Significance
The precise locations of refugia from which temperate species expanded following the Last Glacial Maximum have yet to be precisely identified. We use a novel method that harnesses genomic footprints of past geography to estimate the latitude and longitude of these refugia. We detect expansion from a northern source near the edge of glaciation in one eastern North American tree species and from a southern source in a second codistributed species. The inferred northern expansion origin in a climatically inhospitable region provides strong support for the existence of elusive microrefugia. Our approach addresses classic questions about the nature of population persistence under different climatic conditions and demonstrates how leveraging genomic data allows statistical validation of controversial historical paradigms.
Keywords: approximate Bayesian computation, eastern North America, glacial refugia, microrefugia, temperate trees
Abstract
Temperate species experienced dramatic range reductions during the Last Glacial Maximum, yet refugial populations from which modern populations are descended have never been precisely located. Climate-based models identify only broad areas of potential habitat, traditional phylogeographic studies provide poor spatial resolution, and pollen records for temperate forest communities are difficult to interpret and do not provide species-level taxonomic resolution. Here we harness signals of range expansion from large genomic datasets, using a simulation-based framework to infer the precise latitude and longitude of glacial refugia in two widespread, codistributed hickories (Carya spp.) and to quantify uncertainty in these estimates. We show that one species likely expanded from close to ice sheet margins near the site of a previously described macrofossil for the genus, highlighting support for the controversial notion of northern microrefugia. In contrast, the expansion origin inferred for the second species is compatible with classic hypotheses of distant displacement into southern refugia. Our statistically rigorous, powerful approach demonstrates how refugia can be located from genomic data with high precision and accuracy, addressing fundamental questions about long-term responses to changing climates and providing statistical insight into longstanding questions that have previously been addressed primarily qualitatively.
Locations in which temperate taxa from eastern North America might have persisted during the dramatic climatic changes of the Last Glacial Maximum (LGM; ca. 21.5 ka) (1) remain largely unknown, even though there is no question that temperate species across the globe have repeatedly experienced major range shifts in response to Pleistocene glacial cycles (2–4). Identifying the locations of glacial refugia has been a focus of many phylogeographic studies, in part because of the importance of refugial histories for explaining contemporary population genetic structures (5), defining regions of long-term conservation priority (6), predicting migration capacity given future climate change (7), and inferring how dispersal limitation has impacted extinction rates and species richness patterns (8, 9).
Numerous refugial regions have been proposed in eastern North America, including the Gulf and Atlantic Coastal Plains, the Lower Mississippi River Valley, the Southern Appalachians, peninsular Florida, and central Texas (10–14), and opinions on the extent of such refugia vary widely. For example, proposals for some species include widespread distributions throughout unglaciated eastern North America during the LGM (15–17), some of which include areas not far from the southern edge of the Laurentide Ice Sheet (18). Moreover, most pollen assemblages across this region suggest nonanalog conifer-dominated communities with only trace to low levels of temperate deciduous tree pollen or communities with species characteristic of more open vegetation (1, 15, 19, 20), fueling the ongoing debate about the nature of glacial refugia for temperate taxa.
Likewise, the discovery of macrofossils of several mesic cool-temperate deciduous taxa in the northern Lower Mississippi River Valley (ca. 35°N) (1, 21) suggests that some species might have survived in localized microrefugia. Microrefugia are sites in which unusual local microclimates may have allowed persistence of isolated populations within a broader region of generally inhospitable climate (22–26). However, the dominance of Picea, Larix, and other boreal taxa in regional pollen assemblages (1, 19) implies that such microrefugia would have supported only a select set of temperate species. Although regional climates were cold and dry, some local sites along the edge of the valley likely had a cool, humid climate, possibly due to frequent fogs that transported moisture from nearby glacial meltwater (21, 27). However, even if some species might have been able to survive in these microrefugia given their ecological characteristics and specific habitat affinity, it is not clear whether these microrefugia persisted throughout the entire LGM, and, consequently, whether they were likely sources of postglacial recolonization. In other words, even if cool, mesic microsites along the northern Lower Mississippi River Valley may have in theory provided opportunities for persistence of temperate species adapted to such conditions, whether they were actually a source of expansion of species that now populate previously glaciated regions is far from clear. As such, classical views of displacement to more distant, southern geographic regions may still be most likely for many taxa (3, 4, 13, 28).
Here we address this debate by using genomic data to infer the actual latitude and longitude of postglacial range expansion in two broadly distributed hickory species: bitternut hickory, Carya cordiformis (Wangenh.) K. Koch, (Juglandaceae) and shagbark hickory, Carya ovata, (Mill.) K. Koch from eastern North America. Although they are codistributed, the species differ in their respective habitat affinities, with C. cordiformis found predominantly in mesic sites and bottomlands, whereas C. ovata is more abundant on dry sites in upland habitats (29–31). Leveraging the resolution of genomic data, combined with analytical advances for estimating the latitude and longitude from which range expansion was initiated and quantifying statistical uncertainty in these estimates, our work tackles classic questions about the nature of population persistence under different climatic conditions and demonstrates how genomic data can provide statistical insight into controversial historical scenarios.
Results and Discussion
Locating Geographic Coordinates of Refugia with Genomic Data.
The ability to extract detailed spatial and demographic information about range expansion from population genomic data (32) provides a powerful, statistically rigorous means of identifying the geographic coordinates of glacial refugia from which range expansion proceeded when species were displaced by changing climatic conditions. Here we infer different expansion origins for the two codistributed hickory species and show the inferred origins to be both precisely and accurately estimated.
