Significance
DNA-reactive compounds target actively proliferating cells. Therefore, they are presupposed to kill cancer cells selectively, and many of them are used as chemotherapeutic agents. In this study, we have discovered a cell death pathway involving nc886 and PKR as another mechanism for the cytotoxicity. Our study provides an insight how a proapoptotic protein responds to a DNA-reactive compound via a regulatory noncoding RNA (ncRNA) as a molecular signal. Since the nc886/PKR pathway operates in most normal cells including nonproliferating ones, our finding may answer to a conundrum why a DNA-damaging compound harms quiescent cells and is of future clinical utility by considering nc886/PKR when designing a chemotherapeutic regimen with minimal side effects on normal cells.
Keywords: nc886, protein kinase R, doxorubicin, cytotoxicity, RNA polymerase III
Abstract
DNA-reactive compounds are harnessed for cancer chemotherapy. Their genotoxic effects are considered to be the main mechanism for the cytotoxicity to date. Because this mechanism preferentially affects actively proliferating cells, it is postulated that the cytotoxicity is specific to cancer cells. Nonetheless, they do harm normal quiescent cells, suggesting that there are other cytotoxic mechanisms to be uncovered. By employing doxorubicin as a representative DNA-reactive compound, we have discovered a cytotoxic mechanism that involves a cellular noncoding RNA (ncRNA) nc886 and protein kinase R (PKR) that is a proapoptotic protein. nc886 is transcribed by RNA polymerase III (Pol III), binds to PKR, and prevents it from aberrant activation in most normal cells. We have shown here that doxorubicin evicts Pol III from DNA and, thereby, shuts down nc886 transcription. Consequently, the instantaneous depletion of nc886 provokes PKR and leads to apoptosis. In a short-pulse treatment of doxorubicin, these events are the main cause of cytotoxicity preceding the DNA damage response in a 3D culture system as well as the monolayer cultures. By identifying nc886 as a molecular signal for PKR to sense doxorubicin, we have provided an explanation for the conundrum why DNA-damaging drugs can be cytotoxic to quiescent cells that have the competent nc886/PKR pathway.
DNA-reactive compounds have been widely used for cancer chemotherapy for a long time. They impair various aspects of DNA metabolism by alkylating DNA, intercalating into DNA, interfering with the action of topoisomerases, and evicting histones from DNA (1). All these mechanisms have a preferential impact on actively proliferating cells that need DNA replication, which is a basis for cancer-specific cytotoxicity. On this assumption, concern about side effects is confined in a subset of normal cells that are proliferating. Nonetheless, there are cases in which DNA-reactive anticancer drugs are toxic to nonproliferating normal cells (1). This cannot be explained by their intrinsic genotoxic effect and the underlying mechanism remains to be elusive.
Protein kinase R (PKR) is an interferon-inducible serine/threonine kinase. It is present in most mammalian cells in a latent state and typically is activated by double-stranded RNA (dsRNA) (2). Upon binding to dsRNA, PKR undergoes dimerization and autophosphorylation. The phosphorylated PKR (phospho-PKR) is an active kinase that phosphorylates eukaryotic initiation factor 2 α subunit (eIF2α), leading to the shutdown of global protein synthesis and consequently to apoptosis. However, the PKR pathway is far more complicated than indicated by the description above. In addition to dsRNA, PKR is controlled by a number of cellular and external factors to play critical roles in diverse signaling pathways including nuclear factor-κB (NF-κB) and p53 (3). As these cellular pathways are important in determining cell death versus cell proliferation, PKR is implicated, not surprisingly, in cancer pathology. Dysregulation of PKR-mediated apoptosis is a critical component in tumor development itself as well as in the resistance of cancer cells to chemotherapeutic drugs.
Several reports indicated PKR’s role in the cytotoxicity of doxorubicin, a DNA-reactive compound that have been used clinically for a long time (4–8). PKR seems to be activated by doxorubicin; in this case, PKR should somehow sense doxorubicin (or a molecular signal induced by it). However, this mechanism is completely unknown and is even puzzling, given that doxorubicin induces genotoxic stresses in the nucleus, but PKR-mediated apoptotic signaling occurs mainly in the cytoplasm. Even if doxorubicin provokes PKR and the resultant apoptosis, the significance of these events is questionable because doxorubicin’s genotoxic effect will lead to cell death anyway independently of PKR.
In this study, we have solved those questions by a noncoding RNA (ncRNA) that binds to PKR and suppresses its activation (9). It is nc886 (also known as VTRNA2-1 or pre-miR-886), a 101-nucleotide (nt)-long ncRNA transcribed by RNA polymerase III (Pol III) and ubiquitously expressed in normal human tissues. The expression level of nc886 is elevated in some cancer cells (10, 11) but is completely silenced by CpG DNA hypermethylation in other malignancies (12–17). Another interesting feature of nc886 is its short half-life (∼1 h; refs. 9 and 18), which makes it suitable as a candidate signaling molecule that quickly responds to stimuli. Here, we have found nc886 to be the key signaling molecule that links a DNA-reactive compound and PKR to therapeutic apoptosis.
Results
PKR Contributes to Doxorubicin-Induced Apoptosis.
