Significance
Photosynthetic organisms utilize sunlight as a form of energy. Under low light, they maximize their capacity to harvest photons; however, under excess light, they dissipate part of the harvested energy to prevent photodamage, at the expense of light-use efficiency. Optimally balancing light harvesting and energy dissipation in natural (fluctuating light) conditions is considered a target for improving the productivity of both algae and plants. Here we have studied the energy dissipation process in the green alga Chlamydomonas reinhardtii in vivo. We found that it is remarkably different from that of higher plants, highlighting the need of developing tailor-made strategies to optimize the light harvesting–energy dissipation balance in different organisms.
Keywords: photosynthesis, low pH, photosystem II, fluorescence, ultrafast spectroscopy
Abstract
Sunlight drives photosynthesis but can also cause photodamage. To protect themselves, photosynthetic organisms dissipate the excess absorbed energy as heat, in a process known as nonphotochemical quenching (NPQ). In green algae, diatoms, and mosses, NPQ depends on the light-harvesting complex stress-related (LHCSR) proteins. Here we investigated NPQ in Chlamydomonas reinhardtii using an approach that maintains the cells in a stable quenched state. We show that in the presence of LHCSR3, all of the photosystem (PS) II complexes are quenched and the LHCs are the site of quenching, which occurs at a rate of ∼150 ps−1 and is not induced by LHCII aggregation. The effective light-harvesting capacity of PSII decreases upon NPQ, and the NPQ rate is independent of the redox state of the reaction center. Finally, we could measure the pH dependence of NPQ, showing that the luminal pH is always above 5.5 in vivo and highlighting the role of LHCSR3 as an ultrasensitive pH sensor.
Photosynthetic organisms have evolved large antenna systems comprising many pigment–protein complexes to maximize light harvesting of photosystem (PS) I and II reaction centers (RCs) (1). However, under high light intensity, this natural design results in a number of absorbed photons that exceeds the capacity of the photochemical reactions and can damage the photosynthetic apparatus (2). To avoid photodamage, nearly all photosynthetic species have developed a variety of photoprotective strategies, collectively termed nonphotochemical quenching (NPQ). In NPQ, a rapid and reversible excited energy quenching is the dominant process that dissipates the excess-absorbed energy as heat, before the excitation energy from the antennae reaches the RCs (3). The NPQ processes ensure healthy growth of photosynthetic organisms but also reduce their overall photosynthetic efficiency. Recently, manipulations of NPQ were successfully shown to improve biomass production (4, 5). Therefore, a molecular understanding of NPQ mechanisms is of paramount importance also for improving photosynthesis (6).
In the green alga Chlamydomonas reinhardtii, NPQ depends on the light-harvesting complex stress-related (LHCSR) proteins (7, 8) LHCSR1 and LHCSR3. Both are capable of triggering efficient energy quenching (7, 9, 10). In contrast to PSBS, the protein involved in NPQ in land plants (11), the LHCSRs are pigment-binding proteins (8). A unique property of the LHCSR proteins in vitro, compared with other pigment-binding antennae, is their pH sensitivity (8, 12, 13): At pH 4.4 to 5.5, the fluorescence of LHCSR3 is severely quenched compared with pH 7.5. This pH-induced quenching has been proposed to be the result of a conformational change of the protein triggered by the protonation of residues of the C-terminal domain (12). Since in vivo the C terminus is exposed to the thylakoid lumen, it is conceivable that the pH sensitivity of the LHCSRs enables them to switch to the quenching state in response to the lumen acidification. Clearly, the luminal pH and the dependence of NPQ on it in vivo are critical factors in this photoregulatory process. However, thus far, they have not been determined.
Over the past decades, the molecular mechanisms of NPQ have been extensively studied in land plants (see ref. 3 and references therein), whereas in green algae they remain elusive. In plants, the fluorescence quenching in LHCII aggregates in vitro—that has a signature similar to that of the quenching in vivo (14)—has been used as a basis for the development of the so-called LHCII aggregation model (15). This model proposes that LHCII undergoes a conformational change under low pH, thus forming a dissipative state upon aggregation. This proposal was further supported by in vivo studies with either time-resolved fluorescence (16) or freeze-fracture electron microscopy (17). Although there is no consensus on the underlying physical mechanism of NPQ in land plants (for a recent review, see ref. 18), the involvement of LHCII aggregation and membrane reorganization is generally accepted. A central question appearing in NPQ studies on green algae is whether the LHCII aggregation model is also responsible for the LHCSR-dependent NPQ process. To answer this question as well as to gain insight into the ultrafast kinetics of NPQ, we performed time-resolved fluorescence measurements with both high temporal and spectral resolutions. One of the main limitations is the difficulty in maintaining the cells in a stable quenched state during the measurements, since NPQ recovers rapidly after the light is switched off [by a significant fraction in a few seconds and almost entirely in a few minutes (7)]. In this work, we have used an approach in which acetic acid was used to maintain the cells in a stable quenched state to perform the ultrafast fluorescence measurements.
This approach also allowed us to disclose the pH dependence of NPQ and to quantify the change of the light-limited charge separation rate in PSII—sometimes labeled the functional antenna size—upon quenching in vivo.
Results
Acetic Acid-Induced NPQ and Its pH Dependence.
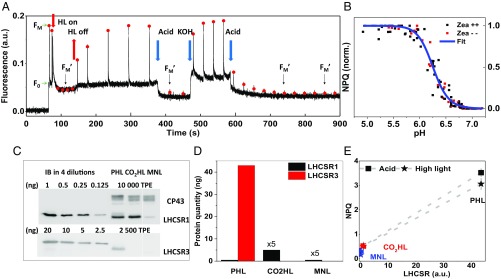
The most frequently used medium to grow C. reinhardtii is TAP (Tris-acetate-phosphate) (19), which contains 17.4 mM acetic acid that serves as a source of carbon and energy. However, acetic acid was also proven to induce quenching when added to the cell solution (9, 10, 20). This approach was employed in this work to lock the cells in their quenched state. A state transition-deficient mutant, stt7-9 (21), was used, which permits us to separate the effect of NPQ from that of state transitions. Indeed, upon addition of acetic acid, the chlorophyll (Chl) fluorescence of C. reinhardtii decreases in a few seconds and fully recovers by titrating the pH back to 7.0 (Fig. 1A). The high sensitivity of Chl fluorescence to environmental pH allows us to explore the pH dependence of the NPQ process; note that the ionophore nigericin was added during the pH titration to ensure complete pH equilibrium within the cells (22). The pH titration curve (Fig. 1B) shows that the fluorescence quenching starts already at pH values around 6.7 and saturates at pH 5.5, well above the suggested cytotoxic pH <5 (23). In higher plants, zeaxanthin (Zea) has been reported to significantly affect the pH dependence of LHCII fluorescence quenching (24). This is not the case in C. reinhardtii, as the level of NPQ in high light (HL)-grown cells, which contain Zea [Zea/violaxanthin (Vio) ratio of 0.41 ± 0.07; Z++], and in dark-adapted cells, in which the amount of Zea is reduced by 75% (Zea/Vio ratio of 0.10 ± 0.01; Z−), is similar and shows an identical pH dependence (Fig. 1B).
