Abstract
The last two steps of the purine biosynthetic pathway may be catalyzed by different enzymes in prokaryotes. The genes that encode these enzymes include homologs of purH, purP, purO and those encoding the AICARFT and IMPCH domains of PurH, here named purV and purJ, respectively. In Bacteria, these reactions are mainly catalyzed by the domains AICARFT and IMPCH of PurH. In Archaea, these reactions may be carried out by PurH and also by PurP and PurO, both considered signatures of this domain and analogous to the AICARFT and IMPCH domains of PurH, respectively. These genes were searched for in 1,403 completely sequenced prokaryotic genomes publicly available. Our analyses revealed taxonomic patterns for the distribution of these genes and anticorrelations in their occurrence. The analyses of bacterial genomes revealed the existence of genes coding for PurV, PurJ, and PurO, which may no longer be considered signatures of the domain Archaea. Although highly divergent, the PurOs of Archaea and Bacteria show a high level of conservation in the amino acids of the active sites of the protein, allowing us to infer that these enzymes are analogs. Based on the results, we propose that the gene purO was present in the common ancestor of all living beings, whereas the gene encoding PurP emerged after the divergence of Archaea and Bacteria and their isoforms originated in duplication events in the common ancestor of phyla Crenarchaeota and Euryarchaeota. The results reported here expand our understanding of the diversity and evolution of the last two steps of the purine biosynthetic pathway in prokaryotes.
Keywords: Archaea, Bacteria, bioinformatics, comparative genomics, evolution, phylogeny
Introduction
Purines and their derivatives are biomolecules essential to all living organisms as they play important roles in signaling pathways, carbohydrate metabolism, and as precursors of nucleic acids (Smith and Atkins 2002). Additionally, the enzymes that catalyze their synthesis are important targets for antimicrobial and anticancer compounds (Kirsch and Whitney 1991). The enzymatic reactions involved in the de novo biosynthesis of purines were elucidated in the 1950s (Buchanan and Hartman 1959) and all genes that encode these enzymes were identified and their products biochemically characterized by the year 2000 (Zalkin 1983; Parker 1984; Schrimsher et al. 1986; Aiba and Mizobuchi 1989; Watanabe et al. 1989; Cheng et al. 1990; Inglese et al. 1990; Gu et al. 1992; He et al. 1992; Marolewski et al. 1994; Graupner et al. 2002; Hoskins et al. 2004; Ownby et al. 2005).
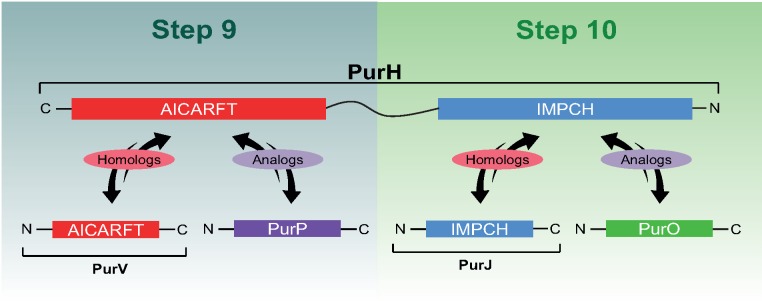
There are generally ten pur genes in the following order: purF, purD, purN, purL, purM, purE, purK, purC, purB, and purH, each encoding an enzyme responsible for a step in the purine biosynthetic pathway (PBP). The majority of the pur genes were initially described in Escherichia coli and later, orthologs were found in other prokaryotes and eukaryotes, suggesting that these are the canonical genes encoding enzymes of the PBP in different evolutionary lineages (Chopra et al. 1991; Ni et al. 1991; Gu et al. 1992; Rayl et al. 1996; Nilsson and Kilstrup 1998; Peltonen and Mäntsälä 1999; Sampei et al. 2010; Liu et al. 2014). This scenario changed when some authors showed that the enzymatic reactions of three of the 10–11 steps of the PBP may be catalyzed by different nonhomologous enzymes in distinct microorganisms. For example, PurT, a novel enzyme involved in the PBP was described in 1994 as an analog of the already known PurN that catalyses the third step of the PBP (Marolewski et al. 1994). Two new enzymes, PurP and PurO were later described in the PBP of Archaea as analogs of PurH, until then considered as the canonical enzyme catalyzing the two last steps of the PBP (Graupner et al. 2002; Ownby et al. 2005). PurP and PurO are currently considered signatures of the Archaea domain (Graupner et al. 2002; Ownby et al. 2005; Zhang, Morar, et al. 2008; Zhang, White, et al. 2008; Armenta-Medina et al. 2014). These two separate enzymes are analogous to the domains AICARFT and IMPCH of PurH, which contains them fused (fig. 1). The domain AICARFT catalyses the penultimate reaction of the PBP, converting AICAR in FAICAR and the domain IMPCH catalyses the last reaction of the PBP, converting FAICAR into IMP, the final product of the pathway (Zhang, Morar, et al. 2008).
Fig. 1.
—Nomenclature and homology/analogy relationships among the proteins involved in the last two steps of the purine biosynthetic pathway. PurH is composed of two domains, IMPCH and AICARFT. The relationships of homology and analogy among the domains of PurH and other proteins are shown. PurH is shown in an inverted position to match the steps of the purine biosynthetic pathway.
A comparative genomic analysis of the Archaea domain showed that in some free-living species of the phylum Euryarchaeota the domains AICARFT and IMPCH of PurH are encoded by distinct genes. These archaeal species do not contain genes encoding PurH nor its analogs PurP and PurO (Brown et al. 2011). In this study, the authors also showed that species of the phylum Crenarchaeota do not possess the genes coding for PurH nor homologs of its domains or PurO, but PurP was found (Brown et al. 2011). This study indicates that the diversity of enzymes involved in the PBP of Archaea is higher than previously thought. In their study, Brown et al. (2011) did not include the domain Bacteria, where most of the prokaryotic diversity resides.
Purine biosynthesis is among the most ancient metabolic pathways and probably evolved in the LUCA (Caetano-Anollés et al. 2007). According to the hypothesis of Horowitz (1945), enzymes in the last steps of biosynthetic pathways are the first to be recruited. Curiously, the last two steps of the PBP show the highest variation.
Nowadays, the availability of completely sequenced genomes in public databases representing most of the diversity of prokaryotic higher taxa potentially provide a comprehensive picture of the diversity and evolution of biological processes. In this study, we report on a genomic analysis concerning the diversity and evolution of the two last steps of the PBP in prokaryotic lineages. The results are presented in a taxonomical framework that includes the currently accepted phylogenetic classification of the prokaryotes.
Materials and Methods
Searches for Purine Biosynthetic Genes (pur) in Prokaryotic Genomes
A total of 1,403 nonredundant fully sequenced prokaryotic genomes deposited in the NCBI (National Centre for Biotechnology Information) database until July of 2014 were used in this study. Bacterial and archaeal strains identified at the genus or species levels were treated as distinct Operational Taxonomical Units (OTUs) in the analyses. The program TBLASTN was used to perform searches for purH, purP and purO and for genes that code for the domains AICARFT and IMPCH in the nucleotide collection (nr/nt), RefSeq Representative genomes (refseq_representative_genomes) and ReFseq Genome (refseq_genomes) databases. The genes coding for the domains AICARFT and IMPCH were named hereafter as purV and purJ, respectively (fig. 1). Additionally, the program BLASTP was used to perform searches for PurH, PurP, PurO, PurV, and PurJ in the nonredundant protein sequences (nr) database. All TBLASTN and BLASTP searches (Altschul et al. 1990) were performed with the default parameters of the programs, except for the filters and masking options that were disabled.
BLAST searches were done individually in each completely sequenced genome and the presence of conserved domains typical of the searched proteins was used as the homology criterion to recover the sequences. All sequences containing the searched domains were recovered and included in the analyses. Amino acid sequences of PurH (GI: 16131836) of Escherichia coli and PurP (GI: 15668306) and PurO (GI: 34588137) of Methanocaldococcus jannaschii were used as queries. The amino acid sequence of PurH was used as query to perform searches for PurH, PurV, and PurJ and for the genes that encode these proteins.
Diversity of the Last Two Steps of the PBP
During the BLAST searches, the presence or absence and the number of copies of purH, purV, purJ, purP, and purO in all the genomes analyzed as well as their genomic context in relation to the other genes of the PBP were registered. Taxonomic patterns of occurrence of these genes in prokaryotic genomes were registered in all categories, from domain to species.
