Abstract
External photocontrol over RNA function has emerged as a useful tool for studying nucleic acid biology. Most current methods rely on fully synthetic nucleic acids with photocaged nucleobases, limiting application to relatively short synthetic RNAs. Here we report a method to gain photocontrol over RNA by postsynthetic acylation of 2′-hydroxyls with photoprotecting groups. One-step introduction of these groups efficiently blocks hybridization, which is restored after light exposure. Polyacylation (termed cloaking) enables control over a hammerhead ribozyme, illustrating optical control of RNA catalytic function. Use of the new approach on a transcribed 237 nt RNA aptamer demonstrates the utility of this method to switch on RNA folding in a cellular context, and underlines the potential for application in biological studies.
RNA is a highly versatile biological macromolecule, carrying out a diverse range of cellular functions including gene expression, catalysis, and cell signaling among other functions.1In recent years, research has uncovered many additional properties and applications of RNA,2 including modulation of signaling pathways by circular RNAs,3 RNA epigenetics,2b and RNA nanobiology.4 Due to the complexity of RNA biology, there is a high demand for chemical tools to study functions and properties of RNA.5
One important way to gather insight into RNA’s biological role is by exerting external control over its function,6 allowing researchers to study downstream effects. External control can be obtained by introducing labels that perturb its natural state and function and that can be removed on demand. This has been achieved in multiple laboratories by introducing photoreleasable protecting groups (PPGs or photocaging groups) into RNA that can be removed by light exposure.7 For example, time-resolved RNA folding kinetics were analyzed in detail by employing an RNA strand that contained photoprotected guanosine,8 and embryonic development has been studied using RNAs with photocleavable backbones.9
The chief method for obtaining caged RNA is by use of phosphoramidite reagents that are incorporated by solid-phase synthesis.7b,c,k,m,10 While useful in the hands of chemists, synthesis of the modified nucleotides is specialized, and requires DNA synthesizers for their incorporation into RNA; thus nonspecialist laboratories often cannot access this technology easily or cheaply. Further, limitations of solid-phase synthesis require that the PPGs be applied in relatively short RNAs, precluding their use with most biological RNAs, which are commonly much longer. One approach to overcoming this limitation involves diazoketones that react with RNA phosphodiesters.11 However, the resulting phosphotriester adducts can hydrolyze to cause RNA strand cleavage.12
To address these obstacles, we set out to develop a general and accessible method that can be applied postsynthetically to synthetic and native RNAs regardless of length. Inspired by previous studies showing that 2′-OH groups of RNA can be selectively acylated in aqueous buffers with activated acyl compounds in structure mapping experiments,13,14 we imagined a strategy (Figure 1) in which PPGs are reacted with 2′ hydroxyls postsynthetically to block structure and interactions, but are rendered photoresponsive by including photocleavable bonds. Addition of several such blocking groups to the RNA (“cloaking”) would in principle cover and protect it from folding and interacting with other molecules. Subsequently, the acylation could be reversed by exposing RNA to light, removing the blocking groups and switching on activity.
Figure 1.
Modular design of photocloaking agents and cloaking strategy. (A) Schematic of the photocloaking strategy; polyacylation of 2′-OH groups in an existing RNA blocks RNA structure and function, while photoexposure (365 nm) removes these blocking groups. (B) Structures of PCA reagents 1 and 2. (C) Gel electrophoretic analysis of untreated, cloaked, and uncloaked RNA.
Design of our photocloaking agents (PCAs) began with the knowledge that the 2′-OH groups of RNA exhibit relatively high nucleophilicity due to their low pKa.13,14 For reaction with these groups, we envisioned the combination of an active acyl group with an o-nitroveratryl photoreactive group15 (Figure 1). We chose unmethylated and methylated veratryl groups to test (PCAs 1 and 2) (Figure 1B); PCA 1 is less sterically hindered, while 2 is expected to have higher efficiency in photocleavage.16We also included a dimethylaminoethyl group to enhance solubility. Finally, testing a range of leaving groups (imidazoles and triazoles (see Supporting Information (SI)), led to the choice of 2-chloroimidazole as having the ideal level of reactivity.