Fig. 1.
(A) A typical PAM Chl fluorometer trace of steady-state fluorescence in stt7-9. NPQ can be induced with high light (80 to 130 s), as well as by acetic acid (4.5 mM) (370 to 470 s). The quenching is fully reversible after KOH addition setting the pH back to 7 (470 to 500 s). FM and FM′ are indicated by red dots. Cells were grown photoautotrophically in high light. (B) pH titration curve of NPQ in cells obtained by sequentially adding ∼2 µL acetic acid (1 M stock) with nigericin (100 µM) to the same cell culture. NPQ levels were measured within ∼2 min after adding acetic acid; six and three biological replicas were measured for cells with and without zeaxanthin, respectively. For each replica, the NPQ values were normalized to 0 (NPQ_min) and 1 (NPQ_max). The fitted curve is a logistic function (y = 1/[1 + (x/x0)^p]), yielding x0 (the midpoint pH) of 6.25 ± 0.09. (C) Determination of the expression level of LHCSR1 and LHCSR3 in cells grown in high light photoautotrophically (PHL), photoautotrophically plus 5% CO2 (CO2HL), and in normal light mixotrophically (MNL) by immunoblots on the total protein extracts. For stoichiometry assessment, different amounts of inclusion bodies (IBs) of LHCSR1 and LHCSR3 were loaded on the gel. CP43 was used as a control. (D) The relative content of LHCSR1 and LHCSR3 proteins in 10 µg of TPE is shown. (E) Correlations between the total amount of LHCSR proteins (LHCSR1+LHCSR3) and the NPQ levels (connected by dashed lines) induced in the cells by high light and acetic acid. SDs of the NPQ values are included (n = 3).
In higher plants, it was shown that without PSBS, the LHC antennae can still be quenched by low luminal pH (25), and thus it is essential to verify whether the acetic acid-induced quenching is LHCSR-dependent. To do so, we compared the NPQ levels induced by either high light or acetic acid in cells with different amounts of LHCSRs. In wild-type C. reinhardtii, the expression level of LHCSR1 and LHCSR3 changes dramatically under different growth conditions, leading to different NPQ levels (26–28). The immunoblot results in Fig. 1 C and D show that this is also the case in the stt7-9 mutant. We found that the NPQ levels caused by acetic acid positively correlate with the amount of LHCSRs (Fig. 1E), suggesting that it is indeed LHCSR-dependent. Note that PsbS is hardly expressed in the cells (SI Appendix, Fig. S1A) and does not trigger a significant amount of NPQ either, as tested by using a double mutant of npq4/stt7-9 (SI Appendix, Fig. S5 and more discussion in SI Appendix).
A question that follows is whether the acid-induced and the high light-induced fluorescence quenching mechanisms are the same. Fig. 1E also shows that the NPQ levels induced by HL and acetic acid are very similar, showing the same dependence on LHCSRs. The similarity is also observed in the low-temperature time- and spectrally resolved fluorescence kinetics (SI Appendix, Figs. S6 and S7 and Table S1). This indicates that the quenching induced by HL and by acetic acid proceeds via the same mechanism. Therefore, acetic acid can be used to study NPQ at a physiologically relevant temperature.
Ultrafast Fluorescence Quenching at Physiological Temperature.
To investigate the mechanism of quenching, the time- and wavelength-resolved fluorescence of the cells was measured before and after adding acetic acid. The use of acid is key for this type of measurement, as it offers several advantages: (i) no distortion of the kinetics from any intermediate NPQ state; (ii) stable luminal pH; and (iii) absence of PSII heterogeneity, as all of the measurements can be done with closed PSII RCs. Indeed, independent of the state [unquenched (UQ) or quenched (Q)], all of the PSII RCs are closed by preilluminating the cells in the presence of DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea] and HA (hydroxylamine) (29). It should be emphasized that this is possible for cells in the quenched state, only thanks to the use of acid. In the case of HL-induced quenching, DCMU and HA would block the linear electron flow, inhibiting the lumen acidification and consequently NPQ.
