Significance
Multiciliated cells (MCCs) have core roles in organ formation and function, where they control fluid flow and particle displacement. MCCs direct fluid movement in the brain and spinal cord, clearance of respiratory mucus, and ovum transport from the ovary to the uterus. Deficiencies in MCC functionality lead to hydrocephalus, chronic respiratory infections, and infertility. Prostaglandins are lipids that are used to coordinate cellular functions. Here, we discovered that prostaglandin signaling is required for MCC development in the embryonic zebrafish kidney. Understanding renal MCC genesis can lend insights into the puzzling origins of MCCs in several chronic kidney diseases, where it is unclear whether MCCs are a cause or phenotypic outcome of the condition.
Keywords: kidney, prostaglandin, zebrafish, pronephros, chemical screen
Abstract
Multiciliated cells (MCCs) are specialized epithelia with apical bundles of motile cilia that direct fluid flow. MCC dysfunction is associated with human diseases of the respiratory, reproductive, and central nervous systems. Further, the appearance of renal MCCs has been cataloged in several kidney conditions, where their function is unknown. Despite their pivotal health importance, many aspects of MCC development remain poorly understood. Here, we utilized a chemical screen to identify molecules that affect MCC ontogeny in the zebrafish embryo kidney, and found prostaglandin signaling is essential both for renal MCC progenitor formation and terminal differentiation. Moreover, we show that prostaglandin activity is required downstream of the transcription factor ets variant 5a (etv5a) during MCC fate choice, where modulating prostaglandin E2 (PGE2) levels rescued MCC number. The discovery that prostaglandin signaling mediates renal MCC development has broad implications for other tissues, and could provide insight into a multitude of pathological states.
Multiciliated cells (MCCs) project bundles of motile cilia on their apical surface that control fluid flow and particle displacement. In mammals, MCCs direct cerebrospinal fluid circulation in the brain and spinal cord, propel respiratory mucus, and facilitate ovum transport (1). Deficiencies in MCC function lead to hydrocephalus, chronic respiratory infections, and infertility (1). MCCs are also found transiently during human organogenesis, such as in the esophagus (2) and kidney tubules (3, 4). In lower vertebrates, MCCs in the embryonic kidney are necessary for excretory flow (5). MCCs are not present in healthy human adult kidneys, but have been noted in biopsies from patients with chronic renal diseases, where it is hypothesized that they reflect a reversion to a more primitive state to facilitate excretion (1, 5).
Due to the roles of MCCs in fluid regulation and disease, examining MCC development in various animal and cell-culture models has been a popular topic in recent years. Discoveries from several avenues now suggest a conserved pathway of MCC genesis (5). However, many questions remain about the mechanisms that predicate MCC fate choice during organogenesis and the processes that orchestrate MCC differentiation. The simplicity and experimental tractability of the zebrafish embryo kidney offer a platform to study MCC ontogeny in vivo. Vertebrate kidneys have segmented functional units termed nephrons, which typically have a blood filter, tubule, and duct (6). The kidney originates from the intermediate mesoderm and undergoes successive generation of multiple forms (7). In zebrafish, the first form, or pronephros, has two segmented nephrons (8), where MCCs drive fluid movement and are intercalated with transporter cells that modify the filtrate (Fig. 1A). MCC fate in the zebrafish pronephros is reliant on the transcription factor ets variant 5a (etv5a), but its targets have not been identified (9).
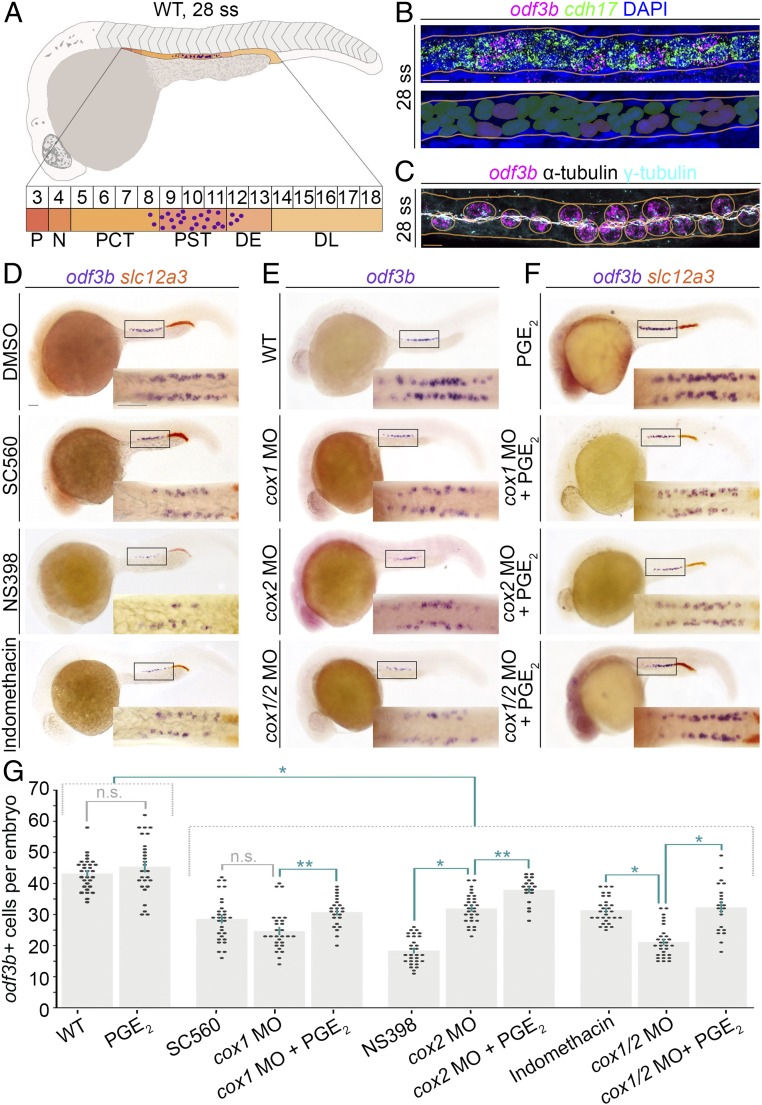
Fig. 1.
Chemical screen reveals prostaglandin signaling mediates renal MCC development in the zebrafish embryonic kidney, the pronephros. (A, Top) Schematic of a WT zebrafish embryo and pronephros at the 28 ss (lateral view) that is composed of a series of distinct functional segments with transporter cells (shades of orange) and MCCs (purple dots). The pronephros is situated along the trunk, lateral with respect to the adjacent somitic mesoderm and dorsal to the yolk and yolk sac extension. (A, Bottom) Somite map (indicated by the numbered box sequence) demarcates the location of nephron segments with respect to somites numbered 3 to 18. N, neck; P, podocyte. The formation of MCCs (purple dots) occurs in the central region of the nephron tubule, mostly located in the PST, with a few cells in the PCT and DE segments as well. (B, Top) Whole-mount smiFISH on a representative WT 28 ss embryo showing labeling for the presence of odf3b transcripts (magenta) and cdh17 transcripts (green). Nuclei are labeled with DAPI (blue), and the nephron is outlined by orange lines. (B, Bottom) Pseudocolored nuclei indicate scoring as transporter cell with cdh17+ only (green) or MCC with coexpression of cdh17+/odf3b+ (purple). (Scale bar, 10 μm.) (C) MCCs of a representative 28 ss WT embryo visualized by FISH, to label odf3b transcripts (magenta), and IF to label cilia with antiacetylated α-tubulin (white) and basal bodies with γ-tubulin (cyan). MCCs (circled in orange) exhibit odf3b expression and are distinguished by multiple ciliated basal bodies. (Scale bar, 10 μm.) (D–F) Embryos (lateral view) were labeled by WISH for the MCC marker odf3b (purple), and in D and F for the DL marker slc12a3 (orange). (D–F, Insets) Magnified dorsal views of the areas demarcated by the black boxes. (Scale bar, 50 μm.) (G) Quantification of odf3b+ cells. Each dot on the graph represents the total odf3b+ MCC number in one embryo. Data are represented as mean ± SEM; significance was determined by ANOVA, where *P < 0.0001, **P < 0.05, n.s., not significant.
Zebrafish fecundity, rapid external development, small size, and transparency have enabled researchers to employ chemical screens to study embryogenesis. This has illuminated roles of prostaglandin signaling in hematopoietic stem cell regulation (10) and endoderm (11) and nephron formation (12). Prostaglandins are functionally diverse lipid mediators that signal through G protein-coupled receptors (13). Prostaglandins are synthesized from arachidonic acid by a cyclooxygenase (COX1, COX2) into an intermediate that is metabolized into an active prostanoid, like prostaglandin E2 (PGE2). Prostaglandins exit the cell to interact with receptors on receiving cells. Interestingly, PGE2 affects ciliogenesis by regulating intraflagellar transport (14). However, the role of prostaglandin signaling in renal MCC formation is unknown. Defects in prostaglandin biosynthesis or receptors affect fate choice in the zebrafish embryo kidney, leading to an expanded distal early (DE) tubule segment and a reduced distal late (DL) tubule segment (12). By comparison, the proximal convoluted tubule (PCT) and proximal straight tubule (PST) segments were unchanged based on assessment of genes that mark transporter cells, but MCCs were not analyzed (12).
