Abstract
Background:
Circulating tumor DNA analysis is an emerging genotyping strategy that can identify tumor-specific genetic alterations in plasma including mutations and rearrangements. Detection of ROS1 fusions in plasma requires genotyping approaches that cover multiple breakpoints and target a variety of fusion partners. Compared to other molecular subsets of non-small cell lung cancer (NSCLC), experience with detecting ROS1 genetic alterations in plasma is limited.
Patients and Methods:
To describe the spectrum of ROS1 fusions in NSCLC and determine sensitivity for detecting ROS1 fusions in plasma, we queried the Guardant Health plasma dataset and an institutional tissue database and compared plasma findings to tissue results. In addition, we used the Guardant360 NGS assay to detect potential genetic mediators of resistance in plasma from patients with ROS1-positive NSCLC who were relapsing on crizotinib.
Results:
We detected seven distinct fusion partners in plasma, most of which (n=6/7) were also represented in the tissue dataset. Fusions pairing CD74 with ROS1 predominated in both cohorts (plasma: n=35/56, 63%; tissue: n=26/52, 50%). There was 100% concordance between the specific tissue- and plasma-detected ROS1 fusion for seven patients genotyped with both methods. Sensitivity for detecting ROS1 fusions in plasma at relapse on ROS1-directed therapy was 50%. Six (33%) of 18 post-crizotinib plasma specimens harbored ROS1 kinase domain mutations, five of which were ROS1 G2032R. Two (11%) post-crizotinib plasma specimens had genetic alterations (n=1 each BRAF V600E and PIK3CA E545K) potentially associated with ROS1-independent signaling.
Conclusions:
Plasma genotyping captures the spectrum of ROS1 fusions observed in tissue. Plasma genotyping is a promising approach to detecting mutations that drive resistance to ROS1-directed therapies.
Keywords: ROS1, Lung Cancer, Liquid Biopsy, Targeted Therapy
INTRODUCTION
ROS1-rearranged (ROS1-positive) non-small cell lung (NSCLC) is a subset of NSCLC characterized by dependency on ROS1 signaling and marked sensitivity to ROS1 tyrosine kinase inhibitors (TKIs).1 Despite biological and clinicopathologic similarities between ROS1-positive and anaplastic lymphoma kinase (ALK)-positive NSCLC, multiple studies have established these diseases as separate entities.2,3 For example, structural differences between the two kinases render some ALK-directed therapies such as alectinib ineffective against ROS1-positive NSCLC.3 There are also differences in the characteristic rearrangements that define these two molecular subgroups. Most ALK-positive NSCLCs result from the juxtaposition of EML4 to ALK and ALK fusions primarily occur at a highly conserved breakpoint in intron 19.4 In contrast, a multitude of ROS1 breakpoints and fusion partners have been described in ROS1-positive NSCLC.1,4 Diagnostic approaches that can reliably identify ROS1 fusions are as critical as therapies that selectively and effectively target ROS1.
ROS1 fusions are most often detected in tissue specimens using fluorescence in-situ hybridization (FISH) or next-generation sequencing (NGS).1 Compared to RNA-based genotyping, DNA-based NGS assays are more prone to false-negatives or failure to detect a ROS1 rearrangement due to incomplete ROS1 intronic coverage.5 Analysis of circulating tumor DNA (ctDNA) is a promising strategy for identifying cancer-related molecular alterations, including mutations and rearrangements, in plasma. Plasma genotyping has several potential advantages over tissue genotyping including better resolution of spatial and temporal intratumoral heterogeneity.6 Liquid biopsies are increasingly used for selection of EGFR-directed therapies based on studies confirming the utility of plasma genotyping in this setting.7 However, detection of rearrangements in plasma is more challenging than identifying short variants such as point mutations and insertions/deletions.8
Early studies suggest that plasma genotyping assays detect ALK fusions and ALK kinase domain mutations with a high degree of concordance with tissue genotyping. 9,10 However, few studies have assessed feasibility of plasma genotyping in ROS1-positive NSCLC.11,12 The variety of breakpoints and fusion partners in ROS1-positive NSCLC may pose unique challenges for DNA hybrid capture-based plasma genotyping. 8 Here, we performed molecular analysis of plasma from patients with ROS1-positive NSCLC to describe the spectrum of ROS1 alterations in plasma, compare findings in plasma and tissue, and evaluate the potential role of liquid biopsies in management of this disease.
