Abstract
Background:
Current standard-of-care technologies such as imaging and cyst fluid analysis are unable to consistently distinguish intraductal papillary mucinous neoplasms of the pancreas (IPMN) at high-risk of pancreatic cancer from low-risk IPMN. The objective was to create a single-platform assay to identify IPMN that are at high-risk for malignant progression.
Study Design:
Building on the Verona International Consensus Conference BD-IPMN biomarker study, specific protein, cytokine, mucin, DNA, and miRNA cyst fluid targets were identified for creation of a q-PCR based assay as we have previously published. This included mRNA markers: ERBB2, GNAS, IL1b, KRAS, MUCs1, 2, 4, 5AC, 7, PGE2R, PTGER2, PTGES2, PTGES1, TP63; miRNA targets: miRs 101, 106b, 10a, 142, 155, 17, 18a, 21, 217, 24, 30a, 342, 532, 92a, and 99b; and GNAS and KRAS mutational analysis. A multi-institutional international collaborative contributed IPMN cyst fluid samples to validate this platform. Cyst fluid gene expression levels were normalized, z-transformed, and utilized in classification and regression analysis by a support vector machine (SVM) training algorithm.
Results:
From fifty-nine IPMN patient cyst fluids, principal component analysis confirmed no institutional bias/clustering. Lasso-penalized logistic regression with binary classification and 5-fold cross validation utilized AUC as evaluation criteria to create the optimal signature to discriminate IPMN into low-risk (low/moderate dysplasia) or high-risk (high-grade dysplasia/invasive cancer). The most predictive signature was achieved with IL1β, MUC4, and PTGES2 to accurately discriminate high from low-risk cysts with up to an AUC of 0.86, p=0.002.
Conclusions:
We have identified a single-platform PCR-based assay of cyst fluid to accurately predict IPMN with high-malignant potential for further studies.
Keywords: Intraductal papillary mucinous neoplasm, IPMN, pancreas, cyst, biosignature, biomarker, signature, gene expression, miRNA, molecular marker, pancreatic cancer, dysplasia
Precis
Current standard-of-care clinical guidelines are unable to accurately identify patients with intraductal papillary mucinous neoplasms of the pancreas at high risk of pancreatic cancer. Therefore, a single platform biosignature to accurately predict intraductal papillary mucinous neoplasms with high malignant potential was created.
Introduction:
Intraductal papillary mucinous neoplasms (IPMN) are pancreatic cysts with adenomatous proliferation of ductal epithelium causing mucin production and dilatation of the pancreatic ductal system. The incidence of asymptomatic pancreatic cysts, including IPMN, is 3–15% of the population and is increasing secondary to ubiquitous cross-sectional imaging and advancements in imaging quality.1, 2 Up to 24% of patients at autopsy will have a pancreatic cyst, of which 20% may contain atypia or high-grade dysplasia.3 As a result, IPMN has become the most common cystic precursor lesion of pancreatic adenocarcinoma (PDAC), representing 10–25% of resected pancreatic neoplasms.4, 5
The main challenge in treating IPMN is in accurately predicting malignant potential and thus determining the risk-benefit of a surgical resection. Studies on the clinical signs and imaging characteristics of the disease have evolved to form the basis of multiple clinical consensus guidelines.6, 7 Though the sensitivity of the current treatment guidelines is satisfactory, the specificity remains poor. As a result, the vast majority of resected IPMNs will be low-risk and we have reported to contain only low or moderate grade dysplasia on final pathology.5, 8 Surgical intervention often involves major pancreatic resection which carries significant risk of mortality and morbidity.9 Thus, particularly for small BD-IPMN, enhanced tests with improved negative predictive value are desired to avoid the potential complications of surgical resection for the cysts that will prove to have low malignant potential and may otherwise have been treated with surveillance alone.
Current clinical guidelines lack accuracy in determining the level of cyst dysplasia6, 10–13, and additional molecular diagnostic data to distinguish high from low-risk cysts are desperately needed. We have previously shown that EUS-FNA cytology has limited utility in surgical decision making for IPMN, and have identified multiple prognostic molecular biomarkers within the cyst fluid.14–18 Cyst fluid is easily and safely accessible preoperatively and contains shed genetic material from the cyst wall that is representative of the entire cyst.14 Though previous biomarker studies have focused on proteins, RNA, DNA, miRNA, cytokines, glycoproteins, or mucins in the cyst fluid, we endeavored to combine the most predicitive markers from each of these classes using the coding genes in one comprehensive assay. Thus, the objective of this study was to create a gene expression based assay encorporating multiple IPMN biomarkers into a signature that could accurately stratify IPMN as low or high-risk for malignant progression.
