The majority of the RNAP-binding genes or up- and down-regulated genes are still under σ70 control (Figure 2E; Supplementary Figure S3C and D), indicating that other mechanisms related to σ70 exist to redistribute RNAP.
Analysis using an information-theory based σ70 model (39) predicted that σ70 promoter-like sites in intergenic regions have high density, and the number of these sites is greatly larger than the number of actively initiating promoters in the genome (51,52), consistent with a previous report (42). Our study found that the mean promoter density of ‘Shock All’ intergenic regions is much higher than the mean promoter densities of ‘LB only’ and ‘Continuous’ intergenic regions. Then we compared the previously reported 78 regions containing promoter islands (53,54) with the ‘LB only’, ‘Shock All’ and ‘Continuous’ intergenic regions. Interestingly, we found that more regions of the 78 promoter islands were included in ‘Shock All’ subset (12/78), compared with ‘LB only’ (1/78) and ‘Continuous’ (3/78). The result suggests that the regions with promoter islands tend to have higher density of promoter-like sites, even if different methods were used to detect the promoters. The obtained dynamic maps of the location of genome-wide RNAP-binding sites during the osmotic stress response showed a dynamic redistribution of RNAP across the genome from ‘LB only’ to ‘Shock All’ genes. Additionally, we did not find possible conserved DNA elements responsible for osmotic adaptation in the promoter regions of ‘Shock All’ genes. Next, we explored the function of promoter density in the intergenic regions in redistributing RNAP. Our study demonstrated a positive linear correlation between the density of σ70 promoter-like sites in the intergenic region and RNAP-binding ratio, even if the promoter-like sites may be not evenly distributed in each intergenic region. One possible explanation is that the RNAP binding before transcription initiation is a dynamic balance between RNAP–DNA association and dissociation. RNAP primarily searches for promoter-like sequences by 3D diffusion (55,56), and different promoter-like sites will not influence RNAP binding to each other, on account of the relatively low RNAP concentration (3600–6000/cell).
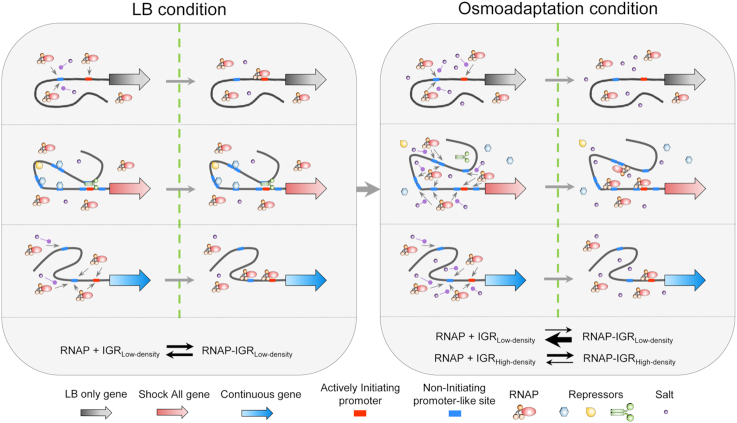
Based on the above property of promoter density, we propose a model for RNAP redistribution during osmotic response (Figure 8). In the LB condition before osmotic stress, most RNAPs mediated by σ70 locate at the intergenic regions of ‘LB only’ and ‘Continuous’, including growth-related genes (e.g. ATP synthesis, lipopolysaccharide synthesis and translational machineries). When the cytoplasmic salt concentration increases transiently, RNAP shows a genome-wide dynamic dissociation with the nucleoid. Because salt can neutralize the electrostatic interaction between RNAP and DNA (57,58), all intergenic regions show decreased RNAP binding. Due to the low and moderate densities of σ70 promoter-like sites in the intergenic regions of ‘LB only’ and ‘Continuous’ genes, RNAP is completely and partially released from these intergenic regions, respectively. In contrast, free RNAP released from above mentioned regions could re-associate to the intergenic regions of ‘Shock All’ genes, because of their high densities of σ70 promoter-like sites, resulting in a relatively high RNAP binding at high salt. This whole process shows a kinetics of RNAP dissociation from the intergenic regions with low promoter density and re-association to those with high promoter density. Once RNAP binds to a promoter-like site, it could reach the real promoter by facilitated diffusion, including 1D sliding or hopping along the DNA (55,56). Thus, there are more ways for RNAP to reach the actively initiating promoter with the help of the high promoter density and subsequently to orchestrate the expression of stress-responsive genes, which also coincides with the appearance of new RNA synthesis in the cell (Figure 1B). This increased RNAP binding leads to the transcription of genes related to osmotic response (e.g. osmC, osmB and osmY), as well as the betaine and proline biosynthetic processes (betAB, betT; proVWX), which could increase the cytoplasmic osmolytes amount. These enriched processes could protect the cell against osmotic challenge (Supplementary Table S3).