Here, we report a chemical screen for regulators of MCC formation in the zebrafish pronephros. Our studies identified essential roles for prostaglandin signaling in renal MCC specification and terminal differentiation. Prostaglandin-deficient embryos formed fewer renal MCC progenitors and instead developed more transporter cells. Further, the loss of prostaglandin activity affected renal MCC differentiation, evident by disrupted apical docking of basal bodies and reduced ciliogenesis. Finally, we show that 16,16-dimethyl prostaglandin E2 (dmPGE2) partially rescued MCCs in etv5a-deficient embryos, placing prostaglandin signaling downstream of a critical MCC gene. Taken together, our study uncovers fundamental roles for prostaglandin signaling in renal MCC ontogeny. These discoveries have important implications for understanding MCC genesis in development and in the context of renal pathological conditions.
Results
Chemical Screen Identifies Candidate Modulators of Renal MCC Development.
Many components and molecular interactions that control MCC genesis remain unknown. The zebrafish pronephros is useful to study renal MCC ontogeny, because this organ and its constituent cells form quickly. By the 28 somite stage (ss), or 24 h post fertilization (hpf), renal progenitors in a wild-type (WT) embryo complete a mesenchymal transition into a polarized epithelium with discrete segment domains (Fig. 1A) that express cadherin 17 (cdh17) (Fig. 1B) (15, 16). The epithelial MCC populace is dispersed in a “salt-and-pepper” pattern in the central region (Fig. 1A), expresses outer dense fiber of sperm tails 3b (odf3b), and possesses bundles of apically localized cilia (Fig. 1 B and C) (17). Given the utility of chemical screens to study developmental processes in the zebrafish, we hypothesized that this approach could be used to gain new insights into MCC development by assessing odf3b expression.
We utilized the SCREEN-WELL Harvard Institute of Chemistry and Cell Biology (ICCB) Known Bioactives Library. At 2 hpf, WT embryos were arrayed in 96-well plates, and at 4 hpf they were treated with dimethyl sulfoxide (DMSO) vehicle control, a blinded chemical from a library well, or left untreated (18). At the 28 ss, the drugs were removed and the embryos were fixed. Three biological replicates were performed on each library component. For the analysis of MCC development, whole-mount in situ hybridization (WISH) was performed on the fixed embryo samples using the differentiating MCC marker odf3b (SI Appendix, Fig. S1A). Following WISH, blinded scoring was used to assess the resultant phenotypes. Of the compounds screened, 34% were lethal, 57% were associated with normal expression of odf3b, and 9% had an MCC phenotype categorized as either more or less odf3b+ pronephros cells (SI Appendix, Figs. S1B and S2 and Dataset S1). Based on the ICCB library categories of classifications, the 9% of chemicals that affected MCCs were 32% bioactive lipids, 29% inhibitors, 10% kinase inhibitors, 10% nuclear receptor ligands, 7% CNS receptor ligands, 7% ion-channel ligands, and 5% endocannabinoids (SI Appendix, Figs. S1B and S2 and Dataset S1). Several hits were prostaglandin pathway agonists. These included a bioactive prostaglandin, receptor agonists, and an arachidonic acid metabolite, which all caused reduced MCC numbers at the screen dosage and treatment window (SI Appendix, Figs. S1C and S2). Also, we noted that a variety of prostaglandin modulators were lethal at the dosage and developmental time tested in the screen (SI Appendix, Fig. S1C).
The observation of prostaglandin screen hits was surprising. Prior research had not detected a role for prostaglandin signaling in renal MCC formation, despite being required to drive ciliogenesis by regulating intraflagellar transport in other developing tissues (14). In this study, researchers examined cilia in the leaky tail (lkt) zebrafish mutant, which encodes a defect in an ATP-binding cassette transporter (ABCC4) that can export PGE2 (14). lkt mutants evinced no change in renal cilia (14). However, transcripts encoding Lkt/ABCC4 were not expressed in the developing kidney (14). We wondered if a role for prostaglandin signaling in renal MCC ontogeny was not discerned because this transporter is not involved in supplying kidney progenitors with prostaglandins, and as PGs can exit cells with or without such facilitated transport. Therefore, we sought to further investigate the possible role of prostaglandins in renal MCC ontogeny.
Prostaglandin Signaling Is Required for the Development of Renal MCCs.
To explore how prostaglandin signaling influences renal MCC genesis, we performed COX loss-of-function studies. At the 60% epiboly stage, we treated WT embryos with vehicle or one of the following pharmacological compounds: the COX1 inhibitor SC-560, COX2 inhibitor NS-398, or nonselective COX inhibitor indomethacin (10–12). We treated at a later stage than in the screen to minimize the disruption of cell movements by prostaglandins during early development (19, 20), and selected compound concentrations based on previous effective dosages in zebrafish (10–12). At the 28 ss, the embryos were fixed for WISH analysis using the MCC marker odf3b and the specific DL marker solute carrier family 12, member 3 (slc12a3) (8). After treating with 50 μM SC-560, 50 μM NS-398, or 30 μM indomethacin (12), embryos with a smaller DL segment, a benchmark of prostaglandin deficiency, had a significant decrease in renal MCCs (Fig. 1 D and G). Of note, drug treatments were ∼70% penetrant, consistent with prior work (12). Knockdown of cox1 or cox2 with validated morpholino (MO) reagents (12) also significantly decreased MCC number, which was further exacerbated by cox1/2 knockdown (Fig. 1 E and G). Analysis of live cox1, cox2, and cox1/2 morphants revealed pericardial edema at 48 hpf, suggesting fluid imbalance and kidney dysfunction (21–23) as well as hydrocephaly, as noted previously in lkt mutants and cox1 morphants (SI Appendix, Fig. S3) (14).
Next, we performed studies to test if provision of a prostaglandin ligand could rescue MCCs in cox morphants. PGE2 is abundant in zebrafish embryos and functions in gastrulation and organogenesis (10–12, 19, 20). Further, two PGE2 receptors, prostaglandin E receptor 2a and 4a (Ptger2a, Ptger4a; also known as EP2 and EP4, respectively), are expressed in the renal progenitors that give rise to the zebrafish pronephros (12). While prostanoids have a short half-life, leading to challenges with PGE2 treatment, a long-acting derivative, dmPGE2, has been effectively used in vivo (10–12). Thus, we treated control embryos and cox morphants with 100 μM dmPGE2 from 60% epiboly to the 28 ss, and MCCs were assessed by WISH for odf3b. While dmPGE2 treatment alone had no effect on MCC number in WT embryos, dmPGE2 was sufficient to rescue MCC number in cox morphants (Fig. 1 F and G). The finding that dmPGE2 had no effect on MCC development in WT embryos was contradictory to the screen results (SI Appendix, Fig. S1C), which was possibly due to the different treatment window. Moreover, library well contamination and/or compound degradation can lead to variations. Nevertheless, the sum of our pharmacological and genetic studies establishes that prostaglandin biosynthesis is required for renal MCC development.
Prostaglandins Specify the MCC Progenitor Pool in the Embryonic Kidney.
MCC development is progressive and begins with progenitor specification, followed by differentiation events including centriole duplication, apical docking of basal bodies, and ciliary outgrowth (1). There is rich evidence for prostaglandin signaling in cell fate specification, including liver versus pancreas fate (11) and pronephros segment fate choice (12). To examine if prostaglandin signaling regulates MCC progenitor specification, we first determined a suite of markers to enable their identification and quantification. One promising candidate was the transcription factor paired box 2a (pax2a). Similar to Pax2 in other vertebrates, Pax2a is expressed in the zebrafish pronephric mesoderm (24). At the 28 ss, pax2a transcripts were expressed in a similar pattern of cells compared with those expressing odf3b within the nephrons (Fig. 2 A and B). Further, whole-mount fluorescence in situ hybridization (FISH) analysis on WT embryos revealed that cells with pax2a transcripts also expressed odf3b transcripts at the 28 ss (Fig. 2A). These data led us to conclude that pax2a marks the renal MCC population. Given the early expression of pax2a before more mature markers such as odf3b (9) and the similar number of pax2a+ cells to odf3b+ cells at the 28 ss (SI Appendix, Fig. S4C), we hypothesized that pax2a may mark MCC progenitors as well. Additionally, the Notch ligand encoded by jagged 2b (jag2b) was a second candidate for identifying MCC precursors. Previous research has demonstrated that Jag2b+ renal progenitors interact with their neighboring Notch-expressing cells to inhibit transcription of ciliogenesis genes and promote transporter cell fate (25, 26). Indeed, jag2b and odf3b transcripts colocalize in pronephros cells characterized by bundles of multiple cilia, indicating they are labeling maturing or terminally differentiated MCCs (26).
