Abstract
Background
Canine parvovirus type 2 (CPV-2) was first identified in the late 1970s; it causes intestinal hemorrhage with severe bloody diarrhea in kennels and dog shelters worldwide. Since its emergence, CPV-2 has been replaced with new genetic variants (CPV-2a, CPV-2b, and CPV-2c). Currently, information about the genotype prevalence of CPV-2 in Vietnam is limited. In the present study, we investigated the genotype prevalence and distribution of CPV-2 in the three regions of Vietnam.
Methods
Rectal swabs were collected from 260 dogs with suspected CPV-2 infection from northern, central, and southern Vietnam from November 2016 to February 2018. All samples were identified as parvovirus positive by real-time PCR, and further genotyping was performed using a SimpleProbe® real-time PCR assay.
Results
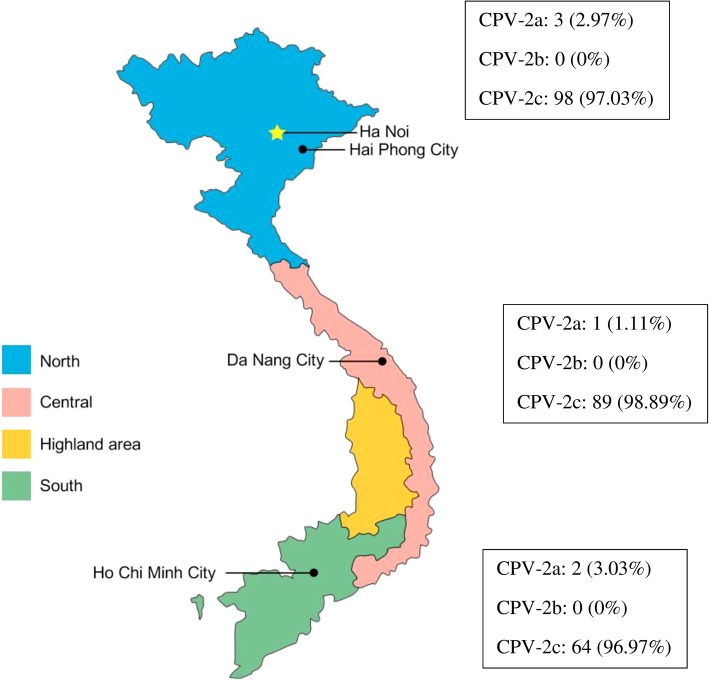
Of the 260 Vietnamese CPV-2 isolates, 6 isolates (2.31%) were identified as CPV-2a, 251 isolates (96.54%) were identified as CPV-2c and 3 isolates (1.15%) were untypable using the SimpleProbe® real-time PCR assay. In northern Vietnam, the percentages of CPV-2a and CPV-2c were 2.97% (3/101) and 97.3% (98/101), respectively. In central Vietnam, the percentages of CPV-2a and CPV-2c were 1.11% (1/90) and 98.89% (89/90), respectively. In southern Vietnam, the percentages of CPV-2a and CPV-2c were 3.03% (2/66) and 96.97% (64/66), respectively. CPV-2b was not observed in this study. The VP2 genes of CPV-2c in Vietnam are more genetically similar to those of CPV-2c strains in China and Taiwan than to those of prototype CPV-2c strains (FJ222821) or the first Vietnamese CPV-2c (AB120727).
Conclusion
The present study provides evidence that CPV-2c is the most prevalent variant in Vietnam. Phylogenetic analysis demonstrated that the recent Vietnamese CPV-2c isolates share a common evolutionary origin with Asian CPV-2c strains.
Electronic supplementary material
The online version of this article (10.1186/s12985-019-1159-z) contains supplementary material, which is available to authorized users.
Keywords: Canine parvovirus type 2, Genotype, SimpleProbe® real-time PCR, Vietnam, SimpleProbe®
Background
Canine parvovirus type 2 (CPV-2) is one of the most dangerous enteropathogens, causing fatal disease in dogs and puppies worldwide [1]. CPV-2 is a nonenveloped, small DNA virus with a diameter of approximately 25 nm and a single-stranded DNA genome of approximately 5 kb [2]. CPV-2 belongs to the genus Parvovirus in the family Parvoviridae, which includes feline panleukopenia virus (FPV), mink enteritis virus, raccoon parvovirus, and porcine parvovirus [3]. Clinical manifestations of CPV-2 infection are characterized by intestinal hemorrhage with severe bloody diarrhea; other clinical signs include anorexia, depression and vomiting [1, 3, 4].
CPV-2 is a rapidly evolving virus, leading to the mutation of novel variants since its first recognition in the late 1970s [5]. For example, CPV-2a was approximately discovered in 1980 in the USA, and CPV-2b and 2c were identified in 1984 and 2000 in the USA and Italy, respectively, based on residue 426 (Asn in 2a, Asp in 2b, and Glu in 2c) in the VP2 protein of the parvovirus [4, 6]. When a novel CPV-2 variant appears, it very rapidly replaces old variants [7, 8]. In recent decades, CPV-2c has been found to be widespread in European countries [1, 9], the USA [10], South America [11–13] and Africa [14, 15]. The first reported occurrence of the CPV-2c variant in Vietnam was in 2004 [16]; however, since then, this strain has not been prevalent in Asia [17]. Surprisingly, novel Asian CPV-2c isolates were identified in China [18–21], Taiwan [17, 22], Laos [23] and Thailand [24] in the past few years.
In Vietnam, CPV-2 infection was first observed as sporadic cases in 1994 (unpublished data). Subsequently, after its first emergence, there were widespread outbreaks of canine hemorrhagic enteritis with high morbidity and mortality occurring across the whole country [16]. Along with the increasing number of pet dogs in Vietnam, CPV-2 infection has emerged as a veterinary public health concern that greatly affects puppies because of its high mortality and morbidity. However, current information related to the antigenic types of CPV prevailing in Vietnam is poorly understood. Thus, in the present study, we investigated the genotype prevalence and distribution of CPV-2 from naturally infected dogs in three regions of Vietnam using a SimpleProbe® real-time polymerase chain reaction (PCR) assay.
