Significance
Proteorhodopsin (PR) is found in marine bacteria in various ecosystems and is one of the most abundant photoreceptors. It converts light into a transmembrane, electrochemical gradient as a source of energy for the cell. PR, like many microbial rhodopsins, forms functionally unresolved higher oligomers. Here, experimental evidence for functionally relevant cross-protomer interactions within the PR pentamer is presented. An Asp–His–Trp triad is formed across the oligomerization interface, participates in the photocycle, and potentially acts as a pH sensor. These findings provide a potentially paradigm shifting aspect for the mechanism of microbial rhodopsins. Such an insight could only be obtained by combining solid-state NMR spectroscopy with dynamic nuclear polarization and light-induced cryotrapping of photointermediates.
Keywords: proteorhodopsin, NMR, DNP, oligomer, photocycle
Abstract
Proteorhodopsin (PR) is a highly abundant, pentameric, light-driven proton pump. Proton transfer is linked to a canonical photocycle typical for microbial ion pumps. Although the PR monomer is able to undergo a full photocycle, the question arises whether the pentameric complex formed in the membrane via specific cross-protomer interactions plays a role in its functional mechanism. Here, we use dynamic nuclear polarization (DNP)-enhanced solid-state magic-angle spinning (MAS) NMR in combination with light-induced cryotrapping of photointermediates to address this topic. The highly conserved residue H75 is located at the protomer interface. We show that it switches from the (τ)- to the (π)-tautomer and changes its ring orientation in the M state. It couples to W34 across the oligomerization interface based on specific His/Trp ring orientations while stabilizing the pKa of the primary proton acceptor D97 within the same protomer. We further show that specific W34 mutations have a drastic effect on D97 and proton transfer mediated through H75. The residue H75 defines a cross-protomer Asp–His–Trp triad, which potentially serves as a pH-dependent regulator for proton transfer. Our data represent light-dependent, functionally relevant cross talk between protomers of a microbial rhodopsin homo-oligomer.
Proteorhodopsin (PR) is a heptahelical retinal-binding membrane protein that was discovered through metagenomic screens of seawater samples in the SAR86 group of uncultured γ-proteobacteria (1). It functions as a light-dependent proton pump, establishing a proton gradient across the bacterial membrane. Bacteria may use this gradient for retinal-based phototrophy (2). Numerous PR-like sequences have been found in different seas of the world with specific properties such as green proteorhodopsin (GPR) and blue proteorhodopsin (BPR), which are color tuned depending on the depth of seawater in which they occur (3). A single residue substitution (L105Q) within the retinal-binding pocket is strongly correlated with shifting the retinal absorption maximum from 520 nm in GPR to 490 nm in BPR (4, 5).
Green and blue proteorhodopsin variants form higher homo-oligomeric complexes (Fig. 1) in which specific cross-protomer salt bridge contacts were identified to control the formation of predominantly pentamers and a small fraction of hexamers (6–8). Protein oligomerization may be advantageous for structural stability of individual subunits as well as for their functionality. However, it is unknown whether the pentameric and hexameric PR arrangements have a direct functional relevance for proton transport (7, 9). Specific interactions across the protomer interface with mechanistic relevance for the photocycle have not been demonstrated yet.
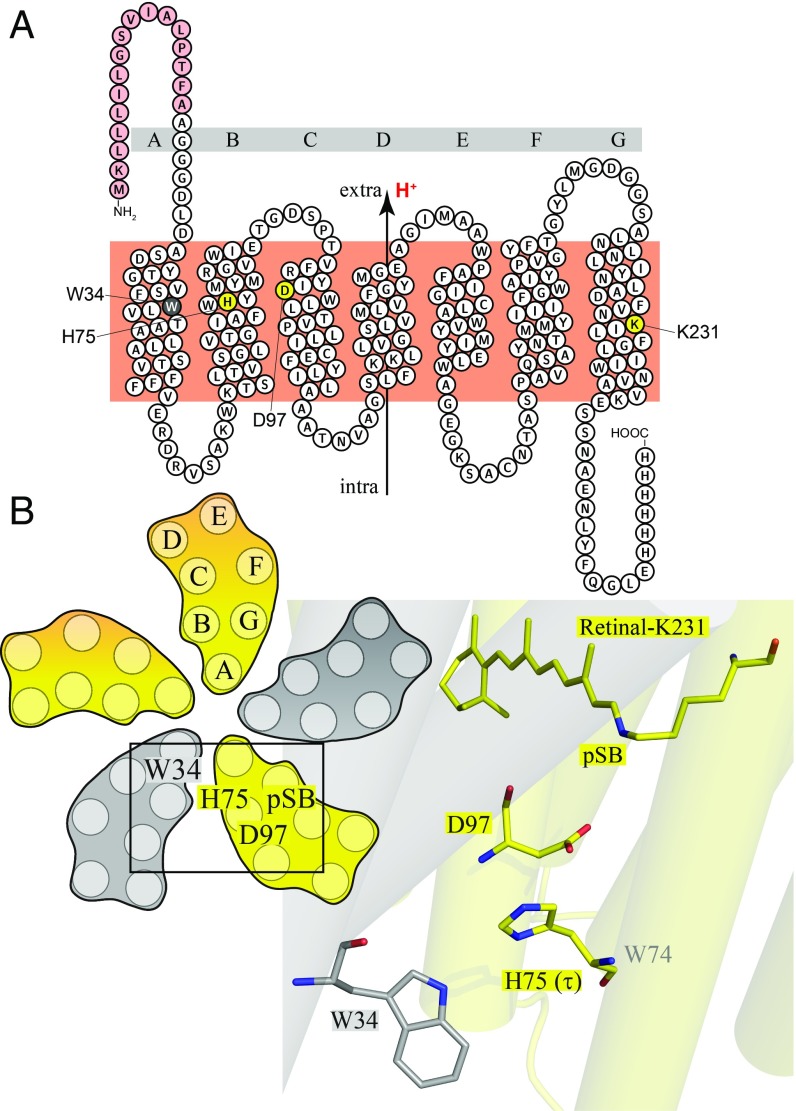
Fig. 1.
Overview of the GPR oligomeric structure and residues present at the cross-protomer interface. (A) Topological plot of GPR. The seven transmembrane helices (A-G), the signal sequence (red), and the positions of W34 (gray), H75, D97 (proton acceptor), and K231 (Schiff base; yellow) are labeled. Protons are transported from the intracellular to the extracellular side. (B) Overview of the residue positions in the pentamer and detailed structural arrangements of the interprotomer cluster consisting of W34 in one protomer (gray) and H75, D97, and pSB in the neighboring protomer (yellow). Data from PDB ID: 4JQ6 (7).
The oligomerization of PR as well as its 3D structure has been extensively studied. Atomic force microscopy (6) and EPR distance measurements (10) first revealed the higher oligomeric state and orientation of protomers toward each other. Native mass spectrometry demonstrated pentamer formation in detergent micelles (11). X-ray crystallography of two BPR variants provided well-resolved pentamer and hexamer structures, enabling a precise mapping of the retinal-binding pocket and also the oligomeric interface (7). A cross-protomer hydrogen bond network was discovered, involving residues W34, H75, and D97. This network was proposed to be involved in the photocycle and proton transport mechanism. As BPR and GPR share roughly 80% sequence identity, it can be supposed that their protein structures have high similarity. W34 and H75 are in close proximity across the protomer interface. H75, on the other hand, also forms an intraprotomer contact to the primary proton acceptor D97 (12) (Fig. 1). Solid-state NMR spectroscopy has been extensively used for the analysis of structural and mechanistic details of GPR complexes in the membrane (5, 8, 12–16). Solution-state NMR yielded a low-resolution structure model of a GPR monomer (17).
