Summary
When studying microtubules in vitro, label free imaging of single microtubules is necessary when the quantity of purified tubulin is too low for efficient fluorescent labelling or there is concern that labelling will disrupt function. Commonly used techniques for observing unlabelled microtubules, such as video enhanced differential interference contrast, dark-field and more recently laser-based interferometric scattering microscopy, suffer from a number of drawbacks. The contrast of differential interference contrast images depends on the orientation of the microtubules, dark-field is highly sensitive to impurities and optical misalignments. In addition, all of these techniques require costly optical components such as Nomarski prisms, dark-field condensers, lasers and laser scanners. Here we show that single microtubules can be imaged at high speed and with high contrast using interference reflection microscopy without the aforementioned drawbacks. Interference reflection microscopy is simple to implement, requiring only the incorporation of a 50/50 mirror instead of a dichroic in a fluorescence microscope, and with appropriate microscope settings has a similar signal-to-noise ratio to differential interference contrast and fluorescence. We demonstrated the utility of interference reflection microscopy by high-speed imaging and tracking of dynamic microtubules at 100 frames per second. In conclusion, the optical quality of interference reflection microscopy falls within the range of other microscope techniques, being inferior to some and superior to others, depending on the metric used and, with minimal microscope modification, can be used to study the dynamics of unlabelled microtubules.
Keywords: Interference reflection microscopy, label free imaging, microtubules
Introduction
In vitro imaging of single microtubules has proved to be an important tool to study microtubule dynamics (Hotani & Horio, 1988; Walker et al., 1988), mechanics (Gittes et al., 1993; Dogterom & Yurke, 1997), and regulation (Gell et al., 2010). Typically, fluorescence microscopy, in particular total internal reflection fluorescence (TIRF) microscopy, is used for imaging microtubule dynamics and gliding assays due to its high signal-to-background noise ratio (SBR; a measure of the optical contrast of the microtubule) and the wide range of available fluorescent labels (Gell et al., 2010). However, efficient fluorescent labelling requires large quantity of protein, which is difficult to obtain for some sources of tubulin (Widlund et al., 2012). Furthermore, fluorescence-induced photo-damage may be a concern (Vemu et al., 2016) especially if oxygen scavengers can react with the molecules of interest. In such cases, label free imaging is required.
There are many label-free imaging techniques available for imaging microtubules. Video-enhanced differential interference microscopy (DIC) and dark-field microscopy (DF) have been regularly used for almost 30 years (Hotani & Horio, 1988; Walker et al., 1988; Gittes et al., 1993; Bormuth et al., 2007). With DIC it is possible to image a large field of view at high frame rates. The drawback of using DIC is that the contrast of microtubules depends on their orientation with respect to the axis of the Nomarski prims. Although this issue can be solved (Shribak et al., 2008), the solution adds extra layers of complexity to the set up. Dark-field can visualise microtubule at a high SBR assuming very clean solutions and surfaces. Otherwise, stray light caused by misalignment or light scattered by impurities will overwhelm the microtubule signal. Another technical inconvenience is that both techniques require the use of high numerical-aperture (NA) condensers that limit access to the sample during experiments. Other than DIC and dark-field, a number of techniques have emerged over the years. For example, Amos & Amos (1991) imaged single microtubules by confocal reflection microscopy, requiring minimal modification to a confocal setup. Medina et al. (2010) visualised microtubules by defocusing the bright-field microscope, though the contrast was low even after image processing. Recently, Andrecka et al. (2016) used interferometric scattering (iSCAT) microscopy to image microtubule dynamics at high frame rates. Although iSCAT is capable of label-free imaging of single proteins (Young et al., 2018), it requires a fast laser scanning system (Ortega Arroyo et al., 2016). Rotating-coherent-scattering microscopy (ROCS) which is also based on laser scattering was used to image microtubules far from the surface (Koch & Rohrbach, 2018). Just like iSCAT, ROSC requires the use of a laser and scanners. Another recent technique uses spatial light interference microscopy (SLIM); although capable of imaging a large field of view (200 × 200 μm2), the frame rate is low (6.25 fps) (Kandel et al., 2017).
