Significance
Polycomb Repressor Complex 2 (PRC2) is an epigenetic effector enzyme with key roles in gene silencing and establishment of facultative heterochromatin. Mutations of this histone lysine methyltransferase and its substrate H3K27 are linked to multiple human pathologies, and PRC2 has been identified as a therapeutic target in cancer treatment. The H3K27 methylation activity of PRC2 is sensitive to the chromatin environment, including the methylation state of H3K36 on the same histone tail. The mechanistic basis of this negative cross-talk is poorly understood. The present work sheds light on this question by identifying a regulatory feature on the catalytic subunit of PRC2, EZH2, which acts as a specific sensor for H3K36 methylation state. The work reveals an unusual mode of enzyme modulation by a substrate molecule.
Keywords: PRC2 methyltransferase, designer chromatin, chemical biology
Abstract
Enhancer of Zeste Homolog 2 (EZH2) is the catalytic subunit of Polycomb Repressor Complex 2 (PRC2), the enzyme that catalyzes monomethylation, dimethylation, and trimethylation of lysine 27 on histone H3 (H3K27). Trimethylation at H3K27 (H3K27me3) is associated with transcriptional silencing of developmentally important genes. Intriguingly, H3K27me3 is mutually exclusive with H3K36 trimethylation on the same histone tail. Disruptions in this cross-talk result in aberrant H3K27/H3K36 methylation patterns and altered transcriptional profiles that have been implicated in tumorigenesis and other disease states. Despite their importance, the molecular details of how PRC2 “senses” H3K36 methylation are unclear. We demonstrate that PRC2 is activated in cis by the unmodified side chain of H3K36, and that this activation results in a fivefold increase in the kcat of its enzymatic activity catalyzing H3K27 methylation compared with activity on a substrate methylated at H3K36. Using a photo-cross-linking MS strategy and histone methyltransferase activity assays on PRC2 mutants, we find that EZH2 contains a specific sensing pocket for the H3K36 methylation state that allows the complex to distinguish between modified and unmodified H3K36 residues, altering enzymatic activity accordingly to preferentially methylate the unmodified nucleosome substrate. We also present evidence that this process may be disrupted in some cases of Weaver syndrome.
DNA is packaged in the eukaryotic cell nucleus in one of two general chromatin states: transcriptionally active (structurally open) euchromatin or transcriptionally silent (condensed) heterochromatin. Heterochromatin can be further divided into a constitutive form, which cannot convert to euchromatin, and a facultative form, which retains that potential (1). The histone proteins involved in packaging the DNA in each of these states are decorated with complex, characteristic patterns of posttranslational modifications (PTMs) that contribute to the local structure of chromatin (2, 3).
Histone H3 contains several key lysine residues on its N-terminal tail, including H3K27 and H3K36, which are the sites of various regulatory PTMs. H3K27me3, catalyzed by PRC2, is known to be important to the establishment of gene silencing through the recruitment of the PRC1 silencing complex that is ultimately responsible for the establishment of facultative heterochromatin (4–6), while H3K36me3 is commonly associated with active transcription within euchromatic regions (7). Notably, both these PTMs are dynamic, and dedicated demethylase enzymes are responsible for their removal (8–10).
The delicate balance between H3K27me3 and H3K36me2/3 is critical to the establishment and maintenance of cellular identity (11–13). Indeed, disruption of these methylation patterns is associated with several pathologies. Mutations in the histone methyltransferase (HMT) enzymes responsible for installing methylation at both the H3K27 and H3K36 positions (i.e., PRC2 and NSD1-3/SETD2, respectively) have been identified in numerous cancers and overgrowth disorders (7, 14, 15). Both activating and inactivating mutations of EZH2, the SET domain-containing catalytic subunit of PRC2, have been identified in hematologic malignancies, lymphomas, myeloproliferative neoplasms, and myelodysplastic syndromes (15). NSD1 mutations have been identified in pediatric acute myelogenous leukemia, neuroblastoma, gliomas, and carcinomas of the gastrointestinal tract, as well as head and neck cancers (7, 16, 17) and SETD2 mutations in clear cell renal cell carcinomas (18). PRC2 mutations have also been described in patients with Weaver syndrome (19, 20), while NSD1 loss of function has been identified in patients with Sotos syndrome (21). Further underscoring the importance of an H3K27/H3K36 methylation balance, lysine-to-methionine mutations at these positions lead to epigenetic dysregulation that is thought to drive the progression of rare pediatric cancers (11, 12, 17, 22–25).
PRC2 is a large molecular machine whose HMT activity is tuned by a variety of inputs associated with the local chromatin environment (26, 27). The minimal active version of the enzyme comprises EZH2 as well as three other proteins: EED, SUZ12, and RbAp46/48 (28). The core architecture of PRC2 is important for sensing the chromatin state. Catalytic stimulation of the complex is carried out via a positive feedback mechanism through the binding of EED to H3K27me3 on a neighboring nucleosome (29, 30). This acts to boost PRC2 activity locally within heterochromatic regions, leading to so-called “spreading” (29). The core PRC2 complex is further elaborated by a series of ancillary subunits that have various roles in regulating enzyme activity in time and space (31, 32). For example, the JARID2 binding partner can allosterically stimulate PRC2 activity in a manner analogous to H3K27me3 binding to EED (31) and has a role in targeting the core enzyme to relevant genomic loci along with other factors, such as AEBP2 and PCLs (26, 33).
