Abstract
Sex chromosomes play a central role in genetics of speciation and their turnover was suggested to promote divergence. In vertebrates, sex chromosome–autosome fusions resulting in neo-sex chromosomes occur frequently in male heterogametic taxa (XX/XY), but are rare in groups with female heterogamety (WZ/ZZ). We examined sex chromosomes of seven pests of the diverse lepidopteran superfamily Gelechioidea and confirmed the presence of neo-sex chromosomes in their karyotypes. Two synteny blocks, which correspond to autosomes 7 (LG7) and 27 (LG27) in the ancestral lepidopteran karyotype exemplified by the linkage map of Biston betularia (Geometridae), were identified as sex-linked in the tomato leafminer, Tuta absoluta (Gelechiidae). Testing for sex-linkage performed in other species revealed that while LG7 fused to sex chromosomes in a common ancestor of all Gelechioidea, the second fusion between the resulting neo-sex chromosome and the other autosome is confined to the tribe Gnoreschemini (Gelechiinae). Our data accentuate an emerging pattern of high incidence of neo-sex chromosomes in Lepidoptera, the largest clade with WZ/ZZ sex chromosome system, which suggest that the paucity of neo-sex chromosomes is not an intrinsic feature of female heterogamety. Furthermore, LG7 contains one of the major clusters of UDP-glucosyltransferases, which are involved in the detoxification of plant secondary metabolites. Sex chromosome evolution in Gelechioidea thus supports an earlier hypothesis postulating that lepidopteran sex chromosome–autosome fusions can be driven by selection for association of Z-linked preference or host-independent isolation genes with larval performance and thus can contribute to ecological specialization and speciation of moths.
Keywords: Coleophora, Depressaria, Hofmannophila, Opisina, Phthorimaea, Sitotroga
Introduction
Sex chromosomes represent intriguing portions of the genome which play an important role in many evolutionary processes including sexual and intragenomic conflict and speciation (Masly and Presgraves 2007; Mank et al. 2014). Indeed, the formation of postzygotic isolation can be characterized by two empirical rules, both involving sex chromosomes, inferred from analyses of hybrid fitness. The first of these known as the large-X effect refers to the disproportionately large effect of the X chromosome compared with autosomes in introgression analyses of hybrid incompatibilities (Masly and Presgraves 2007; Dufresnes et al. 2016). The second, Haldane’s rule, which has proved to be one of the most robust generalizations in evolutionary biology, states that when in the F1 offspring of two different animal races one sex is absent, rare, or sterile, that sex is the heterogametic sex (Haldane 1922; Delph and Demuth 2016).
It was shown that larger and more heteromorphic sex chromosomes were associated with faster evolution of postzygotic isolation (Turelli and Begun 1997; Lima 2014). Sex chromosome size can increase via sex chromosome–autosome fusions, which result in so-called neo-sex chromosomes. These have been suggested to promote divergence in fish (Kitano et al. 2009; Kitano and Peichel 2012), mammals (Graves 2016), and moths (Nguyen et al. 2013; Nguyen and Carabajal Paladino 2016), although little is known about their functional role in this process. Neo-sex chromosomes also provide insight into the evolution of animal sex chromosomes (Pala et al. 2012; Bachtrog 2013; Natri et al. 2013), which are much older than the sex chromosome systems examined in plants (Charlesworth 2015). To identify the evolutionary forces driving sex chromosome–autosome fusions, the occurrence of derived multiple sex chromosome systems was recently analyzed in vertebrates (Pokorná et al. 2014; Pennell et al. 2015). These analyses yielded a striking pattern of a higher incidence of fusions in male heterogametic (♀XX, ♂XY) than female heterogametic (♀WZ, ♂ZZ) taxa. Moreover, it was shown that Y–autosome fusions occur most frequently. Theoretical models suggested that a combination of two or more evolutionary forces, such as underdominance of the fusions, male-biased mutation rates for fusions, and female-biased reproductive sex ratio, is needed to explain the asymmetry between the Y and W chromosomes (Pennell et al. 2015; Kirkpatrick 2017).
Moths and butterflies (Lepidoptera), together with their sister order caddisflies (Trichoptera), constitute the most speciose lineage with female heterogamety. In their overview of 40 lepidopteran species with identified sex chromosomes, Traut et al. (2007) listed 12 moths with multiple sex chromosomes. Since then, more neo-sex chromosome systems have been reported in this order (Nguyen et al. 2013; Šíchová et al. 2013, 2015, 2016; Smith et al. 2016; Fraïsse et al. 2017; Mongue et al. 2017; Traut et al. 2017; Picq et al. 2018).
Some of the derived sex chromosome systems correspond to a conspicuously large sex chromosome pair (Nguyen et al. 2013; Šíchová et al. 2013; Mongue et al. 2017; Picq et al. 2018), which suggests that both W and Z sex chromosomes fused with an autosome. Similar large chromosome pairs were also observed in representatives of the families Pyralidae, Oecophoridae, and Gelechiidae with reduced chromosome numbers, but were considered autosomal fusion products (Ennis 1976). Carabajal Paladino et al. (2016), however, showed that the large chromosome pair corresponds to sex chromosomes in an invasive gelechiid pest, the tomato leafminer Tuta absoluta (Gelechiidae).
To test for the presence of neo-sex chromosomes in their genomes, we examined the karyotypes of several pests of the diverse superfamily Gelechioidea, which contains ∼18,500 species (van Nieukerken et al. 2011) and comprises among others the above-mentioned Oecophoridae and Gelechiidae families. Our results confirmed a sex chromosome–autosome fusion, which occurred in a common ancestor of all three main lineages of Gelechioidea, namely the Gelechiid, Scythridid, and Depressariid assemblages (Sohn et al. 2016). A synteny block involved in the fusion was identified as an autosome homoeologous to the chromosome 7 of the ancestral karyotype represented by the peppered moth Biston betularia (Geometridae) (cf. Van’t Hof 2013). Furthermore, we discovered another fusion between the neo-sex chromosomes and homoeologue of the B. betularia chromosome 27 within the tribe Gnorimoschemini (Gelechiinae). A potential role of the sex chromosome turnover in the divergence of Gelechioidea is discussed.
Materials and Methods
Insects
Representatives of five families within Gelechioidea were either obtained from laboratory stocks or collected from natural populations. A laboratory stock of the potato tuber moth, Phthorimaea operculella (Gelechiidae), was provided by the Atomic Energy Commission of Syria (Damascus, Syria). Larvae were reared on wax-coated potato slices as described in Saour and Makee (1997). Cultures of the Angoumois grain moth, Sitotroga cerealella (Gelechiidae), from the Instituto de Microbiología y Zoología Agrícola (IMYZA), Instituto Nacional de Tecnología Agropecuaria (INTA) (Buenos Aires, Argentina), and the Institute for Biological Control JKI, Federal Research Centre for Cultivated Plants (Darmstadt, Germany) were kept on wheat grains (Méndez et al. 2016). A laboratory colony of the tomato leafminer, T.absoluta (Gelechiidae), from IMYZA, INTA was maintained on potted tomato plants under the conditions detailed in Cagnotti et al. (2012). Specimens of the coconut black-headed caterpillar, Opisina arenosella (Xylorictidae), were obtained from the colony maintained on coconut leaflets at the Crop Protection Division of the Coconut Research Institute of Sri Lanka (Lunuwila, Sri Lanka). The larch case-bearer Coleophora laricella (Coleophoridae) and the brown house-moth Hofmannophila pseudospretella (Oecophoridae) were collected as larvae from wild populations in Levín (Lišov, Czech Republic). The dingy flat-body moth Depressaria daucella (Depressaridae) was collected as larvae and pupae in Slapy u Tábora (Tábor, Czech Republic). The material obtained in the field was immediately processed for its future analysis, and barcoded using a fragment of the cytochrome c oxidase subunit I (COI) gene as described in Hebert et al. (2004). The sequences obtained were checked in the BOLD animal identification database (Ratnasingham and Hebert 2007) to confirm the identity of the specimens (for accession numbers of the sequences, see supplementary table S1, Supplementary Material online).
