Abstract
Plants respond to herbivore attack with a dramatic functional reorganization that involves the activation of direct and indirect defenses and tolerance, which in turn make large demands on primary metabolism. Here we provide the first characterization of the transcriptional reorganization that occurs after insect attack in a model plant-herbivore system: Nicotiana attenuata Torr. ex Wats.-Manduca sexta. We used mRNA differential display to characterize one-twentieth of the insect-responsive transcriptome of N. attenuata and verified differential expression for 27 cDNAs. Northern analyses were used to study the effects of folivory and exposure to airborne methyl jasmonate and for kinetic analyses throughout a 16-h- light/8-h-dark cycle. Sequence similarity searches allowed putative functions to be assigned to 15 transcripts. Genes were related to photosynthesis, electron transport, cytoskeleton, carbon and nitrogen metabolism, signaling, and a group responding to stress, wounding, or invasion of pathogens. Overall, transcripts involved in photosynthesis were strongly down-regulated, whereas those responding to stress, wounding, and pathogens and involved in shifting carbon and nitrogen to defense were strongly up-regulated. The majority of transcripts responded similarly to airborne methyl jasmonate and folivory, and had tissue- and diurnal-specific patterns of expression. Transcripts encoding Thr deaminase (TD) and a putative retrotransposon were absent in control plants, but were strongly induced after herbivory. Full-length sequences were obtained for TD and the pathogen-inducible α-dioxygenase, PIOX. Effects of abiotic and biotic stimuli were investigated for transcripts encoding TD, importin α, PIOX, and a GAL83-like kinase cofactor.
Progress in the molecular biological understanding of plant defense responses against pathogens is considerable and a detailed biochemical and genetic understanding of the signal cascades mediating local, as well as systemic responses is rapidly unfolding (for review, see Baker et al., 1997; Hammond-Kosack and Jones, 1997; Somssich and Hahlbrock, 1998). In contrast, our understanding of the molecular mechanisms of plant-insect interactions is in its infancy. With the exception of two notable advances, namely the characterization of the first herbivore-specific elicitors (Alborn et al., 1997; Pohnert et al., 1999) and the demonstration of the central role of the octadecanoid pathway for resistance in three plant-insect systems (Howe et al., 1996; McConn et al., 1997; Baldwin, 1998), knowledge about the molecular basis of herbivore resistance is only beginning to emerge (Korth and Dixon, 1997; Stotz et al., 1999; Reymond et al., 2000; Ryan, 2000).
Overlap in the signaling pathways regulating pathogen- and insect-plant interactions has been reported (for review, see Hammerschmidt and Schultz, 1996; Reymond and Farmer, 1998) and should be expected. Herbivores force their way past a plant's protective barriers with a diverse array of feeding apparatuses and in doing so, cause wounds that can be opportunistically used as points of entry for microbes. Moreover, many plant-herbivore associations require a microbial partner (Berenbaum, 1988) and plant resistance traits against herbivores may target the microbial fauna, as well as the herbivores. Some herbivores (e.g. aphids) vector pathogenic organisms during feeding (Carter, 1973), which should select for defense responses that do not distinguish between microbes and herbivores.
The interaction between the specialist lepidopteran herbivore Manduca sexta and its natural host Nicotiana attenuata has the essential elements of a model system for plant-herbivore interactions: strong induced resistance (Baldwin, 1998; van Dam et al., 2000) mediated by rapidly activated direct (nicotine: Baldwin, 1999) and indirect (volatile emission: Halitschke et al., 2000, 2001; Kahl et al., 2000) defenses that are activated by products of the octadecanoid cascade (for review, see Baldwin and Preston, 1999). In nature N. attenuata is attacked by a suite of specialist and generalist herbivores (Baldwin and Ohnmeiss, 1993) and the feeding damage of one of these herbivores, namely M. sexta, is “recognized” by the plant, as evidenced by an endogenous jasmonic acid (JA) burst (McCloud and Baldwin, 1997). Attack by these larvae, or simply the addition of their oral secretions to mechanical wounds, results in an apparent change in defensive strategies: it switches from deploying a direct defense (nicotine induction) to a putative indirect defense (volatile emission; Kahl et al., 2000). This switching is mediated by yet another herbivore-specific hormonal response: an ethylene burst (Kahl et al., 2000). Moreover, the JA-induced resistance in this species comes at a substantial fitness costs (Baldwin, 1998), which in turn results from decreased intra-specific competitive ability (van Dam and Baldwin, 1998). In short, this model system exhibits a rich suite of phenotypic responses that are known to be important for plant-herbivore interactions.
Here we present the first installment of the molecular characterization of the N. attenuata-M. sexta interaction in analyzing one-twentieth of the insect-responsive transcriptome of N. attenuata by mRNA differential display (DDRT-PCR). By applying the DDRT-PCR methodology to hydroponically cultivated host plants continuously attacked by larvae for 24 h, we were able to simultaneously detect up- and down-regulated mRNAs, early and late responses, local and systemic responses, as well as shoot- and root-specific responses. Given the importance of the octadecanoid cascade in resistance in this species (Baldwin, 1998), and the role of methyl jasmonate (MeJA) in intra- and inter-plant signaling (Farmer and Ryan, 1990; Karban et al., 2000), we additionally examined the effects of airborne MeJA on mRNA accumulation. Our analysis provides the first glimpse into the large transcriptional changes that occur in N. attenuata after herbivore attack. We consider the similarities between the responses to pathogens and herbivores and the metabolic reorganization demanded by these defense responses.
RESULTS
We used DDRT-PCR to monitor changes in transcript accumulation in plants of N. attenuata exposed to folivory by first-instar larvae of M. sexta. With the goal of characterizing local and systemic responses, we pooled equal amounts of RNA extracted from shoots and roots for reverse transcription. Hydroponic culture provided standardized shoot tissue, as well as root tissue devoid of soil contamination. The expression window of the DDRT-PCR analysis presented here was defined by the combination of a single arbitrary primer R1 (selected from a complete set of 20 primers) with a complete set of 10 anchor primers and represents about one-twentieth of the insect-responsive transcriptome of N. attenuata (Bauer et al., 1993).
A total of 79 cDNA fragments exhibited differential expression on the display gel. All of the cDNAs were re-amplified by PCR, subcloned, and sequenced in sets of three to five per transformation. The analysis of 320 single-sequence runs allowed us to assign the 79 cDNAs to 53 contigs, revealing that several cDNAs had been multiply primed. Of the 53 individual sequences, 49 transcripts were detectable on northern blots. Herbivore-induced changes in transcript accumulation were verified for 27 cDNAs (Table I). Clone pDH64.4, encoding 18S rRNA, was not differential, but was added, since it served to standardize RNA blots. Sequence similarity searches allowed us to assign presumptive functions to 15 of the 27 differentially accumulating cDNAs. According to their functions, the genes can be divided into seven groups related to photosynthesis, electron transport, cytoskeleton, carbon metabolism, nitrogen metabolism, signaling, and a group of genes responding to stress, wounding, or invasion of pathogens.
Table I.
Summary of pDH clones containing cDNA inserts isolated by DDRT-PCR
| Clone | Length | Anchor Primer | GenBank Accession No. | BLAST Results
|
|
|---|---|---|---|---|---|
| Sequence similarity | E value | ||||
| bp | |||||
| pDH6.1 | 210 | A1 | AW191805 | Bell pepper Sn-1 gene (X79230) | 2 × 10−20 |
| pDH9.4 | 645 | A1 | AW191806 | G. coronatum β-tubulin (AAF08229) | 6 × 10−82 |
| pDH9.8 | 618 | A1 | AW191807 | No similarities found | – |
| pDH10.4 | 582 | A1 | AW191808 | Caenorhabditis elegans cosmid F54B3 (Z48583) | 7 × 10−4 |
| pDH10.7 | 571 | A1 | AW191809 | Arabidopsis unknown protein (AAC14534) | 4 × 10−32 |
| pDH11.1 | 711 | A2 | AW191810 | B. japonicum FixO protein homolog (B47468) | 7 × 10−25 |
| pDH14.2 | 289 | A2 | AW191811 | Tomato threonine deaminase gene (M61915) | 6 × 10−6 |
| pDH19.3 | 485 | A2 | AW191812 | Arabidopsis dehydration-induced protein RD22 (Q08298) | 2 × 10−22 |
| pDH20.2 | 273 | A3 | AW191813 | Potato germin-like protein mRNA (AF067731) | 9 × 10−14 |
| pDH21.4 | 347 | A3 | AW191814 | No similarities found | – |
| pDH23.5 | 252 | A3 | AW191815 | Human chromosome 4 clone B366O24 (AC004067) | 5 × 10−3 |
| pDH25.4 | 172 | A3 | AW191816 | Rice protein similar to retrotransposon RIRE1 (BAA88170) | 6 × 10−6 |
| pDH30.4 | 163 | A3 | AW191817 | No similarities found | – |
| pDH31.5 | 123 | A3 | AW191818 | No similarities found | – |
| pDH39.1 | 672 | A4 | AW191819 | Tomato cDNA clone cLEB8F13 (AI483151) | 1 × 10−138 |
| pDH40.1 | 615 | A4 | AW191820 | Tomato importin α-subunit (O22478) | 7 × 10−6 |
| pDH41.6 | 433 | A5 | AW191821 | Tobacco mRNA for pathogen-induced oxygenase (AJ007630) | 1 × 10−123 |
| pDH46.4 | 289 | A6 | AW191822 | Tobacco blp5 mRNA for luminal-binding protein BiP (X60058) | 4 × 10−53 |
| pDH48.4 | 604 | A6 | AW191823 | Tomato cDNA clone cLER17A21 (AI775929) | 1 × 10−14 |
| pDH55.2 | 164 | A6 | AW191824 | Arabidopsis ferredoxin-dependent glutamate synthase GLU2 (AAC78549) | 4 × 10−19 |
| pDH55.7 | 163 | A6 | AW191825 | Wild tobacco unknown gene upstream of cnx5 (AF124162) | 5 × 10−48 |
| pDH61.1 | 270 | A7 | AW191826 | Tomato lhbC1 gene for LHCII type III (X60275) | 8 × 10−39 |
| pDH63.5 | 374 | A8 | AW191827 | Potato mRNA for GAL83 protein (AJ012215) | 8 × 10−86 |
| pDH64.4 | 74 | A8 | AW191828 | Tetrameles nudiflora 18S ribosomal RNA gene (U41502) | 2 × 10−23 |
| pDH64.7 | 566 | A8 | AW191829 | Nicotiana sylvestris mRNA for the small subunit of Rubisco (X01722) | 1 × 10−146 |
| pDH68.1 | 152 | A9 | AW191830 | No similarities found | – |
| pDH69.3 | 162 | A9 | AW191831 | Tobacco Mg protoporphyrin IX chelatase (CHL H) mRNA (AF014052) | 8 × 10−81 |
| pDH73.2 | 276 | A10 | AW191832 | No similarities found | – |
The length of the inserts is given in base pairs excluding the 3′-poly(A) stretch. In addition, anchor primers used in combination with the single random primer R1 for DDRT-PCR and the GenBank accession nos. are listed. Sequence similarities and expectation (E) values obtained from BLAST searches correspond with the highest scoring matches with nucleotide or protein sequences found in the GenBank or The Arabidopsis Information Resource databases. Accession nos. of the heterologous sequences are given in parentheses.
