Significance
Ascofuranone (AF) and ascochlorin (AC) are fungal natural products with similar chemical structures, originally isolated from Acremonium egyptiacum. Both have many useful biological properties; in particular, AF is a promising drug candidate against the tropical disease, African trypanosomiasis. However, the difficulty of the synthetic method and the inaccessibility of bioengineering methods have inhibited industrial production. This study identified all of the genes required for the branched biosynthetic pathways of AF/AC, which are clustered at two separate loci in the genome. In addition, we established the A. egyptiacum strain selectively producing AF, by genetically blocking the AC biosynthetic pathway. This study benefits the field of combinatorial biosynthesis through presenting biocatalysts and paves the way to cost-effective AF production with bioengineering.
Keywords: ascofuranone, ascochlorin, secondary metabolism, terpene cyclase, African trypanosomiasis
Abstract
Ascofuranone (AF) and ascochlorin (AC) are meroterpenoids produced by various filamentous fungi, including Acremonium egyptiacum (synonym: Acremonium sclerotigenum), and exhibit diverse physiological activities. In particular, AF is a promising drug candidate against African trypanosomiasis and a potential anticancer lead compound. These compounds are supposedly biosynthesized through farnesylation of orsellinic acid, but the details have not been established. In this study, we present all of the reactions and responsible genes for AF and AC biosyntheses in A. egyptiacum, identified by heterologous expression, in vitro reconstruction, and gene deletion experiments with the aid of a genome-wide differential expression analysis. Both pathways share the common precursor, ilicicolin A epoxide, which is processed by the membrane-bound terpene cyclase (TPC) AscF in AC biosynthesis. AF biosynthesis branches from the precursor by hydroxylation at C-16 by the P450 monooxygenase AscH, followed by cyclization by a membrane-bound TPC AscI. All genes required for AC biosynthesis (ascABCDEFG) and a transcriptional factor (ascR) form a functional gene cluster, whereas those involved in the late steps of AF biosynthesis (ascHIJ) are present in another distantly located cluster. AF is therefore a rare example of fungal secondary metabolites requiring multilocus biosynthetic clusters, which are likely to be controlled by the single regulator, AscR. Finally, we achieved the selective production of AF in A. egyptiacum by genetically blocking the AC biosynthetic pathway; further manipulation of the strain will lead to the cost-effective mass production required for the clinical use of AF.
Ascofuranone (1) and ascochlorin (2) (Fig. 1) are fungal secondary metabolites with polyketide-terpene hybrid origins (meroterpenoids) that exhibit diverse physiological activities, including antivirus, antitumor, antiinflammatory, and hypolipidemic activities (1–4). Recent reports indicated that 1 is a potential lead candidate for drug development against cancer (5) and alveolar echinococcosis (6). Most notably, 1 is a strong inhibitor of cyanide-insensitive alternative oxidases (7, 8) and a promising drug candidate against African trypanosomiasis (9). This disease, also known as African sleeping sickness in humans and nagana in livestock, is caused by the protozoan parasite Trypanosoma brucei. T. brucei depends exclusively on glycolysis as an energy source in the mammalian bloodstream and relies on trypanosome alternative oxidase (TAO) for reoxidizing NADH to continue glycolysis (9). Mammalian hosts lack this protein, and thus TAO is considered to be a unique target for antitrypanosomal drugs (9); indeed, 1 showed therapeutic efficacy against a rodent infection model of African trypanosomiasis (10). However, the investigation of 1 as a treatment against African trypanosomiasis, a neglected tropical disease that affects economically disadvantaged patients, was inhibited by the complexity of the stereoselective synthesis of 1 (11, 12). Therefore, the identification of the biosynthetic genes has been awaited for cost-effective industrial production, through engineering of the pathway.
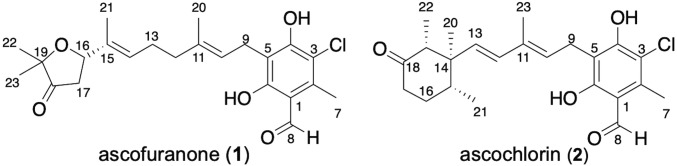
Fig. 1.
Chemical structures of ascofuranone (1) and ascochlorin (2).
Ascofuranone (1) and ascochlorin (2) were originally isolated from the filamentous fungus Acremonium egyptiacum (synonym: Acremonium sclerotigenum), which has been known as Ascochyta viciae (1, 2, 13, 14). Interestingly, these compounds have been isolated from various fungi; for example, 2 from Cylindrocladium ilicicola (15, 16), Cylindrocarpon sp. (17), Fusarium sp. (18), Microcera sp. (19), and Nectria coccinea (20), and 1 from Paecilomyces variotii (21) and Verticillium hemipterigenum (22), indicating the broad distribution of the biosynthetic pathways. Compounds related to 1 and 2 are considered to be synthesized through the prenylation of orsellinic acid (3) and terminal cyclization via epoxidation (17, 18, 23) (Fig. 2A), but the details of their biosyntheses have not been established. Recently, we reported that Stachybotrys bisbyi PYH05-7 encodes the 3-producing polyketide synthase (PKS) StbA, the UbiA-family prenyltransferase StbC, which produces ilicicolinic acid B (4) (Fig. 2A), and the nonribosomal peptide synthetase (NRPS)-like reductase StbB for the synthesis of LL-Z1272β (ilicicolin B) (5), a putative precursor of 1 and 2 (24). By analogy to the biosynthetic pathways of other fungal meroterpenoids, such as pyripyropene, paxilline, and aflatrem (25), the terminal olefin of the prenyl chain of 5 is thought to be epoxidized by a flavin monooxygenase (FMO), and then cyclized by a membrane-bound terpene cyclase (TPC). Considering the different cyclization patterns of 1 and 2, two distinct TPCs were assumed to exist in A. egyptiacum.
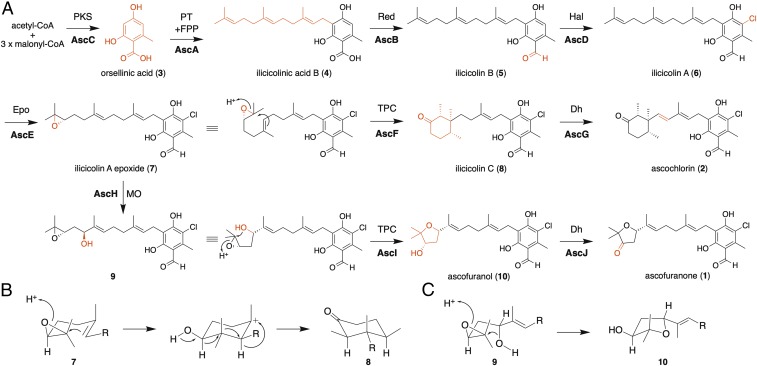
Fig. 2.
