Significance
Our results provide an example of reactivation of a signaling pathway important in embryogenesis that can be exploited to promote tissue repair and help host recovery from bacterial pneumonia. Although this approach of regenerative medicine will not curtail infection per se, it reduces suffering and shortens recovery time by fostering tissue repair, thus complementing antibiotic treatment and improving patient outcomes from serious infections. Our findings open up a frontier in the treatment of microbial infections using microRNA-based regenerative therapy.
Keywords: pneumonia, bacteria, alveolus, regeneration, miRNA
Abstract
Bacterial pneumonia remains a leading cause of morbidity and mortality worldwide. A defining feature of pneumonia is lung injury, leading to protracted suffering and vulnerability long after bacterial clearance. Little is known about which cells are damaged during bacterial pneumonia and if the regenerative process can be harnessed to promote tissue repair and host recovery. Here, we show that infection of mice with Streptococcus pneumoniae (Sp) caused substantial damage to alveolar epithelial cells (AEC), followed by a slow process of regeneration. Concurrent with AEC regeneration, the expression of miRNA-302 is elevated in AEC. Treatment of Sp-infected mice with miRNA-302 mimics improved lung functions, host recovery, and survival. miRNA-302 mediated its therapeutic effects, not by inhibiting apoptosis and preventing damage, but by promoting proliferation of local epithelial progenitor cells to regenerate AEC. These results demonstrate the ability of microRNA-based therapy to promote AEC regeneration and enhance host recovery from bacterial pneumonia.
Pneumonia is a major cause of severe morbidity, often resulting in hospitalization, admission to intensive care units, long recovery, and a high rate of mortality (1). Streptococcus pneumoniae (Sp) is the leading cause of bacterial pneumonia and secondary pneumonia following flu infection (2). The pathobiology of pneumonia is characterized by robust host immune responses that cause lung damage (2–4). Studies of microbial infection have mostly focused on bacterial virulence and host immune responses, with the goal of developing interventions based on antimicrobials or vaccines. However, full recovery from bacterial pneumonia is dependent not only on clearance of microbial pathogens but also on regeneration of the damaged airway epithelium. Failure to repair epithelial damage can disrupt the epithelial barrier that protects the lung from external insults, leading to susceptibility to recurring infections and development of chronic and progressive lung diseases, which include chronic obstructive pulmonary disease (COPD), idiopathic pulmonary fibrosis, and emphysema (5, 6).
It is now well established that regulatory pathways involved in tissue growth and differentiation during embryogenesis are reactivated in the process of regeneration following injury in adults (7, 8). microRNAs (miRNAs) have emerged as key modulators of the regeneration process by controlling expression of signaling and transcription factors involved in multiple facets of tissue development and regeneration (9, 10). The miRNA cluster miR-302–367, known as the miR-302 family, is highly expressed at early stages of fetal mouse lung development and contributes to enhanced proliferation of lung progenitor cells during embryogenesis (11). However, the miR-302–367 gene cluster is not actively expressed in healthy adult lungs (https://lungmap.net/). Studies using mouse models with physical and chemical injuries have shown that the regenerative process in the lung parenchyma depends on proliferation and differentiation of local stem or progenitor cell populations, which include the alveolar epithelial cells (AEC) and bronchiolar Club cells (12–14). It remains largely unknown what role the miR-302 family and epithelial regeneration play in repair and recovery from lung injuries due to infection by microbial pathogens.
In this study, we established a murine pneumonia model with diffuse bacterial infection of alveolar spaces, resulting in acute inflammation and substantial damage to AEC, followed by a slow process of regeneration over an extended period (>30 d). We found the expression of miR-302 was up-regulated in AEC and coincided with AEC regeneration. We hypothesized that transient expression of miR-302 genes critical for fetal lung development would stimulate local stem cells in the lung parenchyma to regenerate epithelium and promote animal recovery following Sp infection. Here, we showed that treatment of Sp-infected mice with miR-302 mimics improved lung functions, host recovery, and survival by promoting proliferation of alveolar epithelial progenitor cells to regenerate AEC and repair damaged alveolar epithelium. These findings suggest that miRNA-based therapy may be used as a powerful therapeutic to promote AEC regeneration and enhance host recovery from bacterial pneumonia.
Results
AEC Damage and Regeneration in Sp-Infected Mouse Lung.
C57BL/6 mice were infected intranasally (i.n.) under anesthesia with ∼5 × 106 colony-forming units (cfu) of the Sp TIGR4 strain (SpT4), resulting in direct infection of the lower respiratory tract and acute bacterial pneumonia with an ∼40% mortality rate (15, 16). Histology of lung sections of surviving mice showed numerous foci of inflammatory lesions starting at 2 d post infection (dpi), peaking at 7 dpi, and returning to normal state by 30 dpi (Fig. 1A). Bacterial loads in the lung were determined by cfu plating and visualized by immunostaining of lung sections with antibodies specific to the type 4 capsule on the surface of SpT4 (Fig. 1 B and C). Both cfu plating and immunostaining showed high levels of bacteria in the lung at 2 dpi that were cleared by 7 dpi, while immunostaining showed that most bacteria were localized to the alveolar spaces. Thus, this infection model recapitulates pathological and clinical features of severe acute pneumonia with extensive inflammation in the lung and infected mice either succumbing to infection or clearing bacteria.
Fig. 1.