The photosynthesis-related clones pDH61.1, pDH64.7, and pDH69.3 encode the LHB C1 subunit of the peripheral light-harvesting antenna (LHC II) of photosystem II (Schwartz et al., 1991), the small subunit (RBC S) of Rubisco (Pinck et al., 1984), and the subunit CHL H of the Mg protoporphyrin IX chelatase (Kruse et al., 1997), respectively. The Mg chelatase catalyzes the committed step in chlorophyll biosynthesis by inserting Mg2+ into protoporphyrin IX, thereby directing tetrapyrrols to the chlorophyll rather than the heme pathway (for review, see Grimm, 1998). Clone pDH11.1 encodes a protein with homology to the FixO protein of Bradyrhizobium japonicum and is therefore likely to possess a cytochrome c-type function relating the gene to the respiratory electron transport chain (Preisig et al., 1993). A cytoskeletal compound is represented by pDH9.4, which is similar to β-tubulin of the fungus Glomus coronatum (M. Stommel and P. Franken, unpublished data). Carbon metabolism is represented by pDH14.2 encoding biosynthetic Thr deaminase (TD), which catalyzes the committed step in iso-Leu biosynthesis by converting Thr to 2-ketobutyrate (Samach et al., 1991). As indicated by the similarity to the Arabidopsis ferredoxin-dependent Glu synthase (Fd-GOGAT; Lin et al., 1999), the gene product encoded by pDH55.2 is involved in nitrogen metabolism. Signaling, sensu lato, is represented by pDH40.1 and pDH63.5, which share similarities with importin α of tomato (T. Kunik, L. Mizrachy, V. Citovsky, and Y. Gafni, unpublished data) and the potato kinase cofactor StubGAL83 (Lakatos et al., 1999), respectively.
Responses to stress, wounding, or pathogens are found in a group of genes encoded by cDNAs with similarity to the wound-inducible Sn-1 gene from bell pepper (pDH6.1; Pozueta-Romero et al., 1995), the dehydration-responsive protein RD22 from Arabidopsis (pDH19.3; Yamaguchi-Shinozaki and Shinozaki, 1993), a potato germin (pDH20.2; Campbell et al., 1998), a rice retrotransposon (pDH25.4; T. Sasaki, T. Matsumoto, and K. Yamamoto, unpublished data), the pathogen-inducible oxygenase PIOX from tobacco (pDH41.6; Sanz et al., 1998), and a luminal-binding protein (BiP) from tobacco (pDH46.4; Denecke et al., 1991).
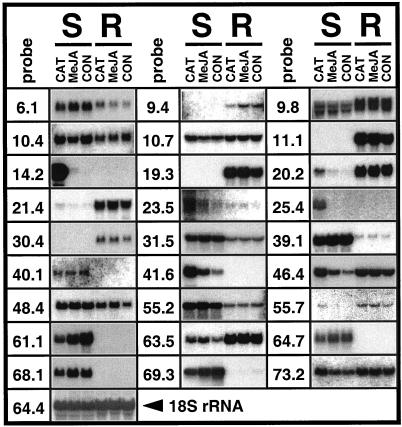
Northern analyses were conducted to determine the effects of folivory and exposure to airborne MeJA, respectively, on transcript accumulation in shoots and roots compared with untreated control plants (Fig. 1). Herbivore attack increased the accumulation of 16 transcripts: 12 in shoots (pDH9.8, pDH10.7, pDH14.2, pDH20.2, pDH21.4, pDH23.5, pDH25.4, pDH41.6, pDH46.4, pDH55.2, pDH63.5, and pDH73.2), three in roots (pDH11.1, pDH19.3, and pDH30.4), and one in shoots and roots (pDH55.7). Herbivore attack decreased the accumulation of nine transcripts: seven in shoots (pDH31.5, pDH39.1, pDH40.1, pDH61.1, pDH64.7, pDH68.1, and pDH69.3), one in roots (pDH9.4), and one in shoots and roots (pDH10.4; see also Fig. 2). The accumulation of transcripts encoded by pDH6.1 and pDH48.4 simultaneously decreased in shoots and increased in roots. In contrast to the majority of transcripts that were detectable in shoot or root tissue of untreated control plants, transcripts encoded by pDH14.2 and pDH25.4 were absent in controls, but strongly induced in shoots after herbivory.
Figure 1.
Accumulation of transcripts in shoots (S) and roots (R) of plants exposed for 24 h to folivory by caterpillars (CAT) and airborne methyl jasmonate (MeJA), respectively, in comparison to untreated control plants (CON). Northern blots were hybridized with the indicated pDH gene probes. The probe pDH64.4 encoding N. attenuata 18S rRNA served as control for equal blotting.
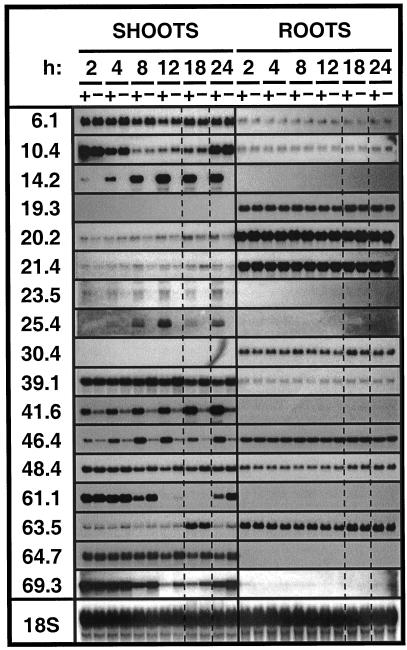
Figure 2.
Time course of transcript accumulation in shoots and roots of plants exposed to folivory (+) for the indicated time intervals. Undamaged control plants (−) were harvested simultaneously. Plants were grown under a 16-h-light regime with the 2-h sample taken after 5 h of light and the 18-h sample taken after 5 h of darkness (lanes bordered by dashed lines). Blots are indexed with the corresponding gene probes including the 18S-rRNA probe as control.
Exposure of plants to airborne MeJA resulted in qualitatively similar patterns of changes in transcript accumulation as did folivory for 15 of differentially expressed transcripts (Fig. 1; pDH9.8, pDH10.4, pDH11.1, pDH14.2, pDH19.3, pDH23.5, pDH30.4, pDH40.1, pDH41.6, pDH46.4, pDH55.2, pDH55.7, pDH61.1, pDH63.5, and pDH69.3; for effects of high concentrations of MeJA on pDH14.2, pDH40.1, pDH41.6, and pDH63.5, see Fig. 3). With the exception of pDH10.4 (decrease in shoots), caterpillar attack resulted in stronger effects compared with the exposure of airborne MeJA. Compared with control plants, weak or no effect of airborne MeJA could be seen for 12 clones (pDH6.1, pDH9.4, pDH10.7, pDH20.2, pDH21.4, pDH25.4, pDH31.5, pDH39.1, pDH48.4, pDH64.7, pDH68.1, and pDH73.2). We did not observe antagonistic effects of herbivory and MeJA exposure.
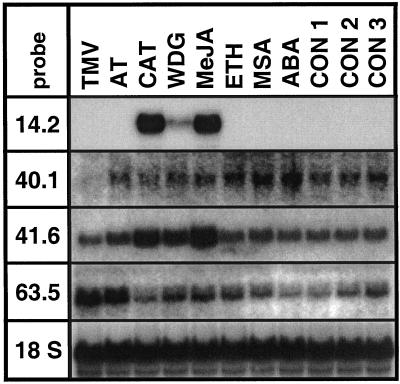
Figure 3.
Accumulation of transcripts encoding TD (pDH14.2), importin-α (pDH40.1), PIOX (pDH41.6), and the putative kinase cofactor pDH63.5 in response to biotic and abiotic stimuli. RNA was extracted from shoots 72 h after inoculation with TMV with carborundum powder, 24 h after inoculation with the disarmed strain LBA4404 of A. tumefaciens (AT), and 12 h after folivory by caterpillars (CAT), repeated wounding in 30-min intervals (WDG), application of MeJA to leaves (MeJA), exposure to ethylene (ETH), application of MSA to leaves (MSA), and of ABA to roots (ABA), respectively. The control lanes correspond to untreated plants (CON 1), plants treated with lanolin paste (CON 2), and plants treated with carborundum powder (CON 3), respectively. Equal blotting was verified by hybridization with the 18S-rRNA probe.
Considering the data obtained from these treatments (Fig. 1) and the putative function of particular genes (Table I), we selected a subset of 17 cDNAs for a more detailed kinetic analysis. For this analysis the accumulation of transcripts in roots and shoots after herbivore attack was determined by northern analysis of untreated control and attacked plants harvested six times throughout a 16-h light/8-h dark cycle (Fig. 2). This sampling design allowed us to simultaneously monitor diurnal changes in transcript accumulation. Diurnal effects on transcript accumulation were pronounced for pDH10.4, pDH21.4, pDH25.4, pDH46.4, pD61.1, pDH63.5, and pDH69.3 and weak for pDH6.1, pDH14.2, pDH23.5, and pDH48.4. Changes in the amounts of transcripts found in roots were weak or absent and did not show pronounced diurnal effects (Fig. 2). With respect to herbivory, transcripts of pDH63.5 displayed an inconsistent expression pattern in independent experiments (Figs. 1–3). The fastest responses to herbivory were found for pDH14.2, pDH41.6, and pDH46.4 with increases in transcripts already visible 2 h after larvae had been placed on the plants.
The effects of abiotic and biotic stimuli on mRNA accumulation were investigated for transcripts encoding TD (pDH14.2), importin-α (pDH40.1), PIOX (pDH41.6), and the putative kinase cofactor pDH63.5 (Fig. 3). Accumulation of TD-specific transcripts increased dramatically upon herbivory and application of MeJA to leaves, whereas the response to wounding was weak and treatments with tobacco mosaic virus (TMV), the disarmed strain of Agrobacterium tumefaciens, ethylene, methyl salicylate (MSA), and abscisic acid (ABA) did not show any effects compared with the corresponding controls. Transcripts of pDH40.1 accumulated slightly in response to ethylene, MSA, and ABA, were slightly reduced upon herbivory, and were absent in plants inoculated with TMV. Larval feeding, wounding, and MeJA all showed strong effects on the accumulation of PIOX transcripts, whereas A. tumefaciens, ethylene, and MSA were much less effective. Amounts of transcripts encoded by pDH63.5 strongly accumulated after inoculation with TMV and A. tumefaciens and weakly in response to wounding, MeJA, ethylene, and MSA. Herbivory did not evoke changes compared with the control, but the wound-response was suppressed during larval feeding (Fig. 3).