The GPR photocycle can be divided into a series of photointermediates, during which a proton is transported across the bacterial plasma membrane (SI Appendix, Fig. S1A). The first step of the photocycle is the formation of the K state where the retinal isomerizes from all-trans to 13-cis after photon absorption. Next, a proton is transferred from the protonated Schiff base (pSB) to D97, forming the M state photointermediate, which shows a characteristic retinal absorption blue shift to ∼400 nm. In the following steps of the N and O states, the SB is reprotonated by the proton donor E108, and D97 releases the proton to the extracellular side. The retinal returns to the ground state by isomerization back to the all-trans conformation (18).
H75 is a highly conserved and functionally important residue throughout the proteorhodopsin family, and it was subjected to detailed studies in GPR (12, 19–21). By forming a pH-dependent H bond with the primary proton acceptor (D97), H75 contributes to the unusually high pKa value of D97. Surprisingly, H75 seems to delay the photocycle (12). The direct interaction of H75 with the primary proton acceptor raises the question whether it plays a direct role during the photocycle since D97 becomes protonated in the M state (SI Appendix, Fig. S1A). Being located at the protomer interface, H75 could also be responsible for intersubunit cross talk during the photocycle through direct contact with W34, which was proposed in the BPR crystal structure (7).
Solid-state NMR, especially in combination with dynamic nuclear polarization (DNP), provides an ideal approach to address these questions as light-induced and cryotrapped photointermediate states could be probed directly within the lipid bilayer (13, 22, 23). DNP enables new perspectives for such studies since sensitivity can be boosted by orders of magnitude by magnetization transfer from suitable polarizing agents containing stable radicals to the nuclei of interest (24). Such an approach is of emerging importance for the detection of subpopulations of cryotrapped photointermediate states as demonstrated for bacteriorhodopsin and channelrhodopsin-2 (22, 25).
For our study, GPR was reconstituted into a synthetic lipid environment in which it retains its native higher oligomeric composition. Further, proteoliposomes were doped with the radical-based polarizing agent AMUPol (26). By using cryogenic temperatures, GPR was trapped in different photointermediate states. K state trapping was achieved by illuminating GPRWT directly in the DNP-NMR probe at 100 K. For M state trapping, the GPRE108Q mutant was used, which shows an elongated lifetime of the M state (13). It was illuminated at room temperature, subsequently flash frozen in liquid nitrogen, and quickly transferred into the DNP magic-angle spinning (MAS) probe (13, 22) (SI Appendix, Fig. S1B). Due to the DNP sensitivity enhancement, different H75 tautomers and rotamers can be observed. We are able to show that H75 undergoes drastic conformational changes during the photocycle. H75 forms a cluster with W34 across the protomer interface and therefore might be responsible for intersubunit cross talk. By specific mutations of W34, we reveal that homo-oligomerization of GPR not only contributes to its structural stability but also plays a functional role, as mutations in the protomer interface drastically alter its activity.
Results
H75 Changes Its Conformation During the Photocycle.
Uniformly 13C-15N-His-labeled green proteorhodopsin [(13C6-15N3-His)-GPR] samples (SI Appendix, Fig. S1C) were prepared to follow changes in the H75 conformation and its interaction pattern during the photocycle. The GPR construct used here included a tobacco etch virus (TEV) protease cleavage site located before the purification 6xHis tag. TEV protease cleavage ensured elimination of the 6xHis tag, leaving H75 as the sole histidine residue in the protein. Therefore, all detected NMR signals of (13C6-15N3-His)-GPR can be attributed to H75. Protein expression, purification, TEV protease cleavage of the 6xHis tag, and reconstitution were successfully performed and confirmed by SDS/PAGE, Western blot, blue native PAGE, and size exclusion chromatography (SI Appendix, Fig. S2).
Histidine residues often play an important role in enzymatic mechanisms (27–30) since the imidazole side chain can adopt three different states: At a pH below its pKa value of ∼6.0, both Nε2 and Nδ1 nitrogens are protonated, resulting in the positively charged (+) state. Above the pKa, a tautomeric equilibrium between the protonated Nε2 (τ)-state and protonated Nδ1 (π)-state is present, which is usually shifted toward the (τ)-state in a 4:1 ratio (in aqueous solutions) if no additional interactions stabilize the (π)-state (31).
Here, we have monitored H75 in different GPR photointermediates by DNP-enhanced MAS-NMR. Signal enhancement was achieved by doping the reconstituted GPR sample with the biradical polarizing agent AMUPol (26), leading to a 40- to 60-fold signal increase, as previously observed (5, 8, 13) (SI Appendix, Fig. S1B). Under cryogenic DNP conditions different photocycle intermediates can be cryotrapped by following specific sample preparations and illumination protocols, as previously described by us for GPR and channelrhodopsin-2 (13, 22). Low sample amounts (∼2 mg protein) in the NMR rotor and a 1:2 (wt/wt) lipid to protein ratio provide ideal illumination conditions but also make the use of DNP enhancement mandatory.
(13C6-15N3-His)-GPRWT (wild-type) signals of H75 were assigned from 13C double-quantum–single-quantum (DQSQ) (SI Appendix, Fig. S3A) (32) and 15N-13C transferred-echo double resonance (TEDOR) correlation spectra (SI Appendix, Fig. S3C) (33). It has been shown that the 13C chemical shifts of Cγ and Cδ2 are sensitive indicators of the tautomeric state of histidine (34–36). Therefore, we can conclude that H75 is present primarily as a (τ)-tautomer in the ground state. A small (π)-state subpopulation is detected as well (see also below).
For K state trapping, (13C6-15N3-His)-GPRWT was illuminated under DNP conditions at 100 K, directly inside the MAS probe for ∼40 min. In previous studies (13) it was shown that ∼30% K state could be achieved by applying this thermal trapping procedure, resulting in significant chemical shift perturbations in the retinal chromophore. Here, the comparison between dark and illuminated 13C DQSQ spectra shows no differences between ground state and K state H75 signals, indicating that the first step of retinal 13-cis isomerization does not influence H75 (SI Appendix, Fig. S4).
The next step in the GPR photocycle is the M state, which can be trapped by illumination at room temperature directly inside the MAS rotor and subsequent fast freezing in liquid nitrogen (13). These frozen samples can then be transferred into the precooled DNP probe. Unfortunately, the M state in GPRWT only shows a low population, but we were able to show a drastic enhancement by introducing the primary proton donor mutation E108Q, as shown before (13). This mutation elongates the M state by preventing reprotonation of the Schiff base. It was shown that it does not induce structural changes or chemical shift changes within the retinal chromophore. The mutant displays the typical predominant pentameric and hexameric oligomerization profile after reconstitution (SI Appendix, Fig. S5A). One-dimensional double-quantum-filtered (DQF) and 2D TEDOR DNP spectra show identical H75 signals in (13C6-15N3-His)-GPRE108Q and (13C6-15N3-His)-GPRWT, respectively (SI Appendix, Fig. S5 B and C). Therefore, it can be confirmed that GPRE108Q behaves similarly to GPRWT. The Schiff base becomes deprotonated in the M state. To prove successful trapping, additional lysine 15Nε labeling in addition to 13C6-15N3-His was employed, so that the occurrence of the deprotonated SB signal and a decrease in the pSB resonance could be observed in (13C6-15N3-His, 15Nε-Lys)-GPRE108Q. Successful M state cryotrapping is demonstrated in SI Appendix, Fig. S6. Since a complete conversion to the M state cannot be achieved by thermal trapping, ground state species are still present after illumination. They can be quantified by the intensity of the pSB resonance in the M state spectrum. Here, a conversion of ∼50% is achieved.
