Significance
Light-chain amyloidosis (AL) is a progressive and often fatal degenerative disease caused by conformational changes within Ig light chains that generally lead to aggregation. Current chemotherapy treatments aim to eliminate the clonal plasma cells that secrete full-length light chains; however, complete eradication is achieved in only 30–40% of the patients. The kinetic stabilizer strategy introduced herein does not require an understanding of the nonnative light-chain structure–proteotoxicity relationships driving organ degeneration in AL to be successful at stopping disease progression because it stops light-chain conformational excursions at the beginning of the aggregation cascade. The small-molecule kinetic stabilizers identified bind to conserved residues at the variable-domain–variable-domain interface in the native dimer, stabilizing this putative nontoxic structure.
Keywords: kinetic stabilizer, high-throughput screen, dimerization, structural biology, proteotoxicity
Abstract
In Ig light-chain (LC) amyloidosis (AL), the unique antibody LC protein that is secreted by monoclonal plasma cells in each patient misfolds and/or aggregates, a process leading to organ degeneration. As a step toward developing treatments for AL patients with substantial cardiac involvement who have difficulty tolerating existing chemotherapy regimens, we introduce small-molecule kinetic stabilizers of the native dimeric structure of full-length LCs, which can slow or stop the amyloidogenicity cascade at its origin. A protease-coupled fluorescence polarization-based high-throughput screen was employed to identify small molecules that kinetically stabilize LCs. NMR and X-ray crystallographic data demonstrate that at least one structural family of hits bind at the LC–LC dimerization interface within full-length LCs, utilizing variable-domain residues that are highly conserved in most AL patients. Stopping the amyloidogenesis cascade at the beginning is a proven strategy to ameliorate postmitotic tissue degeneration.
Secretion of an Ig light chain (LC) by a clonally expanded plasma cell population can lead to the disease LC amyloidosis (AL)—both a cancer and a proteinopathy (1, 2). “Free” LCs secreted without an associated antibody heavy chain (HC) initially adopt a well-defined homodimeric structure, wherein the monomers may be covalently linked by an interchain disulfide bond (Fig. 1A) (3). LC monomers comprise an N-terminal variable (V) domain attached to a C-terminal constant (C) domain (SI Appendix, Fig. S1). Each patient’s clonal plasma cells secrete a single, unique LC sequence. Most LCs are rapidly removed by the kidney.
Fig. 1.
High-throughput screen for LC kinetic stabilizers. (A) LCs consist of variable (V) (yellow) and constant (C) (gray) Ig domains, which form homodimers linked by a disulfide bond between C214 residues (black line). Cleavage of dye-labeled protein by protease leads to a decrease in FP in the resulting peptides. The rate of proteolysis depends on a LC’s kinetic stability, which can be enhanced by the binding of a stabilizing small molecule. (B) Proteolysis of fluorescein-labeled and unlabeled LCs, coincubated with 100 nM PK and visualized by SDS/PAGE. Symbols show normalized band intensities, and lines show fits to a single-exponential decay model; n = 3. Black, Coomassie-stained total LC (10 µM); green, fluorescence of labeled LC (20 nM). (C) Kinetics of fluorescein-labeled LC proteolysis measured by FP at 37 °C in a fluorimeter cuvette. Lines show fits to a single-exponential decay model. (D) Kinetics of proteolysis measured by FP in a 384-well plate at 22 °C. Points show data for individual wells, and lines show the mean; n = 16. (E) The extent of proteolysis after 24 h at 22 °C depends on the concentration of the LC-stabilizing kosmotrope Na2SO4, but not on the concentration of protease. (F) Postproteolysis compound addition to LCs identifies false-positive screen hits. Each point represents the mean measurement (n = 3) for a single compound. Green shaded areas indicate compounds considered to be hits. (G) Compounds that provide protection from both serine protease PK and metalloprotease thermolysin (green area) are likely to be stabilizers of LCs, rather than protease inhibitors.
However, since amyloidogenic full-length (FL) LCs are generally less stable than nonamyloidogenic FL LCs, they can misfold, or misfold and misassemble, into nonnative species including cross–β-sheet amyloid fibrils, which are a hallmark of AL (4–8). Sequence also seems to play a role, as not all destabilized FL LCs aggregate in patients (4–8). How aggregation occurs in patients is not known, but several processes have been described in vitro, including destabilization-dependent endoproteolysis that releases amyloidogenic LC fragments (4, 9, 10). LC fragments including V domains are observed in patient deposits alongside FL LCs (11–13).
Since we do not understand the structure–proteotoxicity relationships driving AL, a conservative strategy is to block FL LC misfolding at its origin by stabilizing the FL LC native state. Such a strategy has been effective at ameliorating the transthyretin amyloidoses (14–19). A small molecule that stabilizes FL LC dimers should prevent any misfolding and/or endoproteolysis that lead to LC aggregation and organ toxicity. We refer to such molecules as kinetic stabilizers, since they reduce the rate at which LCs transiently visit nonnative, aggregation-prone, and protease-sensitive conformations (20). The interfaces between the domains of the LC dimer are an important determinant of stability and aggregation propensity that we and others have identified as potential targets for stabilization (21, 22). We therefore seek treatments that prevent newly synthesized and secreted FL LCs from misfolding and aggregating, thus reducing organ proteotoxicity, such that patients can eventually tolerate effective chemotherapy regimens.
Using a protease-coupled fluorescence polarization (PCFP) assay that assesses LC kinetic stability, we screened 650,000 small molecules and identified FL LC kinetic stabilizers in four structural classes that protect recombinant and plasma cell-secreted λ6a LCs from endoproteolysis. NMR and X-ray crystallography reveal that these small molecules bind to the V-domain–V-domain interface in the FL LC dimer and in isolated V domains, utilizing conserved residues found in most patient-derived LCs. Identifying these small-molecule LC kinetic stabilizers and their binding site represents a step toward developing an eventual kinetic stabilizer drug for AL.
Results
A PCFP Assay of LC Stability.
To identify FL LC kinetic stabilizers, we developed a PCFP assay (Fig. 1A). This assay measures the protease resistance of FL LCs—a reflection of the kinetic stability imparted by small-molecule binding (4, 5). Cleavage of the AL patient-derived λ6–57 FL LC known as WIL (23) to smaller fragments requires conformational excursions (4) and leads to reduced fluorescence polarization (FP) of the LC-conjugated fluorescein fluorophore (Fig. 1A) (24). We introduced a surface-exposed K79C V-domain mutation [numbered according to the Kabat (25) scheme] into FL WIL and into the non-AL FL LC JTO (23), to which we conjugated fluorescein via an attached maleimide (SI Appendix, Fig. S1). The resulting conjugates, referred to as WIL-FL* and JTO-FL*, were purified by ion-exchange chromatography. Both WIL-FL* and JTO-FL* are obligate dimers because of the disulfide bond between monomers. Proteinase K (PK), a broad-spectrum serine protease that efficiently cleaves transiently unfolded LCs under physiological conditions (4) was used for the PCFP assay to minimize any sequence-specific cleavage site differences between LCs.
We measured PK proteolysis of WIL-FL* and JTO-FL* (20 nM) spiked into unlabeled WIL-FL and JTO-FL (10 µM), respectively, at 37 °C in PBS (pH 7.4). Both labeled LCs are degraded at increased rates compared with the parent sequences based on the disappearance of the LC band by SDS/PAGE (Fig. 1B; kobs = 7.6 ± 0.3 × 10−4 s−1 for WIL-FL* versus 2.4 ± 0.9 × 10−4 s−1 for WIL-FL; 9.5 ± 3.5 × 10−5 s−1 for JTO-FL* versus no measurable decrease for JTO-FL; mean ± SD; n = 3). The K-to-C mutation and subsequent fluorescein conjugation reduces the kinetic stability of both LCs, possibly by decreasing the solvent entropy change upon folding in the region displaying the solvated dye, that is, by attenuating the hydrophobic effect. Importantly, endoproteolysis of AL-associated WIL-FL* is significantly faster than that of the more kinetically stable JTO-FL* (Fig. 1B). Treatment of WIL-FL* (20 nM) with PK (500 nM) at 37 °C in PBS leads to reduction of FP (λex = 488 nm; λem = 520 nm) at a rate (kobs = 9.9 × 10−4 s−1) comparable to that measured by SDS/PAGE, whereas the FP of JTO-FL* is reduced more slowly (kobs = 1.8 × 10−4 s−1), as expected (Fig. 1C).
