Significance
RNA and protein molecules interact to perform translation, splicing, and other fundamental processes. These interactions are defined by their strength and specificity, but it remains infeasible to experimentally measure these properties for all biologically important RNA–protein complexes. Development of computational strategies for calculating RNA–protein energetics has been hindered by unique complexities of RNA–protein binding, particularly the propensity of RNA to adopt multiple conformations in the unbound state. We describe a method, Rosetta-Vienna RNP-ΔΔG, combining 3D structure modeling with RNA secondary structure-based energetic calculations to predict RNA–protein relative binding affinities. For several diverse systems and in rigorous blind tests, the accuracy of Rosetta-Vienna RNP-ΔΔG compared with experimental measurements is significantly better than that of prior approaches.
Keywords: RNA–protein complex, conformational change, binding affinity, blind prediction, energetic prediction
Abstract
Interactions between RNA and proteins are pervasive in biology, driving fundamental processes such as protein translation and participating in the regulation of gene expression. Modeling the energies of RNA–protein interactions is therefore critical for understanding and repurposing living systems but has been hindered by complexities unique to RNA–protein binding. Here, we bring together several advances to complete a calculation framework for RNA–protein binding affinities, including a unified free energy function for bound complexes, automated Rosetta modeling of mutations, and use of secondary structure-based energetic calculations to model unbound RNA states. The resulting Rosetta-Vienna RNP-ΔΔG method achieves root-mean-squared errors (RMSEs) of 1.3 kcal/mol on high-throughput MS2 coat protein–RNA measurements and 1.5 kcal/mol on an independent test set involving the signal recognition particle, human U1A, PUM1, and FOX-1. As a stringent test, the method achieves RMSE accuracy of 1.4 kcal/mol in blind predictions of hundreds of human PUM2–RNA relative binding affinities. Overall, these RMSE accuracies are significantly better than those attained by prior structure-based approaches applied to the same systems. Importantly, Rosetta-Vienna RNP-ΔΔG establishes a framework for further improvements in modeling RNA–protein binding that can be tested by prospective high-throughput measurements on new systems.
RNA binding proteins (RBPs) affect nearly all aspects of RNA biology, including alternative splicing, localization, translation, and stability (1, 2), and novel RNA–protein biophysical phenomena, ranging from in vivo phase separations to helicase-induced rearrangements, are being discovered at a rapid pace (3, 4). The function of an RBP depends on its ability to identify a specific target RNA sequence and structure, a process governed by the energetics of the interactions between each RNA and every RBP in its biological milieu (5). Recently developed high-throughput experimental methods have been used to quantitatively characterize the binding landscapes of a handful of RBPs (6–11), improving our understanding of the relationship between RNA sequence, structure, and binding affinity. However, these empirically derived landscapes are limited to specific systems with solubilities and affinities within the concentration windows accessible to these methods. To understand RNA–protein systems inaccessible to experimental characterization and to rationally design new RNA–protein interactions, a general physical model is needed to predict RNA–protein binding energies. Physical models have proved useful for predicting changes in binding free energies (ΔΔG) for macromolecular interactions that do not involve RNA, including protein–protein, protein–small molecule, and protein–DNA interactions (12–16). The best methods for these other macromolecular interactions report accuracies of between 1 and 2 kcal/mol and include rigorous blind studies, validating their use for applications that range from drug discovery to protein–protein interface design (14, 16, 17). However, the accuracy of these methods deteriorates when molecules are highly flexible or undergo large conformational changes (18, 19), factors common in RNA–protein binding events (20, 21). Accurate prediction of RNA–protein binding affinities is therefore challenging, and a complete prediction framework for RNA–protein complexes has yet to be developed and systematically tested.
Existing computational methods that attempt to quantitatively predict relative RNA–protein binding affinities have achieved limited success, likely as a result of neglecting key features of the binding process such as intramolecular interactions and the unbound states. A previously developed method to predict relative RNA–protein binding affinities from a database-derived statistical potential produced significant correlations with experimental measurements for protein mutants, but for RNA mutants the calculations exhibited no detectable correlation with experiment (22). Another approach used molecular dynamics simulations in combination with a nonlinear Poisson Boltzmann model and linear response approximation. Despite the complexity of the method and computation exerted on a single model system, statistical uncertainties in relative binding affinity calculations were reported to be 1 to 3 kcal/mol (23). More recently, a machine-learning method, GLM-Score, developed to predict absolute nucleic acid–protein binding affinities from structures of bound complexes reported excellent accuracies (R2 = 0.75), but has not been tested in its ability to predict relative binding affinities on independent RNA–protein complexes (14).
The propensity of RNA to adopt multiple stable conformations in the unbound state makes it problematic to predict binding affinities with standard approaches that typically neglect the unbound state altogether. However, RNA is also distinctive from other molecules in that its unbound energetics can be predicted from a simple secondary structure-based model derived from dozens of optical melting experiments (24–26). One straightforward but previously untested solution for the treatment of unbound RNA energetics in RNA–protein binding affinity calculations is to use these secondary structure-based calculations. RNA secondary and tertiary structure modeling are commonly integrated to increase the accuracy of RNA 3D structure prediction (27), but this combination has yet to be tested for quantitatively predicting binding affinities.
Here we present a complete structure-based computational framework, Rosetta-Vienna RNP-ΔΔG, for predicting RNA–protein relative binding affinities, bringing together secondary structure-based energetic calculations of unbound RNA free energies and a unified energy function for bound RNA–protein complexes. Rosetta-Vienna RNP-ΔΔG achieves root-mean-squared errors (RMSEs) of 1.3 kcal/mol on a dataset of binding affinities of the MS2 coat protein with thousands of variants of its partner RNA hairpin (6) and 1.5 kcal/mol on a diverse, independent set of RNA–protein complexes. Additionally, we rigorously evaluated the accuracy of the method through a blind challenge that involved making predictions and separately measuring binding affinities of the human PUF family protein PUM2 with hundreds of RNA sequences using the high-throughput RNA MaP technology (28). On all tests, the prediction accuracy of Rosetta-Vienna RNP-ΔΔG appreciably exceeds that of previous structure-based energetic calculation methods and approaches the kilocalorie-per-mole accuracy seen for protein–protein and other well-studied complexes. Rosetta-Vienna RNP-ΔΔG establishes a framework for using high-throughput experimental data that will likely continue to be collected in the next several years to test further improvements in modeling RNA–protein binding.