Materials and Methods:
Biological Samples
The International IPMN Cyst Fluid Collaborative was created of groups from high-volume pancreatic surgery centers with an expertise in IPMN across Europe and the United States that was born out of the Verona Consensus Conference.11, 14 IPMN cyst fluid samples were obtained from prospectively maintained institutional databases/repositories after approval by the Institutional Review Board of the University of Illinois at Chicago. Only samples with a confirmed diagnosis of IPMN on final pathology and with the specific grade of dysplasia determined by an expert pancreatic pathologist were included in the study. Analysis evaluated samples by “low-risk” (low and moderate-grade dysplasia) or “high-risk” (high-grade dysplasia and invasive cancer) pathology for the purpose of risk-stratification, as has been used in multiple other biomarker studies in this field due to it’s clinical applicabiity.15, 16, 19
Quantitative analysis of messenger RNAs and microRNAs
Total RNA was extracted from 100–400 μl of IPMN fluid using Quick-RNA™ MicroPrep R1050/R1051 (Irvine, CA), implemented on a Maxwell16 instrument. DNAse treatment was performed according to the manufacturer’s instructions. Subsequently, total RNA was split into two paths for messenger RNA (mRNA) and micro RNA (miRNA) analysis using quantitative PCR.
For analysis of mRNA, total RNA was reverse transcribed using random primers and the High Capacity cDNA reverse transcription kit (#4368814; Thermo Fisher Scientific), according to the manufacturer’s instructions. cDNA was prepared for quantitative PCR (qPCR) using a pre-amplification step, with the Taqman PreAmp master mix kit (#4384267; Thermo Fisher Scientific). Taqman gene expression assays were pooled to serve as primers for the pre-amplification step according to the manufacturer’s instructions. Assays included IL1b(Hs01555410_m1), muc-1(Hs00159357_m1), muc-2(Hs00894025_m1), muc-4(Hs00366414_m1), muc-5ac(Hs01365616_m1), muc-7(Hs00379529_m1), PTGER2(Hs04183523_m1), PTGS1(Hs00377726_m1), PGE2-R(Hs00168755_m1), KRAS(Hs00364282_m1), GNAS(Hs00255603_m1), GAPDH(Hs99999905_m1), RPLP0(Hs99999902_m1), TP63(Hs00978341_m1), ERBB2(Hs01001580_m1), PTGES2(Hs00228159_m1). qPCR reactions were performed using Taqman Fast Advanced master mix (#4444556; Thermo Fisher Scientific) in 384-well plates using a ViiA7 real-time PCR instrument (Life Technologies). All reactions were performed in triplicate and in volumes of 10 μl. Real-time data were processed using the comparative C(t) method.20 The chosen endogenous control gene was RPLP0, based on performance across the entire dataset. Reverse transcription of miRNA was performed using the Taqman microRNA reverse transcription kit (#4366596), with Taqman miRNA assays in place of random primers. The assays used for this study included miR17–3p, miR142–3p, miR532–3p, miR342–3p, miR30a-3p, miR21, miR155, mir101, mir10a, miR106b, miR18a, miR217, miR24, miR92a, miR99b, and RNU6B. Real-time data were processed using the comparative C(t) method, using the RPLP0 gene as an endogenous control.20
PCR amplification and Sequencing of GNAS and KRAS mutation sites
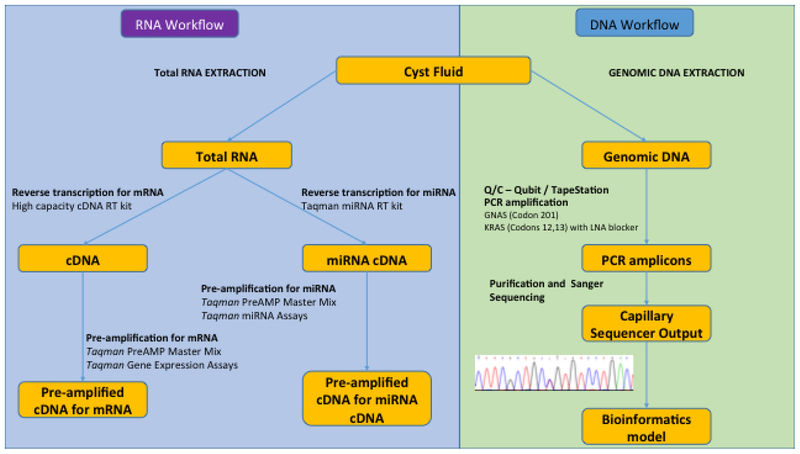
Genomic DNA was extracted from IPMN fluid using the Maxwell16 Tissue DNA kit (AS1030; Promega, Madison, WI). Mutation analysis of codons 12 and 13 in KRAS and codon 201 in GNAS were performed by PCR followed by Sanger sequencing. Each 50 μl PCR reaction contained 1– PCR buffer with 1.5 mM MgCl2, 0.5 μl HotStarTaq DNA polymerase (203203, Qiagen, Germantown, MD), 0.2 mM dNTP mix (D7295, Sigma-Aldrich Corp., St Louis, MO), 20 pmols of forward and reverse primers and 5 μl DNA template. The KRAS PCR reaction in addition contained 25 pmols of a LNA oligo (5’ GC+T+G+G+T+G+G+C+GTA/3’invdT 3’) to suppress wild type amplification (Exiqon, Woburn, MA). Amplification products were purified and bi-directionally sequenced on an ABI3130XL genetic analyzer using the PCR primers and the BigDye 3.1 terminator cycle sequencing kit. Sequence chromatograms were visualized manually to determine if a mutation was present. The analytical sensitivity is 1% mutant sequence for KRAS codons 12 and 13 and 15% mutant sequence for GNAS codon 201. Appropriate positive and contamination controls were included. Mutation nomenclature was according to standard guidelines (http://varnomen.hgvs.org/recommendations/DNA/variant/substitution/). Sample workflow is outlined in Figure 1.
