Abstract
Rapid and accurate immune monitoring plays a decisive role in effectively treating immune-related diseases especially at point-of-care, where an immediate decision on treatment is needed upon precise determination of the patient immune status. Derived from the emerging clinical demands, there is an urgent need for a cytokine immunoassay that offers unprecedented sensor performance with high sensitivity, throughput and multiplexing capability, as well as short turnaround time at low system complexity, manufacturability and scalability. In this paper, we developed a label-free, high throughput cytokine immunoassay based on a magnet patterned Fe3O4/Au core-shell nanoparticle (FACSNP) sensing array. By exploiting the unique superparamagnetic and plasmonic properties of the core-shell nanomaterials, we established a facile microarray patterning technique that allows the fabrication of uniform, self-assembled microarray in a large surface area with remarkable tunability and scalability. The sensing performance of the FACSNP microarray was validated by real-time detection of four cytokines in complex biological samples, showing high sensitivity (~ 20 pg/mL), selectivity and throughput with excellent statistical accuracy. The developed immunoassay was successfully applied for rapid determination of functional immunophenotype of leukemia tumor associated macrophages, manifesting its potential clinical applications for real-time immune monitoring, early cancer detection, and therapeutic drug stratification towards personalized medicine.
Introduction
The ongoing revolution in fundamental immunology and clinical discovery critically hinges on the availability of diagnostic tools capable of decentralized point-of-care measurements [1] to provide immediate quantitative information at the bedside or in the clinic.[2] In particular the case of immune monitoring for practical medical treatment,[3] fast, accurate and high throughput analysis of multiple immune cells secreted cytokines using a small sample volume[4] is highly required to precisely determine the rapidly changing immune status of patients in different inflammatory disease conditions.[5] The current “gold standard” clinical technology is mainly based on Enzyme-linked immunosorbent assay (ELISA). The complex labeling and washing processes require a total assay time up to more than 72 hours and a sample volume of 0.5-2 mL per test per patient, which greatly hinders its application for immune monitoring at the point of care.[6]
Label-free optical biosensing platforms, where the optical responses are measured at real-time without the need for secondary labeling, offer unique advantages in rapid analysis of complex biological samples.[7] Among these techniques, inclusive of photonic crystal,[8] optical ring resonator,[9] surface plasmon resonance (SPR),[10] fiber optics and interferometry,[11] the nanoplasmonic biosensing based on the localized surface plasmon resonance (LSPR) of noble metal nanoparticles (NPs) has shown exquisite levels of sensing performance.[12] Recent advances in nanomaterials and nanotechnology have spurred the design and fabrication of next-generation nanoplasmonic biosensors with various nanostructures,[13] such as nanorod,[14] nano-bipyramid,[15] nano-flower,[16] nano core-shell structure[17], and nanohole arrays.[18] The nanoplasmonic structures offer remarkable potential in sensor sensitivity, tunability, miniaturization, high throughput capability, and large-scale fabrication.[19] The integration of these platforms into highly functional microfluidic devices has provided novel biological interfacing opportunities and promising features for practical biomarker detection.[20] However, the implementation of such devices for real clinical and pharmaceutical settings has still been prohibited due to the deficiency in throughput and manufacturability without necessarily compromising the desirable sensitivity, multiplicity and reliability.[21] Various microarray nanoplasmonic sensing platforms are flourishing owing to the rapid technology evolution in nanofabrication.[22] They hold great promise in massively parallel quantification of multiple analytes by immobilizing specific antibodies in separate spots on a single array. The versatility of parallel detection has significantly increased the throughput of the nanoplasmonic biosensors for multiplex label-free analysis.[23] Yet, majority of these sensing arrays were fabricated using electron beam lithography,[24] direct laser writing,[25] chemical electrodeposition[26], and dip pen nanolithography,[27] which often require dedicated instrumentations, labor intensive fabrication procedures and are extremely costly for large scale production.[28] While there are a few multi-analyte high-throughput nanoplasmonic sensing platforms being developed at a fast pace, it has become clear that the cost, operation complexity, sensing performance, throughput, and scalability are equally challenging issues that must be addressed before the technology can be widely accepted in medical practice.[29]
Here, we developed a high-throughput, label-free, multiplex LSPR immunoassay based on a facile magnet assisted fabrication method for Fe3O4/Au core-shell nanoparticles (FACSNPs) microarrays. By harnessing the unique superparamagnetic property of the iron oxide nanocore, we demonstrated an easy-to-implement, scalable nanoparticle surface patterning technique for generating regular-shape, well dispersed, individual sensing spots over a large area. Moreover, the strong plasmonic coupling afforded by the decorated gold nanoparticles (AuNPs) on the FACSNPs exhibit superior sensitivity to the local refractive index change upon cytokine binding. Incorporating our previous developed LSPR dark-field imaging technique, the FACSNPs microarray biosensors can conduct 384 of test on four different cytokines for each sample with 16 replicates per cytokine-test. The integration of the FACSNPs microarray sensors into a simple optofluidic device allows real-time, massively parallel detection of multiple cytokines with a low limit of detection to ~ 20 pg/mL using 1 μL of real biological samples. The stability, accuracy and reproducibility of the immunoassay are further confirmed by standard ELISA and validated through successful demonstration of its practical use for functional immunophenotyping of leukemia tumor-associated macrophage (TAM) phenotypic polarization.
