See Bateman et al. (doi:10.1093/brain/awz082) for a scientific commentary on this article.
The forms of Aβ that contribute to Alzheimer’s disease remain poorly defined. Brinkmalm and Hong et al. report the discovery of covalently cross-linked dimers formed from a large range of Aβ monomers. The identification of these dimers may open up a new phase of Alzheimer’s disease research.
Keywords: amyloid-β-protein, human neurons, long-term potentiation, mass spectrometry, video microscopy
Abstract
The primary structure of canonical amyloid-β-protein was elucidated more than 30 years ago, yet the forms of amyloid-β that play a role in Alzheimer’s disease pathogenesis remain poorly defined. Studies of Alzheimer’s disease brain extracts suggest that amyloid-β, which migrates on sodium dodecyl sulphate polyacrylamide gel electrophoresis with a molecular weight of ∼7 kDa (7kDa-Aβ), is particularly toxic; however, the nature of this species has been controversial. Using sophisticated mass spectrometry and sensitive assays of disease-relevant toxicity we show that brain-derived bioactive 7kDa-Aβ contains a heterogeneous mixture of covalently cross-linked dimers in the absence of any other detectable proteins. The identification of amyloid-β dimers may open a new phase of Alzheimer’s research and allow a better understanding of Alzheimer’s disease, and how to monitor and treat this devastating disorder. Future studies investigating the bioactivity of individual dimers cross-linked at known sites will be critical to this effort.
See Bateman et al. (doi:10.1093/brain/awz082) for a scientific commentary on this article.
Introduction
Although the precise cause of Alzheimer’s disease remains obscure, evidence from multiple sources indicate that the amyloid-β-protein plays a key role (Masters et al., 2015; Karran and De Strooper, 2016). There is general agreement that amyloid-β monomer is innocuous (Walsh and Teplow, 2012) or may even have a physiological function (Pearson and Peers, 2006; Puzzo et al., 2008) and that aggregation and further assembly is required for toxicity (Yankner and Lu, 2009). Both intrinsic and extrinsic factors may influence how and why wild-type amyloid-β folds to form toxic assemblies and it is likely that certain post-translational modifications of amyloid-β may facilitate formation of toxic structures.
Aqueous extracts of Alzheimer’s disease brain potently disrupt hippocampal long-term potentiation (LTP), alter synaptic form and number, and impair memory consolidation, and these effects are reversed when amyloid-β is depleted by anti-amyloid-β antibodies (Shankar et al., 2008; Barry et al., 2011; Freir et al., 2011; Borlikova et al., 2013; Wang et al., 2017; Hong et al., 2018). The amyloid-β in these samples migrates on sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE) with molecular weights consistent for monomers and dimers (Shankar et al., 2008; Mc Donald et al., 2010, 2012; Wang et al., 2017; Yang et al., 2017; Hong et al., 2018; Jin et al., 2018). The amyloid-β ∼4 kDa species (4kDa-Aβ) appears to be composed of both native monomers and monomers derived from soluble aggregates that are unstable when electrophoresed in SDS (Mc Donald et al., 2015). Characterization of the ∼7 kDa amyloid-β (7kDa-Aβ) species from water soluble extracts of Alzheimer’s disease brain has been challenging because brain extracts are biologically complex and the amount of 7kDa-Aβ is minute. Nonetheless, prior studies that use non-denaturing size separation and a battery of 12 anti-APP/amyloid-β antibodies support the premise that 7kDa-Aβ comprises both native amyloid-β dimers and dimers generated from the denaturation of soluble aggregates (Mc Donald et al., 2015). However, it is not known what sort of bonds hold these dimers together. Moreover, there is continued concern that 7kDa-Aβ could be an artefact of SDS-PAGE (Hepler et al., 2006; Watts et al., 2014).
Here, we isolated 4kDa-Aβ and 7kDa-Aβ from the aqueous phase of brain and by solubilizing purified amyloid plaques, and used video microscopy of human neurons and LTP to assess bioactivity. 7kDa-Aβ from both sources potently disrupted neuritic integrity of induced pluripotent stem cell (iPSC)-derived neurons and blocked hippocampal LTP, whereas 4kDa-Aβ had no effect. While our results demonstrate that at least a subpopulation of 7kDa-Aβ have Alzheimer’s disease-relevant toxic activity, it seems likely that other forms of amyloid-β might also have cognition-disrupting activity. Subsequent analysis using mass spectrometry (MS) revealed that 4kDa-Aβ contained a rich diversity of amyloid-β sequences encompassing a large number of N- and C-termini. Analysis of 7kDa-Aβ identified 11 mass matches consistent with covalently cross-linked amyloid-β dimers including species such as: 1–37 × 1–38, 1–38 × 1–40, 1–40 × 1–40, 2–40 × 1–40, 1–40 × 1–42, and 1–42 × 1–42. MS/MS of trypsin/LysC-digested 7kDa-Aβ identified the most abundant amyloid-β dimer as having a cross-link between Asp1 and Glu22. Collectively, these results recommend the further study of amyloid-β dimers as targets for therapy and as potential biomarkers of disease.
Materials and methods
Reagents and chemicals
Aβ1–40 and Aβ1–42 were synthesized and purified by Dr James I. Elliott at the ERI Amyloid laboratory, Oxford, CT, USA. Peptide mass and purity (>99%) were confirmed by electrospray/ion trap mass spectrometry and reverse-phase HPLC. N-terminally extended (NTE)-amyloid-β−31Aβ1–42 was expressed and purified as described previously (Szczepankiewicz et al., 2015). Recombinant Aη-α (APP505–612, APP695 numbering) was a gift from Drs M. Willem and C. Haass (Ludwig-Maximillian University, Munich), and dityrosine cross-linked dimers were produced from Aβ1–42 (O'Malley et al., 2014). Peptide standards were prepared as described previously (Hong et al., 2018) and were stored frozen at 10 ng/μl in 50 mM ammonium bicarbonate, pH 8.5.
Gel filtration standards were purchased from Bio-Rad (Hercules). Antibodies and their sources are described in Table 1. All other chemicals and reagents were of the highest purity available and unless indicated otherwise were obtained from Sigma-Aldrich.
Table 1.
Antibodies used in this study and their sources
| Antibody | Type | Epitope | Dilution for immunoprecipitation | Concentration for western blot | Source |
|---|---|---|---|---|---|
| 2E9 | Monoclonal | APP545-555 | - | 1 μg/ml | Haass Lab |
| 28D10 | Monoclonal | APP585-600 | - | 1 μg/ml | Haass Lab |
| 3D6 | Monoclonal | Aβ1–5 | - | 1 μg/ml | Elan |
| 6E10 | Monoclonal | Aβ6–10 | - | 1 μg/ml | BioLegend |
| 266 | Monoclonal | Aβ16–23 | - | 1 μg/ml | Janssen |
| 4G8 | Monoclonal | Aβ17–24 | - | 1 μg/ml | BioLegend |
| 2G3 | Monoclonal | Aβ terminating at Val40 | - | 1 μg/ml | Janssen |
| 21F12 | Monoclonal | Aβ terminating at Ala42 | - | 1 μg/ml | Janssen |
| AW7 | Polyclonal | Pan anti-Aβ | 1:50 | - | Walsh Lab |
Aβ = amyloid-β.
Preparation of aqueous extracts from human brain
Frozen brain tissue was provided by the Massachusetts ADRC Neuropathology Core, Massachusetts General Hospital and used in accordance with the Partners Institutional Review Board (Protocol: Walsh BWH 2011). Brain tissue was obtained from nine patients who died with end-stage Alzheimer’s disease, one individual with mild Alzheimer’s disease and one subject free of Alzheimer’s disease (Table 2). Aqueous extracts were prepared as described previously (Wang et al., 2017). Cortical grey matter (10–20 g) was Dounce-homogenized in five volumes of ice-cold artificial CSF base buffer (artificial CSF-B) (124 mM NaCl, 2.8 mM KCl, 1.25 mM NaH2PO4, 26 mM NaHCO3, pH 7.4) supplemented with protease inhibitors (5 mM EDTA, 1 mM EGTA, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 2 μg/ml pepstatin, 120 μg/ml pefabloc and 5 mM NaF]. The resulting homogenates were centrifuged at 200 000g for 110 min and 4°C in a SW41 Ti rotor (Beckman Coulter) and the upper 80% of the supernatant was removed and dialyzed against fresh artificial CSF-B, with three buffer changes over a 72-h period. Dialysates were removed to clean tubes, aliquoted and stored at −80°C until required.
Table 2.
Demographic details of the cases used in this study
| ADRC No. | Age | Gender | PMI (h) | CLDX | NPDX | B&B, CERAD | Use to prepare | |
|---|---|---|---|---|---|---|---|---|
| Aqueous extract | Amyloid plaque | |||||||
| 414 | 75 | F | <48 | AD | AD | NA, NA | + | |
| 453 | 75 | F | <24 | AD | AD | NA, C | + | |
| 1849 | 68 | F | 36 | AD | AD | VI, C | + | |
| 1185 | 89 | F | 14 | AD | AD | VI, C | + | + |
| 1242 | 79 | F | NA | AD | AD | VI, C | + | + |
| 722 | 69 | F | 3 | AD | AD | V, A | + | + |
| 464 | 92 | F | 4 | AD | AD | V, C | + | |
| 1167 | 84 | F | NA | AD | AD | VI, C | + | |
| 1444 | 86 | F | 56 | DLB | AD | V-VI, C | + | |
| 1670a | 87 | M | 18 | Mild AD | AD | IV, A | + | |
| 1821 | 82 | M | 18 | Ctrl | Ctrl | II, C | + | |
AD = Alzheimer’s disease; ADRC = Alzheimer's Disease Research Center; B&B = Braak stage; CERAD = Consortium to Establish a Registry for Alzheimer's disease score; CLDX = clinical diagnosis; DLB = dementia with Lewy bodies; F = female; NA = information not available; NPDX = neuropathology diagnosis; PMI = post-mortem interval.
Indicates brain from a subject who died with mild Alzheimer’s disease (MMSE score 23, examined 9 months prior to death).