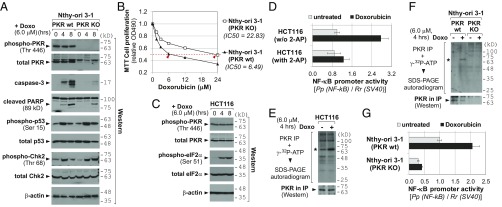
Doxorubicin has been shown to provoke the PKR pathway (4–8). In the case of human cells, the experimental evidence has been mostly based on knockdown (KD) experiments by using small interfering (or hairpin) RNA (siRNA or shRNA) against PKR. We confirmed this with PKR knockout (KO) cells that were generated from a thyroid cell line Nthy-ori 3-1 and a hepatocarcinoma cell line Huh7.5 by the CRISPR-Cas technique (10) (Fig. 1A and SI Appendix, Fig. S1A). Doxorubicin treatment induced apoptotic marker proteins, such as caspase-3 and cleaved Poly (ADP ribose) polymerase (PARP) in parental Nthy-ori 3-1 cells [PKR wild type (WT)], but this induction was significantly attenuated in PKR KO cells (Fig. 1A). The comparable induction of p53 and Chk2 (whose phospho-forms are indicators for DNA damage, Fig. 1A) in PKR WT and KO cells ascertained that doxorubicin’s genotoxic effect was equally efficient between them and that our apoptosis data could not be attributed to a difference in a DNA damage response. In MTT cell proliferation assays, PKR KO cells were more resistant to doxorubicin than PKR WT cells (Fig. 1B and SI Appendix, Fig. S1B). We also performed siRNA-mediated KD of PKR in another cell line, HCT116 from colon cancer, and observed a higher IC50 value in PKR KD than control KD (SI Appendix, Fig. S2), similarly to PKR KO data.
Fig. 1.
Activation of PKR by doxorubicin and its contribution to the doxorubicin cytotoxicity. (A) Western blot of indicated proteins after doxorubicin (Doxo) treatment. Molecular size markers in kilodaltons (kDa) are indicated on the right. (B) MTT cell proliferation assays. Doxorubicin was treated for 24 h. Each data point is an average of triplicate samples. SDs are not shown because they are small (ranging 0.010∼0.030) and obscured by markers in most data points. (C) Western blot of indicated proteins as in A. (D) Luciferase assays measuring NF-κB activity. Luciferase-expressing plasmids (the firefly luciferase from the NF-κB–responsive promoter and the Renilla luciferase from the constitutive the SV40 promoter for normalization) were transfected. At 24 h after transfection, cells were pretreated with 2-AP (2 mM) for 1 h and then replaced with a culture medium containing doxorubicin (6 µM) for 8 h. Firefly values (“Pp”) were divided by Renilla values (“Rr”), and the value of untreated HCT116 was set as 1. (E and F) In vitro kinase assay. The autoradiogram (Upper) and PKR Western (Lower) for equal IP efficiency are shown. Asterisks indicate the PKR band. (G) Luciferase assays, as described in D, except for no treatment of 2-AP.
Doxorubicin Results in PKR Activation.
The above data corroborated that PKR contributes to doxorubicin-induced apoptosis (4–8). Because PKR induces apoptosis when activated (2), we presumed PKR activation upon doxorubicin treatment. However, astonishingly there was no induction of PKR phosphorylation at Thr446, a representative marker for active PKR, in our Nthy-ori 3-1 and HCT116 data (Fig. 1 A and C). To rule out technical issues such as our Western blot procedure or antibody quality, we included a sample treated with Poly(I:C), a known PKR activator, and repeated phospho-PKR Western blot. Of eight antibodies tested, none of them yielded a legitimate band in doxorubicin-treated samples, although three detected phosphorylation at Thr446 or Thr451 in the Poly(I:C)-treated sample (SI Appendix, Fig. S3A). Of note, eIF2α phosphorylation was seen both in doxorubicin and Poly(I:C) samples at a similar level (SI Appendix, Fig. S3B). Having this puzzling result, we carefully assessed the experimental data in previous literatures and found that those literatures did not provide phospho-PKR Western blot as concrete data (elaborated in SI Appendix, Fig. S3 legend).
One conceivable scenario is that PKR per se, not as an active form, is required for doxorubicin-induced apoptosis. However, we disfavored this possibility because eIF2α was phosphorylated (Fig. 1C and SI Appendix, Fig. S3B) and NF-κB was activated in our luciferase reporter assays employing the NF-κB–responsive promoter (Fig. 1D). These two events represent the canonical downstream pathways of active PKR (3). Furthermore, the NF-κB induction upon doxorubicin treatment was negated by the PKR inhibitor 2-aminopurine (2-AP), convincingly showing PKR dependency (Fig. 1D). So, we questioned whether the Western blot of phospho-Thr446 (or Thr451) is a valid indicator in the context of doxorubicin treatment. Thr446 or Thr451 is not the sole residue that undergoes phosphorylation but several other Ser/Thr and also Tyr residues have been identified to be phosphorylated (19, 20). Therefore, we performed an in vitro kinase assay to measure the overall autophosphorylation level. A radiolabeled phospho-PKR was immunoprecipitated and resolved on a SDS-polyacrylamide gel (Fig. 1 E and F). The asterisked band is PKR, because it migrated at the correct position and was not detectable in PKR KO Nthy-ori 3-1 cells (Fig. 1F). Importantly, PKR phosphorylation was increased when treated with doxorubicin. The NF-κB luciferase data in Nthy-ori 3-1 cells were in line with the in vitro kinase assay data; the NF-κB promoter activity was elevated by doxorubicin in PKR WT cells, but was lower and unresponsive to doxorubicin in PKR KO cells (Fig. 1G).