Multiple Alignments and Phylogeny of PurP and PurO
The amino acid sequences of the genes purH, purV, purJ, purP, and purO recovered in the BLAST searches were code-aligned in the Guidance server with the MAFFT algorithm (Penn et al. 2010). The program MEGA 6.0 (Tamura et al. 2013) was used to edit the multiple alignments of the proteins PurP and PurO, choose the substitution matrix, and to perform the phylogenetic analyses with the maximum likelihood (ML) method. The phylogenetic trees were visualized and edited in the program Archaeopterix (Han and Zmasek 2009).
Amino Acid Sequence Analyses
Sliding window plot analyses were done with the program SWAAP 1.0.2 (Pride 2000) using the multiple alignment of proteins. The average identity of the sequences was calculated with the model K2P in a sampling window of ten amino acids with only one amino acid displaced along the multiple alignments. The ConSurf Server (http://consurf.tau.ac.il/2016; last accessed March 14, 2019; Ashkenazy et al. 2016) was used to estimate the evolutionary conservation of amino acid positions in two multiple alignments of PurO: the first alignment contained only archaeal PurOs and the second contained the PurO sequences of Methanothermobacter thermoautotrophicus and the bacterial PurOs. The ConSurf score was calculated using the default parameters with the two multiple alignments described earlier for each amino acid of the tertiary structure of the PurO from M. thermoautotrophicus (PDB ID code 2NTL), which was previously functionally characterized (Kang et al. 2007). The logos were produced in the program Web Logo (Crooks et al. 2004) only with the positions of the active sites of PurOs. The average identity/divergence between archaeal and bacterial proteins was calculated with the software MEGA 6.0 in pairwise comparisons between proteins from these two domains.
Results
Comparative Genomics
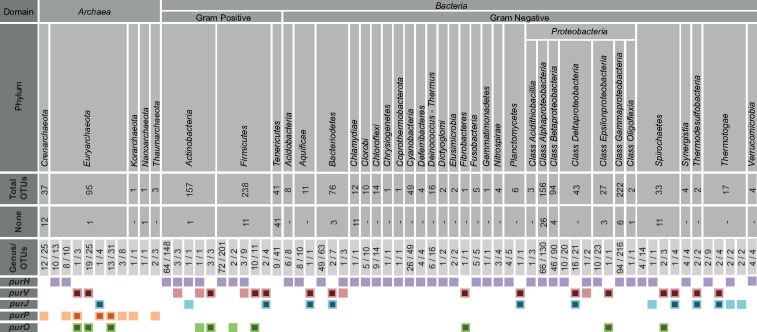
The comparative genomic analysis was carried out with a total of 1,403 completely sequenced genomes available in the NCBI database, representing 1,266 OTUs of the domain Bacteria and 137 OTUs of the domain Archaea. These OTUs represent 27 out of the 34 described phyla of the domain Bacteria and all five phyla of the domain Archaea according to List of Prokaryotic names with Standing in Nomenclature—LPSN (Euzéby 1997). From the 1,403 analyzed genomes, 132 did not have genes coding for PurH, PurO, purP, PurV, and PurJ (fig. 2 and supplementary tables S1 and S2, Supplementary Material online).
Fig. 2.
—Comparative genomic analysis of genes coding for the ninth and tenth steps of the purine biosynthetic pathway in 1,403 complete prokaryotic genomes. Colored boxes represent presence of the gene and smaller black boxes inside the colored ones represent the cases in which purH is replaced by a combination of genes that are functionally equivalent to purH. The numbers below each taxonomical category indicate the number of genera and OTUs analyzed. None indicates the number of OTUs that do not contain any of the genes encoding the last two steps of the PBP.
PurH-coding genes were found in genomes of 23 OTUs of the domain Archaea, all of which are in the class Methanomicrobia and in the families Methanoregulaceae, Methanocorpusculaceae, Methanomicrobiaceae, Methanospirillaceae, and Methanosarcinaceae and in the class Thermoplasmata, families Ferroplasmaceae, Picrophilaceae, and Thermoplasmataceae of the phylum Euryarchaeota (fig. 2 and supplementary table S1, Supplementary Material online). In contrast, PurH-coding genes were found in 1,083 OTUs of the domain Bacteria distributed in most phyla of this domain (fig. 2 and supplementary table S1, Supplementary Material online).
Genes that code for proteins homologous to the domains AICARFT and IMPCH of PurH were found in OTUs of the domains Archaea and Bacteria (fig. 2 and supplementary table S1, Supplementary Material online). The name purJ was used to designate the domain IMPCH of the PurH of Salmonella typhymurium, when it was incorrectly identified as a gene (Gots et al. 1969). Therefore, from this point on, we will use the name purJ for the gene encoding the domain IMPCH in accordance with its original nomenclature (Gots et al. 1969). For the domain AICARFT, we propose the name purV (fig. 1). The majority of the purV and purJ were recovered from bacterial genomes: 109 purVs, 28 from the domain Archaea and 81 from domain Bacteria; and 54 purJs, 4 from the domain Archaea and 50 from domain Bacteria (table 1 and supplementary table S1, Supplementary Material online). The genes purV and/or purJ were found in 10 of the 27 bacterial phyla analyzed, including Gram-positive and Gram-negative OTUs, indicating that they are widely distributed.
Table 1.
Occurrence and Co-Occurrence of the Genes Encoding the Last Two Steps of the Purine Biosynthetic Pathway in Bacteria and Archaea and the Genomic Context They are Found
| Occurrence Per OTU |
Co-Occurrence Per OTU |
In Context with Other Pur Genes |
Not in Context with Other Pur Genes |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Genes | Archaea | Bacteria | Total | Genes | Archaea | Bacteria | Genes | Archaea | Bacteria | Archaea | Bacteria |
| purH | 23 | 1,083 | 1,106 | purH/purV | — | 17 | purV | 25 | 26 | 3 | 55 |
| purV | 28 | 81 | 109 | purH/purJ | — | 4 | purJ | — | 16 | 4 | 34 |
| purJ | 4 | 50 | 54 | purH/purO | — | 3 | purO | 6 | 15 | 53 | 7 |
| purP I | 25 | — | — | purV/purJ | — | 44 | purP I | — | — | 25 | — |
| purP II | 62 | — | — | purV/purO | 25 | 19 | purP II | 25 | — | 37 | — |
| purP III | 61 | — | — | purH/purV/purO | — | 1 | purP III | 32 | — | 29 | — |
| purP IV | 6 | — | — | purP II/purP III | 37 | — | purP IV | 6 | — | — | — |
| purO | 59 | 22 | 81 | purP II/purP III/purJ | 4 | — | |||||
| purP I/purO | 24 | — | |||||||||
| purP I/purP II/purP III/purO | 1 | — | |||||||||
| purP II/purP III/purV/purO | 3 | — | |||||||||
| purP II/purP III/purH | 10 | — | |||||||||
| purP II/purP III/purP IV/purO | 6 | — | |||||||||
Note.—Genes in genomic context are together with other genes of the PBP.
The gene purO was until now considered a signature of the domain Archaea (Graupner et al. 2002; Ownby et al. 2005; Zhang, Morar, et al. 2008; Zhang, White, et al. 2008; Armenta-Medina et al. 2014). Surprisingly, 22 homologs of purO were found in bacterial genomes, whereas 59 purOs were found in archaeal genomes (table 1 and fig. 2). Genes that code for homologs of PurP were only found in genomes of OTUs of the domain Archaea, a total of 154, with the number of copies varying from one to four per genome (fig. 2 and supplementary table S1, Supplementary Material online). Homologs of PurH, PurO, PurV, and PurJ were not found in genomes of the phyla Crenarchaeota, Korarchaeota, and Thaumarchaeota, but genes coding for homologs of PurP were present.
The patterns of presence and absence of the genes purH, purV, purJ, and purO show that the putative new bacterial genes of the PBP, in general, anticorrelate with purH. In other words, the majority of the OTUs that do not have purH possess purV and purO or purV and purJ in the genome (fig. 2). Similarly, archaeal genomes contained the combinations purV/purO, purJ/purP, and purP/purO, but not purV and purJ, the most common in Bacteria, after purH. These results suggest that the combinations of genes mentioned earlier for bacteria and archaea are replacing purH, maintaining the last two steps of the PBP, as already proposed for the domain Archaea (Brown et al. 2011).
Only two assembly/annotation errors in the genes coding for proteins of the last two steps of the PBP were found in the 1,403 analyzed genomes: two truncated PurH sequences (supplementary table S1, Supplementary Material online). These two sequences were included in our analyses. This remarkably low number of misannotations may be due to the fact that these genes are well represented in the biological databases.
