Significance
Cells respond to nutritional challenges by dramatically reprogramming their transcriptomes. In bacteria, transcriptional regulators are often pleiotropic, complicating identification of direct versus indirect effects. Here, we present the genome-wide transcriptional profile resulting from RNA polymerase (RNAP) binding by ppGpp, a transcription regulator found throughout the bacterial domain of life. Most of the more than 750 transcripts identified here were not identified as regulated by ppGpp in previous studies. The excellent correlation between the genes identified as regulated by ppGpp in our RNA-seq analysis and promoters regulated directly by ppGpp in vitro facilitated identification of sequence features distinguishing inhibited, activated, and unregulated promoters from each other, providing tools for future mechanistic studies of ppGpp action.
Keywords: stringent response, ppGpp, DksA, promoter, transcription
Abstract
The second messenger nucleotide ppGpp dramatically alters gene expression in bacteria to adjust cellular metabolism to nutrient availability. ppGpp binds to two sites on RNA polymerase (RNAP) in Escherichia coli, but it has also been reported to bind to many other proteins. To determine the role of the RNAP binding sites in the genome-wide effects of ppGpp on transcription, we used RNA-seq to analyze transcripts produced in response to elevated ppGpp levels in strains with/without the ppGpp binding sites on RNAP. We examined RNAs rapidly after ppGpp production without an accompanying nutrient starvation. This procedure enriched for direct effects of ppGpp on RNAP rather than for indirect effects on transcription resulting from starvation-induced changes in metabolism or on secondary events from the initial effects on RNAP. The transcriptional responses of all 757 genes identified after 5 minutes of ppGpp induction depended on ppGpp binding to RNAP. Most (>75%) were not reported in earlier studies. The regulated transcripts encode products involved not only in translation but also in many other cellular processes. In vitro transcription analysis of more than 100 promoters from the in vivo dataset identified a large collection of directly regulated promoters, unambiguously demonstrated that most effects of ppGpp on transcription in vivo were direct, and allowed comparison of DNA sequences from inhibited, activated, and unaffected promoter classes. Our analysis greatly expands our understanding of the breadth of the stringent response and suggests promoter sequence features that contribute to the specific effects of ppGpp.
The second messengers ppGpp and pppGpp (together abbreviated here as ppGpp) have long been known to alter gene expression after starvation of Escherichia coli for a variety of nutrients, down-regulating stable RNA (rRNA and tRNA) genes and up-regulating stress and starvation-related genes such as those for amino acid biosynthesis (1, 2). ppGpp regulates transcription initiation from specific promoters by binding directly to RNA polymerase (RNAP) at two sites, generally conserved in proteobacteria (3–5). “Site 1” is at the interface of the ω subunit and the β′ subunit (4), and “site 2” is at the interface of the transcription factor DksA and the secondary channel rim helices of β′ (5). DksA is a small RNAP-binding protein present at relatively constant concentrations in E. coli (6, 7). The combined effects of amino acid substitutions in β′, ω, and DksA within the two ppGpp binding sites render RNAP insensitive to effects of ppGpp on transcription initiation in vitro and in vivo (5).
ppGpp can also bind to and affect the activities of a large number of other proteins in addition to RNAP (1, 2), thereby playing roles in processes as diverse as DNA replication (8, 9), translation (10–12), nucleotide metabolism (2, 11–14), ribosome biogenesis (11, 12, 15, 16), amino acid decarboxylation (17), polyphosphate metabolism (18), and ppGpp metabolism itself (11, 19, 20). Our studies were designed to distinguish the effects of ppGpp on gene expression resulting from its binding to RNAP versus other cellular targets.
Global transcriptional effects of ppGpp have been examined previously, utilizing expression microarrays and strains containing or lacking the genes for ppGpp synthesis (relA and spoT) following starvation for serine (21) or isoleucine (22, 23). The transcriptomes of wild-type, ∆relA ∆spoT, and ∆dksA strains were also compared in early stationary phase (24). Several hundred regulated genes were identified, indicating that ppGpp affects the expression of many cellular products. However, these studies did not distinguish effects on transcription resulting from ppGpp binding directly to RNAP from effects of ppGpp binding to other targets or from secondary events subsequent to the direct effects of ppGpp binding to RNAP. Distinguishing direct from indirect effects is necessary for understanding the mechanisms underlying a regulatory network.
High-throughput sequencing technologies (RNA-seq) that have emerged since publication of the initial global studies are more sensitive, reproducible, and versatile than expression microarrays for genome-wide transcription regulation studies (25). To study the global transcriptional effects of direct ppGpp binding to RNAP, we used RNA-seq to identify regulated transcripts genome-wide produced before and after inducing synthesis of ppGpp in strains with or without the ppGpp binding sites on RNAP. Analysis of transcripts produced after only 5 min of ppGpp induction reduced identification of products resulting from secondary events. Comparison of the effects of ppGpp in strains with and without the ppGpp binding sites on RNAP rather than in strains with and without the ability to synthesize ppGpp eliminated potential effects from loss of ppGpp binding to other cellular targets. Furthermore, by producing ppGpp without nutrient starvation, we avoided identification of effects resulting from changes in metabolism independent of ppGpp production.
The results show that all transcriptional responses to ppGpp under these conditions result from binding of ppGpp to RNAP rather than to transcription factors or other potential targets. The much larger number of genes and pathways identified here as regulated by ppGpp indicates that the transcriptional response to ppGpp is considerably broader than realized previously. A large subset of the regulated promoters was analyzed by in vitro transcription, identifying DNA sequence features contributing to the promoter-specific effects of ppGpp and thus to the mechanism of the stringent response.
Results and Discussion
Transcriptional Response to Synthesis of ppGpp by Induction of a relA Expression Plasmid.
To study the transcriptional effects of ppGpp binding to RNAP and to identify which of its effects are direct, we used a two-pronged approach. First, RNA-seq was used to identify regulated transcripts genome-wide after synthesis of ppGpp in strains with and without the ppGpp binding sites on RNAP, and second a large number of the promoters identified in vivo were analyzed by in vitro transcription to unambiguously identify direct effects of ppGpp.
ppGpp was produced without concurrent starvation by conditional expression of a RelA variant from plasmid pALS13 lacking its autoinhibitory domain, allowing continued synthesis of ppGpp when induced with isopropyl β-d-1-thiogalactopyranoside (IPTG) from the tac promoter in strains with or without the two binding sites for ppGpp on RNAP (referred to as 1+2+ or 1−2−) (Fig. 1 A and B). Plasmid pALS14, which codes for an inactive RelA variant, was used as a control for effects of IPTG induction. We previously demonstrated that representative negatively (rRNA) or positively (iraP) regulated promoters were not regulated in the 1−2− strain in vivo following induction of ppGpp by amino acid starvation with serine hydroxamate (5). We evaluated use of the inducible relA plasmid system instead of treatment with serine hydroxamate, first by testing the response of the promoters for transcription of rRNA and the iraP mRNA.
Fig. 1.
Validation of ppGpp overexpression method. (A) Strains. (B) Workflow of the RNA-seq experiment. (C) qPCR analysis of unstable precursor RNA transcripts from chromosomal rRNA operons after induction of RelA from pALS13 and pALS14 with 1 mM IPTG. (D) qPCR analysis of iraP transcripts after IPTG addition as in C. Means and SDs are from three independent experiments.
