Abstract
Mitochondria continually undergo fission and fusion which allow mitochondria to rapidly change their shape, size, and function throughout the cell life cycle. OMA1, a zinc metalloproteinase enzyme, is a key regulator of the mitochondrial fusion machinery. The paucity of information regarding OMA1 regulation and function largely stems from the fact that there is no direct method to quantitatively measure its activity. Using a fluorescence-based reporter assay, we developed a sensitive method to measure OMA1 enzymatic activity in whole cell lysates.
Keywords: Fluorescence-based reporter, Mitochondria, Fusion, Proteases, OMA1
1. Introduction
Mitochondria are dynamic organelles that continuously undergo two opposing processes, fission and fusion, to maintain normal mitochondrial function (Wai and Langer, 2016; Anand et al., 2013; Leonard et al., 2015; Mishra et al., 2015). Impaired mitochondrial function has been linked to numerous pathologies, including neurodegenerative (Chen and Chan, 2009), cardiovascular (Vasquez-Trincado et al., 2016; Ong et al., 2013), and metabolic diseases (Wai and Langer, 2016). During stressful conditions, mitochondrial fusion allows compensatory mixing of functional mitochondrial content with damaged mitochondria, thereby mitigating acute injury. Thus, disrupted fusion can exacerbate acute damage and lead to irreversible loss of respiratory capacity and subsequent cell death (Mishra et al., 2015; Olichon et al., 2003; Olichon et al., 2006; Mishra et al., 2014; Rainbolt et al., 2015). A key regulator in mitochondrial fusion is the optic atrophy protein termed OPA1 (Olichon et al., 2006; Ishihara et al., 2006). Fusion-competent OPA1 requires both the long and short forms and studies show that loss of the long form (L-OPA1) leads to impaired fusion. OPA1 is cleaved to its short form (S-OPA1) by two zinc metalloproteinases, OMA1 and YME1L (Ishihara et al., 2006; Head et al., 2009; Anand et al., 2014), which are located in the mitochondrial inner membrane. OMA1 is an ATP-independent protease with activities overlapping with the m-AAA proteases. Studies show that OMA1 is a stress-induced protease, whereas YME1L, an ATP-dependent protease, is thought to be constitutively active and is required for normal mitochondrial function as demonstrated by its knockout lethality (Anand et al., 2014; Zhang et al., 2014; Wai et al., 2015). Conversely, over-active OMA1 is pathogenic (Parajuli et al., 2017; Korwitz et al., 2016; Xiao et al., 2014; Acin-Perez et al., 2018), presumably by disrupting OPA1-mediated mitochondrial fusion.
Interestingly, OMA1-deficient mice exhibit protection from neurodegeneration (Korwitz et al., 2016), renal ischemia (Xiao et al., 2014), and heart failure (Acin-Perez et al., 2018) which strongly suggest that OMA1 is a promising therapeutic target. However, the lack of quantitative assays to directly measure OMA1 activity is a serious impediment for characterizing OMA1 function during disease. The current gold standard for qualitatively assessing OMA1 activity is via western blot analysis of OPA1 cleavage from long to short form which is not quantitative or temporally correlated, and where specificity is merely inferred. A decrease in the long form of OPA1 suggests activation of OMA1 during cell stress but only via indirect inference.