Based on these conditions, we were able to probe whether and how H75 is affected in the M state. A comparison of 15N and 13C spectra is shown in Fig. 2. The dark-state 15N spectrum of (13C6-15N3-His, 15Nε-Lys)-GPRE108Q (Fig. 2A, black) shows the same spectral features as the wild type as described above with a major (τ) and a minor (π) population. The two main resonances are Nε2(τ) at 163.5 ppm and deprotonated Nδ1(τ) at 250.4 ppm. The smaller (π)-state signals Nδ1(π) and Nε2(π) are found at 169.1 and 255.2 ppm, respectively.
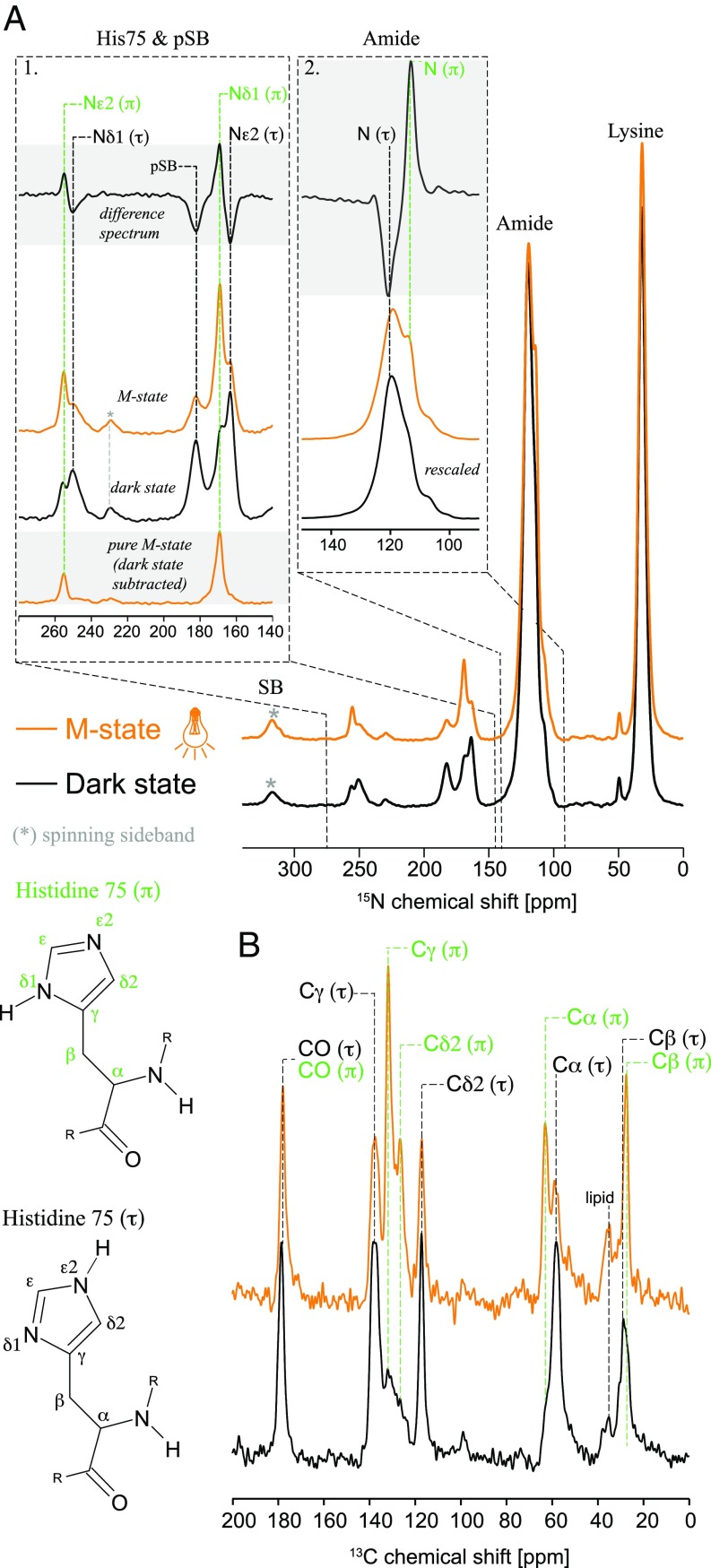
Fig. 2.
Effect of M state trapping on H75 15N and 13C signals. (A) 15N spectra of (13C6-15N3-His, 15Nε-Lys)-GPRE108Q in the dark state (black) and M state (orange). (τ)- and (π)-state resonances are labeled in black and green, respectively. To visualize spectral changes, difference spectra (M state minus dark state) are plotted for regions 1 and 2. The observed intensity changes for Nδ1 and Nε2 show that H75 changes from (τ) as the main population in the dark state to (π) in the M state. The same is reflected in the amide N difference spectrum. *, a spinning sideband from amide N. The pure M state spectrum for region 1 was calculated by subtracting the residual dark-state contribution as quantified via the pSB signal intensity (for details see SI Appendix, Fig. S6C). The spectrum reveals almost 100% (π)-state and no contribution of (τ)-state signals in the M state. (B) 13C 1D DQF spectra of H75 in the dark state (black) and M state (orange). (τ)- and (π)-state resonances are labeled in black and green, respectively. An increase in Cγ(π), Cδ2(π), and Cα(π) populations is observed after M state trapping.
In contrast to the K state, where no changes in H75 could be monitored, the cryotrapped M state shows a drastic effect on the histidine residue (Fig. 2A, orange). After illumination, the (τ)-state is significantly depopulated, and the (π)-state increases. The protonated Nδ1 and deprotonated Nε2 (π)-state signals are now the dominant resonances. Changes are visualized in the difference spectra in Fig. 2 A, 1. An additional shoulder appears also for the backbone signal (Fig. 2 A, 2) that can be attributed to H75 N in the (π)-state.
To estimate how the (τ):(π) ratio changes between dark and M states, peak intensities were analyzed. Therefore, a spectral deconvolution of the overlapping Nδ1(τ) and Nε2(π) resonances in the dark-state spectrum was carried out (SI Appendix, Fig. S6C). The ratio of the integral peak intensities Nδ1(τ):Nε2(π) is approximately 4:1, which is similar to the tautomeric ratio observed for aqueous histidine. The M state spectrum could be simplified by subtracting the residual dark-state contribution quantified by the intensity of the pSB resonance (Fig. 2 A, 1 and SI Appendix, Fig. S6C). The difference spectrum shows that H75 is completely converted into the (π)-state since almost no presence of (τ)-state signals could be monitored.
These observations are also supported by 2D DQSQ spectra, which allow an unambiguous assignment of the 13C H75 chemical shifts (SI Appendix, Fig. S7A), and by 13C-15N TEDOR experiments (SI Appendix, Fig. S7B). The assignment results are plotted onto 13C 1D DQF spectra in Fig. 2B: In the dark state, Cγ(τ) at 137.7 ppm and Cδ2(τ) at 117.2 ppm are the dominant signals. After illumination, these two signals get reduced, and two new dominant resonances occur, corresponding to Cγ(π) at 131.8 ppm and Cδ2(π) at 126.6 ppm (34). The increase in the (π)-state population is also seen by an additional Cα peak at 63 ppm. As observed and discussed for the 15N spectra, the small H75 (τ)-state signals still present after illumination arise from a remaining dark-state population. In summary, our data show unambiguously that H75 swaps its tautomeric state from the (τ)-state to the (π)-state when GPR switches from the ground to the M state. Chemical shift changes from the (τ)-state to the (π)-state are listed in SI Appendix, Table S1.
To exclude the possibility that H75 gets protonated during the M state, we compared the M state 13C DQSQ and 13C-15N TEDOR spectra with data obtained from (13C6-15N3-His)-GPRWT at pH 5 in which H75 is positively charged. As demonstrated in SI Appendix, Fig. S8, the spectral pattern of the M state differs significantly from H75(+), which supports the conclusion that it is indeed the (π)-state that is mainly populated.