Figure 1.
Workflow of cyst fluid preparation for input into the bioinformatics model to predict level of cyst dysplasia.
Statistical Analysis
RQ values were z-transformed, log2 transformed, and scaled (X-mean/standard deviation). Pearson correlation coefficients were utilized to remove highly correlated variables with a cutoff of 0.7. Principal coordinate analysis was then performed. Models were run adding sequencing data from KRAS and GNAS mutational analysis and evaluated as +kras mutation, +gnas mutation, +gnas/+kras mutation, or 0, 1, or 2 mutations. Mutational analysis as an independent variable was appended to the data matrix with 22 markers for learning and utilized in classification and regression analysis by a support vector machine (SVM) training algorithm. The R-package Glmnet21, a package that fits a generalized linear model via penalized maximum likelihood, was used together with logistic regression. Batch effect correction was performed. Highly-corrected markers were removed.
Lasso (Least absolute shrinkage and selection operator) -penalized logistic regression with binary classification and 5-fold cross validation utilized AUC as evaluation criteria to create the optimal signature. A machine learning algorithm identified markers significantly related to the level of dysplasia/risk of pancreatic malignancy. In N patient cyst fluids, each of which consists of p predictive genes and level of dysplasia as single outcome; yi is the classification of dysplasia and xi = (x1, x2, … , xp)T the gene expression (covariate vector) for the ith case. Where the aim is to identify the least number, but optimal subset, of markers which minimize the classification error between high and low-risk IPMN21, 22 the objective of lasso was to solve:
Results:
Selection of targets and cyst fluid
Specific protein, cytokine, mucin, DNA, and miRNA cyst fluid targets were identified from primary research and an extensive literature search of proposed biomarkers in IPMN as previously published.14–16, 18 This included 14 mRNA markers, 15 miRNA targets, as well as GNAS codon 201 and KRAS codons 12 and 13 point mutational analysis. A total of 134 cyst fluid samples were evaluated for inclusion. Sufficient fluid volume of samples with IPMN grade of dysplasia (low (n=18), moderate (16), high-grade (12), invasive (13)) was confirmed for 59 cyst fluid samples. 95% of samples contained sufficient genomic material for further analysis.
Principal component analysis, batch effect correction, removal of confounders
Principal component analysis demonstrated minimal institutional bias/clustering which was batch effect corrected. As highly corrected markers will cause difficulties in machine learning algorithms to identify individual features for the signature, within each group of highly correlated markers (Pearson correlation > 0.7), one representative marker was kept for further analysis using R package caret (https://github.com/topepo/caret/). Thus, confounding genes were removed from the analysis (Figure 2).
Figure 2.
Removal of confounders. A Pearson correlation matrix was constructed between each pair of gene markers. GNAS, miR106B, miR155, miR24, miR92A, and miR532 were removed from the analysis due to high correlation of gene expression that confounded machine learning algorithms in identifying predictive markers for the biosignature.
Specific mutational analysis
Based on previous data,14, 23–25 GNAS codon 201 and KRAS codons 12 and 13 were sequenced for mutational analysis. Of 49 samples with sufficient DNA harvested from the cyst fluid to reliably sequence, 30 (61%) contained a point mutation in GNAS (13/49, 43%) or KRAS (26/49, 53%). For GNAS, seven samples (7/13 (54%) had p.R201H mutations and six (46%) had p.R201C mutations; while for KRAS, seven (27%) had a p.G12R mutation, 14 (54%) had a p.G12V mutation, eight (31%) had a p.G12D mutation, 1 (4%) had a G12F mutation, 1 had a p.G12A mutation, and 1 had a p.G13D mutation. Three samples (3/49, 6%) each had two KRAS codon 12 point mutations. Nine samples (9/49, 18%) contained both GNAS codon 201 and KRAS codon 12 mutations, of which 1 also contained the KRAS codon 13 mutation.