Doxorubicin Suppresses nc886 Expression.
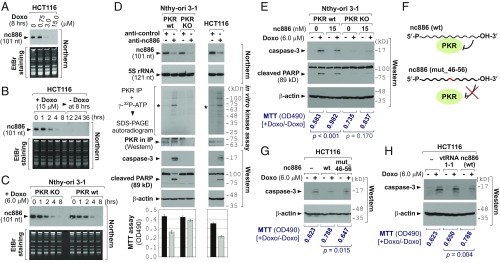
So far we proved that doxorubicin activates PKR. We found a report that doxorubicin inhibits Pol III that transcribes nc886 (21). Our previous study has shown that nc886 is a suppressor of PKR activity (22). All of these inspired us to explore a role of nc886 herein. When treated with titrating amounts of doxorubicin, nc886 disappeared almost completely at or above 3 µM (Fig. 2A). It should be noted that the micromolar range was not unreasonably high compared with clinically relevant concentrations ranging from 5 to 25 µM (23). In our time-course experiments, nc886 was noticeably diminished after 2 h of treatment and was barely seen after 4–8 h in all cell lines we tested (Fig. 2 B and C and SI Appendix, Fig. S4). At these time points, cell proliferation was decreased only modestly (SI Appendix, Fig. S5), indicating that nc886 decreased before doxorubicin elicited its toxic effect on cells. After 8 h, nc886 expression disappeared completely and did not recover after doxorubicin removal (Fig. 2B). Our consistent data across cell lines of diverse tissues origins (colon, thyroid, breast, esophagus, stomach, and lung) suggest that the suppression of nc886 expression by doxorubicin is a ubiquitous phenomenon. nc886 was not decreased by an oxidative stress- or a hypoxia-inducing drug (SI Appendix, Fig. S6), indicating that the doxorubicin effect on nc886 was not simply an end result of any stress condition.
Fig. 2.
nc886’s role in the PKR-mediated cytotoxic effect upon doxorubicin treatment. (A–C) Northern blot of nc886 and EtBr staining as a loading control. (D) Various indicated assays upon transfection of anti-oligos for 24 h. (E) Transfection of the nc886 RNA (an in vitro transcript) combined with doxorubicin treatment. Transfection and treatment were done simultaneously before harvesting cells at 6 h for Western blot. 2 μg of yeast tRNA (the equivalent amount to 15 nM of nc886) was transfected as a negative control (lanes 1–2 and 5–6). (F) A diagram illustrating the feature of the mutant nc886 compared with WT. The mutated portion is highlighted in red. (G and H) The same experiment as E, except that mutant nc886 or vtRNA1-1 (with WT nc886 for comparison) was transfected and cell harvest was at 12 h. Lanes 1–2 are 2 µg of yeast tRNA.
nc886 Suppression Is the Critical Event for Doxorubicin Cytotoxicity Through PKR Activation.
Thus far we have demonstrated concretely that doxorubicin treatment results in PKR activation and also in the suppression of nc886. nc886 is physically associated with PKR (9, 22) (SI Appendix, Fig. S7). Since nc886 inhibits PKR (24), we hypothesized that nc886 suppression is the mechanism for doxorubicin-mediated PKR activation. To prove this hypothesis, we conducted several experiments.
We began by examining whether depletion of nc886 elicits the same cellular response to doxorubicin, by performing nc886 KD using modified antisense oligonucleotides (anti-oligo). The nc886 targeting anti-oligo (anti-nc886) efficiently suppressed nc886 (Northern blot in Fig. 2D) and led to PKR activation (in vitro kinase assay in Fig. 2D). nc886 KD was sufficient to induce apoptosis and to inhibit cell proliferation in PKR WT Nthy-ori 3-1 and HCT116 cells, but not in PKR KO Nthy-ori 3-1 cells (Fig. 2D), assuring that they occurred through PKR. Collectively, nc886 KD emulated doxorubicin’s cytotoxicity through PKR activation.
Doxorubicin causes pleiotropic effects. So we wanted to discern nc886’s contribution and interrogated whether doxorubicin-mediated cytotoxicity was mitigated by ectopic expression of nc886. As a Pol III transcript, nc886 expression is driven by gene-internal promoter elements and, therefore, its expression from an exogenous DNA was also inhibited by doxorubicin (shown later in SI Appendix, Fig. S17). So, our tactic was to deliver nc886 in a form of an in vitro transcript. Doxorubicin-induced apoptosis was attenuated when the nc886 transcript was transfected into PKR WT cells, whereas such attenuation was not seen in PKR KO cells (Fig. 2E and SI Appendix, Figs. S8 and S9). The role of nc886 on PKR was confirmed by testing a mutant nc886 (“mut_46-56”) that is deficient in PKR binding (22) (Fig. 2F). Doxorubicin-induced apoptosis was not mitigated by mut_46-56 (Fig. 2G). This result was further corroborated by a vault RNA (vtRNA), which is a paralog of nc886 but does not bind to PKR (22). A vtRNA did not mitigate the apoptosis either (vtRNA1-1 in Fig. 2H). One of nc886’s aliases is vtRNA2-1, which implies that nc886 might be a component of the vault complex, which has been implicated in cancer drug resistance (25). However, the vault complex did not contain nc886 nor affected doxorubicin sensitivity (SI Appendix, Figs. S10–S13). In the doxorubicin-induced PKR activation, a role of p53 has been suggested but remained unclear due to conflicting results between studies (8, 26). In our data employing p53 WT and null HCT116 cells, p53 barely affected the nc886/PKR pathway (SI Appendix, Fig. S14). All of our data unequivocally demonstrated that nc886 suppression is a key event for PKR activation and apoptosis in response to doxorubicin, regardless of the vault complex or the p53 status.