All the aforementioned techniques are capable of imaging single microtubules, but each fails to meet one or more of the following four criteria: (i) large field of view for higher throughput and to take advantage of the large chips of the new scientific CMOS cameras, (ii) high frame rate to improve temporal resolution, (iii) good signal-to-background noise ratio to achieve high spatial precision and (iv) ease of implementation with low cost. In this work, we demonstrate that interference reflection microscopy (IRM) can image single label-free microtubules and satisfies all criteria. In IRM, the image is formed by the interference between the light reflected from the glass-medium interface and that reflected from the medium-sample interface (Weber, 2003; Limozin & Sengupta, 2009). Since its first use in biology by Curtis (1964), IRM and the more advanced implementation known as reflective interference contrast microscopy have been used intensively to probe cell adhesion to surfaces and other applications (Verschueren, 1985; Barr & Bunnell, 2001). Here we show that it is possible to image single microtubules using IRM in its simplest configuration.
Methods
IRM was implemented by adding a 50/50 mirror (Chroma, Bellows Falls, VT, USA) to a Nikon inverted fluorescence microscope (Ti Eclipse, Nikon, Melville, NY, USA) in the position where usually a dichroic mirror is placed (Fig. S1). No additional excitation or emission filters are needed. The sample was illuminated by a Sola light engine (Lumencore, Beaverton, OR, USA) attached to the microscope’s epi port. The NA of the illumination was set by the aperture iris. The field diaphragm was adjusted to optimise the contrast as presented in the Results section. The illumination light was partially reflected to the objective (100x/1.49 Apochromat, Nikon, Melville, NY, USA) and the light reflected from the sample was collected by the same objective and projected – after passing through the 50/50 mirror – onto a 16 bit sCMOS camera [Zyla 4.2, 6.5 μm pixel size, 2048 × 2048 pixel2 chip size, 72% quantum efficiency, Andor, Belfast, North Ireland]. The illumination was adjusted to nearly saturate the camera’s dynamic range. Experiments were performed in a flow channel formed by two parafilm strips sandwiched between 18 × 18 mm2 and 22 × 22 mm2 coverslips (#1.5H, Marienfeld, Germany) or between a 22 × 22 mm2 coverslip and a 25 × 75 mm2 microscope slide (Thermo Scientific, Waltham, MA, USA) (Gell et al., 2010). The coverslips were cleaned using piranha solution and rendered hydrophobic by silanisation (Gell et al., 2010). In nondynamic assays GMPCPP stabilised, TAMRA labelled (tetramethylrhodamine Ex: 550 nm, Em: 580 nm) bovine microtubules were imaged at room temperature. The microtubules were either passively attached to the hydrophobic surface or fixed by an anti-TAMRA antibody (Life Technologies, Waltham, MA, USA). For dynamic assays, the same configuration was used and microtubule growth was initiated by flowing in polymerisation solution [80 mM PIPES/KOH, pH 6.9, 1 mM ethylene glycol tetra-acetic acid (EGTA), 1 mM MgCl2, 2 mM GTP and 7.5 μM unlabelled bovine tubulin] at 34°C. Dynamic assays were imaged at slow (0.2 fps) and fast (100 fps) frame rates. In some dynamic assays the minus and plus end of the microtubules seeds were labelled using Alexa488 labelled (Ex: 490 nm, Em: 525 nm) tubulin. For all IRM imaging of fluorescently labelled tubulin, a long pass filter (LP 594 nm, Chroma, Bellows Falls, VT, USA) was inserted into the illumination path to avoid exciting the fluorophores. Such a filter is unnecessary if unlabelled tubulin is used. No antifade reagents (Gell et al., 2010) were used.
