Significance
In species with males and females, reproduction requires contributions from both sexes and therefore some degree of cooperation. At the same time, antagonistic interactions can evolve because of the differing goals of males and females. We aligned the interests of the sexes in the naturally promiscuous fruit fly Drosophila melanogaster by enforcing randomized monogamy for more than 150 generations. Males repeatedly evolved to manipulate females less, a pattern visible in both the timing of female reproductive effort and gene expression changes after mating. Male investment in expression of genes encoding seminal fluid proteins, which shape the female postmating response, declined concurrently. Our results confirm the presence of sexually antagonistic selection on postcopulatory interactions that can be reversed by monogamy.
Keywords: sexual selection, sexual conflict, Drosophila melanogaster, experimental evolution, seminal fluid proteins
Abstract
In many animals, females respond to mating with changes in physiology and behavior that are triggered by molecules transferred by males during mating. In Drosophila melanogaster, proteins in the seminal fluid are responsible for important female postmating responses, including temporal changes in egg production, elevated feeding rates and activity levels, reduced sexual receptivity, and activation of the immune system. It is unclear to what extent these changes are mutually beneficial to females and males or instead represent male manipulation. Here we use an experimental evolution approach in which females are randomly paired with a single male each generation, eliminating any opportunity for competition for mates or mate choice and thereby aligning the evolutionary interests of the sexes. After >150 generations of evolution, males from monogamous populations elicited a weaker postmating stimulation of egg production and activity than males from control populations that evolved with a polygamous mating system. Males from monogamous populations did not differ from males from polygamous populations in their ability to induce refractoriness to remating in females, but they were inferior to polygamous males in sperm competition. Mating-responsive genes in both the female abdomen and head showed a dampened response to mating with males from monogamous populations. Males from monogamous populations also exhibited lower expression of genes encoding seminal fluid proteins, which mediate the female response to mating. Together, these results demonstrate that the female postmating response, and the male molecules involved in eliciting this response, are shaped by ongoing sexual conflict.
Sexual reproduction in animals often involves both synergistic and antagonistic interactions between males and females. At a fundamental level, gametes are required from both sexes, and the fitness of both males and females increases with the quantity and quality of offspring produced. There is also scope for antagonism, however, because the evolutionary strategies of the sexes differ (1). Antagonistic interactions can manifest before mating, for example, via male persistence or coercion that harms females, or after mating, by males manipulating female physiology and behavior (2, 3).
Antagonism between the sexes is particularly likely in promiscuous species such as the fruit fly Drosophila melanogaster. In female flies, the transition into a reproductive state requires both the receipt of sperm and the seminal fluid proteins (SFPs) present in the ejaculate (4–8). SFPs modulate gene expression in a time- and tissue-specific manner (9–13), induce oogenesis (14–17), ovulation (18, 19), and sustained egg production (20), and allow sperm storage (21–28). While some of these effects are beneficial to females (29, 30), the action of SFPs has also been suggested to influence the female postmating response in ways that benefit males at the expense of females and therefore constitute manipulation (31). For example, SFPs induce a short-term boost in egg laying (19) that is accompanied by reduced fertilization success (32), refractoriness to remating (14, 15, 28, 33, 34) that could limit the ability of females to replenish sperm reserves or sample higher quality sperm, and a reduction in life span (35, 36). If any of these SFP-mediated effects are favored by selection on males while harming female fitness, then interlocus sexual conflict—conflict occurring through antagonistic interactions between different genetic loci in males and females (37)—should lead to the evolution of female resistance and select for further male manipulation. The observed rapid duplication and sequence-level evolution of SFP genes (38, 39), as well as experiments finding increased male harm to females after female coevolution is arrested (40), are consistent with the possibility of such a conflict-fueled arms race.
To investigate to what extent male effects on the female postmating response are beneficial to males but costly to females (and therefore constitute male manipulation), we evolved three populations of Drosophila melanogaster for >150 generations in a mating system in which each female was randomly paired with a single male every generation, thereby removing all opportunity for mating competition or mate choice. This randomized monogamy manipulation has been employed to remove sexual selection in evolving populations of many different arthropods, including the fruit fly species D. melanogaster (41–45), Drosophila pseudoobscura (46), and Drosophila serrata (47), as well as the dung fly Sepsis cynipsea (48), the beetles Callosobruchus maculatus (49, 50) and Tribolium castaneum (51), and the bulb mite Rhizoglyphus robini (52). Under a monogamous evolutionary regime, interlocus sexual conflict is eliminated because the reproductive interests of males and females are completely tied to one another. In parallel to our three monogamous populations, we established three control populations that evolved under a mating system in which groups of five females and five males were formed each generation, thereby retaining sexual selection and interlocus sexual conflict (the polygamous evolutionary regime). We predicted that, if male effects on the postmating response are costly to females in terms of lifetime reproductive success, or costly to males because investment in reproductive tissues trades off with other components of nonsexual fitness, these effects should decrease in the course of evolution under monogamy. By contrast, if the effects elicited by males are beneficial to females (e.g., by increasing total reproductive output or the quality of offspring), they should be either maintained or elevated during evolution in the monogamous evolutionary regime.
Results and Discussion
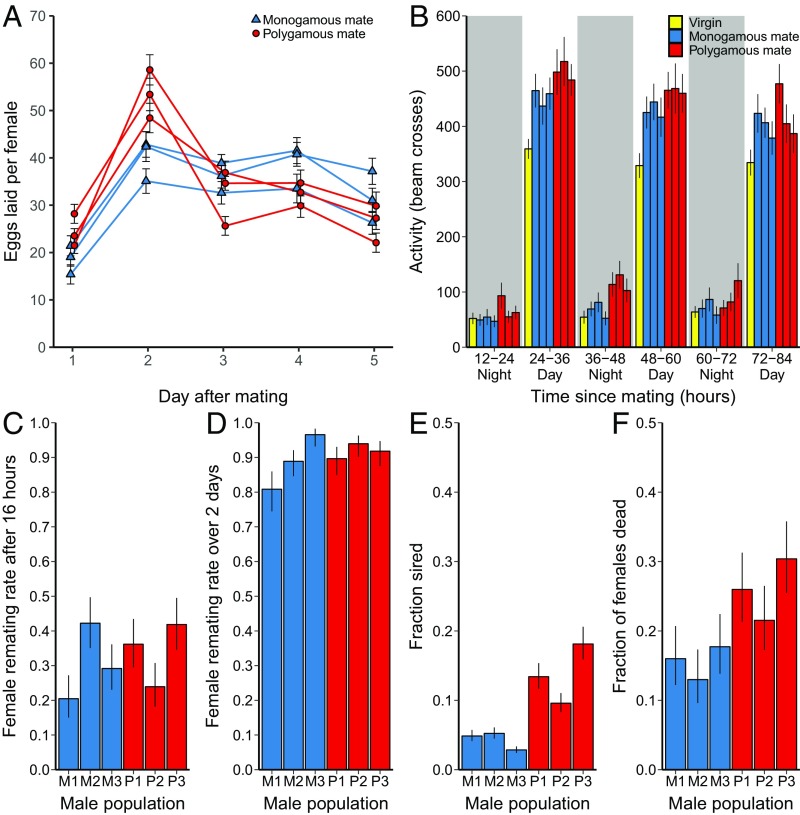
We examined whether males evolved in the monogamous versus polygamous evolutionary regimes (hereafter M males and P males, respectively) differed in their postmating effects on females sampled from the base population from which all evolved populations originated, thereby isolating the effects of males from any differences between evolved females. We first assessed female reproductive investment in the days following mating with evolved males. Wild-type D. melanogaster males stimulate reproduction in females in the first hours after mating via the action of SFPs, an effect that is expected to be beneficial to males in the presence of multiple mating by increasing a male’s share of a given female’s offspring. However, this increased early egg laying might be costly for females if it results in reductions in fertilization rates, offspring quality, or offspring production later in life. Under monogamy, there is no competition from other males, and selection should favor male effects on female reproduction that optimize offspring number and quality. If sexual conflict drives males to stimulate female short-term fecundity beyond the female optimum, this effect on females should therefore be reduced during the course of evolution under monogamy. Consistent with this prediction, M males elicited different female egg-laying patterns from P males over the first 5 d after mating, a window of time that corresponds to the mating and reproductive periods of the evolutionary regime (Fig. 1A, linear mixed model, evolutionary regime × day: F4,15 = 17.0, P < 0.0001). The difference between the effects of the two types of males also changed linearly over time (linear model, F1,3 = 15.1, P = 0.03), with females mated to M males laying on average fewer eggs in the first 2 d after mating and more eggs in the following 3 d. Overall, the total number of eggs laid by females was unaffected by the male evolutionary regime (F1,4 = 0.1, P = 0.75), a result matching what was previously found in D. pseudoobscura (46).