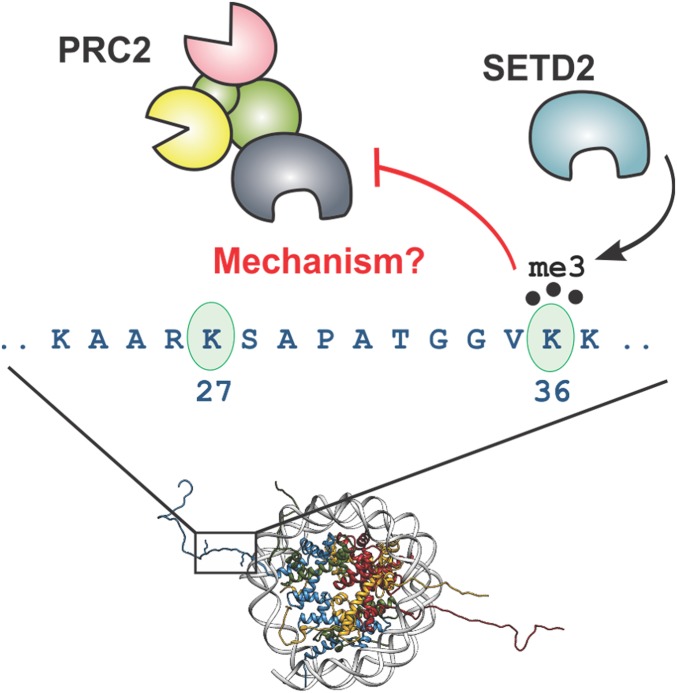
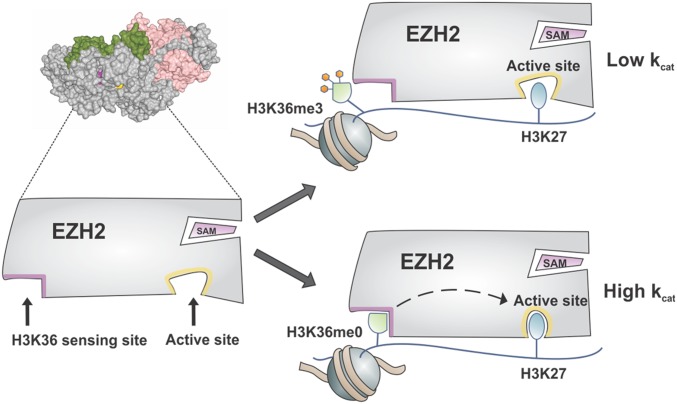
PRC2 activity is known to be sensitive to the modification status of histones, particularly on the H3 tail (Fig. 1). Remarkably, the same PTM chemotype (e.g., lysine methylation) can have opposite effects on activity, depending on the sequence position. Thus, while H3K27me3 leads to stimulation of PRC2, H3K36me3—which marks actively transcribed chromatin—inhibits enzyme activity (34, 35). Notably, the two PTMs are rarely found to coexist on the same histone tail in nature (34–36). Indeed, it is thought that H3K36me3 functions to block heterochromatin spreading by antagonizing PRC2 activity in cis (13, 34, 35). Consistent with this idea, reduction of H3K36 methylation levels in cells leads to a genome-wide increase in H3K27 methylation and an associated derepression of polycomb gene silencing (12, 13). Unlike with H3K27me3, for which the mechanism of PRC2 stimulation is well understood and involves a long-range allosteric activation process (29, 30, 37, 38), the molecular basis of the H3K27me3-H3K36me3 negative cross-talk is unclear. Structural studies on this system have so far failed to resolve the H3K36 side chain. Thus, how PRC2 senses the methylation status of H3K36, and whether the presence of the PTM affects substrate binding and/or enzyme catalysis, are unknown (Fig. 1).
Fig. 1.
Cartoon depicting the functional cross-talk between H3K27 and H3K36. Methylation of H3K27 by PRC2 is inhibited by the presence of H3K36 methylation on the same tail. It is unclear whether this happens through effects on enzyme-substrate binding or catalytic inhibition of the enzyme. Mononucleosome structure (Protein Data Bank ID code 3AV2).
In this study, we used chemical biology methods to shed light on the mechanism of H3K27/H3K36 methylation cross-talk. Photo-cross-linking studies localized the H3K36 sensor to a pocket within the SET domain of EZH2. Our data suggest that this pocket serves as a PTM-gated sensing site to regulate the catalytic activity of the enzyme. Mutagenesis of this region reduces the ability of the enzyme to distinguish between modified and unmodified H3K36 residues. In addition, we investigated the effect of a Weaver syndrome-associated mutation that maps to this region of EZH2 and found biochemical evidence that this cross-talk may be disrupted in certain cases of this disorder.
Results
PRC2 Activity Is Modulated by H3K36.
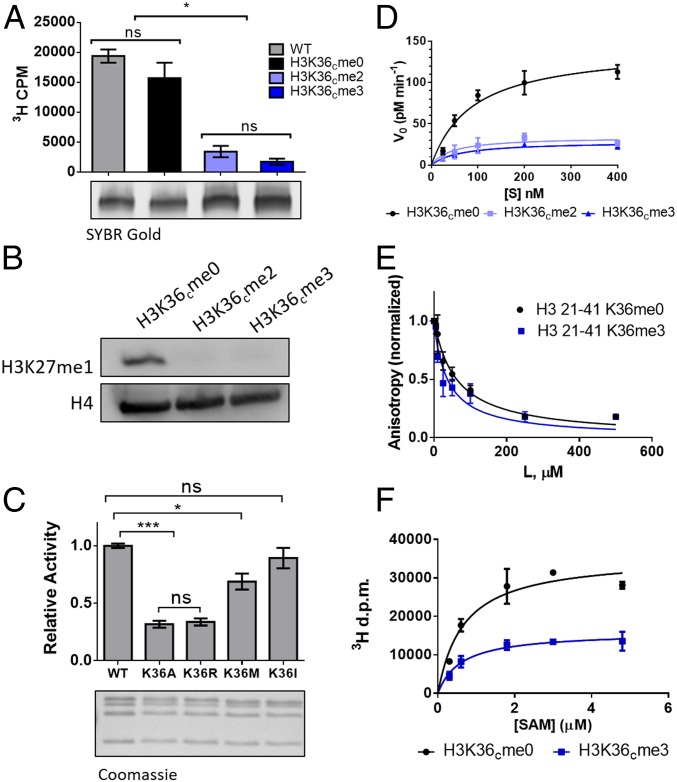
We initiated our studies into the basis of H3K27/H3K36 methylation cross-talk by performing HMT assays using purified recombinant PRC2 complexes and various designer chromatin substrates containing methyl-lysine analogs (SI Appendix, Fig. S1). Consistent with previous reports (34, 35), PRC2 activity was found to be significantly higher on H3K36cme0 (i.e., unmodified) mononucleosome substrates compared with those containing H3K36cme2/3, as measured by scintillation counting using 3H-labeled SAM (Fig. 2A). Similar results were obtained by Western blot analysis using antibodies specific to methylated H3K27 (Fig. 2B). Substitution of H3K36 with alanine or arginine also resulted in diminished H3K27 methylation, while methionine and isoleucine substitutions had a more modest impact on PRC2 function (Fig. 2C).