Summary of ascofuranone (1) and ascochlorin (2) biosyntheses. (A) The overall scheme of 1 and 2 biosyntheses. Enzymes are abbreviated as follows: Dh, dehydrogenase; Epo, epoxidase; Hal, halogenase; MO, monooxygenase; PKS, polyketide synthase; PT, prenyltransferase; Red, reductase; TPC, terpene cyclase. (B) The cyclization reaction catalyzed by AscF. (C) The cyclization reaction catalyzed by AscI.
Developments in genomic resources and bioinformatics tools have accelerated the elucidation of the biosynthetic genes in various organisms (26). In particular, the biosynthetic genes for fungal secondary metabolites often form functional gene clusters (27), which can be predicted by searching the genome for collocated genes with functional motifs for putative biosynthetic enzymes (26). However, it is difficult to predict the relevance of genes with novel functions and to find biosynthetic clusters separated at multiple loci. For plant metabolites, in contrast, coexpression analyses have been effective in identifying nonclustered biosynthetic genes (28, 29); naturally, such a transcriptome-based approach is also promising in the case of fungal multilocus clusters (30). Since previous studies showed that 1 and 2, as well as other related compounds, suppress respiratory chain activities at multiple targets (31, 32), the fungal production of 1 and 2 is likely to be tightly regulated to avoid inhibiting fungal growth. Therefore, we can reasonably anticipate that determining the gene clusters that are differentially expressed in association with 1 production will lead to the identification of the responsible genes.
In this study, we aimed to find the biosynthetic genes for 1 by using a comparative transcription analysis of A. egyptiacum cultures exhibiting all-or-none production. Taking advantage of heterologous expression systems in Aspergillus spp., in vitro reconstruction assays, and a newly established gene disruption method for A. egyptiacum, we identified the complete biosynthetic enzymes of 2 encoded by the most prominently induced gene cluster. We then discovered another gene cluster at a discrete locus involved in the remaining steps of AF production, by virtue of a combination of motif-based and differential expression approaches. In addition, we analyzed the transcriptional factor regulating the biosynthetic genes of both compounds located in separate chromosomal regions.
Results and Discussion
Discovery of the asc-1 Cluster.
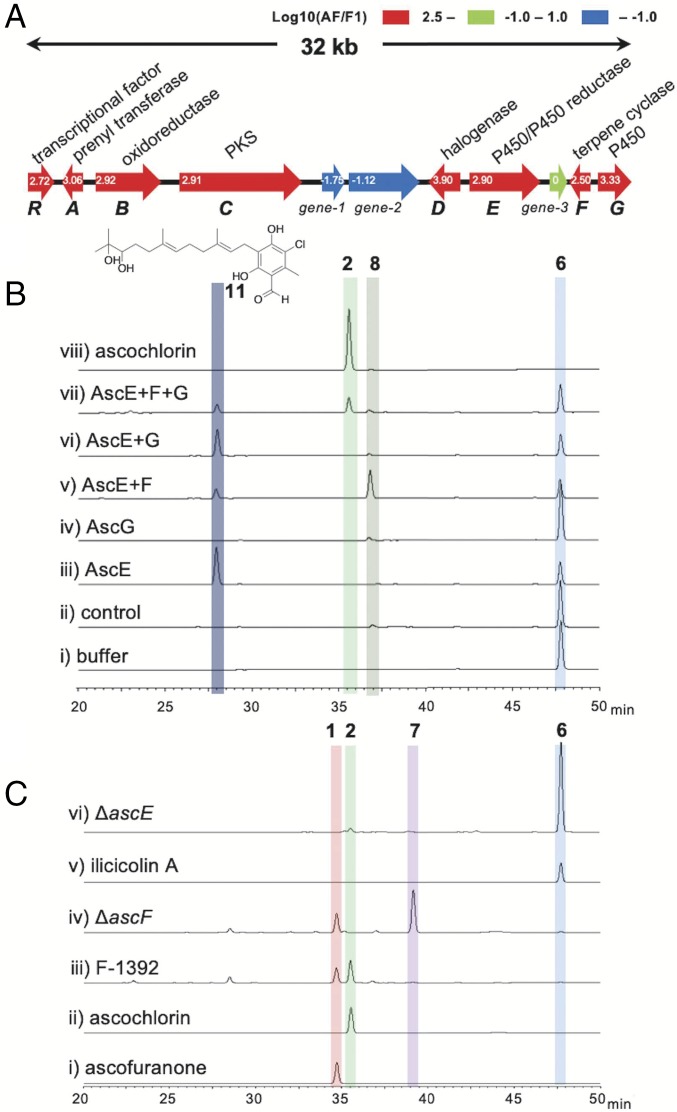
First, we performed a genome-wide transcription analysis using the A. egyptiacum strain F-1392, to identify the differentially expressed gene clusters possibly responsible for the biosynthesis of 1 or 2. The production of 1 and 2 in A. egyptiacum varied dependent on the culture medium, and two media, designated as F1 and AF here, showed the virtual all-or-none difference. We thus prepared poly(A) selected RNAs from mycelia grown in F1 and AF media, which yielded 0.96 and 399 mg of 1 per liter culture, and obtained 611,740 and 846,811 RNA-seq reads, respectively. Although the experiment was preliminary, in terms of read numbers and lacking replicates, mapping the differential expression profiles to the 31-Mb draft genome (SI Appendix, Table S1) shed light on a 32-kb region on scaffold 2 (named the asc-1 cluster; DNA Data Bank of Japan (DDBJ)/European Nucleotide Archive (ENA)/GenBank accession LC406756) (Fig. 3A and SI Appendix, Fig. S1). The transcription of eight genes (ascABCDEFGR) in the asc-1 cluster was more strongly induced (log10 values ≥ 2.5) in AF medium than in F1 medium, and some of them were predicted to encode characteristic enzymes required for the biosyntheses of 1 and 2; that is, PKS, prenyl transferase, and halogenase (SI Appendix, Table S2). Therefore, we named the eight genes ascR and ascA-G as candidates for the 1 or 2 biosynthetic genes (Fig. 3A).