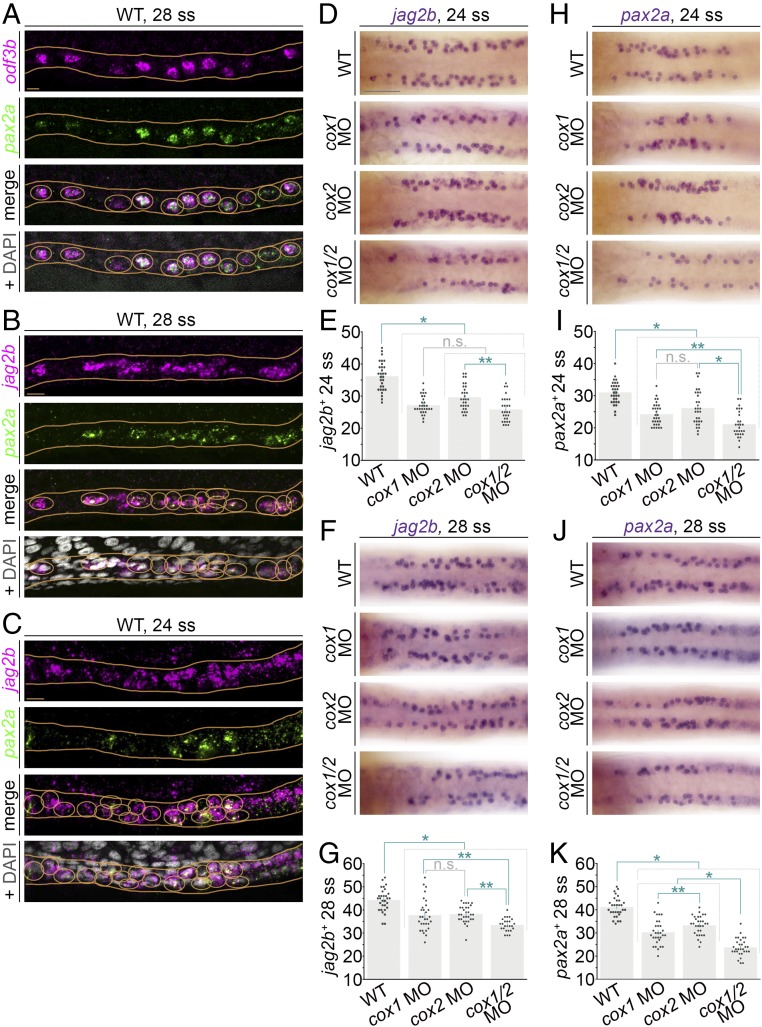
Fig. 2.
Deficiency of prostaglandin biosynthesis leads to decreased renal MCC progenitor number in the zebrafish pronephros. (A–C) FISH in representative WT embryos at the 28 ss reveals (A) colocalization of odf3b transcripts (magenta) and pax2a transcripts (green), and (B and C) that jag2b and pax2a are coexpressed in a salt-and-pepper fashion in the nephron at the (B) 28 ss and (C) 24 ss. Images are maximum image projections. Nephron cells with colocalization of transcripts are circled in orange, and nephron tubules are outlined with orange lines. (A–C, Bottom) Orange circles are around nuclei to indicate the scoring of representative cells that exhibit colabeling of the markers. (Scale bars, 10 μm.) (D and F) jag2b transcript expression at the (D) 24 and (F) 28 ss in representative WT controls and cox-deficient embryos as visualized by WISH. Images are dorsal views of the pronephros. (Scale bar, 50 μm.) (E and G) Quantification of jag2b+ pronephros cells at the 24 and 28 ss. (H and J) Expression of pax2a at the 24 and 28 ss in representative WT controls and cox-deficient embryos as visualized by WISH. Images are dorsal views of the pronephros. (I and K) The average number of pax2a+ cells per pronephros at the (I) 24 and (K) 28 ss is quantified in graphs. For D, F, H, and J, scored cells are indicated by the overlay of an opaque purple dot. For E, G, I, and K, each dot represents the total number of jag2b+ or pax2a+ cells between somites 8 and 12 of one pronephros. Data are represented as mean ± SEM; significance was determined by ANOVA, where *P < 0.0001, **P < 0.05, n.s., not significant.
Using FISH on WT embryos, we found that jag2b and pax2a transcripts were localized within the same pronephros cells at the 28 ss (Fig. 2B), as well as the earlier 24 ss (Fig. 2C). As only weak odf3b expression has been detected in the pronephros at the 24 ss (9), we conclude the jag2b+/pax2a+–coexpressing cells are in a MCC precursor stage. To ascertain if there was an earlier time when jag2b+/pax2a+ cells display the salt-and-pepper MCC pattern in the zebrafish pronephros, we performed FISH at younger stages. At the 20 ss, when the earliest expression of the maturing MCC marker odf3b has been observed in renal progenitors (9), expression of jag2b and pax2a was intermingled throughout renal progenitors (SI Appendix, Fig. S5). Thus, it was difficult to determine whether a specific populace of MCC precursors was present at this stage of development (SI Appendix, Fig. S5). Thus, we selected the 24 ss as the most reliable benchmark for MCC precursor detection by jag2b or pax2a, and concluded that the 28 ss time point was a benchmark for further maturing MCCs.
Next, we interrogated whether there was a role for prostaglandin signaling in MCC progenitor formation. To this end, we inhibited prostaglandin signaling via single or double cox knockdown and quantified jag2b+ or pax2a+ cell number at the 24 and 28 ss with WISH. At both stages, cox-deficient embryos had significantly decreased jag2b+ and pax2a+ cell numbers compared with controls (Fig. 2 D–K and SI Appendix, Fig. S4). Interestingly, we observed subtle but significant decreases in the number of jag2b+ and pax2a+ cells in cox1- and cox2-deficient embryos compared with controls: The average cell-number decreases were about five or six cells (Fig. 2 E, G, I, and K). By comparison, the cox1/2 doubly deficient embryos displayed an average reduction of between 10 and 15 jag2b+ and pax2a+ cells compared with WT control embryos (Fig. 2 E, G, I, and K). Taken together, these results indicate that inhibition of prostaglandin signaling significantly reduces, but does not fully abrogate, the number of jag2b+ or pax2a+ MCC precursors that develop in the pronephros. Thus, we conclude that both cox1 and cox2 have significant roles in MCC precursor ontogeny. It is possible that cox1 and cox2 have the same role(s), and thereby function redundantly. Related to this, the loss of either cox1 or cox2 might lead to up-regulation of the other as a compensatory mechanism to account for the less severe MCC reduction in single knockdowns. Alternatively, cox1 and cox2 may have an independent role(s) in MCC genesis, given the observation that loss of both led to a reduction in the number of pax2a+ cells that was approximately equivalent to the additive loss of cox1 and cox2.
Loss of Prostaglandin Signaling Causes a Switch in Fate Choice Between Transporter Cell and MCC Within the Proximal Tubule Segments.
Next, we explored how the change in MCC number affected pronephros development. MCCs primarily occupy the PST segment in the zebrafish pronephros, where they are intermingled with transporter cells (9). Previous work determined that cox1, cox2, or cox1/2 deficiency has no effect on the absolute length of the PST segment (12). Despite this, it remains unknown if the distribution of MCC and transporter populations within the segment was altered because MCCs were not directly examined (12), and because standard WISH staining methods are less sensitive than FISH with confocal analysis. To examine this aspect, we selected transient receptor potential cation channel, subfamily M, member 7 (trpm7) as a marker to evaluate PST development, as trpm7 is an indicator of progression to the 28 ss (8) and can be utilized to evaluate developmental delay, which is a possible side effect of MO use. Moreover, we observed that the pronephros was composed of distinct subsets of trpm7+ cells and pax2a+ cells, consistent with a distribution of transporters and MCCs, respectively (SI Appendix, Fig. S6A).
To examine PST cellular composition following the loss of prostaglandin activity during nephrogenesis, we performed double FISH in control and cox-deficient embryos with the MCC specific marker odf3b and the specific PST transporter cell marker trpm7 (Fig. 3 and SI Appendix, Fig. S6B). Cell number in the PST was quantified using DAPI as a nuclear marker, and each cell was scored as to whether it expressed trpm7 and/or odf3b transcripts. WT control and experimental groups had no difference in the absolute length of the trpm7+ domain, consistent with prior results (12) (Fig. 3B). Further, pronephros cells were typified by displaying fluorescent signals equivalent to an abundance of either trpm7 or odf3b transcripts, and not a mixture of both transcripts over background signal (Fig. 3A). We observed that control PST segments were composed of ∼50% trpm7+ cells and 50% odf3b+ cells (Fig. 3 B and C). In contrast, cox-deficient embryos had a statistically significant increase in the number of transporter cells in the PST and a statistically significant decrease in MCC number (Fig. 3B). When the percentage of distribution was calculated, these alterations equated to an average range of 70 to 75% trpm7+ cells and 25 to 30% odf3b+ cells in cox-deficient embryos (Fig. 3 B and C).