To enhance contrast and eliminate illumination irregularities and static noise, a background image was subtracted from the acquired images. The background image was generated by averaging 32 or 100 images acquired before flowing in the microtubules or it was the median of 100 images acquired while moving the sample at a high speed after imaging. Further enhancement of images included averaging, Fourier filtering or both (Bormuth et al., 2007). All image processing was performed using Fiji (Schindelin et al., 2012). Tracking of microtubules was done using the tracking software FIESTA (Ruhnow et al., 2011).
For DIC imaging of microtubules, a high NA oil immersion condenser equipped with a high-resolution prism (NA = 1.4, Nikon, Melville, NY, USA) was used for illuminating the sample. To enhance the contrast of the images, a bias retardation of one tenth of the wavelength was applied. Background images were generated by averaging 100 images taken 1–2 μm from the surface. As for IRM images, the images could be further enhanced by averaging and/or filtering.
Fluorescence images were obtained by TIRF microscopy. The microtubules were excited by a 561 nm laser at a power of ~1 mW (at the sample plane) with an exposure time of 100 ms.
As a metric to assess IRM and compare it to different techniques, we measured the signal-to-background noise ratio (SBR), defined by the average intensity of the microtubule signal (intensity of the microtubule minus the intensity of the background) divided by the standard deviation of the back-ground. The background noise was based on a region close to the microtubule, as described below.
For IRM and TIRF images, the signal was obtained by first isolating the microtubule in a rectangular region of interest (ROI) that was slightly longer than the microtubule and 60 pixels in the perpendicular direction. Next, the cross section profile of the microtubule was measured by taking a line scan perpendicular to the microtubule axis. The line width was set to be equal to the microtubule length. In this manner, every point on the cross section profile was an average of all pixels along the microtubule axis. Then, the signal was measured as the difference between the profile peak and the background. The background was measured by thresholding the microtubule image to separate the microtubule from the background and then averaging all pixels below the cut off intensity. In the case of DIC images, the signal-to-background noise ratio was determined as the peak to peak difference of the averaged signal divided by the standard deviation of the background noise (Bormuth et al., 2007).
We also measured the contrast sensitivity coefficient (CSC) defined as the microtubule signal divided by the standard deviation of the microtubule signal. Here, the signal is the mean of all the pixels’ values above the threshold cut off after background subtraction. The noise is the standard deviation of the signal. CSC measurement is applicable to IRM and TIRF microtubule images but not DIC because the standard deviation along the microtubule will be higher than the signal. For DIC, the average signal will nearly vanish because the intensities above and below the background tend to cancel each other.
Results and discussion
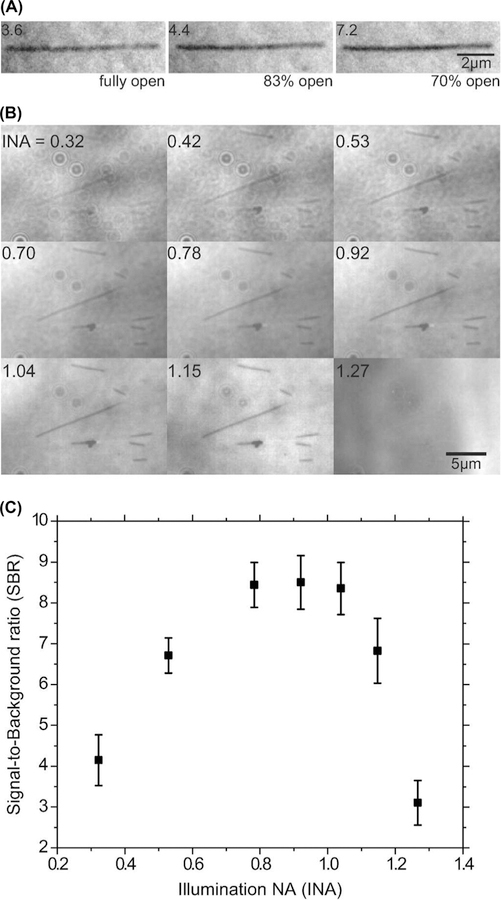
Microtubules are readily visible in IRM without background subtraction. The microtubules appeared dark against a brighter background. Reducing the field of view size by the field diaphragm reduced the amount of stray light reaching the camera and improved the contrast of the microtubule (Fig. 1A). We found that opening the field diaphragm up to 70% of the field of view was a good balance between improving contrast and maintaining a large imaging area. In addition to the field diaphragm, the contrast depended on the illumination NA (Weber, 2003), as shown in Figures 1(B), (C). Qualitatively, microtubule visibility was highest in the NA range 0.7–1.1 (Fig. 1B). This agreed with SBR measurements of background-subtracted images as a function of illumination NA (Fig. 1C). The SBR of a single frame of background-subtracted images was 6.8±0.8 (mean±SD, N=41).