To verify the reversibility of the observed (τ) to (π) conversion during the photocycle, a thermal relaxation experiment was performed, which demonstrates that H75 returns to its initial (τ)-state when the M state is no longer trapped (SI Appendix, Fig. S9). These findings support that the tautomerization of H75 is directly linked to the M state.
Besides the (τ) to (π) conversion, the orientation of the imidazole ring could also change in the M state. Therefore, 15N-15N proton-driven spin diffusion (PDSD) spectra were recorded to monitor intraresidue nitrogen–nitrogen through-space contacts within H75 (Fig. 3A, Left). In the dark-state spectrum, two cross peaks for correlations between Nδ1(τ)-Nε2(τ) and Nδ1(π)-Nε2(π) are observed. In line with the findings described above, the (τ)-state is significantly larger than the (π)-state cross peak. No correlations between imidazole ring 15N nuclei and the backbone N could be detected. In the M state (Fig. 3A, Right), intensities for the Nδ1(τ)-Nε2(τ) and Nδ1(π)-Nε2(π) cross peaks swap as expected since the (π)-state is the dominating population. Furthermore, an additional correlation between Nδ1(π) and N(π) occurs. This means that both nuclei are now closer to each other compared with the ground state, which suggests that H75 adopts a different rotamer configuration, presumably through rotations around the Cα–Cβ and Cβ–Cγ bonds (Fig. 3B). However, a more precise determination of the sidechain orientation via angles χ1 and χ2 would require exact distance measurements (34).
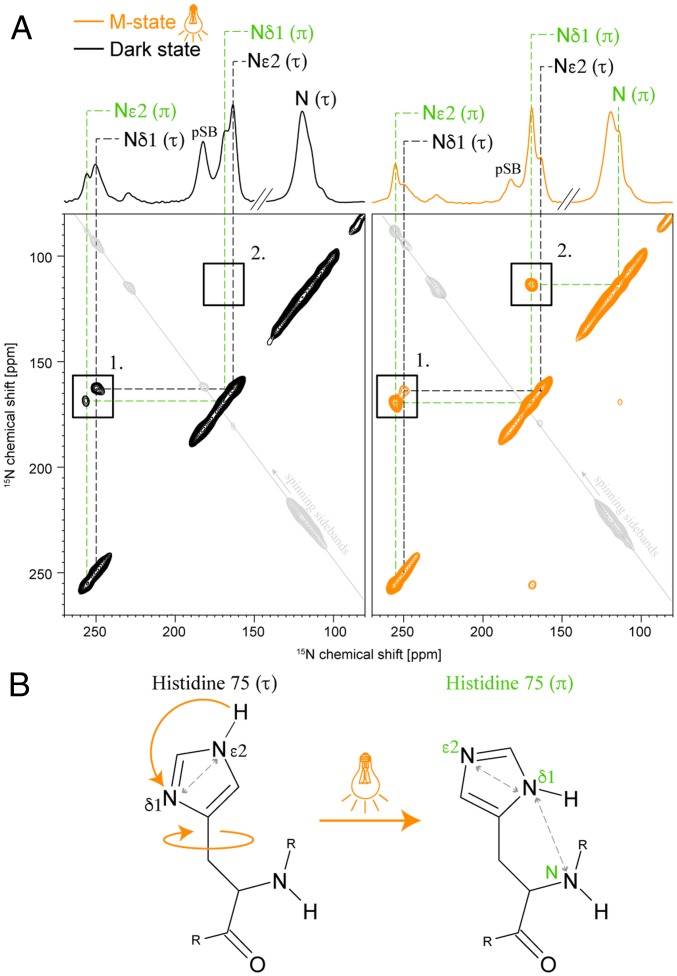
Fig. 3.
Conformational changes of H75 in the M state. (A) 15N-15N PDSD spectra of (13C6-15N3-His, 15Nε-Lys)-GPRE108Q in the dark state (Left, black) and illuminated M state (Right, orange). (τ)-state signals are labeled in black, and (π)-state signals are labeled in green. (1) Short-range contact between Nε2 and Nδ1 within the imidazole ring. The (τ) and (π) peaks swap intensities after illumination, confirming the change to a dominant (π) population in the M state. (2) The occurrence of a Nδ1(π)-N(π) cross peak in the M state indicates the close proximity between these nuclei. (B) 15N-15N PDSD spectra suggest that H75 in the M state changes not only its tautomerization from (τ) to (π) but also the orientation of its side chain. The observed close proximity between Nδ1(π) and N(π) could be explained by a rotation around the Cβ–Cγ bond. Spectra were recorded with a mixing time of 1 s. The asymmetry in cross-peak intensities is caused by longitudinal relaxation differences between the coupled spins, which is slow compared with the recycle delay times used here. Signals in gray are caused by back-folded spinning sidebands due to the reduced spectral width used for spectra acquisition.
Visualizing the W34-H75 Cross-Protomer Contact.
H75 changes its tautomerization state and rotational conformation in the M state, where the proton acceptor D97 receives a proton from the pSB. Previous studies suggested a functional contact between H75 and D97 in GPR and BPR (7, 12, 19). The BPR crystal structure also showed a contact to W34 across the protomer interface, which was suggested to be responsible for intersubunit cross talk during the photocycle (7). In GPR, cross-protomer interactions based on strong salt bridges were discovered by DNP-enhanced solid-state NMR (8). Here, we use this approach to probe whether in GPR a cross-protomer contact between H75 and W34 in the ground and M states occurs.
Therefore, GPR was labeled with 13Cδ1-Trp and 15N3-His (Fig. 4A), so that long-range 13C-15N distances between W34 and H75 could be detected by TEDOR experiments. While H75 is the only histidine residue in a GPR protomer, tryptophan is present 10 times in the primary sequence, which could obscure data interpretation if a tryptophan within the same protomer is adjacent to H75.
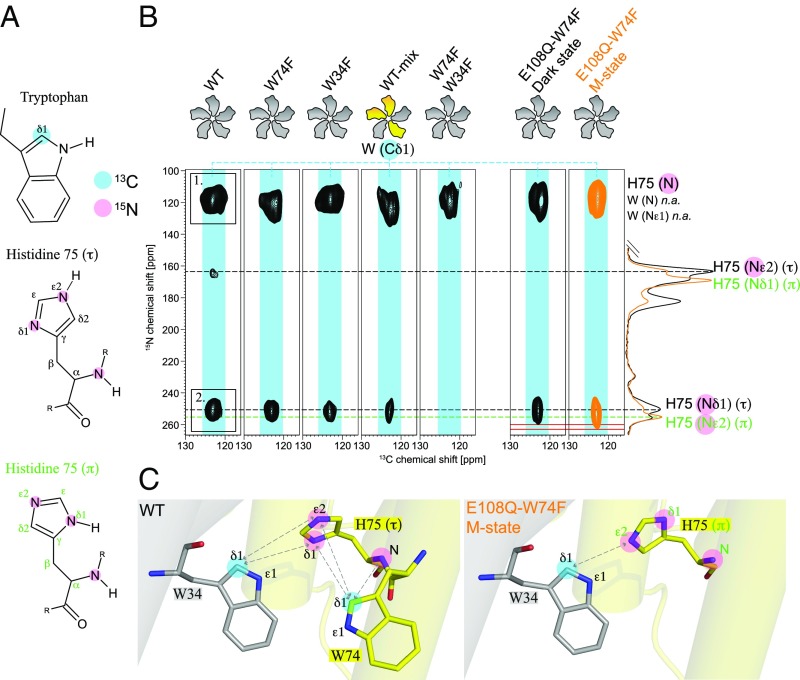
Fig. 4.