Immunoprecipitation/western blot detection of amyloid-β
Aliquots (0.5 ml) of human brain extracts were cleared of antibodies and protein A binding proteins by gently mixing with 15 μl protein A sepharose (PAS) beads for 1 h at 4°C. PAS beads were removed by centrifugation at 6000g for 5 min. The supernatant was transferred to a clean tube and incubated with 10 µl purified AW7 antibody or 10 µl purified pre-immune serum, and 15 μl PAS beads overnight at 4°C with gentle agitation. Amyloid-β-antibody-PAS complexes were collected by centrifugation and washed as previously described (Shankar et al., 2011). Samples treated with AW7 are referred to as immunodepleted and samples treated with pre-immune serum as mock immunodepleted (mock). Beads were eluted by boiling in 15 μl of 2× sample buffer (50 mM Tris, 2% w/v SDS, 12% v/v glycerol with 0.01% phenol red) and samples electrophoresed on hand-cast 15-well 16% polyacrylamide tris-tricine gels. Proteins were transferred to 0.2 µm nitrocellulose at 400 mA and 4°C for 2 h. Blots were microwaved in phosphate-buffered saline (PBS) and amyloid-β detected using anti-amyloid-β monoclonal antibodies (Table 1), and bands visualized using a Li-COR Odyssey infrared imaging system (Li-COR). Synthetic Aβ1–40 and Aβ1–42, recombinant NTE-Aβ and Aη-α peptides were loaded to confirm antibody specificity, and to allow comparison between gels. To allow estimation of the quantitation of 4kDa-Aβ and 7kDa-Aβ, a series of synthetic amyloid-β standards were loaded alongside size exclusion chromatography (SEC) fractions and the content of fractions determined from a standard curve.
Isolation of 4kDa-Aβ and 7kDa-Aβ using immunoprecipitation/size exclusion chromatography
To enrich and purify 4kDa-Aβ and 7kDa-Aβ, artificial CSF extracts were immunoprecipitated with AW7 and the immunoprecipitate eluted in a volatile solvent to allow concentration by lyophilization. Thereafter, formic acid was used to denature the sample so as to reduce any non-covalent structures to their most fundamental building blocks. The denatured material was then size-separated in ammonium bicarbonate—a buffer that allows further concentration by lyophilization and which is suitable for mass spectrometry. Specifically, 10 1-ml aliquots of artificial CSF brain extracts were thawed at room temperature, centrifuged at 13 200g in a benchtop centrifuge and the upper 90% of supernatants removed to clean tubes and each immunoprecipitated with AW7. Immune-complexes were isolated, washed, and bound amyloid-β was eluted by vortexing in 150 µl 500 mM ammonium hydroxide (Welzel et al., 2014). PAS beads were removed by centrifugation and eluted a further two times with 500 mM ammonium hydroxide; the eluates were pooled and lyophilized. The lyophilizate was resuspended in 1 ml 88% formic acid and the entire volume loaded on to a Superdex 75 10/300 GL SEC column. The column was eluted with 50 mM ammonium bicarbonate, pH 8.5 at a flow rate of 0.5 ml/min and 0.5 ml fractions were collected. Prior to application of sample, the column was calibrated using Blue dextran and gel filtration standards. The peak fraction containing Blue dextran was designated as fraction zero. Upon collection, fractions were divided into two aliquots. One 50-µl aliquot was lyophilized and used for western blotting. The remainder was stored frozen at −80°C. Following western blot analysis, fractions that contained 4kDa-Aβ and 7kDa-Aβ were thawed, pooled, exchanged into neuronal medium and added to iPSC-derived neurons.
Live-cell imaging of human iPSC-derived neurons
Human iPSCs-derived neurons were induced and cultured as summarized in Supplementary Fig. 1A (Guix et al., 2018; Jin et al., 2018). At Day 21, neurons were used to investigate the effects of Alzheimer’s disease brain extracts on neuritic integrity. Approximately 7 h prior to addition of sample, images were collected from four fields per well every 2 h for a total of 6 h and baseline neurite length and branch points were calculated. During this time, brain extracts were exchanged into Neurobasal medium supplemented with B27/GlutaMAXTM using PD MidiTrap G-25 columns (GE Healthcare). Following the 6 h period of baseline imaging, half of the medium was removed from each well (leaving ∼100 µl) and 50 µl of exchanged extract or vehicle, added along with 50 µl of fresh medium. Thereafter, images were collected from four fields per well every 2 h for at least 72 h. Phase contrast image sets were analysed using IncuCyte® Zoom 2016A Software (Essen Bioscience). The analysis job Neural Track was used to automatically define neurite processes and cell bodies based on phase contrast images. Typical settings were: segmentation mode, Brightness; segmentation adjustment, 1.2; cell body cluster filter, minimum 500 μm2; neurite filtering, Best; neurite sensitivity, 0.4; neurite width, 2 μm. Total neurite length (in mm) and number of branch points were quantified and normalized to the average value measured during the 6 h period prior to sample addition.
Mouse brain slice preparation and long-term potentiation recording
Animal procedures were performed in accordance with the National Institutes of Health Policy on the Use of Animals in Research and were approved by the Harvard Medical School Standing Committee on Animals. Wild-type C57BL/6 mice were purchased from Jackson Labs and a small colony maintained in-house. Mice (2–3 months old) were anaesthetized with isoflurane and decapitated, then brains were rapidly removed and immediately immersed in ice-cold (0–4°C) artificial CSF. The artificial CSF contained (in mM): 124 NaCl, 3 KCl, 2.4 CaCl2, 2 MgSO4·7H2O, 1.25 NaH2PO4, 26 NaHCO3 and 10 d-glucose, and was equilibrated with 95% O2 and 5% CO2, pH 7.4, 310 mOsm. Coronal brain slices (350 µm) including hippocampus were prepared using a Leica VT1000 S vibratome (Leica Biosystems Inc) and transferred to an interface chamber and incubated at 34 ± 5°C for 20 min and then kept at room temperature for 1 h before recording.
Brain slices were transferred to a submerged recording chamber and perfused (10 ml/min) with oxygenated (95% O2 and 5% CO2) artificial CSF for at least 10 min prior to electrophysiological recordings. Brain slices were visualized using an infrared and differential interference contrast camera (IR-DIC camera, Hitachi) mounted on an upright Olympus microscope. Recording electrodes were pulled from borosilicate glass capillaries (Sutter Instruments) using a micropipette puller (Model P-97; Sutter Instruments) with resistance ∼2 MΩ when filled with artificial CSF. To induce field excitatory postsynaptic potentials (fEPSPs) in the hippocampal CA1, a tungsten wire stimulating electrode (FHC, Inc) was placed on the Schaffer collaterals of the CA3 and a recording electrode was placed at least 300 µm away on the striatum radiatum of the CA1. Test stimuli were delivered once every 20 s (0.05 Hz) and the stimulus intensity was adjusted to produce a baseline fEPSP of 30–40% of the maximal response of the initial slope of fEPSP. Aliquots (0.5 ml) of artificial CSF-B brain extracts or 40 μl of SEC isolated plaque amyloid-β were added to an artificial CSF reservoir to achieve a final volume of 10 ml, and the resulting solutions were perfused over slices for at least 30 min. LTP was induced by theta burst stimulation (TBS), composed of three trains, each of four pulses delivered at 100 Hz, 10 times, with an interburst interval of 200 ms with a 20-s interval between each train. Field potentials were recorded using a Multiclamp amplifier (Multiclamp 700B; Molecular Devices) coupled to a Digidata 1440A digitizer. Signal was sampled at 10 kHz and filtered at 2 kHz and data were analysed using Clampex 10 software (Molecular Devices, Sunnyvale, CA).
Amyloid plaque isolation and amyloid-β purification
Amyloid plaques were isolated from Alzheimer’s disease brain as described previously (Selkoe et al., 1986). Whole hemispheres were thawed on ice and 50–100 g of cortical grey matter isolated. Tissue aliquots of ∼50 g were incubated with five volumes of 2% (w/v) SDS in 50 mM Tris-HCl, pH 7.6, containing 0.1 M β-mercaptoethanol at room temperature for 2 h. Thereafter, the suspension was Dounce homogenized and the resultant homogenate was boiled in a water bath for 15 min. When samples had cooled to room temperature they were passed through a 112-µm nylon mesh and the flow-through collected. This material was centrifuged at 300g for 30 min and the pellet collected and washed three times with 0.1% SDS in 150 mM NaCl. The final pellet was Dounce homogenized and then passed through a 38-µm nylon mesh and the flow-through applied to a preformed sucrose step gradient composed of layers of 1.2 M, 1.4 M, 1.6 M, and 1.8 M sucrose in 1% SDS, 50 mM Tris-HCl, pH 7.6. This preparation was then centrifuged at 72 000g for 1 h at 26°C in a SW28 rotor (Beckman Coulter). Typically, most amyloid plaques are found in the 1.6 M sucrose interface and this material was collected and washed with five volumes of 0.1% SDS in 150 mM NaCl. Plaques were then pelleted by centrifugation at 300g in a benchtop centrifuge for 30 min. To remove SDS, plaques were washed twice with 1 ml Milli-Q® water, and the pelleted plaques were resuspended in 100 µl Milli-Q® water. A small portion of this material (10 µl) was stained with 0.2% Congo red and visualized using polarized microscopy.
The remaining plaque suspension was lyophilized and then incubated in 88% formic acid for 12–14 h with gentle agitation. Insoluble material was removed by centrifugation at 12 000g for 15 min and the upper 90% of supernatant was used for SEC as described above. Fractions (0.5 ml) were collected and a small portion (5 μl) was taken for western blot analysis. Based on western blot results, fractions containing 7kDa-Aβ and amyloid-β monomers were pooled and used for further analysis. For bioactivity experiments approximately equal concentrations (estimated from western blots) of 4kDa-Aβ and 7kDa-Aβ were diluted directly into artificial CSF or neuronal medium and used for LTP and live-neuron video microscopy, respectively. For mass spectrometry, samples were split into 1–12 replicates in 1.5 ml polypropylene tubes, lyophilized and shipped to Gothenburg.
Reconstitution of plaque-derived amyloid-β
Lyophilized 4kDa-Aβ and 7kDa-Aβ were reconstituted in 100 µl (corresponding to the maximum injection volume) 1% ammonium hydroxide/20% acetonitrile in water (v/v/v) vortexed briefly and shaken vigorously for 30–60 min. The resulting solution was then analysed using microflow LC-MS/MS and alkaline mobile phase. The portion of the 4kDa-Aβ and 7kDa-Aβ fractions reconstituted varied from 1/12 to 1/1 of the whole available amount from the SEC fractionations.
In-solution enzymatic digestion of plaque-derived amyloid-β
Shortly before mass spectrometric analysis, samples were digested with Trypsin/Lys-C Mix (Mass Spec Grade, Promega). Dried eluates were dissolved in 20 µl 50 mM ammonium bicarbonate (prepared biweekly by adding 395 mg to 100 ml water) containing 30 mM dithiothreitol (prepared freshly by adding ∼540 µl of 50 mM ammonium bicarbonate to ∼2.5 mg dithiothreitol in a 1.5 ml polypropylene tube) and agitated for 30 min at 60°C. Solutions were then cooled to room temperature and 20 µl 70 mM iodacetamide (prepared freshly by adding 50 mM ammonium bicarbonate to ∼10 mg iodacetamide to ∼770 µl in a 1.5 ml polypropylene tube) was added and the solution agitated in darkness for 30 min. Thereafter, 20 µl 0.08 µg/µl trypsin/Lys-C (prepared freshly by adding 250 µl of 50 mM ammonium bicarbonate to the supplied glass vial containing the enzyme) was added and gently mixed overnight at 37°C. The entire solution (60 µl) was then transferred to a liquid chromatography vial and used for nanoflow LC-MS/MS. Alternatively, 50 mM ammonium bicarbonate was substituted for 0.1% ammonium hydroxide at all stages [prepared daily by adding 4 ml 25% (concentrated) ammonium hydroxide to ∼20 ml water, then adding 20 ml acetonitrile, and finally adding water to a total volume of 100 ml].