Bacterial clearance and alveolar epithelial damage and repair in SpT4-infected mice. Lung tissues were collected on 2, 7, 14, and 30 dpi with SpT4. (A) H&E staining. (B) Immunostaining with antibodies to the type 4 capsular of SpT4 (green). (C) Bacteria loads in lung homogenate measured by cfu plating (LOD: limit of detection). (D and F) Immunostaining with mAb to T1α (D, green), SPC (F, red), and CC10 (F, green). DAPI: blue in B, D, and F. [Scale bar: 500 μm (A); 50 μm (B, D, and F).] (E and G) qRT-PCR of T1α and SPC mRNA in lung tissue. Data are mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001 versus noninfected normal lungs (0 dpi).
Lung alveoli are primarily composed of alveolar epithelial type I cells (AECI) that mediate the key function of gas exchange and alveolar epithelial type II cells (AECII) that produce antimicrobial peptides and surfactant proteins and lipids for reducing alveolar surface tension. Flat-shaped AECI cover more than 90% of the alveolar surface and can be visualized by staining for the cell-type–specific marker T1α. Cuboidal-shaped AECII interdigitate between AECI and can be visualized by staining for the cell-type–specific marker SPC. Substantial destruction of AECI and AECII was observed after SpT4 infection, as evidenced by significant loss of cell-type–specific markers for AECI (T1α) and AECII (SPC) at both mRNA and protein levels (Fig. 1 D–G). The kinetics of AECI and AECII loss and regeneration were similar, with dramatic loss at 2 and 7 dpi and recovery starting after 7 dpi, as visualized by immunostaining of T1α (AECI) and SPC (AECII) (Fig. 1 D and F). Similarly, gene expression analyses of cell-type–specific markers by qRT-PCR for mRNA from the lung tissue showed decreased levels of T1α and SPC at 2, 7, and 14 dpi (Fig. 1 E and G). By 30 dpi, there was substantial recovery of AECI and AECII, although T1α and SPC mRNA levels were still reduced compared with those before infection (Fig. 1 E and G). In contrast to AEC, epithelial cells in the bronchiolar regions, such as Club cells (CC10), were unaffected (Fig. 1F) and neither were other cells in the lung, including basal cells, ciliated cells, smooth muscle and fibroblasts, and vascular endothelial cells at 7 dpi (SI Appendix, Fig. S1A). Modest levels of intra-alveolar fibrotic lesions were observed, as characterized by collagen fiber deposition and inflammatory cell accumulation in alveolar spaces, which subsequently resolved between 7 and 14 dpi (SI Appendix, Fig. S1B). These results showed that mice with SpT4 infection exhibited extensive lung parenchyma injuries that impaired alveolar architecture with specific damage to AECI and AECII, rather than broad injury to all cell types. The regeneration and repair processes were slow and took more than 30 d for full recovery.
miRNA-302 Expression Is Elevated in AEC After Sp Infection.
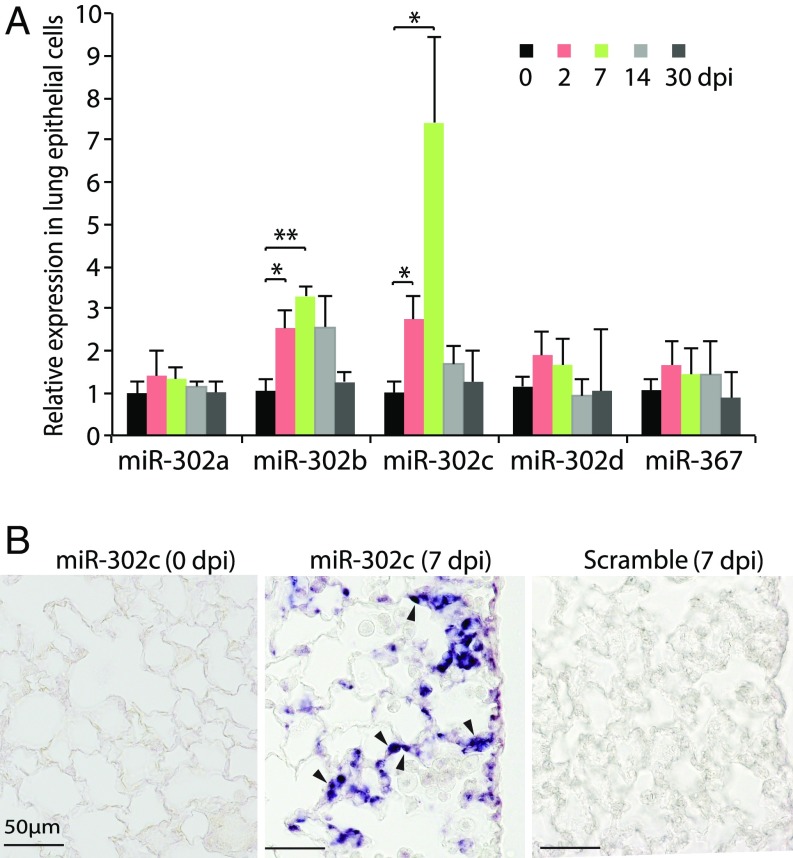
It is known that regulatory pathways important for tissue growth and differentiation during embryonic development can be reactivated in the process of regeneration to promote tissue repair (7, 8). The miRNA cluster miR-302–367 is important for lung epithelial progenitor cell proliferation during embryonic development (11). We explored a potential role of miR-302–367 by asking if their expression is reactivated in the lung epithelium following injuries caused by bacterial pneumonia. While the expression of miR-302–367 was undetectable in the normal adult mouse lung epithelium, two members (miR-302b and miR-302c) of the miR-302–367 cluster were induced by SpT4 infection at 2 dpi, peaked at 7 dpi, and returned to the basal level by 30 dpi (Fig. 2A). Cells expressing miR-302c were evident in the alveolar epithelium by in situ hybridization at 7 dpi, but not before infection (Fig. 2B). Together, these data indicated that expression of miR-302b and miR-302c was reactivated in the adult lung epithelium after SpT4 infection, suggesting a potential role of miR-302b/c during tissue regeneration/repair in response to SpT4 infection-induced lung injury.
