Significance
The human mitochondrial ribosome (mitoribosome) recycling factor (RRFmt) is known to play essential roles in mitochondrial physiology, including protein synthesis, and it has been implicated in human genetic diseases. The RRFmt is among the few protein molecules that carry their N-terminal signal peptide sequence into the mitochondrial matrix that is required for RRFmt’s interaction with the mitoribosome. In this study, we present a cryo-electron microscopic structure of the human mitoribosome in complex with the RRFmt. The structure reveals hitherto unknown features of RRFmt and its interactions with the functionally important regions of the ribosomal RNA that constitute the peptidyl-transferase center and that are linked to the GTPase-associated center of the mitoribosome, shedding light on the mechanism of ribosome recycling in mitochondria.
Keywords: human mitochondrial RRF, mito-specific sequence, 55S–RRFmt complex, cryo-EM structure, mito-specific interactions
Abstract
Mammalian mitochondrial ribosomes (mitoribosomes) are responsible for synthesizing proteins that are essential for oxidative phosphorylation (ATP generation). Despite their common ancestry with bacteria, the composition and structure of the human mitoribosome and its translational factors are significantly different from those of their bacterial counterparts. The mammalian mitoribosome recycling factor (RRFmt) carries a mito-specific N terminus extension (NTE), which is necessary for the function of RRFmt. Here we present a 3.9-Å resolution cryo-electron microscopic (cryo-EM) structure of the human 55S mitoribosome-RRFmt complex, which reveals α-helix and loop structures for the NTE that makes multiple mito-specific interactions with functionally critical regions of the mitoribosome. These include ribosomal RNA segments that constitute the peptidyl transferase center (PTC) and those that connect PTC with the GTPase-associated center and with mitoribosomal proteins L16 and L27. Our structure reveals the presence of a tRNA in the pe/E position and a rotation of the small mitoribosomal subunit on RRFmt binding. In addition, we observe an interaction between the pe/E tRNA and a mito-specific protein, mL64. These findings help understand the unique features of mitoribosome recycling.
Ribosomes are highly complex macromolecular structures that orchestrate the process of protein synthesis in coordination with messenger RNAs (mRNAs), transfer RNAs (tRNAs) and multiple translational factors. Mitochondria are cellular organelles that carry their own genetic material and gene-expression machinery including ribosomes. Mammalian mitochondria synthesize 13 polypeptides that form essential components of the oxidative phosphorylation machinery (1). Although the ribosomal RNA (rRNA) components of the mitochondrial ribosome (mitoribosome) are encoded by mitochondrial (mt)-DNA, all the proteins required for mitochondrial translation including the mt-ribosomal proteins and mt-translation factors are encoded by nuclear genes, translated by the cytoplasmic ribosomes and then transported into the mitochondria (2). The mitoribosomes and their associated translation machinery are distinct from those in the cytoplasm and display features reminiscent of prokaryotic translation (3), in line with the assumption that mitochondria have evolved from endocytosis of an α-proteobacterium by an ancestral eukaryotic cell (3).
The basic features of ribosome recycling are known mostly through biochemical and structural studies of the bacterial system. After the termination step of protein synthesis, the mRNA and a deacylated tRNA in its peptidyl/exit (P/E) state remain associated with the ribosome (4, 5), and the complex is referred to as the posttermination complex (PoTC). To initiate a new round of protein synthesis, the ribosome-bound ligands must be removed from the PoTC and the ribosome must be split into its two subunits. Disassembly of the PoTC requires a concerted action of two protein factors: the ribosome recycling factor (RRF) and the elongation factor G (EF-G) (4, 6, 7). The binding of EF-G in conjugation with guanosine 5′-triphosphate (GTP) to the RRF-bound PoTC dissociates the 70S ribosome into its two subunits on GTP hydrolysis (8).
High-resolution crystallographic studies of RRF from several bacterial species revealed its two-domain structure that adopts nearly an “L” conformation (e.g., refs. 9 and 10). Previous hydroxyl radical probing experiments (11), cryo-electron microscopic (cryo-EM) reconstructions (12–15), and X-ray crystallographic structures (16–18) of RRF-bound ribosome and functional PoTCs have revealed that, although domain I of RRF occupies an almost identical binding position in various RRF-bound 70S structures, domain II adopts different positions in relation to domain I, and has been proposed to play an important role in ribosome recycling (19). Domain I binds to the 50S subunit, spanning over its binding sites for peptidyl (P) and aminoacyl (A) tRNAs and interacting with some key elements of the 23S rRNA, including the P-loop of the peptidyl transferase center (PTC) (20), helices 69 (H69), and 71 (H71), whereas domain II localizes to the intersubunit space in close proximity to the protein L11 stalk-base from the large subunit and protein S12 from the 30S subunit (12–15, 17). Cryo-EM studies of the spinach chloroplast ribosome, in complex with chloroplast-RRF, which possesses an extended N terminus, and hibernation-promoting factor PSRP1, have shown that the overall binding position of chloroplast-RRF on the chlororibosome is similar to that in the bacterial ribosome (21, 22). However, structure of the extended N terminus of the chloroplast-RRF is unknown. Previous studies have inferred RRF-induced disruption of the intersubunit bridges would facilitate subunit separation (12–14, 16, 17).
Despite anticipated similarity between the bacterial and mitochondrial ribosomes, the first mammalian mitoribosome cryo-EM structure had revealed considerable divergence from their bacterial counterparts and several acquired unique features (23) that were subsequently confirmed in higher-resolution studies (24–26). The most striking difference is the reversal in the protein to RNA ratio: Whereas the bacterial ribosomes are high in rRNA, the mitoribosomes are high in protein (27). The overall sedimentation coefficient of the mitoribosome is 55S, and those of its small and large subunits are 28S and 39S, respectively (27). Most mitochondrial translational factors have also acquired insertions and extensions compared with their bacterial counterparts (28, 29). Thus, despite overall resemblances with the prokaryotic system in terms of sequence of events and the accessory protein factors involved, there are considerable differences as well. For example, only two initiation factors, IF2mt and IF3mt, have been identified in mammalian mitochondria, and homolog for IF1 is absent (30–34). Unlike in bacteria, where a single EF-G molecule participates in both the elongation and ribosome recycling steps (35), mammalian mitochondria uses two isoforms of EF-G: EF-G1mt, which functions as a translocase during the polypeptide elongation step (36), and EF-G2mt (37), which works exclusively with RRFmt (38) to catalyze the mitoribosome recycling.