Fig. 3.
Functional characterizations of AscA-G. (A) Schematic representation of the asc-1 cluster, found by the differential expression analysis. The expression change was indicated with log10 value for each gene. (B) HPLC profiles of the in vitro reaction products of ilicicolin A (6) as a substrate when incubated with the buffer (i), the homogenate of the A. sojae wild-type strain (ii), the homogenates containing either AscE (iii) or AscG (iv), and the mixed homogenates containing AscE+AscF (v), AscE+AscG (vi), or AscE+AscF+AscG (vii), and authentic ascochlorin (viii). (C) HPLC profiles of authentic ascofuranone (i), authentic ascochlorin (ii), mycelium extracts of A. egyptiacum F-1392 (iii), ΔascF strain (iv), authentic ilicicolin A (v), and mycelium extract of ΔascE strain (vi). The yields of the compounds are summarized in SI Appendix, Table S8.
The functions of AscR and AscA-G were deduced by bioinformatics analyses, as follows. AscR possessed a Zn2Cys6 binuclear cluster for DNA binding and was presumably a transcription regulator. AscA, AscB, and AscC exhibited more than 50% amino acid identities with the ilicicolin B (5) biosynthetic enzymes (StbABC) in S. bisbyi (24). A flavin-binding enzyme, AscD, was thought to be a halogenase, because it shares 68% amino acid identity with the halogenase that catalyzes the 5-chlorination of ilicicolinic acid B (4) (33). The Pfam motifs indicated that AscG is a membrane-bound P450 monooxygenase, and AscE is a P450 monooxygenase/P450 reductase fusion protein, such as the bacterial soluble P450 monooxygenase BM3 (34). AscF was predicted to be a membrane-bound TPC, sharing 29% amino acid identity to AndB, a TPC in anditomin biosynthesis in Emericella variecolor (35). The multiple alignment of AscF, AndB, and other meroterpenoid TPCs, Pyr4 in pyripyropene biosynthesis (36), and PaxB in paxilline biosynthesis (37), revealed that the active-site E63 and D218 residues of Pyr4 (36) were also conserved in AscF (SI Appendix, Fig. S2). Although three other genes (gene-1, -2, and -3) of unknown function are located within the asc-1 cluster, we excluded them from the analysis since they exhibited little or no expression in the AF medium.
Characterization of the asc-1 Cluster Genes.
To investigate the biosynthesis, we constructed Aspergillus oryzae transformants expressing ascA-D genes under the starch-inducible amyB promoter (38). Comparison of MS and NMR data with literature (24) indicated that 5 (m/z 355, [M−H]−) was produced in the strain expressing ascCAB (yield, 0.71 mg/L) (SI Appendix, Figs. S3 and S4 and Table S3). We further confirmed that AscC, AscA, and AscB in this order catalyzed each reaction as expected from the homology, based on the detection of the specific products 3 and 4 in the transformants expressing ascC and ascCA (SI Appendix, Figs. S4 and S5 and Table S3). Subsequently, the A. oryzae transformant harboring ascCABD yielded an additional compound (SI Appendix, Fig. S4), which was identified as ilicicolin A (6) (0.04 mg/L) by comparison with the authentic standard (SI Appendix, Fig. S6A). These data showed that AscCABD were responsible for the biosynthesis of 6.
Next, we performed an in vitro reconstruction of the succeeding steps, since the meager production of 6 complicated the analysis of the downstream metabolites in A. oryzae transformants. To prepare the AscE-G proteins, we employed an Aspergillus sojae high-copy expression system in which the pyrG selective marker with a truncated promoter enables the transformation of high-copy plasmids (SI Appendix, Fig. S7) (39). Since AscF and AscG are membrane-bound proteins and difficult to purify, a cell-free homogenate was used for the in vitro enzyme reaction. We hypothesized that 6 was epoxidized into ilicicolin A epoxide (7) by AscE or AscG before cyclization by AscF, as in the case of other fungal meroterpenoid biosynthesis in which terpene cyclization follows epoxidation of the prenyl group (25). This is also supported by the previous report of accumulation of 7 in A. egyptiacum mutant strain obtained through random chemical mutagenesis (23). However, the in vitro assay showed that 7 was not detected with either the AscE- or AscG-containing homogenate; alternatively, a compound (11) with an m/z of 423 ([M–H]−) was detected only with the AscE-containing homogenate (Fig. 3B and SI Appendix, Fig. S8 and Table S7). The m/z suggested that 11 was a diol, hydrolyzed from the epoxide 7 (m/z 405, [M−H]−), so we considered that AscE catalyzes the epoxidation to produce 7. Since 11 is unlikely to participate in the production of 1 or 2 in view of the reaction mechanisms, the diol 11 was presumed to be a shunt product formed by endogenous hydrolase in A. sojae, as previously observed in the A. oryzae system (36, 40). These assumptions were later confirmed by our deletion experiment (see below). Another reaction product (8) was detected when a mixed homogenate containing AscE and AscF was incubated with 6 (Fig. 3B). Compound 8 was deduced to be ilicicolin C from the high-resolution (HR)-MS data (SI Appendix, Fig. S8 and Table S7), which was supported by the fact that ilicicolin C was isolated from A. egyptiacum (41). Finally, when the protein extracts containing AscE, AscF, and AscG were incubated with 6, the reaction product was identified as ascochlorin (2) (Fig. 3B). Confirmation of the structure was obtained by direct comparison of the MS/MS fragmentation pattern, and by coinjection with the authentic standard (SI Appendix, Fig. S6 B and C). These results indicated that the AscF-catalyzed terpene cyclization product 8 was oxidized into 2 by the P450 monooxygenase AscG. The absolute configuration of 8 is thought to be identical to that of (14S,15R,19R)-2, which was established by X-ray structure analysis (42).
To experimentally establish the involvement of 7 in the biosynthesis of 1 and/or 2, we constructed A. egyptiacum gene disruptants, using the ku70-deleted strain that we constructed in this study to enhance homologous recombination (43) (SI Appendix, Fig. S9). The ascF-deleted strain (ΔascF) no longer produced 2 (Fig. 3C), but instead accumulated 7 (1.22 g/L; SI Appendix, Table S8), which was isolated and structurally determined by NMR and HR-MS analyses (SI Appendix, Figs. S8 and S10 and Tables S4 and S7). In contrast, the ascE-deleted strain (ΔascE) only accumulated 6 (2.32 g/L, SI Appendix, Table S8), but neither 1 nor 2 (Fig. 3C). These results supported experimental proof that AscE epoxidized the terminal olefin of 6 to produce 7, and AscF cyclized 7 into 8 in AC biosynthesis. For AF biosynthesis, another TPC other than AscF should be involved, since ΔascF still produced 1 (0.50 g/L; SI Appendix, Table S8) (Fig. 3C). Given the fact that ΔascE no longer produced 1, the epoxide 7 was indicated as the last common precursor for the biosyntheses of 1 and 2.