In summary, genes coding for PurH were found in 85% of the bacterial genomes and the combinations purV/purJ and purV/purO were in 63 genomes, representing 5% of the total number of bacterial genomes analyzed. The remaining 10% did not harbor any of the genes for the last two steps of the PBP. In Archaea, genes coding for PurH were in ∼17% of the genomes and the combinations purV/purO, purP/purJ, and purP/purO were found in ∼44% of the genomes, 10% of the genomes did not have the genes for the last two steps of the PBP, and the remaining 29% contained other combinations that did not replace purH completely (fig. 2; table 1; and supplementary table S1, Supplementary Material online).
Genomic Context and Taxonomical Patterns
The results of these studies demonstrated that purP is present only in the domain Archaea, while purO, purV, and purJ were found both in the domains Bacteria and Archaea (fig. 2 and supplementary table S1, Supplementary Material online). The gene combinations purP/purO, purP/purJ, purV/purO, purV/purJ are potentially replacing purH in Bacteria and Archaea, performing the last two steps of the PBP. Approximately 37% of the total number of genes that is potentially replacing purH in Bacteria and Archaea is in genomic context with other genes of the PBP and the remaining 63% is not in context in Bacteria and Archaea (tables 1 and 2). In some bacterial OTUs, the putative new genes, purO, purV, and purJ are in genomic context with themselves or with other genes of the PBP, from which the most frequent are purD and purN (table 2). The arrangements of these genes in Bacteria (table 2) indicate that they are putative operons that are coexpressed and have a role in the PBP.
Table 2.
Bacterial OTUs with purV, purJ, and purO in Genomic Context with Other Genes of the Purine Biosynthetic Pathway
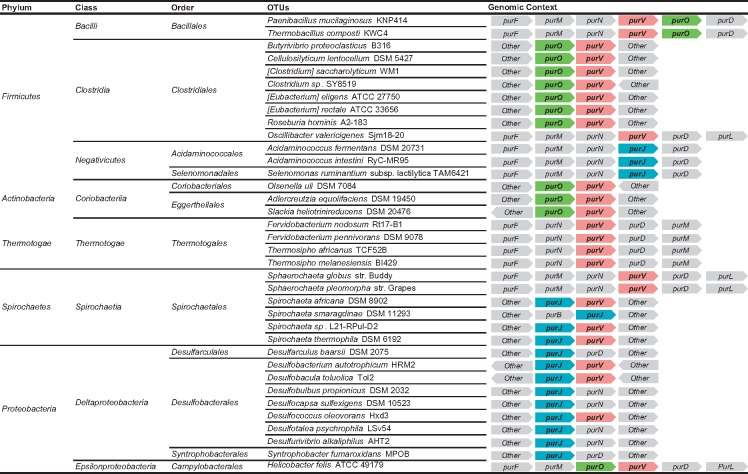 |
Note.—Gray boxes indicate genes that do not participate in last two steps or are not part of the purine biosynthetic pathway.
The comparative genomic analysis shows that there are taxonomical patterns at the genus, family and class levels for the combinations purP, purO, purV, and purJ, which are able to replace purH in Archaea and Bacteria. Thus, the presence of these genes is typical of specific higher taxa of prokaryotes. Some patterns are maintained at the genus, family, class, and at the phylum level (table 3).
Table 3.
Taxonomic Patterns of Ocurrence of purPs, purO, purV, and purJ
| Taxonomic Classification | Taxonomic Patterns | |||
|---|---|---|---|---|
| Archaea | Crenarchaeota | Family Thermoproteaceae | [12] | purP II/purP III |
| Euryarchaeota | Class Halobacteria | [25] | purV-N/purO | |
| Family Methanosaetaceae | [03] | purV/purO/purP II/purP III | ||
| Genus Archaeoglobus | [04] | purJ/purP II/purP III | ||
| Bacteria | Bacteriodetes | Family Prevotellaceae | [06] | purV/purJ |
| Thermotogae | Family Fervidobacteriaceae | [04] | purV/purJ | |
| Thermodesulfobacteria | Phylum Thermodesulfobacteria | [02] | purV/purJ | |
| Proteobacteria | Order Desulfovibrionales | [09] | purV/purJ | |
| Order Desulfobacterales | [08] | purV/purJ | ||
Note.—All OTUs of these taxa, including classes, families, and genera included harbor the gene shown. The numbers between brackets are the amount of OTUs in each group.
The evolutionary history of purH, purV, and purJ is intimately related and these results will be presented elsewhere, whereas the evolution of purO and PurP will be presented in this publication.
Evolution of PurO
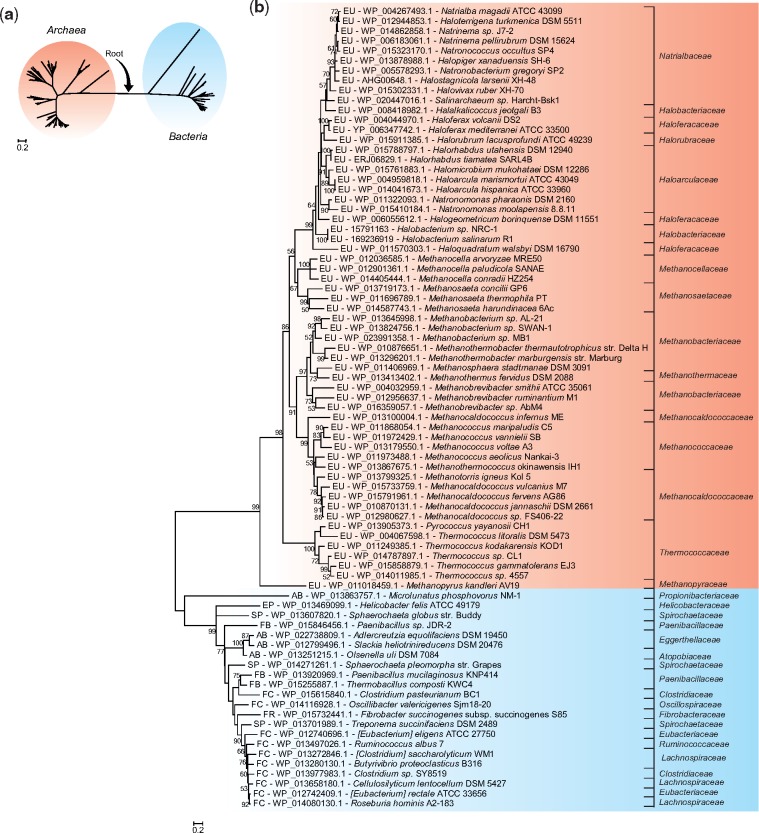
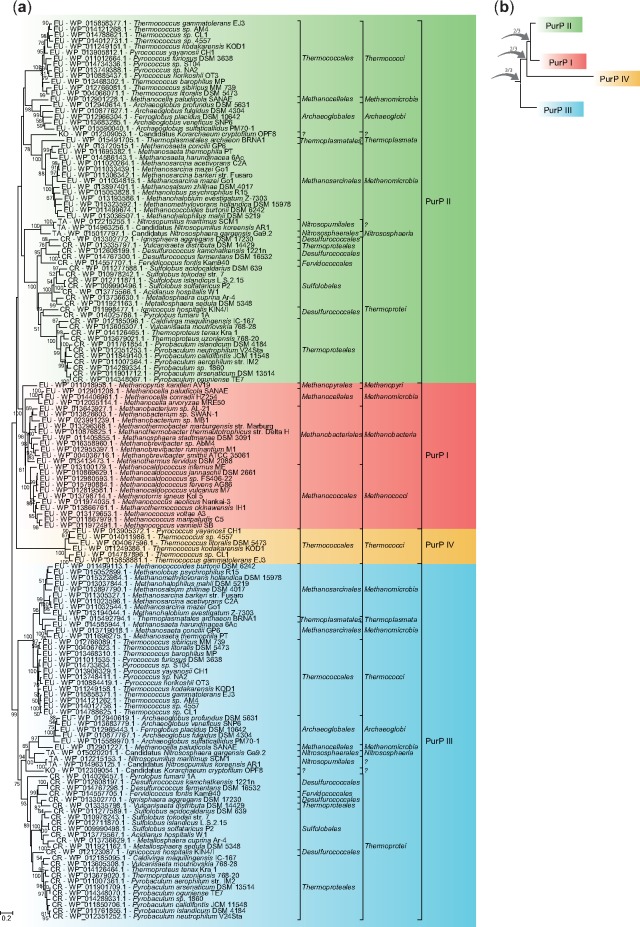
The maximum likelihood (ML) tree of PurO shows two distinct groups, one containing PurOs from OTUs of the domain Archaea and the other one with PurOs of the domain Bacteria (fig. 3a). This topology was obtained both with sequences of amino acids and nucleotides (supplementary fig. S1, Supplementary Material online) and is congruent with the current prokaryotic phylogeny (Woese and Fox 1977) and therefore the root of this tree was placed in the branch that connects these two groups. The topology of the rooted tree of archaeal PurOs agrees with the taxonomy of this domain, at least at the family level (fig. 3b).