LC-MS/MS
Two different liquid chromatography setups were used, one configured for flow rates down to 50 µl/min and one configured for flow rates below 1.5 µl/min. The latter configuration was not suitable for use with alkaline mobile phases, while both worked under acidic conditions.
Analysis of undigested plaque extracts was carried out with alkaline solvents using a microflow LC coupled to electrospray ionization (ESI) quadrupole–orbitrap MS and MS/MS. This was carried out using a Dionex Ultimate 3000 system (solvent rack SRD-3600, pump NCS-3400RS, autosampler WPS 3000TRS, column oven TCC 3000RS) coupled to a Q Exactive (both Thermo Fisher Scientific) equipped with a HESI-II ion source, using a setup based on a reference measurement procedure for amyloid-β quantification (Leinenbach et al., 2014). For analysis with alkaline mobile phases a reverse-phase monolithic ProSwift RP-4H column (length 25 mm, i.d. 1.0 mm, Thermo Fisher Scientific) was used for separation at a flow rate of 300 µl/min and a linear gradient of 0–50% B for 20 min. Mobile phase A consisted of 0.075% ammonium hydroxide/5% acetonitrile in water (v/v/v) while mobile phase B was 0.025% ammonium hydroxide/95% acetonitrile in water (v/v/v); both mobile phases were prepared daily.
Mass spectra of undigested samples (50–100 µl injected) were acquired in positive ion mode with a voltage setting of +4.4 kV and a resolution setting of 70 000; target values were 106 and ion injection times were 250 ms both for MS and MS/MS acquisitions. Acquisitions were performed with one microscan per acquisition. Precursor isolation width was 3 m/z units. Only singly charged ions were deselected for fragmentation. The instrument was operated in data-dependent mode so that each precursor ion scan was followed by five fragment ion scans of the five most intense ions fragmented by so-called higher energy collision induced dissociation (HCD) at a normalized collision energy (NCE) of 25. Minimum AGC target was 2 × 104, peptide match was set to ‘off’, and Exclude isotopes to ‘on’. Selected m/z were excluded for 3 s until eligible again. Other settings depended on sample type. For monomer fractions the m/z range was 400–2000 to cover possible charge states. Monomer fractions from several samples were also analysed under acidic conditions in the same way as described for the digested samples (see below).
The ∼7 kDa fractions were analysed in a variety of ways although most of the settings were the same as for the monomer fractions; the differences are outlined as follows: the standard m/z range was set to 1200–2000 to minimize background ions. The same fragmentation settings as for monomer fractions normally were used, but also other NCE settings as well as a parallel reaction monitoring approach with every second acquisition set to acquire MS/MS data of the possible 6+ ion of Aβ1–42×1–42 with intensity peaking at m/z 1502.76. For this approach the isolation window was four mass units; all other parameters were the same as in the data-dependent mode.
Analysis of digested samples were carried out using acidic solvents and a nanoflow LC coupled to ESI quadrupole–orbitrap MS and MS/MS. In this case a different Dionex Ultimate 3000 system (solvent rack SRD-3400, pump NCS-3500RS, autosampler WPS-3000 TPLRS) was coupled to a Q Exactive™ equipped with a Nanospray Flex™ ion source. Analysis under acidic conditions was carried out with a reversed-phase Acclaim™ PepMap™ C18 (length 20 mm, i.d. 75 µm, particle size 3 µm, pore size 100 Å) trap column used for online desalting and sample clean-up, followed by a reverse-phase Acclaim™ PepMap™ RSLC C18 (length 150 mm, i.d. 75 µm, particle size 2 µm, pore size 100 Å, both Thermo Fisher Scientific) column. Separation was performed at a flow rate of 300 nl/min by applying a linear gradient of 3–40% B for 50 min. Mobile phase A was 0.1% formic acid in water (v/v) and mobile phase B was 0.1% formic acid/84% acetonitrile in water (v/v/v).
Mass spectra of digested samples (6 µl injected) were acquired in positive ion mode with a voltage setting of +1.7 kV and a resolution setting of 70 000. Target values were 106 and ion injection times were 250 ms both for MS and MS/MS acquisitions. Typical acquisitions were performed with an m/z range of 300–1800 with one microscan per acquisition. Precursor isolation width was 3 m/z units. Singly charged ions and ions with undetermined charge state were deselected for fragmentation. The instrument was operated in data-dependent mode so that each MS acquisition was followed by five fragment ion acquisitions of the five most intense ions, which were fragmented by HCD at an NCE of 25. Minimum AGC target was 2 × 104, Peptide match was set to ‘off’, and Exclude isotopes to ‘on’. Selected m/z were excluded for 5 s until eligible again.
Data analysis
Database searches were performed using several different software programs. For spectra from digested samples Proteome Discoverer v2.1 (Thermo Fisher Scientific) and Peaks Studio v8.5 (Bioinformatics Solutions, Inc) were used. For spectra from non-digested samples, which required redetermination of the precursor ion monoisotopic m/z and charge-deconvolution of the peaks, Mascot Daemon v2.6.0 combined with Mascot Distiller v.2.6.3 (both Matrix Science) as well as PEAKS Studio v8.5 were used. For Proteome Discoverer and Mascot Deamon/Distiller, the searches were submitted to the in-house Mascot database server (v2.6.1) while for PEAKS Studio the search feature was built-in. Finally the Kojak feature of the Trans-Proteomic Pipeline (TPP) (Keller et al., 2005; Deutsch et al., 2010) was used in the search of cross-linked species. Parameter settings for the software programs are provided in the Supplementary material .
For intact amyloid-β mass determinations we also used the instrument-supplied analysis software Xcalibur Qual Browser’s built-in deconvolution function, Xtract, and again parameter settings were varied. Typical settings are given in the Supplementary material . Mass lists generated by Xtract and PEAKS Studio 8.5 differed slightly. To generate the deconvoluted spectra and peak lists of the intact dimers presented in the figures and tables the two outputs were combined (see Supplementary material for details).
Data analysis required extensive manual evaluation since we were unable to find a software program that could identify the extensive array of fragment types possible for amyloid-β dimers. Therefore, in-house scripts were used to produce theoretical fragment lists, which were then compared to obtained acquisitions. The very low background in orbitrap data allows for good signal-to-noise ratios even when the signal is low.
Statistical analysis
Electrophysiological data were analysed offline by pCLAMP 10.2 (Molecular Devices) and tested with one-way analysis of variance (ANOVA) with Bonferroni post hoc tests or Student t-tests. For live-cell imaging experiments, differences between groups were tested with ANOVA with Bonferroni post hoc tests or Student t-tests.
Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material .
Results
Numerous studies have shown that solubilized amyloid plaques and aqueous extracts of Alzheimer’s disease brain contain two major amyloid-β bands when analysed by SDS-PAGE (Masters et al., 1985; Enya et al., 1999; McLean et al., 1999; Morishima-Kawashima et al., 2000; Shankar et al., 2008; Mc Donald et al., 2010; Lesne et al., 2013; Watt et al., 2013). The faster migrating species are centred around 4 kDa and the other at ∼7 kDa. In pilot studies we reported that 7kDa-Aβ isolated from the aqueous phase of a single Alzheimer’s disease brain possessed Alzheimer’s disease-relevant toxic activity (Shankar et al., 2008). However, the molecular identity of 7kDa-Aβ is controversial (Watt et al., 2013; Willem et al., 2015) and definitive identification has been hampered due to the low abundance of 7kDa-Aβ. Here, we used a combination of live-cell imaging, LTP and advanced mass spectrometric techniques to characterize the 4kDa-Aβ and 7kDa-Aβ species isolated from the aqueous phase of human brain and from solubilized amyloid plaques.
Bioactive aqueous extracts of Alzheimer brains contain amyloid-β monomers and 7kDa-Aβ
Initial experiments focused on amyloid-β species extracted from the aqueous phase of human brain. In accord with our prior reports (Shankar et al., 2008; Mc Donald et al., 2010, 2015; Hong et al., 2018), immunoprecipitation/western blot analysis of artificial CSF extracts from end-stage (Fig. 1A) and mild Alzheimer’s disease (Supplementary Fig. 2 ) brains revealed two broad bands, one consistent with the 4kDa-Aβ monomer, and the other with 7kDa-Aβ. This pattern is comparable with that seen in a recent study in which we used passive diffusion instead of mechanical homogenization (Hong et al., 2018). When applied to cultured iPSC-derived human neurons, Alzheimer’s disease brain extracts caused a time-dependent loss of neuritic complexity (Fig. 1B, for analysis of data collected over the last 6 h of recordings: brain 1242-mock versus control, P < 0.001; brain 722-mock versus control, P < 0.001; one-way ANOVA; Supplementary Fig. 1 ). Such extracts also impaired LTP (Fig. 1C, statistical comparison for the last 10 min of recording: brain 722-mock versus control, P < 0.001; brain 722-mock versus immunodepleted, P < 0.001; one-way ANOVA), an electrophysiological correlate of learning and memory (Bliss et al., 2003). In both cases depletion of amyloid-β from brain extracts prevented these adverse effects (Fig. 1B and C).
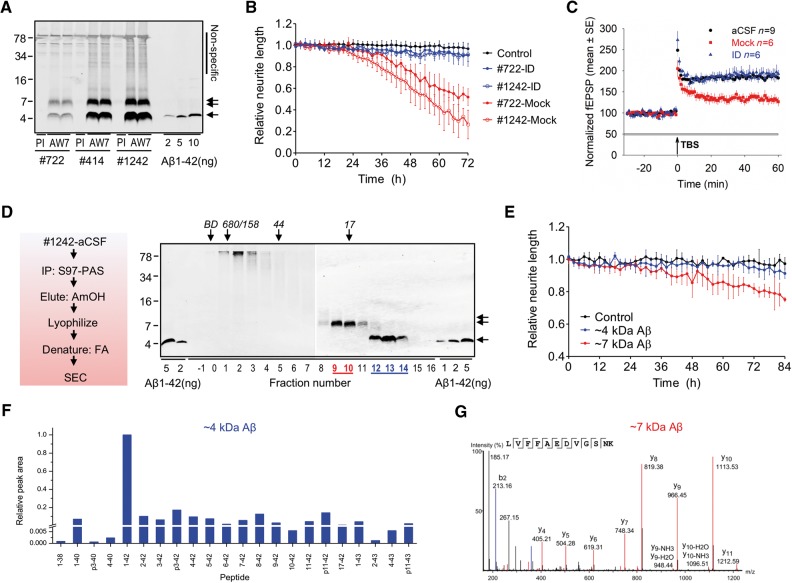
Figure 1.