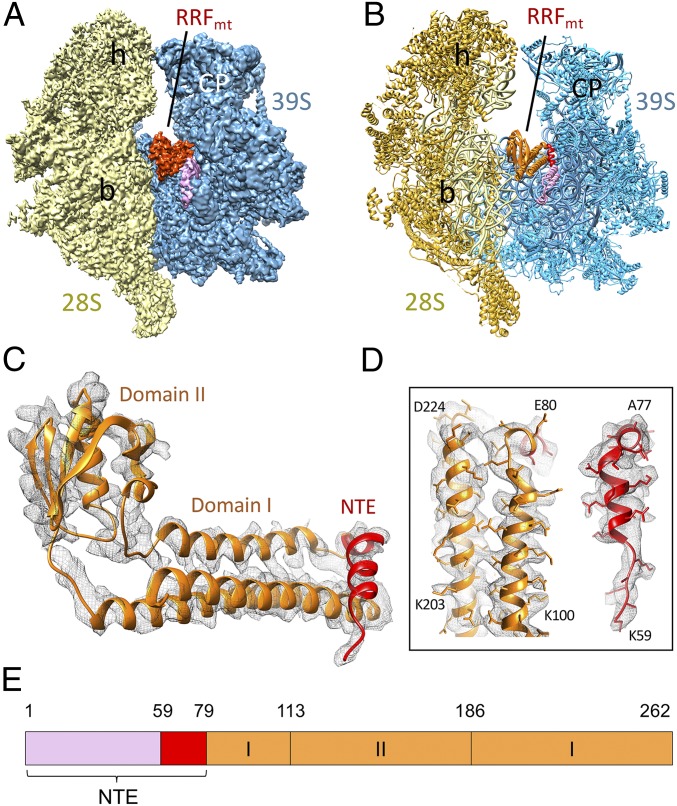
The amino acid (aa) sequence of the human RRFmt is ∼25–30% identical to its bacterial homologs and carries an additional 80-aa-long extension at its N terminus (39). RRFmt was shown to be essential for the viability of human cell lines, and depletion of this factor leads to mitoribosomal aggregation and mitochondrial dysmorphism (40). Here we present a 3.9-Å-resolution cryo-EM structure of the human RRFmt in complex with human 55S mitoribosome and investigate the molecular interactions of RRFmt and its mito-specific N-terminal extension (NTE) with the 55S mitoribosome to gain insights into the process of mitoribosome recycling.
Results and Discussion
RRFmt Binds to the Model Mitoribosome PoTC.
We purified RRFmt carrying the 79-aa NTE, and prepared a model PoTC by treating the mitoribosome with puromycin. The RRFmt binds efficiently to the model mitoribosome PoTC. We obtained an overall 3.7-Å-resolution structure of the mitoribosome–RRFmt complex (SI Appendix, Fig. S1), which showed a clear density for RRFmt, but a relatively weak density for the 28S mitoribosomal subunit, suggesting existence of more than one conformational states for the 28S subunit. Therefore, the dataset was further classified to capture conformational states of the 28S subunit.
RRFmt Preferentially Binds the Rotated 55S Mitoribosome.
After 3D classification, we obtained four subpopulations from a total of 144,051 selected particle images that included three major classes corresponding to intact 55S mitoribosomes with and without bound RRFmt and dissociated 39S mitoribosomal subunit (SI Appendix, Fig. S1C). The 55S classes with and without bound RRFmt were refined to 3.9-Å (Fig. 1) and 4.4-Å resolution, respectively. Superimposition of the RRFmt-bound 55S map (henceforth referred to as Class I) with the published similar resolution cryo-EM maps of the human (24) and porcine (25) mitoribosomes revealed that the conformation of the 28S subunit in our structure is different from the porcine and Class I conformational state of the published human mitoribosomes (24), in both their classical unrotated conformations but similar to the rotated Class II conformation state. This rotation is similar to the ratchet-like intersubunit rotation observed in bacterial ribosome, where the small subunit shows a coordinated 5–10° rotation in a counter clockwise direction with respect to the large subunit (41, 42), with largest and smallest displacements taking place in the peripheral and central regions, respectively, of the small subunit. In our 55S-RRFmt structure, the 28S subunit rotates by ∼8.5° with respect to the 39S subunit (SI Appendix, Fig. S2A), using a pivot point of rotation at the 12S rRNA base A1162 within h27 [all rRNA helices are identified according to bacterial numbering (43, 44)] on the 28S body. This movement results in maximum displacement involving the mito-specific protein S22 by ∼15 Å that lies at the bottom of the 28S-subunit body. In addition to the ratchet-like rotation of the whole 28S subunit, the head of the 28S subunit rotates by ∼5° in an orthogonal direction toward the E site, which is similar to the swiveling movement described previously (45, 46) (SI Appendix, Fig. S2B), resulting in displacement of the 28S head protein S29 further away from the 39S subunit.
Fig. 1.