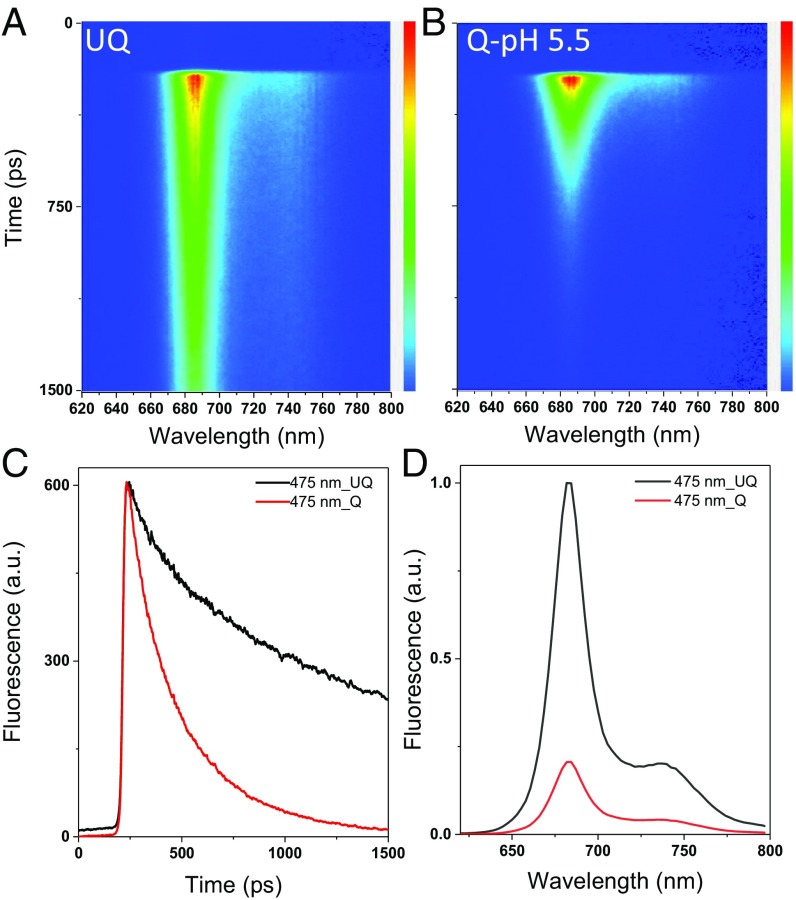
A dramatic shortening of the fluorescence decay at pH 5.5 compared with 7.0 is directly visible from the streak-camera images (Fig. 2 A and B and SI Appendix, Fig. S8), as well as from the fluorescence decay curves at 683 nm (Fig. 2C). Reconstructed steady-state spectra (Fig. 2D) show that the fluorescence quantum yield drops by nearly 80% upon quenching. It corresponds to an NPQ value of 4.0, slightly higher than the value of 3.5 obtained with pulse-amplitude modulation (PAM), which can be explained by the presence of the PSI contribution in the PAM signal (30).
Fig. 2.
Time-resolved fluorescence data at room temperature at 475-nm excitation. (A–C) Streak-camera images of intact cells in the unquenched state, UQ (A), and quenched state, Q (B); their fluorescence decay curves at 683 nm are shown (C). (D) Reconstructed steady-state spectra of UQ and Q cells.
Target Analysis of Time-Resolved Fluorescence Data.
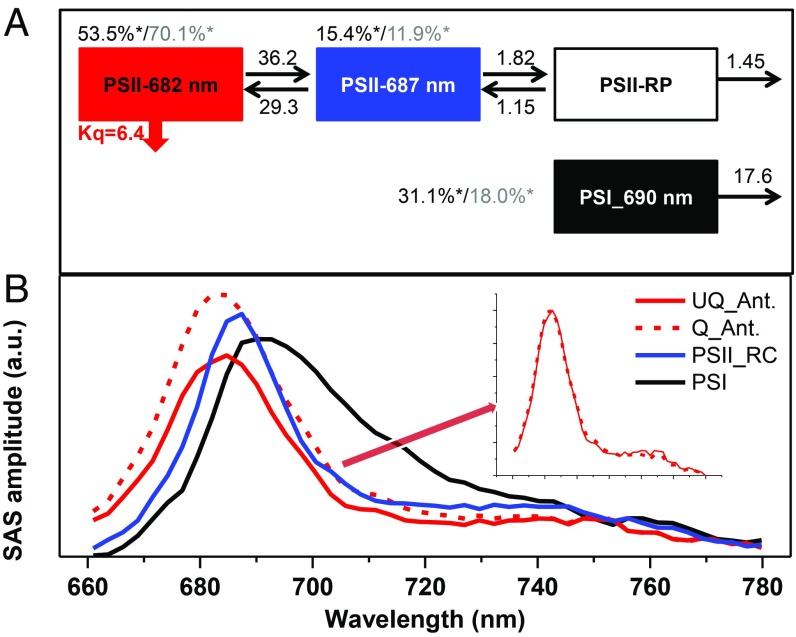
The data of UQ and Q cells upon 400- and 475-nm excitations were fitted simultaneously with a four-compartment model (see fitting qualities in SI Appendix, Fig. S9), in which all of the spectra, energy inputs at time 0, and rate constants are free parameters. In the model, one compartment represents PSI while three were used for PSII (Fig. 3A). The PSI compartment (black in Fig. 3A) has the typical PSI spectrum with a maximum at 690 nm [and emission above 700 nm due to the far-red Chls (31)] and the typical PSI lifetime of ∼56 ps (32). It absorbs ∼31% of the energy upon 400-nm excitation and ∼18% upon 475-nm excitation. As for PSII, the compartment in red (in Fig. 3A), characterized by a blue-shifted spectrum peaking at 682 nm, is assigned to the PSII antenna (PSII-Ant.) and carries most of the initial excitations: 53.5% at 400-nm excitation and 70.1% at 475-nm excitation. A second compartment (blue in Fig. 3A) (PSII-core) has a maximum at 687 nm. This compartment receives 15.4 and 11.9% of the initial excitation at 400 and 475 nm, respectively. Note that the spectra of PSII-Ant and PSII-core were successfully separated at room temperature. These two compartments reach an energetic equilibrium within 15 ps after excitation (see calculated lifetimes and their decay-associated spectra in SI Appendix, Fig. S10), and then the PSII-core transfers an electron to a nonemitting radical pair (RP; white in Fig. 3A), from where the electron is further transferred down or recombines. For the description of the quenched sample, initially we had tried to associate quenching with each compartment except the RP, but we found that the quenching on PSII-Ant. dominates (SI Appendix, Fig. S11), meaning that it is taking place on the antenna. Indeed, the fitting quality of the model in which only PSII-Ant. is quenched remains the same. There is no need to include heterogeneity of PSII in the model to reach a satisfactory fit, meaning that all individual PSII supercomplexes are protected by NPQ. The quenching rate is estimated to be ∼150 ps−1, and the shape of the spectrum of PSII-Ant. remains unchanged (Fig. 3B, Inset) during the NPQ process. In particular, no new species with different spectra are required to fit the fluorescence decay curves of the quenched cells. This conclusion is further supported by the low-temperature time-resolved fluorescence data (see SI Appendix, Figs. S12–S14 and the fitted curves in SI Appendix, Figs. S15–S18), where the differences in spectral shape, if present, are often enhanced.
Fig. 3.
Target analysis results. (A) Schematic model used to describe the time-resolved fluorescence data; rates are given in ns−1, Kq is the rate of quenching, and asterisks indicate the initial excitations at 400 nm (black) and 475 nm (gray). (B) Species-associated spectra (SASs) of the compartments with the same colors as used in A. (B, Inset) Normalized spectra of the UQ_Ant. and Q_Ant. are shown.