Results and Discussion
Synthesis and Characterization of Fe3O4/Au core-shell Nanoparticles
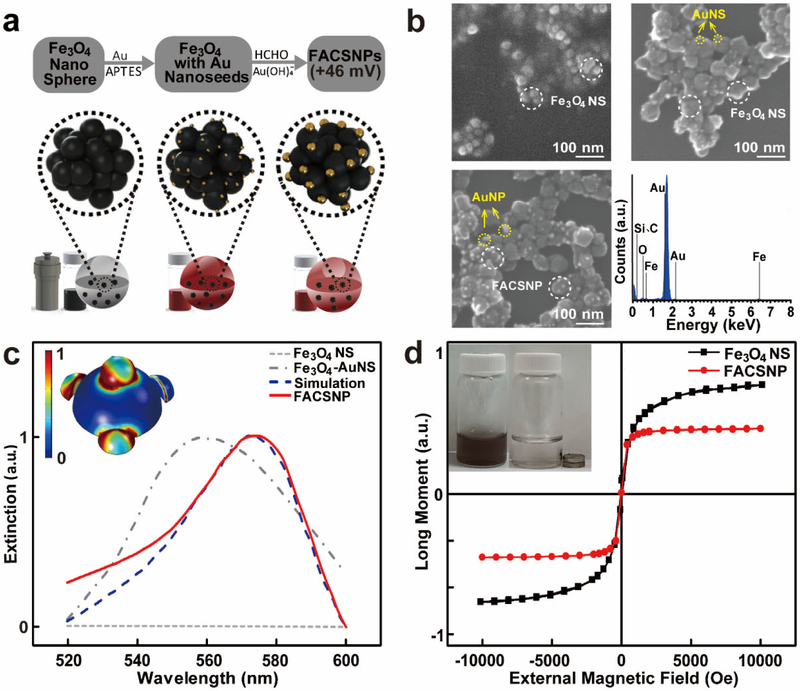
The FACSNPs were synthesized according to the fabrication procedures as illustrated in Figure 1a. (The detailed synthesis processes are described in the Materials and Methods section). Briefly, the monodisperse Fe3O4 nanospheres (Fe3O4 NS) were prepared by a modified solvothermal reduction method.[30] The Fe3O4 NS were then functioned with 3-aminopropyl-triethoxysilane (APTES) to allow the covalent attachment of Au nanoseeds (AuNS) on the Fe3O4 nanocore. Finally, the FACSNPs were formed through in situ seed growth by directly reducing the Au3+ on the Au nanoseeds. We used CTAB to modify the surface of our FACSNPs. This provided the core-shell structured Fe3O4/Au nanoparticles with a positively charged surface showing a zeta potential of +45 mV (Zetasizer Nano ZS90, Malvern). Figure 1b shows the representative scanning electron microscope (SEM) images of the obtained Fe3O4 NS, Fe3O4 NS decorated with AuNS (Fe3O4-AuNS) and FACSNPs. The Fe3O4 nanocore displayed a structure composed of a cluster of many small Fe3O4 nanoparticles with diameters around 20 nm as shown in the high-contrast dark part of the core region. The AuNS and AuNPs (bright dots circled in yellow of the SEM images) were immobilized on the surface of the Fe3O4 nanocore with sizes around 3-5 and 20 nm, respectively. Multiple-crystalline porous structures were observed for the core-shell nanomaterial with an average diameter of 50 – 60 nm. We determined the composition (major elements: Fe, O and Au) of the FACSNPs by energy-dispersive X-ray spectroscopy (EDX, bottom right panel of Figure 1b). Characteristic spectra of Fe, O and Au were observed in the plot, confirming the presence of the major elements in the FACSNPs. Other signature peaks in the EDX spectrum were resulted from the silicon substrate. The morphology, structure, and size of the FACSNPs were further confirmed using transmission electron microscopy (TEM) (Figure S1a). The size distributions of the FACSNPs and the embedded AuNPs were analyzed and shown good agreement with the SEM results (Figure S1b). We also performed the X-ray diffraction (XRD) measurements on the synthesized FACSNPs (Figure S1c). The results clearly show the corresponding spectrum peaks of Fe3O4 phase, in consistent with those reported previously.[30-31]
Figure 1. Synthesis and characterization of the Fe3O4Au core-shell NPs.
(a) Schematics of the fabrication procedures for FACSNPs (b) SEM images of Fe3O4 NS, Fe3O4-AuNS and FACSNPs drop-cast onto a conductive silicon substrate. The element composition of FACSNPs was analyzed by energy-dispersive X-ray spectroscopy shown in the bottom right panel. (c) The UV-Vis spectra of Fe3O4 NS, Fe3O4-AuNSs and FACSNPs. The experiment data shows very good agreement with the simulation results on the extinction spectra of FACSNPs. The inset panel displays the enhanced localized electromagnetic field surrounding the AuNPs immobilized on the Fe3O4 core. (d) Magnetization curves of the Fe3O4 NS and FACSNPs, suggesting the superparamagnetic characteristics of the NPs. The inset figure shows the effect of external magnetic field on FACSNPs in aqueous dispersion.
The UV-Vis spectroscopy on the aqueous dispersions of the Fe3O4 NS, Fe3O4-AuNS and FACSNPs was performed as displayed in Figure 1c. The Fe3O4 NS did not show any characteristic peak in 520-600 nm wavelength range (grey dashed line, Figure 1c). The addition of AuNS on the Fe3O4 resulted in a broad absorbance spectrum with resonance peak at around 550 nm. This can be explained by the large variation of the AuNS size distribution and their random deposition positions on the Fe3O4 NS. The growth of AuNS to AuNPs on the Fe3O4 core gives rise to a resonance peak at around 574 nm (Figure 1c, red line), indicating an absorbance red-shift of the AuNPs through enhanced plasmonic coupling due to the smaller interparticle distances. The much sharper resonance peak suggests a uniform immobilization of AuNPs on the Fe3O4 core. We theoretically calculated (COMSOL Multiphysics) the extinction resonance spectrum of the FACSNPs based on the characteristic parameters from SEM and TEM results (Figure 1c, blue dash line). A highly enhanced localized electromagnetic (EM) field can be observed surrounding the AuNPs with strong coupling between the adjacent nanospheres (inset of Figure 1c). As a result, the optical response of the FACSNPs was determined by the superimposition of the plasmonically coupled neighboring AuNPs, which can be effectively utilized for the detection of local analyte binding events on the nanoparticles.
We further examined the magnetism of Fe3O4 NS and FACSNPs by measuring the magnetic hysteresis loop using a vibrating sample magnetometer (Figure 1d). The remanent magnetizations and coercivities were measured to be close to zero, which demonstrates that both the Fe3O4 NS and FACSNPs exhibited superparamagnetic characteristics. Modification of Fe3O4 NS with AuNPs slightly weakened the magnetization saturation value. But the FACSNPs can still be easily magnetized under external magnetic field, and demagnetized and redispersed immediately in the solution when external magnetic field was removed (inset of Figure 1d). Similar phenomenon was also observed under the dark-field microscope, confirming the superparamagnetism of the FACSNPs (Figure S2). It should be noted that the conventional single-phase solid Fe3O4 NPs also exhibit superparamagnetic properties when the diameters of the NPs are in the range of 8.0-30 nm.[32] However, this smaller NP size makes the growth of 20 nm AuNPs onto the core much less energy favorable as compared to the nanocore of FACSNPs (~60 nm) with a cluster of small Fe3O4 NS. As such, the entire FACSNPs afford both the superparamagnetic and strong plasmonic coupling characteristics, which allows tracking, manipulation and patterning the FACSNPs without losing the advantage of the stable colloidal suspension, rendering them a well-suited material for microarray plasmonic biosensor fabrication as shown below.