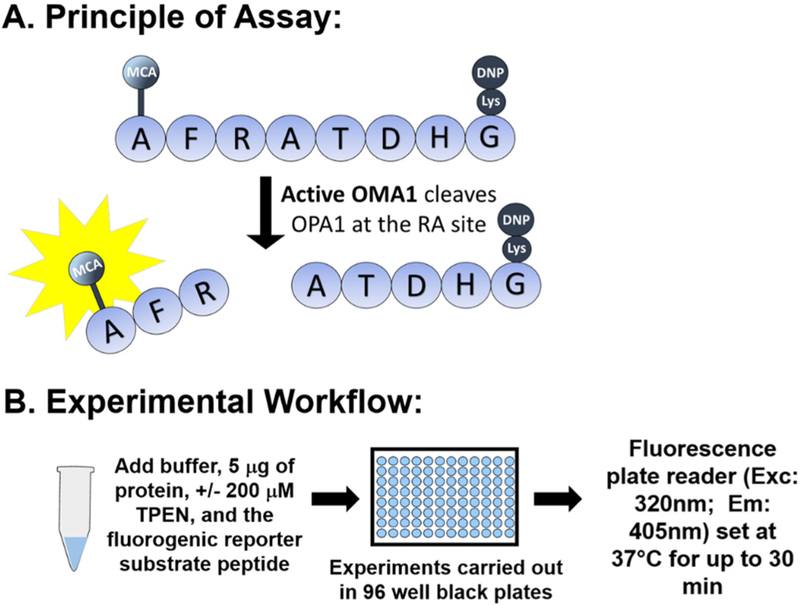
In the present paper, we describe a method which utilizes a fluorescence-based reporter cleavage assay for directly measuring OMA1 activity. Our assay is based on the report by Ishihara et al., showing that OMA1 cleaves OPA1 between the 194 Arginine (R) and 195 Alanine (A) residues, also described as the S1 site (Ishihara et al., 2006). Using this OPA1 S1 cleavage site sequence, an eight amino acid peptide (MCA-AFRATDHG-(lys)DNP) was synthesized, which contains an MCA (7-Methoxycoumarin-4-ylacetyl) fluorophore moiety on the N-terminus and a DNP (2,4-Dinitrophenyl) quencher moiety on the C-terminus. This MCA moiety on the OPA1 fluorogenic reporter substrate has very low intrinsic fluorescence due to the proximity of the DNP quencher group; once cleaved, the quenching is relieved, yielding free MCA fluorescence, which is then measured spectrofluorometrically using a plate reader (Fig. 1.).
Fig. 1.
Basis of the OMA1 activity using fluorescence-based technology. A) Fluorescence is released when OMA1 recognizes and cleaves the OPA1 8-mer peptide (fluorescence reporter) presumably at the RA site. B) Experimental workflow. [Abbreviations: MCA - 7 Methoxycoumarin 4 ylacetyl; DNP - 2,4 Dinitrophenyl; TPEN - N,N,N′,N′-tetrakis(2-pyridinylmethyl)-1,2-ethanediamine.
2. Materials and methods
2.1. Cell model
Normal rat kidney cells (NRK-52E; ATCC No. CRL-1571) were maintained in a humidified incubator with 5% CO2, 95% air at 37 °C in DMEM containing 5% fetal bovine serum. Human kidney cells (HK-2; ATCC No. CRL-2190) were grown in DMEM / F12 (1:1) containing 10% fetal bovine serum. Wild-type (WT) and OMA1-knockout (KO) mouse embryonic fibroblasts (MEF) cells, kindly provided by Dr. Pedro Quirós (Hospital Universitario de Puerto Real, Spain), Clone 9 (ATCC No. CRL- 1439) and NIH/3 T3 (ATCC No. CRL- 1658) were all grown in DMEM and 10% FBS. NRK, MEF, Clone 9, NIH/3 T3 or HK-2 cell lysates were prepared by using radioimmunoprecipitation assay (RIPA) buffer (Pierce) with 1mM PMSF (Sigma), 1.2mM Na3VO4 (Sigma), 2.5mM NaF (Sigma), 1mM DTT (Sigma), 1X of Halt Protease & Phosphatase inhibitor cocktail (Thermo Scientific), and 5mM EDTA (Thermo Scientific). Protein concentrations were determined by Coomassie Plus Protein Assay Reagent (Pierce).
2.2. OMA1 and YME1L siRNA knockdown in NRK cells
50 nM OMA1 siRNA (siGENOME SMARTpool, Dharmacon); target sequences: GUAGGACUCUCAAGAACAA; UUGAAUAGCGUGACCGAUA; GACAUACGCACUUGGGAAA; GGUCAAUGCCUUCGUGCUU and 50 nM YME1L siRNA (Ambion); sequence (5′ ➔ 3′): GUCAUGCUAUUAUUGC AUAtt (sense) and UAUGCAAUAAUAGCAUGACca (antisense) were used to knockdown OMA1 and YME1L in NRK cells, respectively. Scrambled siRNA (ON-TARGETplus Control Pool, Non-Targeting Pool, Dharmacon); target sequences: UGGUUUACAUGUCGACUAA, UGGUUUACAUGUUGUGUGA, UGGUUUACAUGUUUUCUGA; UGGUUUACAUGUUUUCCUA was used as a control. SiRNA was diluted (50 nM final concentration) in OptiMEM 1X (Life Technologies) with Lipofectamine RNAiMAX (Invitrogen) accordingly to manufacturer's instructions. At 50% cell confluency, NRK cells were transfected with the siRNA solution, and lysed 24 h later using RIPA lysis buffer. Successful knockdown of OMA1 and YME1L was confirmed via protein immunoblotting.
