Abstract
Measurement of distances between spectroscopic labels (e.g., spin labels, fluorophores) attached to specific sites of biomolecules is an important method for studying biomolecular complexes. ALLNOX (Addition of Labels and Linkers) has been developed as a program to model interlabel distances based on an input macromolecule structure. Here, we report validation of ALLNOX using measured distances between nitroxide spin labels attached to specific sites of a protein–DNA complex. The results demonstrate that ALLNOX predicts average interspin distances that matched with values measured with pairs of labels attached at the protein and/or DNA. This establishes a solid foundation for using spin labeling in conjunction with ALLNOX to investigate complexes without high-resolution structures. With its high degree of flexibility for the label or the target biomolecule, ALLNOX provides a useful tool for investigating the structure–function relationship in a large variety of biological molecules.
Graphical Abstract

INTRODUCTION
Site-directed spin labeling (SDSL) is a biophysical technique that provides unique information on the structure and dynamics of biomacromolecules.1–3 In SDSL, a spin label, which is a chemically stable radical, is attached to a specific site within a macromolecule. Information is derived by monitoring the behaviors of the spin label using electron paramagnetic resonance (EPR) spectroscopy. The most commonly used spin labels are nitroxide radicals,1–3 although others such as trityl radicals4 and EPR-active metal ions (e.g., Gd3+,5 Cu2+,6) have gained interest recently. The distance between a pair of spin labels can be measured by determining the dipolar interaction strength between the unpaired electrons.1–3 Interspin distances below 2 nm can be determined using continuous-wave (cw-) EPR,7 and with the development of pulsed EPR spectroscopy, the range has been expanded to beyond 10 nm.8,9 The measured distances provide direct structural constraints, which are highly informative in investigating structures of the target molecules and conformational changes in different functional states.
For the majority of spin labels used, the unpaired electron resides at a functional group that is not native to the target macromolecule, and the measured interspin distances differ from the corresponding distances between the points of label attachment. A successful approach for correlating measured interspin distances to a given conformation of the target macromolecule is to model the spin label at the desired sites of a target structure, computationally identify the allowable ensemble of conformers, compute the expected interspin distances, and then compare the expected distances to those measured experimentally. Examples of the first generation of these programs include NASNOX, which computes expected distances for a nitroxide (designated as R5, Figure 1A) attached to DNA or RNA,10,11 and MMM,12 PRONOX,13 and MtsslWizard,14 which compute interlabel distances for a nitroxide (designated as R1) attached on proteins. These programs have been validated by studies on target molecules with reported X-ray or NMR structures10 and have been successfully applied in a number of studies to investigate complexes with no reported high-resolution structures.15,16
Figure 1.
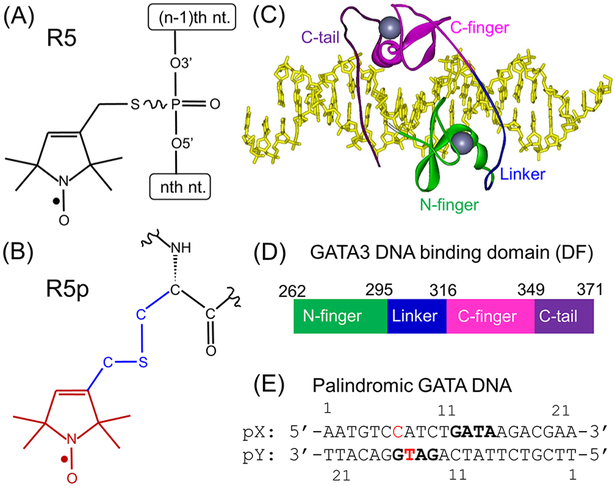
Constructs for spin-labeling studies on GATA3 protein/DNA complex. (A) Chemical structure of the R5 label on DNA. (B) Chemical structure of the R5p label on protein, with the “label” shown in red, “linker” in blue, and “target” in black. (C) A crystal structure (4HCA.pdb) with the “double-finger” fragment of GATA3 bound to a DNA with palindromic GATA sites. The DNA is shown as a stick, the protein is shown as a ribbon with the domains colored, and the two zinc ions are shown in gray. (D) Schematic of the doublefinger GATA3 DNA-binding domain fragment (i.e., “DF”). The two zinc-fingers and the adjacent basic regions are color-coded as in panel (C). The detailed protein sequence is shown in Figure S1. (E) pGATA DNA duplex. The “GATA” motifs are shown in bold, and the pX15 and pY15 nucleotides used for R5 attachment are shown in red.
These first generation programs can only handle one fixed label (i.e., R1 or R5) on a fixed type of biological molecule (i.e., protein or nucleic acids). To overcome these limitations, the ALLNOX (Addition of Labels and Linkers) program was developed17 to permit addition of any label to any site on any target biomolecule. ALLNOX implements a “label-linker-target” strategy to model a desired spectroscopic label at a specific site of a biomacromolecule. A user may upload a “label” moiety as a pdb file (e.g., pyrroline ring of a nitroxide, Figure 1B), define a “linker” (e.g., “–C–S–C–”, Figure 1B), and then specify the “target” for attachment (e.g., “Cα” of an amino acid, Figure 1B). The program models the label onto the target macromolecule structure (as specified by an input pdb file), generates rotamers of the label by stepwise variations of the rotatable torsional angles in the linker, identifies the rotamers without steric collision with the target macromolecule, and then outputs the interlabel distances. ALLNOX can examine a variety of labels (different spin labels, fluorophores, etc.) directly on protein and/or nucleic acids.17 Such flexibility is highly beneficial for evaluation of experimental data in biochemical complexes, especially those containing multiple molecular types.