Magnet Assisted Fabrication of the FACSNP Microarray Biosensor
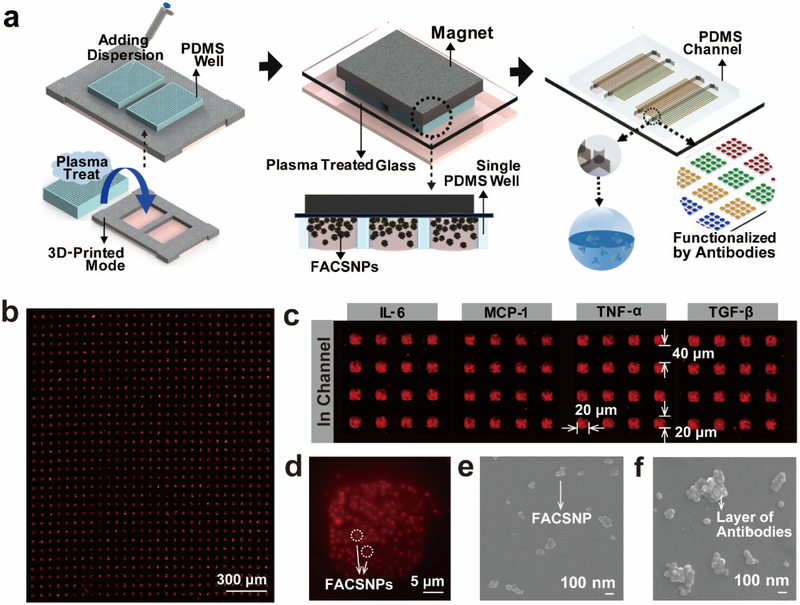
Using the physical and chemical properties of the FACSNPs that arise from both the intrinsic properties of constituent nanoparticles and their interparticle interactions, we adopted a magnet assisted self-assembly process to pattern uniform antibody functioned microarray on a glass substrate. Figure 2a presents the schematics of the fabrication processes (details can be found in Supporting Information Figure S3). We first treated a polydimethylsiloxane (PDMS) microwell mask (20×20 μm well size) and a glass substrate with oxygen plasma to make the surface hydrophilic and negatively charged. Two of the micro well-shape PDMS masks were placed into a 3D-printed plastic frame to fix the pattern positions for subsequent antibody function and sample loading. Ten μL of FACSNPs dispersion was then dropped onto the plasma treated PDMS masks and degassed for 25 min to ensure that all the microwells were fulfilled with the particle dispersion. The treated glass substrate was then attached to the FACSNPs loaded PDMS mask. Ceramic magnets were mounted on the bottom side of the glass substrate to exert a strong magnetic force on the superparamagnetic FACSNPs in the microwells. After overnight incubation, the positively charged FACSNPs were bound to the glass substrate by electrostatic attraction and assembled into uniform square shaped microarrays over a large surface area as presented in the dark-field image (Figure 2b). We compared the patterning results with and without the assistance of the external magnetic field (Figure S4a and b). Here, the magnet served as a concentrator that significantly enriched the local concentration, thus increased the binding possibility of the FACSNPs to the glass substrate and avoided the “coffee ring” effect. The release of the magnetic field allows the redistribution of the FACSNPs owing to the superparamagnetic characteristic. As a result, we obtained a clearly better pattern quality of the FACSNPs with the magnet, showing a much stronger scattering intensity under dark-field imaging with regular microarray shapes (Figure 2c) and well dispersed particle deposition as shown in Figure 2d and 2e.
Figure 2. Schematics of the FACSNP microarray fabrication and characterization of the fabricated microarray chip.
(a) Illustrations of the magnet assisted patterning process of the FACSNP microarray. (b) Dark-field microscopy images of the FACSNP microarray on the glass substrate. The FACSNPs confined in PDMS microwells self-assembled into a series of regular square-shape sensing spot arrays with the assistance of the external magnetic field. (c) After the patterning process, the FACSNP microarrays were functioned with four different antibodies in defined sensing areas for multiplex detection of four cytokines. (d) Dark-field microscopy image of individual FACSNPs biosensing spot at higher magnification, showing the well dispersed FACSNPs immobilized in the sensing spot. (e) SEM image of the FACSNPs before antibody function and (f) after successful antibody attachment.
Following the microarray patterning, we functionalized the FACSNPs with a panel of four cytokine antibodies using parallel microfluidic channels. The detailed function steps based on standard EDC/NHS chemistry are described in the Materials and Methods. The functionalized FACSNPs were imaged under SEM showing a thick layer of antibody coating on the NPs (Figure 2f). We further confirmed the antibody functionalization by measuring the Zeta-potential on the aqueous suspensions of the FACSNPs before and after the process. The functioned FACSNPs showed a neutralized zeta potential of +15 mV, indicating a partially coated surface of the FACSNPs with the antibodies. The successful antibody functionalization yielded four physically separated sensing regions with each consisting of large numbers of microarray sensors targeting specific cytokines (Figure 2c). This permits the multiplex detection of four cytokines in a massively parallel manner with high statistic accuracy. It should be noted that the shape, dimension, periodic distance, pattern area and the number of the target analytes of the microarray biosensor can be readily tuned by changing the design of the PDMS mask (Figure S4c and d), suggesting excellent flexibility, scalability and manufacturability of our fabrication technique.
