Significance
Many infections initiating at mucosal barriers involve polymicrobial communities of organisms within which there are often multiple mechanisms of polymicrobial synergy and antagonism. However, we still understand little about another fundamental aspect of polymicrobial infection: that is, how the interaction of one organism with host cells can modulate the properties of another organism. We examined this topic using a model two-species microbial community and oral epithelial cells. The results show that Streptococcus gordonii can redirect signal transduction pathways within epithelial cells that would otherwise be induced by the keystone pathogen Porphyromonas gingivalis and which converge on a proinflammatory, mesenchymal-like transcriptional program. These observations represent a mechanism by which commensals can maintain homeostasis in the presence of more pathogenic organisms.
Keywords: EMT, commensal, polymicrobial community, FOXO1, periodontal
Abstract
The polymicrobial microbiome of the oral cavity is a direct precursor of periodontal diseases, and changes in microhabitat or shifts in microbial composition may also be linked to oral squamous cell carcinoma. Dysbiotic oral epithelial responses provoked by individual organisms, and which underlie these diseases, are widely studied. However, organisms may influence community partner species through manipulation of epithelial cell responses, an aspect of the host microbiome interaction that is poorly understood. We report here that Porphyromonas gingivalis, a keystone periodontal pathogen, can up-regulate expression of ZEB2, a transcription factor which controls epithelial–mesenchymal transition and inflammatory responses. ZEB2 regulation by P. gingivalis was mediated through pathways involving β-catenin and FOXO1. Among the community partners of P. gingivalis, Streptococcus gordonii was capable of antagonizing ZEB2 expression. Mechanistically, S. gordonii suppressed FOXO1 by activating the TAK1-NLK negative regulatory pathway, even in the presence of P. gingivalis. Collectively, these results establish S. gordonii as homeostatic commensal, capable of mitigating the activity of a more pathogenic organism through modulation of host signaling.
Bacterial colonizers of the skin and mucosal barriers tend to organize into polymicrobial communities. Disease can occur when the homeostatic balance between these communities and the host is disrupted, resulting in dysbiosis. The interbacterial and bacteria–host interactions that dictate homeostasis or dysbiosis are, therefore, a subject of intense scrutiny. In the oral cavity, dysbiotic microbial communities in the gingival compartment can induce periodontal disease, which is the sixth most common infectious condition worldwide (1–5). Subgingival communities provoke a poorly controlled immune response which fails to eliminate the microbial challenge and eventually leads to tissue destruction (2). Polymicrobial synergistic interactions among community inhabitants raise the community pathogenic potential, or nososymbiocity; and species such as Porphyromonas gingivalis that can effect the transition of a commensal community to a pathogenic entity, even at low abundance, are known as keystone pathogens (6). Increasing epidemiological and mechanistic evidence also implicates the oral microbiota in cancers such as oral squamous cell carcinoma (OSCC) (7, 8). Periodontal organisms can contribute to a tumorigenic environment by subverting epithelial cell pathways to reduce apoptosis and increase cell cycle progression, and also by disruption of local inflammatory responses (7–10).
Characterization of the epithelial cell responses to the oral microbiota is imperative for understanding the host–microbe interface that dictates the dysbiotic potential underlying periodontal diseases and potentially OSCC. In addition to its role as a keystone periodontal pathogen, P. gingivalis has also been shown to increase the mesenchymal properties of epithelial cells (11–14). The epithelial–mesenchymal transition (EMT) is an orchestrated process by which epithelial cells change shape and acquire a motile phenotype which can generate self-renewing tumor-initiating cells, and, in malignant tumors, it gives rise to a population of migratory and invasive cancer cells (15). EMT is controlled by a group of transcription factors, including the zinc-finger E-box–binding homeobox proteins (ZEB1 and ZEB2), SNAI1/2, and TWIST1/2 (16, 17). We have found that P. gingivalis can increase expression of ZEB1 in immortalized gingival epithelial cells (11), and others have reported up-regulation of ZEB1 and SNAI1/2 following P. gingivalis challenge of primary cultures of gingival epithelial cells (12). In addition to EMT, ZEB1/2, SNAI1/2, and TWIST1/2 also control inflammatory responses (18–20), which could provide a mechanistic convergence between tumorigenic potential and periodontal diseases.
Organisms that cocolonize the oral epithelium with P. gingivalis include Fusobacterium nucleatum and oral streptococcal species of the mitis group (21–23). These organisms can interact with each other in biofilms and can also modulate epithelial responses to partner species in mixed infections. For example, both P. gingivalis and streptococcal species can inhibit F. nucleatum-induced epithelial IL-8 expression through blocking NF-κB nuclear translocation (24, 25). Moreover, Streptococcus gordonii can reprogram epithelial cell global transcriptional patterns such that the subsequent response to P. gingivalis is diminished (26), and S. gordonii can prevent P. gingivalis-induced gingival epithelial cell (GEC) proliferation (26). The mechanistic details of the influence of S. gordonii on epithelial cell transcriptional responses has yet to be addressed.
In this study, we focus on the ZEB2 transcription factor and show that expression of ZEB2 is induced specifically by P. gingivalis through pathways involving β-catenin and FOXO1. While neither S. gordonii nor F. nucleatum regulated ZEB2, S. gordonii antagonized ZEB2 induction by P. gingivalis through blocking the dephosphorylation and activation of FOXO1. The results position S. gordonii as a homeostatic commensal which operates via host cell manipulation to mitigate the action of a pathogen.
Results
Regulation of EMT-Inducing Transcription Factors by P. gingivalis Alone and in Dual Species Conglomerates.
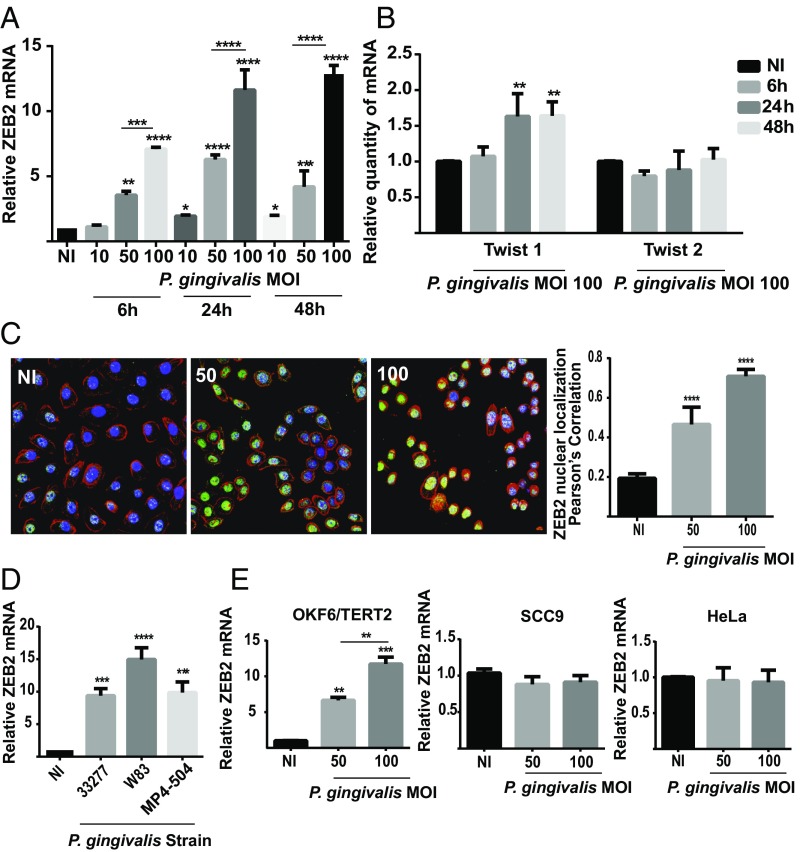
P. gingivalis has been shown to induce a partial or transition EMT phenotype in gingival epithelial cells and to increase the activity of transcription factors that control EMT, including ZEB1 and SNAI1/2 (11, 12). However, regulation of EMT is multifactorial and involves a number of additional transcription factors (15, 17). Hence, we examined the impact of challenge with P. gingivalis on the transcription factors ZEB2 and TWIST1/2. As shown in Fig. 1A, P. gingivalis increased ZEB2 mRNA levels in telomerase immortalized gingival keratinocyte (TIGK) cells in a time-dependent (up to 24 h) and dose-dependent manner, with a maximal 13-fold induction occurring after 48-h infection with a multiplicity of infection (MOI) of 100. In contrast, P. gingivalis had a less pronounced effect on TWIST1/2 mRNA, with a statistically significant increase of under twofold only observed for TWIST1 at 24 and 48 h at an MOI of 100 (Fig. 1B). To corroborate the quantitative reverse transcription-PCR (qRT-PCR) data and begin to test functional relevance, the expression and location of ZEB2 protein were examined. P. gingivalis challenge increased both the amount of ZEB2 protein expression and localization in the nucleus where it is functionally active (Fig. 1C).
Fig. 1.
P. gingivalis up-regulates transcription factors controlling EMT. (A and B) TIGK cells were infected with P. gingivalis 33277 at the times and MOIs indicated. ZEB2 (A) or TWIST1/2 (B) mRNA levels were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. (C) Fluorescent confocal microscopy of TIGK cells infected with P. gingivalis 33277 (Pg) at the MOI indicated for 24 h. Control cells were noninfected (NI). Cells were fixed and probed with ZEB2 antibodies (green). Actin (red) was stained with Texas Red-phalloidin, and nuclei (blue) were stained with DAPI. Cells were imaged at magnification 63×. Shown are merged images of projections of z-stacks (Left) and Pearson’s correlation coefficient of ZEB2 with nuclei (Right) obtained with Volocity software. (D) TIGK cells were infected with P. gingivalis strains at MOI:100 for 24 h. ZEB2 mRNA levels were determined as in A. (E) ZEB2 mRNA levels in different cell types following P. gingivalis 33277 infection for 24 h. Quantitative data represent three independent experiments with three replicates. Error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001, compared with NI unless indicated. Images are representative of three independent experiments.
