Abstract
Inflammation plays an important role in the pathogenesis of atherosclerosis and coronary syndromes; moreover, various lines of evidence suggest that genetic factors do contribute to the risk of coronary artery disease (CAD). The proinflammatory cytokine IL-6 is a central mediator of inflammation associated with CAD. The present study is aimed to investigate the association of single nucleotide polymorphism in the promoter region of the IL-6 gene (-174 G > C) and methylation with the susceptibility of CAD. Genotyping of IL-6 -174 G/C polymorphism was performed by PCR–RFLP. Methylation-specific PCR method was used to study the IL-6 gene promoter methylation. Analysis of 470 subjects (265 CAD patients and 205 controls) showed association of the -174 G/C variant with the CAD risk in dominant model (OR 1.58, 95% CI, 1.024–2.23, P = 0.04). Further, the analysis of the distribution of genotypes and alleles of -174 G > C polymorphism according to clinical features of CAD, revealed significant association of genotype and allele (OR 1.86, 95% CI 1.18–2.84 P = 0.01, and OR 1.71, 95% CI 1.09–2.23 P = 0.02 respectively) with diabetes, and we found no association with hypertension (OR 0.95, 95% CI 0.57–1.59, P = 0.8). We also analyzed the methylation status of IL-6 promoter region between cases and controls showed significant hypo methylation in CAD subjects (OR 2.36, 95% CI 1.51–4.259, P = 0.006). Additionally, GC, CC genotypes and C allele carriers show hypomethylation in CAD cases compared to controls (54.58 vs. 76.85%, 29.83 vs. 40% respectively). In conclusion, the promoter polymorphism -174 G/C is associated with CAD risk and further carriers of ‘C’ allele at -174 locus showed significant hypo methylation which could contribute to increased risk of CAD. The present study highlights the association of allele and genotypes with differential DNA methylation of CpG islands in the IL-6 promoter region which may affect IL-6 gene regulation.
Keywords: Coronary artery disease, IL-6, Polymorphism, Methylation
Introduction
Coronary artery disease (CAD) is the leading cause of mortality and morbidity worldwide [1]. CAD is a major cause of death and disability in developed countries accounting for over one-third of total deaths and now became more epidemic in India [2]. The global burden of disease study estimate of age-standardized CAD death rate of 272 per 100,000 population in India is higher than the global average of 235 per 100,000 population [3]. CAD is a complex, multifactorial disorder that results from major risk factors such as dyslipidemias, diabetes, hypertension, obesity, smoking, stress, unhealthy diet and physical inactivity [4, 5]. In general, individuals with these risk factors are considered at high risk for development of CAD [6].
Atherosclerosis is considered as state of chronic low-grade inflammation occurring within the arterial wall [7], in which onset or progression of atherosclerotic manifestations is contributed by the pro-inflammatory cytokines including IL-6, IL-1, IL-8, IL-10 and TNF-α [8]. Cytokines play an important role in the inflammatory response and are involved in acute phase and chronic phase of the disease. Both the phases are characterized by increased blood flow, vascular permeability along with the accumulation of fluid, leukocytes, and inflammatory mediators such as cytokines finally promotes the pathogenesis of atherosclerosis [9].
The Interleukin-6 (IL-6) is a 26-kDa pleotropic inflammatory cytokine produced by many cell types, including fibroblasts, monocytes, adipocytes and endothelial cells and altered levels are associated with endothelial damage leading to initiation of atherosclerotic events [10]. The IL-6 contributes to CAD progression by affecting metabolic, endothelial and coagulant events, and is viewed as a local and circulating marker of coronary plaque inflammation [11]. The IL-6 was implicated in the pathogenesis of ischemic cardiovascular events, including unstable angina [11] and ACS [12]. IL-6 induces the expression of tissue factor, monocyte chemotactic protein-1, matrix-degrading enzyme, low-density lipoprotein receptors in macrophages which stimulates the aggregation of platelets, proliferation of vascular smooth muscle cells and production of C-reactive protein and fibrinogen that results in the plaque stability [13].
The IL-6 gene, located at chromosome 7p21–24 is composed of 4 introns and 5 exons. Polymorphic variants in the IL-6 gene promoter may affect the expression and secretion of IL-6 and subsequently altered circulating levels might result in relevant biological responses, that leads to the pathogenesis of various diseases such as CAD, myocardial infarction (MI) and ischemic stroke [14]. Polymorphisms in the promoter region (G-597A, G-572C, G-174C) of IL-6 gene have been well studied [15–17], among which, the single nucleotide change from G to C at position -174 (rs1800795) is a crucial polymorphism that affects IL-6 production, therefore predispose an individual to cardiovascular events [18, 19]. Several studies have reported that IL-6 -174 polymorphism has been associated with CAD [18, 19], type II diabetes mellitus [17]. In a meta-analysis, reported the significant association between the incidence of stroke and IL-6 -174G/C polymorphism [20, 21].