FACSNP Microarray Imaging and Calibration for Label-Free High Throughput Cytokine Detection
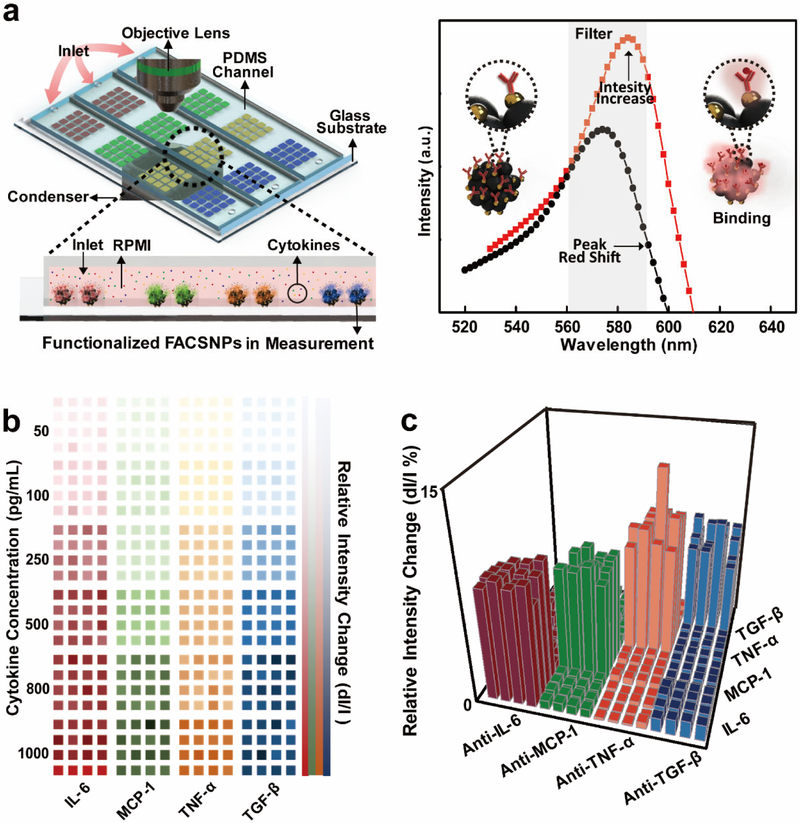
The clinical and immunological relevance of cytokine detection requires real-time, high throughput, sample efficient analysis while simultaneously achieving a low limit of detection (LOD). For instance, the early diagnosis and screening of inflammatory diseases, such as sepsis, cancer and graft-versus-host disease (GVHD), would need to identify multiple serum cytokine profiles at cutoff values less than 100 pg/mL.[33] To provide such discriminatory power for valuable clinical outcomes, we integrated our previously developed LSPR dark-field imaging technique[34] with the FACSNP microarray biosensor into an optofluidic immunoassay for rapid, sensitive and high throughput detection of cytokines in real biological samples. The optical setup and the principle of LSPR microarray imaging are illustrated in Figure 3a. Briefly, the prepared FACSNP microarray chip was mounted on a standard dark-field microscope (Nikon Eclipse Ni-U). Binding of the analyte cytokines onto the FACSNPs induces an increase in the intensity and a spectral red-shift of the scattering light. The collective light intensity shift was then recorded in real-time by an electron-multiplying charge-coupled device (EMCCD) camera. A customized Matlab program was used to automatically select the regions of interest through an edge detection/background subtraction algorithm and quantify the intensity change for each microarray sensing spots.
Figure 3. FACSNP microarray imaging schematics and intensity mapping of massively on-chip biosensing of multiple cytokines,
(a) Schematic of the dark-field microscope setup for FACSNP microarray imaging. The prepared microarray chip was fixed on the motorized stage with the other side in contact with the dark-field condenser via silicon oil. A sample was injected from the inlet of the PDMS channel covered on the prepared pattern, flown through the sample channel, and collected from the outlet. Different cytokines in the sample were captured by the antibody-conjugated FACSNPs on microarray. The light scattered from the FACSNPs was collected by 20X objective lens and imaged by the EMCCD. The right panel shows the principle of the FACSNP microarray imaging. The binding of specific cytokines onto the FACSNP results in an intensity increase and a spectrum red-shift, (b) Mapping of intensity variations of FACSNP microarray for four different types of cytokines (IL-6, MCP-1, TNF-α and TGF-β) at different concentrations. (c) Bar graph showing the scattering intensity variations obtained by the FACSNPs microarray sensing spots within each microfluidic detection channel (horizontal direction) when loaded with one specific recombinant cytokines spiked to the device. The concentration of the specific cytokine in the detection channel is 1000 pg/mL.
The intensity based LSPR microarray imaging provides unique advantages over the traditional plasmonic biosensors based on the spectrum-shift detection schemes. The obtained optical signals from the assembled microarrays contain statistic information over a large amount of the FACSNPs, which minimizes the variances in particle structure and spatial distribution. This distinct feature of LSPR microarray imaging offers unprecedented opportunities for high throughput immunoassay with inherently excellent statistic accuracy. As such, we first performed the calibration of the FACSNPs microarray chip through parallel detection of four different cytokines: interleukin-6 (IL-6), monocyte chemoattractant protein 1 (MCP1), tumor-necrosis-factor alpha (TNF-α) and transforming growth factor beta (TGF-β) in Roswell Park Memorial Institute (RPMI) cell culture medium. The spiked in samples with known concentrations (50, 100, 250, 500, 800, 1000 pg/mL) of cytokines were loaded into the sample loading channels and the quantified signal changes of the microarrays were translated into intensity maps as shown in Figure 3b. Here, we selected a 4 × 4 sensing array for each cytokine and recorded the real time intensity shift of all the sensing spots in one sample channel (Figure S5a). Thus, we can simultaneously acquire 64 real-time binding curves for four cytokines in one loading channel and achieve a total number of 384 (64 × 6) measurements for the whole immunoassay. The sensing matrix can be easily scaled up by increasing the numbers of sensing arrays and sample loading channels. We established the calibration curves for each cytokines based on the concentration dependent fractional intensity change(ΔI/I0) (Figure 3c , S5b). The intensity shift (ΔI/I0) was averaged over the 16 LSPR sensing microarrays by calculating the signal difference before (I0) and after (I0 + ΔI) the sample incubation. We further determined the LOD as defined by 3σ/kslope, where σ is the standard derivation of background signal of the control medium and kslopeis the slope of the linear regression of each calibration curve. The calculated LODs for the four cytokines were 18.96 pg/mL for IL-6, 14.57 pg/mL for MCP-1, 32.62 pg/mL for TNF-α and 22.08 pg/mL for TGF-β. As a result, our FACSNP microarray immunoassay shows comparable sensing characteristics in terms of sensitivity, assay time and sample volume to the most state-of-art LSPR biosensors,[35] while offering unique advantages in selectivity, throughput and manufacturability towards practical applications.