How Does the NPQ Process Affect PSII Functional Antenna Size in Vivo?
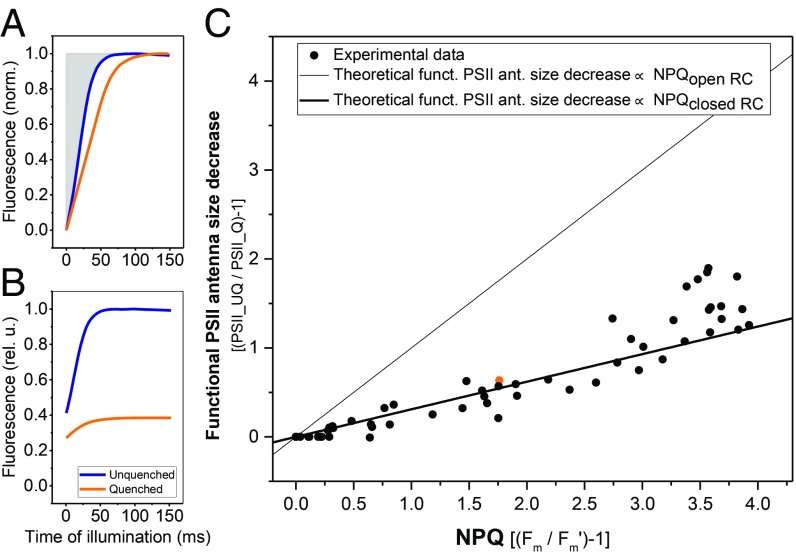
The approach of acid-induced quenching is well-suited for estimating the functional effect of NPQ on the PSII electron transfer rate, as the acid keeps the cells in a constant quenched state without the necessity of sample illumination and thus light-induced RC closure. This permitted us to measure the functional antenna size of PSII upon NPQ in vivo by fluorescence induction in the presence of DCMU (33) (Methods and SI Appendix, Text 2): The comparison of the fluorescence value at the plateau in the presence and absence of acid represents the NPQ value (FM − FM′)/FM′; Fig. 4B), while the area above the normalized induction curve (Fig. 4A) represents the functional antenna size (see SI Appendix, Text 2 for more details). Since the fluorescence induction curve in the presence of DCMU provides the PSII rate before the RC closes (FM), it gives access to information about the competition between NPQ and photochemistry. Our data show a linear decrease in functional antenna size concomitant with increasing NPQ (Fig. 4C), yet to a significantly lower extent than suggested by the NPQ value, demonstrating that NPQ competes for the excitation with the photochemical traps.
Fig. 4.
Functional antenna size of PSII decreases upon NPQ. (A) Typical fluorescence induction traces of cells in the unquenched (blue) and quenched state (orange; NPQ ∼ 1.8). Both curves are normalized to 1 at FM and to 0 at F0. (B) Same curves as shown in A but without normalization. (C) The relative increase in the area above the fluorescence induction curve in the presence of DCMU at increasing concentrations of acetic acid is plotted as a function of NPQ (see Methods for area calculation details). Solid lines depict theoretical scenarios where the functional antenna size decreases proportionally to the NPQ (thin line; NPQclosed RC = kNPQ/kclosed) and proportionally to the photochemical quenching + NPQ (thick line; NPQopen RC = kNPQ/kopen = NPQclosed RC × kclosed/kopen = NPQclosed RC × F0/FM, assuming kNPQ is the same for open and closed RCs); k represents the overall chlorophyll deexcitation rate of PSII supercomplexes. Each experiment is shown as a separate circle, and the specific data point that corresponds to A and B is highlighted in orange. The measurements were performed on four batches of high light-grown algae.
Finally, we investigated the slope of the dependence of the functional antenna size vs. NPQ to verify whether the rate of NPQ, kNPQ, remains similar with open (QA) and closed (QA−) RCs. Within the precision of our measurements, the data closely follow the theoretical scenario where kNPQ is identical with open and closed RCs (Fig. 4C), resulting in a different value of NPQ in the two situations. This indicates that in Chlamydomonas, NPQ decreases the functional antenna size of PSII, and that its rate does not depend on the redox state of the PSII RC.
Discussion
The Dynamic Range of NPQ in Chlamydomonas Is 1.2 pH Units.
Our NPQ titration curve shows that the quenching is activated at pH 6.7 and saturates at pH 5.5, meaning that it only covers ∼1.2 pH units, whereas for higher plants it was reported to attain ∼4 pH units (25). This difference is most likely related to the different properties of the pH sensors in plants and green algae, PsbS and LHCSR, respectively. The midpoint of the pH titration curve is at about 6.25, which indicates that the pKa of the pH-sensing residues must be close to neutral. For LHCSR3, it has been shown that the protonation of Asp and Glu residues at its C terminus triggers quenching (12, 34). Since these residues have a side chain with a pKa as low as ∼4.0 in water, the present results suggest that the environment strongly modulates their pKa. In higher plants, it is zeaxanthin that modulates the pH sensitivity of NPQ (24, 25). This is not the case in C. reinhardtii, since we do not observe any influence of zeaxanthin on the NPQ response to pH. The lack of a zeaxanthin effect on NPQ agrees with the observation that a Zea-lacking mutant of C. reinhardtii has a similar NPQ level as wild-type C. reinhardtii (8).
NPQ as pH Probe in Vivo: The pH of the Lumen Does Not Go Below 5.5.
The value of the luminal pH is a matter of debate, as its determination is technically challenging (23). Several approaches have been developed that either use pH-sensitive dyes (25) or pH-sensitive GFPs (35). However, these approaches require labeling, which is often complicated for in vivo systems. Here, we show that the LHCSR-dependent NPQ can be used as a natural sensor to determine the pH of the lumen. Using the pH calibration curve and the value of NPQ obtained on the same cells after high light exposure, we can conclude that the pH of the lumen does not go below 5.5, even in very high light conditions (1,000 µmol photons·m−2·s−1). Indeed, the NPQ level observed upon HL exposure is always a bit lower than the value obtained with acetic acid at pH 5.5. This result is in agreement with a previous proposal that the luminal pH does not go below 5, as lower values would damage the Mn cluster in PSII (36).