Processing of the Insects
Spread chromosome preparations were made from wing imaginal discs, testes, or ovaries of the last instar larvae of all species using the method of Traut (1976) with slight modifications detailed in Šíchová et al. (2013). For D. daucella, preparations were also made from ovaries of female pupae. The preparations were dehydrated in an ethanol series (70%, 80%, and 100%, 30 s each) and stored at −20 °C.
Nucleic acids were isolated from larvae or pupae. Given the size of the specimens, total RNA was recovered using the NucleoSpin RNA II (Macherey-Nagel, Düren, Germany) kit, RNA blue (Top-Bio, Prague, Czech Republic), or RNAzol (Sigma–Aldrich, St. Louis, MO). The first-strand cDNA was then synthesized by random or oligo-dT primed SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA). Genomic DNA (gDNA) was extracted either by the NucleoSpin Tissue kit (Macherey-Nagel) or the MagAttract HMW DNA Kit (Qiagen, Hilden, Germany) and if needed, amplified by illustra GenomiPhi HY DNA Amplification Kit (GE Healthcare, Milwaukee, WI).
Fluorescence In Situ Hybridization Experiments
To identify sex chromosomes genomic in situ hybridization (GISH) was performed as described in Yoshido et al. (2005). Amplified male gDNA was fragmented by heating to 99 °C for 10 min in a TProfessional TRIO thermocycler (Biometra, Göttingen, Germany), and used as a species-specific competitor DNA (Šíchová et al. 2013). Female gDNA was labeled with fluorescein-12-dUTP (Jena Bioscience, Jena, Germany) using the nick translation protocol of Kato et al. (2006) with 3.5-h incubation at 15 °C. To accurately determine chromosome numbers, fluorescence in situ hybridization (FISH) with (TTAGG)n telomeric probes (tel-FISH) was performed either alone or in combination with GISH as described in Yoshido et al. (2005) and Šíchová et al. (2015). Unlabeled (TTAGG)n telomeric probes were prepared by nontemplate PCR according to Sahara et al. (1999) and labeled with Cy3-dUTP (Jena Bioscience) using the same nick translation protocol as above, but with 1-h incubation at 15 °C. For each slide, the hybridization mixture contained unlabeled fragmented male gDNA (3 µg) and female fluorescein-labeled gDNA (500 ng), and/or Cy3-labeled telomeric probe (200 ng), and sonicated salmon sperm DNA (25 µg). The preparations were counterstained with 0.5 mg/ml DAPI (4’,6-diamidino-2-phenylindole; Sigma–Aldrich) in antifade based on DABCO (1,4-diazabicyclo[2.2.2]octane; Sigma–Aldrich) (for composition, see Traut et al. 1999).
Preparations from FISH experiments were observed in a Zeiss Axioplan 2 microscope (Carl Zeiss, Jena, Germany) equipped with appropriate fluorescence filter sets. Black-and-white images were captured with an Olympus CCD monochrome camera XM10 equipped with cellSens 1.9 digital imaging software (Olympus Europa Holding, Hamburg, Germany). The images were pseudocolored and superimposed with Adobe Photoshop CS6 (Adobe Systems, San Jose, CA).
Screening for T. absoluta Sex-Linked Genes
The sex-linkage of selected genes was tested by means of quantitative PCR (qPCR) using male and female gDNA as template and autosomal gene as a reference (Nguyen et al. 2013; Dalíková et al. 2017). The selected genes were orthologous to markers for all the chromosomes of the ancestral karyotype represented by the B.betularia (Geometridae) linkage map (Van’t Hof 2013) and the Melitaea cinxia (Nymphalidae) genome (Ahola et al. 2014) (supplementary table S2, Supplementary Material online). Primers were designed using available T. absoluta transcriptome sequences (Berger et al. 2016). The 1:1 (female:male) ratio of the used autosomal reference genes, elongation factor 1 alpha (EF-1a) and acetylcholinesterase 1 (Ace-1) (using Ace-1 as target and EF-1a as reference), and the 1:2 ratio of the Z-linked control gene kettin (ket) (using Ace-1 as reference) were verified before analyzing other markers. The genes were tested in triplicates of three independent samples of both male and female gDNAs. Amplification efficiencies (E) of primer pairs were determined from the slope of the standard curve generated by plotting the threshold cycle (Ct) values against the log-concentrations of serial dilutions of male and female gDNAs. The female-to-male (F:M) ratio for each gene was calculated for each female as [(1 + Etarget) ^ (Average_Cttarget_male − Cttarget_female)] / [(1 + Ereference) ^ (Average_Ctreference_male − Ctreference_female)], and then compared with the expected values of 1 and 0.5 corresponding to autosomal position and sex-linkage, respectively, by means of one-sample t-test using R (R Core Team 2013) (supplementary table S3, Supplementary Material online). Composition of the reaction, cycling conditions, and sequences of forward and reverse primers are detailed in supplementary tables S3 and S4, Supplementary Material online.
Once these control genes were validated, the marker genes (supplementary table S2, Supplementary Material online) were analyzed using one biological replicate per sex with three technical replicates per gDNA sample. In this case, the F:M ratio was calculated using the delta delta Ct method as 2 ^ [(Cttarget_female − Ctreference_female) − (Cttarget_male − Ctreference_male)], which is a simplified version of the aforementioned formula that assumes E = 1 for all genes. The obtained values were considered for analysis only if ket and/or the reference genes (EF-1a and Ace-1) compared with each other provided the expected and previously corroborated 0.5 and 1 values, respectively. The experiments were carried out at least three times, using both reference genes, EF-1a and Ace-1. All reactions were performed in a final volume of 25 µl using SYBR Premix Ex Taq II (Perfect Real Time) (TaKaRa, Otsu, Japan) and a final concentration of primers of 0.2 mM for both the target and reference genes (except for pixie ATP-binding cassette subfamily E member 1 [Pix] when a final concentration of 0.3 mM was used for the primers of the target gene). The cycling conditions included an initial denaturation at 95 °C for 3 min, then 45 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, a final denaturation of 95 °C for 15 s, and then an increase of temperature from 65 to 95 °C with increments of 0.5 °C for 5 s for the generation of melting curves. The sequences of forward and reverse primers are detailed in supplementary table S2, Supplementary Material online.
All qPCR experiments were performed in FrameStar 96 well plates (Institute of Applied Biotechnologies [IAB], Prague, Czech Republic) covered by µltraAmp Plate Sealers (Sorenson BioScience, Salt Lake City, UT) or qPCR adhesive foil (IAB) using a C1000 Thermal cycler CFX96 Real-Time System (Bio-Rad, Hercules, CA).
Cloning of Genes of Interest in Other Gelechioid Species
The genes of interest included the reference genes EF-1a and Ace-1, together with the markers proven to be sex-linked in T. absoluta, namely Pix and chitinase h (Chit) for the chromosome homoeologous to B. betularia linkage group (BbLG) 7, and 90-kDa heat shock protein (Hsp90) and twitchin (Tw) for the chromosome homoeologous to BbLG27 (see results for details). Degenerate primers (supplementary table S5, Supplementary Material online) were designed for regions of coding sequences conserved between Lepidoptera and other insect species, and used for RT-PCR amplification of partial sequences with the first-strand cDNA as a template. Amplified fragments were cloned using pGEM-T Easy Vector System (Promega, Madison, WI) or CloneJET PCR cloning kit (Thermo Fisher Scientific, Waltham, MA), and confirmed by Sanger sequencing. The obtained sequences were deposited in GenBank (for accession numbers, see supplementary table S1, Supplementary Material online) and used for the design of species-specific primers for qPCR experiments (supplementary table S4, Supplementary Material online).