Selectivity and Stability of FACSNP Microarray Immunoassay
To validate that our microarray immunoassay can specifically detect target cytokines in a complex biological medium, we performed measurements on a set of samples with each containing only one specific type of the cytokines (IL-6, MCP-1, TNF-α and TGF-β) at the concentration of 1000 pg/mL. Figure 3c presents the fractional intensity changes of FACSNP microarrays after incubating with the prepared spiked-in samples in RPMI. With the presence of the target cytokines, only the corresponding antibody functioned sensing spots yielded significantly increased optical responses, proving the excellent selectivity of the microarray immunoassay. The intensity shift was then converted to analyte concentration based on the calibration curves (Figure S5c), showing no significant difference with the expected concentration of 1000 pg/mL. The microarrays targeting unpresented cytokines in the cell culture medium exhibited signals below the LOD as anticipated (Figure S5c). These results suggest that our multiplex immunoassay demonstrated minimum cross-reactivity and negligible non-specific adsorption among the four cytokine biosensor arrays. We believe that the spatial confinement of the antibody functioned sensing area and physically separated individual sensing arrays are the key factors that contribute to the pronounced selectivity as compared to conventional multiplex immunoassays. In this study, all the microarray immunoassay were used within the same day after antibody functionalization. We anticipate that the functioned chips in proper storage condition at 4 °C could last for 1-3 weeks and believe that the stability of the immunoassay is highly dependent on the chemistry and physics of the antibody and the storage condition.[36] Since the FACSNP microarrays without antibody can be stored for months and the antibody functionalization can be readily achieved within 2 hrs. Thus, it would be more preferable to use the functionalized FACSNP microarray on an as-needed basis for practical application, which has already been widely implemented in commercial immunoassays.
FACSNP Microarray Immunoassay for Functional Immunophenotyping of Tumor-Associated Macrophage
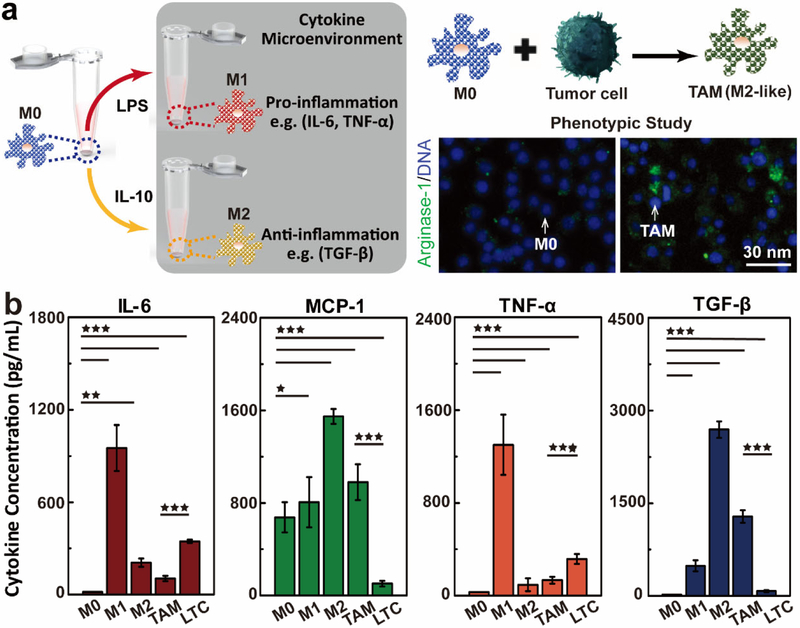
TAMs are the most prominent immune cells in the tumor microenvironment composed of leukocytes, fibroblasts and vascular endothelial cells. They play a vital role in non-resolving inflammation in tumor microenvironment, which is known as a hallmark of cancer.[37] In general, macrophages show a high degree of plasticity in response to local environments and can be polarized to pro-inflammatory (M1) or anti-inflammatory (M2) macrophages. The M1 phenotype are well adapted to promote a strong immune response by secreting high levels of pro-inflammatory cytokines. In contrast, the M2 phenotype are activated by T helper cell 2 and tumor derived cytokines (IL-4, IL-10 and IL-13), which are well-suited for the promotion of proliferation, invasion and angiogenesis of tumor cells and thus the tumor development (Figure 4a). With the unraveling relationship of the apparent dual nature of macrophages to the tumor development, the M2-like TAMs are now being recognized as potential diagnostic biomarkers and therapeutic targets for cancer.[38]
Figure 4. FACSNP microarray immunoassay for functional immunophenotyping of macrophages in biomimetic tumor microenvironment.
(a) The schematics of macrophage polarization under different stimulation conditions. The right panel shows the transformation of the macrophages into M2-like TAMs after exposure to a biomimetic tumor microenvironment. (b) Cytokine secretion profiles of macrophages after treated in different polarization conditions. M0 denotes the unpolarized, original macrophages. M1 is the original macrophages treated by 100ng/mL of LPS for 48 hrs. M2 is the original macrophages treated by 100ng/mL of IL-10 for 48 hrs. TAM presents the macrophages polarized by the leukemia tumor condition (LTC) culture medium. The cytokine concentrations were quantified by measuring the cell medium samples (n=3) using the FACSNP microarray immunoassay. Data were presented as mean ± SD. P-values were calculated using the One-Way ANOVA,* P < 0.05, **P<0.01, ***P<0.0001.
To explore the practical use of our FACSNP microarray immunoassay for clinical diagnosis, we performed the functional immunophenotyping of macrophages exposed to leukemia tumor cell (LTC) microenvironment. Here, we measured the cytokine secretion profiles of macrophages under stimulation or treated using LTC conditioned media (Figure 4b). The original unpolarized macrophages (M0) express no significant secretion for pro-inflammatory cytokines (IL-6 and TNF-α) and anti-inflammatory cytokine TGF-β. The stimulation with lipopolysaccharides (LPS) turned M0 into the M1 phenotype macrophages, where strong IL-6 and TNF-α expressions were observed in the cell culture medium with a negligible amount of TGF-β released. In contrast, the M2 phenotype macrophages polarized by IL-10 showed a significantly increased concentration of anti-inflammatory cytokine (TGF-β) expression, while the inflammatory responses by IL-6 and TNF-α secretion were largely suppressed. The macrophages treated with LTC conditioned media displayed a similar cytokine secretion profile with that of M2 macrophages, indicating their transformation into M2-like TAMs. The measurements of LTC conditioned media showing low levels of all cytokines further confirm that the measured TGF-β and MCP-1 were secreted by the polarized TAMs. Here, the MCP-1 is known as a potent chemotactic factor for monocyte trafficking,[39] thus was detected in all the macrophage culture media. The relatively higher level of MCP-1 released by M2 and TAMs could be attributed to the promoted macrophage recruitment in the tumor microenvironment.[40] All the results obtained from the FACSNP microarray immunoassay were validated by the singleplex ELISA for all four cytokines across the cell medium samples prepared above. An excellent linear correlation (R2=0.9252) was obtained between the results measured by both methods (Figure S6). As such, the FACSNP microarray immunoassay showed discriminative power for immunophenotyping of macrophages in a biomimetic tumor environment, which could be potentially applied as a rapid and high throughput method for point-of-care clinical cancer diagnosis.