Results
Rosetta-Vienna RNP-ΔΔG: A Framework for RNA–Protein Relative Binding Affinity Calculation.
Before developing a framework for calculating relative RNA–protein binding affinities, we first tested a previously published structure-based machine learning method, GLM-Score (14). This method was specifically developed to calculate absolute binding affinities for which it was reported to achieve strong correlation with experimental measurements (R2 = 0.75). This method has not, however, been systematically tested in its ability to calculate relative RNA–protein binding affinities (i.e., affinity predictions have not been made for multiple mutations of a single system). As an initial test, we used the MS2 coat protein system because experimental binding affinities of MS2 for thousands of RNA mutants have recently been measured (6) and crystal structures of the protein with several mutant RNA hairpins have been determined (29). These crystal structures are between 0.37 and 0.87 Å rmsd of each other (SI Appendix, Fig. S1A), suggesting that structures of the mutants are highly similar to the wild-type structure. We then used the wild-type MS2 coat protein–RNA hairpin crystal structure as a template to generate complex structures (Fig. 1B) for 74 RNA mutants that preserve the wild-type RNA secondary structure (canonical mutants) and 660 mutants that introduce a single noncanonical base pair into the RNA hairpin (single noncanonical mutants; Methods). The GLM-Score calculations exhibited RMSEs of 2.52 kcal/mol for the canonical mutants (R2 = 0.04) and 2.35 kcal/mol for the single noncanonical mutants (R2 = 0.07; Fig. 2B). We reasoned that the substantially worse accuracy of the relative binding affinity calculations compared with the previously reported absolute binding affinity calculations may be due to the fact that GLM-Score neglects intramolecular interactions and does not model the unbound states, factors which can play a large role in determining the relative favorability of RNA binding. We did not test other structure-based methods that have previously been shown to have poor accuracy for predicting relative RNA–protein binding affinities (22, 23).
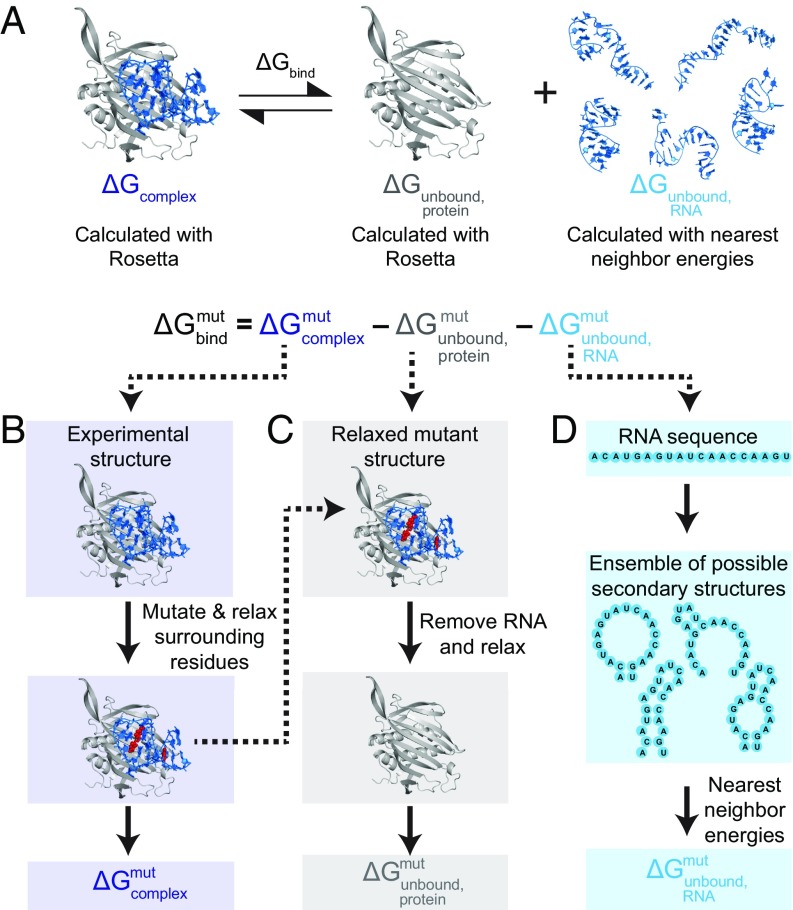
Fig. 1.
Rosetta-Vienna RNP-ΔΔG framework for calculating relative binding affinities for RNA–protein complexes. (A) Schematic overview of relative binding affinity calculation (RNA colored blue, protein colored gray, MS2 coat protein/RNA hairpin complex shown here). The right panel depicts the conformational ensemble of the unbound RNA. All free energies were calculated at standard state. (B) Free energies of the complex (ΔGcomplex) are calculated by first relaxing the wild-type experimental structure in Rosetta to generate an ensemble of 100 highly similar conformations. The 20 lowest-scoring conformations are then mutated (mutated residues shown as red spheres) as directed by a user-inputted list of mutants then relaxed and scored. ΔGmutcomplex is the average of these 20 scores. (C) ΔGmutunbound protein is calculated by removing the RNA from the mutant structure, then relaxing and scoring the protein structure in Rosetta. (D) The free energy of the unbound RNA (ΔGmutunbound RNA) is calculated as the Boltzmann sum over all possible nonpseudoknotted secondary structures, with energies calculated using the nearest-neighbor energy model in Vienna RNA. The reference states for each of the components ΔGmutcomplex, ΔGmutunbound protein, and ΔGmutunbound RNA are fully unfolded states of the macromolecules, as assumed in Rosetta and Vienna RNA packages.