The DNA methylation is an epigenetic modification that plays a crucial role in controlling gene expression in the genome [22] and may significantly contribute to the risks of many complex diseases, including cardiovascular, cancer, and metabolic diseases [22, 23]. Aberrant promoter methylation of several genes has been associated with the development and progression of coronary heart disease (CHD) [24]. As with many inflammatory mediators, IL-6 is also known to be regulated through epigenetic mechanisms. Several studies have been reported the correlation between the methylation of different CpG sites in the promoter region of IL-6 and its mRNA expression [25, 26]. Abnormal methylation patterns in the CpG islands of disease-associated genes might be involved in CAD pathogenesis [27]. Hence the present study is aimed to investigate the association of SNPs in the promoter region of the IL-6 gene (174 G > C) and effect of methylation with the susceptibility to CAD.
Methods
Study Subjects and Sampling
We have enrolled 470 subjects, which includes 265 angiographically documented CAD patients and 205 age, gender and ethnicity-matched healthy controls from Nizam’s Institute of Medical Sciences (NIMS), Hyderabad, India. Patients who had malignancies, myocardial spasms, myocardial bridges, as well as those suffering from autoimmune diseases, congenital heart diseases, or end-stage kidney or liver diseases were excluded from the study. Healthy control subjects were hospitals staff and voluntary blood donors who were screened for dyslipidemia, liver and kidney function and other investigations to demonstrate CAD risk and found to be normal. Baseline characteristics of all of the patients and controls were obtained using a self-designed questionnaire and medical records. Data on gender, age, body mass index, alcohol consumption, tobacco consumption, and hypertension and diabetes were collected from the self-designed questionnaire. Total cholesterol, triglycerides, low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C) levels were obtained from medical records. Institutional Ethical committee of Nizam’s Institute of Medical Sciences, Hyderabad, India has approved the study. Informed consent was obtained from all the study subjects.
Genotyping of -174 G/C Polymorphism
Five milliliters (5 ml) of blood was collected from each subject in an EDTA vacutainer. The genomic DNA was extracted from whole blood by using the phenol–chloroform extraction protocol. Genotyping for IL-6 -174G/C polymorphism was performed by PCR–RFLP. Primers used for PCR amplification was shown in Table 1. PCR was carried out on an Eppendorf thermol cycler under the following conditions: 94 °C for 5 min, followed by 40 cycles of 30 s at 94 °C, 30 s at 60 °C and 1 min at 72 °C for extension, followed by a final extension of 10 min at 72 °C. The PCR products were then digested with 1 U of NlaIII restriction enzyme (reaction volume 10 μl) at 37 °C, for 4 h. The enzyme cut the 198 bp PCR product into four fragments 168, 119, 49 and 30 bp in length. Fragments size of 119 and 49 bp indicates the presence of a mutant homozygous CC genotype, two 168 bp and 30 bp fragments displayed the presence of homozygous GG genotype and three fragments of 168, 119 and 49 bp indicated the presence of heterozygous GC genotype. The resulting products were separated by 3.0% agarose gel electrophoresis. The products were visualized with Ultraviolet light by using Gel documentation system.
Table 1.
Primer sequence information used for IL-6 -174 G/C polymorphism and methylation
Analysis of IL-6 DNA Methylation
Sodium bisulfite conversion procedure was done using the EpiMark bisulfite conversion kit (NEW ENGLAND BioLABS) according to the manufacture’s protocol. Subsequently, 10–60 ng of treated DNA was used for methylation specific PCR (MSP) reaction. The primers for MSP reaction was shown in Table 1. Each PCR reaction was performed with a total volume of 10 μl, which contained 5 μl of Hot-Star Taq Master Mix (Ampliqon), 4 μl of bisulfite- treated DNA template, and 0.3 μl of each primer pair. The reaction mixture was incubated at 95 °C for 5 min, followed by 40 cycles of 95 °C for 45 s, 54 °C for 45 s, 72 °C for 40 s and a final extension at 72 °C for 10 min. The PCR products were checked on 2.5% agarose gels with ethidium bromide staining. MSP products were analyzed for methylation index.
Statistical Analysis
Allele and genotype frequencies between CAD cases and controls were compared in a 2 × 2 contingency table using Fisher’s exact test. All P values were two-sided and differences were considered statistically significant for P < 0.05. Odds ratio (OR) at 95% confidence intervals (CI) was determined to describe the strength of association. Logistic regression analysis was used to assess the independent effect of each risk factor on CAD.
Results
Demographic Characteristics of the Study Population
A total of 470 subjects are recruited for the current study, among which 265 (193 male and 72 female) are CAD patients and 205 (105 male and 100 female) are control group. Mean age of the CAD group is 56.0 ± 12.0 years with 25.26 ± 4.66 BMI kg/m2. Out of 265 CAD patients, 144 (55.3%) had diabetes and 168 (63.39%) had hypertension. The demographic characteristics of the study population are given in Table 2.
