Abstract
The microRNA (miR)-183-5p is expressed at high level in the majority of cancer. The purpose of present study was to investigate the role of oncogenic miR-183-5p in prostate cancer (PCa) as biomarker. We carried out our experiment in 50 prostate cancer patients and 40 patients of benign prostatic hyperplasia (BPH) and 40 adjacent controls tissue. The expression of miR-183-5p was evaluated through reverse transcription qualitative polymerase chain reaction. We found that the expression of miR-183-5p in PCa tissue was significantly up regulated as compared to BPH patients and adjacent normal tissues as control. Additionally, miR-183 expression was correlated with higher prostate-specific antigen, higher Gleason Score and metastatic condition. A receiver operating characteristic curve analysis revealed that miR-183-5p distinguished PCa patients from BPH patients and also from control. In conclusion, our data suggest that oncogenic miR-183-5p may be useful as a new tissue specific diagnostic biomarker in prostate cancer.
Keywords: Prostate cancer, BPH, MicroRNA, RT-PCR and diagnosis
Introduction
Prostate cancer (PCa) is the most commonly diagnosed carcinoma after the age of 50 years in men. It is leading cause of cancer-associated deaths in western countries as well as metropolitan cities of India [1, 2]. At present, serum prostate-specific antigen (PSA) is the standard diagnostic biomarker for PCa, increased levels of which are observed in patients with PCa. However, the level of serum PSA could be also elevated in other disease conditions such as, trauma, prostatitis and BPH [3] which generally leads to over diagnosis and overtreatment [4]. The PSA measurement is insufficient to identify prostate cancer. Furthermore, urinary prostate cancer antigen 3 (PCA3) has been use diagnostic biomarker for PCa screening, but it is not sufficient to distinguish PCa patients from healthy individuals [5, 6]. Therefore, there is an urgent need to identify more effective biomarkers for screening of prostate cancer.
MiRNAs, a novel class of conserved short non-coding RNA molecules, regulate gene expression by binding to the 3′ untranslated region (3′ UTR) of target messenger RNAs (mRNAs) [7, 8]. MicroRNAs participate in crucial roles through post transcriptional regulation of gene expression in almost all biological functions, such as cell proliferation, apoptosis differentiation, development and many more. Many miRNA profiling studies found the specific correlation between miRNAs expression and human cancers [9]. Several differentially dysregulated miRNAs have been identified by high-throughput technologies across various normal and cancer tissues, which are involved in prostate cancer [10, 11]. Oncogenic miR-21 over expression is significantly correlated in hormonal therapy, castration resistance and metastatic disease as well as clinical parameters and predictive biomarker in cancer progression [12, 13]. Many other miRs have role in prostate cancer progression for instance; miR-375, miR-141 and miR-449a are involved in apoptosis, differentiation and development of PCa [14, 15]. Thus, miRNAs have been considered as potential biomarkers for diagnosis and prognosis as well as in therapeutic purpose in PCa [16].
In particular, miR-183-5p is one of the most common cancer-associated miRNAs, which has been reported to have oncogenic properties in numerous malignancies, such as colorectal cancer [17], breast carcinoma [18], lung carcinoma [19], pancreatic carcinoma [20] and so on. MiR-183-5p directly or indirectly participates in PCa development by regulating several tumor suppressors which are involved in prostate cancer progression.
Therefore, the present study was aimed to evaluate the expression of miR-183-5p in the tissue of PCa, BPH patients and adjacent normal control, thereby investigating the potential use of tissue miR-183-5p as a putative PCa diagnostic biomarker.
Materials and Methods
Patient Tissue Samples Collection
All tissue samples of PCa, BPH patients, and adjacent normal tissue as controls were collected in RNA later and stored at − 80 °C, from the Department of Urology, King George’s Medical University Lucknow, India between Nov 2013 and Nov 2016. The BPH tissue samples (N = 40) were obtained from TURP method, whereas PCa tissues (N = 50) were recovered through TRUS biopsy and channel TURP and adjacent control tissues (N = 40). The study was approved by the Research Ethics Committee of King George’s Medical University and written informed consent was obtained from all patients. At the time of samples collection, patients did not receive androgen deprivation therapy or radiotherapy. The clinicopathological characteristics of the patients were revealed in Table 1.
