Significance
For achieving efficient entry into host cells, viruses utilize multiple host factors for mediating the stepwise entry process. Sole expression of sodium taurocholate cotransporting polypeptide (NTCP), a cellular receptor for hepatitis B virus (HBV), is not sufficient for the efficient viral internalization into hepatocytes. Here, we identify epidermal growth factor receptor (EGFR) as a host factor that interacts with NTCP and mediates HBV internalization. Dissociation of the NTCP–EGFR interaction or functional depletion of EGFR attenuated the viral internalization to NTCP-expressing cells and infection. Our data suggest that HBV enters human hepatocytes using an intrinsic NTCP–EGFR complex as a driving force. EGFR is thus an entry cofactor required for HBV infection of the human liver.
Keywords: HBV, NTCP, EGFR, entry, transporter
Abstract
Sodium taurocholate cotransporting polypeptide (NTCP) is a host cell receptor required for hepatitis B virus (HBV) entry. However, the susceptibility of NTCP-expressing cells to HBV is diverse depending on the culture condition. Stimulation with epidermal growth factor (EGF) was found to potentiate cell susceptibility to HBV infection. Here, we show that EGF receptor (EGFR) plays a critical role in HBV virion internalization. In EGFR-knockdown cells, HBV or its preS1-specific fluorescence peptide attached to the cell surface, but its internalization was attenuated. PreS1 internalization and HBV infection could be rescued by complementation with functional EGFR. Interestingly, the HBV/preS1–NTCP complex at the cell surface was internalized concomitant with the endocytotic relocalization of EGFR. Molecular interaction between NTCP and EGFR was documented by immunoprecipitation assay. Upon dissociation from functional EGFR, NTCP no longer functioned to support viral infection, as demonstrated by either (i) the introduction of NTCP point mutation that disrupted its interaction with EGFR, (ii) the detrimental effect of decoy peptide interrupting the NTCP–EGFR interaction, or (iii) the pharmacological inactivation of EGFR. Together, these data support the crucial role of EGFR in mediating HBV–NTCP internalization into susceptible cells. EGFR thus provides a yet unidentified missing link from the cell-surface HBV–NTCP attachment to the viral invasion beyond the host cell membrane.
Hepatitis B virus (HBV), an enveloped virus carrying three envelope proteins [small, middle, and large surface proteins (LHBs)], infects humans, chimpanzees, and tupaias with a selective tropism to the liver (1). HBV infection is initiated by interaction between the viral envelope proteins and host factors on a cell surface through heparan sulfate proteoglycan, followed by specific virus attachment to its host receptor, sodium taurocholate cotransporting polypeptide (NTCP). Subsequent viral internalization into cells and membrane fusion proceed through an endocytosis-dependent manner. However, it has been largely unknown how virus–cell attachment triggers internalization and which host factor (or factors) mediates this process. Clarification of this entry process is essential for a better understanding of the HBV life cycle, its host tropism, and the eventual formation of covalently closed circular DNA (cccDNA) in the nucleus, which serves as a template for HBV replication (2–4).
NTCP was identified as a host receptor essential for HBV entry (5). Expression of NTCP confers HBV susceptibility to human hepatic cell lines such as HepG2, Huh7, or undifferentiated HepaRG cells, which are originally nonsusceptible to infection (6, 7). Nonetheless, some human-derived hepatic or nonhepatic cell lines remain resistant to HBV entry, even though NTCP overexpression enabled viral attachment to the cell surface (8–10). Furthermore, NTCP-complemented HepG2 cells were diversely susceptible to HBV infection, depending on the culture condition, despite sustained levels of NTCP expression (SI Appendix, Fig. S1B). These observations likely indicate that NTCP expression is not sufficient for supporting robust HBV infection. In this study, we identify that epidermal growth factor (EGF) receptor (EGFR) is a host cofactor that is required for NTCP’s viral receptor function and determines the host cell’s ability to support viral internalization.
Results
EGFR Is Critically Involved in HBV/HDV Infection.