P. gingivalis is a host adapted organism with a nonclonal population structure, and isolates from different individuals can vary extensively (27–29). Hence, we next examined the ability of different strains of P. gingivalis to enhance ZEB2 mRNA levels. As shown in Fig. 1D, the commonly used laboratory strain W83, along with the low passage clinical isolate MP4-504, induced ZEB2 expression to a similar degree as did 33277. The property of ZEB2 induction by P. gingivalis thus spans both fimbriated/nonencapsulated (33277) and nonfimbriated/capsulated (W83) lineages. Regulation of ZEB2 by W83 contrasts to ZEB1, which is not induced by W83 (11), indicating that differential pathways can control ZEB1 and ZEB2. Interestingly, however, P. gingivalis did not increase ZEB2 mRNA amounts in the SCC9 line, which are squamous carcinoma cells derived from the tongue, or in the cervical adenocarcinoma HeLa epithelial cell line (Fig. 1E). ZEB2 was regulated by P. gingivalis, however, in another nontumorigenic oral line, OKF6/TERT2, derived from the oral mucosa. Context-dependent regulation of ZEB2 has been observed previously, and ZEB2 can be regulated by Akt in gastric cancer cells (30), but not in OSCC cells (31). The development of a mesenchymal and tumorigenic phenotype is complex, and these data would indicate that P. gingivalis could influence the early stages of the process. However, prolonged bacterial–host cell challenge may also induce a mesenchymal phenotype in OSCC cells (14).
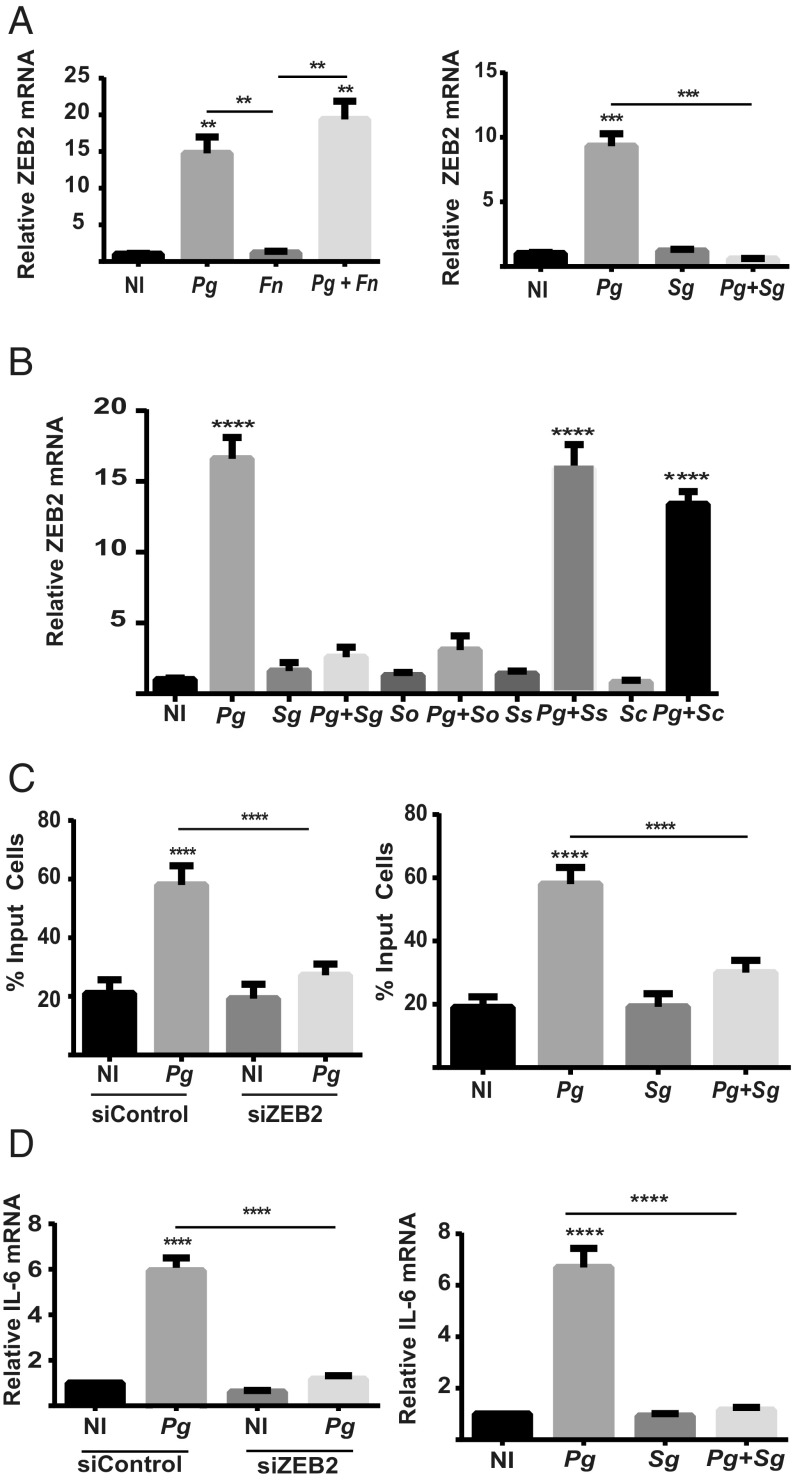
In the oral cavity, P. gingivalis is a component of polymicrobial communities, and it is these communities that constitute the etiological unit in periodontal diseases (6). However, the ability of other community inhabitants to modulate the action of P. gingivalis at the epithelial interface is poorly understood. Organisms such as F. nucleatum and oral streptococci are often coisolated with P. gingivalis and are associated with epithelial cells (21–23). Neither F. nucleatum nor S. gordonii were capable of regulating ZEB2 mRNA levels as monoinfections (Fig. 2A), indicating a restricted distribution of the ZEB2-inducing phenotype among bacterial inhabitants of the oral cavity. While P. gingivalis remained capable of increasing ZEB2 transcripts in the presence F. nucleatum, coinfection with S. gordonii antagonized induction of ZEB2 by P. gingivalis (Fig. 2A). To begin to examine the distribution of ZEB2 antagonism among oral streptococci, other common species were tested. Streptococcus oralis also was capable of suppressing ZEB2 induction; however, Streptococcus sanguinis and Streptococcus cristatus were not (Fig. 2B), and, hence, ZEB2 antagonism is not universal among oral streptococcal species. ZEB2 can control cell migration and also production of inflammatory mediators such as IL-6 (20). As both TIGK cell migration and IL-6 secretion can be modulated by P. gingivalis (11, 32), we examined the influence of ZEB2 on these processes. Fig. 2C shows that knockdown of ZEB2 with siRNA (SI Appendix, Fig. S1) reduced TIGK migration into matrigel in response to P. gingivalis. Similarly, IL-6 mRNA production in response to P. gingivalis infection was reduced in the ZEB2 knockdown cells (Fig. 2D). Thus, ZEB2 can control phenotypic properties of TIGKs with relevance to maintenance of tissue homeostasis and innate immune responses. Consistent with the antagonistic effect on ZEB2 activation, S. gordonii diminished both TIGK migration and IL-6 production in response to P. gingivalis. Collectively, these results show that common oral bacteria display a range of properties with regard to influencing ZEB2 activity in epithelial cells. In some polymicrobial contexts, the P. gingivalis phenotype can prevail while, in other cases, streptococci such as S. gordonii and S. oralis can antagonize ZEB2 induction, even in the presence of an otherwise stimulatory organism.
Fig. 2.
Impact of dual species challenge on ZEB2 mRNA and associated phenotypic properties. (A and B) ZEB2 mRNA levels measured by qRT-PCR in TIGK cells infected with P. gingivalis 33277 (Pg) alone or together with F. nucleatum (Fn) or with oral streptococcal species. Sc, S. cristatus; Sg, S. gordonii; So, S. oralis; Ss, S. sanguinis. Monoinfection was MOI:100 for 24 h. Dual species infection was MOI:100 for each strain for 24 h. (C) Quantitative analysis of TIGK migration through matrigel-coated transwells. TIGK cells were transiently transfected with siRNA to ZEB2 (siZEB2) or scrambled siRNA (siControl) (Left) or nontransfected (Right). TIGKs were challenged with P. gingivalis 33277 and/or S. gordonii at MOI:50 for each strain for 24 h. Control cells were not infected (NI). Data are presented as the mean number of cells invading through the transwell. (D) ZEB2 was silenced with siRNA (Left), and TIGKs were challenged with bacteria as in C for 2 h. IL-6 mRNA levels were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Quantitative data represent three independent experiments with three replicates. Error bars represent the SEM. **P < 0.01, ***P < 0.005, and ****P < 0.001 compared with NI unless indicated.
Wnt/β-Catenin and FOXO1 Signaling Controls ZEB2 Expression in Response to P. gingivalis.
To begin to functionally dissect the mechanism of streptococcal antagonism of P. gingivalis-induced ZEB2 regulation, we first examined the signaling pathways modulated by P. gingivalis to control ZEB2. The TGF-β pathway is the predominant mechanism which controls expression of ZEB2, and thus the role of TGF-β signaling in P. gingivalis-dependent control of ZEB2 was investigated through pharmacological inhibition. Treating TIGK cells with LY-364947, which is a selective, ATP-competitive inhibitor of TGF-β Type I receptor kinase, had no effect on up-regulation of ZEB2 mRNA by P. gingivalis (SI Appendix, Fig. S2A). In the TGF-β pathway, proximate control of ZEB2 transcription is mediated by SMADs, in particular SMAD3, but also SMAD2 and the common-mediator SMAD, SMAD4 (33–35). To further examine the role of the TGF-β pathway, we therefore used siRNA to knockdown SMADs (SI Appendix, Fig. S1). Reduction of SMAD3 alone or of SMAD2, -3, and -4 simultaneously did not impede up-regulation of ZEB2 by P. gingivalis (SI Appendix, Fig. S2 B and C). Taken together, these results indicate that regulation of ZEB2 by P. gingivalis is neither mediated by the TGF-β signal transduction pathway, nor by a parallel pathway that converges on SMAD3.