Fig. 3.
—Phylogenetic relationships among archaeal PurOs and their bacterial counterparts. (a) Maximum likelihood tree (ML) of PurOs showing the root placement at the branch that connects PurOs from Archaea and Bacteria. (b) Rooted ML tree showing PurOs from Archaea and their homologs in Bacteria in distinct groups. The multiple alignment utilized in these reconstructions contained 81 sequences of amino acids with 134 sites. The tree was constructed with the model LG+G + I and bootstrap was performed with 1,000 resamplings. The OTUs are identified by abbreviations that represent their phylum and class, accession number of the protein and species name. The abbreviations are: EU, phylum Euryarchaeota; FC, phylum Firmicutes class Clostridia; FB, phylum Firmicutes class Bacillales; AB, phylum Actinobacteria; SP, phylum Spirochaetes; FR, phylum Fibrobacteres; EP, phylum Proteobacteria classe Epsilonproteobacteria. The scale indicates the number of substitutions per site.
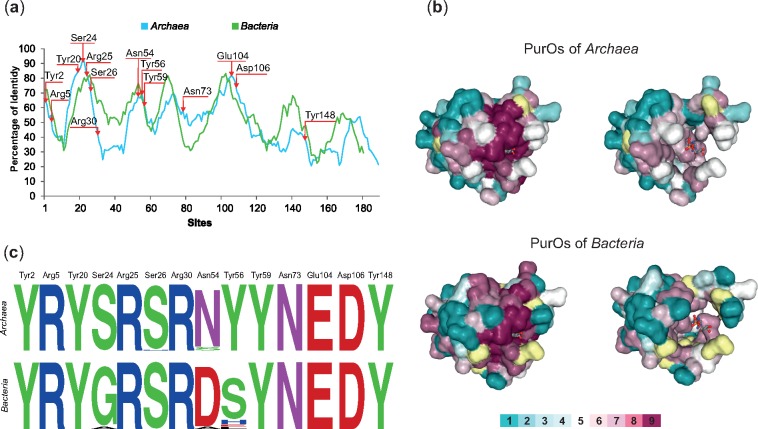
The average divergence of purO homologs in Archaea and Bacteria is 74%, indicating that they are highly divergent. However, the results of the genomic analyses previously shown (fig. 2; tables 1 and 2) suggest that the bacterial homologs of purOs are analogs of their archaeal counterparts. To test whether the archaeal and bacterial PurOs are indeed analogs, additional analyses were performed. The PurO from Methanothermobacter thermoautotrophicus was functionally characterized previously. This enzyme possesses a tetrameric tertiary structure (PDB ID code 2NTL) and its active site comprises 14 amino acid residues, all of them in the same monomer (Kang et al. 2007). The sliding window plot analysis showed that the conservation in the primary structure of the archaeal PurOs and their bacterial counterparts is similar (fig. 4a). The amino acids of the active site of PurO are distributed in the first 3/4 of the primary structure and concentrated in the N-terminal (fig. 4a).
Fig. 4.
—Conservation of PurOs from Archaea and Bacteria. (a) Sliding window plot analysis of the multiple alignments of PurOs in Archaea and Bacteria show that the conservation in the primary structure is similar in these domains. The arrows indicate the positions of the active sites of PurO from Methanothermobacter thermoautotrophicus. (b) Amino acids residues of the 3D structure of PurO from M. thermoautotrophicus complexed with AICAR, its substrate (PDB ID code 2NTL) as visualized in ConSurf. The tertiary structure is presented using a surface-filled model and the AICAR in a ball-and-stick model. The amino acids residues are colored according to their ConSurf conservation scores calculated on the basis of multiple alignments of archaeal and bacterial PurOs. The color-coding bar varies from 1 to 9, where 9 is the most conserved and 1 most variable. Amino acid positions with low confidence are marked in yellow. Tertiary structures on the left show amino acids with scores 1 to 9 and the ones on the right show amino acids with scores 1 to 7. The figure reveals that most highly conserved amino acids (scores 8 and 9) compose or surround the active site of the enzyme. (c) Sequence logos of the positions in the multiple alignment that correspond to the active sites of PurO from M. thermoautotrophicus showing that most amino acids are conserved in Archaea and Bacteria.
The conservation score for each amino acid of PurO from M. thermoautotrophicus (mthPurO) was calculated with ConSurf on the basis of their homologs in multiple alignments of both archaeal and bacterial PurOs (fig. 4b). The results showed that 53 amino acid residues of the mthPurO (26%) are highly conserved in archaeal PurOs (scores 8 and 9) and 35 of these amino acid residues also have ConSurf scores 8 and 9 in bacterial PurOs (supplementary table S3, Supplementary Material online). Most of these highly conserved amino acid residues compose or surround the active site of the enzyme in the tertiary structure (fig. 4b). All amino acids of the PurO active site possess conservation score 9, except for ser24, asn54, and tyr56, that had scores 7 or 8 based on the bacterial multiple alignment.
The logos constructed with the 14 positions of the active site in multiple alignments of purOs from Archaea and Bacteria showed that ten of these amino acids are identical (fig. 4c). The only exceptions are Ser24, Ser26, Asn54, and Tyr56. The Ser24 is 100% conserved in archaeal PurOs and varies in bacterial PurOs, whereas Ser26 is 100% conserved in bacterial PurOs and varies in archaeal PurOs (fig. 4c). The Asn54 is the only amino acid position that varies in both archaeal and bacterial PurOs (fig. 4c). Most of the archaeal PurOs analyzed possess asparagine in this position and from the four that possess serine, three were recovered from OTUs in the Family Methanocellaceae, suggesting that this variation is characteristic of PurOs from this family. The Tyr56 is 100% conserved in archaeal PurOs, but it is replaced mainly by serine in bacterial PurOs, however, glutamic acid, histidine, and leucine were also present at this position (fig. 4c). In general, most of the variations described earlier are chemically equivalent in PurOs of both Archaea and Bacteria. These results are congruent with the hypothesis that purOs in Archaea and Bacteria are analogous.
Evolution of PurP
The diversity of PurPs was investigated in previous studies. Zhang, White, et al. (2008) analyzed PurPs from 22 archaeal species and proposed the existence of three groups: PurP I, PurP II, and PurP III, but they did not explore the evolutionary relationships among them. Brown et al. (2011) analyzed 76 PurPs from archaeal genomes and found similar groups, but proposed the clusters Ia/Ib and II (named by Zhang, White, et al. 2008 as groups I, II, and III, respectively). They hypothesized that clusters Ia/Ib and II originated in an ancient gene duplication event that occurred before the divergence of the Archaea taxa analyzed. Brown et al. (2011) also reported that the cluster Ib (PurP II by Zhang, White, et al. 2008) was monophyletic in some analyses, but in others the cluster Ia fell inside cluster Ib. Besides that, it was not clear if the highly divergent PurP from Thermococcus species was a distinct isoform of PurP or if it was acquired by a lateral gene transfer event from a Crenarchaeota. In this study, we analyzed 154 PurPs from archaeal genomes, including higher taxa not sampled in previous studies and revisited their phylogenetic relationships.
The number of copies of purP in OTUs of the domain Archaea that contain this gene varied from one to three. The ML phylogenetic tree of PurP showed four distinct groups (fig. 5a) that were enumerated according to Zhang, White, et al. (2008) as PurP I, II, and III. The group PurP IV is being reported in this study. The groups I, II, and III were also recovered by Brown et al. (2011) but the internal topologies were distinct from the groups presented in our study. The indels in the PurP alignment (fig. 5b) were used to root the phylogenetic tree (fig. 5a) because indels are rarely fixed and are highly conserved evolutionary events that do not revert easily as compared with nucleotide substitutions (Rokas and Holland 2000). Therefore, genes or proteins that share indels are considered phylogenetically more related (Chan et al. 2007). Indels are used as molecular markers in studies as diverse as protein evolution and taxonomy of microorganisms (Rokas and Holland 2000; Chan et al. 2007; Ajawatanawong and Baldauf 2013; Naushad et al. 2014). The PurPs I, II, and IV share two indels (fig. 5b), indicating that they are phylogenetically more related with each other than with PurP III. These indels were utilized as a nonarbitrary criterion to root the phylogenetic tree of PurP in the branch that connects PurP III with the other groups (fig. 6). The topology of the rooted tree indicates that PurPs I and II are themselves more related than they are with PurP IV (fig. 6a).