Table 2.
Demographic details of CAD subjects
| Variables | CAD patients (n = 265) |
|---|---|
| Age (mean ± SD), years | 56.00 ± 12.00 |
| Gender | |
| Male | 193 (72.83) |
| Female | 72 (27.16) |
| BMI | |
| Male (kg/m2) | 25.01 ± 4.64 |
| Female (kg/m2) | 25.95 ± 4.66 |
| Diabetes mellitus | |
| Yes (%) | 144 (55.3) |
| No (%) | 121 (45.66) |
| Hypertension | |
| Yes (%) | 168 (63.39) |
| No (%) | 97 (36.6) |
Association of -174 G > C Polymorphism with CAD
The frequency distribution of the genotype and alleles of IL-6 in the CAD and healthy control groups are shown in Table 3. The distribution of genotypes of IL-6 gene polymorphism at -174 G > C among healthy controls was in accordance with of Hardy–Weinberg equilibrium. The statistical analysis revealed that differences in the distribution of the IL-6 gene -174 G > C polymorphism between the CAD patients and healthy control groups (OR 1.58, 95% CI 1.024–2.23, = 0.04). And also higher frequency of allele “C” allele at -174 G > C was found in patients with CAD compared to the controls (19.81 vs. 15.36), but failed to reach statistical significance (P = 0.08, OR 1.36, 95% CI 0.96–1.91).
Table 3.
The genotype and allele frequencies of promoter polymorphisms in IL-6 gene in CAD patients and healthy control subjects
| IL-6 SNP | Genotype and allele | Cases (n = 265) | Controls (n = 205) | OR | 95% CI | P value |
|---|---|---|---|---|---|---|
| rs1800795 (G/C) | GG | 163 (61.50%) | 145 (70.73%) | 1.58 | 1.024–2.23 | 0.04* |
| GC | 99 (37.35%) | 57 (27.80%) | ||||
| CC | 03 (1.13%) | 03 (1.46%) | ||||
| G | 425 (80.18%) | 347 (84.63%) | 1.36 | 0.96–1.91 | 0.08 | |
| C | 105 (19.81%) | 63 (15.36%) |
*P value < 0.05 is considered as significant
Further, we analyzed the distribution of genotypes and alleles of -174 G > C polymorphism in association with clinical features of CAD. The genotype frequency distribution between CAD with diabetes and without diabetes revealed the significant distribution with allele and genotypes (OR 1.86, 95% CI 1.18–2.84, P = 0.01 and, OR 1.71, 95% CI 1.09–2.23, P = 0.02 respectively). However, -174 G > C polymorphism did not show association with hypertension (OR 0.95, 95% CI 0.57–1.59 P = 0.8) and with other clinical features (Table 4). Further, logistic regression analysis also revealed association of CAD with type 2 diabetes (OD: 1.99, CI at 95%:1.10–3.58, P = 0.02) and no association with other variables such as age (P = 0.99), sex (P = 0.42), BMI (P = 0.50) and hypertension (P = 0.20) (Table 5).
Table 4.
The genotype and allele frequencies of promoter polymorphisms in the IL-6 gene in CAD patients with and without risk factors
| Clinical variable | Genotype | OR (95% CI) | P | Allele | OR (95% CI) | P | |||
|---|---|---|---|---|---|---|---|---|---|
| GG | GC | CC | G | C | |||||
| CAD T2DM(+) (n = 144) | 79 (54.86%) | 62 (43.04%) | 03 (2.08%) | 1.86 (1.01–2.84) | 0.01* | 220 (76.36%) | 68 (23.64%) | 1.71 (1.09–2.66) | 0.02* |
| CAD T2DM(−) (n = 121) | 84 (69.49%) | 37 (30.5%) | 0 (0%) | 205 (84.715) | 37 (15.285) | ||||
| CAD HTN(+) (n = 168) | 104 (61.9%) | 61 (36.3%) | 03 (1.78%) | 0.95 (0.5–1.5) | 0.8 | 269 (80.05) | 67 (19.94) | 1.02 (0.65–1.59) | 1.00 |
| CAD HTN(−) (n = 97) | 59 (60.83%) | 38 (39.17%) | 0 (0%) | 156 (80.41%) | 38 (19.58%) | ||||
*P value < 0.05 is considered as significant
Table 5.