Fig. 2.
H3K36 methylation inhibits PRC2 catalysis. (A, Top) PRC2 core HMT activity as measured by scintillation counting on mononucleosome substrates containing recombinantly produced unmodified histones (WT) or histones alkylated with methyl-lysine analogs at the K36 position (H3K36cme0-3). Error bars represent SEM (n = 3). *P ≤ 0.05; ns, not significant. (A, Bottom) Analysis of mononucleosome substrates using a native gel stained for DNA using SYBR Gold as a loading control. (B) PRC2 core HMT activity measured by Western blot analysis for methylated H3K27 on unmodified on H3K36cme2/3 12mer nucleosome array substrates. Histone H4 serves as a loading control. (C, Top) PRC2 core HMT activity using 25 nM enzyme on 400 nM unmodified H3K36 mutant 12mer nucleosome array substrates. Data are shown relative to the WT. Error bars represent SEM (n = 4). *P ≤ 0.05; ***P ≤ 0.001; ns, not significant. (C, Bottom) Analysis of 12mer nucleosome arrays with mutations at H3K36 using SDS/PAGE and Coomassie blue staining as a loading control. (D) Kinetic analysis of WT PRC2 HMT activity using 25 nM enzyme on increasing concentrations of unmodified or H3K36cme2/3 12mer nucleosome array substrates. Error bars represent SEM (n = 3). Table 1 presents associated kinetic values. (E) Binding between the PRC2 core and H3 peptides (residues 21–41) modified or unmodified at H3K36. Shown is the change in steady-state anisotropy of a fluorescein-labeled H3 peptide on being displaced from the PRC2 core by added unlabeled substrate peptide. Note that the experiment was performed in the presence of SAM cofactor and H3K27me3-stimulating peptide. Negligible de novo methylation of H3K27 occurs under the conditions used in this assay (SI Appendix, Fig. S13). Error bars represent SEM (n = 3). (F) PRC2 (with JARID2 subunit present) HMT activity as measured by scintillation counting on 400 nM H3K36cme0 or H3K36cme3 12mer nucleosome array substrates in the presence of increasing concentrations of SAM cofactor. Error bars represent SEM (n = 3). Table 1 presents associated Km values.
To better understand the origins of these effects, we turned to more detailed kinetic analyses. In preliminary studies, we found that the use of 12mer nucleosome array substrates allowed for the measurement of enzymatic activity under conditions appropriate for the application of Michaelis–Menten kinetic analysis for the PRC2 core complex, which could not be accomplished using peptide, histone, or mononucleosome substrates (SI Appendix, Fig. S2). Moreover, the use of 12mer arrays overcame the need to include exogenous H2K27me3 peptide in the assay system (a requirement for robust HMT activity when using mononucleosome substrates). These kinetic results are in line with recent structural and biochemical studies indicating that PRC2 can engage multiple neighboring nucleosomes (30, 39). Using this experimental system, we were able to show that the difference in PRC2 activity on H3K36 methylated versus nonmethylated chromatin is primarily a kcat effect (Fig. 2D and Table 1).
Table 1.
Summary of thermodynamic and kinetic data generated in this study
| Substrate | SAM Km†, µM | Kdapp‡, µM peptide | WTEZH2* | M700VEZH2* | ||||
| kcat, min−1 × 10−5 | Km, nM, 601 sites | kcat/Km, min−1 nM−1 × 10−5 | kcat, min−1 × 10−5 | Km, nM, 601 sites | kcat/Km, min−1 nM−1 × 10−5 | |||
| H3K36me0 | 0.62 ± 0.2 | 29.4 ± 6.5 | 571 ± 46 | 89 ± 20 | 6.4 ± 1.4 | 277 ± 21 | 42 ± 12 | 6.6 ± 1.8 |
| H3K36me2 | 136 ± 19 | 44 ± 22 | 3.1 ± 1.5 | 152 ± 9 | 19 ± 6 | 7.8 ± 2.3 | ||
| H3K36me3 | 0.67 ± 0.2 | 18.5 ± 9.1 | 115 ± 9 | 63 ± 15 | 1.8 ± 0.4 | 160 ± 8 | 25 ± 5 | 6.3 ± 1.3 |
| H3K36R | 32.5 ± 2.1 | |||||||
Kinetic studies used the PRC2 core complex (WT or mutant EZH2) using 12mer nucleosome arrays as substrates. Errors = SEM (n = 3).
The Km for SAM was obtained using WT PRC2 core + JARID2 and 12mer nucleosome arrays as substrate. Errors represent SEM (n = 3).
Apparent dissociation constants are reported for the PRC2 core complex using histone peptide substrates. Errors = SEM (n = 3).