Quantitative Analysis of Gene Dose in the Other Gelechioid Species
Quantitative PCR experiments using male and female gDNAs as template were conducted in S. cerealella, P. operculella, C. laricella, O. arenosella, H. pseudospretella, and D. daucella to test for the sex-linkage of Pix, Chit, Hsp90, and Tw. Male and female gene doses of the target genes were compared with EF-1a and/or Ace-1. Three technical and three biological replicates were used per experiment. Composition of the reactions, cycling conditions, and sequences of forward and reverse species-specific primers are detailed in supplementary tables S3 and S4, Supplementary Material online. The F:M ratio was calculated including the E value of the primers, according to the formula mentioned earlier, and then compared with the expected values of 1 and 0.5 corresponding to autosomal position and sex-linkage, respectively, by means of one-sample t-test using R.
Results
Barcoding of Collected Specimens
The field collected larvae used for chromosome preparations were barcoded using a partial sequence of COI. The sequences confirmed the classification of H.pseudospretella (Oecophoridae) and D.daucella (Depressaridae) with 100% identity with their respective records in the BOLD database. In the case of the Coleophoridae specimens, our search retrieved matches with C.laricella and Coleophorasibiricella. Since the geographical distribution of both species does not overlap in the Czech Republic (Laštůvka and Liška 2011), we considered the samples as C. laricella in our analysis. The consensus sequences of the COI fragments for all species examined were deposited in the GenBank database under accession numbers detailed in supplementary table S1, Supplementary Material online.
Karyotype Analyses
In comparison with the most common and ancestral lepidopteran chromosome number n = 31 (see Discussion for details), all species of Gelechioidea studied herein showed a reduced chromosome number ranging from n = 28 to n = 30. These values are in concordance with those observed in other representatives of the superfamily, which shows a modal chromosome number of n = 29 in 15 out of 33 studied species (supplementary table S6, Supplementary Material online). FISH with the telomeric probe marking chromosome ends was used to accurately count chromosome numbers in some of the examined species (cf. Šíchová et al. 2015; not shown).
In the Gelechiid assemblage, a complete karyotype analysis including the identification of sex chromosome constitution analysis has not been performed except for P.operculella (Gelechiidae) (Bedo 1984; Makee and Tafesh 2006; supplementary table S6, Supplementary Material online). In the present study, we analyzed two representatives of the family Gelechiidae, namely T.absoluta and S.cerealella.
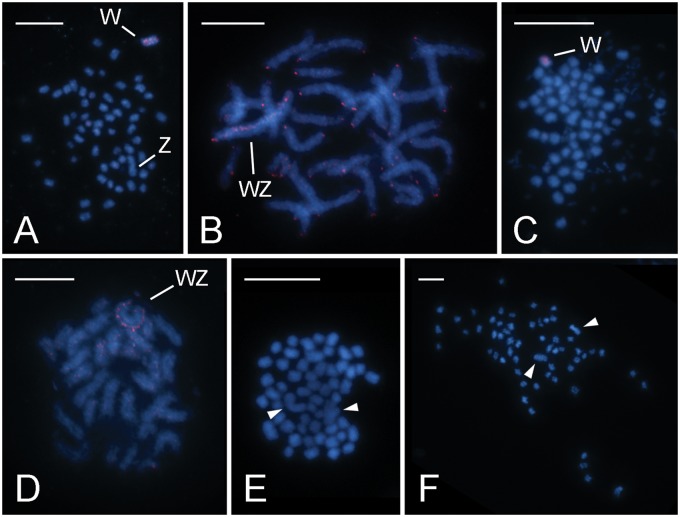
In T. absoluta, Carabajal Paladino et al. (2016) determined the haploid chromosome number of n = 29 and identified the largest elements as sex chromosomes morphometrically. In the present study, we identified the W chromosome by means of GISH, in which the labeled female gDNA-derived probe was hybridized to chromosomes in excess of unlabeled male competitor DNA. In mitotic complements, hybridization signals clearly highlighted one chromosome of the large pair. GISH thus confirmed that this is the W chromosome and implied that the other large element represents the Z chromosome (fig. 1A). The probe produced signals scattered along the W chromosome with notable exception of one subtelomeric and one interstitial gap in pachytene nuclei, and in some experiments also highlighted the chromosome ends (fig. 1B).
Fig. 1.
—Cytogenetic analysis of representatives of the Gelechiid and Scythridid assemblages. Chromosomes were counterstained with DAPI (blue); female derived genomic probes (A–D) were labeled by Cy3 (red). (A and B) GISH in Tuta absoluta (Gelechiidae, Gelechiid assembl.): (A) female mitotic metaphase consisting of 2n = 58 elements; note that the W chromosome is one of the two largest chromosomes in the complement; (B) female pachytene nucleus; the probe labeled the W chromosome in the WZ bivalent and chromosome ends of most bivalents. (C and D) GISH in Sitotroga cerealella (Gelechiidae, Gelechiid assembl.): (C) female mitotic metaphase consisting of 2n = 60 chromosomes; the W chromosome is not conspicuously larger than the other chromosomes; note DAPI-stained small rod-shaped bodies, probably corresponding to bacteria; (D) late pachytene female nucleus; the probe identified the W chromosome in the WZ bivalent. (E and F) Mitotic complements of Coleophora laricella (Coleophoridae, Scythridid assembl.) stained with DAPI: (E) male mitotic metaphase consisting of 2n = 58 chromosomes; note a pair of large chromosomes (arrowheads); (F) female mitotic metaphase comprising 2n = 58 chromosomes; note a pair of large chromosomes (arrowheads). Bar = 10 µm.
The haploid chromosome number of n = 30 was previously described for males of S. cerealella (Lukhtanov and Kuznetsova 1989). We confirmed the chromosome number in mitotic complements, 2n = 60, in males (not shown) as well as in females (fig. 1C). Furthermore, we used GISH to identify the female-specific W chromosome in mitotic complements (fig. 1C). In most mitotic metaphases, the W chromosome was not clearly discernible by size. In order to improve the resolution, GISH experiments were performed on female preparations of elongated pachytene bivalents. These experiments provided a more informative labeling pattern of the female genomic probe on the W chromosome. Hybridization signals of the probe were scattered along the entire W chromosome (fig. 1D). Interestingly, chromosome preparations obtained from the Argentinian S. cerealella females were contaminated with small DAPI-positive bodies, most likely corresponding to some bacteria present in the ovaries (fig. 1C).
Coleophora laricella (Coleophoridae) was the only representative of the Scythridid assemblage examined in this study. Mitotic metaphase complements consisted of n = 29 in both males and females of this species. The karyotype of both sexes comprised a conspicuously large chromosome pair (fig. 1E and F). Surprisingly, GISH provided weak or no hybridization signals in mitotic nuclei (not shown). GISH carried out on less condensed female pachytene chromosomes failed to identify a W-chromosome as well (not shown). Telomeric FISH combined with GISH was used as a control and yielded clear telomeric but no GISH signals (not shown). So it seems that our negative GISH results are not artifactual but rather point to an exceptional molecular composition of the C. laricella W chromosome. The W chromosome of C. laricella presumably does not differ from the rest of the genome in that it comprises a diverse spectrum of ubiquitous repeats present at low abundance.
