Abstract
Many patients with opioid use disorder do not have successful outcomes during treatment but the underlying reasons are not well understood. An OPRD1 variant (rs678849) was previously associated with methadone and buprenorphine efficacy in African-Americans with opioid use disorder. The objective of this study was to determine if the effect of rs678849 on opioid use disorder treatment outcome could be replicated in an independent population. Participants were recruited from African-American patients who had participated in previous studies of methadone or buprenorphine treatment at the outpatient treatment research clinic of the NIDA Intramural Research Program in Baltimore, MD, USA between 2000 and 2017. Rs678849 was genotyped retrospectively and genotypes were compared to urine drug screen results from the previous studies for opioids other than the one prescribed for treatment. Genotypes were available for 24 methadone patients and 55 buprenorphine patients. After controlling for demographics, the effect of rs678849 genotype was significant in the buprenorphine treatment group (RR = 1.69, 95% confidence interval (CI) 1.59–1.79, p = 0.021). Buprenorphine patients with the C/C genotype were more likely to have opioid-positive drug screens than individuals with the C/T or T/T genotypes, replicating the original pharmacogenetic finding. The effect of genotype was not significant in the methadone group (p = 0.087). Thus, genotype at rs678849 is associated with buprenorphine efficacy in African-Americans being treated for opioid use disorder. This replication suggests that rs678849 genotype may be a valuable pharmacogenetic marker for deciding which opioid use disorder medication to prescribe in this population.
Introduction
Opioid use disorder (OUD) includes dependence on heroin and prescription opioid analgesics. The disorder affects millions of people worldwide and has grown into an epidemic in the United States, where the National Survey on Drug Use and Health suggested that ~2.6 million people were suffering from OUD in 2015 1. Society pays a significant price for this ongoing problem in the forms of health care costs, missed work, criminal activity, and premature mortality. Overdose deaths in the United States have reached historically high rates; more than 50,000 Americans died of opioid overdoses in 2016 according to the Centers for Disease Control and Prevention.
Opioids of abuse are primarily agonists of the mu-opioid receptor (MOR). Methadone, a MOR agonist, and buprenorphine, a MOR partial agonist and kappa-opioid receptor antagonist, are FDA approved for OUD treatment. Both medications have been repeatedly proven to be effective at reducing illicit opioid use when compared with placebo 2. However, individual patients may have varying levels of OUD treatment success and many people will not reduce illicit opioid use on methadone or buprenorphine 2.
Variation in treatment efficacy is affected by genetic factors specific to individual patients the (i.e. pharmacogenetics). Pharmacogenetic findings have been described for therapies for alcohol and tobacco use disorders 3–6 However, similar findings are limited with regard to OUD and often focus on dose or serum levels rather than treatment outcome (reviewed in 7). The cytochrome P450 family of genes encodes enzymes that metabolize a large number of molecules, including methadone and buprenorphine 8–10. Pharmacokinetic status of family members such as CYP2B6 and CYP3A4 have been associated with plasma concentrations of methadone but connections to dose requirements have been more equivocal 7, 11–14. Genotypes at ABCB1, a gene encoding a transmembrane efflux pump, have also been linked to methadone serum levels and dose 15–17. As with the CYP genes, a number of studies have failed to replicate the ABCB1 associations with dose (reviewed in 7). Recent genome-wide association studies (GWAS) have implicated other genes potentially relevant to dose or serum levels of methadone, including SPON1 and GSG1L 18 and the region upstream of OPRM1 19.
Pharmacogenetic effects on OUD treatment efficacy have also been identified, although confirmation in additional studies has not occurred. Polymorphisms in ARRB2, DRD2, and BDNF have all been associated with methadone outcome when patients are classified as responders or non-responders 20–22. A small study found patients with the ultra-rapid metabolizer phenotype for CYP2D6 to have a lower rate of successful methadone treatment than those with the poor metabolizer phenotype 23.
Our group has previously studied pharmacogenetics in the Starting Treatment with Agonist Replacement Therapy (START) trial, which was a randomized, open label trial of OUD treatment funded by the National Institute on Drug Abuse (NIDA) in response to a report of liver toxicity problems with buprenorphine 24. Patients were randomized to either methadone or buprenorphine for a 24 week course of treatment. In addition to a liver enzyme analysis, blood samples were collected to study the pharmacogenetics of the two medications. In this sample set, a 3’ untranslated region variant in the mu-opioid receptor gene (OPRM1) predicted methadone efficacy in European-Americans and a polymorphism in the delta-opioid receptor gene (OPRD1) was associated with buprenorphine efficacy in woman of that ethnic group 25, 26. The most significant finding, however, was rs678849 in OPRD1 27. This SNP was originally chosen for study due to its presence in a haplotype previously associated with opioid dependence 28. This intronic variant was associated with both methadone and buprenorphine efficacy in African-Americans and had potential clinical significance due to the large size of the pharmacogenetic effect. Patients with the C/C genotype at rs678849 did worse on buprenorphine than those with the C/T or T/T genotypes. The opposite pharmacogenetic effect was observed in patients receiving methadone.