Unlike the mature rRNAs, the rRNA precursors are highly unstable, and the level of the leader RNA can serve as an estimate of rRNA promoter activity. ppGpp induction inhibited rRNA leader synthesis approximately fourfold in the wild-type strain in the first 5 min, but there was little or no effect in the 1−2− strain or in the strains with the inactive ppGpp production plasmid (Fig. 1C). The positively regulated iraP mRNA increased approximately fourfold within 5 min of ppGpp induction in the 1+2+ strain but not in the 1−2− strain or in the strains with pALS14 (Fig. 1D). The fold effects of ppGpp were qualitatively consistent with, although somewhat smaller than, the results obtained after treatment with serine hydroxamate (5), suggesting that the level of ppGpp produced in our inducible system might be somewhat lower than that produced by wild-type relA during a severe amino acid starvation. Residual activity of the chromosomally encoded relA product could also limit the fold increase in ppGpp concentration after IPTG induction (and thus the observed magnitude of the transcriptional response).
We compared ppGpp and relA mRNA levels in the 1+2+ and 1−2− strains after conditional expression of the active RelA variant without concurrent starvation (SI Appendix, Figs. S1 and S2). ppGpp increased only in the strains containing pALS13 and not with the control plasmid pALS14 (SI Appendix, Fig. S1 A and B). Maximum ppGpp levels in the 1−2− strain were ∼70% as high as those in the 1+2+ strain, correlating qualitatively with relA mRNA levels in the two strains (SI Appendix, Fig. S2 A and B). The slight reduction in ppGpp production in the 1−2− strain could not account for the complete loss of regulation observed, given the reported IC50 for ppGpp in vitro (5) and the threshold concentration of ppGpp required for its effects on rRNA transcription in vivo (26).
Few, if Any, Effects of ppGpp on Transcription Result from ppGpp Binding to Targets Other than RNAP During the Initial Phase of the Response.
RNA was sequenced from four strains (1+2+ and 1−2−, each with pALS13 or pALS14) before (time 0) and at 5 and 10 min after addition of IPTG to induce ppGpp synthesis. Three biological replicates were measured at each time point (Fig. 1 A and B).
The RNA-seq results are displayed as a heatmap (Fig. 2A) comparing RNA transcripts that changed in a statistically significant manner by more than twofold to those at time 0 in response to ppGpp synthesis (log2 ≥ 1.0 or −1; adjusted P value < 0.05; false discovery rate controlled). The results for individual genes are provided in three different formats. In general, fold effects are reported in the text, whereas log2 values are reported in Datasets S1 and S2. A spreadsheet listing the log2 responses to ppGpp for all detected transcripts is provided as Dataset S1, grouped by statistical category, as well as gene identifier information (including b number and gene coordinates). Dataset S2 lists all genes alphabetically, including those for which no transcripts were detected or for which the effect was not statistically significant. SI Appendix, Table S1 ranks the magnitudes of the effects for those genes most affected by ppGpp (i.e., the top 10% negatively regulated and positively regulated genes) and indicates the putative function of the gene product.
Fig. 2.
Global gene expression changes in response to ppGpp induction. (A) Heatmap shows log2 changes in the 1+2+ and 1−2− strains. (B) Venn diagram showing number of genes inhibited or activated at least twofold in response to ppGpp in the 1+2+ and 1−2− strains. Small solid red circle (1−2−, 5 min) in the activated genes diagram represents two genes, one unique to the 1−2− strain at 5 min and one shared with the other strains.
The number of genes whose expression was inhibited or activated after ppGpp production in the 1+2+ (wild-type) or 1−2− (mutant) strains is shown in the Venn diagram in Fig. 2B. In the 1+2+ strain, expression of 757 genes changed twofold or more (log2 ≥ 1.0 or −1) by 5 min (inhibited and activated genes combined), and expression of 1,224 genes changed by 10 min. In contrast, expression of only three genes changed twofold or more in the 1−2− strain in the first 5 min and 52 genes by 10 min. Two of the three transcripts that changed in the first 5 min in the 1−2− strain changed only slightly (ydgI, 2.3-fold inhibited; phoH, 2.6-fold activated in Dataset S1). Even though these transcripts met our arbitrary twofold cutoff for biological significance, they changed much more in the 1+2+ strain (15.6-fold decrease and 9.9-fold increase, respectively). Thus, whatever mechanism accounted for the effects of ppGpp on these transcripts in the 1−2− strain, it accounted for only a minor fraction of the regulation observed in the strain with wild-type RNAP. The third transcript, yeeD, remained unchanged in the 1+2+ strain, although it increased slightly in the mutant strain. We conclude that there are very few, if any, effects of ppGpp on transcription that result from ppGpp binding to targets other than RNAP, ruling out a major contribution of ppGpp to effects on transcription from binding to transcription factors (or other proteins) during the initial phase of the stringent response.
In the 1+2+ host strain without IPTG, the level of ppGpp at time 0 (i.e., without induction) was the same with the plasmids encoding the active and inactive relA. Thus, the basal ppGpp level was determined by the wild-type relA and spoT genes on the chromosome, not by potential leaky expression from the plasmid-encoded relA (SI Appendix, Expanded Materials and Methods). Interestingly, ppGpp was higher in the 1−2− strain than in the 1+2+ strain (SI Appendix, Fig. S1 A and B). Although our understanding of the mechanisms controlling basal ppGpp levels is far from complete, there is an inverse correlation between ppGpp levels and the steady-state growth rate (26). Since the 1−2− strain grew significantly slower than the 1+2+ strain under these growth conditions (30-min versus 20-min doubling time), probably because of its altered regulation of transcription genome-wide even in the relatively rich medium used here, the higher basal levels of ppGpp could result from the reduced growth rate. Alternatively, the higher levels of ppGpp could cause the decreased growth rate, or there could be feedback systems controlling ppGpp synthesis and/or degradation that are affected by ppGpp binding to RNAP. We return later in the manuscript to the genes whose expression differs in the 1−2− and 1+2+ strains in the absence of induction of the relA plasmid.
The Breadth of the Transcriptional Response to ppGpp Is Greater than Recognized Previously.
A much larger number of genes responded to ppGpp in our study than in a previous expression microarray study in which serine hydroxamate was used to induce ppGpp synthesis (21). Because the timescales were the same, we were able to compare our results with those in that study. Expression of 104 genes changed within 5 min in the previous report, whereas 757 genes responded to ppGpp induction within 5 min in our study. Ten minutes after ppGpp induction, 264 genes changed in the previous study, whereas 1,224 genes changed in our study. The previous study used a cutoff of 2.8-fold (log2 = 1.5) versus the twofold cutoff used in our study. When we used a log2 = 1.5 cutoff, approximately threefold more genes were still identified in our study (325 and 754 genes at 5 and 10 min, respectively, versus the 104 and 264 genes at 5 and 10 min, respectively, in the previous study). Not all of the genes regulated by ppGpp in the previous report were also identified as regulated in our study. In summary, more than 75% of the genes identified as regulated by ppGpp in our study were not identified as regulated by ppGpp previously. The genes we identified probably still represent a lower limit of the number of transcripts affected by ppGpp, since ppGpp synthesis from the chromosomally encoded relA or spoT genes would likely obscure changes in transcription that might occur at very low ppGpp concentrations.
There were a number of differences in experimental design that could account for the differences in the results obtained in the different studies. First, ppGpp was induced by amino acid starvation in the reports cited above rather than by induction of relA with IPTG. We suggest that many of the transcripts reported previously might have resulted from changes in metabolism, independent of ppGpp binding to RNAP. Second, use of ppGpp binding-deficient mutant strains (4, 5) made it possible for us to eliminate ppGpp effects on RNAP without eliminating ppGpp production, retaining ppGpp binding to other cellular targets and thus their effects on metabolism. Third, many secondary effects were probably eliminated by our focus on genes responding within the first 5 min of ppGpp induction. Finally, the previous studies all employed expression microarrays, the best technology available for genome-wide analysis at the time. However, this technique has largely been superseded by the much more sensitive and reproducible RNA-seq technology (25), allowing analysis of regulation of lower abundance transcripts.