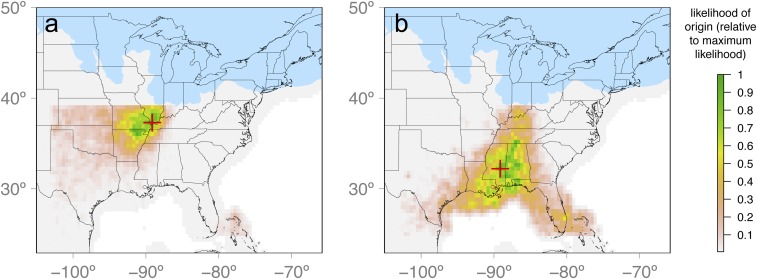
To estimate the geographic expansion origins of each hickory species, we used an approximate Bayesian computation (ABC) framework (33) in which the latitude and longitude of the expansion origin is a model parameter (32) estimated by comparing empirical genetic summary statistics to summary statistics generated from spatially explicit demographic and coalescent simulations initiated from a broad geographic prior for different potential origins of population expansion (Fig. 1 and Table 1). Because it is not necessary to define specific refugial scenarios to test a priori (e.g., refs. 34 and 35), expansion origins can be identified de novo and independently from alternative sources of inference, such as the fossil record, which may then serve as a source of corroboration. For example, our genomic results for C. cordiformis (Fig. 2A) converge with independent macrofossil evidence (1, 21) in identifying a likely location of microrefugia for mesic cool-temperate deciduous trees. Specifically, the latitude and longitude of the estimated expansion origin (Ω) for C. cordiformis was located near the confluence of the Mississippi and Ohio Rivers (37.3°N, 89.1°W), with high-likelihood areas covering the northern Lower Mississippi River Valley (Fig. 2A). This inferred origin close to the LGM ice sheet margin is consistent with controversial proposals for expansion from microrefugia and with macrofossil evidence of the presence of some mesic cool-temperate deciduous trees in this region (1, 21). In contrast, for C. ovata, the expansion origin corresponds to more traditional proposals of a southern refuge, in this case in central Mississippi in the eastern Gulf Coastal Plain (32.2°N, 89.1°W), with high-likelihood areas covering most of Alabama, Mississippi, and southeastern Louisiana (Fig. 2B).
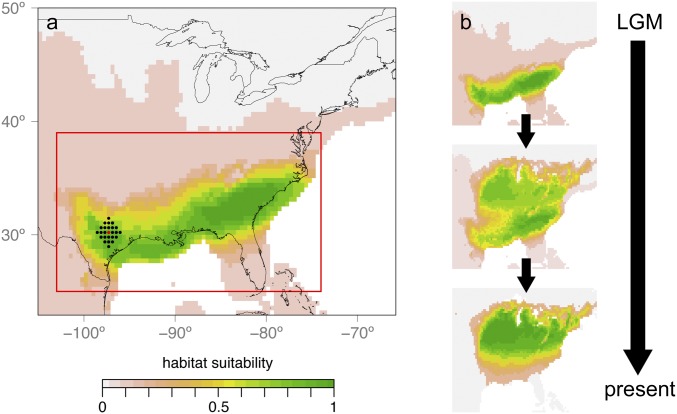
Fig. 1.
Schematic overview of demographic simulations. (A) Simulations were initiated in the LGM landscape (shown here for C. cordiformis) from a central deme (see red dot as an example) plus an area extending three additional demes (black dots) in all directions. Different geographic sources of expansion were modeled as selected from a uniform geographic prior (red rectangle). Per deme carrying capacities were scaled relative to maximum carrying capacity (Kmax) according to habitat suitability from an SDM. (B) Individuals were allowed to colonize the landscape with relative carrying capacities shifting from the LGM to present.
Table 1.
Priors on model parameters
| Parameter | Description | Distribution | Minimum | Maximum |
| Latitude | Latitude of Ω (°) | Uniform | 25 | 39 |
| Longitude | Longitude of Ω (°) | Uniform | −103 | −74 |
| Nanc | Ancestral population size before initiating expansion (number of individuals) | Log-uniform | 103.3 | 105.0 |
| Kmax | Maximum carrying capacity of a deme (number of individuals) | Log-uniform | 103.3 | 105.0 |
| m | Migration rate between neighboring demes (proportion of individuals migrating per generation) | Log-uniform | 10−2.0 | 10−0.3 |
The same priors were used in both species.
Fig. 2.
Estimated expansion origins (Ω; red cross) in C. cordiformis (A) and C. ovata (B). The shading of pixels depicts a probability surface (kernel density) showing the likelihood that each pixel served as the expansion origin relative to the pixel with the highest likelihood (i.e., Ω). Glaciated regions are shown in blue. The results presented in A and B are based on retention of four and three PC axes of variation in genetic summary statistics, respectively. Results based on retaining additional PC axes are presented in SI Appendix, Figs. S2 and S3.
In addition to being corroborated by independent data, our results are strongly statistically supported, with clearly distinguished regions that differ in their relative likelihoods of serving as the expansion origin (Fig. 2), low errors associated with the estimated expansion origin overall (Fig. 3), and a low probability that the expansion origin was misidentified given our specific empirical datasets (SI Appendix, Fig. S8). Comparisons of prior and posterior distributions show that demographic model parameters were mostly estimated with good precision (e.g., the per deme maximum carrying capacity, Kmax, and migration rate, m), with the exception of the ancestral population size, Nanc (SI Appendix, Fig. S4). Latitude and longitude were also estimated with good precision (SI Appendix, Fig. S5), although estimates of these variables considered in isolation should be interpreted with caution due to the 2D nature of the simulations; the kernel density of retained simulations provides a more powerful approach to their joint inference (Fig. 2).