Visualizing the cross-protomer H75-W34 contact. (A) For the detection of the cross-protomer H75-W34 contact, (13Cδ1-Trp, 15N3-His)-GPRWT (wild type) and various mutants have been prepared. (B) Two-dimensional TEDOR spectra of GPRWT, GPRW74F, GPRW34F, GPRWT-mix, GPRW74F-W34F, and GPRE108Q-W74F, all with (13Cδ1-Trp, 15N3-His) labeling. The 1D spectra on the Right are from (13C6-15N3-His, 15Nε-Lys)-GPRE108Q (Fig. 3A) in the dark state (black) and M state (orange) and have been plotted to identify the H75(τ) and (π) resonances. Cross peak 1 in GPRWT (Left) is observed in all spectra and corresponds to a mixture of correlations between W34/W74-Cδ1 and H75-N as well as natural abundance correlations between Trp-Cδ1 and Trp-N or Trp-Nε1. Cross peak 2 can be attributed to a correlation between W74 or W34-Cδ1 and H75-Nδ1(τ). Spectra of GPRW74F and GPRW34F confirm that both tryptophan residues contribute to the interaction. The mixed-labeled sample GPRWT-mix consisting of (13Cδ1-Trp)-GPRWT and (15N3-His)-GPRWT proves that a W34-H75 interprotomer contact exists. M state trapping of (13Cδ1-Trp, 15N3-His)-GPRE108Q-W74F shows a stretching of signal 2 toward the Nε2(π) resonance. (C) The GPRWT sample shows close proximity of H75-Nδ1(τ) to W34-Cδ1 and an intraprotomer contact to W74-Cδ1. The GPRE108Q-W74F sample indicates a turn of H75 so that Nε1(π) and W34-Cδ1 occur in close proximity. Data from PDB ID: 4JQ6 (7).
The residues H75 and W34 could only come in close proximity through oligomeric contacts since they are 18 Å apart within one protomer, which is beyond the detection radius of a 13C-15N TEDOR experiment of up to 6 Å. Eight of the remaining nine tryptophan Cδ1 nuclei are at least 9 Å away from the H75 imidazole ring (SI Appendix, Fig. S10). However, W74, which is located next to H75, could potentially give rise to intraprotomer cross peaks.
A 13C-15N TEDOR spectrum of (13Cδ1-Trp, 15N3-His)-GPRWT (wild type) is shown in Fig. 4B, Left. Two dominant cross peaks are observed. Peak 1 corresponds to a correlation between H75-N and Cδ1 of an adjacent Trp (W34 or W74), but it also contains contributions from intraresidue contacts between Trp-Cδ1 and Trp-N or Trp-Nε1, which occur at natural abundance (0.4%). Cross peak 2 represents a correlation between W34/W74-Cδ1 and H75-Nδ1(τ). The remaining smaller cross peak arises from W34/W74-Cδ1 and H75-Nε2(τ) correlation.
To distinguish between interprotomer W34-H75 and intraprotomer W74-H75 contacts, the effects of W34F and W74F mutations on the TEDOR spectra were analyzed. None of these mutations affected the oligomeric state of GPR (SI Appendix, Fig. S11). Since peak 2 can be detected in the TEDOR spectra of both GPRW34F and GPRW74F (Fig. 4B), it can be concluded that both tryptophan contacts contribute to the signal seen in GPRWT.
For further validation, a mixed sample (GPRWT-mix) was produced. In this sample, the GPR pentamer consists of a mixture of (13Cδ1-Trp)-GPRWT and (15N3-His)-GPRWT protomers, which can be achieved by disassembling and reassembling GPR complexes as demonstrated by us previously (8). In such a mixed-labeled pentamer, all intraprotomer 13C-15N correlations are suppressed. The signal observed at the position of cross peak 2 therefore belongs unambiguously to the interprotomer correlation between W34-Cδ1 and H75-Nδ1(τ) (Fig. 4B). A drawback of the mixing procedure is the drastic reduction of the number of 13C-15N contacts. Hence, the spectrum had to be acquired with five times more scans to gain a similar signal-to-noise ratio compared with the other samples. Cross peak 2 vanishes in the double mutant GPRW74F-W34F, demonstrating that no other tryptophan residues (e.g., W98, SI Appendix, Fig. S10) are involved in the H75 correlations.
Our experiments demonstrate a cross-protomer contact between W34 and H75. Both residues must be oriented to each other in a way that W34-Cδ1 and H75-Nδ1(τ) are in close proximity. Such an orientation also allows for the intraprotomer contact between W74-Cδ1 and H75-Nδ1(τ), as seen in the BPR crystal structure (Fig. 4C, WT).
Next, we analyzed whether this interprotomer interactions is affected in the M state. Therefore, a double mutant GPRE108Q-W74F was prepared, which is M state trappable (E108Q) and does not contain the intraprotomer H75-W74 contact (W74F). TEDOR spectra of (13Cδ1-Trp, 15N3-His)-GPRE108Q-W74F in the dark and M states are shown in Fig. 4B, Right. The dark-state spectrum is comparable to GPRW74F; the interprotomer W34-H75 correlation can be detected. After successful M state trapping, the W34-Cδ1-H75-Nδ1(τ) cross peak becomes broader and is stretched by ∼3 ppm toward H75-Nε2(π). This additional peak intensity indicates a correlation between W34-Cδ1 and H75-Nε3 (π) and close proximity between both nuclei. No correlation is observed for H75-Nδ1 (π) (along 169.1 ppm). Such an observation supports a rotation and a reorientation of H75 toward W34 in the M state (Fig. 4C, E108Q-W74F M state), as shown in Fig. 3A.
W34 Mutations Alter the Functionality of GPR.