Methods
Specimen collection
Rectal swabs were collected from 260 dogs with suspected CPV-2 infection from northern, central, and southern Vietnam from November 2016 to February 2018. These dogs had displayed clinical symptoms of CPV-2 infection, including diarrhea or bloody diarrhea. Fecal samples were collected from dogs with alimentary signs (diarrhea or bloody diarrhea) by inserting a swab into the rectum (~ 2 cm), rotating the swab and then resuspending the swab in 1 ml of phosphate-buffered saline. All specimens were transported on ice and stored at -20 °C at the key laboratory for biotechnology and animal disease, Vietnam National University of Agriculture, Hanoi, Vietnam. Information including the sampling year, age, clinical history, and CPV-2 types of the sampled dogs is summarized in Additional file 1.
DNA extraction and parvovirus screening
Total DNA was extracted using a Genomic DNA Mini Kit (Geneaid Biotech, Ltd., Taipei, Taiwan) following the manufacturer’s protocol. All of the clinical specimens were confirmed to be infected with parvovirus by real-time PCR, as described by Lin et al. [25].
SimpleProbe® assay for CPV-2 genotyping
Parvovirus-positive samples were further characterized by a SimpleProbe® real-time PCR assay, following the method developed by Hoang et al. [26]. First, the reaction was carried out in a final volume of 10 μl containing 3 mM MgCl2, 3 pM SimpleProbe® (TIB MOLBIOLGmbH, Berlin, Germany), 5 pM and 2 pM each of primers Parvo-F (5′-ACA CCT gAg AgA TTT ACA TAT ATA gCA CA-3′) and Parvo-A (5′-ATT AgT ATA gTT AAT TCC TgT TTT ACC TCC-3′), 1 μl 10x Light Cycler 480 Genotyping master mix and 1 μl 10x diluted template DNA. Real-time PCR was cycled under the same thermal conditions as those previously described for the SimpleProbe® assay [26].
VP2 gene amplification, sequencing and sequence analyses
To clone full length VP2, we performed PCR amplification using the primer pair VP2F (5′- CGGTGCAGGACAAGTAAAA -3′)/VP2R (5′-GGTGCTAGTTGATATGTAATA -3′) that amplified a 1755 bp fragment of the gene encoding the capsid protein. The PCR conditions were as follows: denaturation at 94 °C for 2 min; 40 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 1 min, and extension at 72 °C for 2 min; and a final extension step at 72 °C for 5 min. PCR products were electrophoresed, followed by the purification of electrophoresis products by a Wizard® SV Gel and PCR clean-up system (Promega Corporation, USA). The PCR products obtained from the previous step were cloned by a T&A® Cloning kit before being sent for automated sequencing (MB mission biotech, Inc., Taiwan). The resulting sequences were compared with reference FPV (M38246), CPV-2 (M38245), CPV-2a (M24003, and M24000), new CPV-2a (AY742953, AB054213, EU009200, JQ686671, KF676668, and KR611488), CPV-2b (M74852 and M74849), new CPV-2b (AB120720, AB120721, AB120722, AB120723, AB120724, AB120725, AB120726, AY742955, AY869724, JQ268284, KR611459, and KR611461), CPV-2c (FJ222821, AB120727, FJ005235, KM236569, KR611522, KT156832, KT162005, KY937650, MF467229, and KU244254), CPV-2a vaccine strain (FJ197847) and previous Vietnamese strains (AB120720, AB120721, AB120722, AB120723, AB120724, AB120725, AB120726). The Clustal W method and MegAlign program (DNASTAR, Madison, WI, USA) were used for multiple alignments of the nucleic acid and amino acid sequences. Phylogenetic analyses were conducted by the maximum likelihood methods using MEGA 6 based on the Tamura-Nei model [27].
Results
SYBR-based real-time PCR amplification
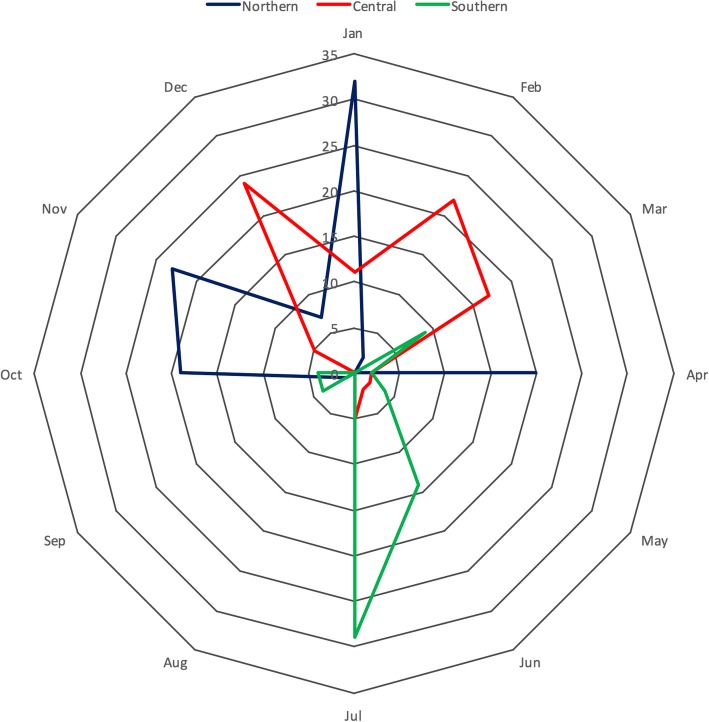
A total of 260 specimens were collected from suspected parvovirus-infected dogs in three regions of Vietnam, including northern, central and southern Vietnam. All 260 samples were positive for parvovirus DNA. The age of the sampled dogs ranged from 1 month to 12 months old. The demographic summary of puppies that tested positive for parvovirus by real-time PCR is also presented in Additional file 1. The number of puppies less than 3 months old accounted for 26.92% (70/260) of the sample, dogs of 3 to 6 months of age accounted for 49.62% (129/260) of the sample, and dogs older than 6 months accounted for 8.85% (23/260) of the sample. For 38 samples, the age group was unspecified. In terms of gender, 51.15% (133/260) of the sample was male dogs, and 36.15% (94/260) was female dogs. For 33 samples, the gender was not reported. In the northern areas of Vietnam, CPV-2 cases often peak in November and January, while the peak time for central areas is from December to March, and the peak time for southern areas is in June and July (Fig. 1).