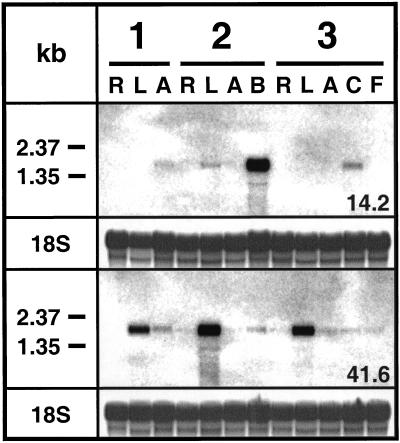
Tissue-specific expression of TD and PIOX was investigated in undamaged plants during vegetative growth, bolting, and flowering (Fig. 4). TD-specific transcripts were readily detected in apical buds of the developing axis, barely detectable in the stems of the vegetative stage, the leaves of bolting plants, and the calyx of flowering plants, and not detectable in all other tissues. PIOX-specific transcripts were abundant in the leaves of vegetative, bolting-, and flowering-stage plants. Small amounts of transcripts were detectable in all other tissues. It is interesting that the highest expression for TD- and PIOX-specific transcripts was found in bolting plants, albeit in different plant organs.
Figure 4.
Tissue-specific accumulation of transcripts encoding TD (probe pDH14.2) and PIOX (probe pDH41.6) in undamaged plants during vegetative growth (1), bolting (2), and flowering (3). Total cellular RNA was extracted from roots (R), leaves (L), axis (A), apical buds (B), calyx (C), and flowers without calyx (F), respectively. Hybridizations with 18S rRNA are provided as controls for equal blotting.
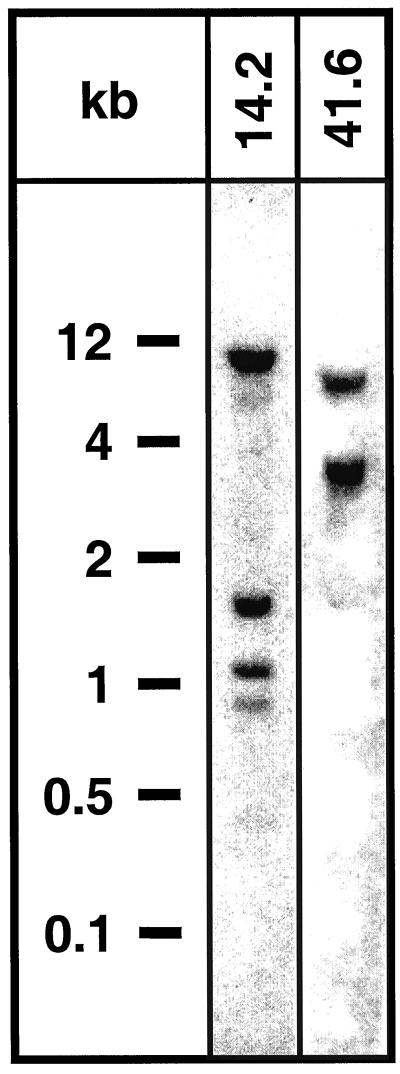
To estimate the copy number of TD and PIOX genes in the genome, Southern blots were prepared from genomic DNA digested with EcoRV. Blots were probed for TD and PIOX using the 3′-specific gene probes pDH14.2 and pDH41.6, respectively, which did not contain internal Eco-RV sites (Fig. 5). Three pronounced bands and one weak band were detected for TD and two bands of different intensity for PIOX.
Figure 5.
Southern blots of genomic DNA of diploid N. attenuata restricted with EcoRV and hybridized with the gene probes pDH14.2 (TD) and pDH41.6 (PIOX). The gene probes did not contain Eco-RV sites.
Full-length sequences of PIOX and TD were isolated from a cDNA library as described in “Materials and Methods.” The clone p51A1 (PIOX) contained a 2,178-bp cDNA insert consisting of a 54-bp 5′-untranslated region (UTR), a 1,932-bp open reading frame (ORF) terminated by a TAA stop codon, and a 167-bp 3′-UTR followed by a 25-bp poly(A) stretch (Fig. 6). The sequence of the cDNA insert of the clone pDH41.6 (which was generated by DDRT-PCR) extends beyond the 3′ end of p51A1, indicating variable processing of 3′ ends. In accordance with the notion that plant 3′-UTRs contain multiple polyadenylation signals similar to the mammalian consensus sequence AATAAA (Joshi, 1987; Wu et al., 1995), we found putative signals at the positions indicated in Figure 6. The nucleotide sequence of p51A1 showed 94% identity to tobacco PIOX (Sanz et al., 1998). The ORF of p51A1 translated into a polypeptide of 643 amino acids with a predicted molecular mass of 73.8 kD and 97% similarity to the deduced amino acid sequence of tobacco PIOX. Database searches revealed additional similarities of 85% to a FEEBLY-like protein from Arabidopsis (X. Lin, S. Kaul, C.D. Town, M. Benito, T.H. Creasy, B. Haas, D. Wu, C.M. Ronning, H. Koo, C.Y. Fujii, T.R. Utterback, M.E. Barnstead, C.L. Bowman, O. White, W.C. Nierman, and C.M. Fraser, unpublished data), of 71% to the tomato FEEBLY protein (van der Biezen et al., 1996), and of 37% to the chicken prostaglandin G/H synthase 2 precursor (Xie et al., 1991). Alignments of the amino acid sequences show high similarity throughout the entire length between the sequences of p51A1, tobacco PIOX, and the Arabidopsis FEEBLY-like protein (Fig. 7).
Figure 6.
Nucleotide and deduced amino acid sequence of clone p51A1 encoding full-length N. attenuata PIOX. The stop codon is marked with a star and putative polyadenylation signals within the 3′-UTR are underlined. Within the 5′-UTR two nonsense codons out-of-frame followed by one in-frame are indicated by dots. The clone pDH41.6, generated by DDRT-PCR, starts at position 1,792 (arrow) and is identical in sequence to p51A1 except for a 71-bp extension beyond the 3′ end of p51A1. Numbers refer to the nucleotide sequence not counting the poly(A) stretches. The GenBank accession number of p51A1 is AF229926.
Figure 7.
Alignment of deduced amino acid sequences of N. attenuata PIOX (att_piox; GenBank accession no. AF229926), tobacco PIOX (tob_piox; GenBank accession no. AJ007630), Arabidopsis FEEBLY-like protein (ara_feebly; GenBank accession no. AAF24612), tomato FEEBLY protein (tom_feebly; GenBank accession no. U35643), and chicken prostaglandin G/H synthase 2 precursor (chick_pgh2; GenBank accession no. P27607). Amino acid identity is indicated by black shading and similarity by gray shading. Uppercase letters in the consensus sequence indicate fully conserved residues, lowercase letters indicate partial conserved residues.
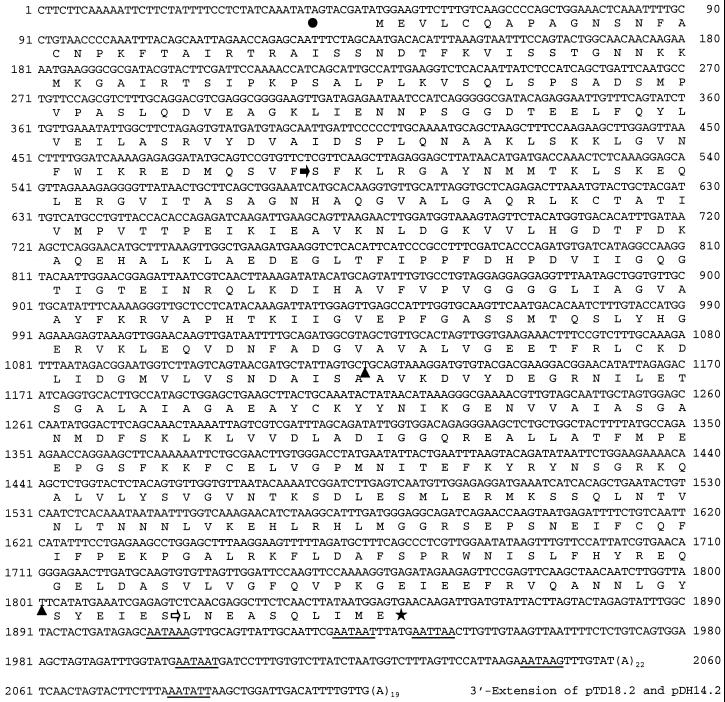
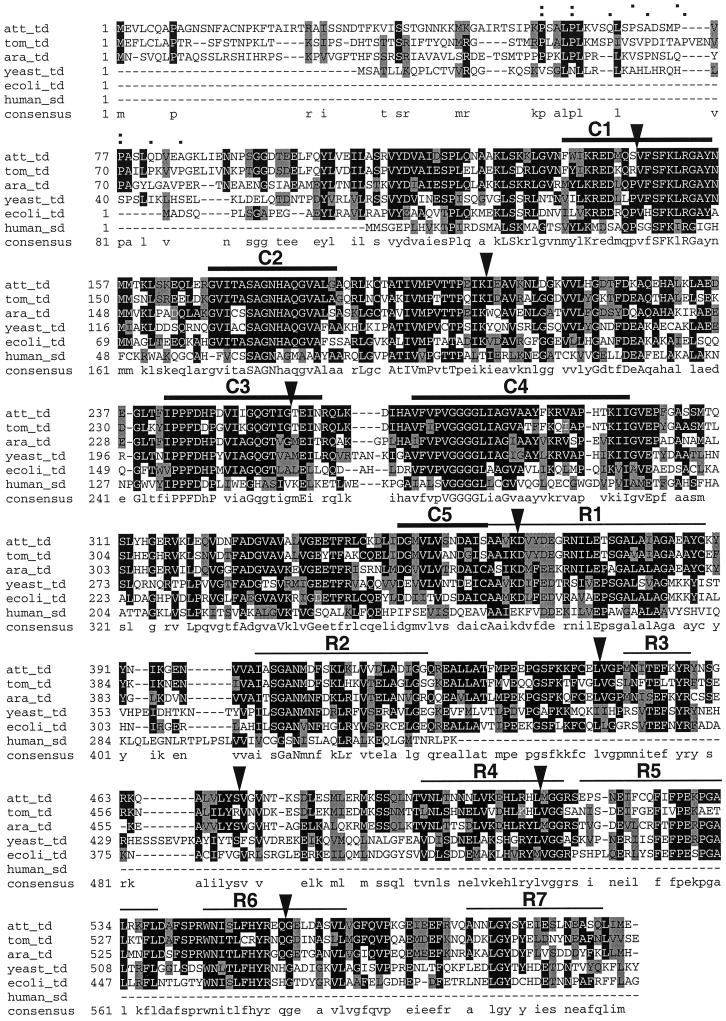
Screening with the pDH14.2 gene probe for TD clones provided a 5′-truncated sequence, designated as pTD18.2, which in turn served to generate a 5′ gene probe as described in “Materials and Methods.” Using the 5′ gene probe we isolated the clone pTD13, which harbored a 2,082-bp cDNA insert encoding full-length TD. The cDNA featured a 5′-UTR of 46 bp, an ORF of 1,806 bp, and a 3′-UTR of 208 bp followed by a poly(A) stretch of 22 bp (Fig. 8). Compared with pTD13, the 3′-UTRs of pTD18.2 and pDH14.2 were extended, indicating variable 3′ processing of the mRNAs as observed for PIOX. The ORF of the pTD13 insert encodes a polypeptide of 601 amino acids with a calculated molecular mass of 65.6 kD. Alignments of the amino acid sequence of N. attenuata with other sequences available in databases revealed similarities of 73% to tomato TD (Samach et al., 1991), 73% to Arabidopsis TD (X. Lin, S. Kaul, C.D. Town, M. Benito, T.H. Creasy, B. Haas, D. Wu, C.M. Ronning, H. Koo, C.Y. Fujii, T.R. Utterback, M.E. Barnstead, C.L. Bowman, O. White, W.C. Nierman, and C.M. Fraser, unpublished data), 59% to yeast TD (Kielland-Brandt et al., 1984), 58% to Escherichia coli TD (Cox et al., 1987), and 46% to human Ser deaminase (Ogawa et al., 1989). Catalytic domains, regulatory domains, and exon-intron boundaries are highly conserved, whereas the putative chloroplast transit peptide shows only moderate similarity between the three plant species (Fig. 9).