Cryo-EM structure of the human 55S mitochondrial ribosome in complex with RRFmt. (A) 3D cryo-EM map of the 55S mitoribosome-RRFmt complex as seen from the subunit–subunit interface side, with segmented densities corresponding to the small subunit (28S, yellow), large subunit (39S, blue), and RRFmt (orange, red, and pink). (B) Molecular interpretation of the cryo-EM map shown in A. A darker shade of yellow differentiates the 28S ribosomal proteins from the 12S rRNA, whereas a lighter shade of blue differentiates the 39S ribosomal proteins from the 16S rRNA. Landmarks of the 28S subunit: h, head; b, body. Landmarks of the 39S subunit: CP, central protuberance. (C and D) Molecular model of RRFmt as derived from the cryo-EM map, showing well-resolved densities for both the conserved domains I and II (orange) and the NTE (red). The better-resolved segments of RRFmt with sidechains are shown in D. (E) Domain organization of the human RRFmt. The structurally resolved and unresolved portions of the NTE are depicted in red and pink colors, respectively (SI Appendix, Fig. S8).
Interestingly, the conformation of the empty 55S cryo-EM map (henceforth referred to as Class II) is not only different from RRFmt-bound 55S map (Class I), but also from the published bovine (23) and porcine (25) 55S mitoribosome structures. This conformational change involves a ∼8° rotation of the 28S subunit around a long axis of the subunit, such that its shoulder side moves toward 39S subunit while its platform side moves away from the 39S subunit (SI Appendix, Fig. S2C). Previous cryo-EM studies of the empty human 55S mitoribosome (24) and posttranslocational-state mammalian 80S ribosome (47) have also reported the existence of similar conformation that is termed subunit rolling (47). The fact that the 55S ribosome in the unrotated state is empty (i.e., not bound by RRFmt) suggests that the rotated state is the preferred substate for RRFmt binding. It should be noted that the ribosome in eubacterial ribosome-RRF complex is almost invariably captured in a rotated state (14–16); however, it is not clear whether RRF binding induces such a state or whether it binds to a prerotated state ribosome. In both class I and II maps, we find weak mass of densities corresponding to indigenously bound mRNA (SI Appendix, Fig. S3 E and F) and a tRNA in the pe/E state (SI Appendix, Fig. S3 A–C) (45), the anticodon of which is situated toward the P site but closer to the E site (24, 48). The anticodon of pe/E tRNA moves by ∼10 Å along with the 12S rRNA segment G1419-G1421 of the 28S subunit head in the RRFmt-bound state (SI Appendix, Supplementary Results and Figs. S3D and S4). However, we do not observe any change in the mRNA density between the two classes. In both classes, the anticodon stem-loop of the tRNA interacts with the mito-specific protein mL64 (SI Appendix, Fig. S3 A and B), suggesting a direct role of this protein in tRNA movement and stability in the pe/E state. This finding also reveals that the deacylated tRNA remains bound to the mitoribosome upon RRFmt binding, and its position is significantly different from the usual P/E state deacylated-tRNA present in the analogous eubacterial complexes (15).
Bridges between the two ribosomal subunits are essential for a functionally intact ribosome (23–25, 49, 50) that allow dynamic movements required to facilitate various steps of the translation cycle. There are about 15 intersubunit bridges that have been identified in the mammalian 55S ribosome (23, 24), and seven of them are either destabilized or completely broken during ratchet-like intersubunit rotation (24). Interestingly, three (mB1a, mB1b, and mB2) of these seven bridges are specific to the mitoribosomes, involving mito-specific proteins S29, L40, L46, and L48 (24, 25). The RRFmt binding could stabilize the rotated conformational state of the mitoribosome with multiple weakened mito-specific intersubunit bridges to prepare the complex for the subsequent step of subunit dissociation on EF-G2mt binding.
Molecular Interactions of RRFmt with Components of the 55S Ribosome.
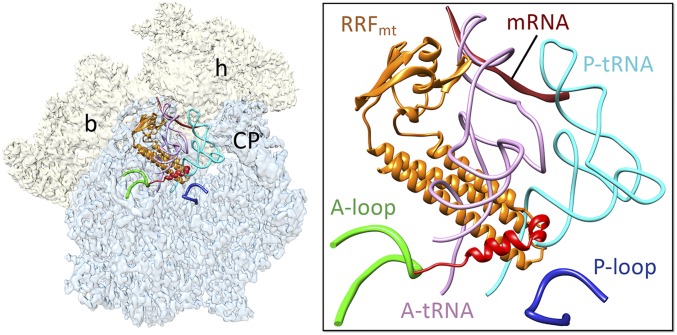
In our cryo-EM map of the 55S-RRFmt complex, a well-resolved “L”-shaped density readily attributable to RRFmt is visible in the intersubunit space of the 55S mitoribosome (Fig. 1 A and B and SI Appendix, Fig. S5), matching the overall size and domain composition of the conserved portion of RRFmt to that of bacterial RRF on (12–17) and off (9, 10) the 70S ribosome. The long arm of the l-shaped density is represented by three long α-helices that run parallel to each other and constitute the larger domain I, with the mito-specific N-terminal sequence extending from the tip of domain I in an obliquely perpendicular direction, whereas the smaller domain II is composed of an α/β motif (Fig. 1C). As in bacterial ribosome–RRF complexes, RRFmt in mitoribosome is located on the cleft of the 39S subunit and is positioned such that its domain I is oriented toward the PTC and overlaps with the binding positions of the A- and P-site tRNAs, whereas domain II is oriented toward the intersubunit space and is positioned close to protein S12. In its current position, domain I will preclude the binding of both A- and P-site tRNAs in their classical A/A and P/P binding states (Fig. 2), but it would allow the binding of P-site tRNA in its P/E hybrid state. Thus, the overall binding positions of structurally conserved regions of domains I and II of RRFmt in our structure is almost identical to those of domain I of RRF in the bacterial (12, 14–17) and chloroplast ribosomes (22).
Fig. 2.