The results on the flagella regulon illustrate how induction of ppGpp from the relA plasmid and analysis within 5 min resulted in identification of genes only directly regulated by ppGpp. The flagella regulon is a three-tiered transcriptional cascade that consists of FhlDC, a master regulator that turns on expression of genes in a second level of the cascade including the flagellar sigma factor σ28 encoded by fliA. In the third level of the cascade, σ28 activates a multigene operon that includes fliC, the gene coding for the major flagellin (27). Our RNA-seq data indicate that ppGpp directly inhibits transcription of fhlDC, but not the genes in the third level of the cascade, consistent with our previous conclusions (28) and in contrast to the conclusions from an expression microarray study that reported that all but one of the genes in the flagellar cascade were inhibited by ppGpp (22).
We used a version of the “Pathway Tools Omics Dashboard” slightly modified from the one available on the EcoCyc webserver (29) to visualize our transcriptomic data. The results are presented in a series of panels, each containing genes broadly related to a cell function defined by the Omics Dashboard tool (Fig. 3, SI Appendix, Fig. S3, and legends). Although there is some arbitrariness in the classifications, the representation provides a snapshot of the breadth of the effects of ppGpp on transcription. We note that this display does not take into account the magnitude of the changes within a particular category, only the percentages of genes whose expression increased or decreased at least twofold in response to ppGpp production. For brevity, most of the discussion below is limited to the results at 5 min after IPTG addition, although the data at 10 min are also presented in Datasets S1 and S2.
Fig. 3.
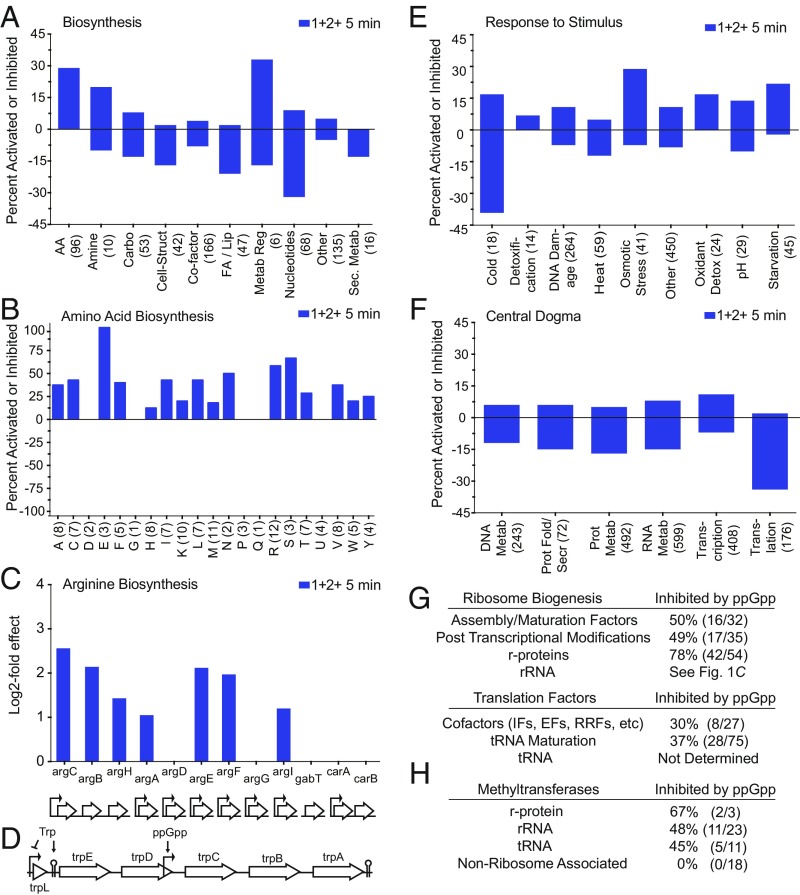
Visualization of transcriptomics data in representative categories. Bars above the line represent percentages of genes increased by ppGpp, and bars below the line represent percentages of genes inhibited by ppGpp. Number of genes in the category is indicated beneath its name on the x axis. Categories are in alphabetical order. (A) EcoCyc dashboard “Biosynthesis” category. (B) Amino acid biosynthesis pathways whose transcription is regulated by ppGpp. (C) Arginine biosynthesis-related operons regulated by ppGpp. (D) Tryptophan biosynthesis operon regulation by ppGpp. (E) ppGpp-responsive genes in the EcoCyc Dashboard “Response to Stimulus” category. (F) ppGpp-responsive genes in the EcoCyc Dashboard “Central Dogma” category. (G) ppGpp-responsive genes related to translation (SI Appendix, Table S2). (H) ppGpp-responsive genes coding for methyltransferases. ppGpp-responsive genes in additional EcoCyc Dashboard categories are in SI Appendix, Fig. S3.
Biosynthesis.
In general, biosynthesis genes [including amino acid biosynthesis (AA), fatty acid biosynthesis, and nucleotide biosynthesis genes] responded to ppGpp induction (Fig. 3A). Many amino acid biosynthesis pathways increased even though all 20 aa were provided in the culture medium, but ppGpp did not increase expression of all amino acid biosynthesis pathways the same (Fig. 3B). Furthermore, not all genes within a regulon for a given amino acid responded the same, for example, expression of at least 7 of the 12 genes in the arginine biosynthesis pathway (R) increased after ppGpp induction, even though they might have been expected to have been repressed by ArgR in the presence of arginine (Fig. 3C).
The analysis also identified a previously unsuspected ppGpp-responsive promoter in the tryptophan biosynthetic operon (trpLEDCBA). Transcription from the trp operon is regulated by both the trp repressor, TrpR, and by a transcriptional attenuator responsive to translation of the leader peptide, trpL (30). Since tryptophan was present in the medium, transcription of trpLED was turned off. However, transcription from the downstream part of the operon, trpCBA, increased within 5 min in response to ppGpp (>2-fold for trpC and 1.9-fold for trpB and trpA). Our data suggest that a previously identified promoter just upstream of trpC (31) is ppGpp responsive (Fig. 3D), perhaps increasing tryptophan levels by allowing trpCBA to synthesize tryptophan from serine and indole when trpLED transcription is off (32).
Response to stimulus.
Fig. 3E indicates that many genes in this diverse category, which includes genes for the responses to DNA damage, osmotic stress, oxidants, and environmental treatments, unexpectedly responded to ppGpp. These results suggest that ppGpp senses not only nutrition-based stresses. For example, a high percentage of cold response genes appear to be sensitive to ppGpp, as predicted previously (33).
Central dogma: RNA, protein, and nucleotide synthesis.
A large number of genes related to nucleotide, protein, and RNA metabolism, translation, and DNA synthesis are negatively regulated by ppGpp (Fig. 3F), consistent with many previous reports (21, 23, 34). Our data are consistent with the general picture that cells go into survival mode in response to production of ppGpp, turning down synthesis of products required for growth and division.
ppGpp has long been known to regulate ribosomal RNA synthesis directly, whereas effects of ppGpp on ribosomal protein synthesis have generally been ascribed to posttranscriptional regulation in which most r-protein mRNAs are translationally inhibited and destabilized by binding specific translational repressor r-proteins. The activities of these r-proteins as translational repressors are ultimately determined by whether they are titrated away by binding to rRNAs whose synthesis is controlled directly by ppGpp (35). Some fraction of this regulation of r-protein mRNA stability likely occurs very rapidly, within just a minute or two of ppGpp induction (36), but our results along with those from some earlier studies indicate that there are also direct effects of ppGpp on r-protein promoters (37, 38). Although the mRNAs for some r-proteins were too short to be retained in our RNA preparation, all measurable r-protein mRNAs (corresponding to 78% of the total number of r-protein genes) were inhibited by ppGpp within 10 min. Nevertheless, during a stringent response, the translational feedback mechanism rather than the direct effect on promoter activity is still likely to be responsible for the majority of inhibition of r-protein mRNAs by ppGpp (39).