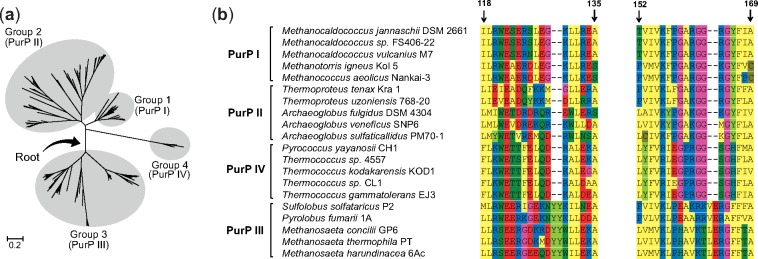
Fig. 5.
—Groups of PurP defined by phylogenetic analysis and detail of the multiple alignment. (a) Unrooted maximum likelihood tree of PurP constructed with 154 amino acid sequences containing 399 sites, the model LG+G + I and bootstrap with 1,000 resamplings. Root placement is indicated. (b) Indels shared by PurPs I, II and IV that were used to root the phylogenetic tree of PurP shown in (a). Five PurPs of each group are represented in the alignment.
Fig. 6.
—Phylogenetic trees of the PurPs and the proposed events of duplication that gave rise to the different groups. (a) Phylogenetic tree rooted on the branch that connects PurP III with the other PurPs. This tree is shown unrooted in figure 5a. (b) Proposed evolutionary history of PurPs. Arrows indicate duplication events and the numbers on the arrows indicate the number of times the branch is recovered in phylogenetic analyses performed with different settings (supplementary fig. S2, Supplementary Material online). The abbreviations indicate the phylum of each sequence: EU, phylum Euryarchaeota; CR, phylum Crenarchaeota; TA, phylum Thaumarchaeota; KO, phylum Korarchaeota. The scale indicates the number of substitutions per site.
Group I includes PurPs I from the methanogenic archaea Class I, Orders Methanobacteriales, Methanococcales, and Methanopyrales, and PurPs from the Order Methanocellales, which are methanogenic archaea Class II (fig. 6a). Methanogenic Archaea Class I and II are not phylogenetically related (Petitjean et al. 2015). The topology of this group is similar to the one obtained by Brown et al. (2011) and is congruent with the archaeal phylogeny, except for the PurP I from Methanocellales, grouped as a sister of the subgroup containing PurPs I from Methanobacteriales and Methanococcales, both methanogenic archaea Class I. It indicates that the Methanocellales, that are methanogenic archaea Class II, acquired its PurP I in an event of horizontal gene transfer from a methanogenic archaea Class I. All OTUs from these classes contain only one copy of purP, the only exception is Methanocella paludicola SANAE, with three copies of the gene, but only one copy is classified in group I (supplementary table S4, Supplementary Material online). Methanogenic archaea Class I are phylogenetically related to the Class Thermococci, however, the PurPs from all methanogenic archaea Class I analyzed fell into the group PurP I, which is not phylogenetically related to any PurP of Thermococci. Thus, the relationship of PurPs I with the other PurPs is incongruent with the archaeal phylogeny (Petitjean et al. 2015; Hug et al. 2016). The group PurP I includes the PurP from Methanocaldococcus jannaschii, the only PurP that had its FAICAR synthase activity experimentally characterized (Zhang, White, et al. 2008).
The group II encompasses the PurPs II and PurPs III from the same 61 OTUs of the phyla Crenarchaeota, Euryarchaeota, Thaumarchaeota, and Korarchaeota (supplementary table S4, Supplementary Material online). The composition of these groups is similar to what was reported by Zhang, White, et al. (2008) and Brown et al. (2011) but the topologies of these groups are not. The group PurP II is composed of 62 proteins, one in each OTU, except for Methanosarcina mazei Go1 that harbors two PurPs II. The group PurP III is composed of 61 proteins, one copy in each OTU. In some OTUs, the genes that encode the PurP II and III are in genomic context with other pur genes, what suggests that they are functionally related with the PBP (supplementary table S5, Supplementary Material online). The group PurP II possesses two subgroups with several cases of horizontal gene transfer. These inferences were made because the groups of PurPs formed are not congruent with the archaeal phylogeny (Koonin 2015; Petitjean et al. 2015; Hug et al. 2016). For example: 1) PurPs from Euryarchaeota are together with one PurP from an OTU belonging in the phylum Korarchaeota, indicating that it was acquired by a horizontal gene transfer event from an Euryarchaeota (fig. 6a); 2) the PurP II from a Methanocellales is more related to PurPs II from Archaeoglobales than to PurPs II from Methanosarcinales, while phylogenetically, Archaeoglobales are more distantly related than the other two orders; 3) PurPs II from Thermoplasmatales are more related to PurPs II from Methanosarcinales, while Archaeoglobales are known to be phyllogenetically closer to Thermoplasmatales; 4) The second subgroup contained PurPs II from Thaumarchaeota and Crenarchaeota, where Thermoproteales formed a sister subgroup with the Desulfurococcales, whereas it is known from archaeal phylogenies that Desulfurococcales is closer to Sulfobolales than to Thermoproteales; 5) the PurPs II from Desulfurococcales do not form a monophyletic group (fig 6a). Similar phylogenetic incongruences were found among the PurPs III (fig. 6a).
The group PurP IV is composed of PurP homologs encoded in the genome of six out of 13 OTUs analyzed in the family Thermococcaceae (supplementary table S4, Supplementary Material online). These six OTUs also harbor PurPs II and III (supplementary table S4, Supplementary Material online). The PurPs IV are highly divergent from the others PurPs but highly similar among them, with identities varying from 70% to 87%. In our analyses, the PurP IV always formed a distinct group, in contrast to what was reported by Brown et al. (2011), where the two representatives of these PurPs emerged inside PurP II as a basal group in Crenarchaeota (cluster Ib by Brown et al. 2011). The genes coding PurPs IV are in the same genomic context with other pur genes (supplementary table S5, Supplementary Material online). The PurPs IV are being described for the first time in this study as a novel putative isoform of PurP (supplementary table S4, Supplementary Material online).
The internal topology of the groups I, II, and III of the PurP tree is generally not congruent with the phylogenetic relationships among orders, classes, and phyla of the Archaea, with few exceptions. PurPs encoded in the same genomes fall in different groups. For instance, the groups PurPs II and III composed by PurPs from de same OTUs, representing four archaeal phyla, and the three PurPs from M. paludicola SANAE fall in groups I, II, and III. Finally, the overall topology of the PurP tree is incongruent with the archaeal phylogeny, but congruent with the hypothesis that the main PurP groups are composed by paralogs that originated in gene duplication events. Thus, based on the finding presented earlier, we propose that groups I, II, III, and IV are paralogs that originated in three events of gene duplication that occurred in the ancestral of the Archaea domain (fig. 6b).
The first duplication event, also proposed by Brown et al. (2011), originated the PurP III and the ancestral of PurPs IV, I, and II. Here, we proposed two additional subsequent duplications events that originated the PurPs IV, I, and II. These isoforms of PurP were selectively maintained or lost during the taxonomic diversification of the domain Archaea. The internal nodes representing the duplication events that originated the paralogs I, II, and IV do not have enough bootstrap support in the protein ML tree (fig. 6a) similar to what was found by Brown et al. (2011). Therefore, we constructed other ML trees with the multiple alignment of nucleotides containing only unambiguous sites as determined in the Guidance server (Penn et al. 2010). The alternative ML trees were inferred with different nucleotide positions (first and second or all positions of the codon) and with the amino acid positions of the protein multiple alignment. Only the ML tree obtained with the protein multiple alignment yielded an alternative evolutionary history (supplementary fig. S2, Supplementary Material online) that was not similar to the tree proposed for the PurPs in this study (fig. 6b).