Conclusion
In conclusion, we have demonstrated a high-throughput, label-free, multiplex immunoassay that enables the analysis of multiple immune biomarkers in a rapid, accurate and sensitive manner. The key to the success of this platform is by synergistically utilizing both the superparamagnetic and nanoplasmonic properties of the FACSNPs for large scale array patterning and high throughput sensing. The magnet assisted patterning approach shows great advantages over many other technologies for large-scale array fabrication in terms of feasibility, flexibility, scalability and cost-effectiveness. The multiplex immunoassay based on the patterned microarray can generally serve as a powerful tool for routinely monitoring a wide variety of biomarkers with rapid turn-around time and high statistic accuracy, which can be readily implemented for point-of-care clinical diagnosis. This platform possesses unique characteristics that do not exist with currently available technologies in clinical settings, which can be further expanded by integrating with nano- and microfluidic systems to provide multiscale measurements from whole-blood level to single-cell level for comprehensive functional analysis of the immune system.
Materials and Method
Fe3O4/Au core-shell Nanoparticle Synthesis
Firstly, 0.15 g of FeCl3·6H2O was added into a 4 mL mixed solution of 3 mL ethylene glycol and 1 mL ethanolamine under vigorous magnetic stirring to form a stable light brown solution. 0.1 g of polyethylene glycol 2000 and 0.4 g of NaAc was added into the above solution. After 30 minutes of vigorous magnetic stirring, the solution was transferred to a Teflon-lined stainless-steel autoclave and was heated to 200 0C for 8 hrs. The products (Fe3O4 NS) were collected and washed with ethanol and deionized water three times and dispersed in ethanol. Then, 40 μL of APTES was added into the dispersion under continuous magnetic stirring for 3 hrs at room temperature. The APTES modified Fe3O4 NS were harvested and were washed with ethanol and deionized water three times. The Au nanoseeds were prepared by adding 0.6 mL ice-cold and fresh 0.1 M NaBH4 solution into 10 mL aqueous solution of 2.5×10−4 M HAuCl4 and 2.5×10−4 M trisodium citrate. The solution color changed into pink immediately. After that, 5 mL of APTES-modified Fe3O4 NS aqueous dispersion was mixed with 3 mL of gold nanoseeds solution. The mixture was quickly ultrasonicated and then stayed under magnetic stirring for 2 hrs. After washing with ethanol and deionized water, the products (Fe3O4 NS-AuNS) were dispersed in deionized water. 150 μL of 1% HAuCl4 solution was added to 10 mL of 2mM potassium carbonate solution under vigorous magnetic stirring. After 30 minutes, the solution aged at 4 0C overnight to form Au(OH)4− ions solution. To further grow the AuNS to AuNPs on the surface of the Fe3O4 NS, 2 mL of Au(OH)4− ions solution and 8 μL of formaldehyde was added to the Fe3O4 NS-AuNS aqueous dispersion. The FACSNPs were harvested by a magnet and incubated with 0.1 M CTAB solution at 18 0C for 48 hrs. The final products were washed with ethanol and deionized water.
FACSNP Microarray Fabrication and Functionalization
The FACSNP microarray was fabricated based on a magnet assisted patterning technique. The detailed fabrication procedures are described in Supporting Information Section 4. To functionalize the FACSNP microarray, we fabricated a PDMS layer with microfluidic flow channels. The microfluidic channels can be easily aligned onto the FACSNP microarray as we used a plastic mold to carefully confine the patterning area. After placing the microfluidic channel layer on the FACSNP microarray chip, the microarray sensing spots were functionalized by forming a self-assembled monolayer (SAM) through simple ligand exchange. A stock solution of thiolated alkane 10-Carboxy-1-decanethiol (HS-(CH2)10-COOH) was diluted to 1 mM in the mixed solution consisted of 10% ethanol and 90% deionized water and injected through the microfluidic channel on the glass substrate. The stronger affinity of the thiol anchor group with the gold surface enabled the thiolated alkane to replace the CTAB coating and serve as a linker to probe antibodies. The antibody was linked to −COOH functional group through standard 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide/N-hydroxysuccinimide (EDC/NHS) coupling chemistry. Briefly, we injected a mixture of 0.4 M EDC (Tokyo chemical Industry) and 0.1 M NHS at a 1:1 volume ratio in 2-(N-morpholino) ethanesulfonic acid (MES) buffer through the microfluidic channels and activated −COOH on the gold surfaces of the FACSNPs. After the surface activation, we diluted primary cytokine antibodies (eBioscience) from 500 μg/mL to 50 μg/mL in 1x PBS, loaded them into individual channels and incubated them at room temperature for 60 min. Each channel will cover an area of 500 μm × 2.5 cm FACSNP microarray with hundreds of sensing spots functionalized with distinct antibody molecules. Therefore, the sensing spots arrays can afford multiplex detection of four different cytokines. During all the process steps, we loaded the reagent solutions using a syringe pump (LEGATO101, Kd Scientific) at 1.5pμL/min. Between every step, the microarray sensing spots were thoroughly washed to remove any excessive solutions or molecules using 20 μL of deionized water or 1x PBS (depends on various conditions) at 2 μL/min.