We miniaturized the PCFP assay to a 384-well microplate format and verified that we observed similar patterns of proteolysis under conditions suitable for high-throughput screening. In PBS at 22 °C, WIL-FL* (20 nM) is extensively cleaved by PK (500 nM) over 24 h, whereas JTO-FL* is cleaved much more slowly (Fig. 1D). We previously observed correlations between rates of denaturant-induced unfolding and rates of endoproteolysis (4). Initial experiments showed that the FP of both WIL-FL* and JTO-FL* decreased at a similar rate in the presence of PK and increased in the absence of PK. This appears to be due to denaturation of the LCs on the plate surface (SI Appendix, Fig. S2). Addition of Pluronic F-127 detergent to untreated plates (Greiner; no. 655076) prevented the increase in FP in the absence of PK and yielded proteolysis kinetics similar to those in cuvettes with PK. Employing low-binding plates with or without Pluronic F-127 detergent (0.02%) does not alter the rate of WIL-FL* endoproteolysis. Thus, we added Pluronic F-127 detergent to untreated plates for the high-throughput screen and subsequent experiments due to the increased cost of low-binding plates. This detergent may have the additional benefit of reducing colloidal aggregation of small molecules (26). Addition of the kosmotropic salt sodium sulfate reduces the extent of WIL-FL* proteolysis after 24 h, consistent with its stabilizing effect (Fig. 1E). The extent of WIL-FL* endoproteolysis is not influenced by the concentration of PK, consistent with FL LC conformational excursions from the native state being rate limiting for proteolysis (4), a condition referred to as EX1 kinetics (Fig. 1E). The assay exhibited a robust Z′ score of 0.8 (27).
Identification of Small Molecules That Protect LCs from Proteolysis.
We screened the Maybridge HitFinder Library (16,000 small molecules), employing the 384-well PCFP assay with WIL-FL*. The results guided further miniaturization of the assay to 1,536-well plates, optimizing the concentrations of WIL-FL* (10 nM), PK (250 nM), and kinetic stabilizer candidate (6.75 µM). We screened the Scripps Florida library (653,085 small molecules), identifying 2,777 primary hits exhibiting >20.6% (mean + 3SDs) retention of WIL-FL* FP [relative to 0.68% vehicle (DMSO) control assigned 0%, with 100% corresponding to the FP of JTO-FL* without PK; SI Appendix, SI Materials and Methods]. The PCFP assay was repeated in triplicate on the 2,777 primary screening hits, of which 1,422 molecules had >20.6% mean retention of WIL-FL* FP.
To identify and exclude molecules that interfered with FP measurements, the assay was repeated, but candidate stabilizers were not added until after 24 h of PK treatment (Fig. 1F). Compounds that artifactually increase FP would result in similar apparent retention of FP in both assay configurations. Six hundred sixty-two molecules had >1.25-fold greater mean activity (n = 3) in the primary screen than in this counterscreen (2,115 molecules artifactually increased FP). To eliminate PK inhibitors, the PCFP screen was rerun in triplicate on the 2,777 hits using the protease thermolysin (200 nM; candidate stabilizer concentration, 6.75 µM; Fig. 1G). Thermolysin is a broad-spectrum metalloprotease that is likely not inhibited by compounds that inhibit PK, a serine protease. Plotting the thermolysin versus PK data identified 1,243 compounds on the diagonal that were not protease inhibitors (Fig. 1G). The 269 small molecules that fulfilled all three assessment criteria [exhibited PCFP activity in triplicate, passed the FP artifact counterscreen (Fig. 1F), and exhibited >20% efficacy in the PCFP thermolysin counterscreen] were retained.
The potency of these surviving 269 LC kinetic stabilizers was measured by recording concentration dependence data in the three PCFP configurations covered in the preceding paragraph, along with 147 candidates that appeared to inhibit one or both proteases (Dataset S1). The 128 compounds exhibiting a dose–response curve were chosen for further evaluation. We further verified that these 128 compounds do not inhibit PK by measuring their ability to inhibit proteolysis of a fluorogenic peptide substrate (SI Appendix, Fig. S3). To exclude the possibility that differences in the initial FP amplitude could lead to misinterpretation of the endpoint FP measurement, we measured FP kinetics upon PK treatment. Ninety of the 128 compounds reduced the rate and amplitude of FP change relative to vehicle, consistent with LC kinetic stabilization (SI Appendix, Fig. S4). Finally, to verify that the FP measurements were reporting on FL LC proteolysis sensitivity, we measured protection of WIL-FL* from PK proteolysis imparted by the 128 compounds mentioned above using SDS/PAGE. Twenty-six molecules stabilize FL LCs by this assay (SI Appendix, Figs. S4 and S5). Sixteen molecules showed clear activity in both the kinetic FP and SDS/PAGE assays (SI Appendix, Fig. S4 and Table S1).
The 16 hit molecules fall into four classes (compounds 1–16, SI Appendix, Table S1): coumarins (1–3, 9–15), an aryl cyanoacrylamide (4), biaryl hydrazones (5–6), and hydantoins (7–8). Two sulfones (17–18) derived from the Maybridge library were also validated as hits (SI Appendix, Table S1). We purchased eight representative small molecules and determined their midpoint effective concentration (EC50) values in the PCFP assay by titration (Table 1). Dissociation constants (KD ligand) measured for each compound using equilibrium dialysis are similar to the EC50 values, indicating that the small-molecule–bound FL LC is highly resistant to proteolysis (Table 1).
Table 1.
Small molecules that stabilize LCs
 |
7-Amino-Coumarins Are Fluorogenic Kinetic Stabilizers of λ6a LCs.
Ten of the 16 hits and a further 14 of the 128 screening hits surviving the counterscreens are coumarins modified at the 7-position by either a diethylamino group (21 molecules; e.g., 1 and 2; Table 1) or a 1-phenylethoxy group (three molecules; e.g., 3; Table 1). Coumarins are often regarded as promiscuous binders; however, 7,839 other coumarin-containing molecules in the library, of which 432 are modified at the 7-position, were not identified as hits after counterscreening. We initially focused on compound 1, 7-diethylamino-4-methylcoumarin, a commercially available dye known as “Coumarin 1.”
To verify the activity of 1 and investigate the structural requirements at the 7-position of coumarins, we measured protection from PK proteolysis of unlabeled WIL-FL. Incubation of WIL-FL (10 µM) with 1 (100 µM; Table 1 and Fig. 2A), but not the related compounds 19, 20, and 21 (100 µM; Fig. 2A and SI Appendix, Table S1), protects this FL LC against PK proteolysis (t = 2 h). Compound 9, which lacks a methyl group at the 4-position, also stabilizes WIL-FL, but is less efficacious than 1 (Fig. 2A and SI Appendix, Table S1). Incubation of WIL-FL (20 µM) with 1 (1 µM) results in a large fluorescence emission increase of 1 relative to the weaker, red-shifted fluorescence of 1 in PBS (Fig. 2B; λex = 373 nm), suggesting that the altered environment of 1 associated with binding and the restricted mobility of the 7-diethylamino group contribute to its fluorogenicity (28). The FP of 1 increases from 0.05 in buffer to 0.35 in the WIL-FL•1 complex, consistent with an increase in rotational correlation time. In contrast, 19 (1 µM) is fluorescent in PBS, but its emission intensity is reduced in the presence of WIL-FL (20 µM; Fig. 2B).
Fig. 2.
