Abstract
The template-directed formation of phosphodiester bonds between two nucleic acid components is a pivotal process in biology. To induce such a reaction in the absence of enzymes is a challenge. This challenge has been met for the extension of a primer with mononucleotides, but the ligation of short oligonucleotides (dimers or trimers) has proven difficult. Here we report a method for ligating dimers and trimers of ribonucleotides using in situ activation in aqueous buffer. All 16 different dimers and two trimers were tested. Binding studies by NMR showed low millimolar dissociation constants for complexes between representative dimers and hairpins mimicking primer–template duplexes, confirming that a weak template effect is not the cause of the poor ligating properties of these short oligomers. Rather, cyclization was found to compete with ligation, with up to 90% of dimer being converted to the cyclic form during the course of an assay. This side reaction is strongly sequence dependent and more pronounced for dimers than for trimers. Under optimized reaction conditions, high yields were observed with strongly pairing purines at the 3′-terminus. These results show that short oligomers of ribonucleotides are competent reactants in enzyme-free copying.
INTRODUCTION
The transmission of genetic information in the cell involves the formation of phosphodiester bonds between a primer and nucleotide building blocks, directed by a template sequence. The synthesis of the complementary strand is called ‘copying', and two rounds of copying produce a full replica of the original genetic polymer. The enzymatic version of the copying reaction, catalyzed by polymerases, is well known (1), but simpler versions of this process, directed solely by intermolecular forces and chemical reactivity also exist (2). In most instances, enzyme-free genetic copying was studied in an attempt to re-enact what may have happened in the prebiotic phase of evolution, using either DNA or RNA templates. Still, important questions remain open as to how this process may have led to an early version of replication (3,4).
The most likely nucleic acid to have undergone polymerase-free replication is RNA, and the term ‘RNA world' has been coined to describe a scenario, in which this biopolymer acted both as genetic material and as biocatalyst (5–7). The RNA world hypothesis assumes that the transmission of genetic information in a prebiotic world was driven by RNA replication, rather than replication of DNA, today's prime carrier of genetic information in cells. The hypothesis is corroborated by the activity of ribozymes with activity in polymerization, ligation, and splicing, as well as other findings that suggest that a ribonucleotide-based system was an early precursor of today's biochemistry (8–12).
The most pristine form of genetic copying is enzyme- and ribozyme-free copying, i.e. copying in the absence of any biopolymer catalyst. This form of copying is not found in the cell today, but it has been observed experimentally in vitro, using activated nucleotides. The simplest version of this process is oligomerization of activated ribonucleotides on a template (13–15). The more common version is template-directed primer extension, which starts from an existing template-primer duplex or hairpin (16–20). In either case, the activated ribonucleotides have organic leaving groups at their 5′-phosphates, rather than the pyrophosphate leaving group of polymerase substrates. Under typical reaction conditions, elongation of strands takes hours or days, unless modified primers and/or nucleotides are used (21,22). Further, yields are often low (4,23), and the partial hydrolysis of monomers makes it difficult to copy longer sequences (24), unless primer and template are immobilized and spent monomers are removed periodically (25).
Non-enzymatic ligation of RNA strands on a template is a method for copying genetic information via a ‘block condensation' approach. If performed with a short splint strand, rather than a long template, the chemical ligation is a method for synthesizing longer RNA constructs in enzyme-free fashion. Early experiments on chemical ligation were performed by Naylor and Gilham. In 1966, they reported the condensation of two hexathymidilates to the corresponding dodecadeoxynucleotide in the presence of polyadenylic acid as template (26). Later, non-enzymatic replication systems were reported using modified (27) or unmodified, triplex-forming DNA (28). However, the ligation of RNA strands was found to be low yielding in many sequence contexts. Shabarova et al. found that both cyanogen bromide- and carbodiimide-induced ligation reactions are lower yielding for RNA than for DNA (29). Successful, but often incomplete chemical ligations using either of these reagents or cyanogen imidazole were later reported by Sawai (30,31), and Damha for DNA dumbbell ligations involving a ribonucleotide (32), as well as Sutherland for oligoribonucleotide ligations with acetylated species (33). Slow reactions of diphosphates and imidazolides of oligoribonucleotides had been found by Szostak et al. (34), and the same group recently reported that the ligation of preactivated trimers is 100-fold slower than that of monomers (35). This low reactivity is quite surprising, as the template effect should be stronger for oligomers than for monomers, and the bond forming mechanism should be the same.
We became interested in studying enzyme-free ligation of short RNA strands because we recently found reaction conditions that induce the simultaneous oligomerization of ribonucleotides and primer extension with monomers (36). The oligomerization produces a statistical distribution of chain lengths, and not just a primer and a single template, and it is interesting to ask whether all strands produced in the process can be part of a self-replicating system. This prompted us to ask whether very short sequences, namely dimers and trimers, would react under the reaction conditions mentioned above, which also support ribonucleotide-induced peptide chain formation (37) and the formation of cofactors from nucleotide precursors (38). If so, the results may help to bridge the gap between oligomerization of monomers and copying with prebiotically plausible strand mixtures. Further, a methodology for ligating RNA strands would also be useful for practical applications, including the synthesis of circular RNA (39), labeled siRNA (40), modified mRNA (41) and functional nanostructures (42,43).
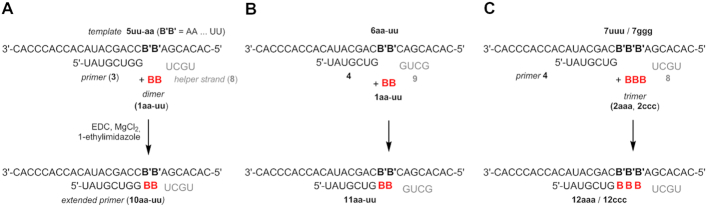
Here we present the results of enzyme-free template-directed ligation of each of the 16 possible dinucleotides at the 3′-terminus of a primer, using an aqueous solution of a carbodiimide and an organocatalyst. The general reaction scheme for dimers is shown in Figure 1. Optimization of the reaction was performed for dimers, and all 16 possible dinucleotide sequences were tested to reveal the effect of the dimer sequence on the yield of ligation. The optimized protocol gave yields exceeding 80% for some sequences, despite the tendency of dimers to cyclize. Exploratory experiments with trimers indicated that they suffer from cyclization to a lesser extent than dimer, resulting in higher yields for difficult-to-ligate sequences. Still, even for a poorly ligating dimer, the yield of ligation was found to be higher than that for extending by a monomer in a competition assay.
Figure 1.
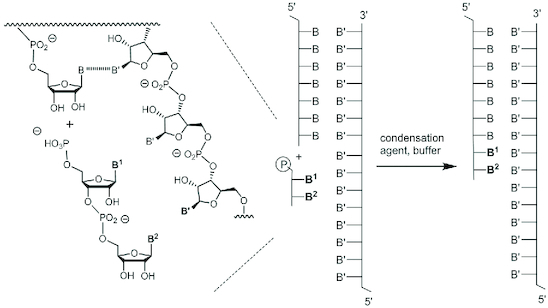
Enzyme-free template-directed ligation of a dinucleotide at the terminus of a primer; B and B' are nucleobases.