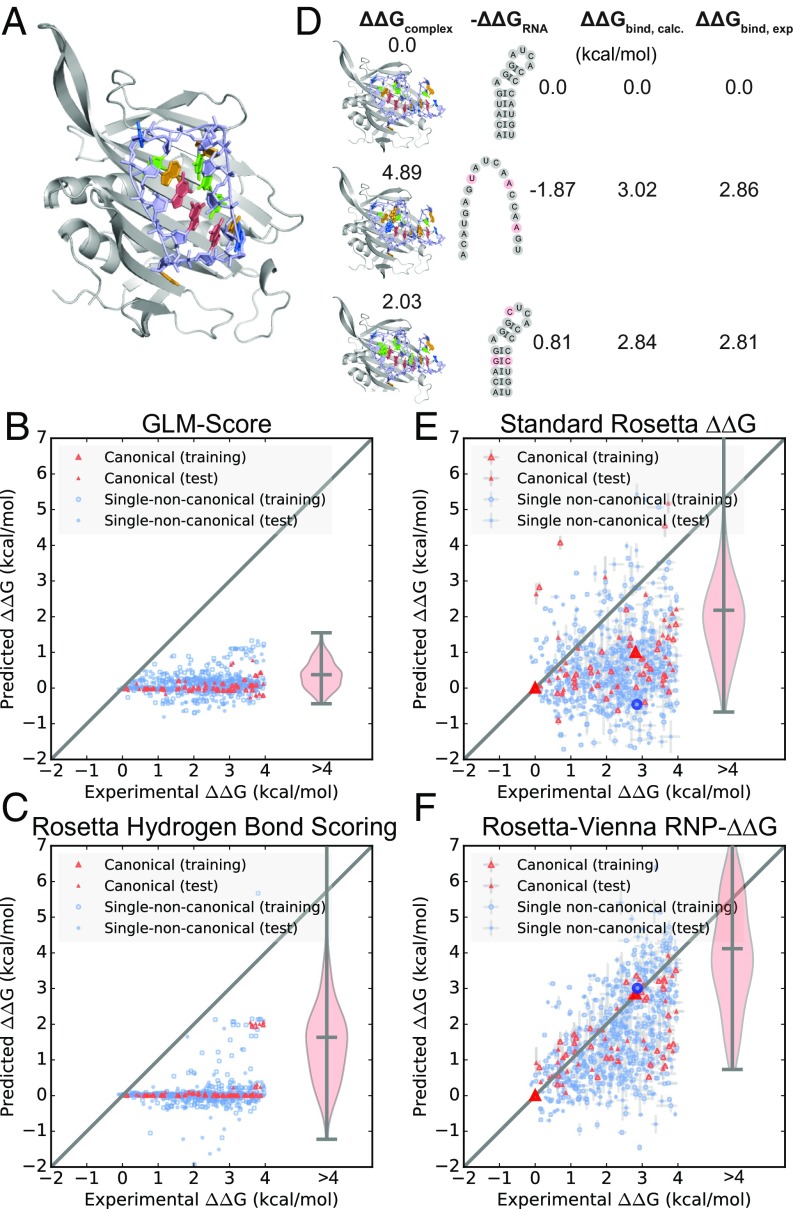
Fig. 2.
Calculation of relative RNA–protein binding affinities for 734 mutants of the MS2 coat protein RNA hairpin. (A) Crystal structure of the MS2 coat protein–RNA hairpin complex [Protein Data Bank (PDB) ID code 1ZDH]. Experimental ΔΔG versus ΔΔG calculations for canonical and single-noncanonical RNA hairpin mutants using (B) GLM-Score and (C) the Rosetta hydrogen bond scoring model. Note that the reference states for ΔGcomplex, ΔGunbound RNA, and ΔGunbound protein are the fully unfolded states and that ΔΔG refers to the free energy of a mutant relative to wild type. Positive ΔΔG indicates a mutant with a free energy that is less stable than the wild type, while negative ΔΔG indicates a mutant with more stability than the wild type. (D) Decomposition of the binding affinity calculation for the wild-type complex (top row) and two example mutants (-12G,-7C,0C mutant, middle row; -8U,-3A,1A mutant, bottom row). The first column shows the mutated complex structures, with mutated residues shown with spheres, and calculated values. The second column shows calculated values and the predicted secondary structures of the unbound RNA, with mutated residues colored red, the third column shows the final calculated , and the fourth column shows the experimental values. (E) Calculations for RNA hairpin mutants using the standard Rosetta ΔΔG approach for unbound RNA free energies. (F) Calculations for the same RNA hairpin mutants with Rosetta-Vienna RNP-ΔΔG, in which unbound RNA free energies were computed from the partition function of all possible secondary structures. The violin plots show calculations for the canonical hairpins that have experimental relative binding free energies greater than 4 kcal/mol. Pearson correlation coefficients for all calculations are given in SI Appendix, Table S3.
To further assess whether a simple model that considers only intermolecular interactions is insufficient to accurately model relative binding affinities, we tested a simple hydrogen bond scoring model in Rosetta (30). This model considers only the hydrogen bonding scores between the RNA and the protein in the bound complexes (Methods). This is not intended to be a test of the Rosetta hydrogen bonding score term but rather a test of a simple calculation framework that considers only a single type of intermolecular interaction. For the MS2 canonical and single noncanonical mutants, these calculations gave RMSEs of 2.31 kcal/mol and 2.44 kcal/mol, respectively (Fig. 2C). These calculations are less accurate than the GLM-Score calculations of absolute binding affinities and are outside the 1 to 2 kcal/mol accuracy range that methods for other macromolecular systems have achieved.