Fig. 1.
Monthly distribution of CPV-2 isolates in three regions of Vietnam. The numbers of positive cases in each of the three regions were individually calculated according to the sample collection date to analyze the monthly distribution
Genotype analysis and geographical distribution of CPV-2 variants
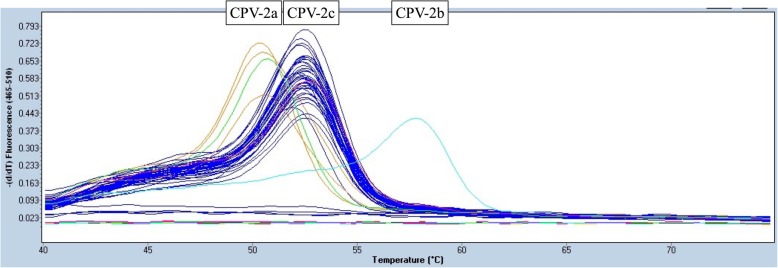
Among the 260 samples collected from the three regions in Vietnam, 257 samples (98.85%) were clearly genotyped according to melting temperature analysis using a SimpleProbe® real-time PCR assay (Table 1). However, there were three cases (1.15%) from northern Vietnam for which differentiation was not possible by the SimpleProbe® real-time PCR assay (Additional file 1). Among 257 samples, six isolates (2.31%) were identified as CPV-2a and 251 isolates (96.54%) were identified as CPV-2c. In northern Vietnam, the percentages of CPV-2a and CPV-2c were 2.97% (3/101) and 97.03% (98/101), respectively. In central Vietnam, the percentages of CPV-2a and CPV-2c were 1.11% (1/90) and 98.89% (89/90), respectively. In southern Vietnam, the percentages of CPV-2a and CPV-2c were 3.03% (2/66) and 96.97% (64/66), respectively (Fig. 2). CPV-2b was not observed in this study (Fig. 3). Our results showed that CPV-2c is currently the most prevalent CPV-2 field strain circulating in Vietnam.
Table 1.
Melting temperature of Vietnam samples
| CPV-2 genotype | SimpleProbe® analysis | Reference melting temperature (CI95%) [26] | |
|---|---|---|---|
| Number | Melting temperature of Vietnamese samples (CI95%) | ||
| 2a | 6 | 50.23 (50.2–50.3) | 50.2 (50.1–50.5) |
| 2b | 0 | N/Aa | 57.8 (57.7–58.5) |
| 2c | 251 | 52.75 (52.7–52.8) | 52.3 (52.2–53.2) |
aN/A not available
Fig. 2.
Geographical distribution of CPV-2 variants in three regions of Vietnam. Of a total of 257 genotyped CPV-2 isolates, 101 isolates, 90 isolates and 66 isolates were collected from northern, central and southern Vietnam, respectively. The percentage of CPV-2 variants was independently calculated in each region
Fig. 3.
Melting curve analysis of clinical CPV-2 isolates using a SimpleProbe® real-time PCR assay
Genotype of CPV-2 vaccinated dogs
A total of 44 dogs (16.92%) were confirmed to be vaccinated against CPV-2. Among 44 CPV-2 vaccinated dogs, 6 were younger than 3 months old, and 32 were 3 months old or older; the age of the remaining 6 dogs could not be accurately determined. Interestingly, all vaccinated dogs were identified as having CPV-2c infection using the SimpleProbe® real-time PCR assay.
Amino acid and nucleotide sequence analysis of VP2
To investigate the association and diversity of CPV-2 strains circulating in Vietnam, we sequenced and analyzed 16 complete VP2 genes of CPV-2 strains collected from the three regions of Vietnam. Phylogenetic analysis based on the nucleotide sequences of the complete VP2 gene revealed two clusters that were prototype CPV-2c strains and Asian CPV-2c clusters (15 isolates from this study together with the Chinese strain [KR611522, KT156832, KT162005, KY937650, and MF467229], and Taiwanese strain [KU244254]) (Fig. 4). The VP2 genes of CPV-2c in Vietnam are more genetically similar to CPV-2c strains in China and Taiwan than to prototype CPV-2c strains (FJ222821) or first Vietnamese CPV-2c (AB120727) (Fig. 4). In addition, one Vietnamese strain (DN33) in this study was clustered with a CPV-2a vaccine strain (FJ197847) (Fig. 4).
Fig. 4.