Bioactive aqueous extracts of Alzheimer’s disease brains contain amyloid-β monomers and 7kDa-Aβ species. (A) Artificial CSF extracts of Alzheimer’s disease brains (brain 722, 414 and 1242) contain amyloid-β species that migrate on SDS-PAGE at ∼4 kDa (single arrow) and ∼7 kDa (double arrow). Brains were immunoprecipitated with the pan anti-amyloid-β antiserum, AW7, or pre-immune serum (PI) and blotted with the mid-region anti-amyloid-β monoclonal antibody, 4G8. Non-specific bands detected when pre-immune serum was used are indicated on the right, and migration of molecular weight marker is on the left. (B) Time-course plots show that amyloid-β-containing artificial CSF extracts of brains 1242 (red open circles) and 722 (red filled circles) cause neurito-toxicity compared to the medium-alone control (black) (last 6 h; brain 1242-mock versus control, P <0.001; brain 722-mock versus control, P < 0.001; one-way ANOVA), or brain extracts immunodepleted (ID) of amyloid-β with purified AW7 antiserum; brain 1242-ID (blue open circles) and brain 722-ID (blue filled circles) (last 6 h; brain 1242-mock versus immunodepleted, P < 0.001; brain 722-mock versus immunodepleted, P < 0.001; one-way ANOVA). (C) The amyloid-β-containing artificial CSF extract of brain 722 (red) blocks LTP compared to artificial CSF vehicle control (black) or brain 722-ID (blue) (last 10 min; brain 722-mock versus control, P < 0.001; brain 722-mock versus immunodepleted, P < 0.001; one-way ANOVA). (D) Artificial CSF extract of brain 1242 was immunoprecipitated with AW7 beads, antigen eluted with 500 mM ammonium hydroxide, the eluate lyophilized, the lyophilizate dissolved in formic acid and the resulting solution subjected to SEC, and fractions used for western blotting with 4G8. (E) The amyloid-β monomer and 7kDa-Aβ fractions shown in D were exchanged into iPSC-derived human neuron culture medium and added to iPSC-derived human neuron cells. 7kDa-Aβ, but not amyloid-β monomer, caused time-dependent neurito-toxicity (last 6 h; 7kDa-Aβ versus control, P < 0.001; 4kDa-Aβ versus control, P > 0.05; one-way ANOVA). (F) Liquid chromatography (LC)-mass spectrometry (MS) analysis of SEC fractions containing the 4kDa-Aβ species detected 22 different amyloid-β primary structures. Data are shown as relative abundance of detected peaks, with the most abundant peak (Aβ1–42) set to a value of 1 (see Supplementary Fig. 6 for an LC-MS trace). (G) MS/MS analysis of trypsin/LysC-digested SEC fractions containing the ∼7 kDa species confirmed the presence of amyloid-β residues 17–28.
Using the same antibody that was effective in removing bioactivity from brain extracts we isolated 4kDa-Aβ and 7kDa-Aβ and tested their activity. Artificial CSF-B extract of Alzheimer’s disease brain 1242 was immunoprecipitated with AW7 and the antibody-bound material eluted with ammonium hydroxide. The elute was then lyophilized, denatured with formic acid and the resulting material size-separated using SEC. A portion of each fraction was used for western blotting. Three 4G8-reactive bands were detected: (i) material that eluted just after the void (fractions 1–3) and migrated at high molecular weight on 16% polyacrylamide SDS gels; (ii) SEC low molecular weight species (fractions 9–10) that migrated at ∼7 kDa on 16% polyacrylamide SDS gels; and (iii) SEC low molecular weight species (fractions 12–14), which migrated at ∼4 kDa on 16% polyacrylamide SDS gels (Fig. 1D).
Importantly, when SEC-isolated synthetic Aβ1–42 was incubated in either artificial CSF-B buffer or artificial CSF-B extract from a control brain and subjected to our immunoprecipitation/lyophilization/SEC purification process, peptide was recovered only in fractions 11–14 (Supplementary Fig. 3 ). These data demonstrate that significant amounts of 7kDa-Aβ are not generated as a result of the immunopurification procedure, and that this control brain lacks detectable 7kDa-Aβ.
Fractions containing peak 4kDa-Aβ and 7kDa-Aβ species were each pooled and exchanged into iPSC-derived human neuron medium. The amount of 4G8-reactive 4kDa-Aβ and 7kDa-Aβ species was quantified by LiCor imaging and equal concentrations (∼100 ng/ml) of 4kDa-Aβ and 7kDa-Aβ were applied to iPSC-derived human neuron cells and the effects of treatments monitored using video microscopy. Neurons treated with 7kDa-Aβ (red) caused a time-dependent decrease of neurite length, whereas amyloid-β monomers (blue) had no effect (Fig. 1E). When neuritic complexity was monitored by quantifying the number of neuritic branch points, again only the 7kDa-Aβ (red) had a negative effect (Supplementary Fig. 4A ).
To confirm that toxicity mediated by 7kDa-Aβ fractions was indeed attributable to amyloid-β and not a non-amyloid-β species, we tested the effect of ∼7 kDa fraction ± immunodepleted with AW7. Reassuringly, the mock-treated ∼7 kDa fraction impaired neuritic integrity, whereas the AW7-treated fraction did not (Supplementary Fig. 5 ).
In parallel experiments, large volumes (15 ml) of artificial CSF-B extracts of brain 1185 were immunoprecipitated with AW7 and size separated as in Fig. 1D. The resulting 4kDa-Aβ and 7kDa-Aβ (Supplementary Fig. 6A ) were then used for LC-MS. Preliminary experiments using a covalently cross-linked dimer and SEC-isolated brain-derived 4kDa-Aβ revealed that a weakly alkaline water-acetonitrile LC gradient allowed better recovery of amyloid-β than using mobile phases with acidic solvents. Application of this approach to SEC-isolated 4kDa-Aβ allowed detection of a large array of amyloid-β primary structures. For the ∼4 kDa fraction of Alzheimer’s disease brain 1185 (Supplementary Fig. 6 ) a total of 22 different primary structures were detected (Fig. 1F). However, when 7kDa-Aβ was analysed neither MS nor MS/MS allowed detection of amyloid-β.
We hypothesized that the difficulty in detecting 7kDa-Aβ by LC-MS could reflect a high level of molecular heterogeneity (Roberts et al., 2012), i.e. the ∼7 kDa band detected by western blot may be composed of multiple primary structures such that the amounts of individual components are below the level of detection by LC-MS. If this were the case, we reasoned that digesting 7kDa-Aβ with trypsin/Lys-C would generate common fragments that would be at higher concentrations than their individual precursors. Importantly, when 7kDa-Aβ from brain 1185 was digested with trypsin/Lys-C and analysed using LC-MS/MS a peptide fragment corresponding to Aβ17–28 was detected (Fig. 1G). These data are consistent with the recognition of 7kDa-Aβ by a variety of anti-amyloid-β antibodies (Mc Donald et al., 2015) (Fig. 2) and provide the first mass spectrometric evidence that 7kDa-Aβ contains at least the mid- and C-terminal region sequence of amyloid-β. However, the anticipated tryptic fragments from the N-terminal half (Aβ1–5 and Aβ6–16) were not well detected.
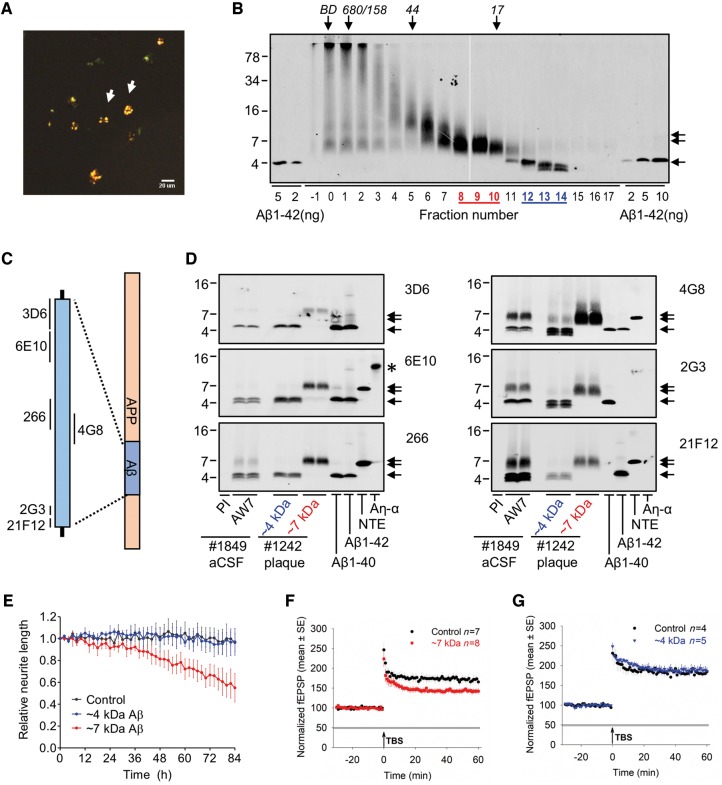
Figure 2.
Aqueous extracts of Alzheimer’s disease brains and solubilized amyloid plaques contain similar bioactive 7kDa-Aβ species. (A) Congo red-positive amyloid plaques were isolated from Alzheimer’s disease brain 1242, and (B) dissolved in formic acid, chromatographed on SEC and used for western blot with 4G8. The elution of Blue dextran (BD) and globular protein standards is indicated by downward arrows and SDS-PAGE molecular weight standards are on the left. (C and D) Plaque SEC-isolated ∼4 and ∼7 kDa fractions, and AW7 immunoprecipitation of artificial CSF extracts were used for western blotting using six different anti-amyloid-β monoclonal antibodies. Synthetic Aβ1–40, Aβ1–42 and recombinant NTE-Aβ (−31A-β1–42) and Aη-α peptides were loaded as controls. Single arrow and double arrows indicate the 4kDa-Aβ and 7kDa-Aβ species, respectively, and an asterisk marks the position of Aη-α. The 7kDa-Aβ species from artificial CSF extracts and solubilized plaques co-migrate and exhibit similar immunoreactivity anti-amyloid-β monoclocal antibodies. (E) Time-course plots show that brain 1242 plaque-derived 7kDa-Aβ species (red) causes neurito-toxicity, whereas brain 1242 plaque-derived amyloid-β monomer (blue) is indistinguishable from the medium alone control (black) (last 6 h; 7kDa-Aβ versus control, P < 0.001; 4kDa-Aβ versus control, P > 0.05; one-way ANOVA). (F and G) Time-course plots show that brain 1444 plaque-derived 7kDa-Aβ (red), but not amyloid-β monomer (blue), blocks LTP compared to artificial CSF control (black) (last 10 min; 7kDa-Aβ versus control, P < 0.05; 4kDa-Aβ versus control, P > 0.05; one-way ANOVA).