Fig. 1.
Female postmating response when mated to males from six populations with differing evolutionary histories (monogamous in blue, polygamous in red). (A) Female egg production (population mean ± SE, n = 19–20 groups of females per male population) across the first 5 d after mating. (B) Female activity levels, measured as the number of beam crosses in 12-h intervals in the first 3.5 d after mating (population mean ± SE, n = 21–24 females per male population). Virgin activity levels are indicated with yellow bars for comparison. (C) Female remating rate (population mean ± SE, n = 43–48 females per male population) when placed with a new male from the ancestral population 16 h after a first mating with males from the evolved populations and observed for 4 h. (D) Female remating rate (population mean ± SE, n = 58–72 females per male population) when females were first mated to males from the evolved populations and then housed with four ebony males continuously from 4 to 48 h after first mating. (E) Sperm competition outcomes from the same experiment (population mean ± SE, n = 38–64 females per male population), assessing the offspring of all doubly mated females. (F) Female death in the same experiment, 4 d after mating with males (population mean ± SE, n = 75–79 females per male population).
We next examined whether the evolutionary regime of males differentially induced postmating activity in females. Within hours after mating, activity of D. melanogaster females greatly increases as they dedicate more time to searching for suitable oviposition sites, foraging, and feeding. This change is driven by receipt of SFPs (53) and is expected to benefit males by shifting female reproductive effort toward the period of time when paternity share is highest. However, early investment would come at a cost to females if it trades off with reproductive success (54) within the experimental regimes. We therefore continuously monitored female locomotor activity levels after mating to test whether females mated with M males exhibited different locomotor activity than females mated with P males. As predicted, if male-induced hyperactivity is costly to females, females mated to M males exhibited overall lower locomotor activity than females mated to P males (Fig. 1B, linear mixed model, evolutionary regime: F1,4 = 19.5, P = 0.01, evolutionary regime × time: F5,20 = 0.6, P = 0.67). As a consequence, females mated to M males had locomotor activity profiles shifted in the direction of virgin females (Fig. 1B, virgins indicated with yellow bars).
We then tested whether M males were also less effective than P males in eliciting female refractoriness to remating. Preventing female remating is an important component of male postcopulatory competitive success and is also mediated by SFPs (14, 15). However, reluctance to remate might be costly to females if it limits their ability to replenish sperm reserves or mate with higher-quality males. Inducing refractoriness in females has no obvious advantage to males under monogamy, and therefore male effects on refractoriness are predicted to decline if the SFPs responsible for inducing refractoriness carry a physiological cost to females (36). We measured male effects on female refractoriness in our populations by first mating females to males from each of the evolved populations and then presenting the females with a new male from the ancestral population 16 h later. Contrary to our prediction, there was no difference between M males and P males in eliciting female refractoriness (Fig. 1C, generalized linear mixed model, evolutionary regime: χ21 = 0.2, P = 0.63). Tests of this prediction have yielded mixed results in other studies. D. melanogaster males evolved in the absence of sexual competition have sometimes been found to suppress female remating less than polygamous males (55), an effect also observed in D. pseudoobscura (46), but a more recent study found no difference between D. melanogaster males evolved under monogamy and polygamy (45). Some of the differences between experimental outcomes might be explained by differences in how female remating rates were measured (e.g., the amount of time elapsed between females encountering first and second males, the number of males provided to the females, or the amount of time males spent with females). Because remating rates were low in the assay that we employed, and the opportunity for remating was limited to a 6-h window on the morning after the first mating, it is possible that our design could have missed male effects occurring earlier or later after mating (56). We therefore performed an additional experiment, using a 2-d interaction period between males and females that corresponded to the time window when mating occurred during the course of experimental evolution. In this design, we first allowed males from the evolved populations to mate with females carrying a recessive ebony marker and then housed these females continuously with four ebony male competitors. This allowed us to determine whether females remated during a 2-d period and, if so, how many of the offspring were sired by the focal males. Remating rates of females were much higher in this experiment, but there was again no significant effect of the evolutionary regime of males on their ability to induce refractoriness (generalized linear mixed model, evolutionary regime: χ21 = 0.6, P = 0.45, Fig. 1D). This suggests that male effects on female refractoriness may not be costly to females under our evolutionary regimes.
Beyond inducing refractoriness to remating, males can also increase their fitness by outperforming other males when there is competition between sperm for fertilization. We therefore also assessed sperm competition outcomes (sperm defense, or P1) in all doubly mated females in the experiment measuring female refractoriness. We found that P males had almost a three times higher success in sperm competition when mating first compared with M males (Fig. 1E, generalized linear mixed model, evolutionary regime: χ21 = 9.8, P = 0.002), an effect not observed in previous experimental evolution with D. melanogaster that spanned fewer generations (45, 57). This advantage in sperm competition occurred despite the fact that the increased relative egg laying of females induced by P males should result in these females having less sperm from P males in the spermatheca by the time of the second mating. Females mated first to P males also suffered higher mortality than those mated to M males, despite the short window of time (4 d) over which the experiment was conducted (Fig. 1F, generalized linear mixed model, χ21 = 6.7, P = 0.01). The evolution of reduced male harm to females by M males suggests that the wild-type cost of mating (35, 58, 59) to females represents a true fitness cost that is not offset by gains in offspring quantity or quality (60) and therefore was selected against under monogamy.
Postmating changes in female physiology and behavior like those that we observed are mediated by and/or coincident with changes in patterns of gene expression that vary between the level of the whole fly (9, 10, 61), the reproductive tract (62), and the head (11, 63). Because of the central role of the nervous system in behavior, we measured gene expression not only in abdomens, which house the reproductive tissues, but also in the heads of females both before mating and 24 h after mating with either M males or P males. As expected, mating resulted in more genes changing expression in the abdomen [6,206 or 71.7% of all genes at 10% false discovery rate (FDR)] than in the head (1,485 or 16.0% of all genes) [see Datasets S1and S2 for gene identities and SI Appendix, Fig. S1, for gene ontology (GO) enrichment]. Interestingly, however, the evolutionary regime of males had a greater effect on the expression of genes in the female head than abdomen. In the abdomen, only 45 genes responded differentially depending on whether females mated with M or P males. In contrast, the male selection regime had a significantly greater impact on expression of genes in the head (3.8 versus 0.5% in the abdomen, Fisher’s exact test, P < 0.0001), with 351 genes differentially expressed (see SI Appendix, Fig. S1A, for GO enrichment in the abdomen and SI Appendix, Fig. S1B, for GO enrichment in the head).
