Significance
Despite the importance of immune receptors in the biology of plants and animals, the precise biochemical mechanisms leading to the biosynthesis and secretion of the peptide activators of these immune receptors remain unexplored. Here, we describe the biosynthesis, processing, and secretion of a peptide activator that binds a rice immune receptor. Our studies also reveal that this peptide activator is a member of an important class of peptides present in both eukaryotic and prokaryotic species. Thus, these results provide a major advance in our knowledge of the biology of peptide ligands.
Keywords: peptidase, ABC transporter, RIPP, sulfation, immunogen
Abstract
The rice immune receptor XA21 is activated by the sulfated microbial peptide required for activation of XA21-mediated immunity X (RaxX) produced by Xanthomonas oryzae pv. oryzae (Xoo). Mutational studies and targeted proteomics revealed that the RaxX precursor peptide (proRaxX) is processed and secreted by the protease/transporter RaxB, the function of which can be partially fulfilled by a noncognate peptidase-containing transporter component B (PctB). proRaxX is cleaved at a Gly–Gly motif, yielding a mature peptide that retains the necessary elements for RaxX function as an immunogen and host peptide hormone mimic. These results indicate that RaxX is a prokaryotic member of a previously unclassified and understudied group of eukaryotic tyrosine sulfated ribosomally synthesized, posttranslationally modified peptides (RiPPs). We further demonstrate that sulfated RaxX directly binds XA21 with high affinity. This work reveals a complete, previously uncharacterized biological process: bacterial RiPP biosynthesis, secretion, binding to a eukaryotic receptor, and triggering of a robust host immune response.
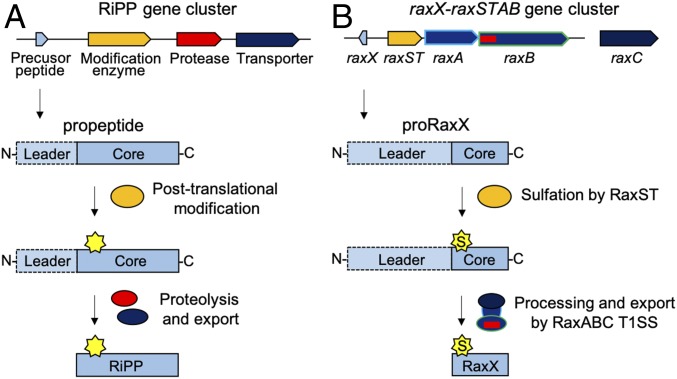
Ribosomally synthesized and posttranslationally modified peptides (RiPPs) include antimicrobial, anticancer, insecticidal, and quorum sensing peptides (1). RiPPs are structurally and functionally diverse but share commonalities. They are ribosomally synthesized as a precursor peptide (propeptide) with a cleavable N-terminal leader and a posttranslationally modified core that becomes the final secreted bioactive RiPP (Fig. 1A) (1–3).
Fig. 1.
Biosynthetic pathways of RiPPs and RaxX. (A) General RiPP biosynthetic pathway. The RiPP precursor (propeptide) and biosynthetic proteins are ribosomally synthesized. The core, which becomes the final RiPP product, is posttranslationally modified by enzyme(s) encoded in the same genomic region. Multiple posttranslational modifications can take place on a single propeptide. The N-terminal leader is enzymatically removed by a protease, and the modified core is exported by a transporter, releasing the mature bioactive RiPP. (B) RaxX biosynthetic pathway. proRaxX is ribosomally synthesized, and the core is sulfated by the sulfotransferase RaxST encoded upstream. We hypothesize that the PCAT RaxB removes the N-terminal leader and transports the sulfated mature RaxX peptide through the T1SS composed of RaxB, the periplasmic adaptor protein RaxA, and the genetically unlinked outer membrane protein RaxC.
RiPPs achieve substantial chemical diversity through extensive posttranslational modifications and are divided into over 20 groups (1). RiPPs that have not been well studied or formally categorized are tyrosine sulfated peptides.
Tyrosine sulfation is a posttranslational modification that influences receptor–ligand binding in diverse host–microbe interactions (4). For example, in humans, tyrosine sulfation of the integral membrane protein C-C chemokine receptor type 5 (CCR5) significantly enhances binding of the HIV envelope glycoprotein, facilitating HIV entry (5).
In rice, the transmembrane immune receptor XA21, which shares similarities to animal Toll-like receptors and the Arabidopsis flagellin-sensitive 2 and EF-Tu receptors (6), responds to sulfated derivatives of the microbial peptide required for activation of XA21-mediated immunity X (RaxX) produced by the Gram-negative pathogenic bacterium Xanthomonas oryzae pv. oryzae (Xoo) (7–10). The XA21/RaxX interaction conforms to the “gene-for-gene relationship” initially described by Flor (11) in the 1940s. The rice XA21 immune receptor, which is encoded by a specific resistance gene, recognizes Xoo strains that produce the cognate microbial molecule, RaxX. This recognition leads to a host immune response. Thus, plants lacking the XA21 immune receptor are susceptible to RaxX-producing Xoo strains, and conversely, plants carrying XA21 are unable to recognize and respond to Xoo strains lacking RaxX (7–10). Xoo strains lacking RaxX are also compromised in virulence of rice plants lacking XA21 (10). These results suggest a role for RaxX in bacterial virulence.
Tyrosine sulfated RaxX16, a 16-residue synthetic peptide derived from residues 40–55 of the 60-residue RaxX precursor peptide (proRaxX), is the shortest characterized immunogenic derivative of RaxX (10). RaxX16 shares a 13-residue sequence with high similarity to the 18-residue plant peptide containing sulfated tyrosine (PSY1) hormone (10). PSY1 promotes cellular proliferation and expansion in planta (12) and increases root growth in both Arabidopsis and rice (10). Exogenous application of sulfated RaxX16 also promotes root growth (10), which is consistent with the hypothesis that RaxX mimics host PSY1 and PSY1-like proteins. These observations indicate that RaxX16 contains the biologically active domain of RaxX.
In Arabidopsis, PSY1 is ribosomally synthesized as a 75-residue propeptide and proteolytically processed into the mature secreted 18-residue sulfated and glycosylated peptide (12). Based on similarities between PSY1 and RaxX16, we hypothesized that sulfated proRaxX is also processed and secreted to produce an extracellular molecule that can interact with host receptors.
The raxX gene is adjacent to the raxSTAB operon, encoding the RaxST tyrosylprotein sulfotransferase, which catalyzes sulfation of proRaxX tyrosine residue 41, and two components of a predicted type I secretion system (T1SS): the RaxA periplasmic adaptor protein and the RaxB peptidase-containing ATP-binding cassette transporter (PCAT) (Fig. 1B) (7, 13). In other characterized PCATs, the N-terminal peptidase domain cleaves the substrate leader immediately after a Gly–Gly (GG) motif, and the processed substrate is secreted through the PCAT, periplasmic adaptor protein, and TolC, an outer membrane channel protein (14).
Previously, we reported that derivatives of the immunogenic Xoo strain PXO99 containing null alleles of raxX, raxST, or raxC (encoding the Xoo TolC ortholog) evade XA21-mediated recognition, but strains with null alleles of raxA or raxB elicit a partial XA21-mediated immune response (7, 13). These results suggest that RaxA and RaxB are important for RaxX secretion, but RaxX can also be released outside the cell independent of raxA/raxB.
Here, we report that a synthetic RaxX peptide derivative directly binds the extracellular domain (ECD) of the XA21 immune receptor (XA21ECD), an interaction enhanced by tyrosine sulfation of RaxX. We also demonstrate that RaxB is required for proteolytic processing and secretion of proRaxX and identify a second PCAT, peptidase-containing transporter component B (PctB), that can partially compensate for deletion of raxB. Our genetics and targeted proteomics data indicate that proRaxX is ribosomally synthesized and cleaved downstream of a GG motif in an RaxB-dependent manner. This proteolytic event releases the mature RaxX core peptide containing the sulfated Tyr and the minimal 16-residue active region (Fig. 1B). These results indicate that RaxX is a tyrosine sulfated RiPP, a previously undescribed class of RiPPs that mediate intercellular interactions.
Results
Sulfated RaxX Peptide Binds XA21 with High Affinity.
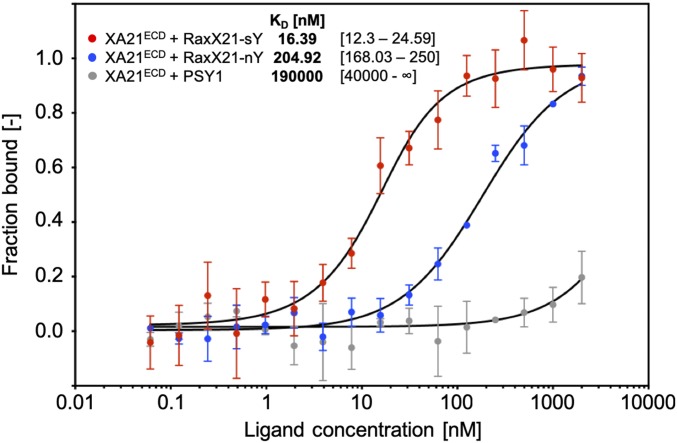
We previously demonstrated that in vivo tyrosine sulfation of RaxX by RaxST is required to activate XA21-dependent immune responses (7), suggesting that sulfated, but not nonsulfated, RaxX is recognized by XA21. To test if RaxX directly binds to the XA21 immune receptor, we used microscale thermophoresis (MST), an optical tool that analyzes protein and small molecule interactions (15). The MST assays were conducted using XA21ECD (XA21 residues 23–649) with varying concentrations of peptide to measure the dissociation constant (Kd). Because we were unable to purify sufficient levels of Xoo-produced RaxX, we used a synthetic 21-residue derivative (RaxX21: proRaxX residues 35–55), which has previously been shown to activate XA21-mediated immune responses in a sulfation-dependent manner (7). When we titrated RaxX21 in the presence of XA21ECD, we observed a Kd of ∼205 nM with the nonsulfated form (RaxX21-nY) (Fig. 2). This Kd is 12.5-fold higher than the sulfated form (RaxX21-sY), which had a Kd of ∼16 nM (Fig. 2), indicating that sulfated RaxX binds XA21 with higher affinity than nonsulfated RaxX.