Considering these results, we concluded that the seven genes, ascA-G, in the asc-1 cluster encode the biosynthetic enzymes for the production of 2. AscE is a P450 monooxygenase that catalyzes stereoselective epoxidation of the terminal double bond of the prenyl group (44). In contrast, in most of the cases, fungal meroterpenoid biosynthesis employs FAD-dependent monooxygenases for the formation of epoxide (25). Notably, AscF is the rare meroterpenoid TPC that produces a monocyclic terpene, and the cyclization reaction is proposed to be initiated by protonation of the terminal epoxide of 7 to generate a monocyclic tertiary cation, which is followed by a series of hydride and methyl shifts with abstraction of proton, leading to the formation of the (14S,15R,19R)-trimethylcyclohexanone ring structure of 8 (Fig. 2B). AscA-E are also involved in AF biosynthesis, but additional enzymes are required for the later steps, after the common precursor 7, which should be encoded outside of the asc-1 cluster.
Identification of the Ascofuranone Biosynthetic Genes.
Once again, the differential expression analysis was exploited to identify the genes responsible for the late stage of AF biosynthesis. Since one additional oxygen atom must be incorporated into the side chain of 7, we focused on P450s, the most abundant oxygenases in fungi. Among the gene clusters induced at least 10-fold in AF medium, only three harbored P450 monooxygenase genes other than the asc-1 cluster (SI Appendix, Table S5). One cluster on scaffold 6 encodes homologs of desmethylbassianin biosynthesis enzymes, DmbS, DmbC, and DmbA (45), and thus was not likely to be involved in AF biosynthesis. Another cluster on scaffold 3 was excluded since it encodes an acetyltransferase and a peptidase, which both seemed irrelevant to AF biosynthesis. Combining the function prediction (see below), we targeted the other cluster on scaffold 3 (asc-2 cluster; DDBJ/ENA/GenBank accession no. LC406757) located on the different scaffold from asc-1 cluster (SI Appendix, Fig. S1).
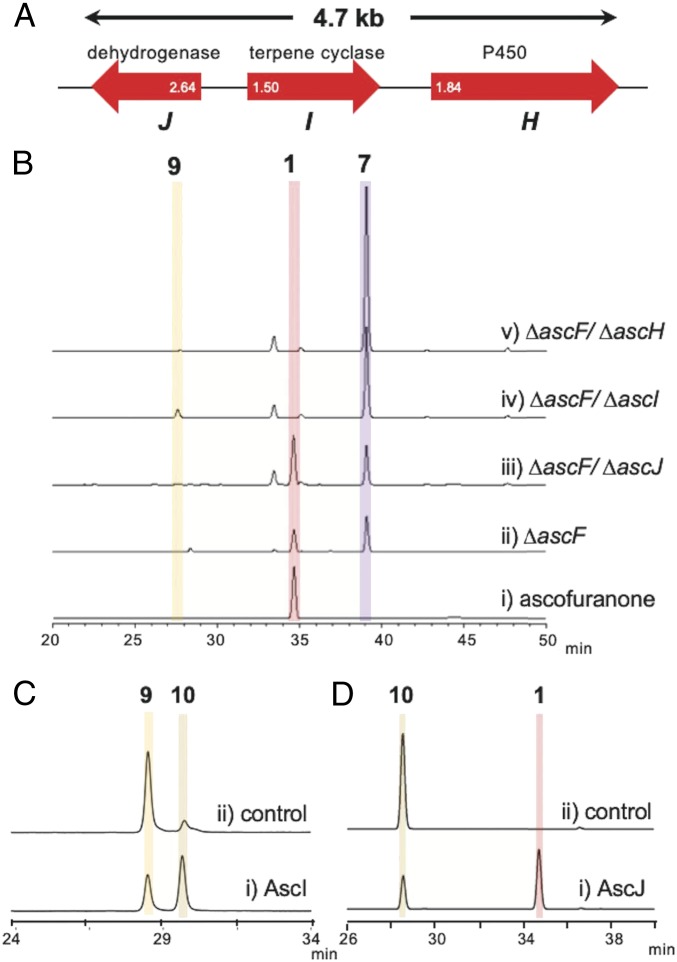
The asc-2 cluster is composed of three genes, designated as ascHIJ (Fig. 4A). All three genes exhibited higher (log10 values ≥ 1.5) expression in AF medium (SI Appendix, Table S2). AscH is a P450 sharing 38% amino acid identity with the hexadecane hydroxylase P450-ALK2-A from Candida tropicalis (46). AscJ is classified into the NAD(P)-dependent, short-chain alcohol dehydrogenase family, sharing 33% identity with the aflatoxin biosynthesis protein AflH (47). Finally, AscI is an eight-transmembrane protein deduced by SOSUI program (48) without any Pfam motif; however, it is similar to the citreoviridin biosynthetic protein CtvD (49) and AurD in aurovertin biosynthesis (50), although it shares only 27% and 29% identities, respectively. Both enzymes are regioselective hydrolases for cyclizing a bisepoxide to give a 3,4-dihydroxyl-tetrahydrofuran structure, suggesting that AscI is the TPC for AF biosynthesis.
Fig. 4.
Functional analyses of AscH, AscI, and AscJ. (A) Schematic representation of the asc-2 cluster, found by the differential expression analysis. The expression change was indicated with log10 value for each gene. (B) HPLC profile of authentic ascofuranone (i) and mycelium extracts of A. egyptiacum disruptants ΔascF (ii), ΔascF/ΔascJ (iii), ΔascF/ΔascI (iv), and ΔascF/ΔascH (v). (C) HPLC profile of in vitro reaction products of 9 as a substrate when incubated with microsomal fractions of the yeast harboring the ascI-expressing vector (i) and the void vector (ii). Note that 10 detected in the control is the uncontrollable product generated nonenzymatically in the preparation of the substrate 9. (D) HPLC profile of in vitro reaction products of 10 as a substrate when incubated with the homogenates of the AscJ-overexpressing strain of A. sojae (i) and wild type (ii). The yields of compounds for B are summarized in SI Appendix, Table S8.