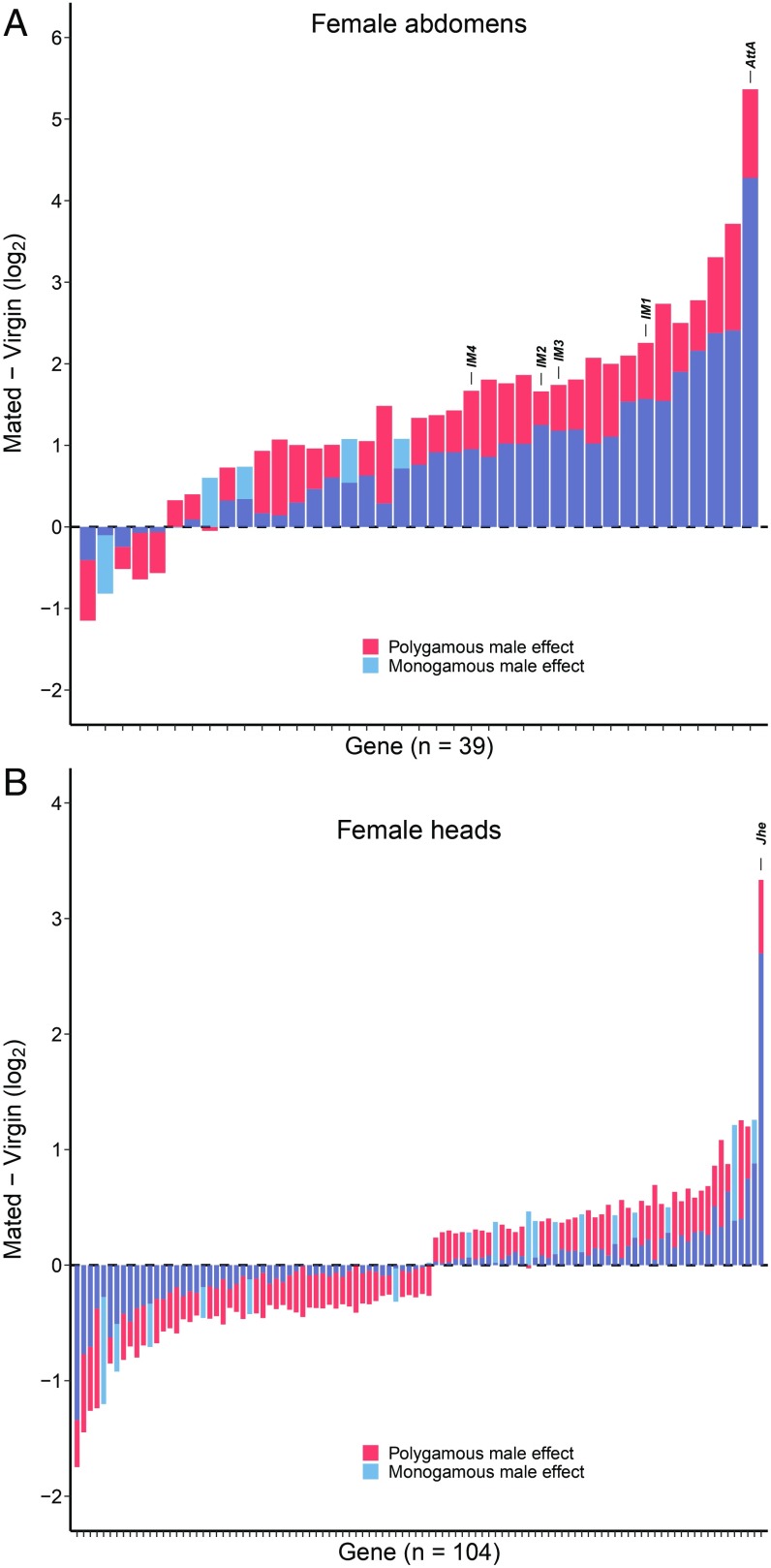
Given that mating with M males induced a weaker response in female reproductive investment and locomotor activity than mating with P males, we predicted that M males would also elicit a weaker female gene expression response. To test this prediction, we examined genes that were differentially elicited depending on male evolutionary regime and responded to mating in the same direction regardless of male identity (i.e., those with a significant main effect of mating). In the abdomen, 34 of 39 (87.2%) of these genes responded less strongly when females mated with M males, significantly more than would be expected if up and down changes were equally likely (Fig. 2A, binomial test, P < 0.0001). Similarly, in the head, 87 of 104 (83.7%) of these genes responded less strongly when females mated with M males (Fig. 2B, binomial test, P < 0.0001), demonstrating that M males indeed had a dampened effect on female postmating gene expression compared with P males.
Fig. 2.
Change in gene expression 24 h after mating (log2 mated–log2 virgin) in the female abdomen (A) and female head (B) for mating-responsive genes that are significantly differentially affected by polygamous versus monogamous male mates. Genes encoding immune-induced peptides (A) and Juvenile hormone esterase (B) are labeled. In both female abdomens and heads, monogamous males elicit a weaker transcriptional response.
The greater difference in male effects on female gene expression in the head than in the abdomen might be explained by the fact that changes in female behavior are regulated by the effects of SFPs on both the nervous and the endocrine systems. For example, receptors for sex peptide, which enters the hemolymph soon after mating (64, 65), are located in both the female reproductive tract and the brain (64, 66), where they are necessary for sex peptide to elicit egg laying. Sex peptide elevates egg production by increasing juvenile hormone production (67) in the corpus allatum, an endocrine gland located in the head. Elevated juvenile hormone titer is responsible for increased early life reproduction, and juvenile hormone mediates key trade-offs between reproduction, metabolism, survival, and stress resistance (68, 69). Interestingly, the differentially induced gene with the largest change in female heads was Juvenile hormone esterase (Jhe), which is responsible for the breakdown of juvenile hormone and shows a nearly twofold higher expression in females mated to P males. Juvenile hormone esterase might therefore serve to limit male manipulative effects on female reproductive investment through its degradation of juvenile hormone.
Four lines of evidence led us to hypothesize that the different effects reported in this study might be due to a lower investment in SFPs by M males than by P males. First, in a previous study carried out earlier in the course of experimental evolution, we found a signal of reduced expression of SFP genes as a group (although no individual SFP genes were differentially expressed) in M males when examining whole-fly gene expression (70). Second, M males stimulated female reproduction less than P males in the first 2 d following mating, a period when the SFPs sex peptide and ovulin are more important than the presence of sperm in eliciting oogenesis and ovulation (15, 19, 20). Sex peptide is also responsible for an increase in locomotor activity levels in mated females (53), and we observed lower activity levels in females mated to M males. Third, SFPs are known to modulate sperm competition success (71–75), and M males had lower sperm competition success. SFPs are also responsible for reduced female survival [the well-established “cost of mating” (35, 36)], and females mated to M males survived better than females mated to P males, a result sometimes (41), but not always (45, 76), found after evolution under monogamy. Finally, SFPs are directly responsible for activation of the immune system, in particular induction of expression of antimicrobial peptide genes, which occurs after mating (9–13). This activation of the immune system in females was visible in the female abdomen transcriptome and markedly weaker when females mated to M males versus P males (Fig. 2A). The top overall candidate for differential male effects in the female abdomen was Attacin-A (more than twofold higher expression when females mated to P males), a gene that codes for an antimicrobial peptide. The postmating change in expression of Attacin-A was previously shown to be exclusively regulated by male SFPs (10). Four other genes encoding immune-induced peptides (IM1, IM2, IM3, and IM4) were also differentially affected, with P males inducing 33–64% higher postmating expression of these genes than M males. Like Attacin-A, two of these genes (IM1 and IM2) were previously identified as responding to mating only via the action of SFPs (10). Together, these results suggest a role for SFPs in the differential response of females mated to M versus P males.