Discussion
The PBP is ancient and its derivatives are involved in the synthesis of nucleic acid precursors, carbohydrate metabolism, and in several cellular signaling pathways. Information on the diversity of the genes encoding the enzymes of the PBP is important to many biotechnological applications, such as the development of anticancer and antimicrobial drugs. In this study, we expand the scientific knowledge on the diversity in the last two steps of the PBP in prokaryotic lineages. To accomplish this, an extensive genome analysis was performed with 1,403 completely sequenced and annotated prokaryotic genomes. Comparative genomics and genomic context analyses showed that the diversity of PBP in the domain Bacteria is higher than previously reported. For example, the genes purV, purJ, and purO, initially reported only from Archaea, were found in this study to be relatively common in Bacteria. The gene purO was until now, considered a signature of the domain Archaea.
The occurrence of genes coding for PurH in Bacteria and Archaea was previously reported (Brown et al. 2011; Armenta-Medina et al. 2014). However, our results showed that contrary to what occurs in the domain Archaea, the enzyme PurH seems to be the preferred evolutionary alternative selected by most bacterial phyla to catalyze the last two steps in the purine biosynthesis pathway. Genes encoding PurH were found in 17% of the archaeal genomes analyzed and in 85% of the bacterial genomes (fig. 2). This difference may be due to the fact that the domain AICARFT from the PurHs uses tetrahydrofolate (THF) as a donor of formyl and in bacteria, in contrast to archaea, THF is preferentially used as the C1 donor (Maden 2000). A similar observation was made by Brown et al. (2011), when they reported that the few archaeal OTUs that could synthesize folates also possessed PurH or PurV encoded in their genomes. These included OTUs from the Class Halobacteria and from the Order Methanomicrobiales. On the other hand, Archaea that do not possess PurH nor PurV do not synthesize THF (White 1988, 1997; Choquet et al. 1994).
The genomic analyses performed in this study, as well as the taxonomical patterns of gene occurrence indicate that the combinations purV/purO, purV/purJ are able to replace purH in members of the domain Bacteria that lack this gene, similar to what was proposed for Archaea (Brown et al. 2011). Genes that replace purH occurred in 73% of the archaeal genomes and in 5% of the bacterial genomes included in our analyses (fig. 2). Approximately 37% of the purO, purV, and purJ were in genomic context with other genes of the PBP in both bacterial and archaeal genomes and 63% were not in genomic context (table 1). The fact that part of the genes coding for enzymes in the PBP are in genomic context is expected because genes functionally related tend to be in the same operon in prokaryotic genomes (Korbel et al. 2004). The genomic context is commonly used in the prediction of the functional interactions among genes (Huynen et al. 2000).
No homologs of genes that code for PurH, PurO, PurV, and PurJ were found in genomes studied of OTUs in the phyla Crenarchaeota, Korarchaeota, and Thaumarchaeota, but genes that code for homologs of PurP were found and they may be involved in the PBP, as will be discussed below. A total of 132 genomes did not harbor any of the genes coding for the last two steps of the PBP nor their homologs. These OTUs, 14 Archaea and 118 Bacteria, representing ∼10% of each domain are not able to synthesize purines de novo. All these 132 OTUs that lacked the genes coding for the last two steps of the PBP also lacked the other genes of the PBP, with the exception of 54 OTUs that possessed one or a few genes of the pathway, from which purB or purC were the most frequently encountered (supplementary table S2, Supplementary Material online). Parasites, such as many members of the family Rickettsiaceae, Helicobacter pylori, Borrelia burgdorferi, and symbionts such as Nanoarchaeum equitans and Serratia symbiotica acquire purines by recycling pre-existing purines found in the growth substrate through one the salvage purine pathways (Waters et al. 2003; Jewett et al. 2009; Liechti and Goldberg 2012). However, approximately one-third (43 OTUs) of these 132 OTUs lacked the four salvage pathways described in prokaryotes, indicating that they acquire purines from their hosts. Most of these OTUs are obligatory endosymbionts or intracellular parasites (supplementary table S2, Supplementary Material online).
According to Xu et al. (2007) the fusion of the genes that code for the domains AICARFT and IMPCH originating the PurH was favored during evolution once FAICAR, product of the AICARFT domain of PurH, is spontaneously converted into its precursor, AICAR. Therefore, the origin of PurH contributed to accelerate the conversion of FAICAR into IMP by the domain IMPCH. The hypothesis proposed by Xu et al. (2007) for the origin of PurH, implies that the domains AICARFT and IMPCH coded by two distinct genes was the ancestral condition of the PBP and therefore, organisms with the PurH in their genomes would have an adaptive advantage. Zhang, Morar, et al. (2008) speculated that although there is no tunnel or channel between the domains AICARFT and IMPCH of PurH, there is a predominance of positive charges between them, what favors the transfer of FAICAR from the domain AICARFT to the domain IMPCH, corroborating the suggestions made by Xu et al. (2007) for the origin of PurH. However, our results show that purV and purJ are functionally involved in the PBP in bacteria, like was proposed by Brown et al. (2011) for Archaea. The domains AICARFT and IMPCH of the human PurH produce peptides able to catalyze their respective enzymatic reactions (Rayl et al. 1996). These results indicate that AICARFT and IMPCH do not need to be fused to perform their catalytic reactions. Therefore, the fact that the domains AICARFT and IMPCH are coded by distinct genes in many living OTUs of the domains Archaea and Bacteria does not implicate that these two genes cannot catalyze the last two steps of the PBP. The experimental characterization of the peptides encoded by these genes will certainly bring more information on their catalytic mechanisms and evolution.
The molecular phylogeny of the PurOs indicates that the ancestral form of this enzyme was present in the common ancestor of the domains Archaea and Bacteria. An alternative hypothesis to the origin of bacterial PurOs would be the lateral transfer of an archaeal PurO to an ancient bacterium. But, in this case, the bacterial PurOs would have emerged within the group of archaeal PurOs in the phylogenetic tree, which was not the case. Based on these considerations, we inferred that the ancestral PurO was present in the common ancestor of Archaea and Bacteria. The absence of purO in the majority of the analyzed bacterial genomes would be the result of losses of this gene during species diversification in this domain. Taking these results into consideration, purO can no longer be considered a signature of the domain Archaea, as previously suggested (Graupner et al. 2002; Ownby et al. 2005; Zhang, Morar, et al. 2008; Zhang, White, et al. 2008; Armenta-Medina et al. 2014).
The maintenance of basic biochemical functions in homologous protein domains that experienced drastic structural divergence is a recently described phenomenon (Zhang et al. 2014). Therefore, the divergence at the primary structure level observed among bacterial and archaeal PurOs does not necessarily imply in functional differences even when the divergences result in changes in the tertiary structure. This phenomenon was also observed with PurOs from Bacteria and Archaea, where the amino acids from the catalytic site as well as the ones surrounding it are highly conserved (fig. 4b) as opposed to the majority of the other amino acids that are highly variable (supplementary table S3, Supplementary Material online). The variation in the conservation of the primary structure of the PurO of Archaea and Bacteria is coincident (fig 4a), indicating that the selection pressure was similar on these proteins after the divergence of the domains Archaea and Bacteria. In addition, the conservation of the amino acid residues of the active site is not related to the conservation of the region of the primary structure in which they are located, even at positions where amino acids residues vary (fig. 4a). It suggests that they were conserved in the PurOs of archaeas and their bacterial counterparts despite the evolutionary divergence that resulted from speciation events. Further evidences that support the hypothesis that bacterial and archaeal PurOs are analogous include the anticorrelation between the occurrence of the genes purO/purV and purH (fig. 2) and the fact that part of the bacterial homologs of purO are in genomic context with purV and other genes of the PBP (table 1).
The PurPs are only present in archaeal genomes and the phylogenetic tree of these enzymes shows a division in four groups. Due to the different evolutionary histories recovered in the ML tree constructed with a protein multiple alignment containing only amino acid positions with high levels of reliability, the relationships among these PurP paralogs proposed here must be viewed with caution. Perhaps, phylogenetic analyses including a greater number of PurP sequences or other methodological approaches, such as the use of complex networks, can help to solve this problem (Andrade 2011; Carvalho et al. 2015).
The collective analyses of PurPs in archaeal genomes indicate that PurP IV is a new isoform, distinct from the ones previously described in the PBP (Zhang, White, et al. 2008; Brown et al. 2011). The tertiary structure of the PurP IV from Thermococcus kodakarensis was resolved, however, it was not enzymatically characterized (Zhang, White, et al. 2008; Brown et al. 2011).
The incongruences between the phylogeny inferred with the PurPs in this study and the phylogeny of the domain Archaea may be the result of both the absence of PurPs in the genome of some OTUs (e.g., in the Class Halobacteria; supplementary text) or events of horizontal gene transfer (e.g., transference of PurP I among unrelated methanogenic Archaea—fig. 6).