Quenching Site, Rate, and Mechanism.
Electron microscopy on isolated PSII–LHCII–LHCSR3 supercomplexes has shown that LHCSR3 binds to the LHCs at different positions (37). In agreement with this, fluorescence upconversion experiments on isolated supercomplexes have suggested that under low pH, LHCSR3 quenches LHCs and consequently the excitation energy within the PSII supercomplex (38). Our data indicate that this is also happening in vivo. Based on our target model (Fig. 3), LHCSR directly binds and quenches the PSII-Ant. and concomitantly the PSII-core but not PSI. The quenching rate at physiological temperature is estimated to be ∼150 ps−1, which is surprisingly fast considering that there are almost 200 Chls in a supercomplex (∼4.6 LHCII trimers per RC according to our estimations; see SI Appendix for details). With this rate, the quenching is even very competitive when the PSII reaction center is running at full capacity, in agreement with our observation that F0 was severely quenched (Fig. 1A) and that the light-limited charge separation rate of PSII decreases upon quenching (Fig. 4). A much faster quenching rate of 18 ps−1 has been reported by Kim et al. (38) for isolated supercomplexes. However, since there is no energy equilibrium between the two compartments implemented in their model, this quenching rate is dramatically overestimated. Moreover, in their measurements, the open/closed state of PSII, which severely influences the kinetics, is not defined. In contrast, PSIIs in our measurements are fully closed in both UQ and Q states. This maximizes the time the excitation spends on the antenna, so that we could precisely fit both the energy equilibrium within PSII and the quenching rate.
Importantly, because of the high spectral resolution of our data, we could show that there is not a spectral signature associated with NPQ at both room temperature (Fig. 3) and 77 K (SI Appendix, Figs. S12–S14). This conclusion is further supported by the indistinguishability of the spectral shape of UQ and Q in the original data after 100 ps, when the PSI contribution is minimized (see also the time-gated spectra in SI Appendix, Fig. S19). This is an important observation since LHCII aggregates, which have a specific spectral feature even at room temperature (39, 40), serve as a well-received model for NPQ in higher plants (14). In our data, we did not observe the appearance at any time point during NPQ of a red shoulder that would suggest induction of LHCII aggregation, even though the cells have a substantial NPQ (a value of ∼3.5). This observation is consistent with our previous results on LHCSR1-induced quenching (10), suggesting that the mechanism of quenching in C. reinhardtii is different from what has been proposed for higher plants (14, 16, 41, 42).
Finally, our data allow us to separate PSI and PSII in the model because of their significant differences in both kinetics and spectra (32). We found that both the lifetime and amplitude of PSI are not affected by acid, indicating that the proposed mechanism of acid-induced quenching in vascular plants does not occur in C. reinhardtii (43).
The Effects of NPQ on PSII Light Harvesting.
It was recently concluded that upon NPQ in plant thylakoids, the functional antenna size of PSII increases (44). Here we demonstrate that this is not the case in C. reinhardtii cells, for which we observed that NPQ induces a decrease of the PSII functional antenna size (Fig. 4). Moreover, we show that this decrease follows the expected value in the scenario where NPQ competes with photochemical quenching without any change in the physical antenna size. Interestingly, our results also indicate that the rate of quenching in C. reinhardtii is identical at F0 and at FM, although small changes of kNPQ on the order of ∼10% could not be excluded. This is again different from what was recently observed in higher plants, where the quenching rate was found to be lower in the case of open RCs (45). The lack of the QA redox state-dependent NPQ regulation in C. reinhardtii results in a larger loss of energy in the presence of the open reaction center than in plants, meaning that NPQ lowers the quantum yield of photochemistry.
Conclusions: Plants vs. Green Algae.
Our results show that substantial differences in the mechanism of NPQ exist between C. reinhardtii and vascular plants. It is likely that the mode of quenching, LHCSR- vs. PSBS-dependent, is the reason for these differences. Indeed, while LHCSR is a pigment-binding protein (8), this does not seem to be the case for PSBS (46), implying that the former can in principle directly act as a quencher while the latter cannot. This also means that while in C. reinhardtii the quenchers only need to be activated by the low luminal pH, in plants they have to be created during the NPQ induction. This might explain why LHCII aggregation, which was shown to generate quenchers (47), is present in plants but does not occur in C. reinhardtii. In this respect, it should be noticed that in higher plants the promotion of LHCII aggregation/clustering (48) is expected to be Zea-dependent. The absence of Zea effect on C. reinhardtii NPQ is thus in line with the fact that a reorganization of the antenna proteins within the membrane is not required for NPQ induction in Chlamydomonas.
The absence in C. reinhardtii of the NPQ regulation by the RC redox state might also be determined by the properties of PSBS and LHCSR and especially by their docking site, which is suggested to be the core complex for PSBS (49, 50) and the outer antenna for LHCSR (37). Binding to the core is a prerequisite for sensing the redox state of the RC, and thus regulating the quenching in accordance with it, as observed in plants (45). It is therefore tempting to speculate that the evolution of NPQ from the LHCSR-dependent process in green algae and mosses to the PSBS-dependent process in higher plants is functional in maintaining a higher quantum yield of charge separation in the presence of open reaction centers under NPQ conditions.
Our results demonstrate that even though the pH dependence is evolutionarily conserved from green algae to higher plants irrespective of the different sensor proteins involved, the quenching process is far from identical. In green algae, LHCII aggregation is absent and the additional mechanism, which regulates the quenching rate depending on the redox state of the reaction center, is lacking. Taken together, our results suggest that the LHCSR-dependent quenching is an on-site and solely pH-driven protective mechanism which does not require the aggregation of the LHC proteins as well as any additional feedback loop.
Methods
Cell Growth Conditions.
To eliminate the effects of antenna redistribution and focus exclusively on NPQ, we have used the C. reinhardtii stt7-9 mutant, which is impaired in state transitions (21). Cells were grown under three different light/carbon supply conditions: high-light photoautotrophic growth in high-salt medium (HSM) (51) (∼500 μmol photons·m−2·s−1) or in the presence of 5% CO2, and normal-light mixotrophic growth in TAP (∼50 μmol photons·m−2·s−1) as previously described (26). The light source was made of fluorescence tubes (Philips; MASTER PL-L, 55W/840), the spectrum of which is shown in SI Appendix, Fig. S2.