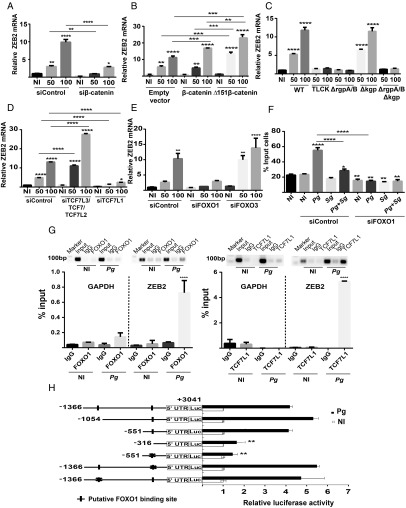
The Wnt/β-catenin signaling pathway can also control EMT, along with the activity of the EMT-related transcription factors SNAI and TWIST (36), and P. gingivalis has been shown to activate the Wnt/β-catenin pathway in epithelial cells (12, 37). When β-catenin was suppressed with siRNA (SI Appendix, Fig. S1), the up-regulation of ZEB2 by P. gingivalis was suppressed (Fig. 3A). Further support for a role for β-catenin came from exogenous overexpression of native β-catenin or a constitutively active mutant lacking the N-terminal 151 amino acids which comprise the degradation complex stabilization domain (38) (SI Appendix, Fig. S1). Ectopic expression of native β-catenin caused an increase in ZEB2 mRNA upon a P. gingivalis challenge of MOI 100, and exogenous Δ151 β-catenin expression further increased ZEB2 mRNA levels at MOI 50 and 100 (Fig. 3B). One mechanism by which P. gingivalis stimulates Wnt/β-catenin signaling is by proteolytic degradation of the β-catenin destruction complex (37). Mutants of P. gingivalis which lack the arginine-specific gingipains RgpA and RgpB therefore are unable to stabilize β-catenin. As shown in Fig. 3C, a ΔrgpAB mutant of P. gingivalis failed to up-regulate ZEB2 mRNA levels whereas loss of the lysine-specific gingipain Kgp had no effect on ZEB2 induction by P. gingivalis. The involvement of arginine-gingipain processing of β-catenin was corroborated by preincubation of P. gingivalis WT with TLCK, a gingipain inhibitor, which also prevented elevation of ZEB2 mRNA (Fig. 3C).
Fig. 3.
ZEB2 responses to P. gingivalis are controlled by β-catenin and FOXO1 pathways. (A) TIGK cells were transiently transfected with siRNA to β-catenin or scrambled siRNA (siControl) and infected with P. gingivalis 33277 for 24 h at the MOI indicated. ZEB2 mRNA levels were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. (B) TIGK cells were transiently transfected with plasmid expressing full-length β-catenin, a Δ151 truncation derivative, or with empty vector. Cells were challenged with P. gingivalis and ZEB2 mRNA measured as described in A. (C) TIGK cells were challenged with P. gingivalis strains for 24 h at the MOI indicated. WT, P. gingivalis 33277; ΔrgpA/B, deletion mutant of the rgpA and rgpB arginine gingipain genes; Δkgp, deletion mutant of the kgp lysine gingipain gene; ΔrgpAB Δkgp, triple gingipain deletion mutant; TLCK, WT preincubated with the protease inhibitor TLCK (100 µM, 2 h). ZEB2 mRNA was measured as described in A. (D and E) TIGK cells were transiently transfected with siRNA to TCF7L2/TCF7L3/TCF7, or TCF7L1 (D), FOXO1 or FOXO3 (E), or control scrambled siRNA. Cells were challenged with P. gingivalis, and ZEB2 mRNA was measured as described in A. (F) TIGK cells were transiently transfected with siRNA to FOXO1 or control scrambled siRNA. Cells were challenged with P. gingivalis 33277 and/or S. gordonii at MOI:50 for each strain for 24 h. Quantitative analysis of TIGK migration through matrigel-coated transwells is presented as the mean number of migrated cells. (G) TIGK cells were challenged with P. gingivalis 33277 MOI:100 for 24 h, or left uninfected (NI), and subjected to chromatin immunoprecipitation (ChIP) using anti-FOXO1 IgG, anti-TCF7L1 IgG, or preimmune IgG. The precipitated DNA was subsequently analyzed by end point PCR and by qPCR with primers to the ZEB2 promoter region or the GAPDH promoter as a control. qPCR was expressed relative to the input DNA. (H) Luciferase assay for ZEB2 promoter activity in TIGKs challenged with P. gingivalis 33277 MOI:100 for 30 min, or left uninfected (NI). Cells were transiently transfected with a ZEB2 promoter–luciferase reporter plasmid, or a constitutively expressing Renilla luciferase reporter. Derivatives of the ZEB2 promoter included serial deletions and site-specific mutations (denoted X) in the FOXO1 binding sites. FOXO luciferase activity was normalized to the level of Renilla luciferase. Quantitative data represent three independent experiments with three replicates. Error bars represent the SEM. *P > 0.05, **P < 0.01, ***P < 0.005, and ****P < 0.001 compared with NI unless indicated.
β-Catenin does not bind directly to DNA (39) but is a coactivator of transcription factors, mainly TCF7L3, TCF7, TCF7L1, and TCF7L2 (40). The role of these transcription factors was investigated by an siRNA approach. Knockdown of TCF7L3, TCF7, and TCF7L2 (SI Appendix, Fig. S1) had no effect on ZEB2 induction by P. gingivalis (Fig. 3D). However, knockdown of TCF7L1 abrogated the ability of P. gingivalis to increase ZEB2 levels (Fig. 3D), indicating a role for this transcription factor in P. gingivalis-induced ZEB2 mRNA expression.
P. gingivalis can also activate the FOXO1 transcription factor, and FOXO1 has been found to regulate ZEB2 production (41). To assess the participation of FOXO1 in the regulation of ZEB2 mediated by P. gingivalis, we used siRNA knockdowns. Reduction of FOXO1, but not FOXO3 (SI Appendix, Fig. S1), suppressed P. gingivalis induction of ZEB2 (Fig. 3E). We also confirmed that FOXO1 activity can control phenotypic outcomes dependent on ZEB2 as knockdown of FOXO1 reduced the amount of TIGK invasion of matrigel in response to P. gingivalis (Fig. 3F).
Next, we sought to determine whether TCF7L1 and FOXO1 regulate ZEB2 by binding directly to the promoter region and initiating transcription. As shown in Fig. 3G, following challenge of TIGK cells with P. gingivalis, ChIP with antibodies specific for TCF7L1 or FOXO1 immunoprecipitated fragments of genomic DNA that could be amplified using primers specific to the ZEB2 promoter by end-point PCR. Quantitative (q) PCR also showed enrichment of the ZEB2 promoters in the TCF7L1 and FOXO1 pulled-down conditions, compared with control IgG. The control GAPDH promoter did not precipitate with TCF7L1 or FOXO1 antibodies. Moreover, consistent with the siRNA data, antibodies to TCF7L2 did not immunoprecipitate the ZEB2 promoter (SI Appendix, Fig. S3). As previous reports have found that FOXO1 suppresses ZEB2 expression in different contexts (41, 42), we corroborated the role of FOXO1 in inducing ZEB2 in response to P. gingivalis infection using promoter–luciferase reporters. Two putative consensus FOXO1 binding domains (43) were identified upstream of ZEB2, and we cloned a fragment spanning −1366 to +3041 relative to the transcriptional start site (44, 45) next to a promoterless luc gene. As shown in Fig. 3H, the full-length construct responded to P. gingivalis challenge with increased transcriptional activity, and serial deletion of the promoter construct revealed that loss of both putative FOXO1 binding sites was required to reduce ZEB2 transcription. Promoter constructs harboring three point mutations in the center of the putative FOXO1 response element at either the proximal (−352 to −345) or distal (−1070 to −1063) position showed no significant reduction of transcription in the presence of P. gingivalis, and mutation of the proximal site in an otherwise functional truncated construct abrogated transcription. Together, these data show that both potential FOXO1 binding sites are redundantly functional and can promote transcription in the presence of P. gingivalis.
S. gordonii Modulates Activation of FOXO1 by P. gingivalis.
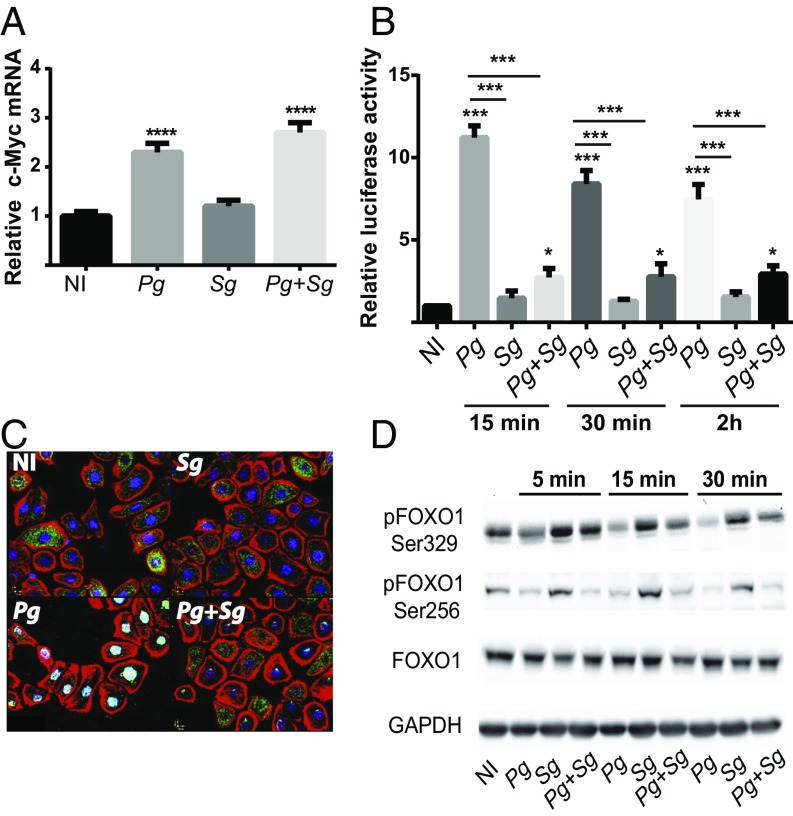
Next, we investigated whether S. gordonii modulates activation of β-catenin and/or FOXO1 to impede up-regulation of ZEB2 by P. gingivalis. As shown in Fig. 4A, coinfection of S. gordonii with P. gingivalis had no impact on mRNA levels of c-Myc, a gene regulated by β-catenin. However, challenge with S. gordonii prevented activation of FOXO in the luciferase reporter assay (Fig. 4B), and S. gordonii blocked P. gingivalis-induced translocation of FOXO1 to the nucleus (Fig. 4C). Nuclear localization of FOXO1 in resting cells was at very low levels (Fig. 4C); thus, despite its antagonistic action, S. gordonii did not cause a further decrease in FOXO1 activation. P. gingivalis can activate FOXO1 through a pathway involving reactive oxygen species (ROS)-mediated activation of JNK (46). Hence, we first examined the impact of S. gordonii on ROS and JNK activation by P. gingivalis. S. gordonii had no effect on either ROS or JNK activation by P. gingivalis (SI Appendix, Fig. S4), indicating that S. gordonii acts on a different FOXO1 control pathway. A wide range of posttranslational modifications exert control over the activity and nuclear localization of FOXO1 (42, 47), and, to begin to decipher the mode of action of S. gordonii, we began a screen for specific amino acid phosphorylations. P. gingivalis challenge induced dephosphorylation of Ser256, as reported previously (46); however, this was unaffected by coinfection with S. gordonii (Fig. 4D). Dephosphorylation of Ser329 was also induced by P. gingivalis, and this was reduced by S. gordonii coinfection. Dephosphorylation of Ser329 inhibits shuttling of FOXO1 out of the nucleus, thus increasing activity (42). Hence, preventing Ser329 dephosphorylation could represent a mechanism by which S. gordonii antagonizes P. gingivalis action.