7-Diethylamino-4-methylcoumarin (1) is an environment-sensitive fluorophore that stabilizes LCs. (A) SDS/PAGE gel showing proteolysis of unlabeled WIL-FL (10 µM) in the presence of 1% DMSO vehicle and 100 µM small molecule; t = 2 h. Modifications to the core coumarin structure (21) are shown in red for each small molecule. (B) Fluorescence emission (λex = 373 nm) of 1 (1 µM), but not 19 (1 µM), increases in the presence of unlabeled WIL-FL (20 µM). (C) Titrations of LC constructs into 1 (1 µM); n = 3. Lines indicate fits to a one-site binding model. (D) Dissociation constants of 1 for recombinant LC variants (n = 3), measured as for C. (E) SV-AUC of WIL-FL C214S (20 µM) and WIL-V (20 µM) in the absence (red) or presence (blue) of 1 (100 µM). Data for dimeric WIL-FL (20 µM) are shown in gray. (F) Titration of κ I O18/O8-FL (blue) and AL12-FL (red) into 1 (1 µM). Data for WIL-FL (gray) are shown for comparison. (Inset) Fluorescence spectra of 1 (1 µM) in PBS (dashed gray line) or the presence of 20 µM κ I O18/O8-FL (blue) or AL12-FL (red). (G) Competition of 1 (1 µM) for the WIL-FL binding site indicates that the presence of other hit molecules (10 µM; numbering as per SI Appendix, Table S1) reduces binding of 1 (1 µM). The unrelated molecule diflunisal (Dif) does not compete with 1 for LC binding. (H) Binding of 1 to human serum albumin (HSA) in the presence or absence of diflunisal determined by fluorescence of 1, measured as for C. (I) Fluorescence of 1 in the presence of the LC homodimer (black) or LC:HC heterodimer (Fab, red) of the human IgG antibody 5J8.
The fluorogenicity of 1 makes it a convenient probe to investigate whether LCs other than WIL-FL can be kinetically stabilized and whether other nonfluorescent candidate kinetic stabilizers bind LCs at the same site. Isothermal titration calorimetry measurements indicate that 1 binds to WIL-FL with a KD ligand of 3.08 ± 0.52 µM (mean ± SD; n = 3; SI Appendix, Fig. S6). To measure binding, we titrated different WIL constructs into 1 in PBS containing 0.02% Pluronic F-127 and measured the fluorescence emission of the LC•1 complex in a 384-well plate format (Fig. 2C; λex = 373 nm, λem = 445 nm). We used a low concentration of 1 (1 µM) and varied LC concentration (serial dilution from 133 µM to 18 nM in 1.5-fold decrements) to minimize background fluorescence and simplify the analysis. Binding of compound 1 to WIL-FL is fit well by a 1-site model with an apparent KD ligand of 3.14 ± 0.3 µM (mean ± SD; n = 5), whereas binding to the WIL V domain has a steeper dependence on LC concentration and is fit less well by a 1:1 binding model (apparent KD ligand of 79.3 ± 5.2 µM). No binding of 1 (1 µM) was detected to the isolated λ3 C domain (Fig. 2C). Recombinant FL λ6–57 LCs form disulfide-linked dimers (4), but their isolated V domains populate an equilibrium between monomers and dimers (4, 22, 29). The apparent positive cooperativity in the binding of 1 to WIL-V is consistent with induced dimerization of the LC V domain.
Concentration-dependent NMR chemical shift data indicate that WIL-V has a dimerization equilibrium constant (KD dimer) of ∼5 mM (SI Appendix, Table S2) (22). Fitting the fluorescence data to a model of binding-induced dimerization constrained by the NMR-derived KD dimer (SI Appendix, SI Materials and Methods) yields an apparent KD ligand of 3.0 ± 1.2 µM for WIL-V2•1 (SI Appendix, Fig. S6), indicating that the apparent low affinity for WIL-V compared with WIL-FL is due to a low concentration of WIL-V dimer at equilibrium. Since the affinities for WIL-FL and dimeric WIL-V are indistinguishable, we can assume that 1 binds only to dimeric WIL-V and, hence, estimate the apparent KD dimer from the fluorogenic binding data in the presence of 1 (1 µM) to be 1.45 mM, compared with 5 mM in the absence of 1.
A successful kinetic stabilizer must bind to and stabilize patient LCs with a vast range of sequences (30, 31). To determine whether 1 can stabilize other FL LC sequences, we measured binding of 1 to three more recombinant λ6–57 FL LC disulfide-linked dimers: JTO-FL; ALMC2-FL, an AL-associated LC sequence derived from the ALMC2 cell line (32); and the germline sequence 6aJL2-FL. Serial dilution of these LCs from 133 µM to 18 nM in 1.5-fold decrements into 1 (1 µM) in PBS yields KD ligand values of between 1 and 20 µM (Fig. 2D, Left). The isolated V domains from WIL, ALMC2, and 6aJL2 bind to 1 (1 µM) with reduced affinity (Fig. 2D, Left), while JTO-V has KD ligand of 16.5 ± 0.4 µM (mean ± SD; n = 3), similar to that of JTO-FL (20.3 ± 1 µM), consistent with JTO-V being mainly dimeric at this concentration (SI Appendix, Table S2) (22). Eliminating the interchain disulfide bond between the C domains by the C214S mutation reduces the affinity of LCs for 1 (1 µM) (Fig. 2D), consistent with this mutation’s destabilization of the dimeric interface (4).
To further test the hypothesis that 1 binds at the interface between the V domains, we measured binding of 1 to WIL-FL and JTO-FL variants with mutations designed to disrupt the dimer (22). The F98D mutation, which disrupts the V-domain–V-domain interface, rendered binding of 1 (1 µM) undetectable in the presence or absence of the C214 interchain disulfide bond (Fig. 2D, Right). The F118D mutation, which destabilizes the C-domain–C-domain interface, or the L117G or P141A mutations, which destabilize the hydrophobic core of the C domain, resulted in reduced binding affinity of 1 (1 µM) to WIL-FL C214S and, to a lesser extent, JTO-FL C214S (Fig. 2D, Right), with KD values similar to those exhibited by the isolated V domains (∼20–50 µM).
To further investigate the ability of small molecules to induce dimerization of LCs, we performed sedimentation velocity analytical ultracentrifugation (SV-AUC) analysis of WIL-V and WIL-FL C214S (20 µM) in the presence or absence of 1 (100 µM). Continuous sedimentation distributions (Fig. 2E) show the apparent size of the sedimenting species. Deviations from the expected size indicate exchange between species, in this case monomer and dimer. The size distributions of both LCs (20 µM) increase upon binding 1 (100 µM). WIL-FL C214S has a KD dimer of ∼16 µM, estimated by concentration-dependent SV-AUC experiments (SI Appendix, Fig. S7).
Of the two classes of human LCs, λ and κ, λ LCs are more commonly observed in AL, but a fraction of patients secrete κ1–33 LCs (12, 33). To ask whether κ LCs possess a similar small-molecule binding site as λ LCs, we incubated 1 (1 µM) with two κ1–33 LCs (serial dilution from 133 µM to 18 nM in 1.5-fold decrements), which represent the patient-derived sequence AL12-FL (34) and its germline precursor κ I O18/O8-FL (7). Recombinant expression of these κ1–33 LCs using a similar protocol to that used for λ6–57 LCs yielded both monomeric and dimeric LCs, as shown by SV-AUC (SI Appendix, Fig. S7). Consistent with this reduced propensity to dimerize, binding of 1 to κ1–33 LCs is weaker than binding to λ6–57 LCs (Fig. 2F). Binding of an excess of κ1–33 LCs (20 µM) to 1 (1 µM) afforded FL LC•1 fluorescence (Fig. 2F, Inset). This result indicates that a binding site for 1 is present in dimers of both λ6–57 and κ1–33 LCs.
To ask whether the noncoumarin small molecules in Table 1 bind to FL LCs at the same or an overlapping site, we measured binding of 1 (1 µM) to WIL-FL (serial dilution from 150 µM to 20 nM in 1.5-fold decrements) in the presence of the other screening hits (10 µM). The apparent binding affinity of LCs for 1 was reduced by six nonfluorescent molecules identified in the screen, but not by the negative control diflunisal, an antiinflammatory drug that stabilizes transthyretin (15, 35) (Fig. 2G). These data suggest that the hits 4–8 have binding sites that overlap with that of 1, or exhibit negatively cooperative binding with 1 to WIL-FL.