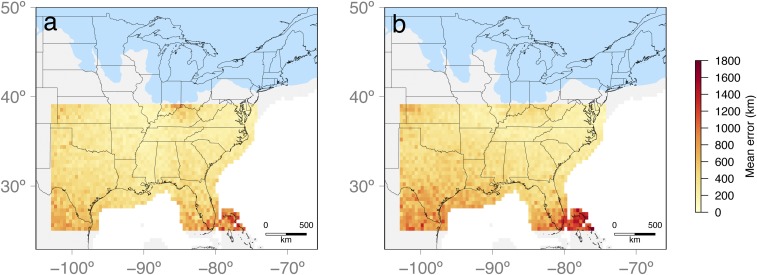
Fig. 3.
Mean error in expansion origins (Ω) estimated from PODs for C. cordiformis (A) and C. ovata (B). Colors of each pixel show the mean geographic distance between the inferred Ω and the true expansion origin, when the true origin was located in that pixel. Mean errors of each pixel are calculated from a total of 10 PODs.
A set of complementary power analyses validated the accuracy of our estimates, which are based on analysis of >1,000 SNPs in approximately 150 individuals from across each species’ range (36). Specifically, the average error in estimating the expansion origin from pseudo-observed datasets (PODs) simulated with known expansion origins (Fig. 3) was low in both species (C. cordiformis: median, 250 km; mean, 338 km; C. ovata: median, 325 km; mean, 421 km). Likewise, it is highly unlikely that the empirical expansion origins estimated for each species would have been incorrectly inferred if the true expansion were actually from another geographic region (SI Appendix, Fig. S8). The only region with a high average error is located in the Bahamas and southern Florida far from the inferred expansion origin coordinates (Fig. 3), and only C. ovata has a low probability of failure to detect true expansion from this region (SI Appendix, Fig. S8D), which most likely reflects the high overall error for this region, given that it is not hypothesized to be an expansion source for cool-temperate trees (1, 11, 13). Importantly, however, it is particularly unlikely for C. cordiformis that a northern expansion origin would be inferred, given expansion from a southern source (SI Appendix, Fig. S8 A and B), providing further confidence in the inferred existence of putative microrefugia in this region (1, 21).
Our analyses also show that our models are capable of generating data consistent with aspects of the empirical data in both species. Specifically, we estimated Wegmann’s P value, defined as the proportion of retained simulations with a smaller likelihood than that of the empirical data (37). Wegmann’s P value was high (0.86 for C. cordiformis and 1.0 for C. ovata), suggesting that the models were able to generate simulated datasets that closely matched the empirical data in terms of the summary statistics we used. Direct comparison of the empirical, simulated, and retained summary statistics also suggests that the retained simulations closely matched the empirical data (SI Appendix, Figs. S6 and S7). However, much lower Wegmann’s P values for higher dimensions of the data (i.e., for principal component axes of variation in summary statistics beyond those retained in the presented analysis; SI Appendix, Figs. S2 and S3 and Table S3), suggest there are some aspects of our datasets for which the models are a poor fit.
As with any model-based inference, our models contain a number of simplifications that affect their scope of inference. Previous analysis of the species (17) revealed no signatures of multiple genetically isolated refugia in either species, except for a genetically distinct Texas population of C. ovata. This population was not included here because ignoring its distinctive phylogeographic history would invalidate any inference based on allele frequency gradients arising due to range expansion from the same ancestral population, which is the foundation of the summary statistics that we used (32, 38). We expect that if expansion of the populations studied here had occurred from multiple refugia or a broad, diffuse area, we might have obtained results with high error or with signal of expansion from multiple regions, yet we obtained a strong signal for a high likelihood of expansion from a single, geographically well-defined region (Fig. 2).
It is also worth considering whether the inferred expansion origins can be confidently dated to the LGM. If range-wide genetic structure in the empirical dataset corresponds to events that occurred during a different time period, this would confound inferences based on comparisons with simulations initiated during the LGM. However, previous analysis found no evidence of genetic structure that likely predates the LGM (17). The divergence of the genetically distinct Texas population of C. ovata may be an exception, but as noted, its distinctive history is separate from and thus not part of the expansion origins estimated here. Demographic events that occurred more recently than postglacial expansion could affect local populations but would not be expected to fundamentally alter range-wide genetic structure. Likewise, high gene flow could homogenize populations and erode expansion signals over time, yet the empirical data retain clear expansion signals (SI Appendix, Tables S1 and S2).
In addition, we note that X-ORIGIN predicts the origin of lineages that genetically contributed to postglacial recolonization (32), not the complete LGM geographic distribution of the entire species, as with inferences based on ecological niche models (e.g., ref. 17). Additional populations that made little or no contribution to postglacial expansion may have existed in areas identified as having a low likelihood of serving as the expansion origin. For example, some southern populations of C. cordiformis (SI Appendix, Fig. S1A) may have persisted since the LGM, and the genetic distinctiveness of the excluded Texas population of C. ovata (17) strongly suggests that C. ovata was also present west of the Mississippi River. In this sense, our models do not capture the entire LGM and postglacial history of either species. Our models are also unable to generate data consistent with the higher dimensions of the observed data (SI Appendix, Table S3), which may reflect the effects of locally distributed alleles in the empirical data that are found at high frequency only in a few populations, or the effects of unusual patterns of gene flow between particular populations, which our range-wide models are not designed to accommodate.