Fig. 2.
miR-302 expression in AEC after SpT4 infection. (A) qRT-PCR analysis of miR-302–367 polycistron (miR-302b/c/a/d family and miR-367) from isolated lung epithelial cells of mouse distal lungs at 0, 2, 7, 14, and 30 dpi. (B) In situ hybridization of digoxigenin-labeled LNA miR-302c antisense probe and LNA-scrambled control probe (Exiqon) on lung tissue sections at 0 and 7 dpi. Arrowheads point to the alveolar epithelial cells. (Scale bars: 50 μm.) Data shown are means ± SEM (A, n = 4 per group). *P < 0.05; **P < 0.01.
miRNA-302 Mimic Treatment Improves Lung Function, Host Recovery, and AEC Regeneration in Sp-Infected Mice.
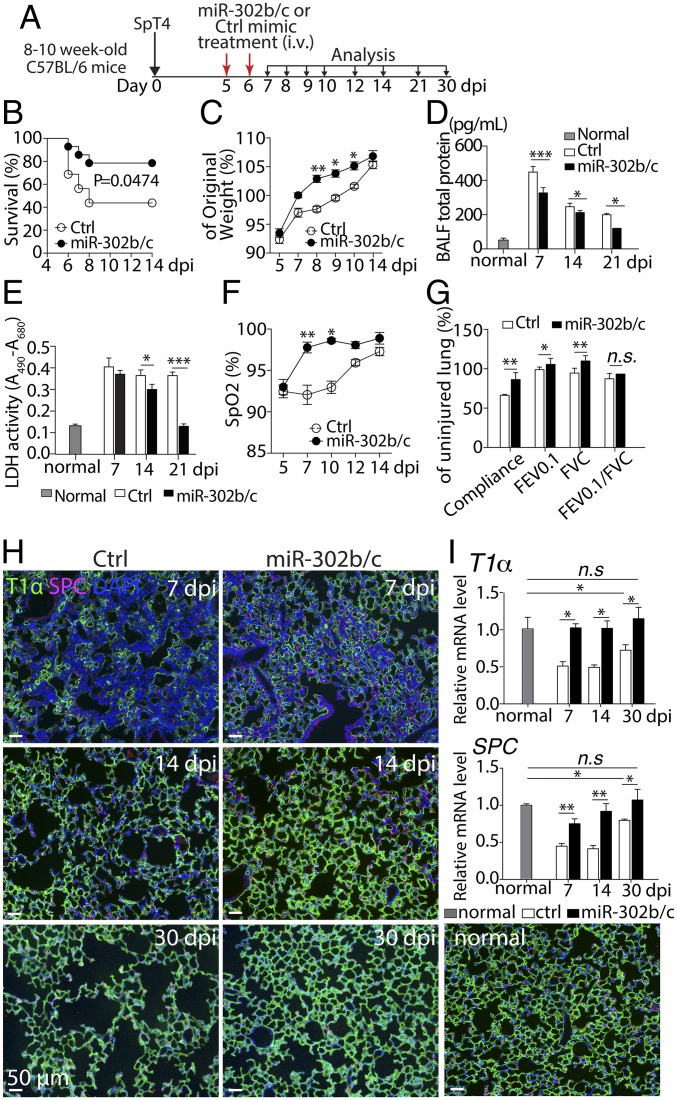
We next tested the role of miR-302b/c using in vivo administration of miRNA mimics with the goal of developing novel therapeutics to improve recovery from bacterial pneumonia. To determine whether i.v. administration of miR-302b/c mimics led to accumulation of these miRNAs in the lung, we examined lung tissues and observed that miR-302b/c levels peaked at 4 h and returned to baseline 24 h after injection (SI Appendix, Fig. S2A). We then treated SpT4-infected mice at 5 and 6 dpi with miR-302b/c mimics (miR-302b/c) or negative control mimics (Ctrl) by tail-vein injections (Fig. 3A). Treatment with miR-302b/c increased survival following infection compared with untreated, infected mice (Fig. 3B), and surviving treated mice regained body weight more rapidly than surviving untreated controls (Fig. 3C). Mice treated with miR-302b/c had lower levels of total protein and lactate dehydrogenase (LDH) activities in their bronchoalveolar lavage fluid (BALF) relative to the Ctrl group (Fig. 3 D and E), indicating less damage to lung tissue integrity in miR-302b/c–treated mice. Assessment of gas exchange by pulse oximetry (SpO2) showed accelerated recovery of oxygenation in the miR-302b/c-treated compared with the Ctrl-treated group (Fig. 3F). Analysis of lung mechanics by flexiVent revealed significant improvements in compliance, forced expiratory volume (FEV0.1), and forced vital capacity (FVC) in miR-302b/c–treated animals compared with Ctrl (Fig. 3G). The FEV0.1/FVC ratio was not significantly affected, consistent with only modest levels of lung fibrotic lesion formation induced by SpT4 infection that was not affected by miR-302b/c treatment (SI Appendix, Fig. S2B). Collectively, these data show that treatment with miR-302b/c mimics improved pulmonary function and recovery from bacterial pneumonia.
Fig. 3.