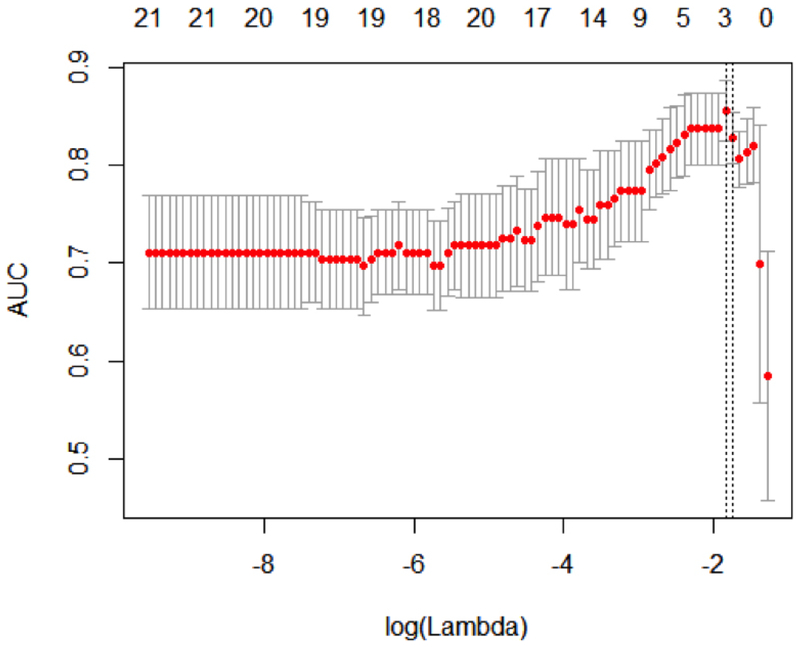
Lasso Regression results
In a binomial logistic regression model with area under the curve (AUC) as the objective function, the maximum AUC was achieved with miR21, miR342, IL1β, KRAS, MUC4, and PTGES2 resulting in an AUC of 0.82 (p=0.003) to differentiate low from high-risk cysts. Subset analysis including iterations involving GNAS and KRAS point mutation analysis were performed to determine the most accurate predictive biosignature. When a mutation in either GNAS or KRAS was considered, the most predictive signature was achieved with IL1β, MUC4, and PTGES2 to construct the equation of:
(Figure 3). Thus, utilizing PCR data, the level of expression of IL1β, MUC4, and PTGES2 in IPMN cyst fluid can be entered into this equation and enabled accurate discrimination of IPMN with low or moderate grade dysplasia (low-risk) from high grade dysplasia or invasive cancer (high-risk).
Figure 3.
Intraductal papillary mucinous neoplasm (IPMN) cyst fluid biosignature can differentiate high from low-risk cysts. Lasso penalized logistic regression with cross validation identified a 3-gene cyst fluid signature with optimal accuracy to predict the risk of pancreatic malignancy in IPMN. In this model, low-risk (low and moderate grade dysplasia) vs high-risk (high-grade dysplasia and invasive cancer) cysts were predicted with an accuracy, as measured by AUC, of 86%; y=0.36+ (−0.06IL1β) + (−0.17MUC4) + (−0.50PTGES2); AUC=0.86, p-value=0.002.
Discussion:
Evidence supports a progression model for IPMN from low grade dysplasia to adenocarcinoma, however, the time frame for this transformation is unknown.26, 27 Currently many cysts at low-risk of malignant transformation are being removed at the expense of mental, physical, and financial cost for patients and society, with the added risk of up to 2% mortality and approximately 40% morbidity post-operatively. Balancing the risks and benefits of resection is the crux of the challenge in surgical decision making for this disease, where the ramifications of missing an occult pancreatic adenocarcinoma, or delay in resection that allows progression, may result in significant cancer-related mortality. In the United States, the vast majority of patients currently undergoing surgical resection for IPMN will have low-risk cysts determined on final pathology despite multiple U.S., European, and international guidelines intended to direct patient selection towards high-risk lesions.5 It has been demonstrated that up to 65% of lesions predicted by the guidelines to be high-risk for high-grade dysplasia or invasive cancer are found to be low-risk on final pathology28, while other small BD-IPMN predicted to have a low-risk of malignancy with the same guidelines will demonstrate high-risk pathology up to 25% of the time.29
The two most commonly used guidelines for clinical decision making in the United States are the revised Sendai (Fukuoka) and American Gastroenterological Association (AGA) guidelines. The Fukuoka guidelines have been found to have a high false positive rate with 21% specificity for malignancy. The same study found the AGA guidelines to have a lower false positive rate with 44% specificity, but with a higher false-negative rate and 12% more of malignancies overlooked.30 Similar analysis supported that the Fukuoka guidelines had a 65–72% false negative rate to identify high-risk cysts while the AGA guidelines misidentified 45% of high-risk IPMN.31, 32 Thus, the field is in need of novel and reliable biomarkers that will be able to differentiate between cysts with minimal risk of malignant transformation from those with high-risk pathology or occult malignancy.33 For this reason, a biosignature that utilizes the inherent molecular makeup of the cyst, as opposed to only size or radiographic findings alone, has great practical and clinical value.34 Preoperative next-generation sequencing of cyst fluid has shown the ability to differentiate mucinous from non-mucinous cysts through mutational analysis of KRAS and GNAS, and in the same study, a combination of mutations or deletions in TP53/PIK3CA/PTEN served as a marker of advanced neoplasia.35 Additional studies evaluating cyst fluid for subtle mutations, loss of heterozygosity, and aneuploidy in eleven genes aided in identification of pancreatic cyst type and histologies for which surgery may be recommended.24 Thus DNA-based testing of genetic material shed into the cyst fluid is a reliable tool through which to study the biology of IPMN disease.