Fig. 1.
Optimal microscope settings for IRM. (A) The same microtubule imaged at three different field diaphragm openings, without background subtraction. The illumination was adjusted such that the average image intensity is nearly the same in all three images. Numbers at the upper corner are the measured SBRs of the microtubule. (B) The same field of view was imaged at different illumination NAs (INA, top left corner), without background subtraction. Again, the intensity was adjusted to give nearly equal illumination levels. (C) SBR as a function of illumination NA for the same field of view as in B, calculated after background subtraction.
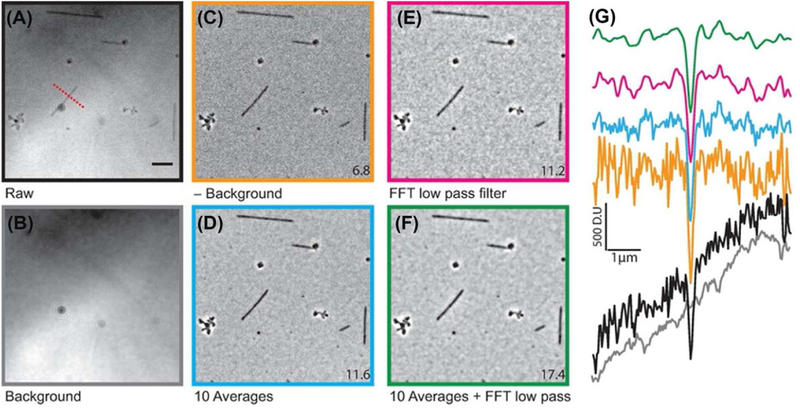
To further improve the SBR, images were either averaged or filtered or both. Upon averaging, the SBR initially increased with a power law of about 0.5, as expected for a photon-shot-noise limited image (dashed line in Fig. S2A). Beyond 10 frame averages, there was little additional increase in SBR (Fig. S2A) due to the presence of other sources of noise that are insensitive to averaging, such as dirt and other inhomogeneities in the image. We found that taking 100 averages for the background was sufficient to obtain a high SBR (Fig. S2B). By averaging ten background-subtracted images, the SBR increased to 11.6 ± 1.6 (Fig. 2D). A comparable increase to 11.2±1.2 (Fig. 2E) was obtained by using a low-pass Fourier filter. Filtering the averaged images results in a final SBR of 17.4±2.4 (Fig. 2G).
Fig. 2.
Image acquisition and processing. Raw images (A) were acquired at intensities that nearly saturated the camera. The background (B, average of 32 images) was subtracted to yield the background-subtracted image (C). The images are either averaged (D), filtered (E) or both (F). The numerical values at the corner of the images are the average SBRs calculated as described in the Methods section. Scale bar is 5 μm. (G) Typical line scans across a microtubule (dashed red line in A) for all conditions (scans matched to respective images using the colour of the image frame). DU corresponds to digital unit, the raw signal obtained from each pixel of the sCMOS camera.