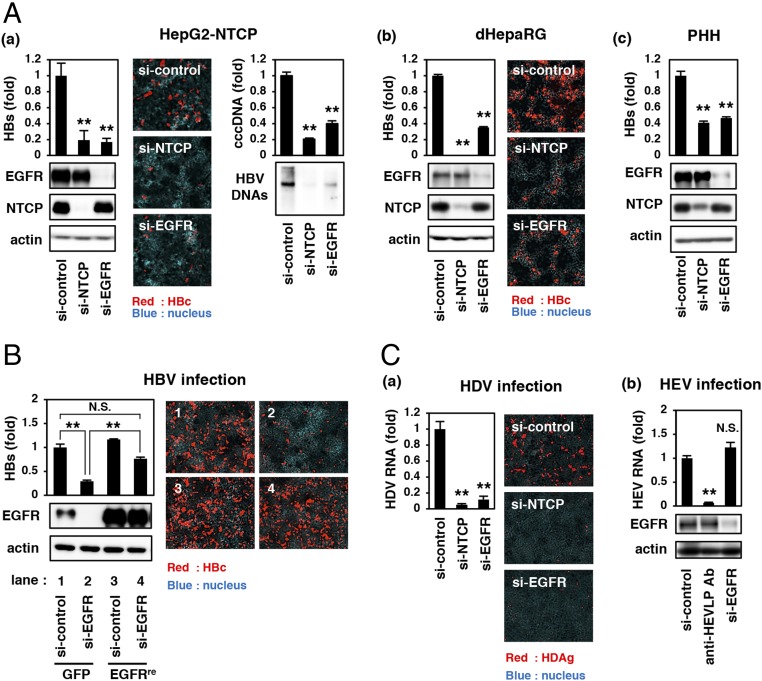
From a screening of bioactive ligands (Materials and Methods), we found that EGF clearly augmented the susceptibility of NTCP-complemented cells (HepG2-NTCP cells) to HBV infection, although the expression of NTCP was not affected (SI Appendix, Fig. S1B). We then examined the role of EGF-related host factor(s) in HBV infection. Contribution of EGFR on HBV infection was evaluated by using siRNA knockdown. As shown in Fig. 1A, siRNA-mediated knockdown of endogenous EGFR drastically reduced HBV infection to HepG2-NTCP cells (Fig. 1 A, a). A similar observation was obtained in differentiated HepaRG (dHepaRG) cells and primary human hepatocytes (PHHs) (Fig. 1 A, b and c). Cell viability, NTCP transporter activity, and cell-surface NTCP expression were not reduced by EGFR knockdown (SI Appendix, Fig. S2). To exclude the possibility of an siRNA off-target effect, we transduced EGFR cDNA resistant to si-EGFR (EGFRre) by lentivirus vector and found that the HBV susceptibility of si-EGFR–treated HepG2-NTCP cells was significantly rescued by complementation with EGFRre (Fig. 1B, lane 2 vs. lane 4), which reached no significant difference in HBV infection of the original cells (Fig. 1B, lane 1 vs. lane 4). These results suggest that EGFR is critically involved in the process of HBV infection. To address the specific role of the EGFR in viral infection, we examined the contribution of EGFR to the infection by other two viruses, hepatitis D and E viruses (HDV and HEV, respectively). As shown in Fig. 1C, knockdown of EGFR significantly reduced the infection of HDV, but not HEV (Fig. 1C).
Fig. 1.
Critical role of EGFR in supporting the infection by HBV and HDV. (A, a–c) HepG2-NTCP cells (a), dHepaRG cells (b), and PHHs (c) transfected with siRNA against NTCP (si-NTCP), siRNA against EGFR (si-EGFR), or nonrelevant siRNA (si-control) were used for HBV infection assay and examined by detecting extracellular HBs (Upper Left graphs in a–c), intracellular HBc (Center images in a and b), cccDNA (Upper Right graph in a), and HBV DNAs (Lower Right in a). Protein production of EGFR, NTCP, and actin are also shown (Lower Left in a–c). (B) HepG2-NTCP cells that were transduced with a lentiviral vector expressing siRNA-resistant EGFR (EGFRre) or GFP as a control and then transfected with si-control or si-EGFR were evaluated for HBV infection by detecting extracellular HBs (Upper Left graph) and intracellular HBc (Right images). EGFR and actin proteins are also shown (Lower Left). (C, a and b) HepG2-NTCP cells (prepared as in A, a) (a) or PLC/PRF/5 cells treated with the indicated agents (b) were subjected to the infection assay for HDV (a) or HEV (b). An anti-HEVLP antibody was used for a positive control for HEV entry inhibition. Statistical significance was determined using Student’s t test, **P < 0.01. N.S., not significant.
EGFR Knockdown Attenuates Virus Internalization.