Our results greatly expand the number of genes related to the translation machinery that are regulated by ppGpp. Fig. 3G and SI Appendix, Table S2 indicate that about one-half of the genes related to translation were inhibited by ppGpp induction, including genes coding for r-proteins, rRNA processing, ribosome maturation and modification enzymes, and ribosome assembly, initiation, elongation, and termination factors. Consistent with a recent proposal that SuhB, a protein originally annotated as an inositol monophosphatase, plays a role in transcription antitermination and/or ribosome assembly (40), we found that SuhB is one of the most strongly ppGpp-inhibited transcripts (14-fold; SI Appendix, Table S1).
tRNAs were not included in our RNA-seq study, because they were too short to be retained quantitatively during RNA preparation (Materials and Methods). However, 12 of the 86 tRNA genes are within rRNA operons and are thus directly regulated by ppGpp (35), and promoters for many other tRNAs are also likely to be regulated directly by ppGpp (e.g., ref. 41). The RNA-seq analysis showed that some genes coding for tRNA processing and modifying enzymes (e.g., rnpA, encoding the protein component of RNase P; and rluB, encoding a tRNA pseudouridine synthase) are also among the most strongly ppGpp inhibited genes (SI Appendix, Table S2 and Datasets S1 and S2).
Other affected transcripts in the “central dogma” category include those encoding rRNA helicases, GTPases, and chaperones needed for ribosome assembly (DeaD, SrmB, RimM, RimP, Era, ObgE), RNases (RNase III, RNase T), as well as polyamine transporters and RNA-modifying enzymes (methyltransferases, pseudouridylases, and acetyltransferases). Although transcription of some components of the ribosome has long been implicated as a target of ppGpp, the number of genes in the translation category directly regulated more than twofold within 5 min by ppGpp is far greater than recognized previously (at least 115 genes; SI Appendix, Table S2). For example, to our knowledge, methyltransferases have not been identified previously as part of the ppGpp regulon (SI Appendix, Table S3). Our data indicate that only the methyltransferases that modify substrates involved in translation are regulated by ppGpp (Fig. 3H and SI Appendix, Table S3).
As noted above, a major goal of our study was to determine whether ppGpp binding to proteins other than RNAP played a direct role in the effects of ppGpp on transcription. Previous screens have identified ∼70 E. coli proteins that bind ppGpp directly in vitro, and many of the activities of these enzymes are inhibited (11, 12). In addition to altering transcription from many genes whose products participate in nucleotide biosynthesis (SI Appendix, Table S2), ppGpp binds directly to at least 11 nucleotide biosynthesis-related enzymes, reducing NTP pools (2, 11, 12). Effects of ppGpp on NTP levels are critical components of the regulation of transcription, including rRNA transcription, in E. coli as well as in other bacteria (3, 11, 12, 14). Changes in NTP pools in the 1−2− strain are apparent even in the thin-layer chromatograms (SI Appendix, Fig. S1).
At least 13 of the targets that ppGpp binds to directly are related to translation, including GTPases required for ribosome assembly, initiation, or elongation. The reduction in NTP synthesis and thereby NTP levels (especially GTP) by ppGpp would make ppGpp more effective as a competitive inhibitor of the GTPases. Direct inhibition of transcription of mRNAs responsible for synthesis of nucleotides and the translation apparatus, as well as direct ppGpp inhibition of nucleotide synthesis and translation-related enzymes themselves, are likely to synergize to tightly control nucleotide metabolism and translation during a stringent response.
Other pathways regulated by ppGpp.
The omics dashboard analysis also identified some classes of genes whose regulation by ppGpp was not expected. As illustrated in SI Appendix, Fig. S3D, transcription from many genes in the “Energy” category was activated by ppGpp. Many genes in the large “Cell Exterior” category were also regulated by ppGpp (SI Appendix, Fig. S3F), including a large number of genes in the cell wall, plasma membrane, transport, and LPS metabolism groups. Eight of the 12 most inhibited genes and 5 of the 8 most activated genes are annotated as coding for importers, transporters, permeases, or outer membrane proteins (SI Appendix, Table S1). Previous reports have implicated ppGpp as a regulator of fatty acid enzyme expression or enzyme activity (e.g., ref. 42).
Regulation by ppGpp Under Steady-State Conditions.
As indicated above, there were differences in steady-state transcription in the 1+2+ and 1−2− strains (i.e., at time 0, before IPTG addition). Many of the 223 genes that were expressed at least twofold differently in the two strains were in pathways whose expression also changed in response to ppGpp induction (compare SI Appendix, Table S4 and SI Appendix, Fig. S4 with Dataset S1 and Fig. 3). A substantial number of the transcripts that were differentially expressed in steady state were in amino acid biosynthetic pathways and were lower in the 1−2− than in the 1+2+ strain, consistent with the requirement for ppGpp for activation of amino acid biosynthetic pathway expression. Similarly, transcripts from nucleotide biosynthesis genes were higher in the 1−2− than in the 1+2+ strain in the absence of ppGpp induction, consistent with their inhibition by ppGpp. However, some of the differences between the transcriptomes of the wild-type and 1−2− strains, like those from comparison of the wild-type and ∆relA ∆spoT (or ∆dksA) strains, may reflect transcripts that changed to compensate for defects in cellular metabolism in the mutant strains and not promoters that are regulated directly by ppGpp.
The RNA-Seq Dataset Is a Good Predictor of Promoters Negatively Regulated by ppGpp and an Even Better Predictor of Promoters Positively Regulated by ppGpp.
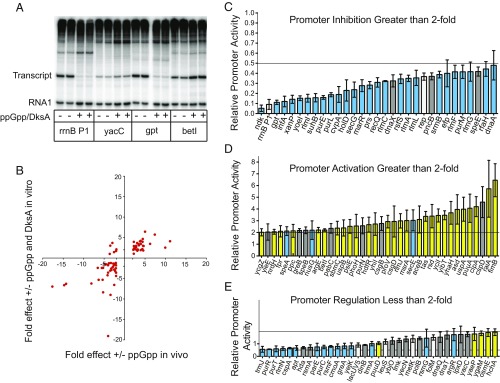
The 757 genes whose transcripts were regulated within 5 min by ppGpp represent 586 operons. To ask whether the promoters for these genes are regulated directly by ppGpp, 134 promoters from the RNA-seq dataset were chosen for analysis in vitro in a purified system, either because they were affected strongly in vivo, the function of the product was of interest to us, or both. DNA fragments containing the promoters were cloned into pRLG770, a plasmid containing transcription terminators downstream of the promoter fragment insertion site (4). In vitro transcription analysis was performed on reactions containing ppGpp and DksA (since DksA forms part of ppGpp binding site 2) (5). Transcripts were detected from 104 promoters, including 77 that were regulated at least twofold by ppGpp within 5 min in the RNA-seq analysis described above.
A representative gel showing effects of ppGpp and DksA on three of the promoters (as well as on rrnB P1) is shown in Fig. 4A. For those promoters analyzed both in vitro and in vivo, the effects in vivo were generally somewhat larger than the effects observed in vitro (not surprising since solution conditions were not optimized for individual promoters). Thirty-eight of the 44 genes whose transcription was inhibited at least twofold by ppGpp in vivo after 5 min of ppGpp induction (86%) were also inhibited in vitro, and 33 of the 34 genes that were activated in vivo were also activated in vitro (97%). The effect of ppGpp/DksA on each of these 77 transcripts is compared side-by-side in vivo and in vitro in SI Appendix, Table S6, and the overall correlation is illustrated by a scatterplot (Fig. 4B, plotted on a linear scale to match that in SI Appendix, Table S6). As indicated above, discrepancies in the responses to ppGpp in vitro versus in vivo were primarily quantitative, not qualitative. A small number of cases in which genes were inhibited slightly in vivo but activated slightly in vitro (upper left quadrant of the plot) will require further analysis.