Aqueous extracts of Alzheimer brains and solubilized plaques contain similar bioactive 7kDa-Aβ species
As plaques are known to contain 7kDa-Aβ, but at much higher quantities than 7kDa-Aβ in aqueous Alzheimer’s disease brain extracts, we sought to systematically compare the properties and activity of 7kDa-Aβ from these two sources. Using a well-established method for isolating plaques from human cortex (Selkoe et al., 1986), we obtained microgram quantities of Congo red-positive plaques (Fig. 2A) from six Alzheimer’s disease brains (Table 2). Plaques were then solubilized with formic acid, and size-separated using SEC. A portion of each fraction was used for western blotting and the identified 7kDa-Aβ (fractions 8–10; Fig. 2B) and monomer fractions (12–14; Fig. 2B) were each pooled and further analysed alongside artificial CSF extracts of Alzheimer’s disease brain 1849 (Fig. 2C and D).
Plaque-derived material from brains 1242 (Fig. 2B) and 722 (Supplementary Fig. 7A ) were mixed with 2× SDS-PAGE sample buffer and electrophoresed alongside AW7 immunoprecipitation of artificial CSF extracts of brains 1849 and 453 (Fig. 2D, Supplementary Fig. 8B and C) and a battery of six different anti-amyloid-β antibodies was used to identify epitopes present on plaque-derived and aqueous-soluble 7kDa-Aβ. Synthetic Aβ1–40 and Aβ1–42, and recombinant NTE-Aβ (−31Aβ1–42) and Aη-α peptides were included to verify the specificity of antibodies and to provide relevant molecular weight markers (Fig. 2D, Supplementary Fig. 8B and C). Plaque-derived and aqueous-soluble ∼4 kDa bands each resolved into three different components, the recognition of which depended on the antibody used (Fig. 2D and Supplementary Fig. 8B ). Monoclonal antibody 3D6 (which requires a free Asp1 on amyloid-β) detected a single band in both sources of monomer, whereas the mid-region antibody, 4G8 and the C-terminal antibodies, 2G3 and 21F12 often detected two additional faster migrating bands. Monomer from plaques and aqueous brain showed similar immunoreactivity with the antibodies tested, but the amounts differed.
7kDa-Aβ from aqueous extracts and plaques also appeared highly similar, and was best recognized by mid-region (266 and 4G8) and C-terminal (2G3 and 21F12) antibodies (Fig. 2D and Supplementary Fig. 8B ). To investigate whether 7kDa-Aβ contained APP sequence N-terminal of amyloid-β Asp1 we used the anti-APP antibodies 28D10 and 2E9. 28D10 and 2E9 recognize APP sequences 585–600 and 545–555 (695 numbering), respectively. Both antibodies detected recombinant Aη-α, but not brain-derived 4kDa-Aβ or 7kDa-Aβ, although these species were readily detected when blots were reprobed with 4G8 (Supplementary Fig. 8C ).
Having shown that 7kDa-Aβ from aqueous extracts and plaques co-migrate on SDS-PAGE, and share several common epitopes, we proceeded to investigate their bioactivity. When applied to iPSC-derived human neurons at a concentration comparable to that used for aqueous 7kDa-Aβ (∼100 ng/ml; Fig. 1E) plaque-derived 7kDa-Aβ from two different Alzheimer’s disease brains (brains 1242 and 722) decreased neurite length by at least 20% (Fig. 2E and Supplementary Fig. 7B ). Specifically, over the last 6 h of recording 7kDa-Aβ caused a significant decrease in neurite length relative to pretreatment values, and compared to the vehicle control (P < 0.001). In contrast, concentration-matched plaque-derived amyloid-β monomer had no effect (Fig. 2E and Supplementary Fig. 7B ). Similar results were obtained when branch points were analysed (Supplementary Figs 4B and 7C ). LTP experiments require much larger sample volumes than used for video microscopy, therefore we specifically generated large batches of plaque-derived 4kDa-Aβ and 7kDa-Aβ (Supplementary Fig. 9 ) to test on LTP. When used at ∼72 ng/ml brain 1444 plaque-derived 7kDa-Aβ significantly blocked LTP, whereas concentration-matched amyloid-β monomer had no effect (7kDa-Aβ versus control, P < 0.05; 4kDa-Aβ versus control, P > 0.05; one-way ANOVA; Fig. 2F and G). Collectively, these results demonstrate that plaque-derived 7kDa-Aβ and aqueous 7kDa-Aβ have similar biochemical, immunological, and toxic properties, thus indicating that their molecular structure is the same, or highly similar.
Solubilized plaques contain covalently cross-linked amyloid-β heterodimers and a diverse mixture of amyloid-β monomers
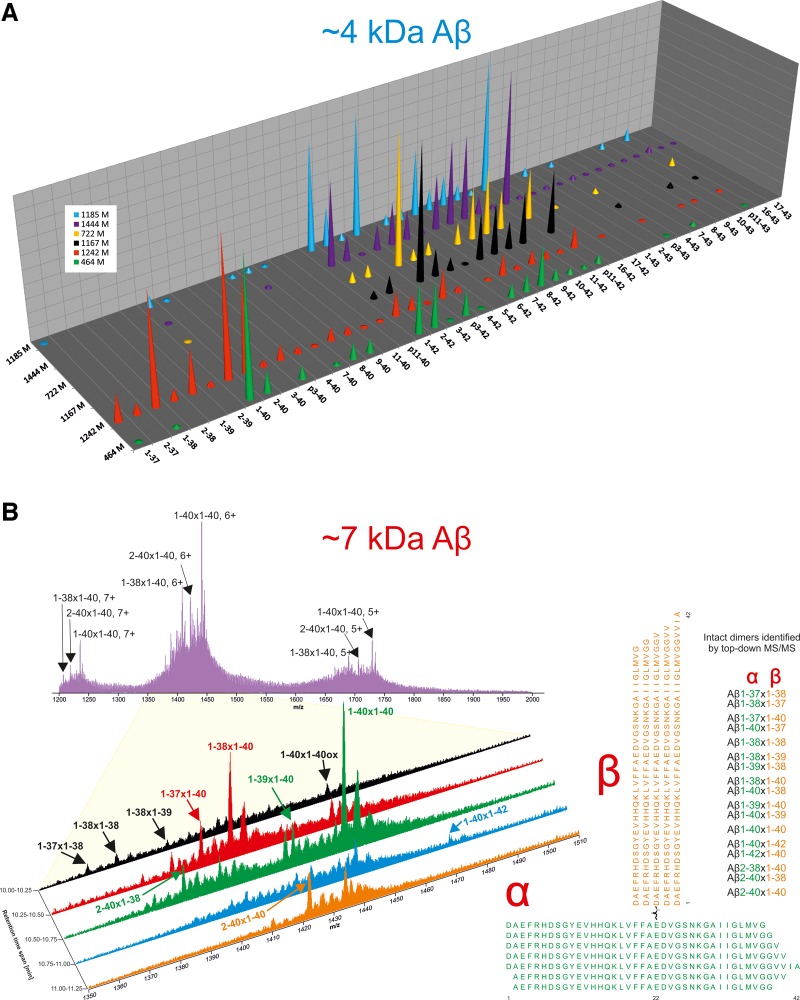
As it had been relatively straight forward to obtain mass estimates for undigested aqueous 7kDa-Aβ using LC-MS/MS and mildly alkaline solvents (Fig. 1F), we began our analysis of plaque-derived amyloid-β using the same approach. MS/MS analysis of SEC-isolated 7kDa-Aβ from six different brains identified a total of 36 amyloid-β primary structures. Their relative abundance is shown in Fig. 3A and expanded LC-MS traces are shown in Supplementary Fig. 10 . Many peptides were detected in all brains, but there were notable differences between samples. Specifically, there was clear segregation between brains in which the predominant peptides terminated at Ala42 versus Val40. In four brains (brains 1185, 722, 1167 and 1444) the most abundant peptides terminated at Ala42, whereas in the other two brains (brains 1242 and 464) peptides terminating at Val40 were more common. In these latter two brains, peptides terminating at Val39, Gly38 and Gly37 were also evident. These differences in Ala42/Val40 content are also reflected in western blot analysis of brains 1242 and 722 (Fig. 2 and Supplementary Fig. 8 ). We identified amyloid-β peptides with 15 different N-termini, and many of the detected peptides, except the previously elusive p3 fragment (Aβ17–42), had been reported in prior studies (Mori et al., 1992; Miller et al., 1993; Portelius et al., 2010). In addition, peptides terminating at Thr43, oxidized peptides, as well as occasional shorter peptides were also detected (Supplementary Table 1 ).
Figure 3.
Solubilized amyloid plaques contain covalently cross-linked amyloid-β heterodimers and a diverse mixture of amyloid-β monomers. (A) LC-MS detects a rich array of amyloid-β primary structures in SEC-isolated monomer fractions of solubilized plaque. Results are for monomers isolated from six brains (brains 1185, 1444, 722, 1167, 1242 and 464). The height of cones corresponds to the relative abundance of detected peaks, with the most abundant peak in each sample set to a value of 1. The identity of the peaks shown were confirmed by tandem MS/MS (Supplementary Table 1 ). (B) LC-MS of the ∼7 kDa fractions from brain 1242 reveals the presence of amyloid-β species consistent with covalently cross-linked heterodimers. The upper panel shows the average data for the retention time span 10–12 min and the magnified panel shows five 0.25-min retention time spans. The right panel illustrates structures of amyloid-β dimers consistent with the identified masses. Approximately 10% of the respective SEC-isolated fractions was used for LC-MS and MS/MS analysis.