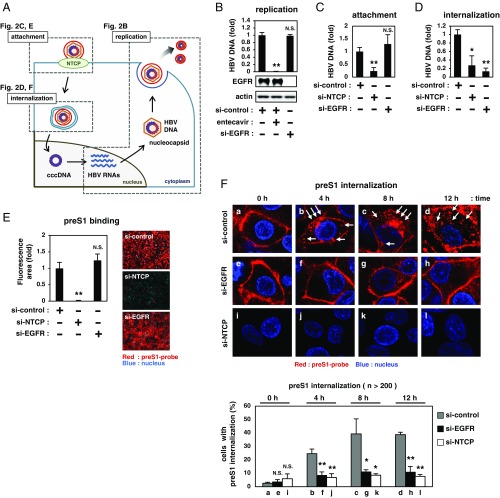
We next investigated which step in the HBV life cycle was affected by depletion of EGFR. HBV attaches to hepatocytes and subsequently internalizes into cells through endocytosis, which eventually leads to the formation of cccDNA in the nucleus; cccDNA produces HBV RNAs and drives HBV replication to produce HBV particles (Fig. 2A) (4). During the life cycle, the activity of HBV replication was not affected by depletion of EGFR (Fig. 2B), as demonstrated using HepG2.2.15.7 cells (which autonomously replicate HBV but do not support HBV attachment and internalization) (11, 12). While viral attachment was not decreased in EGFR-knockdown cells (Fig. 2C), the level of HBV DNA internalized into cells was significantly reduced by knockdown of EGFR (Fig. 2D). These data are consistent with the results that EGFR depletion blocked the infection by both HBV and HDV (Fig. 1C), two viruses that share the same mechanisms during the viral internalization process. The amino-terminal preS1 region of the LHBs, especially amino acids 2 to 48, is essential for HBV entry through NTCP attachment, and a peptide consisting of this region labeled with fluorescence is frequently used for the analysis of HBV/HDV entry (5, 13, 14). This tetramethylrhodamine-labeled myristoylated preS1 (amino acids 2 to 48) peptide (preS1-probe) still bound to the cells under depletion of EGFR, in contrast to its complete loss of binding to the NTCP-knockdown cells (Fig. 2E), in agreement with the result of the attachment assay (Fig. 2C). To examine the process after attachment, we observed incorporation of preS1-probe using high-magnification confocal microscopy, as previously reported (15). After preS1-probe attachment to the cell surface at 4 °C and washing out free probe (indicated as 0 h in Fig. 2F), the cells were transferred to 37 °C to allow preS1-probe incorporation up to 12 h. In HepG2-NTCP cells transfected with nonrelevant siRNA (si-control), preS1-probe was incorporated inside the cells, with the formation of speckled patterns increasing with incubation time (Fig. 2 F, Upper, a–d) as reported (8, 15). In contrast, EGFR-knockdown cells showed a much lower frequency of preS1 incorporation, with retaining the plasma membrane localization at least up to 12 h (Fig. 2 F, Upper, e–h). The quantified number of cells showing preS1 incorporation was significantly decreased upon EGFR depletion (Fig. 2 F, Lower). These results clearly suggest that EGFR plays a significant role in HBV internalization.
Fig. 2.
EGFR is involved in HBV internalization. (A) HBV life cycle. HBV attaches to the cell surface through NTCP (attachment), followed by internalization inside cells (internalization) and translocation into the nucleus to form cccDNA. cccDNA drives HBV replication by transcription of HBV RNAs, nucleocapsid formation, and secretion of HBV particles (replication). Evaluation of the activity of each step is shown in C and E (attachment), D and F (internalization), and B (replication). (B) HBV replication was evaluated by quantifying extracellular HBV DNA from HepG2.2.15.7 cells transfected with the indicated siRNA at 9 d posttransfection. Entecavir was used as a positive control to suppress HBV replication. (C) For evaluating viral attachment, HepG2-NTCP cells transfected with siRNAs (as indicated in Fig. 1 A, a) were exposed to HBV at 4 °C to allow viral attachment to the cell surface. After washing, cell-surface HBV DNA was quantified. (D) For quantifying the internalized HBV, HBV-attached cells (prepared as in C) were transferred to 37 °C to allow viral internalization into the cells and to quantify intracellular HBV DNA. (E) HepG2-NTCP cells transfected with siRNAs (as in Fig. 1 A, a) were exposed to preS1-probe (Right images). The fluorescence intensities are shown in the graph (Left). (F) siRNA-transfected HepG2-NTCP cells were attached to preS1-probe at 4 °C for 1 h, washed of free preS1-probe, and then transferred to 37 °C to allow incorporation of preS1-probe up to 12 h. Cells were observed using high-magnification confocal microscopy for preS1-probe internalization (red) after 0, 4, 8, and 12 h of incubation at 37 °C (Upper). White arrows show the internalized preS1 speckles (b–d). Percentages of cells exhibiting preS1 internalization are indicated in the graph (Lower) (see Materials and Methods).