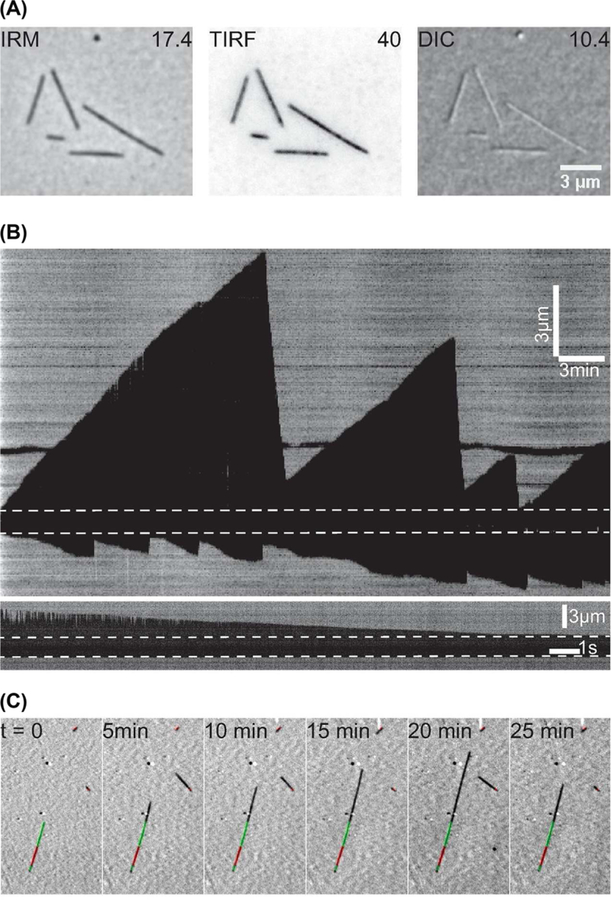
To compare the performance of IRM to other techniques, the same field of view of microtubules was imaged by IRM, TIRF and DIC and the SBR for each image was measured (Fig. 3A). For TIRF of highly labelled microtubules (30%, to reduce speckles; Waterman-Storer et al., 1998), the SBR was 40 ± 10 (mean±SD, N=41), prior to photobleaching. In terms of CSC, both IRM and TIRF images were similar, CSCTIRF = 2.64 ± 0.13 (mean ± SD) compared to CSCIRM = 2.7 ± 0.18. Although the variation in intensity along the microtubule in fluorescence images is caused by the fluorophore distribution within the microtubule, for IRM it is a result of microtubule-to-surface distance variation as well as noise sources such as dirt. DIC images had a lower SBR (10 ± 2) than IRM (after averaging and filtering). Aside from the higher SBR, the advantage of using IRM over DIC is that the contrast is independent of microtubule orientation as can be seen in Figure 3(A). Such dependence has an impact when tracking microtubules imaged in DIC (Danuser et al., 2000; Janson & Dogterom, 2004).
Fig. 3.
IRM performance. (A) The same field of view was imaged using IRM, TIRF and DIC. The IRM and DIC images were averaged 10× and low-pass filtered for a final frame rate of 10 fps. TIRF images were single snaps of 100 ms exposure time (10 fps) and the contrast inverted. The numerical values are the average SBRs. (B) Example kymographs of dynamic microtubules imaged using IRM at two frame rates: 0.2 fps (upper) and 100 fps (lower). The dashed line represents the GMPCPP seed. The lower kymograph shows a single shrinking event. Just after catastrophe, the microtubule extension beyond the seed is relatively long, and as a result the tip fluctuates upwards causing the change in contrast. (C) Example of combining IRM and fluorescence microscopy. Stabilised GMPCPP Alexa488-labelled extensions were grown (green) from TAMRA labelled seeds (red) to mark the plus end (long green extension) and minus end (short green extension) of the microtubule. Such tripartite fluorescent microtubules allow unambiguous identification and facilitate the study of drugs or MAPs.