Logistic regression analysis of association between the IL-6-174 G/C genotypes and risk of coronary artery disease
| Variable | B | SE | Wald | Odds ratio | 95% CI | P |
|---|---|---|---|---|---|---|
| Age | − 0.00001 | 0.011 | 0.000001 | 1.0 | 0.97–1.02 | 0.99 |
| Sex | − 0.247 | 0.308 | 0.642 | 0.78 | 0.42–1.42 | 0.42 |
| BMI | 0.017 | 0.026 | 0.435 | 1.0 | 0.96–1.07 | 0.50 |
| HTN | − 0.403 | 0.315 | 1.636 | 0.66 | 0.36–1.23 | 0.20 |
| DM | 0.688 | 0.299 | 5.271 | 1.99 | 1.10–3.58 | 0.021* |
B = coefficient, SE std. error, HTN hypertension, DM diabetes mellitus
DNA Methylation of IL-6 Promoter
The percentage of methylation of IL-6 promoter was quantified using the MSP method in 97 CAD cases and 81 controls. Methylation status between cases and controls revealed significant hypomethylation in CAD subjects (OR 2.36, 95% CI 1.51–4.259, P = 0.006). Regardless of case–control status, carrier of homozygous “GG” had 45.16%, “GC” had 50% and individuals homozygous for the variant genotype “CC” had 4.8% methylation. We also found “C allele” carriers showed hypomethylation in both cases and control group (Cases G: 70.16% vs. C 29.83%, Controls G: 60% vs. C: 40%) (Fig. 1).
Fig. 1.
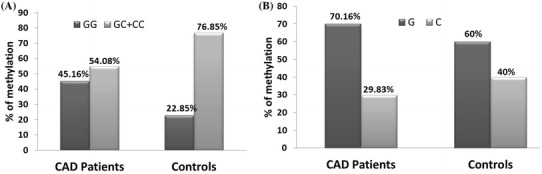
Extent of IL-6 promoter methylation levels according to rs1800795 -174 G/C. IL-6 promoter methylation was determined using methylation-specific PCR. DNA methylation results are sorted according to rs1800795 -174 G/C genotype and alleles. a The rs1800795 GC + CC genotypes show hypomethylation in CAD cases compared to controls (54.58 vs. 76.85%). b Similarly, methylation results are sorted based on the rs1800795 alleles. “C allele” carriers also displayed hypomethylation in CAD subjects compared to the controls (29.83 vs. 40%). The data shows significant hypomethylation in IL-6 promoter in carriers of “GC” and “CC” genotypes in CAD cases when compared to controls (n = 97 CAD cases and 81 controls)
Discussion
In recent years, the disease susceptibility has been increased when genomic factors combined with the environmental factors [18, 19]. IL-6 is a pleiotropic cytokine with a broad range of humoral and cellular immune effects relating to inflammation, host defense, and tissue injury [28].The SNPs associated with the disease might alter the gene transcription and expression by modulating transcription factor (TF) binding site [29, 30]. Functional variation in the promoter region of the IL-6 gene can influence gene transcription [31]. This would affect plasma or tissue levels of IL-6 which in turn would influence plasma levels of cardiovascular disease risk factors and thus leads to cardiovascular disease [31]. The G (-174) C SNP in the promoter of IL-6 gene is considered a key expression regulator of the gene [32]. Several studies have assessed the relationship between the IL-6 gene polymorphisms and pathogenesis of CAD and showed significant association with the CAD susceptibility [33–38]. The SNPs in the promoter region and intron 3 of the IL-6 gene have shown to play a role in the blood pressure regulation and progression of atherosclerosis in the Japanese [28]. Galimudi et al. [39] reported that CC genotype of IL-6 -174G/C gene was associated with development of CAD. In present study, we also found that IL-6 -174G/C was associated with CAD in dominant model with 1.58-fold risk for developing CAD. However, few studies have reported that IL-6 rs1800795 is not associated with an increased risk of CAD in Tunisians, Chinese population and in Isfahan population [40–42]. But in a recent meta-analysis on 50 studies suggested that the IL-6 − 174 G > C polymorphism was positively associated with susceptibility to CAD [18], which is in line with the results of our study. Hou et al. conducted a meta-analysis with 42 studies including 15,145 cases and 21,496 controls, and they reported that C allele of IL-6 -174G/C was correlated with an increased risk of CAD in Caucasians [43]. Song et al. [44] found that the CC genotype of IL-6 -174G/C was related to the onset of cardiovascular events. The C allele of G (− 174) C polymorphism was determined as a risk factor for MI [45]. In patients with type 2 diabetes, rs1800795 SNP of the IL-6 gene is significantly related to increased risk of CVD [46]. In the sub group analysis, we also observed that the significant association of CC genotype and C allele in CAD patients with diabetes. The carriers of the C allele were at increased risk of developing CAD in patients with type 2 diabetes. Based on these evidences, we speculate that polymorphisms in the promoter region may result in variation in transcriptional regulation and altered gene expression which in turn could affect the disease phenotypes and influence an individual’s risk of disease.