We next asked whether trimethylation of H3K36 perturbs the interaction of PRC2 with its substrate. Fluorescence anisotropy experiments demonstrated that PRC2 binds similarly to H3K36me0, H3K36me3, and H3K36R peptide substrates in the presence of SAM, with Kdapp values of ∼18–32 µM (Fig. 2E, Table 1, and SI Appendix, Fig. S3). This is consistent with pulldown experiments using chromatin array substrates with variations at position H3K36 (SI Appendix, Fig. S5) and with the kinetic data discussed above showing that Km values on chromatin array substrates were also similar to one another (Table 1). To determine whether the affinity for the cofactor changed as a function of H3K36 methylation state, we performed a titration of SAM at a saturating concentration of substrate. These studies necessitated the use of a PRC2 complex containing the JARID2 subunit to obtain a sufficient signal on the H3K36cme3 substrate for curve fitting and accurate determination of the Km for SAM. Importantly, the activity of JARID2-PRC2, like the core complex, is sensitive to methylation at H3K36 (SI Appendix, Fig. S6). However, this sensitivity was apparently not related to altered affinity for the cofactor, since the Km for SAM was unaffected by the methylation state of H3K36 (Fig. 2F and Table 1).
Taken together, our biochemical data supports a kinetic scheme in which overall PRC2 affinity for its substrate and cofactor is unchanged by methylation at H3K36, but enzyme association with a favorable substrate (S1) results in an activated complex with an increased kcat value, indicated by the activation coefficient γ (for WT PRC2, γ = 5; Eq. 1). In contrast, association of PRC2 with a poor substrate (S2) results in the formation of a less active complex (Eq. 2):
| [1] |
| [2] |
Photo-Cross-Linking Reveals a Sensing Pocket for H3K36 in EZH2.
The precise location of the putative H3K36 sensing pocket on PRC2 remains unknown. Initial biochemical work implicated the Suz12 subunit in the sensing mechanism (34), while structural studies, including a recent cryo-EM structural model of PRC2 bound to a dinucleosome (30), suggest that H3K36 is more likely to engage the EZH2 subunit than the Suz12 subunit (SI Appendix, Fig. S7). To map the H3K36-sensing region of PRC2 in an unbiased fashion, we used a photo-cross-linking strategy. To this end, we generated 12-mer nucleosome arrays in which a diazirine probe was site-specifically incorporated at position 36 in histone H3 (SI Appendix, Fig. S8). We imagined that the small size of this photoactivatable group and our observation that K36M nucleosomes acted as PRC2 substrates (Fig. 1) would facilitate cross-linking to the H3K36 sensing pocket on PRC2. Incubation of these arrays with PRC2 core complex led to UV-dependent generation of a major cross-linked species between H3 and EZH2, as determined by Western blot analysis (SI Appendix, Fig. S8 F–H). Cross-links to the other subunits were not observed.
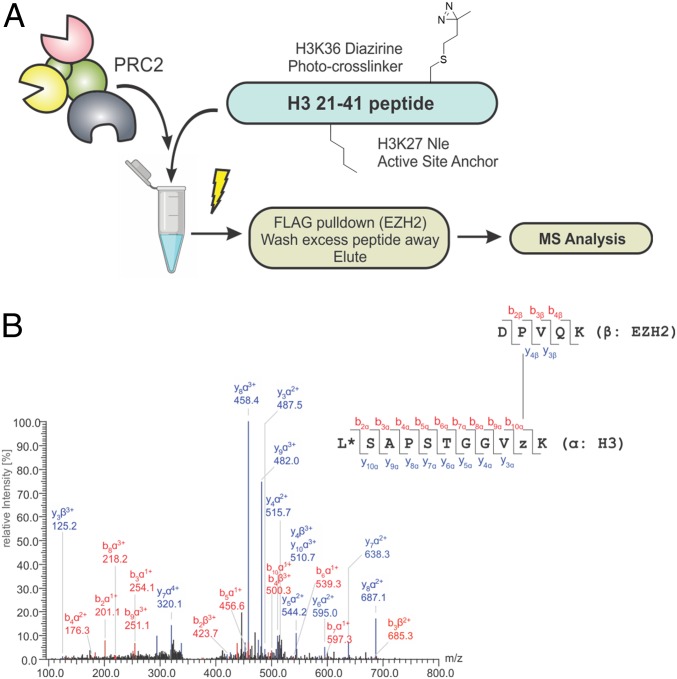
Building on this result, we next set out to localize the H3K36-binding site on EZH2 by incorporating an MS readout into the workflow (Fig. 3A). To simplify the analytics, we designed a photoactivatable peptide probe encompassing residues 21–41 of the histone H3.3 sequence. The probe contained two key features: a norleucine (Nle) residue at position 27 and, as before, a diazirine moiety at position 36 (Fig. 3A and SI Appendix, Fig. S9 A–C). H3 peptides containing Nle in place of Lys-27 are known to be potent orthosteric inhibitors of PRC2 (40). Thus, we expected that installation of this unnatural amino acid would serve to anchor the peptide to the EZH2 active site, thereby improving cross-linking efficiency. Consistent with our design, the synthetic probe peptide was found to be a potent competitive inhibitor of PRC2 activity (SI Appendix, Fig. S9D).
Fig. 3.
Mapping the H3K36 sensing site using photo-cross-linking MS. (A) In the experimental workflow, a diazirine-containing probe peptide is incubated with PRC2 and the mixture is UV-irradiated. Following enrichment and digestion, MS is used to map the H3K36 sensing site. Details of probe synthesis are provided in SI Appendix, Fig. S9. (B) Representative MS/MS spectrum corresponding to a cross-link between the probe peptide and the EZH2 subunit. (SI Appendix, Table S1 identifies all cross-links.)