Characterization of the asc-2 Cluster Genes.
To link the candidate genes ascHIJ to AF biosynthesis, each candidate gene was disrupted in the A. egyptiacum ΔascF strain accumulating the precursor 7. HPLC analyses of these double disruptants showed that ΔascF/ΔascH and ΔascF/ΔascI could not produce 1, indicating that AscH and AscI are essential for AF biosynthesis, although ΔascF/ΔascJ could produce it (0.41 g/L; SI Appendix, Table S8) (Fig. 4B). In contrast, the cell-free homogenate from the A. sojae strain expressing ascE, H, and I did not convert 6 into 1, while the addition of the ascJ homogenate to the mixture sufficed to produce 1 (SI Appendix, Fig. S11). Taken together, AscJ is also required for the biosynthesis, although its function is compensated by the endogenous dehydrogenase in A. egyptiacum. These results clearly demonstrated that AscHIJ are responsible for AF biosynthesis.
The double disruptant ΔascF/ΔascI specifically accumulated a new compound 9 (0.05 g/L, SI Appendix, Table S8) (Fig. 4B), which was isolated from the large-scale culture, and analyzed by HR-MS and NMR. Its molecular formula was established as C23H31ClO5 from the HR-MS data (m/z 421.1799, calc. 421.1787; SI Appendix, Fig. S8 and Table S7), indicating that it contains an additional oxygen compared with 7. The NMR data revealed that 7 and 9 are very similar to each other, but one methylene signal (δ 1H: 2.01/2.05 ppm; 13C: 36.4 ppm) disappeared and one oxymethine signal (δ 1H: 4.20 ppm; 13C: 75.5 ppm) appeared in 9 (SI Appendix, Fig. S12 and Table S4). Based on the association with the spin system of H-17/H-18, we assigned this signal as H-16. Notably, under acidic condition, 9 was nonenzymatically converted into ascofuranol (10), whose planar structure was identified by HR-MS and NMR analyses, and its absolute configuration was determined as (16S,18S)-10 by the Mosher and NOESY analyses (SI Appendix, Figs. S13 and S14 and Table S6). The absolute configurations of 7 and 9 were thus thought to be (16S)-7 and (16S,18S)-9, respectively.
We further conducted in vitro assays to attribute the succeeding reactions to the candidate genes. The microsomal fraction of the yeast expressing ascI efficiently converted 9 into 10, while that from the yeast harboring a blank vector did not afford any product (Fig. 4C). The Km and Vmax values of AscI were 50.4 ± 11.1 µM and 129 ± 13 nmol/min, respectively. In addition, we found that the cell-free homogenate of A. sojae expressing ascJ dehydrogenizes 10 into 1 (Fig. 4D). Thus, we demonstrated that 7 is hydroxylated by the P450 monooxygenase AscH, and the resultant 9 is cyclized by AscI to 10, which is oxidized into 1 by AscJ (Fig. 2A).
Next, we investigated the reaction mechanism of the meroterpenoid TPC AscI, which lacks significant sequence similarity to any known TPCs. Sequence alignment revealed that AscI also harbors several conserved acidic residues that are thought to be essential for the catalysis (36) (SI Appendix, Fig. S15). Indeed, the alanine substitutions of AscI D61, E103, D296, E353, and D355 resulted in significant loss of activities (SI Appendix, Fig. S16). Notably, D61, D296, and E353 are also conserved in AurD and CtvD (SI Appendix, Fig. S15), suggesting that these residues are important for the protonation-initiated epoxide ring opening, which facilitates the 6-endo-tet cyclization to form the tetrahydrofuran ring of 10 (Fig. 2C).
Collectively, the three genes, ascH-J, in the asc-2 cluster encode the late-step biosynthetic enzymes, indicating that AF biosynthesis represents another rare example of a fungal multilocus biosynthetic cluster. Although there have been a few reports (51–54), there is still no established strategy for exploring such clusters. In the cases of prenyl xanthone and austinol, seeking the gene candidates based on the homology of the characteristic enzymes, a nonreducing PKS and a prenyltransferase, successfully led to the identification of the separate clusters, because there were only a few paralogs in the genome (52, 54). This was not the case for 1, since more than 100 genes encode P450 monooxygenases in the genome of A. egyptiacum, and the TPC, which is usually considered as a characteristic core enzyme, has not been known. The expression analysis was shown to be useful for the motif-independent identification of the fungal biosynthetic gene clusters, although previous studies exploited many sample conditions for the expression analysis and dedicated algorithms (55, 56). This study indicated that, even with a simple comparison of producing and nonproducing conditions, the expression analysis is a powerful method to identify the split gene clusters when combined with a motif-based approach.
Coregulation of the Multilocus Gene Clusters by AscR.
Last, we analyzed the coregulation of the multilocus AF biosynthetic genes. Since the putative DNA-binding protein AscR was highly expressed in AF medium (SI Appendix, Table S2), it is predicted to be a transcription factor positively regulating the expression of the asc-1 cluster genes. An A. egyptiacum strain constitutively expressing AscR was constructed to verify this, and it produced both 1 and 2 in F1 medium (SI Appendix, Fig. S17A), suggesting the coregulation of the asc-1 and asc-2 clusters by AscR. This was further corroborated by the fact that all of the promoters of ascA-J possessed the shared sequence motif of TCGGYGNNTTW detected by MEME (SI Appendix, Fig. S17B), containing the CGG nucleotide triplet essential for the DNA binding of transcription factors with the same Zn2Cys6 binuclear cluster (57). In fact, this motif was not present in the promoters of the three intervening genes in the asc-1 cluster, which showed little or no expression in AF medium. We thus consider that AscR positively regulates the expression of both the asc-1 and -2 cluster genes by binding to this motif.