MATERIALS AND METHODS
General
Anhydrous solvents were from Sigma/Aldrich/Fluka (Deisenhofen, Germany), Merck (Darmstadt, Germany), VWR Chemicals (Darmstadt, Germany), or Acros (Geel, Belgium) and were stored over molecular sieves. Methoxytrimethylsilane was from TCI (Zwijndrecht, Belgium). The controlled pore glass (cpg) for RNA synthesis loaded with the 3′-terminal nucleoside as well as the 2′-TBDMS-protected phosphoramidites were from Sigma (Deisenhofen, Germany) and had the usual DMT group at their 5′-position, a tert-butyldimethylsilyl (TBDMS) group as 2′-protecting group, and a tert-butylphenoxyacetyl group (tac) as nucleobase protecting group for A, C and G. Triethylamine trihydrofluoride 98%, Dowex 50 WX8-200 cation exchange resin, N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide (EDC), 1-ethylimidazole, MOPS and HEPES were also from Sigma, as well as the reagents for oligonucleotide synthesis, including Deblock solution (3% tricholoracetic acid in CH2Cl2), Cap A (10% tert-butylphenoxyacteyl anhydride in THF), Cap B (16% 1-methylimidazole in THF), Activator (0.25 M 4,5-dicyanoimidazole in MeCN) and Oxidizer (THF/H2O/pyridine/iodine; 66/12/22/0.6, v/v/v/w). Chemical phosphorylation reagent 2-[2-(4,4′-dimethoxytrityloxy)ethylsulfonyl]ethyl-(2-cyanoethyl)-(N,N-diisopropyl) phosphoramidite was from Glen Research or was synthesized in-house following a known procedure (44). Hairpin oligonucleotides (13aa, 13ag) were purchased from Biospring (Frankfurt a. M., Germany) in HPLC-purified form and were used without modification. Tetramers were prepared as described in chapter 3 of the Supplementary Information. All other oligonucleotides, except for dimers and trimers, were from Biomers (Ulm, Germany). Deuterated water (99.9%) was from Euriso-Top (Saclay Gif/Yvette, France). The NMR standard 3-(trimethylsilyl)propionic acid-d4 (TPS) was from Merck (Darmstadt) and SepPak RP-C18 cartridges were from Waters (Eschborn, Germany).
Dinucleotides
The following protocol is for 1ag and is representative. Dinucleotides were synthesized manually, using columns for automated synthesis connected to two syringes. The cpg loaded with the first guanosine nucleoside (80 mg, 2.5 μmol loading) was placed in the synthesis column and was then treated with Deblock solution for 2 min via the syringe. The solution was flushed out via the syringes, and the cpg was washed with dry MeCN (2 mL). The phosphoramidite solution for the adenosine building block (0.12 M in MeCN, 0.4 mL, 20 eq) was mixed with Activator solution (0.4 mL, 40 eq), the mixture was added to the cpg and was allowed to react for 20 min with mixing via the syringes every 2 min. The solution was removed and replaced by a fresh coupling mixture of the same phosphoramidite and Activator. The second coupling was again performed for 20 min to ensure high yields. The cpg was washed with MeCN (2 mL), followed by capping with a mixture of Cap A and Cap B solutions (2 mL, 1:1, v/v) for 1 min. The cpg was washed with MeCN (2 mL), and the phosphite linkage was then oxidized with Oxidizer solution (3 mL), added over the course of 3 min, followed by washing with dry MeCN (2 mL). The DMT groups were removed with Deblock solution for 2 min. A second coupling cycle was performed with Chemical Phosphorylation Reagent without capping, otherwise following the same protocol. The cpg was removed from the column, dried at 1 mbar for 15 min, and was then treated with concentrated aqueous ammonia (1 mL, 30%) for 16 h at 35°C. The supernatant was collected, and the cpg was washed with water (3 × 1 mL). The combined aqueous solutions were freed of excess ammonia with a gentle flow of nitrogen, directed onto the surface of the solution, for approx. 1 h. The remaining solution was then lyophilized, producing a colorless solid. The partially protected dimer was dissolved in triethylamine trihydrofluoride 98% (50 μL, 900 μmol, 450 eq) for 10 h at 25°C. Remaining fluoride was quenched with methoxytrimethylsilane (260 μL), leading to a precipitate after 10 h. If the precipitation was incomplete, acetone (50 μL) was added to induce further precipitation of the desired dinucleotide. The supernatant was removed, and the solid was washed with a cold solution of NaClO4 in acetone (50 mM, 3 × 500 μL). The crude product was dried at 0.1 mbar for 3 h, dissolved in water and lyophilized. Purification on a C-18 SepPak cartridge (1.0 g) involved rinsing the cartridge with MeCN (2 mL), demineralized water (20 mL), and brine (0.25 M NaCl, 5 mL). The dimer (2 μmol) was dissolved in brine (0.25 M, 250 μL) and loaded on the cartridge, followed by washing with brine (0.25 M, 5 mL) and water (5 mL). The dimer was eluted with MeCN (5% in H2O, 6 mL). Fractions (1.5 mL) were analyzed via UV absorption. The fractions containing the dimer were collected and lyophilized. Dimers were analyzed by 1H- and 31P-NMR spectroscopy and MALDI-TOF MS, and yields were determined by UV absorption. Yields ranged from 25 to 50% for the different dimers. The synthesis of trimers followed similar procedures, except that the first coupling step was followed by a second coupling step, which was then followed by the phosphorylation. A list of yields, as well as analytical data can be found in the Supplementary Information.
Ligation
The following protocol is for the ligation with dimer 1ag and is representative. A polypropylene vessel was rinsed with aqueous ammonia (25%) and dried at 80°C for 1 h. Stock solutions of oligonucleotides in deionized water were added to the vial to give concentrations of 40 μM primer (3) 60 μM template (5uc), 60 μM helper (8) and 2 mM dimer (1ag) in the final volume. The strands were annealed by heating to 80°C for 10 min and cooling to room temperature in 20 min. Then, a stock solution of cold condensation buffer (2.5-fold concentrated) and water were added to give the final volume of 10 μL, with the following concentration of buffer components: 500 mM MOPS, 150 mM 1-EtIm, 80 mM MgCl2 and 800 mM EDC. If necessary, the pH was adjusted to a value of 6.0 using 10 M NaOH or 20 M HCl. Ligation was allowed to occur at 4°C for at least 7 days and was monitored by drawing samples (0.7 μL) after stated intervals that were analyzed by MALDI-TOF mass spectrometry, as described below. Primer extension with monomers or trimers in condensation buffer followed the same procedure.
Kinetics of cyclization
Cyclization of dinucleotide 1aa was monitored via MALDI-TOF mass spectrometry during primer extension assays in condensation buffer. The ratio of signal intensities of the dimer 1aa and the cyclic dimer cyc-1aa, with its lower mass, were plotted over time. The rate constant of cyclization was determined by fitting the equation y = y0 exp(–kt), where y is the fraction of dimer, k is the rate constant and t is time, to the experimental data, for a first order reaction. The same procedure was applied to determine the rate constant of cyclization for trimer 2aaa.
Mass spectrometry
Mass spectra were measured on Microflex MALDI-TOF spectrometer (Bruker Daltonics) with a N2 laser (337 nm), using the Flex Control software in linear negative mode. For samples with a mass higher than 1000 Da, the laser was set at 70% of its maximal intensity. One spectrum was the sum of 200 laser shots at 1.5 Hz frequency. For target compounds with a mass lower than 1000 Da, laser intensity was dropped to 50%, with a frequency of 2 Hz and accumulation of 150 shots. Aliquots of assay solutions (0.7 μL) were desalted with the ammonium form of cation exchange resin Dowex 50 WX8-200 in 25 μL volume for 20 min. Sample of 0.5 μL were dried on the MALDI target, and a mixture (0.4 μL) of matrix and co-matrix solutions (0.3 M trihydroxyacetophenone in EtOH, and 0.1 M aqueous ammonium citrate; 2:1) was placed on the spot. Analysis was performed on the [M–H]− ions.
RESULTS
Figure 2 shows the sequences employed in our study. Either dinucleotides with a 5′-terminal phosphate (1) or trimers with a 5′-terminal phosphate (2) were employed. The ligation experiments with dimers were performed in two different sequence contexts, using either primer 3 or primer 4, and combined with either template 5aa–5uu or template 6aa–6uu, where the two letters indicate the sequence of the two templating bases in 3′ to 5′ direction. All 16 different dinucleotides 1aa–uu were prepared by manual solid-phase synthesis with chemical phosphorylation of the 5′-terminus, as described in the Materials and Methods section, and were obtained in pure form after cartridge purification. Analytical data are listed in the Supplementary Information. The two trimers tested, 2aaa and 2ccc were prepared similarly, using solid-phase synthesis with 2′-TBDMS protected phosphoramidites.
Figure 2.
Sequences and ligation reactions studied.