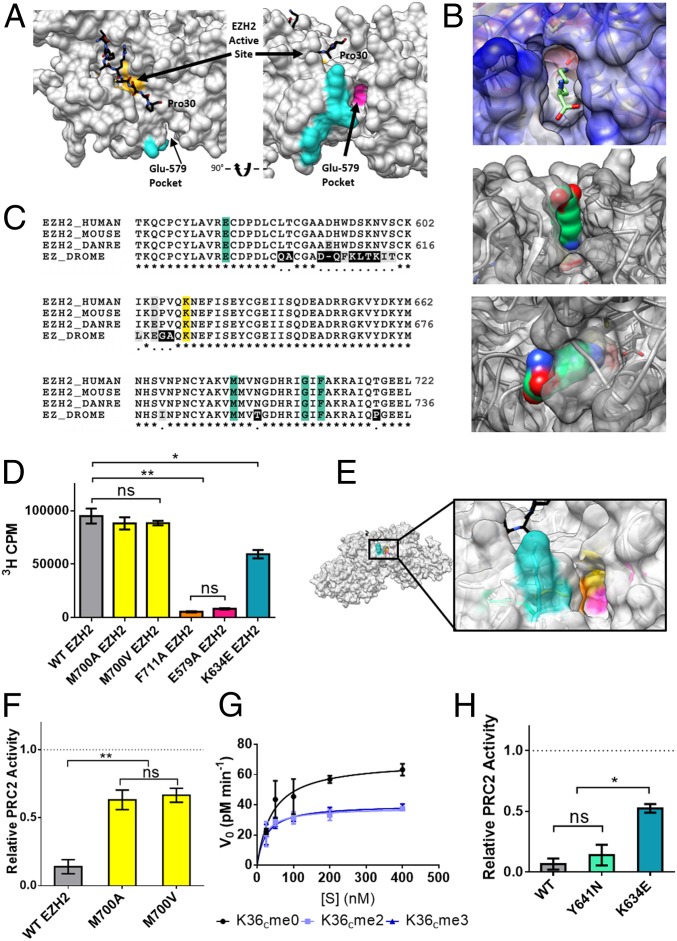
PRC2 core complex was incubated with excess probe peptide and then irradiated with UV light. The complex was then isolated by immunoprecipitation to remove excess peptide (Fig. 3A), digested with trypsin, and analyzed by liquid chromatography coupled to tandem MS (LC-MS/MS). Analysis of the LC-MS/MS data using the StavroX software tool (41) led to the identification of four unique cross-linked peptides with a score >100, indicating high probability of the cross-linked product (41). All these identified cross-linked peptides map to the same tryptic fragment from the EZH2 subunit, namely D630PVQK634 (Fig. 3B and SI Appendix, Table S1). Inspection of available PRC2 structural data shows that this peptide is surface-exposed and proximal to the predicted H3 substrate-binding channel on EZH2 (Fig. 4A and SI Appendix, Fig. S10A). Analysis of the topological and electrostatic properties of this region revealed a solvent-accessible pocket with the appropriate properties to accommodate an unmodified lysine side chain (Fig. 4B). Specifically, the walls of this pocket are lined with hydrophobic side chains (e.g., Met-700 and Phe-711), while the base contains a negatively charged glutamic acid residue (Glu-579). Notably, these residues in EZH2 are conserved from fruit fly to human (Fig. 4C). We refer to this region of EZH2 as the Glu-579 pocket.
Fig. 4.
Role of the Glu-579 pocket on EZH2. (A) View of PRC2 with substrate H3 tail (black) bound in the active site (yellow). The cross-linked tryptic peptide is highlighted in cyan and the adjacent Glu-579 pocket highlighted in magenta (Protein Data Bank ID code 5HYN). (B, Top) an unmodified lysine side chain depicted as sticks modeled into the Glu-579 pocket in EZH2. (B, Middle and Bottom) a lysine side chain in space-filling view is shown modeled into the same pocket. (C) Sequence alignment of EZH2 shows conservation of pocket residues (teal) from fruit fly to human. DANRE, Danio rerio; DROME, Drosophila melanogaster. Cross-links were observed to the K634 residue of EZH2 (yellow), a residue notably mutated in some cases of Weaver syndrome. (D) WT or mutant PRC2 HMT activity as measured by scintillation counting on 200 nM unmodified mononucleosomes. Error bars represent SEM (n = 3). **P ≤ 0.01; *P ≤ 0.05; ns, not significant. (E) Zoom-in view of the Glu-579 pocket in EZH2 (Protein Data Bank ID code 5HYN), with pocket residues color-coded as follows: K634 in teal, M700 in yellow, F711 in orange, and E579 in pink. (F) WT or mutant PRC2 HMT activity as measured by scintillation counting on H3K36cme3 mononucleosomes, normalized to each enzyme’s activity on H3K36cme0 mononucleosome substrates (dotted line). Error bars represent SEM (n = 3). **P ≤ 0.01. (G) Kinetic analysis of M700V-EZH2 mutant on unmodified or H3K36cme2/3 12mer array substrates. Error bars represent SEM (n = 3). Table 1 presents associated kinetic values. (H) WT or disease-associated mutant EZH2-containing PRC2 HMT activity on H3K36cme3 mononucleosomes, normalized to each enzyme’s activity on H3K36cme0 mononucleosome substrates (dotted line). Error bars represent SEM (n = 3). *P ≤ 0.05; ns, not significant.
Mutagenesis of the Putative H3K36 Sensing Pocket Increases Tolerance of PRC2 to H3K36 Methylation.