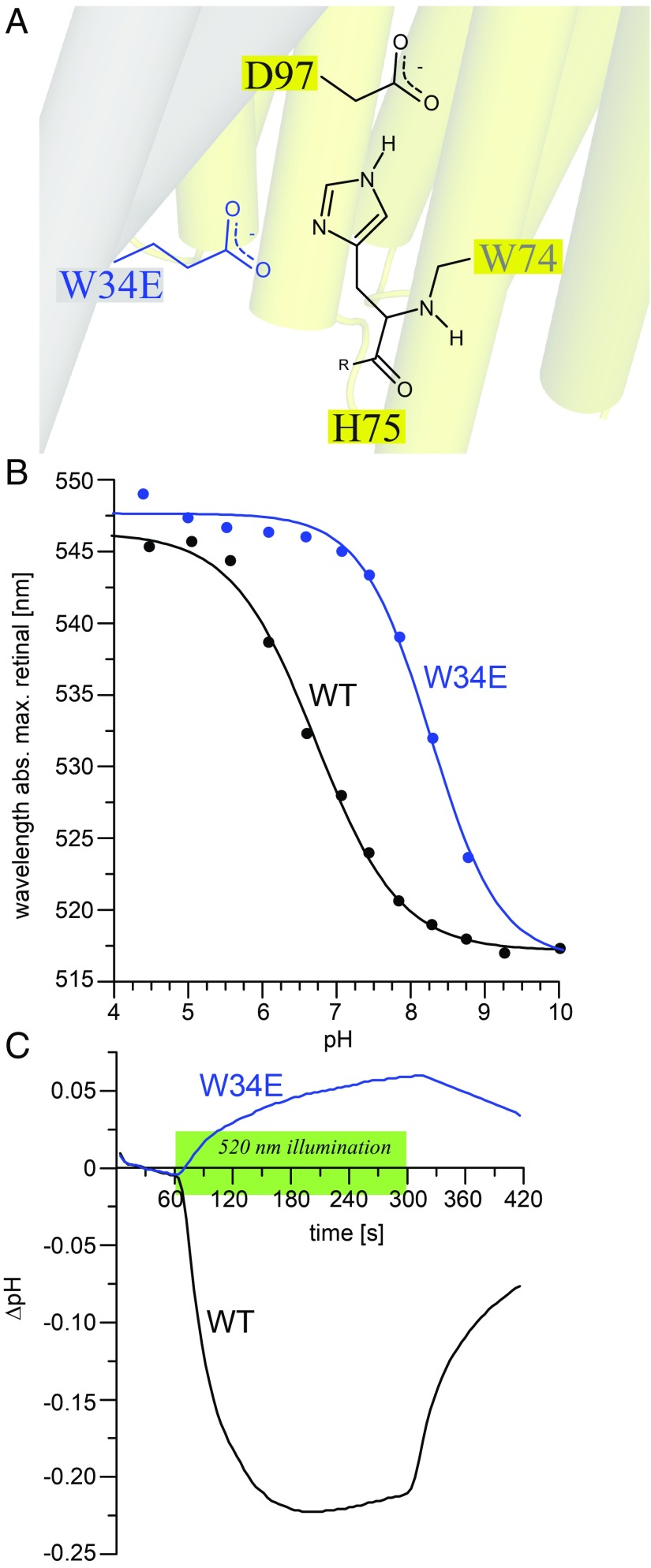
Our DNP-NMR results show that W34 and H75 have close contact across the protomer interface in GPR, which raises the question of its functional role. Therefore, the effects of replacing W34 with positively and negatively charged residues on the pKa of the primary proton acceptor D97 and on proton transport were tested. It was found that positively charged mutations lower the pKa value (GPRWT pKa is ∼7), while negative charges cause an increase (SI Appendix, Fig. S12A). It can therefore be concluded that the primary proton acceptor is affected by mutations at W34 across the protomer interface. The largest effect was caused by W34E, in which the D97 pKa was shifted to 8.1 (Fig. 5 A and B). Therefore, D97 is more likely to be found protonated in this mutant compared to the wild type under our experimental conditions.
Fig. 5.
Effects of the W34E mutation on GPR properties. (A) W34E forming a close contact with H75 across the protomer interface. (B) pH titrations of GPRWT and GPRW34E. The mutation causes a pKa shift of the primary proton acceptor D97 from ∼6.9 (GPRWT, black) to ∼8.3 (GPRW34E, blue). (C) Proton transport measurements of GPRWT (black) and GPRW34E (blue). WT shows a decrease of pH during illumination, corresponding to an outward proton transport. W34E shows a reverse effect, corresponding to an inward-driven proton pumping.
Light-induced proton transport was probed in live E. coli cells containing heterologous expressed W34E mutants. The cell suspension was subjected to 520-nm illumination for 2 min, during which the pH was constantly monitored (37–39). Cells expressing GPRWT show a pH drop during the illumination period, which can be explained by light-induced, outward-driven proton transport across the bacterial membrane (Fig. 5C). Positively charged GPRW34K and GPRW34R mutants do not drastically affect proton transport (SI Appendix, Fig. S12B). In contrast, the negatively charged GPRW34E mutant causes a pH increase upon illumination (Fig. 5C) indicative of reverse, inward-driven proton transport. The same observation is made for GPRW34D (SI Appendix, Fig. S12B). Similar results were seen before for BPR (7). A double mutation of W34E and H75N abolishes the negative mutation effect and again shows a normal outward proton transport (SI Appendix, Fig. S12B). It can be concluded that these functional modifications are mediated by H75 located in between W34E across the protomer interface and D97, responsible for proton transport.
To further evaluate how H75 is affected by the W34E mutation, 2D DQSQ spectra of (13C6-15N3-His)-GPRW34E were recorded and compared with the wild type (SI Appendix, Fig. S13). Mutation-induced chemical shift changes are observed for Cγ and Cδ2. These changes could reflect alterations of the H75 structure and orientation in GPRW34E, which might influence the protonation state of neighboring D97, causing its drastic pKa shift and a reverse proton transport across the bacterial membrane.
Applying the M state trapping protocol described above to the double mutant (13C6-15N3-His, 15Nε-Lys)-GPRE108Q-W34E showed no effect on the H75 tautomerization or the Schiff base resonances (SI Appendix, Fig. S14), which means that the M state did not form. The W34E-induced increase of the D97 pKa indicates a favored protonated state of the primary proton acceptor, which cannot accept another proton from the pSB during the photocycle. This might explain the reversed proton transfer by GPRW34E, which was also observed before in GPRWT at acidic conditions (<pH 5) (18). Under these circumstances the pSB could transfer its proton to the proton donor E108 and be reprotonated by the protonated D97. In the double mutant GPRE108Q-W34E, the proton donor E108 is replaced, and the proton acceptor D97 is strongly affected. The proton of the pSB is trapped as it cannot be transferred to either one of the residues, explaining why no M state formation can be detected for the double mutant after illumination.
Discussion
Our studies present an Asp–His–Trp triad in a retinal protein and cross-protomer interactions involving noncanonical residues that are part of the photocycle. The trapped M state of the photocycle results in changes in the tautomerization state and rotational conformation of H75.
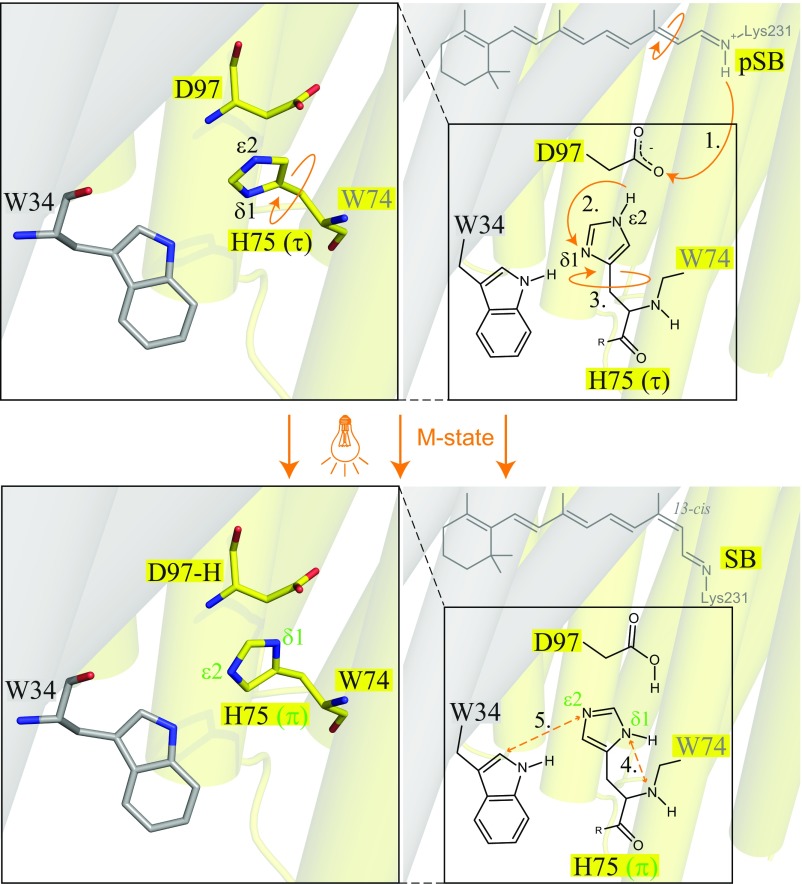
Fig. 6 summarizes all information we gain from dark- and M state histidine spectra. During illumination and trapping of GPRE108Q in the M state, a proton is transferred from the protonated Schiff base to the primary proton acceptor D97 (Fig. 6, step 1). The protonated D97 influences the neighboring H75 to change its tautomerization from the (τ)-state to the (π)-state (Fig. 6, step 2). The cross-protomer contact with W34 additionally forces (π)-state H75 to perform a rotation, presumably around the Cβ–Cγ bond (Fig. 6, step 3), leading to the close proximity of H75-Nδ1(π) and H75-N(π) and W34-Cδ1 and H75-Nε2(π) in the M state (Fig. 6, steps 4 and 5). In this way, the unfavorable arrangement of two protons facing each other between W34 and H75 is avoided. The exact interaction and orientation of H75 with D97 remain undetermined.