Fig. 4.
S. gordonii impedes P. gingivalis-induced FOXO activation. (A) TIGK cells were infected with P. gingivalis 33277 (Pg) and/or S. gordonii (Sg) at MOI:50 for each strain for 30 min. c-Myc mRNA levels were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. (B) TIGKs were transiently transfected with the FOXO promoter–luciferase reporter plasmid, or a constitutively expressing Renilla luciferase reporter, and FOXO luciferase activity was normalized to the level of Renilla luciferase. Cells were challenged with P. gingivalis 33277 (Pg) and/or S. gordonii (Sg) at MOI:50 for each strain for the times indicated. NI, no infection control. (C) Fluorescent confocal microscopy of TIGK cells infected with P. gingivalis 33277 (Pg) and/or S. gordonii (Sg). Control cells were noninfected (NI). Cells were fixed and probed with FOXO1 antibodies (green). Actin (red) was stained with Texas Red-phalloidin, and nuclei (blue) were stained with DAPI. Cells were imaged at magnification 63×, and shown are merged images of projections of z-stacks obtained with Volocity software. (D) Immunoblot of lysates of TIGK cells challenged with P. gingivalis 33277 (Pg) and/or S. gordonii (Sg) at MOI:50 for each strain for the times shown, and probed with the antibodies indicated. GAPDH was used as a loading control. Quantitative data represent three independent experiments with three replicates. Error bars represent the SEM. *P > 0.05, ***P < 0.005, and ****P < 0.001 compared with NI unless indicated. Images are representative of three independent experiments.
S. gordonii Antagonism Is Mediated Through the TAK1-NLK Pathway.
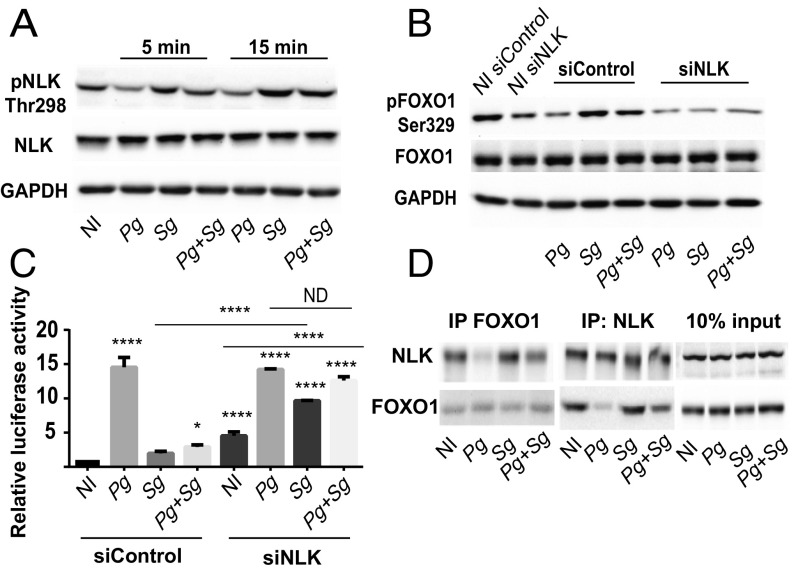
FOXO1 Ser329 is a target of Nemo-like Kinase (NLK) (48), and thus we examined the effect of P. gingivalis and S. gordonii on the activation of NLK. Fig. 5A shows that S. gordonii, but not P. gingivalis, can induce the phosphorylation and activation of NLK, and coinfection with S. gordonii and P. gingivalis maintained the increased phosphorylation state of NLK. Further, siRNA knockdown of NLK (SI Appendix, Fig. S1) prevented S. gordonii from inducing phosphorylation of FOXO1 at Ser329 (Fig. 5B). To test the functional relevance of NLK with regard to FOXO1 activity, we used the luciferase reporter assay. Silencing NLK caused an increase in FOXO1 activity in both the no-infection and S. gordonii-infection conditions (Fig. 5C), consistent with the function of NLK as an inhibitor of FOXO (48) and as a target of S. gordonii. Additionally, in the NLK knockdown cells, S. gordonii was no longer able to antagonize P. gingivalis-induced activation of FOXO, implicating NLK as an effector of S. gordonii-mediated antagonism. In contrast, knockdown of the dual specificity tyrosine-phosphorylated and -regulated kinase 1A (DYRK1A), which can also phosphorylate FOXO1 at Ser329 (49), had no effect on antagonism of P. gingivalis-induced activation of the FOXO promoter (SI Appendix, Fig. S5). To examine the association between NLK and FOXO1 in bacterially infected cells, immunoprecipitation was performed. Precipitation with either NLK or FOXO1 antibodies revealed a physical association between the two proteins in S. gordonii-challenged cells (Fig. 5D).
Fig. 5.
S. gordonii activates NLK to inhibit FOXO1. (A) TIGK cells were challenged with P. gingivalis (Pg) and/or S. gordonii (Sg) at MOI:50 for each strain for the times indicated. Control cells were noninfected (NI). Lysates of TIGK cells were immunoblotted and probed with the antibodies indicated. GAPDH was used as a loading control. (B) TIGK cells were transiently transfected dually with siRNA to NLK (siNLK) or scrambled siRNA (siControl) and challenged with bacteria as in A for 30 min. Control cells were noninfected (NI). Lysates of TIGK cells were immunoblotted and probed with the antibodies indicated. GAPDH was used as a loading control. (C) TIGK cells were transiently transfected dually with siRNA to NLK (siNLK) or scrambled siRNA (siControl), along with the FOXO promoter–luciferase reporter plasmid, or a constitutively expressing Renilla luciferase reporter. FOXO luciferase activity after 15 min of bacterial challenge as in A was normalized to the level of Renilla luciferase. (D) TIGK cells were challenged with bacteria as in A for 15 min and cross-linked with dithiobis(succinimidyl propionate), and lysates were immunoprecipitated with either NLK or FOXO1 antibodies. Immunoblots show cell lysates before immunoprecipitation (10% input) and immunoprecipitates (IP) with FOXO1 or NLK antibodies. Quantitative data represent three independent experiments with three replicates. Error bars represent the SEM. *P > 0.05 and ****P < 0.001 compared with NI unless indicated. ND, no statistical difference. Images are representative of three independent experiments.
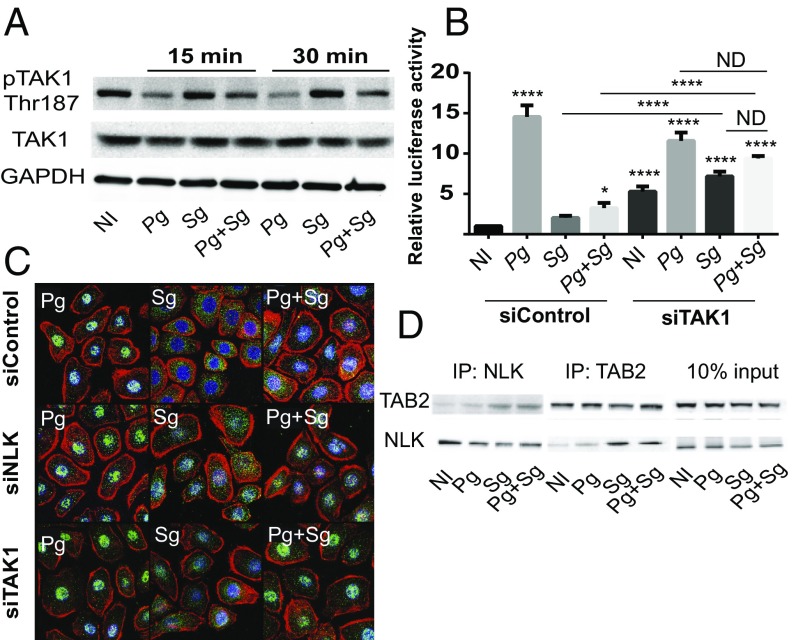
Upstream of NLK in the regulation of FOXO1 is TAK1, which can phosphorylate and activate NLK (48). Thus, the activation of TAK1 by P. gingivalis and S. gordonii was examined by immunoblotting. S. gordonii alone and in combination with P. gingivalis increased the phosphorylation levels of TAK1 compared with P. gingivalis alone (Fig. 6A), consistent with the involvement of the TAK1-NLK pathway in S. gordonii antagonistic activity. Interestingly, challenge of TIGKs with P. gingivalis alone results in dephosphorylation of the Thr-187 residue of TAK1 compared with the no-infection condition, most notably at 30 min, indicating the existence of an additional pathway by which P. gingivalis can impact FOXO1 activity. Functional relevance of TAK1 was corroborated by siRNA knockdown (SI Appendix, Fig. S1), which prevented S. gordonii-mediated antagonism of P. gingivalis-induced FOXO promoter activity (Fig. 6B). Resting activity of FOXO and S. gordonii-induced activation were also increased, due to reduction of the antagonistic TAK1-NLK axis. To examine nuclear location of FOXO1 directly, immunofluorescent microscopy was utilized. Fig. 6C shows increased nuclear localization of FOXO1 with TAK1 or NLK knockdown in the S. gordonii infection conditions and the inability of S. gordonii to antagonize nuclear localization in response to P. gingivalis, thus confirming the results of the reporter assays. TAK1-binding protein 2 (TAB2) is a TAK1-interacting protein that can also bind to NLK and function as a scaffold protein to facilitate the interaction between TAK1 and NLK (50). Therefore, we investigated binding of TAB2 to NLK using immunoprecipitation. As shown in Fig. 6D, challenge of TIGK cells with S. gordonii alone or in combination with P. gingivalis increased the recovery of coprecipitates of TAB2 and NLK, compared with no infection or P. gingivalis infection alone.