The evolutionary origin of fungal multilocus biosynthetic clusters has also been a matter of debate. In the cases of the biosyntheses of aflatrem and austinol, ancestrally single-locus clusters were presumably divided into two separated loci by chromosomal rearrangement, supported by the presence of the relict copy in one cluster and the functional gene in the other cluster (51, 52). However, phylogeny and synteny analyses of trichothecene biosynthesis genes revealed that ancestrally separated genes could work with the major biosynthetic cluster as a coregulated pathway and, depending on the species, were later merged into the major cluster (53). For AF biosynthesis, there was no trace of past duplication suggesting a split into the asc-1 and -2 clusters. BLAST searches revealed partially syntenic clusters of the asc-1 cluster in several related fungi of the class Sordariomycetes, but there is no asc-2 syntenic cluster. Interestingly, the asc-1 syntenic cluster in Coniella lustricola also contains the ascH ortholog (PSR83571; 50% amino acid identity), whereas the fungus does not encode a plausible ascI ortholog. Since the orthologs of the ascH and asc-1 cluster genes are widely distributed among Sordariomycetes, ascH is likely to play another role in the biosynthesis of AC-related compounds. Given that the report of AF-producing fungi (13) and the distribution of ascI orthologs were biased to the class Eurotiomycetes, we assume that ascI was horizontally transferred and grafted upon the preexisting ascH to make a coregulated asc-2 cluster, although the elucidation will require further detailed investigation including synteny and phylogeny analyses using a wide range of fungal species.
Conclusions
We clarified the entire biosynthetic pathways of ascofuranone (1) and ascochlorin (2) in A. egyptiacum (Fig. 2). The biosyntheses of 1 and 2 share the common pathway up to the generation of ilicicolin A epoxide (7). Notably, the biosynthetic genes of 1 and 2 are localized at distinct chromosomal regions, but all of their promoters share a common conserved motif, and they are probably regulated by the transcriptional factor, AscR. This study thus contributes to increasing the knowledge on meroterpenoid biosynthesis and demonstrates the power of a differential expression analysis for exploring multilocus biosynthetic clusters. From a clinical viewpoint, the elucidated genes, as well as the established method for the genetic manipulation of the strain F-1392, will be the keys for the drug development of 1. We have already established the ascF-deleted strain producing exclusively 1, but not 2 (Fig. 3C), and further manipulation will lead to the cost-effective industrial production of ascofuranone.
Materials and Methods
A. egyptiacum (synonym: A. sclerotigenum) strain F-1392 (13, 14) is a descendant of the nitrosoguanidine-induced mutant no. 34, characterized in the original paper reporting 1 (2). The differential expression analysis was performed with an Ion PGM system, and the raw sequence reads and the expression profile per gene were deposited in the DDBJ under the accession nos. DRA006136 and E-GEAD-282, respectively. A. egyptiacum gene disruptants were obtained from the newly established Δku70/ΔpyrG strain by homologous recombination, after transformation by the protoplast–polyethylene glycol method (58). Aspergillus oryzae NSAR1 (59) (niaD−, sC−, ΔargB, adeA−) was used as the host for the heterologous coexpression of ascA-D, and Aspergillus sojae P6-1 (ΔpyrG) (39) was used as the host for the high-copy heterologous expression of ascE-J. The products from mycelia of each transformant were extracted with acetone, and the in vitro reaction products were extracted with ethyl acetate. The extracts were analyzed by UV-HPLC and liquid chromatography–MS, using ODS columns with standard chromatographic methods. The purified products were monitored by NMR analyses, including 1HNMR, 13CNMR, heteronuclear multiple bond coherence, heteronuclear multiple quantum coherence, and COSY. Full experimental procedures are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Prof. Katsuya Gomi (Tohoku University) and Prof. Katsuhiko Kitamoto (The University of Tokyo) for kindly providing the expression vectors and the fungal strain. We also thank Dr. Keiko Gomi and Dr. Ryoichi Sakaue (Kikkoman Corporation) for their helpful advice. This work is financially supported by Ministry of Education, Culture, Sports, Science, and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI Grants JP17H04763 (to T.A.), JP17KT0095 (to T.A.), JP17K15679 (to Y.K.), 23117004 (to M.M.), JP16H06443 (to I.A.), JP18K19139 (to I.A.), and 26253025 (to K.K.); Japanese Initiative for Progress of Research on Infectious Disease for Global Epidemic [Grant JP18fm0208020 (to Y.K.)]; Japan Science and Technology Agency/National Natural Science Foundation of China Strategic International Collaborative Research Program (to I.A.); and Research Program on Emerging and Reemerging Infectious Diseases Grant 17fk0108119j0001 (to K.K.). We also acknowledge support from the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry and from the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries and Food Industry.
Footnotes
Conflict of interest statement: Y.A., Y.S., and K.I. are employees of Kikkoman Corporation. Y.A., Y.S., and K.K. are inventors on the patent application PCT/JP2018/18405. Y.A., T.A., Y.S., I.A., and K.K. are inventors on the Japanese patent application 2017-220055. M.Y. and Y.K. are executive directors of Institute of Mitochondrial Science Company, Ltd. (IMS). M.Y. and Y.K. received financial interests from IMS. M.Y., Y.K., D.K.I., and K.K. hold equity of IMS. K.K. received research funding from Kikkoman Corporation.
A portion of this manuscript was presented as the master’s degree thesis of R.C. at Graduate School of Medicine, the University of Tokyo, in February 2015.
This article is a PNAS Direct Submission. C.A.T. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the DNA Data Bank of Japan (accession nos. DRA006136, E-GEAD-282, LC406756, and LC406757).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819254116/-/DCSupplemental.