EGFR Relocalization Triggers the preS1–NTCP Internalization.
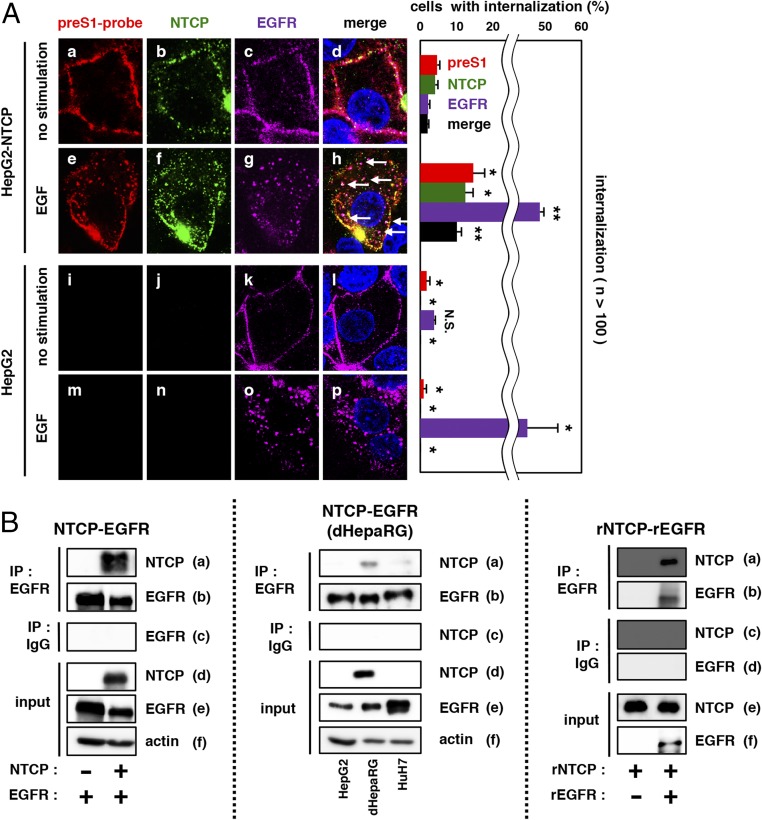
EGFR is a receptor tyrosine kinase functionally regulated by its phosphorylation that is induced by ligands such as EGF (16). EGF stimulation induces EGFR activation through its dimerization and autophosphorylation, triggering downstream signaling such as the mitogen-activated protein kinase and phosphatidylinositol 3-kinase (PI3K) pathways and inducing the endocytosis of EGFR itself (17). In preS1-attached cells, EGF also triggered EGFR relocalization with speckled patterns within as early as 30 min after stimulation (Fig. 3 A, Left, c vs. g). Interestingly, EGF stimulation also induced a rapid relocalization of preS1-probe and NTCP to the speckled distribution in parallel with EGFR endocytosis such that preS1-probe, NTCP, and EGFR were colocalized (Fig. 3 A, Left, h, white arrows and Right). A coimmunoprecipitation assay showed that NTCP overproduced in 293T cells was copurified with EGFR (Fig. 3 B, Left, a). Coprecipitation of endogenous NTCP with EGFR was also observed in HepaRG cells (Fig. 3 B, Center, a). Recombinant NTCP (18) also coprecipitated with recombinant EGFR in vitro (Fig. 3 B, Right, a). Wang et al. (19) reported that NTCP colocalized and comigrated with EGFR during EGF-induced trafficking in the absence of HBV, consistent with our data. These results raise the possibility that HBV utilizes the intrinsic EGFR endocytosis pathway for viral internalization, which should be further analyzed in the future. The above data suggest that EGFR relocalization triggers the preS1–NTCP internalization.
Fig. 3.