Fig. 2.
Sulfated RaxX peptide binds XA21ECD with high affinity. Quantification of binding between sY (red) and nY (blue) RaxX21 and sulfated PSY1 (gray) peptides with the XA21 ectodomain (XA21ECD) by MST. Data points indicate the fraction of fluorescently labeled XA21 bound to the peptides during the assay (fraction bound [−]). The Kd and confidence intervals shown in brackets are indicated in nanomolar. SE bars are representative of at least two independent measurements performed with independent protein preparations.
Although RaxX shares similarities with PSY1 in both sequence and root growth-promoting activities, synthetic sulfated PSY1 peptides derived from Arabidopsis and rice fail to activate XA21-dependent immune responses (10). Consistent with these reports, sulfated PSY1 failed to specifically bind XA21ECD (Fig. 2). These results indicate that RaxX binding to XA21ECD is a specific interaction, which is enhanced by sulfation.
Selected Reaction Monitoring MS Detects RaxX as a Secreted Mature Peptide.
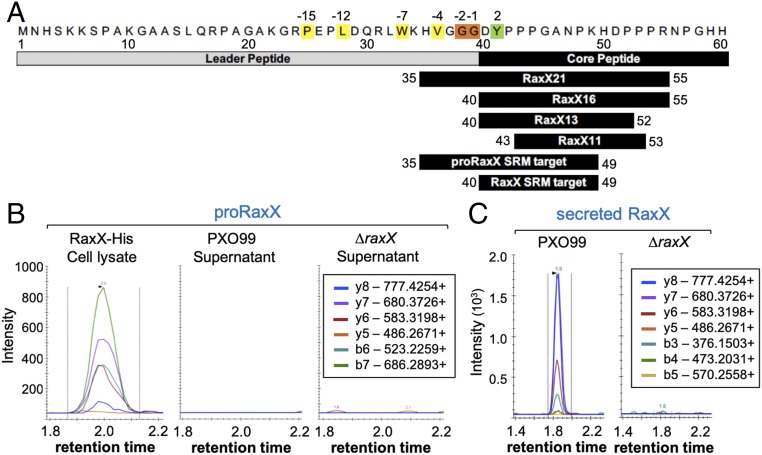
Based on its similarity to PSY1 and its direct binding to XA21ECD, we hypothesized that sulfated RaxX interacts with host receptors as a secreted proteolytically processed, mature peptide. To test this hypothesis, we assessed the presence of secreted RaxX in the extracellular milieu. Secreted proteins were harvested from Xoo strains grown in plant-mimicking media, concentrated, and digested with trypsin. Because the N-terminal 39 residues of proRaxX are dispensable for RaxX immunogenicity and root growth-promoting activity (10), we hypothesized that proRaxX is processed downstream of Gly39 (Fig. 3A). The predicted mature RaxX and nonprocessed proRaxX peptides (represented by the DYPPPGANPK and HVGGGDYPPPGANPK tryptic peptides, respectively) were specifically detected by selected reaction monitoring (SRM)–MS, which was calibrated using tryptic digests of RaxX16 (DYPPPGANPKHDPPPR) synthetic peptide and purified recombinant full-length proRaxX (RaxX60) (Fig. 3A and SI Appendix, Fig. S1 A and B).
Fig. 3.
Mature RaxX is detected in Xoo supernatants. (A) proRaxX sequence. Numbers above refer to the residue position relative to the predicted cleavage site; numbers below are relative to propeptide. Cleavage of proRaxX after Gly38, Gly39 (orange) would result in a leader with hydrophobic residues (yellow) at conserved locations typical of PCAT substrates and a core containing Tyr41 (green) sulfated by RaxST. Shown below this precursor are the synthetic RaxX peptide derivatives (RaxX21, RaxX16, RaxX13, and RaxX11) and the two tryptic peptide targets detected by SRM-MS. (B and C) SRM-MS chromatograms of nonprocessed proRaxX (B) and the predicted mature RaxX (C) tryptic peptides detected from ΔraxX(praxX-His) cell lysates or from PXO99 or ΔraxX supernatants. Lines correspond to the individual SRM transitions monitored. The legends indicate the detected peptide b- and y-series fragment ions.
Similar to previous reports (7), we detected nonprocessed proRaxX in the cell lysate (Fig. 3B). However, only mature, processed RaxX was detected in the supernatant of the wild-type PXO99 strain (Fig. 3C). RaxX was not detected in the ΔraxX negative control (Fig. 3C), indicating that the observed peaks are specific for RaxX. Because trypsin is not known to cleave after glycine, which precedes the detected DYPPPGANPK tryptic peptide, these results suggest that proRaxX is cleaved by an endogenous Xoo protease after Gly39 and secreted outside the cell as a mature peptide (Fig. 1B).
To characterize the full sequence of mature RaxX, we attempted to detect tryptic peptides located at the C-terminal end of RaxX. However, SRM-MS analysis of supernatants from the wild-type strain failed to detect the HDPPPR tryptic peptide (corresponding to proRaxX residues 50–55) (SI Appendix, Fig. S1C) adjacent to the DYPPPGANPK peptide. As an alternative approach to elucidate the nature of the RaxX C terminus, we performed shotgun liquid chromatography (LC)–MS to analyze the presence of RaxX from Xoo supernatants. Although we detected the same DYPPPGANPK tryptic peptide as observed in the SRM-MS analysis (SI Appendix, Fig. S1 D and E), we did not detect tryptic peptides located C-terminal to this peptide. Additionally, we did not detect RaxX from Xoo supernatants that were processed without trypsin digestion. We were, therefore, unable to confirm the identity of the C terminus of the processed RaxX using our MS approaches.
We next attempted immunoblot analysis to assess the RaxX protein. However, an antibody generated against RaxX failed to detect RaxX16 or RaxX from Xoo supernatants (SI Appendix, Fig. S1 F and G). We also performed immunoblot analysis of C-terminal HA-tagged RaxX pulled down using an anti-HA resin. Immunoblots with an anti-HA antibody detected proRaxX-HA pulled down from the cell lysate, but we did not detect a band from supernatants (SI Appendix, Fig. S1H). Based on these results, we hypothesize that proRaxX is further processed at its C terminus.
Sequence Analysis Identifies a Candidate Secondary RaxX Maturation and Secretion System: PctAB.
Prokaryotic RiPPs typically encode the maturation and secretion proteins in the same genomic region as the peptide (Fig. 1A). We previously demonstrated that a strain carrying a null allele of raxC, encoding the predicted T1SS outer membrane protein, fails to activate XA21-mediated immunity, supporting a role for raxC in RaxX secretion (13). In contrast, strains carrying null alleles of raxB or raxA, encoding the putative RaxX-associated PCAT and periplasmic adaptor proteins, respectively, maintain the ability to partially activate XA21-mediated immunity (13). These results suggest that RaxX can be released in an raxA/raxB-independent manner.
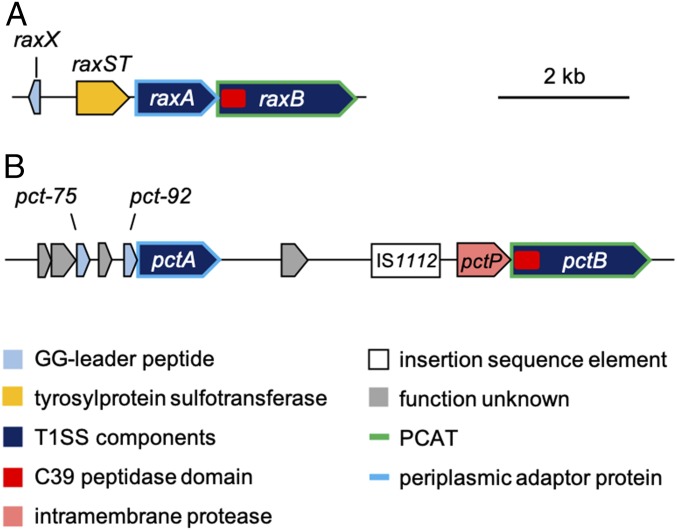
The RaxB sequence contains an N-terminal C39 peptidase domain characteristic of PCATs [e.g., Escherichia coli colicin V (ColV) transporter CvaB (16)], which have dual functionality as both a transporter and a protease (Fig. 4A) (13, 17, 18). A query of the RaxB C39 peptidase domain sequence in BLAST searches of the PXO99 genome identified only one other candidate PCAT, which shares 50% sequence identity with RaxB (41% identity with E. coli CvaB). We named this gene (PXO_RS14825) pctB (Fig. 4B).
Fig. 4.
Genetic map of candidate RaxX biosynthetic proteins. (A) raxX is encoded upstream of the raxSTAB operon, containing genes for the RaxST sulfotransferase and components of a T1SS: the RaxA periplasmic adaptor protein and the RaxB PCAT. (B) Genes for components of a second peptidase-containing transporter system are not genetically linked to raxX: pctA and pctB encoding a periplasmic adaptor protein and PCAT, respectively.