The hypomethylation is a feature of transcriptionally active genes [47]. The expression of IL-6 gene is regulated by both transcriptional and post-transcriptional mechanisms through epigenetics which leads to the pathogenesis of inflammatory diseases including CAD [48]. The methylation level of IL-6 promoter has been associated with air pollution exposure, [49] which has been known to increase cardiovascular morbidity and mortality. In addition, serum IL-6 level has been associated with increased risk of mortality in patients with CHD [50]. Zuo et al. [51] have observed that hypomethylation in IL-6 promoter was strongly associated with risk estimation for AMI. Several studies have observed the correlations of DNA methylation in IL-6 promoter with diet and environmental factors, which would confound the associations between IL-6 promoter hypo methylation and risk for CHD [48, 52–54]. The implication of IL-6 in the pathogenesis of CHD and the inverse correlation of IL-6 promoter methylation with CHD risk factors have been demonstrated [55]. Previous studies have been reported that hypomethylation of IL-6 promoter was associated with the pathogenesis of systemic lupus erythematous, rheumatoid arthritis, chronic periodontitis and CHD [56–60]. Gene-specific DNA methylation profiles and LINE-1 hypo methylation are associated with myocardial infarction risk [61]. In the present study, the methylation in the promoter region of the IL-6 gene showed significant association with the CAD, particularly in patients with CC genotype and C allele have shown hypomethylation in cases which may lead to increased transcription activity and subsequently up regulation of IL-6 production in targeted tissues finally contribute to the susceptibility to CAD. These observations are consistent with the results of other studies [57–59]. Suggesting that hypomethylation status is associated with the over transcription of genes that is related to the pathogenesis of inflammatory diseases.
Conclusion
In conclusion, our results revealed that IL-6 rs1800795 –174G (G/C) might contribute to the risk of CAD. IL-6 hypo methylation was observed in CAD cases which may help in identifying individuals at risk of developing CAD. Further − 174 G/C polymorphism have influence on CAD patients with diabetes. Our study demonstrates that genetic and epigenetic variability plays an important role in the disease pathogenesis.
Author Contributions
L.K performed clinical investigations and contributed samples. V.K.K provided research material. B.I and S.K.K conducted experiments and analyzed the data. B.I, S.K.K and V.K.K wrote and reviewed the manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors have no conflicts of interest to declare.
References
- 1.Sanchis-Gomar F, Perez-Quilis C, Leischik R, Lucia A. Epidemiology of coronary heart disease and acute coronary syndrome. Ann Transl Med. 2016;13:256. doi: 10.21037/atm.2016.06.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreatsoulas C, Sloane D, Pogue J, Velianou JL, Anand SS. Referrals in acute coronary events for CARdiac catheterization The RACE CAR trial. Can J Cardiol. 2010;8:e290–e296. doi: 10.1016/S0828-282X(10)70436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prabhakaran D, Jeemon P, Roy A. Cardiovascular diseases in India. Current epidemiology and future directions. Circulation. 2016;16:1605–1620. doi: 10.1161/CIRCULATIONAHA.114.008729. [DOI] [PubMed] [Google Scholar]
- 4.Gensini GF, Comeglio M, Colella A. Classical risk factors and emerging elements in the risk profile for coronary artery disease. Eur Heart J. 1998;19:53–61. [PubMed] [Google Scholar]
- 5.Gupta R, Mohan I, Narula J. Trends in coronary heart disease epidemiology in India. Ann Glob Health. 2016;82:307–315. doi: 10.1016/j.aogh.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 6.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 7.Paoletti R, Gotto AM, Jr, Hajjar DP. Inflammation in atherosclerosis and implications for therapy. Circulation. 2004;109:20–26. doi: 10.1161/01.CIR.0000131514.71167.2e. [DOI] [PubMed] [Google Scholar]
- 8.Alexander RW. Inflammation and coronary artery disease. N Engl J Med. 1994;331:468–469. doi: 10.1056/NEJM199408183310709. [DOI] [PubMed] [Google Scholar]
- 9.Georges JL, Loukaci V, Poirier O, Evans A, Luc G, Arveiler D. Interleukin-6 gene polymorphisms and susceptibility to myocardial infarction: the ECTIM study. J Mol Med (Berl) 2001;79:300–305. doi: 10.1007/s001090100209. [DOI] [PubMed] [Google Scholar]