EGFR relocalization triggers the preS1–NTCP internalization. (A) HepG2-NTCP or HepG2 cells attached with preS1-probe at 4 °C were stimulated with EGF or left unstimulated (no stimulation) at 37 °C for 30 min and observed by confocal microscopy (preS1-probe, red; NTCP, green; EGFR, purple; nucleus, blue). White arrows indicate the colocalization of preS1, NTCP, and EGFR (Left, d and h). The percentages of cells showing the internalization for preS1, NTCP, and EGFR are indicated in the graph (Right). **P < 0.01; *P < 0.05. N.S., not significant. (B, Left) 293T cells overexpressing EGFR together with or without NTCP were harvested to immunoprecipitate with anti-EGFR antibody (IP: EGFR) or normal mouse IgG as a negative control (IP: IgG) and to detect NTCP or EGFR in the precipitates. NTCP, EGFR, and actin in the total cell lysate were also detected (input). (B, Center) HepG2, dHepaRG, and Huh7 cells were subjected to coimmunoprecipitation assay for endogenous protein interaction. (B, Right) Recombinant NTCP (rNTCP) was incubated with or without recombinant EGFR (rEGRF) in vitro, which was subjected to coimmunoprecipitation analysis as shown above.
Functional Interaction with EGFR Is Required for NTCP Receptor Function.
Based on the above results, we hypothesized that EGFR mediates HBV–NTCP internalization after HBV attachment to the cell surface. To address whether EGFR is essential for HBV internalization, we performed loss-of-function analyses using three different approaches: mutation, competition, and pharmacological inhibition.
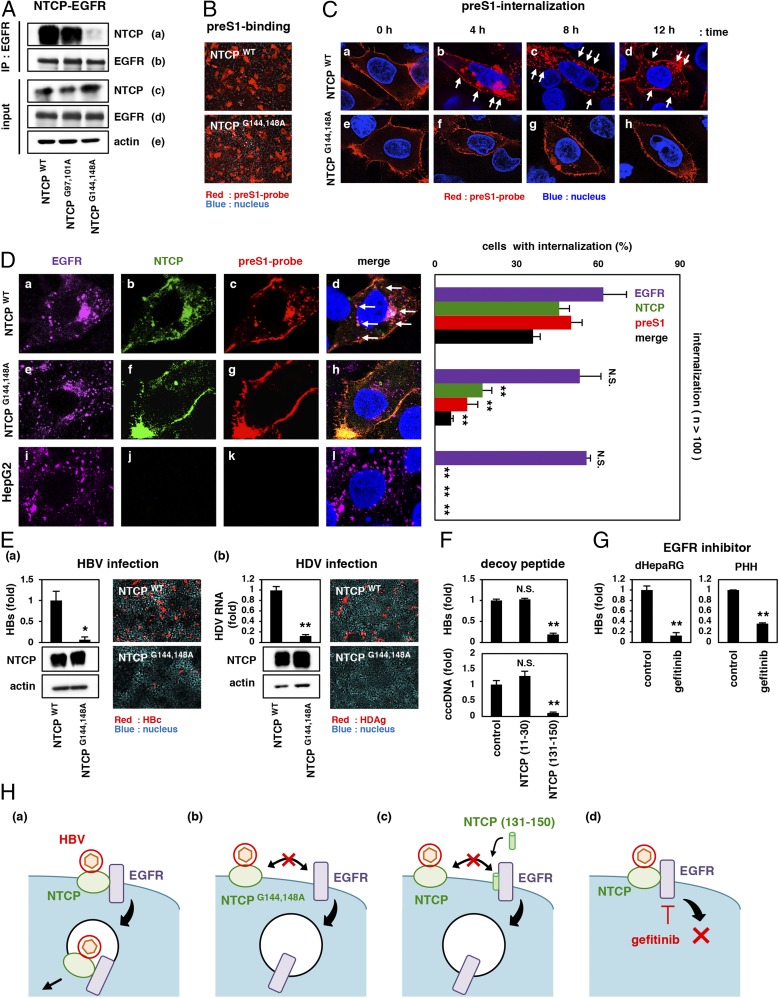
First, we determined the region of NTCP responsible for the interaction with EGFR (Materials and Methods and SI Appendix, Fig. S3A). By screening the NTCP fragment peptides consisting of 20 aa, we found that the amino acid 131 to 150 region [NTCP (131–150)] showed the highest binding signal to EGFR (SI Appendix, Fig. S3 A and B). Further mutagenesis analysis revealed that upon replacement of Gly with Ala at amino acids 144 and 148 of NTCP (NTCPG144,148A), NTCP lost the ability to bind to EGFR (Fig. 4 A, a). Notably, expression of NTCPG144,148A rendered the level of preS1 binding (Fig. 4B) and bile acid transport to cells (SI Appendix, Fig. S4) equivalent to that of NTCPWT. However, preS1 internalization into NTCPG144,148A-expressing cells was less observed than that in NTCPWT-producing cells (Fig. 4 C, b–d vs. f–h). In both of these cells, EGF stimulation induced the endocytosis of EGFR at 30 min after stimulation (Fig. 4 D, a and e), but the internalization of preS1 and NTCP was significantly attenuated in NTCPG144,148A-expressing cells (Fig. 4 D, f and g), in contrast to the colocalization of EGFR, NTCP, and preS1 in speckles in NTCPWT-expressing cells (Fig. 4 D, d, white arrows). This loss of EGFR–NTCP interaction, which dissociated preS1–NTCP localization from EGFR, resulted in the reduction in the infection by both HBV and HDV (Fig. 4 E). This result was