PATIENTS AND METHODS
Study Population
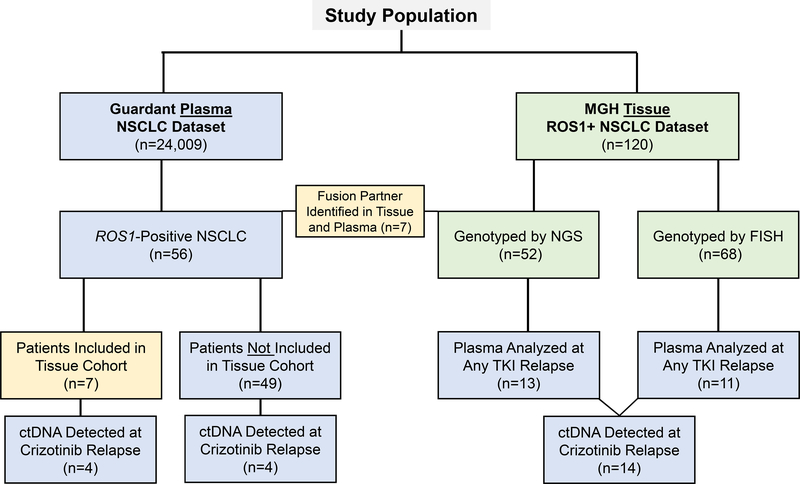
To evaluate the spectrum of ROS1 fusions in plasma, we assembled a cohort of patients (n=56) with ROS1-positive NSCLC from a deidentified dataset containing molecular testing results from 24,009 lung cancer patients who underwent plasma genotyping using the Guardant360 assay between February 2015 and February 2018. We also reviewed a clinical database of patients (n=120) with ROS1-positive NSCLC seen at Massachusetts General Hospital (MGH) between 2011 and 2018 to assess the spectrum of ROS1 fusions in tissue. Twenty-four (20%) patients in the tissue cohort also underwent plasma genotyping using the Guardant360 assay during their disease course. Seven (35%) of these patients were part of the 24,009 patient Guardant dataset. The remaining 17 patients underwent plasma genotyping with Guardant360 after the plasma database used for this study was assembled. We performed molecular analysis of plasma specimens from 18 patients with ROS1-positive NSCLC who were relapsing on crizotinib to detect genetic alterations potentially contributing to relapse. Figure 1 provides a schematic of the study population. This study was approved by the Massachusetts General Hospital Institutional Review Board.
Figure 1. Study Population.
The schematic represents the population of patients studied in this analysis. Plasma specimens are indicated in blue. Tissue specimens are identified by green rectangles. Yellow boxes refer to overlap between two methods. NSCLC: non-small cell lung cancer; NGS: next-generation sequencing; FISH: fluorescence in-situ hybridization; TKI: tyrosine kinase inhibitor.
Data Collection
Medical records were retrospectively reviewed to extract data on clinical and molecular characteristics of patients in the tissue cohort. For patients in the plasma genotyping cohort, data were gathered from information provided to Guardant Health at the time that plasma testing was requested. Data were updated as of September 30, 2018.
Genetic Assessment
All plasma specimens were genotyped using the hybrid-capture based Guardant360 NGS assay as previously described.13 To detect diverse ROS1 fusions, the Guardant360 assay includes capture probes targeting known fusion partners and reported ROS1 breakpoints. ROS1 rearrangements were detected in tissue specimens in the MGH cohort using NGS (n=52) or FISH (n=68). Several NGS platforms were utilized to identify ROS1 rearrangements in tissue: the MGH Solid Fusion Assay (n= 23),14 Foundation One (n=19),15 DFCI Oncopanel (n=6),16 MSK Impact (n=3),17 or Oncoplex (n=1).18 Crizotinib-resistant tissue specimens (n=21) were analyzed using SNaPshot NGS (n=16),14 Foundation One (n=2),15 DFCI Oncopanel (n=1),16 MSK Impact (n=1),17 and a targeted NGS panel developed at the University of Vermont (n=1). All patients included in this study provided consent for molecular testing.