Doxorubicin Inhibits Pol III Transcription.
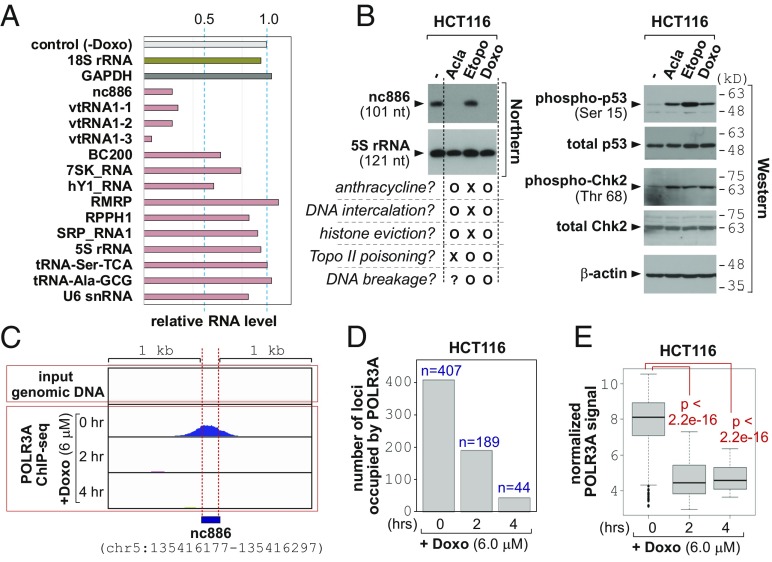
Next, we investigated the mechanism for nc886 suppression by doxorubicin. A classic study had shown that doxorubicin inhibits Pol III transcription when introduced into in vitro transcription assays (21). Pol III transcribes medium-sized ncRNAs, many of which are stable and abundant RNAs with fundamental cellular roles (designated collectively as “Pol III genes”; see Fig. 3A and Dataset S1). Upon doxorubicin treatment, the expression of most Pol III genes was decreased with varying degrees (Fig. 3A and SI Appendix, Figs. S15 and S16). The kinetics of nc886 reduction indicated that its transcription was shut down almost immediately by doxorubicin, given that nc886’s half-life is ∼1 h (9, 18). This notion was corroborated by measurement of two precursor tRNAs (pre-tRNAs), which disappeared within an hour (SI Appendix, Fig. S16 N and P). Pre-tRNAs are regarded as the most sensitive indicator to assess Pol III transcription rates, because they are processing intermediates. Collectively, Pol III transcription appeared to be inhibited readily upon doxorubicin treatment. It should be underscored that most Pol III genes were not so severely (<twofold) affected until 4 h (Fig. 3A and SI Appendix, Figs. S15 and S16), when doxorubicin had already elicited PKR-dependent apoptosis (Fig. 1A).
Fig. 3.
The eviction of Pol III and inhibition of transcription by doxorubicin. (A) Expression levels of Pol III genes at 4 h after doxorubicin treatment, relative to 0 h, which is designated as “control (-Doxo)”. Full data and detailed description are in SI Appendix, Figs. S15 and S16. (B) Northern (Left) and Western (Right) blot after 8 h-treatment of aclarubicin (Acla; 6 µM), etoposide (Etopo; 30 µM), and doxorubicin (Doxo; 6 µM), as well as no treatment control (the leftmost lane). Features of the three drugs are briefly summarized at the bottom of the Northern blot. (C) The POLR3A ChIP-seq peak of nc886. Ten million tags were used for normalization of relative POLR3A signal from ChIP-seq reads. The normalized ChIP-seq density (y axis) is shown along the genomic coordinate (hg19) of nc886 and its flanking 1-kb region at both sides (x axis). IGV 2.3 was used for visualization. (D) Number of POLR3A-occupied Pol III loci. (E) A boxplot of normalized POLR3A occupancy per each locus. The ANOVA test was used to calculate P values.
The next question was the detailed mechanism for doxorubicin to inhibit Pol III. Doxorubicin belongs to the anthracycline family consisting of flat aromatic moieties and imposes stresses on DNA/chromatin in multiple ways. It intercalates into DNA to generate topological tension. Another well-known effect is topoisomerase II (topo II) poisoning by trapping of topo II in the double-strand cleavage form and preventing ligation (27). The cellular outcomes of these effects are DNA strand breakage, eviction of histones from chromatin, and DNA damage responses. To discern which event is causative of the suppression of Pol III transcription, we compared three different compounds (Fig. 3B). Aclarubicin is an anthracycline compound and is a DNA intercalator. However, it is not a topo II poison unlike doxorubicin (27). Etoposide, a nonanthracycline compound, does not intercalate into DNA but acts as a topo II poison (27).