To test if prototypical PCA reagent 1 is effective in reacting with RNA, it was incubated for 4 h at 100 mM at room temperature with a 12 nucleotide (nt) RNA strand (10 μM) in water (see SI). Analysis by denaturing polyacrylamide gel electrophoresis (PAGE) revealed up to five new mobility-shifted bands, suggestive of several adducts per strand, with little unreacted RNA remaining (Figure 1C). The polyacylation was further confirmed by mass spectrometry (Figures S1–S3), although fewer labels were observed, likely due to photo-deprotection caused by the UV laser during the mass spectrometric analysis.
Next, uncloaking of RNA by photoremoval of the PCA labels was investigated. Photocloaked RNA in water (1.0 μM) was exposed to 365 nm light with a UV transilluminator and analyzed with denaturing PAGE gels. The results showed that the majority of shifted bands resolved back to a band having the mobility of untreated RNA. Successful photo-uncloaking was subsequently confirmed by MALDI-TOF, which documented restoration of the mass of the original RNA (Figure S3). As a control, the analogous DNA strand was incubated with PCA 1, and little (<5%) reaction was observed (Figure S4), supporting the conclusion that PCA 1 reacts with the 2′-OH groups of RNA.
Encouraged by the initial results, we next assessed how these multiple carbonate groups affect RNA helix formation and interactions. A16 nt single-stranded RNA (untreated or cloaked) was hybridized with a DNA complement and analyzed by native gel electrophoresis and UV melting (Figures S5 and S6). The gel analysis confirmed near-complete blocking of hybridization, and thermal denaturation showed a 10 °C drop in melting temperature after polyacylation of the RNA (Figure S5). To further test the effect on hybridization, a nucleic-acid-templated reaction was employed with complementary fluorogenic DNA (Q-STAR) probes.17 Figure 2B shows that the cloaked RNA was very inefficient in templating the fluorogenic reaction, suppressing signal to 9% of untreated RNA (Figure 2). After exposure to light, a strong restoration of signal (82% of original) was observed (Figure 2B,C), consistent with uncloaking at relatively high efficiency.
Figure 2.
Control of RNA hybridization by photocloaking, as measured by nucleic-acid-templated probes. (A) Schematic of the RNA-templated Q-STAR reaction. (B) Time course of fluorescence when RNA template has been treated with DMSO (untreated), PCA 1 (cloaked), and exposed to light after PCA 1 treatment (uncloaked). The templated reaction is severely inhibited upon cloaking and restored after photo-uncloaking. (C) Total fluorescence of Q-STAR reactions incubated for 1 h at 25 °C with RNA template 1 that was untreated, cloaked, and uncloaked. Error bars represent the standard deviation of triplicate experiments.
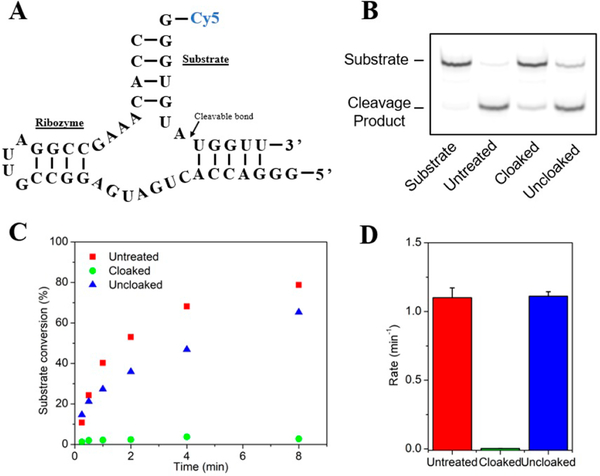
Next, we explored the ability of this photocloaking strategy to control the more complex function of a ribozyme RNA, which must not only hybridize, but also bind metals and form proper tertiary structure to function.18 Several examples of photocontrolled ribozymes have been reported previously;7e,19 however, all prior studies have been performed with modifications incorporated into RNAs during solid-phase synthesis. Thus, we tested whether PCA 1 could be used to block a ribozyme in a single postsynthetic step, and subsequently phototrigger its activity. A 34 nt hammerhead ribozyme RNA19a was cloaked under standard conditions at room temperature and isolated by precipitation, and was then incubated with Cy5-labeled substrate in 50 mM Tris pH = 6.0, 10 mM MgCl2 and analyzed by PAGE (Figure 3B and C). Control ribozyme showed almost full cleavage of the substrate under these conditions as expected, but the cloaked ribozyme exhibited strong abrogation of activity. Importantly, after light exposure, ribozyme function was almost completely restored. Analysis of ribozyme reaction rates (Figure 3D and Figure S9) revealed that cloaking suppressed the rate by 370-fold, showing strong blocking of ribozyme function. This was readily restored in RNA exposed to light, which exhibited a rate the same as that of the untreated ribozyme.