RESULTS
Spectrum of ROS1 Fusion Partners
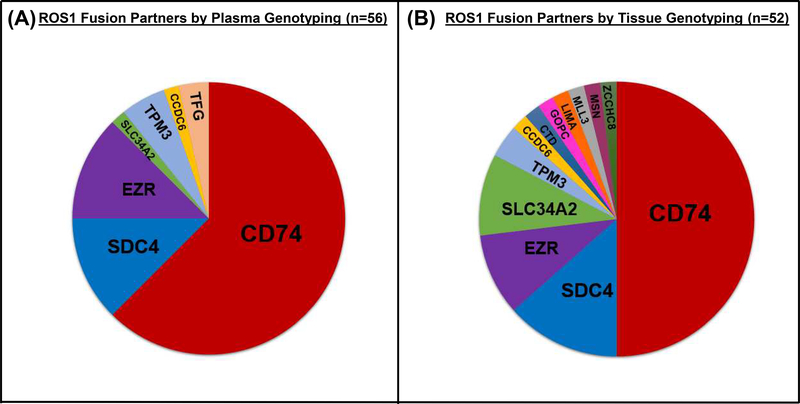
Through review of the Guardant360 dataset, we identified 56 patients with ROS1-positive NSCLC. CD74 was the most common fusion partner (n=35/56, 63%). Six distinct fusion partners were detected in the remaining specimens: EZR (n=7, 12.5%), SDC4 (n=7, 12.5%), TPM3 (n=3, 5%), TFG (n=2, 4%), CCDC6 (n=1, 2%), and SLC34A2 (n=1, 2%). The fusion partners observed in ROS1-positive plasma specimens are depicted in Figure 2A. Our findings confirm that hybrid-capture-based plasma genotyping can detect a variety of ROS1 fusions.
Figure 2. Spectrum of ROS1 Fusion Partners.
The pie charts represent the relative prevalence of distinct fusion partners in plasma (A) and tissue (B).
In the MGH tissue database, the fusion partner was known in 52 (43%) of the 120 patients with ROS1-positive NSCLC. We observed twelve unique fusion partners in tissue specimens, among which CD74-ROS1 fusions were the most common. The spectrum of ROS1 fusion partners in tissue (Figure 2B) was similar to what we observed in plasma based on Guardant testing with the exception that several rare (GOPC, LIMA, MSN, ZCCHC8) or novel (CTD-2021J15.1, MLL3) fusion partners were only identified in tissue.19,20
Tissue-Plasma Concordance and Sensitivity for Detecting ROS1 Fusions
Tissue-Plasma Concordance
We compared tissue and plasma findings for seven patients at our institution who were genotyped using both methods during the study period. For all seven patients, plasma was obtained at relapse on ROS1-directed therapy. The ROS1 fusion partner was detected in tissue either at diagnosis (n=4) or progression on targeted therapy (n=3). Fusions present in these seven specimens included CD74-ROS1 (n=4), EZR-ROS1 (n=1), CCDC6-ROS1 (n=1), and SDC4-ROS1 (n=1). In all cases, the plasma fusion partner was identical to the tissue fusion partner.
Sensitivity for Detecting ROS1 Fusions in Plasma at Relapse
In total, twenty-four patients in the MGH cohort underwent plasma genotyping at relapse on ROS1-directed therapy, including the seven patients discussed above in the tissue-plasma concordance analysis. Seventeen (71%) patients were relapsing on crizotinib at the time plasma was collected. The remaining patients were progressing on investigational next-generation ROS1 inhibitors. Previous studies have demonstrated that sensitivity for detecting genetic alterations in plasma is impacted by disease burden and location of progressing metastatic sites.7,21 At the time that plasma was collected, ten (42%) patients in our cohort were only progressing in the thoracic cavity. Two patients were relapsing in the brain and thoracic cavity. The remaining 12 (50%) patients had progressive disease involving extra-thoracic extra-cranial disease sites.
Circulating tumor DNA was not detected in four (17%) plasma samples, three of which were collected at relapse on crizotinib. These four specimens were obtained from two patients with intrathoracic-only progression, one patient with brain and hilar node relapse, and one patient who had isolated progression of a soft tissue mass involving T1. In total, fourteen (82%) of 17 post-crizotinib plasma specimens from patients at our institution contained ctDNA. The ROS1 fusion was identified in plasma from 10 (50%) of the 20 plasma specimens where circulating tumor DNA was present. Detected fusions included CD74-ROS1 (n=6), CCDC6-ROS1 (n=1), EZR-ROS1 (n=1), SDC4-ROS1 (n=1), and TFG-ROS1 (n=1). Overall, six (60%) of the 10 specimens that lacked plasma ROS1 fusions did not have progression outside of the thorax or central nervous system compared to 3 (30%) of those with detectable fusions. Table 1 summarizes the concordance between each patient’s plasma and tissue ROS1 fusion result and details the sites of progressive disease and preceding ROS1 inhibitor at the time plasma was collected. Patients progressing on crizotinib are highlighted in yellow whereas those relapsing on a next-generation ROS1 inhibitor are captured in blue rows.
Table 1.