In this work, we report validation of ALLNOX using EPR measured distances obtained with a protein–DNA complex with reported crystal structures (Figure 1C). The target macromolecule is a complex of a DNA duplex and the DNA-binding domain of GATA3, which is a member of the mammalian GATA family of transcription factor specifically recognizing DNAs with a consensus sequence of “GATA”.18,19 The DNA-binding domain of GATA3 contains two highly conserved zinc fingers, which are referred to as the N-terminal zinc finger (N-finger) and C-terminal zinc finger (C-finger) (Figure 1C,D). The C-finger and its adjacent basic region are necessary and sufficient for binding to the cognate consensus DNA sequence WGATAR (W = A/T, R = A/G).18,19 However, the N-finger has been shown to participate in interactions with protein partners but can also bind to DNA independently with a slightly different preferred sequence (i.e., GATC).20,21 On certain DNA sequences with two proximal GATA sites, the N-finger can participate in DNA binding together with the C-finger, leading to a greatly increased binding affinity.22,23
Members of the GATA family have been characterized by both X-ray crystallography and NMR.23–27 In particular, the Chen lab reported crystal structures of full GATA3 DNA binding domain (Figure 1C) containing both zinc fingers and the adjacent basic regions (Figure 1D) bound to DNAs containing composite GATA sites (Figure 1E).23 This provides an ideal case to test ALLNOX, as the complex contains different macromolecule types (protein and DNA) that require different labeling chemistry. In this report, we attached the R5 nitroxide label to DNA targets (Figure 1A), and the R5p nitroxide label to the protein fragment (Figure 1B). A pulsed EPR scheme, Double Electron Resonance (DEER)8, was employed to measure interspin distances between pairs of labels attached to protein, to DNA, and one at the protein and the other at the DNA. In parallel, ALLNOX was used to predict expected internitroxide distances using a crystal structure of a DNA-bound GATA3 fragment (pdb id 4HCA,23 Figure 1C). In all data sets examined, ALLNOX predicted average distances within 0.15 nm (1.5 Å) of the measured value. The results validate the accuracy of ALLNOX, thus establishing its utility for SDSL studies.
MATERIALS AND METHODS
Protein Mutagenesis, Expression, and Purification.
To introduce a cysteine for R5p attachment at a desired site, mutagenesis was carried out using vector pET-28a(kanamycin resistant) encoding the “double-finger” GATA3 fragment (designated as “DF”, Figure 1D), which included human GATA3 amino acids 260–371 fused with a 6X His-tag.23 Threonine 280 (N-finger) and isoleucine 362 (C-tail) were individually (i.e., constructs “T280” and “I362”, respectively) or simultaneously (i.e., construct “DM”) mutated to cysteine for subsequent spin label attachment. Site-directed mutagenesis was carried out using the Quikchange kit. Mutant plasmids with sequence verified were transformed into a BL21(DE3)-Gold competent cell for protein expression. The overexpressed protein was purified using affinity chromatography (i.e., Ni-NTA) followed by cation exchange chromatography. Details of mutagenesis, protein expression, and purification are described in the Supporting Information section S1.
Spin Labeling of Proteins.
The R5p label (Figure 1B) is covalently attached to a cysteine with a thioether linkage, which is stable in the presence of certain reducing agents [e.g., TCEP (tris(2-carboxyethyl)phosphine)] used to maintain activities of most nucleic acid-binding proteins.28 Each labeling reaction was carried out in a solution containing 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.5), 500 mM NaCl, 0.5 μM zinc acetate, 0.5 mM TCEP, 0.5 mM 3-iodomethyl-1-oxyl-2,2,5,5-tetramethylpyrroline (i.e., the “R5 precursor”, prepared as reported previously29,30], and 1050 μM protein. The reaction mixture was incubated with constant shaking at 4 °C in the dark overnight. After incubation, the unreacted precursor was removed using cation-exchange chromatography, and the purified labeled protein fractions were concentrated to desired volume. The efficiency of protein spin labeling was estimated using a spin counting procedure described previously31 and was found to be ~75% for the T280, I362, and DM constructs (Supporting Information, section S2.2).
Spin Labeling of DNAs.