Motivated by these results, we sought to develop a more complete framework for relative RNA–protein binding affinity calculations that includes the effects of mutations in the unbound ensembles and treatment of intramolecular interactions. Our method takes as input a structure of an RNA–protein complex and predicts the change in binding affinity that would result from mutating RNA or protein residues in the complex. Briefly, structures of the mutant complexes were generated from the input structure by computationally mutating specified residues and then allowing neighboring residues to relax in response to the mutation in Rosetta (Fig. 1; see Methods for additional details). The free energy of this complex was then approximated with an all-atom energy function that includes terms describing hydrogen bonding, electrostatics, torsional energy, van der Waals interactions, and solvation (31). Details are provided in SI Appendix. The free energy of the unbound protein was similarly calculated from the protein structure in the absence of the RNA (Fig. 1C), while the unbound RNA free energy was calculated either in Rosetta from the RNA structure in the absence of the protein (similar to conventional prediction schemes) or from a secondary structure ensemble-based method (Fig. 1D; discussed below). Relative binding affinity was then computed as the difference in bound and unbound energies relative to this difference for the initial input structure.
Because unbound RNA is highly flexible and often exists in heterogeneous conformational ensembles, we hypothesized that the treatment of unbound RNA energetics would substantially impact the accuracy of our relative binding affinity calculations. We therefore tested two different methods for calculating these free energies. First, unbound RNA free energies were calculated using a 3D structure-based approach that is standard in protein–protein and protein–small molecule relative binding affinity calculations (18, 32). The structure of each partner RNA was taken from the RNA–protein complex and relaxed then scored in the absence of the protein. We hypothesized that this “standard Rosetta ΔΔG” 3D structure-based approach, in which a single 3D structure is used to represent the unbound conformational ensemble, may not work as well for RNA as it does for proteins due to the relative flexibility of RNA in unbound states, and because of the likelihood of mutations to introduce alternative secondary structures. Our second approach, which we called “Rosetta-Vienna RNP-ΔΔG,” was to instead calculate unbound RNA free energies using partition function calculations enumerating all possible unbound secondary structures. These calculations used the nearest-neighbor energy model as encoded in the ViennaRNA package, frequently used to predict RNA secondary structure (24, 25). The nearest-neighbor energy model offers the potential of efficiently capturing the effects of numerous unbound structures whose component free energies have been probed and tested in dozens of empirical studies. The applicability of Rosetta-Vienna RNP-ΔΔG for the unbound RNA state is highlighted by two illustrative examples that show how changes in binding affinity are determined by both stability of the complex and the stability of the unbound RNA (Fig. 2D and SI Appendix).
Over all of the measurements, the Rosetta-Vienna RNP-ΔΔG model significantly outperformed the standard Rosetta ΔΔG approach for calculating unbound RNA free energies (Fig. 2 E and F and SI Appendix, Tables S1–S3). For the 74 canonical mutants and 660 single noncanonical mutants, RMSEs for calculations made with the Rosetta-Vienna RNP-ΔΔG model, which uses a secondary structure ensemble model to calculate unbound RNA free energies, were 1.11 and 1.28 kcal/mol, respectively, compared with 2.03 and 2.15 kcal/mol, respectively, for calculations performed with the standard Rosetta ΔΔG approach, which uses a single 3D structure to represent the unbound RNA conformational ensemble. These Rosetta-Vienna RNP-ΔΔG correlations were statistically significant (P < 0.01; SI Appendix, Table S3). Additionally, calculations that included both ΔΔGcomplex and ΔΔGunbound RNA were more accurate than either term alone (SI Appendix).
Calculations Across Additional Diverse RNA–Protein Systems.
To assess the accuracy and applicability of Rosetta-Vienna RNP-ΔΔG for systems other than the MS2 coat protein, we calculated relative binding affinities for four additional diverse RNA–protein systems: a conserved component of the signal recognition particle (SRP) (Fig. 3A); the A protein of the human U1 snRNP (U1A), a widely studied model system for RNA recognition motifs (Fig. 3C); Pumilio homolog 1 (PUM1), a single-stranded RBP made up of eight repeats that each specifically recognize a single nucleotide (Fig. 3B); and FOX-1 (Fig. 3D), which recognizes a five-nucleotide single-stranded consensus sequence. These systems were chosen because experimental structures and quantitative affinities for protein mutants and for RNA mutants are available in the literature for each of these systems (33–38). We expected that the ΔΔG calculations for these systems would be less accurate than those for the MS2 system because we did not train any part of the method using these systems (discussed further in SI Appendix).
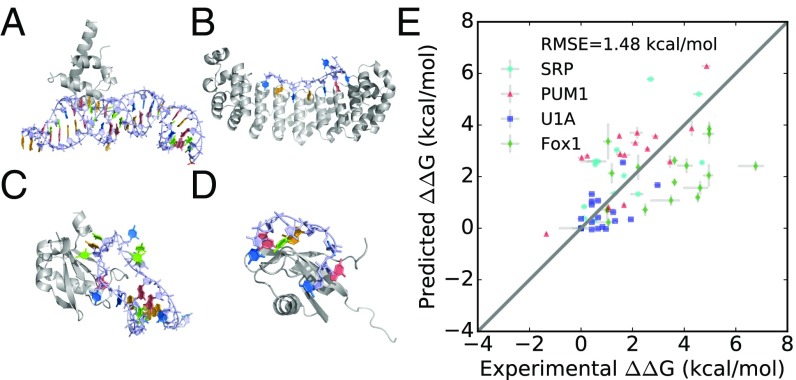
Fig. 3.
Recovery of relative binding affinities in independent RNA–protein systems. Structures of (A) a conserved component of SRP (PDB ID code 1HQ1) (33), (B) PUM1 (PDB ID code 1M8W) (37), (C) U1A (PDB ID code 1URN) (38), and (D) FOX-1 (PDB ID code 2ERR) (35). (E) Calculations of ΔΔGbind using Rosetta-Vienna RNP-ΔΔG.