Effects of miR-302b/c mimics treatment on SpT4-infected mice. (A) Schematic of experimental design. Mice were infected with SpT4 on day 0 and then treated with either miR-302b/c or Ctrl mimics at 5 and 6 dpi and monitored daily for survival (B), gain of body weight (C), and blood oxygen levels (F). Total protein levels (D) and LDH activities (E) in BALF at indicated dpi. Pulmonary functions (G) were analyzed at 21 dpi for compliance, forced expiratory volume in 0.1 s (FEV0.1), and forced vital capacity (FVC). (H) Immunostaining of lung sections with antibodies to T1α and to SPC. (Scale bars: 50 µm.) (I) qRT-PCR of T1α and SPC mRNA in lung tissues. Normal: uninfected/untreated control. Data in B were analyzed using the Gehan–Breslow–Wilcoxon test of cumulative data (n = 14 for miR-302b/c; n = 16 for Ctrl mimics). Data shown are means ± SEM (C–G, n = 4 per group; I, n = 10 per group). *P < 0.05; **P < 0.01; ***P < 0.001.
We next evaluated the effect of miR-302 mimics treatment on the process of lung AEC regeneration. Analyses of lung sections by immunostaining (Fig. 3H) and qRT-PCR (Fig. 3I) of the T1α and SPC markers showed that miR-302b/c mimics-treated mice had a significant increase of AECI (T1α) and AECII (SPC) at 7, 14, and 30 dpi compared with the Ctrl group. By 30 dpi, the Ctrl mice still had lower expression of T1α and SPC markers compared with uninfected controls. In striking contrast, miR-302b/c mimics-treated mice fully recovered as there were no significant differences in expression of T1α and SPC markers between miR-302b/c mimics-treated and uninfected mice (Fig. 3I). Thus, therapeutic treatment with miR-302b/c mimics at 5 and 6 d post SpT4 infection promoted regeneration of AEC and repair of damaged alveolar epithelium, resulting in improved pulmonary function, lower mortality rate, and faster recovery of surviving animals from pneumonia.
miRNA-302 Mimic Treatment Increases AEC Proliferation in Vivo.
To study the mechanisms by which miR-302 mimics treatment might help recovery at cellular and molecular levels, we examined its effect on apoptosis and proliferation of lung epithelial cells and on expression of genes associated with these processes. TUNEL staining of lung sections showed no significant difference in the number of apoptotic cells between miR-302b/c and Ctrl groups at 7, 14, and 21 dpi (SI Appendix, Fig. S3 A and B). Similarly, flow cytometry detecting cleaved caspase 3 from EpCAM+ epithelial cells showed no significant difference (SI Appendix, Fig. S3 C and D). Furthermore, expression of apoptosis-associated genes (Dapk1, Stk17B, Bax) (17–19) in lung epithelial cells as determined by qRT-PCR was not affected at 7, 14, and 21 dpi, except increased level of Stk17B at 7 dpi in miR-302b/c–treated mice (SI Appendix, Fig. S3E). These results indicate that inhibition of apoptosis was unlikely to explain the effect of miR-302b/c mimics in reducing tissue injury during bacterial pneumonia.
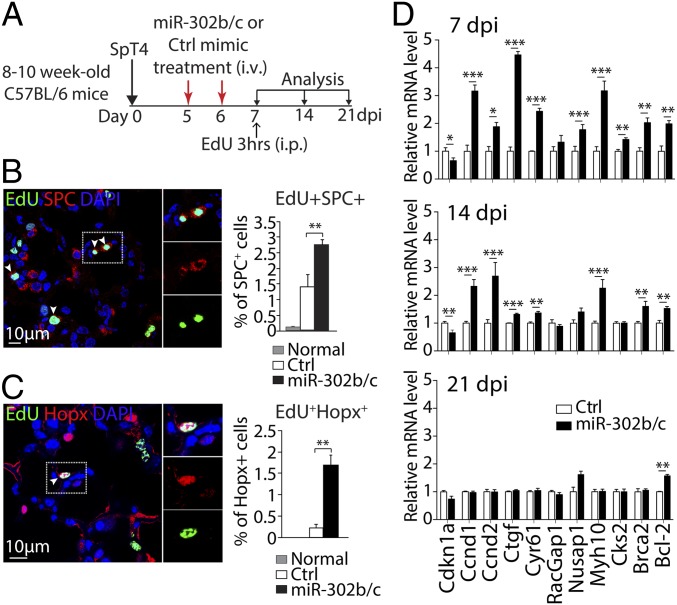
An alternative mechanism of miR-302b/c mimics treatment was stimulating proliferation of local progenitor cells and, thereby, enhancing regeneration of AEC, tissue repair, and recovery (20). To examine cell proliferation in vivo following bacterial pneumonia, SpT4-infected mice, either treated with miR-302b/c or Ctrl mimics at 5 and 6 dpi, were pulsed with 5-ethynyl-2′-deoxyuridine (EdU) for 3 h at 7 dpi (Fig. 4A). Proliferating epithelial cells were quantified by visualizing EdU-labeled cells coimmunostained with markers of AECI (Hopx) and AECII (SPC) (12, 13). There was a significant increase in AECI and AECII proliferation detected in the miR-302b/c–treated compared with the Ctrl-treated group (Fig. 4 B and C). Gene expression analyses using qRT-PCR showed that lung epithelium of miR-302b/c–treated mice had increased expression of genes associated with positive regulation of cell proliferation, including Ccnd1, Ccnd2, Ctgf, Cyr61, Nusap1, Myh10, Cks2, and Brca2 (21–28), compared with Ctrl-treated mice (Fig. 4D). In addition, expression of Cdkn1a, a cell-cycle inhibitor gene (29), was reduced by miR-302b/c treatment (Fig. 4D). These results show that local epithelial cell proliferation, especially AECI and AECII, accounted for the accelerated repair in lung alveoli after miR-302b/c treatment following acute Sp-induced lung injury.
Fig. 4.