A series of PRC2 mutants were generated to test whether the Glu-579 pocket in EZH2 plays a role in H3K36 methylation sensing (SI Appendix, Fig. S1D). Mutation of either Glu-579 or Phe-711 to alanine largely abolished PRC2 activity (Fig. 4D) and cross-linking to chromatin (SI Appendix, Fig. S8J), a result consistent with a functional role for this region of EZH2. Mutation of Met-700, which lies near the base of the Glu-579 pocket (Fig. 4E), was more insightful in terms of exploring the question of H3K36 sensing. Substitution of this residue with either alanine or valine had little impact on the activity of PRC2 toward chromatin substrates containing unmethylated H3K36 (Fig. 4D). Strikingly, however, both mutants were markedly more active toward H3K36cme3-containing substrates compared with the wild-type (WT) enzyme (Fig. 4F). Indeed, the catalytic efficiency of the M700V mutant enzyme (as measured by the kcat/Km ratio) was unchanged as a function of H3K36 methylation state of the substrate (Fig. 4G and Table 1). This result supports a role for the Glu-579 pocket of EZH2 in the H3K36 sensing mechanism. We propose that the M700A/V mutations enlarge the Glu-579 pocket to allow it to accommodate the larger size of the methylated H3K36 side chain.
The EZH2 sequence identified in the cross-linking MS experiment includes lysine 634. This residue is located on the edge of the Glu-579 pocket and, intriguingly, has been found to be mutated to glutamic acid in some patients diagnosed with Weaver syndrome, a pediatric overgrowth disorder (20). We tested the effect of this mutation on PRC2 activity as a function of H3K36 methylation state. Similar to the M700A/V mutations discussed above, this mutant was significantly less sensitive to the H3K36 methylation compared with the WT enzyme (Fig. 4H). As a control, we also tested a known hyperactive, cancer-associated EZH2 mutant, Y641N, located between the H3K36 sensing site and the active site (42). Notably, in this case, we did not observe a change in PRC2 sensitivity to the H3K36 methylation state (Fig. 4H).
Discussion
PRC2 is a critical “writer” of epigenetic information with the ability to integrate multiple signals emanating from its chromatin substrate to regulate its overall HMT activity (26). One such input is methylation of H3K36. The presence of this PTM negatively regulates PRC2 activity, despite being located nine residues away from the substrate lysine (i.e., H3K27). Here we have used purified enzymes and designer chromatin substrates to explore the basis of this negative cross-talk. Detailed kinetic analyses point to a process in which the unmodified H3K36 side chain, but not its methylated derivatives, stimulates enzyme activity primarily through changes in kcat. Photo-cross-linking studies led to the identification of a putative H3K36 sensing pocket on the EZH2 subunit, the mutagenesis of which disrupts the normal negative cross-talk process. Collectively, our data point to a mode of PRC2 regulation in which the catalytic activity of the enzyme is stimulated by binding of unmodified lysine 36 into a conserved structural feature within the SET domain of EZH2.
One key question regarding the mechanism of H3K36 methylation sensing is how the PTM affects the HMT activity of the enzyme. Our data indicate that the H3K36me2/3 does not, strictly speaking, inhibit the enzyme, but rather fails to activate its HMT activity in a manner analogous to the unmodified side chain. Several lines of evidence lead us to this conclusion. First, we observe no difference in the affinity of PRC2 for its substrate (Kdapp and Km) as a function of H3K36 methylation, a finding in agreement with previously studies suggesting that PRC2-substrate association/binding is uncoupled from its activity (39, 43). In contrast, the catalytic activity of the enzyme is strongly affected by the chemical structure of the side chain at position 36. In this regard, the behavior of the H3K36A/R and H3K36M/I mutants is particularly insightful; the former are poor substrates for PRC2, whereas the latter are better tolerated. We propose that the H3K36M/I mutations, because of their size and shape, can bind the sensing pocket in EZH2 in a manner that still activates the enzyme. We note that Lys-to-Met/Ile mutations in histone tails are known to afford tight-binding orthosteric inhibitors of several HMTs (11, 12, 40, 44). Thus, there is precedence for these two side chains being accommodated in lysine binding pockets. In contrast, the corresponding alanine and arginine substitutions do not afford tight-binding orthosteric inhibitors of HMTs (40), which is consistent with our finding that incorporation of these residues at position 36 affords poor PRC2 substrates.
Our sensing model is also supported by mutagenesis of the putative sensing region itself, that is, the Glu-579 pocket in EZH2. Expanding the size of the pocket through substitution of Met-700 with either alanine or valine had the remarkable effect of dampening the sensitivity of the enzyme to the H3K36 methylation state.
Based on the foregoing considerations, we propose that PRC2 makes specific interactions with the unmodified H3K36 side chain, likely via the Glu-579 pocket in EZH2, and that these trigger structural changes in the EZH2 active site (Fig. 5). Methylation of this residue disrupts these contacts, likely for steric reasons.
Fig. 5.
Model of H3K36 sensing by PRC2. Engagement of PRC2 with chromatin containing unmodified H3K36 leads to activation of the EZH2 catalytic subunit, as manifested by an increase in kcat for H3K27 methylation. This process is disrupted on dimethylation or trimethylation of H3K36.
Exactly how ligand (i.e., H3K36) engagement with the Glu-579 pocket leads to PRC2 activation remains unclear. The Glu-579 residue is located within the CXC domain of EZH2, and the pocket is located at the cleft formed by the interface between the CXC and SET domains. Thus, binding of unmodified H3K36 at this region may simply serve to stabilize the SET domain and thereby promote catalysis. We note that stabilization of SET domains via engagement with flanking domains has been suggested previously (45). Alternatively, this activation may occur through effects of the H3K36 modification state on the substrate H3K27 side chain positioning in the active site. An unmodified H3K36 side chain bound in the Glu-579 pocket could serve to “pull” the H3K27 side chain into proper position at the active site, whereas an altered binding mode between H3K36me2/3 and PRC2 may prevent proper engagement of EZH2 with H3K27 through twisting of the histone tail.