The R5 label (Figure 1A) was attached to a DNA strand following protocols published previously.29,30 Briefly, all DNA oligonucleotides were synthesized by solid-phase chemical synthesis (Integrated DNA Technologies, Coralville, IA, USA), and the desired phosphate for spin labeling is chemically modified to a phosphorothioate in this step. The R5 precursor was then reacted with the phosphorothioate group in a solution containing ~0.5 mM crude DNA, 200 mM R5 precursor, 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) (pH 5.8), and 40% (v/v) acetonitrile. The reaction mixture was incubated in the dark at room temperature for 24 h under constant shaking. Labeled DNA was purified by anion-exchange high-pressure liquid chromatography, followed by desalting using a reversed-phase column. Desalted oligonucleotides were lyophilized, resuspended in water, and stored at −20 °C. The final concentrations of each DNA strand were determined by absorbance at 260 nm using extinction coefficients of pX, 228 300 M−1.cm−1; pY, 210 100 M−1.cm−1; sX, 201, 300 M−1. cm−1; and sY, 169 700 M−1.cm−1.
EPR Sample Assembly.
DNA duplexes were prepared following an annealing protocol as reported previously.29,32 Briefly, a duplex stock was prepared by annealing the R5-labeled strand with the complementary strand at a ratio of 1:1.1 or annealing two R5-labeled strands at a ratio of 1:1. After appropriate amounts of the respective strands were mixed, the mixture was heated at 95 °C for 1 min and then cooled at room temperature for 1 min. An appropriate amount of salt was then added to the mixture to reach a final concentration of 50 mM Tris–HCl (tris(hydroxymethyl)-aminomethane hydrochloride) (pH 7.5) and 100 mM NaCl. The solution was left standing at room temperature for >1 h to allow duplex formation.
To assemble a GATA3 DF/DNA complex, appropriate amounts of DNA duplex and protein were mixed in a high salt solution containing 50 mM HEPES, pH 7.5, 500 mM NaCl, 0.5 μM zinc acetate, 0.5 mM TCEP, with the protein concentration ranging from 10 to 50 μM and the total solution volume being >500 μL. The ratio of protein/DNA varied depending on the spin-labeled construct. The mixture was then dialyzed against a low salt solution containing 50 mM HEPES (pH 7.5), 200 mM NaCl, 0.5 μM zinc acetate and 0.5 mM TCEP at 4 °C overnight to allow DNA/protein binding. The dialyzed complex was concentrated to the desired volume with complex concentrations ranging from 200 to 300 μM.
DEER Spectroscopy.
Each final DEER sample contained 50 mM HEPES, pH 7.5, 200 mM NaCl, 0.5 μM zinc acetate, and 40% (v/v) glycerol. The total volume was at least 25 μL, with spin concentrations being ~200 μM. Samples were loaded into round glass capillaries (2.0 mm i.d. × 2.4 mm o.d., Vitrocom, Inc., Mountain Lakes, NJ) sealed at one end, flash-frozen in liquid nitrogen, and used immediately for DEER measurements.
DEER measurements were carried out at 78 K on a Bruker ELEXSYS E580 X-band spectrometer following previously reported procedure and parameters.15 Briefly, a dead time free four-pulse scheme8 was used, with the pump pulse frequency set at the center of the nitroxide spectrum and the observer frequency being approximately 70 MHz higher. The observer π pulse was 32 ns, and the pump π pulse was usually set at 16 ns. The accumulation time in each measurement ranged from 12 to 24 h with 512 shots per point. Interspin distances were computed from the resulting dipolar echo evolution data using the Tikhonov regularization module in DeerAnalysis2013.33 From the computed distance distribution, average distance (⟨Rdeer⟩) and width of distribution (σdeer) of a selected population were calculated as reported previously.10
ALLNOX Computation of Interspin Distances.
Interspin distances were computed using the web-based ALLNOX program.17 A reported GATA3/DNA complex crystal structure (pdb id 4HCA.pdb,23 Figure 1C) was formatted as described17 and used as the input structure. R5 labels attached to the DNA (Figure 1A) were modeled using the standard “R5-CS-nucP” label-linker-target assembly.17 R5p labels attached to the protein (Figure 1B) were modeled with a “Custom” label– linker–target assembly, with the “label selection” set as “R1(R5)”, the “target selection” set as “aaCα”, and the “linker” set as “CSC”.17 The “conformation mode” was set to “automatic”, with each rotatable torsion angle (three for R5 and four for R5p, respectively) varied among 60°, 180°, and 300°. The clash criterion “vdW” was set at the default value of 0.75.
The ALLNOX output file contains a list of accepted and rejected nitroxide rotamers, as well as the ensemble of individual distances between each pair of allowable rotamers.17 Using the output distances between the nitrogen atoms of the nitroxide (i.e., “distance N-N”) as the predicted distances, the average of the ensemble was computed as the predicted distance (⟨Rnox⟩), and the standard deviation of the ensemble was computed as the width of the distribution (σnox).
Note that the current version of ALLNOX program only supports modeling of one R5 diastereomer,17 specifically the R5 attached to the “OP1” atom of the target nucleotide specified in the user input pdb file. In this work a scheme was developed to account for the presence of two R5 diastereomers at a DNA labeling site (Figure 1A) for distances measured between two DNA sites or between protein and DNA. First, the properly formatted 4HCA.pdb file was used as the input structure to obtain the result representing the Sp-R5 diastereomer. Then, a modified pdb file was generated by switching the designations of “OP1” and “OP2” in the target nucleotide while keeping the coordinates of the atoms unchanged. This modified pdb file was used as the input structure to obtain the result representing the Rp-R5 diastereomer. Interspin distances obtained from the Sp-and Rp-R5 were then combined together as the ensemble of ALLNOX predicted distances.