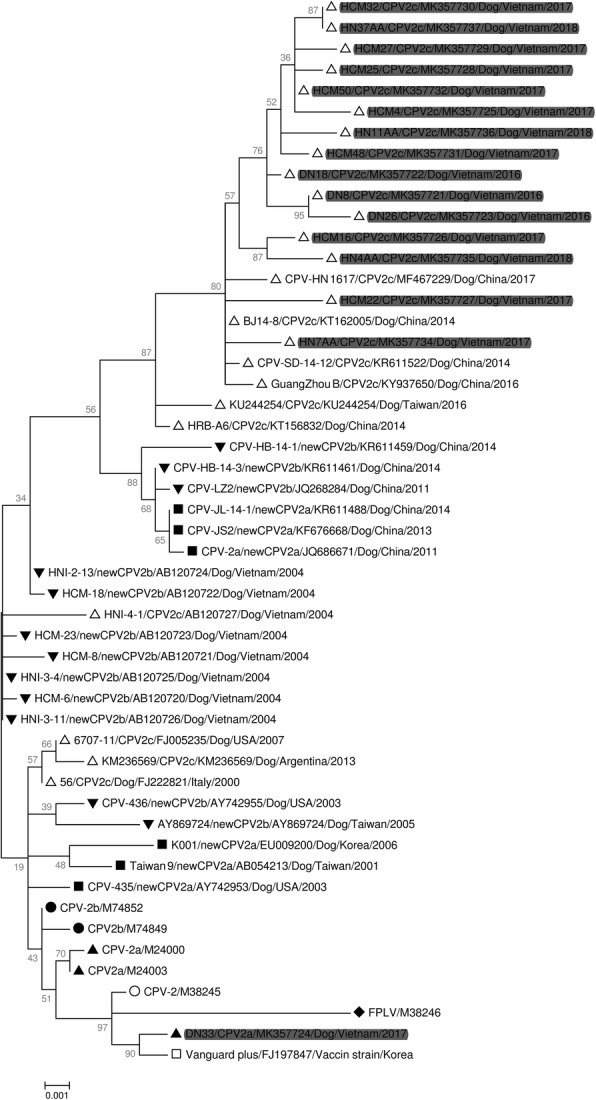
Phylogenetic relationships based on the complete VP2 gene of CPV-2 between. Vietnamese isolates and reference strains. HN: HaNoi (North Vietnam), DN: DaNang (Central Vietnam), HCM: HoChiMinh city (South Vietnam), CPV: Canine parvovirus, FPLV: Feline panleukopenia virus. The analysis was performed employing the maximum likelihood method based on 1000 replicates using MEGA 6 software. CPV variants are indicated by □, ◆, ◯, ▲, ■, ●, ▼ and △ for the vaccine strain, FPLV, CPV-2, CPV-2a, new CPV-2a, CPV-2b, new CPV-2b, and CPV-2c, respectively. Gray underlay represents CPV-2 isolates in the present study
There were several nucleotide mutations in the VP2 gene of all strains (Table 2), but only 6 mutations (nucleotide position 13, 800, 899, 970, 1109, and 1341) resulted in changes in the amino acid sequence. Amino acid sequence comparison among the 11 reference strains and 16 isolates at amino acids 5–555 is illustrated in Table 2. For the complete VP2 analysis, one sequence with an Asn and 15 sequences with a Glu at position 426 were classified as CPV-2a and CPV-2c, respectively. All CPV-2c strains showed substitution at position Ala5Gly, Phe267Tyr, Tyr324Ile, and Gln370Arg (Table 2). Thirteen of the 15 CPV-2c strains obtained showed a unique substitution at position 447 (Ile to Met) caused by mutation of TAT to TGT at nucleotide positions 1340–1342 of the VP2 gene (Table 2).
Table 2.
Nucleotide sequences and amino acid substitution in the full VP2 gene of CPV-2
| Strains | Position of nucleotide sequence (amino acid) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 13–15 (5) | 259–261 (87) | 799–801 (267) | 898–900 (300) | 964–966 (322) | 967–969 (323) | 970–972 (324) | 1000–1002 (334) | 1021–1023 (341) | 1108–1110 (370) | 1276–1278 (426) | 1318–1320 (440) | 1339–1341 (447) | 1663–1665 (555) | |
| Reference strain | ||||||||||||||
| CPV-2 | ||||||||||||||
| M38245 (USA) | GCA (Ala) | ATG (Met) | TTT (Phe) | GCT (Ala) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| CPV-2a | ||||||||||||||
| M24000 (USA) | GCA (Ala) | TTG (Leu) | TTT (Phe) | GGT (Gly) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | ACA (Thr) | ATA (Ile) | ATA (Val) |
| FJ197847 (vaccine) | GCA (Ala) | ATG (Met) | TTT (Phe) | GGT (Gly) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| New CPV-2a | ||||||||||||||
| AY742953 (USA) | GCA (Ala) | TTG (Leu) | TTT(Phe) | GGT (Gly) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| KF676668 (China) | GCA (Ala) | TTG (Leu) | TAT(Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | GCA (Ala) | ATA (Ile) | GTA (Val) |
| KR611488 (China) | GCA (Ala) | TTG (Leu) | TAT(Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | GCA (Ala) | ATA (Ile) | GTA (Val) |
| JQ686671 (China) | GCA (Ala) | TTG (Leu) | TAT(Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | GCA (Ala) | ATA (Ile) | GTA (Val) |
| CPV-2b | ||||||||||||||
| M78452 (USA) | GCA (Ala) | TTG (Leu) | TTT (Phe) | GGT (Gly) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAT (Asp) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| New CPV-2b | ||||||||||||||
| AY742955 (USA) | GCA (Ala) | TTG (Leu) | TTT(Phe) | GGT (Gly) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAT (Asp) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| JQ268284 (China) | GCA (Ala) | TTG (Leu) | TAT(Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAT (Asp) | GCA (Ala) | ATA (Ile) | GTA (Val) |
| KR611459 (China) | GCA (Ala) | TTG (Leu) | TAT(Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAT (Asp) | GCA (Ala) | ATA (Ile) | GTA (Val) |
| KR611461 (China) | GCA (Ala) | TTG (Leu) | TAT(Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAT (Asp) | GCA (Ala) | ATA (Ile) | GTA (Val) |
| CPV-2c | ||||||||||||||
| FJ005235 (USA) | GCA (Ala) | TTG (Leu) | TTT (Phe) | GCT (Ala) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAA (Glu) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| FJ222821 (Italy) | GCA (Ala) | TTG (Leu) | TTT (Phe) | GGT (Gly) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAA (Glu) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| KY937650 (China) | GGA (Gly) | TTG (Leu) | TAT(Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CAA (Gln) | GAA (Glu) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| KU244254 (Taiwan) | GCA (Ala) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| AB120727 (Vietnam) | GCA (Ala) | TTG (Leu) | TTT (Phe) | GGT (Gly) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCG (Pro) | CAA (Gln) | GAA (Glu) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| This study | ||||||||||||||
| CPV-2a | ||||||||||||||
| DN33/MK357724 | GCA (Ala) | ATG (Met) | TTT (Phe) | GCT (Ala) | ACA (Thr) | AAC (Asn) | TAT (Tyr) | GCT (Ala) | CCA (Pro) | CAA (Gln) | AAT (Asn) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| CPV-2c | ||||||||||||||
| HN7AA/MK357734 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| HN4AA/MK357735 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCC (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| N11AA/MK357736 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HN37AA/K357737 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HCM4/MK357725 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HCM16/MK357726 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HCM22/MK357727 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATA (Ile) | GTA (Val) |
| HCM25/MK357728 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HCM27/MK357729 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HCM32/MK357730 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HCM48/MK357731 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| HCM50/MK357732 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| DN8/MK357721 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| DN18/MK357722 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
| DN26/MK357723 | GGA (Gly) | TTG (Leu) | TAT (Tyr) | GGT (Gly) | ACA (Thr) | AAC (Asn) | ATT (Ile) | GCT (Ala) | CCA (Pro) | CGA (Arg) | GAA (Glu) | ACA (Thr) | ATG (Met) | GTA (Val) |
Discussion
CPV-2c is currently the most prevalent genotype in Asian countries [17–22]. The first Asian CPV-2c case was detected in Vietnam in 2004 [16]. However, information regarding the current genotype prevalence of CPV-2 strains in Vietnam is limited. This study is the first to investigate the genotype prevalence of CPV-2 in 3 regions of Vietnam. Similar to other Asian countries [17–22], the CPV-2c variant appears to be the most prevalent genotype in the dog population in all regions of Vietnam. Surprisingly, no CPV-2b cases were found in Vietnam in the present study; however, according to previous reports, CPV-2b had the highest detection rate in Vietnam in 2004 [16] (Table 3). The reasons for this discrepancy may be as follows: i) current CPV-2b vaccines are effective in preventing and controlling the disease in Vietnam; or ii) there is limited or no protection from current vaccines against the CPV-2c variant, and this variant rapidly replaced the previous circulation of CPV-2 strains. As such, we used a SYBR Green-based real-time PCR assay, which was sensitive, specific and reliable for the detection of CPV-2, FPV and porcine parvovirus DNA [25]. However, there were three parvovirus-positive cases from northern Vietnam for which differentiation was not possible by our SimpleProbe® real-time PCR assay. This issue may be due to i) unusual changes in the probe sequence region [26]; or ii) the infection of Vietnamese dogs by other parvovirus species for which the sequence analysis has not been investigated.