To demonstrate the capability of IRM, dynamic microtubules were imaged. Averaged background-subtracted images were taken at a frame rate of 0.2 fps. Growth and shrinkage phases where easily observed in the videos and kymographs (Fig. 3B, upper panel and Supplementary Video 1). Although such a low frame rate is suitable for measuring growth rate and catastrophe frequency, it is more challenging to measure the shrinkage rates, which are an order of magnitude higher than growth rates. For this reason, shrinking microtubules were imaged at 100 fps, which provided detailed dynamics of the shrinking end (Fig. 3B, lower panel). It was possible to track the microtubule ends using the tracking software FIESTA with a length precision of ≈ 20 nm (Supplementary Video 1). Although this precision is less than the one that can be obtained by TIRF (≈10 nm; Ruhnow et al., 2011) or iSCAT (≈1 nm; Andrecka et al., 2016), it is sufficient for measuring shortening speeds. In addition, IRM can be readily combined with fluorescence microscopy (Fig. 3C), thus freeing up one of the fluorescence channels to image, for example, molecular motors, MAPs or specially modified tubulins.
In TIRF and DIC, thermal fluctuations of long microtubules away from the surface lead to loss of contrast. In IRM, such fluctuations lead to a reversal in microtubule contrast; this can be used to measure the height of the microtubule, similar to fluorescence interference contrast microscopy (Lambacher & Fromherz, 1996; Kerssemakers et al., 2006) (Supplementary Video 2). Recently, the ability to measure distances from the surface using IRM was used as a means to calibrate the TIRF evanescent field in a combined IRM-TIRF-optical tweezers setup using a light emitting diode for illumination (Simmert et al., 2018). It is also worth mentioning that IRM is flexible in terms of what objective to use: it was possible to image microtubules using high end and low end objectives as well as phase and DIC objectives (Fig. S3). The presence of a Nomarski prism or the phase ring did not seem to influence the contrast in the images.
In addition to dirt, IRM is sensitive to drift, which is noticeable when imaging for extended periods of time (Fig. S4). Drift can be avoided by thermally stabilising the setup (Mahamdeh & Schäffer, 2009) or correcting the images for drift (Carter et al., 2007; Kim & Saleh, 2008; Ortega Arroyo et al., 2016).
In conclusion, IRM proved to be a powerful tool for imaging unlabelled microtubules in surface assays. IRM only requires a minor, cost-effective modification to any epi-microscope, requires a onetime alignment, is capable of high-speed wide-field [100 fps at full chip (130 × 130 μm2)] imaging of microtubules at a high spatial precision for long periods of time and can be readily combined with other techniques.
Supplementary Material
Fig. S1. Schematic of IRM implementation. Epi-illumination from the light source passes through the aperture diaphragm before reaching the 50/50 mirror. The mirror partially reflects the light up to the objective to illuminate the sample. The reflected light is collected and projected onto the camera chip (by the tube lens) where it interferes to generate the image. Image contrast is the result of the interference between the light reflected form the glass/water interface (I1) and the light reflected from the water/microtubule interface (I2). Depending on the microtubule/surface distance (h), the optical path difference between I1 and I2 will result in a constructive (bright signal) or destructive (dark signal) or anything in between. For example, if light with a wavelength of 600 nm is used for imaging, the contrast will switch between dark and bright when the microtubule height changes by about 100 nm.
Fig. S2. Effect of number of image averages on SBR. (A) SBR as a function of number of averaged frames. The dashed line is a power law of 0.5 which is expected for a shot noise limited imaging. The subtracted background was the average of 100 frames. (B) Effect of the number of averaged background images on noise reduction. An ROI was imaged twice. Each time, 1000 images were collected. The first set was used as background and was averaged 10, 32, 100 and 1000 times. The 2nd set was subtracted from the background images and averaged (1, 2, 5, 10, 20, 50, 100, 200, 500 and 1000 averages). Next, the standard deviation of the averaged background-subtracted images was plotted as a function of the number of averages. The lower the number of background averages the faster the noise saturated. The solid line is a power law of −0.5 calculated over the range of measurements and represents the expected behaviour for a shot noise limited imaging.
Fig. S3. IRM imaging using various objective types. Same stabilised microtubules were imaged using three objectives (not background subtracted). The presence of a phase ring or DIC prisms did not affect the ability to image microtubules.