Figure 8.
Nucleotide and deduced amino acid sequence of clone pTD13 encoding full-length N. attenuata TD. The stop codon is marked with a star, putative polyadenylation signals within the 3′-UTR are underlined, and an in-frame nonsense codon in the 5′-UTR is indicated by a dot. The partial cDNA clone pTD18.2 starts at position 487 (black arrow) of the nucleotide sequence of pTD13 and extends 46 bp beyond the 3′ end of pTD13. In pTD18.2 we found two conservative base substitutions marked with triangles at position 1,126 (T to A) and position 1,801 (T to C), respectively. The cDNA clone pDH14.2 generated by DDRT-PCR starts at position 1,820 (white arrow) and matches in sequence to pTD18.2. Numbers refer to the nucleotide sequence not counting the poly(A) stretches. The GenBank accession number of pTD13 is AF229927.
Figure 9.
Alignment of deduced amino acid sequences of TDs from N. attenuata (att_td; GenBank accession no. AF229927), tomato (tom_td; GenBank accession no. P25306), Arabidopsis (ara_td; GenBank accession no. AAF04418), Saccharomyces cerevisiae (yeast_td; GenBank accession no. P00927), E. coli (ecoli_td; GenBank accession no. P04968), and the Ser deaminase from human (human_sd; GenBank accession no. P20132). Amino acid identity is indicated by black shading and similarity by gray shading. Uppercase letters in the consensus sequence indicate fully conserved residues, lowercase letters indicate partial conserved residues. The five catalytic domains C1 to C5 and the eight regulatory domains R1 to R7 as defined by Taillon et al. (1988) are displayed as bars. Exon-intron boundaries determined for the tomato gene by Samach et al. (1991) are indicated by triangles. Pro residues in a putative chloroplast transit peptide of N. attenuata and tomato are indicated by dots in the upper and lower row, respectively, above the alignment.
DISCUSSION
We employed DDRT-PCR to initiate the global characterization of molecular responses of N. attenuata to folivory by the specialist herbivore M. sexta. Investigating one-twentieth of the transcriptome, we found herbivore-induced changes in the abundance of 27 transcripts of plant origin, which implies that the insect-responsive transcriptome of N. attenuata contains more than 500 different mRNA species. Among those, about 200 genes would be expected to encode unknown functions since BLAST queries of 40% of the cDNAs did not reveal similarities to known genes (Table I).
Down-Regulation of Photosynthesis-Related Genes
Herbivory strongly diminished the abundance of transcripts of the three photosynthesis-related genes lhb C1 (pDH61.1), chl H (pDH69.3), and rbc S (pDH64.7). Airborne MeJA reduced only transcript amounts of lhb C1 and chl H, whereas rbc S was not affected (Fig. 1; Table I). Consistent with the literature, dramatic diurnal effects were seen in the accumulation of transcripts of lhb C1 and chl H, but not of rbc S (Kloppstech, 1985; Gibson et al., 1996; Papenbrock et al., 1999). The diurnal effects were superimposed on the herbivore-induced responses (Fig. 2). Decreased amounts of rbc S transcripts or Rubisco had been observed after treatments with ABA, MeJA, or sugars, and after wounding, stress, or exposure to phytopathogens, respectively. ABA, MeJA, and sugars decreased the amounts of LHC II subunits or transcripts concomitantly (Reinbothe et al., 1994 and refs. therein; Koch, 1996). The expression of photosynthetic and defense genes is known to be inversely correlated, with sugars being substrates and central regulators of the carbon reallocation between sources and sinks (Somssich and Hahlbrock, 1998; Roitsch, 1999). From this respect, the herbivore-induced suppression of the chl H, lhb C1, and rbc S might benefit the plant by redirecting the carbon flux toward defenses.
Other Down-Regulated Genes
The bell pepper gene Sn-1 is homologous to pDH6.1, encodes a protein with similarity to two major latex proteins from opium poppy, and shares motifs with major allergen proteins and PR proteins. SN-1 is wound inducible and is proposed to be a secretory protein likely to participate in the early disease resistance response (Pozueta-Romero et al., 1995; Osmark et al., 1998). Genes encoding proteins with similarity to major latex proteins have also been found to be induced by ethylene (Aggelis et al., 1997) and to be repressed by plant-parasitic nematodes (Hermsmeier et al., 2000). We found herbivory to diminish transcripts of pDH6.1 in leaves (Figs. 1 and 2), indicating that major latex proteins are involved in plant-pathogen and plant-insect interactions.
A cytoskeletal compound is encoded by pDH9.4 and is most similar to β-tubulins from the zygomycete fungus G. coronatum and Arabidopsis. Transcripts of the Arabidopsis homolog accumulate preferentially in roots, at low levels in flowers, and are barely detectable in leaves (Oppenheimer et al., 1988). We found transcripts encoded by pDH9.4 to be restricted to root tissue and to be diminished after herbivory (Fig. 1). Decreases in β-tubulin mRNA have also been reported after treatment of soybeans with elicitors of defense responses (Gianfagna and Lawton, 1995) and infection of parsley cells with Phytophthora infestans (Gross et al., 1993). In both cases the decrease in β-tubulin transcripts was accompanied with an increase in transcripts encoding components of the defense response.
A homolog of importin α, a subunit of the nuclear pore-targeting complex, is encoded by pDH40.1. Importin α mediates the import of cytosolic proteins into the nucleus (Adam, 1995; Görlich, 1997). Plant importin α binds to virulence factors of A. tumefaciens and several phytopathogenic viruses (Ballas and Citovsky, 1997; Kunik et al., 1998). Transcripts of pDH40.1 were shoot specific and amounts were diminished after herbivory, application of MeJA, and inoculation with TMV. Treatments with ABA and MSA induced a slight increase, whereas the disarmed strain of A. tumefaciens did not affect accumulation of transcripts (Figs. 1 and 3). These data suggest that ABA and MSA are not regulating responses of importin α to biotic stimuli.
Clone pDH63.5 is similar to StubGAL83, a cofactor of the potato kinase complex StubSNF1 (Lakatos et al., 1999). SNF1-type complexes regulate carbohydrate and fatty acid metabolism during nutritional or environmental stresses (Hardie et al., 1998; Roitsch, 1999). The GAL83-like protein AKINβ1 from Arabidopsis showed an expression pattern very similar to pDH63.5, which was most abundant in roots, but declined rapidly in shoots within 30 min of illumination (Fig. 2; Bouly et al., 1999). In contrast, transcripts of StubGAL83 were reported to be most abundant in leaves and barely detectable in roots of potato (Lakatos et al., 1999). The evaluation of expression patterns of pDH63.5 derived from three independent experiments was hampered by the low abundance of transcripts in shoots due to the strong suppression by light (Figs. 1–3). Although effects of herbivory compared with controls are ambiguous, the data clearly show decreased transcript levels after herbivory compared with wounding, application of MeJA, MSA, and ethylene, and in particular to treatments with A. tumefaciens and TMV, which caused strong increases in transcript accumulation (Fig. 3). These findings provide evidence that larval cues suppress wound responses during feeding and suggest that SNF1 kinase complexes might be involved in the allocation of resources during exposure of the plants to pathogens or herbivores.
Up-Regulation of TD: Shifting Carbon to Defense?
Biosynthetic TDs are highly conserved among bacteria, fungi, and plants and show significant similarity to human Ser deaminase, as well (Fig. 9). The conservation of catalytic domains (Taillon et al., 1988), exon-intron boundaries, and putative plastid-targeting sequences (Samach et al., 1991) in combination with the similar developmental expression of TD in potato, tomato, and N. attenuata provide strong evidence that pTD13 indeed encodes biosynthetic TD and not biodegradative TD (Szamosi et al., 1993). In undamaged plants, TD transcripts (pDH14.2) accumulated mainly in the apex of the emerging axis, which consists of vegetative and flower buds (Fig. 4). These findings are consistent with data obtained for tomato where the peak enzyme activity was found in sepals from buds and flowers (Szamosi et al., 1993) and for potato where the transcript accumulation was restricted to bud tissue (Hildmann et al., 1992). Among the genes presented in this work, TD showed the strongest and most specific response to insect attack: transcripts encoding TD strongly accumulated after herbivory and application of MeJA to leaves, weakly in response to low concentrations of airborne MeJA and wounding, and not at all after inoculation with TMV, A. tumefaciens, or exposure to ABA, ethylene, and MSA (pDH 14.2; Figs. 1 and 3; see also Schittko et al., 2001). Local induction of TD transcript accumulation after wounding or application of ABA or JA has been reported for potato and tomato, but data on systemic responses are contradictory (Hildmann et al., 1992; Pena-Cortez et al., 1993; Dammann et al., 1997). Discrepancies in our results of the ABA treatment might be due to differences in the applied concentrations of ABA and in the experimental setup. Two possible roles for TD suggest a central role in defense responses against phytophagous insects: the provision of 2-ketobutyrate to fuel the synthesis of defensive compounds found in glandular trichomes or epicuticular waxes, duvatrienediol diterpenes, and sugar esters (Keen and Wagner, 1985; Severson et al., 1985; Chortyk et al., 1996); and the induction of pathogenesis-related (PR) proteins via α-aminobutyric acid, a derivative of 2-ketobutyrate (Lotan and Fluhr, 1990; Eyal et al., 1992). The strong herbivore-induced expression of TD might indeed reflect the high demand in enzyme for the synthesis of defense compound precursors.