Binding positions of bound RRFmt, mRNA, and A- and P-site tRNAs on the mitoribosome. RRFmt binding would be in direct steric clash with the acceptor arm of both aminoacyl- and peptidyl- tRNAs on the large subunit. Coordinates of the mRNA (dark brown) and tRNAs (pink and light blue) were derived from the structure of porcine mitoribosome (25) (PDB ID: 5AJ4). The conserved RRFmt domains and its NTE are colored as in Fig. 1. The NTE of RRFmt lies in close proximity to the functionally important and conserved A (green) and P loops (dark blue). A thumbnail to left depicts an overall orientation of the 55S mitoribosome, with semitransparent 28S (yellow) and 39S (blue) subunits, and overlaid positions of ligands. Landmarks on the thumbnail: h, head, and b, body of the 28S subunit, and CP, central protuberance of the 39S subunit.
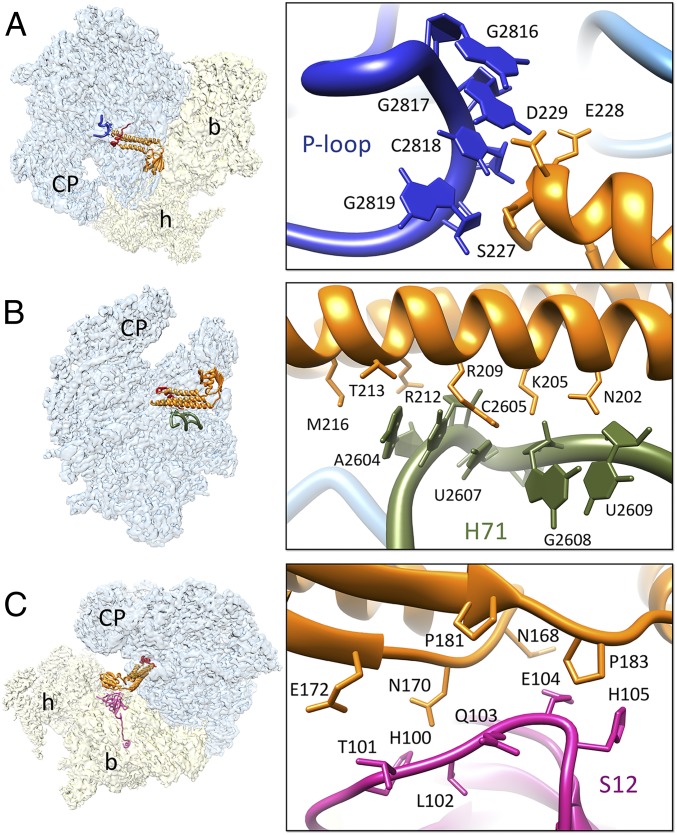
Interactions of domain I.
Although the overall shape and composition are significantly different among the bacterial, mitochondrial, and chloroplast ribosomes, their internal rRNA cores are very much conserved (21–26). The strategic positioning of domain I on the 55S mitoribosome allows RRFmt to interact with several important elements of the large subunit, such as components of the PTC, helix 69 (H69), and helix 71 (H71), which are known to play crucial roles during various phases of the bacterial translational cycle (51). The tip of domain I is positioned very close to the P-loop region (H80) of the PTC, such that its aa residues Ser227–Asp229 make direct contacts with the rRNA bases G2816–G2819 (Fig. 3A). The P-loop region is known to interact with the CCA end of the P-site bound tRNA, and hence it is essential that the peptidyl tRNA is removed from the P site of the 39S subunit before the binding of RRFmt. Interestingly, these interacting residues are conserved among bacterial, mitochondrial, and chloroplast RRFs; accordingly, structural studies of the bacterial and chloroplast 70S-RRF complexes (12–17, 21) have shown similar interactions of domain I with the P-loop. The three long α-helices of domain I run almost parallel to a portion of 16S rRNA helix H71 such that aa residues Asn202, Lys205, Arg209, Arg212, Thr213, and Met216 from α-helix 3 of RRFmt make extensive interactions with the H71 rRNA bases A2604–U2609 (Fig. 3B). Surprisingly, except for the two arginine residues (Arg209 and Arg-212), none of the interacting α-helix 3 residues are conserved between the bacterial and the human mitochondrial RRF, and mutation of Arg209 (Arg132 in Escherichia coli) to other amino acid residues drastically reduces the affinity of RRF to the 70S ribosome (52). This high network of interactions seems to provide the essential anchoring points for RRFmt on the 39S subunit, as mutations in the analogous region of the prokaryotic factor can lead to cell death or generation of temperature-sensitive phenotypes in bacteria (53). Although domain I of RRFmt is positioned in the close vicinity of H69, no specific interaction between domain I and H69 was detected. In the presence of RRFmt, the conformation of H69, a key 16S rRNA helix that partners with the functionally important 12S rRNA helix 44 to form the highly conserved and crucial intersubunit bridge B2a in the 55S ribosome, slightly changes and moves in the direction of the 28S head movement (SI Appendix, Fig. S6), as it has been shown to adopt different conformations in bacterial ribosome on RRF binding (12, 14, 15, 18). Because similar change has been also observed in the rotated 28S subunit in the absence of RRFmt (24), it is likely that the observed change in our RRFmt-bound map is primarily the effect of subunit rotation.
Fig. 3.
Interactions of structurally conserved domains of RRFmt with the 55S mitoribosome. (A) Contacts between the domain I (orange) and the P loop, the 16S rRNA H80 (blue). (B) Interactions between the domain I and H71 (olive green) of the 16S rRNA. (C) Contacts between the ribosomal protein S12 (magenta) and RRFmt domain II. Thumbnails to left depict overall orientations of the 55S mitoribosome, with semitransparent 28S (yellow) and 39S (blue) subunits, and overlaid positions of RRFmt. Landmarks on the thumbnails are same as in Fig. 2.
Interactions of domain II.