Fig. 6.
S. gordonii activates NLK through TAK1. (A) TIGK cells were challenged with P. gingivalis (Pg) and/or S. gordonii (Sg) at MOI:50 for each strain for the times indicated. Control cells were noninfected (NI). Lysates of TIGK cells were immunoblotted and probed with the antibodies indicated. GAPDH was used as a loading control. (B) TIGK cells were transiently transfected dually with siRNA to TAK1 (siTAK) or scrambled siRNA (siControl), along with the FOXO promoter–luciferase reporter plasmid, or a constitutively expressing Renilla luciferase reporter. FOXO luciferase activity after 15 min of bacterial challenge as in A was normalized to the level of Renilla luciferase. (C) Fluorescent confocal microscopy of TIGK cells challenged with bacteria as in A. Cells were fixed and probed with FOXO1 antibodies (green). Actin (red) was stained with Texas Red phalloidin, and nuclei (blue) were stained with DAPI. Cells were imaged at magnification 63×, and shown are representative merged images of projections of z-stacks (10 slices per z stack) obtained with Volocity software. (D) TIGK cells were challenged with bacteria as in A for 15 min and cross-linked with dithiobis(succinimidyl propionate), and lysates were immunoprecipitated with either NLK or TAB2 antibodies. Immunoblots show cell lysates before immunoprecipitation (10% input) and immunoprecipitates (IP) with TAB2 or NLK antibodies. Quantitative data represent three independent experiments with three replicates. Error bars represent the SEM. *P > 0.05 and ****P < 0.001 compared with NI unless indicated. ND, no statistical difference. Images are representative of three independent experiments.
Collectively, these results support the notion that S. gordonii can activate the TAK1-TAB2-NLK pathway to increase Ser329 phosphorylation on FOXO1 and that this inhibitory pathway supersedes the otherwise stimulatory effect of P. gingivalis.
Phosphorylation of FOXO1 Ser329 Is Required for S. gordonii Antagonism.
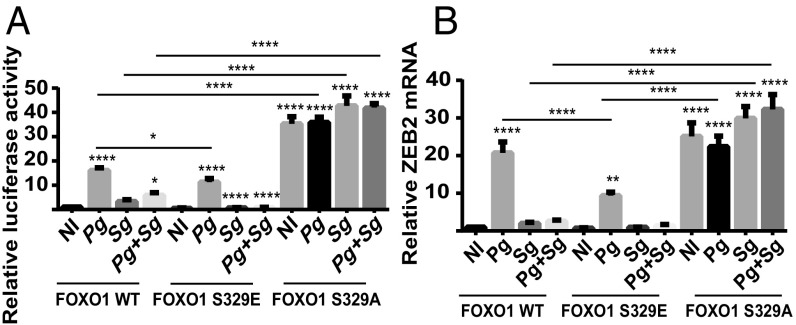
To further verify the role of Ser329 phosphorylation in the antagonistic action of S. gordonii, we generated and tested phosphomimetics of FOXO1. TIGK cells were transfected with FOXO1 WT, FOXO1 S329A, or FOXO1 S329E and challenged with S. gordonii and P. gingivalis alone or in combination. The S329A construct cannot be phosphorylated at amino acid 329 and will thus prevent NLK-dependent exclusion of FOXO1 from the nucleus. Consequently, FOXO activity was higher in the S329A-transfected cells under all-infection and no-infection conditions (Fig. 7A). Consistent with a role for phosphorylation of S329 in S. gordonii antagonism of P. gingivalis-induced FOXO1 activation, coinfection with S. gordonii and P. gingivalis did not result in a decrease in FOXO activity compared with P. gingivalis infection alone. In contrast, transfection with S329E, which, by mimicking the phosphorylated condition, will increase exclusion of FOXO1 from the nucleus, resulted in lower levels of FOXO activity in the no-infection and P. gingivalis- and S. gordonii-infection conditions. Moreover, there was significantly less FOXO activity after coinfection with P. gingivalis and S. gordonii. We also verified the involvement of S329 phosphorylation in the control of ZEB2 expression. Similar to the results with FOXO, ZEB2 mRNA levels were higher in the S329A-transfected cells under no-infection conditions, and antagonism of ZEB2 by S. gordonii was lost (Fig. 7B). Transfection with S329E resulted in lower levels of ZEB2 mRNA in the P. gingivalis-infection condition (Fig. 7B). These results are consistent with the concept that dephosphorylation and activation FOXO1 by P. gingivalis can regulate the expression of ZEB2 and that this is the primary site of intervention by S. gordonii.
Fig. 7.
Phosphomimetic mutant of FOXO1 Ser-329 abrogates S. gordonii-mediated antagonism of FOXO1 activation. (A) TIGK cells were transiently transfected dually with FOXO1, FOXO1S329E, or FOXO1S329A, along with the FOXO promoter–luciferase reporter plasmid, or a constitutively expressing Renilla luciferase reporter. Cells were challenged with P. gingivalis (Pg) and/or S. gordonii (Sg) at MOI:50 for each strain for 15 min. Control cells were noninfected (NI). FOXO luciferase was normalized to the level of Renilla luciferase. (B) TIGK cells were transiently transfected with FOXO1, FOXO1S329E, or FOXO1S329A and challenged with bacteria as in A for 24 h. ZEB2 mRNA levels were measured by qRT-PCR. Data were normalized to GAPDH mRNA and are expressed relative to noninfected (NI) controls. Quantitative data represent three independent experiments with three replicates. Error bars represent the SEM. *P > 0.05, **P < 0.01, and ****P < 0.001 compared with NI unless indicated.
Discussion
Bacteria in the oral cavity engage host epithelial cells in a complex molecular dialogue. The action of specific organisms, such as P. gingivalis, at the epithelial interface has been extensively studied. However, in vivo oral bacteria are conglomerated in polymicrobial communities, which can be synergistically pathogenic (51–53). Polymicrobial communities also contain commensal organisms which can antagonize the action of pathogens. This colonization resistance is traditionally attributed to direct antimicrobial activity: e.g., the production of bacteriocins or competition for niches and nutrients (54). Additionally, commensals regulate basic developmental features and functions of the immune system in a manner that primes it for vigorous defense against overt pathogens, while maintaining tolerance to innocuous antigens (55, 56). Commensals can also induce homeostatic immunity that couples antimicrobial function with tissue repair (57). An emerging component of community–host interaction is the capacity for one organism to mitigate or potentiate the action of another through manipulation of signaling pathways in host epithelial cells (26, 53, 58).
In this study, we show that P. gingivalis up-regulates ZEB2 expression in gingival epithelial cells to a much higher extent compared with TWIST1/2 or, as reported previously, ZEB1 and SNAI1/2 (11, 12). This suggests that ZEB2 represents a major target for P. gingivalis-induced remodeling of the gingival epithelial cell transcriptional program, and this could have major consequences for homeostasis. Increased ZEB2 expression has been reported in esophageal and oral squamous cell cancer and is associated with poor prognosis (59–61). In addition, ZEB2 has recently been shown to control inflammatory responses, including the production of IL-6, a cytokine with a role in periodontal bone resorption (62). IL-6 is also involved in the progression of several types of cancer, including OSCC (63–66). Indeed, P. gingivalis challenge enhanced the migration of TIGK cells into matrigel and increased production of IL-6 in a ZEB2-dependent manner, consistent with a central role for ZEB2 in the epithelial cell response to P. gingivalis, with relevance to both periodontitis and tumorigenesis.
The control mechanisms for ZEB proteins comprise an intricate network of signaling pathways overlaid with noncoding RNA (15), and, in this study, we found that P. gingivalis impinges on Wnt/β-catenin and FOXO1 pathways. Disruption of either of these pathways separately was sufficient to reduce P. gingivalis-induced up-regulation of ZEB2, suggestive of a finely balanced regulatory mechanism. β-Catenin is a coactivator of gene expression, functioning mainly through the TCF7 family of transcription factors. A systematic siRNA inhibition approach identified TCF7L1 as responsible for ZEB2 induction, and this result was corroborated by chromatin immunoprecipitation showing that TCF7L1 binds to the ZEB2 promoter region and stimulates transcription. The TCF7 proteins possess common structural features, are often expressed in overlapping patterns, and display functional redundancy. However, individual family members also exhibit unique features that are not recapitulated by the other proteins (40). TCF7L2 has been shown previously to regulate ZEB1, and here we report that ZEB2 can be regulated by TCF7L1.
P. gingivalis can activate FOXO1 in TIGK cells (46), and, herein, we show the relevance of this pathway to ZEB2 regulation. FOXO activity is controlled by an array of posttranslational modifications (PTMs), including phosphorylation of a number of amino acid residues (42, 67). We found that P. gingivalis can dephosphorylate the serine 256 and serine 329 residues in FOXO1, outcomes that are associated with retention of FOXO1 in the nucleus (42, 43). Moreover, expression of an exogenous phosphomimetic of serine 329 reduced FOXO1 activity and ZEB2 transcriptional levels. The function of FOXO transcription factors is highly context-specific (42), and other studies have reported that FOXO1 can reduce ZEB2 expression in hepatocarcinoma cells (41). However, FOXO3 can switch between a transcriptional repressor and a transcriptional activator according to the phosphorylation state of one serine residue, S574 (68), and, thus, the phosphorylation status of serine 329 may toggle FOXO1 between an activator and repressor of ZEB2 transcription.
F. nucleatum and oral streptococci, such as S. gordonii, S. oralis, S. sanguinis, and S. cristatus, are community partners of P. gingivalis in oral biofilms (69) and major colonizers of epithelial surfaces (21, 22). Individually, none of these species regulated ZEB2 expression, and, further, F. nucleatum, S. sanguinis, and S. cristatus did not impede regulation by P. gingivalis. However, both S. gordonii and S. oralis were capable of antagonizing regulation of ZEB2 by P. gingivalis. Although these streptococcal species are closely related, a number of studies have demonstrated different properties in the context of polymicrobial communities. S. gordonii and P. gingivalis provide mutual physiological support (70) whereas the interaction between P. gingivalis and Streptococcus mitis or S. cristatus is more antagonistic. P. gingivalis induces S. mitis DNA fragmentation and death in an in vitro biofilm system (71), and S. cristatus can inhibit fimbrial production by P. gingivalis, thus impeding colonization and pathogenicity (72). Hence, the composition and ratios of even closely related species in polymicrobial communities may be an underappreciated and key component of community function and pathogenic potential.