Figure 8.
Schematic of osmotic response model based on the density of σ70 promoter-like sites. In each condition, the left section illustrates the potential interaction among RNAP, promoter-like site and salt. The right section represents the equilibrium state of RNAP-σ70 promoter-like sites interaction. The small gray arrow represents the potential RNAP binding to the promoter-like sites. The purple round-head arrow shows the neutralization of the RNAP–DNA interaction by salt. The meaning of each symbol is annotated at the bottom. In LB condition, RNAPs bind to the intergenic regions of ‘LB only’ and ‘Continuous’ genes. The actively initiating promoters of ‘Shock All’ genes may be blocked by the bending, wrapping or bridging of some repressors. In osmoadaptation condition, repressors are released from the nucleoid and RNAPs dissociate with the intergenic regions with low promoter density and RNAPs redistribute to the intergenic regions of ‘Shock All’ genes with high density of σ70 promoter-like sites. IGRLow-density, intergenic region with low density of σ70 promoter-like sites; IGRHigh-density, intergenic region with high density of σ70 promoter-like sites.
Thus, our study provides a mechanism for RNAP redistribution through the reorganized nucleoid during osmotic response. If we further focus on another subgroup of ‘Shock All’ genes, named ‘Shock only’, which show apparent RNAP binding in Na10 and/or Na20 but not in Na45 (Figure 2A), an even higher mean promoter density (8.0 #/100 bp) is identified compared with the mean promoter density of ‘Shock All’ genes (7.4 #/100 bp). The ‘Shock only’ subgroup includes the earliest osmotic stress-response genes, such as betABT and puuR, involved in glycine betaine synthesis (59,60) and regulation of putrescine utilization pathway (45,61,62). These results indicate that the first genes responding to salt shock have an even higher promoter density, which is consistent with the proposed function of promoter-like sites in improving RNAP binding at high salt.
However, one apparent contradiction in the model is that the ‘Shock All’ genes are inactive in fast-growing cells (LB) prior to osmotic stress even though their intergenic regions have higher promoter density. They are activated only after salt shock, indicating that these σ70 promoter-like sites are inaccessible to RNAP prior to salt shock. One possibility is that in fast-growing cells, ‘Shock All’ genes are in spatially remote regions, blocked by nucleoid-associated proteins, such as H-NS, Fis and HU (63–67), relative to RNAP foci. Thus, limited numbers of RNAP could access to those intergenic regions. In contrast, the ‘LB only’ genes are in spatial proximity with RNAP foci readily accessible to RNAP for transcription, even if their intergenic regions have a relatively low promoter density. Upon salt shock, H-NS and other NAPs either dissociate from the nucleoid (33) and/or relocate to other positions in the genome, accompanying by disappeared RNAP foci, thus unmasking the intergenic regions of ‘Shock All’ genes to capture RNAP by the mechanism proposed above. Transcription-coupled supercoiling facilitates the reorganization of the nucleoid so that the stress-responsive genes may be located at the edges (DNA loops) of the hyper-condensed nucleoid 10–20 min after salt shock, as manifested by the RNAP ‘ring’ surrounding the hyper-condensed nucleoid (Figure 1C). Further experiments are warranted to test this possibility.
In conclusion, we identified that hyperdensity of σ70 promoter-like sites in the intergenic regions of salt-responsive genes is the driving force to redistribute RNA polymerase for transcription to adapt to osmotic stress conditions. Our results also suggest that the redistribution of RNAP and the spatial reorganization of the nucleoid orchestrates new programming of genome-wide transcription, which raises a theme of the correlation between the genes’ spatial localization and their transcription level for future investigation.
DATA AVAILABILITY
The Delila programs and models including multiscan and the sigma70 model are available from https://alum.mit.edu/www/toms.
All the ChIP-chip data and transcriptome data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession numbers GSE110831 and GSE110730.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Carmen Mata Martin for SR-SIM imaging, Kevin Franco for providing the promoter-less DNA sequence and Mikhail Kashlev for helpful discussions. We thank Tim Durfee and Fenfei Leng for reading the manuscripts and providing helpful comments. We also thank the NIH Fellows Editorial Board for editorial assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of National Institutes of Health, National Cancer Institute, Center for Cancer Research. Funding for open access charge: Intramural Research Program of National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Conflict of interest statement. None declared.