Figure 3.
Use of photocloaking to control hammerhead ribozyme activity. (A) Sequence of the ribozyme and Cy-5 labeled substrate. (B) Denaturing PAGE analysis of substrate conversion (1 h) after incubation with untreated, cloaked and photo-uncloaked ribozyme. (C) Time course of substrate conversion when incubated with the differently treated ribozymes.(D) Plot showing initial rates of substrate cleavage. Error bars represent the standard deviation of triplicate experiments.
We proceeded to test the ability of PCAs to control longer RNAs produced by transcription. Photocontrolled aptamers have recently gained interest due to their potential for therapeutic and imaging applications.20 We hypothesized that the current postsynthetic polyacylation strategy would offer a convenient and simpler method for preparing photocontrolled aptamers of arbitrary length. To test this, the transcribed 105 nt F30 Broccoli aptamer21 was chosen as a substrate. Broccoli RNA folds into a compact tertiary structure, forming a binding site for the DFHBI dye, resulting in a strong increase in fluorescence.21After cloaking with PCA 1, initial studies revealed strong suppression of fluorescence in Broccoli RNA (Figure S10), but a relatively poor recovery after light exposure. Our hypothesis was that this poor recovery was due to low quantum yields for uncaging of nitrobenzyl carbonates,16 which may be problematic on longer RNAs containing greater numbers of acyl groups. To address this, we prepared PCA 2 (Figure 1B), which bears a methyl group at the benzylic position to improve photocleavage efficiency.16
F30 Broccoli RNA (4 μg) was incubated with PCA 2 (100 mM) in water for 4 h and then isolated by precipitation (Figure S11). Untreated and cloaked Broccoli (500 nM) were dissolved in folding buffer and DFHBI (1 μM) was added and the aptamer was refolded. The cloaked sample exhibited very dim fluorescence, whereas the untreated sample showed pronounced emission (Figure 4). When the cloaked aptamer was exposed to light and incubated with DFHBI, fluorescence was completely restored to the level of the untreated sample. These experiments emphasized the strength of the photocloaking approach, which is carried out in a single step on RNAs considerably longer than are conveniently synthesized by chemical methods. Based on these results, we conclude that both PCA1 and PCA2 can be used to gain photocontrol of short RNAs, and for longer strands PCA2 is more efficient.
Figure 4.