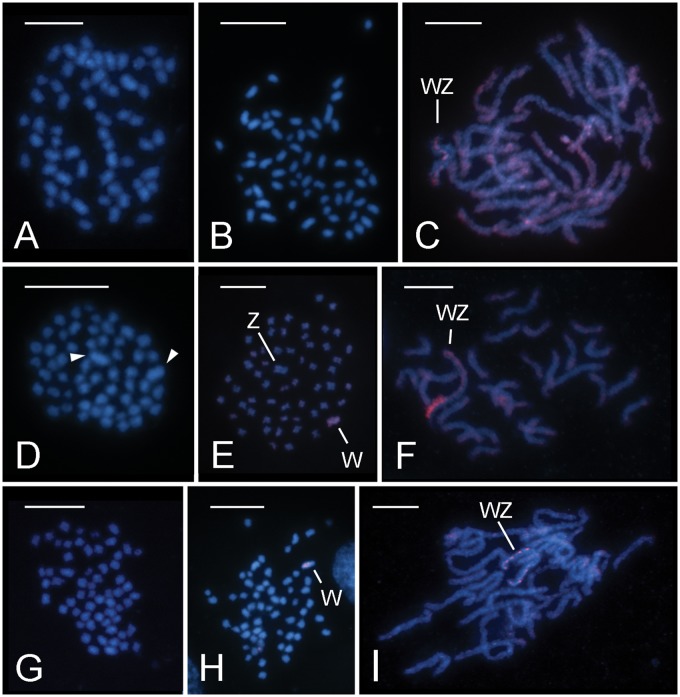
Within the Depressariid assemblage, three species, namely O.arenosella (Xyloryctidae), H. pseudospretella (Oecophoridae), and D. daucella (Depressaridae), were investigated. In O. arenosella, the diploid chromosome complement consisted of 2n = 60 chromosomes in both males (fig. 2A) and females (fig. 2B). No elements showed significant size differences in O. arenosella, with all chromosomes decreasing gradually in size, which is typical for lepidopteran karyotypes (fig. 2A and B). In addition, no mitotic chromosome was reliably discerned by GISH in this species as the female-derived genomic probe labeled all chromosomes more or less with the same intensity (not shown). In pachytene, the probe labeled all bivalents, some along the entire chromosome length and some preferentially in subterminal regions. However, one bivalent was conspicuous by its heteromorphic staining with one of its threads intensively stained while the other was not (fig. 2C). It is reasonable to assume that this bivalent corresponds to the WZ sex chromosome pair. The absence of hybridization signals on the Z chromosome is likely a result of its hemizygosity in females from which the GISH probe was derived. The sex chromosome bivalent identity was further supported by its meiotic pairing pattern, as the signal-free chromosome typically twisted several times around its labeled partner (fig. 2C). This was due to the size difference between the sex chromosomes with the W being much shorter than the Z chromosome (cf. Marec and Traut 1994).
Fig. 2.
—Cytogenetic analysis of representative of the Depressariid assemblage. Chromosomes were counterstained with DAPI (blue); female-derived genomic probes (C, E, F, H, I) were labeled by Cy3 (red). (A–C) Opisina arenosella (Xyloryctidae): (A) male mitotic metaphase consisting of 2n = 60 elements; (B) female mitotic metaphase consisting of 2n = 60 chromosomes; (C) GISH on female pachytene nucleus; note the hybridization signals on all bivalents either along the entire chromosomes or with preference for subterminal regions; the WZ bivalent is identified by the signal intensity that differs between the W and Z chromosome threads, as well as by the characteristic pairing of the longer Z chromosome twisted around the much shorter W chromosome. (D–F) Hofmannophila pseudospretella (Oecophoridae): (D) male mitotic metaphase consisting of 2n = 56 chromosomes; note the two largest chromosomes (arrowheads); (E and F) GISH on female chromosome preparations; (E) female mitotic metaphase comprising 2n = 56 chromosomes; note that GISH identified the W chromosome as one of the two largest chromosomes; (F) female pachytene nucleus; note the size and bipartite organization of the WZ bivalent with about one-third of the W chromosome thread strongly labeled with the probe. (G–I) Depressaria daucella (Depressariidae): (G) male mitotic metaphase comprising 2n = 60 chromosomes; note that there is no conspicuously larger chromosome pair; (G–I) GISH on female chromosome preparations; (H) female mitotic metaphase consisting of 2n = 60 elements with the W chromosome identified by the probe; (I) female pachytene nucleus; note the WZ bivalent showing scattered hybridization signals of the probe on the W chromosome thread. Bar = 10 µm.
In H. pseudospretella, a reduced diploid chromosome number of 2n = 56 with two large chromosomes was observed in mitotic metaphase nuclei of both sexes (fig. 2D and E). The female-derived genomic probe clearly highlighted one of the large chromosomes in female mitotic metaphase complements (fig. 2E). Thus, the largest chromosome pair most likely comprises the sex chromosomes. However, in female pachytene nuclei, a WZ bivalent could not be identified without the use of GISH. This method revealed a bipartite organization of the W chromosome, as it strongly labeled one terminal region corresponding to roughly one-third of the sex chromosome bivalent (fig. 2F).
The diploid chromosome number was 2n = 60 in both sexes of D. daucella. Neither male nor female mitotic complement comprised any notably larger chromosome (fig. 2G and H). GISH identified one of the larger chromosomes as the W chromosome in the D. daucella female mitotic metaphase complements (fig. 2H). In female pachytene nuclei, the WZ bivalent was easily discerned by the heterochromatic W thread (not shown). GISH showed scattered hybridization signals colocalizing with DAPI positive blocks on the W chromosome (fig. 2I).
Identification of Sex-Linked Synteny Blocks in Gelechioidea
To identify sex-linked synteny blocks, the sex-linkage of T. absoluta genes was tested by qPCR using male and female gDNA as template. This method can detect hemizygosity of Z-linked markers caused either by the absence or molecular degradation of their W-linked gene copies (Nguyen et al. 2013; Dalíková et al. 2017). The variable female-to-male (F:M) ratio between the selected reference genes EF-1a and Ace-1, using Ace-1 as target and EF-1a as reference, was 1.000 ± 0.102 (SE), which statistically differed from 0.5 (P < 0.05) but not from 1 (P > 0.05) (supplementary table S3, Supplementary Material online). The F:M ratio for ket, using Ace-1 as reference, gave a value of 0.498 ± 0.090, which significantly differed from 1 (P < 0.05) but not from 0.5 (P > 0.05) (supplementary table S3, Supplementary Material online). These results indicated that females and males had the same copy number of both Ace-1 and EF-1a genes, and that females had half the number of copies of ket with respect to males, which was expected as this gene represents a standard marker for the lepidopteran Z chromosome (cf. Nguyen et al. 2013; Van’t Hof 2013). The analysis thus confirmed that the Ace-1 and EF-1a genes are autosomal and can be used as reference genes for further studies. It also proved ket as a good control gene for the screening of sex-linked markers in T. absoluta.
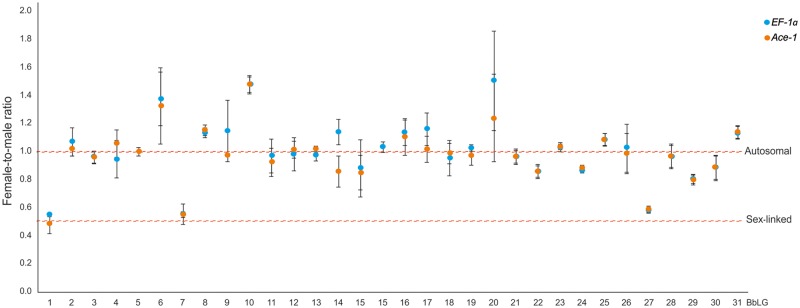
The results of the screening of marker genes in T. absoluta are presented in supplementary table S2, Supplementary Material online and figure 3. Markers orthologous to genes of B.betularia (Geometridae) LG1 (ket), LG7 (Pix) and LG27 (Hsp90) were sex-linked in this species, with F:M ratios ranging from 0.491 (ket) to 0.590 (Hsp90), considering the values obtained with both reference genes (EF-1a and Ace-1). The rest of the markers ranged from 0.800 for ribosomal protein L4 (marker for BbLG29) to 1.508 for 18–56 protein (marker for BbLG20), and were considered autosomal. Deviation of markers from the expected F:M value of 1 could be attributed to differences in primer efficiency, which was not corrected in the initial screening.