The positioning of domain II in the present structure is different compared with the orientation of domain II in the 70S-RRF (17) and the 70S-chlRRF (21) complexes. Although two RRF domains are placed in almost right angles to each other in most bacterial complexes, domain II adopts a more open conformation in the chloroplast complex. In our structure, domain II occupies an intermediate position relative to the positions of domain II in the bacterial and chloroplast complexes (SI Appendix, Fig. S7). None of the ribosomal components from the 39S subunit are found within interacting distance from domain II of RRFmt. The only structural element from the 28S subunit that is found in close proximity to domain II of RRFmt is protein S12. A loop region of protein S12 encompassing His100–His105 is within hydrogen-bonding distance (∼4 Å) from the asparagine and proline-rich segment that involves aa residues Asn168, Asn170, Glu172, Pro181, and Pro183 of the RRFmt domain II (Fig. 3C). Both the prolines and Asn170 are conserved among the bacterial and human mitochondrial RRFs. Although interactions between S12 and bacterial RRF are known to exist in the 70S-RRF structures (12, 14, 16), these interactions are more extensive in our 55S-RRFmt complex.
Structure and interactions of the mito-specific NTE.
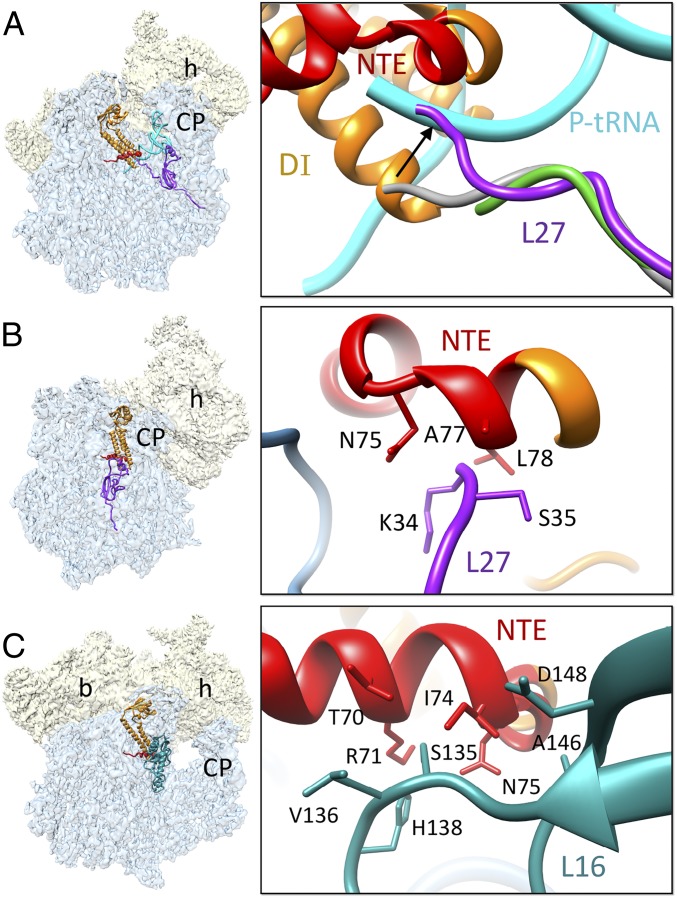
The human RRFmt carries a functionally essential mito-specific 80-aa extension at its N terminus (39, 40) (SI Appendix, Fig. S7). In our map, we find additional density (Fig. 1 C and D) at the apical region of RRFmt domain I that is contiguous with the RRFmt α-helix1 and could be readily assigned to the last 21-aa residues of the NTE. Residues Lys65–Val79 fold into a well-defined α-helix, and residues Lys59–Gly64 constitute a loop region (Fig. 1 C and D). The observed NTE segment runs beyond the C terminus α-helix in an oblique perpendicular direction to rest of the domain I, and is found sandwiched between the 39S subunit and rest of the RRFmt domain I, where it makes several novel interactions with the 39S subunit. The density corresponding to remaining 59-aa residues of the NTE appears to be flexible, as it is somewhat fragmented and is visible only at low-density threshold levels in the low-pass filtered Class I map at ∼7 Å resolution (SI Appendix, Fig. S8A).
The ribosomal elements that are within reach of the resolved 21-aa segment of NTE are 16S rRNA helices H89, H90, and H92 and ribosomal proteins L16 and L27. Protein L27 lies in close proximity to the PTC and was reported to interact with the apex region of domain I in the bacterial recycling complex (15, 17, 18). The N-terminal region of L27 is known to be highly flexible, and hence was disordered in the factor-free (46, 49) as well as RRF-bound 70S ribosomal complexes (17). The mature human mitoribosomal protein L27 is ∼118 aa long, which is slightly larger than its bacterial counterpart. In our structure, we have observed additional density extending from the previously known N terminus of L27, compared with that in empty human 55S ribosome structure (24), suggesting that contacts with the NTE of RRFmt have stabilized the N terminus of the L27 in our map, and therefore, allowed us to model additional aa residues, except for the first three amino acid residues. Compared with the empty human and porcine 55S ribosomal structures (24, 25), the N-terminal region of L27 has undergone a large conformational change, and moves by ∼9 Å toward the P-site tRNA (Fig. 4A), such that it would occlude the binding of the accepter arm of the peptidyl-tRNA on the 39S subunit (Fig. 4A). The N terminus of L27 makes multiple contacts with the mito-specific NTE of RRFmt through its residues Ser35 and Lys34. Ser35 interacts with Ala77 and Leu78, whereas the conserved Lys34 makes close contacts with Asn75 and Leu78 from the NTE of RRFmt (Fig. 4B). Thus, L27 could stabilize the binding of RRFmt through the mito-specific interactions described here. The large subunit protein L16 is known to be in close proximity to bacterial RRF (14, 17, 18), but specific interactions between the two proteins could not be identified in previous studies. L16 and the NTE of RRFmt contact each other at multiple sites. Residues Ser135–His138, Ala146, and Asp148 from the L16 are involved in direct interactions with the NTE residues Thr70, Arg71, Ile74, and Asn75 (Fig. 4C). Outside the NTE, Leu233 from RRFmt domain I also makes close contact with His138 of L16.
Fig. 4.