In response to this need, an IPMN cyst fluid gene biosignature was shown to have the ability to discriminate high from low-risk IPMN with up to 86% accuracy. This is compared the 50%, 76%, and 60% accuracies of the Fukuoka, AGA, and ACR criteria, respectively, in the multiinstitutional 5-year study reported by Xu et al.36 The current study utilizes a unique gene panel heretofore not evaluated as a biosignature and that requires only a single PCR platform to quantify.
A combination of IL1β, MUC4, and PTGES2 were consistent across the predictive models. IL1β is secreted into the extracellular space where it can be measured in pancreatic cyst fluid.37 We previously determined that IL1β was nearly undetectable in the cyst fluid of low grade IPMN and serous cystadenomas and that its presence in dysplastic cysts reflected an inflammatory microenvironment.16 with a likelihood ratio of 17– to distinguish low from high-risk cysts. Further, IL1β is a known mediator of pancreatic cancer cell invasion.37–42 MUC4 is implicated in IPMN development, and increased expression may transform borderline cysts to a malignant phenotype.43 Our previous data identified high cyst fluid expression with high-risk IPMN.15, 44 Prostaglandin E synthase 2 (PTGES2) is an enzyme that is encoded by the PTGES2 gene. It catalyzes the conversion of prostaglandin H2 to prostaglandin E2 (PGE2), which, in excess, is known to contribute to inflammatory diseases and cancer. Elevated PGE2 has been implicated in distinguishing IPMN from other mucinous pancreatic cysts and trends to increase with higher levels of IPMN cyst dysplasia and pancreatic cancer.45, 46
There are limitations to the study. Bias was minimized as much as possible by including an international multi-institutional cohort, running samples in large batches, preselecting candidate biomarkers,14, 18 and through robust statistical methods with 5-fold cross-validation. Nonetheless, the samples were collected prospectively, but compiled and evaluated retrospectively. To become a routine study in this disease it is vital that validation with additional sample sets be performed. Furthermore, as the value of this test is in accurate prediction of the level of IPMN dysplasia preoperatively, additional validation utilizing prospective collection of EUS-FNA cyst fluid compared to final surgical pathology of resected specimens will be necessary in order to be practice changing. Little is known about the genetic features of IPMNs undergoing surveillance, thus, there may be a bias in selection as the lesions selected for operative intervention likely contained high-risk or worrisome features. Thus, future analysis will include IPMN phenotype, including duct type and size, patient demographics, and features that led to surgical intervention; information not included in the current dataset. However, in other excellent analyses of IPMN cyst fluid where clinical nomograms were included in a predictive model, the accuracy was not significantly higher than what was achieved in this analysis.19 Regardless, future validation sets will include prospective collection of detailed cyst phenotype variables and patient demographics in the equation, which when combined with the correct biosignature may enhance the selection of high-risk lesions over clinical nomograms alone, and will also allow for determination of additive predictive value over Fukuoka guideline characteristics alone.19, 24, 47 Certainly, a strength of the current study was inclusion of the gene expression, mutational analysis, and epigenetics of targets selected for the biosignature. Interestingly, when KRAS and GNAS mutational analysis were added to the model, they were not selected as contributing features to the predictive value, possibly because of the high prevalence of these mutations in IPMN overall.48, 49 Though previous studies have focused on signatures made from groups of proteins, RNA, DNA, miRNA, cytokines, glycoproteins, or mucins individually, we endeavored to combine the best predictive markers from each category using only the cyst fluid into one rapid comprehensive assay. Proteins predictive of phenotype that had been identified in our previous analyses14, 18 were represented in this analysis by expression of their coding gene. A strength of the analysis was building the model around samples with pathologically confirmed IPMN, which allowed the signature to focus specifically on low-risk compared to high-risk IPMN, though future validation sets may also evaluate if the signature can also discriminate between other non-IPMN histologies or levels of dysplasia. Further, the number of genes evaluated was limited and highly selected based on our comprehensive review of the literature in order to decrease model overfitting, however, there may be other genes that, if included, may have contributed to the model.