RESULTS AND DISCUSSIONS
Validating ALLNOX Predictions Using a GATA3 Protein–DNA Complex Crystal Structure.
To validate ALLNOX, spin labeling measurements were carried out on a complex with a reported crystal structure (pdb id 4HCA.pdb,23 Figure 1C) that has the GATA3 double-finger fragment (i.e., “DF”, Figure 1D) bound to a DNA duplex containing palindromic GATA recognition sites separated by one base-pair (pGATA, with individual strands designated respectively as pX and pY) (Figure 1E). Four labeling sites were investigated in this work: T280 (N-finger) and I362 (C-tail) on the protein (Figures 1D and S1) and pX15 and pY15 on the DNA (Figure 1E). On the basis of the reported 4HCA crystal structure,23 these sites are solvent accessible and located away from the protein/DNA interface and therefore allow efficient spin labeling and minimize potential disruption of the protein– DNA interactions. Indeed, control studies using gel shift assays (Supporting Information, section S2.1) and continuous-wave EPR spectroscopy (Supporting Information, section S2.2) both confirmed that mutagenesis and spin-labeling did not interfere with formation of the protein–DNA complex, indicating that spin labeling did not significantly perturb folding and function of the GATA3 DF fragment.
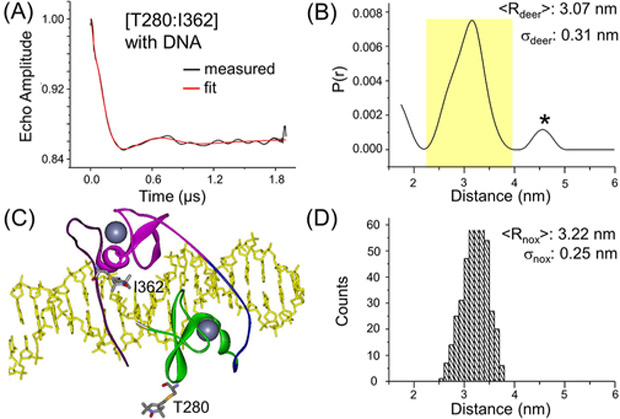
A complex was first assembled using unlabeled pGATA DNA duplex and double-labeled GATA3 fragment, with R5p attached to T280 and I362. The interspin distance was measured using DEER, with the observed echo decay profile clearly showing oscillations (Figure 2A). Analyses of the echo decay profile using the Tikhonov regularization approach yielded a distance distribution profile with the major population having an average distance ⟨Rdeer⟩ = 3.07 nm, with a width of distribution σdeer = 0.31 nm (Figure 2B). Using the reported 4HCA crystal structure, R5p labels were modeled at T280 and I362, and the sterically allowed conformers were identified (Figure 2C). The analyses gave an averaged distance ⟨Rnox⟩ = 3.22 nm and a width of distribution (σnox) of 0.25 nm (Figures 2D). The ⟨Rnox⟩ value differs from ⟨Rdeer⟩ by only 0.15 nm (Table 1), which is below the 0.3 nm (3 Å) range of variation in the measured and predicted averaged distance.17 The predicted width of distribution is also comparable with the measured distribution (Table 2). As such, the T280–I362 measurement indicates that ALLNOX accurately models the distance between a pair of R5p labels attached to the protein within a protein/DNA complex. In ALLNOX, the protein prediction module is derived from the previously developed PRONOX program.13 For the R1 nitroxide label (linker “C– S–S–Cβ”) used most frequently for proteins,1 ALLNOX and PRONOX give similar results: Control studies showed that for the T280–I362 pair, the predicted inter-R1 distance is 3.28 nm with ALLNOX and 3.26 nm with PRONOX. However, R1 cannot be used in the present experiments as the presence of the reducing agent breaks the “S–S” linker, while PRONOX cannot model the R5p label (linker C–S–Cβ, Figure 1B) because it only accepts R1. ALLNOX allows custom label definition, thus overcoming this limitation in PRONOX.
Figure 2.
Assessment of the interspin distance between a pair of R5p attached at T280 and I362 of the protein. (A) DEER measured background corrected echo evolution data on the DF/pGATA complex assembled with a protein/DNA ratio of 1:1.1. (B) Distance distribution profile computed from data shown in (A). The major population is shaded in yellow, and “*” indicates a minor population deemed “significant” on the basis of DEERconstruct analyses (see Supporting Information section S3). (C) ALLNOX generated structure of the spin-labeled DF/pGATA complex. The structure is depicted with the same scheme as described in Figure 1C. At the T280 and I362 site, one of the allowable R5p conformers is shown in stick form. (D) Histogram of ALLNOX predicted distance distribution.
Table 1.