ROS1 Fusions Detected in Plasma at Relapse
| Patient | Plasma Fusion | Tissue Fusion* | ROS1 Inhibitor | Sites of Progression |
|---|---|---|---|---|
| 1 | CD74-ROS1 | CD74-ROS1 | Lorlatinib | Extra-thoracic nodes, Mediastinal nodes |
| 2 | CD74-ROS1 | CD74-ROS1 | Crizotinib | Bone, Liver, Lung, Peritoneum |
| 3 | CD74-ROS1 | N/A—FISH | Crizotinib | Bone, Brain |
| 4 | CD74-ROS1 | CD74-ROS1 | Lorlatinib | Lung |
| 5 | CD74-ROS1 | N/A—FISH | Crizotinib | Lung, Soft Tissue |
| 6 | CD74-ROS1 | CD74-ROS1 | Entrectinib | Extra-thoracic nodes, Liver |
| 7 | CCDC6-ROS1 | CCDC6-ROS1 | Lorlatinib | Liver |
| 8 | EZR-ROS1 | EZR-ROS1 | Crizotinib | Brain, Lung |
| 9 | SDC4-ROS1 | SDC4-ROS1 | Crizotinib | Brain, Lung, Liver, Mediastinal nodes |
| 10 | TFG-ROS1 | N/A—FISH | Crizotinib | Lung |
| 11 | None | N/A—FISH | Crizotinib | Lung |
| 12 | None | N/A—FISH | Crizotinib | Lung |
| 13 | None | SLC34A2-ROS1 | Crizotinib | Pleural effusion |
| 14 | None | SLC34A2-ROS1 | Crizotinib | Lung |
| 15 | None | CD74-ROS1 | Repotrectinib | Lung, Mediastinal lymph nodes |
| 16 | None | N/A—FISH | Crizotinib | Mediastinal lymph nodes |
| 17 | None | N/A—FISH | Crizotinib | Liver, Lung |
| 18 | None | N/A—FISH | Crizotinib | Brain, Mediastinal node, Retroperitoneal node |
| 19 | None | EZR-ROS1 | Crizotinib | Adrenal, Lung, Retroperitoneal node |
| 20 | None | N/A—FISH | Cabozantinib | Liver |
| 21 | No ctDNA | LIMA-ROS1 | Crizotinib | Lung |
| 22 | No ctDNA | SLC34A2-ROS1 | Crizotinib | Pleural effusion |
| 23 | No ctDNA | N/A—FISH | Crizotinib | Brain, Hilar nodes |
| 24 | No ctDNA | N/A—FISH | Lorlatinib | Isolated soft tissue mass |
Tissue fusion partner was determined at any point molecular testing was performed during a patient’s disease course
Molecular Landscape of Crizotinib Resistance
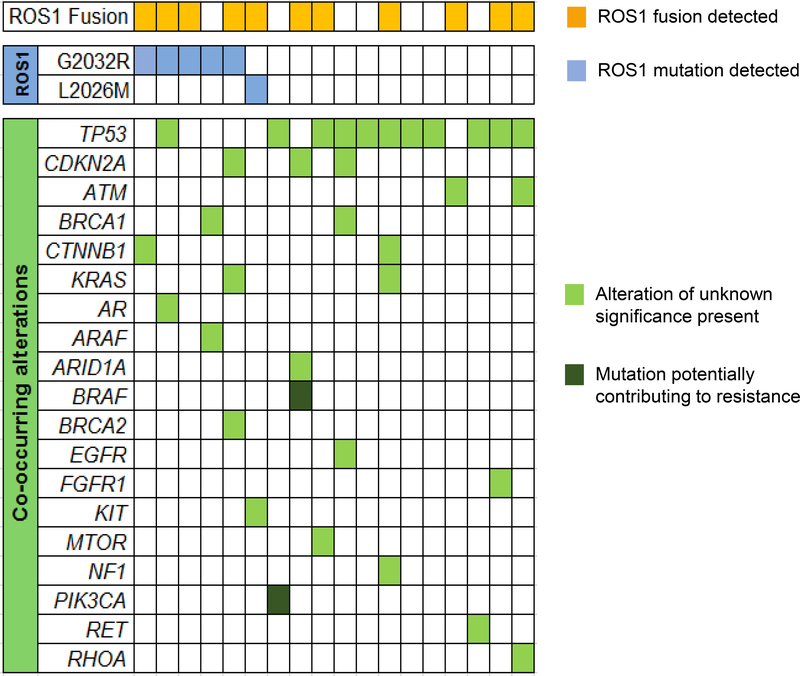
Acquired mutations in the ROS1 kinase domain reactivate ROS1 signaling during treatment with crizotinib by introducing steric interference or altering electrostatic interactions.1 These secondary ROS1 kinase domain mutations drive resistance to ROS1 inhibition.2 To determine the prevalence of ROS1 kinase domain mutations in plasma at relapse and detect mutations in other genes that may contribute to crizotinib relapses, we analyzed plasma from the 18 patients with acquired resistance to crizotinib who had detectable ctDNA in plasma specimens (Figure 3).