The free monoclonal LC concentration in the plasma of AL patients is typically in the range of 1–20 µM (10–100 mg/dL) (36), whereas the concentration of antibodies and albumin is much higher. Therefore, kinetic stabilizer binding selectivity to FL LCs in preference to other plasma proteins is important to minimize the proportion of free FL LC. Albumin binds many small molecules in blood, typically with micromolar KD values. Titration of albumin (serial dilution from 400 µM to 53 nM in 1.5-fold decrements) with 1 (1 µM) results in increased coumarin fluorescence, with an apparent KD of 37 ± 3 µM (Fig. 2H). Affinity of 1 for albumin is reduced in the presence of 100 µM diflunisal (Fig. 2H), consistent with diflunisal binding competitively to albumin.
The heterodimeric interface between LCs and HCs in an antibody is structurally distinct from that of the LC homodimer interface. To verify that our kinetic stabilizers bind FL LC dimers selectively over antibodies, we compared the binding of 1 (1 µM) to the LC and antigen binding fragment (Fab) (the LC:HC dimer) of 5J8, a human anti-influenza hemagglutinin antibody (serial dilution from 120 µM to 16 nM in 1.5-fold decrements) (37). The 5J8 FL LC from the λ3 class (not associated with AL) binds to 1 with an apparent KD of ∼50 µM (Fig. 2I), an event that protects the 5J8 LC from proteolysis by PK (SI Appendix, Fig. S8). In contrast, the 5J8 Fab shows no apparent binding to 1 as measured by fluorescence (Fig. 2I), and 1 does not protect the 5J8 Fab from PK proteolysis (SI Appendix, Fig. S8).
Structural Insight into LC Kinetic Stabilizer Binding.
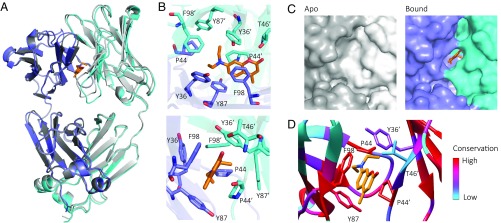
We determined the crystal structures of JTO-FL with and without bound 1 [PDB ID codes 6MG5 (38) and 6MG4 (39), respectively]. Our attempts to crystallize other LC•kinetic stabilizer complexes (e.g., the WIL-FL•1 complex) have so far been unsuccessful. The unbound JTO-FL LC structure is very similar to previously published FL LC structures (3, 5, 40) (Fig. 3A). LC homodimers are covalently linked by an interchain disulfide bond between C214 residues in the C domains of the monomers. In the apo JTO-FL structure, refined at 1.75-Å resolution, the conformation of the V domains is the same as that in the published JTO-V dimer structure (41).
Fig. 3.
Kinetic stabilizer binding to the V-domain–V-domain dimer interface. Crystal structures of JTO-FL with bound 1 (in orange) (LC blue, cyan) and without 1 (LC gray). (A) Overview of structures, aligned by one chain of the dimer. (B) Kinetic stabilizer 1 binds in a pocket formed by the side chains of residues 44, 87, and 98 from one protomer and 36′, 44′, and 46′ from the other protomer. Residues are numbered according to the Kabat system (SI Appendix, Fig. S1). (C) Surface representation of the binding site region. 1 binds in a pocket that is not present in the unliganded LC. (D) Conservation of binding site residues in human germline LC genes (SI Appendix, Fig. S10) mapped onto the JTO-FL•1 structure. Red, highly conserved residues; blue, weakly conserved residues.
The structure of the JTO-FL•1 complex, refined at 1.8-Å resolution, unambiguously shows density for 1 at the interface formed by the two V domains (SI Appendix, Fig. S9). This binding site is consistent with previous reports that the interface between V domains in the dimer can accommodate various aromatic ligands (21, 42), although the exact binding site that we identify below has not been previously observed. The binding site for 1 comprises residues P44, Y87, and F98 from one protomer and Y36ʹ, P44ʹ, and T46ʹ from the other protomer (Fig. 3B), which together form a cavity that is not present in the unbound structure (Fig. 3C). The JTO-FL F98D variant (22) has no measurable affinity for 1 (Fig. 2D), consistent with disruption of the π–π stacking interaction observed in the structure. The 7-diethylamino substructure projects through the V-domain–V-domain interface, interacting with both P44 residues. These interactions may explain the apparent requirement for modification at the 7-position of the coumarin scaffold. The carbonyl oxygen of 1 hydrogen bonds with the side-chain hydroxyl and/or backbone amide of T46ʹ. A CH–π interaction between 1 and Y87 may also contribute to the binding free energy. Binding of 1 causes a rotation of the V domains relative to each other (SI Appendix, Fig. S9), which creates the interface cavity (Fig. 3C). The distance between the Cα residues of F98 and P44ʹ increases from 7.2 to 9.7 Å to accommodate 1. The LC dimer has two pairs of F98 and P44 residues, creating two potential binding sites, but only one molecule of 1 is observed per dimer. Aligning a second molecule of 1 utilizing the same intermolecular interactions results in a steric clash between the diethylamino substructures (SI Appendix, Fig. S9).
The residues used by JTO to bind 1 are highly conserved (Fig. 3D and SI Appendix, Fig. S10). Residues Y36, P44, Y87, and F98 are present in 88%, 99%, 95%, and 100% of all human germline LC genes, respectively, whereas T46 is only present in two functional germline genes. Mutation of residue 46 in WIL-FL and JTO-FL to leucine, which is the residue present in 75% of LC germline genes, slightly enhances the binding affinity of 1 for these FL LCs (SI Appendix, Fig. S11), indicating that the side-chain hydroxyl group of T46′ is not necessary for binding of 1.
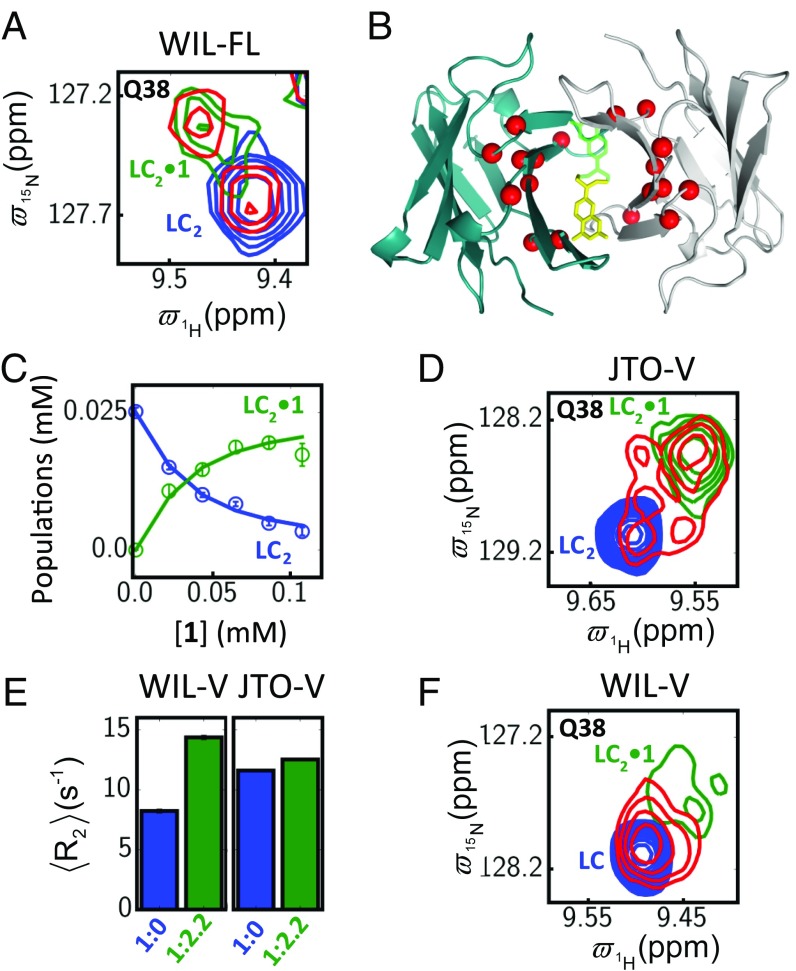
To verify that 1 binds to different LCs in solution at the same site identified in the crystal structure, we recorded the NMR spectra of WIL-FL, WIL-V, and JTO-V in the absence and presence of 1. We used high concentrations of LC (100–500 μM) and solubility-limited concentrations of 1 (up to 500 μM) to maximize the NMR signal from the bound state. Two-dimensional 1H–15N heteronuclear single-quantum coherence (HSQC) experiments, wherein each amide in a protein is represented as a peak in the spectrum, allowed identification of residues perturbed by binding of 1. Chemical shift and intensity perturbations of similar residues in each spectrum occur upon addition of 1 (Fig. 4A). In each case, the perturbed residues are concentrated in the interface between the two V domains, close to the site at which 1 binds according to the crystal structure (Fig. 4B). Upon titration of 1 into WIL-FL, which is an obligate dimer (4), new peaks of the bound state LC2•1 appear for the amides close to the binding site (Fig. 4 A and B and SI Appendix, Fig. S12). The changes in intensity for LC2 and LC2•1 peaks can be fit by a two-state binding model (SI Appendix, SI Materials and Methods) with an apparent KD ligand of 19 ± 2 µM (Fig. 4C).