Immediately upstream of pctB is a gene, which we named pctP (PXO_RS14830), encoding a putative rhomboid family intramembrane serine protease (Fig. 4B) that typically cleaves the transmembrane domain of membrane proteins (19). Farther upstream are an insertion sequence (IS1112), a region of uncertain heritage likely derived from multiple insertion and deletion events, and a predicted gene (PXO_RS14840), which we named pctA, encoding a putative periplasmic adaptor protein (Fig. 4B). PctA shares 32% sequence identity with RaxA [24% identity with E. coli CvaA, the adaptor component of the ColV secretion system (16)]. Given the close proximity of pctA and pctB and the absence of other PCAT-encoding genes in the genome, we hypothesized that PctA and PctB form an alternate T1SS that can secrete sufficient levels of sulfated RaxX to activate XA21.
PctB Partially Compensates for the Loss of RaxB.
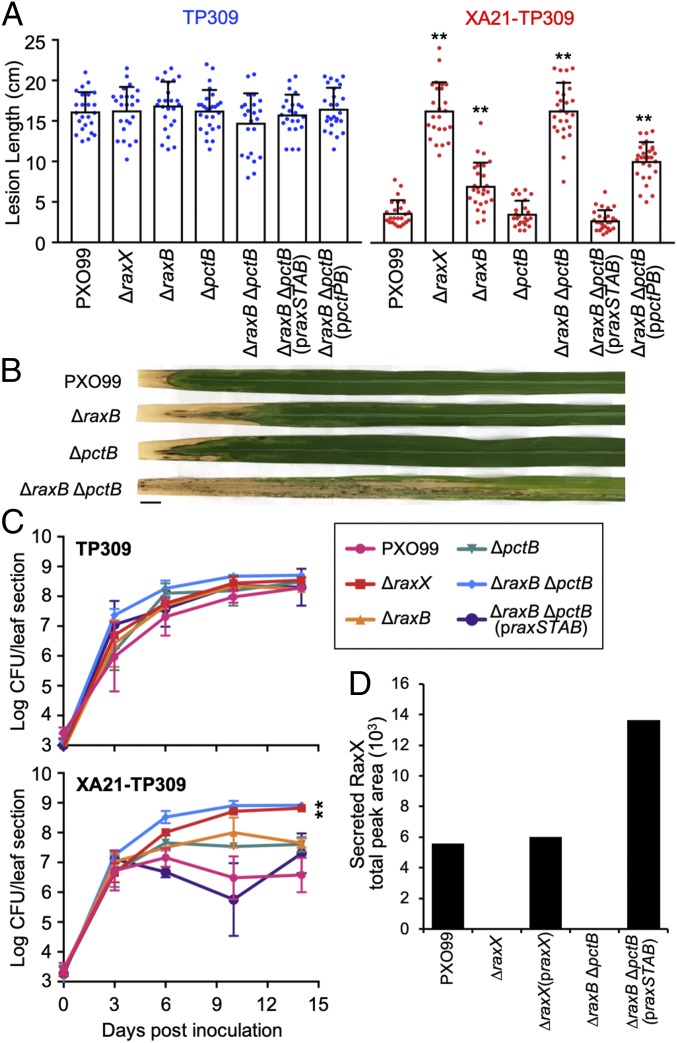
To assess the potential role of PctB in RaxX maturation and secretion, we deleted raxB and pctB singly or together from the PXO99 genome. The resultant mutant strains (designated ΔraxB, ΔpctB, and ΔraxB ΔpctB, respectively) were inoculated onto rice plants by clipping leaf tips with scissors dipped in bacterial suspension. Lesion lengths and bacterial densities in planta measured 14 d after inoculation were comparable with wild-type PXO99 on Taipei 309 (TP309) rice plants, which lack Xa21 (Fig. 5 A and C) (20). This indicates that deletion of genes encoding these putative transporters does not directly compromise Xoo virulence under the tested conditions.
Fig. 5.
The ΔraxB ΔpctB double mutant does not secrete RaxX, but secretion can be restored with the addition of the praxSTAB plasmid. (A–C) Rice plants were inoculated by scissor clipping with PXO99-derived strains. (A) Bars represent the mean + SD of lesions (centimeters) measured 14 d postinoculation (dpi) of TP309 (blue dots) or XA21-TP309 (red dots) plants (n = 21–28). (B) Inoculated XA21-TP309 leaves 14 dpi. (Scale bar: 1 cm.) (C) Bacterial densities in planta. Data points represent the mean log cfu per 10-cm leaf section ± SE (n = 3). (D) SRM-MS was used to detect the presence of RaxX in Xoo supernatants. Bars represent the total peak area of the chromatograms shown in SI Appendix, Fig. S3B. Similar results were observed in five (A) or two (C) other experiments, except that ΔraxB formed lesions comparable with PXO99 in roughly one-half. **P < 0.01 compared with PXO99 using Dunnett’s test.
We next tested the effect of the mutant strains on XA21-TP309 rice, a derivative of TP309 that expresses Xa21 (20), to assess XA21-dependent responses. As previously reported (20), XA21-TP309 rice was resistant to PXO99 infection, and it had short lesions (<5 cm) and low bacterial densities (6.8 × 106 cfu per leaf) in planta (Fig. 5 A–C). We observed that the ΔraxB mutant formed short (<5-cm) to intermediate (6- to 9-cm) lesions on XA21-TP309, which varied between experiments, but the ΔpctB mutant consistently formed short (<5-cm) lesions similar to PXO99 (Fig. 5 A and B). Both ΔraxB and ΔpctB single mutants accumulated to populations of ∼4.5 × 107 cfu per leaf, roughly 6.6-fold higher than PXO99 (Fig. 5C). In contrast, the ΔraxB ΔpctB double mutant formed 4-fold longer lesions (>14 cm) than PXO99 and accumulated to 8.4 × 108 cfu per leaf, over 120-fold higher than the PXO99 population (Fig. 5 A–C). These phenotypes were comparable with the ΔraxX control (Fig. 5 A and C). Similarly, the ΔraxA ΔpctA double mutant, containing mutations in genes for the predicted periplasmic adaptor proteins RaxA and PctA, formed longer lesions than the ΔraxA and ΔpctA single mutants (SI Appendix, Fig. S2). Collectively, these results suggest that sulfated RaxX is not secreted in both the ΔraxA ΔpctA and ΔraxB ΔpctB double mutants, allowing these strains to evade XA21. These results also suggest that PctA and PctB can partially compensate for the loss of RaxA and RaxB, respectively.
RaxB Is the Primary PCAT Required for RaxX Maturation and Secretion.
The ΔraxB ΔpctB double mutant accumulates to high population levels in XA21-TP309 plants (Fig. 5C), indicating that this strain can evade detection by the XA21 immune receptor. This result supports the hypothesis that these PCATs secrete sulfated RaxX. Given that raxB is genetically clustered with raxX (Figs. 1B and 4A), we hypothesized that RaxB functions as the primary transporter of RaxX and that it is sufficient to activate XA21 in the absence of PctB. To test this hypothesis, we complemented the ΔraxB ΔpctB double mutant with praxSTAB, a broad host range vector expressing the entire raxSTAB operon with the presumptive native promoter. On XA21-TP309, the praxSTAB plasmid restored raxB gene expression (SI Appendix, Fig. S3A) as well as activation of XA21-mediated immunity, resulting in short lesions and bacterial densities comparable with wild-type PXO99 (Fig. 5 A and C). Similarly, praxSTAB restored the ability of the ΔraxA ΔpctA double mutant to activate XA21-mediated immunity (SI Appendix, Fig. S2). These results suggest that the RaxAB system is sufficient to secrete RaxX and activate XA21 in the absence of the PctAB system.
As a control, we also expressed praxSTAB in the wild-type PXO99 background. We observed no significant alterations on lesion development with the addition of praxSTAB (SI Appendix, Fig. S2), suggesting that overexpression of raxSTAB did not directly contribute to enhanced activation of the XA21-mediated immune response.
We next assessed, using SRM-MS, if RaxX was secreted in the ΔraxB ΔpctB double mutant. RaxX was detected in the supernatant of the ΔraxB ΔpctB double mutant only in the presence of the praxSTAB plasmid (Fig. 5D). We detected SRM transition peaks of RaxX in the praxSTAB-containing strain at the same retention time as wild-type PXO99 and the ΔraxX mutant complemented with plasmid-expressed raxX (praxX) controls, which were collected and analyzed in the same batch (SI Appendix, Fig. S3B). Together, these results indicate that RaxB is a sufficient PCAT for proRaxX maturation and secretion.
We also tested if PctB is sufficient to restore RaxX secretion in the ΔraxB ΔpctB double mutant. For this purpose, we transformed the ΔraxB ΔpctB double mutant with a broad host range vector expressing the pctPB cluster with its presumptive native promoter (ppctPB). The ppctPB plasmid restored pctB gene expression (SI Appendix, Fig. S3A) but failed to fully restore XA21-mediated recognition of the ΔraxB ΔpctB double mutant (Fig. 5A). The ppctPB-complemented ΔraxB ΔpctB double mutant still formed intermediate to long lesions (∼10 cm) roughly threefold longer than PXO99 (Fig. 5A). This result suggests that PctB alone cannot effectively secrete RaxX to activate XA21-mediated immunity.
RaxB and PctB Carry Conserved Residues Characteristic of Bifunctional PCATs.