Like the other pharmacogenetic associations described above, replication of the rs678849 finding is necessary before the variant can be used to guide treatment decisions in a clinical setting. In this study we attempted to replicate the effect of the SNP in an independent population of African-Americans in Baltimore who had received either methadone or buprenorphine as part of OUD treatment studies. A combined analysis of the replication sample and the START trial population was also performed.
Materials and Methods
Participants and sample collection
Replication Cohort (Baltimore)
Individuals were recruited for one of 4 OUD treatment studies at the outpatient treatment research clinic at the NIDA Intramural Research Program (NIDA IRP) (Baltimore, MD, USA) between 2000 and 2017: Protocol 326 - Combined Behavioral and Pharmacologic Treatment of Polydrug Abuse - Arms 326–1 and 326–2; Protocol 385 - Real-time assessment of drug craving, use, and abstinence during outpatient treatment: A development and feasibility study; Protocol 407 - Clonidine for relapse prevention in buprenorphine-maintenance patients; Protocol 020 - Developing field tools for real-time assessment of exposure to psychosocial stress and drug use in an outpatient treatment population (Arms Methadone (020-MTD), Buprenorphine (020-BUP), and Office-based Buprenorphine (020-OBOT). Methodologies for studies 326–1, 326–2, 385, 407, 020-MTD, and 020-BUP have been previously published 29–33. Methodology for study 020-OBOT is provided in the supplemental material. All patients were at least 18 years of age and had physical dependence on opioids. Cocaine and opioid positive urine samples were also required for inclusion in studies 326–1, 326–2, and 385. Patients were excluded for any of the following reasons: 1) Axis 1 psychiatric disorders (e.g schizophrenia, bipolar disorder, etc); 2) alcohol or sedative dependence; 3) severe medical illness; 4) any condition that would interfere with urine collection; 5) severe cognitive impairment that would prevent informed consent. Ethnicity was self-reported. Treatment consisted of open-label methadone or buprenorphine/naloxone in combination with weekly individual counseling. Urine drug screens were performed three times per week (020-OBOT two times per week). Summarized details for the individual studies are presented in Table 1. For the pharmacogenetic replication study, individuals were re-recruited between 2015 and 2017 and provided a venous blood sample. In total, genotypes were available for 24 methadone patients and 55 buprenorphine patients. The institutional review board for the NIDA Intramural Research Program approved all protocols for the original trials and the protocol of the pharmacogenetic study. All subjects provided written informed consent for the original study and the pharmacogenetic study.
Table 1.
Sample size and methodological details for trials of methadone or buprenorphine for the treatment of opioid dependence
| Study | N | Medication | Length | Abstinence Reinforcement | Dose Limits | Miscellaneous |
|---|---|---|---|---|---|---|
| 326–1 | 1 | Methadone | 25 weeks | Cocaine and/or opioids | 70 mg/day | 7 visits/week |
| 326–2 | 1 | Methadone | 25 weeks | Cocaine or opioids | 100 mg/day | 7 visits/week |
| 385 | 4 | Methadone | 20 weeks | Opioids | 100 mg/day | 7 visits/week |
| 020-MTDa | 18 | Methadone | 46 weeks | None | No ceiling; target dose 100 mg/day | 5–7 visits/week |
| 020-BUPa | 25 | Buprenorphine | 46 weeks | None | No ceiling; target dose 16 mg/day | 5 visits/week |
| 020-OBOTa | 19 | Buprenorphine | 22 weeks | None | 24 mg/day | 2–3 visits/week |
| 407 | 11 | Buprenorphine | 28 weeks | Opioids | 24 mg/day | 7 visits/week. Randomized to clonidine or placebo for weeks 7–20 |
Protocol 020 arms: Methadone Maintenance (020-MTD); Buprenorphine Maintenance (020-BUP; Office-Based Therapy (020-OBOT)
Discovery Cohort (START)
START was a 24 week, randomized, open-label trial of methadone and buprenorphine/naloxone. Methodology and primary outcomes for the trial have been described 24. Recruitment at federally licensed OUD treatment programs in the United States took place between May 2006 and October 2009. All patients met DSM-IV-TR criteria for opioid dependence. Patients were excluded for any of the following reasons: <18 years of age, cardiomyopathy, liver disease, acute psychosis, blood levels of alanine amino transferase or aspartate amino transferase greater than five times the maximum normal level, or poor venous access. Institutional review boards at participating sites approved the study. Oversight was provided by the NIDA Clinical Trials Network Data Safety and Monitoring Board. All patients provided written informed consent.
Genotyping
DNA was extracted from 2uL of whole blood using the DNA Extract All Reagents Kit (ThermoFisher). Genotyping of rs678849 was performed on an ABI 7900 Thermocycler using a Taqman SNP Genotyping Assay (ThermoFisher) as previously described 34.