Comparison of Averaged Distances
| data set | DEER Measureda (nm) |
ALLNOX (nm) |
difference (nm) |
|
|---|---|---|---|---|
| protein–protein | T280 – I362 | 3.07 | 3.22 | −0.15 |
| DNA–DNA | pX15–pY15 | 2.80 | 2.74 | 0.06 |
| protein–DNA | I362–pX15 | 2.90 | 2.95 | −0.05 |
| I362–pY15 | 3.60 | 3.64 | −0.04 | |
| T280–pY15 | 3.32 | 3.23 | 0.09 | |
Major population in the DEER measured distance distribution profile.
Table 2.
Comparison of the Widths of Distance Distributions
| data set | DEER measureda (nm) |
ALLNOX (nm) |
|
|---|---|---|---|
| protein–protein | T280–I362 | 0.31 | 0.25 |
| DNA–DNA | pX15–pY15 | 0.35 | 0.25 |
| protein–DNA | I362–pX15 | 0.28 | 0.20 |
| I362–pY15 | 0.28 | 0.25 | |
| T280–pY15 | 0.24 | 0.34 | |
Major population in the DEER measured distance distribution profile.
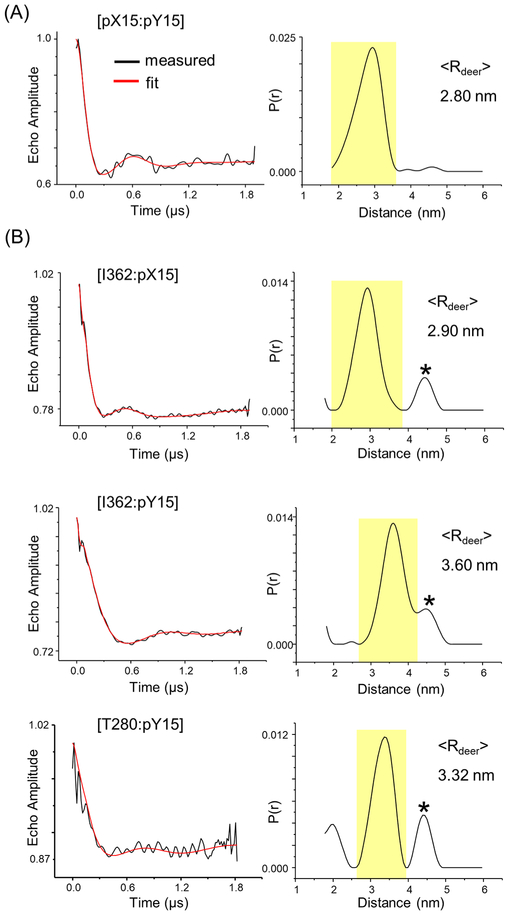
In addition to the protein module, ALLNOX has a module to handle labels attached to nucleic acids, which was built upon the previously validated NASNOX program.10,11 To test the nucleic acid prediction functionality in ALLNOX, we attached a pair of R5 labels at the DNA (pX15 and pY15, Figure 1E), and measured the interspin distance within the DF/pGATA complex. The DEER measurementd yield a major population with ⟨Rdeer⟩ = 2.80 nm (Figure 3A). ALLNOX gave a predicted distance ⟨Rnox⟩ = 2.74 nm (Table 1). The difference of 0.06 nm is well within the 0.3 nm range of variability, and indicates that ALLNOX correctly predicted the DNA–DNA distance.
Figure 3.
DEER data obtained in the DF/pGATA complex. (A) Pair of R5 labels at DNA pX15 and pY15 sites. To ensure all spin-labeled DNA was bound, the complex was assembled with a protein/DNA ratio of 1.1:1. (B) A R5p label at the protein and a R5 label at the DNA. Complexes were assembled with a protein/DNA ratio of 1:1. In each panel, on the left is the background correct echo evolution (black) with the fit trace (red) overlaid and on the right is the distance distribution profile with the major population shaded in yellow. “*” indicates a minor population deemed “significant” on the basis of DEERconstruct analyses (see Supporting Information section S3).
A critical step forward in the ALLNOX program is to predict interspin distances between protein and DNA, which is beyond the ability of either PRONOX or NASNOX. To test this functionality, we measured distances in the DF/pGATA complex with a R5p label attached at the protein fragment and a R5 label attached at the DNA duplex. Three DEER data sets were obtained, I362–pX15, I362–pY15, and T280–pY15, with the major populations giving ⟨Rdeer⟩ of 2.90, 3.60, and 3.32 nm, respectively (Figure 3B). The corresponding ALLNOX predicted ⟨Rnox⟩ values were 2.95, 3.64, and 3.23 nm, respectively (Table 1). The differences between ⟨Rdeer⟩ and ⟨Rnox⟩ range between 0.03 and 0.09 nm, well within the 0.3 nm range of variation. In addition, the predicted widths of distribution (σnox) are comparable with the measured distribution (Table 2). As such, the analyses demonstrated that ALLNOX is able to accurately predict interspin distances between protein and DNA.