REFERENCES
- 1. Bakshi S., Siryaporn A., Goulian M., Weisshaar J.C.. Superresolution imaging of ribosomes and RNA polymerase in live Escherichia coli cells. Mol. Microbiol. 2012; 85:21–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Endesfelder U., Finan K., Holden S.J., Cook P.R., Kapanidis A.N., Heilemann M.. Multiscale spatial organization of RNA polymerase in Escherichia coli. Biophys. J. 2013; 105:172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Riley M., Abe T., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R., Glasner J.D., Horiuchi T., Keseler I.M., Kosuge T. et al.. Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic Acids Res. 2006; 34:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Maeda H., Fujita N., Ishihama A.. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000; 28:3497–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gribskov M., Burgess R.R.. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986; 14:6745–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993; 72:165–168. [DOI] [PubMed] [Google Scholar]
- 7. Raffaelle M., Kanin E.I., Vogt J., Burgess R.R., Ansari A.Z.. Holoenzyme switching and stochastic release of sigma factors from RNA polymerase in vivo. Mol. Cell. 2005; 20:357–366. [DOI] [PubMed] [Google Scholar]
- 8. Gao Y., Yurkovich J.T., Seo S.W., Kabimoldayev I., Drager A., Chen K., Sastry A.V., Fang X., Mih N., Yang L. et al.. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res. 2018; 46:10682–10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Browning D.F., Busby S.J.. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004; 2:57–65. [DOI] [PubMed] [Google Scholar]
- 10. Browning D.F., Busby S.J.. Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. 2016; 14:638–650. [DOI] [PubMed] [Google Scholar]
- 11. Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 2000; 54:499–518. [DOI] [PubMed] [Google Scholar]
- 12. Gourse R.L., Gaal T., Bartlett M.S., Appleman J.A., Ross W.. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 1996; 50:645–677. [DOI] [PubMed] [Google Scholar]
- 13. Paul B.J., Ross W., Gaal T., Gourse R.L.. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 2004; 38:749–770. [DOI] [PubMed] [Google Scholar]
- 14. Cabrera J.E., Jin D.J.. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol. Microbiol. 2003; 50:1493–1505. [DOI] [PubMed] [Google Scholar]
- 15. Cagliero C., Zhou Y.N., Jin D.J.. Spatial organization of transcription machinery and its segregation from the replisome in fast-growing bacterial cells. Nucleic Acids Res. 2014; 42:13696–13705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stracy M., Lesterlin C., Garza de Leon F., Uphoff S., Zawadzki P., Kapanidis A.N.. Live-cell superresolution microscopy reveals the organization of RNA polymerase in the bacterial nucleoid. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4390–E4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin D.J., Mata Martin C., Sun Z., Cagliero C., Zhou Y.N.. Nucleolus-like compartmentalization of the transcription machinery in fast-growing bacterial cells. Crit. Rev. Biochem. Mol. 2016; 52:96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin D.J., Cagliero C., Zhou Y.N.. Role of RNA polymerase and transcription in the organization of the bacterial nucleoid. Chem. Rev. 2013; 113:8662–8682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin D.J., Cagliero C., Zhou Y.N.. Growth rate regulation in Escherichia coli. FEMS Microbiol. Rev. 2012; 36:269–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cagliero C., Grand R.S., Jones M.B., Jin D.J., O'Sullivan J.M.. Genome conformation capture reveals that the Escherichia coli chromosome is organized by replication and transcription. Nucleic Acids Res. 2013; 41:6058–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Herring C.D., Raffaelle M., Allen T.E., Kanin E.I., Landick R., Ansari A.Z., Palsson B.O.. Immobilization of Escherichia coli RNA polymerase and location of binding sites by use of chromatin immunoprecipitation and microarrays. J Bacteriol. 2005; 187:6166–6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Grainger D.C., Hurd D., Harrison M., Holdstock J., Busby S.J.. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:17693–17698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wade J.T., Struhl K.. Association of RNA polymerase with transcribed regions in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:17777–17782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kempf B., Bremer E.. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch. Microbiol. 1998; 170:319–330. [DOI] [PubMed] [Google Scholar]