- 10.Hojo Y, Ikeda U, Takahashi M, Shimada K. Increased levels of monocyte-related cytokines in patients with unstable angina. Atherosclerosis. 2002;161:403–408. doi: 10.1016/S0021-9150(01)00636-0. [DOI] [PubMed] [Google Scholar]
- 11.Plutzky J. Inflammatory pathways in atherosclerosis and acute coronary syndromes. Am J Cardiol. 2001;88:10–15. doi: 10.1016/S0002-9149(01)01924-5. [DOI] [PubMed] [Google Scholar]
- 12.Loppnow H, Werdan K, Buerke M. Vascular cells contribute to atherosclerosis by cytokineandinnate-immunity-related inflammatory mechanisms. Innate Immun. 2008;14:63–87. doi: 10.1177/1753425908091246. [DOI] [PubMed] [Google Scholar]
- 13.Fernandes MT, Fernandes KB, Marquez AS, Cólus IM. Association of interleukin-6 gene polymorphism (rs1800796) with severity and functional status of osteoarthritis in elderly individuals. Cytokine. 2015;75:316–320. doi: 10.1016/j.cyto.2015.07.020. [DOI] [PubMed] [Google Scholar]
- 14.Monaco C, Andreakos E, Kiriakidis S, Feldmann M, Paleolog E. T-cell-mediated signalling in immune, inflammatory and angiogenic processes: the cascade of events leading to inflammatory diseases. Curr Drug Targets Inflamm Allergy. 2004;3:35–42. doi: 10.2174/1568010043483881. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Li E, Zhang LH, Jian LG, Liu HP, Wang T. IL-6 174G/C and IL-6-572C/G polymorphisms are associated with increased risk of coronary artery disease. Genet Mol Res. 2015;14(3):8451–8457. doi: 10.4238/2015.July.28.12. [DOI] [PubMed] [Google Scholar]
- 16.Ghazouani L, Ben HadjKhalifa S, Abboud N, Ben Hamda K, Ben Khalfallah A, Brahim N, et al. TNF-alpha -308G > A and IL-6 -174G > C polymorphisms in Tunisian patients with coronary artery disease. ClinBiochem. 2010;43(13–14):1085–1089. doi: 10.1016/j.clinbiochem.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 17.Libra M, Signorelli SS, Bevelacqua Y, Navolanic PM, Bevelacqua V, Polesel J. Analysis of G (174) C IL-6 polymorphism and plasma concentrations of inflammatory markers in patients with type 2 diabetes and peripheral arterial disease. J ClinPathol. 2006;59:211–215. doi: 10.1136/jcp.2004.025452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yen RW, Vertino PM, Nelkin BD, Yu JJ, El-Deiry W, Cumaraswamy A, et al. Isolation and characterization of the cDNA encoding human DNA methyltransferase. Nucleic Acids Res. 1992;20:2287–2291. doi: 10.1093/nar/20.9.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol Rev. 2006;86:515–581. doi: 10.1152/physrev.00024.2005. [DOI] [PubMed] [Google Scholar]
- 20.Tso AR, Merino JG, Warach S. Interleukin-6 174G/C polymorphism and ischemic stroke: a systematic review. Stroke. 2007;38:3070–3075. doi: 10.1161/STROKEAHA.107.492231. [DOI] [PubMed] [Google Scholar]
- 21.Kim SK, Chung JH, Kwon OY. Promoter polymorphism (− 174, G/C) of interleukin-6 and arterial thromboembolic events: a meta-analysis. Med Sci Monit. 2016;22:4345–4353. doi: 10.12659/MSM.901467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mann DA. Epigenetics in liver disease. Hepatology. 2014;60:1418–1425. doi: 10.1002/hep.27131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozanne SE, Constancia M. Mechanisms of disease. The developmental origins of disease and the role of the epigenotype. Nat. Clin Pract Endocrinol Metab. 2007;3:539–546. doi: 10.1038/ncpendmet0531. [DOI] [PubMed] [Google Scholar]
- 24.Rodenhister D, Mann M. Epigenetics and human disease: translating basic biology into clinical application. CMAJ. 2006;174:341–348. doi: 10.1503/cmaj.050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poplutz MK, Wessels I, Rink L, Uciechowski P. Regulation of the interleukin-6 gene expression during monocytic differentiation of HL-60 cells by chromatin remodeling and methylation. Immunobiology. 2014;219:619–626. doi: 10.1016/j.imbio.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Dandrea M, Donadelli M, Costanzo C, Scarpa A, Palmieri M. MeCP2/H3meK9 are involved in IL-6 gene silencing in pancreatic adenocarcinoma cell lines. Nucleic Acids Res. 2009;37:6681–6690. doi: 10.1093/nar/gkp723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang J, Peng W, Li H, Wang W, Wei Y, Li W. Methylation of p15INK4b and expression of ANRIL on chromosome 9p21 are associated with coronary artery disease. PLoS ONE. 2012;7(10):e47193. doi: 10.1371/journal.pone.0047193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka C, Mannami T, Kamide K. Nucleotide polymorphisms in the interleukin-6 gene associated with blood pressure and atherosclerosis in Japanese general population. Hypertens Res. 2005;28:35–41. doi: 10.1291/hypres.28.35. [DOI] [PubMed] [Google Scholar]