At therapeutic doses (23, 28), doxorubicin and aclarubicin suppressed nc886 expression, whereas etoposide did not (Fig. 3B). In all three drugs, phosphorylation of p53 and Chk2 was seen (Fig. 3B), indicating that all treatments were effective and, more importantly, that nc886 suppression was not due to the aftermath of DNA damage responses. Our etoposide data proved that the suppression of Pol III transcription was not caused by topo II poisoning (or the resultant DNA breakage). Our data from linear and circular plasmid DNAs also suggested that Pol III inhibition by doxorubicin was not caused by intercalation into DNA and the resultant DNA torsion (SI Appendix, Fig. S17) either.
Because doxorubicin and aclarubicin, but not etoposide, are known to evict histones (29, 30), we surmised that these two drugs would lead to Pol III dissociation from DNA. To directly test this idea, we examined bound DNA to POLR3A, the catalytic subunit of Pol III enzyme, by chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) (31). A POLR3A peak was present at the correct position at nc886 and other Pol III loci, but absent in a Pol II gene GAPDH (Fig. 3C and SI Appendix, Fig. S18). Importantly, the peaks in all of the Pol III loci disappeared upon doxorubicin treatment. In the human genome, there are 4,553 Pol III loci that were curated in our previous study (18). When we analyzed the ChIP-seq data comprehensively in these loci (Dataset S1), we found global dissociation of POLR3A to occur upon doxorubicin treatment, as indicated by the number of POLR3A-bound loci (Fig. 3D) and the intensity of POLR3A signal estimated from seq reads (Fig. 3E and SI Appendix, Fig. S16). ChIP-seq data were reconfirmed by ChIP-PCR (SI Appendix, Fig. S19). Collectively, doxorubicin evicts Pol III from DNA.
A Short Pulse Doxorubicin Treatment Provokes the nc886/PKR Pathway but Not DNA Damage Response.
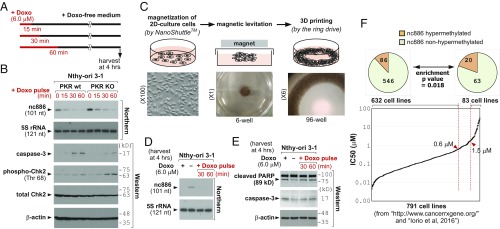
Collectively from our data, it appears that the transcription of nc886 was shut down immediately upon doxorubicin treatment (Figs. 2 and 3) and was not resumed after removal of doxorubicin afterward (Fig. 2B). Hence, a short pulse treatment might be sufficient to activate the nc886/PKR pathway and apoptosis. We tested this possibility by treating cells with doxorubicin and replacing the medium with a doxorubicin-free medium at 15, 30, and 60 min (Fig. 4A). The 30-min pulse of doxorubicin was enough to suppress nc886 and induce apoptosis in a PKR-dependent manner at 4 h (Fig. 4B) but was too short to induce the DNA damage response as measured by phospho-Chk2 (Fig. 4B). So we found a doxorubicin treatment condition in which the nc886/PKR pathway, rather than the DNA damage response, dominantly operated to induce apoptosis.
Fig. 4.
Short pulse treatment of doxorubicin at micromolar concentration: its application in the 3D culture system and survey in various cancer cells. (A) Schematic diagram of pulse-treatment experiments. (B) Northern and Western blots, as performed according to the scheme in A. (C) A brief summary illustrating how to generate the 3D culture system, with schematic (Upper) and actual images (Lower). (D and E) Northern and Western blots described in A–C. Lane 1, continuous doxorubicin treatment; lane 2, no doxorubicin control; lanes 3–4, doxorubicin pulse as described in A. (F) A rank distribution plot of IC50 values of 791 cell lines in ascending order (see the text for detailed description and Dataset S2 for full information). We selected two groups of cells: doxorubicin-sensitive (IC50 < 0.6) and -resistant (IC50 > 1.5). In each group, cells were further classified according to the degree of nc886 CpG methylation (from 10 CpG sites in the nc886 genomic region) and depicted in the pie charts on the top. Hypermethylation was defined as >90% methylation on average. Statistical significance of differential methylation between the two groups was calculated by a χ2 test.
The nc886/PKR Pathway Is Effective also in the 3D Cell Culture System.
So far, our experiments were performed in the conventional 2D monolayer cell culture, which is significantly different from the in vivo tumor environment in many aspects including drug sensitivity. Since cells cultured in 3D show features closer to in vivo physiology (32), we attempted to examine doxorubicin’s effect on nc886 and apoptosis in a 3D culture by using a recently developed protocol. In this technique (33), cells were coated with magnetic nanoparticles and levitated by applying a magnetic field. The levitated cells formed a 3D structure with extracellular matrix and were bioprinted onto a plate (Fig. 4C). Doxorubicin, both in continuous and pulse treatments to a 3D culture, suppressed nc886 and induces apoptosis (Fig. 4 D and E).