AEC proliferation following miR-302b/c mimics treatment of SpT4-infected mice. (A) Schematic of experimental design. (B and C) Confocal images of lung sections at 7 dpi by Click-iT EdU Alexa Fluor 488 imaging and coimmunostaining with antibodies to SPC (AECII, B) and to Hopx (AECI, C), and quantification of EdU+SPC+ and EdU+Hopx+ cells as percentage of total SPC+ and Hopx+ cells, respectively. Arrowheads in B and C point to nucleus of proliferating (EdU+) cells. Quantitative analyses represent counting of multiple fields from five independent samples per group (∼2,200 SPC+ cells and ∼765 Hopx+ cell per sample). Data are means ± SEM (n = 5–6 per group). (D) Expression of indicated genes of interest by qRT-PCR analysis of the mRNA of isolated lung epithelial cells at 7, 14, and 21 dpi. Data are means ± SEM (n = 3 per group and time point). *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t test). (Scale bars: 10 µm.)
To determine the possible effects of miR-302b/c treatment on other cells in the lung, we assessed cell proliferation by coimmunostaining of various cell-type–specific markers and incorporation of EdU among different cell populations. We detected no proliferation in bronchial basal cells (p63) or ciliated cells (β-tubulin IV) in either miR-302b/c or Ctrl groups (SI Appendix, Fig. S4 A and B). On the other hand, we observed proliferating bronchiolar Club cells (CC10), smooth muscle cells (αSMA), vascular endothelial cells (PECAM1), and macrophages (F4/80) at 7 dpi in both groups (SI Appendix, Fig. S4 C–J). Further quantification revealed 3.4-fold more proliferating bronchiolar Club cells in the miR-302b/c–treated group (SI Appendix, Fig. S4D), while proliferation of other three cell populations was not significantly different between the miR-302b/c and Ctrl groups (SI Appendix, Figs. S4F, S4H, and S4J). Moreover, no differences were found in lung fibrotic lesion formation and resolution at 7, 14, and 21 dpi between the miR-302b/c and Ctrl groups (SI Appendix, Fig. S2B). These data indicate that miR-302 mimics treatment had a minimal effect on proliferation of cell types other than bronchiolar and alveolar epithelial cells in the lung.
Discussion
Our results show that bacterial pneumonia causes extensive damage to AEC in lung parenchyma and induces transient expression of miRNA-302 in the lung epithelium. Administration of miR-302 mimics to SpT4-infected mice improves AEC regeneration and lung function and enhances mouse recovery and survival. These results provide an example of a signaling pathway important in embryogenesis that can be reactivated and exploited for regenerative lung therapy following microbial infection.
miRNA mimics are double-stranded RNA molecules intended to “mimic” native miRNAs; they have been used successfully to augment the function of endogenous miRNA in mouse models and are being tested in clinical trials for cancer treatment (30, 31). We show in this study that a miRNA mimics approach can be used as a treatment of microbial infection by accelerating the proliferation of lung progenitor cells and regeneration of AEC to repair lung injury following bacterial pneumonia. These results suggest that the adult lung is capable of initiating a regenerative response after microbial infection to repair tissue injury by utilizing pathways typically expressed during embryogenesis and fetal lung development. However, the natural regenerative process is slow (not fully recovered after 30 d), leaving the host vulnerable to external insults and infections. Our results also showed transient expression of miR-302 in alveolar epithelium following bacterial pneumonia that is concomitant with regeneration of AEC and recovery of lung functions, suggesting that up-regulation of miR-302 may play a role in the regenerative process. Given the slow process of natural regeneration, we reasoned that addition of exogenous miR-302 by miRNA mimics can increase proliferation of lung progenitor cells and accelerate the repair of lung injury and functions during bacterial pneumonia. Like other miRNAs, members of the miR-302 family engage a broad collection of mRNA targets, and the major targets of the miR-302 family are genes involved in the cell cycle and proliferation. Indeed, our results showed that miR-302 mimics resulted in up-regulation of genes associated with promoting cell proliferation, which likely contributed to enhanced proliferation of AECI and AECII observed in miR-302 mimics-treated mice. AECI and AECII are known local progenitor cells in lung alveoli. Injury models using either chemical or mechanical insults show that AECII and AECI increased their proliferation to replace the lost alveolar epithelial cells and contribute to the repair/regeneration of alveolar epithelium (12, 13). Our previous work has shown that miR-302 targets the cell-cycle inhibitor Cdkn1a and that expression of miR-302 is essential for the proliferation of lung epithelial progenitor cells during embryonic development (11, 32). In this study, miR-302b/c mimics treatment led to decreased expression of Cdkn1a in lung epithelium (Fig. 4D). Together, these data suggest that the mechanism by which miR-302b/c mimics promoted mouse lung repair/regeneration and host recovery from bacterial pneumonia was through regulating expression of genes that promote the proliferation of AECI and AECII.
Efficient in vivo delivery of miRNA mimics to targeted cells is the key to the success of this approach. We selected intravenous administration as our first choice because our previous studies with i.v. delivery had shown accumulation of miR-302b/c mimics in the lung (32). In fact, intravenous delivery has proven to be an efficient means to deliver drugs to the lung because the entire right side of the heart is dedicated to pump blood exclusively into the lung. More importantly for our purpose, bacterial pneumonia causes substantial damage to the integrity of lung epithelium, which allows efficient penetration of intravenously delivered drug into lung epithelial cells (33). Systematic administration of miRNA mimics resulted in a rapid therapeutic effect shortly after the second dose, including increased proliferation of AECI and AECII and improved lung functions (Figs. 3 and 4). One downside of this systematic delivery approach is accumulation and possible off-site effects in other organs. Our previous studies have shown that miR-302 mimics delivered by i.v. injection did not cause an adverse effect in other organs including heart, liver, and intestine (32). Furthermore, bacterial pneumonia is an acute infection mostly limited to the lung. In our model, no bacteria or tissue injuries are detected in other organs, and the resulting immune responses are localized to the lung mucosa with minimal responses in the spleen (16). Thus, we expect that miR-302b/c mimics have the most effect in the lungs of SpT4-infected mice and minimal adverse effects in other organs. Additionally, the studies reported here are focused on more targeted approaches, including intranasal delivery and specific targeting of AEC.