Statistical analysis
Rs678849 was in Hardy-Weinberg Equilibrium in the replication cohort as determined by chi-square analysis (p>0.05). The effect of rs678849 genotype on treatment outcome, as defined by all available urine drug screens for opioids other than the one prescribed during treatment (buprenorphine or methadone), was analyzed using a generalized estimating equation (GEE), which can be used for repeated binary measures. The GEE was performed independently for both treatment groups in the replication cohort to analyze the main effects of rs678849 genotype on treatment outcome. To maintain consistency with the original rs678849 pharmacogenetic analysis, individuals with T/T genotype were combined with individuals with the C/T genotype in the GEE and all subsequent analyses. Sex, age, dose, time, study, and cocaine dependence status were included in the model as covariates. Cocaine dependence status was included because of a previous identification of an association between rs678849 genotype and cocaine dependence in African-Americans 34. Given the sample sizes for the two treatment groups and the size of the previously observed pharmacogenetic effect, the power to detect a similar effect in the replication cohort was 58% and 97% for methadone and buprenorphine, respectively. An additional GEE was performed with both medication groups to analyze the interaction between rs678849 genotype and treatment. For all GEEs, an independence correlation structure was assumed and robust variance estimators were used. The estimates produced by the GEE are reported here as relative risks with bootstrapped 95% confidence intervals based on 10,000 replicates. Missing urine drug screens were excluded from all analyses. Patients with the C/C genotype had a higher proportion of missing tests compared to patients in the combined C/T and T/T genotypes group (4.71% vs 3.64%, p = 0.035).
A combined analysis was performed on the combined data from weeks 17–24 of buprenorphine patients in the START and Baltimore cohorts. The proportions of opioid positive urine samples in the C/C genotype group and the C/T+T/T genotypes group during those weeks were compared by chi-square analysis. Response to treatment was measured for the same time period. Non-responders were defined as patients who a) were not retained in treatment until at least week 17 (i.e. dropout) or b) were opioid positive for ≥ 50% of their urine drug screens in weeks 17–24. All other patients were defined as responders. The proportions of responders in the two genotype groups were assessed by chi-square analysis.
Results
Participants and demographics
Table 2 contains information on mean age, mean maximal dose, the percentage of men, and the mean percentage of opioid positive urine drug screens for methadone and buprenorphine patients in the original START trial, the replication cohort, and the combined sample. In the replication cohort, the buprenorphine treatment group had a higher percentage of males than the methadone treatment group (83.6% vs 54.2%, p = 0.006) and had significantly more opioid positive urine drug screens (53.3 ± 37.5% vs 34.8 ± 27.7%, p = 0.035). Buprenorphine patients in the replication sample also had significantly lower maximal doses than buprenorphine patients in the START trial (16.9 ± 3.3 vs 22.5 ± 6.9, p <0.001). In contrast, methadone patients in the replication sample had significantly higher maximal doses than patients in the START trial (97.3 ± 16.1 vs 79.2 ± 26.5, p = 0.005).
Table 2.
Demographic information and treatment outcomes for African-American patients treated with methadone or buprenorphine/naloxone for opiod dependence by rs678849 genotype
| START | Replication | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Treatment | Methadone | Buprenorphine | Methadone | Buprenorphine | Buprenorphine | |||||
| rs678849 | C/C | C/T+T/T | C/C | C/T+T/T | C/C | C/T+T/T | C/C | C/T+T/T | C/C | C/T+T/T |
| Number (% male) | 21 (66.7%) | 15 (80.0%) | 24(62.5) | 17(70.1%) | 13 (46.5%) | 11 (63.6%) | 33 (75.8) | 22 (95.5%) | 57 (70.2%) | 39 (84.6%) |
| Mean age±SD | 48.6±7.9 | 48.5±9.6 | 49.6±8.8 | 44.3±1.3 | 51.5±6.4 | 51.5±6.1 | 50.0±6.5 | 48.9±5.7 | 49.8±7.6 | 46.9±8.3 |
| Mean maximal dose ±SD | 86.0±27.5 | 69.7±21.8 | 22.0±7.2 | 23.3±6.3 | 90.4±17.1 | 105.5±9.9 | 16.6±3.8 | 17.4±2.4 | 18.9±6.1 | 19.9±5.4 |
| Mean % opioid positive UDS± SD | 42.7±30.0% | 64.2±36.1% | 60.1±37.2 | 30.7±32.3 | 29.7±28.8 | 40.8±25.0 | 58.7±38.5 | 45.1±37.4 | 59.6±36.8% | 38.8±36.0% |
Abbreviations: UDS, Urine Drug Screens; SD, Standard Deviation
Replication analysis
A GEE was used to analyze the effect of rs678849 genotype on urinalysis data for each treatment group separately, as well as for a genotype x treatment analysis comparing the genotypic effects in the two medication groups, while accounting for the effects of age, sex, time, dose, study, and cocaine dependence status. Missing tests were excluded from the analysis. As in the START trial, the effect of rs678849 genotype was significant in the buprenorphine treatment group (RR = 1.69, 95% confidence interval (CI) 1.59–1.79, p = 0.021). Buprenorphine patients with the C/C genotype were more likely to have opioid-positive drug screens than individuals with the C/T or T/T genotypes, replicating the original pharmacogenetic finding in the START cohort (Figure 1). The effect of genotype was not significant in the methadone group (p = 0.087), although the direction of the effect matched that observed in the START trial. There was also no significant interaction between genotype and treatment group in the gene x environment analysis (p = 0.076).