Epigenetic Silencing of nc886 Is Related to Cellular Resistance to Doxorubicin.
nc886 expression is silenced by the promoter CpG hypermethylation in a subset of cancer cells (9, 11–15, 18, 34). To implicate this phenomenon in our experimental data here so far, we downloaded doxorubicin IC50 values of 931 cell lines from a cancer drug database (https://www.cancerrxgene.org/) and also obtained CpG methylation values in 957 cell lines from a published study (35). Comparison of the two datasets yielded 791 cell lines with both values (Dataset S2). We sorted IC50 values in ascending order, plotted (Fig. 4F), and found only 27 cell lines to have IC50 > 6 µM (3.4% of 791 cell lines), the concentration used in most of our experiments here. In a lower IC50 cutoff (1.5 µM), 83 cell lines (10.5%) met this criterion. In addition, a majority of tumors that have acquired doxorubicin resistance also had IC50 values < 6 µM (SI Appendix, Fig. S20 and Dataset S3).
As aforementioned, epigenetic silencing of nc886, which is an intriguing and unique feature among Pol III genes (reviewed in ref. 36), occurs in some cancer cells. In theory and also based on our data here, this silencing should have resulted in cell death, because PKR should have been activated. However, nc886-silenced in vitro cell lines and in vivo tumor cells do exist. Our previous studies (15, 37) elucidated this by a tumor surveillance model. In this model, such cell death would have occurred to ensure the elimination of precancerous cells, which would have contributed to the lowering of a cancer incidence rate. However, certain cells acquire genetic/epigenetic alterations, for example overexpression of eIF2B that neutralizes the apoptotic effect of phospho-eIF2α, to bypass the PKR cell death pathway (15). Only those cells could have survived upon nc886 silencing and developed into clinically detectable and in vitro isolatable malignant cells. We hypothesized that those cells lost the PKR pathway for cell death and therefore tended to be resistant to doxorubicin, especially at micromolar concentrations. Because nc886 is a not a microRNA nor mRNA, it is challenging to estimate its expression level from public array or sequencing data. Therefore, we looked into CpG methylation values as a proxy indicator for nc886 silencing. nc886 has a CpG island (−189 to +82, as the 5′-end of the nc886 transcript being the +1 reference point), and we analyzed 10 CpG sites within or close to this island. We calculated an average of the 10 sites and regarded >90% to be hypermethylation. In this analysis (pie charts in Fig. 4F) only 86 of 632 doxorubicin-sensitive (IC50 < 0.6 µM) cell lines were nc886-hypermethylated (12.0%). When we did the same analysis in doxorubicin-resistant (IC50 > 1.5 µM) cell lines, we observed a significant enrichment of hypermethylated cells (20 of 83; 19.4%). These data supported our idea that the PKR cell death pathway has become defective in nc886-silenced cells, which is a contributing factor for their resistance to doxorubicin.
Discussion
In this study, we have found that nc886/PKR signaling is another important pathway in the cytotoxicity of doxorubicin. Our data clearly demonstrated that doxorubicin decreases nc886, thereby activates PKR, and leads to apoptosis. This is a signal transduction pathway in which a genotoxic stress in the nucleus is transmitted to a cytoplasmic protein through an ncRNA. Also, our finding calls a need to reconsider the long-standing tenet that DNA-reactive compounds are selectively toxic to proliferating cells, since nc886 and PKR are expressed in most normal cells regardless of cell proliferation (3, 9).
Our data would be applicable to improve the therapeutic regimen for doxorubicin (and probably all anthracycline drugs) by considering the nc886/PKR mechanism. When we want to exploit doxorubicin’s original genotoxic role to kill cancer cells, combination with a PKR inhibitor is a reasonable formulation to minimize a collateral damage to quiescent normal cells. Alternatively, we could use the nc886/PKR pathway as a target to kill tumor cells. Although one would suspect whether this strategy harms normal cells as well, we anticipate that doxorubicin could trigger the nc886/PKR pathway in tumor cells more proficiently, because the nc886 expression level is high and PKR activity is low usually in tumors compared with normal cells. Global Pol III transcription, accordingly nc886 expression, increases during tumorigenesis (reviewed in ref. 38) (9, 10, 18). Due to PKR’s proapoptotic function, PKR is supposed to be suppressed in malignancies and it has been shown so in a number of literatures (reviewed in ref. 39). When targeting nc886/PKR, a short pulse (30 min) treatment of doxorubicin at a high dose (> several micromolars) will be sufficient to induce apoptosis, with a minimal genotoxic effect adversely on proliferating normal cells such as cells at the hematopoietic lineage, hair follicle cells, and epithelial cells of the intestine. Also, it is obvious that this regimen is inappropriate for nc886-silenced cancer cells.
In addition to DNA-reactive drugs, the PKR pathway also operates in response to various external stimuli such as deprivation of growth factors, heat shock, metabolic stress, and biotoxins. Some PKR upstream regulators, such as PACT/RAX, heat shock proteins, ribosomes, and some cellular ncRNAs are implicated in conveying these stresses to PKR (reviewed in refs. 40 and 41) (42). However, stress-sensing mechanisms have not been elucidated yet in many cases. As mentioned earlier, nc886 has a short half-life and so can respond quickly to external signals. nc886 can be a potent PKR regulator given that its binding affinity to PKR is very high (KD = ∼12 nM, ref. 22) and it is abundantly expressed (105 RNA molecules per cell, ref. 9). Collectively, nc886 is well suited as a molecular signal for PKR and we surmise that nc886 plays this role in other stress conditions besides doxorubicin.