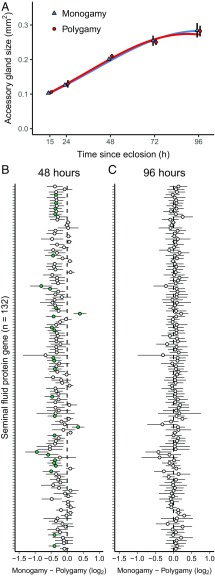
We investigated the possibility that M males invested less in SFPs than P males by measuring the size of the accessory gland, where most SFPs are produced. Previous studies examining mature adults found that D. pseudoobscura males evolved under monogamy invested less in the accessory glands (77), while D. melanogaster males evolving at different intensities of sexual competition did not differ in accessory gland investment (78). Because of the importance of early adult maturation in the sexual success of D. melanogaster males, both in general (79) and within the evolutionary regimes (80), we took a different approach and assessed the growth of the accessory gland by measuring its size across the first 4 d post eclosion. Male accessory glands more than doubled in size in the first few days (age: χ21 = 686.8, P < 0.0001), with growth slowing as males matured (age2: χ21 = 162.2, P < 0.0001). There was no difference, however, between M and P males in either overall size (evolutionary regime: χ21 = 0.1, P = 0.73) or growth trajectory (evolutionary regime × age: χ21 = 0.5, P = 0.46; evolutionary regime × age2: χ21 = 0.8, P = 0.36).
We also assessed gene expression in the male reproductive tract at two time points (48 and 96 h after eclosion) during the same period of early adult maturation. Forty-eight hours after eclosion, 586 genes were differentially expressed between M and P males (see Dataset S2, for gene identities and SI Appendix, Fig. S2, for GO enrichment). SFP genes were overrepresented in the list of differentially expressed genes; of 132 SFP genes, 27 were significantly different between M and P males at 48 h (Fig. 3B; 20.5 versus 5.5% expectation, Fisher’s exact test P < 0.0001). Of the 27 SFP genes that were differentially expressed, 25 (92.6%) showed lower expression in M males (binomial test, P < 0.0001). When considering all SFP genes, M males showed on average 16.6% lower expression than P males. The breadth of this reduced investment in SFP gene expression in M males (113 of the 132 SFP genes show nominally lower expression) is consistent with the idea that these genes are tightly coregulated (81) and suggests that fine-tuned tailoring of the composition of the ejaculate may be to some extent evolutionarily constrained. The reduced investment in SFP gene expression in M males was confirmed by qPCR analyses for the five most well-characterized SFP genes (Acp26Aa, Acp29AB, Acp36DE, Acp62F, and SP) across an extended time course spanning the first 5 d post eclosion (SI Appendix, Fig. S3). By 96 h after eclosion, M males and P males showed similar transcriptional profiles (Fig. 3C) with no genes significantly different in expression between male types. Differences in investment in SFP gene expression between males from the two evolutionary regimes were therefore visible at the time point marking the beginning of the male–female interaction period over the course of experimental evolution but absent at the end of the interaction period. The relative immaturity in the pattern of expression of SFP genes in M males is consistent with what has been previously found when examining gene expression in the heads of M males (80) and suggests the possibility of a generalized maturation deficit under selection in the monogamous regime.
Fig. 3.
(A) Accessory gland size over the first 4 d after eclosion for males from experimentally-evolved monogamous and polygamous populations. Data points are mean (±SE) of three monogamous populations (blue triangles) and three polygamous populations (red circles), and lines indicate predictions from the fit model (n = 8–16 accessory glands per population and age combination). (B) The difference in expression of individual seminal fluid protein genes between males from monogamous and polygamous populations (log2 monogamy–log2 polygamy) at 48 and 96 h post eclosion. Error bars indicate 95% confidence intervals, and green circles indicate genes that are significantly different between monogamy and polygamy at 10% FDR (27 genes at 48 h, 0 genes at 96 h).
We found that M males elicited weaker postmating responses in female reproduction and activity, but did not differentially induce female refractoriness. The evolution of differential male-mating effects that we identified cannot be explained by differences in effective population size (Ne) between the evolutionary regimes. The number of males and females was the same in M and P populations, but there was greater variance in male reproductive success under polygamy. Thus, if male performance was reduced because of inbreeding effects in our experiments, this effect should be greater in P than in M populations. Furthermore, past work directly estimating Ne based on molecular markers in experimental populations with differing intensities of sexual competition found no difference between monogamous and promiscuous populations (82). Diminished M male effects on females could also have arisen if deleterious variation was purged less efficiently from these populations (83, 84), but past work with these populations found no evidence for this (42, 80). Our findings therefore support the hypothesis that a substantial part of male effects on the female postmating response are detrimental to females and/or carry a cost to male nonsexual fitness. As a result, the ability of males to elicit postmating responses in female reproduction and activity diminished over the course of evolution under monogamy because the reproductive interests of males and females were aligned.
We also detected the signature of reduced male manipulation in the gene expression patterns of mated females, where M males induced weaker transcriptional change. Postmating changes in gene expression are particularly profound in the female abdomen, which contains the reproductive tissues as well as the metabolism-regulating fat body. Surprisingly, however, we found that male evolutionary history more strongly affects gene expression in the mated female’s head than in the abdomen. This suggests that, rather than acting directly on female physiology, male manipulation promoted by sexual conflict has targeted the female central nervous system to influence female behavior. Finally, M males showed reduced investment in expression of many genes encoding seminal fluid proteins. This addresses an ongoing controversy about the function of SFPs (31), supporting the notion that these proteins serve as male armaments in male–female coevolution fueled by sexual conflict.
Materials and Methods
Fly Populations and Experimental Evolution Design.
Six experimental populations, each with a census size of 100 males and 100 females, were established from the Ives (IV) population of D. melanogaster (85) in 2007 after a mutagenesis treatment that increased genetic variation and subsequently maintained under either randomized monogamy (populations M1, M2, and M3) or polygamy (populations P1, P2, and P3) (42). In each generation, males and females from monogamous populations were randomly paired in vials and allowed to interact for 2 d. In polygamous populations, groups of five males and five females were placed together in vials and allowed to interact for 2 d. After these 2 d, the males from all populations were discarded and females were placed into new bottles in groups of 50 for 3 d of egg laying, after which the females were also discarded. Virgins collected from these bottles were then used to constitute the next generation. The populations had undergone between 168 and 196 generations of experimental evolution when the measures collected in this study were obtained. Measurements were preceded by one generation in which all populations were mass-reared in the same manner to control for any parental effects, except for the measurements of male accessory gland size and reproductive tract transcriptomes, which were preceded by four generations of controlled rearing. During the course of experimental evolution and common garden rearing before all assays, flies were reared and maintained on either 2% yeast media [water, agar, brewer’s yeast, cornmeal, sucrose, propionic acid, and Nipagin (Sigma-Aldrich)] or, after a move to a new laboratory, a richer 6% yeast media that substituted fruit juice for sucrose. Flies were maintained at 25 °C with a 12 h:12 h light:dark cycle.
Standardized females used in the egg laying, activity, remating, and gene expression experiments were from the IV base population, which is maintained at several thousand individuals with flies mixed and moved to new media on a 14-d schedule. For postcopulatory male competition experiments, virgin ebony females and male competitors were obtained from the IVe population, which originates from and is maintained in the same manner as the IV population. The recessive phenotype of dark-body coloration allowed for offspring of ebony competitors to be easily distinguished from those of focal males that have a wild-type appearance.
Female Egg Laying, Activity Levels, and Refractoriness to Remating.