Pigment Analysis.
Pigments were extracted from the cells with 80% acetone. The relative amount of carotenoids was determined by HPLC as described (48).
Fluorescence Measurements.
High light-adapted cells naturally express zeaxanthin; they were used throughout this work for all fluorescence measurements unless stated otherwise. To check the role of zeaxanthin in NPQ, HL-grown cells were kept in the dark in the growing medium for ∼3 h. Cells with/without zeaxanthin were pelleted down and resuspended in HSM before further experiments. Saturating pulse (2,000 μmol photons·m−2·s−1, 250 ms) and actinic light (1,000 μmol photons·m−2·s−1), both peaking at 630 nm, and a blue measuring beam peaking at 430 nm were used for all of the measurements, and were provided by the Dual-PAM-100 (Walz) fluorimeter.
Acetic Acid-Induced Chlorophyll Fluorescence Quenching.
Cells were washed with fresh HSM (pH 6.9). For Fig. 1A, 4.5 mM acetic acid (1 M stock) was added to decrease the pH to 5.5, and KOH was used to adjust the pH back to 7.0. For the pH titrations in Fig. 1, different amounts of acetic acid were used together with 100 µM nigericin. pH values were recorded directly in the PAM cuvette by a microelectrode (pH-500; Unisense), while NPQ values were measured by the Dual-PAM.
Immunoblots and Total Protein Extract Preparation.
Total protein extract (TPE) preparation was performed as described (26). Immunoblot analyses were performed as described (10); 10 and 2.5 µg of TPEs were loaded for the determination of LHCSR1 and LHCSR3, respectively. CP43 was used as loading control, and the CP43 antibody was used together with the other antibodies. To evaluate the amount of LHCSRs per sample, we used LHCSR3 and LHCSR1 apoprotein overexpressed in Escherichia coli as in refs. 10, 12, and 52. Both LHCSR1- and LHCSR3-overexpressed apoproteins have a His tag at the C terminus, which explains their different mobility in SDS/PAGE compared with the WT proteins. All antibodies used were from Agrisera, but the anti-PsbS was a gift of Stefano Caffarri, Université Aix-Marseille, St. Paul Les Durance, France (53).
Isolation of PSI Complexes.
Photosystem I was purified as in ref. 54, and the isolated complex was used to estimate the PSI spectral shapes in a simultaneous target analysis of the 77 K data.
Time-Resolved Fluorescence.
Measurements were performed with a subpicosecond streak-camera setup (details in ref. 55) which combines a femtosecond laser source (Coherent Vitesse Duo + regenerative amplifier Coherent RegA 9000 + optical parametric amplifier Coherent OPA 9400) with a streak-camera detecting system (Hamamatsu; C5680). Fluorescence emission was collected at a right angle to the incident beam by a spectrograph (Chromex; 250IS; 50 grooves per mm ruling, blaze wavelength 600 nm, spectral width 260 nm), with the central wavelength set at 720 nm. Scattered excitation light was removed with an optical long-pass filter.
Two excitation wavelengths were used, 400 and 475 nm; the former more or less equally excites Chl a and b, and the latter selectively excites Chl b so that more antennae are initially excited. The laser beam was focused to a small spot of ∼50-μm diameter, and laser power was set to 15 µW with a repetition rate of 250 kHz. Cells were pelleted down, resuspended in fresh HSM, and directly measured in a 10-by-10 mm quartz cuvette at room temperature. Magnetic stirring (∼25 Hz) was used to prevent cell sedimentation. Care was taken to minimize the optical path length (<1 mm) to allow measurements on highly concentrated samples (OD750 nm 3) without significant self-absorption. Note that, to keep the cells in a fully quenched state, 4.5 mM acetic acid was added and, to ensure the PSII RCs were in a homogeneous closed state, 20 μM DCMU and 1 mM HA were added for both UQ and Q states. For each fluorescence measurement, a high signal/noise ratio was achieved by averaging a sequence of 200 single images with a CCD exposure time of 10 s for each. All images were background- and shading-corrected before global and target analysis. The laser did not cause damage or actinic effects, as no change in fluorescence decay curves was observed after continuous laser illumination for 30 min; see typical examples in SI Appendix, Fig. S20.
Simultaneous Target Analysis.
Data obtained with the streak-camera setup were globally analyzed with the R package TIMP-based Glotaran (56). The data were fitted with a kinetic model (so-called target analysis). For details of the methodology of target analysis, see ref. 57. With this approach, the spectrum of each resolved species and the energy transfer and quenching rates were estimated.
Determination of the Effect of Quenching on the Functional Antenna Size of PSII in Vivo.
The measurements of the light-limited rate of PSII electron transfer (indicated below as the functional antenna size) were conducted measuring the fluorescence rise in the presence of DCMU. The JTS-10 spectrophotometer (Bio-Logic) was used for all of the measurements in “fluorescence” mode, as described in ref. 58. A detailed description of the experiments and data analysis is provided in SI Appendix, Figs. S21 and S22.
Supplementary Material
Acknowledgments
We thank Jean-David Rochaix (University of Geneva) for providing the stt7-9 mutant, Giovanni Finazzi (CEA Grenoble) for the npq4/stt7-9 double mutant, Stefano Caffarri (Université Aix-Marseille) for the PsbS antibodies, Bart van Oort (VU Amsterdam) for critically reading the manuscript, Lauren Nicol (VU Amsterdam) for the isolated thylakoid membrane of Arabidopsis, and Tom van den Berg (VU Amsterdam) for assistance in the HPLC experiment. W.J.N. was supported by a European Commission Marie Curie Actions Individual Fellowship (799083). This work was supported by the European Research Council via an ERC consolidator grant (214113 to R.C.) and by the Dutch Organization for Scientific Research (NWO) via a Vici grant (to R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817796116/-/DCSupplemental.