Table 3.
Review of CPV-2 genotyping in Vietnam
| Study period | Region of Vietnam | Genotype of CPV-2 | References | ||
|---|---|---|---|---|---|
| 2a | 2b | 2c | |||
| 2003–2004 | Northern and Southern | 0 | 8 | 1 | Nakamura et al., 2004 [16] |
| 2016–2018 | Northern, Central and Southern | 6 | 0 | 251 | This study |
The country of Vietnam is divided into three regions with different climates. Northern Vietnam has a humid subtropical climate, while the central region has a tropical monsoon climate, and southern Vietnam has a tropical savanna climate. Another factor that may affect the study is the humidity in Vietnam, which is 84–100% on average. However, because of differences in latitude and topography, the climate tends to be markedly different across regions, which may cause the difference in the timing of parvovirus outbreaks in specific regions. In the northern part of the country, due to more seasons than other regions (4 seasons in total), the seasonal periods of parvovirus outbreaks are usually from October to November (winter transition time), from December to January (spring transition time), and from March to May, with a peak in April (summer transition time) (Fig. 1). Northern areas show a disease period spread over many months, whereas outbreaks often occur at a more concrete, specific time in the central and southern parts of the country. In fact, in central Vietnam, the period from December to March is the time when parvovirus infection increased rapidly in dogs. In the southern part of the country, small dogs are more likely to be affected during the period from June to August when the weather is beginning to switch from the dry to rainy season. Thus, CPV-2 disease can occur in Vietnam at any time of the year depending on the geographic location; however, when there is a change in climate, temperature and humidity make puppies susceptible to infection and cause CPV-2 to become widespread in the environment due to prolonged survival time. Various contradicting studies emphasize the disease’s seasonality in different specific geographical regions. Some scientists have indicated that the highest incidence of this disease is detected in spring and summer [28, 29], while the opposite is true for other locations [30–33].
Dogs aged 3 months to 6 months accounted for the highest detection rate (129/260; 49.62%), while this rate for the 1 month to 2 month age group was 26.92% (70/260), and the rate for dogs over 6 months old was only 8.85% (23/260). For 38 samples, the age group was not reported. The percentage of dogs under 6 months of age who were greatly affected has been previously reported [29, 34]. The reason dogs from 3 months to 6 months old are easily susceptible to CPV is due to the decrease in maternal antibodies [35, 36]. In addition, the higher incidence of CPV-2 in dogs younger than 6 months old might be due to quicker intestinal crypt cell multiplication with a higher mitotic index by changes in bacterial flora and daily diet during weaning time [1, 36]. Very few cases are recorded over 1 year of age; the cause may be slight exposure to the virus, leading to antibody production in the host; previous vaccination of animals; or another reason not yet identified. Dogs in the 7 to 12 month age group still exhibit CPV disease, which may be because of poor vaccine preservation or incorrect vaccination timing [37].
We found that the proportion of infected females was less than that of males. Of 260 positive cases, 94 were females (36.15%), and 133 (51.15%) were males. Coincidently, the current finding is consistent with studies by Thomas et al. [38] and Gombac et al. [39], as well as other reliable publications [34, 38, 40–42]. Many assumptions could be analyzed; however, the most relevant cause could be that the majority of samples were collections from male dogs during the study. Another explanation for this result could be that male dogs are more likely to be infected; in fact, more male dogs were admitted for treatment than female dogs. However, there is reportedly no influence of sex on the incidence of CPV [43, 44]. The high incidence in male dogs may also be due to some behaviors and habits of pet owners or the hobby of selecting and keeping male dogs [45].
A large number of dogs have been vaccinated against CPV-2 but still suffer from the disease, with most of the cases (32/44; 72.72%) 3 months of age and older. This issue may be related to failure to properly use the vaccine or to preserve the vaccine [11, 46]. Many studies have shown that CPV-2 and CPV-2b vaccines provide protection against the CPV-2a/2b virus and provide complete protection against the new CPV-2c variant in dogs for up to 9 years [47–49]. However, there are some disagreements regarding the use of traditional vaccines against the new antigen variant CPV-2c [50, 51]. Similar to a study conducted in Taiwan, four of 22 CPV-2c-infected dogs died despite vaccination, including one adult dog that completed the vaccination program [17]. This phenomenon has also been seen in Vietnam where 100% of vaccinated positive cases were of genotype 2c. Many CPV-2 vaccines are being used in Vietnam; however, there is little documentation of the effects of CPV-2 vaccination against the current CPV-2c variant. Therefore, the efficacy of the current vaccine against the CPV-2c variant remains to be evaluated.