- 29.Koch W, Kastrati A, Bittiger C. Interleukin-10 and tumor necrosis factor gene polymorphisms and risk of coronary artery disease and myocardial infarction. Atherosclerosis. 2001;159:137–144. doi: 10.1016/S0021-9150(01)00467-1. [DOI] [PubMed] [Google Scholar]
- 30.Sharma S, Rathored J, Ghosh B. Genetic polymorphisms in TNF genes and tuberculosis in North Indians. BMC Infect Dis. 2010;10:165. doi: 10.1186/1471-2334-10-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ridker PM, Rifai N, Stampfer MJ, Hennekens CH. Plasma concentration of interleukin-6 and the risk of future myocardial infarction among apparently healthy men. Circulation. 2000;101:1767–1772. doi: 10.1161/01.CIR.101.15.1767. [DOI] [PubMed] [Google Scholar]
- 32.Buraczynska M, Zukowski P, Drop B, Baranowicz-Gaszczyk I, Ksiazek A. Effect of G (− 174) C polymorphism in interleukin-6 gene on cardiovascular disease in type 2 diabetes patients. Cytokine. 2016;79:7–11. doi: 10.1016/j.cyto.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Rivera-Chavez FA, Peters-Hybki DL, Barber RC, O’Keefe GE. Interleukin-6 promoter haplotypes and interleukin-6 cytokine responses. Shock. 2003;20:218–223. doi: 10.1097/00024382-200309000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mao L, Geng GY, Han WJ, Zhao MH, Wu L, Liu HL. Interleukin-6 (IL-6) -174G/C genomic polymorphism contribution to the risk of coronary artery disease in a Chinese population. Genet Mol Res. 2016;15:2. doi: 10.4238/gmr.15027803. [DOI] [PubMed] [Google Scholar]
- 35.Wang K, Dong PS, Zhang HF, Li ZJ, Yang XM, Liu H. Role of interleukin-6 gene polymorphisms in the risk of coronary artery disease. Genet Mol Res. 2015;14:3177–3183. doi: 10.4238/2015.April.10.29. [DOI] [PubMed] [Google Scholar]
- 36.Sun GQ, Wu GD, Meng Y, Du B, Li YB. IL-6 gene promoter polymorphisms and risk of coronary artery disease in a Chinese population. Genet Mol Res. 2014;26(13):7718–7724. doi: 10.4238/2014.September.26.9. [DOI] [PubMed] [Google Scholar]
- 37.Satti HS, Hussain S, Javed Q. Association of interleukin-6 gene promoter polymorphism with coronary artery disease in Pakistani families. Sci World J. 2013;13:538365. doi: 10.1155/2013/538365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phulukdaree A, Khan S, Ramkaran P, Govender R, Moodley D, Chuturgoon AA. The interleukin-6 -147 g/c polymorphism is associated with increased risk of coronary artery disease in young South African Indian men. Metab Syndr Relat Disord. 2013;11(3):205–209. doi: 10.1089/met.2012.0130. [DOI] [PubMed] [Google Scholar]
- 39.Galimudi RK, Spurthi MK, Padala C, Kumar KG, Mudigonda S, Reddy SG, et al. Interleukin 6 (-174G/C) variant and its circulating levels in coronary artery disease patients and their first degree relatives. Inflammation. 2014;37:314–321. doi: 10.1007/s10753-013-9742-8. [DOI] [PubMed] [Google Scholar]
- 40.Ghazouani L, Abboud N, Ben HadjKhalifa S, Added F, Ben Khalfallah A, Nsiri B, Mediouni M, Mahjoub T. 174G > C interleukin-6 gene polymorphism in Tunisian patients with coronary artery disease. Ann Saudi Med. 2011;31:40–44. doi: 10.4103/0256-4947.75777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong Z, Li Q, Zhang J, Wei Y. Association between interleukin 6 and interleukin 16 gene polymorphisms and coronary heart disease risk in a Chinese population. J Int Med Res. 2013;41:1049–1056. doi: 10.1177/0300060513483405. [DOI] [PubMed] [Google Scholar]
- 42.Meraj P, Reza G, Fereshteh A, Kamran G. Association between rs1800795 (− 174 G/C) polymorphism in the promoter of IL6 gene and risk of relapsing-remitting multiple sclerosis (RRMS) in Isfahan population. Open J Genet. 2014;4:407–413. doi: 10.4236/ojgen.2014.45038. [DOI] [Google Scholar]
- 43.Hou H, Wang C, Sun F, Zhao L, Dun A, Sun Z. Association of interleukin-6 gene polymorphism with coronary artery disease: an updated systematic review and cumulative meta analysis. Inflamm Res. 2015;64:707–720. doi: 10.1007/s00011-015-0850-9. [DOI] [PubMed] [Google Scholar]
- 44.Song Y, Gu HD, He Y, Wang JW. Role of IL-6 polymorphism on the development of cardiovascular events and coronary artery disease in patients receiving hemodialysis. Genet Mol Res. 2015;14:2631–2637. doi: 10.4238/2015.March.30.23. [DOI] [PubMed] [Google Scholar]