References
- 1.Croce R, van Amerongen H. Natural strategies for photosynthetic light harvesting. Nat Chem Biol. 2014;10:492–501. doi: 10.1038/nchembio.1555. [DOI] [PubMed] [Google Scholar]
- 2.Rochaix JD. Regulation and dynamics of the light-harvesting system. Annu Rev Plant Biol. 2014;65:287–309. doi: 10.1146/annurev-arplant-050213-040226. [DOI] [PubMed] [Google Scholar]
- 3.Ruban AV. Nonphotochemical chlorophyll fluorescence quenching: Mechanism and effectiveness in protecting plants from photodamage. Plant Physiol. 2016;170:1903–1916. doi: 10.1104/pp.15.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berteotti S, Ballottari M, Bassi R. Increased biomass productivity in green algae by tuning non-photochemical quenching. Sci Rep. 2016;6:21339. doi: 10.1038/srep21339. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Kromdijk J, et al. Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science. 2016;354:857–861. doi: 10.1126/science.aai8878. [DOI] [PubMed] [Google Scholar]
- 6.Foyer CH, Ruban AV, Nixon PJ. Photosynthesis solutions to enhance productivity. Philos Trans R Soc Lond B Biol Sci. 2017;372:20160374. doi: 10.1098/rstb.2016.0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 8.Bonente G, et al. Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 2011;9:e1000577. doi: 10.1371/journal.pbio.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosuge K, et al. LHCSR1-dependent fluorescence quenching is mediated by excitation energy transfer from LHCII to photosystem I in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2018;115:3722–3727. doi: 10.1073/pnas.1720574115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinc E, et al. LHCSR1 induces a fast and reversible pH-dependent fluorescence quenching in LHCII in Chlamydomonas reinhardtii cells. Proc Natl Acad Sci USA. 2016;113:7673–7678. doi: 10.1073/pnas.1605380113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominici P, et al. Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J Biol Chem. 2002;277:22750–22758. doi: 10.1074/jbc.M200604200. [DOI] [PubMed] [Google Scholar]
- 12.Liguori N, Roy LM, Opacic M, Durand G, Croce R. Regulation of light harvesting in the green alga Chlamydomonas reinhardtii: The C-terminus of LHCSR is the knob of a dimmer switch. J Am Chem Soc. 2013;135:18339–18342. doi: 10.1021/ja4107463. [DOI] [PubMed] [Google Scholar]
- 13.Kondo T, et al. Single-molecule spectroscopy of LHCSR1 protein dynamics identifies two distinct states responsible for multi-timescale photosynthetic photoprotection. Nat Chem. 2017;9:772–778. doi: 10.1038/nchem.2818. [DOI] [PubMed] [Google Scholar]
- 14.Ruban AV, et al. Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature. 2007;450:575–578. doi: 10.1038/nature06262. [DOI] [PubMed] [Google Scholar]
- 15.Horton P, Wentworth M, Ruban A. Control of the light harvesting function of chloroplast membranes: The LHCII-aggregation model for non-photochemical quenching. FEBS Lett. 2005;579:4201–4206. doi: 10.1016/j.febslet.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Holzwarth AR, Miloslavina Y, Nilkens M, Jahns P. Identification of two quenching sites active in the regulation of photosynthetic light-harvesting studied by time-resolved fluorescence. Chem Phys Lett. 2009;483:262–267. [Google Scholar]
- 17.Johnson MP, et al. Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell. 2011;23:1468–1479. doi: 10.1105/tpc.110.081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duffy CD, Ruban AV. Dissipative pathways in the photosystem-II antenna in plants. J Photochem Photobiol B. 2015;152:215–226. doi: 10.1016/j.jphotobiol.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 19.Gorman DS, Levine RP. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Endo T, Asada K. Dark induction of the non-photochemical quenching of chlorophyll fluorescence by acetate in Chlamydomonas reinhardtii. Plant Cell Physiol. 1996;37:551–555. [Google Scholar]
- 21.Lemeille S, et al. Analysis of the chloroplast protein kinase Stt7 during state transitions. PLoS Biol. 2009;7:e45. doi: 10.1371/journal.pbio.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finazzi G, et al. Nonphotochemical quenching of chlorophyll fluorescence in Chlamydomonas reinhardtii. Biochemistry. 2006;45:1490–1498. doi: 10.1021/bi0521588. [DOI] [PubMed] [Google Scholar]
- 23.Kramer DM, Sacksteder CA, Cruz JA. How acidic is the lumen? Photosynth Res. 1999;60:151–163. [Google Scholar]
- 24.Rees D, et al. pH dependent chlorophyll fluorescence quenching in spinach thylakoids from light treated or dark adapted leaves. Photosynth Res. 1992;31:11–19. doi: 10.1007/BF00049532. [DOI] [PubMed] [Google Scholar]
- 25.Johnson MP, Ruban AV. Restoration of rapidly reversible photoprotective energy dissipation in the absence of PsbS protein by enhanced deltapH. J Biol Chem. 2011;286:19973–19981. doi: 10.1074/jbc.M111.237255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polukhina I, Fristedt R, Dinc E, Cardol P, Croce R. Carbon supply and photoacclimation cross talk in the green alga Chlamydomonas reinhardtii. Plant Physiol. 2016;172:1494–1505. doi: 10.1104/pp.16.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niyogi KK, Truong TB. Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr Opin Plant Biol. 2013;16:307–314. doi: 10.1016/j.pbi.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Bonente G, Pippa S, Castellano S, Bassi R, Ballottari M. Acclimation of Chlamydomonas reinhardtii to different growth irradiances. J Biol Chem. 2012;287:5833–5847. doi: 10.1074/jbc.M111.304279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardol P, et al. Impaired respiration discloses the physiological significance of state transitions in Chlamydomonas. Proc Natl Acad Sci USA. 2009;106:15979–15984. doi: 10.1073/pnas.0908111106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalaji HM, et al. Frequently asked questions about chlorophyll fluorescence, the sequel. Photosynth Res. 2017;132:13–66. doi: 10.1007/s11120-016-0318-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le Quiniou C, et al. PSI-LHCI of Chlamydomonas reinhardtii: Increasing the absorption cross section without losing efficiency. Biochim Biophys Acta. 2015;1847:458–467. doi: 10.1016/j.bbabio.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ünlü C, Drop B, Croce R, van Amerongen H. State transitions in Chlamydomonas reinhardtii strongly modulate the functional size of photosystem II but not of photosystem I. Proc Natl Acad Sci USA. 2014;111:3460–3465. doi: 10.1073/pnas.1319164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazár D. Chlorophyll a fluorescence induction. Biochim Biophys Acta. 1999;1412:1–28. doi: 10.1016/s0005-2728(99)00047-x. [DOI] [PubMed] [Google Scholar]