Figure 1.
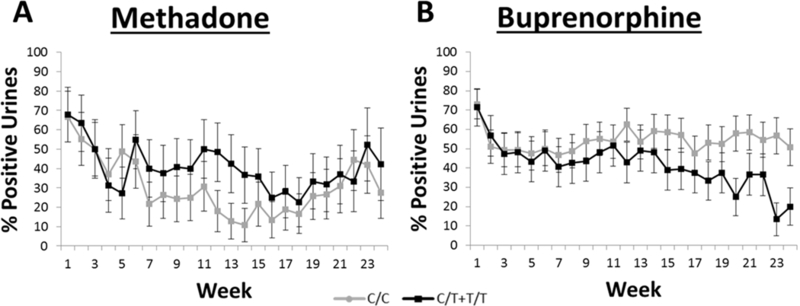
Average percentage of opioid positive urine drug screens for African-Americans based on rs678849 genotype for the first 24 weeks of treatment. Patients were treated for opioid dependence with methadone (A) or buprenorphine (B) as part of four studies at the NIDA Intramural Research Program. The mean percentage of opioid-positive during each week is provided for individuals with either the C/C genotype or the C/T and T/T genotypes. Error bars represent S.E.M. Time, age, sex, dose, study, and cocaine dependence were used as covariates. Buprenorphine patients with the C/C genotype (n = 33) were more likely to submit urines that were positive for opioids than patients in the combined C/T and T/T genotype group (n = 22; RR = 1.69, 95% confidence interval (CI) 1.59–1.79, p = 0.021). No effect of rs678849 genotype was observed in the methadone treatment group (p = 0.087).
Combined analysis
The START trial and replication data sets were combined to analyze urine drug screen results for weeks 17–24, because that time period represents the last two months of treatment in the START trial. In that eight week period African-American buprenorphine patients with the C/C genotype at rs678849 had opioid positive urine samples 56.3% of the time, compared to 30.7% for patients in the combined C/T and T/T genotypes group (p < 0.0001). Patients were also grouped as either “responders” or “non-responders” for further analysis. Non-responders were either not retained in treatment until at least week 17 or submitted ≥50% opioid positive urines in weeks 17–24 of treatment. Only 28.1% (16/57) of African-American buprenorphine patients with the C/C genotype met responder criteria, compared to 61.5% (24/39) for patients in the combined C/T and T/T genotypes group (p = 0.001). The number needed to treat (NNT) was 3.
Discussion
The choice of prescribing methadone or buprenorphine to an individual patient is rarely an evidence-driven decision based on which medication is most likely to provide the better outcome. This situation arises from a lack of knowledge about the factors affecting treatment outcome; there is currently no FDA-approved pharmacogenetic marker for selecting an OUD medication. In this study, we replicated a pharmacogenetic association between an intronic variant in OPRD1 (rs678849) and the efficacy of buprenorphine in treating African-Americans with OUD. This is the first successful replication of a pharmacogenetic effect in OUD treatment to our knowledge and rs678849 has the potential to improve treatment outcomes if used as a biomarker for guiding clinical prescribing practices.
OUD is a strong candidate for the use of pharmacogenetic markers due to two factors. The first is that the available treatments for the disorder work well in reducing or preventing illicit opioid use for many people but are ineffective in a subset of the population. While there are environmental factors that affect patient response to substance use disorder therapies (e.g. psychiatric co-morbidities, personal relationships and support networks) 35, there is also a role for genetic background in altering treatment efficacy through pharmacokinetic or pharmacodynamic mechanisms. Identification of variants associated with treatment outcome could be used to stratify patients based on predicted treatment efficacy.
The second important factor is that OUD has more than one FDA-approved pharmacotherapy. Therefore patients for whom one medication is predicted to be ineffective can be prescribed an alternate option. Patients are often not prescribed the most efficacious medication as a first line therapy and there are significant repercussions to the poor treatment outcomes that result from this issue. Continued use of illicit opioids leads to reduced quality of life, as well as increased risks of disease and overdose. Ineffective OUD treatment also creates monetary burdens for the patients, the healthcare system, and society in general. OUD costs the United States billions of dollars annually in the form of health care and lost productivity 36. The C/C genotype has a frequency of ~54% among African-Americans. Since African-American patients with the C/C genotype at rs678849 have poor outcomes with buprenorphine, they may be better served by the prescription of methadone or naltrexone. Similarly, patients carrying the T allele at the variant might have better outcomes if prescribed buprenorphine rather than other medications. Selecting the medication with the best chance of a successful outcome using a pharmacogenetic biomarker such as rs678849 could help minimize the time between the start of medication and reduced opioid use, improve patient quality of life, and reduce the costs associated with OUD.