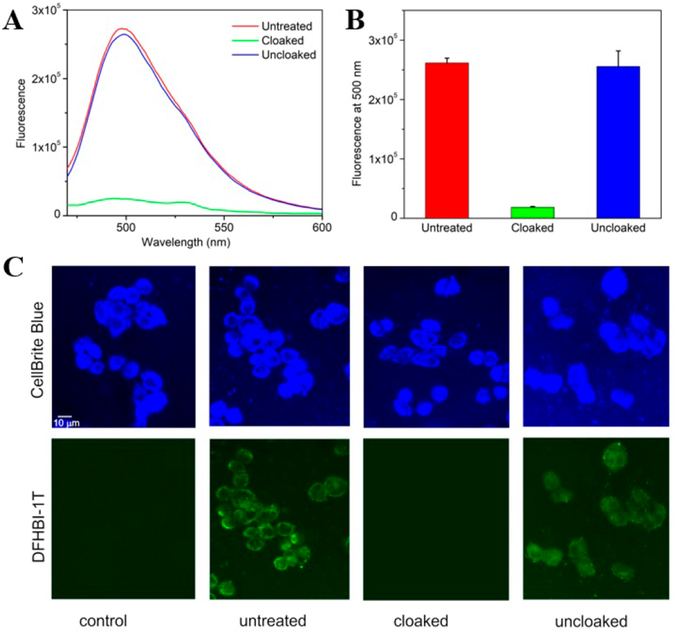
Cloaking effects on 105–237 nt Broccoli aptamer RNAs prepared by in vitro transcription. (A) Fluorescence emission spectra of untreated, cloaked and uncloaked Broccoli 105mer incubated with DFHBI. (B) Emission of differently treated Broccoli aptamers with DFHBI. The PCA 2 cloaked aptamer exhibits a substantially decreased fluorescence which is fully restored after light exposure. Error bars represent the standard deviation of triplicate experiments. λex = 450 nm. (C) Epifluorescence microscopy images of HeLa cells transfected with untreated or cloaked 237 nt Broccoli aptamer.
Finally, the capability to use this photocloaking strategy to switch on RNA function in cells was assessed by transfecting a cloaked Broccoli RNA construct into human cells and then exposing cells to light. For these experiments, we employed a larger 237 nt RNA construct encoding dimeric Broccoli aptamers, which is known to be stable in the intracellular medium.22 In vitro cloaking of this longer RNA was carried out with PCA 2 under standard conditions. The RNA was then transfected into HeLa cells, and after incubating for 5 h, cells were illuminated with 365 nm light under an epifluorescence microscope. As a control, we used the same RNA mock-treated with DMSO alone. The experiments revealed that the untreated RNA showed a positive fluorescence signal after transfection (Figure 4C), consistent with the formation of folded Broccoli structure. In contrast, cloaked RNA showed no measurable signal above background, confirming that cloaking strongly inhibits function in this 237 nt RNA. Finally, experiments with subsequent light exposure revealed the emergence of a visible green fluorescence signal attributable to restored Broccoli RNA (Figure 4C).
An overall assessment of the efficiency of the current RNA cloaking and photo-uncloaking strategy suggests that the new approach compares favorably in performance to previous RNA caging designs. The 10 °C difference in melting temperature upon cloaking is comparable to previously reported values using photocaged nucleobase phosphoramidites.7b,23 The present methodology performed similarly in interfering with hybridization compared to other caging methods.7,24 Using the current methodology on the Broccoli aptamer, we attained marked OFF/ON behavior, comparable or better than previous studies with light controlled DNA aptamers developed using photocaged phosphoramidites.20b However, to the best of our knowledge our data represent the first example of a photocaged RNA aptamer, likely due to size limitations with prior methods.
In summary, our new photocloaking approach is a convenient method for obtaining photoprotected RNA, representing significant advantages over existing technologies. One potential drawback of this new approach is its stochastic nature, which generates RNAs with varying levels of acylation and does not direct PPGs to specific positions. However, our data show that sufficient levels of acylation can be readily achieved to strongly suppress RNA activity. Importantly, a great advantage of this postsynthetic labeling is that it allows for one-step photoprotection of long, biologically relevant RNAs that are difficult or impossible to synthesize via solid-state oligonucleotide synthesis. Since photocloaking can be achieved without specialized equipment, the new method could potentially find widespread use for study of the biology of RNAs.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for support from the U.S. National Institutes of Health (GM068122 and GM127295) and The Netherlands Organization for Scientific Research (RUBICON) and EMBO ALTF 934-2014 to W.A.V.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b12408.