Nevertheless, even though our models admittedly do not capture the entire LGM and postglacial history of the species, our results provide strong statistical support for specific geographic locations of glacial refugia that served as expansion origins in both species, which we were able to corroborate with independent fossil evidence (C. cordiformis) and compare with previous phylogeographic knowledge from other species (C. ovata). As such, our approach highlights the power of harnessing signals from genomic data to infer the locations of postglacial range expansion, either to test hypotheses from the fossil record and climate change models or to identify refugia de novo for taxa lacking detailed species-specific knowledge.
Ecology Impacts Where Species Persist.
The identification of the northern Lower Mississippi River Valley as the likely expansion origin in C. cordiformis with high statistical support provides some of the strongest genetic evidence to date that northern microrefugia, in addition to or instead of classic southern refugial areas, played a role in the LGM survival and postglacial recolonization of some temperate tree species (22, 24–26, 39). Given the dominance of boreal species in the cold, dry climates of this region during the LGM (1, 19, 21), regional climates were unlikely to have been generally suitable for survival of C. cordiformis (Fig. 1) (17) and other temperate species. Evidence of expansion from this region therefore suggests survival in localized habitats best characterized as microrefugia (26). However, the widespread presence of nonanalog communities in eastern North America during the LGM (1, 20) has complicated efforts to understand community-level responses to Pleistocene glaciation. In particular, the extent to which codistributed species shared glacial refugia remains unknown. Although our results provide strong evidence that northern microrefugia existed for some temperate deciduous trees, including C. cordiformis, these microrefugia may have been occupied by only a subset of species adapted to local microhabitats. In particular, the inferred expansion origin for C. ovata in the eastern Gulf Coastal Plain (Fig. 2)—a region previously proposed as a general glacial refugium for many temperate taxa (11, 13) and predicted to have contained suitable regional-scale climates for C. ovata (17)—is compatible with classic phylogeographic paradigms of displacement far from the edges of continental ice sheets.
The different refugial histories inferred for these two codistributed species suggest that contemporary regional communities may have been assembled from populations from diverse geographic sources. The ability of relict populations to persist in local areas during periods of climate change is affected by a variety of abiotic and biotic constraints (39), and species differing in such traits as habitat affinity and dispersal ability often exhibit different phylogeographic patterns and population genetic structure (34, 40–42). Here, consideration of the specific habitat affinities of C. cordiformis and C. ovata compared with the local microhabitats that likely existed in the northern Lower Mississippi River Valley during the LGM may explain why different refugia were inferred. Specifically, the putatively cool, moist microclimates along the edge of the valley (21) were likely suitable for survival of the mesic, primarily bottomland species C. cordiformis (30, 31) but might have been less favorable for the persistence of C. ovata than drier, upland areas (29, 31) that may have existed in the low hills and rolling plains of the eastern Gulf Coastal Plain.
Although the phylogeographic histories of species with differing ecological requirements may be nonconcordant, are any patterns generalizable across taxa? Interestingly, we note an emerging pattern in which tree species hypothesized to have persisted in northern regions are often adapted to mesic or wetter microsites. In both eastern North America and Europe, northern persistence has been inferred for various mesic trees and shrubs [e.g., C. cordiformis (this study), Dirca palustris (43), Fagus grandifolia (18), Fagus sylvatica (44–46)] and habitat generalists [e.g., Acer rubrum (18); Quercus rubra (16)], but not for species adapted to drier microhabitats. It is possible that cool, moist, sheltered microclimates may have been more common near the margins of continental ice sheets than warm, dry, upland microsites, but given that the potential for northern mircorefugia has been explicitly investigated in only a handful of tree species, further research, especially in dry-adapted taxa, is needed.
Conclusions.
Our analyses demonstrate a powerful approach to addressing pressing, unresolved questions about the nature of population persistence under different climatic conditions. The phylogeographic history of temperate tree taxa has been at the center of this debate because of enigmatic aspects of the pollen record for temperate trees. In particular, we have used genetic data to estimate the actual latitude and longitude from which the range expansion of plant taxa was initiated and to quantify relative statistical support for these estimates, as opposed to relying on pollen data, projections from distributional models, or qualitative interpretation of genetic structure. Our results corroborate both classic refugial hypotheses and more controversial proposals concerning the existence of northern microrefugia by showing that even species that are codistributed today may have substantially differing histories as a function of differences in their ecology. Our work points to the utility of population genomic data to resolve longstanding questions in other, ecologically divergent taxa from eastern North America and in taxa from other regions of the world where phylogeographic narratives concerning the regional biota are in flux (2, 4, 25, 28).
Finally, increasing evidence of expansion out of northern microrefugia suggests that conventional wisdom about management of genetic diversity may need to be revised. Northern populations that were recently recolonized are often thought to be unimportant for conservation of genetic diversity and long-term species survival relative to southern populations that are believed to be reservoirs of unique genetic diversity (6, 47). However, especially in eastern North America, where genetic diversity of temperate trees does not typically decline with increasing latitude (17, 48), midlatitude regions that potentially harbored microrefugia, such as the northern Lower Mississippi River Valley, may be equally important reservoirs of genetic diversity and long-term population stability and ought to be additional areas of high conservation priority.
Materials and Methods
Study Species.