Using a single, easily accessible platform, such as PCR, is extremely cost-effective, practical, and proven to be reproducible. This technology is in place in most every institution and accessible for outpatient use throughout the world without requiring complex or expensive equipment and reagents, sequencing technology, or tests of heterozygosity. Signatures using this technology have precedent in other cancer histologies for widespread use and application in patient treatment decision making.50–54 DNA is a very stable molecule for transport and easy to collect since it is shed into the IPMN cyst fluid. Cyst fluid is accessible by EUS-FNA and often collected as part of a standard IPMN clinical workup. The clinical utility of creating such an assay would ultimately be to evaluate small IPMN that do not meet current clinical criteria for resection in order to provide additional quantitative data that can be used potentially for early detection and to further inform the patient and treating physician of the risk of malignant transformation.
In conclusion, utilizing one of the largest multi-national IPMN cyst fluid banks, a biosignature has been identified that predicts IPMN with high-malignant potential utilizing a PCR-based assay.
Support:
Dr Maker is supported by a grant from NIH/NCI (K08-CA190855), Drs Scalpa and Lawlor are supported by a grant from the Associazione Italiana Ricerca Cancro (#12182), and an FP7 European Community Grant Cam-Pac (#602783).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure Information: Nothing to disclose.
Abstract presented at the Americas Hepato-Pancreato-Biliary Association Annual Meeting, Miami, FL, April 2017.
References
- 1.Laffan TA, Horton KM, Klein AP, et al. Prevalence of unsuspected pancreatic cysts on MDCT. AJR Am J Roentgenol 2008; 191(3):802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scheiman JM, Hwang JH, Moayyedi P. American gastroenterological association technical review on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 2015; 148(4):824–48 e22. [DOI] [PubMed] [Google Scholar]
- 3.Kimura W, Nagai H, Kuroda A, et al. Analysis of small cystic lesions of the pancreas. International Journal of Pancreatology 1995; 18(3):197–206. [DOI] [PubMed] [Google Scholar]
- 4.Andrejevic-Blant S, Kosmahl M, Sipos B, et al. Pancreatic intraductal papillarymucinous neoplasms: a new and evolving entity. Virchows Arch 2007; 451(5):863–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khoury RE, Kabir C, Maker VK, et al. What is the Incidence of Malignancy in Resected Intraductal Papillary Mucinous Neoplasms? An Analysis of Over 100 US Institutions in a Single Year. Ann Surg Oncol 2018; 25(6):1746–1751. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka M, Fernandez-del Castillo C, Adsay V, et al. International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas. Pancreatology 2012; 12(3):183–97. [DOI] [PubMed] [Google Scholar]
- 7.Vege SS, Ziring B, Jain R, et al. American gastroenterological association institute guideline on the diagnosis and management of asymptomatic neoplastic pancreatic cysts. Gastroenterology 2015; 148(4):819–22; quize12–3. [DOI] [PubMed] [Google Scholar]
- 8.Vilmann P, Saftoiu A. Endoscopic ultrasound-guided fine needle aspiration biopsy: equipment and technique. J Gastroenterol Hepatol 2006; 21(11):1646–55. [DOI] [PubMed] [Google Scholar]
- 9.Wang H, Chen T, Wang H, et al. A systematic review of the Physiological and Operative Severity Score for the enUmeration of Mortality and morbidity and its Portsmouth modification as predictors of post-operative morbidity and mortality in patients undergoing pancreatic surgery. Am J Surg 2013; 205(4):466–72. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M, Fernandez-del Castillo C, Adsay V, et al. International consensus guidelines 2012 for the management of IPMN and MCN of the pancreas. Pancreatology 2012; 12(3):183–97. [DOI] [PubMed] [Google Scholar]
- 11.Adsay V, Mino-Kenudson M, Furukawa T, et al. Pathologic Evaluation and Reporting of Intraductal Papillary Mucinous Neoplasms of the Pancreas and Other Tumoral Intraepithelial Neoplasms of Pancreatobiliary Tract: Recommendations of Verona Consensus Meeting. Ann Surg 2016; 263(1):162–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basturk O, Hong SM, Wood LD, et al. A Revised Classification System and Recommendations From the Baltimore Consensus Meeting for Neoplastic Precursor Lesions in the Pancreas. Am J Surg Pathol 2015; 39(12):1730–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Haddad MA, Kowalski T, Siddiqui A, et al. Integrated molecular pathology accurately determines the malignant potential of pancreatic cysts. Endoscopy 2015; 47(2):136–42. [DOI] [PubMed] [Google Scholar]