Mito-specific Interactions between the NTE of RRFmt and the 39S mitoribosomal subunit proteins. (A) The N-terminal region of L27 (purple) in the human 55S–RRFmt complex has shifted by ∼9 Å toward the peptidyl tRNA-binding site compared with the positions of L27 in the porcine (gray) (25) (PDB ID: 5AJ4) and empty human (green) (24) (PDB ID: 3J9M) mitoribosomes, to interact with the NTE (red) of RRFmt. In this conformation, L27 would block the binding of tRNA in the 39S P site. Mito-specific interactions between the NTE and mitoribosomal proteins are shown in panels (B) L27 and (C) L16 (dark cyan). Thumbnails to the left depict overall orientations of the 55S mitoribosome, with semitransparent 28S (yellow) and 39S (blue) subunits, and overlaid positions of ligands. Landmarks on the thumbnails are same as in Fig. 2.
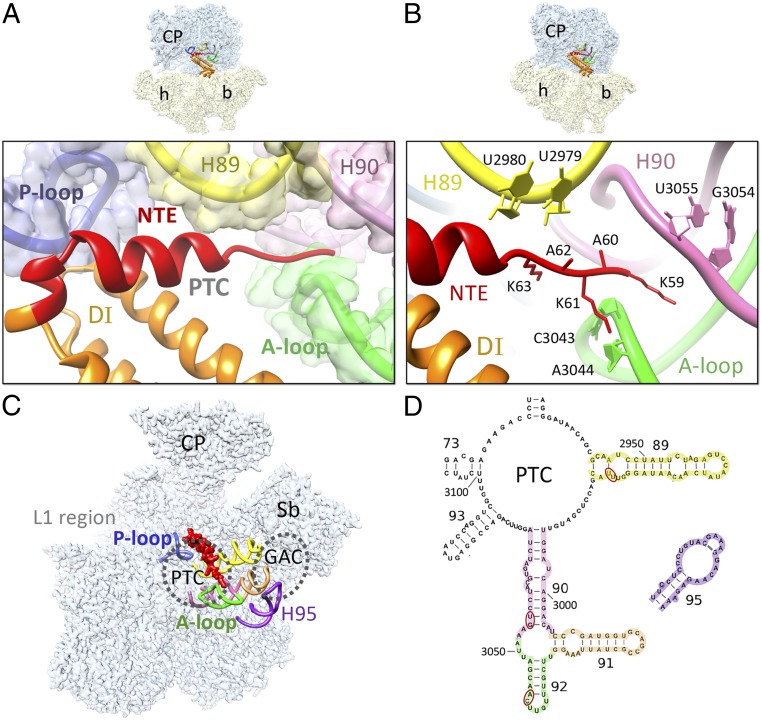
The RRFmt NTE interaction with the 16S rRNA helix 92 (H92) is of particular interest because it has the functionally important A-loop that is known to interact with the CCA end of the A-site tRNA and is thus associated with PTC (Fig. 5A). Lys61 from the RRFmt NTE interacts with nucleotides C3043 and A3044 of the 16S rRNA H92, whereas the neighboring Ala60 also contacts C3043 (Fig. 5B). This stretch of the NTE has unusual presence of multiple lysine residues. RRFmt NTE residues Ala62 and Lys63 interact with the 16S rRNA H89 base U2979 (Fig. 5B), and residues Gly64 and Gln67 is positioned very close to the neighboring base U2980. It should be noted that H89, along with the poorly resolved 59-aa segment of the NTE (SI Appendix, Fig. S8B), extends up to the GTPase-associated center of the L7/L12 stalk base (50), and these interactions might influence the subsequent EF-G2mt-catalyzed GTP hydrolysis step. Unlike the 70S ribosomal recycling, GTP hydrolysis on EF-G2mt is not required for the splitting of the 55S ribosome, and the hydrolysis reaction is only necessary for the release of EF-G2mt from the mitoribosome (38). A strategically placed lysine residue Lys59 from the NTE interacts with the backbone phosphates of bases G3054 and U3055 on the 16S rRNA H90 through charge-based interactions (Fig. 5B). Interestingly, all three rRNA helices are part of the domain V of 23S rRNA that comprise the highly-conserved PTC (Fig. 5 C and D), whereas helices H89-93 constitute the region of domain V that is known for stabilizing the elongation factor-binding region of the 70S ribosome (43). Simultaneous interactions of the RRFmt NTE with three 16S rRNA helices that have been implicated in EF-G binding (H90, H92) and GTP hydrolysis (H89) would have direct effect on the subsequent EF-G2mt–mediated steps during the mitoribosome recycling process.
Fig. 5.
Mito-specific interactions between the NTE of RRFmt and the 16S rRNA components of the 39S subunit. (A) The NTE (red) of RRFmt interacts with several functionally important segments of the peptidyl-transferase center (PTC), including the P loop (blue), H89 (yellow), H90 (pink), and A loop (H92) (green). (B) Mito-specific interactions of the NTE with the nucleotide bases of H89, H90, and the A loop. Color codes are same as in A. Thumbnails on the top depict overall orientations of the 55S mitoribosome in A and B, with semitransparent 28S (yellow) and 39S (blue) subunits, and overlaid positions of RRFmt. Landmarks on the thumbnails are same as in Fig. 2. (C) Overall location of the NTE on the 39S subunit (semitransparent blue), as seen from the subunit’s interface side, with the rRNA components of PTC that extend up to the GTPase-associated center (GAC), which includes the α-sarcin-ricin stem loop (H95). Both PTC and GAC are depicted as dashed circles. (D) Secondary structure of the PTC region of the 16S rRNA, highlighting helices that are color-coded as in A–C, and interacting nucleotides with red ovals.