Optical setup and FACSNP microarray immunoassay
After the FACSNP microarray antibody functionalization, the microfluidic channel layer was removed from the glass substrate and immediately replaced it with another PDMS layer with parallel sample-flow microfluidic channels. The sample-flow microfluidic channels (500 μm (W) × 2.5 cm (L) × 50 μm (H)) were bonded perpendicular to the direction of the antibody functionalization channels. We subsequently mounted the constructed FACSNP microarray chip on a motorized X-Y stage (ProScanIII, Prior Scientific, Rockland, MA), loaded Roswell Park Memorial Institute (RPMI) medium into each of the on-chip flow channels using a syringe pump at 1.5 μL/min to stabilize the initial light intensity of sensing spots. After signal stabilization, ~ 1 μL sample was loaded into each of the sample-flow channels using a syringe pump at 0.1 μL/min and the images of sensing spot arrays were real-time captured by the EMCCD camera and recorded them using NIS-Element BR analysis. A customized Matlab program was used to analyze and quantify the scattering intensity increase for each microarray pattern.
Cytokine measurement with ELISA.
ELISA kits (Mouse IL-6 ELISA MAX™ Deluxe, Mouse IL-10 ELISA MAX™ Deluxe, and Mouse TNF-α ELISA MAX™ Deluxe, Biolegend, Mouse MCP-1/CCL2 Uncoated ELISA kit, Mouse TGF-beta 1 ELISA kit, Invitrogen) were used in this study. According to the manufacturer’s protocol, capture antibody solution was firstly added into all wells of a 96-well plate provided in the kit. The plate was subsequently sealed and incubated at 4°C overnight. After washing and blocking, 100 μL of each standard and cell culture supernatant sample was added into the wells and incubated for 2 hrs at room temperature. The wells were then incubated with detection antibody for 1 hr, followed by washing and incubation with HRP-labeled Avidin for 30 minutes. TMB mixture (1:1) was then added for 20 minutes in the dark to visualize chemiluminescence. Then, 100 μL of 2 M sulfuric acid solution was added in each well to stop the reaction. The reading of ELISA results was carried out using a plate reader (SpectraMax i3, Molecular Devices) by recording the absorbance at 450 nm within 15 mins after adding the stop solution.
Cell culture and macrophage polarization
Mouse macrophage cells (RAW264.7, ATCC) were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (FBS, Gibco), and 1% penicillin/streptomycin (Gibco). All cell lines were grown no more than 20 passages in a 37 °C (5% CO2) incubator. Macrophage polarization was induced in vitro. Briefly, untreated RAW264.7 cells were labeled as M0 macrophages. RAW264.7 cells (106 cells/mL) were seeded in 12-well plates overnight before polarization. M1-like macrophages were obtained by treating RAW264.7 cells with 100 ng/mL lipopolysaccharide (LPS, Sigma-Aldrich) for 24 hrs. M2 macrophages were obtained by stimulating RAW264.7 cells with 50 ng/mL of IL-10 (BioLegend) for 24 hrs. Subsequently, the media were replaced by fresh complete cell culture media for M1 and M2 macrophages. The cell culture supernatants for M1 and M2 macrophages were then collected after another 24 hrs. B-cell acute lymphoblastic leukemia (B-ALL) cells was isolated from a well-characterized model of pediatric Ph+ B-ALL, in which lethally-irradiated C57BL/6 mice are reconstituted with retrovirally-infected hematopoietic stem and progenitor cells ectopically co-expressing the B-ALL-associated P190 BCR-ABL1 isoform, as well as GFP for fluorescent cell tracing. Following the isolation, B-ALL cells were cultured and expanded in Iscove's Modified Dulbecco's Medium (IMDM, Invitrogen) supplemented with 10% FBS, 1% penicillin/streptomycin, 100 μM L-glutamine and 50 μM β-mercaptoethanol in a 37 °C (with 5% CO2) incubator. The culture supernatants of leukemia tumor cells were collected after the concentration of tumor cells reached 13×106 cells/mL and centrifuged at 2000×g for 10 min to remove cell debris. Biomimetic TAMs were obtained by culturing M0 RAW264.7 cells in complete culture media supplemented with conditioned media of leukemia tumor cells (1:1) for 24 hrs.
Statistical Analysis
In Figure 1c, the data of UV-Vis absorbance spectra of Fe3O4-AuNSs, FACSNPs and the simulation results of FACSNPs extinction spectrum were normalized to 0 - 1 range to clearly show the spectrum shift. In Figure 1d, the magnetization curves of the Fe3O4 NS and FACSNPs were also normalized to 0 - 1 range. The normalizations in both figures were processed by Microsoft Office Excel. In Figure 3b, all the measured intensity values from the 16 FACSNP microarray spots for four different types of cytokines at different concentrations were transformed to “Heat Map” by Microsoft Office Excel. All the statistical analysis of the measured cytokine concentrations were obtained by calculating the mean and standard derivation of the signal responses from a set of 16 microarray sensing spots. The standard deviations (SD) were presented as error bars in the bar charts. P-values were calculated using the One-Way ANOVA,* P < 0.05, **P<0.01, ***P<0.0001 using Microsoft Office Excel.
Supplementary Material
Acknowledgement
This work was partially supported by the National Science Foundation (CBET 1701322 to W. C., CBET 1701363 to P.C.) and the National Institute of Health (R21EB025406 to W. C.). The authors would like to acknowledge Prof. Jun Cui and Dr. Xubo Liu for their help with the magnetism measurements.
Footnotes
Supporting Information
Supplementary information and figures S1-S6 on materials characterization, microarray fabrication and the microarray immunoassay.