It is also possible that ligand binding to the Glu-579 pocket activates catalysis through a more subtle allosteric-type process. The Glu-579 pocket is separated from the SET domain active site by ∼21 Å (SI Appendix, Fig. S10B). Thus, in an allosteric process, the ligand state of the H3K36 sensing pocket would be communicated to the active site directly through the SET domain. This is comparable with the canonical allosteric activation process in PRC2, which is initiated by product binding within the EED subunit of the complex at a site located some 30 Å from the active site in the SET domain (SI Appendix, Fig. S10B). Ultimately, high-resolution structural information will be needed to fully understand how the proposed engagement of the Glu-579 pocket by the unmodified H3K36 side chain influences the structure of the SET domain and the active site to promote catalysis.
Finally, the current biochemical studies may have bearing on our understanding of the pediatric overgrowth disorders Weaver syndrome and Sotos syndrome. These disorders phenocopy each other to a remarkably high degree; both are characterized by increased skeletal growth and height during development (14, 46). Sotos syndrome results from loss-of-function mutations or deletions in genes encoding the H3K36-specific HMTs, NSD1, or SETD2 (47, 48). It is known that loss of H3K36 methylation (either by H3K36M mutation or loss of function in NSD1/SETD2) can result in redistribution of PRC2 and PRC1 in the genome, driving aberrant transcriptional profiles in these cases (12, 13). The catalytic subunit of PRC2, EZH2, has an important role in the regulation of skeletal growth and height (49). Mutations in multiple PRC2 subunits have been found in patients with Weaver syndrome (19, 20, 50). Intriguingly, we found that a Weaver mutant that maps to the putative H3K36 sensing region in EZH2 exhibits a reduction of sensitivity to the H3K36 methylation state in vitro. It is thus possible that the overgrowth pathogenesis observed in Weaver syndrome is driven in part by aberrantly localized PRC2 and/or increased PRC2 activity on K36-methylated histone tails in patients with this mutation, contributing to the striking phenotypic overlap observed with Sotos syndrome. Testing this mechanistic idea will be an interesting line of inquiry for future studies.
Methods
Detailed descriptions of the experimental methods used in this study are provided in SI Appendix.
Mononucleosome and Nucleosome Array Reconstitution.
Mononucleosomes and dodecameric array substrates were assembled from purified histone octamers and recombinant DNA in the presence of buffer DNA by salt gradient dialysis as described previously (51). Histone H3 analogs bearing H3K36 methylation states and diazirine photo-cross-linkers were generated using a cysteine alkylation strategy.
Recombinant PRC2.
Recombinant PRC2 was generated by expressing EZH2 (mutant or WT) containing an N-terminal FLAG tag with AEBP2, EED, SUZ12, RbAp48, and, optionally, JARID2 in Spodoptera frugiperda (Sf9) cells using a MultiBac baculovirus expression system (52).
HMT Assays.
The methyltransferase activity of PRC2 was measured using a scintillation-based assay. PRC2 product identity was confirmed by Western blot analysis against methylated H3K27. Typical assay conditions are as follows: unmodified or modified 12mer arrays were incubated with ∼25 nM PRC2 in 10 μL of HMT buffer (50 mM Hepes pH 7.5, 0.5 mM MgCl2, 1 mM DTT) in the presence of 1 μM [3H]-SAM (PerkinElmer) for 45 min at 30 °C. Assays performed with mononucleosome substrates also contained 20 µM H3K27me3-stimulating peptide. Reactions were quenched by spotting on P81 phosphocellulose filter paper (Reaction Biology). After drying, activity was measured using scintillation counting with 1 mL of Ultima Gold scintillation mixture on a MicroBeta2 scintillation counter (PerkinElmer).
Peptide-Binding Studies.
Peptides corresponding to residues 21–41 of the H3 tail, unmodified or trimethylated at H3K36, were used to compete away 100 nM fluorescently labeled H3 peptide with a known Kdapp bound to PRC2. PRC2 concentration was fixed at 400 nM in binding buffer (50 mM Hepes pH 8, 50 nM NaCl, 2 mM MgCls, 1 mM DTT) with 5 µM SAM and 100 µM H3K27me3-stimulating peptide. Fluorescence anisotropy measurements were recorded with a Horiba DM302 fluorimeter.
Synthesis of Photo-Cross-Linker Peptide.
Fmoc-SPPS was used to synthesize a histone peptide containing norleucine at the position corresponding to H3K27 and cysteine at the position corresponding to H3K36. Cysteine alkylation was used to generate the diazirine photo-cross-linker peptide.
Cross-Linking MS.
PRC2 was incubated with a ∼45-fold molar excess of cross-linker peptide and irradiated with UV light to generate covalent cross-links. Cross-linked protein was subjected to trypsin digestion and analyzed by MS. Cross-links were identified using the StavroX software tool (41).
Supplementary Material
Acknowledgments
We thank Dr. Zack Brown and Dr. Galia Debelouchina for their insight and helpful discussions on this project. We also thank Dr. Tharan Srikumar of the Princeton Proteomics and Mass Spectrometry facility. This work was supported by National Institutes of Health Grants R37-GM086868 and P01-CA196539.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819029116/-/DCSupplemental.