Several unanswered questions remain about the underlying mechanism of this pharmacogenetic effect. First, the direct functional consequences of rs678849 genotype have yet to be established. The variant has been associated with opioid dependence 28, 37 and cocaine dependence 34 in the past, along with the pharmacogenetic effect replicated in this study. Genotype at rs678849 was also associated with regional brain volume in individuals of European descent 38, further suggesting that the variant is relevant to human phenotypes. The location of the SNP in intron 1 of OPRD1 might suggest a role in expression or splicing of the gene. ChIP-seq data from the Roadmap Epigenomics Consortium found the rs678849 locus to be associated with epigenetic markers of active enhancers, including H3K4me1 in dorsolateral prefrontal cortex and spleen and H3K27ac in dorsolateral prefrontal cortex and inferior temporal lobe 39, 40. However, to date rs678849 has only been shown to be an expression quantitative trait locus (eQTL) for PHACTR4 and ATPIF1, genes located >300kb upstream of OPRD1 (GTEx Portal), and none of these associations have been found in brain or spleen41. Additional ChIP-analysis for markers of promoters and/or transcription start sites (i.e. H3K4me3 and H3K9ac) also identified the rs678849 region in several tissues, including inferior temporal lobe, anterior caudate, and spleen 39, 40. Although splice variants of OPRD1 have recently been found in human brain, none of the newly identified exons in intron 1 were in the vicinity of rs678849 42. Additional studies will be necessary to determine the functional consequences of rs678849 genotype with regard to OPRD1, PHACTR4, and ATPIF1 and what, if any, role epigenetics may play in this mechanism.
Another issue is that it is unclear why a variant in OPRD1 would affect buprenorphine efficacy, since the medication is thought to function through the mu- and kappa-opioid receptors (MOR and KOR) rather than the delta-opioid receptor (DOR). However, some connections between buprenorphine and DOR have been published. Although buprenorphine has no efficacy at DOR, it does have affinity for the receptor 43, 44, which could allow DOR to act as a sink for the drug. Belcheva et al also demonstrated that DOR, specifically the delta-2 subtype, was upregulated in the frontal and parietal cortexes of rats treated with buprenorphine 45, 46. Further, the effect of this SNP could be related to the formation of MOR-DOR heterodimers, which have been described in the central nervous system 47. The differences in pharmacogenetic effects on buprenorphine and methadone may also be informative in regards to mechanism. Methadone patients demonstrated an opposite effect of rs678849 genotype on efficacy in the original START trial compared to buprenorphine patients. Although this effect was not significant in the current study, the direction of the effect was the same. Chronic treatment of cells with methadone, unlike morphine, results in desensitization of DOR 48. If rs678849 genotype affects OUD treatment outcomes by causing differential expression of DOR, then desensitization of the receptor in methadone would mitigate this effect and might explain the differences in pharmacogenetic effects between the two treatment groups.
Finally, the specificity of the rs678849 effect on buprenorphine to the African-American population has yet to be explained. Variations in OPRD1 haplotype structure between African-Americans and European-Americans could also be relevant if rs678849 is not the causative variant. Other differences in genetic background might result in population-specific epistasis, masking the effects of rs678849 in patients of European descent. Environmental and socioeconomic factors, such as employment and stable housing, vary between ethnic groups and can have significant bearing on treatment outcomes 35, 49. Some studies have also found that European-Americans are more likely to be retained in treatment compared to minority patients 50, 51, although other studies have not found an effect of ethnicity 49. Gene x gene and gene x environment interactions that differ between populations are therefore likely to exist, further emphasizing the need for more detailed analyses of rs678849 in the context of race and socioeconomic status.
There are some limitations of this study that should be noted. First, both the initial analysis and the replication analysis were retrospective. The trials of methadone and buprenorphine from which subjects were recruited were not designed to identify genetic markers of treatment outcome, but for other clinical purposes. Therefore, there is a possibility that the rs678849 effect we have observed is a false positive caused by unknown confounding variables being unequally distributed between the genotype groups. While the replication of the effect in an independent cohort significantly reduces the likelihood of this possibility, a prospective clinical trial will be a necessary next step to verify the suitability of this finding to clinical care. Such a trial would be designed to specifically test the effect of rs678849 genotype on buprenorphine outcome in African-American and randomization would be stratified by genotype and structured to minimize the unequal distribution of confounding variables.
Another limitation of the replication study is the relatively small number of methadone patients that were successfully re-consented for pharmacogenetic analysis. This could potentially explain the inability to replicate the original effect of rs678849 genotype on methadone outcome because we were underpowered to detect it. Other differences between the discovery and replication cohorts could also have affected the methadone replication. The original START study was a nationwide trial run through the NIDA Clinical Trials Network, while the replication samples are exclusively from the Baltimore area. Socioeconomic factors in Baltimore could differ from some or all of the recruitment areas in START and affect outcome. Regional genetic variation in the African-American community could also play an unforeseen role. There are methodological differences between the discovery and replication studies as well. Polydrug abuse and alcohol dependence are both common in opioid dependent patients but patients with alcohol or sedative dependence were excluded from all of the trials represented in the replication cohort. In contrast, there was no such exclusion in the START trial. Finally, the START trial collected only one urine sample per week compared to three in the Baltimore studies, possibly making the replication data a better estimate of illicit opioid use during treatment. While the variations between studies might contribute to the lack of significance in the methadone arm of the replication analysis, the replication of the buprenorphine finding despite these differences suggests that this pharmacogenetic effect for buprenorphine is quite robust.