In conclusion, our study provides direct visualization and structure of the human mitoribosome-bound human RRFmt and its mito-specific NTE, and their interactions with the functionally relevant components of the mitoribosome. These include both A and P loops of the PTC (20) and components of the PTC that directly communicate with the GTPase-associated center of the large ribosomal subunit. Our observation that the mito-specific RRFmt NTE spans across A and P loops, encompassing the entrance of the nascent polypeptide-exit tunnel, suggests that NTE interaction with the mitoribosome ensures a complete inaccessibility to tRNAs and other ligands to both PTC and the entrance-nascent polypeptide-exit tunnel of the mitoribosome. This situation is in sharp contrast to that in bacterial RRF, which directly interacts only with the P loop. Extensive interactions between the RRFmt and mitoribosome are a feature of RRFmt-mediated mitoribosome recycling. The presence of a density corresponding to tRNA in the pe/E state is also in contrast to the situation in analogous eubacterial complex, which carries a deacylated tRNA in the P/E state. Future structural studies involving EF-G2mt should provide further insights into the precise functional roles of the interactions observed in the present study.
Materials and Methods
Referenced details of the Materials and Methods, including mitoribosome isolation, purification, overexpression and purification of RRFmt, preparation of mitoribosome-RRFmt complex, cryo-EM data collection, image processing, and model building are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Linda Spremulli, Zafia Chrzanowska-Lightowlers, and Robert Lightowlers for help in initial stages of the project by providing RRFmt clones. We thank Mona Gupta and Prem Kaushal for their initial contributions to the project, ArDean Leith and Nilesh Banavali for help with computation, and Nilesh Banavali for critical reading of the manuscript. We acknowledge the use of the Wadsworth Center’s Media and Culture core facility for help in producing large volumes of HEK293S GnTI cells, and the Wadsworth Center’s and New York Structural Biology Center’s EM facilities. New York Structural Biology Center EM facilities are supported by grants from the Simons Foundation (349247), NYSTAR, the National Institutes of Health (GM103310), and the Agouron Institute (F00316). This work was supported by National Institutes of Health Grant R01 GM61576 (to R.K.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM maps and atomic coordinates have been deposited in the Electron Microscopy and PDB Data Bank (www.wwpdb.org) under accession codes EMD-0514 and PDB ID 6NU2, respectively, for the RRFmt-bound 55S mitoribosome (Complex I) and EMD-0515 and PDB ID 6NU3, respectively, for the unbound 55S mitoribosome (Complex II).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1815675116/-/DCSupplemental.
References
- 1.O’Brien TW, Denslow ND, Anders JC, Courtney BC. The translation system of mammalian mitochondria. Biochim Biophys Acta. 1990;1050:174–178. doi: 10.1016/0167-4781(90)90162-u. [DOI] [PubMed] [Google Scholar]
- 2.Lightowlers RN, Rozanska A, Chrzanowska-Lightowlers ZM. Mitochondrial protein synthesis: Figuring the fundamentals, complexities and complications, of mammalian mitochondrial translation. FEBS Lett. 2014;588:2496–2503. doi: 10.1016/j.febslet.2014.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pel HJ, Grivell LA. Protein synthesis in mitochondria. Mol Biol Rep. 1994;19:183–194. doi: 10.1007/BF00986960. [DOI] [PubMed] [Google Scholar]
- 4.Kaji A, et al. The fourth step of protein synthesis: Disassembly of the posttermination complex is catalyzed by elongation factor G and ribosome recycling factor, a near-perfect mimic of tRNA. Cold Spring Harb Symp Quant Biol. 2001;66:515–529. doi: 10.1101/sqb.2001.66.515. [DOI] [PubMed] [Google Scholar]
- 5.Karimi R, Pavlov MY, Buckingham RH, Ehrenberg M. Novel roles for classical factors at the interface between translation termination and initiation. Mol Cell. 1999;3:601–609. doi: 10.1016/s1097-2765(00)80353-6. [DOI] [PubMed] [Google Scholar]
- 6.Janosi L, et al. Evidence for in vivo ribosome recycling, the fourth step in protein biosynthesis. EMBO J. 1998;17:1141–1151. doi: 10.1093/emboj/17.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rao AR, Varshney U. Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 2001;20:2977–2986. doi: 10.1093/emboj/20.11.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peske F, Rodnina MV, Wintermeyer W. Sequence of steps in ribosome recycling as defined by kinetic analysis. Mol Cell. 2005;18:403–412. doi: 10.1016/j.molcel.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Saikrishnan K, Kalapala SK, Varshney U, Vijayan M. X-ray structural studies of Mycobacterium tuberculosis RRF and a comparative study of RRFs of known structure. Molecular plasticity and biological implications. J Mol Biol. 2005;345:29–38. doi: 10.1016/j.jmb.2004.10.034. [DOI] [PubMed] [Google Scholar]
- 10.Selmer M, Al-Karadaghi S, Hirokawa G, Kaji A, Liljas A. Crystal structure of Thermotoga maritima ribosome recycling factor: A tRNA mimic. Science. 1999;286:2349–2352. doi: 10.1126/science.286.5448.2349. [DOI] [PubMed] [Google Scholar]
- 11.Lancaster L, Kiel MC, Kaji A, Noller HF. Orientation of ribosome recycling factor in the ribosome from directed hydroxyl radical probing. Cell. 2002;111:129–140. doi: 10.1016/s0092-8674(02)00938-8. [DOI] [PubMed] [Google Scholar]
- 12.Agrawal RK, et al. Visualization of ribosome-recycling factor on the Escherichia coli 70S ribosome: Functional implications. Proc Natl Acad Sci USA. 2004;101:8900–8905. doi: 10.1073/pnas.0401904101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barat C, et al. Progression of the ribosome recycling factor through the ribosome dissociates the two ribosomal subunits. Mol Cell. 2007;27:250–261. doi: 10.1016/j.molcel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Gao N, et al. Mechanism for the disassembly of the posttermination complex inferred from cryo-EM studies. Mol Cell. 2005;18:663–674. doi: 10.1016/j.molcel.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Yokoyama T, et al. Structural insights into initial and intermediate steps of the ribosome-recycling process. EMBO J. 2012;31:1836–1846. doi: 10.1038/emboj.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weixlbaumer A, et al. Crystal structure of the ribosome recycling factor bound to the ribosome. Nat Struct Mol Biol. 2007;14:733–737. doi: 10.1038/nsmb1282. [DOI] [PubMed] [Google Scholar]