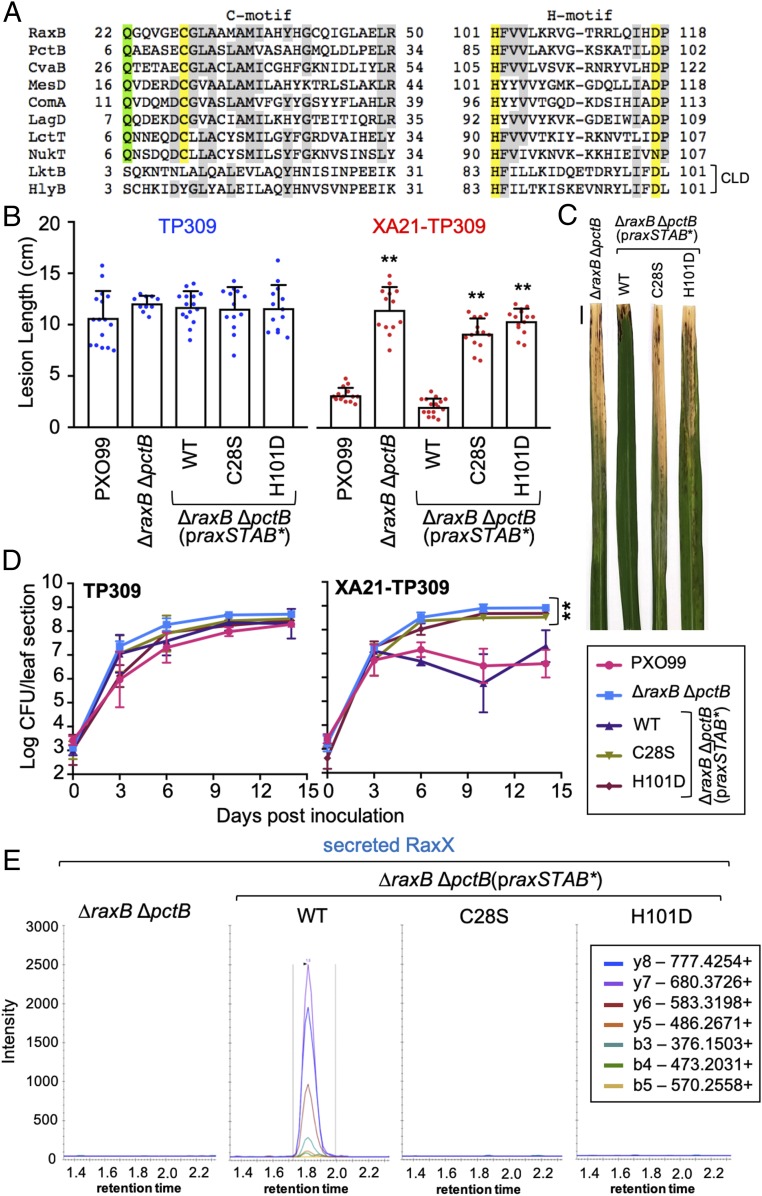
PCATs, such as E. coli CvaB, function as both a transporter and a protease (17, 18). The peptidase domain of this class of ATP-binding cassette (ABC) transporters contains two conserved motifs: a cysteine (C) motif and a histidine (H) motif (Fig. 6A) (17, 21). We found that both RaxB and PctB contain sequences that match the conserved C and H motifs, including the conserved Cys, His, and Asp residues (Cys28/12, His101/85, and Asp117/101 of RaxB/PctB, respectively) predicted to form the active site catalytic triad (Fig. 6A) (18, 22). Additionally, they contain the conserved Gln residue (Gln22/6 of RaxB/PctB, respectively) proposed to form the oxyanion hole that stabilizes a negatively charged tetrahedral intermediate generated during amide/ester hydrolysis (Fig. 6A) (22, 23). In contrast, ABC transporters that have a degenerate C39-like domain (CLD) with no proteolytic activity, such as the E. coli hemolysin transporter HlyB, do not contain the catalytically essential Cys or conserved Gln (Fig. 6A) (24). The presence of the complete Cys–His–Asp/Asn catalytic triad and conserved oxyanion hole Gln characteristic of bifunctional PCATs is consistent with the hypothesis that RaxB and PctB also possess proteolytic activity to process proRaxX.
Fig. 6.
Mutation of the RaxB peptidase catalytic triad impairs proRaxX maturation and secretion. (A) Alignment of the peptidase C and H motifs of RaxB and PctB to select PCATs or ABC transporters with a CLD (listed in Materials and Methods). The Cys–His–Asp/Asn catalytic triad is highlighted in yellow, the oxyanion hole Gln is in green, and residues common in at least six of the transporters are in gray. (B–D) Rice plants were inoculated by scissor clipping with the indicated strains. (B) Bars represent the mean + SD of lesion measurements (centimeters) on TP309 (blue dots) or XA21-TP309 (red dots) plants 14 d postinoculation (dpi; n = 10–16). (C) Inoculated XA21-TP309 leaves 14 dpi. (Scale bar: 1 cm.) (D) Bacterial densities in planta. Data points represent the mean log cfu per 10-cm leaf section ± SE (n = 3). Performed at the same time as Fig. 5C. (E) SRM-MS chromatograms of RaxX tryptic peptide detected in supernatants from the ΔraxB ΔpctB double mutant-derived strains. Similar results were observed in five (B) or two (D) other independent experiments. **P < 0.01 compared with PXO99 using Dunnett’s test.
Mutation of the RaxB Peptidase Domain Catalytic Triad Impairs RaxX Maturation and Secretion.
We next assessed the role of the RaxB peptidase domain in proRaxX maturation. For these experiments, we transformed the ΔraxB ΔpctB double mutant with a derivative of praxSTAB containing site-directed missense substitutions of the active site cysteine (Cys28) or histidine (His101) of the RaxB peptidase domain. Cys28 was mutated to Ser (C28S), and His101 was mutated to Asp (H101D), which we hypothesized would abolish proteolytic activity by disrupting formation of the predicted Cys28, His101, Asp117 catalytic triad as demonstrated with E. coli CvaB (18).
Expression of the C28S and H101D mutant derivatives impaired praxSTAB-complementation of the ΔraxB ΔpctB double-mutant phenotype on XA21-TP309 but had no effect on the TP309 control plants (Fig. 6 B–D). Wild-type praxSTAB fully complemented the ΔraxB ΔpctB double-mutant phenotype on XA21-TP309, resulting in short lesions (<4 cm) and low bacterial densities (3.9 × 107 cfu per leaf). In contrast, expression of the mutated peptidase derivatives of praxSTAB resulted in long lesions (>9 cm) and high bacterial densities (108 cfu per leaf) comparable with the strain without plasmid (Fig. 6 B–D). These results indicate that the predicted RaxB peptidase domain is catalytically active and necessary to process and secrete biologically active RaxX.
To assess if RaxX is secreted into supernatants of the C28S or H101D RaxB peptidase mutants, we carried out SRM-MS analysis. We detected high SRM transition peaks corresponding to RaxX in supernatants of the ΔraxB ΔpctB double-mutant strain carrying wild-type praxSTAB (Fig. 6E). In contrast, SRM transitions corresponding to RaxX were not detected in the ΔraxB ΔpctB double mutant carrying praxSTAB with either the C28S or H101D mutation (Fig. 6E). We also detected no SRM transitions corresponding to RaxX in the ΔraxX negative control and the ΔraxB ΔpctB double mutant without plasmid (Figs. 3C and 6E and SI Appendix, Fig. S3B). The lack of detectable secreted RaxX in the RaxB peptidase mutants supports our inoculation results, providing additional evidence that RaxB serves as the proRaxX maturation protease.
Mutation of the GG Cleavage Site Compromises proRaxX Maturation and Secretion.
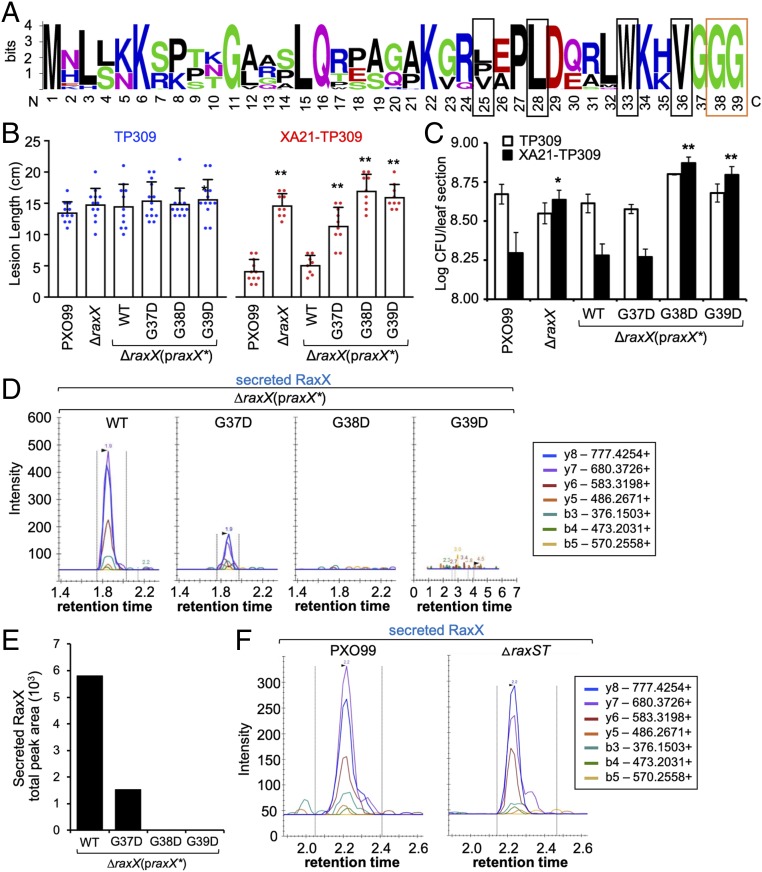
RiPP propeptides typically carry a cleavable N-terminal leader that is removed by a protease (Fig. 1A). PCATs cleave the N-terminal leader of substrates downstream of a GG motif, which contains GG or Gly–Ala immediately preceding the cleavage site (17, 25). Based on the observation that key residues in the RaxB catalytic peptidase domain are required for RaxX secretion (Fig. 6), we hypothesized that proRaxX contains an N-terminal GG motif that is removed by RaxB. In support of this hypothesis, our SRM-MS analysis indicated that the N-terminal 39 residues of proRaxX are removed before secretion (Fig. 3). We noted that this leader peptide ends with three conserved glycines and contains conserved hydrophobic residues at positions −4, −7, −12, and −15 distal to the cleavage site, a pattern typical of PCAT substrates and necessary for interaction with the peptidase (Figs. 3A and 7A and SI Appendix, Fig. S4A) (26–29). These observations indicate that the predicted proRaxX leader resembles the leader of GG motif-containing propeptides typically processed and secreted by PCATs.