supported by a competition analysis using NTCP (131–150). As shown in SI Appendix, Fig. S3C, excess amounts of NTCP (131–150) peptide interfered with the NTCP–EGFR coprecipitation (SI Appendix, Fig. S3 C, a). Introduction of this NTCP (131–150) as a decoy peptide similarly attenuated the EGF-induced internalization of preS1–NTCP (SI Appendix, Fig. S5 B, j and k), without affecting the preS1–cell binding itself (SI Appendix, Fig. S5A). This decoy peptide thereby remarkably reduced the HBV infection (Fig. 4F). Moreover, pharmacological inactivation of EGFR by treatment with a known inhibitor, gefitinib (20), also attenuated the preS1 internalization (SI Appendix, Fig. S6 A, g) and thus reduced HBV infection in both dHepaRG cells and PHHs (Fig. 4G). Based on the above data, we concluded that the functional interaction with EGFR is required for NTCP’s ability to support viral infection.
Fig. 4.
Functional interaction with EGFR is required for NTCP’s ability to mediate viral infection. (A) 293T cells overproducing EGFR and either NTCPWT, NTCPG97,101A, or NTCPG144,148A were lysed and immunoprecipitated with an anti-EGFR antibody (IP: EGFR) or were recovered without immunoprecipitation (input). NTCP, EGFR, and actin were detected. (B and C) Huh7 (B) or HepG2 (C) cells overexpressing either NTCPWT or NTCPG144,148A were examined for preS1 attachment (B) as well as preS1 internalization at the indicated time points (C) as in Fig. 2 E and F. The white arrows show internalized preS1 vesicles in C. (D) EGFR (purple), NTCP (green), preS1-probe (red), and the nucleus (blue) were detected in HepG2-NTCPWT, HepG2-NTCPG144,148A, and HepG2 cells after 30 min of EGF stimulation. White arrows indicate the colocalization of preS1, NTCP, and EGFR (Left, d). The percentages of cells showing the internalization of preS1, NTCP, and EGFR are indicated on the graph (Right). (E, a and b) HepG2-NTCPWT and HepG2-NTCPG144,148A cells (a) as well as Huh7-NTCPWT and Huh7-NTCPG144,148A cells (b) were used for the infection assays with HBV and HDV, respectively, as in Fig. 1. (F and G) HBV infection assay using HepG2-NTCP (F) and dHepaRG cells and PHH (G) upon treatment with or without the indicated compounds [NTCP (11–30) or NTCP (131–150) in F; gefitinib in G]. HBs levels (F and G) and cccDNA levels (F) were detected to evaluate HBV infection. (H, a–d) Proposed model for HBV–NTCP–EGFR internalization. HBV attaches to NTCP on the cell surface and is recruited to the NTCP–EGFR complex, which dynamically translocates from the cell surface to the intramembrane vesicles for viral internalization (a). Dissociation of the NTCP–EGFR interaction either by introducing point mutations in NTCP (b), by binding competition with a decoy peptide (c), or by functional inactivation of EGFR (d), deprives NTCP of the ability to support HBV infection. Thus, EGFR plays a critical role in mediating the entry process after HBV–NTCP attachment. **P < 0.01; *P < 0.05. N.S., not significant.