In conclusion, our results open up a frontier in developing therapies for treating microbial infection by regenerative medicine. Although this regenerative approach does not curtail microbial growth per se, it reduces suffering and shortens recovery time by fostering tissue repair and thus could be used in conjunction with antimicrobial therapy to improve patient outcomes from serious infections. In addition to treating acute pneumonia, this approach may provide a long-term benefit of reducing severe chronic pathological conditions, such as COPD, as repeated lung infections/injuries and defects in tissue regeneration/repair have been implicated in these devastating conditions.
Materials and Methods
Animals.
C57BL/6 mice (8–10-wk old) were purchased from Charles River Laboratories and were housed in a specific pathogen-free environment at the animal facilities of the University of Pennsylvania and Temple University. All animal experiments were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee at University of Pennsylvania and Temple University.
Pathogens and Infections.
Sp strain TIGR4 (serotype 4) (16) was propagated in tryptic soy broth (Difco) at 37 °C and 5% CO2 without shaking until cultures reached log phase, OD620 between 0.8 and 1.0 as determined by Spectronic200 spectrophotometer (Thermo). For lung infection, ∼5 × 106 cfu of TIGR4 in 30 μL PBS was inoculated i.n. in mice that were anesthetized by i.p. injection with 100 μL Ketamine/Xylazine (100 mg/3.8 mg/kg). The dose was confirmed by viable counting via plating of inoculum after infections. This infection resulted in an acute pneumonia with ∼40% mortality rate; mice either succumbed to the infection or cleared bacteria within days. Typically, each experimental group started with 50% more mice than needed, and surviving mice were used for analysis at later time points (n > 3 per time point per group) for injury and repair. Mice were observed for clinical signs of morbidity by monitoring body weights and survival daily. Lung homogenates and BALF were prepared as described (16), and bacterial titers were determined by serial dilutions and plating in triplicate. The limit of detection was 4 cfu/mL of lung homogenates.
RNA Purification and RT-PCR Analysis.
For qRT-PCR of miR-302–367 cluster concentration in epithelial cells, RNA was extracted from isolated epithelial cells using a mirVana miRNA isolation kit (Ambion). For gene expression of targeted genes in lung, total RNA was isolated from lung lobes or lung epithelial cells at the indicated days post infection using TRIzol reagent and reverse-transcribed using High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems). qRT-PCR was performed with primers as described in SI Appendix, Table S1. miRNA qRT-PCR was performed by using the miR-302 LNA PCR primer sets (Exiqon). SYBR green detection of amplification was performed using the StepOne Plus cycler (Applied Biosystems). Transcript expression values were generated with the comparative threshold cycle (Delta CT) method by normalizing to the expression of the GAPDH gene.
Histology.
Lung tissues were inflated and fixed in 4% paraformaldehyde, embedded in paraffin wax, and sectioned at 7-μm intervals. H&E staining was performed using standard procedures. Immunohistochemistry was performed using the following antibodies: type 4 pneumococcal capsular polysaccharides (1: 500; Staten Serum Institut), proSurfactant protein c (1:200; Millipore), CC10 (T-18, 1:500; Santa Cruz), T1α (8.1.1, 1:100; Hybridoma Bank at University of Iowa), Hopx (E-1, 1:200; Santa Cruz), p63 (D2K8X, 1:200; Cell Signaling Technology), β-Tubulin IV (ONS1A6, 1:100; BioGenex), αSMA (1A4, 1:500; Sigma), PECAM1 (MEC13.3, 1:100; BD Pharmingen), F4/80 (BM8, 1:250; eBioscience), and cleaved caspase-3 (Asp175, 5A1E, 1:400; Cell Signaling). Slides were mounted with Vectashield mounting medium containing DAPI (Vector Laboratories). Apoptosis was measured using an In Situ Cell Death Detection Kit (Roche). Cell proliferation was measured using a Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo). The slides were imaged and subjected to an independent blinded analysis, using a Zeiss LSM 710 confocal microscope and ImageJ software. Images shown are representative views of multiple fields from at least five independent samples per group. Quantitation of cell numbers was done using images acquired on confocal microscopy and the ImageJ with the “Cell Counter” plug-in, counting multiple fields from five independent samples per group and ∼2,200 SPC+ cells and ∼765 Hopx+ cells per sample.
Preparation of miRNA Mimics and in Vivo Treatment.
miR-302b/c mimics and negative control mimics were custom-ordered from Dharmacon (GE healthcare), formulated with neutral lipid emulsion (NLE) (BIOO Scientific). To determine the effect of miRNA mimics on respiratory repair after Sp infection, 10 μg of NLE-formulated miR-302b/c mimics or control mimics were administered twice by tail-vein injection at 5 and 6 dpi.
Measurement of Pulse Oximetry.
The MouseOx Pulse-oximeter (Starr Life Sciences) was used to measure blood oxygen saturation (SpO2) in Sp-infected mice. Mice were anesthetized, and neck hairs were removed using an electric trimmer before infection. For readings, the oximeter clip was placed on the neck and the percentage of SpO2 was measured each second over several minutes; data shown are the average of SpO2 readings recorded over 3–5 min per mouse.
Pulmonary Function Testing.