Fig. 7.
Mutation of the GG cleavage site compromises proRaxX maturation and secretion. (A) Sequence logo of the leader from all Xanthomonas proRaxX alleles. Conserved GG preceding the cleavage site is boxed in orange; hydrophobic residues in conserved positions typical of PCAT substrates are boxed in black. (B and C) Rice plants were inoculated by scissor clipping. (B) Bars represent the mean + SD of lesion measurements (centimeters) on TP309 (blue dots) or XA21-TP309 (red dots) plants 14 d postinoculation (dpi; n = 8–13). Similar results were observed in at least six independent experiments. (C) Bars represent the mean log cfu per leaf section ± SE measured 14 dpi (n = 4). (D) SRM-MS chromatograms of RaxX tryptic peptide detected in Xoo supernatants. (E) Quantification of the total peak area shown in D. (F) SRM-MS chromatograms of RaxX tryptic peptide detected in supernatants from the wild-type PXO99 and ΔraxST mutant strains. *P < 0.05 compared with PXO99 using Dunnett’s test; **P < 0.01 compared with PXO99 using Dunnett’s test.
Studies of the propeptide mesentericin Y105 (MesY) produced by Leuconostoc mesenteroides indicate that the GG motif preceding the MesY cleavage site is critical for efficient processing and secretion (26). Based on this observation, we hypothesized that the GG motif in the predicted proRaxX leader sequence is critical for RaxB-mediated processing of proRaxX. To test this hypothesis, we generated site-directed missense substitutions of the conserved glycine residues (Gly37–39) preceding the cleavage site of proRaxX. As Aucher et al. (26) demonstrated that secretion of MesY was significantly reduced or completely abolished when Gly at the −2 position from the cleavage site was mutated to Ala or Asp or when Gly at the −1 position from the cleavage site was mutated to Arg or Asp, we generated homologous missense substitutions of proRaxX Gly37–39. As a control, we also generated missense substitutions of Gly–Ala pairs farther upstream of the cleavage site: Gly11, Ala12 and Gly20, Ala21. These mutant variants of proRaxX were expressed as derivatives of the praxX plasmid in the ΔraxX mutant background.
The resulting mutants were inoculated onto rice plants. All of these mutants formed long (>12-cm) lesions comparable with PXO99 on the TP309 control plants (Fig. 7B, Left and SI Appendix, Fig. S4 B and C). On XA21-TP309, ΔraxX formed long lesions (>15 cm), and this phenotype was complemented to short lesions by praxX as previously reported (Fig. 7B and SI Appendix, Fig. S4 B and C) (7). Similarly, praxX derivatives containing mutations in RaxX residue Gly11, Ala12, Gly20, or Ala21 complemented the ΔraxX mutant phenotype (SI Appendix, Fig. S4 B and C). This result suggests that these residues are not necessary for processing or secretion of RaxX. In contrast, praxX derivatives containing mutations in Gly37–39 were impaired in their ability to complement the ΔraxX mutant phenotype (Fig. 7B and SI Appendix, Fig. S4 B and C). Expression of the G37D and G38A derivatives resulted in intermediate lesions (11–12 cm). In contrast, the G38D, G38R, and G39D derivatives resulted in long lesions (>15 cm) similar to the ΔraxX mutant without plasmid control (Fig. 7B and SI Appendix, Fig. S4 B and C). Bacterial densities were also comparable among the ΔraxX mutant and the G38D- and G39D-expressing strains (Fig. 7C). These results suggest that Gly37–39, which immediately precede the cleavage site, are essential for proRaxX maturation and/or secretion.
We next assessed if we could detect secreted RaxX in supernatants of Xoo strains carrying mutations in the leader peptide. We did not detect SRM transitions of RaxX above background levels in supernatants of the ΔraxX mutant carrying the G38D and G39D derivatives (Fig. 7D), suggesting that proRaxX is neither processed nor secreted when Gly38 or Gly39 is mutated to Asp. In contrast, we detected RaxX in supernatants of the ΔraxX mutant carrying the G37D praxX derivative, although at reduced levels compared with expression of wild-type praxX (1.5 × 103 vs. 5.8 × 103 total peak area, respectively) (Fig. 7E). The accumulation of intermediate amounts of RaxX peptide secreted from the G37D mutant strain is consistent with the formation of intermediate lesion lengths on XA21-TP309 plants inoculated with this strain (Fig. 7 B and E). Taken together, our results suggest that proper proRaxX maturation and secretion, which require the Gly38, Gly39 cleavage site, are necessary steps in the biogenesis of the immunogenic peptide.
Sulfation Is Not Necessary for RaxX Secretion.
Some RiPP proteases process posttranslationally modified propeptides more efficiently than unmodified propeptides (30). We, therefore, assessed if nonsulfated proRaxX could be processed and secreted. We analyzed the presence of RaxX in supernatants of the ΔraxST mutant, a PXO99-derived strain containing a null allele of the sulfotransferase gene raxST (7). We detected RaxX from the ΔraxST supernatant with SRM transitions comparable with those observed from the wild-type PXO99 supernatant (Fig. 7F). This result suggests that sulfation of proRaxX is not required for processing or secretion by the RaxB and PctB transporters.
Discussion
Tyrosine Sulfation of RaxX Enhances XA21 Binding.
RaxX immunogenicity is dependent on Tyr sulfation (7). Our MST results demonstrate that this modification increases the affinity of RaxX binding to XA21ECD (Fig. 2), resulting in a Kd comparable with the RaxX concentration needed to trigger the half-maximal immune response of XA21 (∼20 nM) and induce root growth (7, 10). As we have yet to confirm the identity of the C terminus of RaxX, additional work is needed to determine if the fully processed sulfated peptide binds XA21 more efficiently.
Tyrosine sulfation has also been observed to enhance interaction of the disulfated pentapeptide phytosulfokine (PSK) with its cognate receptor PSKR1 in Arabidopsis and carrot. Based on crystal structure studies, the two sulfate groups on PSK form hydrogen bonds and van der Waals packing with residues on PSKR1 (31). This sulfate group-mediated interaction stabilizes PSKR1, which in turn, allosterically induces heterodimerization with the somatic embryogenesis receptor-like kinase coreceptor, resulting in activation of PSKR1 (31). Analogous structural studies will provide insights into how sulfation of RaxX contributes to the RaxX-XA21 interaction.
RaxX Is a Member of the Tyrosine Sulfated RiPP Family That Spans Bacteria and Plants.
RaxX is predicted to mimic the activity of the plant peptide hormone PSY1 (10). Our results indicate that proRaxX is ribosomally synthesized and then, proteolytically processed by RaxB into a mature peptide (Fig. 1B). This mature RaxX peptide includes the same residues covered by RaxX16 (Fig. 3A), which is sufficient to activate XA21-mediated immunity and promote root growth in rice (10). These residues also precisely overlap the conserved 13-residue region found in mature PSY1 from Arabidopsis and in putative PSY-like proteins from various plant species, including rice (10, 12). Furthermore, RaxX and PSY1 start with the same residues, Asp–Tyr, with the critical sulfated tyrosine at the same position. RaxX, therefore, intimately resembles host PSY-like proteins.
The small size (<10 kDa), presence of a posttranslational modification (sulfated tyrosine in this case), and proteolytic maturation of RaxX and PSY1 are defining characteristics of RiPPs (1). In addition to PSY1, plants produce other secreted tyrosine sulfated peptides that regulate plant growth, including PSK, root meristem growth factor (RFG), and Casparian strip integrity factor (CIF) (32–35). Similar to RaxX and PSY1, PSK, RFG, and CIF are proteolytically processed from larger propeptides and secreted.
Given the similarities of RaxX, PSY1, PSK, RFG, and CIF, we propose that these bacterial and plant peptides be classified as a group of RiPPs defined by the presence of the sulfated tyrosine.
Does PctB Process and Secrete Other RiPPs?
Our results indicate that RaxAB is the cognate proRaxX maturation and secretion system and that it can be partially complemented by PctAB. This functional redundancy is rather unusual given the low sequence identity shared between the two systems, which are also independently distributed among Xanthomonads. For instance, the Xanthomonas albilineans genome contains pctAPB but not raxX or raxSTAB, whereas the Xanthomonas euvesicatoria genome contains raxX and raxSTAB but not pctAPB. These observations suggest that RaxX is not the cognate substrate for PctB.
Because PctB is predicted to have a functional C39 peptidase domain (Fig. 6A), we hypothesize that its cognate substrate is a GG motif-containing peptide. Based on the annotated PXO99 sequence (NC_010717.2), two hypothetical genes, PXO_RS14845 and PXO_RS14850 (here referred to as pct-92 and pct-75, respectively), are encoded upstream of pctA (Fig. 4B). pct-92 and pct-75 are predicted to encode 92- and 75-residue propeptides, respectively, that contain a GG-like leader of 28 and 19 aa, respectively (SI Appendix, Fig. S4A). If Pct-92 and Pct-75 are both PctB substrates, then this would suggest that PctB can tolerate leaders that vary in sequence and length. Such substrate tolerance has also been observed in other PCATs, such as LagD, HalT, and LicT from Lactococcus lactis, Bacillus halodurans, and Bacillus licheniformis, respectively, which process and transport two cognate peptides that function together as a two-component antimicrobial molecule (17, 36, 37). Because the leader of the two propeptide components can vary in sequence and length (SI Appendix, Fig. S4A), these transporters exhibit a relaxed specificity and sometimes tolerate noncognate substrates (37). This may explain why PctB is able to tolerate RaxX, at least to some extent. However, we have yet to confirm that pct-92 and pct-75 encode actual peptides. It is also unclear if these putative peptides are posttranslationally modified, because no known modification enzymes are encoded in the same genomic region (Fig. 4B). Therefore, additional studies are needed to analyze these putative PctB substrates and identify essential determinants that allow PctB to distinguish substrates.