2.3. Western blot analysis
NRK cell lysates (25 μg) were resolved on Bolt 4–12% Bis-Tris Plus SDS precast gels (Invitrogen) and then transferred to PVDF membrane. Western blot analysis was performed using antibodies against proteins: YME1L (1:1000 in T/TBS, Abgent, #AP4882a); OMA1 (1:500 in 5% BSA, Santa Cruz, # sc-515788); and β-actin (1:1000, Sigma, # A5441). Actin served as the loading control for all western blot experiments. Probed membranes were washed three times in T/TBS and immune-reactive proteins were detected using horseradish peroxidase conjugated secondary antibodies (1:30000 dilution) (Seracare KPL, USA) and enhanced with chemiluminescence (Thermoscientific, USA).
2.4. OMA1 activity assay
Our OMA1 activity assay utilizes an 8-mer peptide derived from the cleavage sequence of OPA1, which is reported to be specific for OMA1 (Ishihara et al., 2006) and is referred to as the OPA1 fluorogenic reporter substrate in this study. The peptide contains a fluorogenic MCA moiety on the N-terminus and a DNP quencher moiety on the C-terminus (Fig. 1A), which was custom-synthesized by LifeTein LLC and handled accordingly to the manufacturer's instructions. Using a final 100 μl reaction volume and black, opaque 96 well plates (Costar), the reagents were added quickly in the following order as shown in Fig. 1B: 1) OMA1 activity assay buffer (50mM of Tris/HCl, pH 7.5, 40mM of KCl), 2) 5 μg of protein sample, 3) +/− 200 μM of the zinc chelator N,N,N′,N′-Tetrakis (2-pyridylmethyl)ethylenediamine (TPEN), which is a 1:250 dilution of stock TPEN dissolved in ethanol and lastly, 4) the OPA1 fluorogenic reporter substrate (5 μM final concentration, which is 1:200 dilution of stock peptide dissolved in DMSO). Importantly, TPEN was not added to living cells, only to the isolated protein lysates immediately prior to adding the fluorogenic reporter substrate (Fig. 1B). Relative fluorescence (RFU) was recorded (excitation/emission of 320/405 nm) every five minutes for 30 min at 37 °C using a fluorescent plate reader (SpectraMax M2e, Molecular Devices equipped with SoftMax Pro v5 software).
For the statistical analysis, the average fluorescence of the OPA1 fluorogenic reporter substrate alone was measured (< 200 relative fluorescence units) and that value was subtracted from the sample values. The values were reported as a difference of TPEN inhibition [(sample X) - (sample X + TPEN)] where the slopes between 0 and 30 min were calculated by linear regression and each treatment was compared to the appropriate controls. Thus, OMA1 activity was calculated by subtracting TPEN-inhibitable fluorescence from total fluorescence to enhance specificity.
2.5. Statistical analysis
Results are presented as mean ± standard error of the mean (S.E.M.) using Graph Pad Prism 7 software. At least 3–4 independent biological replicates were carried out for each experiment and treatment, with three replicates of each individual sample. Km (substrate concentration that gives half-maximal enzyme velocity) and Vmax (maximum velocity extrapolated to infinite substrate concentration) were analyzed using Michaelis-Menten (nonlinear regression) built-in analysis in Graph Pad Prism 7. Depending on the number of conditions, either a one-way ANOVA followed by the appropriate post hoc comparisons or an unpaired students t-test were performed (unless otherwise stated). All differences were judged to be significant at p < .05.