To measure egg-laying rates of females mated to males with different evolutionary histories, virgin males from the six populations and virgin standard females from the IV base population were collected and kept in same-sex groups of ∼20 individuals. At 2 d of age, groups of 10 females were placed with 20 males from each of the evolved populations at lights-on for 2 h to allow mating. After the 2-h mass-mating period, males were removed and females were collected and placed in new vials in groups of three and allowed to lay eggs for the remainder of the light cycle. Females were then moved to holding vials overnight before being placed into new vials at lights-on for another day of egg laying. This was repeated for 5 d and counts were made of the number of eggs laid by each group on each day. Any groups in which a female died during the experiment were discarded. Daily egg counts were modeled as the response variable in a linear mixed model (LMM) with male evolutionary regime and the day of egg laying, along with the interaction, as fixed effects and replicate population as a random effect using lme4 (86) in R (87). We also fit a linear model with relative egg laying (females mated to polygamous males/females mated to monogamous males) as the response variable and day as a continuous predictor to test whether the magnitude of the relative difference declined or increased linearly across the 5 d. Significance was assessed with F tests.
For measurement of activity levels, males from the evolved populations and IV females were collected, aged, and mated in the same manner as above. We included virgin females to assess the overall direction and magnitude of changes in activity after mating. After the 2-h mating period, flies were briefly anesthetized with CO2, males were removed, and females were placed individually into 5-mm glass tubes with simple media (2% agar, 5% sucrose), plugged with a small piece of yarn. These tubes were placed in DAM2 Drosophila Activity Monitors (Trikinetics Inc.) that detect movement along the glass tube by counting infrared beam breaks. Total beam breaks per animal for each 12-h light:dark time interval were analyzed. To allow flies to recover from anesthesia, the first 6 h of recording were discarded, so data begins during the first night period after the morning mating and spans three nights and 3 d. Data for flies that died during the measurement period or in the day immediately following were discarded. Activity was modeled with male evolutionary regime and time in 12-h bins, along with the interaction, as fixed effects and replicate population as a random effect using lme4 (86) in R (87). Significance was assessed with F tests.
To measure female willingness to remate, evolved males along with IV male competitors and IV females were collected as virgins and kept in same-sex groups for 4 d. Individual males from the evolved populations were then placed into vials along with one virgin IV female, separated by a cardboard divider. On the next afternoon, the dividers were removed and flies were allowed to interact and mate for 1 h. After mating, males were removed from vials and replaced with new males from the IV population with a cardboard divider again separating the two flies. On the next morning, 16 h after the first mating session began, dividers were removed and whether or not a female remated in the next 6 h was recorded. The experiment was performed across 4 d in balanced blocks. Female willingness to remate was modeled with male evolutionary regime as a fixed effect and replicate population as a random effect with a binomial error distribution and logit link function using lme4 (86) in R (87). Females that did not mate with the first male were excluded from the analysis.
Male Postcopulatory Competition and Harm to Females.
An additional measure of female remating rate, along with the outcomes of sperm competition and male harm to females, was measured together with a design matching the timing of mating and egg production in the experimental evolution regimes. Males from the evolved populations and standard ebony females and males were collected as virgins and kept in same-sex groups of ∼20 for 2 d. Females were then individually placed with a male from one of the evolved populations to allow mating. After 4 h, the males were discarded and females placed individually in new vials along with four ebony males. Females stayed with these males for 44 more hours (i.e., until 2 full days had passed since first encountering males, matching the duration of the interaction period during the course of experimental evolution). At this point, males were removed and females were again placed individually in new vials for 2 d of egg laying. A female remated if she produced at least one wild-type offspring (sired by the evolved male that she first encountered) and at least one ebony offspring (sired by one of the four ebony males) and did not remate if she produced only wild-type offspring. Sperm defense (P1) was assessed by examining in remated females the proportion of offspring in the final vial that were wild type in appearance. Female death was also recorded at the end of the experiment, providing a measure of evolved male harm to females. Remating rate, death, and sperm competition outcomes were all modeled with generalized linear mixed models. Male evolutionary regime was included as a fixed effect and replicate population as a random effect with a binomial error distribution and logit link function in lme4 (86) within R (87). For sperm competition, an additional random effect was included for each female to account for covariance between the multiple offspring in a single female’s brood. Significance was assessed with likelihood-ratio tests.
Female Transcriptomic Response to Mating with Evolved Males.
We measured the gene expression profiles of standard IV females as either virgins or 24 h after mating with males from each of the three monogamous and three polygamous populations. Males from the evolved populations and standard females were first collected as virgins and kept in same-sex groups of ∼20 individuals for 2 d. Shortly after lights-on, females were individually paired with males from all of the evolved populations and watched for 2 h. After copulation occurred in a vial, males were discarded and newly mated females were placed in a holding vial. Any vials in which copulation did not occur after 2 h were discarded. For virgin samples, females were moved between vials similarly, except without the presence of a male. The next morning, all females were snap-frozen in liquid nitrogen. For each treatment, pools of 20 abdomens and 20 heads were dissected and transferred to −80 °C until RNA extraction. At the end of this process, there were 18 samples in total (one sample after mating with males from each of the six evolved populations and three virgin samples of matched age for each tissue). For details on the subsequent RNA extraction, library preparation, sequencing, quality checking, mapping, and read counting, see SI Appendix. Sequence data is available at NCBI GEO under accession no. GSE128404 (88). Analyses of abdomen and head datasets were conducted separately. Genes with less than one count per million in more than 2/3 of the libraries were removed before further analysis, a filter aimed at retaining genes that are not expressed at all in one condition (e.g., virgin) but expressed in the remaining samples and therefore of biological interest. This left 8,661 genes in the abdomen dataset and 9,293 genes in the head dataset. Normalization factors were calculated with the trimmed mean of M-values normalization (TMM) method (89) and observational- and sample-specific weights were used with the voomWithQualityWeights function in the limma package (90–92). The duplicateCorrelation function (93) from the limma package modeled repeated measurements at the sample level because each sample was sequenced in two independent runs. We fit gene-level linear models with a main effect of treatment (three levels: virgin, mated to a monogamous male, or mated to a polygamous male). We then performed two contrasts within this framework: virgin versus mated, to test for a significant overall effect of mating, and mated to a polygamous male − virgin versus mated to a monogamous male − virgin, to identify genes responding differently to mating depending on male evolutionary history. For both contrasts, we report genes detected with a 10% FDR (94). We used gene-set enrichment analysis in the R package tmod (95) to test for enrichment of all “biological process” GO terms in absolute log2 fold change-sorted lists.
Accessory Gland Size.
To track accessory gland development for the first 96 h after eclosion, virgin males from all six experimental populations were collected within 2 h of eclosion and allowed to age for 15, 24, 48, 72, and 96 h in vials of 20 males each. Males were then snap-frozen and stored at −80 °C until dissection. Accessory glands (8–16) and wings per population and age combination were then dissected and measured using ImageJ (96) as described in Ruhmann et al. (79). Samples for two populations (M2 and P2) at one time point post eclosion (72 h) were lost due to a technical problem. Accessory gland size was modeled with male evolutionary regime, age, age2 (to model slowing growth as flies mature), and all two-way interactions as fixed effects and replicate population as a random effect using a gamma error distribution and log link function with lme4 (86) in R (87). The natural log of wing size, measured as the L3 vein length, was included as a covariate to control for differences in overall size between males. Significance was assessed with likelihood-ratio tests.
Gene Expression in Evolved Male Reproductive Tracts.