Consistent with our analysis of aqueous 7kDa-Aβ (Fig. 1), mass spectrometry analysis of plaque-derived 7kDa-Aβ proved challenging. However, it was possible to distinguish several features matching potential amyloid-β dimers. Figure 3B shows an average trace for brain 1242 over the whole m/z range and an expanded LC-MS trace shows dimers identified by MS/MS (Supplementary Table 2 ). Dimers composed of monomers terminating at Val40 were the most abundant species, but dimers containing amyloid-β terminating at Gly37, Gly38, Val39, and Ala42 were also detected. Excluding oxidized species, 10 different variants were confirmed, seven of which may occur in two orientations. The fragment pattern from these dimers are very similar to that of monomeric Aβ1–40/42 (Brinkmalm et al., 2012) long b-ions dominating the fragment ion spectrum (Supplementary Figs 11–13 and Supplementary Table 3 ), but only one of the two amyloid-β chains appeared to undergo fragmentation. For Aβ1–40×1–40 only one ladder of long b-ions is formed (Supplementary Fig. 11 ) and for Aβ1–38×1–40 two such ladders can be seen (Supplementary Fig. 12 ). This is quite different from the pattern seen with synthetically produced (Aβ1 − 42)DiY dimer, where multiple ladders were observed showing a fragmentation pattern involving both chains (Supplementary Fig. 14 and Supplementary Table 3 ). In the other brains, the mass spectrometry signals obtained were too low to give MS/MS data in the regular data-dependent mode, for which single scan MS/MS data were acquired for precursor signals over a preset threshold. However, a small but distinct increase in the 6+ ion signal at m/z 1502.76 was observed in all samples examined (Supplementary Fig. 13 ). The theoretical mono-isotopic mass (i.e. the mass of the lightest isotope combination) of an uncharged Aβ1–42×1–42 dimer formed by a loss of H2O is: 2 × 4511.270 – 18.011 Da = 9004.529 Da. Taking into account that the seventh isotope, which is ∼6 Da heavier, is one of the most abundant for this relatively large compound the predicted ion mass, including the six protons giving the charge, is ∼9016.57 Da, i.e. a perfect match for a 6+ ion with an m/z = 1502.76. To verify the potential Aβ1–42×1–42 dimer, the instrument was set to acquire MS/MS data from a four mass unit wide window centred at m/z 1502.76. Thereafter, multiple MS/MS acquisitions were summed and 7kDa-Aβ (from brain 1185) was identified as being Aβ1–42×1–42 (Supplementary Fig. 13 ). For the other four brains investigated no MS/MS data were obtained, but the LC-MS traces nevertheless provided information on the most abundant dimers. Interestingly, in brains where Aβ40 was the dominant monomer (brains 464 and 1242), the Aβ1–40×1–40 dimer was also the most prominent 7kDa-Aβ species.
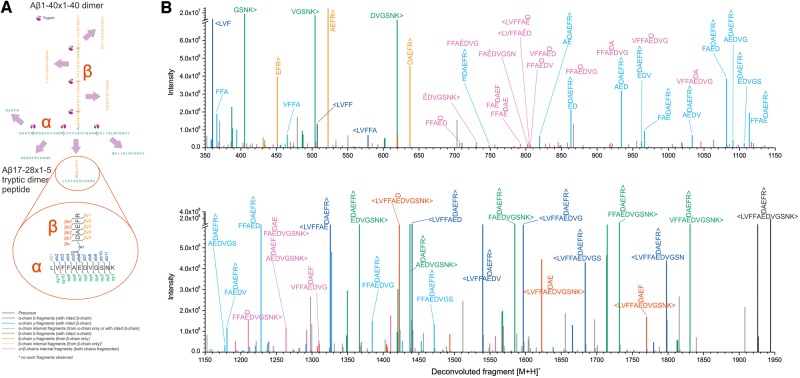
LC-MS/MS analysis of digested 7kDa-Aβ identifies a covalent link between Asp1 and Glu22
Having proven that 7kDa-Aβ is a covalently linked amyloid-β dimer, we were anxious to determine the position(s) at which the component monomers are linked. Given the highly heterogeneous primary structures evident in mass spectra for ∼7 kDa (Fig. 3B), we investigated whether proteolytic digestion might be useful to enrich common peptide fragments that contain cross-linked amino acids. When plaque-derived 7kDa-Aβ from brain 1242 was subjected to digestion with trypsin/Lys-C the dimer-specific fragment, Aβ17–28×1–5, was readily detected (see Fig. 4A for a schematic). Importantly, this fragment was found in multiple different digestion experiments using ammonium bicarbonate, and in separate digestions in ammonium hydroxide. The deconvoluted fragment spectrum shown in Fig. 4B provides compelling evidence that 7kDa-Aβ is composed of two amyloid-β chains linked between Glu22 and Asp1 (see Supplementary Fig. 15 containing the fully annotated spectrum and Supplementary Table 4 ). Analysis using three different software programs (PEAKS Studio, Mascot/Distiller, and Trans-Proteomic Pipeline/Kojak) all corroborate this conclusion. However, in view of the facts that: (i) we did not observe the Aβ17–28×1–5 fragment in samples that contained a verifiable intact dimer (e.g. brain 1185); and (ii) 7kDa-Aβ is highly heterogeneous, it seems likely that dimers may arise due to linkages at other sites besides the Glu22×Asp1 link we have confirmed. In future studies it will be important to generate and investigate the bioactivity of synthetically produced Glu22-Asp1 dimers.
Figure 4.
Amyloid-β heterodimers are covalently cross-linked and the most abundant species is linked between Asp1 and Glu22. (A) Schematic of tryptic peptides released from a putative Aβ1–40×1–40 dimer crosslinked at D1 and E22, and Aβ17–28 x 1–5 related fragments detected in B. (B) Deconvoluted fragment ion spectrum obtained from MS/MS of the tryptic peptide Aβ17–28 x 1–5 from the ∼7 kDa plaque-derived fraction of brain 1242 shows peaks from several different fragment ion types. Peaks corresponding to identified amyloid-β sequence are coloured and unassigned peaks are in grey. Approximately 1% of the SEC-isolated fraction was used for this analysis (10% for enzymatic digestion of which 10% was analysed by LC-MS and MS/MS).
Discussion
Prior work from our laboratory and others indicated that human brain extracts that contain SDS-stable 7kDa-Aβ exert a range of Alzheimer’s disease-relevant effects (Shankar et al., 2008; Freir et al., 2011; Jin et al., 2011; Larson et al., 2012; Borlikova et al., 2013; Yang et al., 2017; Hong et al., 2018). In a pilot study, we found that native 7kDa-Aβ size-isolated from the aqueous phase of a single Alzheimer’s disease brain blocked LTP (Shankar et al., 2008), and in a separate study, that immunopurified aqueous 7kDa-Aβ induced aberrant phosphorylation of tau and neuritic degeneration (Jin et al., 2011). Yet, until now the molecular composition of bioactive 7kDa-Aβ was unknown.
Here, our initial efforts focused on analysis of 4kDa-Aβ and 7kDa-Aβ that had been immuno-isolated and size-separated from bioactive aqueous Alzheimer’s disease brain extracts. LC-MS and MS/MS readily identified an array of primary structures in the amyloid-β monomer fraction, but failed to identify any interpretable signal in the 7kDa-Aβ fraction. Given our demonstration that 4kDa-Aβ contained at least 36 different primary structures (not counting oxidized variants) it is evident that a very large number of molecularly distinct dimers could be formed (Roberts et al., 2012), and that this may explain why it was not possible to detect any signal for 7kDa-Aβ. Following this logic, we reasoned that proteolytic digestion of 7kDa-Aβ would generate common fragments at concentrations approaching the sum of their individual precursors. Moreover, proteolytic peptides typically generate better signals in LC-MS than the longer endogenous peptides. Trypsin/Lys-C cleaves C-terminal of Lys and Arg residues, such that digestion of Aβ1–40 is predicted to generate four fragments: 1–5; 6–16; 17–28; and 29–40. However, cleavage of amyloid-β sequences with variable N- and C-termini will give rise to fragments with variable N- and C-termini, whereas the internal 17–28 fragment should be common to all amyloid-β species and therefore should be the most readily detected. When 7kDa-Aβ was digested with trypsin/Lys-C peptide fragments corresponding to Aβ17–28 and Aβ29–42 were indeed detected, but N-terminal fragments were not well detected. These data confirm that 7kDa-Aβ contains at least the mid-region of amyloid-β, and are consistent with 7kDa-Aβ having heterogeneous N- and C-termini, but no obvious covalent cross-links involving residues 17–28.
The difficulties to identify some of the expected fragments, e.g. 6–16, indicate that we were working close to the limit of reliable detection. Thus, the unambiguous identification of covalent links in aqueous 7kDa-Aβ will likely require improvement in the sensitivity of current mass spectrometry approaches and/or much higher concentrations of 7kDa-Aβ. As 7kDa-Aβ is present in aqueous extracts at <200 ng/g of wet weight grey matter (Mc Donald et al., 2010, 2012, 2015), we turned to another more concentrated source of 7kDa-Aβ–amyloid plaques (Masters et al., 1985; Roher et al., 1996; Sergeant et al., 2003; Shankar et al., 2008). Before pursuing the identification of plaque-derived 7kDa-Aβ, we directly compared the biochemical, immunological, and bioactive properties of this material, versus aqueous 7kDa-Aβ derived from both mild Alzheimer’s disease and end-stage Alzheimer’s disease brains. By every parameter tested 7kDa-Aβ from these three sources behaved highly similarly. Therefore, we determined that elucidating the structure of plaque-derived 7kDa-Aβ would provide insight about aqueous 7kDa-Aβ.
LC-MS analysis of plaque-derived 7kDa-Aβ from six different Alzheimer’s disease brains unequivocally demonstrated the presence of a family of covalently cross-linked amyloid-β dimers that included: 1–42 × 1–42, 1–40 × 1–40 and 1–40 × 1–38. However, because of the complexity of the spectra and the potential for confounding post-translational modifications it was not possible to unambiguously identify the sites at which the component monomers were cross-linked. Thus, we again turned to tryptic digestion to simplify the study of these heterogeneous samples. Careful analysis of MS/MS spectra identified a branched fragment in which amino acids 17–28 were linked to residues 1–5 at Glu22 and Asp1. Given that the intact mass of dimers was 18 Da less than the sum of the mass of two component monomers, it appears that the most common point of linkage arises from a condensation reaction between the alpha nitrogen of Asp1 and the carboxylic acid group of Glu22. Such cross links are uncommon, but certain conformations in which a H-bond donor or Lewis acid group are positioned near the carboxylate group may favour their formation. It is noteworthy that LC-MS analysis of undigested plaque-derived 7kDa-Aβ samples from five other brains also confirmed the presence of covalent amyloid-β dimers. However, none of these (including brain 1185 from which the identity of 1–42 × 1–42 was confirmed by MS/MS) yielded a signal corresponding to the 17–28 × 1–5 fragment. In brain 1185 a dimer formed with a net mass loss of 17 Da fits slightly better than an 18 Da loss (typical of the elimination of H2O) (Supplementary Fig. 13 and Supplementary Table 3 ). These results imply that 7kDa-Aβ is heterogeneous both with regard to the component monomers that contribute to dimers and the linkages that hold dimers together. Indeed, it has not escaped our attention that aqueous 7kDa-Aβ did not yield a 17–28 × 1–5 tryptic fragment.
Covalent dimers appear to exist both as native dimers and incorporated in fibrils (Mc Donald et al., 2015), but whether dimers are formed from free monomers and/or from neighbouring monomers packed together in the same fibril is not known. Given the disordered state of amyloid-β monomers (Baumketner et al., 2006; O'Malley et al., 2014) a richer diversity of residues would be capable of forming cross-links than in structurally restrained monomers present in fibrils (Lu et al., 2013; Colvin et al., 2016; Qiang et al., 2017). Moreover, multiple studies suggest that the formation of intramolecular anti-parallel β-sheet structures, at the mid-region and/or C-terminus of the amyloid-β molecule, may be critical to amyloid-β aggregation and that a subpopulation of β-hairpin-containing amyloid-β molecules may act as a seed or nucleus and facilitate aggregation of intrinsically disordered amyloid-β peptides (Hoyer et al., 2008; Sandberg et al., 2010; Cruz et al., 2012; Roychaudhuri et al., 2013). Thus, covalent bonds between residues at these sites might be expected to preclude fibril formation, whereas bond formation within these stretches of amyloid-β sequence would be unlikely in monomers present in fibrils. In both monomers and fibrils the N-terminus of amyloid-β is highly flexible and thus linkages within this domain might be particularly favoured. However, our data do not shed light on which residues beyond Asp1 and Glu22 may be involved.