We also note that the measured distance distribution profiles presented multiple populations (Figures 2 and 3). For each data set, the “major” population with the highest occupancy (i.e., highest probability) correlated nicely with the values predicted by ALLNOX (Table 1). Additional analyses were carried out on the minor population(s) using the DEERcon-struct program developed by the Fanucci group34 to determine whether the population was “insignificant” (i.e., representing artifacts arising from DEER data analyses as constrained by the experimental setup and signal-to-noise ratio) or “significant” (i.e., representing additional distances presented in the sample) (Supporting Information, section S3). The DEERconstruct analyses indicated that the minor populations presented in the protein–protein measurement (Figure 2) and protein–DNA measurements (Figure 3B) were “significant” (Supporting Information, Table S1), indicating additional distances in the samples measured. These minor distance populations likely represent conformations of the DF/pGATA complex that differ from the 4HCA crystal structure, although the details of these conformations are not known, and it is not clear whether these conformations are related to other DF structures obtained in crystallography studies.23
GATA 3 C-Finger Binds to a Single DNA Site in a Similar Fashion to That with pGATA.
As an application of ALLNOX, we examined the solution conformation of the DF fragment bound to a DNA duplex containing only one “GATA” sequence (designated as sGATA, Figure 4A). Previous studies indicate that the GATA3 C-finger prefers a WGATAR (W = A/T; R = A/G) sequence while the N-finger favors GATC.19,21 Thus, it is expected that the sGATA construct will engage the C-finger of the DF fragment. In addition, multiple crystal or NMR structures have been published on the local binding mode between C-finger and GATA DNA.23– 27 Although the DNA sequence and the protein construct used in those structures vary, the interface remains highly conserved. As such, one would expect that the conformation of C-finger bound to one GATA site would be highly similar to that observed in the crystal structures of the double-finger protein.
Figure 4.
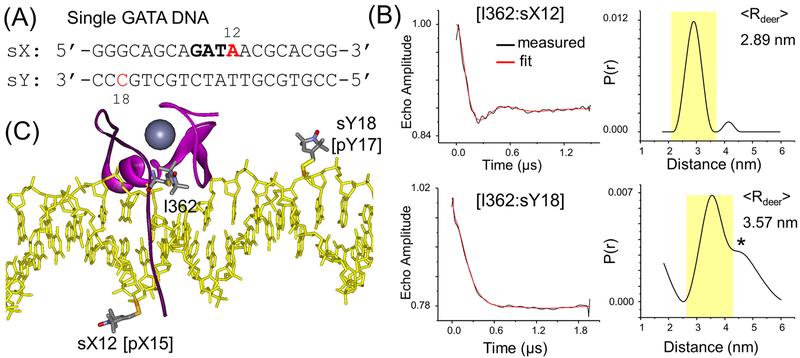
GATA3 double-finger fragment binding to a DNA with a single GATA site. (A) sGATA DNA duplex. The “GATA” motif is in bold, and the R5 labeling sites are in red. (B) DEER result on I362–sX12 (top) and I362-sY18 (bottom). The complexes were assembled with a protein/DNA ratio of 1:1. In each data set, on the left is the background correct echo evolution (black) with the fit trace (red) overlaid and on the right is the distance distribution profile with the major population shaded in yellow. “*” indicates a minor population deemed “significant” on the basis of DEERconstruct analyses (see Supporting Information section S3). (C) ALLNOX generated spin-labeled complex structure. The structure is depicted with the same scheme as described in Figure 1C. One each of the allowable spin label conformers is shown at I362, sX12, and sY18. To mimic C-finger binding to one GATA site, the N-finger and the linker of GATA3 are not shown.
Two sites at sGATA were labeled, one at a time, with R5 (sX12 and sY18, Figure 4A). Complexes were assembled with the single R5-labeled DNA and a DF fragment with an R5p attached at I362, and the interspin interaction was measured by DEER. The data set for I362–sX12 showed a clear oscillation in the echo evolution trace, and the resulting distance distribution showed one dominant distance population with ⟨Rdeer⟩ = 2.89 nm (Figure 4B). sX12, which is located between T and A on the GATA site, is the equivalent site of pX15 in the 4HCA crystal structure (Figures 1E and 4C). This measured I362-sX12 distance is the same as that of I362–pX15 measured with the pGATA DNA (Figure 3B), and differs by 0.06 nm from the ALLNOX predicted value obtained from equivalent sites within the 4HCA crystal structure (“I362–pX15”, 2.95 nm, Table 1). This supports the notion that the C-finger binds to the single GATA construct with a similar conformation as that observed in the crystal structure of the complex with palindromic GATA sites.
A second DEER data set was obtained between I362 and sY18, which yielded a major population with ⟨Rdeer⟩ = 3.57 nm (Figure 4B). sY18, located eight base-pairs away from sX12 on the complementary strand, is equivalent to pY17 in the 4HCA crystal structure (Figures 1E and 4C). Using the 4HCA structure as input, ALLNOX predicted ⟨Rnox⟩ = 3.66 nm for I362–sY18. This very nice agreement between ⟨Rdeer⟩ and ⟨Rnox⟩ further supports the conclusion that the C-finger binds with a similar conformation to DNA containing single or composite GATA sites.