Further experimental details, spectra, Figure S1–S9 (PDF)
REFERENCES
- (1).Armitage BA Curr. Opin. Chem. Biol 2011, 15 (6), 806. [DOI] [PubMed] [Google Scholar]
- (2).(a) Cech TR; Steitz JA Cell 2014, 157 (1), 77. [DOI] [PubMed] [Google Scholar]; (b) He C Nat. Chem. Biol 2010, 6 (12), 863. [DOI] [PubMed] [Google Scholar]
- (3).Chen L Nat. Rev. Mol. Cell Biol 2016, 17 (4), 205. [DOI] [PubMed] [Google Scholar]
- (4).Delebecque CJ; Lindner AB; Silver PA; Aldaye FA Science 2011, 333 (6041), 470. [DOI] [PubMed] [Google Scholar]
- (5).Li F; Dong J; Hu X; Gong W; Li J; Shen J; Tian H; Wang J Angew. Chem., Int. Ed 2015, 54 (15), 4597. [DOI] [PubMed] [Google Scholar]
- (6).(a) Lubbe AS; Szymanski W; Feringa BL Chem. Soc. Rev 2017, 46 (4), 1052. [DOI] [PubMed] [Google Scholar]; (b) Brieke C; Rohrbach F; Gottschalk A; Mayer G; Heckel A Angew. Chem., Int. Ed 2012, 51, 8446. [DOI] [PubMed] [Google Scholar]
- (7).(a) Ankenbruck N; Courtney T; Naro Y; Deiters A Angew. Chem., Int. Ed 2018, DOI: 10.1002/anie.201700171. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Govan JM; Young DD; Lusic H; Liu Q; Lively MO; Deiters A Nucleic Acids Res 2013, 41 (22), 10518. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Mikat V; Heckel A RNA 2007, 13 (12), 2341. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Lucas T; Schafer F; Muller P; Eming SA; Heckel A; Dimmeler S Nat. Commun 2017, 8 (May), 15162. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Chaulk SG; MacMillan AM Nat. Protoc 2007, 2 (5), 1052. [DOI] [PubMed] [Google Scholar]; (f) Matsushita-Ishiodori Y; Ohtsuki T Acc. Chem. Res 2012, 45 (7), 1039. [DOI] [PubMed] [Google Scholar]; (g) Shah S; Rangarajan S; Friedman SH Angew. Chem 2005, 117 (9), 1352. [DOI] [PubMed] [Google Scholar]; (h) Meyer A; Mokhir A Angew. Chem., Int. Ed 2014, 53 (47), 12840. [DOI] [PubMed] [Google Scholar]; (j) Lu J; Koo SC; Li N-S; Piccirilli JA Nucleosides, Nucleotides Nucleic Acids 2015, 34 (2), 114. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Resendiz MJE; Schön A; Freire E; Greenberg MM J. Am. Chem. Soc 2012, 134 (30), 12478. [DOI] [PMC free article] [PubMed] [Google Scholar]; (l) Panja S; Paul R; Greenberg MM; Woodson SA Angew. Chem., Int. Ed 2015, 54 (25), 7281. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Höbartner C; Silverman SK Angew. Chem., Int. Ed 2005, 44 (44), 7305. [DOI] [PubMed] [Google Scholar]; (n) Gautier A; Gauron C; Volovitch M; Bensimon D; Jullien L; Vriz S Nat. Chem. Biol 2014, 10 (7), 533. [DOI] [PubMed] [Google Scholar]
- (8).(a) Wenter P; Fürtig B; Hainard A; Schwalbe H; Pitsch S Angew. Chem., Int. Ed 2005, 44 (17), 2600. [DOI] [PubMed] [Google Scholar]; (b) Furtig B; Wenter P; Pitsch S; Schwalbe H ACS Chem. Biol 2010, 5 (8), 753. [DOI] [PubMed] [Google Scholar]
- (9).(a) Shestopalov IA; Sinha S; Chen JK Nat. Chem. Biol 2007, 3 (10), 650. [DOI] [PubMed] [Google Scholar]; (b) Tang X; Maegawa S; Weinberg ES; Dmochowski IJ J. Am. Chem. Soc 2007, 129 (36), 11000. [DOI] [PubMed] [Google Scholar]
- (10).Liu Q; Deiters A Acc. Chem. Res 2014, 47 (1), 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Ando H; Furuta T; Tsien RY; Okamoto H Nat. Genet 2001, 28 (4), 317. [DOI] [PubMed] [Google Scholar]