Currently, there are many publications regarding the circulation of the CPV-2c strain in many countries in Asia [17–21, 23–25, 52, 53] Although the first publication on CPV-2c was in Vietnam in 2004 [16], the CPV-2c variant has not been prevalent in Asia. Surprisingly, novel Asian CPV-2c isolates were reported in China [18–21], Taiwan [17, 22], Laos [23], and Thailand [24]. This finding suggests that novel CPV-2c is now becoming increasingly prevalent in Asian countries. Based on the results of the phylogenetic analysis, the cause for the large amount of CPV-2c detected in Vietnam is the invasion of a foreign strain from 2016 to 2018. In addition, there was no phylogenetical relation between Vietnamese CPV-2c and prototype CPV-2c strains (European or American strains) or the past Vietnamese CPV-2c in 2004, while these strains are similar to the recent Asian CPV-2c strains (Fig. 4). Phylogenetic analysis revealed that the recent Vietnamese CPV-2c strains share a common evolutionary origin with Asian CPV-2c strains. This finding could be explained by dog imports from China and other neighboring nations with a lack of oversight due to a long shared border with these countries and the effort needed from authorities to apply restrictions.
Amino acid substitutions of Ala5Gly, Phe267Tyr, Tyr324Ile and Gln370Arg of CPV-2c have been observed in China [18, 20, 21] and Taiwan [17, 22]. However, the functions of CPV-2 residues 5, 267, 324 and 370 are still unknown and remain to be elucidated. The substitution of Ile447Met is unique to the Vietnamese CPV-2c strains. Residue 447 is not exposed on the capsid surface [54], and substitutions in this position may not affect the antigenicity of the virus. However, Agbandie’s study showed that the binding of DNA within the internal surface of the parvovirus protein shell may cause specific conformational changes in the protein [54]. Therefore, the function of residue 447 remains to be elucidated.
Conclusions
This study is the first CPV-2 epidemiological survey in Vietnam to document the presence and relative distribution of CPV-2 variants in three regions of Vietnam over the past few years. Phylogenetic analysis demonstrated that the recent Vietnamese CPV-2c isolates share a common evolutionary origin with Asian CPV-2c strains. It is important for veterinarians and dog owners to understand the current geno-prevalence of CPV-2 and effective prevention methods based on outbreak periods.
Additional file
The genotypes of 260 canine parvovirus type 2 isolates collected from Vietnamese dogs. (DOCX 76 kb)
Acknowledgments
Funding
This work was also supported by the Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106.05-2018.337.
Availability of data and materials
Not applicable.
Abbreviations
- CPV-2
Canine parvovirus type 2
- FPV
Feline panleukopenia virus
- PCR
Polymerase chain reaction
Authors’ contributions
MH and WHL performed the molecular data analysis and wrote the manuscript. BTTN performed the cloning and sequence analysis. VPL and MTC managed the study, provided materials and reagents, contributed to the interpretation of the data, and cowrote the manuscript. CNL designed and analyzed the experimental data and wrote the manuscript. All of the authors read and approved the final manuscript.
Ethics approval and consent to participate
The study did not involve any animal experiments. The Institutional Animal Care and Use Committee (IACUC) of Vietnam National University of Agriculture did not deem it necessary for this research group to obtain formal approval to conduct this study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Minh Hoang, Email: kira2108.hua@gmail.com.
Wei-Hao Lin, Email: j7967633@gmail.com.
Van Phan Le, Email: letranphan@gmail.com.
Bui Thi To Nga, Email: buitonga@gmail.com.
Ming-Tang Chiou, Email: mtchiou@mail.npust.edu.tw.
Chao-Nan Lin, Email: cnlin6@mail.npust.edu.tw.
References
- 1.Decaro N, Buonavoglia C. Canine parvovirus: a review of epidemiological and diagnostic aspects, with emphasis on type 2c. Vet Microbiol. 2012;155:1–12. doi: 10.1016/j.vetmic.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reed AP, Jones EV, Miller TJ. Nucleotide sequence and genome organization of canine parvovirus. J Virol. 1988;62:266–276. doi: 10.1128/jvi.62.1.266-276.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoelzer K, Parrish CR. The emergence of parvoviruses of carnivores. Vet Res. 2010;41:39. doi: 10.1051/vetres/2010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parrish CR, Aquadro CF, Strassheim ML, Evermann JF, Sgro JY, Mohammed HO. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J Virol. 1991;65:6544–6552. doi: 10.1128/jvi.65.12.6544-6552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appel MJ, Scott FW, Carmichael LE. Isolation and immunisation studies of a canine parco-like virus from dogs with haemorrhagic enteritis. Vet Rec. 1979;105:156–159. doi: 10.1136/vr.105.8.156. [DOI] [PubMed] [Google Scholar]
- 6.Parrish CR, O'Connell PH, Evermann JF, Carmichael LE. Natural variation of canine parvovirus. Science. 1985;230:1046–1048. doi: 10.1126/science.4059921. [DOI] [PubMed] [Google Scholar]
- 7.Buonavoglia C, Martella V, Pratelli A, Tempesta M, Cavalli A, Buonavoglia D, Bozzo G, Elia G, Decaro N, Carmichael L. Evidence for evolution of canine parvovirus type 2 in Italy. J Gen Virol. 2001;82:3021–3025. doi: 10.1099/0022-1317-82-12-3021. [DOI] [PubMed] [Google Scholar]
- 8.Martella V, Cavalli A, Pratelli A, Bozzo G, Camero M, Buonavoglia D, Narcisi D, Tempesta M, Buonavoglia C. A canine parvovirus mutant is spreading in Italy. J Clin Microbiol. 2004;42:1333–1336. doi: 10.1128/JCM.42.3.1333-1336.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decaro N, Elia G, Martella V, Campolo M, Desario C, Camero M, Cirone F, Lorusso E, Lucente MS, Narcisi D, et al. Characterisation of the canine parvovirus type 2 variants using minor groove binder probe technology. J Virol Methods. 2006;133:92–99. doi: 10.1016/j.jviromet.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Hong C, Decaro N, Desario C, Tanner P, Pardo MC, Sanchez S, Buonavoglia C, Saliki JT. Occurrence of canine parvovirus type 2c in the United States. J Vet Diagn Investig. 2007;19:535–539. doi: 10.1177/104063870701900512. [DOI] [PubMed] [Google Scholar]