We reasoned that in situ activation would overcome the difficulties with preparing and handling activated strands. We chose the system shown in Figure 2A for the initial experiments. The starting point for the optimization of the reaction conditions was the ‘General Condensation Buffer’ described by Jauker et al. for primer extension with monomers (36). This is an aqueous buffer containing EDC as water-soluble carbodiimide, 1-ethylimidazole (1-EtIm) as organocatalyst, MgCl2 and HEPES. In some experiments, a two-fold diluted version was used to lower the likelihood of side reactions between the strands and the condensing agent. Further, a short downstream-binding strand dubbed ‘helper' (8) was added to provide additional stacking interactions to the incoming dinucleotide (45). In the strand system of Figure 2B, this was tetramer 9. The short helper strands bind transiently and are readily removed in washing steps, when immobilized templates are used (25). Finally, we tested either room temperature or 4°C as reaction temperature, the latter to favor binding of the dimer to the template.
While monomers had been employed at a concentration of 50 mM, dimers were employed at 10 mM. Ligation was monitored via MALDI-TOF mass spectrometry. Except for reactions with 1gg, extended primers were found to have only slightly lower desorption and ionization yields than 3, as shown in calibration plots with representative synthetic control sequences (Supplementary Figure S18, SI). The four different dimers of the sequence NG, where N is any of the four nucleotides (A, C, G or U), were tested. Figure 3 shows a representative spectrum. Mass spectrometry does not provide information on whether a 3′,5′- or 2′,5′-diester has been formed, but it is reasonable to assume that either of two hydroxy groups of the 3′-terminus of the primer can attack as nucleophile. The maximum conversion, observed after a reaction time of one week or more, was between 9% for 1ug and 46% for 1cg. So, significant but not quantitative extension was found with the initial buffer composition. Because the highest conversion yield was not far from yields for extension with pre-activated monomers, such as OAt-CMP (24), this finding suggested that dinucleotides are indeed suitable for ligation with in situ activation. Still, optimization of the reaction conditions was called for.
Figure 3.
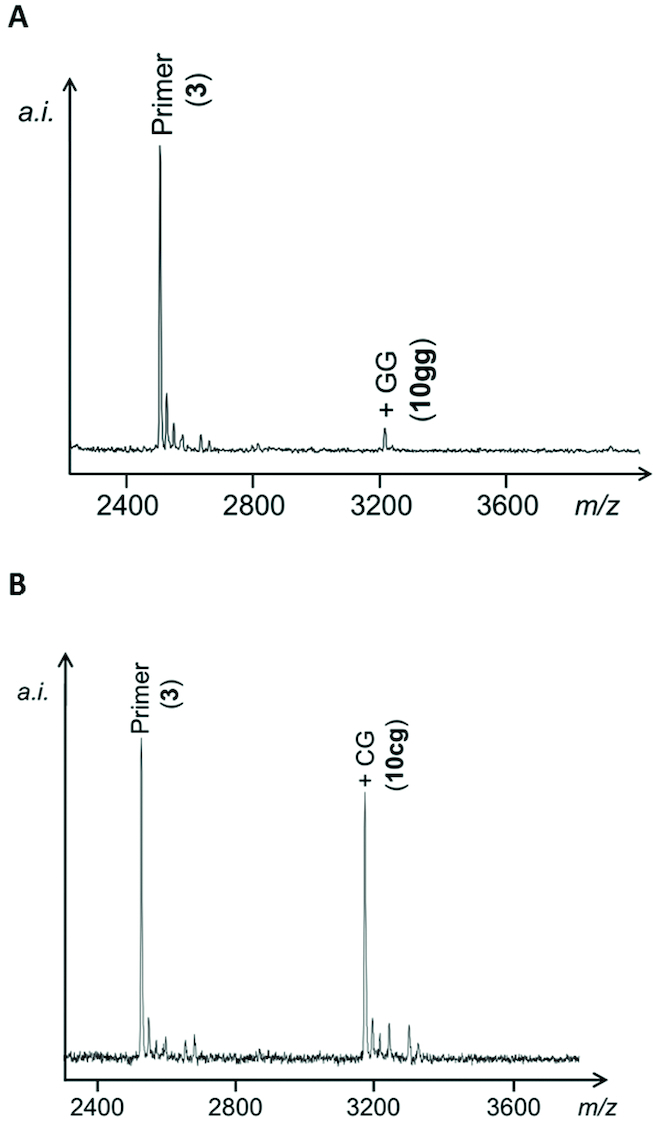
MALDI-TOF mass spectra from ligation assays (A) with dimer 1gg and the duplex of 3 on template 5cc, or (B) with dimer 1cg on template 5gc after 16 days at 4°C. Conditions: 10 mM dimer, 40 μM primer, 44 μM template in half-concentrated condensation buffer (250 mM HEPES, 400 mM EDC, 75 mM 1-ethylimidazole, 40 mM MgCl2 at pH 7.5). No further conversion was observed after longer reaction times.
Binding strength of dimers
First, we studied the binding equilibrium between dimers and templates to ascertain whether higher concentrations may be required for high yielding ligations. Two different methods were employed. The first used hairpins 13aa/ag (Figure 4) as models for primer–template duplexes and chemical shift changes in NMR spectra upon addition of the dimer to measure binding of the dimer 1cu or 1uu. The NMR-based methodology has previously been established for studying the analogous binding equilibrium for monomers (24,46). Representative NMR data are shown in the Supplementary Information. Using this method, dissociation constants of 2 mM (13aa/1uu) and 5–8 mM (13ag/1cu) were found (Supplementary Figure S19, SI). In the latter case, two independent titrations over different concentration ranges were performed. Compared to the Kd values for monomers UMP (500 mM) and CMP (20 mM) determined in a similar hairpin system (46), these values are one to two orders of magnitude lower, confirming the gain in affinity expected for the second nucleotide.
Figure 4.
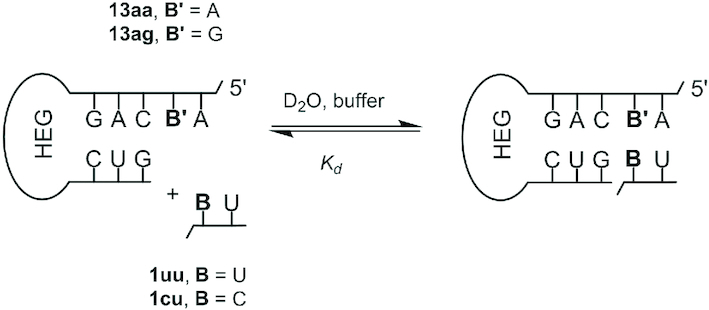
Binding equilibrium between hairpin and dimer to determine a dissociation constant (Kd). HEG stands for a hexaethyleneglycol linker (47).
The second method tested the inhibitory effect of unactivated dimer on the ligation of a pre-activated dimer to estimate binding strength. A similar approach has previously been used by us to measure the binding of monomers (46). Figure 5 shows kinetics for ligation with the oxyazabenzotriazole (OAt) ester 14ag, which had been prepared from 1ag and HOAt, as described in the Supplementary Information. In the absence of inhibitor, ∼15% conversion to 10ag was observed. Upon addition of half an equivalent of 1ag, the yield dropped, and it dropped further, when a full equivalent of the inhibitor was added in a subsequent assay. At 5 mM concentration of 1ag, no ligation product was detectable, indicating <2% conversion to the extended primer or >90% suppression of the reaction at 2.5 equivalents over 14ag. There is too little conversion for a full quantitative treatment (46), but the available data are consistent with the conclusion that the dissociation constant for the complex of the strands with 1ag must be ≤1 mM. This value is smaller than the ones obtained in the hairpin system, as expected for binding in the interior of a longer sequence with a neighboring helper strand, rather than to the dangling residues of a hairpin. Further, 1ag contains two purines that will stack better than the two pyrimidines of 1uu. We also note that the upper limit of the Kd value is similar to the dissociation constants of 0.2 mM recently reported by Szostak et al. for a derivative of a dinucleotide (GpppG) binding to a primer:template duplex (48).
Figure 5.