- (12).Blidner RA; Svoboda KR; Hammer RP; Monroe WT Mol. BioSyst 2008, 4 (5), 431. [DOI] [PubMed] [Google Scholar]
- (13).(a) Spitale RC; Flynn RA; Zhang QC; Crisalli P; Lee B; Jung J-W; Kuchelmeister HY; Batista PJ; Torre EA; Kool ET; Chang HY Nature 2015, 519 (7544), 486. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Spitale RC; Crisalli P; Flynn RA; Torre EA; Kool ET; Chang HY Nat. Chem. Biol 2013, 9, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).(a) Merino EJ; Wilkinson KA; Coughlan JL; Weeks KM J. Am. Chem. Soc 2005, 127 (12), 4223. [DOI] [PubMed] [Google Scholar]; (b) Low JT; Weeks KM Methods 2010, 52 (2), 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).(a) Matsuo K; Kioi Y; Yasui R; Takaoka Y; Miki T; Fujishima S; Hamachi I Chem. Sci 2013, 4 (6), 2573. [Google Scholar]; (b) Knežević NŽ; Trewyn BG; Lin VSY Chem. - Eur. J 2011, 17 (12), 3338. [DOI] [PubMed] [Google Scholar]
- (16).Klán P; Šolomek T; Bochet CG; Blanc A; Givens R; Rubina M; Popik V; Kostikov A; Wirz J Chem. Rev 2013, 113 (1), 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).(a) Franzini RM; Kool ET J. Am. Chem. Soc 2009, 131, 16021. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Velema WA; Kool ET J. Am. Chem. Soc 2017, 139 (15), 5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Liu Y; Wilson TJ; Lilley DM J. Nat. Chem. Biol 2017, 13, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).(a) Chaulk SG; MacMillan AM Nucleic Acids Res. 1998, 26 (13), 3173. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Nierth A; Singer M; Jäschke A Chem. Commun. (Cambridge, U. K.) 2010, 46 (0), 7975. [DOI] [PubMed] [Google Scholar]; (c) Young DD; Deiters A Bioorg. Med. Chem. Lett 2006, 16 (10), 2658. [DOI] [PubMed] [Google Scholar]
- (20).(a) Buff MCR; Schäfer F; Wulffen B; Müller J; Pötzsch B; Heckel A; Mayer G Nucleic Acids Res. 2010, 38 (6), 2111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Li L; Tong R; Chu H; Wang W; Langer R; Kohane DS Proc. Natl. Acad. Sci. U. S.A 2014, 111 (48), 17099. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Qiu L; Wu C; You M; Han D; Chen T; Zhu G; Jiang J; Yu R; Tan W J. Am. Chem. Soc 2013, 135 (35), 12952. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Heckel A; Mayer G J. Am. Chem. Soc 2005, 127 (3), 822. [DOI] [PubMed] [Google Scholar]; (e) Pinto A; Lennarz S; Rodrigues-Correia A; Heckel A; O’Sullivan CK; Mayer G ACS Chem. Biol 2012, 7 (2), 360. [DOI] [PubMed] [Google Scholar]
- (21).Filonov GS; Moon JD; Svensen N; Jaffrey S R J. Am. Chem. Soc 2014, 136 (46), 16299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Filonov GS; Kam CW; Song W; Jaffrey SR Chem. Biol 2015, 22 (5), 649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Deiters A; Garner RA; Lusic H; Govan JM; Dush M; Nascone-Yoder NM; Yoder JA J. Am. Chem. Soc 2010, 132 (44), 15644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wu L; Wang Y; Wu J; Lv C; Wang J; Tang X Nucleic Acids Res. 2013, 41 (l), 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.