Figure 3. Landscape of Genetic Alterations in Crizotinib-Resistant Plasma Specimens.
The grid depicts genetic alterations detected in plasma collected from patients with ROS1-positive lung cancer at relapse on crizotinib. In some cases, multiple genetic alterations involving a single gene were identified but this is not captured in the heatmap. Apart from CDKN2A which includes copy number changes, all other genetic alterations were single nucleotide variants.
As described in the section above, fourteen of the 18 patients were receiving care at our institution when plasma was sent. The other four specimens represented cases from the Guardant de-identified dataset where the clinical history provided by the ordering provier indicated recurrence on crizotinib. Findings from the 14 patients from our institution are listed in Table 1 alongside plasma results from patients with recurrence after treatment with other ROS1 inhibitors. Of the 18 plasma specimens analyzed at crizotinib relapse, six (33%) had ROS1 kinase domain mutations, including five specimens with the G2032R solvent front mutation and one with an L2026M gatekeeper mutation. Two of the twelve specimens lacking on-target mutations had other mutations (PIK3CA E545K and BRAF V600E) potentially associated with ROS1-independent resistance. As paired tissue biopsies were not available for these two patients, we could not confirm whether the PIK3CA and BRAF mutations arose in the context of ROS1-positive NSCLC. We detected genetic alterations in other cancer-associated genes in plasma from the remaining 10 patients including ARAF R211C, FGFR1 I538I, KIT E88K, KRAS I24N and V8I, MTOR Y64C, NF1 E1516D, and RET P695S. However, these mutations were variants of unknown significance. Overall, we identified potential genetic mediators of resistance in 44% of post-crizotinib plasma specimens using the 73-gene panel.
We have previously reported results from analysis of repeat tissue biopsies from 14 patients with ROS1-positive NSCLC who were progressing at extracranial sites during treatment with crizotinib.2 Since the original study, an additional 11 patients have undergone tissue genotyping at crizotinib relapse. We compared the frequency of ROS1 mutations in post-crizotinib tissue and plasma specimens. Of note, four patients underwent paired plasma and tissue sampling at progression. The proportion of tissue specimens with ROS1 kinase domain mutations (n=9/25, 36%) was comparable to our observations from plasma analysis.
Tissue-Plasma Concordance
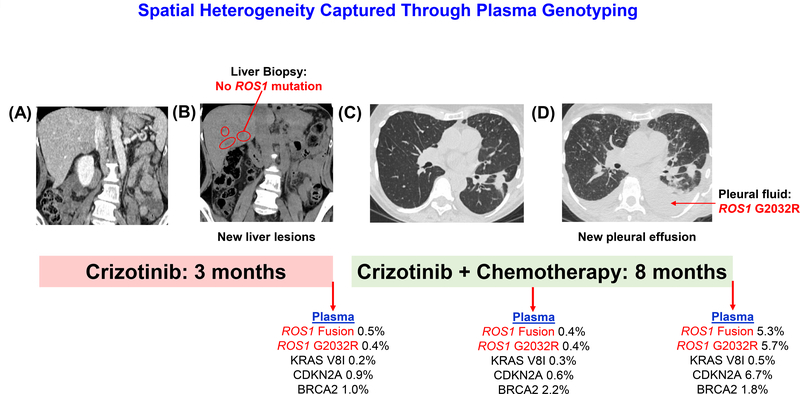
Tissue and plasma were simultaneously analyzed for four of the 18 patients relapsing on crizotinib. The interval between tissue and plasma collection was between 1–28 days. The remaining patients either did not undergo tissue biopsy at progression (n=10) or information regarding tissue sampling was not provided when plasma testing was requested (n=4). In two instances, neither tissue nor plasma contained ROS1 mutations or non-ROS1 genetic alterations previously implicated in activation of bypass pathways. ROS1 G2032R was detected in tissue and plasma specimens collected 15 days apart from Patient #13 in Table 1. In the remaining case (Patient #2 in Table 1), ROS1 G2032R was detected in plasma but was not identified in a biopsy obtained from an enlarging liver lesion which was performed the day before plasma collection. Due to widespread progression, the patient was subsequently treated with the combination of carboplatin, pemetrexed, and crizotinib with response lasting eight months. At progression on combination therapy, she underwent sampling of pleural fluid and plasma, both of which demonstrated ROS1 G2032R, confirming the initial plasma results (Figure 4).