Materials and Methods
Cell lines, antibodies, plasmids, anti-oligos and other reagents are described in refs. 9, 10, and 15 and in SI Appendix. Sequences for primers (for PCR and Northern blot) are summarized in Dataset S4. Luciferase assays, cell proliferation assays, and in vitro kinase assays were performed as described in refs. 9 and 15. POLR3A ChIP-seq was done as described (18), and the raw files are available (31). Three-dimensional cell culture was done as described in SI Appendix. Unless otherwise specified, assays were done in triplicate to calculate an average and SD.
Supplementary Material
Acknowledgments
We thank Drs. Anindya Dutta and Etsuko Shibata (University of Virginia) for helpful discussion and Drs. Glauco R. Souza and Kim Brath (n3D Biosciences and Greiner Bio-One) and Ms. Lauren S. Richardson for assistance with the 3D culture. This work was supported by National Cancer Center, Korea Grants NCC-1810071-1 (to Y.S.L.) and NCC-1810072-1 (to I.-H.K.), the Collaborative Genome Program for Fostering New Post-Genome Industry of the National Research Foundation funded by Ministry of Science and ICT Grant NRF-2017M3C9A6044517 (to Y.-S.L.), and Thailand Research Fund Grant MRG5980034 (to N.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The ChIP-seq data has been deposited in the NCBI Sequence Read Archive (SRA), https://www.ncbi.nlm.nih.gov/sra/ (accession no. PRJNA522927).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814510116/-/DCSupplemental.
References
- 1.Cheung-Ong K, Giaever G, Nislow C. DNA-damaging agents in cancer chemotherapy: Serendipity and chemical biology. Chem Biol. 2013;20:648–659. doi: 10.1016/j.chembiol.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Ortega MB, et al. Clinical and therapeutic potential of protein kinase PKR in cancer and metabolism. Expert Rev Mol Med. 2017;19:e9. doi: 10.1017/erm.2017.11. [DOI] [PubMed] [Google Scholar]
- 3.Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–6120. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 4.Cheng X, Bennett RL, Liu X, Byrne M, Stratford May W. PKR negatively regulates leukemia progression in association with PP2A activation, Bcl-2 inhibition and increased apoptosis. Blood Cancer J. 2013;3:e144. doi: 10.1038/bcj.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bennett RL, et al. Increased expression of the dsRNA-activated protein kinase PKR in breast cancer promotes sensitivity to doxorubicin. PLoS One. 2012;7:e46040. doi: 10.1371/journal.pone.0046040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peidis P, Papadakis AI, Muaddi H, Richard S, Koromilas AE. Doxorubicin bypasses the cytoprotective effects of eIF2α phosphorylation and promotes PKR-mediated cell death. Cell Death Differ. 2011;18:145–154. doi: 10.1038/cdd.2010.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tu YC, et al. Blocking double-stranded RNA-activated protein kinase PKR by Japanese encephalitis virus nonstructural protein 2A. J Virol. 2012;86:10347–10358. doi: 10.1128/JVI.00525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon CH, Lee ES, Lim DS, Bae YS. PKR, a p53 target gene, plays a crucial role in the tumor-suppressor function of p53. Proc Natl Acad Sci USA. 2009;106:7852–7857. doi: 10.1073/pnas.0812148106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K, et al. Precursor miR-886, a novel noncoding RNA repressed in cancer, associates with PKR and modulates its activity. RNA. 2011;17:1076–1089. doi: 10.1261/rna.2701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee EK, et al. nc886, a non-coding RNA and suppressor of PKR, exerts an oncogenic function in thyroid cancer. Oncotarget. 2016;7:75000–75012. doi: 10.18632/oncotarget.11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahn JH, et al. nc886 is induced by TGF-β and suppresses the microRNA pathway in ovarian cancer. Nat Commun. 2018;9:1166. doi: 10.1038/s41467-018-03556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang HJ, et al. Integrated genomic analysis of recurrence-associated small non-coding RNAs in oesophageal cancer. Gut. 2016;66:215–225. doi: 10.1136/gutjnl-2015-311238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KS, et al. nc886, a non-coding RNA of anti-proliferative role, is suppressed by CpG DNA methylation in human gastric cancer. Oncotarget. 2014;5:3944–3955. doi: 10.18632/oncotarget.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee HS, et al. Epigenetic silencing of the non-coding RNA nc886 provokes oncogenes during human esophageal tumorigenesis. Oncotarget. 2014;5:3472–3481. doi: 10.18632/oncotarget.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kunkeaw N, et al. Cell death/proliferation roles for nc886, a non-coding RNA, in the protein kinase R pathway in cholangiocarcinoma. Oncogene. 2013;32:3722–3731. doi: 10.1038/onc.2012.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treppendahl MB, et al. Allelic methylation levels of the noncoding VTRNA2-1 located on chromosome 5q31.1 predict outcome in AML. Blood. 2012;119:206–216. doi: 10.1182/blood-2011-06-362541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao J, et al. DNA methylation-mediated repression of miR-886-3p predicts poor outcome of human small cell lung cancer. Cancer Res. 2013;73:3326–3335. doi: 10.1158/0008-5472.CAN-12-3055. [DOI] [PubMed] [Google Scholar]