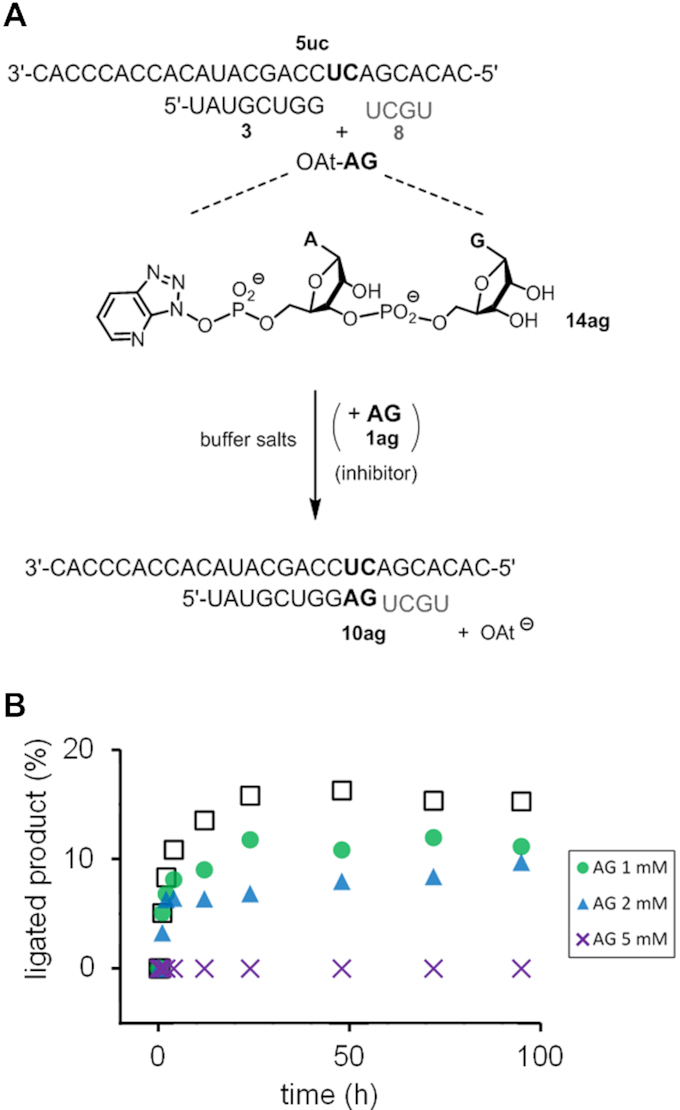
Inhibitory effect of unactivated dimer 1ag on the ligation of activated dimer 14ag to 3 on template 5uc with helper 8. (A) Reaction scheme, and (B) kinetics of ligation at 2 mM 14ag in the absence (open squares) or in the presence of 1ag as inhibitor at the concentrations given. Conditions: 200 mM HEPES, 400 mM NaCl, 80 mM MgCl2, pH 8.9, 22°C.
Taken together, the affinity study confirmed that the additional nucleotide of dimers adds at least one to two orders of magnitude to the binding constants known for monomers (46), particularly when additional stacking interactions with a helper strand are accessible. For the assays of Figure 2 this means that the occupancy of the ligation site at low millimolar concentrations of dimers will usually be at least 50%. This is so high that there is little reason to work at a concentration higher than 10 mM for the dimer starting materials. Rather, other parameters affecting the ligation reaction seemed more promising targets for optimization.
Optimization of reaction conditions
To this end, we performed a screen of reaction conditions using template/primer combination 5ac/3 and 1ug, a dimer sequence poorly incorporated in unmodified General Condensation Buffer, giving just 9% conversion after one week. The results of representative assays are shown in Figure 6 and Table 1. First, the pH was varied. An improvement in primer conversion was found when the pH was adjusted to 6.0, i.e. conditions that favor protonation and thus the electrophilicity of the carbodiimide condensing agent. Second, tetramer helper 8 was added to provide stacking interactions downstream of the ligation site (49), as mentioned in the context of the binding study, above. The additional short strand induced a modest increase in yield. Downstream-binding strands can also suppress secondary structures in the template that interfere with primer extension (50).
Figure 6.
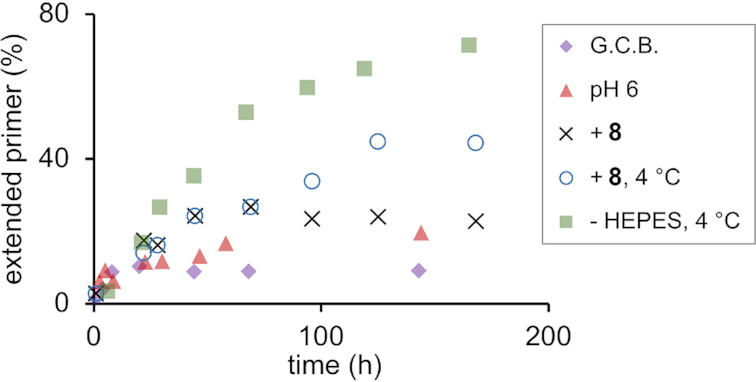
Kinetics of ligation of dimer 1ug (10 mM) to primer 3 (40 μM) on template 5ac (60 μM) under different reaction conditions. The legend to the right of the plot lists changes in the reaction buffer: G.C.B. = general condensation buffer (500 mM HEPES, 800 mM EDC, 150 mM 1-ethylimidazole, 80 mM MgCl2, pH 7.4), + 8 denotes assay solutions with helper 8, the solution lacking HEPES contains all other components. In all cases, except G.C.B., the pH was 6.0.
Table 1.
Results of ligations of dimer 1ug to primer 3 on template 5ac, with or without helper 8 and different reaction conditionsa
| Oligomers | pH | T [°C] | Buffer | Conversion [%]b |
|---|---|---|---|---|
| 5ac/3 | 7.5 | 22 | HEPES | 9 |
| 5ac/3 | 6.0 | 22 | HEPES | 19 |
| 5ac/3/8 | 6.0 | 22 | HEPES | 24 |
| 5ac/3/8 | 6.0 | 4 | HEPES | 49 |
| 5ac/3/8 | 6.0 | 4 | MOPS | 68 |
| 5ac/3/8 | 6.0c | 4 | None | 71 |
aConditions: 10 mM dimer, 40±10 μM primer, 60±15 μM template, 60±15 μM helper, 500 mM HEPES or MOPS, 800 mM EDC, 150 mM 1-ethylimidazole and 80 mM MgCl2.
bAs determined after ≤6 days.
cStarting value.
A significant improvement resulted from lowering the temperature. At 4 °C, the conversion reached twice the value measured at room temperature, possibly because cold temperatures favor binding and reduce hydrolysis of carbodiimide and activated species. Another significant increase in conversion was found when the buffer salt was changed from HEPES to MOPS, i.e. a compound whose pKa is better matched than that of the former and that does not contain a hydroxy group, which may exhibit reactivity toward active species. A further screen revealed that omitting the buffer salt entirely further improved the yield slightly, leading to 71% conversion after 7 days at 4 °C. In the absence of a buffer, the pH usually shifts to more basic values over time, favoring formation of phosphodiesters, but disfavoring activation with EDC. For most subsequent assays, we opted for the version with MOPS as buffer to ensure a stable pH. Overall, at the end of the optimization study, the ligation of 1ug to primer 3 was 7-fold higher yielding than under the initial, unoptimized conditions.
Effect of dimer sequence
We then turned to testing the effect of dimer sequence on the ligation reaction. For this, the full set of 16 different dimers (1aa–uu) and the primer–template combination 6aa–uu/4 was used (Figure 2B). The yield of primer extension was found to strongly depend on the dimer employed. Figure 7 shows a MALDI-TOF mass spectrum for ligation of dimer 1cg with two strongly pairing bases. Even though this is a self-complementary sequence, which may suffer from partial self-pairing of the dimer, a high-yielding reaction was observed with minimal side reactions with the excess of the electrophilic carbodiimide. Figure 8 shows kinetics for four representative dimers, including two that performed well (1gg and 1cg) and two that gave much less conversion (1uc and 1au).
Figure 7.