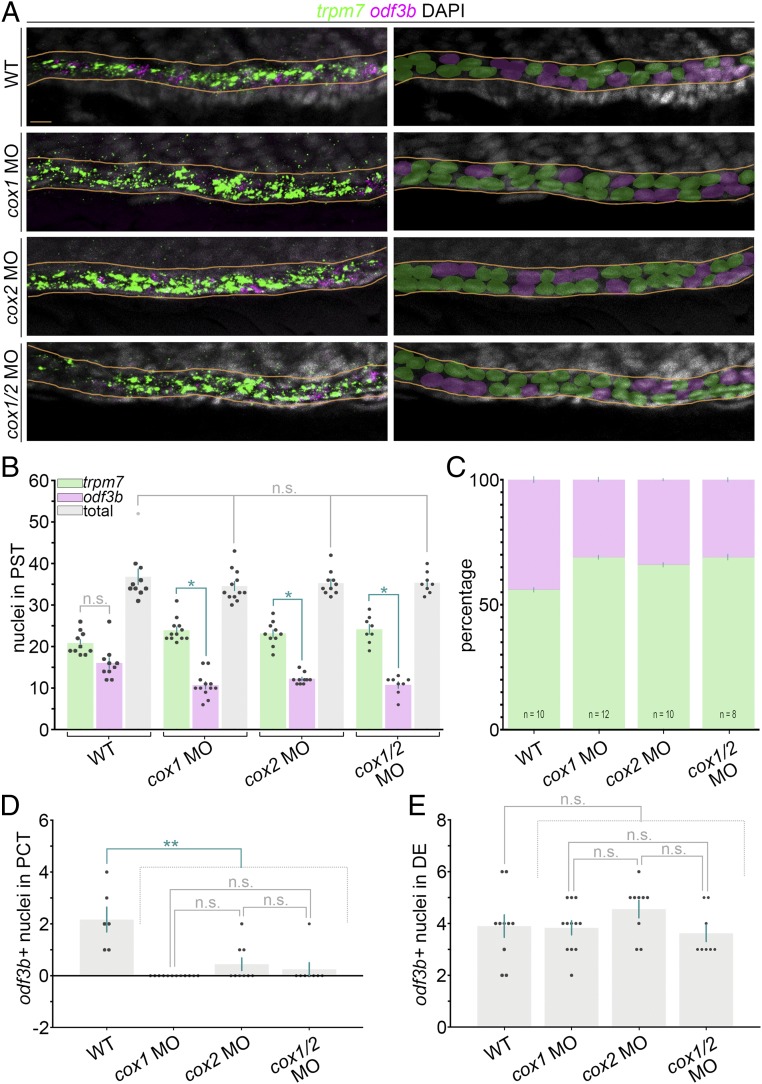
Fig. 3.
Prostaglandin signaling influences MCC versus transporter cell fate in the zebrafish pronephros. (A, Left) FISH analysis at the 28 ss in representative WT control and cox-deficient embryos for odf3b (magenta), trpm7 (green), and DAPI to label nuclei (gray). Images are maximum image projections. (A, Right) Nuclei were pseudocolored to show scoring as MCC (magenta) or PST transporter cell (green) based on odf3b or trpm7 transcript localization, respectively. Nephron tubules are outlined in orange lines. (Scale bar, 10 μm.) (B) Quantification of MCC and transporter cell number within the PST segment. (C) Percentage of MCC and transporter cell numbers within the PST segment. (D) Quantification of MCC number in the PCT segment. (E) Quantification of MCC number in the DE segment. Data are represented as mean ± SEM, and each dot on the graphs represents one embryo; significance was determined by ANOVA, *P < 0.0001, **P < 0.0002, n.s., not significant.
We also assessed MCC numbers in the adjacent segments, where the PCT is located rostrally and the DE segment is located caudally with respect to the trpm7+ PST segment in both WT and cox-deficient embryos (12). Prostaglandin deficiency affected MCC fate in the PCT of cox-deficient embryos (Fig. 3D), while MCC numbers in the DE were unchanged in cox-deficient embryos compared with WT controls (Fig. 3E). Taken together, these results indicate that proximal pronephros cells elect a transporter cell fate when there is a loss of prostaglandin signaling.
Cilia Outgrowth on Maturing MCCs Is Mediated by Prostaglandin Signaling.
To become fully differentiated, an MCC progenitor must undergo maturation events including basal body duplication, apical docking, and cilia outgrowth (1). Prostaglandin signaling is known to regulate cilia outgrowth through intraflagellar transport (14), discovered when ciliogenesis defects in the zebrafish lkt mutant were linked to a disruption of the Lkt/ABCC4T804M transporter. Upon PGE2 export by ABCC4, PGE2 binds the EP4 receptor to activate cAMP and ciliogenesis (14). Intriguingly, lkt mutants do not have a pronephros ciliogenesis defect (14). Additionally, abcc4 transcripts were not detected in the pronephros (14), presenting the possibility that prostaglandin signaling might function with other molecular machinery to regulate renal MCC differentiation. Thus, we hypothesized that prostaglandin signaling might have additional, as-of-yet undetected roles in maturation events of pronephros cells, particularly ciliogenesis.
To examine kidney cilia in prostaglandin-deficient embryos, we performed whole-mount FISH in combination with immunofluorescence (IF) (17). In embryos that experienced a loss of prostaglandin signaling through cox gene knockdown, we saw a notable decrease in pronephric cilia compared with WT control embryos (Fig. 4A and SI Appendix, Fig. S7A). To quantify this, we used the freehand line tool in Fiji to trace the acetylated α-tubulin+ band of cilia in the nephron tubule and measure fluorescence intensity (Fig. 4 B and C). We found that cilia intensity was statistically decreased in cox-deficient embryos compared with WT controls (Fig. 4 B and C). These results indicate that prostaglandin signaling is required for ciliogenesis in the embryonic nephron.
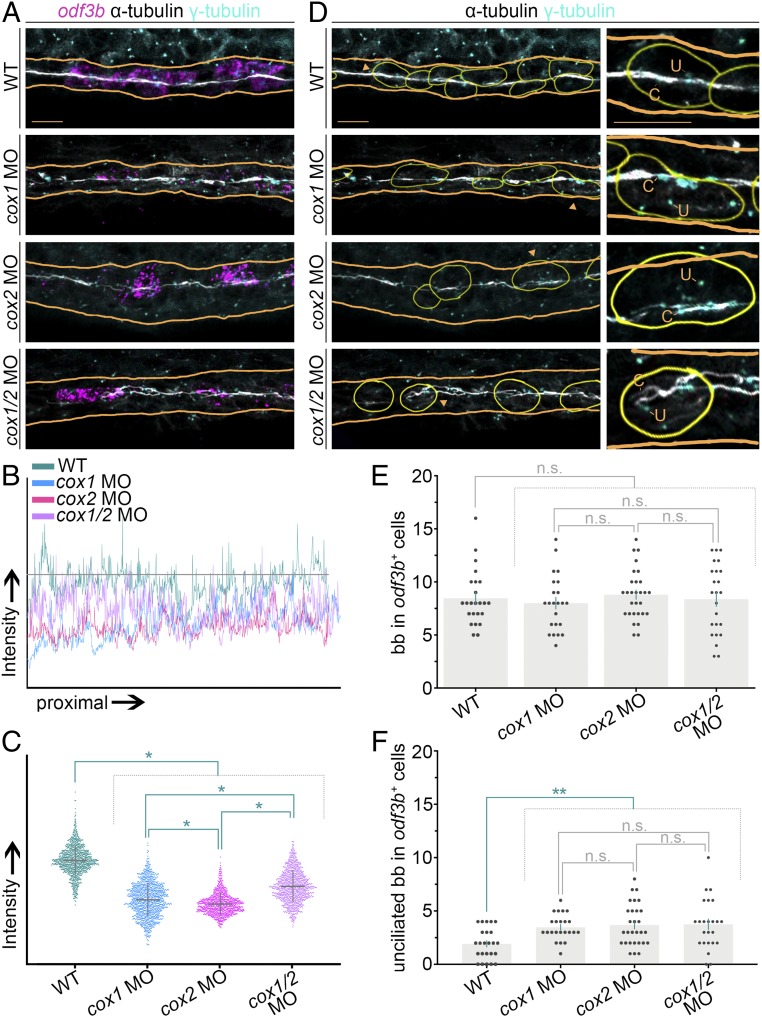
Fig. 4.
Ciliary outgrowth and basal body docking in renal MCCs are influenced by prostaglandin deficiency. (A) FISH and IF at the 28 ss for odf3b transcripts (magenta), acetylated α-tubulin (white), and γ-tubulin (cyan) for representative WT embryos and cox1, cox2, and cox1/2 morphants. Orange lines outline the respective nephrons. Images are maximum image projections. (Scale bar, 10 μm.) (B) Quantification of cilia intensity with a line surface plot of signal intensity in control and cox-deficient samples. The grey threshold line is the difference of the highest- and lowest-intensity points for the WT control. (C) Distribution of individual intensity values along the length of the cilia band in control and cox-deficient samples. (D) Identification of ciliated and unciliated basal bodies by analysis of cilia with antiacetylated α-tubulin (white) and basal bodies with γ-tubulin (cyan). Yellow outlines demarcate odf3b+ cells from A. (D, Insets) Magnified views of the cells labeled by the orange arrowheads. Each “u” demarcates an unciliated basal body, and each “c” demarcates a ciliated basal body. (Scale bar, 10 μm.) (E and F) Quantification of (E) basal bodies, labeled by γ-tubulin, in odf3b+ cells and (F) unciliated basal bodies in odf3b+ cells. For both graphs, each dot represents a single odf3b+ cell, and four or five cells were analyzed in each embryo. Data are represented as mean ± SEM; significance was determined by ANOVA, where *P < 0.0001, **P < 0.02, n.s., not significant.