We also note that crystal structures have been reported that show the C-finger of the DF fragment binds to a GATA site at one DNA while the N-finger binds to another site at a separate DNA.23 In the sGATA DNA experiments, the DNA:protein ratio was 1:1, and the C-finger is expected to bind to the single GATA site, consequently excluding the N-finger from specific binding to the DNA. DEER measurements in complexes assembled with R5p labeled at T280 of the DF fragment (located at the N-finger) and R5-labeled sGATA DNA revealed dipolar interactions between the spin labels, but the resulting distance distribution profile did not reveal a well-defined distance population (Supporting Information, section S4). This indicates that the N-finger is located in the proximity of the sGATA DNA but does not engage the DNA in a specific conformation.
CONCLUSIONS
Studies of the GATA3 protein/DNA complex demonstrated that ALLNOX predicts average interspin distances that match with values measured between sites on protein, on DNA, and between protein and DNA. This establishes a solid foundation for using SDSL in conjunction with ALLNOX to investigate complexes without high-resolution structures, such as that of the double-finger GATA3 protein bound to a single GATA site. This is highly beneficial for SDSL studies on structure– function relationships in protein–nucleic acid complexes, such as the CRISPR–Cas systems being developed for genome editing.28,35 Furthermore, while work reported here focused on spin labeling, ALLNOX allows great flexibility for addressing various labels, linkers, and targets and should be valuable in integrative structural modeling based on information derived from spectroscopic labels (e.g., fluorophores, spin labels).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Lin Chen for advice on the GATA3 system and Brian Sutch and Jaime Avila for assistance on the web-based ALLNOX modeling. Work reported was supported in part by grants from NSF (CHE-1213673; MCB-1818107; P.Z.Q.) and NIH (R01-GM124413; P.Z.Q.).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jpca.9b01027.
Additional descriptions on procedure for protein expression and purification, characterization of protein-DNA binding, analysis of distribution profiles, and measurement on complexes with a single GATA site (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Hubbell WL; Altenbach C Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Curr. Opin. Struct. Biol 1994, 4, 566–573. [Google Scholar]
- (2).Ding Y; Nguyen P; Tangprasertchai NS; Reyes CV; Zhang X; Qin PZ Nucleic acid structure and dynamics: perspectives from site-directed spin labeling In Electron Paramagnetic Resonance; The Royal Society of Chemistry, 2015; Vol. 24, pp 122–147. [Google Scholar]
- (3).Sahu ID; Lorigan GA Site-directed spin labeling EPR for studying membrane proteins. BioMed Res. Int 2018, 2018, 3248289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Bagryanskaya EG; Krumkacheva OA; Fedin MV; Marque SRA Development and application of spin traps, spin probes, and spin labels In Methods Enzymol; Qin PZ, Warncke K, Eds.; Academic Press, 2015; Vol. 563, pp 365–396. [DOI] [PubMed] [Google Scholar]
- (5).Feintuch A; Otting G; Goldfarb D Gd3+ spin labeling for measuring distances in biomacromolecules: why and how? In Methods Enzymol; Qin PZ, Warncke K, Eds.; Academic Press, 2015; Vol. 563, pp 415–457. [DOI] [PubMed] [Google Scholar]
- (6).Yang Z; Ji M; Cunningham TF; Saxena S Cu2+ as an ESR probe of protein structure and function In Methods Enzymol; Qin PZ, Warncke K, Eds.; Academic Press, 2015; Vol. 563, pp 459–481. [DOI] [PubMed] [Google Scholar]
- (7).Altenbach C; Oh KJ; Trabanino RJ; Hideg K; Hubbell WL Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry 2001, 40, 15471–82. [DOI] [PubMed] [Google Scholar]
- (8).Pannier M; Veit S; Godt A; Jeschke G; Spiess HW Deadtime free measurement of dipole-dipole interactions between electron spins. J. Magn. Reson. 2000, 142, 331–40. [DOI] [PubMed] [Google Scholar]
- (9).Schmidt T; Walti MA; Baber JL; Hustedt EJ; Clore GM Long distance measurements up to 160 A in the GroEL tetradecamer using Q-band DEER EPR spectroscopy. Angew. Chem 2016, 128, 16137–16141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Cai Q; Kusnetzow AK; Hubbell WL; Haworth IS; Gacho GP; Van Eps N; Hideg K; Chambers EJ; Qin PZ Site-directed spin labeling measurements of nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nucleic Acids Res 2006, 34, 4722–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Price EA; Sutch BT; Cai Q; Qin PZ; Haworth IS Computation of nitroxide-nitroxide distances for spin-labeled DNA duplexes. Biopolymers 2007, 87, 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Polyhach Y; Bordignon E; Jeschke G Rotamer libraries of spin labelled cysteines for protein studies. Phys. Chem. Chem. Phys 2011, 13, 2356–2366. [DOI] [PubMed] [Google Scholar]