Fig. 4.
Transcription regulation by ppGpp/DksA in vitro. (A) Representative transcriptional responses to ppGpp/DksA in vitro (rrnB P1 and gpt, inhibited more than twofold; yacC, regulated less than twofold; betI, activated more than twofold). Some unrelated gel lanes between the rrnB P1 and yacC reactions were cropped out. RNA transcripts (labeled “transcript”) are ∼200 nt, ∼50 nt (+1 to approximately +50) from the DNA fragment containing the promoter, and ∼150 nt (approximately +50 to approximately +200) from the rrnB T1 terminator fragment in the plasmid. The RNA 1 transcript (∼110 nt) encoded by the plasmid served as an internal control, indicating that the transcription reaction was active even when no transcript was obtained from the test promoter. (B) Scatterplot comparing transcriptional responses to ppGpp/DksA in vitro (y axis) with responses after 5 min of ppGpp induction in the RNA-seq analysis (x axis). Fold effects in the scatterplot are displayed on a linear scale to match the fold effects in SI Appendix, Table S6. Points represent effects on the 77 individual genes whose expression increased more than twofold (data points to the right of the y axis) or decreased more than twofold (data points to the left of the y axis) in vivo and that were also analyzed by transcription in vitro. The six transcripts that increased in vitro but decreased in vivo were argR, speA, rsmG, secE, nusG, and trmA (SI Appendix, Table S6). (C–E) Responses to ppGpp and DksA of the 104 promoters for which transcripts were obtained in vitro. Promoters are named by the gene immediately downstream. (C) Promoters inhibited more than twofold (transcription <0.5 in vitro compared with without ppGpp/DksA). (D) Promoters activated more than twofold by ppGpp/DksA. (E) Promoters affected less than twofold in vitro. (1 on the y axis = no effect.) Colors indicate responses of the same genes in the RNA-seq dataset. Blue, inhibited; yellow, activated; gray, regulated less than twofold; white, promoters that changed in the presence of IPTG in the strain containing pALS14 (i.e., without ppGpp induction).
Fig. 4 C–E illustrates effects of ppGpp/DksA in vitro on all 104 promoters for which transcripts were obtained. Thirty-one promoters were inhibited from ∼2-fold (dnaA) to as much as ∼20-fold (ndk; Fig. 4C), and 38 promoters were activated from ∼2-fold (ycgZ) to ∼6.5-fold (fimB; Fig. 4D). Effects on the 35 promoters that were affected less than twofold in vitro are illustrated in Fig. 4E. Even though the magnitudes of the effects of ppGpp/DksA in vitro on these promoters in the “unregulated” class were small, most still correlated well with the results obtained in vivo. Positive regulation in vitro was reported previously for only a few of the promoters included in Fig. 4 and SI Appendix, Table S6 (e.g., refs. 43 and 44). The fold effect comparisons of ppGpp/DksA on several amino acid biosynthetic promoters reported previously were not included here, because the solution conditions used in vitro were different (45). However, these promoters are included in the list of promoters that have been tested in vitro (SI Appendix, Table S5).
In the bar graphs, the inhibited promoters are ordered from largest to smallest effect of ppGpp/DksA on transcription, and the activated promoters are ordered from smallest to largest effect on transcription. The colors of the bars match those in the heatmap in Fig. 2A (inhibition in vivo is shown as a blue bar, and activation in vivo is shown as a yellow bar), again illustrating the excellent correlation between the effects observed in the RNA-seq and in vitro transcription analyses, even for many promoters in the “unregulated” class (genes regulated twofold or less in vitro).
There are multiple potential explanations for promoters that were active in the RNA-seq dataset but were inactive in vitro. For example, the in vitro transcription reactions might lack a transcription factor, the promoter might utilize a holoenzyme other than Eσ70, or the solution conditions might be inappropriate. In summary, although our results may be biased somewhat by nonrandom sampling, we conclude that the RNA-seq results are an excellent predictor of direct inhibition in vitro and an even better predictor of direct activation in vitro.
We predicted originally that positive regulation by ppGpp in vivo might be indirect, since amino acid biosynthetic promoters were not activated by ppGpp in vitro (46). It was proposed that, in the cell, positive regulation might arise indirectly from inhibition of rRNA transcription, which in turn would make more RNAP available for promoters that recruited RNAP weakly and thus whose activities could be improved by increasing the RNAP concentration (46, 47). However, the discovery of DksA as a transcription factor and of its role in formation of ppGpp binding site 2 demonstrated that ppGpp can activate amino acid biosynthetic promoters directly (45). Although there may be cases where regulation by ppGpp/DksA in vivo may contain a component that is indirect through reduction in rRNA transcription (e.g., ref. 48), the data presented here suggest that the activation observed for most ppGpp-activated promoters in vivo is likely to be direct.
Identification of Sequence Motifs Associated with Promoters Inhibited or Activated by ppGpp/DksA.
Attempts to identify promoter motifs responsible for regulation by ppGpp/DksA have had two major limitations. First, unlike classical transcription factors that bind to specific DNA sequences, ppGpp and DksA bind to RNAP but do not bind DNA. Rather, the sequence signatures responsible for promoter regulation by ppGpp/DksA are based on specific kinetic properties that result from multiple RNAP–promoter interactions, effects of sequence on DNA trajectory or curvature, and ease of strand separation (3, 49). At this time, it is not possible to predict the kinetic properties of a promoter solely from inspection of its sequence. Second, previous promoter sequence comparisons were compromised because the datasets of ppGpp/DksA-regulated promoters included many promoters that are regulated indirectly.
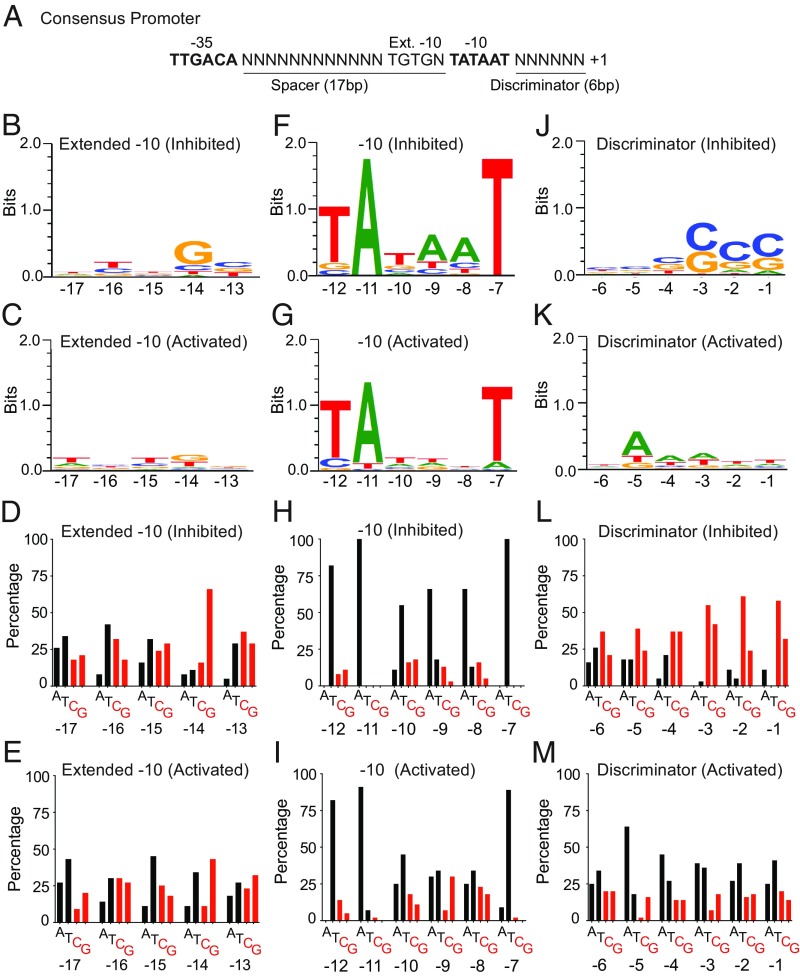
The much larger number of promoters for which there is now evidence for direct regulation by ppGpp both in vitro and in vivo (Fig. 4 and SI Appendix, Table S6) allowed us to begin to search for conserved sequence motifs. The results are displayed as sequence logos (50, 51) and as base distribution histograms (Figs. 5 and 6). In the sequence logos, the overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of individual letter symbols within the stack reflects the relative frequency of the corresponding base at that position. A sequence logo emphasizes the nucleotides shared among promoter sequences, whereas a base distribution histogram emphasizes the enrichment or depletion of each base at a given position (which is sometimes not obvious from a sequence logo). Although 118 promoter sequences were used for this analysis, the three subclasses (inhibited, unregulated, and activated) are still small enough that additional sequences would improve the predictive power of the data. Nevertheless, a few sequence signatures are already apparent.