- 34.Ballottari M, et al. Identification of pH-sensing sites in the light harvesting complex stress-related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii. J Biol Chem. 2016;291:7334–7346. doi: 10.1074/jbc.M115.704601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H, Pu X, Wang L, Liu L, Theg SM. A new fluorescence-based method to monitor the pH in the thylakoid lumen using GFP variants. Biochem Biophys Res Commun. 2017;486:1–5. doi: 10.1016/j.bbrc.2016.12.032. [DOI] [PubMed] [Google Scholar]
- 36.Kramer DM, Cruz JA, Kanazawa A. Balancing the central roles of the thylakoid proton gradient. Trends Plant Sci. 2003;8:27–32. doi: 10.1016/s1360-1385(02)00010-9. [DOI] [PubMed] [Google Scholar]
- 37.Semchonok DA, et al. Interaction between the photoprotective protein LHCSR3 and C2S2 photosystem II supercomplex in Chlamydomonas reinhardtii. Biochim Biophys Acta Bioenerg. 2017;1858:379–385. doi: 10.1016/j.bbabio.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 38.Kim E, Akimoto S, Tokutsu R, Yokono M, Minagawa J. Fluorescence lifetime analyses reveal how the high light-responsive protein LHCSR3 transforms PSII light-harvesting complexes into an energy-dissipative state. J Biol Chem. 2017;292:18951–18960. doi: 10.1074/jbc.M117.805192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miloslavina Y, et al. Far-red fluorescence: A direct spectroscopic marker for LHCII oligomer formation in non-photochemical quenching. FEBS Lett. 2008;582:3625–3631. doi: 10.1016/j.febslet.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 40.van Oort B, et al. Different crystal morphologies lead to slightly different conformations of light-harvesting complex II as monitored by variations of the intrinsic fluorescence lifetime. Phys Chem Chem Phys. 2011;13:12614–12622. doi: 10.1039/c1cp20331b. [DOI] [PubMed] [Google Scholar]
- 41.Dall’Osto L, et al. Two mechanisms for dissipation of excess light in monomeric and trimeric light-harvesting complexes. Nat Plants. 2017;3:17033. doi: 10.1038/nplants.2017.33. [DOI] [PubMed] [Google Scholar]
- 42.Johnson MP, Ruban AV. Photoprotective energy dissipation in higher plants involves alteration of the excited state energy of the emitting chlorophyll(s) in the light harvesting antenna II (LHCII) J Biol Chem. 2009;284:23592–23601. doi: 10.1074/jbc.M109.013557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jajoo A, et al. Low pH-induced regulation of excitation energy between the two photosystems. FEBS Lett. 2014;588:970–974. doi: 10.1016/j.febslet.2014.01.056. [DOI] [PubMed] [Google Scholar]
- 44.Belgio E, et al. Economic photoprotection in photosystem II that retains a complete light-harvesting system with slow energy traps. Nat Commun. 2014;5:4433. doi: 10.1038/ncomms5433. [DOI] [PubMed] [Google Scholar]
- 45.Farooq S, et al. Dynamic feedback of the photosystem II reaction centre on photoprotection in plants. Nat Plants. 2018;4:225–231. doi: 10.1038/s41477-018-0127-8. [DOI] [PubMed] [Google Scholar]
- 46.Fan M, et al. Crystal structures of the PsbS protein essential for photoprotection in plants. Nat Struct Mol Biol. 2015;22:729–735. doi: 10.1038/nsmb.3068. [DOI] [PubMed] [Google Scholar]
- 47.van Oort B, van Hoek A, Ruban AV, van Amerongen H. Aggregation of light-harvesting complex II leads to formation of efficient excitation energy traps in monomeric and trimeric complexes. FEBS Lett. 2007;581:3528–3532. doi: 10.1016/j.febslet.2007.06.070. [DOI] [PubMed] [Google Scholar]
- 48.Xu P, Tian L, Kloz M, Croce R. Molecular insights into zeaxanthin-dependent quenching in higher plants. Sci Rep. 2015;5:13679. doi: 10.1038/srep13679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fey H, et al. Isolation of highly active photosystem II core complexes with a His-tagged Cyt b559 subunit from transplastomic tobacco plants. Biochim Biophys Acta. 2008;1777:1501–1509. doi: 10.1016/j.bbabio.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 50.Su X, et al. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science. 2017;357:815–820. doi: 10.1126/science.aan0327. [DOI] [PubMed] [Google Scholar]
- 51.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1960;46:83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Natali A, Roy LM, Croce R. In vitro reconstitution of light-harvesting complexes of plants and green algae. J Vis Exp. 2014;(92):e51852. doi: 10.3791/51852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tibiletti T, Auroy P, Peltier G, Caffarri S. Chlamydomonas reinhardtii PsbS protein is functional and accumulates rapidly and transiently under high light. Plant Physiol. 2016;171:2717–2730. doi: 10.1104/pp.16.00572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drop B, et al. Photosystem I of Chlamydomonas reinhardtii contains nine light-harvesting complexes (Lhca) located on one side of the core. J Biol Chem. 2011;286:44878–44887. doi: 10.1074/jbc.M111.301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Stokkum IHM, van Oort B, van Mourik F, Gobets B, van Amerongen H. (Sub)-Picosecond spectral evolution of fluorescence studied with a synchroscan streak-camera system and target analysis. In: Aartsma TJ, Matysik J, editors. Biophysical Techniques in Photosynthesis. Springer; Dordrecht, The Netherlands: 2008. pp. 223–240. [Google Scholar]
- 56.Snellenburg JJ, Laptenok SP, Seger R, Mullen KM, van Stokkum IHM. Glotaran: A Java-based graphical user interface for the R package TIMP. J Stat Softw. 2012;49:1–22. [Google Scholar]
- 57.van Stokkum IH, Larsen DS, van Grondelle R. Global and target analysis of time-resolved spectra. Biochim Biophys Acta. 2004;1657:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 58.Nawrocki WJ, Santabarbara S, Mosebach L, Wollman FA, Rappaport F. State transitions redistribute rather than dissipate energy between the two photosystems in Chlamydomonas. Nat Plants. 2016;2:16031. doi: 10.1038/nplants.2016.31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.