Successful replication of the effect of rs678849 genotype on buprenorphine efficacy in African-Americans suggests that the SNP may be an important biomarker of OUD treatment outcome in this population. Prescreening African-American OUD patients for rs678849 genotype prior to the start of medication could have significant benefits. However, more information on the variant and the pharmacogenetic effect are necessary to apply the marker to clinical care. A prospective clinical trial of rs678849 in a population of OUD patients randomized to methadone or buprenorphine will be needed. The goal of such a trial would be to move rs678849 towards FDA approval as a pharmacogenetic marker for selecting an OUD medication. There is also currently minimal information on the functional consequences of rs678849 genotype and how those consequences directly or indirectly affect buprenorphine efficacy. Understanding these biological mechanisms may provide other intermediate phenotypes that predict outcome in buprenorphine treatment or identify additional pharmaceutical targets that are relevant to OUD.
Supplementary Material
Funding and Disclosure
START study funding came from the National Institute on Drug Abuse through the Clinical Trials Network (CTN) through a series of grants provided to each participating node: the Pacific Northwest Node (U10 DA01714), the Oregon Hawaii Node (U10 DA013036), the California/Arizona Node (U10 DA015815), the New England Node (U10 DA13038), the Delaware Valley Node (U10 DA13043), the Pacific Region Node (U10 DA13045), and the New York Node (U10 DA013046). Protocol 326, Protocol 385, Protocol 407, and Protocol 020 were supported by the Intramural Research Program of the NIH National Institute on Drug Abuse (NIDA). Dr. Berrettini was supported by the Delaware Valley Node of the CTN (U10 DA13043) and by NIDA grant R21 DA036808. Dr. Crist was supported by NIDA grant K01 DA036751. Drs. Crist, Berrettini and Doyle were supported by NIDA grant R01 DA044015. The funders had no role in design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Dr. Kampman has received consulting fees from Alkermes and Opiant Pharmaceuticals. He has also received grant funding from Alkermes, Opiant Pharmaceuticals, and Indivior. Dr. Berrettini has received consulting fees from Mundipharma and Geisinger Health Systems and grant support from Saniona. All other authors report no conflicts of interest.
References
- 1.2015. National Survey on Drug Use and Health: Detailed Tables. Substance Abuse and Mental Health Services Administration, Rockville, MD. [Google Scholar]
- 2.Mattick RP, Breen C, Kimber J, Davoli M. Buprenorphine maintenance versus placebo or methadone maintenance for opioid dependence. Cochrane Database Syst Rev 2014;(2): CD002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kranzler HR, Covault J, Feinn R, Armeli S, Tennen H, Arias AJ , et al. Topiramate treatment for heavy drinkers: moderation by a GRIK1 polymorphism. Am J Psychiatry 2014; 171(4): 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lerman C, Wileyto EP, Patterson F, Rukstalis M, Audrain-McGovern J, Restine S , et al. The functional mu opioid receptor (OPRM1) Asn40Asp variant predicts short-term response to nicotine replacement therapy in a clinical trial. Pharmacogenomics J 2004; 4(3): 184–192. [DOI] [PubMed] [Google Scholar]
- 5.Anton RF, Oroszi G, O’Malley S, Couper D, Swift R, Pettinati H , et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry 2008; 65(2): 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chamorro AJ, Marcos M, Miron-Canelo JA, Pastor I, Gonzalez-Sarmiento R, Laso FJ. Association of micro-opioid receptor (OPRM1) gene polymorphism with response to naltrexone in alcohol dependence: a systematic review and meta-analysis. Addict Biol 2012; 17(3): 505–512. [DOI] [PubMed] [Google Scholar]
- 7.Dennis BB, Bawor M, Thabane L, Sohani Z, Samaan Z. Impact of ABCB1 and CYP2B6 genetic polymorphisms on methadone metabolism, dose and treatment response in patients with opioid addiction: a systematic review and meta-analysis. PLoS One 2014; 9(1): e86114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kharasch ED, Stubbert K. Role of cytochrome P4502B6 in methadone metabolism and clearance. J Clin Pharmacol 2013; 53(3): 305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iribarne C, Picart D, Dreano Y, Bail JP, Berthou F. Involvement of cytochrome P450 3A4 in N-dealkylation of buprenorphine in human liver microsomes. Life Sci 1997; 60(22): 1953–1964. [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi K, Yamamoto T, Chiba K, Tani M, Shimada N, Ishizaki T , et al. Human buprenorphine N-dealkylation is catalyzed by cytochrome P450 3A4. Drug Metab Dispos 1998; 26(8): 818–821. [PubMed] [Google Scholar]
- 11.Totah RA, Sheffels P, Roberts T, Whittington D, Thummel K, Kharasch ED. Role of CYP2B6 in stereoselective human methadone metabolism. Anesthesiology 2008; 108(3): 363–374. [DOI] [PubMed] [Google Scholar]