Next, we sought to determine if the odf3b+ renal MCCs in cox-deficient embryos exhibited normal basal body progression. We reasoned that a net decrease in pronephric cilia could be explained by a decrease in the amplification of basal bodies or the number of apically docked basal bodies within MCCs. Therefore, we scrutinized both the number and location of basal bodies by modifying a previously described approach (27). In this analysis, we outlined odf3b+ cells in silico by combining odf3b and DAPI channels. The outlines were then transferred to a composite of acetylated α-tubulin and γ-tubulin channels from the same embryo. By outlining odf3b expression, we were able to subtract odf3b transcripts from the image and visualize the γ-tubulin expression of basal bodies in the MCC population (Fig. 4D and SI Appendix, Fig. S7B). Quantification of absolute basal body number in odf3b+ renal cells revealed that there was not a statistically significant difference between control and cox1, cox2, and cox1/2 knockdowns (Fig. 4E), indicating that prostaglandin deficiency does not affect normal basal body duplication in the pronephros. Next, we scored apical docking of basal bodies in MCCs by assessing if they were ciliated or not in odf3b+ cells (Fig. 4 D and F). This analysis revealed a statistically significant increase of unciliated basal bodies in odf3b+ cells of cox1-, cox2-, and cox1/2-deficient embryos compared with WT controls (Fig. 4 D and F). These data suggest prostaglandin signaling promotes basal body trafficking to the apical surface in renal MCCs.
Prostaglandin Signaling Acts Downstream of etv5a During Renal MCC Development.
We next sought to define how prostaglandin signaling relates to the known genetic hierarchy of zebrafish renal MCC ontogeny, which is reliant on the core transcription factor etv5a (9). etv5a is a member of the Pea3 subfamily of E26 transformation specific (ETS) transcription factors, where it regulates target genes by binding to an ∼10-bp element known as the Ets-binding site. Etv5a and its mammalian ortholog are known to exert diverse roles in embryogenesis. For example, Etv5a is important for cilia formation in Kupffer’s vesicle and subsequent left/right patterning (21), as well as ventral mesoderm production of hematopoietic precursors in zebrafish (28). Etv5, the mammalian ortholog, is required for such processes as branching morphogenesis and ureteric bud tip formation during metanephric kidney formation (29, 30), limb outgrowth (31), neuronal development (32), self-renewal of spermatogonial stem cells (33), and maintenance of cell identity in the lung (34). In the mouse ovary, Etv5 is expressed in cumulus and granulosa cells (35). Interestingly, COX2 is critical for oocyte maturation, and in vitro studies in mice have demonstrated that Etv5 increases the transcriptional activity of the Cox2 promoter (35). During pronephros development, etv5a transcripts localize to the renal progenitor domain, where pax2a+/jag2b+ MCC precursors emerge, and knockdown of etv5a or overexpression of a dominant-negative etv5a diminishes MCC number (9).
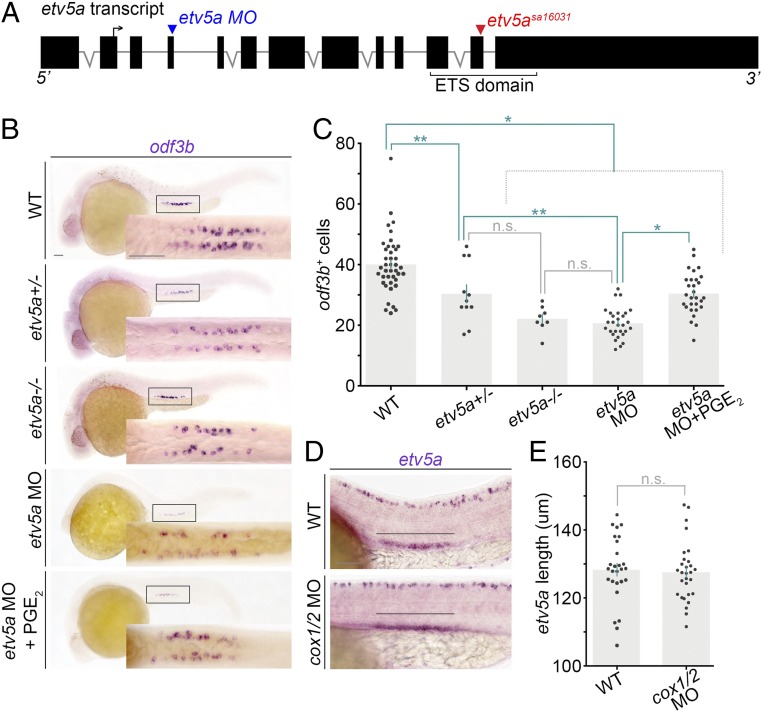
To further assess the requirement of etv5a during MCC development, we conducted genetic studies on etv5a. We obtained the etv5asa16031 mutant, which contains a G→T substitution that encodes a premature stop mutation in the ETS DNA-binding domain of Etv5a (Fig. 5A) (36). etv5asa16031−/− mutant embryos had reduced MCC numbers that were statistically equivalent to etv5a morphants (Fig. 5 B and C). Interestingly, etv5asa10316+/− embryos had a significant decrease in MCC number compared with controls, but had significantly more MCCs compared with the etv5a morphant embryos (Fig. 5 B and C). Based on phenotypic congruence between etv5a mutants and knockdowns, we employed the latter to study prostaglandin signaling in the context of etv5a deficiency, namely to explore whether there was a connection between Etv5a and prostaglandin signaling during MCC development.
Fig. 5.
Prostaglandin signaling acts downstream of etv5a during MCC genesis. (A) Schematic of the etv5a locus with locations of the splice MO (blue arrowhead) and the etv5asa16031 mutation (red arrowhead). The forward arrow in exon 2 depicts the ATG start site. (B) WISH at the 28 ss on representative genotype-confirmed WT embryos, etv5a morphants, and etv5a morphants treated with dmPGE2. Embryos are imaged in a lateral view, and the Insets correspond to a dorsal view of the pronephros. (Scale bar, 50 μm.) (C) Quantification of odf3b+ cells at the 28 ss. Each dot represents the total number of maturing renal MCCs in one embryo. Data are represented as mean ± SEM; significance was determined by ANOVA. (D) Lateral view of the etv5a domain (black line) in the pronephros of representative WT and cox1/2 morphants. (Scale bar, 50 μm.) (E) Quantification of absolute etv5a length in WT versus cox1/2 morphants. Each dot represents the etv5a domain length in one nephron. Data are represented as mean ± SEM; significance was determined by Student’s t test, where *P < 0.0001, **P < 0.006, n.s., not significant.
Given the ability of mammalian Etv5 to influence the transcriptional activity of Cox2 (35), we hypothesized that Etv5a might indeed operate to regulate prostaglandin biosynthesis. To explore the relationship between etv5a and prostaglandin signaling, we injected WT embryos with an etv5a splice MO (9) and treated with vehicle or 100 μM dmPGE2 at 60% epiboly. WISH at the 28 ss revealed that odf3b expression was partially rescued in etv5a-deficient embryos treated with dmPGE2 compared with controls (Fig. 5 B and C). Further, renal etv5a expression was unchanged in cox1/2 morphants (Fig. 5 D and E) and dmPGE2-treated WT embryos (SI Appendix, Fig. S8). Additionally, examination of the DNA sequence 1 kb upstream of zebrafish cox1 and cox2 revealed multiple putative Etv5-binding sites for each gene (SI Appendix, Fig. S9), suggesting the possibility that there could be direct regulation, as reported from in vitro studies with the murine orthologs (35). Taken together, our results suggest that prostaglandins act downstream of etv5a to promote renal MCC genesis.
Discussion
MCCs are an important cell type that is necessary for directing fluid flow during the formation and function of many tissues and organs and also drives locomotion in a variety of animals (37). A comprehensive understanding of the biological processes that regulate MCC specification and maturation can provide insights into known MCC-related conditions in humans, including hydrocephalus, chronic respiratory infections, and female infertility (38). Additionally, knowing the genes that mediate the stages of MCC fate choice and differentiation can help to illuminate the mysterious origins of MCCs in diseased tissues, like in the adult human kidney, where they are not typically present in healthy conditions (1, 5). In the last decade there have been major advances in identifying the genetic components of MCC genesis and ciliary genes, as well as cataloging the protein localization and functions of many factors in MCCs (e.g., refs. 39–46). This and other ongoing work are transforming our understanding of MCC development and function, but more research is still needed to fully decipher the underlying molecular mechanisms of these events.
In lieu of the inaccessibility of mammalian MCCs, the zebrafish pronephros is one useful genetic tool to examine MCC ontogeny and the cell biology of multiciliogenesis in vivo due to its simple anatomy and experimental tractability. Further, the existence of MCCs in the zebrafish pronephros (25, 26) provides a context to examine renal MCC genesis. Our previous studies have discovered an essential role for etv5a in renal MCC specification and for prostaglandin signaling in regulating nephron patterning during embryonic kidney formation (9, 12). Regarding the latter, we found that prostaglandin signaling was required for the proper balance of distal segment fate choice in the zebrafish pronephros, while proximal segment boundaries developed normally (12). The genesis of MCCs was overlooked because the PST, where most MCCs reside, had a normal overall segment size (12).