Figure 4: Spatial Heterogeneity Captured Through Plasma Genotyping.
The figures illustrate the disease course of a patient with ROS1-positive NSCLC sequentially treated with crizotinib followed by crizotinib combined with chemotherapy. The patient did not have liver metastases at diagnosis (A) but developed new liver lesions (B) after 3 months on crizotinib at which time only plasma detected ROS1 G2032R. At subsequent progression on combination therapy, G2032R was detected in pleural fluid (pleural fluid is absent pre-treatment in Panel C and present at relapse in Panel D) and plasma.
DISCUSSION
In this manuscript, we present the first comprehensive analysis of plasma from patients with ROS1-positive NSCLC. Our findings demonstrate that plasma genotyping can detect the same spectrum of ROS1 fusions observed in tissue. In our study, plasma genotyping identified genetic alterations mediating resistance in 8 of 18 (44%) crizotinib-resistant patients, including six with ROS1 kinase domain mutations. The frequency of secondary ROS1 mutations in plasma was similar to that observed in tumor tissue, suggesting that plasma genotyping may have utility in the diagnostic evaluation of patients with resistant disease.
At relapse on ROS1 targeted therapy, we found that the sensitivity of plasma genotyping for detecting ROS1 fusions was 50%. In comparison, sensitivity for detecting ALK fusions was 86% in a similarly-sized cohort of patients relapsing on targeted therapy.9 In EGFR-mutant NSCLC, the sensitivity for detecting known sensitizing EGFR mutations (i.e., exon 19 deletion and L858R) and the T790M resistance mutation was 82–86% and 70%, respectively, in an analysis of over 200 patients.7 The relatively low sensitivity for detecting ROS1 fusions is consistent with previous studies showing that detection of ROS1 rearrangements is more challenging than identifying activating mutations or other oncogenic fusions (e.g. ALK rearrangements).5 For tissue genotyping, several strategies have been employed to improve ROS1 fusion detection, including extensive tiling of introns, greater probe coverage of the more common fusion partner genes, and creation of bioinformatics programs to optimize fusion calls. Applying these approaches to plasma NGS testing could potentially improve ROS1 fusion detection in plasma. For now, given the limitations of current plasma diagnostics, confirmatory tissue testing is recommended to exclude the possibility of a false negative result in cases where plasma fails to uncover an oncogenic driver alteration. In contrast to initial diagnosis where high sensitivity for detecting ROS1 fusions is critical, clinical decision-making in the relapse setting may depend more on identifying other alterations than finding the ROS1 fusion. Specifically, when a diagnosis of ROS1-positive NSCLC has been established, detecting ROS1 kinase domain mutations in plasma at high sensitivity at relapse is more informative for selection of subsequent therapies than improving ROS1 fusion calls.
In addition to the limitations of plasma genotyping assays, intrinsic disease characteristics can compromise the utility of plasma genotyping. For example, studies have shown that ctDNA yield is lowest when metastatic sites are limited to the thoracic cavity or central nervous system.7,22 A sizeable subset of patients with ROS1-positive NSCLC present with lung-only metastases (M1a disease).2 Furthermore, pharmacokinetic limitations of crizotinib result in a high incidence of CNS relapses.2 These disease features may contribute to the limited sensitivity of plasma genotyping for detection of ROS1 fusions. Indeed, nine (45%) of 20 specimens that were analyzed for presence of ROS1 fusion at relapse were collected from patients who were only progressing in the chest or brain. Notably, six (60%) of the 10 specimens that lacked plasma ROS1 fusions represented intrathoracic or brain-only progression compared to 3 (30%) of those with detectable fusions. Larger tissue-plasma concordance studies are needed to determine whether the relatively low sensitivity observed in our study is truly reflective of all-comers with ROS1-positive NSCLC or rather represents a population inadvertently enriched for those with lower disease burden.