Other possible cross-links consistent with our results from undigested 7kDa-Aβ could arise from condensation reactions involving alternate amino terminal residues besides Asp1 and the involvement of carboxylates from amino acids other than Glu22. Indeed, if dimers are formed from two monomers with N-termini beginning after Asp1 these could not contain an Asp1-Glu22 link and therefore would have to be held together by a covalent bond between other amyloid-β residues. While the mass assignment of the trypsin/Lys-C generated 17–28 × 1–5 is undisputable, this is not the case for intact dimers. A precise mass assignment relies on determination of the mono-isotopic mass either from direct measurement of the actual monoisotopic peak, or by using the distribution of the isotopic envelope. Since the spectra from undigested 7kDa-Aβ are highly complex with overlapping isotopic envelopes this cannot be done and hence there is a 1 or 2 Da uncertainty in our precursor mass estimates.
In vitro studies have shown that amyloid-β monomers can be induced to form covalent dimers by the phenolic coupling of tyrosine residues (Galeazzi et al., 1999; Al-Hilaly et al., 2013; O'Malley et al., 2014, 2016; Vazquez de la Torre et al., 2018) and the action of the enzyme transglutaminase, which can catalyse the formation of an isopeptide bond between Gln15 and Lys16 (Hartley et al., 2008; O'Malley et al., 2016). Importantly, in Alzheimer’s disease there is evidence of increased protein cross-linking involving both oxidative phenolic coupling (Hensley et al., 1998; Al-Hilaly et al., 2013) and transglutaminase-mediated isopeptide formation (Wilhelmus et al., 2009). However, mass spectrometry of digested 7kDa-Aβ did not identify the 6–16 × 6–16 fragments that should be derived from diytrosine or isopeptide cross-linked amyloid-β species. Of course, the absence of proof is not the proof of absence, and further investigations using a larger collection of brains will be required to define the array of cross-links that facilitate amyloid-β dimer formation.
Prior studies that used synthetic amyloid-β cross-linked at different sites suggest that different dimers may share some properties, but also exhibit important differences (reviewed in Vazquez de la Torre et al., 2018). For instance, dimers tend to aggregate more rapidly and form more intermediate-sized aggregates than monomers, but the type of aggregates formed are quite different (O'Malley et al., 2016). How these properties affect bioactivity has not been thoroughly investigated and whether all molecularly distinct dimers, or only a subpopulation are toxic will require further study. Moreover, it is clear that under certain circumstances, bands migrating at ∼7 kDa can arise as artefacts (Hepler et al., 2006; Watts et al., 2014). Thus, at this stage we cannot determine which specific component(s) of 7kDa-Aβ fractions is/are neurotoxic.
Our unambiguous demonstration that covalent amyloid-β dimers are present in human brain and that at least some fraction of this material has disease-relevant bioactivity opens up a completely new area of Alzheimer’s disease research. For instance, while there have been some suggestions that the levels of amyloid-β dimers, or antibodies that recognize amyloid-β dimers are elevated in Alzheimer’s disease CSF and blood (Moir et al., 2005; Klyubin et al., 2008; Villemagne et al., 2010), measurement of amyloid-β dimers in human biofluids has not been systematically investigated. Clearly it will be important to generate dimer-specific antibodies with a preference for those dimers proven to be toxic. Similarly, while recent results from anti-amyloid-β immunotherapy trials have been encouraging (Sevigny et al., 2016; Swanson et al., 2018), it will be important to assess whether test antibodies can engage with toxic amyloid-β dimers. Moreover, effort should be devoted to understand the process(es) by which amyloid-β dimers are formed and whether these can be modulated.
Supplementary Material
Acknowledgements
We thank Dr Tracy Young-Pearse for the gift of virally infected iPSCs, and Drs Frederique Bard and Christian Haass for providing antibodies. We thank Dr Jessica Mc Donald for conducting preliminary experiments and Ms Ali Desousa for helping establish the plaque purification procedure. We also thank Dr Malgorzata Rozga for valuable discussions regarding possible peptide links and Dr Magnus Palmblad for introducing G.B. to TPP/Kojak software package.
Glossary
Abbreviations
- 4/7kDa-Aβ =
4/7 kDa amyloid-β; iPSC = induced pluripotent stem cell
- LTP =
long-term potentiation
- SDS-PAGE =
sodium dodecyl sulphate polyacrylamide gel electrophoresis
- SEC =
size exclusion chromatography
Funding
This work was supported by grants to D.M.W. from Science Foundation Ireland (Grant 08/1N.1/B2033), the National Institute on Aging (AG046275), Bright Focus, the Swedish Alzheimer Foundation, the Swedish Brain Foundation, the Swedish Research Council and the European Research Council. H.Z. is a Wallenberg Academy Fellow. K.B. holds the Torsten Söderberg Professorship at the Royal Swedish Academy of Sciences. D.M.W. is an Alzheimer Association Zenith Fellow. G.B. was funded by Old Maidservant Foundation for this project.
Competing interests
None of the authors have biomedical financial interests or potential conflicts of interest related to the work performed in the present study. Unrelated to the current study, K.B. has served as a consultant or at advisory boards for Alzheon, BioArctic, Biogen, Eli Lilly, Fujirebio Europe, IBL International, Merck, Novartis, Pfizer, and Roche Diagnostics, and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg. Unrelated to this study, H.Z. has served at scientific advisory boards for Eli Lilly, Roche Diagnostics, Wave, Samumed and CogRx, has received travel support from Teva and is a co-founder of Brain Biomarker Solutions in Gothenburg AB, a GU Ventures-based platform company at the University of Gothenburg. D.M.W. is an advisor to CogRx and Regeneron, and has active collaborations with Medimmune, Sanofi, Gen2 and Roche. Since submission of this manuscript D.M.W. joined Biogen.
References
- Al-Hilaly YK, Williams TL, Stewart-Parker M, Ford L, Skaria E, Cole M, et al. A central role for dityrosine crosslinking of Amyloid-beta in Alzheimer's disease. Acta Neuropathologica Commun 2013; 1: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry AE, Klyubin I, Mc Donald JM, Mably AJ, Farrell MA, Scott M, et al. Alzheimer's disease brain-derived amyloid-beta-mediated inhibition of LTP in vivo is prevented by immunotargeting cellular prion protein. J Neurosci 2011; 31: 7259–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumketner A, Bernstein SL, Wyttenbach T, Bitan G, Teplow DB, Bowers MT, et al. Amyloid beta-protein monomer structure: a computational and experimental study. Protein Sci 2006; 15(3): 420–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TV, Collingridge GL, Morris RG. Introduction. Long-term potentiation and structure of the issue. Philos Trans R Soc Lond Ser B, Biol Sci 2003; 358: 607–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borlikova GG, Trejo M, Mably AJ, Mc Donald JM, Sala Frigerio C, Regan CM, et al. Alzheimer brain-derived amyloid beta-protein impairs synaptic remodeling and memory consolidation. Neurobiol Aging 2013; 34: 1315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmalm G, Portelius E, Ohrfelt A, Mattsson N, Persson R, Gustavsson MK, et al. An online nano-LC-ESI-FTICR-MS method for comprehensive characterization of endogenous fragments from amyloid beta and amyloid precursor protein in human and cat cerebrospinal fluid. J Mass Spectrom JMS 2012; 47(5): 591–603. [DOI] [PubMed] [Google Scholar]
- Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, et al. Atomic resolution structure of monomorphic Abeta42 amyloid fibrils. J Am Chem Soc 2016; 138: 9663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz L, Rao JS, Teplow DB, Urbanc B. Dynamics of metastable beta-hairpin structures in the folding nucleus of amyloid beta-protein. J Phys Chem B 2012; 116: 6311–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch EW, Mendoza L, Shteynberg D, Farrah T, Lam H, Tasman N, et al. A guided tour of the trans-proteomic pipeline. Proteomics 2010; 10: 1150–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enya M, Morishima-Kawashima M, Yoshimura M, Shinkai Y, Kusui K, Khan K, et al. Appearance of sodium dodecyl sulfate-stable amyloid beta-protein (Abeta) dimer in the cortex during aging. Am J Pathol 1999; 154: 271–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freir DB, Nicoll AJ, Klyubin I, Panico S, Mc Donald JM, Risse E, et al. Interaction between prion protein and toxic amyloid beta assemblies can be therapeutically targeted at multiple sites. Nat Commun 2011; 2: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeazzi L, Ronchi P, Franceschi C, Giunta S. In vitro peroxidase oxidation induces stable dimers of beta-amyloid (1-42) through dityrosine bridge formation. Amyloid 1999; 6: 7–13. [DOI] [PubMed] [Google Scholar]
- Guix FX, Corbett GT, Cha DJ, Mustapic M, Liu W, Mengel D, et al. Detection of aggregation-competent tau in neuron-derived extracellular vesicles. Int J Mol Sci 2018; 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley DM, Zhao C, Speier AC, Woodard GA, Li S, Li Z, et al. Transglutaminase induces protofibril-like amyloid beta-protein assemblies that are protease-resistant and inhibit long-term potentiation. J Biol Chem 2008; 283: 16790–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley K, Maidt ML, Yu Z, Sang H, Markesbery WR, Floyd RA. Electrochemical analysis of protein nitrotyrosine and dityrosine in the Alzheimer brain indicates region-specific accumulation. J Neurosci 1998; 18: 8126–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepler RW, Grimm KM, Nahas DD, Breese R, Dodson EC, Acton P, et al. Solution state characterization of amyloid beta-derived diffusible ligands. Biochemistry 2006; 45: 15157–67. [DOI] [PubMed] [Google Scholar]
- Hong W, Wang Z, Liu W, O'Malley TT, Jin M, Willem M, et al. Diffusible, highly bioactive oligomers represent a critical minority of soluble Abeta in Alzheimer's disease brain. Acta Neuropathol 2018; 136: 19–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer W, Gronwall C, Jonsson A, Stahl S, Hard T.. Stabilization of a beta-hairpin in monomeric Alzheimer's amyloid-beta peptide inhibits amyloid formation. Proc Natl Acad Sci USA 2008; 105: 5099–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, O'Nuallain B, Hong W, Boyd J, Lagomarsino VN, O'Malley TT, et al. An in vitro paradigm to assess potential anti-Abeta antibodies for Alzheimer's disease. Nat Commun 2018; 9: 2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin M, Shepardson N, Yang T, Chen G, Walsh D, Selkoe DJ.. Soluble amyloid beta-protein dimers isolated from Alzheimer cortex directly induce Tau hyperphosphorylation and neuritic degeneration. Proc Natl Acad Sci USA 2011; 108: 5819–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran E, De Strooper B.. The amyloid cascade hypothesis: are we poised for success or failure? J Neurochem 2016; 139 (Suppl 2): 237–52. [DOI] [PubMed] [Google Scholar]