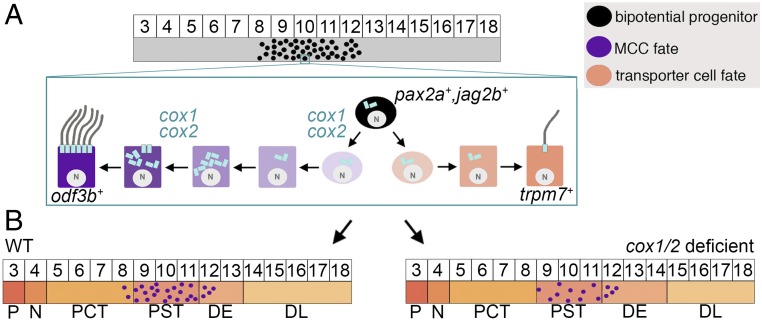
Here, we have delineated critical roles for prostaglandin signaling during renal MCC development in specification and differentiation. Indeed, the requirement for COX1/COX2 activity at sequential stages of MCC genesis is a fascinating example of a stage-dependent modularity of prostaglandin signaling. These results led us to propose an integrated model based on this work and our previous results (12) whereby prostaglandin activity influences the choice between MCC and transporter cell fate, as well as nephron distal segment fate commitment (Fig. 6). Additionally, our work has revealed that prostaglandin signaling is required for crucial aspects of terminal MCC differentiation to proceed, where its deficiency impacts basal body docking and subsequently reduces cilia formation (Fig. 6A).
Fig. 6.
Integrated working model depicts the proposed roles for cox1/2 during renal MCC fate choice and differentiation in the zebrafish pronephros. (A) The intermediate mesoderm (gray), which gives rise to the embryonic kidney, includes a populace of bipotential renal progenitors (black circles) that can form MCCs or transporter cells. Expression of cox1/2 supports adoption of MCC fate, and subsequently promotes basal body docking during MCC differentiation. (B) Prostaglandin signaling in renal progenitors is essential for MCC versus transporter fate in the proximal segments and also modulates distal segment fate, where cox1/2 deficiency is associated with altered DE/DL domains.
The discovery that prostaglandin inhibition reduces the number of MCC progenitors and, in the proximal tubule nephron segments, inversely increases the number of transporter cells supports a mechanism whereby prostaglandin signaling regulates a binary fate choice within bipotential renal progenitors (Fig. 6A). Further, we have placed prostaglandin signaling downstream of the ETS transcription factor etv5a (9) in renal MCC development, as dmPGE2 partially rescued MCC number in etv5a-deficient embryos. How this relates to the set of genes that regulate proximal segment identity (47, 48) is an interesting topic for future studies. Delineating the relationship between prostaglandin signaling and other MCC regulators is another important area for continued study as well. For example, elegant studies have identified Notch signaling as a common negative regulator of MCC fate across various tissues (25, 26, 39, 49). Renal MCC development in the zebrafish pronephros is also mediated by the transcription factor MDS1 and EVI1 complex locus (mecom), which acts upstream of Notch signaling and downstream of retinoic signaling to control MCC fate choice (22). Assembling the higher-order genetic networks that control MCC development will involve determining the relationships between these known players and the definition of their functional activities.
Another aspect of future interest will be elucidating the relationship of prostaglandin signaling with other known factors of MCC differentiation, like geminin coiled-coil domain containing (gmnc) (40), v-myb avian myeloblastosis viral oncogene homolog (myb) (41), and multicilin (46). With regard to MCC terminal differentiation, the question of why there are more unciliated basal bodies in the pronephros of prostaglandin-deficient embryos is an intriguing aspect for cell biology studies. The miR-34b-myb pathway regulates basal body docking to the cell membrane (27). Evaluating this pathway in the background of prostaglandin deficiency is just one place to start that may possibly shed light on the underlying molecular mechanisms of multiciliogenesis. Defective ciliary transition-zone formation can also affect basal body apical docking. The transition zone forms at the early stages of ciliogenesis and may stabilize the basal body connection to the epithelial cell membrane. A newly discovered transition-zone protein, Dzip1L, was found to affect only MCC motile cilia bundles during zebrafish pronephros development, where genetic disruption caused cystic kidneys in mice and was linked to human autosomal-dominant polycystic kidney disease (50). Evaluating whether prostaglandin signaling affects the formation of the transition-zone machinery (e.g., Dzip1L) in MCCs is thus one such area for exploration.
With regard to the prostaglandin signaling pathway, much work is needed to identify the components that are relevant to MCC development, as the current report has only examined the contributions of cox1, cox2, and PGE2. For example, PGE2 is not the only prostanoid in zebrafish embryos, where PGF2α, PGI2, and TXA2 have been detected (19). Testing whether other prostanoids rescue renal MCC specification, basal body docking, and/or cilia outgrowth may provide additional insights into the mechanisms of multiciliogenesis events. Additionally, as Lkt/ABCC4 mutants do not evince renal cilia phenotypes, future work is needed to identify the relevant transporter(s) that supports MCC genesis in the pronephros as well as prostaglandin receptors.
The important roles of MCCs in physiology and disease underscore the need to fully elucidate their ontogeny. Understanding renal MCC specification and maturation may lend insights into the puzzling origins of MCC emergence in chronic kidney conditions, where it is unclear whether MCCs are a cause or phenotypic outcome of disease. The mysterious appearance of MCCs in the adult human kidney has been reported within nephron tubules in the context of several pathological states, including hypercalcemia, congenital nephrosis, and glomerulonephritis (4, 51–55). This indicates that human renal tissues have the capacity for multiciliation, consistent with the observation of MCCs in fetal human kidney samples (3, 4), though MCCs are absent under healthy conditions.
Innovative research is needed to establish experimental models for aberrant MCC generation that can be utilized to delineate how and why these cells are induced to form in various kidney disease states, as well as their effects during the pathology of such conditions. One interesting possible avenue to explore MCC generation subsequent to kidney damage may in fact be the zebrafish kidney. In response to some types of damage, nephrons in the embryonic and adult zebrafish kidney can undergo robust epithelial regeneration (56–62). Exploring if the response to kidney damage entails formation of more MCCs might provide a way to better understand the reparative pathways that are stimulated in injury states.
In conclusion, our work has identified roles for prostaglandin signaling in renal MCC formation, a discovery that may have important implications for MCCs in other tissues, and by extension MCC-related disorders in these locales. Moreover, in addition to prostaglandin signaling, our chemical screen identified other molecules that influence renal MCC development. Further analysis of these hits, by our group and others, can be leveraged to gain more powerful insights into MCC genesis. Given the conservation of MCC development across tissues, we speculate that our dataset contains inroads to appreciate a number of other fundamental aspects of MCC formation, as well as unique aspects relevant to the intermediate mesoderm and renal lineages.
Materials and Methods
Animal Studies.
Zebrafish were maintained in the Center for Zebrafish Research, and experimental studies were approved by the University of Notre Dame Institutional Animal Care and Use Committee under protocol 16-025, in accordance with the Guide for the Care and Use of Laboratory Animals (63). Embryos were produced by pairwise matings of adult zebrafish between 3 and 12 mo of age, with populations including a mixture of both sexes collected, raised at 28.5 °C in embryo E3 media, and staged by somite number for these studies.
Chemical Screen and Other Pharmacological Treatments.
Tübingen strain fish were used for the studies and staged as described (64). For screening, WT zebrafish embryos were arrayed in 96-well plates at 2 h post fertilization. At 4 hpf the embryo media were replaced with 1:100 dilutions of drugs from the ICCB Known Bioactives Library (Enzo Life Sciences) (18). For treatments with 50 μM SC-560, 50 μM NS-398, 30 μM indomethacin, or 100 μM dmPGE2 (American Bioanalytical, Enzo Life Sciences, Santa Cruz), embryos were arrayed in 12-well plates and treated protected from light at 60% epiboly. Drugs were removed at the 28 ss and embryos were fixed in 4% paraformaldehyde.
Expression Analysis and Image Acquisition.