C. cordiformis and C. ovata are widespread, wind-pollinated and animal-dispersed trees native to the temperate deciduous forest biome of eastern North America (SI Appendix, Fig. S1) (49). Both species inhabit a range of different sites (29, 30); however, C. cordiformis is a more mesic species common in fertile bottomlands, whereas C. ovata is more abundant in dry, upland habitats (31). Carya fossil pollen from the LGM is broadly distributed longitudinally across southeastern North America (1, 50), while macrofossils are known from just two sites along the Lower Mississippi River Valley (1, 21, 51). A lack of strong genetic structure characterizes both species, except for a phylogeographic break between a single Texas population and all other populations of C. ovata (17), which suggests that both species were fairly geographically widespread over southeastern North America during the LGM. The Texas population of C. ovata was excluded here because it likely has a separate history from the rest of the population and did not make a major contribution to postglacial recolonization of the study area (17); ignoring phylogeographic breaks within the species to infer a single expansion history for the entire species would invalidate any inference based on allele frequency gradients (32, 38).
Empirical Genetic Data.
Previously published datasets of putatively unlinked SNPs identified without ascertainment bias from double-digest restriction site-associated DNA sequencing (17, 36) were obtained for both species. Populations represented by fewer than five individuals per species were excluded because they contain limited information on allele frequency differences among populations. The minimum minor allele frequency was set to 3.3% (17). This resulted in final datasets of 1,046 SNPs from 20 populations (168 individuals total) for C. cordiformis and 1,018 SNPs from 17 populations (148 individuals) for C. ovata, with an overall genotyping rate of 88% in both species. The high precision and accuracy inherent in our power analyses (Fig. 3 and SI Appendix, Fig. S8) indicates that 1,000 SNPs was an adequate number for distinguishing among scenarios. Details of SNP discovery and filtering are provided in SI Appendix.
Demographic and Coalescent Simulations.
The X-ORIGIN pipeline (32) was used to conduct demographic and coalescent simulations and to estimate the expansion origin of each species. The key components of X-ORIGIN are to (i) simulate a spatially explicit demographic model of population expansion, with carrying capacities of demes and migration rates scaled relative to habitat suitability from species distribution models (SDMs); (ii) to simulate genetic data for each set of simulated demographic conditions (i.e., different carrying capacities and migration rates) under a spatially explicit coalescent model; and (iii) to use summary statistics and ABC to estimate model parameters from the empirical genetic data, including the latitude and longitude of population expansion (Ω). We describe key details here; a complete overview of the pipeline is provided elsewhere (32).
To generate the dynamic landscapes required for demographic simulations (Fig. 1), previously published SDMs (17) (Datasets S1–S4) were converted into downscaled raster landscapes at 25-arcmin resolution (∼46.3 km) with 10 habitat-suitability bins for the LGM and current time period. The landscape for the intermediate time period was generated by averaging carrying capacities during the LGM and current time period, as described previously (52). All unglaciated areas with zero habitat suitability in the LGM landscape were converted to the lowest nonzero habitat-suitability bin, so that it was possible to initiate simulations from regions that may have contained climatic microrefugia. A broad, uniform prior on the latitude and longitude of range expansion was chosen (Fig. 1 and Table 1), because SDMs predicted a wide area to have contained potentially suitable habitat for both species during the LGM (Fig. 1).
For each species, 106 time-forward demographic simulations were generated in SPLATCHE2 (53) and were initiated from a source population with latitude and longitude of the central deme chosen from the geographic prior using the X-ORIGIN pipeline (32). Initial source populations extended three additional demes in all directions beyond the central deme, to avoid enforcing a genetic bottleneck by collapsing individuals into a single deme. This created an initial area of expansion seven demes wide (175 arcmin, or ∼324 km; Fig. 1). Simulations were initiated at 21.5 ka, and individuals were allowed to colonize the landscape according to the carrying capacity of each deme (K; scaled relative to Kmax based on habitat suitability of the dynamic SDM) and the proportional migration rate among neighboring demes (m). Priors on Kmax and m followed a log-uniform distribution (Table 1) and were selected to ensure full recolonization of the species range within the time frame of the simulation, but not so rapidly as to allow nearly instantaneous recolonization after the onset of the simulation (an extremely unlikely scenario). Habitat suitability maps representing the LGM, intermediate time period, and current conditions were each successively used for one-third of the total generations (52). Generation time was 50 y, corresponding roughly to the minimum time to reach peak reproductive age (29, 30).
After the demographic portion of each simulation, genetic data were simulated to match the format of the empirical data. Input parameters to SPLATCHE2 specified that the same number of SNPs as in the empirical data should be simulated, along with the same number of individuals per population and same geographic locations of populations. Coalescent genetic simulations were performed with the ancestry of alleles traced from the present backward to the initial demes at 21.5 ka. At this time, all initial demes were collapsed into a single ancestral source population of size Nanc (following a log-uniform prior; Table 1) in which all alleles were allowed to coalesce. The SNP mutation model in SPLATCHE2 was used to generate genotypes for each individual according to the simulated pattern of coalescence, with a minimum minor allele frequency fixed at 3.3% to match that of the empirical data (17).
Estimating the Expansion Origin.
Spatial summary statistics (population pairwise directionality index, Ψ, and pairwise FST) (32) were generated for both the empirical data (SI Appendix, Tables S1 and S2) and all simulated datasets in Arlequin v.3.5 (54) using scripts provided with X-ORIGIN. The directionality index (Ψ) is a statistic used the infer the direction of range expansion between two populations based on allele frequency gradients that arise due to genetic drift during expansion (38). For the purpose of calculating pairwise Ψ (38) in the empirical data, ancestral alleles for each SNP were defined by the allele with the highest frequency in populations from areas predicted by the SDMs to have contained suitable habitat during the LGM. Principal component (PC) analysis was performed on all pairwise summary statistics (Ψ and FST) to reduce the dimensionality of the summary statistic datasets while still retaining information about the majority of variation in the simulated data. An optimization based on Wegmann’s P values was used to select the optimal number of PC axes to retain (see below for details).