Up-Regulation of Genes Responding to Stress, Wounding, and Pathogens
Transcripts of a BiP accumulate constitutively in roots, but exert a marked responsiveness to herbivory in shoot tissues (pDH46.4; Figs. 1 and 2). BiPs are molecular chaperones that mediate import and maturation of secretory proteins in the endoplasmic reticulum (ER), but also remove malfolded proteins during ER stress (Jelitto-Van Dooren et al., 1999). ER stress is for instance encountered after a burst of PR gene induction, since PR proteins are manufactured and delivered via the secretory pathway. Since BiPs accumulate locally and systemically with identical timing, gaseous signals, in particular nitric oxide, ethylene, and MeJA are considered candidate compounds (Jelitto-Van Dooren et al., 1999). Ethylene and MeJA also play important roles in insect-plant interactions (Fig. 1; Stotz et al., 2000; Kahl et al., 2000) and massive overexpression of BiP mRNA has been reported to follow persistent wounding of soybean leaves by whiteflies (Kalinski et al., 1995). The insect-induced overexpression of BiP could be linked to the overexpression of TD via the TD-mediated induction of PR proteins discussed above.
Clone pDH25.4 shares the highest similarity with a gene from rice, which is similar to the copia-like retrotransposon RIRE 1 from Oryza australiensis (Noma et al., 1997). Several types of plant retrotransposons have been shown to be transcriptionally activated by mechanical stimuli, wounding, MeJA, SA, and pathogens (for review, see Kumar and Bennetzen, 1999). The insect-induced accumulations of leaf-specific transcripts encoding the putative retrotransposon, BiP (pDH46.4) and TD (pDH14.2) are strikingly similar over a day/night cycle (Fig. 2). This might indicate a coordinated regulation e.g. via a common trans-acting factor that binds to identical cis-acting elements of the corresponding promoters.
PIOX catalyzes the first step in α-oxidation of linolenic, linoleic, and oleic acid, respectively, to the corresponding 2(R)-hydroperoxy fatty acids. Suggested functions of PIOX are the synthesis of signaling compounds belonging to the oxylipin pathway, e.g. precursors of JA, or the production of substances toxic to pathogens (Sanz et al., 1998; Hamberg et al., 1999). In undamaged plants, transcripts of PIOX accumulate in photosynthetic tissues (pDH41.6; Fig. 4) and amounts increase substantially after herbivory, wounding, and application of MeJA, whereas airborne MeJA, A. tumefaciens, and MSA were less effective (pDH41.6; Figs. 1 and 3). Sanz et al. (1998) found the expression of tobacco PIOX also to be induced by wounding and JA. Compared with the effect of herbivory in our experiments (Fig. 2), the response reported by Sanz et al. (1998) was equally fast during the incompatible plant-pathogen interaction, but about 14 h delayed during the compatible interaction. It is obvious that PIOX (pDH 41.6) confers less insect specificity than TD (pDH 14.2) and is also not expressed ectopically after herbivory (Figs. 3 and 4). The FEEBLY-like protein of Arabidopsis displays 85% similarity to N. attenuata PIOX, but only 71% similarity to the tomato FEEBLY protein, not considering the C-terminal extension of the tomato protein (Fig. 7). Therefore, the Arabidopsis protein is more likely to exert the function of PIOX than of FEEBLY, with FEEBLY presumably playing a role in nitrogen metabolism according to its mutant phenotype (van der Biezen et al., 1996).
Clone pDH19.3 encodes a homolog of RD22, a seed-specific protein of Arabidopsis that is induced in vegetative-stage plants during drought, salt stress, or application of ABA (Yamaguchi-Shinozaki and Shinozaki, 1993). In contrast to RD22, transcripts of pDH19.3 were only detectable in roots and increased slightly after herbivory and exposure to airborne MeJA (Fig. 1). In spite of the higher BLAST scores for RD22, the amino acid sequence of pDH19.3 shares 65% similarity with the RD22 protein and the auxin down-regulated protein ADR6 from soybean (Datta et al., 1993). Amounts of ADR6 mRNA are strongly diminished in soybean roots parasitized by the soybean cyst nematode (Hermsmeier et al., 1998), indicating that the functions of proteins with similarity to RD22 or ADR6 are not restricted to developmental and environmental stimuli.
Germin-like proteins (GLPs) constitute a superfamily of enzymes that are functionally diverse and involved in many developmental and stress-related processes (Dunwell and Gane, 1998). Among the plant GLPs, oxalate oxidase, an H2O2-producing enzyme, is understood best (Berna and Bernier, 1999). We isolated a homolog (pDH20.2) of a GLP from potato (Campbell et al., 1998), which is constitutively expressed in roots and induced in leaves after herbivory (Figs. 1 and 2). Among the numerous stimuli promoting the accumulation of GLPs, induction by pathogens has been reported in many cases (Vallelian-Bindschedler et al., 1998; Berna and Bernier, 1999 and refs. therein) and our results demonstrate that plants utilize GLPs for responses to herbivory, as well.
Other Up-Regulated Genes
Fd-GOGAT catalyzes the assimilation of nitrogen into Glu that provides, together with Gln, the substrate for the biosynthesis of the main products of nitrate assimilation (Temple et al., 1998). The enzyme is also essential for the refixation of photorespiratory ammonium liberated from Gly (Somerville and Ogren, 1980). The partial clone pDH55.2 shares 86% and 93% amino acid similarity with two forms of the Arabidopsis Fd-GOGAT, designated GLU1 and GLU2, respectively (Lam et al., 1995). Expression of GLU1 is restricted to leaves, whereas transcripts of GLU2 are abundant in leaves and roots. GLU1 is assumed to play a major role in photorespiration and primary nitrogen assimilation, whereas GLU2 drives nitrogen assimilation in roots and maintains a basal assimilation in leaves (Coschigano et al., 1998). Although pDH55.2 is more similar to the root-specific GLU2, we found pDH55.2 primarily expressed in leaves (Fig. 1). Herbivory and airborne MeJA induced accumulation of pDH55.2 transcripts, which implies that up-regulation of Fd-GOGAT might be involved in the herbivore-induced allocation of nitrogen to defense. Baldwin et al. (1994a) and Baldwin (1998) demonstrated that 15NO3 was rapidly assimilated, reduced, and used to synthesize nicotine after MeJA induction and herbivory, a process that has to involve Fd-GOGAT.
The deduced amino acid sequence of clone pDH11.1 shares 58% similarity with the cytochrome c-type FixO protein from B. japonicum, which operates in the bacterial respiratory chain and is essential for root nodulation and symbiotic nitrogen fixation (Preisig et al., 1993). Transcripts of pDH11.1 were absent from shoots, but very abundant in roots and increased slightly after herbivory and exposure to airborne MeJA (Fig. 1). The gene product of pDH11.1 presumably has a vital function since transcripts accumulate constitutively to high amounts, but the current lack of information on eukaryotic homologs of FixO does not allow one to draw conclusions beyond its general role in oxidation. The herbivore-induced increase could be indicative of an increased catabolism.
Concluding Remarks
The estimate that more than 500 genes of N. attenuata respond to herbivory, together with the fact that about one-half of the known genes are involved in plant-pathogen interactions provides evidence that plant-insect interactions reach comparable complexities and specificities. A high complexity is also evident from the data obtained for Arabidopsis (Reymond et al., 2000; Stotz et al., 2000). None of the genes presented in this work share similarity with insect-responsive genes hitherto isolated from Arabidopsis. The finding that five genes (encoding β-tubulin, germin, an LHC II subunit, PIOX, and RBC S) that showed pronounced responses in the Nicotiana-Manduca system did not respond in Arabidopsis exposed to folivory by Pieris rapae (Reymond et al., 2000) demonstrates that species specificity adds to an even higher complexity. The ”ask-the-plant-approach” via DDRT-PCR allowed to isolate genes, which would have been difficult to predict to be elicited or repressed by herbivore attack a priori, but from the vantage of hindsight, their responses are reasonable. The recruitment of pathogen-responsive transcripts and the repression of photosynthetic genes were expected responses (Batz et al., 1998), but the insights into how a plant recruits nitrogen and carbon from primary metabolism for defense responses are unexpected. The patterns of transcript accumulations in herbivore-attacked plants and those treated with JA were largely similar, consistent with a central role played by JA in herbivore resistance in this species (Baldwin, 1998). Fifteen of the 27 genes responded to airborne MeJA, with signaling being particular effective for BiP (pDH46.4) and PIOX (pDH41.6; Fig. 1). N. attenuata co-occurs in its native habitat with sagebrush (Artemesia tridentata), a species that releases MeJA into the atmosphere (Farmer and Ryan, 1990; Karban et al., 2000), hence the responsiveness of BiP and PIOX may contribute to interactions between these two species. The analysis provided evidence for exclusively insect-responsive promotors (in pDH25.4 and pDH14.2). The response of these genes in addition to five additional insect-elicited transcripts are characterized more fully in a second companion paper (Schittko et al., 2001) and the active components responsible for the insect-specific elicitation are described in a third companion paper (Halitschke et al., 2001). Insect-responsive promoters will not only allow researchers to engineer insect-resistance, but also to analyze the contribution of individual transgenes to the Darwinian fitness of natural plant populations exposed to herbivory.
MATERIALS AND METHODS
Rearing of Insects and Cultivation of Plants
Larvae of the tobacco hornworm (Manduca sexta L., Lepidoptera, Sphingidae) were hatched overnight at 28°C from eggs received from Carolina Biological Supply (Burlington, NC). Seeds of the inbred line DI-92 of Nicotiana attenuata Torr. ex Wats. had been collected from T40S R19W section 10 of southwest Utah in 1988 and were germinated in smoke-treated soil (Baldwin et al., 1994b). One-week-old seedlings were then adapted to hydroponic growth conditions by transferring them to communal 25-L hydroponic boxes containing 0.292 g/L of Peter's hydrosol (W.R. Grace, Fogelsville, PA) and 0.192 g/L of Ca(NO3)2. The day/night cycle was set to 16 h of illumination at 1,000 to 1,500 μmol m−2 s−1 photosynthetically active radiation from high-pressure Na lights at 32°C and 8 h of darkness at 28°C with 65% constant relative humidity. After 5 d, plants were placed individually into culture vessels each containing 1 L of no-nitrogen hydroponic solution (Baldwin and Schmelz, 1994). Nitrogen was supplied by adding 2 mL of 1 m KNO3 after transferring to the individual vessels and another 1 mL after 7 to 10 d. Treatments were initiated 18 h after the second addition of nitrogen with plants in the vegetative stage of growth with approximately 12-cm rosette diameters (21 d post-germination, dpg). Untreated plants harvested during vegetative growth (21 dpg), bolting (25 dpg), and flowering (50 dpg) were used in the analyses of tissue-specific gene expression.