3. Results and discussion
Our study offers compelling evidence for the development and feasibility of a novel OMA1 activity assay (Patent Pending). Using fluorescence-based technology, this assay employs a custom peptide, derived from the protein sequence for the OMA1-specific cleavage site within OPA1 (Ishihara et al., 2006) flanked by the fluorophore (MCA) and quencher (DNP), which is referred to as an OPA1 fluorogenic reporter substrate (Fig. 1A).
Michaelis–Menten kinetics was used to determine the Km and Vmax for cleavage of the OPA1 fluorogenic reporter substrate. Six concentrations (1, 5, 10, 15, 20, and 30 μM) of the OPA1 fluorogenic reporter substrate were evaluated using untreated NRK cell lysates (5 μg) in the activity assay (Fig. 2A). The Vmax was calculated to be 34.6 relative fluorescence (RFU)/min and the Km 7.1 μM. Next, we selected 5 μM of the OPA1 fluorogenic reporter substrate in the OMA1 activity assay to ensure we have a sensitive assay when using a relatively small amount of NRK cell lysates (5 μg). Generally, it is preferable to use a substrate concentration at least double its Km value to avoid rate changes due to substrate depletion. However, the linear response over the 30 min reaction period (Fig. 2B) strongly indicates that substrate depletion is negligible during this time – not surprising given the small amounts of OMA1 enzyme present in the lysate aliquots. OMA1 belongs to a unique subset of proteases that are zinc dependent and ATP independent. Numerous studies have documented that the metal chelator, o-Phenanthroline, inhibits OMA1 dependent cleavage of OPA1 when added to living cells (0.5–1mM for 6h) (Ishihara et al., 2006; Rainbolt et al., 2016; Kaser et al., 2003). We first tested o-Phenanthroline in our OMA1 activity assay by adding this compound to protein lysates (not cells) immediately prior to initiation of the assay; however, the compound had intrinsic fluorescence that interfered with the fluorescence-based assay. Therefore, we next tested another zinc chelator, TPEN, which dose-dependently significantly inhibited OMA1 activity in NRK cell lysates compared to NRK cell lysates without TPEN, suggesting that zinc is necessary for enzymatic activity (Fig. 2C). Given the profound inhibition (81%) using 200 μM TPEN, this dose was selected for all subsequent OMA1 activity assay studies. OMA1 activity will be calculated by subtracting TPEN -inhibitable fluorescence from total fluorescence.
Fig. 2.
Kinetic parameters of OMA1 activity assay A). Michalis-Menten kinetics using a dose response of OPA1 fluorogenic reporter substrate concentrations (1, 5, 10, 15, 20 and 30 μM) on the rate of the reaction resulted in values of Km = 7.1 uM and Vmax = 34.6. B). Time course of OMA1 activity assay using untreated NRK cells with 5 μM of OPA1 fluorogenic reporter substrate resulted in a linear increase in relative fluorescence (RFU) over 30 min. TPEN (200 μM) inhibited ~80% of relative fluorescence. C). TPEN dose response showing that TPEN significantly (****p < .0001) reduces OMA1 activity at all doses tested using a one-way ANOVA followed by a Sidak's posthoc tests. Data is expressed as mean ± SEM; n = 3–5.
It is important to note that prior studies have reported aspartic protease inhibitors (e.g. pepstatin A), cysteine protease inhibitors (e.g. E64D) and serine protease inhibitor (e.g. pefabloc/TPCK) do not prevent OMA1-dependent OPA1 cleavage (Ishihara et al., 2006; Baker et al., 2014a, 2014b). In our assay, we lysed cells with RIPA lysis buffer containing numerous protease inhibitors (including cysteine, serine, aspartic, trypsin-like inhibitors-all present in HALT cocktail). No statistical significance was seen in OMA1 activity in cells lysed with RIPA supplemented with a protease inhibitor cocktail compared to with RIPA without it (data not shown).