We first calculated relative binding affinities for these four systems using GLM-Score, the Rosetta hydrogen bond scoring model, and the standard Rosetta ΔΔG approach described previously. The RMSEs for the GLM-Score calculations ranged from 1.09 to 4.47 kcal/mol, the hydrogen bond scoring model RMSEs ranged from 0.82 to 3.28 kcal/mol, and the standard Rosetta ΔΔG method gave RMSEs ranging from 0.89 to 4.17 kcal/mol across the four systems (SI Appendix, Fig. S3 and Tables S1 and S3).
The Rosetta-Vienna RNP-ΔΔG method gave significantly better accuracies, recovering relative binding affinities with RMSE accuracies of 1.47 kcal/mol for the 14 SRP mutants, 0.75 kcal/mol for the 19 U1A mutants, 1.35 kcal/mol for the 17 PUM1 mutants, and 2.13 kcal/mol for the 17 FOX-1 mutants. These RMSE accuracies are better than all prior approaches for each of the four tested systems. Additionally, the correlations between Rosetta-Vienna RNP-ΔΔG calculations and experimentally measured values were statistically significant (P < 0.01; SI Appendix, Table S3). The RMSE over all systems was 1.48 kcal/mol, with overall protein mutant RMSE of 1.21 kcal/mol (36 sequences) and overall RNA mutant RMSE of 1.73 kcal/mol (37 sequences) (Fig. 3E and Table 1). Decomposing the calculations into the contributions from ΔΔGcomplex and ΔΔGunbound RNA suggested again that the combination of these terms provides the most accurate results, as expected from basic thermodynamic principles (Fig. 1 and SI Appendix, Table S2). These calculations suggest the range of accuracies that the Rosetta-Vienna RNP-ΔΔG method will give for arbitrary RNA–protein systems and suggest that the accuracy of these calculations will be worse for systems with flexible bound ensembles, like FOX-1. A modified framework that allowed significant structural changes in the RNA–protein complex upon mutation did not further improve the accuracy of the calculations (SI Appendix).
Table 1.
Modeling accuracies across multiple systems
| System | No. of data points | RNA length (no. of nucleotides) | Rosetta-Vienna RNP-ΔΔG RMSE, kcal/mol |
| MS2 canonical (training) | 37 | 19 | 1.22 |
| MS2 canonical (test) | 37 | 19 | 0.99 |
| MS2 single-noncanonical (training) | 330 | 19 | 1.27 |
| MS2 single-noncanonical (test) | 330 | 19 | 1.28 |
| PUM1 | 17 | 8 | 1.35 |
| SRP | 14 | 47 | 1.47 |
| U1A | 19 | 21 | 0.75 |
| FOX-1 | 17 | 7 | 2.13 |
| PUM2 (round 1) | 509 | 20–62 | 1.50* |
| PUM2, no fifth-position mutants (round 1) | 363 | 20–62 | 1.32* |
| PUM2 (round 2, blind) | 528 | 20–62 | 1.60 |
| PUM2, no fifth-positions mutants (round 2, blind) | 385 | 20–62 | 1.40 |
| Overall | 1,838 | 1.44 |
Pearson correlation coefficients are given in SI Appendix, Table S3.
These predictions were made using a PUM1 crystal structure in addition to the three PUM2 crystal structures. The RMSE accuracy of the initial blind predictions in round 1 was 1.94 kcal/mol and 1.47 kcal/mol without fifth-position mutants.
Blind Predictions of PUM2 Binding Affinities.
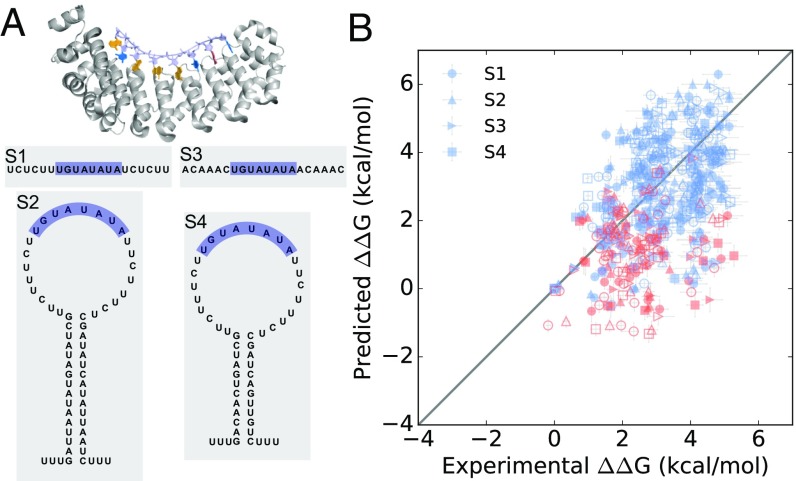
To evaluate the predictive power of this method, we made blind predictions of PUM2–RNA binding affinities. PUM2 is a single-stranded RBP that binds an eight-nucleotide consensus sequence (Fig. 4A). PUM2 is homologous to PUM1, which was included in our previous tests. However, at the time that these predictions were made, the detailed binding preferences of PUM1 versus PUM2 were unknown. Additionally, our blind tests included substantially more sequences than the previous tests on PUM1 (17 data points for previous PUM1 tests and >1,000 data points for PUM2 blind tests). To ensure that the tests were blind, the predictions and measurements were carried out separately by different authors (SI Appendix, Fig. S4). While K.K. made Rosetta-Vienna RNP-ΔΔG predictions, I.J. and P.P.V. used the high-throughput RNA MaP platform to independently measure binding affinities for PUM2 with single and double mutants of the consensus sequence in the context of four different scaffolds. In scaffolds S1 and S3 the PUM2 binding sequence was flanked by short single-stranded sequences, while in scaffolds S2 and S4 the PUM2 sequence was embedded inside long hairpin loops (Fig. 4A).