Mechanistically, antagonism of ZEB2 by S. gordonii involved inhibition of FOXO1. Several lines of evidence indicate that this antagonistic effect of S. gordonii on FOXO1 is mediated by the TAK1-NLK pathway. This evidence includes the following: Challenge of epithelial cells with S. gordonii prevented P. gingivalis-induced dephosphorylation of the serine329 residue of FOXO1, a target of NLK; S. gordonii and S. gordonii in combination with P. gingivalis, but not P. gingivalis alone, activated NLK and increased binding of NLK to FOXO1, and silencing of NLK abrogated the antagonistic effect of S. gordonii; TAK1, which is upstream of NLK, was phosphorylated following challenge with S. gordonii and S. gordonii in combination with P. gingivalis, but not P. gingivalis alone, and silencing of TAK1 also reduced the antagonistic effect of S. gordonii; TAB2, a scaffolding protein which facilitates binding of TAK1 to NLK, showed an increased association with NLK following infection with S. gordonii, and S. gordonii in combination with P. gingivalis, but not P. gingivalis alone; and exogenous expression of a nonphosphorylatable Ser329A mutant of FOXO1 prevented antagonism by S. gordonii. NLK is a negative regulator of FOXO activity as phosphorylation at Ser329 increases binding to the 14-3-3 protein and shuttling of FOXO out of the nucleus (48, 73). Our finding that exogenous expression of a FOXO1 Ser329A mutant significantly increased FOXO1 activity in noninfected cells and also prevented S. gordonii antagonism would tend to support the importance of NLK-mediated phosphorylation in the control of FOXO1 but does not exclude a role for additional mechanisms. S. gordonii did not have an effect on the pathways by which P. gingivalis as a monospecies infection activates FOXO1: i.e., up-regulation of ROS and activation of JNK, and dephosphorylation of Ser256. Thus, the TAK1-NLK pathways would appear to supersede these pathways, consistent with the observation that NLK can modulate FOXO1 regardless of PI3/Akt activity (which targets Ser256) (48). NLK is emerging as a regulator of a number of signaling molecules and transcription factors, including PPAR-γ, Myb proteins, Notch1 (74–76), YAP (77), SMADs (78), NF-κB (79), C/EBPs (80), and Wnt/β-catenin (81). The extent to which S. gordonii can impact epithelial responses through these regulators requires further investigation. Additionally, the means by which P. gingivalis and S. gordonii initially transmit information, along with the host cell sensing mechanisms, are unknown and represent a significant area for future studies. P. gingivalis can engage integrin receptors on epithelial surfaces through the FimA component fimbriae (82) and also secretes a serine phosphatase intracellularly (83, 84), and both of these are potential candidate effectors for inciting signaling events. S. gordonii can adhere to carbohydrate receptors on epithelial cells (85, 86), which, similarly, could stimulate signal transduction events. Moreover, oral streptococci, such as S. gordonii, produce hydrogen peroxide, which is capable of impacting host cell physiology (87).
Epithelial surfaces of the oral cavity are colonized immediately after birth and perform several important defense functions. These include barrier function to prevent penetration of bacteria into underlying tissue, the production of antimicrobial molecules to control colonization, and the secretion of inflammatory mediators (53, 58, 88). Disruption of epithelial cell function is thus a component of periodontal disease, as well as the hallmark of OSCC. Taken together, the results of the current study show that the presence of S. gordonii and related organisms can divert epithelial cell responses to P. gingivalis that are transduced through FOXO1 and consequently diminish inflammatory and EMT properties that are dependent on ZEB2. Oral streptococci, such as S. gordonii, are major components of the oral microbiome. Originally considered passive early colonizers of tooth surfaces, the role of these organisms is now realized to be more nuanced and context-dependent. In infective endocarditis, streptococci originating from the oral cavity are clearly overt pathogens (86). In subgingival biofilms, S. gordonii can enhance community pathogenicity, and, in this context, it is considered an accessory pathogen (89). On the basis of our results here, we propose a designation for S. gordonii as a homeostatic commensal at the oral epithelial interface. The features underlying this designation are an ability to inhibit the activity of a more pathogenic organism (in this case, P. gingivalis) and maintain a homeostatic host–community interaction. Intriguingly, S. gordonii does not directly impede P. gingivalis, but, rather, it is modulation of host signaling that dictates outcome.
Materials and Methods
Bacterial Strains, Eukaryotic Cells, and Growth Conditions.
P. gingivalis ATCC 33277, W83, low passage strain MP4-504, ΔrgpAB, Δkgp, and ΔrgpAB/kgp (90) were cultured in trypticase soy broth (TSB) supplemented with yeast extract (1 mg/mL), hemin (5 μg/mL), and menadione (1 μg/mL). S. gordonii strain DL1, S. oralis strain 10557, S. sanguinis strain 10556, and S. cristatus strain CC5A were grown in Todd–Hewitt broth. F. nucleatum strain ATCC 25586 was cultured in brain heart infusion (BHI) broth supplemented with hemin (5 μg/mL) and menadione (1 μg/mL). All bacterial strains were cultured anaerobically to midlog phase as described previously (91).
Human telomerase immortalized gingival keratinocytes (TIGKs) are derived from gingival epithelium and are maintained in our laboratory (92). OKF6/TERT2 keratinocytes are derived from the oral mucosa and were originally established in the Rheinwald laboratory (93). SCC9 and HeLa cells were obtained from the ATCC. TIGK and OKF6/TERT2 keratinocytes were maintained at 37 °C and 5% CO2 in Dermalife-K serum-free culture medium (Lifeline Cell Technology). SCC9 and HeLa cells were cultured in DMEM supplemented with 10% FBS. Epithelial cells at 80% confluence were stimulated with bacteria as described for individual experiments. Cells remained attached to the substrate at the end of the bacterial challenge, and, in parallel experiments, no loss of cell viability was observed by trypan blue staining following bacterial challenge.
Antibodies and Reagents.
Antibodies to FOXO1 (for blots), phospho-FOXO1 (Ser256, -329), β-catenin, JNK, phospho-JNK (Thr183/Tyr185), NLK, phospho-NLK (Thr298), TAK and phospho-TAK (Thr187), TAB2, and GAPDH, along with horseradish peroxidase (HRP)-conjugated secondary antibodies, and Cell Lysis buffer were from Cell Signaling Technology. TCF7L1 (for blots), TCF7L2, and Alexa Fluor 488-conjugated anti-rabbit secondary antibodies were from ThermoFisher. ZEB2 antibodies were from Novus. For ChIP, FOXO1 antibodies were from Abcam, and TCF7L1 antibodies were from Santa Cruz. Small interfering RNA (siRNA), against β-catenin, NLK, TAK, and SMAD4, was from Santa Cruz; SMAD2, SMAD3, and LEF1 were from OriGene; ZEB2, FOXO3, TCF7L1, TCF7L2, and DYRK1 were from Thermo Fisher; FOXO1 was from Cell Signaling; and TCF7 was from Dharmacon. Flag antibody, TGF-β receptor inhibitor LY-364947, and TLCK were from Sigma-Aldrich. Texas Red-phalloidin was from Life Technologies. Aminophenyl fluorescein (APF) was from Sigma-Aldrich.
Quantitative Reverse Transcription-PCR.
Total mRNA from TIGK cells was isolated and purified with an RNeasy Mini Kit (Qiagen). RNA concentrations were determined by spectrophotometry on a NanoDrop 2000 (ThermoFisher). cDNA from total RNA was synthesized (2 μg RNA per reaction volume) using a High Capacity cDNA reverse transcription kit (Applied Biosystems). Real-time RT-PCR used TaqMan Fast Universal PCR Master Mix and gene expression assays for ZEB2, TWIST1, TWIST2, SMAD2, SMAD3, SMAD4, FOXO1, FOXO3, TCF7L3, NLK, TAK, c-Myc, and GAPDH (Applied Biosystems). For β-catenin, TCF7, TCF7L1, TCF7L2, and GAPDH, Power SYBR Green PCR Master Mix, and primers were used (SI Appendix, Table S1). Real-time PCR was performed on an Applied Biosystems StepOne Plus cycler with StepOne software V2.2.2 and the auto-calculated threshold cycle selected. The cycle threshold (Ct) values were determined, and mRNA expression levels were normalized to GAPDH and expressed relative to controls following the 2−ΔΔCT method.
Immunoblotting.
TIGKs were lysed with cold cell lysis buffer containing PhosSTOP Phosphatase Inhibitor and Protease Inhibitor (Roche). Lysates (20 ng of protein) were separated by 10% SDS-polyacrylamide gel electrophoresis, blotted onto a PVDF membrane, and blocked with 5% skim milk in Tris-buffered saline (TBS) containing 0.1% Tween 20. Blots were probed at 4 °C overnight with primary antibodies, followed by 1 h with HRP-conjugated secondary antibody at room temperature. Membranes were developed using ECL detection (ThermoFisher), and images were collected using a ChemiDoc XRS Plus (Bio-Rad).
Plasmid Construction, RNA Interference, Transfections, and Luciferase Assay.
WT FOXO1 plasmid (pcDNA3 Flag FKHR) was a gift from Kunliang Guan, University of California, San Diego, La Jolla, CA (Addgene plasmid #13507; RRID:Addgene_13507; n2t.net/addgene:13507) (94). S329E and S329A mutations were generated by PCR-based site-directed mutagenesis using the primers described in SI Appendix, Table S1. Mutations were confirmed by sequencing. To construct the mammalian expression vectors for β-catenin and TCF7L1, cDNA fragments were amplified using primer sets listed in SI Appendix, Table S1. CDNA fragments were ligated into the pcDNA3 plasmid to create N-terminal Flag-tagged proteins. To construct ZEB2 promoter-firefly luciferase reporters, DNA fragments were amplified from human genomic DNA using the primers listed in SI Appendix, Table S1. DNA fragments were cloned into the pGL3-Basic plasmid. A series of ZEB2 promoter fragments containing mutations were generated by PCR mutagenesis (Q5 Site-Directed Mutagenesis Kit from NEB). All constructs were confirmed by sequencing. The FOXO Reporter and Negative Control Reporter were from BPS Bioscience. For siRNA, TIGKs were transfected with plasmids or siRNA for 24 h using LipoJet transfection agent (SignaGen). At 48 h after transfection, the medium was replaced, and cells were stimulated with bacteria. Luciferase activity was measured with the Dual-Glo Luciferase Assay system (Promega) and normalized to the Renilla internal control.