ABC (35) was used to compare empirical and simulated summary statistics and infer the latitude and longitude of population expansion (Ω) and other demographic parameters. The 5,000 simulations (0.5%) with transformed PCs of summary statistics most closely matching the empirical data were retained using ABCtoolbox (37). A 2D kernel density of the expansion origin was calculated from the retained simulations using scripts provided with X-ORIGIN, except that we applied a log10 transformation to the default weighting values for each simulation (based on distance between the empirical and simulated datasets) before calculating the kernel density, to reduce the effect of a few extreme outliers with very small distances. Posterior distributions of other model parameters (Kmax, Nanc, m) were estimated from the retained simulations using ABC-generalized linear model adjustment (55) implemented in ABCtoolbox (37).
To determine how well the models are able to generate the observed data, we calculated Wegmann’s P value (37), which is the proportion of retained simulations with a smaller likelihood than that of the empirical data based on the transformed PCs of spatial summary statistics (Ψ and FST). Small P values indicate that a model is unable to generate the observed data (37). We also examined Wegmann’s P values across different numbers of retained PC axes for spatial summary statistics, to determine the optimal number of PC axes to retain for estimating the expansion origin and other model parameters. Based on this optimization, we retained four PC axes in C. cordiformis that together explained 67.7% of the overall variation and three PC axes in C. ovata that together explained 63.4% of the overall variation.
In addition, we calculated the average error in estimating the expansion origin, by estimating this parameter from PODs with known expansion origins using the same estimation procedure as for the empirical data and a subset of 5 × 105 simulations. Ten PODs were generated for each possible expansion origin, and the geographic distance between the inferred expansion origin and the true expansion origin simulated for each POD was calculated using the R package “geosphere” (56). We also used these PODs to test whether the estimated empirical expansion origins are likely to have been incorrectly inferred, by plotting the true expansion origin of all PODs with an inferred expansion origin matching the high-likelihood area estimated for the empirical data (defined as all areas with a likelihood ≥0.5 relative to the highest observed likelihood).
Data Accessibility
Empirical genetic datasets (17) are available from Deep Blue Data (DOI: 10.7302/Z2JS9NNG; ref. 36). Projected SDMs are provided in Datasets S1–S4. Scripts for simulations and data analysis are distributed with X-ORIGIN (32).
Supplementary Material
Acknowledgments
We thank B. J. Belcher, P. Cousineau, R. D’Andrea, J. Ronson, D. Saenz, and P. Tichenor for assistance with fieldwork; Q. He for guidance implementing the X-ORIGIN pipeline; and the National Science Foundation (Graduate Research Fellowship Program Award DDIG 1501159), the University of Michigan Department of Ecology and Evolutionary Biology, and Rackham Graduate School for graduate student support and research funding. We also thank the following organizations for assistance in coordinating fieldwork: Berea College Forest, Connecticut State Forests (SFs), Daniel Boone National Forest (NF), Davy Crockett NF, George Washington NF, Holly Springs NF, Hoosier NF, Jefferson NF, Kisatchie NF, Land Between the Lakes National Recreation Area, Lower Wisconsin State Riverway, Mark Twain NF, Matthaei Botanical Gardens, Monongahela NF, Murray State University Hancock Biological Station, Nantahala NF, Oconee NF, Ozark NF, Prentice Cooper SF, Shimek SF, Society of Ontario Nut Growers, Stephen F. Austin Experimental Forest, Sumter NF, Tombigbee NF, the University of Michigan E. S. George Reserve, and Uwharrie NF.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. V.L.S. is a guest editor invited by the Editorial Board.
Data deposition: Empirical genetic datasets are available from Deep Blue Data (DOI: 10.7302/Z2JS9NNG).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901656116/-/DCSupplemental.
References
- 1.Jackson ST, et al. Vegetation and environment in eastern North America during the last glacial maximum. Quat Sci Rev. 2000;19:489–508. [Google Scholar]
- 2.Shafer ABA, Cullingham CI, Côté SD, Coltman DW. Of glaciers and refugia: A decade of study sheds new light on the phylogeography of northwestern North America. Mol Ecol. 2010;19:4589–4621. doi: 10.1111/j.1365-294X.2010.04828.x. [DOI] [PubMed] [Google Scholar]
- 3.Hewitt G. Post-glacial re-colonization of European biota. Biol J Linn Soc Lond. 1999;68:87–112. [Google Scholar]
- 4.Qiu YX, Fu CX, Comes HP. Plant molecular phylogeography in China and adjacent regions: Tracing the genetic imprints of quaternary climate and environmental change in the world’s most diverse temperate flora. Mol Phylogenet Evol. 2011;59:225–244. doi: 10.1016/j.ympev.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Hewitt G. The genetic legacy of the quaternary ice ages. Nature. 2000;405:907–913. doi: 10.1038/35016000. [DOI] [PubMed] [Google Scholar]
- 6.Hampe A, Petit RJ. Conserving biodiversity under climate change: The rear edge matters. Ecol Lett. 2005;8:461–467. doi: 10.1111/j.1461-0248.2005.00739.x. [DOI] [PubMed] [Google Scholar]
- 7.Feurdean A, et al. Tree migration rates: Narrowing the gap between inferred post-glacial rates and projected rates. PLoS One. 2013;8:e71797. doi: 10.1371/journal.pone.0071797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Svenning JC, Skov F. Could the tree diversity pattern in Europe be generated by postglacial dispersal limitation? Ecol Lett. 2007;10:453–460. doi: 10.1111/j.1461-0248.2007.01038.x. [DOI] [PubMed] [Google Scholar]