WISH was conducted as described (65, 66). RNA probes were digoxigenin- (odf3b, jag2b, etv5a) or fluorescein-labeled (cdh17, slc12a3, pax2a), and generated by in vitro transcription using plasmid templates (8, 67, 68). Embryos were placed in glycerol and images were taken using a Nikon Eclipse Ni with a DS-Fi2 camera. FISH was conducted as described (69, 70), using the identical probes for WISH. Stains were developed with TSA Plus Cy3 and Fluorescein Kits (PerkinElmer Life Sciences). For single-molecule fluorescence in situ hybridization (smiFISH), embryos were fixed in fresh 4% formaldehyde (17) and then washed in 1× PBS with 0.1% Tween (PBST). PBST was replaced with 50% wash buffer (2× SSC, 10% formamide) diluted in 1× PBST for 5 to 10 min. Embryos were quickly washed in 100% wash buffer, followed by two washes at 37 °C for 30 min. Samples were incubated in probe mixture in Hyb buffer (2× SSC, 10% formamide, 1 g/10 mL dextran sulfate) at 37 °C overnight. Probe mixture was removed and the embryos were washed for 1 min with prewarmed (37 °C) wash buffer, followed by four washes in wash buffer at 37 °C for 15 min each, and finally washed in 1× PBST. Twenty-nucleotide probes were designed against the zebrafish odf3b and cdh17 mRNA transcripts with the Stellaris Probe Designer, version 4.2 (Biosearch Technologies). A FLAP sequence (5′-CCTCCTAAGTTTCGAGCTGGACTCAGTG-3′) was added to the 5′ end of each individual probe. The sequences were synthesized (Invitrogen, Integrated DNA Technologies) in a 96-well plate, and 100 μM working stocks were stored at −20 °C. Individual probes for odf3b were combined to make an odf3b probe mix, and cdh17 probes were combined to make a cdh17 probe mix. The probe mixes were annealed to 5 μM Quasar 670 FLAP (odf3b) or Quasar 570 FLAP (cdh17) at 85 °C for 3 min, 65 °C for 3 min, and 25 °C for 5 min. The final probe/FLAP mixture was added to embryos at 240 nM in Hyb buffer. Embryos were mounted (17) and imaged on a Leica SP8 confocal microscope. For imaging cilia and basal bodies, FISH was combined with IF with previously reported antibodies (17). For FISH and IF data analysis, embryos were placed in Aqua-Poly/Mount and imaged on a Nikon C2 confocal microscope. Z stacks were processed with Fiji (https://fiji.sc/) into maximum image projections, and all figures were assembled using Adobe Photoshop CS5.
Gene Knockdown.
Antisense morpholino oligonucleotides were purchased from Gene Tools and solubilized in DNase/RNase–free water to create 4 mM stocks and stored at −20 °C. The sequences and dosages used were cox1 5′-TCAGCAAAAAGTTACACTCTCTCAT-3′, 0.4 mM (10, 12); cox2 5′-AACCAGTTTATTCATTCCAGAAGTG-3′, 0.8 mM (10, 12); and etv5a 5′-ATACATTAGGGAGTACCTGTAGCTG-3′, 0.22 mM (9, 28).
Genotyping of etv5asa16031 Allele.
Genomic DNA was isolated from individual adult fins or embryos as described (23). The PCR amplification was performed using the following primers that flank the mutation site: 5′-GTGGCTCGACGCTGGGGCATCCAGAAGAACAGACCCGCCATGAACTACGACAAGCTGAGT-3′ and 5′-CATGGAGAAGAGCGCTTCAGGATCGCACACAAATTTGTAGACGTACCTCTCGCCGGCAAC-3′. The cycling conditions were a 4-min denaturation step at 94 °C, 45 cycles of 94 °C for 30 s, annealing for 30 s at 68 °C, and 72 °C for 1 min, and a final extension step at 72 °C for 10 min. PCR products were purified using a Qiagen PCR Purification Kit and sequenced.
Quantification and Statistical Analysis.
Quantification of cell number and absolute expression domain length were performed as described (9) and in a blinded fashion to eliminate bias. For FISH colocalization studies, the signal intensity was normalized against the background signal in the Nikon software. The nuclear area of pronephros MCCs was outlined in Photoshop or Fiji, and then scored for the presence or absence of each respective marker in the appropriate channels. Cilia and basal body quantification was done on maximum image projections from a Nikon C2 confocal microscope at 60× magnification. Z stacks were processed in Fiji, and composites of different channel combinations were saved. Using the odf3b/DAPI composite for each image, odf3b+ cells were outlined on a new layer in Photoshop. The new layer was then duplicated and copied to the antiacetylated α/γ-tubulin composite to quantify basal body number in odf3b+ cells. For cilia quantification, the freehand line tool in Fiji was used to trace the entire cilia bundle on the antiacetylated α-tubulin channel, and then the fluorescence intensity for points along this line was obtained. The fluorescence intensity was averaged across the different embryo images for each experimental condition and displayed in both an intensity plot and a dot plot. For the intensity plot, a threshold was determined by subtracting the lowest point of intensity from the highest point of intensity for the control. All experiments were completed in biological triplicate, with group sizes of at least n = 10 embryos. Confocal imaging was performed with group sizes of at least n = 3 representative embryos, with at least one representative embryo from each biological set, but typically two to five samples per replicate were imaged and quantified and, as with our other studies, in a blinded fashion to minimize bias. ANOVA and Student’s t tests were used to assess statistical significance. GraphPad Prism software was used to perform statistics and generate graphical depictions. A detailed catalog of all statistical values is provided in Dataset S2.
Supplementary Material
Acknowledgments
We thank the Gallagher family for their generous support of stem cell research. We thank the Department of Biological Sciences for their dedicated assistance, and all staff of the Center for Zebrafish Research for the care of our aquarium. We especially thank our lab and members of the Smith lab for helpful discussions, critiques, and insights about this work. Funding was provided by NIH Grant R01DK100237 (to R.A.W.), NSF Graduate Research Fellowships (to A.N.M. and B.E.D.), and the Marine Biological Laboratory.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813492116/-/DCSupplemental.
References
- 1.Spassky N, Meunier A. The development and functions of multiciliated epithelia. Nat Rev Mol Cell Biol. 2017;18:423–436. doi: 10.1038/nrm.2017.21. [DOI] [PubMed] [Google Scholar]
- 2.Ménard D. Morphological studies of the developing human esophageal epithelium. Microsc Res Tech. 1995;31:215–225. doi: 10.1002/jemt.1070310305. [DOI] [PubMed] [Google Scholar]
- 3.Zimmermann HD. Cilia in the fetal kidney of man. Beitr Pathol. 1971;143:227–240. German. [PubMed] [Google Scholar]
- 4.Katz SM, Morgan JJ. Cilia in the human kidney. Ultrastruct Pathol. 1984;6:285–294. doi: 10.3109/01913128409018587. [DOI] [PubMed] [Google Scholar]
- 5.Marra AN, Li Y, Wingert RA. Antennas of organ morphogenesis: The roles of cilia in vertebrate kidney development. Genesis. 2016;54:457–469. doi: 10.1002/dvg.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poureetezadi SJ, Wingert RA. Little fish, big catch: Zebrafish as a model for kidney disease. Kidney Int. 2016;89:1204–1210. doi: 10.1016/j.kint.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: Insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip Rev Dev Biol. 2013;2:559–585. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wingert RA, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:1922–1938. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marra AN, Wingert RA. Epithelial cell fate in the nephron tubule is mediated by the ETS transcription factors etv5a and etv4 during zebrafish kidney development. Dev Biol. 2016;411:231–245. doi: 10.1016/j.ydbio.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.North TE, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nissim S, et al. Prostaglandin E2 regulates liver versus pancreas cell-fate decisions and endodermal outgrowth. Dev Cell. 2014;28:423–437. doi: 10.1016/j.devcel.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poureetezadi SJ, Cheng CN, Chambers JM, Drummond BE, Wingert RA. Prostaglandin signaling regulates nephron segment patterning of renal progenitors during zebrafish kidney development. eLife. 2016;5:e17551. doi: 10.7554/eLife.17551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tootle TL. Genetic insights into the in vivo functions of prostaglandin signaling. Int J Biochem Cell Biol. 2013;45:1629–1632. doi: 10.1016/j.biocel.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Jin D, et al. Prostaglandin signalling regulates ciliogenesis by modulating intraflagellar transport. Nat Cell Biol. 2014;16:841–851. doi: 10.1038/ncb3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerlach GF, Wingert RA. Zebrafish pronephros tubulogenesis and epithelial identity maintenance are reliant on the polarity proteins Prkc iota and zeta. Dev Biol. 2014;396:183–200. doi: 10.1016/j.ydbio.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKee R, Gerlach GF, Jou J, Cheng CN, Wingert RA. Temporal and spatial expression of tight junction genes during zebrafish pronephros development. Gene Expr Patterns. 2014;16:104–113. doi: 10.1016/j.gep.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marra AN, Ulrich M, White A, Springer M, Wingert RA. Visualizing multiciliated cells in the zebrafish through a combined protocol of whole mount fluorescent in situ hybridization and immunofluorescence. J Vis Exp. 2017;129:e56261. doi: 10.3791/56261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poureetezadi SJ, Donahue EK, Wingert RA. A manual small molecule screen approaching high-throughput using zebrafish embryos. J Vis Exp. 2014;93:e52063. doi: 10.3791/52063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cha YI, Kim SH, Solnica-Krezel L, Dubois RN. Cyclooxygenase-1 signaling is required for vascular tube formation during development. Dev Biol. 2005;282:274–283. doi: 10.1016/j.ydbio.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Cha YI, et al. Cyclooxygenase-1-derived PGE2 promotes cell motility via the G-protein-coupled EP4 receptor during vertebrate gastrulation. Genes Dev. 2006;20:77–86. doi: 10.1101/gad.1374506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Znosko WA, et al. Overlapping functions of Pea3 ETS transcription factors in FGF signaling during zebrafish development. Dev Biol. 2010;342:11–25. doi: 10.1016/j.ydbio.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Cheng CN, Verdun VA, Wingert RA. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling. Dev Biol. 2014;386:111–122. doi: 10.1016/j.ydbio.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kroeger PT, Jr, et al. The zebrafish kidney mutant zeppelin reveals that brca2/fancd1 is essential for pronephros development. Dev Biol. 2017;428:148–163. doi: 10.1016/j.ydbio.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M. Characterization of three novel members of the zebrafish Pax2/5/8 family: Dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063–3074. doi: 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- 25.Ma M, Jiang YJ. Jagged2a-Notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS Genet. 2007;3:e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Pathak N, Kramer-Zucker A, Drummond IA. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134:1111–1122. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, et al. miR-34b regulates multiciliogenesis during organ formation in zebrafish. Development. 2013;140:2755–2764. doi: 10.1242/dev.092825. [DOI] [PubMed] [Google Scholar]