Mice were anesthetized (4% isofluorane), tracheostomized (18 g), placed on the flexiVent system (SCIREQ), and ventilated with a tidal volume (7 mL/kg) at a frequency of 150 breaths/min and a positive end expiratory pressure of 3 cm H2O. Anesthesia was titrated between 2 and 4% isofluorane to prevent spontaneous breathing; then, all pulmonary function measurements were performed at the same level of anesthesia and repeated in triplicate. After each measurement, the lung was conditioned to total lung capacity (i.e., 30 cm H2O). Whole-lung dynamic respiratory mechanics were measured including pulmonary compliance, and resistance was determined by fitting the linear single-compartment model using a multiple linear regression, followed by the forced oscillation technique. In addition, negative-pressure–forced expirations were performed using the forced expiration extension for mice of the flexivent system to measure FEV at specific time points during expiration (i.e., FEV 0.1 s), forced vital capacity (FVC), and subsequent for calculations of the ratio of forced expiratory volumes to forced vital capacity (i.e., FEV 0.1 s/FVC), as previously described (34). Total time of measurement was <15 min. All data were analyzed using FlexiVent software (version 7.5, service pack 4).
Quantification of Total Proteins and LDH Activity in BALF.
BALF was collected and centrifuged at 1,650 × g for 5 min at 4 °C, and supernatant was stored at −80 °C until detection. Total protein concentration was measured using a Bradford protein assay kit (Bio-Rad), and LDH activity was assessed using the enzymatic detection of the CytoTox 96 nonradioactive cytotoxicity assay (Promega) according to the manufacturer’s protocol and read on a PerkinElmer plate reader. Three to five individual mice were measured and pooled together at each time point.
Epithelial Cell Isolation and Flow Cytometry.
Lung epithelial cells were isolated as previously described (35). For FACS analysis, single-cell preparations were incubated for 30–45 min at 4 °C with the following primary antibodies: EpCAM (G8.8; eBioscience) and Streptavidin, R-Phycoerythrin Conjugate (SAPE), CD31 (1:500, 390; eBioscience), and CD45 (1:500, 30-F11; BioLegend). Cleaved Caspase-3 (1:400, 5A1E; Cell Signaling Technology) and allophycocyanin-conjugated secondary antibody. Incubations were done in PBS (without phenol red) plus 2% FBS, and samples were collected on a BD FACSCanto Flow Cytometer and analyzed using FlowJo.
miRNA in Situ Hybridization.
In situ hybridizations were performed on 6-μm paraffin-embedded lung sections. Sections were deparaffinized, rehydrated in graded ethanols, and pretreated with 10 μg/mL of proteinase K (Roche) for 10 min at room temperature. After protease digestion, the digoxigenin-labeled Locked Nucleic Acid (LNA) miR-302c antisense probe or LNA-scrambled control probe (Exiqon) was hybridized to the slides in a humidified chamber at 60 °C overnight at a concentration of 20 nM in the hybridization buffer of 5× SSC, 50% formamide, 0.1% Tween-20, 500 μg/mL yeast RNA, and 9.2 mM citric acid. Posthybridization washes were performed three times for 30 min each at 60 °C in 2× SSC 2× SSC, 50% formamide; five times for 5 min each at 25 °C in PBS with 0.05% Tween-20 (PBST). Slides were blocked with 2% normal goat serum and 2 mg/mL BSA in PBST for 1 h at 25 °C and incubated with anti-digoxigenin alkaline phosphatase-conjugated antibody (Roche). Slides were rinsed in PBST and developed with nitro blue tetrazolium/5-bromo-4-chloro-3- indolyl-phosphate (NBT/BCIP) (Roche) to yield a blue-purple precipitate. Sections were mounted for viewing.
Statistical Analyses.
Data are presented as mean ± SEM. Unpaired, one-tailed, Student’s t test was used to calculate statistical significance between two groups, and a one-way ANOVA was used for multiple group comparison followed by Bonferroni correction unless stated otherwise. All statistical tests were performed using Prism software (GraphPad Software). Data shown are means ± SEM. P values are depicted as follows: *P < 0.05, **P < 0.01, and ***P < 0.001. Results with P > 0.05 were considered not significant (n.s.).
Supplementary Material
Acknowledgments
This work was supported by National Institutes for Health Grants R00-HL111348 and R01- HL132115 (to Y.T.); R01-AI038446 and R01-AI105168 (to J.N.W.); and R01-AI095740, R01-AI105431, and R21-AI128569 (to H.S.). We acknowledge that we obtained the miR-302 family expression data from the Lung Map database (https://lungmap.net/) on June 23, 2016.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818522116/-/DCSupplemental.