References
- 1.Trojer P, Reinberg D. Facultative heterochromatin: Is there a distinctive molecular signature? Mol Cell. 2007;28:1–13. doi: 10.1016/j.molcel.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- 4.Cao R, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 5.Kirmizis A, et al. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys-27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong S, et al. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 10.Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 11.Lewis PW, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, et al. Histone H3K36 mutations promote sarcomagenesis through altered histone methylation landscape. Science. 2016;352:844–849. doi: 10.1126/science.aac7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streubel G, et al. The H3K36me2 methyltransferase Nsd1 demarcates PRC2-mediated H3K27me2 and H3K27me3 domains in embryonic stem cells. Mol Cell. 2018;70:371–379.e5. doi: 10.1016/j.molcel.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 14.Tatton-Brown K, Rahman N. The NSD1 and EZH2 overgrowth genes, similarities and differences. Am J Med Genet C Semin Med Genet. 2013;163C:86–91. doi: 10.1002/ajmg.c.31359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Comet I, Riising EM, Leblanc B, Helin K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat Rev Cancer. 2016;16:803–810. doi: 10.1038/nrc.2016.83. [DOI] [PubMed] [Google Scholar]
- 16.Ley TJ, et al. Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papillon-Cavanagh S, et al. Impaired H3K36 methylation defines a subset of head and neck squamous cell carcinomas. Nat Genet. 2017;49:180–185. doi: 10.1038/ng.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanu N, et al. SETD2 loss of function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34:5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooney E, Bi W, Schlesinger AE, Vinson S, Potocki L. Novel EED mutation in patient with Weaver syndrome. Am J Med Genet A. 2017;173:541–545. doi: 10.1002/ajmg.a.38055. [DOI] [PubMed] [Google Scholar]
- 20.Tatton-Brown K, et al. Childhood Overgrowth Collaboration Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2:1127–1133. doi: 10.18632/oncotarget.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurotaki N, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat Genet. 2002;30:365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 22.Castel D, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 2015;130:815–827. doi: 10.1007/s00401-015-1478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bender S, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell. 2013;24:660–672. doi: 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 24.Behjati S, et al. Distinct H3F3A and H3F3B driver mutations define chondroblastoma and giant cell tumor of bone. Nat Genet. 2013;45:1479–1482, and erratum (2014) 46:316. doi: 10.1038/ng.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bayliss J, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med. 2016;8:366ra161. doi: 10.1126/scitranslmed.aah6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holoch D, Margueron R. Mechanisms regulating PRC2 recruitment and enzymatic activity. Trends Biochem Sci. 2017;42:531–542. doi: 10.1016/j.tibs.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Xu C, et al. Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2) Proc Natl Acad Sci USA. 2010;107:19266–19271. doi: 10.1073/pnas.1008937107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu H, et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS One. 2013;8:e83737. doi: 10.1371/journal.pone.0083737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margueron R, et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762–767. doi: 10.1038/nature08398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poepsel S, Kasinath V, Nogales E. Cryo-EM structures of PRC2 simultaneously engaged with two functionally distinct nucleosomes. Nat Struct Mol Biol. 2018;25:154–162. doi: 10.1038/s41594-018-0023-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanulli S, et al. Jarid2 methylation via the PRC2 complex regulates H3K27me3 deposition during cell differentiation. Mol Cell. 2015;57:769–783. doi: 10.1016/j.molcel.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CH, et al. Distinct stimulatory mechanisms regulate the catalytic activity of polycomb repressive complex 2. Mol Cell. 2018;70:435–448.e5. doi: 10.1016/j.molcel.2018.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper S, et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between polycomb complexes PRC1 and PRC2. Nat Commun. 2016;7:13661. doi: 10.1038/ncomms13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmitges FW, et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell. 2011;42:330–341. doi: 10.1016/j.molcel.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 35.Yuan W, et al. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Y, et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci USA. 2012;109:13549–13554. doi: 10.1073/pnas.1205707109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Justin N, et al. Structural basis of oncogenic histone H3K27M inhibition of human polycomb repressive complex 2. Nat Commun. 2016;7:11316. doi: 10.1038/ncomms11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiao L, Liu X. Structural basis of histone H3K27 trimethylation by an active polycomb repressive complex 2. Science. 2015;350:aac4383. doi: 10.1126/science.aac4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, et al. Molecular analysis of PRC2 recruitment to DNA in chromatin and its inhibition by RNA. Nat Struct Mol Biol. 2017;24:1028–1038. doi: 10.1038/nsmb.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown ZZ, et al. Strategy for “detoxification” of a cancer-derived histone mutant based on mapping its interaction with the methyltransferase PRC2. J Am Chem Soc. 2014;136:13498–13501. doi: 10.1021/ja5060934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Götze M, et al. StavroX: A software for analyzing crosslinked products in protein interaction studies. J Am Soc Mass Spectrom. 2012;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 42.Sneeringer CJ, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107:20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CH, et al. Allosteric activation dictates PRC2 activity independent of its recruitment to chromatin. Mol Cell. 2018;70:422–434.e6. doi: 10.1016/j.molcel.2018.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jayaram H, et al. S-adenosyl methionine is necessary for inhibition of the methyltransferase G9a by the lysine 9 to methionine mutation on histone H3. Proc Natl Acad Sci USA. 2016;113:6182–6187. doi: 10.1073/pnas.1605523113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson JR, et al. Crystal structure and functional analysis of the histone methyltransferase SET7/9. Cell. 2002;111:105–115. doi: 10.1016/s0092-8674(02)00964-9. [DOI] [PubMed] [Google Scholar]
- 46.Tatton-Brown K, et al. Childhood Overgrowth Collaboration Mutations in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am J Hum Genet. 2017;100:725–736. doi: 10.1016/j.ajhg.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Douglas J, et al. Partial NSD1 deletions cause 5% of Sotos syndrome and are readily identifiable by multiplex ligation dependent probe amplification. J Med Genet. 2005;42:e56. doi: 10.1136/jmg.2005.031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatton-Brown K, Rahman N. Sotos syndrome. Eur J Hum Genet. 2007;15:264–271. doi: 10.1038/sj.ejhg.5201686. [DOI] [PubMed] [Google Scholar]
- 49.Lui JC, et al. EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat Commun. 2016;7:13685. doi: 10.1038/ncomms13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatton-Brown K, et al. Childhood Overgrowth Consortium Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am J Med Genet A. 2013;161A:2972–2980. doi: 10.1002/ajmg.a.36229. [DOI] [PubMed] [Google Scholar]
- 51.Fierz B, et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berger I, Fitzgerald DJ, Richmond TJ. Baculovirus expression system for heterologous multiprotein complexes. Nat Biotechnol. 2004;22:1583–1587. doi: 10.1038/nbt1036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.