Chromatin Immunoprecipitation.
Chromatin was prepared using the ChIP-IT Express Enzymatic Shearing Kit (Active Motif) in accordance with the manufacturer’s instructions. Cells fixed by 1% formaldehyde were lysed and gently homogenized in a Dounce homogenizer to release the nuclei. After centrifugation, the nuclear pellets were resuspended and sheared by Enzymatic Shearing Mixture. Immunoprecipitation was performed overnight at 4 °C with 2 µg of antibody plus 25 μL of protein G magnetic beads. Beads were then washed three times in ChIP buffer. The digested chromatin was eluted, cross-linking was reversed, and proteinase K treatment was performed. The eluted immunoprecipitated DNA and samples of ChIP input DNA were purified with a QIAquick PCR Purification Kit (Qiagen) and then subjected to PCR and qRT-PCR. The GAPDH promoter region was amplified and used to normalize the amount of input material (see SI Appendix, Table S1 for primer sequences).
Immunofluorescence and Confocal Laser Scanning Microscopy.
TIGK cells were grown on glass coverslips, washed twice in PBS, and fixed with 4% paraformaldehyde for 10 min. Permeabilization was with 0.2% Triton X-100 for 10 min at room temperature before blocking in 10% goat serum for 20 min. FOXO1 was detected by reacting with antibody (1:1,000) overnight at 4 °C, followed by Alexa Fluor 488-conjugated secondary antibodies at 1:2,000 for 3 h in the dark. Following a 20-min blocking in 0.1% goat serum, actin was labeled with Texas Red-phalloidin for 2 h at room temperature in the dark. Coverslips were mounted on glass slides using ProLong Gold (Invitrogen) with 4′6-diamidino-2-phenylindole (DAPI) mounting medium before imaging with a Leica SP8 confocal microscope. Images were analyzed using Volocity 6.3 Software (PerkinElmer).
Matrigel Invasion Assay.
Cell motility was measured by assessment of the migration rate of TIGKs using a BD BioCoat Matrigel Invasion Chamber (BD Biosciences). Transfected/control cells (2 × 105) were plated on transwell filters coated with matrigel and reacted with bacteria for 18 h. The lower compartment of the invasion chambers contained cell culture medium. Cells remaining on the upper surface of the filter were removed, and the cells that migrated through the filter were fixed with 1% methanol and stained with toluidine blue. Cells were enumerated from three random 20× fields for each filter using a Nikon Eclipse TS100 microscope.
Statistical Analysis.
Statistical analyses were conducted using GraphPad Prism software. Data were evaluated by ANOVA with Tukey’s multiple comparison test.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants DE011111, DE012505, and DE017921 (to R.J.L.); DE026939 and DE028346 (to D.P.M.); DE028166 (to Z.R.F.); and DE023193 (to M.W.); and by Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research Grant JP18K17066, The Nakatomi Foundation, and The Mochida Memorial Foundation for Medical and Pharmaceutical Research (to J.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microscopy files are available in the BioStudies database (www.ebi.ac.uk/biostudies) under accession no. S-BSST247.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900101116/-/DCSupplemental.
References
- 1.Kassebaum NJ, et al. Global burden of severe periodontitis in 1990-2010: A systematic review and meta-regression. J Dent Res. 2014;93:1045–1053. doi: 10.1177/0022034514552491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hajishengallis G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat Rev Immunol. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajishengallis G, Lamont RJ. Breaking bad: Manipulation of the host response by Porphyromonas gingivalis. Eur J Immunol. 2014;44:328–338. doi: 10.1002/eji.201344202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darveau RP. Periodontitis: A polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 5.Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: Dynamic communities and host interactions. Nat Rev Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hajishengallis G, Lamont RJ. Dancing with the stars: How choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 2016;24:477–489. doi: 10.1016/j.tim.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atanasova KR, Yilmaz O. Looking in the Porphyromonas gingivalis cabinet of curiosities: The microbium, the host and cancer association. Mol Oral Microbiol. 2014;29:55–66. doi: 10.1111/omi.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitmore SE, Lamont RJ. Oral bacteria and cancer. PLoS Pathog. 2014;10:e1003933. doi: 10.1371/journal.ppat.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahingur SE, Yeudall WA. Chemokine function in periodontal disease and oral cavity cancer. Front Immunol. 2015;6:214. doi: 10.3389/fimmu.2015.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE, Davey ME. 2013. Porphyromonas gingivalis: Keeping the pathos out of the biont. J Oral Microbiol, 10.3402/jom.v5i0.19804. [DOI] [PMC free article] [PubMed]
- 11.Sztukowska MN, et al. Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cell Microbiol. 2016;18:844–858. doi: 10.1111/cmi.12554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, et al. Human primary epithelial cells acquire an epithelial-mesenchymal-transition phenotype during long-term infection by the oral opportunistic pathogen, Porphyromonas gingivalis. Front Cell Infect Microbiol. 2017;7:493. doi: 10.3389/fcimb.2017.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdulkareem AA, Shelton RM, Landini G, Cooper PR, Milward MR. Periodontal pathogens promote epithelial-mesenchymal transition in oral squamous carcinoma cells in vitro. Cell Adhes Migr. 2017;12:127–137. doi: 10.1080/19336918.2017.1322253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ha NH, et al. Prolonged and repetitive exposure to Porphyromonas gingivalis increases aggressiveness of oral cancer cells by promoting acquisition of cancer stem cell properties. Tumour Biol. 2015;36:9947–9960. doi: 10.1007/s13277-015-3764-9. [DOI] [PubMed] [Google Scholar]
- 15.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scanlon CS, Van Tubergen EA, Inglehart RC, D’Silva NJ. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J Dent Res. 2013;92:114–121. doi: 10.1177/0022034512467352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goossens S, Vandamme N, Van Vlierberghe P, Berx G. EMT transcription factors in cancer development re-evaluated: Beyond EMT and MET. Biochim Biophys Acta Rev Cancer. 2017;1868:584–591. doi: 10.1016/j.bbcan.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez C, David JM, Palena C. Epithelial-mesenchymal transition and inflammation at the site of the primary tumor. Semin Cancer Biol. 2017;47:177–184. doi: 10.1016/j.semcancer.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katsura A, et al. ZEB1-regulated inflammatory phenotype in breast cancer cells. Mol Oncol. 2017;11:1241–1262. doi: 10.1002/1878-0261.12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudney JD, Chen R, Zhang G. Streptococci dominate the diverse flora within buccal cells. J Dent Res. 2005;84:1165–1171. doi: 10.1177/154405910508401214. [DOI] [PubMed] [Google Scholar]
- 22.Colombo AV, Silva CM, Haffajee A, Colombo AP. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J Med Microbiol. 2006;55:609–615. doi: 10.1099/jmm.0.46417-0. [DOI] [PubMed] [Google Scholar]
- 23.Dzink JL, Gibbons RJ, Childs WC, 3rd, Socransky SS. The predominant cultivable microbiota of crevicular epithelial cells. Oral Microbiol Immunol. 1989;4:1–5. doi: 10.1111/j.1399-302x.1989.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhang G, Rudney JD. Streptococcus cristatus attenuates Fusobacterium nucleatum-induced cytokine expression by influencing pathways converging on nuclear factor-κB. Mol Oral Microbiol. 2011;26:150–163. doi: 10.1111/j.2041-1014.2010.00600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mans JJ, et al. The degree of microbiome complexity influences the epithelial response to infection. BMC Genomics. 2009;10:380. doi: 10.1186/1471-2164-10-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tribble GD, Kerr JE, Wang BY. Genetic diversity in the oral pathogen Porphyromonas gingivalis: Molecular mechanisms and biological consequences. Future Microbiol. 2013;8:607–620. doi: 10.2217/fmb.13.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dashper SG, et al. Porphyromonas gingivalis uses specific domain rearrangements and allelic exchange to generate diversity in surface virulence factors. Front Microbiol. 2017;8:48. doi: 10.3389/fmicb.2017.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olsen I, Chen T, Tribble GD. Genetic exchange and reassignment in Porphyromonas gingivalis. J Oral Microbiol. 2018;10:1457373. doi: 10.1080/20002297.2018.1457373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao L, et al. Src promotes EGF-induced epithelial-to-mesenchymal transition and migration in gastric cancer cells by upregulating ZEB1 and ZEB2 through AKT. Cell Biol Int. 2018;42:294–302. doi: 10.1002/cbin.10894. [DOI] [PubMed] [Google Scholar]
- 31.Hong KO, et al. Inhibition of Akt activity induces the mesenchymal-to-epithelial reverting transition with restoring E-cadherin expression in KB and KOSCC-25B oral squamous cell carcinoma cells. J Exp Clin Cancer Res. 2009;28:28. doi: 10.1186/1756-9966-28-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang H, et al. Porphyromonas gingivalis-induced reactive oxygen species activate JAK2 and regulate production of inflammatory cytokines through c-Jun. Infect Immun. 2014;82:4118–4126. doi: 10.1128/IAI.02000-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegarty SV, Sullivan AM, O’Keeffe GW. Zeb2: A multifunctional regulator of nervous system development. Prog Neurobiol. 2015;132:81–95. doi: 10.1016/j.pneurobio.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Zhao L, et al. Bufalin inhibits TGF-β-induced epithelial-to-mesenchymal transition and migration in human lung cancer A549 cells by downregulating TGF-β receptors. Int J Mol Med. 2015;36:645–652. doi: 10.3892/ijmm.2015.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen F, et al. Baicalein inhibits migration and invasion of gastric cancer cells through suppression of the TGF-β signaling pathway. Mol Med Rep. 2014;10:1999–2003. doi: 10.3892/mmr.2014.2452. [DOI] [PubMed] [Google Scholar]
- 36.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y, et al. Noncanonical activation of β-catenin by Porphyromonas gingivalis. Infect Immun. 2015;83:3195–3203. doi: 10.1128/IAI.00302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Z, et al. Beta-catenin promotes survival of renal epithelial cells by inhibiting Bax. J Am Soc Nephrol. 2009;20:1919–1928. doi: 10.1681/ASN.2009030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valenta T, Hausmann G, Basler K. The many faces and functions of β-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hrckulak D, Kolar M, Strnad H, Korinek V. TCF/LEF transcription factors: An update from the internet resources. Cancers (Basel) 2016 doi: 10.3390/cancers8070070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong T, et al. FOXO1 inhibits the invasion and metastasis of hepatocellular carcinoma by reversing ZEB2-induced epithelial-mesenchymal transition. Oncotarget. 2017;8:1703–1713. doi: 10.18632/oncotarget.13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eijkelenboom A, Burgering BM. FOXOs: Signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol. 2013;14:83–97. doi: 10.1038/nrm3507. [DOI] [PubMed] [Google Scholar]