Fig. 4.
Kinetic stabilizer 1 binds to LCs in solution. (A) Superposition of selected region of 1H–15N HSQC NMR spectra of WIL-FL (0.05 mM monomeric concentration) in the absence (blue contours) or presence of 1 at LC:1 ratios (LC monomer:ligand) of 1:0.8 (red) and 1:2.2 (green). (B) Residues with combined chemical shift changes (SI Appendix, SI Materials and Methods) larger than 0.1 ppm upon binding of 1 are mapped as red spheres onto the structure of the JTO-FL•1 complex; the ligand is shown as yellow or green sticks in the two possible but mutually exclusive binding sites. (C) Intensity of peaks associated with free LC dimer (LC2, blue) and complex (LC2•1, green) as a function of 1 binding to WIL-FL. (D) HSQC spectral region highlighting correlations for residue Q38 of JTO-V. Colors are as for A. Note the presence of exchange peaks connecting free and ligand-bound correlations when approximately equal amounts of LC2 and 1 are added. (E) 15N transverse relaxation rates (R2) for WIL-V (averaged over 29 residues) and JTO-V (32 residues) with (green) or without (blue) addition of 1. (F) Spectra of WIL-V residue Q38, in the absence (blue) or presence of 1, at LC:1 ratios of 1:0.9 (red) and 1:2.2 (green). Note that the cross-peaks from the bound form at the LC:1 ratio of 1:0.9 are broadened. See SI Appendix, Figs. S12–S14, for full HSQC spectra.
Concentration-dependent chemical shift measurements (22) indicate that JTO-V is dimeric at concentrations used for NMR (generally greater than 100 µM) with a dimerization KD of ∼12.5 µM (SI Appendix, Table S2). Binding of 1 to JTO-V (140 µM) at approximately equimolar stoichiometry results in the appearance of exchange cross-peaks between peaks originating from LC2 and LC2•1 (Fig. 4D, red contours, and SI Appendix, Fig. S13) in a regular HSQC experiment that is not designed to measure exchange rates. A ZZ-exchange experiment (43) measured on- and off-rates of 1.0 ± 0.10 µM−1 s−1 and 57 ± 2 s−1, respectively, for the JTO-V2•1 complex, yielding a KD of 57 ± 4 µM for a two-state model (SI Appendix, Supplementary NMR Analysis and Fig. S14), in agreement with the value measured by fluorescence (Fig. 2D).
Binding of 1 to isolated WIL-V (400 µM) is more complex, since this V domain is monomeric in the absence of ligand (KD dimer ∼ 5 mM) (22) and becomes dimeric upon addition of 1. 15N transverse relaxation (R2) rates, which report on overall protein tumbling and hence on molecular size, double for WIL-V upon addition of excess 1 (500 µM), whereas they do not change for JTO-V (140 µM), which is dimeric without and with 1 bound (Fig. 4E). Notably, WIL-V bound to 1 exhibits multiple HSQC peaks for most interfacial residues and may represent alternative conformations of the dimer or of the bound ligand (Fig. 4F and SI Appendix, Fig. S15). The presence of multiple conformations for the LC2•1 complex of WIL-V prevents accurate quantification of KD. Chemical exchange saturation transfer experiments (44) indicate that the KD ligand for WIL-V2•1 is <50 µM (SI Appendix, Fig. S14), consistent with the effective KD ligand of 3 ± 1.6 µM determined from fluorogenic binding titrations (Fig. 2C and SI Appendix, Fig. S6). Removal of 1 from WIL-V by dialysis yielded an HSQC spectrum indistinguishable from that of the starting material (SI Appendix, Fig. S16), indicating that binding is reversible and noncovalent. Thus, the heterogeneity observed in the bound state spectra (Fig. 4F) is likely due to alternate binding modes.
We next asked how binding of kinetic stabilizers would affect the stability of different LCs. In these experiments, we generally used an excess of kinetic stabilizer (limited by small-molecule solubility) to saturate LC binding and thus investigate the properties of ligand-stabilized LCs, as a proxy for the behavior of small molecules that bind to LCs with nanomolar affinity. Ablation of the interchain disulfide bond by the C214S mutation results in reduced LC kinetic stability (4) and reduced small-molecule binding affinity (Fig. 2D). Nonetheless, binding of 1 (100 µM) reduces the rate constant of PK proteolysis of WIL-FL C214S (10 µM) from 6.9 × 10−3 to 2.8 × 10−3 s−1 (Fig. 5A).
Fig. 5.
Small-molecule kinetic stabilizer binding stabilizes LCs against proteolysis, unfolding, and aggregation. (A) Proteolysis of recombinant WIL-FL C214S (10 µM) in the presence of vehicle (red) or 1 (100 µM; blue). Symbols show LC band quantitation, and lines show fits to a single-exponential decay model. (B) Unfolding rates of WIL-FL (5 µM) in the presence of 5 (50 µM; orange) or vehicle (red) as a function of urea concentration. Symbols represent rates calculated for individual kinetic transients, and lines are fits to a two-state unfolding model. (C) Equilibrium urea titrations of WIL-FL (5 μM) in the presence of 1 (100 μM; blue symbols), 5 (50 μM; orange symbols), or vehicle (red symbols). (D) NMR hydrogen–deuterium exchange measurements of JTO-V (100 µM, Left) and WIL-V (200 µM, Right) in the presence (blue) or absence (red) of 1 (400 µM). Exchange rates were converted to free energies as described in SI Appendix, SI Materials and Methods. Symbols show residues for which rates could be measured. Dashed lines indicate the average stability for each V domain. (E) Proteolysis of the plasma cell-secreted LC, ALMC2 (5 µM), in the presence of vehicle (red) or 1 (100 µM; blue). The line represents a fit to a single-exponential decay model. (F) Residual soluble WIL-FL C214S dimer (10 μM) measured by SEC at pH 5, 37 °C, in the presence (blue) or absence (red) of 1 (200 µM), as a function of time to assess aggregation kinetics by starting material disappearance. Individual reactions are shown by dashed lines; curves represent fits of all samples in a category (±1) to a single-exponential decay model.
To confirm that protease resistance is driven by stabilization of the LC native-state ensemble by small-molecule binding, we measured urea-mediated unfolding rates of LCs in solution by tryptophan emission fluorescence (4). The intrinsic fluorescence of 1 precluded its use in these experiments. WIL-FL (5 µM) in the presence of 5 (a nonfluorescent kinetic stabilizer used at a solubility-limited concentration of 50 µM; Table 1) unfolds with a rate constant (extrapolated to the absence of denaturant) of 2.43 s−1 compared with 3.57 s−1 for WIL-FL alone (Fig. 5B). Binding of 5 increases the kinetic stability of the FL LC, even though the >5 M urea concentrations used in this experiment will lower the affinity of 5 for the FL LC. Titration of urea into WIL-FL (5 µM) in the absence or presence of 1 (100 µM) or 5 (50 µM) revealed an increase in the midpoint concentration of urea required to unfold the FL LC (Fig. 5C) from 1.4 to 2.0 M. The magnitude of this stabilization imparted by 1 and 5 was indistinguishable (Fig. 5C), indicating that the ligand-bound states are similarly stable.