- 14.Maker AV, Carrara S, Jamieson NB, et al. Cyst fluid biomarkers for intraductal papillary mucinous neoplasms of the pancreas: a critical review from the international expert meeting on pancreatic branch-duct-intraductal papillary mucinous neoplasms. J Am Coll Surg 2015; 220(2):243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maker AV, Katabi N, Gonen M, et al. Pancreatic cyst fluid and serum mucin levels predict dysplasia in intraductal papillary mucinous neoplasms of the pancreas. Ann Surg Oncol 2011; 18(1):199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maker AV, Katabi N, Qin LX, et al. Cyst fluid interleukin-1beta (IL1beta) levels predict the risk of carcinoma in intraductal papillary mucinous neoplasms of the pancreas. Clin Cancer Res 2011; 17(6):1502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maker AV, Lee LS, Raut CP, et al. Cytology from pancreatic cysts has marginal utility in surgical decision-making. Ann Surg Oncol 2008; 15(11):3187–92. [DOI] [PubMed] [Google Scholar]
- 18.Tulla KA, Maker AV. Can we better predict the biologic behavior of incidental IPMN? A comprehensive analysis of molecular diagnostics and biomarkers in intraductal papillary mucinous neoplasms of the pancreas. Langenbecks Arch Surg 2018; 403(2):151–194. [DOI] [PubMed] [Google Scholar]
- 19.Al Efishat MA, Attiyeh MF, Eaton AA, et al. Multi-Institutional Validation Study of Pancreatic Cyst Fluid Protein Analysis for Prediction of High-Risk Intraductal Papillary Mucinous Neoplasms of the Pancreas. Ann Surg 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3(6):1101–8. [DOI] [PubMed] [Google Scholar]
- 21.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw 2010; 33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 22.Lockhart R, Taylor J, Tibshirani RJ, et al. A Significance Test for the Lasso. Ann Stat 2014; 42(2):413–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanda M, Knight S, Topazian MD, et al. Mutant GNAS detected in duodenal collections of secretin-stimulated pancreatic juice indicates the presence or emergence of pancreatic cysts. Gut 2013; 62(7):1024–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Springer S, Wang Y, Dal Molin M, et al. A combination of molecular markers and clinical features improve the classification of pancreatic cysts. Gastroenterology 2015; 149(6):1501–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J, Matthaei H, Maitra A, et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci Transl Med 2011; 3(92):92ra66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez-del Castillo C, Adsay NV. Intraductal papillary mucinous neoplasms of the pancreas. Gastroenterology 2010; 139(3):708–13, 713 e1–2. [DOI] [PubMed] [Google Scholar]
- 27.Garcea G, Dennison AR. Branch-type intraductal papillary mucinous neoplasms: an update. Pancreatology 2011; 11(3):336–42. [DOI] [PubMed] [Google Scholar]
- 28.Correa-Gallego C, Brennan MF, Fong Y, et al. Liberal resection for (presumed) Sendai negative branch-duct intraductal papillary mucinous neoplasms--also not harmless. Ann Surg 2014; 259(3):e45. [DOI] [PubMed] [Google Scholar]
- 29.Fritz S, Klauss M, Bergmann F, et al. Small (Sendai negative) branch-duct IPMNs: not harmless. Ann Surg 2012; 256(2):313–20. [DOI] [PubMed] [Google Scholar]
- 30.Lekkerkerker SJ, Besselink MG, Busch OR, et al. Comparing 3 guidelines on the management of surgically removed pancreatic cysts with regard to pathological outcome. Gastrointest Endosc 2017; 85(5):1025–1031. [DOI] [PubMed] [Google Scholar]
- 31.Ma GK, Goldberg DS, Thiruvengadam N, et al. Comparing American Gastroenterological Association Pancreatic Cyst Management Guidelines with Fukuoka Consensus Guidelines as Predictors of Advanced Neoplasia in Patients with Suspected Pancreatic Cystic Neoplasms. J Am Coll Surg 2016; 223(5):729–737 e1. [DOI] [PubMed] [Google Scholar]
- 32.Singhi AD, Zeh HJ, Brand RE, et al. American Gastroenterological Association guidelines are inaccurate in detecting pancreatic cysts with advanced neoplasia: a clinicopathologic study of 225 patients with supporting molecular data. Gastrointest Endosc 2016; 83(6):1107–1117 e2. [DOI] [PubMed] [Google Scholar]
- 33.Fritz S, Werner J, Buchler MW. Reply to letter: “Liberal resection for (presumed) Sendai negative branch-duct IPMN--also not harmless”. Ann Surg 2014; 259(3):e46. [DOI] [PubMed] [Google Scholar]