Virgin males from all six experimental evolution populations were collected and aged for 48 and 96 h, as described above. Males were then chilled on ice and reproductive tracts (accessory glands, testes, and ejaculatory duct) dissected in PBS. Thirty reproductive tracts were pooled for each population and time point, snap-frozen in liquid nitrogen, and transferred to −80 °C until RNA extraction. For details on the subsequent RNA extraction, library preparation, sequencing, quality checking, mapping, and read counting, see SI Appendix. Sequence data is available at NCBI GEO under accession no. GSE128404 (88). Genes with less than one count per million in any of the libraries were removed before further analysis, leaving 10,738 genes in the final dataset. Normalization factors were calculated with the TMM method (89) and observational- and sample-specific weights were used with the voomWithQualityWeights function in the limma package (90–92). The duplicateCorrelation function (93) from the limma package modeled repeated measurements at the population level because each population was measured at two time points. We fit gene-level linear models testing for the main effects of evolutionary regime and age, as well as their interaction, on gene expression. Within this model framework, we also contrasted monogamous and polygamous evolutionary regimes at each age. We report genes detected with a 10% FDR (94). To test for enrichment or depletion of SFP genes, we used Fisher’s exact tests with the list of SFPs transferred at mating from Findlay et al. (97). One hundred thirty-two of 142 of the SFP genes were present in our dataset. We used gene-set enrichment analysis in the R package tmod (95) to test for enrichment of all biological process GO terms in absolute log2 fold change-sorted lists.
To gain more detailed insight into SFP gene expression dynamics in maturing accessory glands and validate detected differences in gene expression, we followed up our whole-transcriptome approach with a targeted qPCR approach. Males were collected and aged, reproductive tracts were dissected, and samples were frozen as before, except that in this experiment five ages were assessed (24, 48, 72, 96, and 120 h). We performed qPCR for five SFP genes (Acp26Aa, Acp29AB, Acp36DE, SP, and Acp62F, SI Appendix, Fig. S3) and four reference genes (Ef1α48D, RpL13A, αTub84B, and Act42A). For further details on sample preparation, qPCR, and relative gene expression calculation, see SI Appendix. Relative gene expression was analyzed in R (87) using linear mixed models implemented with package lme4 (86). Log2-transformed relative expression values were analyzed with evolutionary regime, age (treated as categorical because expression dynamics were nonlinear), and their interaction as fixed effects and replicate population included as a random effect. We contrasted polygamous and monogamous evolutionary regimes at each age class and report comparisons both nominally significant (P < 0.05) and significant after Bonferroni correction (P < 0.01).
Supplementary Material
Acknowledgments
We thank Sonja Schindler and Veronika Neumann for assistance with accessory gland size measurements, Christine La Mendola for assistance with RNA extractions, and the Lausanne Genomic Technologies Facility for sequencing support. Computations were performed at the Vital-IT Center (www.vital-it.ch) for high-performance computing of the SIB Swiss Institute of Bioinformatics. This study was supported by Swiss National Science Foundation Grants PZ00P3_161430 (to B.H.), 310030B_176406 (to L.K.), and 31003A_162732 (to T.J.K.); European Research Council Advanced Grant 249375 (to L.K.); a grant from the University of Lausanne (to T.J.K.); and Grant FR 2973/1-1 from the German Research Foundation (to C.F.)
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128404).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1821386116/-/DCSupplemental.
References
- 1.Parker GA. 1979. Sexual Selection and Sexual Conflict. Sexual Selection and Reproductive Competition in Insects (Elsevier, Amsterdam), pp 166.
- 2.Arnqvist G, Rowe L. Sexual Conflict. Princeton Univ Press; Princeton, NJ: 2013. [Google Scholar]
- 3.Chapman T, Arnqvist G, Bangham J, Rowe L. Sexual conflict. Trends Ecol Evol. 2003;18:41–47. [Google Scholar]
- 4.Wolfner MF. Battle and ballet: Molecular interactions between the sexes in Drosophila. J Hered. 2009;100:399–410. doi: 10.1093/jhered/esp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfner MF. Tokens of love: Functions and regulation of Drosophila male accessory gland products. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- 6.Ravi Ram K, Wolfner MF. Seminal influences: Drosophila Acps and the molecular interplay between males and females during reproduction. Integr Comp Biol. 2007;47:427–445. doi: 10.1093/icb/icm046. [DOI] [PubMed] [Google Scholar]
- 7.Chapman T. Seminal fluid-mediated fitness traits in Drosophila. Heredity (Edinb) 2001;87:511–521. doi: 10.1046/j.1365-2540.2001.00961.x. [DOI] [PubMed] [Google Scholar]
- 8.Kubli E. Sex-peptides: Seminal peptides of the Drosophila male. Cell Mol Life Sci. 2003;60:1689–1704. doi: 10.1007/s00018-003-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGraw LA, Clark AG, Wolfner MF. Post-mating gene expression profiles of female Drosophila melanogaster in response to time and to four male accessory gland proteins. Genetics. 2008;179:1395–1408. doi: 10.1534/genetics.108.086934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGraw LA, Gibson G, Clark AG, Wolfner MF. Genes regulated by mating, sperm, or seminal proteins in mated female Drosophila melanogaster. Curr Biol. 2004;14:1509–1514. doi: 10.1016/j.cub.2004.08.028. [DOI] [PubMed] [Google Scholar]
- 11.Gioti A, et al. Sex peptide of Drosophila melanogaster males is a global regulator of reproductive processes in females. Proc Biol Sci. 2012;279:4423–4432. doi: 10.1098/rspb.2012.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng J, Zipperlen P, Kubli E. Drosophila sex-peptide stimulates female innate immune system after mating via the Toll and Imd pathways. Curr Biol. 2005;15:1690–1694. doi: 10.1016/j.cub.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 13.Domanitskaya EV, Liu H, Chen S, Kubli E. The hydroxyproline motif of male sex peptide elicits the innate immune response in Drosophila females. FEBS J. 2007;274:5659–5668. doi: 10.1111/j.1742-4658.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- 14.Aigaki T, Fleischmann I, Chen P-S, Kubli E. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron. 1991;7:557–563. doi: 10.1016/0896-6273(91)90368-a. [DOI] [PubMed] [Google Scholar]
- 15.Chen PS, et al. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell. 1988;54:291–298. doi: 10.1016/0092-8674(88)90192-4. [DOI] [PubMed] [Google Scholar]
- 16.Soller M, Bownes M, Kubli E. Mating and sex peptide stimulate the accumulation of yolk in oocytes of Drosophila melanogaster. Eur J Biochem. 1997;243:732–738. doi: 10.1111/j.1432-1033.1997.00732.x. [DOI] [PubMed] [Google Scholar]
- 17.Heifetz Y, Tram U, Wolfner MF. Male contributions to egg production: The role of accessory gland products and sperm in Drosophila melanogaster. Proc Biol Sci. 2001;268:175–180. doi: 10.1098/rspb.2000.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heifetz Y, Lung O, Frongillo EA, Jr, Wolfner MF. The Drosophila seminal fluid protein Acp26Aa stimulates release of oocytes by the ovary. Curr Biol. 2000;10:99–102. doi: 10.1016/s0960-9822(00)00288-8. [DOI] [PubMed] [Google Scholar]