Fig. S4. 3D drift effect on IRM images. A ROI was imaged 2000 times taking 20 s and the images were averaged. The average image was used as the background. The same ROI was then imaged at three different time points (0, 12 and 30 min). At each time point, 1000 images were collected and averaged. The background was subtracted from the averaged images and the results are shown. Surface structures and illumination irregularities are due to drift.
Footnotes
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- Amos LA & Amos WB (1991) The bending of sliding microtubules imaged by confocal light microscopy and negative stain electron microscopy. J. Cell Sci 1991, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrecka J, Ortega Arroyo J, Lewis K, Cross RA, & Kukura P(2016) Label-free imaging of microtubules with sub-nm precision using interferometric scattering microscopy. Biophys. J 110, 214–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr VA& Bunnell SC (2001) Interference reflection microscopy. Curr. Protoc. Cell Biol. Suppl 45, 2301–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bormuth V, Howard J, & Schäffer E (2007) LED illumination for video-enhanced DIC imaging of single microtubules. J. Microsc 226, 1–5. [DOI] [PubMed] [Google Scholar]
- Carter AR, King GM, Ulrich TA, Halsey W, Alchenberger D & Perkins TT (2007) Stabilization of an optical microscope to 0.1 nm in three dimensions. Appl. Opt 46, 421–427. [DOI] [PubMed] [Google Scholar]
- Curtis ASG (1964) The Mechanism of Adhesion of Cells to Glass. J. Cell Biol 20, 199–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danuser G, Tran PT & Salmon ED (2000) Tracking differential interference contrast diffraction line images with nanometre sensitivity. J. Microsc 198, 34–53. [DOI] [PubMed] [Google Scholar]
- Dogterom M, & Yurke B (1997) Measurement of the Force-Velocity Relation for Growing Microtubules. Science 278, 856–860. [DOI] [PubMed] [Google Scholar]
- Gell C, et al. (2010) Microtubule dynamics reconstituted in vitro and imaged by single-molecule fluorescence microscopy. Methods in Cell Bio 95, 221–245. [DOI] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J & Howard J (1993) Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J. Cell Biol 120, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotani H & Horio T (1988) Dynamics of microtubules visualized by darkfield microscopy: treadmilling and dynamic instability. Cell Motil. Cytoskel 10, 229–236. [DOI] [PubMed] [Google Scholar]
- Janson ME & Dogterom M (2004) A bending mode analysis for growing microtubules: evidence for a velocity-dependent rigidity. Biophys. J 87, 2723–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ME, Teng KW, Selvin PR & Popescu G (2017) Label-free imaging of single microtubule dynamics using spatial light interference microscopy. ACS Nano 11, 647–655. [DOI] [PubMed] [Google Scholar]
- Kerssemakers J, Howard J, Hess H & Diez S (2006) The distance that kinesin-1 holds its cargo from the microtubule surface measured by fluorescence interference contrast microscopy. Proc. Natl. Acad. Sci 103, 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K & Saleh OA (2008) Stabilizing method for reflection interference contrast microscopy. Appl. Opt 47, 2070–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MD & Rohrbach A (2018) Label-free imaging and bending analysis of microtubules by ROCS microscopy and optical trapping. Biophys. J 114, 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambacher A & Fromherz P (1996) Fluorescence interference-contrast microscopy on oxidized silicon using a monomolecular dye layer. Appl. Phys. A 63, 207–216. [Google Scholar]
- Limozin L & Sengupta K (2009) Quantitative reflection interference contrast microscopy (RICM) in soft matter and cell adhesion. ChemPhysChem 10, 2752–2768. [DOI] [PubMed] [Google Scholar]
- Mahamdeh M & Schäffer E (2009) Optical tweezers with millikelvin precision of temperature-controlled objectives and base-pair resolution. Opt. Express 17, 17190–17199. [DOI] [PubMed] [Google Scholar]
- Ortega Arroyo J, Cole D & Kukura P (2016) Interferometric scattering microscopy and its combination with single-molecule fluorescence imaging. Nat. Protoc 11, 617–633. [DOI] [PubMed] [Google Scholar]