Fig. 4.
Blind predictions of PUM2 binding affinities. (A) The structure of PUM2 (PDB ID code 3Q0Q) and the four scaffold sequences in which the consensus sequence mutants were embedded. (B) Calculations of ΔΔGbind using Rosetta-Vienna RNP-ΔΔG. Round 1 predictions including a PUM1 structural template with U at the fifth position are shown as open symbols, and round 2 predictions are filled. Red and blue points indicate sequences with and without mutations at the fifth position, respectively.
Three crystal structures of PUM2 with RNA sequences differing at the positions of the fifth and eighth bases have previously been solved (39). There are large differences in the backbone conformations between these structures at the fifth position (SI Appendix, Fig. S1 B and C and Table S4). Because our prediction method does not account for such large backbone conformational changes between mutants within the bound complex, we used all three of the crystal structures to make the predictions (see Methods for details).
We unblinded the experimental data in two successive rounds, each containing data for approximately half of the single and double mutants of the consensus sequence, to allow potential modifications of the prediction procedure to be evaluated in a separate blind test (SI Appendix, Fig. S4). The RMSE for the first round of predictions for 509 sequences was 1.94 kcal/mol (SI Appendix, Fig. S5 and Table S5). This accuracy suggested that the method had some predictive power for the system and the correlation was statistically significant (P < 0.01), but the accuracy was poorer than the overall RMSE for the previously tested systems. Further analysis of the predictions revealed a common feature of most of the worst prediction outliers. Sequences with mutations at the fifth position, plotted with open markers in SI Appendix, Fig. S5, clustered off the line of equality; removing these variants gave RMSE accuracy of 1.47 kcal/mol for the remaining 363 sequences. We hypothesized that these deviations were the result of high backbone flexibility allowing different base orientations and interactions at the fifth position, an effect that our method would not capture. This hypothesis was supported by the different backbone conformations observed in the three crystal structures with A, C, and G bases at the fifth position (SI Appendix, Fig. S1). To fully account for the conformational flexibility at this position would require sampling a properly Boltzmann-weighted ensemble of all bound conformations, which is not currently feasible. Because the Rosetta-Vienna RNP-ΔΔG method relies instead on one or a few representative conformations, we sought to improve the accuracy of these predictions by including an additional representative structure. We looked to homologous proteins to evaluate whether the backbone might adopt yet another conformation when U is bound at this position. Indeed, a PUM1 structure (91% identity of the RNA-binding domain with PUM2) with a U at the fifth position exhibits alternate conformations of both the base and the backbone (SI Appendix, Fig. S1 and Table S4) (39). When we included this PUM1 structure along with the three PUM2 structures as starting structures in our prediction method, the accuracy of the calculations improved from 2.78 kcal/mol RMSE for sequences with mutations at the fifth position initially to 1.88 kcal/mol RMSE (Fig. 4B, open symbols), although this value still exceeded the RMSE over mutants that preserved the fifth position (1.32 kcal/mol; SI Appendix, Table S6). Even with the inclusion of the PUM1 structure, the Rosetta-Vienna RNP-ΔΔG calculations were significantly more accurate for sequences without mutations at the fifth position (Fig. 4 and SI Appendix, Table S6). We additionally assessed the effect of including alternative structures with different conformations of the fifth position U in our calculations; however, the inclusion of the PUM1 structure gave the most accurate results (SI Appendix, Fig. S8 and Table S5).
For the second round of blind predictions, we used the same Rosetta-Vienna RNP-ΔΔG method and again included the PUM1 “U5” structure because it significantly improved the accuracy of the first round of predictions. Based on the first-round calculations, we also anticipated that the predictions for sequences without mutations at the fifth position would be more accurate than predictions for sequences containing mutations at the fifth position. The RMSE of the predictions for 528 sequences across the four scaffolds in two replicate experiments was 1.60 kcal/mol (Fig. 4B, filled symbols and Table 1). For the 385 sequences without mutations at the fifth position, the RMSE improved to 1.40 kcal/mol (SI Appendix, Table S6). The correlation coefficients for the PUM2 calculations were slightly worse than for the comparisons for other systems (R = 0.51 for round 1 and R = 0.43 for round 2; SI Appendix, Table S3) but remained statistically significant (P < 0.01). Furthermore, as before, the Rosetta-Vienna RNP-ΔΔG calculations were more accurate than calculations made with GLM-Score, the Rosetta hydrogen bond scoring model, and the standard Rosetta ΔΔG approach (SI Appendix, Fig. S3 and Table S1). Overall, these blind tests on PUM2 confirmed the predictive power of our method: The accuracy of the Rosetta-Vienna RNP-ΔΔG method is better than 1.5 kcal/mol when bound complexes do not undergo large conformational changes upon RNA mutation.
Discussion
We report a framework for RNA–protein relative binding affinity calculation that computationally models the energetics of all states in the process (Fig. 1). We have demonstrated that accurate calculations require computing free energies of unbound RNA structural ensembles, here estimated using the nearest-neighbor energy model. The overall RMSE accuracy for Rosetta-Vienna RNP-ΔΔG over all six tested systems was 1.38 kcal/mol, comparable to the 1 to 2 kcal/mol accuracy achieved for protein–protein, protein–small molecule, and protein–DNA systems. The method presented here combines the nearest-neighbor energy framework, developed for secondary structure prediction, with Rosetta 3D structure-based energy calculations for RNA and proteins. The inclusion of RNA secondary structure-based energy calculations is straightforward, but previous work on secondary structure-based energy prediction has advanced independently and it has therefore remained an open question whether the two fields could intersect synergistically.