Fig. 5.
Sequence characteristics of promoters regulated by ppGpp/DksA in vitro (Ext −10, −10, and discriminator elements). (A) Consensus promoter elements (nontemplate strand). Sequence conservation in the extended −10 region (B–E), −10 element (F–I), and discriminator element (J–M). Promoters from the ppGpp/DksA-inhibited class and the ppGpp/DksA-activated class were aligned separately to create sequence logos (50, 51) (B, C, F, G, J, and K) and histograms of base distributions (D, E, H, I, L, and M) for each position. In the histograms, base composition is indicated by black for A or T, and red for C or G. Sequence logos and base distributions from the regulated less than twofold by ppGpp/DksA in vitro promoter class are in SI Appendix, Fig. S5.
Fig. 6.
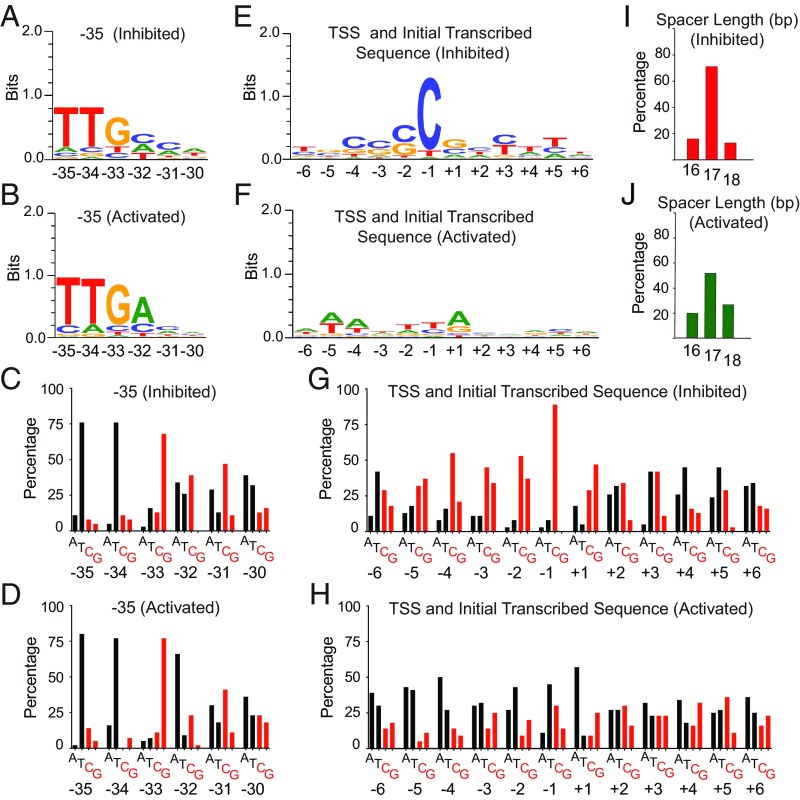
Sequence characteristics of promoters regulated by ppGpp/DksA in vitro (−35 element, TSS region, and −10/−35 spacer length). Sequence conservation in the −35 element (A–D), TSS, and initial transcribed region (E–H) from promoters listed in SI Appendix, Table S5. Sequence logos (50, 51) (A, B, E, and F) and base distribution histograms (C, D, G, and H) are shown for each position as in Fig. 5. (I and J) −10/−35 spacer lengths for the ppGpp/DksA-inhibited and -activated classes.
Bacterial promoters typically contain conserved −10 and −35 elements and two regions of variable sequence and length, the spacer separating the −10 and −35 elements and the discriminator region separating the −10 element from the transcription start site (TSS). The −10/−35 spacer is typically 16–18 bp, and the discriminator is typically 5–8 bp. To accommodate these variable lengths, the −10 element, the −35 element, the extended −10 element, and the discriminator were aligned independent of their exact position relative to the TSS, as were the sequences surrounding the TSS. Figs. 5 and 6 illustrate sequence features of promoters inhibited or activated more than twofold by ppGpp/DksA in vitro, and SI Appendix, Fig. S5 illustrates sequence features in the class regulated by ppGpp/DksA less than twofold in vitro.
−10 element.
Contacts between RNAP and the −10 element have long been recognized as essential for open complex formation (52). The three most highly conserved positions in the −10 element (T-12, A-11, and T-7) (Fig. 5A) matched the consensus sequence in all three promoter classes (inhibited, activated, and unregulated; Fig. 5 F and H, Fig. 5 G and I, and SI Appendix, Fig. S5 C and D, respectively). However, there was a striking difference among the classes at positions −9 and −8, where an A is strongly favored and a G is strongly disfavored in both the inhibited and unregulated classes but not in the activated class (Fig. 5 F–I; the preferences illustrated here are for the nontemplate strand base, but the template strand base could be the one responsible for the effect on transcription). Since A is much preferred over T in the inhibited promoters, A+T content is not an explanation for the preference. Positions −9 and −8 make only backbone contacts to RNAP in the open complex (52), but the preferences could reflect interactions with RNAP in an intermediate not retained in subsequent complexes.
Extended −10 region.
Contacts between RNAP and the extended −10 sequence (Ext −10) immediately upstream of the −10 motif (Fig. 5A) help stabilize the RNAP–promoter interaction. There is little sequence conservation in the Ext −10 region specific to a particular ppGpp/DksA-regulated promoter class (Fig. 5 and SI Appendix, Fig. S5).
Discriminator region.
The sequence between the −10 element and the TSS is often referred to as the discriminator region and has long been implicated in regulation by ppGpp (41). The inhibited promoters have a very high G+C content in the downstream half of the discriminator region, whereas the activated promoters were more A+T-rich in this region (Fig. 5 J–M). The high G+C content presumably makes the double helix in inhibited promoters more difficult to open, and the high A+T content facilitates strand opening of the activated promoters.
Superimposed on the overall G+C versus A+T biases in the discriminators of inhibited versus activated promoters (Fig. 5 J–M), there are specific preferences, most notably a bias for A and against C two positions downstream from the −10 element in activated promoters (Fig. 5M). Although the basis for this sequence preference remains to be determined, we note that the nontemplate strand G at position −5 can make a base-specific contact with σ region 1.2 in the open complex (53, 54), and this interaction interferes with regulation of rRNA promoters by ppGpp (53). The behavior of rRNA promoters indicates that sequence context is critical for regulation by ppGpp/DksA, since changes in several other positions can result in loss of regulation, even in the context of optimal discriminator sequences (53, 55, 56).
TSS region.