There are evidences suggesting that the PurPs II, III, and IV are functionally linked to the PBP. For example, most Archaea included in this study that harbor PurPs II and III or II, III, and IV do not contain the PurP I or analogous enzymes such as PurH or PurV encoded in their genomes (supplementary table S1, Supplementary Material online). In general, these archaeal OTUs are free-living (supplementary table S4, Supplementary Material online) and the genes encoding PurPs II, III, and IV of several distantly related Archaea are in genomic context or in putative operons with other genes of the PBP (supplementary table S5, Supplementary Material online). These OTUs are widespread in the phyla Crenarchaeota, Euryarchaeota, Thaumarchaeota, and Korarchaeota and represent more than one-third of the Archaea analyzed in this study. In some cases, the conservation of the genomic context of the purPs extends to OTUs of different genera, such as the purPs from Desulfurococcus kamchatkensis 1221n and Ignisphaera aggregans DSM 17230 or to all OTUs of one order, such as in Sulfolobales (supplementary table S5, Supplementary Material online). These are ecological and genomic evidences of the involvement of the PurPs II, III, and IV in the PBP.
The PurP I of Methanocaldococcus jannaschii was shown to have FAICAR synthase activity, which is the ninth step of the PBP (Ownby et al. 2005). In contrast, neither Pyrococcus furiosus PurP II and PurP III have showed any detectable FAICAR synthase activity (Zhang, White, et al. 2008). However, the tertiary and quaternary structures of PurP I of M. jannaschii and PurP II of P. furiosus revealed that their active sites were highly conserved (Zhang, White, et al. 2008). Additionally, the PurP II of P. furiosus binds to both ATP and AICAR (Zhang, White, et al. 2008). Based on these findings, Zhang, White, et al. (2008) speculated that the PurP II of P. furiosus could utilize an alternative source of formyl to catalyze the conversion of AICAR to FAICAR while the PurP III would have a distinct catalytic activity or no catalytic function once it is highly divergent from PurP I and II.
The PurP I of M. jannaschii and PurP II of P. furiosus have an hexameric quaternary structure formed by the interaction between two trimers. However, PurP I has a more compact structure, with ∼2.5× more buried surface area between the two trimers than PurP II (Zhang, White, et al. 2008). This weaker interaction between the trimers of PurP II of P. furiosus was attributed to a possible crystallization artifact rather than biologically relevant (Zhang, White, et al. 2008). However, every Archaea analyzed in this study that contain a PurP II also has a PurP III, except for some OTUs of the class Thermococci, which harbor the PurPs II, III, and IV. Therefore, it is possible that this weaker interaction between the trimers of PurP II of P. furiosus is because in its biologically active form, the PurP II and III form heterohexamers composed of trimers with the same isoform. Therefore, we speculate that in this arrangement the PurPs II and III would be able to catalyze the ninth reaction of the PBP, the conversion of AICAR into FAICAR.
The results of this study contribute to a better understanding of the diversity of the PBP in prokaryotes. The taxonomic patterns of occurrence of the genes of the last two steps of the PBP are molecular signatures of certain groups of prokaryotes that could be used as markers in genetic diversity or on functional metagenomic studies. The genes purV, purJ, and purO, previously reported only in the domain Archaea were also found in Bacteria. In light of these results, purO cannot be considered a signature of the domain Archaea, as previously reported. Bacterial PurOs were inferred to have catalytic activity due to the conservation of amino acids in its active site and to participate in the ninth step of the PBP. The findings reported here on the conservation of the active sites of PurOs in Bacteria and Archaea could be used as a guide to select positions to engineer this protein either to gain more insight in its mode of action or modify its activity by mutations as reported for glycerol dehydratase (Maddock et al. 2017).
Some bacterial OTUs containing purV, purJ, and purO are probiotics or biomarkers for human or animal pathologies, such as Roseburia spp., Helicobacter felis, and Prevotella spp. (Fritz et al. 2006; Larsen 2017; Tamanai-Shacoori et al. 2017). Thus, these genes may be used as markers to identify the OTUs or, alternatively, may be used in experiments of molecular docking to develop drugs that specifically inhibit pathogenic species containing these proteins (Meng et al. 2011). The experimental characterization of the activity of the different isoforms of PurP from the Archaea domain ought to be pursued in future studies.
According to the hypothesis of Horowitz (1945), enzymes of the last stages of biosynthetic pathways were the first to be recruited during their origin and Woese (1998) proposed that the Cenancestor was a diverse community of cells that survived and evolved as a biological unit rather than a discrete entity. The last two steps of the PBP show the highest variation when compared with the other steps of this pathway and indeed, this variation is found in both Archaea and Bacteria. This diversity observed in the first recruited enzymes probably resulted from the functional redundancy in ancient times, in accordance with Woese’s proposition. Therefore, our results combined with the propositions presented earlier, are congruent with the hypothesis that the last steps of PBP evolved in the Cenancestor.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors acknowledge the financial support provided by the Brazilian organizations Fundação de Amparo à Pesquisa do Estado da Bahia (FAPESB), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Literature Cited
- Aiba A, Mizobuchi K.. 1989. Nucleotide sequence analysis of genes purH and purD involved in the de novo purine nucleotide biosynthesis of Escherichia coli. J Biol Chem. 26435:21239–21246. [PubMed] [Google Scholar]
- Ajawatanawong P, Baldauf SL.. 2013. Evolution of protein indels in plants, animals and fungi. BMC Evol Biol. 13:140.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ.. 1990. Basic local alignment search tool. J Mol Biol. 2153:403–410. [DOI] [PubMed] [Google Scholar]
- Andrade RFS. 2011. Detecting network communities: an application to phylogenetic analysis. PLoS Comput Biol. 75:e1001131.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armenta-Medina D, Segovia L, Perez-Rueda E.. 2014. Comparative genomics of nucleotide metabolism: a tour to the past of the three cellular domains of life. BMC Genomics 15:800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazy H, et al. 2016. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules Nucleic Acids Res. 44:344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AM, Hoopes SL, White RH, Sarisky CA.. 2011. Purine biosynthesis in archaea: variations on a theme. Biol Direct. 6:63.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan JM, Hartman SC.. 1959. Enzymatic reactions in the synthesis of purines. Adv Enzymol. 21:199–261. [Google Scholar]
- Caetano-Anollés G, Kim SK, Mittenthal JE.. 2007. The origin of modern metabolic networks inferred from phylogenomic analysis of protein architecture. Proc Natl Acad Sci U S A. 104:9358–9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho DS, et al. 2015. What are the evolutionary origins of mitochondria? A complex network approach. PLoS One 109:e0134988.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SK, Hsing M, Hormozdiari F, Cherkasov A.. 2007. Relationship between insertion/deletion (indel) frequency of proteins and essentiality. BMC Bioinformatics 28:227.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng YS, et al. 1990. Glycinamide ribonucleotide synthetase from Escherichia coli: cloning, overproduction, sequencing, isolation, and characterization. Biochemistry 291:218–227. [DOI] [PubMed] [Google Scholar]
- Chopra AK, Peterson JW, Prasad R.. 1991. Nucleotide sequence analysis of purH and purD genes from Salmonella typhimurium. Biochim Biophys Acta. 10903:351–354. [DOI] [PubMed] [Google Scholar]
- Choquet CG, Richards JC, Patel GB, Sprott GD.. 1994. Purine and pyrimidine biosynthesis in methanogenic bactéria. Arch Microbiol. 1616:471–480. [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE.. 2004. WebLogo: a sequence logo generator. Genome Res. 146:1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euzéby JP. 1997. List of bacterial names with standing in nomenclature: a folder available on the Internet. Int J Syst Bacteriol. 472:590–592. [DOI] [PubMed] [Google Scholar]
- Fritz EL, Slavik T, Delport W, Olivier B, van der Merwe SW.. 2006. Incidence of Helicobacter felis and the effect of coinfection with Helicobacter pylori on the gastric mucosa in the African population. J Clin Microbiol. 445:1692–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gots JS, Dalal FR, Shumas SR.. 1969. Genetic separation of the inosinic acid cyclohydrolase-transformylase complex of Salmonella typhimurium. J Bacteriol. 99:441–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graupner M, Xu H, White RH.. 2002. New class of IMP cyclohydrolase in Methanococcus jannaschii. J Bacteriol. 1845:1471–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu ZM, Martindale DW, Lee BH.. 1992. Isolation and complete sequence of the purL gene encoding FGAM synthase II in Lactobacillus casei. Gene 1191:123–126. [DOI] [PubMed] [Google Scholar]