- Keller A, Eng J, Zhang N, Li XJ, Aebersold R.. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol 2005; 1: 2005.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klyubin I, Betts V, Welzel AT, Blennow K, Zetterberg H, Wallin A, et al. Amyloid beta protein dimer-containing human CSF disrupts synaptic plasticity: prevention by systemic passive immunization. J Neurosci 2008; 28: 4231–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson M, Sherman MA, Amar F, Nuvolone M, Schneider JA, Bennett DA, et al. The complex PrP(c)-Fyn couples human oligomeric Abeta with pathological tau changes in Alzheimer's disease. J Neurosci 2012; 32: 16857–71a. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Leinenbach A, Pannee J, Dulffer T, Huber A, Bittner T, Andreasson U, et al. Mass spectrometry-based candidate reference measurement procedure for quantification of amyloid-beta in cerebrospinal fluid. Clin Chem 2014; 60: 987–94. [DOI] [PubMed] [Google Scholar]
- Lesne SE, Sherman MA, Grant M, Kuskowski M, Schneider JA, Bennett DA, et al. Brain amyloid-beta oligomers in ageing and Alzheimer's disease. Brain 2013; 136 (Pt 5): 1383–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R.. Molecular structure of beta-amyloid fibrils in Alzheimer's disease brain tissue. Cell 2013; 154: 1257–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CL, Bateman R, Blennow K, Rowe CC, Sperling RA, Cummings JL.. Alzheimer's disease. Nat Rev Dis Primers 2015; 1: 15056. [DOI] [PubMed] [Google Scholar]
- Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K.. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA 1985; 82: 4245–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Donald JM, O'Malley TT, Liu W, Mably AJ, Brinkmalm G, Portelius E, et al. The aqueous phase of Alzheimer's disease brain contains assemblies built from approximately 4 and approximately 7 kDa Abeta species. Alzheimer's Dementia 2015; 11: 1286–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Donald JM, Savva GM, Brayne C, Welzel AT, Forster G, Shankar GM, et al. The presence of sodium dodecyl sulphate-stable Abeta dimers is strongly associated with Alzheimer-type dementia. Brain 2010; 133 (Pt 5): 1328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JM, Cairns NJ, Taylor-Reinwald L, Holtzman D, Walsh DM.. The levels of water-soluble and triton-soluble Abeta are increased in Alzheimer's disease brain. Brain Res 2012; 1450: 138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, et al. Soluble pool of Abeta amyloid as a determinant of severity of neurodegeneration in Alzheimer's disease. Ann Neurol 1999; 46: 860–6. [DOI] [PubMed] [Google Scholar]
- Miller DL, Papayannopoulos IA, Styles J, Bobin SA, Lin YY, Biemann K, et al. Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer's disease. Arch Biochem Biophys 1993; 301: 41–52. [DOI] [PubMed] [Google Scholar]
- Moir RD, Tseitlin KA, Soscia S, Hyman BT, Irizarry MC, Tanzi RE.. Autoantibodies to redox-modified oligomeric Abeta are attenuated in the plasma of Alzheimer's disease patients. J Biol Chem 2005; 280: 17458–63. [DOI] [PubMed] [Google Scholar]
- Mori H, Takio K, Ogawara M, Selkoe DJ.. Mass spectrometry of purified amyloid beta protein in Alzheimer's disease. J Biol Chem 1992; 267: 17082–6. [PubMed] [Google Scholar]
- Morishima-Kawashima M, Oshima N, Ogata H, Yamaguchi H, Yoshimura M, Sugihara S, et al. Effect of apolipoprotein E allele epsilon4 on the initial phase of amyloid beta-protein accumulation in the human brain. Am J Pathol 2000; 157: 2093–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley TT, Oktaviani NA, Zhang D, Lomakin A, O'Nuallain B, Linse S, et al. Abeta dimers differ from monomers in structural propensity, aggregation paths and population of synaptotoxic assemblies. Biochem J 2014; 461: 413–26. [DOI] [PubMed] [Google Scholar]
- O'Malley TT, Witbold WM 3rd, Linse S, Walsh DM.. The aggregation paths and products of Abeta42 dimers are distinct from those of the Abeta42 monomer. Biochemistry 2016; 55: 6150–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson HA, Peers C.. Physiological roles for amyloid beta peptides. J Physiol 2006; 575 (Pt 1): 5–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portelius E, Bogdanovic N, Gustavsson MK, Volkmann I, Brinkmalm G, Zetterberg H, et al. Mass spectrometric characterization of brain amyloid beta isoform signatures in familial and sporadic Alzheimer's disease. Acta Neuropathol 2010; 120: 185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzzo D, Privitera L, Leznik E, Fa M, Staniszewski A, Palmeri A, et al. Picomolar amyloid-beta positively modulates synaptic plasticity and memory in hippocampus. J Neurosci 2008; 28: 14537–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Yau WM, Lu JX, Collinge J, Tycko R.. Structural variation in amyloid-beta fibrils from Alzheimer's disease clinical subtypes. Nature 2017; 541: 217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts BR, Ryan TM, Bush AI, Masters CL, Duce JA.. The role of metallobiology and amyloid-beta peptides in Alzheimer's disease. J Neurochem 2012; 120 (Suppl 1): 149–66. [DOI] [PubMed] [Google Scholar]
- Roher AE, Chaney MO, Kuo YM, Webster SD, Stine WB, Haverkamp LJ, et al. Morphology and toxicity of Abeta-(1-42) dimer derived from neuritic and vascular amyloid deposits of Alzheimer's disease. J Biol Chem 1996; 271: 20631–5. [DOI] [PubMed] [Google Scholar]
- Roychaudhuri R, Yang M, Deshpande A, Cole GM, Frautschy S, Lomakin A, et al. C-terminal turn stability determines assembly differences between Abeta40 and Abeta42. J Mol Biol 2013; 425: 292–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg A, Luheshi LM, Sollvander S, Pereira de Barros T, Macao B, Knowles TP, et al. Stabilization of neurotoxic Alzheimer amyloid-beta oligomers by protein engineering. Proc Natl Acad Sci USA 2010; 107: 15595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ, Abraham CR, Podlisny MB, Duffy LK.. Isolation of low-molecular-weight proteins from amyloid plaque fibers in Alzheimer's disease. J Neurochem 1986; 46: 1820–34. [DOI] [PubMed] [Google Scholar]
- Sergeant N, Bombois S, Ghestem A, Drobecq H, Kostanjevecki V, Missiaen C, et al. Truncated beta-amyloid peptide species in pre-clinical Alzheimer's disease as new targets for the vaccination approach. J Neurochem 2003; 85: 1581–91. [DOI] [PubMed] [Google Scholar]
- Sevigny J, Chiao P, Bussiere T, Weinreb PH, Williams L, Maier M, et al. The antibody aducanumab reduces Abeta plaques in Alzheimer's disease. Nature 2016; 537: 50–6. [DOI] [PubMed] [Google Scholar]
- Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, et al. Amyloid-beta protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat Med 2008; 14: 837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar GM, Welzel AT, Mc Donald JM, Selkoe DJ, Walsh DM.. Isolation of low-n amyloid beta-protein oligomers from cultured cells, CSF, and brain. Methods Mol Biol 2011; 670: 33–44. [DOI] [PubMed] [Google Scholar]
- Swanson C, Zhang Y, Dhadda S, Wang J, Kaplow J, Lai R, et al. Treatment of early AD subjects with BAN2401, an antiti-Aß protofibril monoclonnal antibody, singnificantly clears amyloid plaque and reduces clinical decline. In: Alzheimer’s Association International Conference, 2018.
- Szczepankiewicz O, Linse B, Meisl G, Thulin E, Frohm B, Sala Frigerio C, et al. N-Terminal extensions retard Abeta42 fibril formation but allow cross-seeding and coaggregation with Abeta42. J Am Chem Soc 2015; 137: 14673–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez de la Torre A, Gay M, Vilaprinyo-Pascual S, Mazzucato R, Serra-Batiste M, Vilaseca M, et al. Direct evidence of the presence of cross-linked abeta dimers in the brains of Alzheimer's disease patients. Anal Chem 2018; 90: 4552–60. [DOI] [PubMed] [Google Scholar]
- Villemagne VL, Perez KA, Pike KE, Kok WM, Rowe CC, White AR, et al. Blood-borne amyloid-beta dimer correlates with clinical markers of Alzheimer's disease. J Neurosci 2010; 30: 6315–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Teplow DB.. Alzheimer's disease and the amyloid beta-protein. Progr Mol Biol Transl Sci 2012; 107: 101–24. [DOI] [PubMed] [Google Scholar]
- Wang Z, Jackson RJ, Hong W, Taylor WM, Corbett GT, Moreno A, et al. Human brain-derived Abeta oligomers bind to synapses and disrupt synaptic activity in a manner that requires APP. J Neurosci 2017; 37: 11947–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt AD, Perez KA, Rembach A, Sherrat NA, Hung LW, Johanssen T, et al. Oligomers, fact or artefact? SDS-PAGE induces dimerization of beta-amyloid in human brain samples. Acta Neuropathol 2013; 125: 549–64. [DOI] [PubMed] [Google Scholar]
- Watts JC, Condello C, Stohr J, Oehler A, Lee J, DeArmond SJ, et al. Serial propagation of distinct strains of Abeta prions from Alzheimer's disease patients. Proc Natl Acad Sci USA 2014; 111: 10323–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welzel AT, Maggio JE, Shankar GM, Walker DE, Ostaszewski BL, Li S, et al. Secreted amyloid beta-proteins in a cell culture model include N-terminally extended peptides that impair synaptic plasticity. Biochemistry 2014; 53: 3908–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmus MM, Grunberg SC, Bol JG, van Dam AM, Hoozemans JJ, Rozemuller AJ, et al. Transglutaminases and transglutaminase-catalyzed cross-links colocalize with the pathological lesions in Alzheimer's disease brain. Brain Pathol 2009; 19: 612–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willem M, Tahirovic S, Busche MA, Ovsepian SV, Chafai M, Kootar S, et al. eta-Secretase processing of APP inhibits neuronal activity in the hippocampus. Nature 2015; 526: 443–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Li S, Xu H, Walsh DM, Selkoe DJ.. Large soluble oligomers of amyloid beta-protein from Alzheimer brain are far less neuroactive than the smaller oligomers to which they dissociate. J Neurosci 2017; 37: 152–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yankner BA, Lu T.. Amyloid beta-protein toxicity and the pathogenesis of Alzheimer disease. The J Biol Chem 2009; 284: 4755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and its Supplementary material .