Fig. 6.
Summary of structural changes observed for H75 during the transition from the dark state to the trapped M state. After M state trapping of GPRE108Q, a proton is transferred from the protonated Schiff base to the primary proton acceptor D97 (step 1). The protonated D97 influences the neighboring H75 to change its tautomeric form from the (τ)-state to the (π)-state (step 2). Additionally, the cross-protomer contact with W34 forces (π)-state H75 to perform a rotation, presumably around the Cβ–Cγ bond (step 3). This leads to close proximity of Nδ1(π) and N(π) and W34 Cδ1 and Nε2(π) in the illuminated M state (steps 4 and 5). Data from PDB ID: 4JQ6 (7).
Previous studies found that mutations such as H75N lead to a significantly faster photocycle (12), for which the reasons could only be speculated. Results in this study reveal tautomerization and conformational changes of H75 during the M state. Presumably, the conformational changes in the H75 residue need to occur before the photocycle continues. These time-consuming conformational steps would then lead to a slowed-down photocycle. The role that is attributed to the conserved residue H75 is extended in this study. It is not only responsible for the high pKa value through an interaction with D97 (12) but is a dynamic part of the photocycle.
Furthermore, H75 is shown to perform a specific task during the photocycle, as it is able to sense protonation of the primary proton acceptor D97 and switch its conformational state accordingly. On the other hand, the protonation of D97 and, with that, the functionality of GPR seem to be linked to the conformational state of H75. The pKa value of histidine lies in the range of pH 6. At higher pH, H75 is deprotonated and can undergo the conformational change from the (τ)-state to the (π)-state during the photocycle. This would not be possible at lower pH (below pH ∼6) where histidine is present in the protonated positively charged state. H75 is located toward the extracellular side of the protein. In this location, the highly conserved H75 could take a specific physiological role and work as an additional pH sensor and functional regulator. A pH drop on the extracellular side of the bacterial membrane could lead to a protonation of H75. This in turn would hinder the conformational changes during the GPR photocycle and attenuate proton transport across the membrane if a strong proton gradient is already present.
The location of H75 between the primary proton acceptor and the protomer interface makes it potentially important for the formation of specific cross-protomer interactions. This study reveals a close contact between H75 and W34, verifying that it is present not only in BPR but also in GPR. Negatively charged mutations of W34 cause a reverse proton transport that is mediated by sensitive conformational changes in H75. The functional importance of this cross-protomer contact might work in both directions. On the one hand, W34 could sense the conformation of H75, which could lead to a cooperative functionality between the protomers. Although an individual GPR protomer shows photocycle activity (9, 40), the question remains whether the GPR oligomers can function more efficiently in a cooperative manner. This intersubunit cross talk could be mediated by the H75-W34 contact. On the other hand, W34 might be responsible for bringing H75 into the right position to be able to sense the protonation of D97 during the photocycle. This becomes evident in SI Appendix, Fig. S15. The mutation GPRE50A causes a destabilization and disruption of higher oligomeric complexes by showing a larger number of monomers after reconstitution (SI Appendix, Fig. S15A). This disruption leads to broader H75 signals in 13C 1D DQF spectra of (13C6-15N3-His)-GPRE108Q-E50A compared with GPRWT and also causes a much smaller tautomerization efficiency of H75 after illumination (SI Appendix, Fig. S15B). Broader H75 signals indicate a higher flexibility and more possible orientations of the residue. The lack of oligomeric stability through disruption possibly causes H75 to move away from D97 and adopt different orientations. This could mean that GPR oligomerization and, specifically, the bulky residue W34 force H75 into the correct position to be able to sense D97 protonation and potentially influence protein functionality as a pH sensor.
The observation that the negatively charged W34E mutation across the protomer interface causes an inversion of proton transport in bacterial cells further illustrates the functional importance of the cross-protomer Asp–His–Trp triad. DNP-NMR experiments revealed mutation-induced chemical shift changes in H75 (SI Appendix, Fig. S13), which could be linked to its functional effect: The negative charge introduced by W34E alters the environment of H75. It presumably shifts H75 toward a protonated state (indicated by chemical shift changes for Cγ and Cδ2) to compensate the negative charge of W34E. Its intramolecular interaction with the primary proton acceptor D97 increases its pKa, keeping it protonated also at higher pH, for example, by sharing or exchanging a proton with the protonated H75. When D97 is protonated at neutral or slightly basic pH, it cannot accept a proton from the pSB during the M state, which induces a reverse proton flow in the protein, also observed in GPRWT under acidic conditions, where D97 is protonated (18, 41).
Conclusion
Here, experimental evidence for functionally relevant cross-protomer interactions within the microbial rhodopsin was presented. An Asp–His–Trp triad is formed across the oligomerization interface, participates in the photocycle, and potentially acts as a pH sensor. The use of DNP-enhanced solid-state NMR in combination with light-induced cryotrapping of photointermediates was essential to obtain such insight.
Material and Methods
Isotope-labeled proteorhodopsin (wild type and mutants) was expressed in Escherichia coli and, after purification and elimination of the 6xHis tag by TEV cleavage, reconstituted into liposomes. All MAS-NMR experiments were performed under cryogenic conditions (100 K) using cross-effect DNP enhancement provided by doping the proteoliposomes with AMUPol. Trapping of the different photointermediates was achieved using protocols combining illumination, freezing, and thermal relaxation. The SI Appendix has a detailed description of all applied methods.
Supplementary Material
Acknowledgments
Andreas Krommes is acknowledged for assistance with the proton transport experiments. This work was funded by Deutsche Forschungsgemeinschaft (DFG)/SFB807 “Transport and communications across membranes” and DFG Grant GL-307/12-1. DNP experiments were enabled through an equipment grant provided by DFG (Grant GL307/4-1) and Cluster of Excellence Macromolecular Complexes Frankfurt. J.M. gratefully acknowledges support by the International Max Planck Research School for Structure and Function of Biological Membranes, Frankfurt.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.G.G. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1817665116/-/DCSupplemental.