- 34.Maker AV. ASO Author Reflections: Improving Identification of Intraductal Papillary Mucinous Neoplasm Patients at Risk-Current Status and the Role of IPMN Molecular Biomarkers. Ann Surg Oncol 2018; 25(Suppl 3):818–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singhi AD, McGrath K, Brand RE, et al. Preoperative next-generation sequencing of pancreatic cyst fluid is highly accurate in cyst classification and detection of advanced neoplasia. Gut 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu MM, Yin S, Siddiqui AA, et al. Comparison of the diagnostic accuracy of three current guidelines for the evaluation of asymptomatic pancreatic cystic neoplasms. Medicine (Baltimore) 2017; 96(35):e7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinarello CA, Wolff SM. The role of interleukin-1 in disease. N Engl J Med 1993; 328(2):106–13. [DOI] [PubMed] [Google Scholar]
- 38.El-Omar EM, Carrington M, Chow WH, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature 2000; 404(6776):398–402. [DOI] [PubMed] [Google Scholar]
- 39.Goode EL, Maurer MJ, Sellers TA, et al. Inherited determinants of ovarian cancer survival. Clin Cancer Res 2010; 16(3):995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greco E, Basso D, Fogar P, et al. Pancreatic cancer cells invasiveness is mainly affected by interleukin-1beta not by transforming growth factor-beta1. Int J Biol Markers 2005; 20(4):235–41. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Zhai X, Jin G, et al. Functional variants in the promoter of interleukin-1beta are associated with an increased risk of breast cancer: a case-control analysis in a Chinese population. Int J Cancer 2006; 118(10):2554–8. [DOI] [PubMed] [Google Scholar]
- 42.Qian N, Chen X, Han S, et al. Circulating IL-1beta levels, polymorphisms of IL-1B, and risk of cervical cancer in Chinese women. J Cancer Res Clin Oncol 2010; 136(5):709–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanno A, Satoh K, Kimura K, et al. The expression of MUC4 and MUC5AC is related to the biologic malignancy of intraductal papillary mucinous neoplasms of the pancreas. Pancreas 2006; 33(4):391–6. [DOI] [PubMed] [Google Scholar]
- 44.Nagata K, Horinouchi M, Saitou M, et al. Mucin expression profile in pancreatic cancer and the precursor lesions. Journal of Hepato-Biliary-Pancreatic Surgery 2007; 14(3):243–254. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt CM, Yip-Schneider MT, Ralstin MC, et al. PGE(2) in pancreatic cyst fluid helps differentiate IPMN from MCN and predict IPMN dysplasia. J Gastrointest Surg 2008; 12(2):243–9. [DOI] [PubMed] [Google Scholar]
- 46.Yip-Schneider MT, Carr RA, Wu H, et al. Prostaglandin E2: A Pancreatic Fluid Biomarker of Intraductal Papillary Mucinous Neoplasm Dysplasia. J Am Coll Surg 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.M DL, DM M, W CL, et al. A novel approach for selecting combination clinical markers of pathology applied to a large retrospective cohort of surgically resected pancreatic cysts. J Am Med Inform Assoc 2017; 24(1):145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Amato E, Molin MD, Mafficini A, et al. Targeted next-generation sequencing of cancer genes dissects the molecular profiles of intraductal papillary neoplasms of the pancreas. J Pathol 2014; 233(3):217–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nissim S, Idos GE, Wu B. Genetic markers of malignant transformation in intraductal papillary mucinous neoplasm of the pancreas: a meta-analysis. Pancreas 2012; 41(8):1195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerami P, Cook RW, Wilkinson J, et al. Development of a prognostic genetic signature to predict the metastatic risk associated with cutaneous melanoma. Clin Cancer Res 2015; 21(1):175–83. [DOI] [PubMed] [Google Scholar]
- 51.Casadaban L, Maker AV. Can Colon Cancer Recurrence and Metastases Be Determined After Surgical Resection Using a Gene Expression Signature? J Clin Oncol 2017; 35(12):1372–1373. [DOI] [PubMed] [Google Scholar]
- 52.Cullen J, Rosner IL, Brand TC, et al. A Biopsy-based 17-gene Genomic Prostate Score Predicts Recurrence After Radical Prostatectomy and Adverse Surgical Pathology in a Racially Diverse Population of Men with Clinically Low- and Intermediate-risk Prostate Cancer. Eur Urol 2015; 68(1):123–31. [DOI] [PubMed] [Google Scholar]
- 53.Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol 2011; 29(35):4611–9. [DOI] [PubMed] [Google Scholar]
- 54.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004; 351(27):2817–26. [DOI] [PubMed] [Google Scholar]