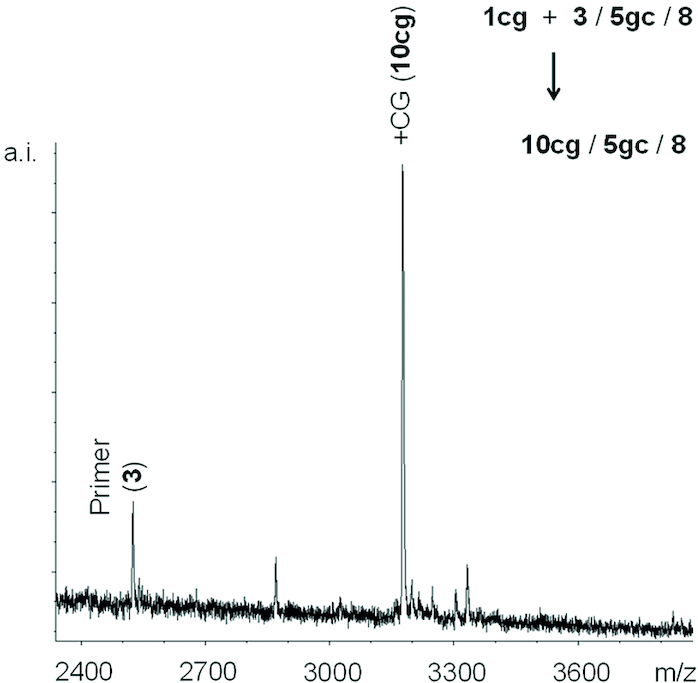
MALDI-TOF spectrum from ligation of dimer 1cg to primer 3 under optimized conditions after 10 d reaction time. Conditions: 30 μM primer, 2 mM dimer, 40 μM template 5gc, 40 μM helper 8, 0.5 M MOPS, 0.8 M EDC, 0.15 M 1-EtIm and 0.08 M MgCl2 at pH 6.0 and 4°C. The low level side product observed at a mass 155 Da above that of the extended primer is probably from a side reaction with EDC.
Figure 8.
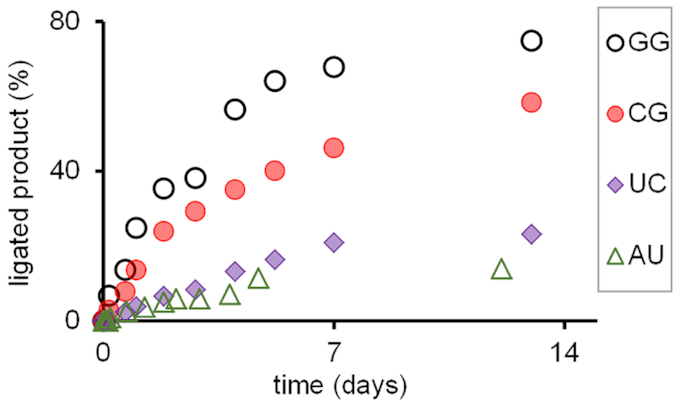
Kinetics of assays of primer extension with dinucleotides 1gg, 1cg, 1uc and 1au in condensation buffer. Conditions identical to those of Figure 7.
Table 2 lists the conversion determined mass spectrometrically for any of the 16 different ligation substrates on their corresponding templates. Extension yield at the end of the assay varied from 3% for 1uu to 83% for 1ug. The average yield for the four dimers with an A residue at the 3′-terminus was 28%, whereas the same value was 19% for those terminating in a C, 68% for those terminating in a G, and just 14% for those with a U as 3′-terminal base, even though the concentration of the dimers had been raised to 5 mM in this case. So, there is a clear order in the observed conversion with 3′-G > 3′-A > 3′-C > 3′-U. For the 5′-terminal base, the average value for the four dimers with an A was 23%, for those with a 5′-C it was 31%, those with a 5′-G it was 34%, and the dimers with 5′-terminal U gave 42% conversion on average, resulting in the order 5′-U > 5′-G ≈ 5′-C > 5′-A. The dimer with the most weakly pairing 5′-base and most strongly pairing 3′-base (1ug) sticks out as the best ligating sequence. The combination of two weakly pairing bases (1uu) gave the poorest performer.
Table 2.
Conversion of the primer in extension assays with dimers of different sequencea
| Sequences | Dimer/trimer | Concentration dimer/trimer [mM] | Conversion [%]b |
|---|---|---|---|
| 6uu/4/9 | 1aa | 2 | 22 |
| 6gu/4/9 | 1ca | 2 | 25 |
| 6cu/4/9 | 1ga | 2 | 10 |
| 6au/4/9 | 1ua | 2 | 56 |
| 6ug/4/9 | 1ac | 2 | 11 |
| 6gg/4/9 | 1cc | 2 | 13 |
| 6cg/4/9 | 1gc | 2 | 28 |
| 6ag/4/9 | 1uc | 2 | 25 |
| 6uc/4/9 | 1ag | 2 | 47 |
| 6gc/4/9 | 1cg | 2 | 65 |
| 6cc/4/9 | 1gg | 2 | 75 |
| 6ac/4/9 | 1ug | 2 | 83 |
| 6ua/4/9 | 1au | 5 | 13 |
| 6ga/4/9 | 1cu | 5 | 19 |
| 6ca/4/9 | 1gu | 5 | 22 |
| 6aa/4/9 | 1uu | 5 | 3 |
| 7uuu/4/8 | 2aaa | 1.0 | 41 |
| 7uuu/4/8 | 2aaa | 0.1 | 17 |
| 7ggg/4/8 | 2ccc | 0.1 | 59 |
aConditions: 30 μM primer, 45 μM template, 45 μM helper, 500 mM MOPS, 800 mM EDC, 150 mM 1-EtIm, 80 mM MgCl2, pH 6.0 and 4°C.
bAs detected by relative peak intensities in MALDI-TOF mass spectra after 20 days, correction factors for lower desorption/ionization yield were applied for dimers with 3′-terminal G and trimers, and data are uncorrected in the other cases.
That the ligating efficiency depends on the sequence and not just the number and type of base pairs can also be seen from the results for isomers in Table 2. For example, 1ua with its 3′-terminal purine is ligated more than four times more efficiently than its isomer 1au with the more weakly pairing pyrimidine terminus. Likewise, 1ga ligates poorly, but the ligation of 1ag, with the strongly pairing G at the 3′-terminal position gives more than four-fold higher conversion. For two strongly pairing bases, the situation is similar but less pronounced, so that 1gc ligates a little less than two-fold more poorly than its isomeric sequence 1cg (28% versus 65%).
To shed more light on the issue and to study the ligation on a broader range of substrates, two different trimers (2aaa and 2ccc) were tested in ligation assays on templates 7uuu and 7ggg (Figure 2C). These were employed at lower concentrations because it was assumed that they bind more strongly to the primer/template/helper complex than their dimer counterparts. As shown in the last three entries of Table 2, at 1 mM concentration, 2aaa ligated to primer 4 giving a conversion of 41%, i.e. approx. twice as high a yield as for the corresponding dimer 1aa employed at twice the concentration. When the concentration was lowered further, to 0.1 M 2aaa, the yield dropped. When trimer 2ccc with its more strongly pairing cytosine bases was employed at 0.1 mM concentration, the ligation gave 59% conversion of the primer at the end of the assay. Again, this is a very significant increase over the 13% conversion measured for 1cc. Together, these results confirmed that longer sequences ligate more readily than dimers under the optimized reaction conditions.
Cyclization of dimers and trimers
As mentioned above, the results of Table 2 suggested that there is an effect that cannot be explained by the base composition of dimers and trimers alone. There appeared to be a competing process that interfered with ligation more readily for some sequences than for others. Symmetrical pyrophosphates or oligomerization products, side products known from assays with monomers (38) were not detected to a significant extent in mass spectra, probably because of the lower concentration of dimers in our assays. Instead, analysis of the low-mass region of mass spectra showed a peak with 18 Da lower mass that grew in intensity over the course of the assays, as well as another side product with a mass 2 Da higher than the dimer starting material (Supplementary Figure S21, SI). While the identity of the latter is currently unclear, the former was identified as the cyclic dimer, most probably resulting from the nucleophilic attack of the 3′-hydroxy group of the dinucleotide added on the activated form of the 5′-phosphate. We cannot exclude that cyclic dimers with a 2′,5′-linkage form, but consider it less likely than the formation of cyclic dimers with 3′,5′-linkages only, due to the slower rate expected for cyclization to 13-membered rings. A cyclization of dinucleotides was previously observed by Ferris when studying the oligomerization of AMP on montmorillonite in presence of EDC (51).