References
- [1].Yager P, Edwards T, Fu E, Helton K, Nelson K, Tam MR, Weigl BH, Nature 2006, 442, 412. [DOI] [PubMed] [Google Scholar]
- [2].Masson J-F, ACS Sensors 2017, 2, 16. [DOI] [PubMed] [Google Scholar]
- [3].Aćimović SS, Ortega MA, Sanz V, Berthelot J, Garcia-Cordero JL, Renger J, Maerkl SJ, Kreuzer MP, Quidant R, Nano Letters 2014, 14, 2636. [DOI] [PubMed] [Google Scholar]
- [4].Dantham VR, Holler S, Barbre C, Keng D, Kolchenko V, Arnold S, Nano Letters 2013, 13, 3347. [DOI] [PubMed] [Google Scholar]
- [5].Yavas O, Aćimović SS, Garcia-Guirado J, Berthelot J, Dobosz P, Sanz V, Quidant R, ACS Sensors 2018, 3, 1376. [DOI] [PubMed] [Google Scholar]
- [6].Hornbeck PV, Current Protocols in Immunology 2015, 110, 2.1.1. [DOI] [PubMed] [Google Scholar]
- [7].Baaske MD, Foreman MR, Vollmer F, Nature Nanotechnology 2014, 9, 933. [DOI] [PubMed] [Google Scholar]
- [8].Vlasov YA, Bo X-Z, Sturm JC, Norris DJ, Nature 2001, 414, 289. [DOI] [PubMed] [Google Scholar]
- [9].Sun Y, Fan X, Analytical and Bioanalytical Chemistry 2011, 399, 205. [DOI] [PubMed] [Google Scholar]
- [10].Homola J, Chemical Reviews 2008, 108, 462. [DOI] [PubMed] [Google Scholar]
- [11].Choi HY, Park KS, Park SJ, Paek U-C, Lee BH, Choi ES, Opt. Lett. 2008, 33, 2455. [DOI] [PubMed] [Google Scholar]
- [12].Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H, Nature Biotechnology 2014, 32, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Eustis S, El-Sayed MA, Chemical Society Reviews 2006, 35, 209. [DOI] [PubMed] [Google Scholar]
- [14].Zijlstra P, Paulo PMR, Orrit M, Nature Nanotechnology 2012, 7, 379. [DOI] [PubMed] [Google Scholar]
- [15].Xu S, Ouyang W, Xie P, Lin Y, Qiu B, Lin Z, Chen G, Guo L, Analytical Chemistry 2017, 89, 1617. [DOI] [PubMed] [Google Scholar]
- [16].Siabi-Gaqan A, Savaloni H, The European Physical Journal B 2013, 86, 257. [Google Scholar]
- [17].Xu Z, Hou Y, Sun S, Journal of the American Chemical Society 2007, 129, 8698. [DOI] [PubMed] [Google Scholar]
- [18].Im H, Lee SH, Wittenberg NJ, Johnson TW, Lindquist NC, Nagpal P, Norris DJ, Oh S-H, ACS Nano 2011, 5, 6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sepúlveda B. Angelomé PC, Lechuga LM, Liz-Marzán LM, Nano Today 2009, 4, 244. [Google Scholar]
- [20] a).Song Y, Chen P, Chung MT, Nidetz R, Park Y, Liu Z, McHugh W, Cornell TT, Fu J, Kurabayashi K, Nano Letters 2017, 17, 2374; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhu J, He J, Verano M, Brimmo AT, Glia A, Qasaimeh MA, Chen P, Aleman JO, Chen W, Lab on a Chip 2018, DOI: 10.1039/C8LC00605A; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Oh B-R, Huang N-T, Chen W, Seo JH, Chen P, Cornell TT, Shanley TP, Fu J, Kurabayashi K, ACS Nano 2014, 8, 2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cetin AE, Coskun AF, Galarreta BC, Huang M, Herman D, Ozcan A, Altug H, Light: Science &Amp; Applications 2014, 3, e122. [Google Scholar]
- [22].Gates BD, Xu Q, Stewart M, Ryan D, Willson CG, Whitesides GM, Chemical Reviews 2005, 105, 1171. [DOI] [PubMed] [Google Scholar]
- [23].Chen P, Chung MT, McHugh W, Nidetz R, Li Y, Fu J, Cornell TT, Shanley TP, Kurabayashi K, ACS Nano 2015, 9, 4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Manfrinato VR, Zhang L, Su D, Duan H, Hobbs RG, Stach EA, Berggren KK, Nano Letters 2013, 13, 1555. [DOI] [PubMed] [Google Scholar]
- [25].Buckmann T, Stenger N, Kadic M, Kaschke J, Frolich A, Kennerknecht T, Eberl C, Thiel M, Wegener M, Advanced Materials 2012, 24, 2710. [DOI] [PubMed] [Google Scholar]
- [26].Claussen JC, Franklin AD, ul Haque A, Porterfield DM, Fisher TS, ACS Nano 2009, 3, 37. [DOI] [PubMed] [Google Scholar]
- [27].Salaita K, Wang Y, Mirkin CA, Nature Nanotechnology 2007, 2, 145. [DOI] [PubMed] [Google Scholar]
- [28].Romanov V, Davidoff SN, Miles AR, Grainger DW, Gale BK, Brooks BD, Analyst 2014, 139, 1303. [DOI] [PubMed] [Google Scholar]
- [29].Oh B-R, Chen P, Nidetz R, McHugh W, Fu J, Shanley TP, Cornell TT, Kurabayashi K, ACS Sensors 2016, 1, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xia Q, Fu S, Ren G, Chai F, Jiang J, Qu F, New Journal of Chemistry 2016, 40, 818. [Google Scholar]
- [31].Zhu Y, Lei J, Tian Y, Dalton Transactions 2014, 43, 7275. [DOI] [PubMed] [Google Scholar]
- [32].Jeong U, Teng X, Wang Y, Yang H, Xia Y, Advanced Materials 2007, 19, 33. [Google Scholar]
- [33].Khan SS, Smith MS, Reda D, Suffredini AF, McCoy JP, Cytometry B Clin Cytom 2004, 61. [DOI] [PubMed] [Google Scholar]
- [34].Jain PK, Huang X, El-Sayed IH, El-Sayed MA, Accounts of Chemical Research 2008, 41, 1578. [DOI] [PubMed] [Google Scholar]
- [35].Chen P, Huang N-T, Chung M-T, Cornell TT, Kurabayashi K, Advanced Drug Delivery Reviews 2015, 95, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36] a).Shimizu M, Nagashima H, Sano K, Hashimoto K, Ozeki M, Tsuda K, Hatta H, Bioscience, Biotechnology, and Biochemistry 1992, 56, 270; [DOI] [PubMed] [Google Scholar]; b) Vermeer AWP, Norde W, Biophysical Journal 2000, 78, 394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37] a).Wang Y, Yu J, Luo X, Wang X, Li M, Wang L, Cellular & molecular immunology 2010, 7; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Caso G, Barry C, Patejunas G, Journal of Hematology & Oncology 2009, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yang L, Zhang Y, Journal of Hematology & Oncology 2017, 10, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Goede V, Brogelli L, Ziche M, Augustin HG, International Journal of Cancer 1999, 82, 765. [DOI] [PubMed] [Google Scholar]
- [40].McClellan JL, Davis JM, Steiner JL, Enos RT, Jung SH, Carson JA, Pena MM, Carnevale KA, Berger FG, Murphy EA, American Journal of Physiology-Gastrointestinal and Liver Physiology 2012, 303, G1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.