Fig. 3.
—Screening of marker genes in Tuta absoluta by means of qPCR. Blue dots represent the average female-to-male ratio values obtained for each marker using EF-1a as the reference gene. Orange dots are the average values for the same variable obtained using Ace-1 as the reference gene. Whiskers show the SE. Red dashed lines are used to show how each value correlates with 1 (autosomal) and 0.5 (sex-linked) expected female-to-male ratios. Note that most of the data points fluctuate ∼1, except for those corresponding to BbLG1, BbLG7, and BbLG27 which are closer to 0.5 than to 1. BbLG, Biston betularia linkage group.
BbLG1 corresponds to the Z chromosomes in the ancestral karyotype of n = 31, while the other two chromosomes (BbLG7 and BbLG27) are autosomes. An extra marker gene was hence considered for further analysis of these autosomes: Chit for BbLG7 and Tw for BbLG27. Orthologs of all four marker and both reference genes were then amplified and cloned from P. operculella, S. cerealella, C. laricella, O. arenosella, H. pseudospretella, and D. daucella. The partial sequences were deposited in NCBI (accession numbers in supplementary table S1, Supplementary Material online) and used for the design of species-specific primers for qPCR experiments (supplementary table S4, Supplementary Material online).
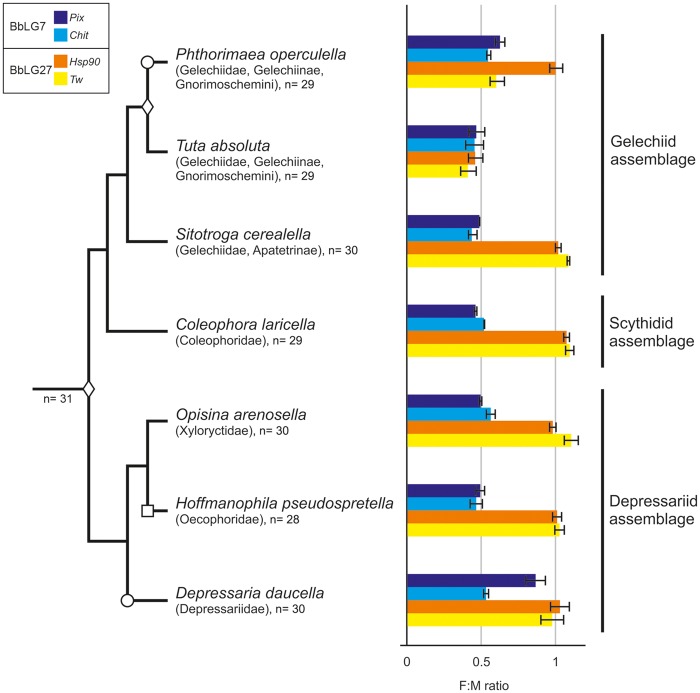
The results in T. absoluta and the other gelechioid species are shown in supplementary table S3, Supplementary Material online and summarized in figure 4. The F:M ratio values for both chromosomal markers corresponding to BbLG7 significantly differed from 1 (P < 0.05) but not from 0.5 (P > 0.05) in all species except for D. daucella, which suggested that the markers were sex-linked. In D. daucella, the F:M ratio of Chit was 0. 528 ± 0.021, while for Pix it was 0. 861 ± 0.074 (EF-1a as the reference gene), which is consistent with sex-linkage of the former and autosomal inheritance of the latter.
Fig. 4.
—Phylogenetic relationship between the species analyzed in this study, including a graphic representation of the results obtained using qPCR for the analysis of selected marker genes. Bar charts show the obtained female-to-male ratios (including SEs) of the copy number of the selected marker genes Pix and Chit for BbLG7, and Hsp90 and Tw for BbLG27, using EF-1a as the reference gene. Values close to 0.5 indicate sex-linkage, while values close to 1 indicate autosomal location of the marker. F:M ratio, female-to-male ratio. Note that the decrease in chromosome numbers coincides with sex chromosome–autosome fusions confirmed by qPCR. Diamond, confirmed fusion; circle, translocation or incomplete degeneration of one marker; square, putative fusion suggested by cytogenetic data.
For the BbLG27 markers, the F:M ratio statistically differed from 0.5 (P < 0.05) but not from 1 (P > 0.05) in S. cerealella, C. laricella, O. arenosella, H. pseudospretella, and D. daucella, indicating that the markers had an autosomal location. The opposite situation was observed in T. absoluta, meaning that both markers were sex-linked in this species. Interesting results were obtained in P. operculella, where Tw was sex-linked but Hsp90 was not (F:M ratios of 0. 578 ± 0. 019 and 0.999 ± 0.047, respectively; EF-1a as the reference gene). These findings, together with the discrepancies found for the markers for BbLG7 in D. daucella, were corroborated using the second reference gene (Ace-1) with a similar outcome (supplementary table S3, Supplementary Material online).
Discussion
In this study, we analyzed the sex chromosomes of seven species sampled across all three major lineages of the superfamily Gelechioidea (cf. Sohn et al. 2016; for phylogenetic relationships, see fig. 4 and supplementary fig. S1, Supplementary Material online). All species under study have a derived chromosome number compared with the ancestral lepidopteran karyotype of n = 31. Our cytogenetic analyses confirmed the expected presence of a large chromosome pair in the karyotypes of T.absoluta (Gelechiidae), C.laricella (Coleophoridae), and H.pseudospretella (Oecophoridae), species with karyotypes reduced to n = 29 in the first two and n = 28 in the latter (figs. 1A, B, E, F and 2D, E). The existence of a conspicuously large chromosome pair was a characteristic feature of the Gelechioidea karyotypes described to date (supplementary table S6, Supplementary Material online) and Ennis (1976) regarded them as autosomal fusion products. The GISH experiments performed in this study, however, confirmed that the largest chromosome pairs are indeed sex chromosomes in T. absoluta and H. pseudospretella (figs. 1A and 2E). In C. laricella, the W chromosome could not be identified (not shown). Thus, our cytogenetic data suggest that the largest chromosome pair corresponds to sex chromosomes only in some gelechioid species. A similar size difference, that is, the largest chromosome pair being about 1.5–2 times larger than the second largest one in a descending size series, was also observed in other Coleophora species (Lukhtanov and Puplesiene 1999) and in P.operculella (Gelechiidae) (Bedo 1984) suggesting chromosome fusions. Interspecific differences were observed in the relative size of the sex chromosomes, which were not so conspicuous in species with n = 30, namely S.cerealella (Gelechiidae), O.arenosella (Xylorictidae), and D.daucella (Depressaridae) (figs. 1C, D and 2A–C, G–I). A larger chromosome pair, which was not detected in our study, was reported for S. cerealella by Lukhtanov and Kuznetsova (1989) based on preparations of metaphase I bivalents from males (supplementary table S6, Supplementary Material online). This inconsistency could be caused by different methods, tissues used for chromosome preparations, and the type of cell division.