Table 1.
Clinicopathological demographic data of the Prostate cancer and BPH patient
| Clinical characteristic’s | Prostate cancer (N = 50) | BPH (N = 40) |
|---|---|---|
| Age | ||
| < 60 | 20 (40%) | 13 (32.5%) |
| ≥ 60 | 30 (60%) | 27 (67.5%) |
| PSA level (ng/ml) | ||
| Median < 20 | 16 (32.00%) | 36 (90%) |
| High ≥ 20 | 34 (68.00%) | 4 (10%) |
| Gleason Score | ||
| ≤ 7a | 29(58.00%) | – |
| > 7b | 21 (42.00%) | – |
| Metastasis | ||
| Mc0 | 33 (66.00%) | – |
| Mxd | 17 (33.00%) | – |
a≤ 7 = Low Gleason Score
b> 7 = High Gleason Score
cM0 = Non metastatic condition
dMx = Metastatic condition
RNA Extraction
The total RNA from prostate tissues, enriched for microRNAs was isolated using the mirVana miRNA Isolation Kit from Ambion (Austin, TX) rigorously according to manufacturer’s recommendations. The concentration and purity of RNA samples were examined by agilent nanodrop on OD260/OD280.
Reverse Transcription (RT) and Quantitative Polymerase Chain Reaction (qPCR)
First, 250 ng of total RNA was reverse transcribed by using the Mir-X miRNA first-stand synthesis kit. The quantitative real time PCR (qRT-PCR) expression analysis of miR-183-5p was done by using the SYBR Advantage qPCR premix kit (Cat no. 639676). The reverse-transcribed miRNAs were used as templates in the qRT-PCR analysis and experiment proceed by using the fast real-time PCR System (7500HT, ABI, Applied Biosystems). The primer used for qPCR of miR-183-5p forward primer, 5′-GTA TGG CAC TGG TAG AAT TCA CT-3′; and mRQ 3′ primer (Universal or Reverse primer) and U6 forward and reverse primer were provided with Reverse transcription kit, (Cat. No. 638313). The reaction volume is 25 µl per reaction where RT-PCR condition where denaturation 95 °C—10 s qPCR × 40 cycles 95 °C—5 s, 60 °C—20 s and dissociation curve 95 °C—60 s, 55 °C—30 s and 95 °C—30 s was performed at this standard condition. Both melting curve analysis and agarose gel run were used to confirm the specificity of the amplification reactions. The relative expression of miR-183-5p was determined using the 2-delta delta Ct Analysis method. We select U6 as the endogenous control/normalization.
Statistical Analysis
All statistical analysis and graphs were performed with the SPSS 21 (SPSS, Inc., Chicago, IL, USA) and graph pad Prism 5 Software. Experimental data are represented as mean ± SEM. Differences were carried out with Student’s t test method and ROC curve was used to assess miR-183-5p as a biomarker, and the AUC was reported. P < 0.05 was considered as statistically significant difference.
Results
Clinicopathological Characteristics of Patients
The demographics and clinical characters of the BPH and PCa patients were briefed in Table 1. Serum PSA levels were significantly elevated in PCa patients when compared with BPH patients (P < 0.001). The subjects age distribution were analogous in these two groups.
High Expression of miR-183-5p in Tissue of PCa Patients Compared with BPH Patients and Adjacent Normal Control
The expression level of miR-183-5p was evaluated in the tissue of PCa, BPH patients and adjacent control subjects by using real-time PCR. The expression level of miR-183-5p was found to be significantly up-regulated in the tissue PCa patients as compared with BPH patients as well as control (P < 0.0001) (Fig. 1a). The relative expression of miR-183-5p was 3.21 ± 0.29 for PCa patients, 2.06 ± 0.17 for BPH patients and 1.4 ± 0.14 for healthy controls.
Fig. 1.