Our results constitute a major improvement over past computational methods, with twofold decreases in RMSE (Table 1). Nevertheless, the tests presented here also highlight several aspects of our method for future improvement. First, our calculations lack explicit treatment of counterions and the electrostatic effects of different salt conditions, effects known to impact RNA structure and protein binding. Second, the nearest-neighbor energies, used to calculate the unbound RNA free energies, introduce potential errors into this method. The accuracy of these energies is on the order of 0.5 kcal/mol for motifs that have been extensively measured and is worse for other motifs such as loops and junctions (40). Unbound RNA free energies for longer RNAs containing many long loops and junctions are therefore likely to be less accurate. Additionally, because the unbound RNA free energies calculated with Vienna do not include pseudoknots, the Rosetta-Vienna RNP-ΔΔG values for complexes containing pseudoknots are likely to be less accurate. Rosetta-Vienna RNP-ΔΔG also requires an experimental structure of the RNA–protein complex of interest and therefore cannot be applied to the many RNA–protein complexes that have not yet been structurally characterized. This limitation could be addressed by computationally predicting structures of RNA–protein complexes de novo, although existing structure prediction methods are not likely to be accurate enough for this approach (41). Finally, while Rosetta-Vienna RNP-ΔΔG models conformational changes in the unbound state, it models only very limited conformational changes in the bound state. Our blind tests on PUM2 binding affinities show that including several conformations as starting structures can partially reduce inaccuracies resulting from nucleotide resolution conformational changes, but treatment of RNA flexibility in the bound complex is an area open for significant improvement. Future work will likely benefit from including conformational ensembles of the bound complexes, including possible register shifts (SI Appendix, Fig. S7) (28), although it will be a challenge to model these bound ensembles with the same computational efficiency as the nearest-neighbor model for unbound RNAs.
We expect the Rosetta-Vienna RNP-ΔΔG method to prove useful for emergent applications such as RNA structure prediction in the context of complex protein mixtures and within concentrated RNA–protein liquid-like phases (1, 4, 42). In cellular contexts, RNA is frequently bound to a multitude of different proteins, and therefore accurate RNA structure prediction for these contexts requires including relevant protein binding energetics (1). While high-throughput methods have been developed for measuring such interactions in vitro, it is unlikely that in the near future we will obtain quantitative experimental data for the binding landscapes of the thousands of RBPs relevant to human biology to all biologically important RNA sequences; computationally predicted protein binding energies may allow initial expansion of RNA structure prediction to include the effects of protein binding. These predictions may also be useful for a variety of applications such as RNP redesign to alter specificity, de novo design of RNA–protein complexes, or design of membraneless RNA–protein bodies (43, 44). Notably, the Rosetta-Vienna RNP-ΔΔG method appears to underpredict the effects of some deleterious sequence mutations, suggesting that these predictions could effectively be used as part of an initial computational filtering procedure before experimentally testing binding. We hope the availability of our method on a freely available server will help accelerate these applications.
Methods
The software used to calculate the relative binding affinities described here is freely available as a ROSIE webserver at rosie.rosettacommons.org/rnp_ddg. Additionally, the software is available to academic users as part of the Rosetta software suite at https://www.rosettacommons.org/. Documentation is available at https://www.rosettacommons.org/docs/latest/application_documentation/rna/rnp-ddg, and a demonstration is available at https://www.rosettacommons.org/demos/latest/public/rnp_ddg/README. Additional details are available in SI Appendix.
Relative binding affinities calculated with Rosetta-Vienna RNP-ΔΔG are available in Datasets S1–S6 for all systems tested here.
Supplementary Material
Acknowledgments
We thank Sarah Denny and members of the R.D. laboratory for useful discussions. Calculations were performed on the Stanford BioX3 cluster, supported by NIH Shared Instrumentation Grant 1S10RR02664701, and the Stanford Sherlock cluster. This work was supported by an NSF Graduate Research Fellowship (K.K.), a Stanford Graduate Fellowship (K.K.), NIH Grants R01GM100953 (to R.D.), R01GM111990 (to W.J.G.), and P01 GM066275 (to R.D., W.J.G.,and D.H.), and a RosettaCommons Grant (to R.D.). W.J.G. is a Chan Zuckerberg Biohub Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819047116/-/DCSupplemental.
References
- 1.Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell. 2014;54:547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 3.Gilman B, Tijerina P, Russell R. Distinct RNA-unwinding mechanisms of DEAD-box and DEAH-box RNA helicase proteins in remodeling structured RNAs and RNPs. Biochem Soc Trans. 2017;45:1313–1321. doi: 10.1042/BST20170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sawyer IA, Sturgill D, Dundr M. Membraneless nuclear organelles and the search for phases within phases. Wiley Interdiscip Rev RNA. 2018;10:e1514. doi: 10.1002/wrna.1514. [DOI] [PubMed] [Google Scholar]
- 5.Jankowsky E, Harris ME. Specificity and nonspecificity in RNA-protein interactions. Nat Rev Mol Cell Biol. 2015;16:533–544. doi: 10.1038/nrm4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buenrostro JD, et al. Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat Biotechnol. 2014;32:562–568. doi: 10.1038/nbt.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin HC, et al. Analysis of the RNA binding specificity landscape of C5 protein reveals structure and sequence preferences that Direct RNase P specificity. Cell Chem Biol. 2016;23:1271–1281. doi: 10.1016/j.chembiol.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert N, et al. RNA bind-n-seq: Quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell. 2014;54:887–900. doi: 10.1016/j.molcel.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tome JM, et al. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nat Methods. 2014;11:683–688. doi: 10.1038/nmeth.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jain N, Lin HC, Morgan CE, Harris ME, Tolbert BS. Rules of RNA specificity of hnRNP A1 revealed by global and quantitative analysis of its affinity distribution. Proc Natl Acad Sci USA. 2017;114:2206–2211. doi: 10.1073/pnas.1616371114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.She R, et al. Comprehensive and quantitative mapping of RNA-protein interactions across a transcribed eukaryotic genome. Proc Natl Acad Sci USA. 2017;114:3619–3624. doi: 10.1073/pnas.1618370114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddy MR, et al. Free energy calculations to estimate ligand-binding affinities in structure-based drug design. Curr Pharm Des. 2014;20:3323–3337. doi: 10.2174/13816128113199990604. [DOI] [PubMed] [Google Scholar]