- 45.Chiappelli M, Tampieri C, Tumini E, Porcellini E, Caldarera CM, Nanni S, et al. Interleukin-6 gene polymorphism is an age dependent risk factor for myocardial infarction in men. Int J Immunogenet. 2005;32:349–353. doi: 10.1111/j.1744-313X.2005.00537.x. [DOI] [PubMed] [Google Scholar]
- 46.Toperoff G, Aran D, Kark JD, Rosenberg M, Dubnikov T, Nissan B, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viana MB, Cardoso FP, Diniz MG, Costa FO, da Costa JE, Gomez RS, et al. Methylation pattern of IFN-c and IL-10 genes in periodontal tissues. Immunobiology. 2011;216:936–941. doi: 10.1016/j.imbio.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 48.Bind MA, Lepeule J, Zanobetti A, Gasparrini A, Baccarelli A, Coull BA. Air pollution and gene-specific methylation in the normative aging study: association, effect modification, and mediation analysis. Epigenetics. 2014;9(3):448–458. doi: 10.4161/epi.27584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 50.Jiang D, Zheng D, Wang L, Huang Y, Liu H, Xu L. Elevated PLA2G7 gene promoter methylation as a gender-specific marker of aging increases the risk of coronary heart disease in females. PLoS ONE. 2013;8(3):e59752. doi: 10.1371/journal.pone.0059752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zuo HP, Guo YY, Che L, Wu XZ. Hypomethylation of interleukin-6 promoter is associated with the risk of coronary heart disease. Arq Bras Cardiol. 2016;107:131–136. doi: 10.5935/abc.20160124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang FF, Santella RM, Wolff M, Kappil MA, Markowitz SB, Morabia A. White blood cell global methylation and IL-6 promoter methylation in association with diet and lifestyle risk factors in a cancer-free population. Epigenetics. 2012;7(6):606–614. doi: 10.4161/epi.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirchner H, Nylen C, Laber S, Barres R, Yan J, Krook A. Altered promoter methylation of PDK4, IL1 B, IL6, and TNF after Roux-en Y gastric bypass. Surg Obes Relat Dis. 2014;10(4):671–678. doi: 10.1016/j.soard.2013.12.019. [DOI] [PubMed] [Google Scholar]
- 54.Peluso M, Bollati V, Munnia A, Srivatanakul P, Jedpiyawongse A, Sangrajrang S. DNA methylation differences in exposed workers and nearby residents of the MaTaPhut industrial estate, Rayong, Thailand. Int J Epidemiol. 2012;41(6):1753–1760. doi: 10.1093/ije/dys129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aumueller E, Remely M, Baeck H, Hippe B, Brath H, Haslberger AG. Interleukin-6 CpG methylation and body weight correlate differently in type 2 Diabetes patients compared to obese and lean controls. J Nutrigenet Nutrigenomics. 2015;8(1):26–35. doi: 10.1159/000381714. [DOI] [PubMed] [Google Scholar]
- 56.Jin Y, Wang Q, Wang G, Zhang X, Yan B, Hu W. Common polymorphisms in the interleukin-6 gene and myocardial infarction risk: a meta-analysis. Genet Test Mol Biomarkers. 2014;18(5):330–340. doi: 10.1089/gtmb.2013.0404. [DOI] [PubMed] [Google Scholar]
- 57.Wilson AG. Epigenetic regulation of gene expression in the inflammatory response and relevance to common diseases. J Periodontol. 2008;79(8):1514–1519. doi: 10.1902/jop.2008.080172. [DOI] [PubMed] [Google Scholar]
- 58.Nile CJ, Read RC, Akil M, Duff GW, Wilson AG. Methylation status of a single CpG site in the IL6 promoter is related to IL6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 2008;58:2686–2693. doi: 10.1002/art.23758. [DOI] [PubMed] [Google Scholar]
- 59.Mi XB, Zeng FQ. Hypomethylation of interleukin-4 and -6 promoters in T cells from systemic lupus erythematosus patients. Acta Pharmacol Sin. 2008;29(1):105–112. doi: 10.1111/j.1745-7254.2008.00739.x. [DOI] [PubMed] [Google Scholar]
- 60.Ishida K, Kobayashi T, Ito S, Komatsu Y, Yokoyama T, Okada M. Interleukin-6 gene promoter methylation in rheumatoid arthritis and chronic periodontitis. J Periodontol. 2012;83:917–925. doi: 10.1902/jop.2011.110356. [DOI] [PubMed] [Google Scholar]
- 61.Guarrera S, Fiorito G, Onland-Moret NC, Russo A, Agnoli C, Allione A, et al. Gene-specific DNA methylation profiles and LINE-1 hypomethylation are associated with myocardial infarction risk. Clin Epigenetics. 2015;7:133. doi: 10.1186/s13148-015-0164-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Na YK, Hong HS, Lee WK, Kim YH, Kim DS. Increased methylation of interleukin 6 gene is associated with obesity in Korean women. Mol Cells. 2015;38:452–460. doi: 10.14348/molcells.2015.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]