- 19.Herndon LA, Wolfner MF. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc Natl Acad Sci USA. 1995;92:10114–10118. doi: 10.1073/pnas.92.22.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng J, et al. Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila. Curr Biol. 2005;15:207–213. doi: 10.1016/j.cub.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 21.Avila FW, Wolfner MF. Acp36DE is required for uterine conformational changes in mated Drosophila females. Proc Natl Acad Sci USA. 2009;106:15796–15800. doi: 10.1073/pnas.0904029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adams EM, Wolfner MF. Seminal proteins but not sperm induce morphological changes in the Drosophila melanogaster female reproductive tract during sperm storage. J Insect Physiol. 2007;53:319–331. doi: 10.1016/j.jinsphys.2006.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertram MJ, Neubaum DM, Wolfner MF. Localization of the Drosophila male accessory gland protein Acp36DE in the mated female suggests a role in sperm storage. Insect Biochem Mol Biol. 1996;26:971–980. doi: 10.1016/s0965-1748(96)00064-1. [DOI] [PubMed] [Google Scholar]
- 24.Bloch Qazi MC, Wolfner MF. An early role for the Drosophila melanogaster male seminal protein Acp36DE in female sperm storage. J Exp Biol. 2003;206:3521–3528. doi: 10.1242/jeb.00585. [DOI] [PubMed] [Google Scholar]
- 25.Neubaum DM, Wolfner MF. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics. 1999;153:845–857. doi: 10.1093/genetics/153.2.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tram U, Wolfner MF. Male seminal fluid proteins are essential for sperm storage in Drosophila melanogaster. Genetics. 1999;153:837–844. doi: 10.1093/genetics/153.2.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong A, et al. A role for Acp29AB, a predicted seminal fluid lectin, in female sperm storage in Drosophila melanogaster. Genetics. 2008;180:921–931. doi: 10.1534/genetics.108.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ram KR, Wolfner MF. Sustained post-mating response in Drosophila melanogaster requires multiple seminal fluid proteins. PLoS Genet. 2007;3:e238. doi: 10.1371/journal.pgen.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Priest NK, Roach DA, Galloway LF. Cross-generational fitness benefits of mating and male seminal fluid. Biol Lett. 2008;4:6–8. doi: 10.1098/rsbl.2007.0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Gonzalez F, Dowling DK. Transgenerational effects of sexual interactions and sexual conflict: Non-sires boost the fecundity of females in the following generation. Biol Lett. 2015;11:20150067. doi: 10.1098/rsbl.2015.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sirot LK, Wong A, Chapman T, Wolfner MF. Sexual conflict and seminal fluid proteins: A dynamic landscape of sexual interactions. Cold Spring Harb Perspect Biol. 2014;7:a017533. doi: 10.1101/cshperspect.a017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapman T, Herndon LA, Heifetz Y, Partridge L, Wolfner MF. The Acp26Aa seminal fluid protein is a modulator of early egg hatchability in Drosophila melanogaster. Proc Biol Sci. 2001;268:1647–1654. doi: 10.1098/rspb.2001.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang C-H, et al. Control of the postmating behavioral switch in Drosophila females by internal sensory neurons. Neuron. 2009;61:519–526. doi: 10.1016/j.neuron.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalb JM, DiBenedetto AJ, Wolfner MF. Probing the function of Drosophila melanogaster accessory glands by directed cell ablation. Proc Natl Acad Sci USA. 1993;90:8093–8097. doi: 10.1073/pnas.90.17.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chapman T, Liddle LF, Kalb JM, Wolfner MF, Partridge L. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature. 1995;373:241–244. doi: 10.1038/373241a0. [DOI] [PubMed] [Google Scholar]
- 36.Wigby S, Chapman T. Sex peptide causes mating costs in female Drosophila melanogaster. Curr Biol. 2005;15:316–321. doi: 10.1016/j.cub.2005.01.051. [DOI] [PubMed] [Google Scholar]
- 37.Rice WR, Holland B. The enemies within: Intergenomic conflict, interlocus contest evolution (ICE), and the intraspecific red queen. Behav Ecol Sociobiol. 1997;41:1–10. [Google Scholar]
- 38.Haerty W, et al. Evolution in the fast lane: Rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–1335. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swanson WJ, Clark AG, Waldrip-Dail HM, Wolfner MF, Aquadro CF. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice WR. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature. 1996;381:232–234. doi: 10.1038/381232a0. [DOI] [PubMed] [Google Scholar]
- 41.Holland B, Rice WR. Experimental removal of sexual selection reverses intersexual antagonistic coevolution and removes a reproductive load. Proc Natl Acad Sci USA. 1999;96:5083–5088. doi: 10.1073/pnas.96.9.5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollis B, Houle D. Populations with elevated mutation load do not benefit from the operation of sexual selection. J Evol Biol. 2011;24:1918–1926. doi: 10.1111/j.1420-9101.2011.02323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holland B. Sexual selection fails to promote adaptation to a new environment. Evolution. 2002;56:721–730. doi: 10.1111/j.0014-3820.2002.tb01383.x. [DOI] [PubMed] [Google Scholar]
- 44.Hollis B, Fierst JL, Houle D. Sexual selection accelerates the elimination of a deleterious mutant in Drosophila melanogaster. Evolution. 2009;63:324–333. doi: 10.1111/j.1558-5646.2008.00551.x. [DOI] [PubMed] [Google Scholar]
- 45.Wensing KU, Koppik M, Fricke C. Precopulatory but not postcopulatory male reproductive traits diverge in response to mating system manipulation in Drosophila melanogaster. Ecol Evol. 2017;7:10361–10378. doi: 10.1002/ece3.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Crudgington HS, Beckerman AP, Brüstle L, Green K, Snook RR. Experimental removal and elevation of sexual selection: Does sexual selection generate manipulative males and resistant females? Am Nat. 2005;165:S72–S87. doi: 10.1086/429353. [DOI] [PubMed] [Google Scholar]
- 47.Rundle HD, Chenoweth SF, Blows MW. The roles of natural and sexual selection during adaptation to a novel environment. Evolution. 2006;60:2218–2225. [PubMed] [Google Scholar]
- 48.Martin OY, Hosken DJ. Costs and benefits of evolving under experimentally enforced polyandry or monogamy. Evolution. 2003;57:2765–2772. doi: 10.1111/j.0014-3820.2003.tb01518.x. [DOI] [PubMed] [Google Scholar]
- 49.Fricke C, Arnqvist G. Rapid adaptation to a novel host in a seed beetle (Callosobruchus maculatus): The role of sexual selection. Evolution. 2007;61:440–454. doi: 10.1111/j.1558-5646.2007.00038.x. [DOI] [PubMed] [Google Scholar]
- 50.Maklakov AA, Bonduriansky R, Brooks RC. Sex differences, sexual selection, and ageing: An experimental evolution approach. Evolution. 2009;63:2491–2503. doi: 10.1111/j.1558-5646.2009.00750.x. [DOI] [PubMed] [Google Scholar]
- 51.Demont M, et al. Experimental removal of sexual selection reveals adaptations to polyandry in both sexes. Evol Biol. 2014;41:62–70. [Google Scholar]
- 52.Radwan J, Unrug J, Snigórska K, Gawrońska K. Effectiveness of sexual selection in preventing fitness deterioration in bulb mite populations under relaxed natural selection. J Evol Biol. 2004;17:94–99. doi: 10.1046/j.1420-9101.2003.00646.x. [DOI] [PubMed] [Google Scholar]
- 53.Isaac RE, Li C, Leedale AE, Shirras AD. Drosophila male sex peptide inhibits siesta sleep and promotes locomotor activity in the post-mated female. Proc Biol Sci. 2010;277:65–70. doi: 10.1098/rspb.2009.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Edward DA, Fricke C, Gerrard DT, Chapman T. Quantifying the life-history response to increased male exposure in female Drosophila melanogaster. Evolution. 2011;65:564–573. doi: 10.1111/j.1558-5646.2010.01151.x. [DOI] [PubMed] [Google Scholar]
- 55.Pitnick S, Brown WD, Miller GT. Evolution of female remating behaviour following experimental removal of sexual selection. Proc Biol Sci. 2001;268:557–563. doi: 10.1098/rspb.2000.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fricke C, Chapman T. Variation in the post-mating fitness landscape in fruit flies. J Evol Biol. 2017;30:1250–1261. doi: 10.1111/jeb.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pitnick S, Miller GT, Reagan J, Holland B. Males’ evolutionary responses to experimental removal of sexual selection. Proc Biol Sci. 2001;268:1071–1080. doi: 10.1098/rspb.2001.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fowler K, Partridge L. A cost of mating in female fruitflies. Nature. 1989;338:760–761. [Google Scholar]