- 12.Shiran MR, Lennard MS, Iqbal MZ, Lagundoye O, Seivewright N, Tucker GT , et al. Pharmacokinetic-pharmacodynamic modeling of mood and withdrawal symptoms in relation to plasma concentrations of methadone in patients undergoing methadone maintenance treatment. J Clin Psychopharmacol 2012; 32(5): 666–671. [DOI] [PubMed] [Google Scholar]
- 13.Tsai HJ, Wang SC, Liu SW, Ho IK, Chang YS, Tsai YT , et al. Assessment of CYP450 genetic variability effect on methadone dose and tolerance. Pharmacogenomics 2014; 15(7): 977–986. [DOI] [PubMed] [Google Scholar]
- 14.Levran O, Peles E, Hamon S, Randesi M, Adelson M, Kreek MJ. CYP2B6 SNPs are associated with methadone dose required for effective treatment of opioid addiction. Addict Biol 2013; 18(4): 709–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levran O, O’Hara K, Peles E, Li D, Barral S, Ray B , et al. ABCB1 (MDR1) genetic variants are associated with methadone doses required for effective treatment of heroin dependence. Hum Mol Genet 2008; 17(14): 2219–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barratt DT, Coller JK, Hallinan R, Byrne A, White JM, Foster DJ , et al. ABCB1 haplotype and OPRM1 118A > G genotype interaction in methadone maintenance treatment pharmacogenetics. Pharmgenomics Pers Med 2012; 5: 53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee HY, Li JH, Sheu YL, Tang HP, Chang WC, Tang TC , et al. Moving toward personalized medicine in the methadone maintenance treatment program: a pilot study on the evaluation of treatment responses in Taiwan. Biomed Res Int 2013; 2013: 741403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang HC, Chu SK, Huang CL, Kuo HW, Wang SC, Liu SW , et al. Genome-Wide Pharmacogenomic Study on Methadone Maintenance Treatment Identifies SNP rs17180299 and Multiple Haplotypes on CYP2B6, SPON1, and GSG1L Associated with Plasma Concentrations of Methadone R- and S-enantiomers in Heroin-Dependent Patients. PLoS Genet 2016; 12(3): e1005910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith AH, Jensen KP, Li J, Nunez Y, Farrer LA, Hakonarson H , et al. Genome-wide association study of therapeutic opioid dosing identifies a novel locus upstream of OPRM1. Mol Psychiatry 2017; 22(3): 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oneda B, Crettol S, Bochud M, Besson J, Croquette-Krokar M, Hammig R , et al. beta-Arrestin2 influences the response to methadone in opioid-dependent patients. Pharmacogenomics J 2011; 11(4): 258–266. [DOI] [PubMed] [Google Scholar]
- 21.Crettol S, Besson J, Croquette-Krokar M, Hammig R, Gothuey I, Monnat M , et al. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog Neuropsychopharmacol Biol Psychiatry 2008; 32(7): 1722–1727. [DOI] [PubMed] [Google Scholar]
- 22.de Cid R, Fonseca F, Gratacos M, Gutierrez F, Martin-Santos R, Estivill X , et al. BDNF variability in opioid addicts and response to methadone treatment: preliminary findings. Genes, brain, and behavior 2008; 7(5): 515–522. [DOI] [PubMed] [Google Scholar]
- 23.Eap CB, Broly F, Mino A, Hammig R, Deglon JJ, Uehlinger C , et al. Cytochrome P450 2D6 genotype and methadone steady-state concentrations. J Clin Psychopharmacol 2001; 21(2): 229–234. [DOI] [PubMed] [Google Scholar]
- 24.Saxon AJ, Ling W, Hillhouse M, Thomas C, Hasson A, Ang A , et al. Buprenorphine/Naloxone and methadone effects on laboratory indices of liver health: a randomized trial. Drug Alcohol Depend 2013; 128(1–2): 71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke TK, Crist RC, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ , et al. Genetic variation in OPRD1 and the response to treatment for opioid dependence with buprenorphine in European-American females. Pharmacogenomics J 2014; 14(3): 303–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crist RC, Doyle GA, Nelson EC, Degenhardt L, Martin NG, Montgomery GW , et al. A polymorphism in the OPRM1 3’-untranslated region is associated with methadone efficacy in treating opioid dependence. Pharmacogenomics J 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crist RC, Clarke TK, Ang A, Ambrose-Lanci LM, Lohoff FW, Saxon AJ , et al. An intronic variant in OPRD1 predicts treatment outcome for opioid dependence in African-Americans. Neuropsychopharmacology 2013; 38(10): 2003–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry 2008; 13(5): 531–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Epstein DH, Schmittner J, Umbricht A, Schroeder JR, Moolchan ET, Preston KL. Promoting abstinence from cocaine and heroin with a methadone dose increase and a novel contingency. Drug Alcohol Depend 2009; 101(1–2): 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epstein DH, Willner-Reid J, Vahabzadeh M, Mezghanni M, Lin JL, Preston KL. Real-time electronic diary reports of cue exposure and mood in the hours before cocaine and heroin craving and use. Arch Gen Psychiatry 2009; 66(1): 