- 28.Chen SY, et al. Etv5a regulates the proliferation of ventral mesoderm cells and the formation of hemato-vascular derivatives. J Cell Sci. 2013;126:5626–5634. doi: 10.1242/jcs.132613. [DOI] [PubMed] [Google Scholar]
- 29.Lu BC, et al. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat Genet. 2009;41:1295–1302. doi: 10.1038/ng.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuure S, Chi X, Lu B, Costantini F. The transcription factors Etv4 and Etv5 mediate formation of the ureteric bud tip domain during kidney development. Development. 2010;137:1975–1979. doi: 10.1242/dev.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mao J, McGlinn E, Huang P, Tabin CJ, McMahon AP. Fgf-dependent Etv4/5 activity is required for posterior restriction of Sonic Hedgehog and promoting outgrowth of the vertebrate limb. Dev Cell. 2009;16:600–606. doi: 10.1016/j.devcel.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fontanet PA, Ríos AS, Alsina FC, Paratcha G, Ledda F. Pea3 transcription factors, Etv4 and Etv5, are required for proper hippocampal dendrite development and plasticity. Cereb Cortex. 2018;28:236–249. doi: 10.1093/cercor/bhw372. [DOI] [PubMed] [Google Scholar]
- 33.Schlesser HN, et al. Effects of ETV5 (ets variant gene 5) on testis and body growth, time course of spermatogonial stem cell loss, and fertility in mice. Biol Reprod. 2008;78:483–489. doi: 10.1095/biolreprod.107.062935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, et al. Transcription factor Etv5 is essential for the maintenance of alveolar type II cells. Proc Natl Acad Sci USA. 2017;114:3903–3908. doi: 10.1073/pnas.1621177114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eo J, Han K, Murphy KM, Song H, Lim HJ. Etv5, an ETS transcription factor, is expressed in granulosa and cumulus cells and serves as a transcriptional regulator of the cyclooxygenase-2. J Endocrinol. 2008;198:281–290. doi: 10.1677/JOE-08-0142. [DOI] [PubMed] [Google Scholar]
- 36.Busch-Nentwich E, et al. 2013 Sanger Institute zebrafish mutation project mutant data submission. ZFIN direct data submission: sa16031 mutant allele. Available at https://zfin.org/ZDB-ALT-130411-4554. Accessed September 20, 2018.
- 37.Brooks ER, Wallingford JB. Multiciliated cells. Curr Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meunier A, Azimzadeh J. Multiciliated cells in animals. Cold Spring Harb Perspect Biol. 2016;8:a028233. doi: 10.1101/cshperspect.a028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsao PN, et al. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136:2297–2307. doi: 10.1242/dev.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou F, et al. Gmnc is a master regulator of the multiciliated cell differentiation program. Curr Biol. 2015;25:3267–3273. doi: 10.1016/j.cub.2015.10.062. [DOI] [PubMed] [Google Scholar]
- 41.Tan FE, et al. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development. 2013;140:4277–4286. doi: 10.1242/dev.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gegg M, et al. Flattop regulates basal body docking and positioning in mono- and multiciliated cells. eLife. 2014;3:e03842. doi: 10.7554/eLife.03842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chung MI, et al. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife. 2014;3:e01439. doi: 10.7554/eLife.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choksi SP, Babu D, Lau D, Yu X, Roy S. Systematic discovery of novel ciliary genes through functional genomics in the zebrafish. Development. 2014;141:3410–3419. doi: 10.1242/dev.108209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tu F, Sedzinski J, Ma Y, Marcotte EM, Wallingford JB. Protein localization screening in vivo reveals novel regulators of multiciliated cell development and function. J Cell Sci. 2018;131:jcs206565. doi: 10.1242/jcs.206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stubbs JL, Vladar EK, Axelrod JD, Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng CN, Wingert RA. Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a transcription factor and retinoic acid in zebrafish. Dev Biol. 2015;399:100–116. doi: 10.1016/j.ydbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chambers JM, Poureetezadi SJ, Addiego A, Lahne M, Wingert RA. ppargc1a controls nephron segmentation during zebrafish embryonic kidney ontogeny. eLife. 2018;7:e40266. doi: 10.7554/eLife.40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development. 1999;126:4715–4728. doi: 10.1242/dev.126.21.4715. [DOI] [PubMed] [Google Scholar]
- 50.Lu H, et al. Mutations in DZIP1L, which encodes a ciliary-transition-zone protein, cause autosomal recessive polycystic kidney disease. Nat Genet. 2017;49:1025–1034. doi: 10.1038/ng.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duffy JL, Suzuki Y. Ciliated human renal proximal tubular cells. Observations in three cases of hypercalcemia. Am J Pathol. 1968;53:609–616. [PMC free article] [PubMed] [Google Scholar]
- 52.Datsis SA, Boman IA. Ciliated renal tubular epithelium in congenital nephrosis. Beitr Pathol. 1974;151:297–303. doi: 10.1016/s0005-8165(74)80006-5. [DOI] [PubMed] [Google Scholar]
- 53.Larsen TE, Ghadially FN. Cilia in lupus nephritis. J Pathol. 1974;114:69–73. doi: 10.1002/path.1711140203. [DOI] [PubMed] [Google Scholar]
- 54.Lungarella G, de Santi MM, Tosi P. Ultrastructural study of the ciliated cells from renal tubular epithelium in acute progressive glomerulonephritis. Ultrastruct Pathol. 1984;6:1–7. doi: 10.3109/01913128409016659. [DOI] [PubMed] [Google Scholar]
- 55.Ong AC, Wagner B. Detection of proximal tubular motile cilia in a patient with renal sarcoidosis associated with hypercalcemia. Am J Kidney Dis. 2005;45:1096–1099. doi: 10.1053/j.ajkd.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 56.Johnson CS, Holzemer NF, Wingert RA. Laser ablation of the zebrafish pronephros to study renal epithelial regeneration. J Vis Exp. 2011;54:e2845. doi: 10.3791/2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Palmyre A, et al. Collective epithelial migration drives kidney repair after acute injury. PLoS One. 2014;9:e101304. doi: 10.1371/journal.pone.0101304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou W, Boucher RC, Bollig F, Englert C, Hildebrandt F. Characterization of mesonephric development and regeneration using transgenic zebrafish. Am J Physiol Renal Physiol. 2010;299:F1040–F1047. doi: 10.1152/ajprenal.00394.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diep CQ, et al. Identification of adult nephron progenitors capable of kidney regeneration in zebrafish. Nature. 2011;470:95–100. doi: 10.1038/nature09669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kroeger PT, Jr, Wingert RA. Using zebrafish to study podocyte genesis during kidney development and regeneration. Genesis. 2014;52:771–792. doi: 10.1002/dvg.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCampbell KK, Springer KN, Wingert RA. Atlas of cellular dynamics during zebrafish adult kidney regeneration. Stem Cells Int. 2015;2015:547636. doi: 10.1155/2015/547636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yakulov TA, et al. CXCL12 and MYC control energy metabolism to support adaptive responses after kidney injury. Nat Commun. 2018;9:3660. doi: 10.1038/s41467-018-06094-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed Natl Acad Press; Washington, DC: 2011. [Google Scholar]
- 64.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 65.Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Combinatorial regulation of novel erythroid gene expression in zebrafish. Exp Hematol. 2008;36:424–432. doi: 10.1016/j.exphem.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cheng CN, Li Y, Marra AN, Verdun V, Wingert RA. Flat mount preparation for observation and analysis of zebrafish embryo specimens stained by whole mount in situ hybridization. J Vis Exp. 2014;89:e51604. doi: 10.3791/51604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wingert RA, Davidson AJ. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev Dyn. 2011;240:2011–2027. doi: 10.1002/dvdy.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lengerke C, et al. Interactions between Cdx genes and retinoic acid modulate early cardiogenesis. Dev Biol. 2011;354:134–142. doi: 10.1016/j.ydbio.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brend T, Holley SA. Zebrafish whole mount high-resolution double fluorescent in situ hybridization. J Vis Exp. 2009;25:1229. doi: 10.3791/1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Drummond BE, Li Y, Marra AN, Cheng CN, Wingert RA. The tbx2a/b transcription factors direct pronephros segmentation and corpuscle of Stannius formation in zebrafish. Dev Biol. 2017;421:52–66. doi: 10.1016/j.ydbio.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.