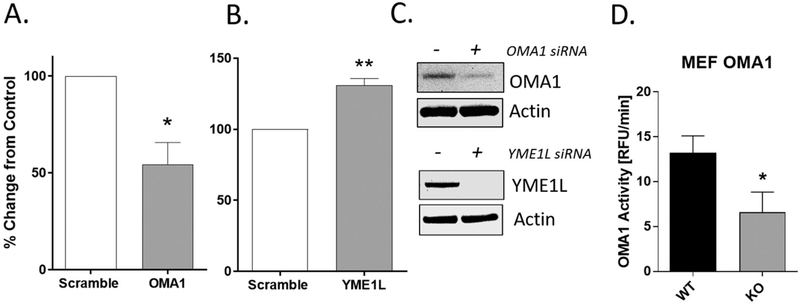
Next, we evaluated whether knockdown of OMA1 in NRK cells reduced apparent enzyme activity in the new assay. Protein lysates harvested from OMA1 siRNA-treated (50 nM; 24 h) NRK cells showed a 45% reduction of fluorescence/min (slope of OMA1 activity) compared to control cells transfected with a scrambled siRNA (Fig. 3A). This reduction in activity is consistent with the extent of lowered OMA1 expression (Fig. 3C inset). The knockdown of OMA1 after 24 h of siRNA transfection was ~ 50%, consistent with the ~ 45% reduction in OMA1 activity. Higher concentrations of OMA1 siRNA or longer time points post-transfection did not improve OMA1 knockdown efficiency (data not shown). Therefore, to confirm that the activity assay was responding to OMA1 protein levels we obtained wild-type (WT) and OMA1-knockout (KO) mouse embryonic fibroblast (MEF) cells kindly provided by Dr. Pedro Quirós. MEF OMA1 KO cells, which have no detectable OMA1 via western blot (not shown), showed a 50% decrease in OMA1 activity compared to WT MEF cells (Fig. 3D). It is also important to point out that another study using shRNA OMA1 knockdown in renal cells showed a similar ~50% knockdown in OPA1 cleavage, which corresponds to our findings with the novel OMA1 activity assay (Xiao et al., 2014). Interestingly, cells treated with YME1L siRNA (50 nM; 24 h) showed a significant increase (**p = .003) in OMA1 activity compared to scrambled siRNA (Fig. 3B). A potential explanation for increase in OMA1 activity in NRK cells treated with YME1L siRNA is that YME1L has been shown to degrade OMA1 (Anand et al., 2014; Rainbolt et al., 2016). In addition, it has been shown that loss of YME1L may act as a cellular stressor leading to increased OMA1 activity (Ruan et al., 2013). Western blots confirmed knockdown of OMA1 and YME1L after transfection with siRNA at the respective time points (24 h for OMA1 and 24 h for YME1L) (Fig. 3C). Taken together, these findings provide strong evidence that the observed protease activity in cell lysates corresponds with OMA1 protein levels and not YME1L. This was not unexpected, as the peptide sequence ‘AFRATDHG’ is specific for the OMA1-dependent OPA1 cleavage site (Site 1), and not the site where YME1L cleaves. The precise site within OPA1 that YME1L cleaves, which is referred to as Site 2, has not been established but it does not overlap and is clearly different than the OMA1 cleavage site (Ishihara et al., 2006). The protease activity remaining in the siRNA knockdown of OMA1 (~50%) is virtually the same as that remaining in the MEF OMA1 KO cells. This suggests that: 1) the small OMA1 fraction remaining after siRNA treatment is inactive, and 2) that roughly 50% of the total activity seen in the assay is due to some other protease(s). Efforts were then made to identify other potential sources of activity including alteration of the peptide sequence and addition of known inhibitors of other metalloproteinases.
Fig. 3.
Assay has specificity toward OMA1, but not YME1L, mediated OPA1 cleavage. A). Protein lysates (5 μg) from NRK cells treated with OMA1 siRNA (50 nM; 24 h; 37 °C) showed reduced OMA1 activity, *p = .015, n = 3. B) Protein lysates from NRK cells treated with YME1L siRNA (50 nM; 24 h; 37 °C) showed an increase in OMA1 activity, **p = .003, n = 3. C) Insets show a representative image of OMA1 and YME1L knockdown using Western blots. D) Protein lysates from OMA1 KO MEF cells showed 50% reduced OMA1 activity compared to wild type MEF cells, *p = .046. Slopes (0–30 min) were converted to percent change from control/scramble rate (RFU/min) values;. n = 6. Data is expressed as mean ± SEM. *p < .05 and **p < 0.01.