- 13.Vangone A, Bonvin AM. Contacts-based prediction of binding affinity in protein-protein complexes. eLife. 2015;4:e07454. doi: 10.7554/eLife.07454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dias R, Kolazckowski B. Different combinations of atomic interactions predict protein-small molecule and protein-DNA/RNA affinities with similar accuracy. Proteins. 2015;83:2100–2114. doi: 10.1002/prot.24928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yan Z, Wang J. Optimizing scoring function of protein-nucleic acid interactions with both affinity and specificity. PLoS One. 2013;8:e74443. doi: 10.1371/journal.pone.0074443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potapov V, Cohen M, Schreiber G. Assessing computational methods for predicting protein stability upon mutation: Good on average but not in the details. Protein Eng Des Sel. 2009;22:553–560. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- 17.Ashtawy HM, Mahapatra NR. A comparative assessment of predictive accuracies of conventional and machine learning scoring functions for protein-ligand binding affinity prediction. IEEE/ACM Trans Comput Biol Bioinformatics. 2015;12:335–347. doi: 10.1109/TCBB.2014.2351824. [DOI] [PubMed] [Google Scholar]
- 18.Moal IH, Agius R, Bates PA. Protein-protein binding affinity prediction on a diverse set of structures. Bioinformatics. 2011;27:3002–3009. doi: 10.1093/bioinformatics/btr513. [DOI] [PubMed] [Google Scholar]
- 19.Yang CY, Sun H, Chen J, Nikolovska-Coleska Z, Wang S. Importance of ligand reorganization free energy in protein-ligand binding-affinity prediction. J Am Chem Soc. 2009;131:13709–13721. doi: 10.1021/ja9039373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rau M, Stump WT, Hall KB. Intrinsic flexibility of snRNA hairpin loops facilitates protein binding. RNA. 2012;18:1984–1995. doi: 10.1261/rna.035006.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Draper DE. Protein-RNA recognition. Annu Rev Biochem. 1995;64:593–620. doi: 10.1146/annurev.bi.64.070195.003113. [DOI] [PubMed] [Google Scholar]
- 22.Zheng S, Robertson TA, Varani G. A knowledge-based potential function predicts the specificity and relative binding energy of RNA-binding proteins. FEBS J. 2007;274:6378–6391. doi: 10.1111/j.1742-4658.2007.06155.x. [DOI] [PubMed] [Google Scholar]
- 23.Olson MA. Calculations of free-energy contributions to protein-RNA complex stabilization. Biophys J. 2001;81:1841–1853. doi: 10.1016/S0006-3495(01)75836-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hofacker IL, et al. Fast folding and comparison of Rna secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- 25.Tinoco I, Jr, et al. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 26.SantaLucia J, Jr, Turner DH. Measuring the thermodynamics of RNA secondary structure formation. Biopolymers. 1997;44:309–319. doi: 10.1002/(SICI)1097-0282(1997)44:3<309::AID-BIP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 27.Miao Z, et al. RNA-puzzles round III: 3D RNA structure prediction of five riboswitches and one ribozyme. RNA. 2017;23:655–672. doi: 10.1261/rna.060368.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jarmoskaite I, et al. 2018. A quantitative and predictive model for RNA binding by human Pumilio proteins. bioRxiv:10.1101/403006.
- 29.Valegârd K, et al. The three-dimensional structures of two complexes between recombinant MS2 capsids and RNA operator fragments reveal sequence-specific protein-RNA interactions. J Mol Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 30.Leaver-Fay A, et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alford RF, et al. The Rosetta all-atom energy function for macromolecular modeling and design. J Chem Theory Comput. 2017;13:3031–3048. doi: 10.1021/acs.jctc.7b00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang N, Kalyanaraman C, Bernacki K, Jacobson MP. Molecular mechanics methods for predicting protein-ligand binding. Phys Chem Chem Phys. 2006;8:5166–5177. doi: 10.1039/b608269f. [DOI] [PubMed] [Google Scholar]
- 33.Batey RT, Sagar MB, Doudna JA. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J Mol Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- 34.Jessen TH, Oubridge C, Teo CH, Pritchard C, Nagai K. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auweter SD, et al. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheong CG, Hall TM. Engineering RNA sequence specificity of Pumilio repeats. Proc Natl Acad Sci USA. 2006;103:13635–13639. doi: 10.1073/pnas.0606294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110:501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- 38.Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 39.Lu G, Hall TM. Alternate modes of cognate RNA recognition by human PUMILIO proteins. Structure. 2011;19:361–367. doi: 10.1016/j.str.2010.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zuber J, Cabral BJ, McFadyen I, Mauger DM, Mathews DH. Analysis of RNA nearest neighbor parameters reveals interdependencies and quantifies the uncertainty in RNA secondary structure prediction. RNA. 2018;24:1568–1582. doi: 10.1261/rna.065102.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kappel K, Das R. Sampling native-like structures of RNA-protein complexes through Rosetta folding and docking. Structure. 2019;27:140–151.e5. doi: 10.1016/j.str.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forties RA, Bundschuh R. Modeling the interplay of single-stranded binding proteins and nucleic acid secondary structure. Bioinformatics. 2010;26:61–67. doi: 10.1093/bioinformatics/btp627. [DOI] [PubMed] [Google Scholar]
- 43.Butterfield GL, et al. Evolution of a designed protein assembly encapsulating its own RNA genome. Nature. 2017;552:415–420. doi: 10.1038/nature25157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Omabegho T, et al. Controllable molecular motors engineered from myosin and RNA. Nat Nanotechnol. 2018;13:34–40. doi: 10.1038/s41565-017-0005-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.