Alignment of the discriminator either with respect to the −10 hexamer or with respect to the TSS showed there was a general preference for G+C-rich sequences in the inhibited promoter population and a general preference for A+T-rich sequences in the activated promoter population (Figs. 5 J–M and 6 E–H). However, because the distance between the −10 hexamer and the TSS can vary, it was not apparent from the alignment with respect to the −10 hexamer that there is very strong conservation of a C at −1 in promoters inhibited by ppGpp/DksA (>85% of the inhibited promoter population) (Fig. 6 E and G). A strong preference for C at −1 in the nontemplate strand has been proposed to reflect stabilization of the incoming NTP (most often ATP) by base stacking with a purine on the template strand (57). We suggest that stabilization of the incoming rNTP by a G on the template strand at −1 may be especially important for promoters negatively regulated by ppGpp/DksA, because they form intrinsically unstable open complexes and require higher initiating rNTP concentrations for transcription initiation (55, 58). This property of rRNA promoter complexes is proposed to facilitate promoter escape (59).
−35 element.
We aligned the promoter sequences by their −35 motif to address its impact on regulation by ppGpp and DksA. In all three promoter classes, there was a strong preference for the most conserved bases, TTG, the first three positions, and a strong bias against a G at the fourth position (Fig. 6 A–D and SI Appendix, Fig. S5 G and H). We conclude that there are no strong −35 element preferences for regulation by ppGpp/DksA.
−10/−35 spacer.
Finally, we tabulated the length of the spacer sequence separating the −10 and −35 hexamers in the inhibited and activated promoter classes. Insertions that increase the rrnB P1 spacer length from 16 to 17 bp reduce regulation of that promoter (53), but Fig. 6 I and J shows that the most frequent spacer length in both the inhibited and activated promoter classes is 17 bp (as it is in the E. coli promoter population as a whole), and the fraction of 16- and 18-bp spacer lengths within a class is roughly equal. Thus, a 16-bp spacer is neither a requirement for, nor a reliable predictor of, inhibition or activation by ppGpp/DksA. Like other determinants of regulation by ppGpp/DksA, the effect of spacer length is context dependent.
Prospect.
We have shown here that genome-wide effects of ppGpp on transcription in E. coli are in large part direct, resulting from ppGpp binding to its sites on RNAP. The physiological impact of ppGpp binding to RNAP was already apparent from previous studies (e.g., refs. 5 and 60), but the number of genes directly regulated by ppGpp is much larger than expected. More than 75% of the genes we identified were not identified as regulated in previous studies. The responses of the unexpected members of the regulon will require further investigation. Although apparently not contributors to transcription regulation by ppGpp in E. coli, transcription factors that bind ppGpp have been reported in another bacterial species (61).
We emphasize that the transcriptional responses that result from direct binding of ppGpp to RNAP are only one part of a broad response that also includes direct binding of ppGpp to many other enzymes. ppGpp regulates some products, for example, many nucleotide synthesis and translation-related enzymes, at both the transcription and activity levels.
The promoter sequence preferences identified here should guide future investigations of the kinetic properties of promoters that account for their regulation by ppGpp and ultimately aid in bioinformatic prediction of the stringent response regulon in species for which biochemical and genetic approaches are not available.
Materials and Methods
Strains, Plasmids, Oligonucleotides.
Strains and plasmids are listed in SI Appendix, Table S7, and oligonucleotide sequences in SI Appendix, Table S8.
Strain Construction.
RLG14535 [MG1655 rpoZ-kanR, rpoC-tetAR (1+2+)] and RLG14538 [MG1655 rpoZ∆2–5-kanR, rpoC R362A R417A K615A N680A K681A-tetAR (1−2−)] were transformed with pALS13 (encodes RelA lacking its autoinhibitory domain) or a control plasmid pALS14 (inactive version of RelA) to create the four strains used in Fig. 1A. See also SI Appendix, Expanded Materials and Methods and Table S7.
Strain Growth, RNA, and ppGpp Analysis.
Briefly, cells were grown at 37 °C in a Mops-based rich defined medium with 100 μg/mL ampicillin, Mops EZ Rich Defined Medium (Teknova; M2105), which contains all 20 aa, bases, vitamins, and glucose (SI Appendix, Expanded Materials and Methods). To evaluate the response of promoters to induction of ppGpp using the relA plasmid system, cells were grown in the medium above, and transcript levels were determined by qPCR after 5, 10, or 15 min of ppGpp induction (5). Strain growth and 32P-labeled ppGpp extraction and quantitation are described in SI Appendix, Expanded Materials and Methods.
RNA Sequencing.
RNA samples were sequenced by the University of Wisconsin–Madison Biotechnology Center using an Illumina HiSeq 2000 set for 50-bp single-end reads (further details in SI Appendix, Expanded Materials and Methods). High-throughput sequencing data have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession no. PRJNA504613.
EcoCyc Omics Dashboard Tool.
Dataset S1 was imported into an EcoCyc (ecocyc.org) “Smart Table” and analyzed using the Omics Dashboard Tool (29) (Fig. 3 and SI Appendix, Figs. S3 and S4). Further details are in SI Appendix, Expanded Materials and Methods.
In Vitro Transcription Analysis.
Promoter fragments chosen for in vitro analysis were amplified from chromosomal DNA and inserted into plasmid pRLG770, which contains transcription termination sequences ∼140 bp downstream from the site of insertion of the promoter fragment. Choice of promoters, cloning into the plasmid vector, multiple round in vitro transcription conditions, and sequence analysis are described in SI Appendix, Expanded Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of our laboratory for support and comments, K. Myers for help with data analysis, and J. Schroeder for help with RNA-seq data submission. Research in the R.L.G. laboratory is supported by NIH Grant R01 GM37048. C.N.D. was supported in part by the Clinical and Translational Science Award program through NIH National Center for Advancing Translational Sciences Grant UL1TR002373. P.S.-V. was supported in part by an NIH Molecular Biosciences Training Grant, and N.K. was supported in part by a Hilldale undergraduate fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The high-throughput sequencing data reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (SRA) (BioProject accession no. PRJNA504613).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1819682116/-/DCSupplemental.