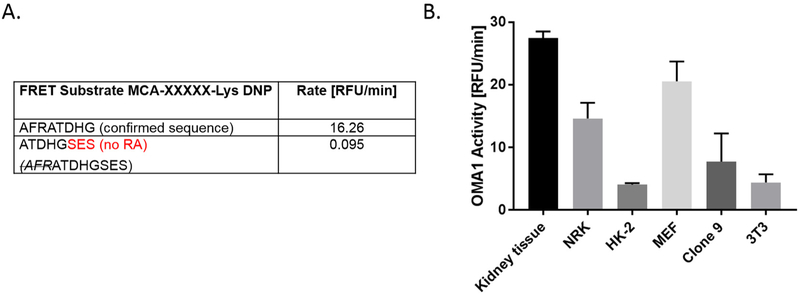
The eight amino acid OPA1 fluorogenic reporter substrate (AFRATDHG) used in the OMA1 activity assay was derived from the rat OPA1 protein sequence (residues 192–199). According to Ishihara et al., the OMA1-dependent OPA1 processing has been reported to occur between the 194 Arginine (R) and the 195 Alanine (A) residues (Ishihara et al., 2006). To test whether the RA sequence is critical for measuring OMA1 activity in our assay, we synthesized an eight amino acid fluorogenic reporter substrate peptide (ATDHGSES) based on a the OPA1 sequence, but a few amino acids downstream of the cleavage site, such that it does not contain the RA sequence. This new OPA1 fluorogenic reporter peptide showed no fluorescent changes in our assay, suggesting that the “RA” residues are required for OMA1 recognition and/or cleavage, as well as cleavage by the other non-specific component (Fig. 4A). Further studies are underway to determine whether OMA1 specificity might be enhanced using an alternate peptide sequences or size.
Fig. 4.
Substrate specificity and applicability of assay across species. A). RA sequence is essential for OPA1 recognition. OMA1 activity was calculated as an average slope over 30 min compared to AFRATDHG substrate. The ‘no RA’ sequence was calculated as a percent difference from the confirmed sequence; n = 3–4. B). Application of OMA1 assay across different species. Untreated rat kidney tissue, NRK (normal rat kidney), human kidney cells (HK-2), and MEF (mouse embryonic fibroblasts), rat liver Clone 9 cells, and mouse 3 T3 fibroblasts were used to show applicability of the OMA1 activity assay across species (mouse, rat, human). Data is expressed as mean ± SEM. n = 2–4.
Next, to determine the applicability of the OMA1 assay across different species, we tested rat cells and kidney tissue, along with human and mouse cell lysates. As shown in Fig. 4B we observed OMA1 activity in rat kidney homogenates, rat NRK cells, rat liver Clone 9 cells, mouse (MEF), mouse 3 T3 fibroblast cell lines, as well as human kidney (HK –2) proximal tubule cells. Despite the variability in basal OMA1 activity, all samples were effectively (~75%) inhibited by the addition of TPEN (data not shown).
To further validate the assay's specificity, we tested whether classic zinc metalloproteinase (MMP) inhibitors could inhibit OMA1 activity. We found that neither the broad-spectrum MMP inhibitor, batimastat (0.5–1 μM), or the meprin A inhibitor, actinonin (0.5–1 μM), altered OMA1 activity (Fig. 5.), suggesting that the OMA1 activity assay does not detect MMP activity. We selected these doses based on the known IC50 concentrations of these agents toward MMP-like activity (Cayman Chemical Product sheets). It is important to remember that these agents were not added to living cells, only to the isolated protein immediately prior to adding the fluorogenic reporter substrate. While the source of the non-OMA1 protease activity remains to be identified, these data strongly suggest that it is not due to meprin A or other matrix metalloproteinases inhibited by batimastat.
Fig. 5.
Classical MMP inhibitors fail to inhibit OMA1 activity. NRK cell lysates were incubated with Actinonin or Batimastat and the designated concentrations. Data is expressed as mean ± SEM; n = 4.