- 9.Qian H, Ricklefs RE. Large-scale processes and the Asian bias in species diversity of temperate plants. Nature. 2000;407:180–182. doi: 10.1038/35025052. [DOI] [PubMed] [Google Scholar]
- 10.Shaw J, Small RL. Chloroplast DNA phylogeny and phylogeography of the North American plums (Prunus subgenus Prunus section Prunocerasus, Rosaceae) Am J Bot. 2005;92:2011–2030. doi: 10.3732/ajb.92.12.2011. [DOI] [PubMed] [Google Scholar]
- 11.Soltis DE, Morris AB, McLachlan JS, Manos PS, Soltis PS. Comparative phylogeography of unglaciated eastern North America. Mol Ecol. 2006;15:4261–4293. doi: 10.1111/j.1365-294X.2006.03061.x. [DOI] [PubMed] [Google Scholar]
- 12.Potter K, et al. Allozyme variation and recent evolutionary history of eastern hemlock (Tsuga canadensis) in the southeastern United States. New For. 2008;35:131–145. [Google Scholar]
- 13.Jaramillo-Correa JP, Beaulieu J, Khasa DP, Bosquet J. Inferring the past from the present phylogeographic structure of North American forest trees: Seeing the forest for the genes. Can J Res. 2009;39:286–307. [Google Scholar]
- 14.Morris AB, Graham CH, Soltis DE, Soltis PS. Reassessment of phylogeographical structure in an eastern North American tree using Monmonier’s algorithm and ecological niche modelling. J Biogeogr. 2010;37:1657–1667. [Google Scholar]
- 15.Bennett KD. The spread of Fagus grandifolia across eastern North America during the last 18 000 years. J Biogeogr. 1985;12:147–164. [Google Scholar]
- 16.Magni CR, Ducousso A, Caron H, Petit RJ, Kremer A. Chloroplast DNA variation of Quercus rubra L. in North America and comparison with other Fagaceae. Mol Ecol. 2005;14:513–524. doi: 10.1111/j.1365-294X.2005.02400.x. [DOI] [PubMed] [Google Scholar]
- 17.Bemmels JB, Dick CW. Genomic evidence of a widespread southern distribution during the last glacial maximum for two eastern North American hickory species. J Biogeogr. 2018;45:1739–1750. [Google Scholar]
- 18.McLachlan JS, Clark JS, Manos PS. Molecular indicators of tree migration capacity under rapid climate change. Ecology. 2005;86:2088–2098. [Google Scholar]
- 19.Davis MB. Quaternary history of deciduous forests of eastern North America and Europe. Ann Mo Bot Gard. 1983;70:550–563. [Google Scholar]
- 20.Jackson ST, Williams JW. Modern analogs in quaternary paleoecology: Here today, gone yesterday, gone tomorrow? Annu Rev Earth Planet Sci. 2004;32:495–537. [Google Scholar]
- 21.Delcourt PA, Delcourt HR, Brister RC, Lackey LE. Quaternary vegetation history of the Mississippi embayment. Quat Res. 1980;13:111–132. [Google Scholar]
- 22.Stewart JR, Lister AM. Cryptic northern refugia and the origins of the modern biota. Trends Ecol Evol. 2001;16:608–613. [Google Scholar]
- 23.Rull V. Macrorefugia and microrefugia: A response to Tzedakis et al. Trends Ecol Evol. 2014;29:243–244. doi: 10.1016/j.tree.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 24.Willis KJ, Van Andel TH. Trees or no trees? The environments of central and eastern Europe during the last glaciation. Quat Sci Rev. 2004;23:2369–2387. [Google Scholar]
- 25.Provan J, Bennett KD. Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol. 2008;23:564–571. doi: 10.1016/j.tree.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Rull V. Microrefugia. J Biogeogr. 2009;36:481–484. [Google Scholar]
- 27.Smith LM. Fluvial geomorphic features of the Lower Mississippi alluvial valley. Eng Geol. 1996;45:139–165. [Google Scholar]
- 28.Tzedakis PC, Emerson BC, Hewitt GM. Cryptic or mystic? Glacial tree refugia in northern Europe. Trends Ecol Evol. 2013;28:696–704. doi: 10.1016/j.tree.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 29.Graney DL. Carya ovata (Mill.) K. Koch. In: Burns RM, Honkala BH, editors. Silvics of North America: 2. Hardwoods. Agricultural Handbook 654. 2nd Ed US Department of Agriculture, Forest Service; Washington, DC: 1990. [Google Scholar]
- 30.Smith HC. Carya cordiformis (Wangenh.) K. Koch. In: Burns RM, Honkala BH, editors. Silvics of North America: 2. Hardwoods. Agricultural Handbook 654. 2nd Ed US Department of Agriculture, Forest Service; Washington, DC: 1990. [Google Scholar]
- 31.Barnes BV, Wagner WH. Michigan Trees. University of Michigan Press; Ann Arbor, MI: 2004. [Google Scholar]
- 32.He Q, Prado JR, Knowles LL. Inferring the geographic origin of a range expansion: Latitudinal and longitudinal coordinates inferred from genomic data in an ABC framework with the program X-ORIGIN. Mol Ecol. 2017;26:6908–6920. doi: 10.1111/mec.14380. [DOI] [PubMed] [Google Scholar]
- 33.Beaumont MA, Zhang W, Balding DJ. Approximate Bayesian computation in population genetics. Genetics. 2002;162:2025–2035. doi: 10.1093/genetics/162.4.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bemmels JB, Title PO, Ortego J, Knowles LL. Tests of species-specific models reveal the importance of drought in postglacial range shifts of a Mediterranean-climate tree: Insights from integrative distributional, demographic and coalescent modelling and ABC model selection. Mol Ecol. 2016;25:4889–4906. doi: 10.1111/mec.13804. [DOI] [PubMed] [Google Scholar]
- 35.Massatti R, Knowles LL. Contrasting support for alternative models of genomic variation based on microhabitat preference: Species-specific effects of climate change in alpine sedges. Mol Ecol. 2016;25:3974–3986. doi: 10.1111/mec.13735. [DOI] [PubMed] [Google Scholar]
- 36.Bemmels JB, Dick CW. 2018 Data from “Genomic evidence of a widespread southern distribution during the Last Glacial Maximum for two eastern North American hickory species.” Deep Blue Data. Available at https://deepblue.lib.umich.edu/data/concern/data_sets/4b29b664s. Deposited May 18, 2018.