Discussion
EGFR shuttles between the cell-surface plasma membrane and the endosomes through membrane trafficking machinery (17). In this study, the HBV preS1 ligand that was bound to the HBV receptor NTCP at the cell surface was shown to be internalized along with EGFR. We therefore propose a model for internalization of HBV as shown in Fig. 4 H, a, in which HBV interacts with NTCP at the cell surface to be recruited to NTCP–EGFR complex that dynamically translocates between the plasma membrane and intracellular vesicles. By taking advantage of the interaction with EGFR, HBV is internalized into intracellular vesicles, thereby acquiring the opportunity for subsequent acidification-dependent membrane fusion leading to the release of the nucleocapsid into the cytoplasm (Fig. 4 H, a). When NTCP is prevented from interacting with EGFR, either by introducing mutations in NTCP (Fig. 4 H, b), by masking the binding interface with a decoy peptide (Fig. 4 H, c), or by inactivation of EGFR (Fig. 4 H, d), cells no longer support HBV entry. Thus, NTCP’s viral receptor function appears dependent on a functional interaction with EGFR. Activated EGFR triggers the downstream signaling, including Ras and PI3K, and induces the endocytosis of EGFR itself (17). It is of particular importance to understand whether the downstream signaling or the EGFR endocytosis machinery is essential for mediating HBV internalization.
The NTCP–EGFR interaction likely occurs irrespective of the presence of HBV (Fig. 3B) (19). What is the original biological significance of the NTCP–EGFR interaction for uninfected cells? Transporter activity and cell-surface expression of NTCP were not affected by the knockdown of EGFR in the experiments in SI Appendix, Fig. S2 B and C. NTCP plays an important role for constituting the enterohepatic circulation of bile acids through transporting bile acids into hepatocytes (21), a phenomenon that should be tightly and rapidly regulated depending on the concentration of bile acids. It is possible that EGFR contributes to the rapid regulation of cell-surface NTCP levels by responding to the change in serum bile acid levels. It has been reported that the exposure of bile acids at high concentration induced the phosphorylation of EGFR and its relocalization from the cell surface (22, 23). Given that the phosphorylation of EGFR induces its endocytosis as rapidly as within 5 min (24), the NTCP–EGFR association may help rapid down-regulation of cell-surface NTCP by sensing high concentrations of bile acids and preventing excess bile acid transport to cells and subsequent cytotoxicity. Further analysis should shed light on a novel regulation system for bile acid homeostasis that involves EGFR.
In this study, we showed that a decoy peptide [NTCP (131–150)] and gefitinib clearly inhibited HBV infection (Fig. 4 F and G). It is suggested that the functional interaction of EGFR and NTCP represents a target for the development of anti-HBV agents (SI Appendix, Fig. S7). Precise analysis of the mode of the NTCP–EGFR interaction should be further analyzed in the future. Because EGFR has been the target of anticancer agents (25), it would be of interest to analyze the effect of such drugs on HBV/HDV infection. Thus, our findings have a significant impact not only for understanding the molecular basis for HBV/HDV infection, but also for the development of antiviral drugs.
Materials and Methods
HBV used in this study was derived from Hep38.7-Tet cells (genotype D) (6, 11). PHHs, dHepaRG, and HepG2-NTCP were infected with HBV as previously described (6). HBV infection was evaluated by detecting HBV surface protein (HBs), HBV core protein (HBc), cccDNA, and/or HBV DNA (6, 11).
Additional experimental procedures are described in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
Plasmids for the production of lentivirus were kindly provided by Dr. Hiroyuki Miyoshi, RIKEN. This study was supported by the Japan Society for the Promotion of Science KAKENHI (Grants JP17H04085, JP66KT0111, and JP16K19145); the JST CREST program; JST MIRAI program; the Japan Agency for Medical Research and Development, AMED (Grants JP18fk0310114j0002, JP18fk0310101j1002, JP18fk0310103j0202, JP18fm0208019j0002, JP18fk0210036j0001, JP17fk0310103j0001, and JP18fm0208019h0202); Takeda Science Foundation; Pharmacological Research Foundation, Tokyo; and The Japan Food Chemical Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1811064116/-/DCSupplemental.