References
- 1.Tamura G, Suzuki S, Takatsuki A, Ando K, Arima K. Ascochlorin, a new antibiotic, found by the paper-disc agar-diffusion method. I. Isolation, biological and chemical properties of ascochlorin. (Studies on antiviral and antitumor antibiotics. I) J Antibiot (Tokyo) 1968;21:539–544. doi: 10.7164/antibiotics.21.539. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki H, Hosokawa T, Sawada M, Ando K. Isolation and structure of ascofuranone and ascofranol, antibiotics with hypolipidemic activity. J Antibiot (Tokyo) 1973;26:676–680. doi: 10.7164/antibiotics.26.676. [DOI] [PubMed] [Google Scholar]
- 3.Magae J, Hosokawa T, Ando K, Nagai K, Tamura G. Antitumor protective property of an isoprenoid antibiotic, ascofuranone. J Antibiot (Tokyo) 1982;35:1547–1552. doi: 10.7164/antibiotics.35.1547. [DOI] [PubMed] [Google Scholar]
- 4.Lee SH, et al. Anti-inflammatory effect of ascochlorin in LPS-stimulated RAW 264.7 macrophage cells is accompanied with the down-regulation of iNOS, COX-2 and proinflammatory cytokines through NF-κB, ERK1/2, and p38 signaling pathway. J Cell Biochem. 2016;117:978–987. doi: 10.1002/jcb.25383. [DOI] [PubMed] [Google Scholar]
- 5.Miyazaki Y, et al. Selective cytotoxicity of dihydroorotate dehydrogenase inhibitors to human cancer cells under hypoxia and nutrient-deprived conditions. Front Pharmacol. 2018;9:997. doi: 10.3389/fphar.2018.00997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enkai S, et al. Medical treatment of echinococcus multilocularis and new horizons for drug discovery: Characterization of mitochondrial complex ii as a potential drug target. In: Inceboz T, editor. Echinococcosis. IntechOpen; London: 2017. pp. 49–70. [Google Scholar]
- 7.Minagawa N, et al. An antibiotic, ascofuranone, specifically inhibits respiration and in vitro growth of long slender bloodstream forms of Trypanosoma brucei brucei. Mol Biochem Parasitol. 1997;84:271–280. doi: 10.1016/s0166-6851(96)02797-1. [DOI] [PubMed] [Google Scholar]
- 8.Shiba T, et al. Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc Natl Acad Sci USA. 2013;110:4580–4585. doi: 10.1073/pnas.1218386110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nihei C, Fukai Y, Kita K. Trypanosome alternative oxidase as a target of chemotherapy. Biochim Biophys Acta. 2002;1587:234–239. doi: 10.1016/s0925-4439(02)00086-8. [DOI] [PubMed] [Google Scholar]
- 10.Yabu Y, et al. The efficacy of ascofuranone in a consecutive treatment on Trypanosoma brucei brucei in mice. Parasitol Int. 2003;52:155–164. doi: 10.1016/s1383-5769(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 11.Mori K, Takechi S. Synthesis of the natural enantiomers of ascochlorin, ascofuranone and ascofuranol. Tetrahedron. 1985;41:3049–3062. [Google Scholar]
- 12.Haga Y, et al. A short and efficient total synthesis of (±)-ascofuranone. Chem Lett. 2011;39:622–623. [Google Scholar]
- 13.Hijikawa Y, et al. Re-identification of the ascofuranone-producing fungus Ascochyta viciae as Acremonium sclerotigenum. J Antibiot (Tokyo) 2017;70:304–307. doi: 10.1038/ja.2016.132. [DOI] [PubMed] [Google Scholar]
- 14.Summerbell RC, et al. The Protean Acremonium. A. sclerotigenum/egyptiacum: Revision, food contaminant, and human disease. Microorganisms. 2018;6:88. doi: 10.3390/microorganisms6030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayakawa S, Minato H, Katagiri K. The ilicicolins, antibiotics from Cylindrocladium ilicicola. J Antibiot (Tokyo) 1971;24:653–654. doi: 10.7164/antibiotics.24.653. [DOI] [PubMed] [Google Scholar]
- 16.Minato H, Katayama T, Hayakawa S, Katagiri K. Identification of ilicicolins with ascochlorin and LL-Z 1272. J Antibiot (Tokyo) 1972;25:315–316. doi: 10.7164/antibiotics.25.315. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi M, et al. A new ascochlorin derivative from Cylindrocarpon sp. FKI-4602. J Antibiot (Tokyo) 2013;66:23–29. doi: 10.1038/ja.2012.75. [DOI] [PubMed] [Google Scholar]
- 18.Ellestad GA, Evans RH, Jr, Kunstmann MP. Some new terpenoid metabolites from an unidentified fusarium species. Tetrahedron. 1969;25:1323–1334. doi: 10.1016/s0040-4020(01)82703-4. [DOI] [PubMed] [Google Scholar]
- 19.Isaka M, et al. Ascochlorin derivatives from the leafhopper pathogenic fungus Microcera sp. BCC 17074. J Antibiot (Tokyo) 2015;68:47–51. doi: 10.1038/ja.2014.90. [DOI] [PubMed] [Google Scholar]
- 20.Aldridge DC, et al. Metabolites of Nectria coccinea. J Chem Soc Perkin 1. 1972;17:2136–2141. doi: 10.1039/p19720002136. [DOI] [PubMed] [Google Scholar]
- 21.Terekhova L, et al. Biosynthesis of ascofuranone by the fungus Paecilomyces variotii Bainier. Microbiology. 1997;66:510–514. [Google Scholar]
- 22.Seephonkai P, Isaka M, Kittakoop P, Boonudomlap U, Thebtaranonth Y. A novel ascochlorin glycoside from the insect pathogenic fungus Verticillium hemipterigenum BCC 2370. J Antibiot (Tokyo) 2004;57:10–16. doi: 10.7164/antibiotics.57.10. [DOI] [PubMed] [Google Scholar]
- 23.Hosono K, Ogihara J, Ohdake T, Masuda S. LL-Z1272alpha epoxide, a precursor of ascochlorin produced by a mutant of Ascochyta viciae. J Antibiot (Tokyo) 2009;62:571–574. doi: 10.1038/ja.2009.80. [DOI] [PubMed] [Google Scholar]
- 24.Li C, et al. Biosynthesis of LL-Z1272β: Discovery of a new member of NRPS-like enzymes for aryl-aldehyde formation. ChemBioChem. 2016;17:904–907. doi: 10.1002/cbic.201600087. [DOI] [PubMed] [Google Scholar]
- 25.Matsuda Y, Abe I. Biosynthesis of fungal meroterpenoids. Nat Prod Rep. 2016;33:26–53. doi: 10.1039/c5np00090d. [DOI] [PubMed] [Google Scholar]
- 26.Chavali AK, Rhee SY. Bioinformatics tools for the identification of gene clusters that biosynthesize specialized metabolites. Brief Bioinform. 2018;19:1022–1034. doi: 10.1093/bib/bbx020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism—from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 28.Rajniak J, Barco B, Clay NK, Sattely ES. A new cyanogenic metabolite in Arabidopsis required for inducible pathogen defence. Nature. 2015;525:376–379. doi: 10.1038/nature14907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau W, Sattely ES. Six enzymes from mayapple that complete the biosynthetic pathway to the etoposide aglycone. Science. 2015;349:1224–1228. doi: 10.1126/science.aac7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Lee TAJ, Medema MH. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet Biol. 2016;89:29–36. doi: 10.1016/j.fgb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 31.Mogi T, et al. Antibiotics LL-Z1272 identified as novel inhibitors discriminating bacterial and mitochondrial quinol oxidases. Biochim Biophys Acta. 2009;1787:129–133. doi: 10.1016/j.bbabio.2008.11.016. [DOI] [PubMed] [Google Scholar]