To confirm the fusions and identify the synteny blocks involved, we tested selected markers for all chromosomes of the ancestral karyotype with n = 31 (Van’t Hof 2013; Ahola et al. 2014) for their sex-linkage in T. absoluta by means of qPCR. The qPCR results confirmed the sex-linkage of markers located on the Z chromosome in other Lepidoptera (Nguyen et al. 2013; Van’t Hof 2013) and identified synteny blocks homoeologous to B.betularia (Geometridae) linkage group (BbLG) 7 and 27 as candidates for fusions (fig. 3). Testing of two markers for each chromosome, namely Pix and Chit for BbLG7, and Hsp90 and Tw for BbLG27, confirmed their sex-linkage in T. absoluta and thus strongly supported fusions of these synteny blocks with the ancestral Z chromosome (supplementary table S3, Supplementary Material online and fig. 4). qPCR analyses of Pix and Chit in the other species showed a sex-linkage of both markers in all gelechioids but D. daucella, in which only Chit and not Pix was sex-linked (supplementary table S3, Supplementary Material online and fig. 4). Assuming current phylogenetic hypotheses (Heikkilä et al. 2014; Sohn et al. 2016; supplementary fig. S1, Supplementary Material online), the qPCR results suggest that the fusion of the Z chromosome and chromosome homoeologous to BbLG7 [hereinafter F(Z; 7)] occurred in a common ancestor of the superfamily Gelechioidea. Thus, the autosomal location of Pix in D. daucella most likely points to a secondary translocation of this gene to an unidentified autosome (cf. Nguyen et al. 2013) or the W chromosome (Van’t Hof 2013) or to incomplete degeneration of its W-linked copy. The latter, however, seems unlikely in this case, as the F(Z; 7) fusion occurred ∼100 Ma (Wahlberg et al. 2013). Sex-linkage analyses of Hsp90 and Tw revealed that these markers are autosomal in all species but two representatives of the family Gelechiidae, T. absoluta and P. operculella, with Tw sex-linked in the latter but not Hsp90 (supplementary table S3, Supplementary Material online and fig. 4). This, together with the autosomal localization of both markers in S. cerealella, suggests that the neo-Z chromosome formed by the F(Z; 7) fusion further fused with BbLG27 [hereinafter F(neo-Z; 27)] in a common ancestor of the tribe Gnorimoschemini. However, we cannot exclude the possibility that the F(neo-Z; 27) fusion occurred earlier in the subfamily Gelechiinae (cf. Karsholt et al. 2013). Autosomal linkage of Hsp90 in P. operculella can again be explained by its translocation (see above). However, given the relative young age of the F(neo-Z; 27) fusion, we cannot fully rule out the Hsp90 allele persisting on the neo-W chromosome. Further research is needed to trace the exact evolutionary origin and level of differentiation of the F(neo-Z; 27) fusion. Moreover, the reduced chromosome number observed in H. pseudospretella (see above), the large size of its neo-sex chromosome pair along with the partial differentiation of its W chromosome suggest that another fusion between the F(Z; 7) and an autosome occurred independently in the family Oecophoridae.
Our results hence clearly show that at least two sex chromosome–autosome fusions occurred in the evolution of the diverse superfamily Gelechioidea. This finding further adds to the growing list of derived sex chromosome systems recently identified in various lepidopteran taxa, such as leafrollers of the family Tortricidae (Nguyen et al. 2013; Šíchová et al. 2013; Picq et al. 2018), leaf miners of the family Gracillaridae (Dalíková et al. 2017; Fraïsse et al. 2017), and Leptidea wood white (Pieridae) (Šíchová et al. 2015, 2016) and Danaus (Nymphalidae) butterflies (Smith et al. 2016; Mongue et al. 2017; Traut et al. 2017). The latter represent yet another case of repeated sex chromosome–autosome fusions, similar to those reported in this study. All these findings illustrate that neo-sex chromosomes are not exceptional in moths and butterflies. Rather, they appear to be relatively common, not only in terms of number of species, as the Tortricidae and Gelechioidea taxa alone comprise together about 17% of the described lepidopteran biodiversity (Beccaloni et al. 2018) but also in the number of independent origins (Nguyen and Carabajal Paladino 2016; cf. Pokorná et al. 2014). This suggests that the paucity of sex chromosome–autosome fusions is not an intrinsic feature of female heterogamety as previously assumed (Pokorná et al. 2014; Pennell et al. 2015).
Lepidoptera possess holokinetic chromosomes, which attach to kinetochore microtubules along most of the chromosomal surface (Wolf 1994). This reduces the risk of formation of dicentric and acentric chromosomes and hence it is expected to facilitate chromosomal rearrangements (Wrensch et al. 1994). Indeed, high variation in chromosome numbers was observed in moths and butterflies (Blackmon et al. 2017). However, this genome instability is confined only to a few lepidopteran taxa (Robinson 1971; Talavera et al. 2013). Comparative genomic studies have revealed that lepidopteran karyotypes are very stable with the modal chromosome number of n = 31 being the ancestral one. Furthermore, it has been shown that chromosome fusions are not random in this insect order since independent fusions observed in distant species involve the same small and repeat-rich chromosomes (Van’t Hof 2013; Ahola et al. 2014). Reconstructions of karyotype evolution in several lepidopteran clades with derived sex chromosome systems also show that the first large-scale chromosome rearrangements which differentiated the karyotypes of examined taxa from the ancestral n = 31 tend to be sex chromosome–autosome fusions (Nilsson et al. 2008; Nguyen et al. 2013; Šíchová et al. 2013; Dalíková et al. 2017; Mongue et al. 2017). Although the reconstruction of karyotype evolution in a group so diverse as Gelechioidea is challenging due to the scarcity of available data (supplementary table S6, Supplementary Material online), the reduced chromosome number of n = 30 in families Gelechiidae, Elachistidae, Xyloryctidae, and Depressariidae suggests that the F(Z; 7) fusion occurred early in the karyotype evolution of gelechioids.
This propensity of lepidopteran sex chromosomes for fusions could shed light on the evolutionary forces driving chromosomal change. The higher rate of sex chromosome–autosome fusions in XX/XY than in WZ/ZZ systems observed in vertebrates (Pokorná et al. 2014; Pennell et al. 2015) led to the conclusion that fusions must be driven by two or more evolutionary forces (Pennell et al. 2015; Kirkpatrick 2017). A simpler explanation for the higher rate of Y-autosome fusions in vertebrates, random genetic drift (Kirkpatrick 2017), was dismissed due to the lack of multiple sex chromosomes in female heterogametic groups (Pennell et al. 2015; Kirkpatrick 2017). Genetic drift, however, can be invoked to explain the high incidence of neo-sex chromosomes in Lepidoptera. In such case, the same pattern observed in vertebrates (a higher incidence of W–autosome than Z–autosome fusions) is expected for lepidopteran multiple sex chromosome systems. However, the W–autosome and Z–autosome fusions resulting in multiple sex chromosome constitutions WZ1Z2 and W1W2Z, respectively, observed so far in Lepidoptera are tied (Traut et al. 2007; Šíchová et al. 2015, 2016; Smith et al. 2016). Furthermore, many of the other recently reported neo-sex chromosomes systems are not informative as males and females exhibit the same chromosome number (Nguyen et al. 2013; Dalíková et al. 2017; Fraïsse et al. 2017; Mongue et al. 2017; this study). Available data thus do not allow us to evaluate the role of genetic drift in sex chromosome–autosome fusions in Lepidoptera.
Chromosome rearrangements such as fusions or inversions affect linkage relationships and thus can play an important role in adaptation and speciation (Yeaman 2013; Charlesworth 2015; Ortiz-Barrientos et al. 2016). In leafrollers of the family Tortricidae, Nguyen et al. (2013) reported the fusion of the Z chromosome with an autosome homoeologous to BbLG15. This chromosome is enriched in genes involved in detoxification and regulated absorption of plant secondary metabolites, namely esterases and ABC transporters, which are crucial for the performance of lepidopteran larvae on their host plants. The fusion thus linked these performance genes together with sex-linked female preference or host-independent isolation genes, which can facilitate adaptation and speciation in the presence of gene flow (Matsubayashi et al. 2010). Furthermore, it was hypothesized that the neo-Z-linked performance genes got amplified to make up for their nonrecombining and thus gradually degenerating maternally inherited gametologues (Nguyen et al. 2013). Following functional divergence of the new performance gene copies supposedly contributed to adaptation to new hosts which could eventually result in the formation of new species (cf. Li et al. 2003). Interestingly, BbLG7, which is involved in the F(Z; 7) fusion shared by all gelechioids, comprises the largest cluster of UDP-glycosyltransferases (UGTs). Enzymes encoded by the UGT gene family catalyze the glycosylation of small lipophilic compounds, turning them into water-soluble and thus more easily excreted products (Ahn et al. 2012). Although UGTs have been considerably understudied compared with other detoxification families, evidence supporting their role in detoxification of plant secondary metabolites and insecticides in Lepidoptera has been growing (Ahn et al. 2011; Wouters et al. 2014; Krempl et al. 2016; Li et al. 2017). Therefore, we hypothesize that the sex chromosome–autosome fusions may indeed contribute to ecological specialization and speciation in moths.