Does the proRaxX Leader Peptide Have Functional Roles in RiPP Biosynthesis Other than Peptidase Recognition?
In the proRaxX leader peptide, residues proximal to the peptidase cleavage site are well conserved in raxX alleles from all Xanthomonas species (Fig. 7A). These include Gly38 and Gly39 preceding the cleavage site, which are critical for proper proRaxX maturation and secretion (Fig. 7), as well as the hydrophobic residues that occupy positions −4, −7, and −12 from the cleavage site (Val36, Trp33, and Leu28 of proRaxX, respectively; Fig. 3A), which are proposed to position the GG cleavage site in the enzyme’s active site (28, 29). These features are also conserved in leaders of PCAT substrates, including RiPPs and unmodified propeptides (SI Appendix, Fig. S4A), suggesting that this region of the proRaxX leader is important for recognition by the RaxB and PctB peptidase domain. Here, we also show that sulfation of RaxX, which occurs one residue from the cleavage site, is not required for processing and secretion (Fig. 7F).
The leader peptide of RiPPs is hypothesized to serve as a recognition motif for posttranslational modification enzymes (2). We previously detected sulfated nonprocessed proRaxX in Xoo cell lysates (7), suggesting that sulfation occurs while the leader is attached. However, under in vitro conditions, RaxST can sulfate a peptide derivative of human CCR5, containing only an aspartate preceding the tyrosine (38). This result suggests that the proRaxX leader is not required for RaxST function. Similarly, the lacticin 481 propeptide LctA, which is posttranslationally modified by dehydration and cyclization reactions catalyzed by LctM, does not require the leader for its modification (39). Instead, the presence of the leader peptide promotes the efficiency of LctM modification of the LctA core peptide even when supplemented in trans as a synthetic peptide (39) or covalently fused to LctM (40). Additional studies are needed to determine if the proRaxX leader helps recruit RaxST or enhance its sulfotransferase activity in an analogous manner.
Knowledge of RiPP Biosynthesis Can Be Used to Engineer a Diversity of Tyrosine Sulfated Molecules.
The simplicity of the RaxX biosynthetic machinery makes it an ideal system to study how sulfation affects substrate specificity and activity of the biosynthetic proteins. This information can provide insights into the development of strategies to engineer tyrosine sulfated molecules, such as hybrid RiPP products.
Conclusion
Here, we show that PCAT processing is required for both the maturation and export of bacterial RaxX, an immunogen that binds a eukaryotic receptor. These results suggest that RaxX is a tyrosine sulfated RiPP, a previously undescribed group of RIPPs that span both plants and bacteria. We further identified and defined the proRaxX leader, which contains residues that are critical for its maturation and secretion. These results set the stage for leader peptide-guided biosynthetic strategies to extend the chemical diversity of hybrid RiPPs.
Materials and Methods
Peptides.
Tyrosine sY and/or nY versions of RaxX13 (DYPPPGANPKHDP), RaxX16 (DYPPPGANPKHDPPPR), and RaxX21 (HVGGGDYPPPGANPKHDPPPR) were synthesized. The synthetic peptides were ordered from Pacific Immunology, with the exception of RaxX16, which was ordered from Peptide 2.0. Tyrosine sulfated PSY1 was synthesized by the Protein Chemistry Facility at the Gregor Mendel Institute. All of the peptides were resuspended in double-distilled water. Recombinant full-length proRaxX (RaxX60-sY and RaxX60-nY) was isolated from an engineered E. coli strain using an expanded genetic code approach as previously described (7).
XA21ECD Expression and Purification.
The ECD of XA21 (amino acids 23–649) was amplified using primers listed in SI Appendix, Table S2 and inserted into the baculovirus transfer vector pMeIBac B1 (Invitrogen) using RecA-mediated sequence and ligation independent cloning strategy (41). A C-terminal Strep II-9xHis tag was fused to XA21ECD and verified by Sanger sequencing. XA21ECD -StrepII-9xHis was produced by secreted expression in baculovirus-infected High Five insect cells and harvested 72 h postinfection. Subsequently, the protein was purified by nickel affinity chromatography (Ni Sepharose excel; GE Healthcare) and subjected to size exclusion chromatography column (Superdex 200 16/60; GE Healthcare) preequilibrated with 50 mM NaH2PO4/Na2HPO4, pH 7.5, 200 mM NaCl, and 5% glycerol. All of the purification steps were checked by SDS/PAGE.
MST.
Purified XA21ECD was labeled with a fluorescent dye using Monolith Protein Labeling Kit RED-NHS (Amine Reactive; NanoTemper Technologies). Fluorescently labeled XA21ECD (at constant concentration 0.0133 μM) was mixed with varying peptide concentrations (ranging from 60 pM to 2 µM) in buffer containing 50 mM NaH2PO4/Na2HPO4, pH 7.5, 200 mM NaCl, 5% glycerol, and 0.001% Tween. Approximately 4–6 μL of each sample was loaded in a fused silica capillary (NanoTemper Technologies). Measurements were performed at room temperature in a Monolith NT.115 instrument at a constant LED power of 65% and MST power of 60%. Measurements were performed repeatedly on independent protein preparations to ensure reproducibility. The data were analyzed by plotting peptide concentrations against percentage changes of normalized fluorescence. Curve fitting was performed using PALMIST software (42).
Bacterial Strains and Culture.
A list of bacterial strains used in this study is provided in SI Appendix, Table S1. E. coli DH5α, which was used for general cloning, was cultured in LB with the appropriate antibiotics at 37 °C (shaking at 230 rpm for liquid cultures). Xoo strains were routinely cultured at 28 °C on peptone sucrose agar (PSA) with the appropriate antibiotics unless otherwise indicated. Nutrient broth or agar was initially used to generate the mutant strains. When needed, sucrose was added to a final concentration of 5%, and antibiotics were added. For proteomic analysis, cells were grown on XOM2 (43) agar, a plant-mimicking medium. Cephalexin was used at 20 μg/mL, kanamycin was used at 50 μg/mL, and geneticin was used at 50 μg/mL.
Protein Extraction.
Xoo proteins were prepared from strains grown on XOM2 agar plates. After incubation at 28 °C for 2 d, the fully grown cells were scraped from the plates and resuspended in 30 mL of water to loosen the extracellular polysaccharide matrix containing the secretome. The cells were pelleted by centrifugation (17,210 × g for 45 min at 4 °C). The supernatants, which contained the secreted proteins, were harvested and concentrated to 0.5–1 mL using Amicon Ultra Centrifugal Filters Ultracel-3K (Millipore). Total protein was also extracted from the cell pellet. The cells were resuspended and lysed by sonication in lysis buffer (25 mM Tris, pH 8, 300 mM NaCl, 1 mM PMSF, 2.5 mM EDTA, 1 mg/mL lysozyme). Soluble proteins were separated by centrifugation (17,210 × g for 45 min at 4 °C). RaxX-HA was pulled down from cell lysates and supernatants using the μMACS HA isolation kit (Miltenyi Biotec).
All of the samples for MS were digested with 1 μg of trypsin (Promega) in the presence of 5 mM DTT. The tryptic peptides were desalted with a C18 column (Harvard apparatus) and eluted with 80% acetonitrile and 0.1% formic acid.
LC-SRM-MS Analysis.
The SRM-targeted proteomic assays were performed on an Agilent 6460 QQQ mass spectrometer system coupled with an Agilent 1290 UHPLC system as previously described (44). Details are provided in SI Appendix, SI Materials and Methods. The data were acquired using Agilent MassHunter version B.08.02 and processed using Skyline software version 3.70 (MacCoss Lab Software).
Shotgun LC-MS Analysis.
Samples were analyzed using an Agilent 6550 iFunnel Q-TOF mass spectrometer coupled to an Agilent 1290 UHPLC system as previously described (45). Details are provided in SI Appendix, SI Materials and Methods.
Immunoblotting.
Immunoblotting is described in SI Appendix, SI Materials and Methods.
Identification of the pct Locus.
PctB (PXO_RS14825) was identified by National Center for Biotechnology Information (NCBI) Protein BLAST analysis of the PXO99 genome (NC_010717.2) using the RaxB (PXO_RS06015) N-terminal 150 residues as the query under the default settings. PctP (PXO_RS14830) and PctA (PXO_RS14840) were identified by algorithms from the NCBI in the rhomboid family (CDD:304416) and the type_I_hylD (CDD:330454) domain classification, respectively. Pct-92 (PXO_RS14845) and Pct-75 (PXO_RS14850) were annotated as hypothetical proteins.
Xoo Mutation and Complementation.