- 11.Perez R, Francia L, Romero V, Maya L, Lopez I, Hernandez M. First detection of canine parvovirus type 2c in South America. Vet Microbiol. 2007;124:147–152. doi: 10.1016/j.vetmic.2007.04.028. [DOI] [PubMed] [Google Scholar]
- 12.Calderon MG, Romanutti C, A DA, Keller L, Mattion N, La Torre J. Evolution of canine parvovirus in Argentina between years 2003 and 2010: CPV2c has become the predominant variant affecting the domestic dog population. Virus Res. 2011;157:106–110. doi: 10.1016/j.virusres.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pedroza-Roldan C, Paez-Magallan V, Charles-Nino C, Elizondo-Quiroga D, Cervantes-Mireles RL, Lopez-Amezcua MA. Genotyping of canine parvovirus in western Mexico. J Vet Diagn Investig. 2015;27:107–111. doi: 10.1177/1040638714559969. [DOI] [PubMed] [Google Scholar]
- 14.Amrani N, Desario C, Kadiri A, Cavalli A, Berrada J, Zro K, Sebbar G, Colaianni ML, Parisi A, Elia G, et al. Molecular epidemiology of canine parvovirus in Morocco. Infect Genet Evol. 2016;41:201–206. doi: 10.1016/j.meegid.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Touihri L, Bouzid I, Daoud R, Desario C, El Goulli AF, Decaro N, Ghorbel A, Buonavoglia C, Bahloul C. Molecular characterization of canine parvovirus-2 variants circulating in Tunisia. Virus Genes. 2009;38:249–258. doi: 10.1007/s11262-008-0314-1. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura M, Tohya Y, Miyazawa T, Mochizuki M, Phung HT, Nguyen NH, Huynh LM, Nguyen LT, Nguyen PN, Nguyen PV. A novel antigenic variant of canine parvovirus from a Vietnamese dog. Arch Virol. 2004;149:2261–2269. doi: 10.1007/s00705-004-0367-y. [DOI] [PubMed] [Google Scholar]
- 17.Chiang SY, Wu HY, Chiou MT, Chang MC, Lin CN. Identification of a novel canine parvovirus type 2c in Taiwan. Virol J. 2016;13:160. doi: 10.1186/s12985-016-0620-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geng Y, Guo D, Li C, Wang E, Wei S, Wang Z, Yao S, Zhao X, Su M, Wang X. Co-circulation of the rare CPV-2c with unique Gln370Arg substitution, new CPV-2b with unique Thr440Ala substitution, and new CPV-2a with high prevalence and variation in Heilongjiang Province, Northeast China. PLoS One. 2015;10:e0137288. doi: 10.1371/journal.pone.0137288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao HWJ, Jiang Y, Cheng Y, Lin P, Zhu H, Han G, Yi L, Zhang S, Guo L, Cheng S. Typing of canine parvovirus strains circulating in North-East China. Transbound Emerg Dis. 2015;64:495–503. doi: 10.1111/tbed.12390. [DOI] [PubMed] [Google Scholar]
- 20.Zhao H, Wang J, Jiang Y, Cheng Y, Lin P, Zhu H, Han G, Yi L, Zhang S, Guo L, Cheng S. Typing of canine parvovirus strains circulating in North-East China. Transbound Emerg Dis. 2017;64:495–503. doi: 10.1111/tbed.12390. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Lin P, Zhao H, Cheng Y, Jiang Z, Zhu H, Wu H, Cheng S. Continuing evolution of canine parvovirus in China: isolation of novel variants with an Ala5Gly mutation in the VP2 protein. Infect Genet Evol. 2016;38:73–78. doi: 10.1016/j.meegid.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Lin YC, Chiang SY, Wu HY, Lin JH, Chiou MT, Liu HF, Lin CN. Phylodynamic and genetic diversity of canine parvovirus type 2c in Taiwan. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed]
- 23.Vannamahaxay S, Vongkhamchanh S, Intanon M, Tangtrongsup S, Tiwananthagorn S, Pringproa K, Chuammitri P. Molecular characterization of canine parvovirus in Vientiane, Laos. Arch Virol. 2017;162:1355–1361. doi: 10.1007/s00705-016-3212-1. [DOI] [PubMed] [Google Scholar]
- 24.Charoenkul K, Tangwangvivat R, Janetanakit T, Boonyapisitsopa S, Bunpapong N, Chaiyawong S, Amonsin A. Emergence of canine parvovirus type 2c in domestic dogs and cats from Thailand. Transbound Emerg Dis. 2019. 10.1111/tbed.13177. [DOI] [PMC free article] [PubMed]
- 25.Lin CN, Chien CH, Chiou MT, Wang JW, Lin YL, Xu YM. Development of SYBR green-based real-time PCR for the detection of canine, feline and porcine parvoviruses. Taiwan Vet J. 2014;40:1–9. [Google Scholar]
- 26.Hoang M, Wu HY, Lien YX, Chiou MT, Lin CN. A SimpleProbe® real-time PCR assay for differentiating the canine parvovirus type 2 genotypereal-time PCR assay for differentiating the canine parvovirus type 2 genotype. J Clin Lab Anal. 2018:e22654. [DOI] [PMC free article] [PubMed]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horner GW. Canine parvovirus in New Zealand: epidemiological features and diagnostic methods. N Z Vet J. 1983;31:164–166. doi: 10.1080/00480169.1983.35010. [DOI] [PubMed] [Google Scholar]
- 29.Houston DM, Ribble CS, Head LL. Risk factors associated with parvovirus enteritis in dogs: 283 cases (1982-1991) J Am Vet Med Assoc. 1996;208:542–546. [PubMed] [Google Scholar]
- 30.Meunier PC, Glickman LT, Appel MJ, Shin SJ. Canine parvovirus in a commercial kennel: epidemiologic and pathologic findings. Cornell Vet. 1981;71:96–110. [PubMed] [Google Scholar]
- 31.Studdert MJ, Oda C, Riegl CA, Roston RP. Aspects of the diagnosis, pathogenesis and epidemiology of canine parvovirus. Aust Vet J. 1983;60:197–200. doi: 10.1111/j.1751-0813.1983.tb09581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pospischil A, Yamaho H. Parvovirus enteritis in dogs based on autopsy statistics 1978-1985. Tierarztl Prax. 1987;15:67–71. [PubMed] [Google Scholar]
- 33.Castro T, Miranda SC, Labarthe N, Silva LE, Cubel Garcia RC: Clinical and epidemiological aspects of canine parvovirus (CPV) enteritis in the State of Rio de Janeiro: 1995 - 2004. 2007.