- 59.Partridge L, Fowler K. Non-mating costs of exposure to males in female Drosophila melanogaster. J Insect Physiol. 1990;36:419–425. [Google Scholar]
- 60.Brommer JE, Fricke C, Edward DA, Chapman T. Interactions between genotype and sexual conflict environment influence transgenerational fitness in Drosophila melanogaster. Evolution. 2012;66:517–531. doi: 10.1111/j.1558-5646.2011.01449.x. [DOI] [PubMed] [Google Scholar]
- 61.Innocenti P, Morrow EH. Immunogenic males: A genome-wide analysis of reproduction and the cost of mating in Drosophila melanogaster females. J Evol Biol. 2009;22:964–973. doi: 10.1111/j.1420-9101.2009.01708.x. [DOI] [PubMed] [Google Scholar]
- 62.Mack PD, Kapelnikov A, Heifetz Y, Bender M. Mating-responsive genes in reproductive tissues of female Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103:10358–10363. doi: 10.1073/pnas.0604046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dalton JE, et al. Dynamic, mating-induced gene expression changes in female head and brain tissues of Drosophila melanogaster. BMC Genomics. 2010;11:541. doi: 10.1186/1471-2164-11-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ottiger M, Soller M, Stocker RF, Kubli E. Binding sites of Drosophila melanogaster sex peptide pheromones. J Neurobiol. 2000;44:57–71. [PubMed] [Google Scholar]
- 65.Ding Z, Haussmann I, Ottiger M, Kubli E. Sex-peptides bind to two molecularly different targets in Drosophila melanogaster females. J Neurobiol. 2003;55:372–384. doi: 10.1002/neu.10218. [DOI] [PubMed] [Google Scholar]
- 66.Yapici N, Kim Y-J, Ribeiro C, Dickson BJ. A receptor that mediates the post-mating switch in Drosophila reproductive behaviour. Nature. 2008;451:33–37. doi: 10.1038/nature06483. [DOI] [PubMed] [Google Scholar]
- 67.Moshitzky P, et al. Sex-peptide activates juvenile hormone biosynthesis in the Drosophila melanogaster corpus allatum. Arch Insect Biochem Physiol. 1996;32:363–374. doi: 10.1002/(SICI)1520-6327(1996)32:3/4<363::AID-ARCH9>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 68.Flatt T, Kawecki TJ. Juvenile hormone as a regulator of the trade-off between reproduction and life span in Drosophila melanogaster. Evolution. 2007;61:1980–1991. doi: 10.1111/j.1558-5646.2007.00151.x. [DOI] [PubMed] [Google Scholar]
- 69.Flatt T, Tu M-P, Tatar M. Hormonal pleiotropy and the juvenile hormone regulation of Drosophila development and life history. BioEssays. 2005;27:999–1010. doi: 10.1002/bies.20290. [DOI] [PubMed] [Google Scholar]
- 70.Hollis B, Houle D, Kawecki TJ. Evolution of reduced post-copulatory molecular interactions in Drosophila populations lacking sperm competition. J Evol Biol. 2016;29:77–85. doi: 10.1111/jeb.12763. [DOI] [PubMed] [Google Scholar]
- 71.Clark AG, Aguadé M, Prout T, Harshman LG, Langley CH. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics. 1995;139:189–201. doi: 10.1093/genetics/139.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fiumera AC, Dumont BL, Clark AG. Associations between sperm competition and natural variation in male reproductive genes on the third chromosome of Drosophila melanogaster. Genetics. 2007;176:1245–1260. doi: 10.1534/genetics.106.064915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fiumera AC, Dumont BL, Clark AG. Sperm competitive ability in Drosophila melanogaster associated with variation in male reproductive proteins. Genetics. 2005;169:243–257. doi: 10.1534/genetics.104.032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chapman T, Neubaum DM, Wolfner MF, Partridge L. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc Biol Sci. 2000;267:1097–1105. doi: 10.1098/rspb.2000.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wigby S, et al. Seminal fluid protein allocation and male reproductive success. Curr Biol. 2009;19:751–757. doi: 10.1016/j.cub.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Crudgington HS, Fellows S, Snook RR. Increased opportunity for sexual conflict promotes harmful males with elevated courtship frequencies. J Evol Biol. 2010;23:440–446. doi: 10.1111/j.1420-9101.2009.01907.x. [DOI] [PubMed] [Google Scholar]
- 77.Crudgington HS, Fellows S, Badcock NS, Snook RR. Experimental manipulation of sexual selection promotes greater male mating capacity but does not alter sperm investment. Evolution. 2009;63:926–938. doi: 10.1111/j.1558-5646.2008.00601.x. [DOI] [PubMed] [Google Scholar]
- 78.Linklater JR, Wertheim B, Wigby S, Chapman T. Ejaculate depletion patterns evolve in response to experimental manipulation of sex ratio in Drosophila melanogaster. Evolution. 2007;61:2027–2034. doi: 10.1111/j.1558-5646.2007.00157.x. [DOI] [PubMed] [Google Scholar]
- 79.Ruhmann H, Wensing KU, Neuhalfen N, Specker J-H, Fricke C. Early reproductive success in Drosophila males is dependent on maturity of the accessory gland. Behav Ecol. 2016;27:1859–1868. [Google Scholar]
- 80.Hollis B, Keller L, Kawecki TJ. Sexual selection shapes development and maturation rates in Drosophila. Evolution. 2017;71:304–314. doi: 10.1111/evo.13115. [DOI] [PubMed] [Google Scholar]
- 81.Mohorianu I, Fowler EK, Dalmay T, Chapman T. Control of seminal fluid protein expression via regulatory hubs in Drosophila melanogaster. Proc Biol Sci. 2018;285:20181681. doi: 10.1098/rspb.2018.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Snook RR, Brüstle L, Slate J. A test and review of the role of effective population size on experimental sexual selection patterns. Evolution. 2009;63:1923–1933. doi: 10.1111/j.1558-5646.2009.00682.x. [DOI] [PubMed] [Google Scholar]
- 83.Long TA, Agrawal AF, Rowe L. The effect of sexual selection on offspring fitness depends on the nature of genetic variation. Curr Biol. 2012;22:204–208. doi: 10.1016/j.cub.2011.12.020. [DOI] [PubMed] [Google Scholar]
- 84.Whitlock MC, Agrawal AF. Purging the genome with sexual selection: Reducing mutation load through selection on males. Evolution. 2009;63:569–582. doi: 10.1111/j.1558-5646.2008.00558.x. [DOI] [PubMed] [Google Scholar]
- 85.Charlesworth B, Charlesworth D. Genetic variation in recombination in Drosophila. I. Responses to selection and preliminary genetic analysis. Heredity. 1985;54:71–83. [Google Scholar]
- 86.Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:1–48. [Google Scholar]
- 87.Team RC. 2018 R: A Language and Environment for Statistical Computing, R Foundation for Statistical Computing. R Version 3.5.1. Available at www.R-project.org. Accessed July 2, 2018.
- 88.Brian Hollis. 2019 Male reproductive tract gene expression and male effects on female gene expression in populations of Drosophila melanogaster evolving in manipulated mating systems. Gene Expression Omnibus (GEO). Available at https:// www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE128404. Deposited March 16, 2019.
- 89.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu R, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res. 2015;43:e97. doi: 10.1093/nar/gkv412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ritchie ME, et al. Empirical array quality weights in the analysis of microarray data. BMC Bioinformatics. 2006;7:261. doi: 10.1186/1471-2105-7-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 94.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 95.Weiner J, III, Domaszewska T. 2016. tmod: An R Package for General and Multivariate Enrichment Analysis. PeerJ Preprints 4:e2420v1. Preprint, posted September 4, 2016.
- 96.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Findlay GD, Yi X, Maccoss MJ, Swanson WJ. Proteomics reveals novel Drosophila seminal fluid proteins transferred at mating. PLoS Biol. 2008;6:e178. doi: 10.1371/journal.pbio.0060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.