- (13).Hatmal M. m. M.; Li Y; Hegde BG; Hegde PB; Jao CC; Langen R; Haworth IS Computer modeling of nitroxide spin labels on proteins. Biopolymers 2012, 97, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hagelueken G; Ward R; Naismith JH; Schiemann O MtsslWizard: In Silico spin-labeling and generation of distance distributions in PyMOL. Appl Magn. Reson. 2012, 42, 377–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Zhang X; Tung C-S; Sowa GZ; Hatmal M. m. M.; Haworth IS; Qin PZ Global structure of a three-way junction in a phi29 packaging RNA dimer determined using site-directed spin labeling. J. Am. Chem. Soc 2012, 134, 2644–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Zhang X; Dantas Machado AC; Ding Y; Chen Y; Lu Y; Duan Y; Tham KW; Chen L; Rohs R; Qin PZ Conformations of p53 response elements in solution deduced using site-directed spin labeling and Monte Carlo sampling. Nucleic Acids Res 2014, 42, 2789–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Beasley KN; Sutch BT; Hatmal M. m. M.; Langen R; Qin PZ; Haworth IS, Computer modeling of spin labels: NASNOX, PRONOX, and ALLNOX In Methods Enzymol; Qin PZ, Warncke K, Eds.; Academic Press, 2015; Vol. 563, pp 569–593. [DOI] [PubMed] [Google Scholar]
- (18).Ko LJ; Engel JD DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol 1993, 13, 4011–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Merika M; Orkin SH DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol 1993, 13, 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pedone PV; Omichinski JG; Nony P; Trainor C; Gronenborn AM; Clore GM; Felsenfeld G The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J. 1997, 16, 2874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Newton A; Mackay J; Crossley M The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J. Biol. Chem 2001, 276, 35794–801. [DOI] [PubMed] [Google Scholar]
- (22).Trainor CD; Omichinski JG; Vandergon TL; Gronenborn AM; Clore GM; Felsenfeld G A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol. Cell. Biol 1996, 16, 2238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Chen Y; Bates DL; Dey R; Chen PH; Machado AC; Laird-Offringa IA; Rohs R; Chen L DNA binding by GATA transcription factor suggests mechanisms of DNA looping and long-range gene regulation. Cell Rep 2012, 2, 1197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Bates DL; Chen Y; Kim G; Guo L; Chen L Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J. Mol. Biol 2008, 381, 1292–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Omichinski JG; Clore GM; Schaad O; Felsenfeld G; Trainor C; Appella E; Stahl SJ; Gronenborn AM NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science 1993, 261, 438–46. [DOI] [PubMed] [Google Scholar]
- (26).Starich MR; Wikstrom M; Arst HN Jr.; Clore GM; Gronenborn AM The solution structure of a fungal AREA protein-DNA complex: an alternative binding mode for the basic carboxyl tail of GATA factors. J. Mol. Biol 1998, 277, 605–20. [DOI] [PubMed] [Google Scholar]
- (27).Liew CK; Simpson RJ; Kwan AH; Crofts LA; Loughlin FE; Matthews JM; Crossley M; Mackay JP Zinc fingers as protein recognition motifs: structural basis for the GATA-1/friend of GATA interaction. Proc. Natl. Acad. Sci U. S. A 2005, 102, 583–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Vazquez Reyes C; Tangprasertchai NS; Yogesha SD; Nguyen RH; Zhang X; Rajan R; Qin PZ Nucleic acid-dependent conformational changes in CRISPR-Cas9 revealed by site-directed spin labeling. Cell Biochem. Biophys 2017, 75, 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Qin PZ; Haworth IS; Cai Q; Kusnetzow AK; Grant GP; Price EA; Sowa GZ; Popova A; Herreros B; He H Measuring nanometer distances in nucleic acids using a sequence-independent nitroxide probe. Nat. Protoc 2007, 2, 2354–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tangprasertchai NS; Zhang X; Ding Y; Tham K; Rohs R; Haworth IS; Qin PZ, An integrated spin-labeling/computational-modeling approach for mapping global structures of nucleic acids In Methods Enzymol; Qin PZ, Warncke K, Eds.; Academic Press, 2015; Vol. 564, pp 427–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Zhang X; Cekan P; Sigurdsson ST; Qin PZ Studying RNA using site-directed spin-labeling and continuous-wave electron paramagnetic resonance spectroscopy In Methods Enzymol; Herschlag D, Ed.; Academic Press, 2009; Vol. 469, pp 303–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Ding Y; Zhang X; Tham KW; Qin PZ Experimental mapping of DNA duplex shape enabled by global lineshape analyses of a nucleotide-independent nitroxide probe. Nucleic Acids Res 2014, 42, No. e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Jeschke G; Chechik V; Ionita P; Godt A; Zimmermann H; Banham J; Timmel CR; Hilger D; Jung H DeerAnalysis2006 - a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 2006, 30, 473–498. [Google Scholar]
- (34).Casey TM; Fanucci GE Spin labeling and Double Electron-Electron Resonance (DEER) to deconstruct conformational ensembles of HIV protease In Methods Enzymol; Qin PZ, Warncke K, Eds.; Academic Press, 2015; Vol. 564, pp 153–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tangprasertchai NS; Di Felice R; Zhang X; Slaymaker IM; Vazquez Reyes C; Jiang W; Rohs R; Qin PZ CRISPR-Cas9 mediated DNA unwinding detected using site-directed spin labeling. ACS Chem. Biol 2017, 12, 1489–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.