Xoo mutants were generated in the Philippine race 6 strain PXO99A (referred to as PXO99 in this work) (46). The mutant strains ΔraxX and ΔraxST and the complemented strain ΔraxX(praxX) used in this study were previously reported (7). The rest of the mutant strains (ΔraxB, ΔpctB, ΔraxB ΔpctB, ΔraxA, ΔpctA, and ΔraxA ΔpctA) were generated by double homologous recombination using the pUFR80 suicide vector (47). Briefly, two flanking sequences upstream and downstream of the target gene were independently PCR amplified with their respective primer sets A, B and C, D (SI Appendix, Table S2). Both PCR products were fused together by overlap extension PCR (48) using external primers A and D. This PCR product was then cloned into pUFR80 using the sites EcoRI/HindIII or BamHI/HindIII. The resultant plasmids, which were verified by sequencing, were electroporated into PXO99 (ΔraxB or ΔraxA was used instead for generation of the ΔraxB ΔpctB or ΔraxA ΔpctA double mutant, respectively). The electroporated cells were then plated onto nutrient agar (NA) with kanamycin (50 μg/mL) and grown at 28 °C for 5–6 d. Positive integrants were screened by PCR, grown in nutrient broth without kanamycin overnight, and then plated onto NA containing 5% sucrose to select for the second cross-over event. Colonies were analyzed by PCR to identify deletion mutants and verified by sequencing the PCR product.
For complementation, all of the plasmids (listed in SI Appendix, Table S3) were constructed using standard recombinant DNA techniques using the primers listed in SI Appendix, Table S2. praxX-ha was generated by cloning the raxX coding sequence into pENTR/D-TOPO (Invitrogen), which was then recombined into pLN615, a gateway destination vector containing an HA tag (49). praxSTAB and ppctPB were derived from pVSP61 (50), into which the indicated gene clusters and 0.4 kb of the upstream sequence, presumed to contain the native promoter, were cloned. Expression of raxSTAB and pctPB was chosen instead of independent expression of raxB and pctB, respectively, to account for potential translational coupling with raxA and pctP, respectively. Site-directed mutagenesis of catalytic residues of the RaxB peptidase domain and residues of the proRaxX leader were generated in praxSTAB and praxX, respectively. All of the plasmids were verified by sequencing. The plasmids were transformed into Xoo-competent cells by electroporation, plated onto PSA with the appropriate antibiotic, and grown at 28 °C. Colonies were analyzed by PCR and verified by sequencing the PCR product.
Rice Plants.
The Oryza sativa ssp. japonica rice variety TP309 and a transgenic TP309 line derived from I106-17 (XA21-TP309), which carries the Xa21 gene driven by its own promoter (20), were used for rice inoculations. Native TP309 does not contain Xa21. TP309 and XA21-TP309 rice plants were grown as previously described (7). Briefly, seeds were germinated in distilled water at 28 °C for 1 wk and then, transplanted to sandy soil in 5.5-inch square pots (three seedlings per pot). Plants were grown in tubs filled with fertilizer water in a greenhouse; 6-wk-old plants were then transferred to a growth chamber at least 2 d before inoculation. The growth chamber was set to 28 °C/24 °C, 80/85% humidity, and 14/10-h lighting for the day/night cycle, respectively.
Rice Inoculation.
Xoo strains were cultured on PSA plates with appropriate antibiotics at 28 °C. Bacteria from the plates were resuspended in sterile water at a density of 108 cfu/mL and then, inoculated onto rice plants using the scissor clipping method (51). Briefly, surgical scissors were dipped in bacterial suspension and used to clip the tips of the two uppermost expanded leaves in each tiller, representing one biological replicate. The lengths (centimeters) of the water-soaked lesions extending from the site of inoculation were measured 14 d after inoculation. The lesions of at least eight biological replicates, each a mean of the lesions formed on the two leaves, were measured. At least three independent experiments were performed with similar results unless indicated otherwise.
Analysis of bacterial growth in planta was performed as previously described (52) with minor modifications. Briefly, leaves (a 10-cm section from the site of inoculation) were harvested for each sample set at each time point. The two leaf sections from each tiller, representing one biological replicate, were cut into 2-mm pieces and incubated in sterile water for 2 h at 28 °C with shaking at 230 rpm. This suspension was serially diluted, plated onto PSA plates with cephalexin (20 μg/mL), and incubated at 28 °C. Colonies were counted 2 d later, and the cell number per 10-cm leaf section was calculated. Three to four biological replicates, each a mean number of colonies from four technical replicates, were measured for each data point. Three independent experiments were performed with similar results.
For in planta bacterial gene expression analysis, TP309 leaves were harvested 6 d after inoculation. The leaves were cut 4 cm from the site of inoculation, and the 4-cm section from two leaves of each tiller, representing one biological replicate, was snap frozen in liquid nitrogen and processed as described below.
RNA Extraction and qRT-PCR.
Total RNA was extracted from homogenized rice leaf tissue using TRIzol reagent (Invitrogen) and treated with the TURBO DNA-free kit (Ambion) to remove residual genomic DNA. cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The cycle threshold (Ct) value was measured on a Bio-Rad CFX96 Real-Time System coupled to a C1000 Thermal Cycler (Bio-Rad) using the iTaq Universal SYBR Green Supermix (Bio-Rad) and 100 ng cDNA. The primers used are listed in SI Appendix, Table S2. The 2(−ΔΔCt) method was used to calculate the relative changes in gene expression normalized to the ampC2 (PXO_RS14730) reference gene control and the PXO99 wild-type strain. Three biological replicates, each a mean of two technical replicates, were analyzed. Three independent experiments were performed with similar results.
Sequence Analysis and Visualization.
The C and H motifs of the RaxB and PctB peptidase domains were identified from alignments of the peptidase domain of the E. coli ColV transporter CvaB (CAA40744.1) (18), the L. mesenteroides mesentericin B105 and Y105 transporter MesD (CAA57402.1) (26, 53), the Streptococcus pneumoniae competence-stimulating peptide transporter ComA (AAA69510.1) (54), the Lactococcus lactis lactococcin G transporter LagD (ACR43772.1) (17), the L. lactis lacticin 481 transporter LctT (AAC72259.1) (55), and the Staphylococcus warneri nukacin ISK-1 transporter NukT (NP_940774.2) (56) as well as the peptidase-like domain of the Aggregatibacter actinomycetemcomitans leukotoxin transporter LktB (CAA37906.1) (57) and the E. coli alpha-hemolysin transporter HlyB (KKA61973.1) (24). The alignment was performed using the default settings in Clustal Omega (European Bioinformatics Institute; EMBL-EBI) (58).
The sequence logo of the proRaxX leader was constructed using WebLogo (59) with the predicted leader sequence of proRaxX alleles from various Xanthomonas species identified by Pruitt et al. (10). The RaxX leader was aligned with the predicted leader of the candidate PctB substrates Pct-92 and Pct-75 as well as the experimentally verified leader sequences from the E. coli ColV propeptide CvaC (AAX22078.1) (25); the L. mesenteroides mesentericin B105 and Y105 propeptides MesB and MesY (AAD54223.1 and AAP37395.1, respectively) (26, 53); the S. pneumoniae competence-stimulating peptide propeptide ComC (AAC44895.1) (54); the L. lactis lactococcin alpha and beta propeptides LagA and LagB, respectively (ACR43769.1 and ACR43770.1, respectively) (17); the L. lactis lacticin 481 propeptide LctA (AAC72257.1) (55); and the S. warneri nukacin ISK-1 propeptide NukA (NP_940772.1) (56).
Statistical Analysis.
Statistical analysis was performed using JMP Pro-13 (SAS Institute Inc.) or GraphPad Prism 7 (GraphPad Software) and is specified in the figures. Differences were considered to be significant when P < 0.05.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants GM59962 and GM122968 and NSF Plant Genome Research Program Grant IOS-1237975 (to P.C.R.). This work was also partially supported by the Office of Biological and Environmental Research’s Genomic 275 Science Program within the US Department of Energy Office of Science Award DE-AC02-05CH11231. K.P. was supported by the Vienna Science and Technology Fund Project LS17-047.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818275116/-/DCSupplemental.