- Ruhnow F, Zwicker D & Diez S (2011) Tracking single particles and elongated filaments with nanometer precision. Biophys. J 100, 2820–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shribak M, LaFountain J, Biggs D & Inouè S (2008) Orientation-independent differential interference contrast microscopy and its combination with an orientation-independent polarization system. J. Biomed. Opt 13, 014011–014011−10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmert S, Abdosamadi MK, Hermsdorf G & Schäffer E (2018) LED-based interference-reflection microscopy combined with optical tweezers for quantitative three-dimensional microtubule imaging. Opt. Express 26, 14499. [DOI] [PubMed] [Google Scholar]
- Vemu A, Atherton J, Spector JO, Szyk A, Moores CA & Roll-Mecak A (2016) Structure and dynamics of single-isoform recombinant neuronal human tubulin. J. Biol. Chem 291, 12907–12915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschueren H (1985) Interference reflection microscopy in cell biology: methodology and applications. J. Cell Sci 75, 279–301. [DOI] [PubMed] [Google Scholar]
- Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP & Salmon ED (1988) Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J. Cell Biol 107, 1437–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer CM, Desai A, Chloe Bulinski J& Salmon ED(1998) Fluorescent speckle microscopy, a method to visualize the dynamics of protein assemblies in living cells. Curr. Biol 8, 1227–S1. [DOI] [PubMed] [Google Scholar]
- Weber I (2003) [2]Reflection interference contrast microscopy. Methods Enzymol 361, 34–47. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Podolski M, Reber S, Alper J, Storch M, Hyman AA, Howard J & Drechsel DN (2012) One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol. Biol. Cell 23, 4393–4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G, Hundt N, Cole D et al. (2018) Quantitative mass imaging of single biological macromolecules. Science 360, 423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Schematic of IRM implementation. Epi-illumination from the light source passes through the aperture diaphragm before reaching the 50/50 mirror. The mirror partially reflects the light up to the objective to illuminate the sample. The reflected light is collected and projected onto the camera chip (by the tube lens) where it interferes to generate the image. Image contrast is the result of the interference between the light reflected form the glass/water interface (I1) and the light reflected from the water/microtubule interface (I2). Depending on the microtubule/surface distance (h), the optical path difference between I1 and I2 will result in a constructive (bright signal) or destructive (dark signal) or anything in between. For example, if light with a wavelength of 600 nm is used for imaging, the contrast will switch between dark and bright when the microtubule height changes by about 100 nm.
Fig. S2. Effect of number of image averages on SBR. (A) SBR as a function of number of averaged frames. The dashed line is a power law of 0.5 which is expected for a shot noise limited imaging. The subtracted background was the average of 100 frames. (B) Effect of the number of averaged background images on noise reduction. An ROI was imaged twice. Each time, 1000 images were collected. The first set was used as background and was averaged 10, 32, 100 and 1000 times. The 2nd set was subtracted from the background images and averaged (1, 2, 5, 10, 20, 50, 100, 200, 500 and 1000 averages). Next, the standard deviation of the averaged background-subtracted images was plotted as a function of the number of averages. The lower the number of background averages the faster the noise saturated. The solid line is a power law of −0.5 calculated over the range of measurements and represents the expected behaviour for a shot noise limited imaging.
Fig. S3. IRM imaging using various objective types. Same stabilised microtubules were imaged using three objectives (not background subtracted). The presence of a phase ring or DIC prisms did not affect the ability to image microtubules.
Fig. S4. 3D drift effect on IRM images. A ROI was imaged 2000 times taking 20 s and the images were averaged. The average image was used as the background. The same ROI was then imaged at three different time points (0, 12 and 30 min). At each time point, 1000 images were collected and averaged. The background was subtracted from the averaged images and the results are shown. Surface structures and illumination irregularities are due to drift.