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kowalczyk WJ, Phillips KA, Jobes ML, Kennedy AP, Ghitza UE, Agage DA , et al. Clonidine Maintenance Prolongs Opioid Abstinence and Decouples Stress From Craving in Daily Life: A Randomized Controlled Trial With Ecological Momentary Assessment. Am J Psychiatry 2015; 172(8): 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Preston KL, Ghitza UE, Schmittner JP, Schroeder JR, Epstein DH. Randomized trial comparing two treatment strategies using prize-based reinforcement of abstinence in cocaine and opiate users. J Appl Behav Anal 2008; 41(4): 551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Preston KL, Kowalczyk WJ, Phillips KA, Jobes ML, Vahabzadeh M, Lin JL , et al. Context and craving during stressful events in the daily lives of drug-dependent patients. Psychopharmacology (Berl) 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crist RC, Ambrose-Lanci LM, Vaswani M, Clarke TK, Zeng A, Yuan C , et al. Case-control association analysis of polymorphisms in the delta-opioid receptor, OPRD1, with cocaine and opioid addicted populations. Drug Alcohol Depend 2013; 127(1–3): 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brewer DD, Catalano RF, Haggerty K, Gainey RR, Fleming CB. A meta-analysis of predictors of continued drug use during and after treatment for opiate addiction. Addiction 1998; 93(1): 73–92. [PubMed] [Google Scholar]
- 36.Birnbaum HG, White AG, Schiller M, Waldman T, Cleveland JM, Roland CL. Societal costs of prescription opioid abuse, dependence, and misuse in the United States. Pain Med 2011; 12(4): 657–667. [DOI] [PubMed] [Google Scholar]
- 37.Sharafshah A, Fazel H, Albonaim A, Omarmeli V, Rezaei S, Mirzajani E , et al. Association of OPRD1 Gene Variants with Opioid Dependence in Addicted Male Individuals Undergoing Methadone Treatment in the North of Iran. J Psychoactive Drugs 2017; 49(3): 242–251. [DOI] [PubMed] [Google Scholar]
- 38.Roussotte FF, Jahanshad N, Hibar DP, Sowell ER, Kohannim O, Barysheva M , et al. A commonly carried genetic variant in the delta opioid receptor gene, OPRD1, is associated with smaller regional brain volumes: replication in elderly and young populations. Hum Brain Mapp 2014; 35(4): 1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chadwick LH. The NIH Roadmap Epigenomics Program data resource. Epigenomics 2012; 4(3): 317–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 2016; 44(D1): D877–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015; 348(6235): 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piltonen M, Parisien M, Gregoire S, Chabot-Dore AJ, Jafarnejad SM, Berube P , et al. Alternative Splicing of the Delta-Opioid Receptor Gene Suggests Existence of New Functional Isoforms. Mol Neurobiol 2018. [DOI] [PubMed] [Google Scholar]
- 43.Negus SS, Bidlack JM, Mello NK, Furness MS, Rice KC, Brandt MR. Delta opioid antagonist effects of buprenorphine in rhesus monkeys. Behav Pharmacol 2002; 13(7): 557–570. [DOI] [PubMed] [Google Scholar]
- 44.Kong H, Raynor K, Yasuda K, Moe ST, Portoghese PS, Bell GI , et al. A single residue, aspartic acid 95, in the delta opioid receptor specifies selective high affinity agonist binding. J Biol Chem 1993; 268(31): 23055–23058. [PubMed] [Google Scholar]
- 45.Belcheva MM, Barg J, McHale RJ, Dawn S, Ho MT, Ignatova E , et al. Differential down- and up-regulation of rat brain opioid receptor types and subtypes by buprenorphine. Mol Pharmacol 1993; 44(1): 173–179. [PMC free article] [PubMed] [Google Scholar]
- 46.Belcheva MM, Ho MT, Ignatova EG, Jefcoat LB, Barg J, Vogel Z , et al. Buprenorphine differentially alters opioid receptor adaptation in rat brain regions. J Pharmacol Exp Ther 1996; 277(3): 1322–1327. [PMC free article] [PubMed] [Google Scholar]
- 47.Law PY, Erickson-Herbrandson LJ, Zha QQ, Solberg J, Chu J, Sarre A , et al. Heterodimerization of mu- and delta-opioid receptors occurs at the cell surface only and requires receptor-G protein interactions. J Biol Chem 2005; 280(12): 11152–11164. [DOI] [PubMed] [Google Scholar]
- 48.Liu JG, Liao XP, Gong ZH, Qin BY. Methadone-induced desensitization of the delta-opioid receptor is mediated by uncoupling of receptor from G protein. Eur J Pharmacol 1999; 374(2): 301–308. [DOI] [PubMed] [Google Scholar]
- 49.Damian AJ, Mendelson T, Agus D. Predictors of buprenorphine treatment success of opioid dependence in two Baltimore City grassroots recovery programs. Addict Behav 2017; 73: 129–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mertens JR, Weisner CM. Predictors of substance abuse treatment retention among women and men in an HMO. Alcoholism, clinical and experimental research 2000; 24(10): 1525–1533. [PubMed] [Google Scholar]
- 51.McCaul ME, Svikis DS, Moore RD. Predictors of outpatient treatment retention: patient versus substance use characteristics. Drug Alcohol Depend 2001; 62(1): 9–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