The MALDI-based kinetics of the formation of the cyclic side product, which should be unable to undergo template-directed ligation, is shown in Figure 9 for 1aa and 1aaa. The cyclization is higher yielding for the dimer, and less pronounced for the trimer with its larger, entropically more difficult-to-form ring. This size dependence may help to explain why others saw lower yields with methylimidazolides of dimers than the corresponding trimers (35) and why ribozyme-catalyzed copying in a eutectic phase was more successful with trimers than with dimers (52).
Figure 9.
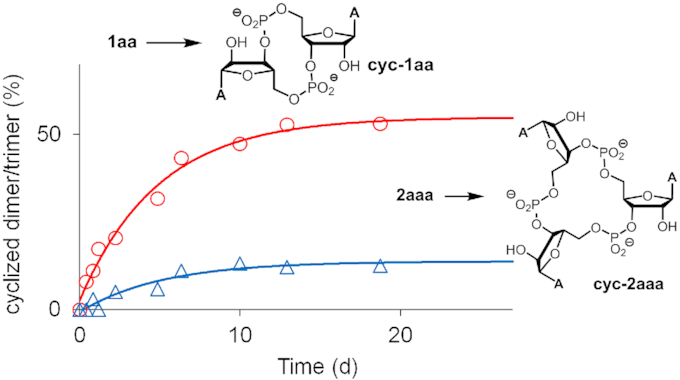
Cyclization of 2aaa and 1aa, as detected in MALDI-TOF mass spectra acquired during assays. Data points are uncorrected signal intensities and lines are monoexponential fits. Conditions: 0.5 M MOPS, 0.8 M EDC, 0.15 M 1-ethylimidazole, 0.08 M MgCl2, 2 mM dimer 1aa or 1 mM trimer 2aaa.
While cyclization is a reaction that interferes with the ligations studied here, it is noteworthy that cyclic dinucleotides have biological significance as signaling molecules (53). This includes cyclic diguanosine monophosphate (c-di-GMP) and cyclic oligoadenylates that are second messengers, acting on enzymes of the CRISPR-Rossman fold family (54). Results from exploratory work suggests that cyclization is also sequence dependent, with pyrimidine dimers 1uu, 1cc, 1cu and 1uc cyclizing to more than 70% during assays, and up to 90% for 1uu (Supplementary Table S2, SI), further corroborating the conclusion that cyclization is one of the causes of incomplete ligation for these sequences (yield ≤ 25% for all four of these sequences, Table 2).
Competition experiment
In a final set of experiments, we tested a more complex mixture, containing a mixture of both CMP as monomer (20 mM) and dimer 1cg (2 mM) as reactants for the extension of primer 3 on template 5gc. The monomer was employed at ten-fold higher concentration than the dimer to compensate somewhat for poorer binding. Figure 10 shows the kinetics of primer extension for either of the two reaction channels. Dimer 1cg is incorporated to a larger extent than the monomer 1c, resulting in an eightfold higher yield of the ligation product. This result confirms that even though monomers react well when they are present as the only reactant, dimers can outcompete them under proper in situ activation conditions. This is significant for prebiotic scenarios that produce mixtures of strands, rather than just monomers as reaction partners for primer extension.
Figure 10.
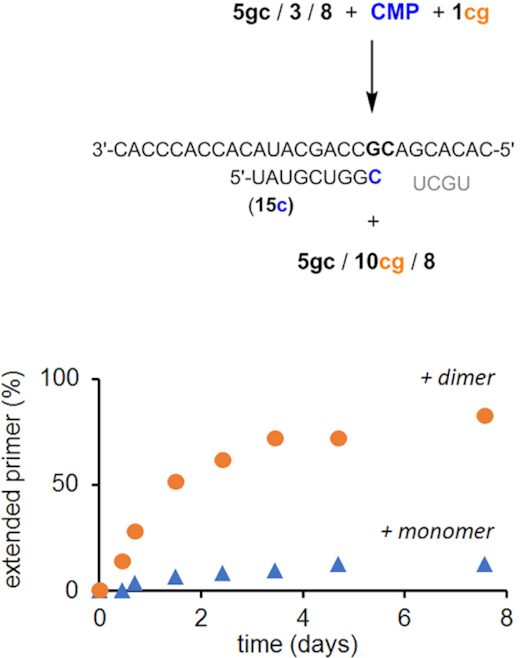
Competition experiment with monomer and dimer in the same solution. Reaction scheme and kinetics of primer extensions at 2 mM 1cg and 20 mM 1c in condensation buffer (500 mM HEPES, 150 mM EtIm, 800 mM EDC, 80 mM MgCl2) at pH 6.0 and 4°C.
DISCUSSION
The ligation of DNA strands is a common process in molecular biology and in the cell, but the ligation of RNA strands is not. While DNA ligases are widely used to link strands, RNA ligases are rare (55). Even during the early studies on enzyme-free ligation, it was noted that for the same sequence and condensing agents, the yields were higher for DNA than for RNA (29). In contrast, enzyme-free primer extension with monomers works well for ribonucleotides reacting with RNA primers (2), but poorly, if at all, for deoxynucleotides extending DNA primers (56). It was unclear why this is. The intrinsic reactivity of the functional groups involved, namely the hydroxy groups of ribose or deoxyribose as nucleophile and the activated phosphate of the other reacting strand, should be the same for monomers and longer strands. So, other factors must govern the rate of the ligation reaction.
Naïvely, one would expect the reactivity of short oligomers to be between that of monomers and longer strands. When we started studying the ligation of dimers, the results showed a reactivity lower than that of both monomers and longer oligomers, in agreement with the findings for pre-activated monomers and trimers (35). The synthesis of pre-activated dimers proved difficult, prompting us to adopt an in situ approach for preparing 14ag. The main reason for this was the lability of the activated species, a phenomenon not observed for either monomers or longer strands. It was only later that we realized that cyclization was a significant side reaction for dimers and trimers (Figure 9). Rules for the cyclization of small organic molecules (57) and for long duplex DNA (58) exist, and cyclic tetramers and octamers had been found as relevant species in oligomerization assays (59), but for the 12- and 18-membered rings formed upon cyclizing dimers and trimers we considered the entropic barriers high enough to make cyclization insignificant compared to template-directed ligation (Figure 11).
Figure 11.
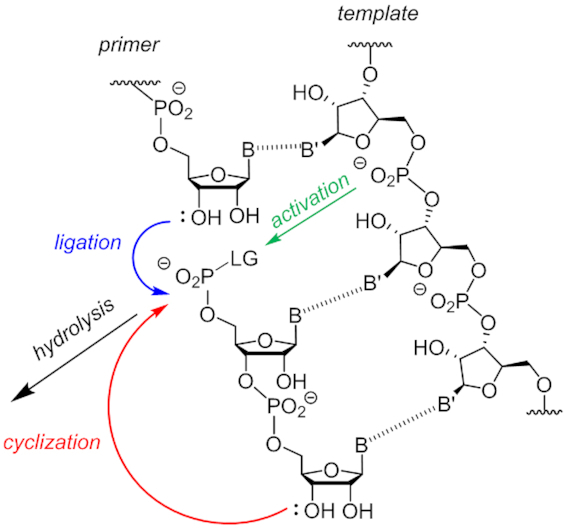
Processes important for the ligation of dimers to primers; LG = leaving group. Either the 3′- or the 2′-hydroxy group may attack the activated phosphate to form a new phosphodiester.
Our results now show that this assumption was not correct. Further, the results from the inhibition study indicate that well binding dimers make efficient inhibitors. Overall, the competition between the different nucleophiles (primer terminus, dimer terminus and water) for the electrophilic phosphorus of the activated phosphate may account for the low yields. The strong dependence of ligation yield on the 3′-terminal nucleobase of dimers (Table 2) corroborates this. Well pairing and well stacking nucleobases at this position favor high yielding ligation, probably by efficiently anchoring the dimer terminus on the template, whereas weakly pairing and weakly stacking bases do not, favoring cyclization instead (shown indicated by the red arrow in Figure 11). Without in situ (re)-activation, too little reactive 5′-phosphate may exist that drives the desired ligation reaction.