Treatments
Twenty freshly hatched larvae were placed on the rosette leaves of individual plants and allowed to feed for 12 or 24 h. Undamaged plants of the same age were harvested as controls for the caterpillar treatment. MeJA (Aldrich, Deisenhofen, Germany), which was close to its thermodynamic equilibrium (90.1% 1R, 2R MeJA and 8.3% 1R, 2S MeJA) for the two naturally occurring epimers, was applied in lanolin paste (Aldrich) to leaves following the procedure of Baldwin et al. (1996). We applied a total of 250 μg of MeJA in four 10-μL droplets of lanolin to four fully expanded leaves of each plant, whereas controls were treated with lanolin paste only. Exposure of plants to airborne MeJA was carried out by placing seven 10-μL droplets of lanolin paste with a total of 437.5 μg of MeJA on the lid of individual hydroponic containers. Ethylene was liberated from 100 μg of ethephon supplied in four 10-μL droplets of lanolin paste on leaves. This quantity of ethephon releases an amount of ethylene comparable with the amount released by a rosette-stage plant under attack by M. sexta larvae (Kahl et al., 2000). MSA was supplied in the same way, but at a concentration of 132 μg per plant, whereas ABA was added to the hydroponic solution to final concentration of 1 μm. Plants were wounded with a fabric pattern wheel in 30-min intervals during a 12-h time course, as described in Baldwin and Schmelz (1994). Inoculation with the disarmed strain LBA4404 of Agrobacterium tumefaciens (Life Technologies, Karlsruhe, Germany) was carried out by dispersing a total of 400 μL of cell suspension (OD600 = 1) to eight rows of wounds generated with the fabric pattern wheel on four fully expanded leaves. For virus treatments, four leaves per plant were wounded using mesh 200 carborundum powder and inoculated with each 1.25 μg of the strain U1 of the TMV suspended in 50 μL of distilled water (Preston et al., 1999).
Extraction of Nucleic Acids, Hybridizations, and Labeling Procedures
Total cellular RNA from four to six plants per treatment was isolated according to Pawlowski et al. (1994). For gel electrophoresis, aliquots of 20 μg of RNA were mixed with 5 volumes of 1× MOPS [3-(N-morpholino)-propanesulfonic acid] buffer, pH 7, containing 48% (v/v) formamide, 5.2% (v/v) formaldehyde, 20 μg ml−1 ethidium bromide, 0.045% (w/v) Orange-G dye (Sigma, Deisenhofen, Germany), and 10% (v/v) glycerol, denatured for 10 min at 68°C, and chilled on ice. Samples were separated alongside RNA size standards (Life Technologies) on gels prepared from 1.2% (w/v) agarose in 1× MOPS buffer, pH 7, containing 2% (v/v) formaldehyde. High-molecular weight genomic DNA was extracted from leaves following the protocol of Richards (1997) and 10 μg of DNA digested with EcoRV was subjected to Southern blotting. High-stringency northern and Southern hybridizations were performed as described in Hermsmeier et al. (1998) with the following modifications. Nucleic acids were blotted onto Nytran SuperCharge membrane (Schleicher & Schuell, Dassel, Germany) using a vacuum blotter (model 785, Bio-Rad, Munich). Size standards were visualized by incubating the membranes 15 min in 5% (v/v) acetic acid, followed by 10 min in 0.5 m sodium phosphate buffer, pH 5.2, containing 0.04% (w/v) methylene blue (Sigma) and 5 to 10 min of repeated rinses in water. The hybridization buffer contained 5× sodium chloride/sodium phosphate/EDTA, 70 mm sodium phosphate buffer, pH 7, 50% (v/v) deionized formamide, 1% (w/v) SDS, 5× Denhardt's solution, and 0.1 mg/mL denatured fish DNA (Acros, Geel, Belgium). Templates for radiolabeling were cDNA inserts PCR amplified from the pCR2.1-TOPO vector (Invitrogen, Groningen, The Netherlands) employing the TOPO-F primer (CTCATCGATGGATATCTGCAGAATTCGCCC) in combination with the TOPO-R primer (CTCATCGATAGTGTGCTGGAATTCGCCC) synthesized by MWG Biotech (Ebersberg, Germany). PCR products were gel purified using the NucleoSpin Extract kit (Machery-Nagel, Düren, Germany). Radiolabeling was carried out in 20-μL PCR reactions containing 1.5 mm MgCl2, 10 μm each dATP, dGTP, and dTTP, 0.5 μm TOPO-F primer, 0.5 μm TOPO-R primer, 0.1 units/μL Platinum Taq DNA polymerase (Life Technologies), 1 μm α-32P-dCTP (NEN, Cologne), and 10 ng of the gel-purified template cDNA in a final 1× concentrated reaction buffer supplied with the polymerase. Unincorporated nucleotides were removed with ProbeQuant G50 spin columns (Amersham-Pharmacia, Braunschweig, Germany). Double-stranded probes were denatured for 30 min at room temperature in final 0.1 m NaOH prior to hybridization. Autoradiographs were recorded on film (Biomax MS, Kodak, Rochester, NY) using a Transscreen for signal amplification (Amersham-Pharmacia).
DDRT-PCR
Total cellular RNA isolated from 20 plants exposed to herbivory for 24 h and from 20 undamaged control plants was used as starting material for DDRT-PCR. RNAs from shoots (whole green parts of the plants) and roots were extracted separately and blended in a 1:1 ratio prior to first-strand cDNA synthesis. The DDRT-PCR procedure was performed according to Hermsmeier et al. (1998) with the following modifications. A single arbitrary primer R1 (TACAACGAGG) was used in combination with the 10 anchor primers A1 (T12AA), A2 (T12AC), A3 (T12AG), A4 (T12CA), A5 (T12CC), A6 (T12CG), A7 (T12GA), A8 (T12GC), A9 (T12GG), and A10 (T12GT), respectively. Primers were synthesized by MWG Biotech. Platinum Taq DNA polymerase (Life Technologies) was employed for intrinsic hot start in DDRT-PCR. Dried display gels were exposed to Kodak Biomax MR film (Amersham-Pharmacia). Re-amplified cDNAs were purified using the NuleoSpin Extract kit (Machery-Nagel) and cloned into the pCR2.1-TOPO vector (Invitrogen) according to the manufacturer's recommendations. Of each transformation, four to eight colonies were screened for plasmid inserts via PCR employing the TOPO-F and TOPO-R primers described above. Depending on the insert sizes, each three to five colonies were selected for plasmid preparation using the Nucleospin Multi 8 kit (Machery-Nagel). Inserts of plasmids were sequenced on an ABI Prism 377 XL DNA sequencer using the BigDye terminator kit (PE-Applied Biosystems, Weiterstadt, Germany) and sequence data were analyzed using the Lasergene software package (DNASTAR, Madison, WI). Sequence similarity searches in the GenBank and The Arabidopsis Information Resource databases were performed using the BLAST algorithms (Altschul et al., 1997). Sequence alignments were generated with the ClustalX program (Thompson et al., 1997).
Screening of cDNA Library
Poly(A)+ RNA was isolated from shoots of soil-grown N. attenuata plants that had been exposed to folivory by M. sexta larvae for 24 h using the Dynabead mRNA purification kit (Dynal, Hamburg). The cDNA library was prepared using the Uni-ZAP XR library kit (Stratagene, Amsterdam). Library construction, screening, and in vivo excision of pBluescript II SK followed the instruction manual supplied with the kit. A partial sequence, designated pTD18.2, was obtained after screening with the 3′ gene probe pDH14.2. The sequence of pTD18.2 was used to derive the sequences of the primers TD-FWD (GCTTATAACATGATGACCAAACTC) and TD-REV (GCACCAAATGGCTCAACTCC) that were employed to generate a 455-bp PCR product from the 5′ end of pTD18.2. The PCR product served as a 5′ gene probe to screen for the corresponding full-length clone pTD13. The gene probe pDH41.6 was used to isolate the full-length clone p51A1. Double-strand sequencing of the excised plasmid clones was conducted by GATC (Konstanz, Germany).
ACKNOWLEDGMENTS
We wish to thank Natalia Sandoval for preparing the N. attenuata cDNA library and Domenica Schnabelrauch for sequencing. We thank editor Carlos Ballaré and two anonymous reviewers for improving the manuscript and apologize to those authors who have not been cited in this work because of space limitations.
Footnotes
This work was supported by the Max Planck Gesellschaft.