- 25. Cheung K.J., Badarinarayana V., Selinger D.W., Janse D., Church G.M.. A microarray-based antibiotic screen identifies a regulatory role for supercoiling in the osmotic stress response of Escherichia coli. Genome Res. 2003; 13:206–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood J.M. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 2011; 65:215–238. [DOI] [PubMed] [Google Scholar]
- 27. Dinnibier U., Limpinsel E., Schmid R., Bakker E.P.. Transient accumulation of potassium glutamate and its replacement by trehalose during adaptation of Growing-Cells of Escherichia-Coli K-12 to elevated Sodium-Chloride concentrations. Arch. Microbiol. 1988; 150:348–357. [DOI] [PubMed] [Google Scholar]
- 28. Cayley S., Record M.T.. Roles of cytoplasmic osmolytes, water, and crowding in the response of Escherichia coli to osmotic stress: Biophysical basis of osmoprotection by glycine betaine. Biochemistry-US. 2003; 42:12596–12609. [DOI] [PubMed] [Google Scholar]
- 29. Cayley S., Lewis B.A., Guttman H.J., Record M.T. Jr. Characterization of the cytoplasm of Escherichia coli K-12 as a function of external osmolarity. Implications for protein-DNA interactions in vivo. J. Mol. Biol. 1991; 222:281–300. [DOI] [PubMed] [Google Scholar]
- 30. Sevin D.C., Sauer U.. Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli. Nat. Chem. Biol. 2014; 10:266–272. [DOI] [PubMed] [Google Scholar]
- 31. Cayley S., Record M.T. Jr. Large changes in cytoplasmic biopolymer concentration with osmolality indicate that macromolecular crowding may regulate protein-DNA interactions and growth rate in osmotically stressed Escherichia coli K-12. J. Mol. Recognit. 2004; 17:488–496. [DOI] [PubMed] [Google Scholar]
- 32. Kontur W.S., Capp M.W., Gries T.J., Saecker R.M., Record M.T. Jr. Probing DNA binding, DNA opening, and assembly of a downstream clamp/jaw in Escherichia coli RNA polymerase-lambdaP(R) promoter complexes using salt and the physiological anion glutamate. Biochemistry-US. 2010; 49:4361–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cagliero C., Jin D.J.. Dissociation and re-association of RNA polymerase with DNA during osmotic stress response in Escherichia coli. Nucleic Acids Res. 2013; 41:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhou Y.N., Jin D.J.. RNA polymerase beta mutations have reduced sigma70 synthesis leading to a hyper-temperature-sensitive phenotype of a sigma70 mutant. J. Bacteriol. 1997; 179:4292–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Toedling J., Sklyar O., Huber W.. Ringo - an R/Bioconductor package for analyzing ChIP-chip readouts. BMC Bioinformatics. 2007; 8:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P.. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003; 19:185–193. [DOI] [PubMed] [Google Scholar]
- 37. Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P.. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003; 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou J., Rudd K.E.. EcoGene 3.0. Nucleic Acids Res. 2013; 41:D613–D624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shultzaberger R.K., Chen Z., Lewis K.A., Schneider T.D.. Anatomy of Escherichia coli sigma70 promoters. Nucleic Acids Res. 2007; 35:771–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. French S.L., Miller O.L. Jr. Transcription mapping of the Escherichia coli chromosome by electron microscopy. J. Bacteriol. 1989; 171:4207–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 42. Huerta A.M., Collado-Vides J.. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003; 333:261–278. [DOI] [PubMed] [Google Scholar]
- 43. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muniz-Rascado L., Garcia-Sotelo J.S., Alquicira-Hernandez K., Martinez-Flores I., Pannier L., Castro-Mondragon J.A. et al.. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keseler I.M., Mackie A., Santos-Zavaleta A., Billington R., Bonavides-Martinez C., Caspi R., Fulcher C., Gama-Castro S., Kothari A., Krummenacker M. et al.. The EcoCyc database: reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017; 45:D543–D550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nemoto N., Kurihara S., Kitahara Y., Asada K., Kato K., Suzuki H.. Mechanism for regulation of the putrescine utilization pathway by the transcription factor PuuR in Escherichia coli K-12. J. Bacteriol. 2012; 194:3437–3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rosenthal A.Z., Hu M., Gralla J.D.. Osmolyte-induced transcription: -35 region elements and recognition by sigma38 (rpoS). Mol. Microbiol. 2006; 59:1052–1061. [DOI] [PubMed] [Google Scholar]
- 47. Lee S.J., Gralla J.D.. Osmo-regulation of bacterial transcription via poised RNA polymerase. Mol. Cell. 2004; 14:153–162. [DOI] [PubMed] [Google Scholar]