To verify that binding of 1 reduces LC unfolding under nondenaturing conditions, we measured residue-specific hydrogen–deuterium exchange rates by NMR of WIL-V and JTO-V (100 µM) in the absence of or with 1 (500 µM). These rates (the decrease in intensity of the HSQC peak associated with each amide) are converted to free energies that report on each amide’s folding equilibrium by normalizing for the published exchange rates of amides in unstructured peptides (SI Appendix, SI Materials and Methods). Both V domains are stabilized by an average of 0.6 kcal/mol in the presence of 1 (500 μM; Fig. 5D), indicating that transient unfolding is reduced by binding of 1.
To verify that the kinetic stabilizers bind LCs secreted from cells analogously to recombinant FL LCs, we measured proteolysis of LCs secreted by ALMC2 cells, an AL patient-derived cell line (32). Purified ALMC2-derived LC (5 μM) is cleaved by PK (100 nM) with a rate constant of 3.83 × 10−4 s−1, whereas no proteolysis was detected after 1 h of incubation by PK in the presence of 1 (100 μM; Fig. 5E).
Inhibition of LC Aggregation.
FL LCs aggregate much less readily than their isolated V domains in vitro (4, 7). Although aggregation can be measured in vitro, it is not clear to what extent these experiments recapitulate processes that occur in patients, where the mechanism of aggregation is unknown. Assessment of the effect of kinetic stabilizers on FL LC aggregation at the hit stage of drug discovery is complicated by the denaturing acidic pH used to destabilize the FL LCs to promote aggregation in vitro, which can also reduce the binding affinity of the kinetic stabilizers, sometimes dramatically.
To assess the influence of 1 on FL LC aggregation, we used WIL-FL C214S, which forms weak dimers (KD = 16 µM; SI Appendix, Fig. S7), increasing its propensity to aggregate compared with WIL-FL, while retaining affinity for 1 (Fig. 2D). WIL-FL C214S aggregates upon vigorous stirring at or below pH 5 to form heterogenous aggregate structures (SI Appendix, Fig. S17). WIL-FL C214S aggregation kinetics assessed by fluorescence of the amyloid-binding dye thioflavin T demonstrates that 1 decreases the rate of aggregation significantly versus vehicle control, but the variability in these experiments is greater than desired (SI Appendix, Fig. S18). This may be because a given LC can stochastically form different aggregate structures with different kinetics.
Thus, we also quantified aggregation by measuring the rate of disappearance of soluble dimeric LC using size exclusion chromatography (SEC) (4). Binding of 1 (200 µM) to WIL-FL C214S (10 µM), which stabilizes the dimer and enhances kinetic stability (Fig. 5A), results in a larger proportion of soluble, dimeric LC eluting from the SEC column after 64 h of shaking at pH 5, which promotes aggregation (SI Appendix, Fig. S19). While individual WIL-FL C214S aggregation reactions exhibit variation in kinetics from run to run, aggregation in the presence of 1 (200 µM; Fig. 5F, dashed blue lines; n = 10) is generally slower than in the absence of 1 (Fig. 5F, red dashed lines; n = 10; SI Appendix, Fig. S19). Fitting all of the aggregation data in Fig. 5F to a single-exponential decay model reveals that WIL-FL C214S aggregates significantly more slowly in the presence of 1 (kagg = 0.011 h−1; Fig. 5F, bold blue curve) than in the absence of 1 (kagg = 0.066 h−1; Fig. 5F, bold red curve; P < 0.001, t test on log-transformed rates).
Discussion
The kinetic stabilizer strategy is a conservative approach, in that it blocks aggregation at the beginning of the amyloidogenicity cascade. Thus, success does not require knowledge about which nonnative LC structure(s) causes proteotoxicity. Through our high-throughput screen and characterization, we have identified several hit molecules that kinetically stabilize LCs by binding at the V-domain–V-domain interface in both FL LCs, and in the more dynamic V domains. In both cases, stabilization of dimeric LCs is achieved. Most, if not all, of our hits bind to a common, conserved site that is not present in the antibody Fab evaluated. FL LC stabilization reduces the rate at which LCs undergo conformational excursions that lead to either aggregation of FL LCs, or aberrant endoproteolysis and aggregation of LC fragments.
Our small-molecule hits exhibit a larger effect on protection against proteolysis, which is rate limited by unfolding and intrinsic protease activity, than on the apparent equilibrium stability or aggregation of LCs as assessed under denaturing conditions that reduce kinetic stabilizer binding affinity. We consider protection from proteolysis under physiological conditions to be a more useful parameter for optimization of more potent kinetic stabilizers than prevention of aggregation, since the relevance of in vitro aggregation to disease-associated aggregation is not yet clear. The identification of a class of fluorogenic kinetic stabilizers allows these tool compounds to be used for other studies on LCs (e.g., quantifying natively folded FL LC concentration in plasma). Optimization of these hit molecules utilizing structure-based design in combination with medicinal chemistry is expected to lead to potent and selective FL LC kinetic stabilizers, which should more dramatically inhibit LC aggregation.
It is not clear how much kinetic stabilization would be needed for a clinically significant outcome. The ability of kinetic stabilizers to reduce LC cardiotoxicity (45) will be explored once more potent kinetic stabilizers become available. We hypothesize that the majority of circulating LCs will need to be bound by small-molecule kinetic stabilizers exhibiting nanomolar affinities to achieve a maximal clinical response, so oral bioavailability and optimized pharmacokinetic and pharmacodynamic properties will likely be critical. The development of FL LC kinetic stabilizers with an excellent safety profile is a priority.
Edmundson et al. (42) identified regions within the interface between the V domains of an amyloidogenic FL LC, known as MCG, that could bind hydrophobic ligands. Brumshtein et al. (21) reported small molecules that bind to the MCG-V domain and prevent its aggregation (21). However, neither of the small molecules for which cocrystal structures were solved (sulfasalazine and methylene blue) were active in our PCFP assay employing WIL-FL* (SI Appendix, Fig. S20). It may be that their effects are specific for MCG-V and/or that their FL LC binding affinity is poor. The conformation of the isolated V-domain dimer in the sulfasalazine and methylene blue cocrystal structures is different from that of the V-domain–V-domain interface in FL MCG and FL JTO, suggesting that sulfasalazine and methylene blue support an alternative dimeric interface structure (SI Appendix, Fig. S20). Our screen did not presuppose a particular binding mode but nevertheless identified kinetic stabilizers that bind at the interface between V domains in FL LCs.
The PCFP assay utilized for our high-throughput screen is a convenient method for measuring protein stability in a high-throughput manner. Because the perturbation applied to the target protein, protease treatment, is irreversible, sample handling requirements are minimized and timing is simplified for FP quantification. To our knowledge, this approach has not been used previously to screen a large library of small molecules. The alternative ThermoFluor or thermal shift assay (46), which measures binding of a fluorogenic dye to partially unfolded conformations upon heating, was pursued. However, we were not able to generate a reliable FL LC screening assay using this methodology, although the ThermoFluor strategy has been used to identify small molecules that stabilize α-crystallin (47). That we were able to screen 650,000 compounds with minimal reagent costs and a straightforward workflow suggests that it should be possible to adapt the PCFP assay to find kinetic stabilizers for other proteinopathies, or small molecules that modulate protein–protein interactions. Use of a conjugation dye with longer excitation and emission wavelengths, and a longer fluorescence lifetime, should reduce assay interference from fluorescent small molecules in the library.