LITERATURE CITED
- Adam SA. The importance of importin. Trends Cell Biol. 1995;5:189–191. doi: 10.1016/s0962-8924(00)88991-6. [DOI] [PubMed] [Google Scholar]
- Aggelis A, John I, Karvouni Z, Grierson D. Characterization of two cDNA clones from mRNAs expressed during ripening of melon (Cucumis meloL.) fruits. Plant Mol Biol. 1997;33:313–322. doi: 10.1023/a:1005701730598. [DOI] [PubMed] [Google Scholar]
- Alborn HT, Turlings TCJ, Jones TH, Stenhagen G, Loughrin JH, Tumlinson JH. An elicitor of plant volatiles from beet armyworm oral secretions. Science. 1997;276:945–949. [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar SP. Signalling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- Baldwin IT. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc Natl Acad Sci USA. 1998;95:8113–8118. doi: 10.1073/pnas.95.14.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT. Inducible nicotine production in native Nicotianaas an example of adaptive phenotypic plasticity. J Chem Ecol. 1999;25:3–30. [Google Scholar]
- Baldwin IT, Karb MJ, Ohnmeiss TE. Allocation of 15N from nitrate to nicotine: production and turnover of a damage-induced mobile defense. Ecology. 1994a;75:1703–1713. [Google Scholar]
- Baldwin IT, Ohnmeiss TE. Alkaloid responses to damage in Nicotiananative to North America. J Chem Ecol. 1993;19:1143–1153. doi: 10.1007/BF00987376. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Preston CA. The eco-physiological complexity of plant responses to insect herbivores. Planta. 1999;208:137–145. [Google Scholar]
- Baldwin IT, Schmelz EA. Constraints on an induced defense: the role of the leaf area. Oecologia. 1994;97:424–430. doi: 10.1007/BF00317335. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Schmelz EA, Zhang Z-P. Effects of octadecanoid metabolites and inhibitors on induced nicotine accumulation in Nicotiana sylvestris. J Chem Ecol. 1996;22:61–73. doi: 10.1007/BF02040200. [DOI] [PubMed] [Google Scholar]
- Baldwin IT, Staszak-Kozinski L, Davidson R. Up in smoke: I. Smoke-derived germination cues for the post-fire annual Nicotiana attenuataTorr. Ex. Watson. J Chem Ecol. 1994b;20:2345–2371. doi: 10.1007/BF02033207. [DOI] [PubMed] [Google Scholar]
- Ballas N, Citovsky V. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of AgrobacteriumVirD2 protein. Proc Natl Acad Sci USA. 1997;94:10723–10728. doi: 10.1073/pnas.94.20.10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batz O, Logemann E, Reinold S, Hahlbrock K. Extensive reprogramming of primary and secondary metabolism by fungal elicitor or infection in parsley cells. Biol Chem. 1998;379:1127–1135. doi: 10.1515/bchm.1998.379.8-9.1127. [DOI] [PubMed] [Google Scholar]
- Bauer D, Müller H, Reich J, Riedel H, Ahrenkiel V, Warthoe P, Strauss M. Identification of differentially expressed mRNA species by an improved display technique (DDRT-PCR) Nucleic Acids Res. 1993;21:4272–4280. doi: 10.1093/nar/21.18.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbaum MR. Allelochemicals in insect-microbe-plant interactions: agents provocateurs in the coevolutionary arms race. In: Barbosa P, editor. Novel Aspects of Insect-Plant Interactions. New York: John Wiley; 1988. pp. 97–123. [Google Scholar]
- Berna A, Bernier F. Regulation by biotic and abiotic stress of a wheat germin gene encoding oxalate oxidase, a H2O2-producing enzyme. Plant Mol Biol. 1999;39:539–549. doi: 10.1023/a:1006123432157. [DOI] [PubMed] [Google Scholar]
- Bouly J-P, Gissot L, Lessard P, Kreis M, Thomas M. Arabidopsis thalianaproteins related to the yeast SIP and SNF4 interact with AKINα1, and SNF1-like protein kinase. Plant J. 1999;18:541–550. doi: 10.1046/j.1365-313x.1999.00476.x. [DOI] [PubMed] [Google Scholar]
- Campbell MA, Herrmann LM, Colvin J. Characterization of a gene encoding a putative germin-like protein from potato (accession no. AF067731) Plant Physiol. 1998;118:711. [Google Scholar]
- Carter W. Insects in Relation to Plant Disease. New York: Wiley Interscience; 1973. [Google Scholar]
- Chortyk OT, Pomonis JG, Johnson AW. Syntheses and characterizations of insecticidal sucrose esters. J Agric Food Chem. 1996;44:1551–1557. [Google Scholar]
- Coschigano KT, Melo-Oliveira R, Lim J, Coruzzi GM. Arabidopsisgls mutants and distinct Fd-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell. 1998;10:741–752. doi: 10.1105/tpc.10.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JL, Cox BJ, FidanzaV, Calhoun DH. The complete nucleotide sequence of the ilvGMEDA cluster of Escherichia coliK-12. Gene. 1987;56:185–198. doi: 10.1016/0378-1119(87)90136-3. [DOI] [PubMed] [Google Scholar]
- Dammann C, Rojo E, Sanchez-Serrano JJ. Abscisic acid and jasmonic acid activate wound-inducible genes in potato through separate, organ-specific signal transduction pathways. Plant J. 1997;11:773–782. doi: 10.1046/j.1365-313x.1997.11040773.x. [DOI] [PubMed] [Google Scholar]
- Datta N, LaFayette PR, Kroner PA, Nagao RT, Key JL. Isolation and characterization of three families of auxin down-regulated cDNA clones. Plant Mol Biol. 1993;21:859–869. doi: 10.1007/BF00027117. [DOI] [PubMed] [Google Scholar]
- Denecke J, Goldman MHS, Demolder J, Seurinck J, Botterman The tobacco luminal binding protein is encoded by a multigene family. Plant Cell. 1991;3:1025–1035. doi: 10.1105/tpc.3.9.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunwell JM, Gane PJ. Microbial relatives of seed storage proteins: conservation of motifs in a functionally diverse superfamily of enzymes. J Mol Evol. 1998;46:147–154. doi: 10.1007/pl00006289. [DOI] [PubMed] [Google Scholar]
- Eyal J, Sagee O, Fluhr R. Dark-induced accumulation of a basic pathogenesis-related (PR-1) transcript and a light requirement for its induction by ethylene. Plant Mol Biol. 1992;19:589–599. doi: 10.1007/BF00026785. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Ryan CA. Interplant communication: airborne methyl jasmonate induces synthesis of proteinase inhibitors in plant leaves. Proc Natl Acad Sci USA. 1990;87:7713–7716. doi: 10.1073/pnas.87.19.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianfagna TJ, Lawton MA. Specific activation of soybean defense genes by the phosphoprotein phosphatase inhibitor okadaic acid. Plant Sci. 1995;109:165–170. [Google Scholar]
- Gibson LCD, Marrison JL, Leech RM, Jensen PE, Bassham DC, Gibson M, Hunter CN. A putative Mg chelatase subunit from Arabidopsis cv C24: sequence and transcript analysis of the gene, import of the protein into chloroplasts, and in-situ localization of the transcript and protein. Plant Physiol. 1996;111:61–71. doi: 10.1104/pp.111.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Grimm B. Novel insights in the control of tretrapyrrole metabolism of higher plants. Curr Opin Plant Biol. 1998;1:245–250. doi: 10.1016/s1369-5266(98)80112-x. [DOI] [PubMed] [Google Scholar]
- Gross P, Julius C, Schmelzer E, Hahlbrock K. Translocation of cytoplasm and nucleus to fungal penetration sites is associated with depolymerization of microtubules and defense gene activation in infected, cultured parsley cells. EMBO J. 1993;12:1735–1744. doi: 10.1002/j.1460-2075.1993.tb05821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke R, Kessler A, Kahl J, Lorenz A, Baldwin IT. Eco-physiological comparison of direct and indirect defenses in Nicotiana attenuata. Oecologia. 2000;124:408–417. doi: 10.1007/s004420000389. [DOI] [PubMed] [Google Scholar]
- Halitschke R, Schittko U, Pohnert G, Boland W, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol. 2001;125:711–717. doi: 10.1104/pp.125.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castarena C. α-Oxidation of fatty acids in higher plants: identification of a pathogen-inducible oxygenase (PIOX) as an α-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem. 1999;274:24503–24513. doi: 10.1074/jbc.274.35.24503. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt R, Schultz JC. Multiple defenses and signals in plant defense against pathogens and herbivores. In: Romeo JT, Saunders J, Barbosa P, editors. Phytochemical Defenses and Signals in Plant Defense against Pathogens and Herbivores. New York: Plenum Press; 1996. pp. 121–154. [Google Scholar]
- Hammond-Kosack KE, Jones JDG. Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Hart JK, Byzova M, Rodermel SR, Baum TJ. Changes in mRNA abundance within Heterodera schachtii-infected roots of Arabidopsis thaliana. Mol Plant-Microbe Interact. 2000;13:309–315. doi: 10.1094/MPMI.2000.13.3.309. [DOI] [PubMed] [Google Scholar]
- Hermsmeier D, Mazarei M, Baum TJ. Differential display analysis of the early compatible interaction between soybean and the soybean cyst nematode. Mol Plant-Microbe Interact. 1998;11:1258–1263. [Google Scholar]
- Hildmann T, Ebneth M, Pena-Cortez H, Sanchez-Serrano JJ, Willmitzer L, Prat S. General roles of abscisic and jasmonic acids in gene activation as a result of mechanical wounding. Plant Cell. 1992;4:1157–1170. doi: 10.1105/tpc.4.9.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelitto-Van Dooren EPWM, Vidal S, Denecke J. Anticipating endoplasmic reticulum stress: a novel early response before pathogenesis-related gene induction. Plant Cell. 1999;11:1935–1943. doi: 10.1105/tpc.11.10.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi CP. Putative polyadenylation signals in nuclear genes of higher plants: a compilation and analysis. Nucleic Acids Res. 1987;15:9627–9640. doi: 10.1093/nar/15.23.9627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl J, Siemens DH, Aerts RJ, Gabler R, Kuhnemann F, Preston CA, Baldwin IT. Herbivore-induced ethylene suppresses a direct defense but not a putative indirect defense against an adapted herbivore. Planta. 2000;210:336–342. doi: 10.1007/PL00008142. [DOI] [PubMed] [Google Scholar]
- Kalinski A, Rowley DL, Loer DS, Foley C, Buta G, Herman EM. Binding-protein expression is subject to temporal, developmental and stress-induced regulation in terminally differentiated soybean organs. Planta. 1995;195:611–621. doi: 10.1007/BF00195722. [DOI] [PubMed] [Google Scholar]
- Karban R, Baldwin IT, Baxter KJ, Laue G, Felton G. Communication between plants: induced resistance in wild tobacco plants following clipping of neighboring sagebrush. Oecologia. 2000;125:66–71. doi: 10.1007/PL00008892. [DOI] [PubMed] [Google Scholar]
- Keen CK, Wagner GJ. Direct demonstration of duvatrienediol biosynthesis in glandular heads of tobacco trichomes. Plant Physiol. 1985;79:1026–1032. doi: 10.1104/pp.79.4.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielland-Brandt MC, Holmberg S, Petersen JGL, Nilsson-Tillgren T. Nucleotide sequence of the gene for threonine deaminase (ILV1) of Saccharomyces cerevisiae. Carlsberg Res Commun. 1984;49:567–575. [Google Scholar]
- Kloppstech K. Diurnal and circadian rhythmicity in the expression of light-induced plant nuclear messenger RNAs. Planta. 1985;165:502–506. doi: 10.1007/BF00398095. [DOI] [PubMed] [Google Scholar]