- 48. Weber A., Kogl S.A., Jung K.. Time-dependent proteome alterations under osmotic stress during aerobic and anaerobic growth in Escherichia coli. J. Bacteriol. 2006; 188:7165–7175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Weber H., Polen T., Heuveling J., Wendisch V.F., Hengge R.. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 2005; 187:1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hengge-Aronis R., Lange R., Henneberg N., Fischer D.. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J. Bacteriol. 1993; 175:259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shimada T., Yamazaki Y., Tanaka K., Ishihama A.. The whole set of constitutive promoters recognized by RNA polymerase RpoD holoenzyme of Escherichia coli. PLoS One. 2014; 9:e90447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Conway T., Creecy J.P., Maddox S.M., Grissom J.E., Conkle T.L., Shadid T.M., Teramoto J., San Miguel P., Shimada T., Ishihama A. et al.. Unprecedented High-Resolution view of bacterial operon architecture revealed by RNA sequencing. mBio. 2014; 5:e01442-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shavkunov K.S., Masulis I.S., Tutukina M.N., Deev A.A., Ozoline O.N.. Gains and unexpected lessons from genome-scale promoter mapping. Nucleic Acids Res. 2009; 37:4919–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Panyukov V.V., Ozoline O.N.. Promoters of Escherichia coli versus promoter islands: function and structure comparison. PLoS One. 2013; 8:e62601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang F., Redding S., Finkelstein I.J., Gorman J., Reichman D.R., Greene E.C.. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat. Struct. Mol. Biol. 2013; 20:174–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Friedman L.J., Mumm J.P., Gelles J.. RNA polymerase approaches its promoter without long-range sliding along DNA. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:9740–9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shaner S.L., Melancon P., Lee K.S., Burgess R.R., Record M.T. Jr. Ion effects on the aggregation and DNA-binding reactions of Escherichia coli RNA polymerase. Cold Spring Harb. Symp. Quant. Biol. 1983; 47 Pt 1:463–472. [DOI] [PubMed] [Google Scholar]
- 58. Roe J.H., Record M.T. Jr. Regulation of the kinetics of the interaction of Escherichia coli RNA polymerase with the lambda PR promoter by salt concentration. Biochemistry-US. 1985; 24:4721–4726. [DOI] [PubMed] [Google Scholar]
- 59. Tondervik A., Strom A.R.. Membrane topology and mutational analysis of the osmotically activated BetT choline transporter of Escherichia coli. Microbiology. 2007; 153:803–813. [DOI] [PubMed] [Google Scholar]
- 60. Andresen P.A., Kaasen I., Styrvold O.B., Boulnois G., Strom A.R.. Molecular cloning, physical mapping and expression of the bet genes governing the osmoregulatory choline-glycine betaine pathway of Escherichia coli. J. Gen. Microbiol. 1988; 134:1737–1746. [DOI] [PubMed] [Google Scholar]
- 61. Capp M.W., Cayley D.S., Zhang W., Guttman H.J., Melcher S.E., Saecker R.M., Anderson C.F., Record M.T. Jr. Compensating effects of opposing changes in putrescine (2+) and K+ concentrations on lac repressor-lac operator binding: in vitro thermodynamic analysis and in vivo relevance. J. Mol. Biol. 1996; 258:25–36. [DOI] [PubMed] [Google Scholar]
- 62. Record M.T. Jr, Courtenay E.S., Cayley D.S., Guttman H.J.. Responses of E. coli to osmotic stress: large changes in amounts of cytoplasmic solutes and water. Trends Biochem. Sci. 1998; 23:143–148. [DOI] [PubMed] [Google Scholar]
- 63. Gao Y., Foo Y.H., Winardhi R.S., Tang Q., Yan J., Kenney L.J.. Charged residues in the H-NS linker drive DNA binding and gene silencing in single cells. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:12560–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lang B., Blot N., Bouffartigues E., Buckle M., Geertz M., Gualerzi C.O., Mavathur R., Muskhelishvili G., Pon C.L., Rimsky S. et al.. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007; 35:6330–6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Singh S.S., Singh N., Bonocora R.P., Fitzgerald D.M., Wade J.T., Grainger D.C.. Widespread suppression of intragenic transcription initiation by H-NS. Genes Dev. 2014; 28:214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kahramanoglou C., Seshasayee A.S.N., Prieto A.I., Ibberson D., Schmidt S., Zimmermann J., Benes V., Fraser G.M., Luscombe N.M.. Direct and indirect effects of H-NS and Fis on global gene expression control in Escherichia coli. Nucleic Acids Res. 2011; 39:2073–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dillon S.C., Dorman C.J.. Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 2010; 8:185–195. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Delila programs and models including multiscan and the sigma70 model are available from https://alum.mit.edu/www/toms.
All the ChIP-chip data and transcriptome data have been deposited in the NCBI Gene Expression Omnibus (GEO) under accession numbers GSE110831 and GSE110730.