In AL, the current treatments aim to eradicate the clonal plasma cells that secrete FL LCs (2, 48). However, complete suppression of the production of the amyloidogenic LC (a complete hematologic response) is achieved in only 30–40% of the patients and most eventually relapse. Kinetic stabilization of LCs is unlikely to contribute to plasma cell death but could reduce organ proteotoxicity and the progression of AL. Patients with prominent cardiac involvement currently have few available options for treatment and represent an urgent unmet medical need, as they are often too sick to tolerate chemotherapy. Kinetic stabilizer pretreatment may allow these patients to tolerate chemotherapy. FL LC stabilization could also become a maintenance therapy upon recurrence of AL after treatment. Since reemergence of the clonal plasma cells is generally slow, organ toxicity caused by conformationally unstable circulating LCs can be minimized by kinetic stabilizer treatment (49). Our data suggest that LC kinetic stabilizers should bind to and stabilize the FL LCs of most patients because conserved residues comprise the kinetic stabilizer binding site. We envision that we can identify patients whose LCs are amenable to kinetic stabilization by measuring susceptibility of their LC in plasma to PK endoproteolysis in the presence and absence of the kinetic stabilizer, allowing for preselection of the patients most likely to benefit.
Materials and Methods
Detailed experimental procedures are provided in SI Appendix, SI Materials and Methods. Recombinant proteins were expressed in Escherichia coli and purified as previously described (4). WIL-FL K83C and JTO-FL K83C were conjugated with fluorescein maleimide (Vector Laboratories) and the labeled proteins, WIL-FL* and JTO-FL*, respectively, purified by ion exchange chromatography. From 400 mL of bacterial culture, we obtained ∼40 mg (1.7-µmol monomer equivalent) of pure, natively folded WIL-FL K79C. A total of 500 nmol of this LC was reacted with 2 equivalents of fluorescein maleimide, yielding 420 nmol of WIL-FL* after repurification. A total of 75 nmol of JTO-FL* was produced at similar overall yield. The screening campaign required 223 nmol of WIL-FL* and 40 nmol of JTO-FL*. Proteolysis assays were generally carried out by incubating LC (10 µM for unlabeled LCs, 20 nM for fluorescein-labeled LCs) with PK (200 nM; Thermo Fisher), in PBS at 37 °C or ambient temperature (22 °C) for screening. FP was measured using either a Jasco 8600 fluorimeter (λex = 488 nm; λem = 520 nm) or PerkinElmer EnVision plate reader equipped with polarizing excitation and emission filters (λex = 485 ± 20 nm; λem = 535 ± 20 nm) and a 505-nm dichroic mirror. FP values were calculated using the following formula:
where P is the polarization, FS and FP are the emission intensities of parallel and perpendicular-polarized light, respectively, and G is a dye- and instrument-specific correction factor. Values are reported as millipolarization units. For the high-throughput screen, 653,085 small molecules from the Scripps Florida screening library were assayed in 1,536-well plates. Final LC concentration was 10 nM in 5 µL. Small molecules were added by pintool from DMSO stock plates to a final concentration of 6.8 µM compound and 0.68% DMSO. Fluorogenic binding of 1 to LCs was measured using a Molecular Devices Gemini plate reader (λex = 373 nm; λem = 480 nm). Crystals of JTO-FL were grown via sitting-drop vapor diffusion using a crystallization buffer consisting of 20% PEG 3350 and 0.2 M NH4H2PO4 at 23 °C. To generate JTO-FL•1 complexes, crystals of apo JTO-FL were soaked with a 10-fold molar excess of 1 for 10 d. Diffraction data were collected at 80 K and a wavelength of 1.0332 Å at beamline 23-ID-D (for apo JTO-FL) or 23-ID-B (for JTO-FL•1) at the Advanced Photon Source. The refined models were deposited in the Protein Data Bank under accession codes 6MG4 for apo JTO-FL and 6MG5 for JTO-FL•1. All samples for NMR were buffered in 50 mM Bis Tris, pH 6.4, 1 mM EDTA, 10% D2O. 1H,15N experiments were recorded at 37 °C on a 14.1-T Bruker AVANCE III HD spectrometer equipped with a cryogenically cooled x,y,z gradient probe. SV-AUC, unfolding, and aggregation experiments were performed as previously described (4).
Supplementary Material
Acknowledgments
We thank Dr. Marina Ramirez-Alvarado for κ LC expression constructs and advice on measuring LC aggregation. This work was supported by the NIH Grants DK46335 and UL1TR002550 (to J.W.K.), the Skaggs Institute for Chemical Biology, and the Lita Annenberg Hazen Foundation. D.E.M. was supported by the George E. Hewitt Foundation for Medical Research. The National Institute of General Medical Sciences and National Cancer Institute Structural Biology Facility at the Advanced Photon Source (GM/CA@APS) has been funded in whole or in part with federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006). This research used resources of the Advanced Photon Source, a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357. The Eiger 16M detector was funded by NIH–Office of Research Infrastructure Programs, High-End Instrumentation Grant 1S10OD012289-01A1. L.E.K. acknowledges funding from the Canadian Institutes of Health Research and the Natural Science and Engineering Research Foundation of Canada. L.E.K. holds a Canada Research Chair in Biochemistry.
Footnotes
Conflict of interest statement: G.J.M., N.L.Y., and J.W.K. have submitted a patent application for the small-molecule kinetic stabilizers of Ig light chains.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 6MG4 (apo JTO) and 6MG5 (JTO•1)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817567116/-/DCSupplemental.
References
- 1.Sipe JD, et al. Amyloid fibril proteins and amyloidosis: Chemical identification and clinical classification International Society of Amyloidosis 2016 Nomenclature Guidelines. Amyloid. 2016;23:209–213. doi: 10.1080/13506129.2016.1257986. [DOI] [PubMed] [Google Scholar]
- 2.Merlini G, et al. Systemic immunoglobulin light chain amyloidosis. Nat Rev Dis Primers. 2018;4:38. doi: 10.1038/s41572-018-0034-3. [DOI] [PubMed] [Google Scholar]
- 3.Schiffer M, Girling RL, Ely KR, Edmundson AB. Structure of a λ-type Bence-Jones protein at 3.5-Å resolution. Biochemistry. 1973;12:4620–4631. doi: 10.1021/bi00747a013. [DOI] [PubMed] [Google Scholar]
- 4.Morgan GJ, Kelly JW. The kinetic stability of a full-length antibody light chain dimer determines whether endoproteolysis can release amyloidogenic variable domains. J Mol Biol. 2016;428:4280–4297. doi: 10.1016/j.jmb.2016.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberti L, et al. Concurrent structural and biophysical traits link with immunoglobulin light chains amyloid propensity. Sci Rep. 2017;7:16809. doi: 10.1038/s41598-017-16953-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez-Alvarado M. Amyloid formation in light chain amyloidosis. Curr Top Med Chem. 2012;12:2523–2533. doi: 10.2174/1568026611212220007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blancas-Mejía LM, et al. Thermodynamic and fibril formation studies of full length immunoglobulin light chain AL-09 and its germline protein using scan rate dependent thermal unfolding. Biophys Chem. 2015;207:13–20. doi: 10.1016/j.bpc.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurle MR, Helms LR, Li L, Chan W, Wetzel R. A role for destabilizing amino acid replacements in light-chain amyloidosis. Proc Natl Acad Sci USA. 1994;91:5446–5450. doi: 10.1073/pnas.91.12.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan GJ, Usher GA, Kelly JW. Incomplete refolding of antibody light chains to non-native, protease-sensitive conformations leads to aggregation: A mechanism of amyloidogenesis in patients? Biochemistry. 2017;56:6597–6614. doi: 10.1021/acs.biochem.7b00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glenner GG, et al. Creation of “amyloid” fibrils from Bence Jones proteins in vitro. Science. 1971;174:712–714. doi: 10.1126/science.174.4010.712. [DOI] [PubMed] [Google Scholar]