References
- 1.Cashel M, Gentry D, Hernandez VJ, Vinella D. The stringent response. In: Neidhardt FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 1458–1496. [Google Scholar]
- 2.Gallant JA. Stringent control in E. coli. Annu Rev Genet. 1979;13:393–415. doi: 10.1146/annurev.ge.13.120179.002141. [DOI] [PubMed] [Google Scholar]
- 3.Gourse RL, et al. Transcriptional responses to ppGpp and DksA. Annu Rev Microbiol. 2018;72:163–184. doi: 10.1146/annurev-micro-090817-062444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50:420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ross W, et al. ppGpp binding to a site at the RNAP-DksA interface accounts for its dramatic effects on transcription initiation during the stringent response. Mol Cell. 2016;62:811–823. doi: 10.1016/j.molcel.2016.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandrangsu P, Lemke JJ, Gourse RL. The dksA promoter is negatively feedback regulated by DksA and ppGpp. Mol Microbiol. 2011;80:1337–1348. doi: 10.1111/j.1365-2958.2011.07649.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutherford ST, et al. Effects of DksA, GreA, and GreB on transcription initiation: Insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase. J Mol Biol. 2007;366:1243–1257. doi: 10.1016/j.jmb.2006.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maciąg M, Kochanowska M, Lyzeń R, Węgrzyn G, Szalewska-Pałasz A. ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid. 2010;63:61–67. doi: 10.1016/j.plasmid.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 9.Rymer RU, et al. Binding mechanism of metal⋅NTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure. 2012;20:1478–1489. doi: 10.1016/j.str.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kihira K, et al. Crystal structure analysis of the translation factor RF3 (release factor 3) FEBS Lett. 2012;586:3705–3709. doi: 10.1016/j.febslet.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Zborníková E, Rejman D, Gerdes K. Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. MBio. 2018;9:e02188-17. doi: 10.1128/mBio.02188-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang B, et al. Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat Chem Biol. 2019;15:141–150. doi: 10.1038/s41589-018-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hou Z, Cashel M, Fromm HJ, Honzatko RB. Effectors of the stringent response target the active site of Escherichia coli adenylosuccinate synthetase. J Biol Chem. 1999;274:17505–17510. doi: 10.1074/jbc.274.25.17505. [DOI] [PubMed] [Google Scholar]
- 14.Kriel A, et al. Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol Cell. 2012;48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng B, et al. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 2014;12:e1001866. doi: 10.1371/journal.pbio.1001866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan H, Hahm J, Diggs S, Perry JJP, Blaha G. Structural and functional analysis of BipA, a regulator of virulence in enteropathogenic Escherichia coli. J Biol Chem. 2015;290:20856–20864. doi: 10.1074/jbc.M115.659136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanjee U, et al. Linkage between the bacterial acid stress and stringent responses: The structure of the inducible lysine decarboxylase. EMBO J. 2011;30:931–944. doi: 10.1038/emboj.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroda A, Murphy H, Cashel M, Kornberg A. Guanosine tetra- and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J Biol Chem. 1997;272:21240–21243. doi: 10.1074/jbc.272.34.21240. [DOI] [PubMed] [Google Scholar]
- 19.Shyp V, et al. Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 2012;13:835–839. doi: 10.1038/embor.2012.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durfee T, Hansen A-M, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Traxler MF, et al. Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the “feast to famine” gradient in Escherichia coli. Mol Microbiol. 2011;79:830–845. doi: 10.1111/j.1365-2958.2010.07498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aberg A, Fernández-Vázquez J, Cabrer-Panes JD, Sánchez A, Balsalobre C. Similar and divergent effects of ppGpp and DksA deficiencies on transcription in Escherichia coli. J Bacteriol. 2009;191:3226–3236. doi: 10.1128/JB.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Z, Gerstein M, Snyder M. RNA-seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryals J, Little R, Bremer H. Control of rRNA and tRNA syntheses in Escherichia coli by guanosine tetraphosphate. J Bacteriol. 1982;151:1261–1268. doi: 10.1128/jb.151.3.1261-1268.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemke JJ, Durfee T, Gourse RL. DksA and ppGpp directly regulate transcription of the Escherichia coli flagellar cascade. Mol Microbiol. 2009;74:1368–1379. doi: 10.1111/j.1365-2958.2009.06939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keseler IM, et al. The EcoCyc database: Reflecting new knowledge about Escherichia coli K-12. Nucleic Acids Res. 2017;45:D543–D550. doi: 10.1093/nar/gkw1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Landick R, Turnbough CL, Jr, Yanofsky C. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. Transcription attenuation; pp. 1263–1286. [Google Scholar]
- 31.Horowitz H, Platt T. Initiation in vivo at the internal trp p2 promoter of Escherichia coli. J Biol Chem. 1983;258:7890–7893. [PubMed] [Google Scholar]
- 32.Kaufmann M, et al. Limited proteolysis of the beta 2-dimer of tryptophan synthase yields an enzymatically active derivative that binds alpha-subunits. Biochemistry. 1991;30:4173–4179. doi: 10.1021/bi00231a010. [DOI] [PubMed] [Google Scholar]
- 33.Jones PG, Cashel M, Glaser G, Neidhardt FC. Function of a relaxed-like state following temperature downshifts in Escherichia coli. J Bacteriol. 1992;174:3903–3914. doi: 10.1128/jb.174.12.3903-3914.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donahue JP, Turnbough CL., Jr Characterization of transcriptional initiation from promoters P1 and P2 of the pyrBI operon of Escherichia coli K12. J Biol Chem. 1990;265:19091–19099. [PubMed] [Google Scholar]
- 35.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 36.Cole JR, Nomura M. Changes in the half-life of ribosomal protein messenger RNA caused by translational repression. J Mol Biol. 1986;188:383–392. doi: 10.1016/0022-2836(86)90162-2. [DOI] [PubMed] [Google Scholar]
- 37.Lindahl L, Post L, Nomura M. DNA-dependent in vitro synthesis of fibosomal proteins, protein elongation factors, and RNA polymerase subunit α: Inhibition by ppGpp. Cell. 1976;9:439–448. doi: 10.1016/0092-8674(76)90089-1. [DOI] [PubMed] [Google Scholar]
- 38.Lemke JJ, et al. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci USA. 2011;108:5712–5717. doi: 10.1073/pnas.1019383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgos HL, O’Connor K, Sanchez-Vazquez P, Gourse RL. Roles of transcriptional and translational control mechanisms in regulation of ribosomal protein synthesis in Escherichia coli. J Bacteriol. 2017;199:e00407-17. doi: 10.1128/JB.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh N, et al. SuhB associates with Nus factors to facilitate 30S ribosome biogenesis in Escherichia coli. MBio. 2016;7:e00114. doi: 10.1128/mBio.00114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Travers AA. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980;141:973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heath RJ, Jackowski S, Rock CO. Guanosine tetraphosphate inhibition of fatty acid and phospholipid synthesis in Escherichia coli is relieved by overexpression of glycerol-3-phosphate acyltransferase (plsB) J Biol Chem. 1994;269:26584–26590. [PubMed] [Google Scholar]
- 43.Aberg A, Shingler V, Balsalobre C. Regulation of the fimB promoter: A case of differential regulation by ppGpp and DksA in vivo. Mol Microbiol. 2008;67:1223–1241. doi: 10.1111/j.1365-2958.2008.06115.x. [DOI] [PubMed] [Google Scholar]
- 44.Gummesson B, Lovmar M, Nyström T. A proximal promoter element required for positive transcriptional control by guanosine tetraphosphate and DksA protein during the stringent response. J Biol Chem. 2013;288:21055–21064. doi: 10.1074/jbc.M113.479998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 47.Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Girard ME, et al. DksA and ppGpp regulate the σS stress response by activating promoters for the small RNA DsrA and the anti-adapter protein IraP. J Bacteriol. 2017;200:e00463-17. doi: 10.1128/JB.00463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haugen SP, Ross W, Gourse RL. Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat Rev Microbiol. 2008;6:507–519. doi: 10.1038/nrmicro1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase σ subunit. Cell. 2011;147:1257–1269. doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haugen SP, et al. rRNA promoter regulation by nonoptimal binding of sigma region 1.2: An additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. doi: 10.1016/j.cell.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 54.Feklistov A, et al. A basal promoter element recognized by free RNA polymerase sigma subunit determines promoter recognition by RNA polymerase holoenzyme. Mol Cell. 2006;23:97–107. doi: 10.1016/j.molcel.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 55.Barker MM, Gourse RL. Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J Bacteriol. 2001;183:6315–6323. doi: 10.1128/JB.183.21.6315-6323.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 57.Gleghorn ML, Davydova EK, Basu R, Rothman-Denes LB, Murakami KS. X-ray crystal structures elucidate the nucleotidyl transfer reaction of transcript initiation using two nucleotides. Proc Natl Acad Sci USA. 2011;108:3566–3571. doi: 10.1073/pnas.1016691108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- 59.Winkelman JT, Chandrangsu P, Ross W, Gourse RL. Open complex scrunching before nucleotide addition accounts for the unusual transcription start site of E. coli ribosomal RNA promoters. Proc Natl Acad Sci USA. 2016;113:E1787–E1795. doi: 10.1073/pnas.1522159113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul BJ, et al. DksA: A critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 61.Cuthbert BJ, et al. Dissection of the molecular circuitry controlling virulence in Francisella tularensis. Genes Dev. 2017;31:1549–1560. doi: 10.1101/gad.303701.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.