References
- 1.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30:108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oman TJ, van der Donk WA. Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6:9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burkhart BJ, Kakkar N, Hudson GA, van der Donk WA, Mitchell DA. Chimeric leader peptides for the generation of non-natural hybrid RiPP products. ACS Cent Sci. 2017;3:629–638. doi: 10.1021/acscentsci.7b00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stone MJ, Chuang S, Hou X, Shoham M, Zhu JZ. Tyrosine sulfation: An increasingly recognised post-translational modification of secreted proteins. N Biotechnol. 2009;25:299–317. doi: 10.1016/j.nbt.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Farzan M, et al. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 6.Dardick C, Schwessinger B, Ronald P. Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr Opin Plant Biol. 2012;15:358–366. doi: 10.1016/j.pbi.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Pruitt RN, et al. The rice immune receptor XA21 recognizes a tyrosine-sulfated protein from a Gram-negative bacterium. Sci Adv. 2015;1:e1500245. doi: 10.1126/sciadv.1500245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwessinger B, et al. A second-generation expression system for tyrosine-sulfated proteins and its application in crop protection. Integr Biol. 2016;8:542–545. doi: 10.1039/c5ib00232j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei T, Chern M, Liu F, Ronald PC. Suppression of bacterial infection in rice by treatment with a sulfated peptide. Mol Plant Pathol. 2016;17:1493–1498. doi: 10.1111/mpp.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pruitt RN, et al. A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone. New Phytol. 2017;215:725–736. doi: 10.1111/nph.14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flor HH. Inheritance of pathogenicity in Melampsora lini. Phytopathology. 1942;32:653–669. [Google Scholar]
- 12.Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:18333–18338. doi: 10.1073/pnas.0706403104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.da Silva FG, et al. Bacterial genes involved in type I secretion and sulfation are required to elicit the rice Xa21-mediated innate immune response. Mol Plant Microbe Interact. 2004;17:593–601. doi: 10.1094/MPMI.2004.17.6.593. [DOI] [PubMed] [Google Scholar]
- 14.Kanonenberg K, Schwarz CKW, Schmitt L. Type I secretion systems–A story of appendices. Res Microbiol. 2013;164:596–604. doi: 10.1016/j.resmic.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 15.Wienken CJ, Baaske P, Rothbauer U, Braun D, Duhr S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat Commun. 2010;1:100. doi: 10.1038/ncomms1093. [DOI] [PubMed] [Google Scholar]
- 16.Zhang LH, Fath MJ, Mahanty HK, Tai PC, Kolter R. Genetic analysis of the colicin V secretion pathway. Genetics. 1995;141:25–32. doi: 10.1093/genetics/141.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Håvarstein LS, Diep DB, Nes IF. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 18.Wu KH, Tai PC. Cys32 and His105 are the critical residues for the calcium-dependent cysteine proteolytic activity of CvaB, an ATP-binding cassette transporter. J Biol Chem. 2004;279:901–909. doi: 10.1074/jbc.M308296200. [DOI] [PubMed] [Google Scholar]
- 19.Freeman M. The rhomboid-like superfamily: Molecular mechanisms and biological roles. Annu Rev Cell Dev Biol. 2014;30:235–254. doi: 10.1146/annurev-cellbio-100913-012944. [DOI] [PubMed] [Google Scholar]
- 20.Song WY, et al. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 21.Dirix G, et al. Peptide signal molecules and bacteriocins in Gram-negative bacteria: A genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides. 2004;25:1425–1440. doi: 10.1016/j.peptides.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 22.Wu KH, Hsieh YH, Tai PC. Mutational analysis of Cvab, an ABC transporter involved in the secretion of active colicin V. PLoS One. 2012;7:e35382. doi: 10.1371/journal.pone.0035382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ménard R, et al. Contribution of the glutamine 19 side chain to transition-state stabilization in the oxyanion hole of papain. Biochemistry. 1991;30:8924–8928. doi: 10.1021/bi00101a002. [DOI] [PubMed] [Google Scholar]
- 24.Lecher J, et al. An RTX transporter tethers its unfolded substrate during secretion via a unique N-terminal domain. Structure. 2012;20:1778–1787. doi: 10.1016/j.str.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Håvarstein LS, Holo H, Nes IF. The leader peptide of colicin V shares consensus sequences with leader peptides that are common among peptide bacteriocins produced by Gram-positive bacteria. Microbiology. 1994;140:2383–2389. doi: 10.1099/13500872-140-9-2383. [DOI] [PubMed] [Google Scholar]
- 26.Aucher W, Lacombe C, Héquet A, Frère J, Berjeaud JM. Influence of amino acid substitutions in the leader peptide on maturation and secretion of mesentericin Y105 by Leuconostoc mesenteroides. J Bacteriol. 2005;187:2218–2223. doi: 10.1128/JB.187.6.2218-2223.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotake Y, Ishii S, Yano T, Katsuoka Y, Hayashi H. Substrate recognition mechanism of the peptidase domain of the quorum-sensing-signal-producing ABC transporter ComA from Streptococcus. Biochemistry. 2008;47:2531–2538. doi: 10.1021/bi702253n. [DOI] [PubMed] [Google Scholar]
- 28.Ishii S, et al. Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway. J Biol Chem. 2010;285:10777–10785. doi: 10.1074/jbc.M109.093781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bobeica SC, et al. Insights into AMS/PCAT transporters from biochemical and structural characterization of a double Glycine motif protease. eLife. 2019;8:e42305. doi: 10.7554/eLife.42305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagedroste M, Smits SHJ, Schmitt L. Substrate specificity of the secreted nisin leader peptidase NisP. Biochemistry. 2017;56:4005–4014. doi: 10.1021/acs.biochem.7b00524. [DOI] [PubMed] [Google Scholar]
- 31.Wang J, et al. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature. 2015;525:265–268. doi: 10.1038/nature14858. [DOI] [PubMed] [Google Scholar]
- 32.Matsubayashi Y, Sakagami Y. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc Natl Acad Sci USA. 1996;93:7623–7627. doi: 10.1073/pnas.93.15.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 34.Doblas VG, et al. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science. 2017;355:280–284. doi: 10.1126/science.aaj1562. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama T, et al. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science. 2017;355:284–286. doi: 10.1126/science.aai9057. [DOI] [PubMed] [Google Scholar]
- 36.Lawton EM, Cotter PD, Hill C, Ross RP. Identification of a novel two-peptide lantibiotic, haloduracin, produced by the alkaliphile Bacillus halodurans C-125. FEMS Microbiol Lett. 2007;267:64–71. doi: 10.1111/j.1574-6968.2006.00539.x. [DOI] [PubMed] [Google Scholar]
- 37.Caetano T, Barbosa J, Möesker E, Süssmuth RD, Mendo S. Bioengineering of lanthipeptides in Escherichia coli: Assessing the specificity of lichenicidin and haloduracin biosynthetic machinery. Res Microbiol. 2014;165:600–604. doi: 10.1016/j.resmic.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 38.Han S-W, et al. Tyrosine sulfation in a Gram-negative bacterium. Nat Commun. 2012;3:1153. doi: 10.1038/ncomms2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levengood MR, Patton GC, van der Donk WA. The leader peptide is not required for post-translational modification by lacticin 481 synthetase. J Am Chem Soc. 2007;129:10314–10315. doi: 10.1021/ja072967+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oman TJ, Knerr PJ, Bindman NA, Velásquez JE, van der Donk WA. An engineered lantibiotic synthetase that does not require a leader peptide on its substrate. J Am Chem Soc. 2012;134:6952–6955. doi: 10.1021/ja3017297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scholz J, Besir H, Strasser C, Suppmann S. A new method to customize protein expression vectors for fast, efficient and background free parallel cloning. BMC Biotechnol. 2013;13:12. doi: 10.1186/1472-6750-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheuermann TH, Padrick SB, Gardner KH, Brautigam CA. On the acquisition and analysis of microscale thermophoresis data. Anal Biochem. 2016;496:79–93. doi: 10.1016/j.ab.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuge S, et al. Expression of Xanthomonas oryzae pv. oryzae hrp genes in XOM2, a novel synthetic medium. J Gen Plant Pathol. 2002;68:363–371. [Google Scholar]
- 44.Batth TS, et al. A targeted proteomics toolkit for high-throughput absolute quantification of Escherichia coli proteins. Metab Eng. 2014;26:48–56. doi: 10.1016/j.ymben.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 45.González Fernández-Niño SM, et al. Standard flow liquid chromatography for shotgun proteomics in bioenergy research. Front Bioeng Biotechnol. 2015;3:44. doi: 10.3389/fbioe.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hopkins CM, White FF, Choi SH, Guo A, Leach JE. Identification of a family of avirulence genes from Xanthomonas oryzae pv. oryzae. Mol Plant Microbe Interact. 1992;5:451–459. doi: 10.1094/mpmi-5-451. [DOI] [PubMed] [Google Scholar]
- 47.Castañeda A, Reddy JD, El-Yacoubi B, Gabriel DW. Mutagenesis of all eight avr genes in Xanthomonas campestris pv. campestris had no detected effect on pathogenicity, but one avr gene affected race specificity. Mol Plant Microbe Interact. 2005;18:1306–1317. doi: 10.1094/MPMI-18-1306. [DOI] [PubMed] [Google Scholar]
- 48.Horton RM, Hunt HD, Ho SN, Pullen JK, Pease LR. Engineering hybrid genes without the use of restriction enzymes: Gene splicing by overlap extension. Gene. 1989;77:61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- 49.Guo M, Tian F, Wamboldt Y, Alfano JR. The majority of the type III effector inventory of Pseudomonas syringae pv. tomato DC3000 can suppress plant immunity. Mol Plant Microbe Interact. 2009;22:1069–1080. doi: 10.1094/MPMI-22-9-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loper JE, Lindow SE. A biological sensor for iron available to bacteria in their habitats on plant surfaces. Appl Environ Microbiol. 1994;60:1934–1941. doi: 10.1128/aem.60.6.1934-1941.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kauffman HE, Reddy APK, Hsieh SPY, Merca SD. An improved technique for evaluating resistance of rice varieties to Xanthomonas oryzae. Plant Dis Rep. 1973;57:537–541. [Google Scholar]
- 52.Bahar O, et al. The Xanthomonas Ax21 protein is processed by the general secretory system and is secreted in association with outer membrane vesicles. PeerJ. 2014;2:e242. doi: 10.7717/peerj.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morisset D, Frère J. Heterologous expression of bacteriocins using the mesentericin Y105 dedicated transport system by Leuconostoc mesenteroides. Biochimie. 2002;84:569–576. doi: 10.1016/s0300-9084(02)01413-x. [DOI] [PubMed] [Google Scholar]
- 54.Ishii S, Yano T, Hayashi H. Expression and characterization of the peptidase domain of Streptococcus pneumoniae ComA, a bifunctional ATP-binding cassette transporter involved in quorum sensing pathway. J Biol Chem. 2006;281:4726–4731. doi: 10.1074/jbc.M512516200. [DOI] [PubMed] [Google Scholar]
- 55.Furgerson Ihnken LA, Chatterjee C, van der Donk WA. In vitro reconstitution and substrate specificity of a lantibiotic protease. Biochemistry. 2008;47:7352–7363. doi: 10.1021/bi800278n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nishie M, Shioya K, Nagao J, Jikuya H, Sonomoto K. ATP-dependent leader peptide cleavage by NukT, a bifunctional ABC transporter, during lantibiotic biosynthesis. J Biosci Bioeng. 2009;108:460–464. doi: 10.1016/j.jbiosc.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 57.Guthmiller JM, Kolodrubetz D, Kraig E. Mutational analysis of the putative leukotoxin transport genes in Actinobacillus actinomycetemcomitans. Microb Pathog. 1995;18:307–321. doi: 10.1006/mpat.1995.0028. [DOI] [PubMed] [Google Scholar]
- 58.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.