- Koch KE. Carbohydrate-modulated gene expression in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:509–540. doi: 10.1146/annurev.arplant.47.1.509. [DOI] [PubMed] [Google Scholar]
- Korth KL, Dixon RA. Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiol. 1997;115:1299–1305. doi: 10.1104/pp.115.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse E, Mock HP, Grimm B. Isolation and characterization of tobacco (Nicotiana tabacum) cDNA clones encoding proteins involved in magnesium chelation into protoporphyrin IX. Plant Mol Biol. 1997;35:1053–1056. doi: 10.1023/a:1005913315905. [DOI] [PubMed] [Google Scholar]
- Kumar A, Bennetzen JL. Plant retrotransposons. Annu Rev Genet. 1999;33:479–532. doi: 10.1146/annurev.genet.33.1.479. [DOI] [PubMed] [Google Scholar]
- Kunik T, Palanichelvam K, Czosnek H, Citovsky V, Gafni Y. Nuclear import of the capsid protein of tomato yellow leaf curl virus (TYLCV) in plant and insect cells. Plant J. 1998;13:393–399. doi: 10.1046/j.1365-313x.1998.00037.x. [DOI] [PubMed] [Google Scholar]
- Lakatos L, Klein M, Höfgen R, Banfalvi Z. Potato StubSNF1 interacts with StubGAL83: a plant protein kinase complex with yeast and mammalian counterparts. Plant J. 1999;17:569–574. doi: 10.1046/j.1365-313x.1999.00406.x. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Coschigano K, Schultz C, Melo-Oliveira R, Tjaden G, Oliveira I, Ngai N, Hsieh M-H, Coruzzi G. Use of Arabidopsismutants and genes to study amide amino acid biosynthesis. Plant Cell. 1995;7:887–898. doi: 10.1105/tpc.7.7.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Kaul S, Rounsley SD, Shea TP, Benito M-I, Town CD, Fujii CY, Mason TM, Bowman CL, Barnsted ME, Feldblyum TV, Buell CR, Ketchum KA, Lee JJ, Ronning CM, Koo H, Moffat KS, Cronin LA, Shen M, van Aken SE, Umayam L, Tallon LJ, Gill JE, Adams MD, Carrera AJ, Creasy TH, Goodman HM, Somerville CR, Copenhaver GP, Preuss D, Nierman WC, White O, Eisen JA, Salzberg SL, Fraser CM, Venter JC. Sequence and analysis of chromosome II of Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Lotan T, Fluhr R. Xylanase, a novel elicitor of pathogenesis-related proteins in tobacco, uses a non-ethylene pathway for induction. Plant Physiol. 1990;93:811–817. doi: 10.1104/pp.93.2.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloud ES, Baldwin IT. Herbivory and caterpillar regurgitants amplify the wound-induced increases in jasmonic acid but not nicotine in Nicotiana sylvestris. Planta. 1997;203:430–435. [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J. Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA. 1997;94:5473–5477. doi: 10.1073/pnas.94.10.5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Nakajima R, Ohtsubo H, Ohtsubo E. RIRE1, a retrotransposon from wild rice Oryza australiensis. Genes Genet Syst. 1997;72:131–140. doi: 10.1266/ggs.72.131. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Gomi T, Konishi K, Date T, Nakashima H, Nose K, Matsuda Y, Peraino C, Pitot HC, Fujioka M. Human liver serine dehydratase: cDNA cloning and sequence homology with hydroxyamino acid dehydratases from other sources. J Biol Chem. 1989;264:15818–15823. [PubMed] [Google Scholar]
- Oppenheimer DG, Haas N, Silflow CD, Snustad DP. The beta-tubulin gene family of Arabidopsis thaliana: preferential accumulation of the beta 1 transcript in roots. Gene. 1988;63:87–102. doi: 10.1016/0378-1119(88)90548-3. [DOI] [PubMed] [Google Scholar]
- Osmark P, Boyle B, Brisson N. Sequential and structural homology between intracellular pathogenesis-related proteins and a group of latex proteins. Plant Mol Biol. 1998;38:1243–1246. doi: 10.1023/a:1006060224012. [DOI] [PubMed] [Google Scholar]
- Papenbrock J, Mock HP, Kruse E, Grimm B. Expression studies in tetrapyrrole biosynthesis: inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta. 1999;208:264–273. [Google Scholar]
- Pawlowski K, Kunze R, deVries S, Bisseling T. Isolation of total, poly(A) and polysomal RNA from plant tissue. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. , Section D5, pp 1–4. [Google Scholar]
- Peña-Cortéz H, Albrecht T, Prat S, Weiler EW, Willmitzer L. Aspirin prevents wound-induced gene expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta. 1993;191:123–128. [Google Scholar]
- Pinck M, Guilley E, Durr A, Hoff M, Pinck L, Fleck J. Complete sequence of one of the mRNAs coding for the small subunit of ribulose bisphosphate carboxylase of Nicotiana sylvestris. Biochimie. 1984;66:539–545. doi: 10.1016/0300-9084(84)90148-2. [DOI] [PubMed] [Google Scholar]
- Pohnert G, Jung V, Haukioja E, Lempa K, Boland W. New fatty acid amides from regurgitant of lepidopteran (Noctuidae, Geometridae) caterpillars. Tetrahedron. 1999;55:11275–11280. [Google Scholar]
- Pozueta-Romero J, Klein M, Houlne G, Schantz M-L, Meyer B, Schantz R. Characterization of a family of genes encoding a fruit-specific wound-stimulated protein of bell pepper (Capsicum annuum): identification of a new family of transposable elements. Plant Mol Biol. 1995;28:1011–1025. doi: 10.1007/BF00032663. [DOI] [PubMed] [Google Scholar]
- Preston CA, Lewandowski C, Enyedi AJ, Baldwin IT. Tobacco mosaic virus inoculation inhibits wound-induced jasmonic acid-mediated responses within but not between plants. Planta. 1999;209:87–95. doi: 10.1007/s004250050609. [DOI] [PubMed] [Google Scholar]
- Preisig O, Anthamatten D, Hennecke H. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicumare essential for a nitrogen-fixing endosymbiosis. Proc Natl Acad Sci USA. 1993;90:3309–3313. doi: 10.1073/pnas.90.8.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Mollenhauer B, Reinbothe C. JIPs and RIPs: the regulation of plant gene expression by jasmonates in response to environmental cues and pathogens. Plant Cell. 1994;6:1197–1209. doi: 10.1105/tpc.6.9.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE. Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol. 1998;1:404–411. doi: 10.1016/s1369-5266(98)80264-1. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–719. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards EJ. Preparation of plant DNA using CTAB. In: Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Short Protocols in Molecular Biology. New York: Wiley; 1997. pp. 2.10–2.11. [Google Scholar]
- Roitsch T. Source-sink regulation by sugar and stress. Curr Opin Plant Biol. 1999;2:198–206. doi: 10.1016/S1369-5266(99)80036-3. [DOI] [PubMed] [Google Scholar]
- Ryan CA. The systemin signaling pathway: differential activation of plant defensive genes. Biochim Biophys Acta. 2000;1477:112–121. doi: 10.1016/s0167-4838(99)00269-1. [DOI] [PubMed] [Google Scholar]
- Samach A, Hareven D, Gutfinger T, Ken-Dror S, Lifschitz E. Biosynthetic threonine deaminase gene of tomato: isolation, structure, and up-regulation in floral organs. Proc Natl Acad Sci USA. 1991;88:2678–2682. doi: 10.1073/pnas.88.7.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz A, Moreno JI, Castarena C. PIOX, a new pathogen-induced oxygenase with homology to animal cyclooxgenase. Plant Cell. 1998;10:1523–1537. doi: 10.1105/tpc.10.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Hermsmeier D, Baldwin IT. Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata: II. Accumulation of plant mRNAs in response to insect-derived cues. Plant Physiol. 2001;125:701–710. doi: 10.1104/pp.125.2.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E, Stasys R, Aebersold R, McGrath JM, Green BR, Pichersky E. Sequence of a tomato gene encoding a third type of LHCII chlorophyll a/b-binding polypeptide. Plant Mol Biol. 1991;17:923–925. doi: 10.1007/BF00037074. [DOI] [PubMed] [Google Scholar]
- Severson RF, Arrendale RF, Chortyk OT, Green CR, Thome FA, Stewart JL, Johnson AW. Isolation and characterization of the sucrose esters of the cuticular waxes of green tobacco leaf. J Agric Food Chem. 1985;33:870–875. [Google Scholar]
- Somerville CR, Ogren WL. Inhibition of photosynthesis in Arabidopsismutants lacking leaf glutamate synthase activity. Nature. 1980;286:257–259. [Google Scholar]
- Somssich IE, Hahlbrock K. Pathogen defense in plants: a paradigm of biological complexity. Trends Plant Sci. 1998;3:86–90. [Google Scholar]
- Stotz HU, Kroymann J, Mitchell-Olds T. Plant-insect interactions. Curr Opin Plant Biol. 1999;2:268–272. doi: 10.1016/S1369-5266(99)80048-X. [DOI] [PubMed] [Google Scholar]
- Stotz HU, Pittendrigh BR, Kroymann J, Weniger K, Fritsche J, Bauke A, Mitchell-Olds T. Induced plant defense responses against chewing insects: ethylene signaling reduces resistance of Arabidopsis against Spodoptera littoralis But Not Plutella xylostella. Plant Physiol. 2000;124:1007–1018. doi: 10.1104/pp.124.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamosi I, Shaner DL, Singh BK. Identification and characterization of a biodegradative form of threonine dehydratase in senescing tomato (Lycopersicon esculentum) leaf. Plant Physiol. 1993;101:999–1004. doi: 10.1104/pp.101.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon BE, Little R, Lawther RP. Analysis of the functional domains of biosynthetic threonine deaminase by comparison of the amino acid sequences of three wild-type alleles to the amino acid sequence of biodegradative threonine deaminase. Gene. 1988;63:245–252. doi: 10.1016/0378-1119(88)90528-8. [DOI] [PubMed] [Google Scholar]
- Temple SJ, Vance CP, Gantt JS. Glutamate synthase and nitrogen assimilation. Trends Plant Sci. 1998;3:51–56. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallelian-Bindschedler L, Mösinger E, Metraux J-P, Schweizer P. Structure, expression and localization of a germin-like protein in barley (Hordeum vulgareL.) that is insolubilized in stressed leaves. Plant Mol Biol. 1998;37:297–308. doi: 10.1023/a:1005982715972. [DOI] [PubMed] [Google Scholar]
- van Dam NM, Baldwin IT. Costs of jasmonate-induced responses in plants competing for limited resources. Ecol Lett. 1998;1:30–33. [Google Scholar]
- van Dam NM, Hadwich K, Baldwin IT. Induced responses in Nicotiana attenuata affect behavior and growth of the specialist herbivore Manduca sexta. Oecologia. 2000;122:371–379. doi: 10.1007/s004420050043. [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Brandwagt BF, van Leeuwen W, Njikamp HJJ, Hille J. Identification and isolation of the FEEBLYgene from tomato by transposon tagging. Mol Gen Genet. 1996;251:267–280. doi: 10.1007/BF02172517. [DOI] [PubMed] [Google Scholar]
- Wu L, Ueda T, Messing J. The formation of mRNA 3′-ends in plants. Plant J. 1995;8:323–329. doi: 10.1046/j.1365-313x.1995.08030323.x. [DOI] [PubMed] [Google Scholar]
- Xie WL, Chipman JG, Robertson DL, Erikson RL, Simmons DL. Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing. Proc Natl Acad Sci USA. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. The plant hormone abscisic acid mediates the drought-induced expression but not the seed-specific expression of rd22, a gene responsive to dehydration stress in Arabidopsis thaliana. Mol Gen Genet. 1993;238:17–25. doi: 10.1007/BF00279525. [DOI] [PubMed] [Google Scholar]