References
- 1.Pfuntner A, Wier LM, Stocks C. 2013. Most frequent conditions in U.S. hospitals, 2010. Healthcare Cost and Utilization Project Statistical Brief #148 (Agency for Healthcare Research and Quality, Rockville, MD)
- 2.Marriott HM, Dockrell DH. Streptococcus pneumoniae: The role of apoptosis in host defense and pathogenesis. Int J Biochem Cell Biol. 2006;38:1848–1854. doi: 10.1016/j.biocel.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 3.Matuschak GM, Lechner AJ. Acute lung injury and the acute respiratory distress syndrome: Pathophysiology and treatment. Mo Med. 2010;107:252–258. [PMC free article] [PubMed] [Google Scholar]
- 4.Jamieson AM, et al. Role of tissue protection in lethal respiratory viral-bacterial coinfection. Science. 2013;340:1230–1234. doi: 10.1126/science.1233632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: Patterns and perpetrators. J Clin Invest. 2012;122:2756–2762. doi: 10.1172/JCI60323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steele MP, Schwartz DA. Molecular mechanisms in progressive idiopathic pulmonary fibrosis. Annu Rev Med. 2013;64:265–276. doi: 10.1146/annurev-med-042711-142004. [DOI] [PubMed] [Google Scholar]
- 7.Baddour JA, Sousounis K, Tsonis PA. Organ repair and regeneration: An overview. Birth Defects Res C Embryo Today. 2012;96:1–29. doi: 10.1002/bdrc.21006. [DOI] [PubMed] [Google Scholar]
- 8.Johnson R, Halder G. The two faces of Hippo: Targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13:63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Porrello ER. microRNAs in cardiac development and regeneration. Clin Sci (Lond) 2013;125:151–166. doi: 10.1042/CS20130011. [DOI] [PubMed] [Google Scholar]
- 11.Tian Y, et al. Regulation of lung endoderm progenitor cell behavior by miR302/367. Development. 2011;138:1235–1245. doi: 10.1242/dev.061762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain R, et al. Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat Commun. 2015;6:6727. doi: 10.1038/ncomms7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci USA. 2011;108:E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malley R, et al. Antibody-independent, interleukin-17A-mediated, cross-serotype immunity to pneumococci in mice immunized intranasally with the cell wall polysaccharide. Infect Immun. 2006;74:2187–2195. doi: 10.1128/IAI.74.4.2187-2195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, et al. Cross-protective mucosal immunity mediated by memory Th17 cells against Streptococcus pneumoniae lung infection. Mucosal Immunol. 2017;10:250–259. doi: 10.1038/mi.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanjo H, Kawai T, Akira S. DRAKs, novel serine/threonine kinases related to death-associated protein kinase that trigger apoptosis. J Biol Chem. 1998;273:29066–29071. doi: 10.1074/jbc.273.44.29066. [DOI] [PubMed] [Google Scholar]
- 18.Singh P, Ravanan P, Talwar P. Death associated protein kinase 1 (DAPK1): A regulator of apoptosis and autophagy. Front Mol Neurosci. 2016;9:46. doi: 10.3389/fnmol.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pawlowski J, Kraft AS. Bax-induced apoptotic cell death. Proc Natl Acad Sci USA. 2000;97:529–531. doi: 10.1073/pnas.97.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hogan BLM, et al. Repair and regeneration of the respiratory system: Complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell. 2014;15:123–138. doi: 10.1016/j.stem.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley KG, et al. Connective tissue growth factor modulates adult β-cell maturity and proliferation to promote β-cell regeneration in mice. Diabetes. 2015;64:1284–1298. doi: 10.2337/db14-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Emerman AB, Zhang ZR, Chakrabarti O, Hegde RS. Compartment-restricted biotinylation reveals novel features of prion protein metabolism in vivo. Mol Biol Cell. 2010;21:4325–4337. doi: 10.1091/mbc.E10-09-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanden Bosch A, et al. NuSAP is essential for chromatin-induced spindle formation during early embryogenesis. J Cell Sci. 2010;123:3244–3255. doi: 10.1242/jcs.063875. [DOI] [PubMed] [Google Scholar]
- 24.Martinsson-Ahlzén H-S, et al. Cyclin-dependent kinase-associated proteins Cks1 and Cks2 are essential during early embryogenesis and for cell cycle progression in somatic cells. Mol Cell Biol. 2008;28:5698–5709. doi: 10.1128/MCB.01833-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma J, Cui B, Ding X, Wei J, Cui L. Over-expression of cyclin D1 promotes NSCs proliferation and induces the differentiation into astrocytes via Jak-STAT3 pathways. Neurochem Res. 2015;40:1681–1690. doi: 10.1007/s11064-015-1635-9. [DOI] [PubMed] [Google Scholar]
- 26.Khanjyan MV, Yang J, Kayali R, Caldwell T, Bertoni C. A high-content, high-throughput siRNA screen identifies cyclin D2 as a potent regulator of muscle progenitor cell fusion and a target to enhance muscle regeneration. Hum Mol Genet. 2013;22:3283–3295. doi: 10.1093/hmg/ddt184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan KY, Ozçelik H, Cheung AN, Ngan HY, Khoo US. Epigenetic factors controlling the BRCA1 and BRCA2 genes in sporadic ovarian cancer. Cancer Res. 2002;62:4151–4156. [PubMed] [Google Scholar]
- 28.Yeung B, Yu J, Yang X. Roles of the Hippo pathway in lung development and tumorigenesis. Int J Cancer. 2016;138:533–539. doi: 10.1002/ijc.29457. [DOI] [PubMed] [Google Scholar]
- 29.Cazzalini O, Scovassi AI, Savio M, Stivala LA, Prosperi E. Multiple roles of the cell cycle inhibitor p21(CDKN1A) in the DNA damage response. Mutat Res. 2010;704:12–20. doi: 10.1016/j.mrrev.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Trang P, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Z, Rana TM. Therapeutic targeting of microRNAs: Current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 32.Tian Y, et al. A microRNA-Hippo pathway that promotes cardiomyocyte proliferation and cardiac regeneration in mice. Sci Transl Med. 2015;7:279ra38. doi: 10.1126/scitranslmed.3010841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agu RU, Ugwoke MI, Armand M, Kinget R, Verbeke N. The lung as a route for systemic delivery of therapeutic proteins and peptides. Respir Res. 2001;2:198–209. doi: 10.1186/rr58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shalaby KH, Gold LG, Schuessler TF, Martin JG, Robichaud A. Combined forced oscillation and forced expiration measurements in mice for the assessment of airway hyperresponsiveness. Respir Res. 2010;11:82. doi: 10.1186/1465-9921-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapman HA, et al. Integrin α6β4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.