While cyclization provides a plausible explanation to the low reactivity paradox (dimers and trimers being less reactive than either monomers or longer oligomers), it does not explain why RNA ligations are lower yielding than DNA ligations. In their early study mentioned above, Dolinnaya, Shabarova and colleagues suspected that local structure plays the dominant role (29). While these authors wrote of the proximity of the reacting groups, we suspect that the governing factor is the conformational equilibrium between the productive and possible unproductive pentavalent intermediates that result from the nucleophilic attack of the hydroxy group at the primer terminus on the activated phosphate. We have previously found that even for the more reactive aminoterminal primers two different steps can be rate-limiting (60). The first is probably the initial nucleophilic attack. But, if the attack of the nucleophile does not occur in an in-line arrangement with the leaving group, a Berry pseudorotation of the intermediate may be required. This pseudorotation that then puts the leaving group in the mandatory apical position may be harder to perform for reactants in RNA duplexes because they lack the conformational flexibility of DNA duplexes. As for our ligations, a strong dependence on sequence and hence local structure is known for the hydrolysis of RNA. Hydrolysis and ligation are related, and conformational constraints that slow down ligation may be smiliar to those that limit the rate of hydrolysis in RNA duplexes.
Independent of the mechanistic details, our results now show that very short sequences (dimers and trimers) that are hard to ligate can be successfully employed in enzyme-free copying reactions. The key to this appears to be the proper in situ activation conditions. If hydrolyzed reactants do not turn into inhibitors, but can undergo re-activation by the condensing agent/organocatalyst combination, significant yields can be achieved, even for the well-cyclizing dimers. The in situ activation approach is easy to implement, relying on commercially available components only and a single synthetic transformation. When the labile activated species is formed in situ, handling and purification losses are also avoided. The optimized protocol described here may therefore become useful for solving related problems in RNA biotechnology that require the ligation of RNA strands in difficult settings.
CONCLUSIONS
In conclusion, our results show that dimers and trimers can be ligated to primers on RNA templates with an optimized combination of condensing agent and organocatalyst at slightly acidic pH. With or without MOPS buffer, unactivated dimer building blocks, which are shown to be potent inhibitors are (re)-activated and react with primers, mostly in acceptable yields. The sequence dependence of the yield, as well as direct spectrometric observation, indicate that cyclization plays an important role as side reaction in the ligation of dimers and, to a lesser degree, trimers. How easily cyclization occurs strongly depends on the 3′-terminal nucleobase. Weakly pairing pyrimidines favor cyclization, strongly pairing and stacking purines favor ligation. Our results thus provide a basis for choosing the right sequence and reaction conditions for ligation short strands, and help to rationalize why nature performs transcription with mononucleotide building blocks only, avoiding dimers with their more complex reactivity.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Eric Kervio for discussions, Sebastian Motsch and Gabrielle Leveau for the synthesis and characterization of trimers, and Jennifer Bremer for the acquisition of NMR spectra.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [RI 1063/16-1 and CRC TRR 235, project P06]. Funding for open access charge: University of Stuttgart.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kornberg A., Baker A.T.. DNA Replication. 2005; Mill Valley: University Science Books. [Google Scholar]
- 2. Kozlov I.A., Orgel L.E.. Nonenzymatic template-directed synthesis of RNA from monomers. Mol. Biol. 2000; 34:781–789. [PubMed] [Google Scholar]
- 3. Gollihar J., Levy M., Ellington A.D.. Many paths to the origin of life. Science. 2014; 343:259–261. [DOI] [PubMed] [Google Scholar]
- 4. Szostak J.W. The eightfold path to non-enzymatic RNA replication. J. Syst. Chem. 2012; 3:2. [Google Scholar]
- 5. Gilbert W. The RNA world. Nature. 1986; 320:618. [Google Scholar]
- 6. Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R.. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell. 1982; 31:147–157. [DOI] [PubMed] [Google Scholar]
- 7. Orgel L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004; 39:99–123. [DOI] [PubMed] [Google Scholar]
- 8. Meyer A.J., Ellefson J.W., Ellington A.D.. Abiotic self-replication. Acc. Chem. Res. 2012; 45:2097–2105. [DOI] [PubMed] [Google Scholar]
- 9. Attawer J., Wochner A., Holliger P.. In-ice evolution of RNA polymerase ribozyme activity. Nat. Chem. 2013; 5:1011–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ekland E.H., Szostak J.W., Bartel D.P.. Structurally complex and highly active RNA ligases derived from random RNA sequences. Science. 1995; 269:364–370. [DOI] [PubMed] [Google Scholar]
- 11. Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S.. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983; 35:649–857. [DOI] [PubMed] [Google Scholar]
- 12. Benner S.A., Ellington A.D., Tauer A.. Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:7054–7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Inoue T., Orgel L.E.. Oligomerization of (guanosine 5′-phosphor)-2-methylimidazolide on poly(C). An RNA polymerase model. J. Mol. Biol. 1982; 162:201–217. [DOI] [PubMed] [Google Scholar]
- 14. Inoue T., Orgel L.E.. A nonenzymatic RNA polymerase model. Science. 1983; 219:859–862. [DOI] [PubMed] [Google Scholar]
- 15. Kanavarioti A., Bernasconi C.F., Alberas D.J., Baird E.E.. Kinetic dissection of individual steps in the poly(C)-directed oligoguanylate synthesis from guanosine 5′-monophosphate 2-methylimidazolide. J. Am. Chem. Soc. 1993; 115:8537–8546. [DOI] [PubMed] [Google Scholar]
- 16. Wu T., Orgel L.E.. Nonenzymatic template-directed synthesis on hairpin oligonucleotides. 3. Incorporation of adenosine and uridine residues. J. Am. Chem. Soc. 1992; 114:7963–7969. [DOI] [PubMed] [Google Scholar]
- 17. Dörr M., Löffler P.M.G., Monnard P.A.. Non-enzymatic polymerization of nucleic acids from monomers: monomer self-condensation and template-directed reactions. Curr. Org. Synth. 2012; 9:735–763. [Google Scholar]
- 18. Li L., Prywes N., Tam C.P., O’Flaherty D.K., Lelyveld V.S., Cagri Izgu E., Pal A., Szostak J.W.. Enhanced nonenzymatic RNA copying with 2-aminoimidazole activated nucleotides. J. Am. Chem. Soc. 2017; 139:1810–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sosson M., Richert C.. Enzyme-free genetic copying of DNA and RNA sequences. Beilstein J. Org. Chem. 2018; 14:603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. O’Flaherty D.K., Kamat N.P., Mirza F.N., Li L., Prywes N., Szostak J.W.. Copying of mixed-sequence RNA templates inside model protocells. J. Am. Chem. Soc. 2018; 140:5171–5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schrum J.P., Ricardo A., Krishnamurthy M., Blain J.C., Szostak J.W.. Efficient and rapid template-directed nucleic acid copying using 2′-amino-2′, 3′-dideoxyribonucleoside−5′-phosphorimidazolide monomers. J. Am. Chem. Soc. 2009; 131:14560–14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Röthlingshöfer M., Kervio E., Lommel T., Plutowski U., Hochgesand A., Richert C.. Chemical primer extension in seconds. Angew. Chem. Int. Ed. 2008; 47:6065–6068. [DOI] [PubMed] [Google Scholar]
- 23. Hill A.R. Jr, Orgel L.E., Wu T.. The limits of template-directed synthesis with nucleoside-5′-phosphoro(2-methyl)imidazolides. Orig. Life Evol. Biosph. 1993; 23:285–290. [DOI] [PubMed] [Google Scholar]