Current understanding of molecular determinants of resistance to crizotinib is derived from two small series, including one from our group which detected acquired ROS1 kinase domain mutations in 9 (64%) of 14 post-crizotinib biopsies from extracranial sites.2,23 In contrast, only 1 (8%) of 12 biopsies from patients relapsing on crizotinib contained ROS1 mutations in the second series.23 In an updated analysis of our tissue cohort, we found ROS1 mutations in 9 (36%) of 25 post-crizotinib tissue specimens. The contradictory findings in the single institution studies might reflect small sample size. Although our plasma cohort was similarly limited by size, we identified ROS1 resistance mutations in six (33%) crizotinib-resistant plasma specimens, supporting the notion that ROS1 kinase domain mutations are a significant cause of resistance to crizotinib. Beyond secondary ROS1 mutations, off-target mechanisms such as activating mutations in KIT or KRAS are also implicated in crizotinib resistance.24,25 In our plasma cohort, two (11%) specimens harbored genetic alterations (PIK3CA E545K and BRAF V600E) potentially associated with activation of bypass signals. Notably, PIK3CA and BRAF mutations have been described in other malignancies. It is possible that these mutations originated from malignant or premalignant lesions independent of the ROS1-positive NSCLC. As oncogenic alterations in other genes can impact efficacy of ROS1-selective approaches, there is value in developing comprehensive assays that enable real-time assessment of resistance.
Notably, one patient relapsing on crizotinib was found to have ROS1 G2032R in plasma but not in a paired liver biopsy (Figure 4). Interestingly, after the patient developed resistance to subsequent treatment with chemotherapy and crizotinib, both plasma genotyping and molecular profiling of a malignant effusion revealed G2032R, confirming the initial plasma findings and suggesting that the prior discordant results may have been due to spatial heterogeneity. The incidence of spatial heterogeneity is not clearly established but is known to increase in later stages of NSCLC and after multiple lines of treatment have been administered. Multiple reports demonstrate that such heterogeneity may result in mixed or lesion-specific responses when treatment is based on single site biopsy.26,27 This suggests that plasma genotyping may be of even greater value later in the disease course where it can improve upon the ability of tissue analysis to capture the complexities of the heterogeneous resistance landscape.
There are limitations of our study that should be considered when interpreting our findings. The Guardant plasma cohort was assembled from a deidentified database. Because tissue findings are seldom provided at the time blood is submitted to Guardant Health, we could not formally evaluate sensitivity or tissue-plasma concordance. We acknowledge that this is a major limitation. Similarly, we could not report positive and negative predictive value since our cohort was entirely comprised of patients known to have ROS1-positive NSCLC. When able, we reported sensitivity and tissue-plasma concordance for cases submitted from our site. A limited number of plasma specimens was assessed in our analysis of resistance to crizotinib. Although the degree of consistency across plasma and tissue cohorts was encouraging, larger studies are necessary to comprehensively assess genetic mediators of resistance to crizotinib. The absence of on-target and off-target mutations in a significant proportion of specimens highlights the need for preclinical models that enable in-depth characterization of multiple facets of resistance, including non-genetic mechanisms of resistance.
In summary, we have performed the largest study of plasma genotyping in patients with ROS1-positive NSCLC. Our findings demonstrate that NGS-based plasma genotyping is an informative method for identifying ROS1 fusions and detecting molecular alterations that confer resistance to ROS1-directed therapies. In cases where neither a ROS1 genetic alteration nor any other oncogenic driver alteration is detected in plasma, tissue analysis should be performed to identify molecular drivers of resistance to ROS1 targeted therapies.
Acknowledgments
DISCLOSURES:
Dr. Dagogo-Jack reports honoraria from Foundation Medicine, personal fees from Boehringer Ingelheim, research support from Pfizer, outside the submitted work.
Ms. Nagy is an employee of Guardant Health.
Dr. Lin reports personal fees from Boehringer Ingelheim, personal fees from Chugai, outside the submitted work.
Dr. Lanman is an employee of Guardant Health..
Dr. Gainor reports personal fees from Bristol-Myers Squibb, personal fees from Genentech, personal fees from Ariad/Takeda, personal fees from Loxo, personal fees from Pfizer, personal fees from Incyte, personal fees from Novartis, personal fees from Merck, personal fees from Agios, personal fees from Amgen, personal fees from Array, personal fees from Clovis Oncology, personal fees from Jounce, research support from Genentech and Ariad/Takeda, outside the submitted work.
Dr. Shaw reports personal fees from Pfizer, personal fees from Novartis, personal fees from Genentech/Roche, personal fees from Ariad/Takeda, personal fees from Ignyta, personal fees from Loxo, personal fees from Blueprint Medicines, personal fees from KSQ Therapeutics, personal fees from Daiichi Sankyo, personal fees from EMD Serono, personal fees from Taiho Pharmaceutical, personal fees from TP Therapeutics, personal fees from Foundation Medicine, personal fees from Natera, outside the submitted work.