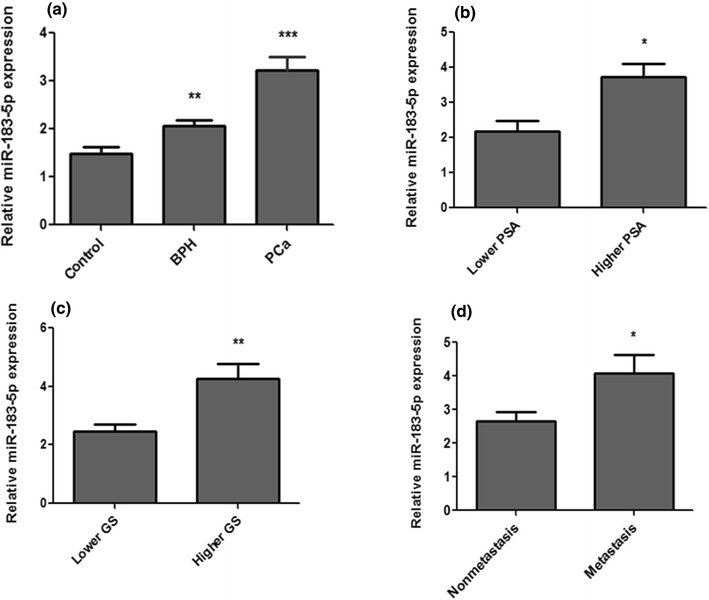
Relative expression levels of miR-183-5p in prostate cancer and BPH. a Relative expression level in PCa, BPH in comparison to control. b Relative expression level in Lower PSA and Higher PSA. c Relative expression level in Lower Gleason Score and Higher Gleason score d Relative expression level in non metastasis (M0) and metastasis (Mx). Data represent in the mean ± SEM. *P < 0.01; **P < 0.001; ***P < 0.0001
Clinicopathological Parameters and Expression of miR-183-5p in Tissue of Prostate Cancer Patient
PCa patients have more aggressive tumors showed significantly higher expression of miR-183-5p as compared to patients have indolent tumors. The mean relative expression level of miR-183-5p in the group of patients have a Gleason Score ≤ 7 was 2.45 ± 0.26, and increased gradually to 4.24 ± 0.51 in patients have a Gleason Score of > 7 (P < 0.0014) (Fig. 1c). The relative expression levels of miR-183-5p in patients have PSA < 20 and PSA ≥ 20 was 2.185 ± 0.29 and 3.728 ± 0.37 respectively (P < 0.010) (Fig. 1b). Furthermore, miR-1183-5p expression level was significantly increased in patients, where, a disease is progress to metastasis condition. Patients have metastasis exhibited a mean relative miR-183-5p expression level was 4.07 ± 0.55, where as non-metastasis condition mean relative expression level was 2.65 ± 0.71 (P < 0.012) (Fig. 1d).
The miR-183-5p May Be Used as a Putative Diagnostic Marker
The miR-183-5p was distinctively expressed in the tissue of PCa patients, BPH patients as well as in control. Thus, the tissue miR-183-5p might be shows potential to use as biomarker. The biomarker potency is evaluated on basis of ROC analysis, tissue miR-139-5p was capable to discriminate PCa patients from control (AUC 0.905; 95% CI 0.847–0.962; Fig. 2a), and it can also discriminate PCa patients from BPH patients (AUC 0.896; 95% CI 0.834–0.957; Fig. 2b).
Fig. 2.