- 11.Glenner GG, Harbaugh J, Ohma JI, Harada M, Cuatrecasas P. An amyloid protein: The amino-terminal variable fragment of an immunoglobulin light chain. Biochem Biophys Res Commun. 1970;41:1287–1289. doi: 10.1016/0006-291x(70)90227-5. [DOI] [PubMed] [Google Scholar]
- 12.Lavatelli F, et al. Amyloidogenic and associated proteins in systemic amyloidosis proteome of adipose tissue. Mol Cell Proteomics. 2008;7:1570–1583. doi: 10.1074/mcp.M700545-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olsen KE, Sletten K, Westermark P. Fragments of the constant region of immunoglobulin light chains are constituents of AL-amyloid proteins. Biochem Biophys Res Commun. 1998;251:642–647. doi: 10.1006/bbrc.1998.9508. [DOI] [PubMed] [Google Scholar]
- 14.Bulawa CE, et al. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci USA. 2012;109:9629–9634. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berk JL, et al. Diflunisal Trial Consortium Repurposing diflunisal for familial amyloid polyneuropathy: A randomized clinical trial. JAMA. 2013;310:2658–2667. doi: 10.1001/jama.2013.283815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coelho T, et al. Tafamidis for transthyretin familial amyloid polyneuropathy: A randomized, controlled trial. Neurology. 2012;79:785–792. doi: 10.1212/WNL.0b013e3182661eb1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenblum H, et al. TTR (transthyretin) stabilizers are associated with improved survival in patients with TTR cardiac amyloidosis. Circ Heart Fail. 2018;11:e004769. doi: 10.1161/CIRCHEARTFAILURE.117.004769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coelho T, et al. Long-term effects of tafamidis for the treatment of transthyretin familial amyloid polyneuropathy. J Neurol. 2013;260:2802–2814. doi: 10.1007/s00415-013-7051-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maurer MS, et al. ATTR-ACT Study Investigators Tafamidis treatment for patients with transthyretin amyloid cardiomyopathy. N Engl J Med. 2018;379:1007–1016. doi: 10.1056/NEJMoa1805689. [DOI] [PubMed] [Google Scholar]
- 20.Hammarström P, Wiseman RL, Powers ET, Kelly JW. Prevention of transthyretin amyloid disease by changing protein misfolding energetics. Science. 2003;299:713–716. doi: 10.1126/science.1079589. [DOI] [PubMed] [Google Scholar]
- 21.Brumshtein B, et al. Inhibition by small-molecule ligands of formation of amyloid fibrils of an immunoglobulin light chain variable domain. eLife. 2015;4:e10935. doi: 10.7554/eLife.10935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rennella E, Morgan GJ, Kelly JW, Kay LE. Role of domain interactions in the aggregation of full-length immunoglobulin light chains. Proc Natl Acad Sci USA. 2019;116:854–863. doi: 10.1073/pnas.1817538116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wall J, et al. Thermodynamic instability of human λ 6 light chains: Correlation with fibrillogenicity. Biochemistry. 1999;38:14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 24.Bolger R, Checovich W. A new protease activity assay using fluorescence polarization. Biotechniques. 1994;17:585–589. [PubMed] [Google Scholar]
- 25.Kabat EA. Sequences of Proteins of Immunological Interest. 5th Ed Department of Health and Human Services, National Institutes of Health; Bethesda: 1991. [Google Scholar]
- 26.McGovern SL, Caselli E, Grigorieff N, Shoichet BK. A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening. J Med Chem. 2002;45:1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]
- 27.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 28.Barik A, Kumbhakar M, Nath S, Pal H. Evidence for the TICT mediated nonradiative deexcitation process for the excited coumarin-1 dye in high polarity protic solvents. Chem Phys. 2005;315:277–285. [Google Scholar]
- 29.Wolwertz ML, Nguyen PT, Quittot N, Bourgault S. Probing the role of λ6 immunoglobulin light chain dimerization in amyloid formation. Biochim Biophys Acta. 2016;1864:409–418. doi: 10.1016/j.bbapap.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 30.Perfetti V, et al. Evidence that amyloidogenic light chains undergo antigen-driven selection. Blood. 1998;91:2948–2954. [PubMed] [Google Scholar]
- 31.Bodi K, et al. AL-base: A visual platform analysis tool for the study of amyloidogenic immunoglobulin light chain sequences. Amyloid. 2009;16:1–8. doi: 10.1080/13506120802676781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arendt BK, et al. Biologic and genetic characterization of the novel amyloidogenic lambda light chain-secreting human cell lines, ALMC-1 and ALMC-2. Blood. 2008;112:1931–1941. doi: 10.1182/blood-2008-03-143040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kourelis TV, et al. Clarifying immunoglobulin gene usage in systemic and localized immunoglobulin light-chain amyloidosis by mass spectrometry. Blood. 2017;129:299–306. doi: 10.1182/blood-2016-10-743997. [DOI] [PubMed] [Google Scholar]
- 34.Sikkink LA, Ramirez-Alvarado M. Salts enhance both protein stability and amyloid formation of an immunoglobulin light chain. Biophys Chem. 2008;135:25–31. doi: 10.1016/j.bpc.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sekijima Y, Dendle MA, Kelly JW. Orally administered diflunisal stabilizes transthyretin against dissociation required for amyloidogenesis. Amyloid. 2006;13:236–249. doi: 10.1080/13506120600960882. [DOI] [PubMed] [Google Scholar]
- 36.Palladini G, et al. New criteria for response to treatment in immunoglobulin light chain amyloidosis based on free light chain measurement and cardiac biomarkers: Impact on survival outcomes. J Clin Oncol. 2012;30:4541–4549. doi: 10.1200/JCO.2011.37.7614. [DOI] [PubMed] [Google Scholar]
- 37.Hong M, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgan GJ, et al. 2019 Structure of full-length human lambda-6A light chain JTO in complex with coumarin 1. Protein Data Bank. Available at https://www.rcsb.org/structure/6MG5. Deposited September 13, 2018.
- 39.Morgan GJ, et al. 2019 Structure of full-length human lambda-6A light chain JTO. Protein Data Bank. Available at https://www.rcsb.org/structure/6MG4. Deposited September 13, 2018.
- 40.Huang DB, Ainsworth C, Solomon A, Schiffer M. Pitfalls of molecular replacement: The structure determination of an immunoglobulin light-chain dimer. Acta Crystallogr D Biol Crystallogr. 1996;52:1058–1066. doi: 10.1107/S090744499600813X. [DOI] [PubMed] [Google Scholar]
- 41.Pokkuluri PR, Solomon A, Weiss DT, Stevens FJ, Schiffer M. Tertiary structure of human lambda 6 light chains. Amyloid. 1999;6:165–171. doi: 10.3109/13506129909007322. [DOI] [PubMed] [Google Scholar]
- 42.Edmundson AB, et al. Binding of 2,4-dinitrophenyl compounds and other small molecules to a crystalline lambda-type Bence-Jones dimer. Biochemistry. 1974;13:3816–3827. doi: 10.1021/bi00715a031. [DOI] [PubMed] [Google Scholar]
- 43.Kloiber K, Spitzer R, Grutsch S, Kreutz C, Tollinger M. Longitudinal exchange: An alternative strategy towards quantification of dynamics parameters in ZZ exchange spectroscopy. J Biomol NMR. 2011;51:123–129. doi: 10.1007/s10858-011-9547-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuwen T, Kay LE, Bouvignies G. Dramatic decrease in CEST measurement times using multi-site excitation. ChemPhysChem. 2018;19:1707–1710. doi: 10.1002/cphc.201800249. [DOI] [PubMed] [Google Scholar]
- 45.Liao R, et al. Infusion of light chains from patients with cardiac amyloidosis causes diastolic dysfunction in isolated mouse hearts. Circulation. 2001;104:1594–1597. [PubMed] [Google Scholar]
- 46.Pantoliano MW, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 47.Makley LN, et al. Pharmacological chaperone for α-crystallin partially restores transparency in cataract models. Science. 2015;350:674–677. doi: 10.1126/science.aac9145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milani P, Merlini G, Palladini G. Novel therapies in light chain amyloidosis. Kidney Int Rep. 2017;3:530–541. doi: 10.1016/j.ekir.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palladini G, et al. Presentation and outcome with second-line treatment in AL amyloidosis previously sensitive to nontransplant therapies. Blood. 2018;131:525–532. doi: 10.1182/blood-2017-04-780544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.