- 18.Park JL, et al. Epigenetic regulation of RNA polymerase III transcription in early breast tumorigenesis. Oncogene. 2017;36:6793–6804. doi: 10.1038/onc.2017.285. [DOI] [PubMed] [Google Scholar]
- 19.Taylor DR, et al. Autophosphorylation sites participate in the activation of the double-stranded-RNA-activated protein kinase PKR. Mol Cell Biol. 1996;16:6295–6302. doi: 10.1128/mcb.16.11.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su Q, et al. Tyrosine phosphorylation acts as a molecular switch to full-scale activation of the eIF2alpha RNA-dependent protein kinase. Proc Natl Acad Sci USA. 2006;103:63–68. doi: 10.1073/pnas.0508207103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logan K, Ackerman S. Effects of antibiotics on RNA polymerase III transcription. DNA. 1988;7:483–491. doi: 10.1089/dna.1.1988.7.483. [DOI] [PubMed] [Google Scholar]
- 22.Jeon SH, et al. Characterization of the direct physical interaction of nc886, a cellular non-coding RNA, and PKR. FEBS Lett. 2012;586:3477–3484. doi: 10.1016/j.febslet.2012.07.076. [DOI] [PubMed] [Google Scholar]
- 23.Mross K, et al. Pharmacokinetics and metabolism of epidoxorubicin and doxorubicin in humans. J Clin Oncol. 1988;6:517–526. doi: 10.1200/JCO.1988.6.3.517. [DOI] [PubMed] [Google Scholar]
- 24.Lee YS. A novel type of non-coding RNA, nc886, implicated in tumor sensing and suppression. Genomics Inform. 2015;13:26–30. doi: 10.5808/GI.2015.13.2.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Zon A, Mossink MH, Scheper RJ, Sonneveld P, Wiemer EA. The vault complex. Cell Mol Life Sci. 2003;60:1828–1837. doi: 10.1007/s00018-003-3030-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman M, Lem C, Muaddi H, Koromilas AE. PKR is not a universal target of tumor suppressor p53 in response to genotoxic stress. Cell Cycle. 2009;8:3606–3607. doi: 10.4161/cc.8.21.9848. [DOI] [PubMed] [Google Scholar]
- 27.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hande KR, et al. Pharmacokinetics of high-dose etoposide (VP-16-213) administered to cancer patients. Cancer Res. 1984;44:379–382. [PubMed] [Google Scholar]
- 29.Pang B, et al. Drug-induced histone eviction from open chromatin contributes to the chemotherapeutic effects of doxorubicin. Nat Commun. 2013;4:1908. doi: 10.1038/ncomms2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang F, Kemp CJ, Henikoff S. Doxorubicin enhances nucleosome turnover around promoters. Curr Biol. 2013;23:782–787. doi: 10.1016/j.cub.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song M-J, et al. 2019 Data from “Polymerase III ChIP-seq in HCT116 cell line.” Sequence Read Archive. Available at https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA522927. Deposited February 28, 2019.
- 32.Ravi M, Paramesh V, Kaviya SR, Anuradha E, Solomon FD. 3D cell culture systems: Advantages and applications. J Cell Physiol. 2015;230:16–26. doi: 10.1002/jcp.24683. [DOI] [PubMed] [Google Scholar]
- 33.Souza GR, et al. Three-dimensional tissue culture based on magnetic cell levitation. Nat Nanotechnol. 2010;5:291–296. doi: 10.1038/nnano.2010.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee BJ, de la Peña P, Tobian JA, Zasloff M, Hatfield D. Unique pathway of expression of an opal suppressor phosphoserine tRNA. Proc Natl Acad Sci USA. 1987;84:6384–6388. doi: 10.1073/pnas.84.18.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iorio F, et al. A landscape of pharmacogenomic interactions in cancer. Cell. 2016;166:740–754. doi: 10.1016/j.cell.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park JL, et al. Epigenetic regulation of noncoding RNA transcription by mammalian RNA polymerase III. Epigenomics. 2017;9:171–187. doi: 10.2217/epi-2016-0108. [DOI] [PubMed] [Google Scholar]
- 37.Jeon SH, Johnson BH, Lee YS. A tumor surveillance model: A non-coding RNA senses neoplastic cells and its protein partner signals cell death. Int J Mol Sci. 2012;13:13134–13139. doi: 10.3390/ijms131013134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall L, White RJ. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer. 2008;8:911–914. doi: 10.1038/nrc2539. [DOI] [PubMed] [Google Scholar]
- 39.Marchal JA, et al. The impact of PKR activation: From neurodegeneration to cancer. FASEB J. 2014;28:1965–1974. doi: 10.1096/fj.13-248294. [DOI] [PubMed] [Google Scholar]
- 40.García MA, et al. Impact of protein kinase PKR in cell biology: From antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moon Y. Ribosomal alteration-derived signals for cytokine induction in mucosal and systemic inflammation: Noncanonical pathways by ribosomal inactivation. Mediators Inflamm. 2014;2014:708193. doi: 10.1155/2014/708193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youssef OA, et al. Potential role for snoRNAs in PKR activation during metabolic stress. Proc Natl Acad Sci USA. 2015;112:5023–5028. doi: 10.1073/pnas.1424044112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.