References
- 1.Verrier ER, Colpitts CC, Schuster C, Zeisel MB, Baumert TF. Cell culture models for the investigation of hepatitis B and D virus infection. Viruses. 2016;8:E261. doi: 10.3390/v8090261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liang TJ. Hepatitis B: The virus and disease. Hepatology. 2009;49(Suppl 5):S13–S21. doi: 10.1002/hep.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas E, Liang TJ. Experimental models of hepatitis B and C–New insights and progress. Nat Rev Gastroenterol Hepatol. 2016;13:362–374. doi: 10.1038/nrgastro.2016.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urban S, Bartenschlager R, Kubitz R, Zoulim F. Strategies to inhibit entry of HBV and HDV into hepatocytes. Gastroenterology. 2014;147:48–64. doi: 10.1053/j.gastro.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 5.Yan H, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwamoto M, et al. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem Biophys Res Commun. 2014;443:808–813. doi: 10.1016/j.bbrc.2013.12.052. [DOI] [PubMed] [Google Scholar]
- 7.Ni Y, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2014;146:1070–1083. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 8.König A, et al. Kinetics of the bile acid transporter and hepatitis B virus receptor Na+/taurocholate cotransporting polypeptide (NTCP) in hepatocytes. J Hepatol. 2014;61:867–875. doi: 10.1016/j.jhep.2014.05.018. [DOI] [PubMed] [Google Scholar]
- 9.Meredith LW, et al. Lentiviral hepatitis B pseudotype entry requires sodium taurocholate co-transporting polypeptide and additional hepatocyte-specific factors. J Gen Virol. 2016;97:121–127. doi: 10.1099/jgv.0.000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishitsuji H, et al. Novel reporter system to monitor early stages of the hepatitis B virus life cycle. Cancer Sci. 2015;106:1616–1624. doi: 10.1111/cas.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwamoto M, et al. Functional association of cellular microtubules with viral capsid assembly supports efficient hepatitis B virus replication. Sci Rep. 2017;7:10620. doi: 10.1038/s41598-017-11015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glebe D, et al. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129:234–245. doi: 10.1053/j.gastro.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 14.Gripon P, Cannie I, Urban S. Efficient inhibition of hepatitis B virus infection by acylated peptides derived from the large viral surface protein. J Virol. 2005;79:1613–1622. doi: 10.1128/JVI.79.3.1613-1622.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fukano K, et al. Troglitazone impedes the oligomerization of sodium taurocholate cotransporting polypeptide and entry of hepatitis B virus into hepatocytes. Front Microbiol. 2019;9:3257. doi: 10.3389/fmicb.2018.03257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bazley LA, Gullick WJ. The epidermal growth factor receptor family. Endocr Relat Cancer. 2005;12(Suppl 1):S17–S27. doi: 10.1677/erc.1.01032. [DOI] [PubMed] [Google Scholar]
- 17.Tomas A, Futter CE, Eden ER. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014;24:26–34. doi: 10.1016/j.tcb.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shimura S, et al. Cyclosporin derivatives inhibit hepatitis B virus entry without interfering with NTCP transporter activity. J Hepatol. 2017;66:685–692. doi: 10.1016/j.jhep.2016.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Wang P, Wang W, Murray JW, Wolkoff AW. The Na(+)-taurocholate cotransporting polypeptide traffics with the epidermal growth factor receptor. Traffic. 2016;17:230–244. doi: 10.1111/tra.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura Y, Bereczky B, Ono M. The EGFR inhibitor gefitinib suppresses ligand-stimulated endocytosis of EGFR via the early/late endocytic pathway in non-small cell lung cancer cell lines. Histochem Cell Biol. 2007;127:541–553. doi: 10.1007/s00418-007-0281-y. [DOI] [PubMed] [Google Scholar]
- 21.Claro da Silva T, Polli JE, Swaan PW. The solute carrier family 10 (SLC10): Beyond bile acid transport. Mol Aspects Med. 2013;34:252–269. doi: 10.1016/j.mam.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao YP, et al. Activation of the Raf-1/MEK/ERK cascade by bile acids occurs via the epidermal growth factor receptor in primary rat hepatocytes. Hepatology. 2002;35:307–314. doi: 10.1053/jhep.2002.31104. [DOI] [PubMed] [Google Scholar]
- 23.Sommerfeld A, Reinehr R, Häussinger D. Bile acid-induced epidermal growth factor receptor activation in quiescent rat hepatic stellate cells can trigger both proliferation and apoptosis. J Biol Chem. 2009;284:22173–22183. doi: 10.1074/jbc.M109.005355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279:16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 25.Singh M, Jadhav HR. Targeting non-small cell lung cancer with small-molecule EGFR tyrosine kinase inhibitors. Drug Discov Today. 2018;23:745–753. doi: 10.1016/j.drudis.2017.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.