- 43.Obsil T, Obsilova V. Structural basis for DNA recognition by FOXO proteins. Biochim Biophys Acta. 2011;1813:1946–1953. doi: 10.1016/j.bbamcr.2010.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Kumar PA, et al. Growth hormone (GH)-dependent expression of a natural antisense transcript induces zinc finger E-box-binding homeobox 2 (ZEB2) in the glomerular podocyte: A novel action of gh with implications for the pathogenesis of diabetic nephropathy. J Biol Chem. 2010;285:31148–31156. doi: 10.1074/jbc.M110.132332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beltran M, et al. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, et al. FOXO responses to Porphyromonas gingivalis in epithelial cells. Cell Microbiol. 2015;17:1605–1617. doi: 10.1111/cmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C, et al. FOXO1 differentially regulates both normal and diabetic wound healing. J Cell Biol. 2015;209:289–303. doi: 10.1083/jcb.201409032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim S, Kim Y, Lee J, Chung J. Regulation of FOXO1 by TAK1-Nemo-like kinase pathway. J Biol Chem. 2010;285:8122–8129. doi: 10.1074/jbc.M110.101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Groote-Bidlingmaier F, et al. DYRK1 is a co-activator of FKHR (FOXO1a)-dependent glucose-6-phosphatase gene expression. Biochem Biophys Res Commun. 2003;300:764–769. doi: 10.1016/s0006-291x(02)02914-5. [DOI] [PubMed] [Google Scholar]
- 50.Li M, et al. TAB2 scaffolds TAK1 and NLK in repressing canonical Wnt signaling. J Biol Chem. 2010;285:13397–13404. doi: 10.1074/jbc.M109.083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lamont RJ, Hajishengallis G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Murray JL, Connell JL, Stacy A, Turner KH, Whiteley M. Mechanisms of synergy in polymicrobial infections. J Microbiol. 2014;52:188–199. doi: 10.1007/s12275-014-4067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ebersole JL, et al. The periodontal war: Microbes and immunity. Periodontol 2000. 2017;75:52–115. doi: 10.1111/prd.12222. [DOI] [PubMed] [Google Scholar]
- 54.Mullineaux-Sanders C, Suez J, Elinav E, Frankel G. Sieving through gut models of colonization resistance. Nat Microbiol. 2018;3:132–140. doi: 10.1038/s41564-017-0095-1. [DOI] [PubMed] [Google Scholar]
- 55.Kamada N, Chen GY, Inohara N, Núñez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belkaid Y, Naik S. Compartmentalized and systemic control of tissue immunity by commensals. Nat Immunol. 2013;14:646–653. doi: 10.1038/ni.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linehan JL, et al. Non-classical immunity controls microbiota impact on skin immunity and tissue repair. Cell. 2018;172:784–796.e18. doi: 10.1016/j.cell.2017.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker JL, Bor B, Agnello M, Shi W, He X. Ecology of the oral microbiome: Beyond bacteria. Trends Microbiol. 2017;25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huo X, et al. Prognostic significance of the epithelial-mesenchymal transition factor zinc finger E-box-binding homeobox 2 in esophageal squamous cell carcinoma. Oncol Lett. 2017;14:2683–2690. doi: 10.3892/ol.2017.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kong YH, et al. Co-expression of TWIST1 and ZEB2 in oral squamous cell carcinoma is associated with poor survival. PLoS One. 2015;10:e0134045. doi: 10.1371/journal.pone.0134045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arunkumar G, et al. Dysregulation of miR-200 family microRNAs and epithelial-mesenchymal transition markers in oral squamous cell carcinoma. Oncol Lett. 2018;15:649–657. doi: 10.3892/ol.2017.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nibali L, Fedele S, D’Aiuto F, Donos N. Interleukin-6 in oral diseases: A review. Oral Dis. 2012;18:236–243. doi: 10.1111/j.1601-0825.2011.01867.x. [DOI] [PubMed] [Google Scholar]
- 63.Shinriki S, et al. Interleukin-6 signalling regulates vascular endothelial growth factor-C synthesis and lymphangiogenesis in human oral squamous cell carcinoma. J Pathol. 2011;225:142–150. doi: 10.1002/path.2935. [DOI] [PubMed] [Google Scholar]
- 64.Nagasaki T, et al. Interleukin-6 released by colon cancer-associated fibroblasts is critical for tumour angiogenesis: Anti-interleukin-6 receptor antibody suppressed angiogenesis and inhibited tumour-stroma interaction. Br J Cancer. 2014;110:469–478. doi: 10.1038/bjc.2013.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Q, et al. IL-6 promotion of glioblastoma cell invasion and angiogenesis in U251 and T98G cell lines. J Neurooncol. 2010;100:165–176. doi: 10.1007/s11060-010-0158-0. [DOI] [PubMed] [Google Scholar]
- 66.Shinriki S, et al. Humanized anti-interleukin-6 receptor antibody suppresses tumor angiogenesis and in vivo growth of human oral squamous cell carcinoma. Clin Cancer Res. 2009;15:5426–5434. doi: 10.1158/1078-0432.CCR-09-0287. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Zhou Y, Graves DT. FOXO transcription factors: Their clinical significance and regulation. BioMed Res Int. 2014;2014:925350. doi: 10.1155/2014/925350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Z, et al. Serine 574 phosphorylation alters transcriptional programming of FOXO3 by selectively enhancing apoptotic gene expression. Cell Death Differ. 2016;23:583–595. doi: 10.1038/cdd.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci USA. 2016;113:E791–E800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuboniwa M, et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat Microbiol. 2017;2:1493–1499. doi: 10.1038/s41564-017-0021-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duran-Pinedo AE, Baker VD, Frias-Lopez J. The periodontal pathogen Porphyromonas gingivalis induces expression of transposases and cell death of Streptococcus mitis in a biofilm model. Infect Immun. 2014;82:3374–3382. doi: 10.1128/IAI.01976-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Xie H, Hong J, Sharma A, Wang BY. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J Periodontal Res. 2012;47:578–583. doi: 10.1111/j.1600-0765.2012.01469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szypowska AA, de Ruiter H, Meijer LA, Smits LM, Burgering BM. Oxidative stress-dependent regulation of Forkhead box O4 activity by Nemo-like kinase. Antioxid Redox Signal. 2011;14:563–578. doi: 10.1089/ars.2010.3243. [DOI] [PubMed] [Google Scholar]
- 74.Ishitani T, et al. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol. 2010;12:278–285. doi: 10.1038/ncb2028. [DOI] [PubMed] [Google Scholar]
- 75.Kanei-Ishii C, et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 2004;18:816–829. doi: 10.1101/gad.1170604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takada I, et al. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–1285. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- 77.Moon S, et al. Phosphorylation by NLK inhibits YAP-14-3-3-interactions and induces its nuclear localization. EMBO Rep. 2017;18:61–71. doi: 10.15252/embr.201642683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeng YA, Rahnama M, Wang S, Sosu-Sedzorme W, Verheyen EM. Drosophila Nemo antagonizes BMP signaling by phosphorylation of Mad and inhibition of its nuclear accumulation. Development. 2007;134:2061–2071. doi: 10.1242/dev.02853. [DOI] [PubMed] [Google Scholar]
- 79.Li SZ, et al. Nemo-like kinase (NLK) negatively regulates NF-kappa B activity through disrupting the interaction of TAK1 with IKKβ. Biochim Biophys Acta. 2014;1843:1365–1372. doi: 10.1016/j.bbamcr.2014.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang ZY, et al. Stabilization of ATF5 by TAK1-Nemo-like kinase critically regulates the interleukin-1β-stimulated C/EBP signaling pathway. Mol Cell Biol. 2015;35:778–788. doi: 10.1128/MCB.01228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Masoumi KC, et al. NLK-mediated phosphorylation of HDAC1 negatively regulates Wnt signaling. Mol Biol Cell. 2017;28:346–355. doi: 10.1091/mbc.E16-07-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yilmaz O, Watanabe K, Lamont RJ. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 2002;4:305–314. doi: 10.1046/j.1462-5822.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 83.Tribble GD, Mao S, James CE, Lamont RJ. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc Natl Acad Sci USA. 2006;103:11027–11032. doi: 10.1073/pnas.0509813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moffatt CE, Inaba H, Hirano T, Lamont RJ. Porphyromonas gingivalis SerB-mediated dephosphorylation of host cell cofilin modulates invasion efficiency. Cell Microbiol. 2012;14:577–588. doi: 10.1111/j.1462-5822.2011.01743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wong A, Grau MA, Singh AK, Woodiga SA, King SJ. Role of neuraminidase-producing bacteria in exposing cryptic carbohydrate receptors for Streptococcus gordonii adherence. Infect Immun. 2018;86:e00068-18. doi: 10.1128/IAI.00068-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Knaus UG, Hertzberger R, Pircalabioru GG, Yousefi SP, Branco Dos Santos F. Pathogen control at the intestinal mucosa–H2O2 to the rescue. Gut Microbes. 2017;8:67–74. doi: 10.1080/19490976.2017.1279378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Feng Z, Weinberg A. Role of bacteria in health and disease of periodontal tissues. Periodontol 2000. 2006;40:50–76. doi: 10.1111/j.1600-0757.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- 89.Whitmore SE, Lamont RJ. The pathogenic persona of community-associated oral streptococci. Mol Microbiol. 2011;81:305–314. doi: 10.1111/j.1365-2958.2011.07707.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lee JY, et al. Maturation of the Mfa1 fimbriae in the oral pathogen Porphyromonas gingivalis. Front Cell Infect Microbiol. 2018;8:137. doi: 10.3389/fcimb.2018.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wright CJ, Wu H, Melander RJ, Melander C, Lamont RJ. Disruption of heterotypic community development by Porphyromonas gingivalis with small molecule inhibitors. Mol Oral Microbiol. 2014;29:185–193. doi: 10.1111/omi.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moffatt-Jauregui CE, et al. Establishment and characterization of a telomerase immortalized human gingival epithelial cell line. J Periodontal Res. 2013;48:713–721. doi: 10.1111/jre.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dickson MA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tang ED, Nuñez G, Barr FG, Guan KL. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.