References
- 1.Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- 2.Gómez-Consarnau L, et al. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol. 2010;8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de la Torre JR, et al. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc Natl Acad Sci USA. 2003;100:12830–12835. doi: 10.1073/pnas.2133554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang WW, Sineshchekov OA, Spudich EN, Spudich JL. Spectroscopic and photochemical characterization of a deep ocean proteorhodopsin. J Biol Chem. 2003;278:33985–33991. doi: 10.1074/jbc.M305716200. [DOI] [PubMed] [Google Scholar]
- 5.Mao J, et al. Structural basis of the green-blue color switching in proteorhodopsin as determined by NMR spectroscopy. J Am Chem Soc. 2014;136:17578–17590. doi: 10.1021/ja5097946. [DOI] [PubMed] [Google Scholar]
- 6.Klyszejko AL, et al. Folding and assembly of proteorhodopsin. J Mol Biol. 2008;376:35–41. doi: 10.1016/j.jmb.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 7.Ran T, et al. Cross-protomer interaction with the photoactive site in oligomeric proteorhodopsin complexes. Acta Crystallogr D Biol Crystallogr. 2013;69:1965–1980. doi: 10.1107/S0907444913017575. [DOI] [PubMed] [Google Scholar]
- 8.Maciejko J, et al. Visualizing specific cross-protomer interactions in the homo-oligomeric membrane protein proteorhodopsin by dynamic-nuclear-polarization-enhanced solid-state NMR. J Am Chem Soc. 2015;137:9032–9043. doi: 10.1021/jacs.5b03606. [DOI] [PubMed] [Google Scholar]
- 9.Hussain S, Kinnebrew M, Schonenbach NS, Aye E, Han S. Functional consequences of the oligomeric assembly of proteorhodopsin. J Mol Biol. 2015;427:1278–1290. doi: 10.1016/j.jmb.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone KM, et al. Structural insight into proteorhodopsin oligomers. Biophys J. 2013;104:472–481. doi: 10.1016/j.bpj.2012.11.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann J, et al. Studying the stoichiometries of membrane proteins by mass spectrometry: Microbial rhodopsins and a potassium ion channel. Phys Chem Chem Phys. 2010;12:3480–3485. doi: 10.1039/b924630d. [DOI] [PubMed] [Google Scholar]
- 12.Hempelmann F, et al. His75-Asp97 cluster in green proteorhodopsin. J Am Chem Soc. 2011;133:4645–4654. doi: 10.1021/ja111116a. [DOI] [PubMed] [Google Scholar]
- 13.Mehler M, et al. Chromophore distortions in photointermediates of proteorhodopsin visualized by dynamic nuclear polarization-enhanced solid-state NMR. J Am Chem Soc. 2017;139:16143–16153. doi: 10.1021/jacs.7b05061. [DOI] [PubMed] [Google Scholar]
- 14.Shi L, et al. Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump–Structural insights. J Mol Biol. 2009;386:1078–1093. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Lake EMR, Ahmed MAM, Brown LS, Ladizhansky V. Solid-state NMR study of proteorhodopsin in the lipid environment: Secondary structure and dynamics. Biochim Biophys Acta. 2009;1788:2563–2574. doi: 10.1016/j.bbamem.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Aslimovska L, Glaubitz C. Molecular dynamics of proteorhodopsin in lipid bilayers by solid-state NMR. J Am Chem Soc. 2011;133:4874–4881. doi: 10.1021/ja109766n. [DOI] [PubMed] [Google Scholar]
- 17.Reckel S, et al. Solution NMR structure of proteorhodopsin. Angew Chem Int Ed Engl. 2011;50:11942–11946. doi: 10.1002/anie.201105648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedrich T, et al. Proteorhodopsin is a light-driven proton pump with variable vectoriality. J Mol Biol. 2002;321:821–838. doi: 10.1016/s0022-2836(02)00696-4. [DOI] [PubMed] [Google Scholar]
- 19.Bergo VB, et al. His-75 in proteorhodopsin, a novel component in light-driven proton translocation by primary pumps. J Biol Chem. 2009;284:2836–2843. doi: 10.1074/jbc.M803792200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfleger N, et al. Solid-state NMR and functional studies on proteorhodopsin. Biochim Biophys Acta. 2009;1787:697–705. doi: 10.1016/j.bbabio.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 21.Rangarajan R, Galan JF, Whited G, Birge RR. Mechanism of spectral tuning in green-absorbing proteorhodopsin. Biochemistry. 2007;46:12679–12686. doi: 10.1021/bi700955x. [DOI] [PubMed] [Google Scholar]
- 22.Becker-Baldus J, et al. Enlightening the photoactive site of channelrhodopsin-2 by DNP-enhanced solid-state NMR spectroscopy. Proc Natl Acad Sci USA. 2015;112:9896–9901. doi: 10.1073/pnas.1507713112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ni QZ, et al. Primary transfer step in the light-driven ion pump bacteriorhodopsin: An irreversible U-turn revealed by dynamic nuclear polarization-enhanced magic angle spinning NMR. J Am Chem Soc. 2018;140:4085–4091. doi: 10.1021/jacs.8b00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ni QZ, et al. High frequency dynamic nuclear polarization. Acc Chem Res. 2013;46:1933–1941. doi: 10.1021/ar300348n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bajaj VS, Mak-Jurkauskas ML, Belenky M, Herzfeld J, Griffin RG. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. Proc Natl Acad Sci USA. 2009;106:9244–9249. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauvée C, et al. Highly efficient, water-soluble polarizing agents for dynamic nuclear polarization at high frequency. Angew Chem Int Ed Engl. 2013;52:10858–10861. doi: 10.1002/anie.201304657. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya S, Sukits SF, MacLaughlin KL, Lecomte JT. The tautomeric state of histidines in myoglobin. Biophys J. 1997;73:3230–3240. doi: 10.1016/S0006-3495(97)78348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bachovchin WW. Contributions of NMR spectroscopy to the study of hydrogen bonds in serine protease active sites. Magn Reson Chem. 2001;39(Suppl 1):S199–S213. [Google Scholar]
- 29.Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen AL, Kay LE. Measurement of histidine pKa values and tautomer populations in invisible protein states. Proc Natl Acad Sci USA. 2014;111:E1705–E1712. doi: 10.1073/pnas.1400577111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blomberg F, Maurer W, Rüterjans H. Nuclear magnetic resonance investigation of 15N-labeled histidine in aqueous solution. J Am Chem Soc. 1977;99:8149–8159. doi: 10.1021/ja00467a005. [DOI] [PubMed] [Google Scholar]
- 32.Hong M. Solid-state dipolar INADEQUATE NMR spectroscopy with a large double-quantum spectral width. J Magn Reson. 1999;136:86–91. doi: 10.1006/jmre.1998.1631. [DOI] [PubMed] [Google Scholar]
- 33.Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. J Am Chem Soc. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Hong M. Protonation, tautomerization, and rotameric structure of histidine: A comprehensive study by magic-angle-spinning solid-state NMR. J Am Chem Soc. 2011;133:1534–1544. doi: 10.1021/ja108943n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goux WJ, Allerhand A. Studies of chemically modified histidine residues of proteins by carbon 13 nuclear magnetic resonance spectroscopy. Reaction of hen egg white lysozyme with iodoacetate. J Biol Chem. 1979;254:2210–2213. [PubMed] [Google Scholar]
- 36.Sudmeier JL, et al. Identification of histidine tautomers in proteins by 2D 1H/13C(delta2) one-bond correlated NMR. J Am Chem Soc. 2003;125:8430–8431. doi: 10.1021/ja034072c. [DOI] [PubMed] [Google Scholar]
- 37.Maresca JA, Keffer JL, Miller KJ. Biochemical analysis of microbial rhodopsins. Curr Protoc Microbiol. 2016;41:1F.4.1–1F.4.18. doi: 10.1002/cpmc.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganapathy S, et al. Modulation of spectral properties and pump activity of proteorhodopsins by retinal analogues. Biochem J. 2015;467:333–343. doi: 10.1042/BJ20141210. [DOI] [PubMed] [Google Scholar]
- 39.Ganapathy S, et al. Retinal-based proton pumping in the near infrared. J Am Chem Soc. 2017;139:2338–2344. doi: 10.1021/jacs.6b11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranaghan MJ, Schwall CT, Alder NN, Birge RR. Green proteorhodopsin reconstituted into nanoscale phospholipid bilayers (nanodiscs) as photoactive monomers. J Am Chem Soc. 2011;133:18318–18327. doi: 10.1021/ja2070957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Verhoefen MK, et al. Low temperature FTIR spectroscopy provides new insights in the pH-dependent proton pathway of proteorhodopsin. Biochim Biophys Acta. 2011;1807:1583–1590. doi: 10.1016/j.bbabio.2011.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.