- 24. Kervio E., Sosson M., Richert C.. The effect of leaving groups on binding and reactivity in enzyme-free copying of DNA and RNA. Nucleic Acids Res. 2016; 44:5504–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Deck C., Jauker M., Richert C.. Efficient enzyme-free copying of all four nucleobases templated by immobilized RNA. Nat. Chem. 2011; 3:603–608. [DOI] [PubMed] [Google Scholar]
- 26. Naylor R., Gilham P.T.. Studies on some interactions and reactions of oligonucleotides in aqueous solution. Biochemistry. 1966; 5:2722–2728. [DOI] [PubMed] [Google Scholar]
- 27. von Kiedrowski G. A self-replicating hexadeoxynucleotide. Angew. Chem. Int. Ed. Engl. 1986; 25:932–935. [Google Scholar]
- 28. Li T., Nicolaou K.C.. Chemical self-replication of palindromic duplex DNA. Nature. 1994; 369:218–221. [DOI] [PubMed] [Google Scholar]
- 29. Dolinnaya N.G., Sokolova N.I., Ashirbekova D.T., Shabarova Z.A.. The use of BrCN for assembling modified DNA duplexes and DNA-RNA hybrids; comparison with water-soluble carbodiimide. Nucleic Acids Res. 1991; 19:3067–3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sawai H., Wada M.. Nonenzymatic template-directed condensation of short-chained oligouridylates on a poly(A) template. Orig. Life. Evol. Biosphere. 2000; 30:503–511. [DOI] [PubMed] [Google Scholar]
- 31. Sawai H., Wada M., Kouda T., Nakamura Ozaki A.. Nonenzymatic ligation of short-chained 2′–5′- or 3′–5′-linked oligoribonucleotides on 2′–5′- or 3′–5′-linked complementary templates. ChemBioChem. 2006; 7:605–611. [DOI] [PubMed] [Google Scholar]
- 32. Carriero S., Damha M.J.. Synthesis of lariat-DNA via the chemical ligation of a dumbbell complex. Org. Lett. 2003; 5:273–276. [DOI] [PubMed] [Google Scholar]
- 33. Bowler F.R., Chan C.K.W., Duffy C.D., Gerland B., Islam S., Powner M.W., Sutherland J.D., Xu J.. Prebiotically plausible oligoribonucleotide ligation facilitated by chemoselective acetylation. Nat. Chem. 2013; 5:383–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rohatgi R., Bartel D.P., Szostak J.W.. Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′-5′ phosphodiester bonds. J. Am. Chem. Soc. 1996; 118:3340–3344. [DOI] [PubMed] [Google Scholar]
- 35. Prywes N., Blain J.C., Del Frate F., Szostak J.W.. Nonenzymatic copying of RNA templates containing all four letters is catalyzed by activated oligonucleotides. eLife. 2016; 5:e17756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jauker M., Griesser H., Richert C.. Copying of RNA sequences without pre‐activation. Angew. Chem. Int. Ed. 2015; 54:14559–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Griesser H., Bechthold M., Tremmel P., Kervio E., Richert C.. Amino acid-specific, ribonucleotide-promoted peptide formation in the absence of enzymes. Angew. Chem. Int. Ed. 2017; 56:1224–1228. [DOI] [PubMed] [Google Scholar]
- 38. Jauker M., Griesser H., Richert C.. Spontaneous formation of RNA strands, peptidyl RNA, and cofactors. Angew. Chem. Int. Ed. 2015; 54:14564–14569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petkovic S., Müller S.. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015; 43:2454–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vogel H., Richert C.. Labeling small RNAs through chemical ligation at the 5′-terminus: enzyme-free or combined with enzymatic 3′-labeling. ChemBioChem. 2012; 13:1474–1482. [DOI] [PubMed] [Google Scholar]
- 41. Mamot A., Sikorski P.J., Warminski M., Kowalska J., Jemielity J.. Azido-functionalized 5′ cap analogues for the preparation of translationally active mRNAs suitable for fluorescent labeling in living cells. Angew. Chem. Int. Ed. 2017; 56:15628–15632. [DOI] [PubMed] [Google Scholar]
- 42. Kalinowski M., Haug R., Said H., Piasecka S., Kramer M., Richert C.. Phosphoramidate ligation of oligonucleotides in nanoscale structures. ChemBioChem. 2016; 17:1150–1155. [DOI] [PubMed] [Google Scholar]
- 43. Kramer M., Richert C.. Enzyme-free ligation of 5′-phosphorylated oligodeoxynucleotides in a DNA nanostructure. Chem. Biodiv. 2017; 14:e1700315. [DOI] [PubMed] [Google Scholar]
- 44. Durand M., Chevrie K., Chassignol M., Thuong N.T., Maurizot J.C.. Circular dichroism studies of an oligodeoxyribonucleotide containing a hairpin loop made of a hexaethylene glycol chain: conformation and stability. Nucleic Acids Res. 1990; 18:6353–6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hagenbuch P., Kervio E., Hochgesand A., Plutowski U., Richert C.. Chemical primer extension: efficiently determining single nucleotides in DNA. Angew. Chem. Int. Ed. 2005; 44:6588–6592. [DOI] [PubMed] [Google Scholar]
- 46. Kervio E., Claasen B., Steiner U.E., Richert C.. The strength of the template effect attracting nucleotides to naked DNA. Nucleic Acids Res. 2014; 42:7409–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Altmann S., Labhardt A.M., Bur D., Lehmann C., Bannwarth W., Billeter M., Wüthrich K., Leupin W.. NMR studies of DNA duplexes singly cross-linked by different synthetic linkers. Nucleic Acids Res. 1995; 23:4827–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhanga W., Tam C.P., Walton T., Fahrenbach A.C., Birraneg G., Szostak JW.. Insight into the mechanism of nonenzymatic RNA primer extension from the structure of an RNA-GpppG complex. Proc. Nat. Acad. Sci. U.S.A. 2017; 114:7659–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vogel S.R., Deck C., Richert C.. Accelerating chemical replication steps of RNA involving activated ribonucleotides and downstream-binding element. Chem. Commun. 2005; 4922–4924. [DOI] [PubMed] [Google Scholar]
- 50. Hänle E., Richert C.. Enzyme-free replication with two or four bases. Angew. Chem. Int. Ed. 2018; 57:8911–8915. [DOI] [PubMed] [Google Scholar]
- 51. Ferris J.P., Ertem G., Agarval V.. Mineral catalysis of the formation of dimers of 5′-AMP in aqueous solution: the possible role of Montmorillonite clays in the prebiotic synthesis of RNA. Origin. Life. Evol. Biosph. 1989; 19:165–178. [DOI] [PubMed] [Google Scholar]
- 52. Attwater J., Raguram A., Morgunov A.S., Gianni E., Holliger P.. Ribozyme-catalysed RNA synthesis using triplet building blocks. eLife. 2018; 7:e35255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ross P., Weinhouse H., Aloni Y., Michaeli D., Weinberger-Ohana P., Mayer R., Braun S., de Vroom E., van der Marel G.A., van Boom J.H., Benziman M.. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature. 1987; 325:279–281. [DOI] [PubMed] [Google Scholar]
- 54. Athukoralage J.S., Rouillon C., Graham S., Grüschow S., White M.F.. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature. 2018; 562:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. El Omari K., Ren J., Bird L.E., Bona M.K., Klarmann G., Legrice S.F.J., Stammers D.K.. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006; 281:1573–1579. [DOI] [PubMed] [Google Scholar]
- 56. Vogel H., Gerlach C., Richert C.. Reactions of buffers in cyanogen bromide-induced ligations. Nucleosides Nucleotides Nucleic Acids. 2013; 32:17–27. [DOI] [PubMed] [Google Scholar]
- 57. Baldwin J.E. Rules for ring closure. J. Chem. Soc., Chem. Commun. 1976; 734–736. [Google Scholar]
- 58. Shore D., Baldwin R.L.. Energetics of DNA twisting I. Relation between twist and cyclization probability. J. Mol. Biol. 1983; 170:957–981. [DOI] [PubMed] [Google Scholar]
- 59. Horowitz E.D., Engelhart A.E., Chen M.C., Quarles K.A., Smith M.W., Lynn D.G., Hud N.V.. Intercalation as a means to suppress cyclization and promote polymerization of base-pairing oligonucleotides in a prebiotic world. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:5288–5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kervio E., Hochgesand A., Steiner U., Richert C.. Templating efficiency of naked DNA. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:12074–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.