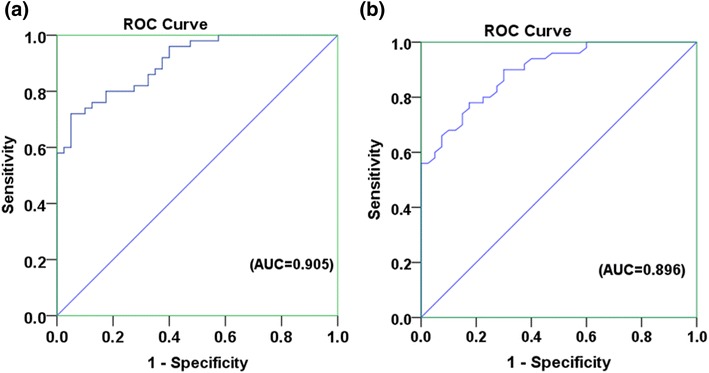
ROC curve analysis. a miR-183-5p can distinguish PCa patients from controls [(AUC = 0.905), (95% CI 0.847–0.96) (P value < 0.001)]. b The expression change of miR-183-5p can distinguish PCa patients from BPH patients [(AUC = 0.896) (95% CI 0.834–0.957) (P value < 0.001)]. ROC receiver operating characteristic, AUC area under the curve, CI confidence interval
Discussion
During the last few years, specific miRNA signatures for PCa have been illustrated in numerous studies suggesting that miRNAs or miRNA profiles can be used as diagnostic markers in cancer including prostate cancer [21–24]. miR-183-5p is the most important among the miRNAs that has been shown to be differentially expression in several cancers, including breast cancer, colorectal cancer, prostate cancer [17, 18, 25]. mir-183 is located on human chromosome 7q32.2 and forms a cluster with miR-96 and miR-182. The miR-183-96-182 cluster could inhibit the invasion and metastasis of lung cancer through directly suppressing the expression of Foxf2 [26]. The expression level of miR-183-5p was showed to be inversely correlated with tumor suppressor SOCS-6 expres-sion in pancreatic cell lines [20]. In the present study, we observed the expression of miR-183-5p in tissue and determined that it was significantly up regulated in PCa patients compared to BPH and control, which revealed a strong association between PCa and miR-183-5p expression. The much higher expression of tissue miR-183-5p was identified in PCa patients with more aggressive tumors (Gleason Score > 7) and with metastasis condition. Therefore, our study point out that tissue miR-183-5p may be tightly associated with progression and risk of prostate cancer.
Most of previous studies also reported that the level of miR-183 expression in prostate cancer was described to be higher than adjacent normal tissues [27]. Furthermore, miR-183 functions as an oncogene by targeting the transcription factor EGR1 and known prostate cancer tumor suppressor PTEN and promoting tumor cell migration in prostate cancer [28]. Mihelich et al. [29] reported that miR-183 was over-expressed in prostate tissue and its expression regulates zinc homeostasis in prostate cells. The oncogenic miR-183 activates the Wnt/b-catenin pathway by directly inhibiting tumour suppressors Dkk-3 and SMAD4 in PC-3 cells [30]. As the SMAD complex directly activates the p21 gene promoter in cooperation with the transcription factor Sp1 [31] and Liu et al. [32] showed that SMAD4 knockdown decreased c-Myc and p21 protein in PC-3 cells. PC-3 cells to look at the role of Dkk-3 on Wnt/b-catenin signalling and b-catenin expression in the nuclear fraction and Tcf transcriptional activity were significantly decreased with ectopic Dkk-3 expression, indicating that over expression of Dkk-3 in PC-3 cells inhibits Wnt/b-catenin signalling.[30]. The statistically significant down-regulation of miR-183-5p in the study group of schizophrenia patients was observed [33].
The major diagnosis tools for PCa are PSA testing, digital rectal examination (DRE), PCA3, Gleason Score and multiparametric magnetic resonance image (mpMRI) [16]. However, presently used diagnostic markers have some limitation due to their wrong prediction and over diagnosis. Therefore, the identification of diagnostic markers for human PCa is required for quality management and treatment strategies. The high stability and easily accessibility of miRNAs make them perfect biomarkers, particularly for surveillance of early stage, presymptomatic diseases in at-risk patients [34, 35]. Therefore, the determination of miRNAs may be a new approach to understanding of PCa pathology and may be also helpful to identify potential biomarkers. Hence, in the present study, we performed the ROC analysis, which revealed that tissue miR-183-5p had quality to discriminate patients of PCa from BPH patients as well as from control tissue. Therefore, these results emphasized a role of miR-183-5p in the pathogenesis of PCa and also suggested that miR-183-5p has quality to be used as biomarker because of its high specificity and sensitivity for PCa detection. Other reports state that higher miR-183 expression was associated with higher PSA at the time of diagnosis, higher prostate tumor and poor overall survival of PCa patients after radical prostatectomy [29]. Larne et al. [36] found that the synthesis and serum levels of PSA are directly affected by miR-183 expression and was found to bind the 3 UTR of PSA directly and increase both protein and messenger RNA levels of PSA.