Footnotes
NOTHING TO DISCLOSE:
Emily Chin
Marguerite Rooney
Jennifer Ackil
Jochen K. Lennerz
Lorin A. Ferris
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lin JJ, Shaw AT. Recent Advances in Targeting ROS1 in Lung Cancer. J Thorac Oncol. 2017;12(11):1611–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gainor JF, Tseng D, Yoda S, et al. Patterns of Metastatic Spread and Mechanisms of Resistance to Crizotinib in ROS1-Positive Non-Small-Cell Lung Cancer. JCO Precis Oncol. 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Facchinetti F, Loriot Y, Kuo MS, et al. Crizotinib-Resistant ROS1 Mutations Reveal a Predictive Kinase Inhibitor Sensitivity Model for ROS1- and ALK-Rearranged Lung Cancers. Clin Cancer Res. 2016;22(24):5983–5991. [DOI] [PubMed] [Google Scholar]
- 4.Ross JS, Ali SM, Fasan O, et al. ALK Fusions in a Wide Variety of Tumor Types Respond to Anti-ALK Targeted Therapy. Oncologist. 2017;22(12):1444–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies KD, Le AT, Sheren J, et al. Comparison of Molecular Testing Modalities for Detection of ROS1 Rearrangements in a Cohort of Positive Patient Samples. J Thorac Oncol. 2018;13(10):1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med. 2018;379(18):1754–1765. [DOI] [PubMed] [Google Scholar]
- 7.Oxnard GR, Thress KS, Alden RS, et al. Association Between Plasma Genotyping and Outcomes of Treatment With Osimertinib (AZD9291) in Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2016;34(28):3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newman AM, Bratman SV, To J, et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat Med. 2014;20(5):548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dagogo-Jack I, Brannon AR, Ferris LA, et al. Tracking the Evolution of Resistance to ALK Tyrosine Kinase Inhibitors through Longitudinal Analysis of Circulating Tumor DNA. JCO Precis Oncol. 2018;2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCoach CE, Blakely CM, Banks KC, et al. Clinical Utility of Cell-Free DNA for the Detection of ALK Fusions and Genomic Mechanisms of ALK Inhibitor Resistance in Non-Small Cell Lung Cancer. Clin Cancer Res. 2018;24(12):2758–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paweletz CP, Sacher AG, Raymond CK, et al. Bias-Corrected Targeted Next-Generation Sequencing for Rapid, Multiplexed Detection of Actionable Alterations in Cell-Free DNA from Advanced Lung Cancer Patients. Clin Cancer Res. 2016;22(4):915–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non-small cell lung cancer. Ann Oncol. 2018;29(4):1049–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanman RB, Mortimer SA, Zill OA, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10(10):e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Z, Liebers M, Zhelyazkova B, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20(12):1479–1484. [DOI] [PubMed] [Google Scholar]
- 15.Frampton GM, Fichtenholtz A, Otto GA, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sholl LM, Do K, Shivdasani P, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight. 2016;1(19):e87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pritchard CC, Salipante SJ, Koehler K, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suehara Y, Arcila M, Wang L, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18(24):6599–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Ahn BC, Lim SW, et al. Characteristics and Outcome of ROS1-Positive Non-Small Cell Lung Cancer Patients in Routine Clinical Practice. J Thorac Oncol. 2018;13(9):1373–1382. [DOI] [PubMed] [Google Scholar]
- 21.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective Validation of Rapid Plasma Genotyping for the Detection of EGFR and KRAS Mutations in Advanced Lung Cancer. JAMA Oncol. 2016;2(8):1014–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol. 2018;29(3):700–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCoach CE, Le AT, Gowan K, et al. Resistance Mechanisms to Targeted Therapies in ROS1(+) and ALK(+) Non-small Cell Lung Cancer. Clin Cancer Res. 2018;24(14):3334–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dziadziuszko R, Le AT, Wrona A, et al. An Activating KIT Mutation Induces Crizotinib Resistance in ROS1-Positive Lung Cancer. J Thorac Oncol. 2016;11(8):1273–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cargnelutti M, Corso S, Pergolizzi M, et al. Activation of RAS family members confers resistance to ROS1 targeting drugs. Oncotarget. 2015;6(7):5182–5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwak EL, Ahronian LG, Siravegna G, et al. Molecular Heterogeneity and Receptor Coamplification Drive Resistance to Targeted Therapy in MET-Amplified Esophagogastric Cancer. Cancer Discov. 2015;5(12):1271–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6(2):147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]