- 34.Vivek Srinivas VM, Mukhopadhyay HK, Antony PX, Pillai RM. Molecular epidemiology of canine parvovirus in Southern India. Vet World. 2013;6:744–749. [Google Scholar]
- 35.Tajpara MM, Jhala MK, Rank DN, Joshi C. Incidence of canine parvovirus in diarrhoeic dogs by polymerase chain reaction. Indian Vet J. 2009;86:238–241. [Google Scholar]
- 36.Stepita ME, Bain MJ, Kass PH. Frequency of CPV infection in vaccinated puppies that attended puppy socialization classes. J Am Anim Hosp Assoc. 2013;49:95–100. doi: 10.5326/JAAHA-MS-5825. [DOI] [PubMed] [Google Scholar]
- 37.Carmichael LE, Joubert JC, Pollock RV. A modified live canine parvovirus vaccine. II. Immune response. Cornell Vet. 1983;73:13–29. [PubMed] [Google Scholar]
- 38.Thomas JSM, Goswami TK, Verma S, Badasara SK. Polymerase chain reaction based epidemiological investigation of canine parvoviral disease in dogs at Bareilly region. Vet World. 2014;7:929–932. [Google Scholar]
- 39.Gombac M, Švara T, Tadic M, Pogacnic M. Retrospective study of canine parvovirosis in Slovenia. Case report. Slovenian Vet Res. 2008;45:73–78. [Google Scholar]
- 40.Phukan ABB, Deka D, Boro PK. Prevalence of canine parvovirus infection in Assam. Indian Vet J. 2010;87:972–974. [Google Scholar]
- 41.Zhao Z, Liu H, Ding K, Peng C, Xue Q, Yu Z, Xue Y. Occurrence of canine parvovirus in dogs from Henan province of China in 2009–2014. BMC Vet Res. 2016;12:138. doi: 10.1186/s12917-016-0753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kitala P, McDermott J, Kyule M, Gathuma J, Perry B, Wandeler A. Dog ecology and demography information to support the planning of rabies control in Machakos District, Kenya. Acta Trop. 2001;78:217–230. doi: 10.1016/s0001-706x(01)00082-1. [DOI] [PubMed] [Google Scholar]
- 43.Sanjukta RKM, Mandakini R. Epidemiological study of canine parvovirus. Indian Vet J. 2011;88:77–79. [Google Scholar]
- 44.Banja BK, Sahoo N, Panda HK, Ray SK, Das PK. Epizootiological status of canine viral haemorrhagic gastroenteritis in Bhubaneswar city. Indian Vet J. 2002;79:850–851. [Google Scholar]
- 45.Deka DPA, Sarma DK. Epidemiology of parvovirus and coronavirus infections in dogs in Assam. Indian Vet J. 2013;90:49–51. [Google Scholar]
- 46.Truyen U. Evolution of canine parvovirus—A need for new vaccines? Vet Microbiol. 2006;117:9–13. doi: 10.1016/j.vetmic.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Larson LJ, Schultz RD. Do two current canine parvovirus type 2 and 2b vaccines provide protection against the new type 2c variant? Vet Ther. 2008;9(2):94–101. [PubMed] [Google Scholar]
- 48.Siedek EM, Schmidt H, Sture GH, Raue R. Vaccination with canine parvovirus type 2 (CPV-2) protects against challenge with virulent CPV-2b and CPV-2c. Ber Munch Tierarztl Wochenschr. 2011;124:58–64. [PubMed] [Google Scholar]
- 49.Spibey N, Greenwood NM, Sutton D, Chalmers WS, Tarpey I. Canine parvovirus type 2 vaccine protects against virulent challenge with type 2c virus. Vet Microbiol. 2008;128:48–55. doi: 10.1016/j.vetmic.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 50.Hernandez-Blanco B, Catala-Lopez F. Are licensed canine parvovirus (CPV2 and CPV2b) vaccines able to elicit protection against CPV2c subtype in puppies?: A systematic review of controlled clinical trials. Vet Microbiol. 2015;180:1–9. doi: 10.1016/j.vetmic.2015.07.027. [DOI] [PubMed] [Google Scholar]
- 51.Miranda C, Thompson G. Canine parvovirus: the worldwide occurrence of antigenic variants. J Gen Virol. 2016;97:2043–2057. doi: 10.1099/jgv.0.000540. [DOI] [PubMed] [Google Scholar]
- 52.Nandi S, Chidri S, Kumar M, Chauhan RS. Occurrence of canine parvovirus type 2c in the dogs with haemorrhagic enteritis in India. Res Vet Sci. 2010;88:169–171. doi: 10.1016/j.rvsc.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 53.Zhang RZ, Yang ST, Feng H, Cui CS, Xia XZ. The first detection of canine parvovirus type 2c in China. J Pathog Biol. 2010;5:246–249. [Google Scholar]
- 54.Agbandje M, McKenna R, Rossmann MG, Strassheim ML, Parrish CR. Structure determination of feline panleukopenia virus empty particles. Proteins. 1993;16:155–171. doi: 10.1002/prot.340160204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genotypes of 260 canine parvovirus type 2 isolates collected from Vietnamese dogs. (DOCX 76 kb)
Data Availability Statement
Not applicable.