- 37.Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L. ABCtoolbox: A versatile toolkit for approximate Bayesian computations. BMC Bioinformatics. 2010;11:116. doi: 10.1186/1471-2105-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peter BM, Slatkin M. Detecting range expansions from genetic data. Evolution. 2013;67:3274–3289. doi: 10.1111/evo.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hampe A, Jump AS. Climate relicts: Past, present, future. Annu Rev Ecol Evol Syst. 2011;42:313–333. [Google Scholar]
- 40.Correa Ribeiro P, Lemos-Filho JP, de Oliveira Buzatti RS, Lovato MB, Heuertz M. Species-specific phylogeographical patterns and Pleistocene east-west divergence in Annona (Annonaceae) in the Brazilian Cerrado. Bot J Linn Soc. 2016;181:21–36. [Google Scholar]
- 41.Papadopoulou A, Knowles LL. Toward a paradigm shift in comparative phylogeography driven by trait-based hypotheses. Proc Natl Acad Sci USA. 2016;113:8018–8024. doi: 10.1073/pnas.1601069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart JR, Lister AM, Barnes I, Dalen L. Refugia revisited: Individualistic responses of species in space and time. Proc Biol Sci. 2010;277:661–671. doi: 10.1098/rspb.2009.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson BJ, Graves WR. Chloroplast phylogeography of Dirca palustris L. indicates populations near the glacial boundary at the last glacial maximum in eastern North America. J Biogeogr. 2016;43:314–327. [Google Scholar]
- 44.de Lafontaine G, Amasifuen Guerra CA, Ducousso A, Petit RJ. Cryptic no more: Soil macrofossils uncover Pleistocene forest microrefugia within a periglacial desert. New Phytol. 2014;204:715–729. doi: 10.1111/nph.12833. [DOI] [PubMed] [Google Scholar]
- 45.Magri D. Patterns of post-glacial spread and the extent of glacial refugia of European beech (Fagus sylvatica) J Biogeogr. 2008;35:450–463. [Google Scholar]
- 46.Magri D, et al. A new scenario for the quaternary history of European beech populations: Palaeobotanical evidence and genetic consequences. New Phytol. 2006;171:199–221. doi: 10.1111/j.1469-8137.2006.01740.x. [DOI] [PubMed] [Google Scholar]
- 47.Petit R, et al. Glacial refugia: Hotspots but not melting pots of genetic diversity. Science. 2003;300:1563–1565. doi: 10.1126/science.1083264. [DOI] [PubMed] [Google Scholar]
- 48.Lumibao CY, Hoban SM, McLachlan J. Ice ages leave genetic diversity “hotspots” in Europe but not in eastern North America. Ecol Lett. 2017;20:1459–1468. doi: 10.1111/ele.12853. [DOI] [PubMed] [Google Scholar]
- 49.Little EL. 1971. Atlas of United States Trees, Volume 1. Conifers and Important Hardwoods, USDA Miscellaneous Publication 1146 (US Department of Agriculture, Washington, DC)
- 50.Prentice C, Bartlein PJ, Webb T., III Vegetation and climate change in eastern North America since the last glacial maximum. Ecology. 1991;72:2038–2056. [Google Scholar]
- 51.Givens CR, Givens FM. Age and significance of fossil white spruce (Picea glauca), tunica hills, Louisiana-Mississippi. Quat Res. 1987;27:283–296. [Google Scholar]
- 52.He Q, Edwards DL, Knowles LL. Integrative testing of how environments from the past to the present shape genetic structure across landscapes. Evolution. 2013;67:3386–3402. doi: 10.1111/evo.12159. [DOI] [PubMed] [Google Scholar]
- 53.Ray N, Currat M, Foll M, Excoffier L. SPLATCHE2: A spatially explicit simulation framework for complex demography, genetic admixture and recombination. Bioinformatics. 2010;26:2993–2994. doi: 10.1093/bioinformatics/btq579. [DOI] [PubMed] [Google Scholar]
- 54.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 55.Leuenberger C, Wegmann D. Bayesian computation and model selection without likelihoods. Genetics. 2010;184:243–252. doi: 10.1534/genetics.109.109058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hijmans RJ, Williams E, Vennes C. 2017 Package “geosphere”. R package version 1.5-7. Available at https://cran.r-project.org/web/packages/geosphere/index.html. Accessed November 6, 2017.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Empirical genetic datasets (17) are available from Deep Blue Data (DOI: 10.7302/Z2JS9NNG; ref. 36). Projected SDMs are provided in Datasets S1–S4. Scripts for simulations and data analysis are distributed with X-ORIGIN (32).