- Han MV, Zmasek CM.. 2009. phyloXML: xML for evolutionary biology and comparative genomics. BMC Bioinformatics 10:356.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Smith JM, Zalkin H.. 1992. Escherichia coli purB gene: cloning, nucleotide sequence, and regulation by purR. J Bacteriol. 1741:130–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz NH. 1945. On the evolution of biochemical syntheses. Proc Natl Acad Sci U S A. 316:153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Anand R, Ealick SE, Stubbe J.. 2004. The formylglycinamide ribonucleotide amidotransferase complex from Bacillus subtilis: metabolite-mediated complex formation. Biochemistry 4332:10314–10327. [DOI] [PubMed] [Google Scholar]
- Hug LA, et al. 2016. A new view of the tree of life. Nat Microbiol. 15:16048.. [DOI] [PubMed] [Google Scholar]
- Huynen M, Snel B, Lathe W, Bork P.. 2000. Predicting protein function by genomic context: quantitative evaluation and qualitative inferences. Genome Res. 108:1204–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglese J, Johnson DL, Shiau A, Smith JM, Benkovic SJ.. 1990. Subcloning, characterization, and affinity labeling of Escherichia coli glycinamide ribonucleotide transformylase. Biochemistry 296:1436–1443. [DOI] [PubMed] [Google Scholar]
- Jewett MW, et al. 2009. GuaA and GuaB are essential for Borrelia burgdorferi survival in the tick-mouse infection cycle. J Bacteriol. 19120:6231–6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YN, Tran A, White RH, Ealick SE.. 2007. A novel function for the NTN hydrolase fold demonstrated by the structure of an archaeal inosine monophosphate cyclohydrolase. Biochemistry 4617:5050–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch DR, Whitney RR.. 1991. Pathogenicity of Candida albicans auxotrophic mutants in experimental infections. Infect Immun. 599:3297–3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2015. Archaeal ancestors of eukaryotes: not so elusive any more. BMC Biol. 13:84.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Jensen LJ, Mering CV, Bork P.. 2004. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 227:911–917. [DOI] [PubMed] [Google Scholar]
- Larsen JM. 2017. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 1514:363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti G, Goldberg JB.. 2012. Helicobacter pylori relies primarily on the purine salvage pathway for purine nucleotide biosynthesis. J Bacteriol. 1944:839–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. 2014. Modification in de novo purine pathway for adenosine accumulation by Bacillus subtilis. Wei Sheng Wu Xue Bao 546:641–647. [PubMed] [Google Scholar]
- Maddock DJ, Gerth ML, Patrick WM.. 2017. An engineered glycerol dehydratase with improved activity for the conversion of meso-2, 3-butanediol to butanone. Biotechnol J. 1212:1700480.. [DOI] [PubMed] [Google Scholar]
- Maden BE. 2000. Tetrahydrofolate and tetrahydromethanopterin compared: functionally distinct carriers in C1 metabolism. Biochem J. 15(350 Pt 3):609–629. [PMC free article] [PubMed] [Google Scholar]
- Marolewski A, Smith JM, Benkovic SJ.. 1994. Cloning and characterization of a new purine biosynthetic enzyme: a non-folate glycinamide ribonucleotide transformylase from E. coli. Biochemistry 339:2531–2537. [DOI] [PubMed] [Google Scholar]
- Meng XY, Zhang HX, Mezei M, Cui M.. 2011. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2:147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naushad HS, Lee B, Gupta RS.. 2014. Conserved signature indels and signature proteins as novel tools for understanding microbial phylogeny and systematics: identification of molecular signatures that are specific for the phytopathogenic genera Dickeya, Pectobacterium and Brenneria. Int J Syst Evol Microbiol. 64(Pt 2):366–383. [DOI] [PubMed] [Google Scholar]
- Ni L, Guan K, Zalkin H, Dixon JE.. 1991. De novo purine nucleotide biosynthesis: cloning, sequencing and expression of a chicken PurH cDNA encoding 5-aminoimidazole-4-carboxamide-ribonucleotide transformylase-IMP cyclohydrolase. Gene 1062:197–205. [DOI] [PubMed] [Google Scholar]
- Nilsson D, Kilstrup M.. 1998. Cloning and expression of the Lactococcus lactis purDEK genes, required for growth in milk. Appl Environ Microbiol. 6411:4321–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ownby K, Xu H, White RH.. 2005. A Methanocaldococcus jannaschii archaeal signature gene encodes for a 5-Formaminoimidazole-4-carboxamide-1–D-ribofuranosyl 5-Monophosphate synthetase: a new enzyme in purine biosynthesis. J Biol Chem. 28012:10881–10887. [DOI] [PubMed] [Google Scholar]
- Parker J. 1984. Identification of the purC gene product of Escherichia coli. J Bacteriol. 1573:712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltonen T, Mäntsälä P.. 1999. Isolation and characterization of a purC(orf)QLF operon from Lactococcus [correction of Lactobacillus] lactis MG1614. Mol Gen Genet. 2611:31–41. [DOI] [PubMed] [Google Scholar]
- Penn O, et al. 2010. GUIDANCE: a web server for assessing alignment confidence scores. Nucleic Acids Res. 38(Web Server):W23–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean C, Deschamps P, López-García P, Moreira D, Brochier-Armanet C.. 2015. Extending the conserved phylogenetic core of Archaea disentangles the evolution of the third domain of life. Mol Biol Evol. 325:1242–1254. [DOI] [PubMed] [Google Scholar]
- Pride DT. 2000. Swaap: a tool for analyzing substitutions and similarity in multiple alignments. Available from:http://www.thepridelaboratory.org/software.html, last accessed March 14, 2019; distributed by the author.
- Rayl EA, Moroson BA, Beardsley GP.. 1996. The Human purH gene product, 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase: cloning, sequencing, expression, purification, kinetic analysis, and domain mapping. J Biol Chem. 2714:2225–2233. [DOI] [PubMed] [Google Scholar]
- Rokas A, Holland PW.. 2000. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 1511:454–459. [DOI] [PubMed] [Google Scholar]
- Sampei G-I, et al. 2010. Crystal structures of glycinamide ribonucleotide synthetase, PurD, from thermophilic eubacteria. J Biochem. 1484:429–438. [DOI] [PubMed] [Google Scholar]
- Schrimsher JL, Schendel FJ, Stubbe J, Smith JM.. 1986. Purification and characterization of aminoimidazole ribonucleotide synthetase from Escherichia coli. Biochemistry 2515:4366–4371. [DOI] [PubMed] [Google Scholar]
- Smith PMC, Atkins CA.. 2002. Purine biosynthesis: big in cell division, even bigger in nitrogen assimilation. Plant Physiol. 1283:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamanai-Shacoori Z, et al. 2017. Roseburia spp.: a marker of health? Future Microbiol. 12:157–170. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 3012:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe W, Sampei G, Aiba A, Mizobuchi K.. 1989. Identification and sequence analysis of Escherichia coli purE and purK genes encoding 5′-phosphoribosyl-5-amino-4-imidazole carboxylase for de novo purine biosynthesis. J Bacteriol. 1711:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters E, et al. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc Natl Acad Sci U S A. 10022:12984–11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RH. 1988. Analysis and characterization of the folates in the nonmethanogenic archaebacteria. J Bacteriol. 17010:4608–4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RH. 1997. Purine biosynthesis in the domain Archaea without folates or modified folates. J Bacteriol. 17910:3374–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Fox GE.. 1977. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 7411:5088–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. 1998. The universal ancestor. Proc Natl Acad Sci U S A. 9512:6854–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, et al. 2007. Structure-based design, synthesis, evaluation, and crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase. J Biol Chem. 28217:13033–13046. [DOI] [PubMed] [Google Scholar]
- Zalkin H. 1983. Structure, function, and regulation of amidophosphoribosyltransferase from prokaryotes. Adv Enzyme Regul. 21:225–237. [DOI] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, Burroughs AM, Aravind L.. 2014. Resilience of biochemical activity in protein domains in the face of structural divergence. Curr Opin Struct Biol. 26:92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Morar M, Ealick SE.. 2008. Structural biology of the purine biosynthetic pathway. Cell Mol Life Sci. 6523:3699–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, White RH, Ealick SE.. 2008. Crystal structure and function of 5-formaminoimidazole-4-carboxamide-1-β-d-ribofuranosyl 5′-monophosphate synthetase from Methanocaldococcus jannaschii. Biochemistry 471:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.