In conclusion, our findings define that miR-183-5p have distinct expression in PCa patients, BPH patients and control. MiR-183-5p over expression is also increased with disease progression and in metastasis condition of PCa. ROC analyses proved that mir-183-5p have quality to be used as diagnostic marker with good specificity and sensitivity. The only limitation of present report is that it has been carried out in small number of subjects. Therefore, there is a need to evaluate it in large sample size of tissue as well as non-invasive sample such as blood, serum etc. and also evaluate its mRNA target in in-vitro experiment.
Acknowledgements
We would like to thank all the members of Professor A A Mahdi laboratory for the helpful discussion. Mohammad Waseem is supported by Senior research fellowship from Science and Engineering Research Board (SERB), Department of Science and Technology, New Delhi, India.
Funding
Science and Engineering Research Board (SERB), Department of Science and Technology, New Delhi, India (SR/SO/HS/-219/2012)
Compliance with Ethical Standards
Conflict of interest
All authors declared that they have no conflict of interests.
References
- 1.Stuopelyte K, Daniūnaitė K, Jankevičius F, Jarmalaite S. Detection of miRNAs in urine of prostate cancer patients. Medicina. 2016;52:116–124. doi: 10.1016/j.medici.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Jain S, Saxena S, Kumar A. Epidemiology of prostate cancer in India. Meta Gene. 2014;29:596–605. doi: 10.1016/j.mgene.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nadler RB, Humphrey PA, Smith DS, Catalona WJ, Ratliff L. Effect of inflammation and benign prostatic hyperplasia on elevated serum prostate specific antigen levels. J Urol. 1995;154:407–413. doi: 10.1016/S0022-5347(01)67064-2. [DOI] [PubMed] [Google Scholar]
- 4.Draisma G, Etzioni R, Tsodikov A, Mariotto A, Wever E, Gulati R, et al. Lead time and overdiagnosis in prostate-specific antigen screening: importance of methods and context. J Natl Cancer Inst. 2009;101:374–383. doi: 10.1093/jnci/djp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roobol MJ, Schroder FH, Van Leeuwen P, Wolters T, Van den Bergh RC, Van Leenders GJ, et al. Performance of the prostate cancer antigen 3 (PCA3) gene and prostate-specific antigen in prescreened men: exploring the value of PCA3 for a first-line diagnostic test. Eur Urol. 2010;58:475–481. doi: 10.1016/j.eururo.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 6.Jackson BL, Grabowska A, Ratan HL. MicroRNA in prostate cancer: functional importance and potential as circulating biomarkers. BMC Cancer. 2014;14:9–30. doi: 10.1186/1471-2407-14-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;21:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 8.Berindan-Neagoe I, Monroig PD, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA Cancer J Clin. 2014;10:311–336. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sassen S, Miska EA, Caldas C. MicroRNA—implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- 11.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–6135. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 12.Guan Y, Wu Y, Liu Y, Ni J, Nong S. Association of microRNA-21 expression with clinicopathological characteristics and the risk of progression in advanced prostate cancer patients receiving androgen deprivation therapy. Prostate. 2016;76:986–993. doi: 10.1002/pros.23187. [DOI] [PubMed] [Google Scholar]
- 13.Leite KR, Reis ST, Viana N, Morais DR, Moura CM, Silva IA, et al. Controlling RECK miR21 promotes tumor cell invasion and is related to biochemical recurrence in prostate cancer. J Cancer. 2015;6:292–301. doi: 10.7150/jca.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kachakova D, Mitkova A, Popov E, Popov I, Vlahova A, Dikov T, et al. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015;34:189–200. doi: 10.1089/dna.2014.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noonan E, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, et al. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 16.Bertoli G, Cava C, Castiglioni I. MicroRNAs as biomarkers for diagnosis, prognosis and theranostics in prostate cancer. Int J Mol Sci. 2016;17:421. doi: 10.3390/ijms17030421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X, Shi K, Wang Y, Song M, Zhou W, Tu H, et al. Clinical value of integrated-signature miRNAs in colorectal cancer: miRNA expression profiling analysis and experimental validation. Oncotarget. 2015;6:37544–37556. doi: 10.18632/oncotarget.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Y, Xiang G, Meng Y, Dong R. MiRNA-183-5p promotes cell proliferation and inhibits apoptosis in human breast cancer by targeting the PDCD4. Reprod Boil. 2016;16:225–233. doi: 10.1016/j.repbio.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Yin Y, Liu X, Xi X, Xue W, Qu Y. Non-small cell lung cancer associated microRNA expression signature: integrated bioinformatics analysis, validation and clinical significance. Oncotarget. 2017;8:245–264. doi: 10.18632/oncotarget.15596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao F, Zhu J, Chen Y, Tang N, Wang X, Li X. MicroRNA-183-5p promotes the proliferation, invasion and metastasis of human pancreatic adenocarcinoma cells. Oncol Lett. 2016;11:134–140. doi: 10.3892/ol.2015.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 22.Catto JW, Alcaraz A, Bjartell AS, White RD, Evans CP, Fussel S, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 23.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 24.Wach S, Nolte E, Szczyrba J, Stöhr R, Hartmann A, Ørntoft T, et al. MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer. 2012;130:611–621. doi: 10.1002/ijc.26064. [DOI] [PubMed] [Google Scholar]
- 25.Larne O, Martens-Uzunova E, Hagman Z, Edsjö A, Lippolis G, den Berg MS, et al. miQ—a novel microRNA based diagnostic and prognostic tool for prostate cancer. Int J Cancer. 2013;132:2867–2875. doi: 10.1002/ijc.27973. [DOI] [PubMed] [Google Scholar]
- 26.Kundu ST, Byers LA, Peng DH, Roybal JD, Diao L, Wang J, et al. The miR-200 family and the miR-183–96–182 cluster target Foxf2 to inhibit invasion and metastasis in lung cancers. Oncogene. 2016;35:173–186. doi: 10.1038/onc.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, et al. Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer. 2010;126:1166–1176. doi: 10.1002/ijc.24827. [DOI] [PubMed] [Google Scholar]
- 28.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 29.Mihelich BL, Khramtsova EA, Arva N, Vaishnav A, Johnson DN, Giangreco AA, et al. miR-183-96-182 cluster is over-expressed in prostate tissue and regulates zinc homeostasis in prostatecells. J Biol Chem. 2011;286:44503–44511. doi: 10.1074/jbc.M111.262915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueno K, Hirata H, Shahryari V, Deng G, Tanaka Y, Tabatabai ZL, et al. microRNA-183 is an oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer. 2013;108:1659–1667. doi: 10.1038/bjc.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ijichi H, Otsuka M, Tateishi K, Ikenoue T, Kawakami T, Kanai F, et al. Smad4-independent regulation of p21/WAF1 by transforming growth factor-beta. Oncogene. 2004;23:1043–1051. doi: 10.1038/sj.onc.1207222. [DOI] [PubMed] [Google Scholar]
- 32.Liu R, Zhou Z, Huang J, Chen C. PMEPA1 promotes androgen receptor-negative prostate cell proliferation through suppressing the Smad3/4-c-Myc-p21 Cip1 signaling pathway. J Pathol. 2004;223:683–694. doi: 10.1002/path.2834. [DOI] [PubMed] [Google Scholar]
- 33.Rizos E, Siafakas N, Katsantoni E, Skourti E, Salpeas V, Rizos I, et al. Let-7, Mir-98 and Mir-181 as biomarkers for cancer and schizophrenia. PLoS ONE. 2015;10:e0123522. doi: 10.1371/journal.pone.0123522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferracin M, Veronese A, Negrini M. Micro-markers: miRNAs in cancer diagnosis and prognosis. Expert Rev Mol Diagn. 2010;10:297–308. doi: 10.1586/erm.10.11. [DOI] [PubMed] [Google Scholar]
- 35.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50:298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larne O, Östling P, Haflidadóttir BS, Hagman Z, Aakula A, Kohonen PO, et al. miR-183 in prostate cancer cells positively regulates synthesis and serum levels of prostate-specific antigen. Eur Urol. 2015;68:581–588. doi: 10.1016/j.eururo.2014.12.025. [DOI] [PubMed] [Google Scholar]