- 18.Wilson DN, et al. X-ray crystallography study on ribosome recycling: The mechanism of binding and action of RRF on the 50S ribosomal subunit. EMBO J. 2005;24:251–260. doi: 10.1038/sj.emboj.7600525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo P, Zhang L, Zhang H, Feng Y, Jing G. Domain II plays a crucial role in the function of ribosome recycling factor. Biochem J. 2006;393:767–777. doi: 10.1042/BJ20050780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 21.Perez Boerema A, et al. Structure of the chloroplast ribosome with chl-RRF and hibernation-promoting factor. Nat Plants. 2018;4:212–217. doi: 10.1038/s41477-018-0129-6. [DOI] [PubMed] [Google Scholar]
- 22.Sharma MR, et al. Cryo-EM study of the spinach chloroplast ribosome reveals the structural and functional roles of plastid-specific ribosomal proteins. Proc Natl Acad Sci USA. 2007;104:19315–19320. doi: 10.1073/pnas.0709856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma MR, et al. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 24.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. Ribosome. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greber BJ, et al. Ribosome. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 26.Kaushal PS, et al. Cryo-EM structure of the small subunit of the mammalian mitochondrial ribosome. Proc Natl Acad Sci USA. 2014;111:7284–7289. doi: 10.1073/pnas.1401657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: Implications for human genetic disease. Gene. 2002;286:73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 28.Christian BE, Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta. 2012;1819:1035–1054. doi: 10.1016/j.bbagrm.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma MR, Kaushal PS, Gupta M, Banavali NK, Agrawal RK. Insights into structural basis of mammalian mitochondrial translation. In: Duchêne A-M, editor. Translation in Mitochondria and Other Organelles. Springer; Berlin: 2013. pp. 1–28. [Google Scholar]
- 30.Gaur R, et al. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol Cell. 2008;29:180–190. doi: 10.1016/j.molcel.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koc EC, Spremulli LL. Identification of mammalian mitochondrial translational initiation factor 3 and examination of its role in initiation complex formation with natural mRNAs. J Biol Chem. 2002;277:35541–35549. doi: 10.1074/jbc.M202498200. [DOI] [PubMed] [Google Scholar]
- 32.Koripella RK, et al. Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit. iScience. 2019;12:76–86. doi: 10.1016/j.isci.2018.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kummer E, et al. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature. 2018;560:263–267. doi: 10.1038/s41586-018-0373-y. [DOI] [PubMed] [Google Scholar]
- 34.Yassin AS, et al. Insertion domain within mammalian mitochondrial translation initiation factor 2 serves the role of eubacterial initiation factor 1. Proc Natl Acad Sci USA. 2011;108:3918–3923. doi: 10.1073/pnas.1017425108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savelsbergh A, Rodnina MV, Wintermeyer W. Distinct functions of elongation factor G in ribosome recycling and translocation. RNA. 2009;15:772–780. doi: 10.1261/rna.1592509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhargava K, Templeton P, Spremulli LL. Expression and characterization of isoform 1 of human mitochondrial elongation factor G. Protein Expr Purif. 2004;37:368–376. doi: 10.1016/j.pep.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 37.Hammarsund M, et al. Identification and characterization of two novel human mitochondrial elongation factor genes, hEFG2 and hEFG1, phylogenetically conserved through evolution. Hum Genet. 2001;109:542–550. doi: 10.1007/s00439-001-0610-5. [DOI] [PubMed] [Google Scholar]
- 38.Tsuboi M, et al. EF-G2mt is an exclusive recycling factor in mammalian mitochondrial protein synthesis. Mol Cell. 2009;35:502–510. doi: 10.1016/j.molcel.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Spremulli LL. Identification and cloning of human mitochondrial translational release factor 1 and the ribosome recycling factor. Biochim Biophys Acta. 1998;1443:245–250. doi: 10.1016/s0167-4781(98)00223-1. [DOI] [PubMed] [Google Scholar]
- 40.Rorbach J, et al. The human mitochondrial ribosome recycling factor is essential for cell viability. Nucleic Acids Res. 2008;36:5787–5799. doi: 10.1093/nar/gkn576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank J, Agrawal RK. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 42.Frank J, Agrawal RK. Ratchet-like movements between the two ribosomal subunits: Their implications in elongation factor recognition and tRNA translocation. Cold Spring Harb Symp Quant Biol. 2001;66:67–75. doi: 10.1101/sqb.2001.66.67. [DOI] [PubMed] [Google Scholar]
- 43.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 44.Wimberly BT, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 45.Ratje AH, et al. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schuwirth BS, et al. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 47.Budkevich TV, et al. Regulation of the mammalian elongation cycle by subunit rolling: A eukaryotic-specific ribosome rearrangement. Cell. 2014;158:121–131. doi: 10.1016/j.cell.2014.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaushal PS, Sharma MR, Agrawal RK. The 55S mammalian mitochondrial ribosome and its tRNA-exit region. Biochimie. 2015;114:119–126. doi: 10.1016/j.biochi.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 50.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 51.Horan LH, Noller HF. Intersubunit movement is required for ribosomal translocation. Proc Natl Acad Sci USA. 2007;104:4881–4885. doi: 10.1073/pnas.0700762104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishino T, et al. Interaction of ribosome recycling factor and elongation factor EF-G with E. coli ribosomes studied by the surface plasmon resonance technique. Genes Cells. 2000;5:953–963. doi: 10.1046/j.1365-2443.2000.00382.x. [DOI] [PubMed] [Google Scholar]
- 53.Janosi L, et al. Mutations influencing the frr gene coding for ribosome recycling factor (RRF) J Mol Biol. 2000;295:815–829. doi: 10.1006/jmbi.1999.3401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.