Sex chromosome turnover has been shown to predate, so far, two large lepidopteran radiations, Tortricidae and Gelechioidea (Nguyen et al. 2013; this study). The F(Z; 7) fusion observed in gelechioids fits well the scenario drawn by Nguyen et al. (2013) and the enrichment in performance genes of the autosomes involved in fusions in both lineages points to more general aspects of the lepidopteran karyotype evolution. The superfamily Gelechioidea provides an opportunity to test the hypothesis on the role of neo-sex chromosomes in the speciation of Lepidoptera, as sister lineages with and without neo-sex chromosomes of different age can be examined in parallel, along with their diversification rates.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
Our thanks go to Hayat Makee and Noha Tafesh (Damascus, Syria) for cultures of P.operculella, Cornel Adler (Berlin, Germany) for cultures of S.cerealella, Cynthia L. Cagnotti (Castelar, Argentina) for cultures of T.absoluta, and Marie Korchová for rearing insects and technical assistance. This work was supported by the Czech Science Foundation (14-35819 P and 17-17211S to P.N. and 17-13713S to F.M.) and the project Postdok_BIOGLOBE (CZ.1.07/2.3.00/30.0032) cofinanced by the European Social Fund and the state budget of the Czech Republic.
Footnotes
Data deposition: This project has been deposited at GenBank under accession numbers MG265651, MG265652, MG265654–MG265661, MG265664–MG265670, MG265672–MG265675, MG265678–MG265690, and MG265692–MG265694.
Literature Cited
- Ahn S-J, et al. 2011. Metabolic detoxification of capsaicin by UDP-glycosyltransferase in three Helicoverpa species. Arch Insect Biochem Physiol. 782:104–118. [DOI] [PubMed] [Google Scholar]
- Ahn SJ, Vogel H, Heckel DG.. 2012. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem Mol Biol. 422:133–147. [DOI] [PubMed] [Google Scholar]
- Ahola V, et al. 2014. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat Commun. 5:4737.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 142:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beccaloni G, et al. 2018. LepIndex: the global Lepidoptera names index. In: Roskov Y, et al., editors. Species 2000 & ITIS catalogue of life, 2018 annual checklist. Leiden: Species 2000: Naturalis.
- Bedo DG. 1984. Karyotypic and chromosome-banding studies of the potato tuber moth, Phthorimaea operculella (Zeller) (Lepidoptera, Gelechiidae). Can J Genet Cytol. 262:141–145. [Google Scholar]
- Berger M, et al. 2016. Insecticide resistance mediated by an exon skipping event. Mol Ecol. 2522:5692–5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmon H, Ross L, Bachtrog D.. 2017. Sex determination, sex chromosomes, and karyotype evolution in insects. J Hered. 1081:78–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagnotti CL, et al. 2012. Effects of X-rays on Tuta absoluta for use in inherited sterility programmes. J Pest Sci. 854:413–421. [Google Scholar]
- Carabajal Paladino LZ, et al. 2016. The effect of X-rays on cytological traits of Tuta absoluta (Lepidoptera: Gelechiidae). Fla Entomol. 99(sp1):43–53. [Google Scholar]
- Charlesworth D. 2015. Plant contributions to our understanding of sex chromosome evolution. New Phytol. 2081:52–65. [DOI] [PubMed] [Google Scholar]
- Dalíková M, et al. 2017. New insights into the evolution of the W chromosome in Lepidoptera. J Hered. 1087:709–719. [DOI] [PubMed] [Google Scholar]
- Delph LF, Demuth JP.. 2016. Haldane’s Rule: genetic bases and their empirical support. J Hered. 1075:383–391. [DOI] [PubMed] [Google Scholar]
- Dufresnes C, et al. 2016. Empirical evidence for large X-effects in animals with undifferentiated sex chromosomes. Sci Rep. 6:21029.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis TJ. 1976. Sex chromatin and chromosome numbers in Lepidoptera. Can J Genet Cytol. 181:119–130. [DOI] [PubMed] [Google Scholar]
- Fraïsse C, Picard MAL, Vicoso B.. 2017. The deep conservation of the Lepidoptera Z chromosome suggests a non-canonical origin of the W. Nat Commun. 81:1486.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J. 2016. Did sex chromosome turnover promote divergence of the major mammal groups? Bioessays 388:734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane J. 1922. Sex ratio and unisexual sterility in hybrid animals. J Gen. 122:101–109. [Google Scholar]
- Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W.. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci U S A. 1101:4812–14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkilä M, Mutanen M, Kekkonen M, Kaila L.. 2014. Morphology reinforces proposed molecular phylogenetic affinities: a revised classification for Gelechioidea (Lepidoptera). Cladistics 306:563–589. [DOI] [PubMed] [Google Scholar]
- Karsholt O, Mutanen M, Lee S, Kaila L.. 2013. A molecular analysis of the Gelechiidae (Lepidoptera, Gelechioidea) with an interpretative grouping of its taxa. Syst Entomol. 382:334–348. [Google Scholar]
- Kato A, Albert P, Vega J, Birchler J.. 2006. Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem. 81(2–3):71–78. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick M. 2017. The evolution of genome structure by natural and sexual selection. J Hered. 1081:3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Peichel CL.. 2012. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes. 943:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, et al. 2009. A role for a neo-sex chromosome in stickleback speciation. Nature 4617267:1079–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krempl C, et al. 2016. Potential detoxification of gossypol by UDP-glycosyltransferases in the two Heliothine moth species Helicoverpa armigera and Heliothis virescens. Insect Biochem Mol Biol. 71:49–57. [DOI] [PubMed] [Google Scholar]
- Laštůvka Z, Liška J.. 2011. Annotated checklist of moths and butterflies of the Czech Republic (Insecta: Lepidoptera). Brno: Biocont Laboratory. [Google Scholar]
- Li WM, Schuler MA, Berenbaum MR.. 2003. Diversification of furanocoumarin-metabolizing cytochrome P450 monooxygenases in two papilionids: specificity and substrate encounter rate. Proc Natl Acad Sci U S A. 100(Suppl 2):14593–14598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu B, Gao X, Liang P.. 2017. Over-expression of UDP-glycosyltransferase gene UGT2B17 is involved in chlorantraniliprole resistance in Plutella xylostella (L.). Pest Manag Sci. 737:1402–1409. [DOI] [PubMed] [Google Scholar]
- Lima TG. 2014. Higher levels of sex chromosome heteromorphism are associated with markedly stronger reproductive isolation. Nat Commun. 5:4743.. [DOI] [PubMed] [Google Scholar]
- Lukhtanov VA, Kuznetsova VG.. 1989. Karyotype structure in higher Lepidoptera (Papilionomorpha.). Entomol Rev. 68:12–31. [Google Scholar]
- Lukhtanov VA, Puplesiene J.. 1999. Polyploidy in bisexual Lepidoptera species (Insecta: Lepidoptera): old hypotheses and new data. Bonn Zool Beitr. 48:313–328. [Google Scholar]
- Makee H, Tafesh N.. 2006. Sex chromatin body as a marker of radiation-induced sex chromosome aberrations in the potato tuber moth, Phthorimaea operculella (Lepidoptera: Gelechiidae). J Pest Sci. 792:75–82. [Google Scholar]