Much of our knowledge of the function and activation of OMA1 is based on OMA1 knockout yeast, cells, and mouse models (Korwitz et al., 2016; Xiao et al., 2014; Quiros et al., 2012; Kong et al., 2014; Khalimonchuk et al., 2012; Bohovych et al., 2014). These are informative proof-of-concept experiments that have established the critical role of OMA1 in helping regulate mitochondrial quality control. However, further advancements in understanding the role of OMA1 and its potential as a therapeutic target are dependent on a semi-quantitative activity measurement, as knockout models can't assess temporal activation and resulting downstream events. Our study was limited by the lack of a reliable, active source of recombinant OMA1 that could be used to validate our new assay. We did in fact test the only commercial source of OMA1 (Abcam); however, it failed to show any activity in the assay, and was quite impure when resolved on SDS-PAGE (data not shown). Another limitation was the inability to completely inhibit all fluorescence using two methods of OMA1 knockdown (Fig. 3), which suggests that the current assay is not fully specific for OMA1. There is no commercially available source of overexpressing OMA1 that could be readily tested but we have begun the process of generating this type of plasmid so these studies can be performed. Despite these limitations, we conclude that the assay is an improvement over using OPA1 western blot analysis as an indirect measure of OMA1 activity. Even this assay required validation of OMA1 specificity using OMA1 and/or YME1L silencing since both proteases can cleave OPA1. Moreover, the assay is quite sensitive, allowing the use of cell lysates, and has wide species applicability, including mouse, rat, and human cells. Many commercial protease assays lack absolute specificity, and contain a component of non-specific activity which must be taken into account when interpreting results. Efforts to refine our assay to make it more specific are ongoing, via the synthesis and testing of new peptides, as well as via identification of OMA1 inhibitors. In addition, we plan to develop a cell-permeable fluorogenic substrate that could be used to detect OMA1 activity in living cells. In the interim, judicious use of this assay will help establish temporal and functional relationships between OMA1 activation and mitochondrial dynamics in rodent and human tissues.
In summary, this is the first study to demonstrate the feasibility of a fast and inexpensive assay to measure OMA1 activity using a fluorescence-based high throughput assay. This study should accelerate our understanding of the role OMA1 plays in both normal and abnormal pathology, and the conditions under which it is induced or inactivated. The OMA1 activity assay may provide a new tool for researchers to further explore the role of OMA1 in their model systems and to identify new drug targets to treat diseases involving over activation of OMA1.
Acknowledgments
Funding
This work was supported in part by University of Arkansas for Medical Research Pilot Research Funds, as well as funding from NIH R01 GM106419. JT is supported by a PhRMA Foundation Pre Doctoral Fellowship; JT and SS were supported by predoctoral fellowships from NIHT32 GM106999.
References
- Acin-Perez R, Lechuga-Vieco AV, Del Mar Munoz M, Nieto-Arellano R, Torroja C, Sanchez-Cabo F, et al. , 2018. Ablation of the stress protease OMA1 protects against heart failure in mice. Sci. Transl. Med 10 (434). [DOI] [PubMed] [Google Scholar]
- Anand R, Langer T, Baker MJ, 2013. Proteolytic control of mitochondrial function and morphogenesis. Biochim. Biophys. Acta 1833 (1), 195–204. [DOI] [PubMed] [Google Scholar]
- Anand R, Wai T, Baker MJ, Kladt N, Schauss AC, Rugarli E, et al. , 2014. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol 204 (6), 919–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Lampe PA, Stojanovski D, Korwitz A, Anand R, Tatsuta T, et al. , 2014a. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33 (6), 578–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker MJ, Lampe PA, Stojanovski D, Korwitz A, Anand R, Tatsuta T, et al. , 2014b. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 33 (6), 578–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohovych I, Donaldson G, Christianson S, Zahayko N, Khalimonchuk O, 2014. Stress-triggered activation of the metalloprotease Oma1 involves its C-terminal region and is important for mitochondrial stress protection in yeast. J. Biol. Chem 289 (19), 13259–13272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC, 2009. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum. Mol. Genet 18 (R2), R169–R176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM, 2009. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol 187 (7), 959–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K, 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25 (13), 2966–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser M, Kambacheld M, Kisters-Woike B, Langer T, 2003. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem 278 (47), 46414–46423. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O, Jeong MY, Watts T, Ferris E, Winge DR, 2012. Selective Oma1 protease-mediated proteolysis of Cox1 subunit of cytochrome oxidase in assembly mutants. J. Biol. Chem 287 (10), 7289–7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong B, Wang Q, Fung E, Xue K, Tsang BK, 2014. p53 is required for cisplatin-induced processing of the mitochondrial fusion protein L-Opa1 that is mediated by the mitochondrial metallopeptidase Oma1 in gynecologic cancers. J. Biol. Chem 289 (39), 27134–27145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korwitz A, Merkwirth C, Richter-Dennerlein R, Troder SE, Sprenger HG, Quiros PM, et al. , 2016. Loss of OMA1 delays neurodegeneration by preventing stress-induced OPA1 processing in mitochondria. J. Cell Biol 212 (2), 157–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AP, Cameron RB, Speiser JL, Wolf BJ, Peterson YK, Schnellmann RG, et al. , 2015. Quantitative analysis of mitochondrial morphology and membrane potential in living cells using high-content imaging, machine learning, and morphological binning. Biochim. Biophys. Acta 1853 (2), 348–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Carelli V, Manfredi G, Chan DC, 2014. Proteolytic cleavage of Opa1 stimulates mitochondrial inner membrane fusion and couples fusion to oxidative phosphorylation. Cell Metab. 19 (4), 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Varuzhanyan G, Pham AH, Chan DC, 2015. Mitochondrial dynamics is a distinguishing feature of skeletal muscle Fiber types and regulates organellar compartmentalization. Cell Metab. 22 (6), 1033–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olichon A, Baricault L, Gas N, Guillou E, Valette A, Belenguer P, et al. , 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem 278 (10), 7743–7746. [DOI] [PubMed] [Google Scholar]
- Olichon A, Guillou E, Delettre C, Landes T, Arnaune-Pelloquin L, Emorine LJ, et al. , 2006. Mitochondrial dynamics and disease, OPA1. Biochim. Biophys. Acta 1763 (5–6), 500–509. [DOI] [PubMed] [Google Scholar]
- Ong SB, Hall AR, Hausenloy DJ, 2013. Mitochondrial dynamics in cardiovascular health and disease. Antioxid. Redox Signal 19 (4), 400–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parajuli N, Shrum S, Tobacyk J, Harb A, Arthur JM, MacMillan-Crow LA, 2017. Renal cold storage followed by transplantation impairs expression of key mitochondrial fission and fusion proteins. PLoS One 12 (10), e0185542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros PM, Ramsay AJ, Sala D, Fernandez-Vizarra E, Rodriguez F, Peinado JR, et al. , 2012. Loss of mitochondrial protease OMA1 alters processing of the GTPase OPA1 and causes obesity and defective thermogenesis in mice. EMBO J. 31 (9), 2117–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt TK, Saunders JM, Wiseman RL, 2015. YME1L degradation reduces mitochondrial proteolytic capacity during oxidative stress. EMBO Rep. 16 (1), 97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainbolt TK, Lebeau J, Puchades C, Wiseman RL, 2016. Reciprocal degradation of YME1L and OMA1 adapts mitochondrial proteolytic activity during stress. Cell Rep. 14 (9), 2041–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Y, Li H, Zhang K, Jian F, Tang J, Song Z, 2013. Loss of Yme1L perturbates mitochondrial dynamics. Cell Death Dis. 4, e896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez-Trincado C, Garcia-Carvajal I, Pennanen C, Parra V, Hill JA, Rothermel BA, et al. , 2016. Mitochondrial dynamics, mitophagy and cardiovascular disease. J. Physiol 594 (3), 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Langer T, 2016. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab 27 (2), 105–117. [DOI] [PubMed] [Google Scholar]
- Wai T, Garcia-Prieto J, Baker MJ, Merkwirth C, Benit P, Rustin P, et al. , 2015. Imbalanced OPA1 processing and mitochondrial fragmentation cause heart failure in mice. Science. 350 (6265), aad0116. [DOI] [PubMed] [Google Scholar]
- Xiao X, Hu Y, Quiros PM, Wei Q, Lopez-Otin C, Dong Z, 2014. OMA1 mediates OPA1 proteolysis and mitochondrial fragmentation in experimental models of is-chemic kidney injury. Am. J. Physiol. Renal. Physiol 306 (11), F1318–F1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Li H, Song Z, 2014. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep. 15 (5), 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]