- 32.Berry EA, et al. Ascochlorin is a novel, specific inhibitor of the mitochondrial cytochrome bc1 complex. Biochim Biophys Acta. 2010;1797:360–370. doi: 10.1016/j.bbabio.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada M, et al. Combinatorial biosynthesis of (+)-daurichromenic acid and its halogenated analogue. Org Lett. 2017;19:3183–3186. doi: 10.1021/acs.orglett.7b01288. [DOI] [PubMed] [Google Scholar]
- 34.Munro AW, et al. P450 BM3: The very model of a modern flavocytochrome. Trends Biochem Sci. 2002;27:250–257. doi: 10.1016/s0968-0004(02)02086-8. [DOI] [PubMed] [Google Scholar]
- 35.Matsuda Y, Wakimoto T, Mori T, Awakawa T, Abe I. Complete biosynthetic pathway of anditomin: Nature’s sophisticated synthetic route to a complex fungal meroterpenoid. J Am Chem Soc. 2014;136:15326–15336. doi: 10.1021/ja508127q. [DOI] [PubMed] [Google Scholar]
- 36.Itoh T, et al. Reconstitution of a fungal meroterpenoid biosynthesis reveals the involvement of a novel family of terpene cyclases. Nat Chem. 2010;2:858–864. doi: 10.1038/nchem.764. [DOI] [PubMed] [Google Scholar]
- 37.Scott B, et al. Deletion and gene expression analyses define the paxilline biosynthetic gene cluster in Penicillium paxilli. Toxins (Basel) 2013;5:1422–1446. doi: 10.3390/toxins5081422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tada S, et al. Construction of a fusion gene comprising the Taka-amylase A promoter and the Escherichia coli β-glucuronidase gene and analysis of its expression in Aspergillus oryzae. Mol Gen Genet. 1991;229:301–306. doi: 10.1007/BF00272170. [DOI] [PubMed] [Google Scholar]
- 39.Araki Y, Masakari Y, Hara S, Yuzuki M. 2018. Transformation method of filamentous fungi. Japanese Unexamined Patent Application Publication JP2018-068292A. [DOI] [PubMed]
- 40.Tagami K, et al. Reconstitution of biosynthetic machinery for indole-diterpene paxilline in Aspergillus oryzae. J Am Chem Soc. 2013;135:1260–1263. doi: 10.1021/ja3116636. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki H, Hosokawa T, Nawata Y, Ando K. Isolation and structure of ascochlorin and its analogs. Agric Biol Chem. 1974;38:1463–1466. [Google Scholar]
- 42.Nawata Y, Ando K, Tamura G, Arima K, Iitaka Y. The molecular structure of ascochlorin. J Antibiot (Tokyo) 1969;22:511–512. doi: 10.7164/antibiotics.22.511. [DOI] [PubMed] [Google Scholar]
- 43.Krappmann S. Gene targeting in filamentous fungi: The benefits of impaired repair. Fungal Biol Rev. 2007;21:25–29. [Google Scholar]
- 44.Tokai T, et al. Fusarium Tri4 encodes a key multifunctional cytochrome P450 monooxygenase for four consecutive oxygenation steps in trichothecene biosynthesis. Biochem Biophys Res Commun. 2007;353:412–417. doi: 10.1016/j.bbrc.2006.12.033. [DOI] [PubMed] [Google Scholar]
- 45.Heneghan MN, et al. The programming role of trans-acting enoyl reductases during the biosynthesis of highly reduced fungal polyketides. Chem Sci (Camb) 2011;2:972–979. [Google Scholar]
- 46.Seghezzi W, Sanglard D, Fiechter A. Characterization of a second alkane-inducible cytochrome P450-encoding gene, CYP52A2, from Candida tropicalis. Gene. 1991;106:51–60. doi: 10.1016/0378-1119(91)90565-s. [DOI] [PubMed] [Google Scholar]
- 47.Sakuno E, Yabe K, Nakajima H. Involvement of two cytosolic enzymes and a novel intermediate, 5′-oxoaverantin, in the pathway from 5′-hydroxyaverantin to averufin in aflatoxin biosynthesis. Appl Environ Microbiol. 2003;69:6418–6426. doi: 10.1128/AEM.69.11.6418-6426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: Classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–379. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- 49.Lin TS, Chiang YM, Wang CC. Biosynthetic pathway of the reduced polyketide product citreoviridin in Aspergillus terreus var. aureus revealed by heterologous expression in Aspergillus nidulans. Org Lett. 2016;18:1366–1369. doi: 10.1021/acs.orglett.6b00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mao XM, et al. Efficient biosynthesis of fungal polyketides containing the dioxabicyclo-octane ring system. J Am Chem Soc. 2015;137:11904–11907. doi: 10.1021/jacs.5b07816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nicholson MJ, et al. Identification of two aflatrem biosynthesis gene loci in Aspergillus flavus and metabolic engineering of Penicillium paxilli to elucidate their function. Appl Environ Microbiol. 2009;75:7469–7481. doi: 10.1128/AEM.02146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lo HC, et al. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J Am Chem Soc. 2012;134:4709–4720. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Proctor RH, McCormick SP, Alexander NJ, Desjardins AE. Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol Microbiol. 2009;74:1128–1142. doi: 10.1111/j.1365-2958.2009.06927.x. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez JF, et al. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc. 2011;133:4010–4017. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umemura M, et al. MIDDAS-M: Motif-independent de novo detection of secondary metabolite gene clusters through the integration of genome sequencing and transcriptome data. PLoS One. 2013;8:e84028. doi: 10.1371/journal.pone.0084028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andersen MR, et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci USA. 2013;110:E99–E107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: The zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsuzaki M, Tatsumi R, Kita K. Protoplast generation from the ascofuranone-producing fungus Acremonium sclerotigenum. Cytologia (Tokyo) 2017;82:317–320. doi: 10.1038/ja.2016.132. [DOI] [PubMed] [Google Scholar]
- 59.Jin FJ, Maruyama J, Juvvadi PR, Arioka M, Kitamoto K. Development of a novel quadruple auxotrophic host transformation system by argB gene disruption using adeA gene and exploiting adenine auxotrophy in Aspergillus oryzae. FEMS Microbiol Lett. 2004;239:79–85. doi: 10.1016/j.femsle.2004.08.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.