- Mank JE, Hosken DJ, Wedell N.. 2014. Conflict on the sex chromosomes: cause, effect, and complexity. Cold Spring Harb Perspect Biol. 612:a017715.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marec F, Traut W.. 1994. Sex chromosome pairing and sex chromatin bodies in W-Z translocation strains of Ephestia kuehniella (Lepidoptera). Genome 373:426–435. [DOI] [PubMed] [Google Scholar]
- Masly JP, Presgraves DC.. 2007. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 5:1890–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi KW, Ohshima I, Nosil P.. 2010. Ecological speciation in phytophagous insects. Entomol Exp Appl. 1341:1–27. [Google Scholar]
- Méndez LM, et al. 2016. Evaluation of three wheat types for the rearing of Sitotroga cerealella Olivier (Lepidoptera: Gelechiidae) commonly used on natural enemies rearing. Rev Soc Entomol Argent. 75:105–116. [Google Scholar]
- Mongue AJ, Nguyen P, Voleníková A, Walters JR.. 2017. Neo-sex chromosomes in the Monarch butterfly, Danaus plexippus. G3 710:3281–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natri HM, Shikano T, Merila J.. 2013. Progressive recombination suppression and differentiation in recently evolved neo-sex chromosomes. Mol Biol Evol. 305:1131–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P, Carabajal Paladino LZ.. 2016. Evolutionary biology: convergent evolution, evolution of complex traits, concepts and methods In: Pontarotti P, editor. On the neo-sex chromosomes of Lepidoptera Heidelberg: Springer; p. 171–185. [Google Scholar]
- Nguyen P, et al. 2013. Neo-sex chromosomes and adaptive potential in tortricid pests. Proc Natl Acad Sci U S A. 11017:6931–6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson NO, Löfstedt C, Dävring L.. 2008. Unusual sex-chromosome inheritance in six species of small ermine moths (Yponomeuta, Yponomeutidae, Lepidoptera). Hereditas 1082:259–265. [Google Scholar]
- Ortiz-Barrientos D, Engelstädter J, Rieseberg LH.. 2016. Recombination rate evolution and the origin of species. Trends Ecol Evol. 313:226–236. [DOI] [PubMed] [Google Scholar]
- Pala I, Hasselquist D, Bensch S, Hansson B.. 2012. Patterns of molecular evolution of an avian neo-sex chromosome. Mol Biol Evol. 2912:3741–3754. [DOI] [PubMed] [Google Scholar]
- Pennell MW, et al. 2015. Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 115:e1005237.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picq S. et al. 2018. Insights into the structure of the spruce budworm (Choristoneura fumiferana) genome, as revealed by molecular cytogenetic analyses and a high-density linkage map. G3 8:2539–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokorná M, Altmanová M, Kratochvíl L.. 2014. Multiple sex chromosomes in the light of female meiotic drive in amniote vertebrates. Chromosome Res. 221:35–44. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; Available from: http://www.R-project.org/; last accessed April 16, 2019. [Google Scholar]
- Ratnasingham S, Hebert P.. 2007. BOLD: the Barcode of Life Data System. Mol Ecol Notes. 73:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R. 1971. Lepidoptera genetics. Oxford: Pergamon Press. [Google Scholar]
- Sahara K, Marec F, Traut W.. 1999. TTAGG telomeric repeats in chromosomes of some insects and other arthropods. Chromosome Res. 76:449–460. [DOI] [PubMed] [Google Scholar]
- Saour G, Makee H.. 1997. Radiation induced sterility in male potato tuber moth Phthorimaea operculella Zeller (Lep., Gelechiidae). J Appl Entomol. 121(1–5):411–415. [Google Scholar]
- Šíchová J, Nguyen P, Dalíková M, Marec F.. 2013. Chromosomal evolution in tortricid moths: conserved karyotypes with diverged features. PLoS One 85:e64520.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šíchová J, et al. 2016. Fissions, fusions, and translocations shaped the karyotype and multiple sex chromosome constitution of the northeast-Asian wood white butterfly, Leptidea amurensis. Biol J Linn Soc. 1183:457–471. [Google Scholar]
- Šíchová J, et al. 2015. Dynamic karyotype evolution and unique sex determination systems in Leptidea wood white butterflies. BMC Evol Biol. 15:89.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DAS, et al. 2016. A neo-W chromosome in a tropical butterfly links colour pattern, male-killing, and speciation. Proc R Soc Lond B Biol Sci. 2831835:20160821.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn J-C, et al. 2016. Phylogeny and feeding trait evolution of the mega-diverse Gelechioidea (Lepidoptera: Obtectomera): new insight from 19 nuclear genes. Syst Entomol. 411:112–132. [Google Scholar]
- Talavera G, Lukhtanov VA, Rieppel L, Pierce NE, Vila R.. 2013. In the shadow of phylogenetic uncertainty: the recent diversification of Lysandra butterflies through chromosomal change. Mol Phyl Evol. 693:469–478. [DOI] [PubMed] [Google Scholar]
- Traut W. 1976. Pachytene mapping in female silkworm, Bombyx mori L. (Lepidoptera). Chromosoma 583:275–284. [DOI] [PubMed] [Google Scholar]
- Traut W, Ahola V, Smith DAS.. 2017. Karyotypes versus genomes: the nymphalid butterflies Melitaea cinxia, Danaus plexippus, and D. chrysippus. Cytogenet Genome Res. 1531:46–53. [DOI] [PubMed] [Google Scholar]
- Traut W, Sahara K, Marec F.. 2007. Sex chromosomes and sex determination in Lepidoptera. Sex Dev. 16:332–346. [DOI] [PubMed] [Google Scholar]
- Traut W, Sahara K, Otto TD, Marec F.. 1999. Molecular differentiation of sex chromosomes probed by comparative genomic hybridization. Chromosoma 1083:173–180. [DOI] [PubMed] [Google Scholar]
- Turelli M, Begun DJ.. 1997. Haldanes’ rule and X-chromosome size in Drosophila. Genetics 1474:1799–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nieukerken EJ, et al. 2011. Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness. Zootaxa 31481:212–221. [DOI] [PubMed] [Google Scholar]
- Van’t Hof AE. 2013. Linkage map of the peppered moth, Biston betularia (Lepidoptera, Geometridae): a model of industrial melanism. Heredity 110:283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg N, Wheat CW, Peña C.. 2013. Timing and patterns in the taxonomic diversification of Lepidoptera (butterflies and moths). PLoS One 811:e80875.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf KW. 1994. The unique structure of lepidopteran spindles. Int Rev Cytol. 152:1–48. [Google Scholar]
- Wouters FC, et al. 2014. Reglucosylation of the benzoxazinoid DIMBOA with inversion of stereochemical configuration is a detoxification strategy in lepidopteran herbivores. Angew Chem Int Ed Engl. 5342:11320–11324. [DOI] [PubMed] [Google Scholar]
- Wrensch DL, Kethley JB, Norton RA.. 1994. Mites: ecological and evolutionary analyses of life-history patterns. In: Houck MA, editor. Cytogenetics of holokinetic chromosomes and inverted meiosis: keys to the evolutionary success of mites, with generalizations on eukaryotes New York: Chapman and Hall; p. 282–343. [Google Scholar]
- Yeaman S. 2013. Genomic rearrangements and the evolution of clusters of locally adaptive loci. Proc Natl Acad Sci U S A. 11019:E1743–E1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshido A, Marec F, Sahara K.. 2005. Resolution of sex chromosome constitution by genomic in situ hybridization and fluorescence in situ hybridization with (TTAGG)n telomeric probe in some species of Lepidoptera. Chromosoma 1143:193–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.